Significance
d polypeptides represent an attractive platform for biomedical applications because of their resistance to proteolytic degradation. However, the structural principles that underlie associations between L- and D-protein partners remain poorly understood because there has been very little atomic-resolution structural characterization of such heterochiral assemblies. Here we report two X-ray crystal structures of the racemic form of an α-helical peptide derived from the influenza M2 protein. Both structures contain large heterochiral coiled–coil interfaces. The ubiquity and regularity of coiled coils has inspired extensive design effort directed toward homochiral tertiary and quaternary structures, and we anticipate that the insights from these crystal structures will facilitate the design of an analogous rich set of heterochiral proteins and assemblies.
Keywords: d peptides, transmembrane peptides, racemic crystallization, racemic detergent, coiled coil
Abstract
Interactions between polypeptide chains containing amino acid residues with opposite absolute configurations have long been a source of interest and speculation, but there is very little structural information for such heterochiral associations. The need to address this lacuna has grown in recent years because of increasing interest in the use of peptides generated from d amino acids (d peptides) as specific ligands for natural proteins, e.g., to inhibit deleterious protein–protein interactions. Coiled–coil interactions, between or among α-helices, represent the most common tertiary and quaternary packing motif in proteins. Heterochiral coiled–coil interactions were predicted over 50 years ago by Crick, and limited experimental data obtained in solution suggest that such interactions can indeed occur. To address the dearth of atomic-level structural characterization of heterochiral helix pairings, we report two independent crystal structures that elucidate coiled-coil packing between l- and d-peptide helices. Both structures resulted from racemic crystallization of a peptide corresponding to the transmembrane segment of the influenza M2 protein. Networks of canonical knobs-into-holes side-chain packing interactions are observed at each helical interface. However, the underlying patterns for these heterochiral coiled coils seem to deviate from the heptad sequence repeat that is characteristic of most homochiral analogs, with an apparent preference for a hendecad repeat pattern.
Polypeptides comprising d-amino acid residues have been sources of growing interest for biological applications, often for functions that depend on recognition by specific natural proteins (1–3). d peptides offer identical versatility in terms of conformation and side-chain functionality relative to conventional peptides (composed of l-amino acid residues), but d peptides are impervious to the action of proteolytic enzymes, which should improve pharmacokinetic properties in vivo relative to those of conventional peptides. The engineering of d peptides to display defined protein-binding preferences is hindered, however, by the dearth of experimental information available for such complexes. Structural principles that are well-known to govern interactions between two l-polypeptide chains are not directly extensible to pairings between peptides of opposite chirality. Favorable heterochiral interactions (between l- and d peptides) that are analogous to homochiral associations between l peptides were postulated decades ago on the basis of geometrical considerations (4, 5). In a 1953 analysis of structural parameters governing coiled–coil formation between right-handed α-helices formed from l peptides, for example, Crick suggested that analogous assemblies should be accessible to pairs of right- and left-handed helices (4). In the same year, Pauling and Corey postulated that heterochiral peptide mixtures could form “rippled” β-sheet assemblies with backbone hydrogen-bonding patterns resembling those of homochiral β-sheets found in proteins (5). Experimental elucidation of structural principles that underlie heterochiral assembly modes would greatly facilitate the design of d polypeptides for specific biomedical applications.
Several structure-independent approaches have enabled discovery of d polypeptides that can recognize specific l-protein partners. “Mirror-image phage display,” a powerful method pioneered by Kim and co-workers, has identified d peptides that bind tightly to l proteins (6). A few of these heterochiral complexes have been characterized at atomic resolution (7, 8), but no general principles of heterochiral recognition have emerged. Shai and co-workers have reported that d peptides corresponding to transmembrane helices of several integral membrane proteins can pair with the natural l-peptide helix within a lipid bilayer (9–11). The retro-inverso design strategy has been widely used in pursuit of biologically active d peptides; however, results from this approach have been mixed (12–15). Computationally designed d peptides that inhibit formation of amyloid fibrils by a Tau-derived l polypeptide have been reported (16).
A few studies support the existence of structurally regular heterochiral assemblies along the lines proposed by Crick and by Pauling and Corey. Sia and Kim have designed an l-peptide/d-peptide pair that forms a tetramer in solution, behavior that is attributed to leucine zipper-type interactions (17). The thermodynamic stability of this heterochiral tetramer is comparable to that of the diastereomeric all-l-peptide tetramer. In contrast, a study by Chung and Nowick suggests that backbone H-bond–mediated interactions characteristic of antiparallel β-sheet secondary structure are much more favorable for homochiral relative to heterochiral strand pairings (18). However, Nilsson and co-workers report that mixing fibril-forming l- and d peptides leads to preferential formation of heterochiral β-sheet assemblies (19). The 30-mer peptide (l-Pro–l-Pro–Gly)10 forms a collagen-like triple helix that is very soluble in aqueous solution, but Nanda and co-workers have found that mixing solutions of enantiomeric triple helices leads to a supramolecular assembly/precipitation process that apparently involves intimate interactions between enantiomeric trimers (20). None of these intriguing studies has provided atomic-resolution insight on heterochiral associations.
Here we report two crystal structures that illuminate structural parameters for heterochiral coiled–coil assembly. These high-resolution data begin to lay a foundation for understanding how an α-helix formed by l residues can be specifically recognized by a d polypeptide. Because α-helices are prominent in many protein–protein interfaces (21–23), this information should be of broad interest. The remarkable recent success of peptide drugs such as teriparatide (24), enfuvirtide (25), and exenatide (26), which contain exclusively proteinogenic l-α-amino acid residues, suggests that the 21st century will see a growing assortment of therapeutic agents in this class, including examples that contain unnatural building blocks such as d-α-amino acid residues.
