Abstract
The safety of traditional Chinese medicine (TCM) is a major strategic issue that involves human health. With the continuous improvement in disease prevention and treatment, the export of TCM and its related products has increased dramatically in China. However, the frequent safety issues of Chinese medicine have become the ‘bottleneck’ impeding the modernization of TCM. It was proved that mycotoxins seriously affect TCM safety; the pesticide residues of TCM are a key problem in TCM international trade; adulterants have also been detected, which is related to market circulation. These three factors have greatly affected TCM safety. In this study, fast, highly effective, economically-feasible and accurate detection methods concerning TCM safety issues were reviewed, especially on the authenticity, mycotoxins and pesticide residues of medicinal materials.
Key Words: Traditional Chinese medicine, Safety issue, Rapid detection, Mycotoxins, Pesticide residues, Authentication, 2D DNA barcodes, Traceability
Abbreviations: AA, aristolochic acid, Afs, aflatoxins; DON, deoxynivalenol, GICA, gold immunochromatographic assay; LOD, limit of detection, OTA, ochratoxin A; PAs, pyrrolizidine alkaloids, SNP, single nucleotide polymorphism; SSCP, single-strand conformation polymorphism, ZEN, zearalenone
Graphical abstract
Adulterants, mycotoxins pollution and pesticide residues are “bottlenecks” that affect use and export of traditional Chinese medicine (TCM). The current review aims to analyze these problems and summarize rapid detection methods for the safety of TCM.
1. Safety issues of traditional Chinese medicine (TCM)
1.1. Safety issues of adulterants and toxic TCM in the market
Various TCM materials are confusing because of historical and geographical reasons and this confusion brings enormous danger to the TCM safety. Potent toxic substances, including aconite, aristolochic acid, anticholinergic, podophyllin, grayanotoxin, pyrrolizidine alkaloids, matrine, gelsemine, teucvin and strychnine1, 2, 3, 4, were easily misidentified, erroneously substituted with other herbs or intentionally adulterated for greater benefit. Traditional identification methods recognize materials by the morphological characteristics of TCM; such methods mainly depend on the expertise of the person who identifies. Once misidentified, the TCM can cause serious toxicity problems5, 6, 7, 8. The problem is, however, that we are facing the lack of the experts in TCM identification. At present, the most commonly used detection platforms are based on analytical laboratory instruments. These approaches fail to meet the purpose of rapid on-site analysis in the quarantine clearance of quarantine-related departments.
1.2. Mycotoxin-related safety problems of TCM materials
Medicinal plants are the main raw materials in TCM production. These plants may be infected by fungi and mycotoxins during their growth in fields or in the process of harvest and storage, thereby increasing the odds of significant health problems induced by TCM (e.g., teratogenesis, immunotoxicity or even cancer)9. Currently, 500 different mycotoxins have been recognized, among which the most common and of particular interest are aflatoxins (AFs), ochratoxins, fumonisins and deoxynivalenol (DON). Medicinal plants, such as platycladi seed and raw malt, are often infected by AFs10. The positive rate of AFs present in Nelumbo nucifera (Gaertn.) is up to 70%, in which the content of 30% AF B1 samples and the total content of 25% AF samples both exceeds the standard limits of 5 μg/kg and 10 μg/kg, respectively11. Moldy licorice samples are infected by both AFs and ochratoxin A (OTA), and their infection levels are relatively high12, 13. In OTA investigations of 57 medicinal material samples distributed in six regions in China, the results showed that the positive rate of molded samples by storage is 74%, and that of un-molded samples is only 8%. The OTA content of partial samples exceeded the standard limit set by the European Union, implying serious undetected toxicity for clinical drug use14. Furthermore, Semen Coicis listed in the ‘Medicine Food Homology’ could be easily infected by zearalenone (ZEN) besides DON, and Baohe pills made from the powder of Semen Coicis are also easily infected by DON15, 16. In recent years, foreign scholars reported that 5% OTA in licorice root was transferred into boiling tea, and 1% OTA was transferred into impregnated tea17, 18, 19. The above results demonstrate the urgent need for the monitoring of mycotoxin residues during TCM production.
1.3. Pesticide-related safety problems of TCM materials
More than 12,000 pesticides exist throughout the world. The pesticides mainly found in TCM materials include organochlorine, organophosphorus, pyrethroid and carbamate pesticides20, 21. Although organochlorine pesticides have been banned for many years, their residues may still exist in TCM because of their stable nature. Moreover, these residues are uneasy to decompose and can be stored in water, soil or biological organisms for a long time. Long-term use of TCM may lead to exposure to pesticide residues beyond safety limits, resulting in bioaccumulation and poisoning. Some cause-and-effect relationships of pesticides (e.g., arsenide and organochlorine) have been clearly established. Epidemiological investigations showed that the risk of cancer is increasing in rural areas, including leukemia, malignant brain tumor, testicular cancer, multiple myeloma and lymphoma. Washing methods can be used to remove residual water-soluble pesticides from the plants successfully. However, to remove most fat-soluble pesticides, which possess high biological attachment coefficient, strong penetrability, and can easily enter plant, washing is much ineffective.
In view of the above, the establishment of accurate, rapid and simple methods for safety monitoring of TCM materials is urgently needed.
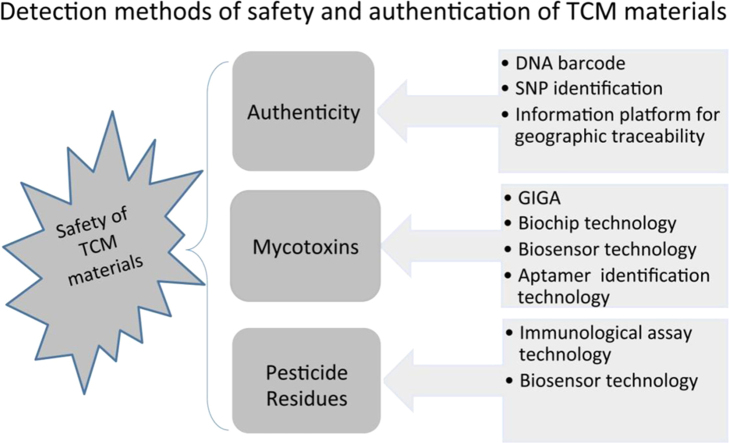
2. New rapid detection methods of the safety monitoring of TCM materials
2.1. Rapid detection of authenticity
2.1.1. DNA barcode
DNA barcode was proposed for the first time by Canadian zoologist Paul Hebert in 200322. It is a new molecular diagnostic technology that identifies species using a recognized standard short sequence in the genome. Gregory23 believed that global DNA barcode innovation research would become a “big science” program after the human genome project. Miller24 explained and popularized DNA barcoding in “the Renaissance of DNA barcode and taxonomy”. DNA barcode has become a global research highlight and direction for biological taxonomy in both academic journals and lay media. DNA barcoding technology has superceded the limitations imposed by traditional morphological identification methods. By establishing a TCM identification database, the digitalized DNA barcode moves TCM identification methods from morphological identification to molecular identification25.
