Abstract
Using high-throughput analyses and the TRANSFAC database, we characterized TF signatures of head and neck squamous cell carcinoma (HNSCC) subgroups by inferential analysis of target gene expression, correcting for the effects of DNA methylation and copy number. Using this discovery pipeline, we determined that human papillomavirus-related (HPV+) and HPV− HNSCC differed significantly based on the activity levels of key TFs including AP1, STATs, NF-κB, and p53. Immunohistochemical analysis confirmed that HPV− HNSCC is characterized by co-activated STAT3 and NF-κB pathways, and functional studies demonstrate that this phenotype can be effectively targeted with combined anti-NF-κB and anti-STAT therapies. These discoveries correlate strongly with previous findings connecting STATs, NF-κB, and AP1 in HNSCC. We identified 5 top-scoring pair biomarkers from STATs, NF-κB and AP1 pathways that distinguish HPV+ from HPV− HNSCC based on TF activity, and validated these biomarkers on TCGA and on independent validation cohorts. We conclude that a novel approach to TF pathway analysis can provide insight into therapeutic targeting of patient subgroup for heterogeneous disease such as HNSCC.
Keywords: HNSCC, HPV, STAT3, NF-κB, Transcription factor
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is the fifth most common cancer worldwide.1 HNSCC has been traditionally associated with tobacco and alcohol exposure. A subset of high-risk human papilloma virus (HPV)-related HNSCC often found in tobacco non-exposed individuals has been described as a distinct clinicopathologic entity.2 HPV− HNSCC is typically associated with poorer outcomes, while HPV+ HNSCC has a more favorable prognosis.3 For patients with locally advanced HNSCC, multi-modality treatments including surgery, radiotherapy and/or cytotoxic chemotherapy have significantly improved, but five-year overall HNSCC survival remains poor.3 Both genetic and epigenetic aberrations have been shown to play a role in HNSCC development, treatment response and survival.4 However, the only targeted agent approved by the FDA for the treatment of HNSCC is cetuximab, a monoclonal antibody directed against epidermal growth factor receptor (EGFR).5
The development of targeted therapy is challenged by diverse genetic and epigenetic alterations heterogeneously distributed in HNSCC. The data suggest that genetic abnormalities affect a limited number of mitogenic pathways, such as PI3K and NOTCH.6-8 However, genome-wide expression array data demonstrate that HNSCC is characterized by expression dysregulation for a majority of genes.9, 10 Transcription factors are a primary determinant in the regulation of gene expression. Unfortunately, direct comprehensive analysis of the dysregulation of key TFs is challenging with existing screening techniques for several reasons: it is difficult to analyze changes in TFs expression due to their transient and low level expression; activation of most TFs requires post-translational modifications, protein cleavage, formation of active transcriptional complex or protein translocation from cytoplasm to nucleus.11 Therapeutic targeting of TF is complicated due to the low level and cycle-dependent expression of TF, diversity of posttranscriptional modifications and redundancy of the downstream gene regulation. Despite that, many laboratories have demonstrated efficiency of several anti-TF therapies, including agents such as bortezomib [targeting NF-κB through proteasome inhibition],12 BEZ-235 [a dual PI3K/mTOR inhibitor],6 and decoy oligonucleotides [STAT3 and NF-κB inhibitors].13, 14
Using high throughput array datasets and the TRANSFAC database, we have developed an inferential method describing TF activity by the gene expression of their targets.15 Using this method we demonstrate that HPV+ and HPV− HNSCC are differentiated by the activity of key TFs including STATs and NF-κB. Approximately 25% of primary HNSCC, almost exclusively HPV−, have these TFs and their targets coordinately upregulated. After validation of this TF alteration pattern in separate clinical cohorts, we further confirmed that HNSCC with key TF pathway alteration are sensitive to therapy directed at these key TF pathways, providing evidence that TF pathway analysis can provide insight into therapeutic targeting.
MATERIALS AND METHODS
The detailed methods can be found in “Supporting Information”
Tissue samples
We used three independent cohorts of HNSCC and normal specimens, overall: 195 HNSCC and 63 non-cancer affected patients (Tables S1, S2, and S3). Every participant signed a written informed consent before participating in this study. This study was approved by Johns Hopkins Medicine Internal Review Board (JHM IRB), and performed under approved research protocol NA_00036235. TMA tissues were collected under ECOG/RTOG-approved protocols. We have also used publicly available data (http://cancergenome.nih.gov/cancersselected/headandneck) for the TCGA-HNSCC cohort that includes 279 HNSCC and 50 control tissues.
DNA and RNA preparation from microdissected tissues
DNA was isolated by phenol-chloroform extraction after proteinase K digestion16 and RNA was isolated with the mirVana miRNA Isolation Kit (Ambion) per manufacturer’s recommendations.
Arrays
Two µg of RNA or DNA were run on Affymetrix HuEx1.0 GeneChips (RNA), Illumina Infinium HumanMethylation27 BeadChips (bisulfite converted DNA), and Affymetrix Genome-wide SNP 6.0 Array (DNA). All high throughput data sets are available in GEO superSeries GSE33232. The data can be freely downloaded from http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33232.
HPV analyses
HPV status of oropharyngeal SCC tumors was tested by in situ hybridization (ISH) for high-risk HPV,17 by quantitative PCR (qPCR).16 P16 expression was checked by immunohistochemical staining.17
Reverse Transcription (RT) and quantitative Real Time PCR (qRT-PCR)
One µg of RNA from the validation cohort was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). QRT-PCR was performed on Taqman 7900HT (Applied Biosystems, Foster City, CA) with gene-specific expression assays (Applied Biosystems) using standard qRT-PCR conditions.
Immunohistochemistry
Immunostaining of 5 µm cuts was carried out on a Bond-Leica autostaining system (Leica Microsystems, Buffalo Grove, IL) using a standard immunohistochemistry (IHC) protocol with 15 min incubation against total NF-κB p65 - RELA (#8242, dilution 1:400) or against total STAT3 (#4904, 1:200 dilution) antibodies (Cell Signaling). Staining was quantified by Aperio software.
Cell Culture
Cell Lines and Cell Culture Conditions
Human HNSCC cell lines Ho1N1, HSC2, and SKN3 were purchased from the Japanese Collection of Research Bioresource. Each cell line was authenticated using a short tandem repeat analysis kit, GenePrint 10 (Promega), as directed at the Johns Hopkins University Core Facility. Cell grew at 37°C in 5% CO2.
Transient Transfection and Cell Proliferation Assay
The expression of STAT1, STAT3 and RELA genes were knocked down by ON-TARGETplus siRNA SMARTpool (Thermo Scientific) using RNAiMAX Transfection reagent (Life Technologies). Cell proliferation was measure by CCK-8 kit (Dojindo).
