Significance
These studies shed light on the role of Mycobacterium tuberculosis (Mtb) 2-ketoglutarate (α-KG) dehydrogenase in fulfillment of noncanonical functions: defense against toxic aldehydes during glutamate anaplerosis and participation by two of its three enzymes in a previously undescribed antinitrosative defense. Analysis of the latter pathway revealed a peroxidase activity, which can be inferred to function as a peroxynitrite reductase as well. Heretofore, antioxidant enzyme systems were known to depend on electrons derived from the oxidant itself in dismutation reactions or derived directly or indirectly from NADPH or NADH. Here, electrons derived from the oxidative decarboxylation of central carbon metabolites α-KG and pyruvate to succinyl CoA (SucCoA) and acetyl CoA (AcCoA), respectively, serve as sources of reducing power.
Keywords: Mycobacterium tuberculosis, α-ketoglutarate dehydrogenase, peroxynitrite reductase, peroxidase, hydroxyoxoadipate synthase
Abstract
Enzymes of central carbon metabolism (CCM) in Mycobacterium tuberculosis (Mtb) make an important contribution to the pathogen’s virulence. Evidence is emerging that some of these enzymes are not simply playing the metabolic roles for which they are annotated, but can protect the pathogen via additional functions. Here, we found that deficiency of 2-hydroxy-3-oxoadipate synthase (HOAS), the E1 component of the α-ketoglutarate (α-KG) dehydrogenase complex (KDHC), did not lead to general metabolic perturbation or growth impairment of Mtb, but only to the specific inability to cope with glutamate anaplerosis and nitroxidative stress. In the former role, HOAS acts to prevent accumulation of aldehydes, including growth-inhibitory succinate semialdehyde (SSA). In the latter role, HOAS can participate in an alternative four-component peroxidase system, HOAS/dihydrolipoyl acetyl transferase (DlaT)/alkylhydroperoxide reductase colorless subunit gene (ahpC)-neighboring subunit (AhpD)/AhpC, using α-KG as a previously undescribed source of electrons for reductase action. Thus, instead of a canonical role in CCM, the E1 component of Mtb’s KDHC serves key roles in situational defense that contribute to its requirement for virulence in the host. We also show that pyruvate decarboxylase (AceE), the E1 component of pyruvate dehydrogenase (PDHC), can participate in AceE/DlaT/AhpD/AhpC, using pyruvate as a source of electrons for reductase action. Identification of these systems leads us to suggest that Mtb can recruit components of its CCM for reactive nitrogen defense using central carbon metabolites.
The bacterium Mycobacterium tuberculosis (Mtb), which causes tuberculosis, has plagued humanity since antiquity (1), is estimated to infect one-third of the population today, and is the leading cause of death by a bacterium. This success as a pathogen reflects Mtb’s metabolic plasticity and resistance to host immunity (2, 3). Recent evidence suggests that certain enzymes of central carbon metabolism (CCM) can mediate both of these facets of Mtb’s adaptation to the host (4–8). Here, we demonstrate that Rv1248c, recently named 2-hydroxy-3-oxoadipate synthase (HOAS) (9), is one such enzyme.
Rv1248c was first annotated as the thiamin diphosphate (ThDP)-dependent E1 component (SucA) of a canonical α-ketoglutarate (α-KG) dehydrogenase complex (KDHC) that produces succinyl CoA (SucCoA) via oxidative decarboxylation of α-KG with concomitant transfer of the resulting succinyl group to CoA (10). Classically, KDHC, composed of three enzymes, joins the oxidative and reductive half-cycles of the TCA cycle (SI Appendix, Fig. S1). The TCA cycle generates high-energy phosphate bonds and biosynthetic precursors of amino acids, nucleotides, and fatty acids (11). However, KDHC activity was not detected in Mtb lysates, and the gene product annotated as the lipoamide-bearing E2 component [Rv2215, dihydrolipoyl succinyl transferase (SucB)] functions as the E2 component dihydrolipoyl acetyl transferase (DlaT) of the pyruvate dehydrogenase complex (PDHC) (5, 12, 13). The Mtb genome does not encode another SucB (12), and KDHC activity could not be demonstrated with purified recombinant Rv1248c plus DlaT and E3 [lipoamide dehydrogenase (Lpd)] in a manner similar to cognate proteins from other actinomycetes (14). Rv1248c by itself produces succinate semialdehyde (SSA) from nonoxidative decarboxylation of α-KG in vitro, and Mtb’s succinate semialdehyde dehydrogenases (SSADHs) can generate succinate from SSA, completing a modified TCA cycle (13). Subsequently, activity-based metabolomic profiling revealed yet another function of Rv1248c that predominated over SSA production: decarboxylation of α-KG, followed by carboligation with glyoxylate to form 2-hydroxy-3-oxoadipate (HOA). Thus, Rv1248c was renamed HOAS (9). Decarboxylation of α-KG is the first step in all three reactions. The Mycobacterium smegmatis α-ketoglutarate decarboxylase (MsKGD) homolog was found to catalyze all three reactions in vitro, with augmentation of catalysis by acetyl CoA (AcCoA)-mediated allosteric modulation (15), whereas Mtb HOAS showed a kinetic preference for the HOAS pathway in vitro (16).
HOAS activity is regulated in Mtb by glycogen accumulation regulator A (GarA), whose activity is controlled, in turn, by Ser/Thr kinases PknG and PknB (SI Appendix, Fig. S1B). GarA also regulates glutamate dehydrogenase (GDH) and glutamate synthase/glutamine oxoglutarate aminotransferase (17). Coregulation of these three enzymes by GarA calls attention to the contribution of HOAS to Mtb’s metabolism of glutamate. Glutamate serves as an anaplerotic substrate entering the TCA cycle as α-KG and is also a key intermediate in nitrogen assimilation and metabolism (18, 19).
Despite extensive studies, the physiological function of HOAS in Mtb and its contribution to virulence remain unknown. Here, we brought genetics, metabolomics, enzymology, and mouse models of infection to bear on that question, using the hoas deletion mutant in Mtb and the deletion mutant complemented in three ways: with the WT allele, with an allele with a point mutation that abrogates catalysis, or with an allele with a point mutation that is insensitive to allosteric regulation by AcCoA. In standard culture conditions, Δhoas showed no defect in growth or changes in levels of CCM metabolites, arguing against Mtb’s reliance on the conventional function of KDHC. However, situational stresses revealed striking phenotypes in Δhoas, indicative of a defensive role of HOAS against products arising from metabolism of glutamate and against reactive nitrogen intermediates (RNIs). Mechanistic analysis of the latter phenotype revealed a previously undescribed route to antioxidant defense mediated by substrates and enzymes of CCM.
Results
HOAS Is Nonessential in Standard Conditions in Vitro.
Transposon site hybridization analyses suggested that HOAS is essential for Mtb to grow under standard conditions in vitro (20). To define the importance of HOAS to Mtb, we generated a monogenic deletion mutant, Δhoas, in virulent H37Rv Mtb, with a hygromycin resistance cassette replacing all of rv1248c except 123 bp of the C-terminal coding region (SI Appendix, Fig. S2A). These nucleotides were retained to avoid disruption of the immediately downstream relB-relE toxin-antitoxin (TA) operon (rv1247c-rv1246c). Southern (SI Appendix, Fig. S2B) and Western (SI Appendix, Fig. S2C) blots confirmed the deletion. Introduction of a full-length chromosomal copy of hoas under the control of a constitutive hsp60 promoter in the strain Δhoas::hoas restored WT protein levels (SI Appendix, Fig. S2C). We also complemented Δhoas with either of two mutant alleles: a variant that is unable to decarboxylate α-KG (Δhoas::hoasE956A) (SI Appendix, Fig. S3A) and a variant insensitive to AcCoA allosteric activation (Δhoas::hoasE1038A) (SI Appendix, Fig. S3B).
Generation of Δhoas established that HOAS is dispensable for in vitro growth. Moreover, Δhoas and Δhoas::E956Ahoas grew at rates comparable to WT in modified Sauton’s minimal medium (SMM) with ammonium sulfate as a nitrogen source and either glycerol or acetate, a glycolytic source and a fatty acid carbon source, respectively (SI Appendix, Fig. S4). Thus, Mtb need not rely on a canonical KDHC for growth. Moreover, the strains did not show the Met, Lys, or heme auxotrophies that result from KDHC deficiency in other bacteria (21).
HOAS Is Required for Mtb Persistence.
