Abstract
Hypoxia-inducible factor-1 (HIF-1) has been recognized as an important cancer drug target. Many recent studies have provided convincing evidences of strong correlation between elevated levels of HIF-1 and tumor metastasis, angiogenesis, poor patient prognosis as well as tumor resistance therapy. It was found that hypoxia (low O2 levels) is a common character in many types of solid tumors. As an adaptive response to hypoxic stress, hypoxic tumor cells activate several survival pathways to carry out their essential biological processes in different ways compared with normal cells. Recent advances in cancer biology at the cellular and molecular levels highlighted the HIF-1α pathway as a crucial survival pathway for which novel strategies of cancer therapy could be developed. However, targeting the HIF-1α pathway has been a challenging but promising progresses have been made in the past twenty years. This review summarizes the role and regulation of the HIF-1α in cancer, and recent therapeutic approaches targeting this important pathway.
Abbreviations: 4E-BP1, eukaryotic translation initiation factor 4E (eIF-4E) binding protein p70 S6 kinase (S6K); ADM, adrenomedullin; AhR, aryl hydrocarbon receptor; AKt, protein kinase B; ARD-1, arrest-defective-1; ARNT, aryl hydrocarbon nuclear translocator; bHLH, basic-helix-loop-helix; CAC, circulating angiogenic cells; ChIP, chromatin immunoprecipitation; C-MYC, myelocytomatosis virus oncogene cellular homolog; CoCl2, cobalt chloride; CPTs, camptothecins; C-TAD, COOH-terminal TAD; DFO, deferoxamine; EGF, epidermal growth factor; eIF-4E, eukaryotic translation initiation factor 4E; ELISA, enzyme-linked immunosorbent assay; EMSA, electrophoretic mobility shift assay; EPO, erythropoietin; ERK, extracellular signal-regulated kinase; FIH-1, factor inhibiting HIF-1; GA, geldanamycin; GAs, geldanamycins; GLUT1, glucose transporter 1; GLUT3, glucose transporter 3; GLUTs, glucose transporters; HDAC, histone deacetylase; HIF-1α, hypoxia-inducible factor-1α; HK1, hexokinase 1; HK2, hexokinase 2; HPH, HIF-1 prolyl hydroxylases; HRE, hypoxia response elements; Hsp90, heat shock protein 90; HTS, high throughput screens; ID2, DNA-binding protein inhibitor; IGF2, insulin-like growth factor 2; IGF-BP2, IGF-factor-binding protein 2; IGF-BP3, IGF-factor-binding protein 3; IPAS, inhibitory PAS; K, lysine residue; LDHA, lactate dehydrogenase; LEP, leptin; LRP1, LDL-receptor-related protein 1; Luc, luciferase; MAPK, mitogen-activated protein kinases; Mdm2, mouse double minute 2 homolog; MEK, MAPK/ERK kinase; MNK, MAP kinase interacting kinase; mTOR, mammalian target of rapamycin; MTs, microtubules; N, asparagine residue; NOS, nitric oxide synthase; N-TAD, NH2-terminal TAD; ODDD, oxygen dependent degradation domain; P, proline residue; PAS, Per and Sim; PCAF, p300/CBP associated factor; PHDs, prolyl-4-hydroxylases; PI3K, phosphatidyl inositol-4,5-bisphosphate-3-kinase; PKM, pyruvate kinase M; pVHL, von Hippel-Lindau protein; Raf, rapidly accelerated fibrosarcoma; Ras, rat sarcoma; RCC, renal cell carcinoma; RT-PCR, reverse transcription polymerase chain reaction; SIRT 1, Sirtuin 1; TAD, transactivation domains; TGF-α, transforming growth factor α; TGF-β3, transforming growth factor beta3; Top I, topoisomerase I; TPT, topotecan; VEGF, vascular endothelial growth factor
KEY WORDS: HIF-1α, HIF-1α inhibitors, Cancer drug discovery and development
Graphical abstract
Hypoxia-inducible factor-1 (HIF-1) has been recognized as an important cancer drug target. This review summarizes the role and regulation of the HIF-1α in cancer and recent therapeutic approaches targeting this important pathway.
1. The role of HIF-1α pathway in cellular adaptation to hypoxic stress
Mammalian cells need to maintain proper oxygen hemostasis in order to execute their aerobic metabolism and energy generation. In cancer, heart diseases, or chronic obstructive pulmonary disorders, the cellular oxygen balance is highly impaired, and cells become hypoxic (having low oxygen (O2) levels)1. Hypoxia is common in many types of solid tumors, where tumor cells proliferate rapidly and form large solid tumor masses, leading to obstruction and compression of the blood vessels surrounding these masses. These abnormal blood vessels often do not function properly and result in poor O2 supply to the center tumor regions2. Tumor cells in this hypoxic region begin to adapt these low oxygen tension conditions by activating several survival pathways. Activation of HIF-1 transcription factor is the most recognized pathway adopted by hypoxic cells in this harsh microenvironment (reviewed in references1, 2, 3, 4, 5, 6, 7, 8, 9, 10).
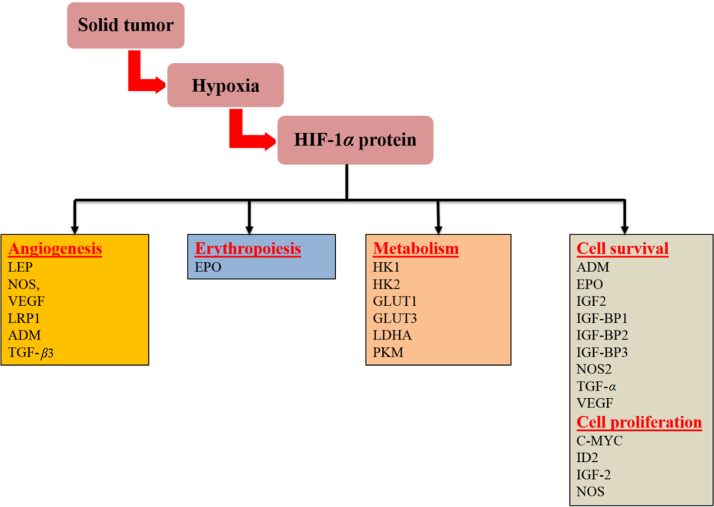
Activated HIF-1 plays a crucial role in adaptive responses of the tumor cells to changes in oxygen through transcriptional activation of over 100 downstream genes which regulate vital biological processes required for tumor survival and progression. Examples include genes involved in glucose metabolism, cell proliferation, migration and angiogenesis11 (Fig. 1). For example, in a rapidly growing tumor tissue, HIF-1 helps hypoxic tumor cells to shift glucose metabolism from the more efficient oxidative phosphorylation to the less efficient glycolytic pathway in order to maintain their energy production (the Warburg effect)12. For this reason, hypoxic cells tend to consume more glucose in order to meet their energy needs. HIF-1 mediates this metabolic conversion through the induction of enzymes involved in the glycolysis pathway and overexpression of glucose transporters (GLUTs) which increase glucose import into tumor cells3. Another example involves the HIF-1 transcriptional induction of several pro-angiogenic factors such as the vascular endothelial growth factor (VEGF), which in turn stimulates the development of new blood vessels to enrich tumor cells with oxygen for their growth13. In addition, HIF-1 promotes tumor metastasis into distant and more oxygenated tissues through the transcriptional activation of oncogenic growth factors such as transforming growth factor beta3 (TGF-β3), epidermal growth factor (EGF) and others13, 14. Altogether, HIF-1 activation in tumor cells is one of the key masters orchestrating their adaptation mechanisms to the hypoxia environment.
Figure 1.
Representative HIF-1α regulatory genes and their effects on cancer progression. LEP, leptin; NOS, nitric oxide synthase; VEGF, vascular endothelial growth factor; LRP1, LDL-receptor-related protein 1; ADM, adrenomedullin; TGF-β3, transforming growth factor-β3; EPO, erythropoietin; HK1, hexokinase 1; HK2, hexokinase 2; GLUT1, glucose transporter 1; GLUT3, glucose transporter 3; LDHA, lactate dehydrogenase; PKM, pyruvate kinase M; IGF2, insulin-like growth factor 2; IGF-BP2, IGF-factor-binding protein 2; IGF-BP3, IGF-factor-binding protein 3; TGF-α, transforming growth factor α; C-MYC, myelocytomatosis virus oncogene cellular homolog; ID2, DNA-binding protein inhibitor7.