Chemical Treatment
Ho1N1, HSC2 and SKN3 cell lines were treated by either 0.1% Dimethyl sulfoxide (DMSO) alone or by 0.1% DMSO with different concentrations of Bortezomib (#681238/8, Kindly provided by NIH, Bethesda, MD), Bay 11-7085 (#B5681, Sigma-Aldrich, St. Louis, MO), Cucurbitacin I (#1571, Tocris, Bristol, UK), or and SH-4-54 (#S7337, Selleckchem, Houston, TX). All cells grew in high glucose DMEM medium with 10% FBS and 1% Penicillin-Streptavidin. Cell proliferation was measure by the CCK-8 kit as described above.
Statistical analyses
Preparation of TF target gene sets
We applied robust multi-array average (RMA) analysis to an expression array dataset for the discovery cohort. We annotated each TF with a list of its experimentally validated targets described in the Transcription Factor Database [TRANSFAC Professional].18 TFs with less than 5 targets were removed. From each TF gene set, we removed genes that were expected to have significantly reduced expression either due to increased methylation (β>0.15) or DNA copy loss (CNV<1.2) on a per tumor sample basis, creating tumor-specific TF gene sets.15 Methylation arrays were preprocessed using R scripts that provided the estimated b value of percentage methylation. SNP-chip arrays were preprocessed using corrected robust linear models with maximum likelihood-based distances (CRLMM) to provide copy number estimates.19 Genes were removed from the TF gene set on a patient-specific basis using the resulting methylation and copy number estimates.
TF activity ranking
The resulting 1,325 tumor-specific TF target gene sets, independently corrected in the discovery and TCGA cohort for DNA-methylation and CNV-dependent expression loss, were used to compare samples from HPV+ and HPV− groups to the pooled values of non-cancer samples for each individual target gene by pseudo-t test, where each individual tumor sample was given a score based on the probability that it came from a normal distribution with mean and standard deviation equal to the expression of that gene in the normal samples. Wilcoxon rank-sum tests were applied to these pseudo-t statistics to compare the expression of targets of each TF relative to the expression of targets not regulated by the TF in each sample relative to normal. Wilcoxon p-values of all TF gene sets were used to rank TFs.
Top Scoring Pair (TSP)
TSP was applied to the combined list of 72 target genes of STAT1, STAT3, NF-κB and AP1 pathways (Table S4). TSP aimed to discover five independent pairs of genes, whose changes in relative level best separate HPV+ and HPV− samples from the discovery cohort. These relative changes were used to score patients in TCGA and a novel validation cohort, where measurements were made by sequencing and RT-PCR methods.
Correlation of IHC staining with clinical data
Protein staining scored by Aperio software was correlated with clinical data of ECOG-TMA cohort. Wilcoxon rank sum tests were used to compare two groups and Kruskal-Wallis tests were used to compare more than 2 groups. Markers’ co-staining was assessed by linear regression.
RESULTS
Cohort assembly and array analysis
To determine gene expression, methylation and copy number in primary HNSCC, we employed Affymetrix HuEx1.0 GeneChips Array, Illumina Infinium HumanMethylation27 BeadChips, and Affymetrix Genome-wide SNP 6.0 Array for 44 HNSCC (multiple primary sites, broad stage distribution) and 25 normal samples from a discovery cohort described earlier [see Table S1 and 8, 9].
Array data annotation and preparation of TF target gene sets
The information regarding the target genes for each TF was obtained from the TRANSFAC 2010.4 Professional database.18 We retained TFs with a minimum of five experimentally validated targets, creating sets of target genes for 1,325 human TFs of the total 2,600 human TF described in TRANSFAC. The decreased expression signals from methylation and copy number-lost target genes would lower the calculated activity of TF, without necessarily lowering the net activity of the TF in a particular tumor sample. To correct for estimated changes in TF activity due to the effects of DNA methylation and copy number changes on target gene expression, we removed genes from each individual sample analysis that had a possibility of reduced expression regardless of TF activity by potential promoter methylation-driven silencing (β>0.15) or by genetic loss of homo- or heterozygosity [Copy Number < 1.2 in Copy Number Variation (CNV) estimation]. These genes were identified by Methylation and SNP arrays used for the same discovery cohort of samples. Methylation and CNV-corrected 1,325 TF target gene sets for each sample were used for further analysis (Figure 1).
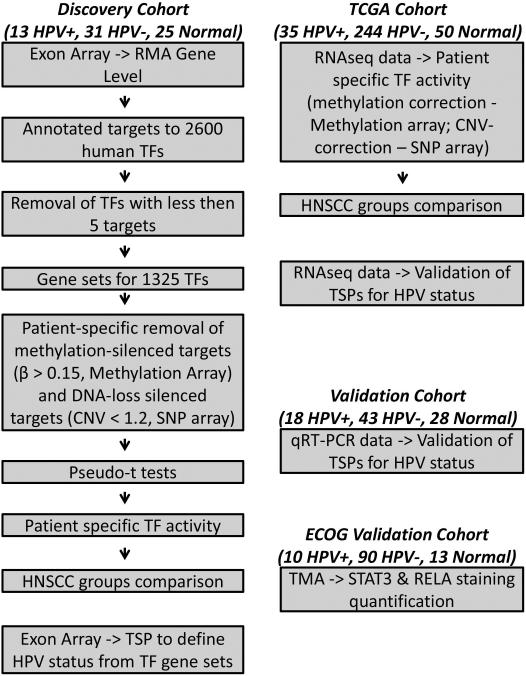
Figure 1.
Experimental set up. Discovery cohort, 44 HNSCC (13 HPV+ and 31 HPV−) and 25 UPPP. Exon Array data for discovery cohort samples was normalized by RMA. 2600 human TF were depicted from TRANSFAC database and were annotated by highly relevant experimentally validated target genes. The list of TF was reduced by removal of TF with less than 5 targets. For the remaining 1325 TFs, the target genes from the TF gene sets were removed if their expression was silenced by hypermethylation or DNA copy loss (using methylation array and SNP assay data) on individual sample basis. Resulting TF target gene sets were then used to compare individual samples to pool of normal by pseudo-t test to evaluate patient specific TF activity. Control-normalized TF gene sets were compared in HPV+ vs HPV− groups. RMA-normalized exon array data alone was used in TSP to define HPV status from TF gene sets for AP1, STAT1, STAT3 and NF-κB pathways. TCGA cohort, 279 HNSCC (35 HPV+ and 244 HPV−) and 50 matched normal. RNA-Seq data was used to evaluate individual gene expression. Target genes from the 1325 TF gene sets were used and were corrected for genes silenced by hypermethylation or DNA copy loss (using methylation array and SNP assay data) and compared for HPV+ and HPV− patients on an individual sample basis similar to the discovery cohort. RNA-Seq was used to validate TSP analysis. Validation cohort, 61 HNSCC (18 HPV+ and 43 HPV−) and 28 normal UPPP, was used for validation of TSP by qRT-PCR. ECOG Validation Cohort, 100 HNSCC (10 HPV+, 90 HPV−) and 13 normal controls, was used in IHC experiments.