Based on standardized DNA barcodes and universal primers, the DNA barcode method is universal. By comparing sequences among species, identification can be easily processed without the taxonomic knowledge of a specialist. DNA barcoding is not restricted by morphological characteristics and physiological conditions. Researchers can accurately determine the information of a species by analyzing DNA sequences. Chen et al.26 first developed ITS2 as a DNA barcoder of medicinal plants, established a plant barcode identification system that was mainly based on ITS2, and used psbA-trnH as a complementary barcode. This system has high identification efficiency in Rosaceae, Asteraceae, and many other families or genera27, 28, 29, 30, 31, 32, 33. Chen et al.34 also developed an animal barcode identification system, which was mainly based on CO1, and used ITS2 as acomplementary barcode. Chen et al. completed the construction of a standardized DNA barcode database of TCM materials and identification website (http://www.tcmbarcode.cn/en/). With this platform, rapid identification of original plants, pills, powder, tissues, or cells can be realized. The TCM barcode database will become permanent data that can be improved by adding new research sequences from taxonomists. Based on the database of the TCM barcode, Chen et al. designed the DNA barcode identification software for many companies to meet their requirements of rapid detection. Chen׳s research35, 36 proposed a new perspective for potential universal barcode sequence identification of all land plants, stimulating wide discussion. The proposed barcode ITS2 has become a hotspot in international plant barcode research.
DNA barcoding can achieve rapid, accurate and automated identification of species without material specificity. In basic laboratories, researchers use DNA barcoding to detect and identify a large number of samples of herbal materials. The whole process of rapid identification can be completed within 4 h, which meets the requirement of entry-exit inspection and quarantine where identification demands a rapid, high-throughput, sensitive, accurate test for TCM materials. DNA barcoding has very good application potential in toxic TCM identification. Our group have developed a special molecular method which could identify all the materials containing aristolochic acid (AA), and we have also designed special primer to identify all the TCM materials containing retronecinetype and otonecine-type pyrrolizidine alkaloids (PAs).
On the other hand, authentication of TCM and their adulterants were widely carried out by using species-specific PCR and microarray37, 38, 39. With the dramatic reduction in the cost of high-throughput sequencing, full-length sequencing of the chloroplast gene sequences can be used to find one suitable sequence as a DNA barcode or the entire chloroplast genome can be used as the ultra-barcode40, 41. Another alternative is the application of the single nucleotide polymorphism (SNP) method42.
2.1.2. SNP identification
Based on DNA barcoding analysis, Chen et al.42 detected two stable SNPs for Panax ginseng and Panax quinquefolius authentication, whereas Liao et al.43 obtained two SNPs for identifying Panax notoginseng. With the development of DNA barcoding, increasing amounts of SNPs have been discovered. A series of detection methods for SNPs has been explored, such as single-strand conformation polymorphism (SSCP) and invader assay with dual-color fluorescence polarization detection. However, all these methods have some shortcomings, such as complex methodology, time-consuming steps, and expensive instruments, all of which discourage wide implementation. Therefore, microarray-in-a-tube, gold nanoparticles (GNPs), and nucleic acid test strips have good potential for fast detection of SNPs.
2.1.2.1. Microarray-in-a-tube
Microarray-in-a-tube is a novel DNA microarray technique in which specific nucleic acid probes are immobilized on the inner surface of a converted Eppendorf tube cap. Different from conventional glass microarray, the probes are arranged on a plastic substrate by agarose film. An inner vessel to store the hybridization solution is placed in the sealed tube. After amplification, the tube can be inverted and hybridization can be performed without contamination44. Liu et al.45 successfully detected single-base mutations of HIV-1 resistance through microarray-in-a-tube. Considering the quenching ratio, single base mismatch discrimination ratio, and time-cost, Wang et al.46 proved that this method has an advantage over the traditional chip. Moreover, Liu et al.47 detected four respiratory tract viruses using microarray-in-a-tube accompanied with reverse transcription-PCR. The sensitivity of the system for virus detection can reach 102 copies/μL. Liu et al.45 prepared kits for several viruses. Microarray-in-a-tube can be used in the detection of similar clinical respiratory viruses, such as distinguishing the SARS virus from other viruses. The major advantages of the method are multi-virus detection and elimination of contamination.
Currently, microarray-in-a-tube is a fast and feasible technology in the detection of SNPs. Based on the theory, SNPs in TCM can be detected through microarray-in-a-tube. Investigations on SNP detection kits for P. ginseng, P. quinquefolius, Ophiocordyceps sinensis, and other expensive medicinal materials are currently underway.
2.1.2.2. GNP technology
GNP is an SNP detection method based on color reaction. It is based on a method in which single- or double-stranded DNA has different electrostatic interactions among the GNPs. GNP is based on the characteristic that DNA bases are strictly complementary paired and hybridized to form double-stranded DNA; it uses the color change or aggregation condition as the signal to determine whether the detected sequence of a target gene has mutations or not48. Given that GNPs have unique physical and chemical properties, this methodology can improve the accuracy and stability of biological detection.
In recent years, the use of GNPs for gene mutation detection and analysis of SNPs has shown rapid development in the research field. Many scholars have contributed to the methods of gene mutation and SNP detection by GNPs49, 50, 51, 52, 53, 54, 55, 56. For example, Rothberg designed a new method according to the dynamics of double-stranded DNA melting, and their results showed that the method can detect a minimum of 100 fmol of target DNA within 5 min. Their probes did not need thiol modification and the PCR products did not require purification, so the detection steps were further simplified and the cost was reduced. Bao׳s studies57 have shown that GNP probes combined with gene chip have many advantages, such as simple operation, short operating time, specificity and high sensitivity. GNP technology is convenient in gene mutation detection and SNP analysis.
GNPs are easily prepared and stored. Their advantages include high detection sensitivity and simple observation. With this technology, the herb-specific detection probes, complementary target sequence, and oligonucleotide DNA with single-base mutation sequences can be designed according to SNP sites. At room temperature, detection probes are hybridized between complementary sequences and single-base mutation sequences in the sequence buffer. With the addition of NaCl solution, the GNP solution produces distinctly different color changes in two hybridization solutions. Thus, the authenticity of medicines can be identified in a fast, effective, and stable manner. GNPs do not require special markers, such as fluorescent dye or expensive equipment, so this technology can achieve low-cost, high-throughput, high sensitivity and high automation detection58. This method can be used for on-site testing of TCM materials.