2. Discovery of the HIF-1 protein
HIF-1 was initially discovered by Semenza and co-workers15 in 1991 during a study conducted on erythropoietin (EPO) gene, a gene responsible for encoding erythropoietin hormone for red blood cells production. Cis-acting DNA sequences (5′–RCGTG-3′) were identified in the 3′-flanking region of this EPO gene. These DNA sequences were found to be essential for the transcriptional activation of EPO gene in response to hypoxia and thus they were named as hypoxia response elements (HRE). Further studies discovered that the transcriptional activation of many regulatory genes is initiated through binding of a specific protein that is induced under hypoxia conditions to the HRE. Later this protein was identified as HIF-116.
3. HIF-1 structure
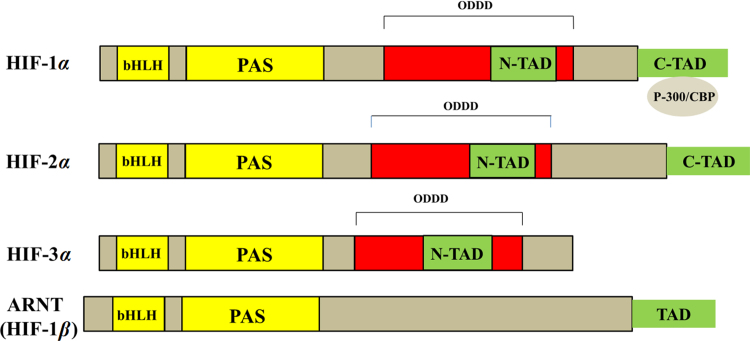
HIF-1 is a heterodimeric transcription factor, composed of two subunits, the HIF-1α (or its analogs HIF-2α and HIF-3α) and HIF-1β subunits17. HIF-1α is an oxygen sensitive subunit and its expression is induced under hypoxic conditions. In contrast, HIF-1β is constitutively expressed. HIF-1β is also known as aryl hydrocarbon nuclear translocator (ARNT), because it was discovered before HIF-1α and was identified as a heterodimeric partner of aryl hydrocarbon receptor (AhR). HIF-1β binds to AhR facilitating its translocation to the nucleus17. These two subunits belong to bHLH-PAS protein family, because their structures are related to two nuclear proteins found in Drosophila (Per and Sim, PAS) which have basic-helix-loop-helix (bHLH) motif18, 19 (Fig. 2). The bHLH proteins are characterized by having recognizable domains (b, HLH, PAS and TAD) which can regulate their own transcriptions as well as expression of other family members. In general, the bHLH-PAS motifs are essential to allow heterodimer formation between HIF-1α and HIF-1β subunits and for binding to the HRE-DNA sequence on the target genes. Base domains were found to have DNA binding properties required for binding the HRE on the gene, while HLH motif is where dimerization with other proteins occurs. Interestingly, PAS was found to be the only domain which is conserved among all members of this protein family (HIF-1α, ARNT, AhR, and PAS) and not found in any other types of proteins.
Figure 2.
Functional domains (bHLH, PAS, TAD) for proteins related to bHLH-PAS family. HIF-1α and HIF-2α share high degree of amino acid sequence similarities and both of them have two distinct TADs (C-TAD and N-TAD). In contrast, HIF-3α only has N-TAD.
The HIF-1α subunit has two transactivation domains (TAD): NH2-terminal (N-TAD) and COOH-terminal (C-TAD). These two domains are responsible for HIF-1α transcriptional activity20. C-TAD interacts with co-activators such as CBP/p300 to modulate gene transcription of HIF-1α under hypoxia. N-TAD is responsible for stabilizing HIF-1α against degradation21. Moreover, all HIF-α subunits are distinct from HIF-1β in that they all have an oxygen-dependent degradation domain (ODDD) overlapping N-TAD in their structures. This ODDD domain is important in mediating O2 regulation stability22 (Fig. 2).
Three closely related isoforms of HIF-α were known: HIF-1α, HIF-2α, and HIF-3α20, 23. Unlike HIF-3α, HIF-2α shows a noticeable degree of amino acid sequence similarity with HIF-1α (48% sequence similarity). This identity resemblance could explain their common capability of hetero-dimerization with HIF-1β and binding with HREs. Interestingly, HIF-1α and HIF-2α show different pattern of tissue distributions. HIF-1α is ubiquitously expressed in the body, but HIF-2α expression is stricter to specific tissues23. HIF-3α was less known until the discovery of the inhibitory PAS (IPAS), a spliced variant of HIF-3α. This domain doesn׳t show any intrinsic transactivation activity in comparison to the C-TAD of HIF-1α and HIF-2α, and it is a dominant-negative regulator over HIF-1/DNA binding ability24.
4. Regulation of HIF-1α pathway
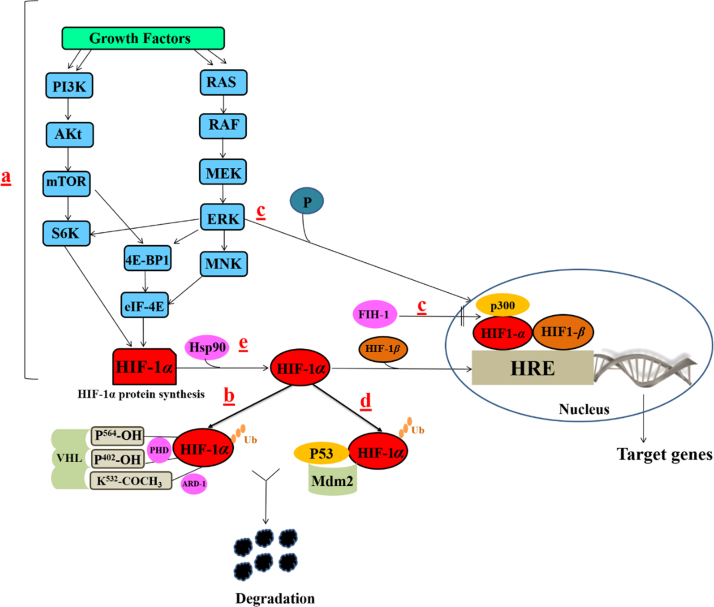
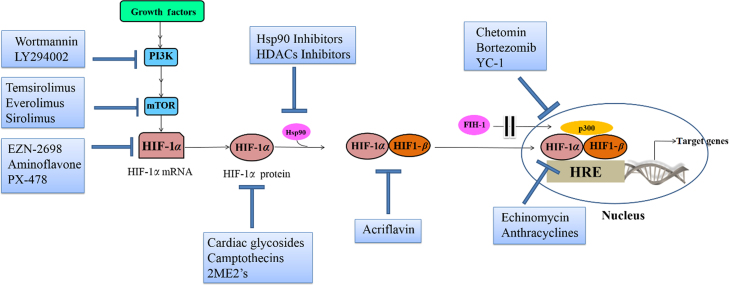
The activity and accumulation of HIF-1α protein were found to be regulated at different levels throughout its life cycle inside the cells. Independently from O2 levels, HIF-1α is constitutively transcribed and synthesized through a series of signaling events involving several growth factors and other signaling molecules25 (Fig. 3a). HIF-1α undergoes quick degradation under normoxic conditions and normally has a very short half-life (about 5 min)26. In contrast, under hypoxic conditions, several pathways have been shown to control HIF-1α stability and transcriptional activity via post-transnational modifications involving hydroxylation, acetylation, ubiquitination, and phosphorylation reactions, as discussed below25.
Figure 3.
Regulation of HIF-1α pathway at different levels. (a) Growth factors related pathways; (b) pVHL related pathways; (c) FIH-1 pathway; (d) Mdm2-p53 mediated ubiquitination and proteasomal degradation pathway; (e) Hsp90. Ras/Raf/MEK: Rat sarcoma/rapidly accelerated fibrosarcoma/MAPK/ERK kinase. These pathways regulate HIF-1α activity by regulating HIF-1α synthesis (a), HIF-1α stability (b, d, e), or HIF-1α transactivation (e, c).