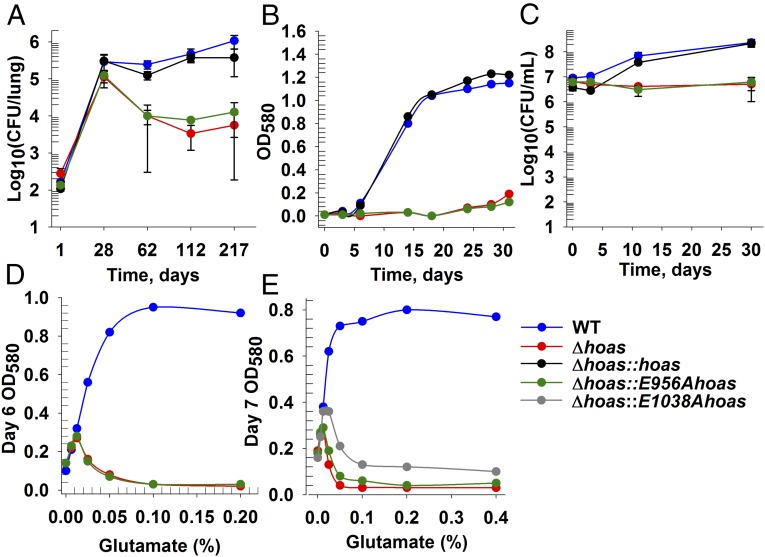
To test whether HOAS is important to Mtb’s ability to cause disease, we infected WT C57BL/6 mice with ∼100 cfu of WT Mtb, Δhoas::hoas, Δhoas, or Δhoas::E956Ahoas. Both Δhoas and Δhoas::E956Ahoas established titers in lungs at levels similar to WT during the acute phase of infection, which is marked by exponential replication of Mtb up to about day 28. About that time, the onset of adaptive immunity, including the induction of inducible nitric oxide synthase (iNOS; NOS2), slows or prevents a further increase in bacterial burden until shortly before death at about a year (22). During the chronic phase of infection, bacillary loads in the lungs infected with Δhoas or Δhoas::E956Ahoas declined to about 2 log10 below the level in mice infected with WT Mtb or Δhoas::hoas (Fig. 1A), with a corresponding reduction in pulmonary pathology (SI Appendix, Fig. S5A). We observed a similar phenotype of impaired survival for the mutant strains in the spleen (SI Appendix, Fig. S5B).
Fig. 1.
Characterization of HOAS-deficient Mtb in vivo and in vitro. (A) Fate of Mtb strains in lungs of C57BL/6 mice after aerosol infection. The mean ± SD for four mice per time point is given. Growth (B) and survival (C) of Mtb strains in vitro in SMM containing glutamate (0.2%) as the sole carbon source. The mean ± SD of triplicate cultures is given. The OD580 at the mid-growth phase of Mtb strains in SMM containing 0.2% glycerol (D) or 0.2% acetate (E) as the sole carbon source, with varying amounts of additional glutamate, is shown. All results are representative of at least two independent experiments.
Mtb Requires KDHC for Glutamate Catabolism.
We probed why both Δhoas and Δhoas::E956Ahoas fail to retain full virulence in mice by first investigating the role of HOAS in Mtb metabolism. We assessed the growth of WT and Δhoas Mtb on 192 individual carbon sources. Glutamate was the only substrate tested on which WT Mtb grew and Δhoas did not (Fig. 1B). Likewise, the active site mutant Δhoas::E956Ahoas was unable to grow in glutamate (Fig. 1B). When we fed the strains U-13C15N glutamate, both Δhoas and Δhoas::E956Ahoas incorporated the labeled isotopomer into their metabolomes like WT (SI Appendix, Fig. S6). Thus, failure to grow was not due to inability to take up glutamate. Despite not growing in glutamate, both Δhoas and Δhoas::E956Ahoas remained viable (Fig. 1C). Similarly, we found that Mtb deficient in the E2 of KDHC, DlaT (23), termed Δdlat, could not grow in the presence of glutamate yet also remained viable (SI Appendix, Fig. S7). In contrast, glutamate supported the growth of Mtb deficient in PdhC (ΔpdhC) (5), the lipoamide-bearing E2 component of the branched chain ketoacid dehydrogenase complex (BCKADHC) (SI Appendix, Fig. S7). Thus, both HOAS and DlaT appear to be essential for Mtb’s ability to grow on glutamate.
Glutamate not only failed to support growth of Δhoas or Δhoas::E956Ahoas but suppressed their growth on glycerol or acetate in a concentration-dependent manner (Fig. 1 D and E). Moreover, basal HOAS activity that was refractory to AcCoA modulation provided only a marginal growth advantage over complete HOAS deficiency when Δhoas::E1038Ahoas was given both glutamate and acetate (Fig. 1E), although this mutant grew like WT when offered combinations of glutamate and glycerol (SI Appendix, Fig. S8). Strikingly, glutamate also suppressed growth of Δdlat in combination with acetate (SI Appendix, Fig. S8). Together, these data reflect that both HOAS and DlaT participate in glutamate metabolism, suggesting that KDHC becomes essential specifically when Mtb is confronted with glutamate.
To confirm that HOAS, DlaT, and Lpd are capable of functioning as a canonical KDHC, we revisited earlier efforts (12) to detect KDHC activity with Mtb lysates and recombinant proteins, now armed with knowledge of the allosteric effect of AcCoA (15). We supplemented Mtb lysates with 1 mM AcCoA, but KDHC activity remained barely detectable. We then reconstituted the complex in vitro by preincubating high concentrations of HOAS (20 μM), DlaT (50 μM), and Lpd (20 μM) on ice for 2–3 h in the presence and absence of AcCoA (200 μM). We used high-resolution MS coupled to a RapidFire device (Agilent Life Sciences) to detect formation of SucCoA as identified by its m/z ratio and isotopomeric envelope compared with a standard (SI Appendix, Fig. S9). Formation of SucCoA was detected, and it was dependent on the presence of all three proteins. Activation by AcCoA (200 μM) enhanced product formation (SI Appendix, Fig. S10), as reported in an NADH formation assay (15). Table 1 lists the steady-state kinetic parameters. Thus, Mtb KDHC can function under certain conditions in vitro, albeit with a low turnover number (kcat = ∼1.9 min−1).
Table 1.
Steady-state kinetic parameters of dehydrogenase and peroxidase complex reactions
| Reaction conditions | 2-Ketoglutarate/pyruvate | CoA | tBuOOH* | NAD+ | |||
| kcat, min−1 | Km, mM | kcat, min−1 | Km, mM | kcat/Km, min⋅mM−1 | kcat, min−1 | Km, mM | |
| KDHC | 0.62 ± 0.05 | 0.10 ± 0.02 | 0.67 ± 0.08 | 0.09 ± 0.04 | — | 0.48 ± 0.05 | 0.04 ± 0.02 |
| KDHC + 200 μM AcCoA | 1.9 ± 0.2 | 0.3 ± 0.1 | 1.5 ± 0.2 | 0.09 ± 0.03 | — | 1.2 ± 0.1 | 0.04 ± 0.02 |
| HOAS peroxidase | 3.9 ± 0.2 | 0.6 ± 0.1 | 3.4 ± 0.4 | 0.4 ± 0.1 | 0.19 ± 0.01 | — | — |
| HOAS peroxidase + 200 μM AcCoA | 3.7 ± 0.2 | 0.4 ± 0.1 | 2.6 ± 0.2 | 0.11 ± 0.02 | 0.16 ± 0.01 | — | — |
| AceE peroxidase | 2.1 ± 0.1 | 0.34 ± 0.04 | 2.7 ± 0.2 | 0.40 ± 0.07 | 0.1 ± 0.01 | — | — |
tBuOOH is not saturating at up to 10 mM, (kcat/Km) determined from linear fit at low substrate concentrations.
Accumulation and Secretion of Four- and Five-Carbon TCA Cycle Intermediates in HOAS and DlaT-Deficient Mtb.