Results and Discussion
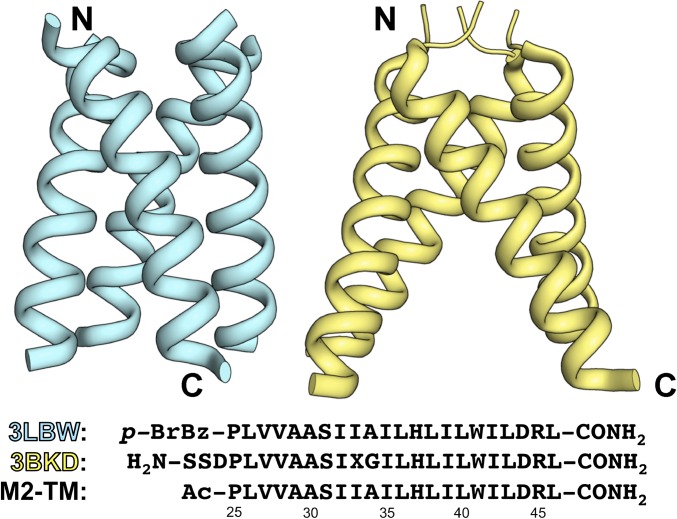
The structures reported here emerged from our application of racemic crystallography to the transmembrane segment of the influenza M2 ion channel [the peptide M2-transmembrane (M2-TM) in Fig. 1]. We selected this system because, although many proteins contain a single membrane-spanning α-helix, M2 is the only case in which an isolated single-pass transmembrane α-helix has been characterized crystallographically (27, 28). Structures for two variants of M2-TM (Fig. 1) show a four-helix quaternary assembly that provides a channel for proton conduction, which is essential for viral propagation. These homochiral assemblies feature N termini clustered on one side and C termini clustered on the other, with substantially nonzero angles between the long axes of each helix and the central axis of the tetramer, 25° for one variant and 33° for the other.
Fig. 1.
X-ray structures of two previously reported sequence variants of M2-TM crystallized from d-OG (Left, 3LBW; Right, 3BKD) and their respective amino acid sequences. p-BrBz refers to the p-bromobenzoyl group, whereas X in the sequence of 3BKD denotes a selenomethionine residue. “M2-TM” refers to the amino acid sequence used in this study. N- and C termini are indicated for both structures shown above.
We wondered whether this tetrameric assembly would be maintained under the conditions of racemic crystallization, a technique first implemented by Zawadzke and Berg (29) that has become increasingly popular in recent years (29–44). It has been proposed that racemates are more prone to crystallization than single enantiomers because racemates can access achiral and centrosymmetric packing arrangements in the solid state (45), arrangements that are not available to a chiral entity. In particular, the availability of space group should result in a greater likelihood of crystal growth from protein racemates relative to single protein enantiomers. For racemic crystallization to be a source of insight on natural protein structure, assembly into the racemate crystal lattice must not significantly alter a polypeptide's conformation or quaternary structure. The available set of racemic and quasiracemic protein crystal structures allows 14 comparisons with chiral crystal counterparts (8, 29, 32, 33, 35–39, 42, 43), and all seem to suggest that native tertiary structure is maintained. Only three racemate structures include specific native-like quaternary contacts (8, 32, 41). In one of these systems (32), native-like quaternary contacts are maintained. In the other two examples (8, 41), there is no chiral crystal structure to which direct comparison could be made; however, indirect evidence suggests in both cases that quaternary associations observed in the solid state are comparable to those that occur in solution. The structures reported here, on the other hand, show that the native tetrameric assembly of homochiral M2-TM is not maintained when the racemate is crystallized. The contrast between previous findings with an α-helical antimicrobial peptide (41) and the current results with the α-helical M2-TM peptide in terms of whether a natural assembly mode is retained upon racemic crystallization suggests that quaternary structures detected in racemic crystals must be interpreted with care.
A peptide closely related to l-M2-TM (Fig. 1, different N-terminal capping group) was previously crystallized from a detergent-solubilized state with octyl-d-glucoside (d-OG) (28); therefore, our efforts to crystallize racemic M2-TM used racemic OG (DL-OG). Racemic M2-TM, prepared by mixing individual enantiomers that were prepared via solid-phase synthesis, was dissolved in a 2% (wt/vol) solution of DL-OG and crystallized using the hanging-drop vapor diffusion method. Crystals were cryoprotected with glycerol and used for synchrotron X-ray data collection to 1.05-Å resolution. In the course of this work, an atomic-resolution structure of the racemic DL-OG alone was determined as well (SI Appendix).
The structure of racemic M2-TM/OG was solved by molecular replacement and refined in space group (see SI Appendix for full details of crystallographic methods). Model refinement for the structure of racemic M2-TM/OG converged at values of R/Rfree of 0.138/0.156. The unit cell of racemic M2-TM/OG contains one l- and one d peptide related by a crystallographic inversion center, whereas the asymmetric unit contains a single peptide (Fig. 2A). One molecule of OG per asymmetric unit was fully resolved in the electron density; the d-glucose component of a detergent molecule packs against a Trp side chain from d-M2-TM as well as engaging in extensive H-bonding interactions (Fig. 2B). Mirror-image versions of these contacts occur as well.