4.1. Oxygen-dependent regulation of HIF-1α pathway ‘hypoxic regulation’
4.1.1. pVHL-dependent pathway
Under normal oxygen tension, HIF-1α protein expression is negatively regulated by proteasomal degradation and ubiquitination in a pathway involving von Hippel−Lindau protein (pVHL), a tumor suppressor protein and one of the recognized components of an E3 ubiquitin protein ligase27. The two proline residues (P402/ P564) located in the LXXLAP amino acid motif in the ODDD of HIF-1α were found to be good substrates for the action of a group of enzymes called prolyl-4-hydroxylases (PHDs) or HIF-1 prolyl hydroxylases (HPH)22, 28. These are 2-OG-dependent dioxygenases enzymes which require oxygen for their hydroxylation action, in addition to other co-factors such as iron and ascorbate. Hence, hydroxylation of proline residues by PHDs occurs only when there is sufficient amount of oxygen7, 28 (Fig. 3b). Moreover, another residue in the ODDD (lysine, K532) could be acetylated by an enzyme called arrest-defective-1 (ARD-1), an acetyl transferase enzyme29 (Fig. 3b). Although ARD activity does not depend on oxygen availability, the expression of ARD is decreased under hypoxia29. Consequently, modified HIF-1α subunits with hydroxylated P402/P564 and acetylated K532 moieties are preferably recognized by pVHL and are tagged for ubiquitination and proteasomal degradation30. Since both the hydroxylation action from PHDs and ARD-1 expression require the presence of oxygen, under hypoxic condition, neither hydroxylation nor acetylation of HIF-1α proline and lysine residues occurs, resulting in stabilization of HIF-1α structure.
4.1.2. pVHL-independent pathway
Another oxygen-dependent major mechanism for negative regulation of HIF-1α pathway under normoxia is through controlling HIF-1α transactivation. This pathway represents another level of post-translational modifications of HIF-1α transactivation domain but does not involve the pVHL protein. The transcriptional activation of HIF-1α target genes is initiated through the cooperative binding of C-TAD in the HIF-1α and the co-activator CBP/p300. In normoxia, oxygen dependent hydroxylation of HIF-1α asparagine (N803) residue by factor inhibiting HIF-1 (FIH-1), also known as asparginyl hydroxylase, blocks the interaction between the two domains, abrogating the subsequent HIF-1α mediated gene transcription31, 32, 33, 34 (Fig. 3c). Alternatively, hypoxia encourages this interaction through the inhibition of oxygen dependent hydroxylation of N803, resulting in transcriptional activation of the target genes32, 33, 34.
In summary, oxygen-dependent regulation of HIF-1α pathway involves a series of post-translational modifications. The pathway involving pVHL regulates HIF-1α stabilization, while that not involving VHL regulates HIF-1α transactivation.
4.2. Oxygen-independent oncogenic regulation of HIF-1α pathway
In addition to the major role of hydroxylases in regulating HIF-1α activity, there are pathways involving oncogenic activation that have considerable contributions in controlling HIF-1α protein levels. In non-hypoxic conditions, growth factors, cytokines and other signaling molecules tend to accumulate HIF-1α protein in cells.
4.2.1. Growth factor signaling pathways
Activation of phosphatidyl inositol-4,5-bisphosphate-3-kinase (PI3K) can upregulate the HIF-1α protein translation35, 36, 37. PI3K regulates protein syntheses through its target protein kinase B (Akt) and downstream component mammalian target of rapamycin (mTOR). mTOR mediates its action via phosphorylation of the eukaryotic translation initiation factor 4E (eIF-4E) binding protein (4E-BP1) disrupting the integrity of these two components, which is essential for inhibiting cap-dependent mRNA translation, resulting in enhanced HIF-1α protein translation38 (Fig. 3a). Additionally, phosphorylation of p70 S6 kinase (S6K) by mTOR promotes the phosphorylation of its substrate, the ribosomal protein S6, and induces protein translation38 (Fig. 3a). This pathway is antagonized by the tumor suppressor protein (PTEN) which reverses the phosphorylation of PI3K products38.
Certain growth factors activate RAS which in turn stimulates RAS/RAF/MEK/ERK kinase cascade36 (Fig. 3a). Activated ERK phosphorylates 4E-BP1, S6K, and MAP kinase interacting kinase (MNK). MNK can also phosphorylate eIF-4E directly39. The net result of these signaling events is the increased rate of mRNA translation into HIF-1α protein. Interestingly, ERK is not only involved in regulation of HIF-1α synthesis but also its transcriptional activation. ERK phosphorylates the co-activator CBP/p300 so it increases HIF-1α/p300 complex formation, and thus stimulats its transcriptional activation function39 (Fig. 3c).
4.2.2. Mdm2 pathway
It was observed that loss of p53 tumor suppressor gene in certain types of tumors is associated with elevated levels of HIF-1α40. This could be explained that in normoxia conditions, HIF-1α binds to the p53 and allows Mdm2 (mouse double minute 2 homolog) mediated ubiquitination and proteasomal degradation of HIF-1α41 (Fig. 3d). In hypoxic tumors, loss or mutations in tumor suppressor genes revokes any chances for Mdm2-mediated degradation of HIF-1α.
4.2.3. Heat shock protein 90 (Hsp90)
It was reported that inhibitors of Hsp90 such as geldanamycin (GA) could nullify HIF-1α levels regardless of the availability of oxygen42. Hsp90 is known to bind directly with HIF-1α inducing some conformational changes in its structure to fit and couple with HIF-1β initiating its transactivation43 (Fig. 3e). In addition, Hsp90 can stabilize HIF-1α against its non-VHL dependent degradation.
5. Targeting HIF-1α pathway for cancer therapy
Considering the multiple roles of HIF-1 in tumor progression and metastasis, there have been great interests in developing inhibitors targeting this pathway. It is clear that HIF-1α regulation pathway is a highly complex network involving several signaling cascades and overlapping mechanisms, each of which could serve as a promising target or step to intervene selectively. It is not surprising that most of the reported HIF-1α inhibitors were originally discovered for targeting other endogenous molecules and later their HIF-1α inhibitory activity was recognized through some empirical testing. For this reason, there are no specific HIF-1α inhibitors currently and developing specific HIF-1α inhibitors in hypoxic tumors represent a challenge.
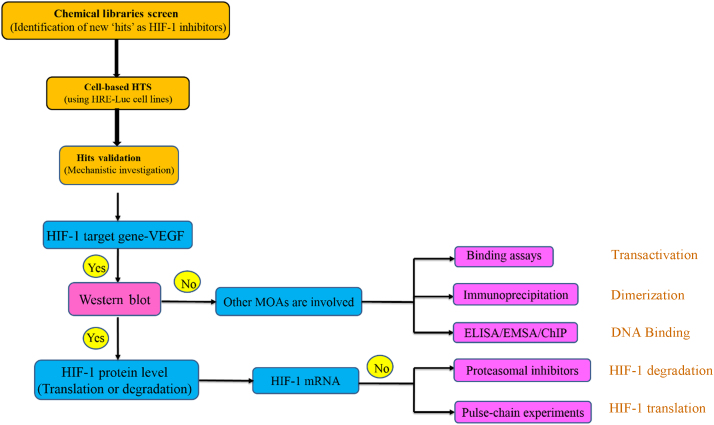
The searching process for novel HIF-1α inhibitors usually starts with chemical libraries screening as a preliminary step to identify novel molecules convenient for HIF-1α inhibition testing. Following this step, cell-based high throughput screens (HTS) are employed to test the identified compounds for their HIF-1α inhibitory action44. Cell-based HTS depends on measuring the level of HIF-1α induced in mammalian cells in response to hypoxia conditions (1% O2) or hypoxia activators, such as deferoxamine (DFO) and cobalt chloride (CoCl2). Stably transfected mammalian cell lines with constructs containing multiple copies of HRE that drives transcription of a reporter gene (luciferase (Luc) reporter gene is the most commonly used one) have been widely used for testing HIF-1α inhibition. HIF-1α inhibition can usually be achieved through modulation of one of the following steps: HIF-1α mRNA expression; HIF-1α protein level (protein translation or degradation); HIF-1α/HIF-1β dimerization; HIF-1α-DNA binding (HIF-1α/HRE); or HIF-1α transcriptional activity (CH-1 of p300/ C-TAD of HIF-1α).
To define which level within HIF-1α pathway is targeted by the identified hit in HTS, Melillo44 described a simple algorithm for validation purposes. This algorithm involves several consequential steps of either cell-based or cell-free assays to consider certain level while ruling out others involved in HIF-1α regulation to precisely dissect the mechanism of action of the tested inhibitor44 (Fig. 4).