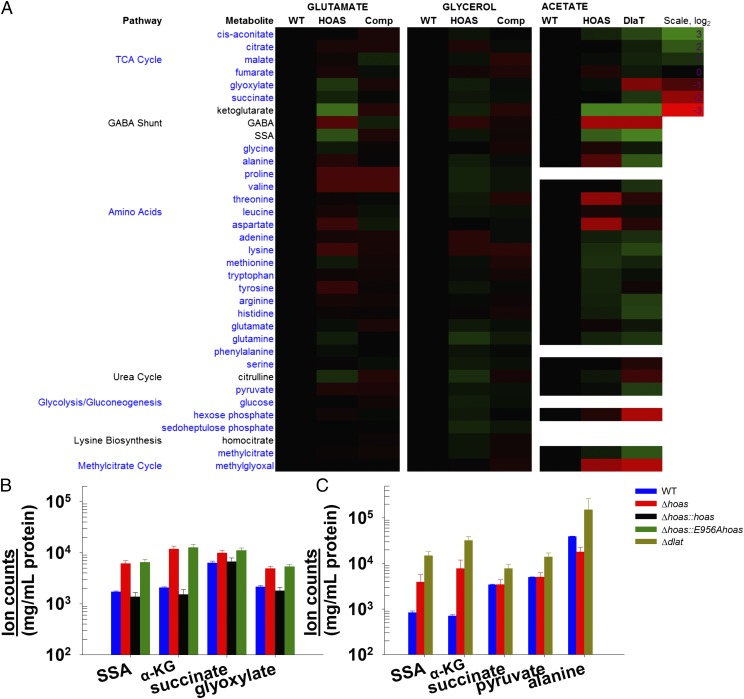
To investigate why the hoas mutants and Δdlat failed to grow, yet survived, when fed glutamate, we measured differences in intrabacterial metabolites between WT, Δhoas, Δhoas::hoas, Δhoas::E956Ahoas, Δhoas::E1038Ahoas, and Δdlat. When strains were fed glycerol, we observed no differences greater than twofold in the levels of TCA cycle, amino acid biosynthesis, or other CCM metabolites between WT, Δhoas, and Δhoas::hoas (Fig. 2A). These observations confirmed that the HOAS-dependent KDHC did not make a measurable contribution to Mtb’s TCA cycle when Mtb was cultured in medium containing glycerol. Similarly, we found no considerable differences between the strains in the abundance of most of the relevant metabolites examined when Mtb was offered glutamate (Fig. 2A). Strikingly, however, both Δhoas and Δhoas::E956Ahoas harbored markedly increased α-KG and SSA levels compared with WT, and slight increases in glyoxylate and succinate (Fig. 2B).
Fig. 2.
Mtb metabolome upon loss of KDHC function under various growth conditions. (A) Heat map profiles of select major pathways when H37Rv Mtb (WT), Δhoas (HOAS), Δhoas::hoas (Comp), or Δdlat (DlaT) was exposed to 0.2% glutamate, 0.2% glycerol, or 0.2% acetate in SMM. Quantitation of metabolites showing major changes when Mtb strains were exposed to 0.2% glutamate (B) or 0.2% acetate (C) is illustrated. The mean ± SD for triplicate experimental samples is given. P < 0.01. In A, the pathway names and corresponding metabolites are shown in fonts of the same color. All results are representative of at least two independent experiments.
We also observed markedly increased α-KG and SSA levels when Δhoas and Δdlat were fed acetate (Fig. 2 A and C) or acetate (0.2%) with glutamate (0.05%) (SI Appendix, Fig. S11A). SSA can arise not only from nonoxidative decarboxylation of α-KG but also from transamination between α-KG and GABA catalyzed by 4-aminobutyrate transaminase (Rv2589, GabT) (SI Appendix, Fig. S1B) Further, when supplied with glutamate, Δhoas and Δhoas::E956Ahoas accumulated intracellular glyoxylate, a cosubstrate of the HOAS carboligation reaction (Fig. 2B).
In parallel, we measured the levels of α-KG, SSA, glyoxylate, and succinate in the medium in which the strains were cultured. With glycerol as the carbon source, we saw no mutant-specific changes in these metabolites (SI Appendix, Fig. S11B). In contrast, with glutamate as the carbon source, Δhoas and Δhoas::E956Ahoas secreted higher levels than WT of the same metabolites (SI Appendix, Fig. S11C). Similarly, increased amounts of α-KG, SSA, glyoxylate, and succinate accumulated in the medium of Δdlat cultured with acetate or a combination of acetate and glutamate (SI Appendix, Fig. S11 D and E). Exogenously added SSA inhibited by >90% the growth of all of the strains cultured in growth-permissive SMM containing acetate ([SSA] = 1.25 mM) and glycerol ([SSA] = 5.0 mM) (SI Appendix, Fig. S12). In contrast, glyoxylate, α-KG, or succinate did not inhibit growth when added at up to 10 mM.
HOAS Activity Is Required for Optimal Mtb Survival in the Presence of RNIs.
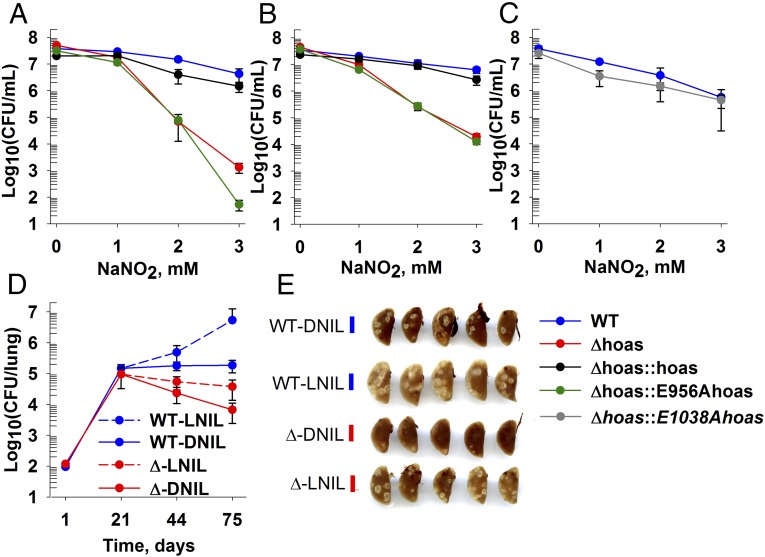
We next investigated the susceptibility of Δhoas to known host-derived bactericidal stresses in an effort to understand its decreased virulence. Several Mtb mutants that are hypersusceptible to RNIs in vitro are attenuated in mice expressing iNOS (23–26). Among those mutants is Δdlat (23). Given the striking metabolic similarities between Δhoas and Δdlat, we hypothesized that iNOS contributed to the attenuation of Δhoas Mtb. First, we tested whether Δhoas, like Δdlat, was hypersusceptible to RNIs in vitro. Mildly acidified nitrite produces small amounts of nitrous acid, whose dismutation gives rise to NO, NO2, and higher nitrogen oxides. NO reacts with superoxide from O2-respiring bacteria to form bactericidal peroxynitrite (5, 27). Mildly acidified nitrite is commonly used to generate fluxes of RNIs similar to those fluxes produced by immunologically activated macrophages (28). Indeed, acidified nitrite was far more bactericidal to Δhoas and Δhoas::E956Ahoas than to WT Mtb with either glutamate or glycerol as the carbon source (Fig. 3 A and B). The regulatory mutant Δhoas::E1038Ahoas survived exposure to RNIs as well as WT Mtb (Fig. 3C). In contrast, Δhoas and Δhoas::E956Ahoas were no more sensitive than WT to acid, starvation, or H2O2 (SI Appendix, Fig. S13 A–C), even in the presence of growth-suppressive concentrations of glutamate. Thus, although Mtb has numerous, independent defenses against H2O2 (29), the enzymatic activity of HOAS serves as a major, nonredundant defense of Mtb against RNIs.
Fig. 3.
Increased sensitivity of Δhoas to RNIs in vitro and partial reversion of its attenuation in vivo upon inhibition of iNOS. Survival of Mtb strains after 4 d of exposure to NaNO2 at pH 5.5 in SMM containing 0.2% glycerol (A and C) or 0.2% glutamate (B). The mean ± SD of triplicate experimental samples is shown from one experiment, representative of at least two experiments. (D) Fate of Mtb strains in lungs of C57BL/6 mice after aerosol infection and start of L-NIL or D-NIL treatment at day 21. The mean ± SD for four mice per point is shown from one experiment, representative of two experiments. P values for differences between L-NIL and D-NIL groups infected with Δhoas at day 44 = 0.056 and at day 75 = 0.048 (two-tailed t test) are shown. (E) Gross pathology of lungs was recorded at day 75 p.i.
HOAS Is Required for Mtb to Persist in iNOS-Expressing Mice.
Considering the extensive killing of Δhoas by RNIs in vitro and the failure of Δhoas to survive during the chronic phase of infection in mice, we tested whether iNOS contributed to attenuation of Δhoas Mtb in vivo. We administered the selective substrate analog iNOS inhibitor L-N6-(1-iminoethyl)Lys (L-NIL) (30) or the inactive enantiomer D-N6-(1-iminoethyl)Lys (D-NIL) to mice infected with either WT or Δhoas beginning on day 22 postinfection (p.i.). By day 75 p.i., lungs from L-NIL–treated mice infected with WT harbored close to 2 log10 more Mtb than lungs from D-NIL–treated mice (Fig. 3D). For mice infected with Δhoas and treated with D-NIL, bacterial numbers fell 1 log10 below WT levels. In contrast, in mice treated with L-NIL, colony-forming units of Δhoas plateaued, failing to match the titers in lungs of L-NIL–treated mice infected with WT. Pulmonary consolidation was correspondingly greater in mice infected with WT or Δhoas and treated with L-NIL than in the mice given D-NIL (Fig. 3E).