2.1.2.3. A nucleic acid amplification test strip method
Isothermal nucleic acid amplification is a methodology that extends the length of target DNA sequences or increases their copy numbers at a specific temperature. Compared with PCR, this technique can conduct amplification in an isothermal period, thereby eliminating the requirements for instruments. Moreover, the temperature control system can be operated by a heating module, water bath, or other simple instruments. Hangzhou USTAR Bio-tech Limited developed a fast detection technique for SNPs. This technique contains one-step PCR and a nucleic acid test strip, which is a detection method for specific extension products. In this technique, the regions containing SNPs are amplified unspecifically at first. The SNP sites are then specifically amplified by allele-specific PCR. Finally, the specific amplification products are detected by a nucleic acid test strip59.
Wang et al.60 designed a suite of loop-mediated isothermal amplification primers for the sequences of the exogenous gene Cry1ab/ac in Bt-transgenic crops. Given the method׳s high reliability, specificity and stability, it can be used in the rapid on-site detection of Bt-transgenic crops. Qin et al.61 established cross-priming amplification for the rapid detection of Vibrio cholerae, and the method was proved to have high specificity and stability. Wang et al.62 developed a new method for the rapid detection of mtDNA G1178A mutation based on the SNP test strip; the result of the SNP test strip was identical to that of DNA sequencing. Zhang et al.63 used this method to detect Mycobacterium tuberculosis in sputum, and found that this method is fast (within 2 h), sensitive, and easy to operate (it does not rely on expensive equipment). Zhang et al.64 developed a nucleic acid test strip method to detect Bursaphelenchus xylophilus, and proved that the method can be applied in the rapid identification of B. xylophilus in entry-exit inspection and quarantine. The test strip technique has also been reported in other fields of biology65, 66, 67, 68.
The author׳s research group has established a sophisticated database of the DNA barcode of TCM plants. More and more SNP(s) have been discovered for identification. Using specific-primer isothermal PCR combined with nucleic acid test strip technique, we can design specific rapid detection kits for some TCM materials, allowing the direct detection of amplification results without electrophoresis, PCR, and other expensive equipment. Therefore, fast on-site detection can be conducted.
2.1.3. Information platform for geographic traceability of TCM
Chinese herbal medicine traceability technology permits circulation information on herbal medicines to be recorded and traced69. This technology ensures the safe use of herbal medicines. Barcode technology is presently the main technology for traceability. A barcode is a set of graphics arranged by certain encoding rules for storing information, and it can be divided into 1D and 2D barcodes. Each code system has its own specific character set and validation functions70. Compared with the 1D barcode, the 2D barcode is extensively used in many fields because it can store large data and encode numbers, letters, and characters. The 2D barcode systems have become popular for media, traceable security system, business cards, social networking, marketing, and electronic payments in China71, 72. Several studies have applied the 2D barcode in food traceability70, 73, 74, 75. 2D barcode not only can be used for the origin traceability of food and herbal medicine, but also can be used in tracking entire production progress76. Yan et al.77 applied the 2D barcode for GAP production progress of herbal medicine, and developed 2D barcode-based GAP production patterns. Jin et al.78 provided a new method of medicinal slice warehousing by 2D barcode-based medicinal slice logistics management. Liu et al.79 successfully converted DNA barcoding information into 2D barcodes. We have developed an automated process of DNA barcode sequences by converting them into colorful 1D barcodes and 2D barcodes. Users can conveniently use mobile terminals, such as mobile phones, to obtain DNA barcode sequences by scanning barcode images, and submit the sequences to the world׳s largest DNA barcode database of TCM (http://www.tcmbarcode.cn/en/) for analysis. Barcode traceability technology is often combined with databases and networks. The barcode acts as a carrier for information transfer and network as a bridge in information flow, and the database acts as a warehouse for traceable information storage. Information of each phase of circulation is stored in a database through the network and converted into a 2D barcode, whereas traceability information can be obtained by scanning a 2D barcode image and searching the database. This combination is convenient for recording and management of information, and can achieve rapid transmission and information retrieval. Considering the popularity of smartphones, 2D barcode traceability technology no longer needs to rely on a specific barcode reading machines or software. The advantages and disadvantages of these fast identification methods were listed in (Table 1).
Table 1.
Comparison of advantages and disadvantages of different detection methods.
| Identification method | Advantage | Disadvantage | Scope of application in TCM |
|---|---|---|---|
| DNA barcoding | Sequence-based | Require advanced equipment | All medicinal plants and animals |
| Very high accuracy | Need comparison in database | ||
| Universal | |||
| Primer-based | |||
| High universality | |||
| Rapid | |||
| Low cost | |||
| Microarray in a tube | SNP detection | Need specific primer | Some closely related species |
| On-site detection | Not universal | ||
| Not require advanced equipment | |||
| After method have been established, single test cost lower than CNY 2.00 | |||
| Database-free | |||
| Can be developed into a species specific kit | |||
| Nano-Au | High accuracy | Need specific primer | Some closely related species |
| SNP detection | Not universal | ||
| On-site detection | |||
| Not require advanced equipment | |||
| Low cost | |||
| Database-free | |||
| Can be developed into a species specific kit | |||
| Nucleic acid amplification test strip | High accuracy | Need specific primer | Some closely related species |
| SNP detection | Not universal | ||
| On-site detection | |||
| Not require advanced equipment | |||
| Low cost | |||
| Database-free | |||
| Can be developed into a species specific kit | |||
| Traceability system | High accuracy | Need database | All medicinal plants and animals |
| High universality | |||
| Very rapid | |||
| Not require advanced equipment | |||
| Low cost |
2.2. Rapid detection technology of mycotoxins
To ensure the quality, safety and efficacy of different products, various analytical techniques have been applied for the detection of mycotoxins in foodstuffs, medicinal plants and their derivative products. Conventional analytical methods of mycotoxin detection involve chromatographic analyses, such as TLC, HPLC, GC, and more recently, techniques such as LC/MS and GC/MS. Most of these methods have high sensitivity, and they were developed for quantitative and qualitative analyses of mycotoxins. To monitor and control the contamination of mycotoxins, rapid detection technology has become the research focus because of its relative simplicity, convenience, accuracy, and efficiency.
2.2.1. Gold immunochromatographic assay (GICA)
GICA is a solid-phase marker immunoassay technique that combines colloidal gold labeling technology and immunoassay, with chromatography analysis technology. The technique not only has the characteristics of good stability and low-cost but also has intuitive and reliable results, suitable for semi-quantitative and quantitative rapid detection of mycotoxins. Nowadays, gold nanoparticles have been extensively employed as immobilizing different biological receptors, e.g., enzyme, DNA, antigen/antibody and other biomolecules. Wang et al.80 found that colloidal gold immunochromatographic dual strip can rapidly and accurately detect samples containing zearalenone and fumonisin B1. The limit of detection (LOD) for fumonisin B1 is 1.0 ng/mL. Multiple testing based on immune colloidal gold test strip has become a new trend for the detection of mycotoxins by immunological methods81.
GIGC is an ideal selection in biotechnological systems because of its inherent advantages (e.g., easy preparation and good biocompatibility). However, owing to the limitation of antigenic epitope on the small biomolecules and a narrow linear range, innovative and powerful techniques are being developed for the amplification of detectable signal. The present limitations of GICA include the small size of the biomolecule as epitope and narrow linear range of assays.