Fig. 2.
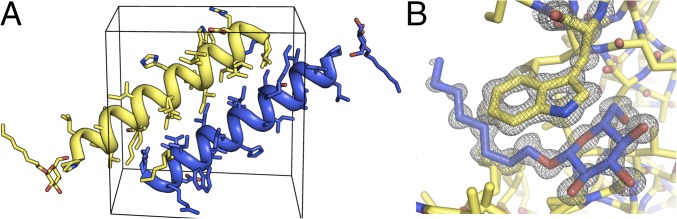
(A) Unit cell of racemic M2-TM/OG in space group . l-M2-TM and d-OG are shown in blue, d-M2-TM and l-OG in yellow. (B) Electron density (2FO-FC map contoured at 1.5σ) surrounding a Trp side chain from d-M2-TM and a molecule of d-OG.
The racemic M2-TM/OG crystal does not contain the homochiral tetrameric assembly seen in the l-peptide crystals (27, 28). Instead, the most intimate interpeptide contacts involve heterochiral pairings, with four distinct interfaces observed between neighboring l- and d peptides. A small knobs-into-holes contact (only one complete “hole”) between enantiomeric α-helices has been observed in a previous X-ray structure of a racemic polypeptide, the designed α-helical peptide Alpha 1 (33); this interface is too small to suggest the principles of heterochiral coiled–coil assembly that are revealed by our structures. In the structure of racemic M2-TM/OG reported here, two of the four unique heterochiral interfaces involve approximately half the length of each interacting helix and bury hydrophobic surfaces from several types of side chain (Ala, Ile, Leu, Pro, and Val), whereas the third interface is more extensive and comprises predominantly Ile side chains (Fig. 3A). All three interfaces correspond to crystallographic inversion operations and thus feature 180° interhelical crossing angles. A fourth contact involves salt-bridging interactions between Arg and Asp side chains on l- and d peptides.
Fig. 3.
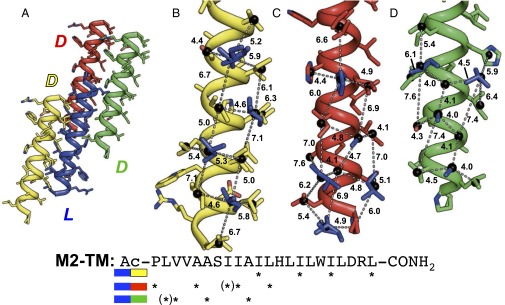
(A) The structure of racemic M2-TM/OG contains three distinct heterochiral interfaces. (B) Distances calculated between side-chain centers-of-mass indicate that the largest interface in the structure of racemic M2-TM/OG contains KIH interactions reflecting a heptad sequence repeat. The remaining two interfaces (C and D) contain spacings between side-chain knobs that are consistent with a nonheptad sequence repeat. (Bottom) Sequence positions of knobs from each heterochiral interface (color-coded) are shown as asterisks below the M2-TM sequence, with marginal interactions (containing a distance ≥7.4 Å) enclosed in parentheses. Distances are reported in units of angstroms (Å). In B–D, side-chain knobs of l-M2-TM are shown in blue.
We analyzed the three heterochiral interfaces (highlighted in Fig. 3 B, C, and D, respectively) involving hydrophobic side chains using side-chain distance calculations identical to those implemented in the SOCKET program (46) to determine whether each interaction mode involved “knobs-into-holes” (KIH) interactions (4) and could therefore be characterized as a coiled coil. The most extensive interface features 12 unique distances among core and flanking side-chain centers-of-mass; these distances range from 4.4 to 7.1 Å and have a mean value of 5.7 Å (Fig. 3B). Walshaw and Woolfson designated side-chain center-of-mass distances of 7.0 and 7.4 Å as stringent and liberal thresholds, respectively, for identifying KIH interactions within coiled coils (46); thus, we conclude that the largest l/d interface in the structure of racemic M2-TM/OG contains true KIH interactions.
The results from these side-chain distance calculations were used to assess possible KIH interactions in the two remaining heterochiral interfaces from the M2-TM/OG structure (Fig. 3 C and D). Intriguingly, this analysis suggested nonheptad repeats of hydrophobic side chains at these heterochiral interfaces. It has previously been noted that l/d-helical assemblies based on a heptad repeat should be limited to interaction surfaces corresponding to about four heptads, because core side chains would stray from the interface over longer interaction surfaces (17). Contact between longer l- and d helices cannot be maintained by supercoiling, as in conventional coiled coils. The interface highlighted in Fig. 3C features a pattern of buried “knob” side chains that is most consistent with a 4,4,3 spacing of hydrophobic side chains, whereas the interface shown in Fig. 3D contains a spacing pattern that could be either 4,4 or 3,4. Consecutive 4,4 spacings are consistent with an 11-residue repeat (“hendecad”), a sequence pattern that has been identified in both native (47–50) and designed (51–53) helical assemblies. In contrast with the conventional heptad repeat, which leads to a left-handed shift in the positions of side chains along the length of a right-handed helix, the hendecad repeat can produce a range of helix geometries that are distinct from that of heptad-based systems. Indeed, for α-helical assemblies in which the sequence periodicity specified by the hendecad repeat (11 residues per 3 helical turns) matches the intrinsic helical periodicity (i.e., 3.67 residues per turn), the positions of every 11th residue are essentially perfectly aligned along the length of the α-helix, and the helical bundle exhibits little to no supercoiling (54).