Thus, inability of Δhoas to persist in the mouse is due, at least in part, to iNOS. Because the Mtb burden in lungs of mice treated with L-NIL was not as high as in mice with iNOS deficiency (22), inhibition of iNOS by L-NIL must not have been complete. Thus, we cannot tell to what degree Δhoas Mtb was attenuated because of hypersusceptibility to the products of iNOS. Nonetheless, we next explored the enzymatic basis of the increased susceptibility of Δhoas.
Oxidative Decarboxylation of 2-Ketoacids Can Provide Reducing Equivalents via DlaT to Support the Alkylhydroperoxide Reductase Colorless Subunit/Alkylhydroperoxide Reductase Colorless Subunit Gene-Dependent Peroxynitrite Reductase/Peroxidase Pathway.
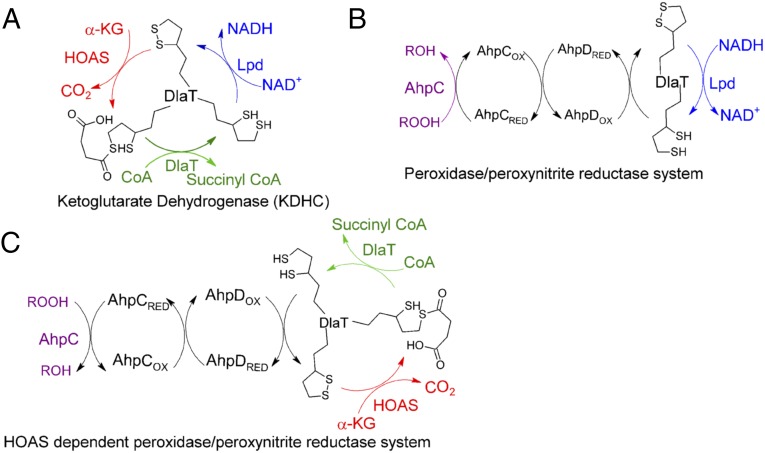
Alkylhydroperoxide reductase colorless subunit (AhpC) and homologous peroxiredoxins are among the most widely distributed antioxidant systems in biology and function as both a peroxidase (P) and a peroxynitrite reductase (PNR) (31). However, species differ in the source of reducing power for the peroxiredoxin-dependent PNR/P. Many use NADPH as the source of electrons. In contrast, Mtb draws on NADH via a four-protein complex in which Lpd acts as E1, running in reverse from its role in KDHC, PDHC, or BCKADHC (32). DlaT serves as E2, the product of a gene neighboring ahpC (AhpD) serves as E3, and AhpC serves as E4 (32). Specifically, Lpd uses NADH to reduce DlaT-bound lipoamide groups, which, in turn, reduce AhpD, a thioredoxin-like protein that reduces the disulfide bonds in AhpC’s catalytic center after AhpC reduces peroxides and peroxynitrite to alcohols, water, and nitrite, respectively (Fig. 4).
Fig. 4.
Redox steps in KDHC (A), Lpd-dependent PNR/P system (B), and HOAS- or AceE-dependent PNR/P system (C) depicting the cycling between oxidized and reduced forms of DlaT-bound lipoamide.
DlaT-bound lipoamide groups are also reduced in the catalytic cycle of canonical 2-ketoacid dehydrogenase complexes to produce AcCoA from 2-ketoacids. In these reactions, NADH is produced by Lpd, not consumed, concomitant with oxidation of the lipoamide groups on DlaT (Fig. 4). Because reduced lipoyl groups are intermediates in both reactions, we hypothesized that in the absence of Lpd and NADH, the DlaT/AhpC/AhpD system could harness reducing equivalents from the oxidative decarboxylation of 2-ketoacids, and thereby reduce peroxides or peroxynitrite. The E1 in this reaction could be either HOAS [acting as α-ketoglutarate decarboxylase (KGD) on α-KG] or pyruvate decarboxylase (AceE; acting as pyruvate decarboxylase on pyruvate) (33, 34), with AcCoA as a product in each case (Fig. 4).
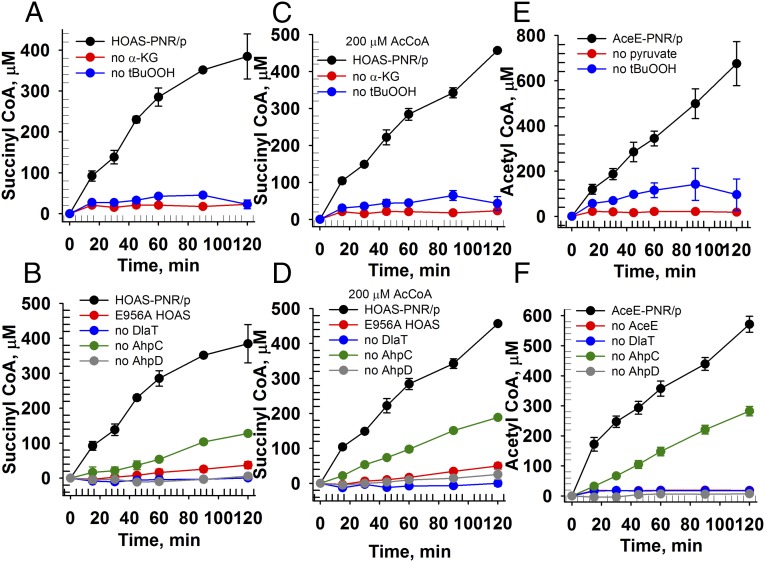
We tested this hypothesis by determining the catalytic production of SucCoA by HOAS, DlaT, AhpC, and AhpD in the presence of CoA, α-KG, and t-butylhydroperoxide [a known substrate for AhpC (32)]. SucCoA was indeed produced, and this process required the presence of each of the foregoing enzymes and reactants (Fig. 5 A–D). The reaction depended on the catalytic activity of HOAS and on the concentrations of α-KG, CoA, and t-butylhydroperoxide, and it followed Michaelis–Menten kinetics (SI Appendix, Figs. S14 and S15). Steady-state kinetic parameters are summarized in Table 1. Similarly, AceE, DlaT, AhpC, and AhpD formed AcCoA in the presence of pyruvate and t-butylhydroperoxide. This reaction likewise depended on the presence of all enzyme components and on the concentrations of pyruvate, CoA, and t-butylhydroperoxide (Fig. 5 E and F), and it followed Michaelis–Menten kinetics (Table 1 and SI Appendix, Figs. S16 and S17).
Fig. 5.
Reconstitution of α-ketoacid–dependent PNR/P systems. SucCoA production by a HOAS-dependent PNR/P system depends on substrates (A) and individual enzyme components (B) in the presence of 200 μM AcCoA (C and D). AcCoA production by an AceE-dependent PNR/P system depends on substrates (E) and individual enzyme components (F). Michaelis–Menten plots and kinetic constants are described in Table 1 and SI Appendix, Figs. S14 and S16. The MS-based assay and data analysis are detailed in Materials and Methods.
Discussion
Enzymes of Mtb’s CCM, including some that subserve glycolysis, gluconeogenesis, the TCA cycle, and the glyoxylate shunt, make an important contribution to the pathogen’s virulence (5–7, 23, 35, 36). Evidence is emerging that some of these enzymes are not simply playing the metabolic roles for which they are annotated, but can protect the pathogen by participating in antioxidant or antinitroxidative defense. Such a role was first seen with DlaT (23, 37) and Lpd (5), and later with the isocitrate lyases (38). Here, we have added HOAS to that list and identified two four-component peroxidase systems, HOAS/DlaT/AhpD/AhpC and AceE/DlaT/AhpD/AhpC, that can sustain peroxidatic action with alternative sources of electrons besides NADPH and NADH, namely, α-KG and pyruvate. Finally, we have helped clarify the role of the KDHC in Mtb.