2.2.2. Biochip technology
Biochip is a new technology developed through life science and microelectronics in recent years. It has incomparable advantages compared with conventional methods including high-throughput, multi-parameter synchronization analysis; fully automatic, rapid analysis; high accuracy and sensitive analysis. In antibody chip technology, various mycotoxins and other hazardous chemicals can be monitored simultaneously, thereby greatly reducing the time of sample extraction and detection and improving efficiency. Wang et al.82 designed an immunochip to simultaneously quantify the contents of AFB1, AFM1, DON, OTA, T-2 toxin, and ZON and the present LODs majorly showed relatively lower.
Although biochip technology has a lot of advantages in advanced optical biosensors for sensing analytes, this technique is expensive in analytical cost, and requires complex labeling process and professional technicians with specialized equipment.
2.2.3. Biosensor technology
Biosensor technology, a prominent technique for preliminary screening of the toxicity of samples, can be defined as having a sensing element for the selective detection of a target and method to transduce the interaction as a measureable signal83. The biosensor׳s high sensitivity, selectivity, low cost, simplicity, miniaturization, portability and integration in automated devices make it a reliable and usable alternative for monitoring mycotoxins84.
Numerous types of biosensors have been developed. Sapsford et al.85 detected the content of AFB1 employed by Array Biosensor, and reported a low LOD of 0.3 ng/mL. Wang et al.86 presented a novel suspension array technology for quantifying AFB1, DON, T-2 toxin and ZEN simultaneously and quantitatively in corn and peanut, and the levels of LOD were better than those obtained using HPLC. Yuan et al.87 designed a surface plasmon resonance biosensor to detect OTA directly, and reported a dramatically improved LOD of 0.042 ng/mL.
In conclusion, these biosensors can achieve the sensitivity and selectivity required for the very strict regulatory limits from legislation. Nowadays, its stability, reproducibility and life-time coupled with the differences in tested samples, less species and instability of antibody, many studies on the biosensors are still at the experimental stage.
2.2.4. Aptamer identification technology
Aptamers, single-stranded oligonucleotides of DNA or RNA sequences, can specifically bind to target molecules in a complex matrix. Aptamer technologies have been widely applied in the analysis of mycotoxins. Yang et al.88 presented the colorimetric detection of OTA using OTA׳s aptamer and unmodified GNPs. In addition, Wang et al.89 developed a new type of structure-switching aptasensor to simultaneously detect OTA and FB1.
Aptamers have the ability under certain physicochemical conditions to fold into defined three-dimensional conformations, which facilitate specific interaction with target molecules having high affinity constants. Based on novel, molecular recognition, aptamer identification technology has great potential for application in the rapid detection of mycotoxins because of high accuracy, precision and specificity. The combination of aptamer recognition techniques and novel nano-materials has been used in various optical and electrochemical analytical methods for mycotoxin analysis.
Consequently, aptamers have emerged owing to inherent advantages compared with antibodies, such as non-requirement of immunization from animals, more chemical and thermal stability, and less variability. Particularly, aptamers are not susceptible to denaturation in the presence of solvents commonly used in the extraction of mycotoxins.
2.3. Rapid detection technology of pesticide residues
Classical instrumental analytical techniques for the determination of pesticides in a large number of samples have been developed. These methods involve GC and LC coupled with various detectors90, 91, 92, GC-MS93, 94, or LC-MS91, 95. In recent years, GC-MS/MS has been intensively used for the determination of pesticides96, 97. Although chromatography-based methods are sensitive and reliable, they require sophisticated equipment, skilled analysts, and time-consuming sample preparation steps. Moreover, organic solvents used in the detection process may lead to environmental pollution.
2.3.1. Immunological assay technology
As a promising method for selective and sensitive analysis, immunoassays have become indispensable analytical tools in a wide range of applications. Immunological methods, which are suitable for both laboratory and field analyses, provide a unique opportunity to screen large numbers of samples quickly and effectively.
Traditional immunoassays, such as enzyme-linked immunosorbent assay (ELISA), are commonly used in the field. ELISAs are invariably considered the gold standard for single analyte measurement. The sensitivity of ELISA is relatively high, but these methods have some drawbacks, including numerous washing and preparation steps, large sample volumes, small surface area, and long diffusion times required for antigen-antibody binding98.
In particular, immunoassay techniques would be a specific, sensitive, rapid and economical analytical tool, but they require extensive pipetting, washing, and incubation. Thus an immunoassay is time-consuming and has limited application in fast detection.
2.3.2. Biosensor technology
Biosensors typically consist of a biological receptor in intimate contact with an electrochemical, optical, gravimetric, or thermal transducer. Biosensors may provide solutions to some of the problems encountered in the measurement of pesticides. As an alternative, a variety of biosensors, biological receptors with selective affinity toward a specific pesticide, have been developed99.
Ferentions et al.100 developed a novel artificial neural network combined with a bioelectric cellular biosensor to detect and classify correctly the presence of investigated pesticide groups with an overall success rate of 83.6%. Qie et al.101 reported that thermometric biosensors evidently simplify sample pretreatment, and exhibit a potentially powerful capability for fast quantitative analysis of pesticide residues. The short assay time per sample of biosensors makes them suitable for the fast detection of large-scale samples.
Biosensor offers a variety of benefits including high selectivity and rapid test. However, in order to get wide use of biosensor technology, its stability, accuracy, and repeatability remains to be further improved.
3. Conclusions and prospects
The quality control of medicinal materials has always been a weak link in the Chinese medicine industry, and it has affected the sustainable development of the TCM industry. Low-cost, on-site fast detection of TCM with the capability of easy operation and large-scale implementation is an urgent must, which depends on not only the availability of portable equipment, but also simplifying the techniques and promoting new detection principles. The future development of the rapid testing of Chinese medicine have three goals: (1) exploring mobile species identification systems and developing kits for SNP detection based on GNPs, microarray-in-a-tube, and nucleic acid amplification test strip; (2) establishing a fast detection system for mycotoxins and pesticide residues of Chinese medicine; (3) establishing a TCM traceability system in China, using some specific kits unlimited from the environment and infrastructure, which can satisfy the requirements for fast detection in remote and undeveloped areas.
3.1. Setting up mobile species identification system (2D DNA barcoding)
Given the rapid development in DNA barcoding, all species are expected to have a unique DNA barcode in the future. Hence, the technique of DNA barcoding has enormous potential applications. The application of handheld rapid species identification equipment will bring great convenience in entry-exit inspection and quarantine work. Compared with traditional DNA sequences (about 200–600 bp) the 2D code has larger information capacity and adjustable size, which is suitable for direct labeling of samples. The 2D code contains more DNA information. Moreover, it will be easier to operate and be more convenient to collect and conduct remote authentication for 2D code using a communication terminal camera with scanner function and wireless communication function. A species identification system within a mobile application will make authentication more convenient and rapid. The DNA barcode for species identification is a global method that enables the identification of TCM to form a unified international standard. It can greatly improve the identification level of Chinese medicinal materials by establishing a Chinese herbal medicine DNA barcode database. Moreover, this global method can speed up the process of modernization of TCM, and provide a scientific basis for the control of raw TCM materials. This method is of great significance to the internationalization of the Chinese herbal medicine identification standard.