We explored an alternative method for crystal growth that has become increasingly popular for integral membrane proteins, crystallization from a lipidic cubic phase (LCP) (55, 56). The lipid most commonly used for LCP crystallization, monoolein, is typically used in racemic form. LCP crystallization involves stabilization of the protein of interest in an ordered lipid bilayer lyotropic phase with percolating aqueous nanochannels. Once exposed to solutions of salts or small-molecule additives, LCPs can produce protein crystals suitable for diffraction measurements. Protein crystal growth from LCP media often results from a transition from a bicontinuous cubic phase to more disordered “swollen” or sponge phases that afford protein molecules increased mobility within the lipid/water network (57). For our crystallization efforts, l- and d-M2-TM polypeptides were grown from a cubic phase derived from the lipid monoolein.
Crystals of racemic M2-TM/LCP generally grew within 3 h, and were vitrified directly and used for synchrotron X-ray data collection to 2.0-Å resolution. Small-angle X-ray scattering (SAXS) analysis of the peptide-doped LCP under the crystallization conditions indicated that crystal formation coincided with a transition from cubic phase with Pn m symmetry to a disordered, fluid l3-sponge phase (SI Appendix). The structure of racemic M2-TM/LCP was solved by molecular replacement and refined in space group P21/c, which is centrosymmetric. The asymmetric unit comprises two molecules of l-M2-TM as well as several lipid fragments that were modeled into elongated regions of electron density between copies of M2-TM. The unit cell of racemic M2-TM/LCP contains eight polypeptides (four l and four d).
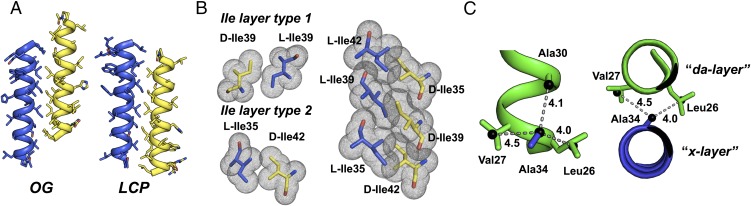
The structure of racemic M2-TM/LCP contains two virtually identical, noncrystallographic symmetry-related heterochiral interfaces that closely resemble the most extensive heterochiral interface in the racemic M2-TM/OG structure (Fig. 4A). In the M2-TM/LCP interfaces, the two enantiomeric peptides are related by a 180° interhelical crossing angle, and contact between l- and d-M2-TM is anchored by a set of interdigitated Ile side chains. The register of this M2-TM/LCP interface matches that of the largest M2-TM/OG interface, but the overlapping surface in the former is shifted by seven amino acid residues (i.e., approximately two helical turns) with respect to the overlapping surface in the latter. The M2-TM/LCP heterochiral pairing features a larger contact surface area than any pairing in the M2-TM/OG structure; five hydrophobic side chains on each helix are buried in KIH interactions at the M2-TM/LCP heterochiral dimer interface.
Fig. 4.
(A) Comparison of heterochiral interfaces in OG- and LCP-derived structures of racemic M2-TM; between the two structures the peptides are shifted with respect to each other by one heptad repeat. (B) Side chains of l- and d-Ile form two distinct layer types that alternate to form a continuous, well-packed interface in both of the interfaces highlighted in A. In both images, l polypeptides are shown in blue and d polypeptides in yellow. Side chains shown in B are derived from the structure of racemic M2-TM/OG. (C) Two orthogonal views of a close-packed “da/x layer”-type interaction found in the structure of racemic M2-TM/OG. In both views, l- and d-M2-TM are shown in blue and green, respectively (color scheme is consistent with that of Fig. 3). Distances are shown in units of angstroms (Å).
Despite differences in the juxtapositions of the l- and d-peptide molecules between the two M2-TM racemate structures (Fig. 4A), these heterochiral dimers share a striking similarity in terms of interactions among mirror-image forms of Ile side chains. In both structures, l-Ile/d-Ile side-chain pairs form two distinct layer types that exhibit local approximate inversion symmetry and accommodate each Ile side chain in its preferred mt conformation (Fig. 4B) (58). As we had observed in the two smaller hydrophobic interfaces in the structure of racemic M2-TM/OG (Fig. 3 C and D), the two noncrystallographic symmetry-related heterochiral interfaces found in the structure of racemic M2-TM/LCP correspond to a hendecad (4, 4, 3) sequence pattern. It is notable that homochiral peptides based on the hendecad sequence motif are considered unlikely to form dimeric coiled coils due to steric clashes within “x layers,” which involve a single side chain from each helix projected directly into the interhelical interface (59). In the two structures of racemic M2-TM, we find side chains projected directly into the interhelical interface in a manner reminiscent of x-layer positions; however, these residues are surrounded by pairs of side chains on the opposing helix that are projected to either side of the interface and resemble the “da layers” described by Lupas et al. (59). This packing motif seems to account for the close inter side-chain distances observed in two of the heterochiral interfaces found in the structure of racemic M2-TM/OG (Fig. 4C). Whether these features are generally characteristic of heterochiral coils, as opposed to being peculiarities of our structures, will not become clear until additional structural data are available.