Figure 4.
Schematic illustration of approaches adopted for discovery of HIF-1α inhibitors and investigation of their underlying inhibitory mechanisms of action (MOAs).
The first step of this validation scheme is to test the effect of the HIF-1α inhibitor on the hypoxic induction of mRNA expression of HIF-1 target genes, such as VEGF, GLUT1, or EPO by reverse transcription polymerase chain reaction (RT-PCR) or enzyme-linked immunosorbent assay (ELISA) method45. If the tested inhibitor showed negative induction effect on the target gene, Western blotting should be the following step to measure the HIF-1α protein levels in tumor cells cultured under hypoxia or in the presence of hypoxia mimics (DFO, CoCl2)46. In case the tested compound impaired the hypoxic induction of HIF-1α protein, the expected mechanism of action would be either hindering HIF-1α protein translation or increasing its degradation. To specify the involvement of protein translation or degradation, experiments can be conducted in the presence of proteasomal inhibitors, such as PS-341 or MG-13247. In addition, pulse-chain experiments are employed for assessment of de novo HIF-1α protein synthesis. The effect of the tested compound on HIF-1α mRNA expression should be excluded by evaluating its level48. If an inhibitor does not negate hypoxic induction of HIF-1α protein, the other mechanisms (DNA binding, protein dimerization, or transcriptional activity) should be considered for further investigation. ELISA, electrophoretic mobility shift assay (EMSA) and chromatin immunoprecipitation (ChIP experiments) are used to test for the inhibition of HIF-1/DNA binding49. Immunoprecipitation experiments are commonly conducted to detect the inhibition of HIF-1α/HIF-1β dimerization50. Unequivocally, the final step in this algorithm would be testing the inhibition of HIF-1α transcriptional activity through testing the interaction between the CH-1 domain of p300 and the C-TAD of HIF-1α peptides in a cell-free binding assay51. Fig. 4 illustrates these steps.
6. Molecules intervening the HIF-1α pathway
There are many small molecules reported as HIF-1α inhibitors in literature. We categorize them in this review based on their targets, structures and putative mechanisms of action (Figure 5, Figure 6, Figure 7 and Table 1).
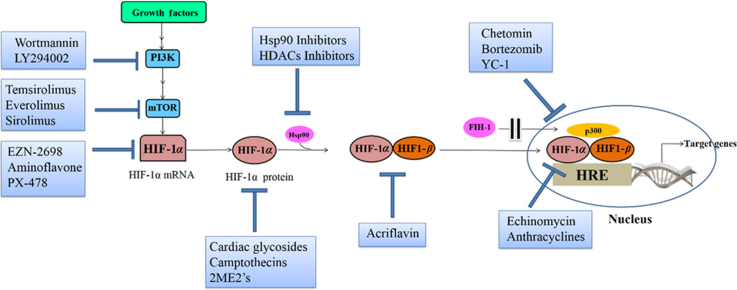
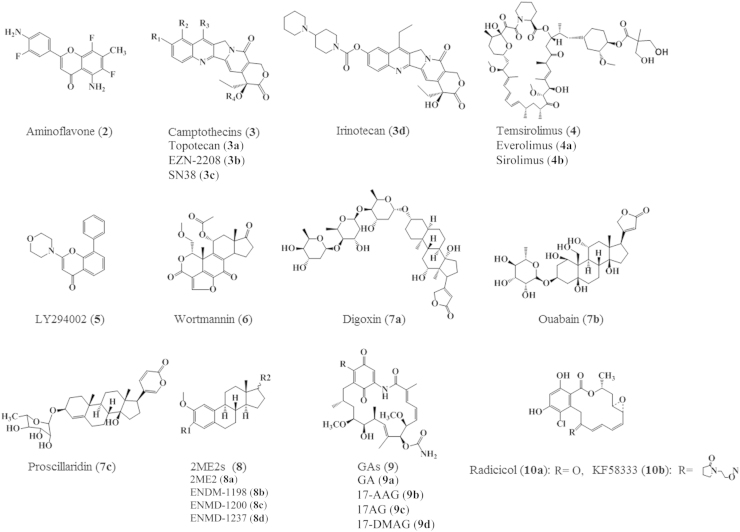
Figure 5.
HIF-1α inhibitors modulate different levels of the HIF-1α activation pathway.
Figure 6.
Chemical structures of molecules inhibiting HIF-1α pathway. Topotecan (3a): R1=OH, R2=CH2N(CH3)2, R3=H, R4=H; EZN-2208 (3b): R1=OH, R2=H, R3=CH2-CH3, R4=CO=CH2NHCOCH2O-(40k 4-arm-PEG); SN38 (3c): R1= OH, R2= H, R3= CH2CH3, R4=H; Everolimus (4a): R1=CH2CH2OH; Sirolimus (4b): R=OCH3; 2ME2 (8a): R1=OH, single bond, R2=OH; ENDM-1198 (8b): R1=CONH2, double bond, R2=H; ENMD-1200 (8c): R1=CONH2, single bond, R2==CH2; ENMD-1237 (8d): R1=CONH2, single bond, R2=H; GA (9a): R=OCH3; 17-AAG (9b): R=–CH2NHCH2CH=CH2; 17AG (9c): R=NH2; 17-DMAG (9d): R=–CH2NH(CH2)2N(CH3)2.
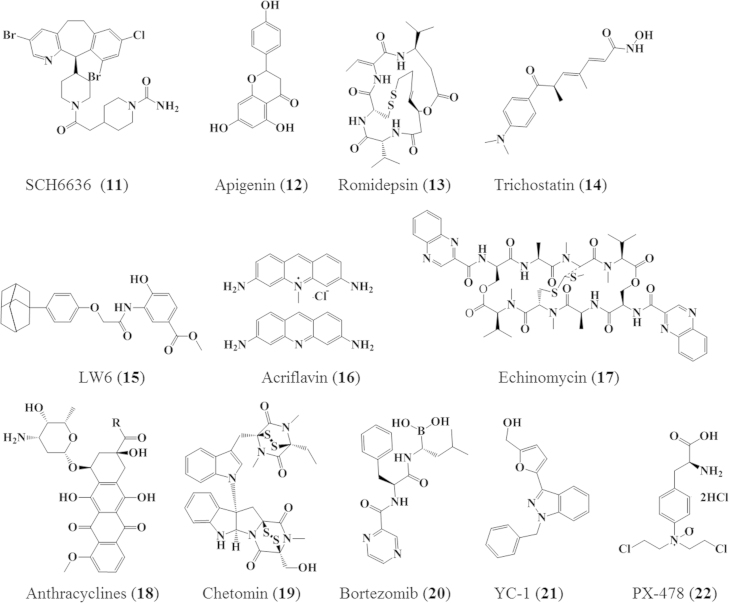
Figure 7.
Chemical structures of molecules inhibiting HIF-1α pathway. Doxorubicin (18a): R=CH2OH; Danuorubicin (18b): R=CH3.
Table 1.
Classification of molecules intervening HIF-1α pathway according to their putative mechanisms of action.