HPV positive and HPV negative patients are different by their TF signature
Methylation and CNV-corrected TF target gene sets were analyzed, where the expression of each target gene was compared for each individual samples to the pool of normal samples. In order to elucidate TF driven differences between HPV+ and HPV− samples, we compared gene expression differences between HPV− and HPV+ samples for the corrected list of gene targets of each TF. We used a Wilcoxon rank sum gene set test to rank the TFs based on the expression changes in their targets. While a single target gene could be either upregulated or downregulated in the particular sample or patient group, the determination of TF activity significance is agnostic to direction of dysregulation in TF gene targets, assuming the presence of both upregulated and downregulated targets in its list. We isolated the fifty TFs with the lowest p-values comparing expression of genes in HPV− and HPV+ groups for further analysis. Of note, due to some overlap in nomenclature of specific TFs annotated in TRANSFAC, some TFs belong to the same TF pathway. Thus, the majority of the top fifty ranked TFs could be assigned to a limited number of key TF pathways, including NF-κB [(RelA−p65)2, NF−kappaB1, NF−kappaB1−p50:RelA-p65], AP-1 [c−Fos:c−Jun, Fra-1, ATF, ATF-1, NF-AT1], retinoic acid signaling [PPARalpha:RXR−alpha, RXR−alpha:PPARalpha, RXR−alpha:PPARgamma, LXR-alpha:RXR−alpha, RORalpha1, RXR−alpha], STAT [STAT1, STAT1alpha, STAT5, STAT6, STAT1:STAT1 and STAT3:STAT3], NOTCH [E12, HES-1], p53 [p53], and RB [E2F-4] pathways (Table S5). Please note, that while we detected canonical NF-κB pathway members, we did not detect any members of NF-κB alternative pathway (NFKB2 or RELB).20 Our data suggest that HPV+ and HPV− HNSCC demonstrate gene dysregulation related specifically to distinct alterations in TF activity (Table S5). These data were recapitulated using TCGA-HNSCC cohort (Table S6). These results support prior reports of overall dysregulation of several common TF pathways in head and neck cancer.21
In order to determine robustness to different CNV and methylation thresholds, we repeated this analysis while varying threshold values, such as β>0.25, β>0.35 and β>0.45 for DNA methylation, and CNV<1, CNV<0.8, and CNV≤0 for DNA copy number. The majority of the top TFs with diverse thresholds were found in the top 50 TF list with originally applied biologically relevant β>0.15 and CNV<1.2 conditions, with most TFs involved in STATs, NF-κB, AP1 and retinoic acid pathways (data not shown). However, we noted that p53 was lost during threshold variations. The p53 pathway is known to be affected in HNSCC via two mechanisms, one – via direct HPV E6 oncoprotein-induced degradation of p53 protein, second – via multiple TP53 mutations.22, 23 Point mutations of TP53 genes as well as chromosomal loss23 and lack of adjustment for CNV correction on p53 itself may have altered the significance of p53 TF pathway dysregulation in this analysis.
Co-activation of NF-κB and STAT3 in HPV− HNSCC
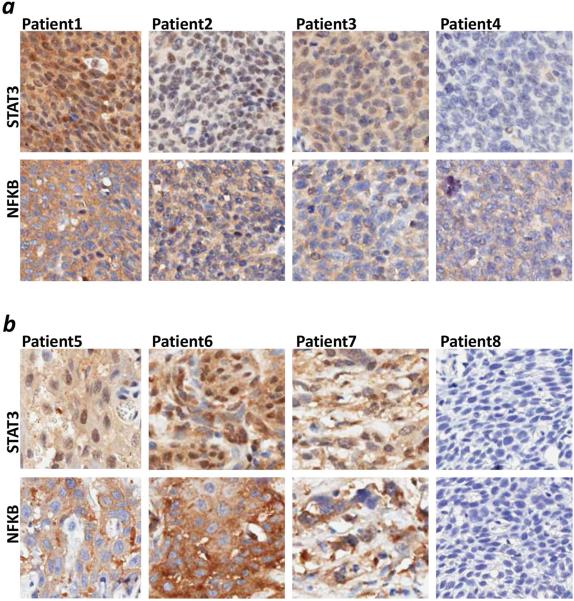
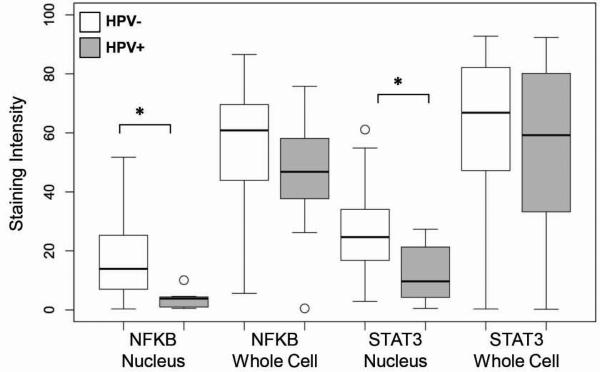
To investigate the coordinated dysregulation and the direction of this dysregulation (upregulation or downregulation) for key TFs, we performed immunohistochemical (IHC) staining experiments on a primary HNSCC tissue microarray (TMA) cohort including 100 HNSCC (consisting of 90 HPV− and 10 HPV+) from a prior clinical trial (ECOG E4393/RTOG 9614) and 13 control samples (Table S2).23 We performed an analysis of STAT3 and NF-κB protein expression for these samples (Figure 2). The protein staining was scored using Aperio software only in the tumor cells in each sample, where staining was independently quantified for the whole cell (reflecting overall protein expression) or for the nuclei (reflecting protein activation and translocation to nuclei) (Table S7, Figure S1). We have evaluated the correlation of protein staining with clinical data, such as age, gender, race, cancer site, as well as with disease, smoking, and HPV statuses. HPV status has the strongest correlation with protein staining. In particular HPV− tumors have increased protein expression and significant protein activation as defined by greater than average cellular and nuclear staining of both STAT3 and NF-κB, as compared to HPV+ (Figure 3), supporting the conclusion that these two groups are differentiated by the activity of TFs such as NF-κB and STATs (Tables S5 and S6). Correlation of nuclear protein staining with HPV was supported by univariate and multivariate analyses (Table S8). The co-staining of NF-κB and STAT3 was evaluated and showed significant co-activation of NF-κB and STAT3 in primary HNSCC (Table S9, Spearman correlation coefficient 0.30, p=0.003). We found significant correlation of STAT3 and NF-κB activation (co-staining in nuclei) (Table S10). We have calculated the median of nuclear and cellular staining of STAT3 and NF-κB for the entire TMA cohort of 100 HNSCC samples (Table S10). We have noticed that 50 of 90 HPV− patients (55.6%) had NF-κB nuclear staining above the median staining intensity (12.85) while none of HPV+ samples had NF-κB staining above the median (Table S10). Similarly, 49 HPV− patients (54.4%) had STAT3 nuclear staining above the median (23.17) while only one HPV+ sample had STAT3 staining above the median (p=0.0012 and 0.0157 for NF-κB and STAT3, Table S10). 30 of 90 HPV− patients (33.3%) had both NF-κB and STAT3 nuclear staining above the median, while no HPV+ patient was found to have this signature (p=0.0302). See table S10 for details.