3.2. Setting up a fast detection system of mycotoxin and pesticide residue in the production of TCM
The design of a rapid detection platform is the primary problem in the quality control of TCM. To set up a fast detection system of mycotoxins and pesticide residues in the production of TCM, the management of TCM supervision departments and other relevant units should select different methods for analyzing various mycotoxins according to their detection technology and detection means to control the quality and safety of TCM. Future research should focus on the identification and quantification of AFs (AFG2, AFG1, AFB2, AFB1), OTA, DON, ZEN, fumonisins (FB1, FB2) and citrinin. According to their detection technology and detection means, the selection of different analytical methods of pesticide residues for the control of the quality and safety of TCM should meet the social demand for rapid identification of medicinal species. Analytical methods must meet the drug test, customs, security functions, and companies׳ demands for rapid identification of medicinal species, which will ensure the safety and efficacy of TCM.
3.3. Setting up the traceability of TCM
Blind transplanting of medicinal materials has led to the decline in medicinal quality and counterfeit medicines in production or sales, which may increase the risk of accidents in TCM102. Establishing herb medicine traceability can not only protect the authenticity of a drug, but also manage the information of seed, planting, processing, and marketing. A traceable system has great application potential in the Chinese herb medicine industry. In consideration of cost and rapidity, barcode traceability can be considered a main technology tool. Once safety problems occur, potential problems can be traced and further characterized by molecular biology or fingerprinting.
Acknowledgments
This project was supported by National Natural Science Foundation of China (No. 81473303) and Grants from the Major Scientific and Technological Special Project for ‘Significant New Drugs Creation’ (No. 2014ZX09304307001).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Contributor Information
Jianping Han, Email: jphan@implad.ac.cn.
Shilin Chen, Email: slchen@implad.ac.cn.
References
- 1.Ng KY, Cheng CL, Xu HX. Safety issues of Chinese medicine: a review of intoxication cases in Hong Kong. Chin Herb Med. 2009;1:29–39. [Google Scholar]
- 2.Dickens P, Tai YT, But PPH, Tomlinson B, Ng HK, Yan KW. Fatal accidental aconitine poisoning following ingestion of Chinese herbal medicine: a report of two cases. Forensic Sci Int. 1994;67:55–58. doi: 10.1016/0379-0738(94)90412-x. [DOI] [PubMed] [Google Scholar]
- 3.Chan TY. Incidence of herb-induced aconitine poisoning in Hong Kong: impact of publicity measures to promote awareness among the herbalists and the public. Drug Saf. 2002;25:823–828. doi: 10.2165/00002018-200225110-00006. [DOI] [PubMed] [Google Scholar]
- 4.Chan TY. Aconitine poisoning: a global perspective. Vet Hum Toxicol. 1994;36:326–328. [PubMed] [Google Scholar]
- 5.Deng JF, Lin TJ, Kao WF, Chen SS. The difficulty in handling poisonings associated with Chinese traditional medicine: a poison control center experience for 1991–1993. Vet Hum Toxicol. 1997;39:106–114. [PubMed] [Google Scholar]
- 6.Poon WT, Lai CK, Ching CK, Tse KY, So YC, Chan YC. Aconite poisoning in camouflage. Hong Kong Med J. 2006;12:456–459. [PubMed] [Google Scholar]
- 7.Lo SHK, Mo KL, Wong KS, Poon SP, Chan CK, Lai CK. Aristolochic acid nephropathy complicating a patient with focal segmental glomerulosclerosis. Nephrol Dial Transplant. 2004;19:1913–1915. doi: 10.1093/ndt/gfh159. [DOI] [PubMed] [Google Scholar]
- 8.Kumana CR, Ng M, Lin HJ, Ko W, Wu PC, Todd D. Hepatic veno-occlusive disease due to toxic alkaloid in herbal tea. Lancet. 1983;322:1360–1361. doi: 10.1016/s0140-6736(83)91112-1. [DOI] [PubMed] [Google Scholar]
- 9.Yang MH. Research progress in fungi and mycotoxin infection of medicinal plants and their products. Guizhou Agric Sci. 2008;36:59–63. [Google Scholar]
- 10.Yang MH, Chen JM, Zhang XH. Immunoaffinity column clean-up and liquid chromatography with post-column derivatization for analysis of aflatoxins in traditional Chinese medicine. Chromatographia. 2005;62:499–504. [Google Scholar]
- 11.Liu SY, Qiu F, Yang MH. Determination of aflatoxins in nelumbinis semen by immunoaffinity column clean-up and HPLC-FLD with on-line post-column photochemical derivatization and LC-MS/MS confirmation. China J Chin Mater Med. 2012;37:305–309. [PubMed] [Google Scholar]
- 12.Wei RW, Yang XL, Qiu F, Yang MH, Qin JP. Simultaneous determination of aflatoxin B1, B2, G1, G2 and ochratoxin A in Glycyrrhiza uralensis by HPLC-fLD after immunoaffinity column with online post-column photochemical derivatization. China J Chin Mater Med. 2011;36:2342–2346. [PubMed] [Google Scholar]
- 13.Wei RW, Qiu F, Kong WJ, Wei JH, Yang MH, Luo ZL. Co-occurrence of aflatoxin B1, B2, G1, G2 and ochrotoxin A in Glycyrrhiza uralensis analyzed by HPLC-MS/MS. Food Control. 2013;32:216–221. [Google Scholar]
- 14.Yang L, Wang L, Pan J, Xiang L, Yang M, Logrieco AF. Determination of ochratoxin A in traditional Chinese medicinal plants by HPLC-FLD. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2010;27:989–997. doi: 10.1080/19440041003647122. [DOI] [PubMed] [Google Scholar]
- 15.Yue YT, Zhang XF, Pan JY, Zhen OY, Wu J, Yang MH. Determination of deoxynivalenol in medicinal herbs and related products by GC-ECD and confirmation by GC-MS. Chromatographia. 2010;71:533–538. [Google Scholar]
- 16.Kong WJ, Li JY, Qiu F, Wei JH, Xiao XH, Zheng Y. Development of a sensitive and reliable high performance liquid chromatography method with fluorescence detection for high-throughput analysis of multi-class mycotoxins in Coix seed. Anal Chim Acta. 2013;799:68–76. doi: 10.1016/j.aca.2013.08.042. [DOI] [PubMed] [Google Scholar]