| Mechanism of inhibition | Inhibitor | Target | ||
|---|---|---|---|---|
| HIF-1α mRNA expression | EZN-2698 (1) | HIF-1α mRNA | ||
| Aminoflavone (2) | ||||
| HIF-1α translation | CPTs (3): | Topoisomerase I | ||
| topotecan (3a), EZN-2208 (3b), SN38 (3c), irinotecan (3d) | ||||
| Temsirolimus (4) | mTOR | |||
| Everolimus (4a) | ||||
| Sirolimus (4b) | ||||
| LY294002 (5) | PI3K | |||
| Wortmannin (6) | ||||
| Cardiac glycosides (7): | HIF-1α protein/HIF-2α mRNA | |||
| digoxin (7a), ouabain (7b), proscillaridin (7c) | ||||
| 2ME2’s (8): | Microtubules | |||
| 2ME2 (8a), ENDM-1198 (8b), ENMD-1200 (8c), ENMD-1237 (8d) | ||||
| HIF-1α stabilization | GAs (9): | Radicicol (10a), KF58333 (10b) | SCH66336 (11), Apigenin (12) | Hsp90 |
| GA (9a), | ||||
| 17-AAG (9b), | ||||
| 17AG (9c), | ||||
| 17-DMAG (9d) | ||||
| Romidepsin (KF228) (13) | HDAC | |||
| Trichostatin (14) | ||||
| LW6 (15) | HDAC/VHL | |||
| HIF-1α dimerization | Acriflavin (16) | PAS-B | ||
| HIF-1/DNA binding | Echinomycin (17) | 5′-CGTG-3′ | ||
| Anthracyclines (18): doxorubicin(18a), danuorubicin(18b) | HRE | |||
| HIF-1 transcriptional activity | Chetomin (19) | CH1 domain of p300 | ||
| Bortezomib (20) | C-TAD of HIF-1α and Asn803 of FIH | |||
| HIF-1α at multiple levels | YC-1 (21) | HIF-1&2 protein/FIH | ||
| PX-478 (22) | HIF-1&2 protein | |||
6.1. Inhibitors of HIF-1α mRNA expression
EZN-2698 (1) is an anti-sense oligonucleotide, an RNA antagonist which specifically binds and inhibits the expression of HIF-1α mRNA as well as HIF-1α target genes. It was reported that inhibition of HIF-1α by EZN-2698 attenuates HIF-1α protein level and tumor progression in various in vitro (human prostate and glioblastoma cell lines) and in vivo studies52. Phase I clinical trials revealed that EZN-2698 is a potential agent in patients with advanced renal cell carcinoma (RCC).
Aminoflavone (AF) (2) is another agent that inhibits HIF-1α mRNA expression. Although it is known as an AhR ligand, its exact mechanism of action as an inhibitor of HIF-1α mRNA expression is still unclear53.
6.2. Inhibitors of HIF-1α translation
Although the nature of the regulating HIF-1α protein translation is still poorly understood, several agents have been progressed that target the rate of HIF-1α protein synthesis, including but not limited to inhibitors of topoisomerase I and II, receptor tyrosine kinase, cyclin-dependent kinase, oncogenic pathways, and microtubule disrupting agents. However, the selectivity of these agents and the contribution of HIF-1α inhibition to their activities have not been fully established.
Camptothecins (CPTs, 3) analogues, which have topoisomerase I (Top I) inhibitory activity, can inhibit HIF-1α protein accumulation in hypoxic U251 human glioma cells. CPTs poison Top 1 by inducing the formation of stable Top 1-DNA cleavage complexes generating double strands DNA breaks48. Among the CPTs is topotecan (Hycamtin®–TPT, 3a) which was originally discovered at the national cancer institute (NCI) in an HTS using a cell-based assay of HIF-1 transcriptional activity45. Though TPT proved to be effective as antitumor agent in xenograft models54, its short half-life motivated the synthesis of other analogues with more favorable pharmacokinetic profiles. EZN-2208 (3b) is a TPT- congener, its high lipid solubility as a PEGylated analog of SN38 (3c), the active metabolite of irinotecan (Camptosar®–CPT-11, 3d)55, have improved its pharmacokinetic and antitumor properties in preclinical models of solid tumors and lymphomas56. A few phase I and II clinical trials have been conducted to provide evidence as for the anticancer activity of this agent.
mTOR Inhibitors (4) can also inhibit HIF-1α translation. The growth factor signaling pathway involving the downstream kinases PI3K/Akt/mTOR represents a major route of HIF-1α translation in over 70% of human cancer cell lines8. Temsirolimus (Torisel®–CCI-779, 4), and its counterparts, everolimus or rad001 (Afinitor®, 4a) and sirolimus (Rapamycin®, 4b) are mTOR inhibitors currently in clinical development for the treatment of solid tumors57. Although mTOR and the overall process of HIF-1α protein synthesis are limited under normoxia, it was found that 75% of sporadic clear cell RCCs are VHL-deficient, thus, in this case HIF-1α translation can proceed smoothly serving as a target for mTOR inhibitors intervention58. Temsirolimus and everolimus have passed phase III clinical trials and FDA approval successfully for treatment of metastatic RRC59. Besides being mTOR inhibitor, temsirolimus was investigated in clinical development for treatment of breast cancer owing to its antiangiogenic activity60. Among all inhibitors of the PI3K/AKT/mTOR pathway, only temsirolimus have gained such success.
PI3K inhibitors LY294002 (5) and wortmannin (6) have been recognized for their anti-HIF activity. High levels of PI3K have been recognized as a common feature of solid tumors, and it is up-regulated by either a gain of function in PI3K itself or in its downstream target Akt. As PI3K-specific inhibitors, LY294002 (5) and wortmannin (6) limited the synthesis of HIF-1α protein in the prostate carcinoma-derived cell lines PC-3 and DU145 in a dose-dependent fashion35.
Cardiac glycosides (7) [digoxin (7a), Ouabain (7b), and proscillaridin (7c)] have been used for the treatment of congestive heart failure for decades. Interestingly, these drugs were identified as inhibitors of HIF-1α protein translation and HIF-2α mRNA expression in a cell-based screen of a library of drugs in clinical trials. Digoxin, which is currently passing phase I clinical trials, was reported to inhibit established tumor xenografts with no effect on the growth of constitutively expressing HIF-1 xenografts. These findings imply the involvement of the HIF-1 in the antitumor activity of digoxin60.
2-Methoxyestradiol (2ME2, 8a), the natural metabolite and its synthetic analogues [ENMD-1198 (8b), ENMD-1200 (8c), and ENMD-1237 (8d)] have been recently recognized as antitumor and antiangiogenic agents in several clinical trials. Mechanistically, these agents bind to the colchicine binding site of tubulin and disrupt tumor interphase microtubules (MTs) which are required for HIF-1α down-regulation. It is worth mentioning that all of these compounds have shown favorable oral bioavailability, metabolic stability and safety profiles61, 62. The promising clinical benefits of cancer treatment with this line of agents will no doubt spur in research to more improved anti-tubulin drugs.
6.3. Inhibitors of HIF-1α stabilization
6.3.1. Hsp90 inhibitors
The binding of HSP90 to HIF-1α promotes HIF-1α activity in two ways: (1) it blocks the VHL-independent proteasomal degradation; and (2) it helps HIF-1α heterodimers acquire the appropriate conformation to recruit p300 and consequently initiate HIF-1 transactivation63. Therefore, Hsp90 antagonists are expected to induce HIF-1α degradation.
Geldanamycins (GAs, 9) are Hsp90 inhibitors. The natural benzoquinone ansamycin antibiotic “geldanamycin” (9a) was the first discovered Hsp90 inhibitor. It disrupts Hsp90 binding to client proteins by competing with its ATP binding site, exposing them to ubiquitination and proteasomal degradation. Geldanamycin semi-synthetic derivatives, 17-demethoxygeldanamycin (17-AAG, 9b) or tanespimycin, 17-dimethylaminoethylamino-17-demethoxygeldanamycin (17-DMAG, 9c) or alvespimycin hydrochloride, IPI-504 or retaspimycin, and 17-AG (IPI-493, 9d), the major metabolite of 17-AAG and IPI-504, have progressed successfully through clinical trials for further clinical development by different pharmaceuticals64. However, specificity of the antitumor activities of this group of agents should be addressed, given the fact that wide range of client proteins, not only HIF-1α may be affected by Hsp90 inhibition.
Radicicols (10) are another chemical class of Hsp90 inhibitors possessing different and overlapping mechanisms for HIF-1α inhibition. For example, radicicol (10a) prevents the interaction between HIF-1α/HIF-1β heterodimers and DNA attenuating the HIF-1 activation process without significant change in HIF-1α protein stability63. KF58333 (10b) down-regulates the HIF-1α and VEGF post-transcriptional expression in different human cancers65.
SCH66336 (11), a farnesyltransferase derivative demonstrating antitumor activity against non-small-cell lung cancer, inhibits the interaction between HIF-1α and Hsp90 promoting HIF-1α proteasomal degradation66.
Apigenin (12) is an antiangiogenic natural flavone. It was reported to block hypoxia-induced up-regulation of HIF-1α, GLUT-1, and VEGF proteins via degradation of HIF-1α through interference with the function of Hsp9067, 68.