Figure 2.
IHC staining of NF-κB and STAT3. TMA containing 100 HNSCC and 13 control tissues was stained against total NF-κB (RELA) and STAT3 antibodies. Representative HNSCC samples of NF-κB and STAT3 co-staining are shown. (a) HPV+ samples. (b) HPV− samples. Samples are arranged from stronger staining and better co-staining on the left to no staining on the right. Both markers co-stain in cellular nuclei of cancer tissues (Spearman correlation 0.30, p=0.003).
Figure 3.
Association of protein level with HPV infection. The level of antibody staining was quantified by the Aperio in the whole cell (total protein expression) or in the nuclei (protein activation) for NF-κB and STAT3. The staining intensity was averaged for three tissue replicates. (* p<0.05).
Target genes of transcription factors STATs, NF-κB and AP1 distinguish HPV+ and HPV− HNSCC
To define expression biomarkers related to TF activity, we defined the TF target genes that reflect the dysregulation of these pathways and examined the ability of these genes to distinguish HPV+ and HPV− HNSCC. We combined the lists of all targets for NF-κB, STAT1, STAT3 and AP1 factors (Table S4) and applied top scoring pair [TSP]24, 25 analysis to this combined list. TSP was applied to the expression array data of 72 combined target genes from the discovery cohort. TSP allowed the discovery of 5 gene pairs among 72 targets that discriminate HPV+ and HPV− (Figure S1), namely: CCND1, CEBPD, ICAM1, IFG1R, IL6ST, IRF1, JAG1, JAK3, NOS3, and SOCS3. Of note, 5 out of 10 of these paired genes belong to more than one of the above dysregulated pathways (Table S4). The five TSPs distinguished HPV+ and HPV− samples from the discovery cohort with a p-value of 9.5 × 10−6 and an Odds Ratio (OR) of 44 [5.0, 2185, 95% CI] (Table 1).
Table 1.
Top 5 pairs of target genes discriminate HPV− and HPV+ patients from different cohorts
| Discovery Cohort | TCGA | Validation Cohort | |
|---|---|---|---|
| p value | 9.5 × 10−6 | 6.7 × 10−8 | 0.0006 |
| odds ratio [95% CI] | 44 [5.0 - 2185] | 8.9 [3.6 26] | 9.6 [2.2 60] |
To validate our discovery of TF differential dysregulation, we adopted RNA-Seq data available for the TCGA HNSCC (The Cancer Genome Atlas) cohort. This cohort consists of 279 tumor samples, including 35 HPV+ and 244 HPV− samples. Validation of these genes as discriminators between HPV+ and HPV− within the TCGA cohort also demonstrated significant separation with a p-value of 6.7 × 10−8 OR = 8.9 [3.6 26, 95% CI] (Table 1).
Validation of transcription factor target genes by quantitative real time PCR (qRT-PCR)
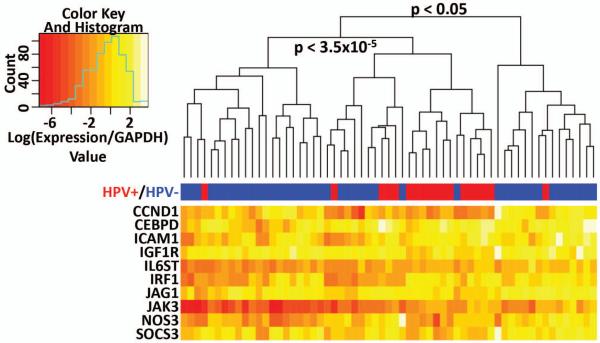
In order to validate our results and evaluate the discriminative ability of the top five gene pairs from the TSP analysis, we assembled an independent validation cohort of 61 HNSCC (including 43 HPV− and 18 HPV+ samples) and 28 control uvulopalatopharyngoplasty (UPPP) samples (Table S3). The TSPs based on expression of the top five gene pairs were analyzed by qRT-PCR, which was able to separate HPV+ and HPV− HNSCC from this validation cohort with a p-value of 0.0006 , OR = 9.6 [2.2 60] (Table 1). Using qRT-PCR data for 5 top scoring gene pairs from TSP, this validation cohort (Table S3) was separated into three groups by unsupervised hierarchical clustering (Figure 4 heat-map was built for the purpose of visualization). We found that 25 out of 61 samples were clustered into a separate group that was enriched by HPV+ patients. That group contains 16 out of 18 HPV+ patients and is distinct from two other primarily HPV− groups by downregulation of CCND1 and partial downregulation of IRF1, ICAM1, IGF1R and CCND1 (p < 3.5 × 10−5). These data suggest that STATs, NF-κB and AP1 pathways are upregulated in HPV− HNSCC patients as compared to HPV+ patients, in concordance with our immunohistochemical data. Two groups of primarily HPV− patients can be distinguished by relative expression of TSP genes, with one group showing higher expression of CCND1, CEBPD, ICAM1, IRF1, IL6ST, JAG1, JAK3, NOS3, and SOCS3. These data suggest that dysregulation of STATs, AP1 and NF-κB pathways is coordinated and that these pathways are co-activated for a subset of HNSCC patients (25%, 15 out of total 61 samples), predominantly HPV− patients (p<0.05 by Fisher’s exact test applied to hierarchically clustered groups, Figure 4). As HPV− tumors have been associated with poor clinical outcome, the detected gene signatures can potentially aid clinical assessment of HNSCC prognostic subgroups.
Figure 4.
Separation of HNSCC by the expression of the five top-scoring gene pairs. The expression of the top five gene pairs discovered by TSP was analyzed by qRT-PCR on 61 HNSCC and 28 UPPP samples from the validation cohort. Logarithm-converted values of GAPDH-relative expression for these genes were used for heatmap preparation in R script. Tumor samples were separated on three groups by unsupervised hierarchical clustering. The middle group of 25 HNSCC contains 16 out of total 18 HPV+ patients. P-values were calculated by Fisher exact test.