HOAS can function in KDHC in vitro. However, KDHC itself seems to play a limited role in Mtb’s TCA cycle during growth on glycolytic or fatty acid carbon sources. In the presence of KDHC, there is little flux of carbon through the node of the TCA cycle that KDHC controls (4) and deletion of its E1 has little or no impact on Mtb’s growth in those conditions, consistent with the low KDHC enzymatic activity observed here and elsewhere (15) and the minimal effect of HOAS deletion on metabolism when bypass pathways like the glyoxylate and GABA shunts are available. Perhaps SucCoA synthetase accounts for the sufficiency of SucCoA for biosynthetic purposes (39). In contrast, we observed drastic metabolic and growth phenotypes when HOAS-deficient Mtb was grown in glutamate. Incomplete metabolism of glutamate/α-KG in HOAS-deficient Mtb led to intracellular and extracellular accumulation of potentially toxic aldehydes, one of which, SSA (40), abrogated growth when added extracellularly.
Why did aldehydes accumulate intracellularly and extracellularly when HOAS-deficient Mtb was presented with extracellular glutamate? WT Mtb can proliferate exponentially with glutamate as its only carbon source. However, extracellular glutamate stopped Mtb from replicating if HOAS was absent or inactive. Glutamate enters the TCA cycle through oxidative deamination to α-KG or as succinate via the GABA shunt. Metabolomic analysis revealed a glutamate-dependent build-up of α-KG and SSA in both Δhoas and Δhoas::E956Ahoas. SSA could arise from HOAS-catalyzed nonoxidative decarboxylation of α-KG or as a product of the GABA shunt in HOAS-deficient strains. SSA can be oxidized to succinate by SSADHs GabD1 and GabD2 (rv0234c and rv1731) (13). GabD1 is inhibited by high concentrations of substrate SSA (41) as well as by glyoxylate (42). Not only could glutamate metabolism drive the GABA shunt but HOAS deficiency could lead to an increase in glyoxylate through a decrease in its disposition through the HOAS reaction. Thus, elevation of SSA could reflect the joint impact of two effects of HOAS deficiency: increased activity of the GABA shunt and inhibition of SSADH activity by increased glyoxylate. The toxic potential of SSA illustrates the principle that intermediary metabolism produces not just molecules that sustain life but life-threatening molecules as well, as shown for Mtb, which also accumulates branched chain α-ketoacids, propionate, maltose 1-phosphate, or glycerol phosphate when the relevant metabolic pathways are disrupted by chemical or genetic means (5, 8, 43–47).
Besides accumulation of growth-inhibitory SSA, other mechanisms may account for or contribute to the suppressive effect of glutamate on growth of HOAS-deficient Mtb, which stands in contrast to the ability of WT Mtb to cocatabolize different substrates, including glutamate, for optimal growth (4). In some bacteria, intracellular metabolites play a signaling role in carbon utilization. For example, in Escherichia coli, elevated α-KG inhibits enzyme I of the phosphotransferase system, blocking glucose uptake (19), and impairs cAMP synthesis, eliciting catabolite repression (48). Moreover, aldehydes acting as electrophiles can form transient adducts with select Lys residues to control enzyme activities and redirect CCM metabolism (49).
Except for dismutation reactions in which H2O2 or O2− serves as both an oxidant and a reductant, previously described systems of enzymatic detoxification of reactive oxygen intermediates and RNIs ultimately depend on reducing equivalents from NADH or NADPH, for which CCM is the major source. However, Lpd, NADH, and NADPH are all known targets of RNIs (50–52). By drawing electrons directly from CCM metabolites, and bypassing Lpd, defense systems that use HOAS or AceE may act as important complements to the Lpd-dependent system.
The turnover number for the PNR/P complexes described here was lower than one might expect for a life-sparing defense. We speculate that the conditions used in vitro did not adequately recapitulate the conditions in the intact cell. We could demonstrate a peroxidase reaction, but reagent peroxynitrite destroyed CoA so rapidly that we could not demonstrate a PNR reaction. Nonetheless, the four-enzyme system containing either HOAS or AceE was capable of cyclic reduction of AhpC, and AhpC was shown to reduce peroxynitrite in an assay system that does not depend on CoA (31).
The peroxidase reaction in which HOAS participates appears to be physiologically relevant during Mtb’s infection of the mouse. The role of HOAS in avoiding toxicity from glutamate anaplerosis may be required for virulence as well, but such a role could not be evaluated by the present studies. By complementing HOAS-deficient Mtb with an active site point mutant, we established that the role of HOAS in both cases is catalytic, not merely structural. HOAS deficiency did not increase susceptibility to the other physiological stresses tested or lead to amino acid auxotrophy (21, 39).
In sum, under the conditions studied here and earlier (23) in vitro and in mice, Mtb uses the E1 and E2 components of its KDHC not for growth but for defense against nutritional imbalance and host immune chemistry.
Materials and Methods
Materials.
β-Mercaptoethanol, β-NADH, bis(2-hydroxyethyl)-amino-Tris(hydroxymethyl)-methane (Bis-Tris methane), Na2EDTA, CoA, AcCoA, SucCoA, pyruvate, NAD+, NADH, t-butylhydroperoxide, hydrogen peroxide, sodium nitrite, potassium phosphate, disodium 2-ketoglutarate, morpholinoethanesulfonic acid, PMSF, potassium ferricyanide, carbenicillin, kanamycin, MgCl2, thiamin hydrochloride, and ThDP were obtained from Sigma Chemical Company. DTT was from USB, and isopropyl-β-d-1-thiogalactopyranoside was from Denville Scientific. QuikChange site-directed mutagenesis kits were from Stratagene.
Media and Culture Conditions.
Mtb was cultured at 37 °C in 7H9 complete medium (Middlebrook 7H9 broth supplemented with 0.2% glycerol, 0.5% BSA fraction V, 0.2% dextrose, 0.085% NaCl, and 0.05% Tween 80). Strains with antibiotic resistance cassettes were grown in the presence of 50 μg/mL hygromycin B (Δhoas, Δdlat), 15 μg/mL streptomycin (ΔpdhC), or a combination of 50 μg/mL hygromycin B and 25 μg/mL zeocin (Δhoas::hoas, Δhoas::E956Ahoas, Δhoas::E1038Ahoas) (5). Modified SMM was used as a base medium for both carbon-defined growth curves and certain survival assays, and it contained 0.05% ammonium sulfate, 0.05% monopotassium phosphate, 0.2% citric acid, 0.005% ferric ammonium citrate, 0.05% magnesium sulfate, 0.01% zinc sulfate, and 0.02% tyloxapol (pH 7.4). For these experiments, indicated concentrations of carbon substrates are expressed as a percentage (vol/vol) for glycerol, a percentage (wt/vol) for acetate, and a percentage (wt/vol) for glutamate. To determine colony-forming units, bacteria were plated on Middlebrook 7H10 agar with 0.5% glycerol and 10% (vol/vol) OADC (Beckon Dickinson).
Mutant Construction.
Native hoas was replaced in H37Rv Mtb with a hygromycin-resistant cassette via allelic exchange using a recombineering approach. A hygromycin resistance gene flanked by ∼800 bp of the genomic region immediately upstream and downstream (123 bp of the C-terminal coding region of hoas included) of hoas was amplified by PCR. The product was electroporated into competent cells from H37Rv Mtb harboring a recombineering plasmid [H37Rv/pNIT(kan)::RecET-SacB] after nitrile induction for 8 h at 37 °C. Transformants were plated onto 7H10 agar supplemented with 50 μg/mL hygromycin B. Candidate clones were screened for kanamycin sensitivity, implicating the loss of the pNIT(kan)::RecET-SacB recombineering plasmid. Deletion mutant colonies were subsequently confirmed by Southern blot. DNA from WT and Δhoas was digested with PvuI (New England Biolabs), and fragments were separated by agarose gel electrophoresis and transferred to a nylon membrane. The membrane was probed with a 1-kb labeled fragment to confirm bands of 3.7 kbp for Δhoas and 1.6 kpb for WT.
Plasmids used to engineer the full-length hoas complementation plasmid (Δhoas::hoas) were made via Gateway Cloning Technology (Invitrogen). The hoas was cloned downstream of an hsp60 promoter into pDE43-MCZ to construct the final construct, pDE43hsp60hoas-MCZ, which integrates into the Mtb genome at the attL5 site. This vector was electroporated into Δhoas competent cells, and the transformation was plated on 7H10 agar supplemented with 50 μg/mL hygromycin B and 25 μg/mL zeocin. The plasmids used for Δhoas and Δhoas::hoas construction were a kind gift from Dirk Schnappinger, Weill Cornell Medical College, and the H37Rv/pNIT(kan)::RecET-SacB strain was a kind gift from Sabine Ehrt, Weill Cornell Medical College. The Δhoas::E956Ahoas and Δhoas::E1038Ahoas were made via site-directed mutagenesis of the pDE43hsp60hoas-MCZ plasmid with primers listed in Site-Directed Mutagenesis. Mutations were confirmed by sequencing. Plasmids were electroporated into Δhoas, and the transformation was plated on 7H10 agar with 50 μg/mL hygromycin B and 25 μg/mL zeocin. Complementation confirmation was assessed by Western blot.