6.3.2. Histone deacetylase (HDAC) inhibitors
Recent evidence indicates that HIF-1α acetylation by p300/CBP associated factor (PCAF) at K674 in an environment of low NAD+ is critical for promoting p300 recruitment and subsequent expression of HIF-1α target genes. Sirtuin 1 (SIRT 1), a redox-sensing deacetylase considered as class III histone deacetylase (HDAC), opposes this reaction by catalyzing HIF-1α deacetylation at high NAD+ levels. However, when hypoxia is induced, the NAD+/NADH levels increase due to glycolysis down-regulating SIRT-1 favoring HIF-1α activation69. The exact mechanism of HDAC inhibitors as HIF-1α inhibitors is still unclear, but they are known to antagonize the HIF-1α transactivation while upregulating ubiquitin-independent degradation pathways.
Romidepsin (13) and trichostatin (14) are known HDAC inhibitors. Romidepsin (Istodax®–FK228-FR901228, 13) is an anticancer agent isolated from the bacterium Chromobacterium violaceum that was first reported by Fujisawa Pharmaceutical Company (now Astellas Pharma) in 199470. Romidepsin is a prodrug with disulphide bond which undergoes reduction in cells by cellular reducing activity involving glutathione releasing a zinc-binding thiol. This thiol blocks class I HDACs activity through reversible interaction with a zinc atom in the binding pocket of Zn-dependent histone deacetylases. The glutathione-mediated activation also implicates its clinical usefulness for counteracting glutathione-mediated drug resistance in chemotherapy71. Romidepsin along with trichostatin (14), an antifungal antibiotic, are antiangiogenic HDAC inhibitors currently evaluated in clinical trials against a number of solid tumors72.
LW6 (15) decreases HIF-1α protein expression through potentiating VHL expression, which in turn interacts with prolyl-hydroxylated HIF-1α for proteasomal degradation. This is demonstrated by its antitumor activity in mice xenografts of human colon cancer HCT116 cells. LW6 is thought to upregulate VHL expression via different proposed mechanisms including demethylation of the VHL promoter, inhibition of HDAC, or suppression of VHL proteasomal degradation73.
6.4. Inhibitors of HIF-1 dimerization
Acriflavin (16) is a mixture of 3,6-diamino-10-methylacridinium chloride (trypaflavin) and 3,6-diaminoacridine (proflavine), which was recently identified as HIF-1α inhibitor in cell-based screening assays. Acriflavin inhibits HIF-1 dimerization by binding to the PAS-B subdomain of HIF-1α and HIF-2α. Its promising safety profile and characteristic ability of binding to both HIF-1α and HIF-2α subunits might distinguish it as a candidate for clinical evaluation especially in cancer subtypes where increased levels of HIF-1α and HIF-2α are the main cause of tumor progress74.
6.5. Inhibitors of HIF-1/DNA binding
Echinomycin (quinomycin A-NSC 526417, 17), a natural cyclic peptide belonging to quinoxaline antibiotic family, was previously identified in a HIF-1-targeted cell-based HTS at the NCI using 140,000 small-molecule library. It binds specifically to the core of the HIF-1 recognition sequence 5′–CGTG-3′ in a sequence-dependent fashion precluding HIF-1 and DNA binding. Although immunoprecipitation experiments proved its ability to selectively inhibit HIF-1/DNA binding, the cytotoxicity encountered during several phase I and II clinical trials limited further clinical evaluation of this agent49.
Anthracyclines (18), the potent well-known chemotherapeutic agents, also act as HIF-1α inhibitors. To determine the molecular basis for their HIF-1 inhibitory action, several mechanistic studies were conducted and demonstrated that anthracyclines; doxorubicin (DXR, 18a), danurubicin (DNR, 18b), epirubicin (EPI), and idarubicin (IDA) disrupt the binding of HIF-1 to DNA in hypoxic human cells. This is attributed to their ability to intercalate DNA and induce topoisomerase II-mediated strand breaks75. Moreover, administration of DXR or DNR to mice bearing human prostate cancer xenografts significantly inhibited tumor growth and vascularization, along with a decrease of circulating angiogenic cells (CAC). These findings imply that anthracyclines inhibition of HIF-1 activity reduced the expression of target genes (e.g., VEGF) responsible for tumor vascularization and growth. For this reason, these two agents attracted considerable interest for further preclinical studies to challenge the current paradigm available for treatment of prostate cancer which is characterized by elevated levels of HIF-1 protein76.
6.6. Inhibitors of HIF-1 transcriptional activity
Interruption of HIF-1α and HIF-1β binding to the co-factor p300 to form an active transcription complex, the final step towards HIF-1 transactivation, represents a potential mechanism by which small molecules may inhibit HIF-1 activity.
Chetomin (19), the antimicrobial dithioketopiperazine fungal metabolite, was identified as a small molecule blocking the transcriptional co-activation of HIF-1 pathway in 2004. Chetomin binds to and disrupts the tertiary structure of the CH1 domain of the co-activator p300, which hinders its interaction with HIF-1 and results in diminished HIF-1 transactivation51. While local toxicity of chetomin restricted its further development, an interesting study was carried out and demonstrated that chetomin HIF-1 inhibition attenuates the expression of HIF-1 target genes and enhances the radiotherapy efficacy in human HT 1080 cells77. Chetomin is one of many compounds indirectly affecting HIF-1α activity that showed great promise but has failed to exhibit safety in preclinical models.
Bortezomib (Velcade®–PS-341, 20), a proteasome inhibitor, inhibits p300-HIF-1α binding at nanomolar concentrations. Paradoxically, at higher concentrations, it increases the level of HIF-1α protein by blocking its proteasomal degradation. At the molecular level, bortezomib stimulates the interaction between C-TAD of HIF-1 and FIH target site (Asn803); thereby enhancing FIH-mediated repression of p300 recruitment. Bortezomib is FDA approved for the treatment of relapsed and refractory multiple myeloma more than 10 years ago78. More recently, the ability of bortezomib to downregulate the PI3K/AKT/mTOR pathway which is upstream of the HIF-1 pathway was demonstrated in prostate cancer cells79. Based on this discovery, recent studies were designed to assess the anticancer spectrum of bortezomib as sole agent or adjuvant therapy for the treatment of different types of cancers80, 81, 82.
6.7. Inhibitors of HIF-1α at multiple levels
YC-1 (21) is an antiplatelet aggregation agent. It was originally discovered as an activator of platelet guanylate cyclase83 and later it was found to have HIF-1 inhibitory activity. YC-1 can enhance FIH binding to HIF-α CAD, which in turn dissociates the binding of the later to p300 co-activator and lead to the functional repression of HIF-1. This mechanism does not involve the FIH active site, Asn80384. YC-1 can also impair HIF-1α and HIF-2α protein accumulation in a dose-dependent manner85. In addition, YC-1 inhibited HIF-1α via the FIH-dependent CAD inactivation as well as via the protein down-regulation in HT1080 and H1299 cancer cell lines84. Literature also revealed the anti-proliferative effect of YC-1 on cancer cells by arresting the cell cycle and promoting cell apoptosis86.
PX-478 (22) is a melphalan derivative which has potent antitumor activity. It was suggested that PX-478 lowers HIF-1α levels by affecting multiple levels in the HIF-1α pathway: (1) inhibition of HIF-1α deubiquitination via p53 and pVHL independent mechanism, resulting in increased levels of polyubiquitinated HIF-1α; (2) reduction of HIF-1α mRNA expression; and (3) inhibition of HIF-1α translation. In this regard, PX-478 was reported active in a panel of cell lines with prominent selectivity towards inhibition of HIF-1α protein translation87.
7. Conclusions
Hypoxia is a prominent micro-environmental feature in many types of solid tumors due to inadequate vascularization. It accounts for developing both radiotherapy and chemotherapy resistance. Tumor cells respond to changes in oxygen via delegating the transcriptional factor HIF-1α for orchestrating vast number of its essential cellular functions. Considering the profound impacts of HIF-1α on cancer progression via gene expression and the unsatisfactory efficacy of chemotherapy, there has been enormous growing interest in the biology of HIF-1α pathway and the development of direct or indirect HIF-1 inhibitors. The identification of novel HIF-1 inhibitors involves panels of cell-based or cell-free screens and assays, but the complexity involved in the regulation of the HIF-1α pathway has made the process of rational design of HIF-1α inhibitors very challenging. To date, no selective HIF-1α inhibitor has been clinically approved, partially due to the requirements of targeting protein-protein interactions without affecting other pathways. Nevertheless, there have been several approved drugs that indirectly affect the HIF-1α pathway and could serve as adjuvant therapy for certain types of cancers along with the existing treatments. Future directions would be directed towards developing drugs that are more specific for HIF-1 inhibition by better understanding of the molecular structure of domains that mediate critical functions of HIF-1. With the rapid advancement of molecular biology and emerging strategies in efficiently disrupting protein-protein interactions, it is very promising that selective HIF-1 inhibitors can be developed in the future.