- 17.Pietri A, Rastelli S, Bertuzzi T. Ochratoxin A and aflatoxins in liquorice products. Toxins. 2010;2:758–770. doi: 10.3390/toxins2040758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrera M, Herrera A, Ariño A. Estimation of dietary intake of ochratoxin A from liquorice confectionery. Food Chem Toxicol. 2009;47:2002–2006. doi: 10.1016/j.fct.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Ariño A, Herrera M, Estopañan G, Juan T. High levels of ochratoxin A in licorice and derived products. Int J Food Microbiol. 2007;114:366–369. doi: 10.1016/j.ijfoodmicro.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Rong WG, Guo H, Yang H. Current research status in China on pesticide contamination of plant material used in making Chinese herbal medicines. Agrochemicals. 2006;45:302–308. [Google Scholar]
- 21.Miao Q, Kong WJ, Wei JH, Yang SH, Yang MH. Analysis and effective control of pesticides residues in traditional Chinese medicine. Chin J Pestic Sci. 2012;14:363–370. [Google Scholar]
- 22.Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schindel DE, Miller SE. DNA barcoding a useful tool for taxonomists. Nature. 2005;435:17. doi: 10.1038/435017b. [DOI] [PubMed] [Google Scholar]
- 24.Miller SE. DNA barcoding and the renaissance of taxonomy. Proc Natl Acad Sci USA. 2007;104:4775–4776. doi: 10.1073/pnas.0700466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen SL, Pang XL, Song JY, Shi LC, Yao H, Han JP. A renaissance in herbal medicine identification: from morphology to DNA. Biotechnol Adv. 2014;32:1237–1244. doi: 10.1016/j.biotechadv.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Chen SL, Yao H, Han JP, Liu C, Song JY, Shi LC. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 2010;5:e8613. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pang XH, Song JY, Zhu YJ, Xie CX, Chen SL. Using DNA barcoding to identify species within Euphorbiaceae. Planta Med. 2010;76:1784–1786. doi: 10.1055/s-0030-1249806. [DOI] [PubMed] [Google Scholar]
- 28.Han JP, Shi LC, Li MH, Yao H, Song JY, Xu HX. Relationship between DNA Barcoding and chemical classification of Salvia L. medicinal herbs. Planta Med. 2009;75:416. [Google Scholar]
- 29.Gao T, Yao H, Song JY, Liu C, Zhu YJ, Ma XY. Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J Ethnopharmacol. 2010;130:116–121. doi: 10.1016/j.jep.2010.04.026. [DOI] [PubMed] [Google Scholar]
- 30.Luo K, Chen SL, Chen KL, Song JY, Yao H, Ma XY. Assessment of candidate plant DNA barcodes using the Rutaceae family. Sci China Life Sci. 2010;53:701–708. doi: 10.1007/s11427-010-4009-1. [DOI] [PubMed] [Google Scholar]
- 31.Pang XH, Song JY, Zhu YJ, Xu HX, Huang LF, Chen SL. Applying plant DNA barcodes for Rosaceae species identification. Cladistics. 2011;27:165–170. doi: 10.1111/j.1096-0031.2010.00328.x. [DOI] [PubMed] [Google Scholar]
- 32.Han JP, Shi LC, Chen XC, Lin YL. Comparison of four DNA barcodes in identifying certain medicinal plants of Lamiaceae. J Syst Evol. 2012;50:227–234. [Google Scholar]
- 33.Gao T, Yao H, Song JY, Zhu YJ, Liu C, Chen SL. Evaluating the feasibility of using candidate DNA barcodes in discriminating species of the large Asteraceae family. BMC Evol Biol. 2010;10:324. doi: 10.1186/1471-2148-10-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao H, Song JY, Liu C, Luo K, Han JP, Li Y. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One. 2010;5:e13102. doi: 10.1371/journal.pone.0013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han JP, Zhu YJ, Chen XC, Liao BS, Yao H, Song JY. The short ITS2 sequence serves as an efficient taxonomic sequence tag in comparison with the full-length ITS. Biomed Res Int. 2013;2013:741476. doi: 10.1155/2013/741476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf M, Chen SL, Song JY, Ankenbrand M, Müller T. Compensatory base changes in ITS2 secondary structures correlate with the biological species concept despite intragenomic variability in ITS2 sequences---a proof of concept. PLoS One. 2013;8:e66726. doi: 10.1371/journal.pone.0066726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carles M, Cheung MKL, Moganti S, Dong TTX, Tsim KW, Ip NY. A DNA microarray for the authentication of toxic traditional Chinese medicinal plants. Planta Med. 2005;71:580–584. doi: 10.1055/s-2005-864166. [DOI] [PubMed] [Google Scholar]
- 38.Tsoi PY, Woo HS, Wong MS, Chen SL, Fong WF, Xiao PG. Genotyping and species identification of Fritillaria by DNA chips. Acta Pharm Sin. 2003;38:185–190. [PubMed] [Google Scholar]
- 39.Che J, Tang L, Liu Y, He W, Chen F. Molecular identity of Crocus sativus and its misused substitutes by ITS sequence. China J Chin Mater Med. 2007;32:668–671. [PubMed] [Google Scholar]
- 40.Li XW, Yang Y, Henry RJ, Rossetto M, Wang YT, Chen SL. Plant DNA barcoding: from gene to genome. Biol Rev. 2014;90:157–166. doi: 10.1111/brv.12104. [DOI] [PubMed] [Google Scholar]
- 41.Li QS, Li Y, Song JY, Xu HB, Xu J, Zhu YJ. High-accuracy de novo assembly and SNP detection of chloroplast genomes using a SMRT circular consensus sequencing strategy. New Phytol. 2014;204:1041–1049. doi: 10.1111/nph.12966. [DOI] [PubMed] [Google Scholar]
- 42.Chen XC, Liao BS, Song JY, Pang XH, Han JP, Chen SL. A fast SNP identification and analysis of intraspecific variation in the medicinal Panax species based on DNA barcoding. Gene. 2013;530:39–43. doi: 10.1016/j.gene.2013.07.097. [DOI] [PubMed] [Google Scholar]
- 43.Liao BS, Han JP, Chen XC, Chen SL, inventors; Chinese Academy of Medical Sciences Institute of Medicinal Plant Development, assignee. Method for rapid identification of Ophiocordyceps Sinensis. China Patent CN102888456A. 2013 Jan 23.
- 44.Lu ZH, Liu QJ, Wang H, inventors; Southeast University, assignee. Flushing-free PCR amplification tube capable of directly detecting gene. China patent CN1448500. 2003 Oct 15.