Screening for Δhoas Growth Phenotype.
A carbon source-dependent growth phenotype of Δhoas was assessed on 192 carbon sources using Biolog PM microplates PM1 and PM2A. SMM (100 μL) was added to each well. Next, 100 μL of H37Rv and Δhoas at OD580 = 0.02 was added to the plates. Plates were incubated at 37 °C for 10 d, and culture densities were measured by absorbance. OD580 values for H37Rv and Δhoas were compared with assessed carbon source-dependent growth defects.
Survival in Glutamate.
Cultures were grown to mid-log phase in 7H9 complete medium and washed twice with modified SMM with 0.2% glutamate. Single-cell suspensions were made in the same medium by centrifuging cultures at 120 × g for 8 min to remove clumps. The inoculum was adjusted to OD580 = 0.01. Survival was determined by plating bacteria for colony-forming units.
Metabolomics.
Cultures were preconditioned in modified SMM with 0.2% glycerol for one full growth cycle. One milliliter of culture at OD580 = 1.0 was filtered through a nitrocellulose membrane (0.22 μM; Millipore GSWP 02500). Mtb-laden membranes were placed atop “swimming pools” constructed from inverted 15-mL falcon tube caps filled with modified SMM + 0.2% glycerol. Samples were incubated at 37 °C at 5% CO2 for 7 d to accumulate biomass, and checked daily to ensure that membranes were in constant contact with growth medium. Membranes were then transferred to fresh SMM-based pools with either 0.2% glycerol, 0.2% glutamate, and 0.2% acetate or 0.2% acetate + 0.05% glutamate for 48 h and were quenched with 700 μL of 40% (vol/vol) acetonitrile, 40% (vol/vol) methanol, and 20% water solution precooled to −40 °C. To analyze glutamate isotopomer uptake, U-13C15N glutamate replaced the unlabeled substrate in SMM. Bacteria were scraped off membranes and lysed by bead beating three times to obtain a small-molecule extract (SME). SME samples were passed through a 0.22 μM Spin X column (Corning). To analyze secreted metabolites, 800 μL of swimming pool medium was extracted from each pool and passed through a Spin X column (53). For MS analysis, both SME and secretome aliquots were mixed in a 1:1 ratio of acetonitrile and 0.2% formic acid and analyzed on an Agilent Accurate Mass 6220 TOF system coupled to an Agilent 1200 LC system using a Cogent Diamond hybrid type C column (Microsolve Technologies). SME concentrations were normalized to protein concentration as quantified via the Pierce BCA Protein Assay Kit (Thermo Scientific) (53).
Minimum Inhibitory Concentration Determination for Metabolites.
Cultures were preconditioned in modified SMM with 0.2% acetate for one full growth cycle. Cultures (OD580 = 0.02) were aliquoted into 96-well plates, and α-KG, SSA, glyoxylate, or succinate was added to the wells (0–10 mM) in a twofold dilution. Plates were incubated at 37 °C in 5% CO2 for 10 d. Growth was measured as change in OD580 at day 10. All experiments were carried out in triplicate.
Nitrosative Stress in Defined Carbon Sources.
Mtb strains were grown to mid-log phase in 7H9 complete medium and washed twice with SMM (acidified to pH 5.5) supplemented with either 0.2% glycerol or 0.2% glutamate. Samples were centrifuged at 120 × g for 8 min to remove clumps. The input inoculum was adjusted to an OD580 = 0.1 in the respective media. Experiments were conducted in 96-well plates. Aliquots of 100× NaNO2 (or water as a control) were added for final concentrations of 1, 3, and 5 mM NaNO2. Bacteria survival was determined by colony-forming units after a 5-d exposure.
Oxidative Stress.
Mtb strains were grown to mid-log phase in 7H9 complete medium and washed twice. Samples were centrifuged at 120 × g for 8 min, and the single-cell inoculum was adjusted to OD580 = 0.1 in the same medium. Bacteria were added to wells of a 96-well plate, and aliquots of 100× H2O2 stocks were added for a final H2O2 concentration of 1, 3, or 5 mM. Water was added to control wells. Bacteria were plated after 4 h for determination of colony-forming units.
Acid Stress.
Mtb strains were grown to mid-log phase in 7H9 complete medium and washed twice with either 7H9 complete medium acidified to pH 4.5 or with nonacidified 7H9 complete medium at pH 6.6. Samples were centrifuged at 120 × g for 8 min, and single-cell inocula were adjusted to OD580 = 0.1 in the respective media. Bacterial numbers were determined by colony-forming units for both input inoculum and after a 6-d exposure.
PBS Starvation.
Mtb strains were gown to mid-log phase in 7H9 complete medium and washed twice with PBS + 0.02% tyloxapol. Single-cell inocula were adjusted to OD580 = 0.1 in PBS + 0.02% tyloxapol. Survival was determined over time by plating bacteria for determination of colony-forming units at various time points.
Mouse Infections.
Female 6- to 8-wk-old C57BL/6 WT mice were infected with Mtb via an Inhalation Exposure System (Model 099C A4212; Glas-Col). Mtb strains were grown to mid-log phase in 7H9 complete medium and washed once with PBS + 0.05% Tween 80 (PBST). Single-cell suspensions were adjusted to OD580 = 0.2 in 6 mL of PBS. Lungs were homogenized in PBS, serially diluted in PBST, and plated on 7H10 agar for determination of colony-forming units. On days 28, 62, 112, and 217, the upper left lobes of the lungs were reserved and fixed in 10% (vol/vol) formalin for pathology. For NIL experiments, 4 mM L-NIL or inactive enantiomer D-NIL (30) was freshly administered in acidified (pH 2.7) drinking water every 48 h beginning on day 22 p.i. (22). On days 21, 45, and 75, the upper left lobes of the lungs were reserved and fixed in 10% (vol/vol) formalin for pathology. This protocol was approved by the Weill Cornell Medical College Institutional Animal Care and Use Committee.
Immunoblotting.
Mtb was grown in 7H9 complete medium to mid-log phase, washed twice in PBST, and resuspended in PBS with 1 mM PMSF. Samples were lysed by bead beating three times and centrifuged at 20,000 × g to obtain supernatants. Protein was diluted in reducing sample buffer to 10 μg, boiled, separated by a 10% SDS/PAGE gel, and transferred to a PVDF membrane. The membrane was probed with antiserum against KGD (1:10,000), now called HOAS (12). Antiserum against DlaT was used as a loading control (1;10,000) (5), and donkey anti-rabbit IgG (1:10,000; Amersham) was used for detection.
Cloning, Overexpression, and Purification.
HOAS was overexpressed and purified as described earlier (16). AceE, DlaT, LPD, AhpC, and AhpD were overexpressed and purified as described earlier (12, 31, 32).
Site-Directed Mutagenesis.
Site directed mutagenesis was performed using the QuikChange kit.
The following primers were used: (E956A) 5′-GCCACTGTCGGCGTACGCCGCCG-3′ (forward) and 5′-CGGCGGCGTACGCCGACAGTGGC-3′ (reverse) and (E1038A) 5′-GCAGTTGTGGGCGGCAGGTTCGATGACCA-3′ (forward) and 5′-TGGTCATCGAACCTGCCGCCCACAACTGC-3′ (reverse). All variants were expressed and purified as described earlier (16).
Enzyme Assays.
Protein concentrations were determined using the Bradford assay method. Ferricyanide reductase assays were performed as described earlier (16). A typical ferricyanide reductase assay reaction mixture (200 μL) in 20 mM Bis-Tris (pH 6.5) contained 5 mM MgCl2; 1.6 mM K3Fe(CN)6; 200–5,000 nM HOAS; and varying amounts of ThDP (0–500 μM), α-KG (0–20 mM), and AcCoA (0–2,000 μM). The time-dependent decrease in absorbance at 420 nm was monitored over 20 min at 37 °C. The linear region of the progress curves was used to calculate the steady-state velocities. All assays were carried out in triplicate in Corning 96-well transparent clear-bottom plates using a Spectramax M5 plate reader (Molecular Devices).
Multienzyme Complex Assays.
Experimental observations suggested that incubation at high concentrations and for prolonged periods on ice lead to the reconstitution of HOAS- and DlaT-dependent enzyme activities, in contrast to other bacterial KDHCs. Hence, multienzyme complexes were reconstituted on ice as high-concentration stocks (100–800 μL total volume) freshly before use and diluted 10-fold into the desired reaction mixture.