NF-κB and STAT co-activated HPV− HNSCC cells are growth-inhibited by combined targeted therapy
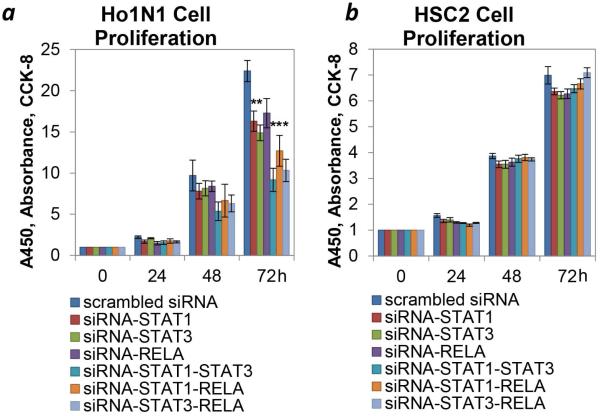
We chose Ho1N1 and HSC2 from total 34 cell lines described in 26 after expression analysis of key TFs and their significant targets (detected via TSP analysis); these cell lines were selected as representative of STAT3–NF-κB over-expressing or base-line expressing tumors based on expression pattern. To evaluate the clinical relevance and potential therapeutic susceptibility of NF-κB and STATs co-activation in HPV− HNSCC, we knocked down the expression of these TFs in vitro by RNAi techniques. . Ho1N1 cells are characterized by over-expression of STAT1, STAT3, RELA and NFΚB1 and their signature targets as defined by our TSP analysis (IRF1, CEBPD, CCND1, ICAM1, JAG1, JAK3, and NOS3). Since our data demonstrate that expression of signature target genes discovered as five TSPs reliably describes the activity of key TF pathways, we conclude that Ho1N1 has hyper-activated STAT3 and NF-κB pathways. Conversely HSC2 showed low baseline expression of STATs, NF-κB genes and their significant targets. Of note, JAK3 expression was not detected in HSC2 and the expression of JAG1 and NOS3 was not significantly different in these two cell lines (Figure S2).
The siRNA SMARTpools for STAT1, STAT3 and RELA allowed significant inhibition of target gene expression in each cell line (Figure S3). Growth inhibition was associated with induction of the cell death by downregulation of NF-κB, shown for wt TP53 head and neck cell lines (HPV−).27 Interestingly, significant growth inhibition was noticed only in the STAT and NF-κB activated Ho1N1 cell line, but not in the HSC2 cell line (Figure 5). Combined downregulation of two genes had additive effect on cell growth, suggesting that these pathways were downregulated independently. The combined inhibition of STAT1 and STAT3 expression had the strongest effect on Ho1N1 cell line proliferation (62% cell growth inhibition, as compared to control scrambled siRNA pool); and second strongest growth inhibition was archived with double STAT3–RELA inhibition (56% cell growth inhibition). The Ho1N1 cell line was the most sensitive to chemical NF-κB and STAT3 inhibitors (Bortezomib, Bay 11-7085, Cucurbitacin and SH-4-54; Figures S4 and S5, and References12, 28-30), supporting our siRNA experiments. NF-κB and STAT3 inhibitors were not used in combination due to the high cellular toxicity of the individual agents. No effect of siRNA inhibition was noted in the low STAT–NF-κB expressing HSC2 cell line (Figure 5b). HSC2 was the most resistant to chemical NF-κB and STAT3 inhibitors (Figures S4 and S5), supporting our siRNA experiments. SKN3 cells, characterized by intermediate expression of STATs, NF-κB and their targets (Figure S2), were used as a validating control. This cell line had intermediate cell growth inhibitory effect of STAT and NF-κB downregulation (Figure S6), with maximal cell growth inhibition with use of combined STAT3 and RELA siRNA (42% cell growth inhibition, Figure S6a). SKN3 has an intermediate response to both NF-κB inhibitors (Figure S4). As for STAT3 inhibitors, SKN3 was sensitive to Cucurbitacin and resistant to SH-4-54 (Figure S5), suggesting an overall intermediate response to STAT3 inhibitors. The diverse response of SKN3 to STAT3 inhibitors may be explained by high toxicity of Cucurbitacin and low specificity of SH-4-54.
Figure 5.
Cooperative downregulation of STATs and NF-κB pathways in HNSCC cell lines. Ho1N1 (a, over-expresser) and HSC2 (b, baseline) cell lines were used in experiments with single and combined downregulation of STAT1, STAT3 and NF-κB subunit – RELA. Cell proliferation of Ho1N1 and HSC2 with or without of gene specific knockdown of STAT1, STAT3 and RELA was measured by CCK8. Values are mean ± SEM, the experiments were performed in pentaplicates. (* indicates a significant difference (p<0.05) between the control (scrambled) and targeted RNAi experiments.
DISCUSSION
Over the past decade, there has been recognition that HNSCC includes favorable prognosis HPV+ and poor prognosis HPV− patients.31 The discovery of novel therapeutic agents for poor prognosis HPV− HNSCC has been challenging, despite use of high throughput sequencing efforts to define novel therapeutic targets. TF are significant drivers of gene expression variation, and characterization of TF alterations facilitates insight into HNSCC biology; however, reliable changes in TF activity are hard to directly detect due to the complexity of TF signaling. Using three high throughput platforms and an innovative integrative strategy that compensates for genetic and epigenetic changes in TF targets, we were able to annotate highly specific target genes to each known TF and infer whole-genome TF activity by the expression of those target genes in each particular tumor sample.
We have found that HPV− patients have strong coordinated dysregulation of several pathways, including coordinated activation of NF-κB and STATs pathways. Our results are supported by similar results shown by other investigators using a cDNA microarray analysis of 10 HPV− HNSCC cell lines.32, 33 We confirmed our results on multiple validation datasets, including an ECOG TMA cohort, a TCGA-HNSCC cohort and independent HNSCC validation cohort, by protein and expression assays, as well as by confirmation of expression alteration for direct targets of specific TFs. The discovered co–activation of NF-κB and STAT3 in primary HNSCC tissue is also in agreement with prior published IHC data.34
We have also shown that co-activation of NF-κB and STAT3 provide an avenue for selective therapeutic targeting in cell line models. We have demonstrated that these alterations occur in a substantial subset of HNSCCs. For example, 15 out of total 61 HNSCC patients (25%) or 14 out of total 43 HPV− HNSCC patients (33%) have higher expression of direct targets of STATs, NF-κB and AP1 TFs, such as CCND1, CEBPD, ICAM1, IRF1, JAG1, JAK3 and NOS3 (Figure 4). The direct dependence of these targets on TFs activity strongly correlates with the previous data showing NF-κB target activation in HNSCC.27
Prior reports have described changes in the activity of the several TFs, like STAT1, STAT3, NF-κB and AP1, in HPV− HNSCC, as well as in HPV mixed HNSCC population and HPV+ cervix patients.35-39 For example, HPV16 E7 was shown to inhibit NF-κB activity.40 Additional TF pathway activity alterations in HNSCC have been shown for p53,22 RB1,23 and retinoic X receptor41 cascades. There are many possibilities for these pathways to interact in HNSCC. Many of these TFs have common upstream regulators: including dependence of NF-κB, STATs and AP1 activation on IKKα, IKKβ, and on cytokine signals.20, 42. They also share common downstream targets: for example, STAT1 and NF-κB regulate CD40 expression43, 44; AP1 and NF-κB regulate IL6, IL8 and VEGF expression.34 Finally, these TFs can also affect their own expression and expression of each other: NF-κB plays a role in paracrine-dependent activation of STAT3 pathway.34, 42 More shared targets can be found in Table S4 and in Reference32. Prior publications also suggest that co-activation of NF-κB and STAT3, especially in the context of dysfunctional p53 will enhance BCL-XL expression and lead to HNSCC cell survival.45 While direct ChIP-Seq experiments are not feasible to perform on the primary HNSCC samples due to the limited tumor availability, we expect overlap in TF binding to their target genes, inconsistent with the literature cited above.