Acknowledgments
This work was partially supported by the NIH grant 1R01CA148706 and funds from the University of Tennessee Health Science Center, College of Pharmacy. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
References
- 1.Semenza GL. HIF-1 and human disease: one highly involved factor. Genes Dev. 2000;14:1983–1991. [PubMed] [Google Scholar]
- 2.Powis G, Kirkpatrick L. Hypoxia inducible factor-1alpha as a cancer drug target. Mol Cancer Ther. 2004;3:647–654. [PubMed] [Google Scholar]
- 3.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 4.Ke QD, Costa M. Hypoxia-inducible factor-1 (HIF-1) Mol Pharmacol. 2006;70:1469–1480. doi: 10.1124/mol.106.027029. [DOI] [PubMed] [Google Scholar]
- 5.Patiar S, Harris AL. Role of hypoxia-inducible factor-1alpha as a cancer therapy target. Endocr Relat Cancer. 2006;13(Suppl 1):61–75. doi: 10.1677/erc.1.01290. [DOI] [PubMed] [Google Scholar]
- 6.Semenza GL. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol Med. 2002;8:S62–S67. doi: 10.1016/s1471-4914(02)02317-1. [DOI] [PubMed] [Google Scholar]
- 7.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 8.Xia Y, Choi H-K, Lee K. Recent advances in hypoxia-inducible factor (HIF)-1 inhibitors. Eur J Med Chem. 2012;49:24–40. doi: 10.1016/j.ejmech.2012.01.033. [DOI] [PubMed] [Google Scholar]
- 9.Bárdos JI, Ashcroft M. Negative and positive regulation of HIF-1: a complex network. Biochim Biophys Acta. 2005;1755:107–120. doi: 10.1016/j.bbcan.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Minet E, Michel G, Mottet D, Raes M, Michiels C. Transduction pathways involved in hypoxia-inducible factor-1 phosphorylation and activation. Free Radic Biol Med. 2001;31:847–855. doi: 10.1016/s0891-5849(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 11.Semenza GL. HIF-1 and mechanisms of hypoxia sensing. Curr Opin Cell Biol. 2001;13:167–171. doi: 10.1016/s0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 12.Weinhouse S, Warburg O, Burk D, Schade AL. On respiratory impairment in cancer cells. Science. 1956;124:267–272. doi: 10.1126/science.124.3215.267. [DOI] [PubMed] [Google Scholar]
- 13.Conway EM, Collen D, Carmeliet P. Molecular mechanisms of blood vessel growth. Cardiovasc Res. 2001;49:507–521. doi: 10.1016/s0008-6363(00)00281-9. [DOI] [PubMed] [Google Scholar]
- 14.Laderoute KR, Calaoagan JM, Gustafson-Brown C, Knapp AM, Li GC, Mendonca HL. The response of c-Jun/AP-1 to chronic hypoxia is hypoxia-inducible factor 1α dependent. Mol Cell Biol. 2002;22:2515–2523. doi: 10.1128/MCB.22.8.2515-2523.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semenza GL, Nejfelt MK, Chi SM, Antonarakis SE. Hypoxia-inducible nuclear factors bind to an enhancer element located 3ʹ to the human erythropoietin gene. Proc Natl Acad Sci USA. 1991;88:5680–5684. doi: 10.1073/pnas.88.13.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldberg MA, Dunning SP, Bunn HF. Regulation of the erythropoietin gene: evidence that the oxygen sensor is a heme protein. Science. 1988;242:1412–1415. doi: 10.1126/science.2849206. [DOI] [PubMed] [Google Scholar]
- 17.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang BH, Rue E, Wang GL, Roe R, Semenza GL. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J Biol Chem. 1996;271:17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- 19.Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Ko HP, Whitlock JP. Induction of phosphoglycerate kinase 1 gene expression by hypoxia. Roles of Arnt and HIF1α. J Biol Chem. 1996;271:21262–21267. doi: 10.1074/jbc.271.35.21262. [DOI] [PubMed] [Google Scholar]
- 21.Yamashita K, Discher DJ, Hu J, Bishopric NH, Webster KA. Molecular regulation of the endothelin-1 gene by hypoxia. Contributions of hypoxia-inducible factor-1, activator protein-1, GATA-2, AND p300/CBP. J Biol Chem. 2001;276:12645–12653. doi: 10.1074/jbc.M011344200. [DOI] [PubMed] [Google Scholar]
- 22.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 23.Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1α regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci USA. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makino Y, Cao RH, Svensson K, Bertilsson G, Asman M, Tanaka H. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414:550–554. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- 25.Kallio PJ, Pongratz I, Gradin K, McGuire J, Poellinger L. Activation of hypoxia-inducible factor 1α: posttranscriptional regulation and conformational change by recruitment of the Arnt transcription factor. Proc Natl Acad Sci USA. 1997;94:5667–5672. doi: 10.1073/pnas.94.11.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salceda S, Caro J. Hypoxia-inducible factor 1α (HIF-1α) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 27.Iwai K, Yamanaka K, Kamura T, Minato N, Conaway RC, Conaway JW. Identification of the von Hippel–Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc Natl Acad Sci USA. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Epstein ACR, Gleadle JM, McNeill LA, Hewitson KS, O׳Rourke J, Mole DR. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 29.Jeong J-W, Bae M-K, Ahn M-Y, Kim S-H, Sohn T-K, Bae M-H. Regulation and destabilization of HIF-1α by ARD1-mediated acetylation. Cell. 2002;111:709–720. doi: 10.1016/s0092-8674(02)01085-1. [DOI] [PubMed] [Google Scholar]
- 30.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 31.Block KM, Wang H, Szabó LZ, Polaske NW, Henchey LK, Dubey R. Direct inhibition of hypoxia-inducible transcription factor complex with designed dimeric epidithiodiketopiperazine. J Am Chem Soc. 2009;131:18078–18088. doi: 10.1021/ja807601b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dann CE, III, Bruick RK, Deisenhofer J. Structure of factor-inhibiting hypoxia-inducible factor 1: an asparaginyl hydroxylase involved in the hypoxic response pathway. Proc Natl Acad Sci USA. 2002;99:15351–15356. doi: 10.1073/pnas.202614999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNeill LA, Hewitson KS, Claridge TD, Seibel JF, Horsfall LE, Schofield CJ. Hypoxia-inducible factor asparaginyl hydroxylase (FIH-1) catalyses hydroxylation at the β-carbon of asparagine-803. Biochem J. 2002;367:571–575. doi: 10.1042/BJ20021162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang BH, Jiang G, Zheng JZ, Lu Z, Hunter T, Vogt PK. Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor 1. Cell Growth Differ. 2001;12:363–369. [PubMed] [Google Scholar]
- 36.Semenza G. Signal transduction to hypoxia-inducible factor 1. Biochem Pharmacol. 2002;64:993–998. doi: 10.1016/s0006-2952(02)01168-1. [DOI] [PubMed] [Google Scholar]
- 37.Conrad PW, Freeman TL, Beitner-Johnson D, Millhorn DE. EPAS1 trans-activation during hypoxia requires p42/p44 MAPK. J Biol Chem. 1999;274:33709–33713. doi: 10.1074/jbc.274.47.33709. [DOI] [PubMed] [Google Scholar]
- 38.Gingras A-C, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 39.Sang N, Stiehl DP, Bohensky J, Leshchinsky I, Srinivas V, Caro J. MAPK signaling up-regulates the activity of hypoxia-inducible factors by its effects on p300. J Biol Chem. 2003;278:14013–14019. doi: 10.1074/jbc.M209702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bae M-K, Ahn M-Y, Jeong J-W, Bae M-H, Lee YM, Bae S-K. Jab1 interacts directly with HIF-1α and regulates its stability. J Biol Chem. 2002;277:9–12. doi: 10.1074/jbc.C100442200. [DOI] [PubMed] [Google Scholar]