HOAS (20 μM), DlaT (50 μM), and LPD (20 μM) were incubated in 20 mM KH2PO4 (pH 7.0) containing additional 1 mM ThDP and 5 mM MgCl2 (incubation buffer) for 4 h on ice for KDHC assays. AcCoA (200 μM) was included in the incubation buffer and in the reaction buffer for the activated KDHC.
HOAS (20 μM), DlaT (50 μM), AhpC (20 μM), and AhpD (50 μM) were incubated in incubation buffer for 4 h on ice for HOAS-dependent peroxidase complex assays. AcCoA (200 μM) was included in the incubation buffer and in the reaction buffer for the activated complex.
AceE (20 μM), DlaT (50 μM), AhpC (20 μM), and AhpD (50 μM) were incubated in incubation buffer for >2 h on ice for AceE-dependent peroxidase complex assays.
A typical KDHC assay reaction mixture (25 μL) in 20 mM KH2PO4 (pH 7.0) contained 1 mM ThDP, 5 mM MgCl2, α-KG (0–5.0 mM), CoA (0–1.0 mM), NAD+ (0–1.0 mM), and 10× stock multienzyme complex (2.5 μL). The final concentrations in the reactions of individual enzymes were HOAS (2 μM), DlaT (5 μM), and LPD (2 μM). For progress curve experiments, the substrates were added at the highest concentrations and individual reaction mixtures were quenched with 75 μL of 0.1% formic acid at the desired times (0, 15, 30, 45, 60, 90, and 120 min). For Michaelis–Menten kinetics, individual substrates were varied as indicated. The constant substrates were added at their respective highest concentrations. Individual reaction mixtures were quenched with 75 μL of 0.1% formic acid at 60 min. All assays were carried out in triplicate in Corning 96-well transparent clear-bottom plates. Activated KDHC assay reaction mixtures contained AcCoA (200 μM) in addition.
A typical HOAS-dependent peroxidase assay reaction mixture (25 μL) in 20 mM KH2PO4 (pH 7.0) contained 1 mM ThDP, 5 mM MgCl2, α-KG (0–5.0 mM), CoA (0–1.0 mM), tert-butyl hydroperoxide (tBuOOH) (0–10.0 mM), and 10× stock multienzyme complex (2.5 μL). The final concentrations in the reaction of individual enzymes were HOAS (2 μM), DlaT (5 μM), AhpC (2 μM), and AhpD (5 μM). For progress curve experiments, the substrates were added at the highest concentrations and individual reaction mixtures were quenched with 75 μL of 0.1% formic acid at the desired times (0, 15, 30, 45, 60, 90, and 120 min). For Michaelis–Menten kinetics, individual substrates were varied as indicated. The constant substrates were added at their respective highest concentrations. Individual reaction mixtures were quenched with 75 μL of 0.1% formic acid at 60 min. All assays were carried out in triplicate in Corning 96-well transparent clear-bottom plates. Activated HOAS-dependent peroxidase assay reaction mixtures (25 μL) contained AcCoA (200 μM) in addition.
A typical AceE-dependent peroxidase assay reaction mixture (25 μL) in 20 mM KH2PO4 (pH 7.0) contained 1 mM ThDP, 5 mM MgCl2, pyruvate (0–5.0 mM), CoA (0–1.0 mM), tBuOOH (0–10.0 mM), and 10× stock multienzyme complex (2.5 μL). The final concentrations in the reaction of individual enzymes were AceE (2 μM), DlaT (5 μM), AhpC (2 μM), and AhpD (5 μM). For progress curve experiments, the substrates were added at the highest concentrations and individual reaction mixtures were quenched with 75 μL of 0.1% formic acid at the desired times (0, 15, 30, 45, 60, 90, and 120 min). For Michaelis–Menten kinetics, individual substrates were varied as indicated. The constant substrates were added at their respective highest concentrations. Individual reaction mixtures were quenched with 75 μL of 0.1% formic acid at 60 min. All assays were carried out in triplicate in Corning 96-well transparent clear-bottom plates.
RapidFire MS was used for the detection and quantitation of CoA, AcCoA, and SucCoA.
Samples (80 μL) were transferred to 384-well polypropylene plates for RapidFire MS analyses, and the plates were centrifuged at 180 × g for 30 s. A 10-point twofold dilution series (1,000 μM − 1.97 μM) standard curve in triplicates of CoA, SucCoA, or AcCoA was also transferred to the analyses plates. The standards were made in the reaction buffer (25 μL) and quenched with 0.1% formic acid (75 μL).
All of the samples were analyzed on an Agilent Life Sciences RapidFire 365 System using a 6230 TOF mass spectrometer fitted with a Jet Stream electrospray ionization source (Agilent). The packing material of the solid phase extraction cartridge used was hypercarb made of graphite carbon-based material that retains highly polar compounds. Thirty-five microliters of sample was aspirated in 600 ms; loaded onto the cartridge; washed with liquid chromatography (LC)-MS grade H2O supplemented with 5 mM ammonium acetate (pH 10) for 4,000 ms with a flow rate of 1.5 mL/min; and eluted with a mixture of LC-MS solvents, 50% H2O, 25% (vol/vol) acetone, and 25% (vol/vol) acetonitrile supplemented with 5 mM ammonium acetate (pH 10) for 5,000 ms with a flow rate of 1.25 mL/min. Finally, the cartridge was equilibrated for 5,000 ms. The mass spectrometer was operated in negative mode, and m/z transitions for CoA, SucCoA, and AcCoA were monitored. To determine the area under the curve (AUC), RapidFire Integrator software was used.
Data Analyses.
Standard curves for CoA, SucCoA, and AcCoA were determined by plotting the AUC against concentration and fitting to a linear equation. The standard curves were used to determine the concentrations of product SucCoA or AcCoA in the samples.
Kinetic data were fitted using the nonlinear, least square, curve-fitting program of SigmaPlot v.12.0 for Windows (Systat Software, Inc.). The data points are the means of experimental triplicates, and the error bars are the SDs. The solid lines are the regression fits. SEs are reported with the fitted constants.
Product formation (SucCoA or AcCoA) for a fixed time was plotted (Michaelis–Menten plots) against varying α-KG, CoA, NAD+, or tBuOOH, and the data were fit to a hyperbolic increase (Eq. 1). Here, p represents the concentration of product at time t, S is the concentration of substrate, V is the maximal velocity, and E is the concentration of enzymes. The fits yielded the kinetic parameters kcat, Km, and kcat/Km:
| [1] |
Supplementary Material
Acknowledgments
We thank Hyungjin Eoh, Madhumitha Nandakumar, Gregory Petsko, and Kristin Burns-Huang for advice and Xiuju Jiang for assistance (Weill Cornell Medical College). This work was supported by NIH Grant RO1AI64768. The Department of Microbiology and Immunology is supported by the William Randolph Hearst Foundation.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 13135.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510932112/-/DCSupplemental.