The overlapping gene targets of p53, p63 and p73 TFs and their ability to distinguish HPV+ and HPV− patients is summarized in Table S11. Notably, the mutation status of TP53 gene has a mixed downstream effect on the activity of its target genes (Table S12). The TP53 mutation status was available for 37 out of 44 HNSCC samples in the discovery cohort from previous deep-sequencing study46, where 25 samples were found to have wild-type TP53 and 7 samples were found to have disruptive TP53 mutations (2 samples with nonsense mutations and 5 samples with missense hot spot mutations: R175H, Y220C, R248Q, R273C, and R282W). The diverse effect of disruptive TP53 mutations can be explained by downregulating effect of nonsense mutations and activating effect of gain-of-function hot spot mutations. Such results correlate with previously published data, summarized in Reference47.
In general, TFs were considered “undruggable” for many years, but with improved understanding of transcription factor biology and new insight into pathway analysis, as well as novel technologies in the development of targeting agents and agent delivery, there is a broad venue for treatment based on TFs and their pathways.48, 49 We have demonstrated that therapeutic agents directed against STATs and NF-κB, such as Cucurbitacin29, Bortezomib12, Bay 11-708528 and SH-4-5430 have high toxicity and limited specificity (See Figures S4 and S5 for details). However, small peptides and peptidomimetics have shown lower cellular toxicity and better specificity to decrease transcription activity of STAT3, NF-κB, MYC and others.50 Oligonucleotide-based therapy, including RNA interference agents and single-stranded decoys have demonstrated specific downregulation of targeted pathways, and are actively used in preclinical and early clinical studies to facilitate AP1, RB1, ERs, NF-κB, and STATs downregulation.13, 14, 51
The correlation of our findings with previous published data suggests that the inferential technique we have developed to define TF activity is robust and a useful tool for discovery of genome-wide changes in TF activity. While we evaluated only the most prominent TF pathways, such as STATs, NF-κB and AP1, many more TFs were found to be significantly and differentially activated in two HNSCC groups. The use of an inferential strategy to deduce activity of specific TFs also has the potential for application in therapeutic targeting, as we were able to use a list of ten target genes that helped us to validate TF signatures experimentally. This strategy could potentially be adapted for clinical practice to identify patients with dysregulated (upregulated or downregulated) pathways. Characterization of a limited list of TF target genes allowed us to accurately characterize TF signatures experimentally, and could potentially be applied to define patients with tumors that are susceptible to specific TF pathway targeting agents as we have done in cell line systems, albeit with potential limitations in translating cell line responses to primary tumor responses to therapy. In addition, we used a correction strategy to infer TF signatures despite heterogeneity of genetic and epigenetic alterations that could potentially mask TF pathway alteration. This strategy has potential to clarify pathway alterations in specific individual tumors within tumor types that exhibit significant heterogeneity in terms of genetic and epigenetic alterations.
In summary, the use of high throughput integrated expression, methylation, and genome copy number analysis to infer TF activity in HNSCC provides insight into biologic variability of behavior and treatment response for HPV+ and HPV− HNSCC patients, and may potentially be used to direct targeted therapy based on TF pathway analysis.
Supplementary Material
WHAT’S NEW.
This is the first comprehensive genome wide analysis of transcription factors (TFs) in primary head and neck squamous cell carcinoma (HNSCC). TF activity was estimated globally using a novel inferential approach that accounts for gene silencing and loss of hetero- and homo-zygosity. This approach also generated biomarkers of TF activity linked to HPV status, which was validated in two additional cohorts using different molecular measurement technologies. Using the employed inferential approach we developed a top-scoring-pair biomarkers based panel. Using this panel we defined therapeutic response in cell line models. These data indicate that TF based analyses of HNSCC have potential for defining therapeutic responsiveness to TF based targeted therapies.
ACKNOWLEDGEMENT
This manuscript is based on a web database application provided by Research Information Technology Systems (RITS)—https://www.rits.onc.jhmi.edu/.
FINANCIAL SUPPORT:
The work is supported by American Head and Neck Society (AHNS) 306481 (DAG), NIDCR/NIH Challenge Grant RC1DE020324 (JAC), NIDCR/NCI P50DE019032 Head and Neck Cancer SPORE (JAC), NIH/NLM R01 LM011000 (MFO), and NIDCR/NIH R01 DE013152 (WMK).