- 45.Liu QJ. Development of three novel integrated DNA microarrays and detection instruments [dissertation]. Nanjing: Southeast University; 2006
- 46.Wang H, Li J, Liu HP, Liu QJ, Mei Q, Wang YJ. Label-free hybridization detection of a single nucleotide mismatch by immobilization of molecular beacons on an agarose film. Nucleic Acids Res. 2002;30:e61. doi: 10.1093/nar/gnf061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu QJ, Bai YF, Ge QY, Zhou SX, Wen T, Lu ZH. Microarray-in-a-tube for detection of multiple viruses. Clin Chem. 2007;53:188–194. doi: 10.1373/clinchem.2006.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang JF, Sun LP, Li H, Wang XY, Zhang QQ. Gold nanoparticles application in gene mutation detection and SNP analysis. Chin J Biochem Mol Biol. 2008;24:489–495. [Google Scholar]
- 49.Bao YP, Huber M, Wei TF, Marla SS, Storhoff JJ, Müller UR. SNP identification in unamplified human genomic DNA with gold nanoparticle probes. Nucleic Acids Res. 2005;33:e15. doi: 10.1093/nar/gni017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charrier A, Candoni N, Liachenko N, Thibaudau F. 2D aggregation and selective desorption of nanoparticle probes: a new method to probe DNA mismatches and damages. Biosens Bioelectron. 2007;22:1881–1886. doi: 10.1016/j.bios.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 51.Li JS, Chu X, Liu YL, Jiang JH, He ZM, Zhang ZW. A colorimetric method for point mutation detection using high-fidelity DNA ligase. Nucleic Acids Res. 2005;33:e168. doi: 10.1093/nar/gni163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pang LL, Li JS, Jiang JH, Shen GL, Yu RQ. DNA point mutation detection based on DNA ligase reaction and nano-Au amplification: a piezoelectric approach. Anal Biochem. 2006;358:99–103. doi: 10.1016/j.ab.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 53.Zhao WA, Chiuman W, Brook MA, Li YF. Simple and rapid colorimetric biosensors based on DNA aptamer and noncrosslinking gold nanoparticle aggregation. ChemBioChem. 2007;8:727–731. doi: 10.1002/cbic.200700014. [DOI] [PubMed] [Google Scholar]
- 54.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 55.Mao X, Xu H, Zeng QX, Zeng LW, Liu GD. Molecular beacon-functionalized gold nanoparticles as probes in dry-reagent strip biosensor for DNA analysis. Chem Commun. 2009;21:3065–3067. doi: 10.1039/b822582f. [DOI] [PubMed] [Google Scholar]
- 56.Xia F, Zuo XL, Yang RQ, Xiao Y, Kang D, Vallée-Bélisle A. Colorimetric detection of DNA, small molecules, proteins, and ions using unmodified gold nanoparticles and conjugated polyelectrolytes. Proc Natl Acad Sci USA. 2010;107:10837–10841. doi: 10.1073/pnas.1005632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bao H. Detection of gene mutation using DNA-gold nanoparticle probes and gene chips [dissertation]. Shanghai: East China Normal University; 2009
- 58.Guo QC, Wang X, Lou XH, Mao HJ, Jia J, Jin QH. Label-free colorimetric assay based on gold nanoparticles in detection of NOS1AP with single nucleotide polymorphism. Chem J Chin Univ. 2010;31:1965–1969. [Google Scholar]
- 59.Hu L, Xu GL, You QM, Wang HY, Gao Q, Zhang WH, et al., inventors; Ustar Biotechnologies (Hangzhou) Ltd., assignee. Method and kit for rapidly detecting single nucleotide polymorphisms (SNPs). China Patent CN102618626A. 2012 Aug 1.
- 60.Wang L, Zhao YZ, Luo Y, Zhou Q, Lai PA, Zhang XD. Rapid detection of transgenic Bt crops by isothermal nucleic amplification. J Inspect Quar. 2011;21:11–15. [Google Scholar]
- 61.Qin Q, Zhu JL. Establishment and optimization of CPA-nucleic acid test strip for rapid detection of vibrio cholerae. Biotechnol Bull. 2013;7:167–171. [Google Scholar]
- 62.Wang H, Hu L, You Q, Zhou XT, Lü XS, Zhai J. A rapid detection of mtDNA G11778A mutation by a SNP-Strip. Chin J Opt Ophthalmol. 2006;8:356–366. [Google Scholar]
- 63.Zhang JL, Li GG, Dong B, Bai QC, Qiao LL. Isothermal amplification test strips for rapid detection of Mycobacterium tuberculosis in sputum specimens. J Med Pest Control. 2013;29:1093–1094. [Google Scholar]
- 64.Zhang YJ, Wang JC, Wei YD. Visual nucleic acid test strips for rapid detection of Bursaphelenchus xylophilus. Plant Prot. 2013;39:94–98. [Google Scholar]
- 65.Fang R, Li X, Hu L, You Q, Li J, Wu J. Cross-priming amplification for rapid detection of Mycobacterium tuberculosis in sputum specimens. J Clin Microbiol. 2009;47:845–847. doi: 10.1128/JCM.01528-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu LT, Curran MD, Ellis JS, Parmar S, Ritchie AV, Sharma PI. Nucleic acid dipstick test for molecular diagnosis of pandemic H1N1. J Clin Microbiol. 2010;48:3608–3613. doi: 10.1128/JCM.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang YL, Li XY, Pan KY, Zhong HY. Development and application of a rapid nucleic acid diagnostic strip for influenza A (H1N1) virus. Chin J Nosocomiol. 2011;21:2871–2873. [Google Scholar]
- 68.Wang L, Luo Y, Zhou Q, Lai PA, Zhang XD, Bai YD. Application of nucleic acid strips in the detection of transgenic EPSPS crops. Lett Biotechnol. 2011;22:238–242. [Google Scholar]
- 69.Liao BS, Song JY, Xie CX, Han JP, Chen SL. Study on traceability system of genuine medicinal materials. China J Chin Mater Med. 2014;39:3881–3888. [PubMed] [Google Scholar]
- 70.Sheng LM, Wei XT. Application of two-dimensional code in the traceability of agricultural products. Mod Agric Sci Technol. 2013;18:330–332. [Google Scholar]
- 71.Wang Y. Application and analysis of two-dimensional code dissemination of information [dissertation]. Taiyuan: Shanxi University; 2013
- 72.Jin K. Study on two-dimensional code cousumers behavior [dissertation]. Shanghai: Shanghai Normal University; 2013
- 73.Shi LM, Guo CZ, Gai ZH, Chen ZF. Design and implementation of green food traceability system based on two-dimension code. Manuf Autom. 2013;16:144–146. [Google Scholar]
- 74.Lu CH, Wang LF, Hu YN, Bai YF, Bai HW, Wang R. Identification and traceability in animals and animal products. Jiangsu J Agric Sci. 2009;25:197–202. [Google Scholar]
- 75.Fan WC. Research on information identification application in milk product traceability system [dissertation]. Nanjing: Nanjing University; 2012