Assignment of a hendecad repeat to the M2-TM sequence produces a close match with the structure of racemic M2-TM/LCP in terms of core and flanking side-chain identities (Fig. 5A), whereas the alternative assignment of a heptad repeat fails because of the drift of side chains designated as “core” and “flanking” away from the helical interface (Fig. 5B). Given that helical supercoiling is not expected to be operative for heterochiral helical assemblies, our findings suggest that nonheptad sequence patterns may be intrinsically more compatible with heterochiral interfaces than is the conventional heptad. A previous study of heterochiral helix assembly in solution was designed based on the assumption that such interactions are governed by the heptad repeat (17), but future heterochiral helix design efforts may be more effective if founded on a hendecad repeat.
Fig. 5.
Assignment of a hendecad repeat (A) to the M2-TM sequence based on the structure of racemic M2-TM/LCP produces a close match with observed core and flanking side chains, whereas assignment of a heptad repeat (B) to the same sequence causes core positions to drift away from the interhelical interface. Distances between side-chain centers-of-mass are reported in angstroms (Å). Sequence positions assigned according to hendecad and heptad repeats are denoted by asterisks below the M2-TM sequence. Core positions at either end of the antiparallel interface are highlighted in both A and B, and below the M2-TM sequence.
Conclusions
Sequences that evolved to reside in membranes may provide a favorable context for discovery of characteristic interaction modes available to l-peptide/d-peptide pairs because the role of van der Waals complementarity is likely to be maximized in such systems. For water-soluble helices, forces other than van der Waals contacts, such as interhelical salt bridging, can influence pairing specificity; in contrast, Coulombic interactions are uncommon for membrane-embedded segments. Despite the differences between bilayer and aqueous environments, packing motifs identified from racemic crystallization of membrane polypeptides are likely to be useful for the design of soluble heterochiral assemblies. It is possible that the identity of hydrophobic side chains at core positions could be used to control assembly preferences in designed heterochiral coiled coils, as is widely practiced in homochiral systems (60–63). For instance, placement of Ile residues at interface positions can strongly influence the oligomerization state of homochiral helix-bundles, apparently by favoring assemblies that accommodate the preferred rotameric state of the Ile side chain (60). Similar criteria are likely to govern oligomerization specificity in heterochiral coiled coils; further investigation will be necessary for elucidation of such factors.
The X-ray structures reported here suggest that nonheptad sequence repeats are important for maintaining contact between l- and d-peptide helices over long interfaces. This consideration is likely to be critical for the design of extended heterochiral coiled coils. It has previously been suggested that heterochiral α-helical interfaces can occur over only about four heptad repeats before core side chains diverge from the interhelical interface; however, we speculate that longer and likely more stable interfaces could be formed between l- and d polypeptides containing nonheptad sequence repeats. Recent work has shown that native and designed l polypeptides harboring a hendecad sequence pattern, either alone or in combination with the heptad repeat, can form homochiral quaternary assemblies with little or no supercoiling (50, 53). This trend is significant in the context of heterochiral assemblies because oligomers formed between l- and d polypeptides are expected to be incompatible with α-helical supercoiling. We therefore speculate that extended hendecad-based heterochiral helical assemblies are attainable.
An antiparallel heterochiral coiled coil can be seen as a manifestation of crystallographic inversion symmetry between neighboring enantiomeric helices, and, in analogous fashion, a rippled β-sheet as a manifestation of crystallographic inversion symmetry between neighboring enantiomeric β-strands (5). The recently reported structure of racemic ester insulin exhibits such a heterochiral β-sheet–like interaction between l- and d strands (64), whereas the racemic structures reported here exemplify this inversion relationship between mirror-image α-helices.
Our findings suggest that racemic crystallography can provide fundamental insights on heterochiral polypeptide recognition. High-resolution data of the type reported here provide a foundation for future efforts to design d polypeptides that can engage in specific and potentially useful interactions with partners comprised of l residues. Given the clinical successes that have been achieved with peptides containing exclusively proteinogenic l-α-amino acid residues (24–26), and the improvements that can result from minor extensions beyond these 20 building blocks (65), it seems likely that peptidic oligomers with unnatural backbones, such as might be derived from d-α-amino acids, will provide pharmaceutically valuable agents in the future.
Materials and Methods
Polypeptide Synthesis.
l- and d-M2-TM polypeptides were prepared using Fmoc-based solid-phase synthesis protocols, and were purified by HPLC. Detailed methods used for the solid-phase synthesis and purification of l- and d-M2-TM can be found in SI Appendix.
Crystallization, Diffraction Data Collection, Structural Solution, and Refinement.
Crystals of racemic M2-TM/OG were grown via hanging-drop vapor diffusion. Several screening conditions yielded crystals overnight; the best were derived from a precipitant solution containing 0.1 M N-(2-acetamido)iminiodiacetic acid (ADA) pH 6.5 and 1.0 M ammonium sulfate. These crystals were briefly treated with glycerol before vitrification in liquid N2.
For preparation of crystals of racemic M2-TM/LCP, l- and d-M2-TM polypeptides were combined with the lipid monoolein by codissolution in 2,2,2-trifluoroethanol followed by removal of the solvent first under a stream of N gas and then under vacuum. The peptide-doped lipid was then mixed with water in a syringe-coupler apparatus, and the resulting gel was dispensed onto glass sandwich plates prepared as described previously (56) for combination with precipitant solutions. Crystals of racemic M2-TM/LCP grew from 18 of 48 precipitant conditions in the Hampton MembFac screen. Optimization of the best condition led to a precipitant solution containing 0.1 M ADA pH 6.5 and 24% (vol/vol) (±)-2-methyl-2,4-pentanediol, which afforded single crystals suitable for X-ray analysis.