Footnotes
CONFLICT OF INTEREST:
The authors declare no conflict of interest
REFERENCES
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Psyrri A, Boutati E, Karageorgopoulou S. Human papillomavirus in head and neck cancers: biology, prognosis, hope of treatment, and vaccines. Anticancer Drugs. 2011;22:586–90. doi: 10.1097/CAD.0b013e328344ec44. [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, Liu L, Lynch CF, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:4294–301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith IM, Mydlarz WK, Mithani SK, Califano JA. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. International journal of cancer Journal international du cancer. 2007;121:1724–8. doi: 10.1002/ijc.22889. [DOI] [PubMed] [Google Scholar]
- 5.Bonner JA, Harari PM, Giralt J, Cohen RB, Jones CU, Sur RK, Raben D, Baselga J, Spencer SA, Zhu J, Youssoufian H, Rowinsky EK, et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. The lancet oncology. 2010;11:21–8. doi: 10.1016/S1470-2045(09)70311-0. [DOI] [PubMed] [Google Scholar]
- 6.Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, Lu Y, Zhang Q, Du Y, Gilbert BR, Freilino M, Sauerwein S, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer discovery. 2013;3:761–9. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun W, Gaykalova DA, Ochs MF, Mambo E, Arnaoutakis D, Liu Y, Loyo M, Agrawal N, Howard J, Li R, Ahn S, Fertig E, et al. Activation of the NOTCH pathway in head and neck cancer. Cancer research. 2014;74:1091–104. doi: 10.1158/0008-5472.CAN-13-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fertig EJ, Markovic A, Danilova LV, Gaykalova DA, Cope L, Chung CH, Ochs MF, Califano JA. Preferential activation of the hedgehog pathway by epigenetic modulations in HPV negative HNSCC identified with meta-pathway analysis. PloS one. 2013;8:e78127. doi: 10.1371/journal.pone.0078127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D, Butterfoss D, Xiang D, Zanation A, Yin X, Shockley WW, Weissler MC, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer cell. 2004;5:489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 11.Tootle TL, Rebay I. Post-translational modifications influence transcription factor activity: a view from the ETS superfamily. Bioessays. 2005;27:285–98. doi: 10.1002/bies.20198. [DOI] [PubMed] [Google Scholar]
- 12.Allen C, Saigal K, Nottingham L, Arun P, Chen Z, Van Waes C. Bortezomib-induced apoptosis with limited clinical response is accompanied by inhibition of canonical but not alternative nuclear factor-{kappa}B subunits in head and neck cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:4175–85. doi: 10.1158/1078-0432.CCR-07-4470. [DOI] [PubMed] [Google Scholar]
- 13.Gill JS, Zhu X, Moore MJ, Lu L, Yaszemski MJ, Windebank AJ. Effects of NFkappaB decoy oligonucleotides released from biodegradable polymer microparticles on a glioblastoma cell line. Biomaterials. 2002;23:2773–81. doi: 10.1016/s0142-9612(02)00013-3. [DOI] [PubMed] [Google Scholar]
- 14.Sen M, Paul K, Freilino ML, Li H, Li C, Johnson DE, Wang L, Eiseman J, Grandis JR. Systemic Administration of a Cyclic Signal Transducer and Activator of Transcription 3 (STAT3) Decoy Oligonucleotide Inhibits Tumor Growth without Inducing Toxicological Effects. Molecular medicine. 2014;20:46–56. doi: 10.2119/molmed.2013.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rathi KS, Gaykalova DA, Hennessey P, Califano JA, Ochs MF. Correcting Transcription Factor Gene Sets for Copy Number and Promoter Methylation Variations. Drug Dev Res. 2014 doi: 10.1002/ddr.21220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho AL, Henrique R, Jeronimo C, Nayak CS, Reddy AN, Hoque MO, Chang S, Brait M, Jiang WW, Kim MM, Claybourne Q, Goldenberg D, et al. Detection of promoter hypermethylation in salivary rinses as a biomarker for head and neck squamous cell carcinoma surveillance. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:4782–9. doi: 10.1158/1078-0432.CCR-11-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singhi AD, Califano J, Westra WH. High-risk human papillomavirus in nasopharyngeal carcinoma. Head Neck. 2012;34:213–8. doi: 10.1002/hed.21714. [DOI] [PubMed] [Google Scholar]
- 18.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–8. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scharpf RB, Irizarry RA, Ritchie ME, Carvalho B, Ruczinski I. Using the R Package crlmm for Genotyping and Copy Number Estimation. J Stat Softw. 2011;40:1–32. [PMC free article] [PubMed] [Google Scholar]
- 20.Nottingham LK, Yan CH, Yang X, Si H, Coupar J, Bian Y, Cheng TF, Allen C, Arun P, Gius D, Dang L, Van Waes C, et al. Aberrant IKKalpha and IKKbeta cooperatively activate NF-kappaB and induce EGFR/AP1 signaling to promote survival and migration of head and neck cancer. Oncogene. 2014;33:1135–47. doi: 10.1038/onc.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rampias T, Sasaki C, Weinberger P, Psyrri A. E6 and E7 gene silencing and transformed phenotype of human papillomavirus 16-positive oropharyngeal cancer cells. J Natl Cancer Inst. 2009;101:412–23. doi: 10.1093/jnci/djp017. [DOI] [PubMed] [Google Scholar]
- 22.Haraf DJ, Nodzenski E, Brachman D, Mick R, Montag A, Graves D, Vokes EE, Weichselbaum RR. Human papilloma virus and p53 in head and neck cancer: clinical correlates and survival. Clinical cancer research : an official journal of the American Association for Cancer Research. 1996;2:755–62. [PubMed] [Google Scholar]
- 23.Poeta ML, Manola J, Goldwasser MA, Forastiere A, Benoit N, Califano JA, Ridge JA, Goodwin J, Kenady D, Saunders J, Westra W, Sidransky D, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552–61. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geman D, d'Avignon C, Naiman DQ, Winslow RL. Classifying gene expression profiles from pairwise mRNA comparisons. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leek JT. The tspair package for finding top scoring pair classifiers in R. Bioinformatics. 2009;25:1203–4. doi: 10.1093/bioinformatics/btp126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatakeyama H, Cheng H, Wirth P, Counsell A, Marcrom SR, Wood CB, Pohlmann PR, Gilbert J, Murphy B, Yarbrough WG, Wheeler DL, Harari PM, et al. Regulation of heparin-binding EGF-like growth factor by miR-212 and acquired cetuximab-resistance in head and neck squamous cell carcinoma. PloS one. 2010;5:e12702. doi: 10.1371/journal.pone.0012702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee TL, Yang XP, Yan B, Friedman J, Duggal P, Bagain L, Dong G, Yeh NT, Wang J, Zhou J, Elkahloun A, Van Waes C, et al. A novel nuclear factor-kappaB gene signature is differentially expressed in head and neck squamous cell carcinomas in association with TP53 status. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:5680–91. doi: 10.1158/1078-0432.CCR-07-0670. [DOI] [PubMed] [Google Scholar]