- 76.Xie M. Grape traceability system for the production process based on Android operating system [dissertation]. Hangzhou: Zhejiang Univesity; 2013
- 77.Yan LH, Luo ZH, Yang JY. Applications of Chinese herbal medicines GAP mode of production based on two-dimensional code technology. Chin J Tradit Med Sci Technol. 2014;2014:286–287. [Google Scholar]
- 78.Jin L, Zhang J, Shen F, Huang Y, Wu YK. Application of ePS platform in logistics management of small package of TCM decoction pieces storehouse. China Pharm. 2013;24:271–272. [Google Scholar]
- 79.Liu C, Shi LC, Xu X, Li H, Xing H, Liang D. DNA barcode goes two-dimensions: DNA QR code web server. PLoS One. 2012;7:e35146. doi: 10.1371/journal.pone.0035146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang YK, Shi YB, Zou Q, Sun JH, Chen ZF, Wang HA. Development of a rapid and simultaneous immunochromatographic assay for the determination of zearalenone and fumonisin B1 in corn, wheat and feedstuff samples. Food Control. 2013;31:180–188. [Google Scholar]
- 81.Feng X, Kong WJ, Yang MH, Ouyang Z. Latest advancement for detection methods of mycotoxins in traditional Chinese medicine. World Sci Technol. 2012;14:1944–1952. [Google Scholar]
- 82.Wang Y, Liu N, Ning BA, Liu M, Lv ZQ, Sun ZY. Simultaneous and rapid detection of six different mycotoxins using an immunochip. Biosens Bioelectron. 2012;34:44–50. doi: 10.1016/j.bios.2011.12.057. [DOI] [PubMed] [Google Scholar]
- 83.Campàs M, Prieto-Simón B, Marty JL. Biosensors to detect marine toxins: assessing seafood safety. Talanta. 2007;72:884–895. doi: 10.1016/j.talanta.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 84.Yeni F, Acar S, Polat ÖG, Soyer Y, Alpas H. Rapid and standardized methods for detection of foodborne pathogens and mycotoxins on fresh produce. Food Control. 2014;40:359–367. [Google Scholar]
- 85.Sapsford KE, Ngundi MM, Moore MH, Lassman ME, Shriver-Lake LC, Taitt CR. Rapid detection of foodborne contaminants using an Array Biosensor. Sens Actuators B:Chem. 2006;113:599–607. [Google Scholar]
- 86.Wang Y, Ning BA, Peng Y, Bai JL, Liu M, Fan XJ. Application of suspension array for simultaneous detection of four different mycotoxins in corn and peanut. Biosens Bioelectron. 2013;41:391–396. doi: 10.1016/j.bios.2012.08.057. [DOI] [PubMed] [Google Scholar]
- 87.Yuan J, Deng DW, Lauren DR, Aguilar MI, Wu YQ. Surface plasmon resonance biosensor for the detection of ochratoxin A in cereals and beverages. Anal Chim Acta. 2009;656:63–71. doi: 10.1016/j.aca.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 88.Yang C, Lates V, Prieto-Simón B, Marty JL, Yang XR. Aptamer-DNAzyme hairpins for biosensing of Ochratoxin A. Biosens Bioelectron. 2012;32:208–212. doi: 10.1016/j.bios.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 89.Wu SJ, Duan N, Ma XY, Xia Y, Wang HX, Wang ZP. Multiplexed fluorescence resonance energy transfer aptasensor between upconversion nanoparticles and graphene oxide for the simultaneous determination of mycotoxins. Anal Chem. 2012;84:6263–6270. doi: 10.1021/ac301534w. [DOI] [PubMed] [Google Scholar]
- 90.Liu HM, Kong WJ, Qi Y, Gong B, Miao Q, Wei JH. Streamlined pretreatment and GC-FPD analysis of multi-pesticide residues in perennial Morinda roots: a tropical or subtropical plant. Chemosphere. 2014;95:33–40. doi: 10.1016/j.chemosphere.2013.07.085. [DOI] [PubMed] [Google Scholar]
- 91.Miao Q, Kong WJ, Yang SH, Yang MH. Comparison of sample preparation methods combined with gas chromatography with electron-capture detection for the analysis of multipesticide residues in lotus seeds. J Sep Sci. 2013;36:2010–2019. doi: 10.1002/jssc.201300032. [DOI] [PubMed] [Google Scholar]
- 92.Liu QZ, Kong WJ, Qiu F, Wei JH, Yang SH, Zheng YG. One-step extraction for gas chromatography with flame photometric detection of 18 organophosphorus pesticides in Chinese medicine health wines. J Chromatogr B. 2012;885–886:90–96. doi: 10.1016/j.jchromb.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 93.Hu YC, Wan L, Zhang JM, Yang F, Cao JL. Rapid determination of pesticide residues in Chinese materia medica using QuEChERS sample preparation followed by gas chromatography–mass spectrometry. Acta Pharm Sin B. 2012;2:286–293. [Google Scholar]
- 94.Lee KG, Jo EK. Multiresidue pesticide analysis in Korean ginseng by gas chromatography-triple quadrupole tandem mass spectrometry. Food Chem. 2012;134:2497–2503. doi: 10.1016/j.foodchem.2012.04.094. [DOI] [PubMed] [Google Scholar]
- 95.Chen LN, Song FR, Liu ZQ, Zheng Z, Xing JP, Liu SY. Multi-residue method for fast determination of pesticide residues in plants used in traditional Chinese medicine by ultra-high-performance liquid chromatography coupled to tandem mass spectrometry. J Chromatogr A. 2012;1225:132–140. doi: 10.1016/j.chroma.2011.12.071. [DOI] [PubMed] [Google Scholar]
- 96.Pareja L, Fernández-Alba AR, Cesio V, Heinzen H. Analytical methods for pesticide residues in rice. TrAC, Trends Anal Chem. 2011;30:270–291. [Google Scholar]
- 97.Wilkowska A, Biziuk M. Determination of pesticide residues in food matrices using the QuEChERS methodology. Food Chem. 2011;125:803–812. [Google Scholar]
- 98.Zhang Q, Sun Q, Hu BS, Shen Q, Yang G, Liang X. Development of a sensitive ELISA for the analysis of the organophosphorous insecticide fenthion in fruit samples. Food Chem. 2008;106:1278–1284. [Google Scholar]
- 99.Marty JL, Garcia D, Rouillon R. Biosensors: potential in pesticide detection. TrAC, Trends Anal Chem. 1995;14:329–333. [Google Scholar]
- 100.Ferentinos KP, Yialouris CP, Blouchos P, Moschopoulou G, Kintzios S. Pesticide residue screening using a novel artificial neural network combined with a bioelectric cellular biosensor. BioMed Res Int. 2013;2013:813519. doi: 10.1155/2013/813519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qie ZW, Ning BA, Liu M, Bai JL, Peng Y, Song N. Fast detection of atrazine in corn using thermometric biosensors. Analyst. 2013;138:5151–5156. doi: 10.1039/c3an00490b. [DOI] [PubMed] [Google Scholar]
- 102.Hu SL. Genuine traditional Chinese medicine. Harbin: Heilongjiang Science and Technology Press; 1989