References
- 1.Donoghue HD, et al. Tuberculosis in Dr Granville’s mummy: A molecular re-examination of the earliest known Egyptian mummy to be scientifically examined and given a medical diagnosis. Proc Biol Sci. 2010;277(1678):51–56. doi: 10.1098/rspb.2009.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell DG, Barry CE, 3rd, Flynn JL. Tuberculosis: What we don’t know can, and does, hurt us. Science. 2010;328(5980):852–856. doi: 10.1126/science.1184784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubin EJ. The granuloma in tuberculosis--friend or foe? N Engl J Med. 2009;360(23):2471–2473. doi: 10.1056/NEJMcibr0902539. [DOI] [PubMed] [Google Scholar]
- 4.de Carvalho LP, et al. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol. 2010;17(10):1122–1131. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Venugopal A, et al. Virulence of Mycobacterium tuberculosis depends on lipoamide dehydrogenase, a member of three multienzyme complexes. Cell Host Microbe. 2011;9(1):21–31. doi: 10.1016/j.chom.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marrero J, Trujillo C, Rhee KY, Ehrt S. Glucose phosphorylation is required for Mycobacterium tuberculosis persistence in mice. PLoS Pathog. 2013;9(1):e1003116. doi: 10.1371/journal.ppat.1003116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puckett S, et al. Inactivation of fructose-1,6-bisphosphate aldolase prevents optimal co-catabolism of glycolytic and gluconeogenic carbon substrates in Mycobacterium tuberculosis. PLoS Pathog. 2014;10(5):e1004144. doi: 10.1371/journal.ppat.1004144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muñoz-Elías EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11(6):638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Carvalho LP, et al. Activity-based metabolomic profiling of enzymatic function: Identification of Rv1248c as a mycobacterial 2-hydroxy-3-oxoadipate synthase. Chem Biol. 2010;17(4):323–332. doi: 10.1016/j.chembiol.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole ST, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393(6685):537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 11.Bunik VI, Fernie AR. Metabolic control exerted by the 2-oxoglutarate dehydrogenase reaction: A cross-kingdom comparison of the crossroad between energy production and nitrogen assimilation. Biochem J. 2009;422(3):405–421. doi: 10.1042/BJ20090722. [DOI] [PubMed] [Google Scholar]
- 12.Tian J, et al. Mycobacterium tuberculosis appears to lack alpha-ketoglutarate dehydrogenase and encodes pyruvate dehydrogenase in widely separated genes. Mol Microbiol. 2005;57(3):859–868. doi: 10.1111/j.1365-2958.2005.04741.x. [DOI] [PubMed] [Google Scholar]
- 13.Tian J, Bryk R, Itoh M, Suematsu M, Nathan C. Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: Identification of alpha-ketoglutarate decarboxylase. Proc Natl Acad Sci USA. 2005;102(30):10670–10675. doi: 10.1073/pnas.0501605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffelder M, Raasch K, van Ooyen J, Eggeling L. The E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl residues of the acetyltransferase AceF. J Bacteriol. 2010;192(19):5203–5211. doi: 10.1128/JB.00597-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner T, Bellinzoni M, Wehenkel A, O’Hare HM, Alzari PM. Functional plasticity and allosteric regulation of α-ketoglutarate decarboxylase in central mycobacterial metabolism. Chem Biol. 2011;18(8):1011–1020. doi: 10.1016/j.chembiol.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Balakrishnan A, Jordan F, Nathan CF. Influence of allosteric regulators on individual steps in the reaction catalyzed by Mycobacterium tuberculosis 2-hydroxy-3-oxoadipate synthase. J Biol Chem. 2013;288(30):21688–21702. doi: 10.1074/jbc.M113.465419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Hare HM, et al. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol Microbiol. 2008;70(6):1408–1423. doi: 10.1111/j.1365-2958.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- 18.Amon J, Titgemeyer F, Burkovski A. A genomic view on nitrogen metabolism and nitrogen control in mycobacteria. J Mol Microbiol Biotechnol. 2009;17(1):20–29. doi: 10.1159/000159195. [DOI] [PubMed] [Google Scholar]
- 19.Doucette CD, Schwab DJ, Wingreen NS, Rabinowitz JD. α-Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat Chem Biol. 2011;7(12):894–901. doi: 10.1038/nchembio.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48(1):77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 21.Carrillo-Castañeda G, Ortega MV. Mutants of Salmonella typhimurium lacking phosphoenolpyruvate carboxykinase and alpha-ketoglutarate dehydrogenase activities. J Bacteriol. 1970;102(2):524–530. doi: 10.1128/jb.102.2.524-530.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacMicking JD, et al. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci USA. 1997;94(10):5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi S, Ehrt S. Dihydrolipoamide acyltransferase is critical for Mycobacterium tuberculosis pathogenesis. Infect Immun. 2006;74(1):56–63. doi: 10.1128/IAI.74.1.56-63.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302(5652):1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- 25.Darwin KH, Lin G, Chen Z, Li H, Nathan CF. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol. 2005;55(2):561–571. doi: 10.1111/j.1365-2958.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- 26.Darwin KH, Nathan CF. Role for nucleotide excision repair in virulence of Mycobacterium tuberculosis. Infect Immun. 2005;73(8):4581–4587. doi: 10.1128/IAI.73.8.4581-4587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci USA. 2000;97(16):8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gold B, et al. Nonsteroidal anti-inflammatory drug sensitizes Mycobacterium tuberculosis to endogenous and exogenous antimicrobials. Proc Natl Acad Sci USA. 2012;109(40):16004–16011. doi: 10.1073/pnas.1214188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.St John G, et al. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc Natl Acad Sci USA. 2001;98(17):9901–9906. doi: 10.1073/pnas.161295398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore WM, et al. L-N6-(1-iminoethyl)lysine: A selective inhibitor of inducible nitric oxide synthase. J Med Chem. 1994;37(23):3886–3888. doi: 10.1021/jm00049a007. [DOI] [PubMed] [Google Scholar]
- 31.Bryk R, Griffin P, Nathan C. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature. 2000;407(6801):211–215. doi: 10.1038/35025109. [DOI] [PubMed] [Google Scholar]
- 32.Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295(5557):1073–1077. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- 33.Feeney MA, et al. Repurposing lipoic acid changes electron flow in two important metabolic pathways of Escherichia coli. Proc Natl Acad Sci USA. 2011;108(19):7991–7996. doi: 10.1073/pnas.1105429108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bunik V, Follmann H. Thioredoxin reduction dependent on alpha-ketoacid oxidation by alpha-ketoacid dehydrogenase complexes. FEBS Lett. 1993;336(2):197–200. doi: 10.1016/0014-5793(93)80801-z. [DOI] [PubMed] [Google Scholar]
- 35.Trujillo C, et al. Triosephosphate isomerase is dispensable in vitro yet essential for Mycobacterium tuberculosis to establish infection. MBio. 2014;5(2):e00085. doi: 10.1128/mBio.00085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marrero J, Rhee KY, Schnappinger D, Pethe K, Ehrt S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc Natl Acad Sci USA. 2010;107(21):9819–9824. doi: 10.1073/pnas.1000715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryk R, et al. Selective killing of nonreplicating mycobacteria. Cell Host Microbe. 2008;3(3):137–145. doi: 10.1016/j.chom.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nandakumar M, Nathan C, Rhee KY. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun. 2014;5:4306. doi: 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- 39.Yu BJ, et al. sucAB and sucCD are mutually essential genes in Escherichia coli. FEMS Microbiol Lett. 2006;254(2):245–250. doi: 10.1111/j.1574-6968.2005.00026.x. [DOI] [PubMed] [Google Scholar]
- 40.Saito N, et al. Metabolite profiling reveals YihU as a novel hydroxybutyrate dehydrogenase for alternative succinic semialdehyde metabolism in Escherichia coli. J Biol Chem. 2009;284(24):16442–16451. doi: 10.1074/jbc.M109.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Carvalho LP, Ling Y, Shen C, Warren JD, Rhee KY. On the chemical mechanism of succinic semialdehyde dehydrogenase (GabD1) from Mycobacterium tuberculosis. Arch Biochem Biophys. 2011;509(1):90–99. doi: 10.1016/j.abb.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jakoby WB, Scott EM. Aldehyde oxidation. III. Succinic semialdehyde dehydrogenase. J Biol Chem. 1959;234(4):937–940. [PubMed] [Google Scholar]
- 43.Eoh H, Rhee KY. Methylcitrate cycle defines the bactericidal essentiality of isocitrate lyase for survival of Mycobacterium tuberculosis on fatty acids. Proc Natl Acad Sci USA. 2014;111(13):4976–4981. doi: 10.1073/pnas.1400390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Savvi S, et al. Functional characterization of a vitamin B12-dependent methylmalonyl pathway in Mycobacterium tuberculosis: Implications for propionate metabolism during growth on fatty acids. J Bacteriol. 2008;190(11):3886–3895. doi: 10.1128/JB.01767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griffin JE, et al. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem Biol. 2012;19(2):218–227. doi: 10.1016/j.chembiol.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalscheuer R, et al. Self-poisoning of Mycobacterium tuberculosis by targeting GlgE in an alpha-glucan pathway. Nat Chem Biol. 2010;6(5):376–384. doi: 10.1038/nchembio.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pethe K, et al. A chemical genetic screen in Mycobacterium tuberculosis identifies carbon-source-dependent growth inhibitors devoid of in vivo efficacy. Nat Commun. 2010;1:57. doi: 10.1038/ncomms1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.You C, et al. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature. 2013;500(7462):301–306. doi: 10.1038/nature12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moellering RE, Cravatt BF. Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science. 2013;341(6145):549–553. doi: 10.1126/science.1238327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richardson AR, et al. Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar typhimurium. Cell Host Microbe. 2011;10(1):33–43. doi: 10.1016/j.chom.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rhee KY, Erdjument-Bromage H, Tempst P, Nathan CF. S-nitroso proteome of Mycobacterium tuberculosis: Enzymes of intermediary metabolism and antioxidant defense. Proc Natl Acad Sci USA. 2005;102(2):467–472. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirsch M, de Groot H. Reaction of peroxynitrite with reduced nicotinamide nucleotides, the formation of hydrogen peroxide. J Biol Chem. 1999;274(35):24664–24670. doi: 10.1074/jbc.274.35.24664. [DOI] [PubMed] [Google Scholar]
- 53.Eoh H, Rhee KY. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2013;110(16):6554–6559. doi: 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.