- 28.Wang Q, Huber N, Noel G, Haar L, Shan Y, Pritts TA, Ogle CK. NF-kappaBeta inhibition is ineffective in blocking cytokine-induced IL-8 production but P38 and STAT1 inhibitors are effective. Inflamm Res. 2012;61:977–85. doi: 10.1007/s00011-012-0490-2. [DOI] [PubMed] [Google Scholar]
- 29.Sun J, Blaskovich MA, Jove R, Livingston SK, Coppola D, Sebti SM. Cucurbitacin Q: a selective STAT3 activation inhibitor with potent antitumor activity. Oncogene. 2005;24:3236–45. doi: 10.1038/sj.onc.1208470. [DOI] [PubMed] [Google Scholar]
- 30.Haftchenary S, Luchman HA, Jouk AO, Veloso AJ, Page BD, Cheng XR, Dawson SS, Grinshtein N, Shahani VM, Kerman K, Kaplan DR, Griffin C, et al. Potent Targeting of the STAT3 Protein in Brain Cancer Stem Cells: A Promising Route for Treating Glioblastoma. ACS medicinal chemistry letters. 2013;4:1102–7. doi: 10.1021/ml4003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinberger PM, Yu Z, Haffty BG, Kowalski D, Harigopal M, Brandsma J, Sasaki C, Joe J, Camp RL, Rimm DL, Psyrri A. Molecular classification identifies a subset of human papillomavirus--associated oropharyngeal cancers with favorable prognosis. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:736–47. doi: 10.1200/JCO.2004.00.3335. [DOI] [PubMed] [Google Scholar]
- 32.Yan B, Chen G, Saigal K, Yang X, Jensen ST, Van Waes C, Stoeckert CJ, Chen Z. Systems biology-defined NF-kappaB regulons, interacting signal pathways and networks are implicated in the malignant phenotype of head and neck cancer cell lines differing in p53 status. Genome biology. 2008;9:R53. doi: 10.1186/gb-2008-9-3-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan B, Yang X, Lee TL, Friedman J, Tang J, Van Waes C, Chen Z. Genome-wide identification of novel expression signatures reveal distinct patterns and prevalence of binding motifs for p53, nuclear factor-kappaB and other signal transcription factors in head and neck squamous cell carcinoma. Genome biology. 2007;8:R78. doi: 10.1186/gb-2007-8-5-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Squarize CH, Castilho RM, Sriuranpong V, Pinto DS, Jr., Gutkind JS. Molecular cross-talk between the NFkappaB and STAT3 signaling pathways in head and neck squamous cell carcinoma. Neoplasia. 2006;8:733–46. doi: 10.1593/neo.06274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–34. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 36.Dauer DJ, Ferraro B, Song L, Yu B, Mora L, Buettner R, Enkemann S, Jove R, Haura EB. Stat3 regulates genes common to both wound healing and cancer. Oncogene. 2005;24:3397–408. doi: 10.1038/sj.onc.1208469. [DOI] [PubMed] [Google Scholar]
- 37.Cho HJ, Kang JH, Kwak JY, Lee TS, Lee IS, Park NG, Nakajima H, Magae J, Chang YC. Ascofuranone suppresses PMA-mediated matrix metalloproteinase-9 gene activation through the Ras/Raf/MEK/ERK- and Ap1-dependent mechanisms. Carcinogenesis. 2007;28:1104–10. doi: 10.1093/carcin/bgl217. [DOI] [PubMed] [Google Scholar]
- 38.Ondrey FG, Dong G, Sunwoo J, Chen Z, Wolf JS, Crowl-Bancroft CV, Mukaida N, Van Waes C. Constitutive activation of transcription factors NF-(kappa)B, AP-1, and NF-IL6 in human head and neck squamous cell carcinoma cell lines that express pro-inflammatory and pro-angiogenic cytokines. Molecular carcinogenesis. 1999;26:119–29. doi: 10.1002/(sici)1098-2744(199910)26:2<119::aid-mc6>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 39.Song JI, Grandis JR. STAT signaling in head and neck cancer. Oncogene. 2000;19:2489–95. doi: 10.1038/sj.onc.1203483. [DOI] [PubMed] [Google Scholar]
- 40.Vandermark ER, Deluca KA, Gardner CR, Marker DF, Schreiner CN, Strickland DA, Wilton KM, Mondal S, Woodworth CD. Human papillomavirus type 16 E6 and E 7 proteins alter NF-kB in cultured cervical epithelial cells and inhibition of NF-kB promotes cell growth and immortalization. Virology. 2012;425:53–60. doi: 10.1016/j.virol.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato O, Kuriki C, Fukui Y, Motojima K. Dual promoter structure of mouse and human fatty acid translocase/CD36 genes and unique transcriptional activation by peroxisome proliferator-activated receptor alpha and gamma ligands. J Biol Chem. 2002;277:15703–11. doi: 10.1074/jbc.M110158200. [DOI] [PubMed] [Google Scholar]
- 42.Bancroft CC, Chen Z, Yeh J, Sunwoo JB, Yeh NT, Jackson S, Jackson C, Van Waes C. Effects of pharmacologic antagonists of epidermal growth factor receptor, PI3K and MEK signal kinases on NF-kappaB and AP-1 activation and IL-8 and VEGF expression in human head and neck squamous cell carcinoma lines. International journal of cancer Journal international du cancer. 2002;99:538–48. doi: 10.1002/ijc.10398. [DOI] [PubMed] [Google Scholar]
- 43.Lee SJ, Qin H, Benveniste EN. Simvastatin inhibits IFN-gamma-induced CD40 gene expression by suppressing STAT-1alpha. J Leukoc Biol. 2007;82:436–47. doi: 10.1189/jlb.1206739. [DOI] [PubMed] [Google Scholar]
- 44.McArdle MA, Finucane OM, Connaughton RM, McMorrow AM, Roche HM. Mechanisms of obesity-induced inflammation and insulin resistance: insights into the emerging role of nutritional strategies. Frontiers in endocrinology (Lausanne) 2013;4:52. doi: 10.3389/fendo.2013.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee TL, Yeh J, Friedman J, Yan B, Yang X, Yeh NT, Van Waes C, Chen Z. A signal network involving coactivated NF-kappaB and STAT3 and altered p53 modulates BAX/BCL-XL expression and promotes cell survival of head and neck squamous cell carcinomas. International journal of cancer Journal international du cancer. 2008;122:1987–98. doi: 10.1002/ijc.23324. [DOI] [PubMed] [Google Scholar]
- 46.Gaykalova DA, Mambo E, Choudhary A, Houghton J, Buddavarapu K, Sanford T, Darden W, Adai A, Hadd A, Latham G, Danilova LV, Bishop J, et al. Novel insight into mutational landscape of head and neck squamous cell carcinoma. PloS one. 2014;9:e93102. doi: 10.1371/journal.pone.0093102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes & development. 2012;26:1268–86. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Redmond AM, Carroll JS. Defining and targeting transcription factors in cancer. Genome biology. 200910:311. doi: 10.1186/gb-2009-10-7-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeh JE, Toniolo PA, Frank DA. Targeting transcription factors: promising new strategies for cancer therapy. Curr Opin Oncol. 2013;25:652–8. doi: 10.1097/01.cco.0000432528.88101.1a. [DOI] [PubMed] [Google Scholar]
- 50.Turkson J, Kim JS, Zhang S, Yuan J, Huang M, Glenn M, Haura E, Sebti S, Hamilton AD, Jove R. Novel peptidomimetic inhibitors of signal transducer and activator of transcription 3 dimerization and biological activity. Mol Cancer Ther. 2004;3:261–9. [PubMed] [Google Scholar]
- 51.Sen M, Thomas SM, Kim S, Yeh JI, Ferris RL, Johnson JT, Duvvuri U, Lee J, Sahu N, Joyce S, Freilino ML, Shi H, et al. First-in-human trial of a STAT3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer discovery. 2012;2:694–705. doi: 10.1158/2159-8290.CD-12-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.