X-ray diffraction data derived from both crystal forms were collected at the Advanced Photon Source (21-ID-F; λ = 0.97872 Å) at Argonne National Laboratory. Data were integrated and scaled using the XDS package (66). Both structures of racemic M2-TM were solved by molecular replacement in Phaser (67), and were refined via maximum likelihood methods using Refmac5 (68). Detailed methods can be found in SI Appendix. The structures have been deposited in the Protein Data Bank with accession codes 4RWB and 4RWC for racemic M2-TM/LCP and M2-TM/OG, respectively.
Supplementary Material
Acknowledgments
We are grateful to Profs. Brian Kobilka, Søren Rasmussen, and Andrew Kruse for sharing their expertise with LCP protein crystallization, to Dr. Ilia Guzei for assistance with X-ray data collection, and to Prof. Dek Woolfson for helpful discussions. This research was supported by NIH Grant GM061238 (to S.H.G.), National Oceanic and Atmospheric Administration Grant NA14OAR4170092 (to K.T.F.), and US National Science Foundation Grant CHE-1152347 (to M.K.M.). D.E.M. was supported in part by an NIH Molecular Biophysics Training Grant (T32 GM08293). D.F.K. was supported in part by an NIH Biotechnology Training Grant (T32 GM008349). G.P.S. gratefully acknowledges partial support through a 3M Corporation Graduate Fellowship. Lab source SAXS measurements at UW-Madison used core facilities that are partially supported by the UW-Madison Nanoscale Science and Engineering Center (DMR-0832760) and Center of Excellence for Materials Research and Innovation (DMR-1121288). Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the US DOE under Contract DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4RWB (racemic M2-TM/LCP) and 4RWC (racemic M2-TM/OG)] and the Cambridge Crystallographic Data Centre, www.ccdc.cam.ac.uk [CSD reference no. 1058798 (racemic OG)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507918112/-/DCSupplemental.
References
- 1.Eckert DM, Malashkevich VN, Hong LH, Carr PA, Kim PS. Inhibiting HIV-1 entry: Discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell. 1999;99(1):103–115. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 2.Weinstock MT, Jacobsen MT, Kay MS. Synthesis and folding of a mirror-image enzyme reveals ambidextrous chaperone activity. Proc Natl Acad Sci USA. 2014;111(32):11679–11684. doi: 10.1073/pnas.1410900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabideau AE, Liao X, Pentelute BL. Delivery of mirror image polypeptides into cells. Chem Sci (Camb) 2015;6(1):648–653. doi: 10.1039/c4sc02078b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crick FHC. The packing of α-helices: Simple coiled-coils. Acta Crystallogr. 1953;6(8-9):689–697. [Google Scholar]
- 5.Pauling L, Corey RB. Two rippled-sheet configurations of polypeptide chains, and a note about pleated sheets. Proc Natl Acad Sci USA. 1953;39(4):253–256. doi: 10.1073/pnas.39.4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schumacher TNM, et al. Identification of D-peptide ligands through mirror-image phage display. Science. 1996;271(5257):1854–1857. doi: 10.1126/science.271.5257.1854. [DOI] [PubMed] [Google Scholar]
- 7.Liu M, et al. D-peptide inhibitors of the p53-MDM2 interaction for targeted molecular therapy of malignant neoplasms. Proc Natl Acad Sci USA. 2010;107(32):14321–14326. doi: 10.1073/pnas.1008930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandal K, et al. Chemical synthesis and X-ray structure of a heterochiral D-protein antagonist plus vascular endothelial growth factor protein complex by racemic crystallography. Proc Natl Acad Sci USA. 2012;109(37):14779–14784. doi: 10.1073/pnas.1210483109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerber D, Shai Y. Chirality-independent protein-protein recognition between transmembrane domains in vivo. J Mol Biol. 2002;322(3):491–495. doi: 10.1016/s0022-2836(02)00807-0. [DOI] [PubMed] [Google Scholar]
- 10.Sal-Man N, Gerber D, Shai Y. Hetero-assembly between all-L- and all-D-amino acid transmembrane domains: Forces involved and implication for inactivation of membrane proteins. J Mol Biol. 2004;344(3):855–864. doi: 10.1016/j.jmb.2004.09.066. [DOI] [PubMed] [Google Scholar]
- 11.Gerber D, Quintana FJ, Bloch I, Cohen IR, Shai Y. D-enantiomer peptide of the TCRalpha transmembrane domain inhibits T-cell activation in vitro and in vivo. FASEB J. 2005;19(9):1190–1192. doi: 10.1096/fj.04-3498fje. [DOI] [PubMed] [Google Scholar]
- 12.Guichard G, et al. Antigenic mimicry of natural L-peptides with retro-inverso-peptidomimetics. Proc Natl Acad Sci USA. 1994;91(21):9765–9769. doi: 10.1073/pnas.91.21.9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, et al. Limitations of peptide retro-inverso isomerization in molecular mimicry. J Biol Chem. 2010;285(25):19572–19581. doi: 10.1074/jbc.M110.116814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, et al. Functional consequences of retro-inverso isomerization of a miniature protein inhibitor of the p53-MDM2 interaction. Bioorg Med Chem. 2013;21(14):4045–4050. doi: 10.1016/j.bmc.2013.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atzori A, Baker AE, Chiu M, Bryce RA, Bonnet P. Effect of sequence and stereochemistry reversal on p53 peptide mimicry. PLoS One. 2013;8(7):e68723. doi: 10.1371/journal.pone.0068723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sievers SA, et al. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature. 2011;475(7354):96–100. doi: 10.1038/nature10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sia SK, Kim PS. A designed protein with packing between left-handed and right-handed helices. Biochemistry. 2001;40(30):8981–8989. doi: 10.1021/bi010725v. [DOI] [PubMed] [Google Scholar]
- 18.Chung DM, Nowick JS. Enantioselective molecular recognition between β-sheets. J Am Chem Soc. 2004;126(10):3062–3063. doi: 10.1021/ja031632z. [DOI] [PubMed] [Google Scholar]
- 19.Swanekamp RJ, DiMaio JTM, Bowerman CJ, Nilsson BL. Coassembly of enantiomeric amphipathic peptides into amyloid-inspired rippled β-sheet fibrils. J Am Chem Soc. 2012;134(12):5556–5559. doi: 10.1021/ja301642c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu F, et al. Self-assembly of left- and right-handed molecular screws. J Am Chem Soc. 2013;135(50):18762–18765. doi: 10.1021/ja4106545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jochim AL, Arora PS. Assessment of helical interfaces in protein-protein interactions. Mol Biosyst. 2009;5(9):924–926. doi: 10.1039/b903202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bullock BN, Jochim AL, Arora PS. Assessing helical protein interfaces for inhibitor design. J Am Chem Soc. 2011;133(36):14220–14223. doi: 10.1021/ja206074j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raj M, Bullock BN, Arora PS. Plucking the high hanging fruit: A systematic approach for targeting protein-protein interactions. Bioorg Med Chem. 2013;21(14):4051–4057. doi: 10.1016/j.bmc.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saag KG, et al. Teriparatide or alendronate in glucocorticoid-induced osteoporosis. N Engl J Med. 2007;357(20):2028–2039. doi: 10.1056/NEJMoa071408. [DOI] [PubMed] [Google Scholar]
- 25.Lalezari JP, et al. A phase II clinical study of the long-term safety and antiviral activity of enfuvirtide-based antiretroviral therapy. AIDS. 2003;17(5):691–698. doi: 10.1097/00002030-200303280-00007. [DOI] [PubMed] [Google Scholar]
- 26.Drucker DJ, et al. DURATION-1 Study Group Exenatide once weekly versus twice daily for the treatment of type 2 diabetes: A randomised, open-label, non-inferiority study. Lancet. 2008;372(9645):1240–1250. doi: 10.1016/S0140-6736(08)61206-4. [DOI] [PubMed] [Google Scholar]
- 27.Stouffer AL, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451(7178):596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Acharya R, et al. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc Natl Acad Sci USA. 2010;107(34):15075–15080. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zawadzke LE, Berg JM. The structure of a centrosymmetric protein crystal. Proteins. 1993;16(3):301–305. doi: 10.1002/prot.340160308. [DOI] [PubMed] [Google Scholar]
- 30.Doi M, et al. Structural characteristics of enantiomorphic DNA: Crystal analysis of racemates of the d(CGCGCG) duplex. J Am Chem Soc. 1993;115(22):10432–10433. [Google Scholar]
- 31.Toniolo C, et al. Structure determination of racemic trichogin A IV using centrosymmetric crystals. Nat Struct Biol. 1994;1(12):908–914. doi: 10.1038/nsb1294-908. [DOI] [PubMed] [Google Scholar]
- 32.Hung LW, Kohmura M, Ariyoshi Y, Kim SH. Structural differences in D and L-monellin in the crystals of racemic mixture. J Mol Biol. 1999;285(1):311–321. doi: 10.1006/jmbi.1998.2308. [DOI] [PubMed] [Google Scholar]
- 33.Patterson WR, Anderson DH, DeGrado WF, Cascio D, Eisenberg D. Centrosymmetric bilayers in the 0.75 A resolution structure of a designed alpha-helical peptide, D,L-Alpha-1. Protein Sci. 1999;8(7):1410–1422. doi: 10.1110/ps.8.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rypniewski W, et al. The first crystal structure of an RNA racemate. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 6):659–664. doi: 10.1107/S090744490601359X. [DOI] [PubMed] [Google Scholar]
- 35.Pentelute BL, et al. X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers. J Am Chem Soc. 2008;130(30):9695–9701. doi: 10.1021/ja8013538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandal K, et al. Racemic crystallography of synthetic protein enantiomers used to determine the X-ray structure of plectasin by direct methods. Protein Sci. 2009;18(6):1146–1154. doi: 10.1002/pro.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandal K, Pentelute BL, Tereshko V, Kossiakoff AA, Kent SB. X-ray structure of native scorpion toxin BmBKTx1 by racemic protein crystallography using direct methods. J Am Chem Soc. 2009;131(4):1362–1363. doi: 10.1021/ja8077973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandal K, et al. Design, total chemical synthesis, and X-ray structure of a protein having a novel linear-loop polypeptide chain topology. Angew Chem Int Ed Engl. 2012;51(6):1481–1486. doi: 10.1002/anie.201107846. [DOI] [PubMed] [Google Scholar]
- 39.Mortenson DE, Satyshur KA, Guzei IA, Forest KT, Gellman SH. Quasiracemic crystallization as a tool to assess the accommodation of noncanonical residues in nativelike protein conformations. J Am Chem Soc. 2012;134(5):2473–2476. doi: 10.1021/ja210045s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee M, Shim J, Kang P, Guzei IA, Choi SH. Structural characterization of α/β-peptides having alternating residues: X-ray structures of the 11/9-helix from crystals of racemic mixtures. Angew Chem Int Ed Engl. 2013;52(48):12564–12567. doi: 10.1002/anie.201306404. [DOI] [PubMed] [Google Scholar]
- 41.Hayouka Z, et al. Evidence for phenylalanine zipper-mediated dimerization in the X-ray crystal structure of a magainin 2 analogue. J Am Chem Soc. 2013;135(42):15738–15741. doi: 10.1021/ja409082w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dang B, Kubota T, Mandal K, Bezanilla F, Kent SB. Native chemical ligation at Asx-Cys, Glx-Cys: Chemical synthesis and high-resolution X-ray structure of ShK toxin by racemic protein crystallography. J Am Chem Soc. 2013;135(32):11911–11919. doi: 10.1021/ja4046795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang CK, King GJ, Northfield SE, Ojeda PG, Craik DJ. Racemic and quasi-racemic X-ray structures of cyclic disulfide-rich peptide drug scaffolds. Angew Chem Int Ed Engl. 2014;53(42):11236–11241. doi: 10.1002/anie.201406563. [DOI] [PubMed] [Google Scholar]
- 44.Mandal PK, Collie GW, Kauffmann B, Huc I. Racemic DNA crystallography. Angew Chem Int Ed Engl. 2014;53(52):14424–14427. doi: 10.1002/anie.201409014. [DOI] [PubMed] [Google Scholar]
- 45.Wukovitz SW, Yeates TO. Why protein crystals favour some space-groups over others. Nat Struct Biol. 1995;2(12):1062–1067. doi: 10.1038/nsb1295-1062. [DOI] [PubMed] [Google Scholar]
- 46.Walshaw J, Woolfson DN. Socket: A program for identifying and analysing coiled-coil motifs within protein structures. J Mol Biol. 2001;307(5):1427–1450. doi: 10.1006/jmbi.2001.4545. [DOI] [PubMed] [Google Scholar]
- 47.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371(6492):37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 48.Brown JH, Cohen C, Parry DAD. Heptad breaks in α-helical coiled coils: stutters and stammers. Proteins. 1996;26(2):134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 49.Peters J, Baumeister W, Lupas A. Hyperthermostable surface layer protein tetrabrachion from the archaebacterium Staphylothermus marinus: Evidence for the presence of a right-handed coiled coil derived from the primary structure. J Mol Biol. 1996;257(5):1031–1041. doi: 10.1006/jmbi.1996.0221. [DOI] [PubMed] [Google Scholar]
- 50.Stetefeld J, et al. Crystal structure of a naturally occurring parallel right-handed coiled coil tetramer. Nat Struct Biol. 2000;7(9):772–776. doi: 10.1038/79006. [DOI] [PubMed] [Google Scholar]
- 51.Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS. High-resolution protein design with backbone freedom. Science. 1998;282(5393):1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 52.Hicks MR, Walshaw J, Woolfson DN. Investigating the tolerance of coiled-coil peptides to nonheptad sequence inserts. J Struct Biol. 2002;137(1-2):73–81. doi: 10.1006/jsbi.2002.4462. [DOI] [PubMed] [Google Scholar]
- 53.Huang P-S, et al. High thermodynamic stability of parametrically designed helical bundles. Science. 2014;346(6208):481–485. doi: 10.1126/science.1257481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strelkov SV, Burkhard P. Analysis of α-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J Struct Biol. 2002;137(1-2):54–64. doi: 10.1006/jsbi.2002.4454. [DOI] [PubMed] [Google Scholar]
- 55.Landau EM, Rosenbusch JP. Lipidic cubic phases: A novel concept for the crystallization of membrane proteins. Proc Natl Acad Sci USA. 1996;93(25):14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4(5):706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cherezov V, Clogston J, Papiz MZ, Caffrey M. Room to move: Crystallizing membrane proteins in swollen lipidic mesophases. J Mol Biol. 2006;357(5):1605–1618. doi: 10.1016/j.jmb.2006.01.049. [DOI] [PubMed] [Google Scholar]
- 58.Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins. 2000;40(3):389–408. [PubMed] [Google Scholar]
- 59.Lupas A, et al. Model structure of the Omp α rod, a parallel four-stranded coiled coil from the hyperthermophilic eubacterium Thermotoga maritima. J Mol Biol. 1995;248(1):180–189. doi: 10.1006/jmbi.1995.0210. [DOI] [PubMed] [Google Scholar]
- 60.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262(5138):1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 61.Gurnon DG, Whitaker JA, Oakley MG. Design and characterization of a homodimeric antiparallel coiled coil. J Am Chem Soc. 2003;125(25):7518–7519. doi: 10.1021/ja0357590. [DOI] [PubMed] [Google Scholar]
- 62.Schnarr NA, Kennan AJ. Strand orientation by steric matching: A designed antiparallel coiled-coil trimer. J Am Chem Soc. 2004;126(44):14447–14451. doi: 10.1021/ja047496v. [DOI] [PubMed] [Google Scholar]
- 63.Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458(7240):859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avital-Shmilovici M, et al. Fully convergent chemical synthesis of ester insulin: Determination of the high resolution X-ray structure by racemic protein crystallography. J Am Chem Soc. 2013;135(8):3173–3185. doi: 10.1021/ja311408y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Checco JW, et al. Targeting diverse protein-protein interaction interfaces with α/β-peptides derived from the Z-domain scaffold. Proc Natl Acad Sci USA. 2015;112(15):4552–4557. doi: 10.1073/pnas.1420380112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.