- 41.Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- 42.Isaacs JS, Jung YJ, Mimnaugh EG, Martinez A, Cuttitta F, Neckers LM. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1α-degradative pathway. J Biol Chem. 2002;277:29936–29944. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]
- 43.Gradin K, McGuire J, Wenger RH, Kvietikova I, fhitelaw ML, Toftgård R. Functional interference between hypoxia and dioxin signal transduction pathways: competition for recruitment of the Arnt transcription factor. Mol Cell Biol. 1996;16:5221–5231. doi: 10.1128/mcb.16.10.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melillo G. Hypoxia-inducible factor 1 inhibitors. Methods Enzymol. 2007;435:385–402. doi: 10.1016/S0076-6879(07)35020-9. [DOI] [PubMed] [Google Scholar]
- 45.Rapisarda A, Uranchimeg B, Scudiero DA, Selby M, Sausville EA, Shoemaker RH. Identification of small molecule inhibitors of hypoxia-inducible factor 1 transcriptional activation pathway. Cancer Res. 2002;62:4316–4324. [PubMed] [Google Scholar]
- 46.Chau N-M, Rogers P, Aherne W, Carroll V, Collins I, McDonald E. Identification of novel small molecule inhibitors of hypoxia-inducible factor-1 that differentially block hypoxia-inducible factor-1 activity and hypoxia-inducible factor-1α induction in response to hypoxic stress and growth factors. Cancer Res. 2005;65:4918–4928. doi: 10.1158/0008-5472.CAN-04-4453. [DOI] [PubMed] [Google Scholar]
- 47.Kaluz S, Kaluzová M, Stanbridge EJ. Proteasomal inhibition attenuates transcriptional activity of hypoxia-inducible factor 1 (HIF-1) via specific effect on the HIF-1α C-terminal activation domain. Mol Cell Biol. 2006;26:5895–5907. doi: 10.1128/MCB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rapisarda A, Uranchimeg B, Sordet O, Pommier Y, Shoemaker RH, Melillo G. Topoisomerase I-mediated inhibition of hypoxia-inducible factor 1: mechanism and therapeutic implications. Cancer Res. 2004;64:1475–1482. doi: 10.1158/0008-5472.can-03-3139. [DOI] [PubMed] [Google Scholar]
- 49.Kong DH, Park EJ, Stephen AG, Calvani M, Cardellina JH, Monks A. Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 DNA-binding activity. Cancer Res. 2005;65:9047–9055. doi: 10.1158/0008-5472.CAN-05-1235. [DOI] [PubMed] [Google Scholar]
- 50.Park J-E, Kong DH, Fisher R, Cardellina J, Shoemaker RH, Melillo G. Targeting the PAS-A domain of HIF-1alpha for development of small molecule inhibitors of HIF-1. Cell Cycle. 2006;5:1847–1853. doi: 10.4161/cc.5.16.3019. [DOI] [PubMed] [Google Scholar]
- 51.Kung AL, Zabludoff SD, France DS, Freedman SJ, Tanner EA, Vieira A. Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer Cell. 2004;6:33–43. doi: 10.1016/j.ccr.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 52.Greenberger LM, Horak ID, Filpula D, Sapra P, Westergaard M, Frydenlund HF. A RNA antagonist of hypoxia-inducible factor-1α, EZN-2968, inhibits tumor cell growth. Mol Cancer Ther. 2008;7:3598–3608. doi: 10.1158/1535-7163.MCT-08-0510. [DOI] [PubMed] [Google Scholar]
- 53.Onnis B, Rapisarda A, Melillo G. Development of HIF-1 inhibitors for cancer therapy. J Cell Mol Med. 2009;13:2780–2786. doi: 10.1111/j.1582-4934.2009.00876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rapisarda A, Hollingshead M, Uranchimeg B, Bonomi CA, Borgel SD, Carter JP. Increased antitumor activity of bevacizumab in combination with hypoxia inducible factor-1 inhibition. Mol Cancer Ther. 2009;8:1867–1877. doi: 10.1158/1535-7163.MCT-09-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 56.Sapra P, Zhao H, Mehlig M, Malaby J, Kraft P, Longley C. Novel delivery of SN38 markedly inhibits tumor growth in xenografts, including a camptothecin-11-refractory model. Clin Cancer Res. 2008;14:1888–1896. doi: 10.1158/1078-0432.CCR-07-4456. [DOI] [PubMed] [Google Scholar]
- 57.Del Bufalo D, Ciuffreda L, Trisciuoglio D, Desideri M, Cognetti F, Zupi G. Antiangiogenic potential of the mammalian target of rapamycin inhibitor temsirolimus. Cancer Res. 2006;66:5549–5554. doi: 10.1158/0008-5472.CAN-05-2825. [DOI] [PubMed] [Google Scholar]
- 58.Kaelin WG., Jr. The von Hippel−Lindau tumor suppressor protein and clear cell renal carcinoma. Clin Cancer Res. 2007;13 doi: 10.1158/1078-0432.CCR-06-1865. 680–4s. [DOI] [PubMed] [Google Scholar]
- 59.Ravaud A, Bernhard JC, Gross-Goupil M, Digue L, Ferriere JM. mTOR inhibitors: temsirolimus and everolimus in the treatment of renal cell carcinoma. Bull Cancer. 2010;97:45–51. doi: 10.1684/bdc.2010.1069. [DOI] [PubMed] [Google Scholar]
- 60.Zhang HF, Qian DZ, Tan YS, Lee K, Gao P, Ren YR. Digoxin and other cardiac glycosides inhibit HIF-1α synthesis and block tumor growth. Proc Natl Acad Sci USA. 2008;105:19579–19586. doi: 10.1073/pnas.0809763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mabjeesh NJ, Escuin D, LaVallee TM, Pribluda VS, Swartz GM, Johnson MS. 2ME2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating HIF. Cancer Cell. 2003;3:363–375. doi: 10.1016/s1535-6108(03)00077-1. [DOI] [PubMed] [Google Scholar]
- 62.LaVallee TM, Burke PA, Swartz GM, Hamel E, Agoston GE, Shah J. Significant antitumor activity in vivo following treatment with the microtubule agent ENMD-1198. Mol Cancer Ther. 2008;7:1472–1482. doi: 10.1158/1535-7163.MCT-08-0107. [DOI] [PubMed] [Google Scholar]
- 63.Hur E, Kim HH, Choi SM, Kim JH, Yim S, Kwon HJ. Reduction of hypoxia-induced transcription through the repression of hypoxia-inducible factor-1α/aryl hydrocarbon receptor nuclear translocator DNA binding by the 90-kDa heat-shock protein inhibitor radicicol. Mol Pharmacol. 2002;62:975–982. doi: 10.1124/mol.62.5.975. [DOI] [PubMed] [Google Scholar]
- 64.Porter JR, Ge J, Lee J, Normant E, West K. Ansamycin inhibitors of Hsp90: nature׳s prototype for anti-chaperone therapy. Curr Top Med Chem. 2009;9:1386–1418. doi: 10.2174/156802609789895719. [DOI] [PubMed] [Google Scholar]
- 65.Kurebayashi J, Otsuki T, Kurosumi M, Soga S, Akinaga S, Sonoo H. A radicicol derivative, KF58333, inhibits expression of hypoxia-inducible factor-1alpha and vascular endothelial growth factor, angiogenesis and growth of human breast cancer xenografts. Jpn J Cancer Res. 2001;92:1342–1351. doi: 10.1111/j.1349-7006.2001.tb02159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han JY, Oh SH, Morgillo F, Myers JN, Kim E, Hong WK. Hypoxia-inducible factor 1α and antiangiogenic activity of farnesyltransferase inhibitor SCH66336 in human aerodigestive tract cancer. J Natl Cancer Inst. 2005;97:1272–1286. doi: 10.1093/jnci/dji251. [DOI] [PubMed] [Google Scholar]
- 67.Osada M, Imaoka S, Funae Y. Apigenin suppresses the expression of VEGF, an important factor for angiogenesis, in endothelial cells via degradation of HIF-1α protein. FEBS Lett. 2004;575:59–63. doi: 10.1016/j.febslet.2004.08.036. [DOI] [PubMed] [Google Scholar]
- 68.Melstrom LG, Salabat MR, Ding XZ, Strouch MJ, Grippo PJ, Mirzoeva S. Apigenin down-regulates the hypoxia response genes: HIF-1α, GLUT-1, and VEGF in human pancreatic cancer cells. J Surg Res. 2011;167:173–181. doi: 10.1016/j.jss.2010.10.041. [DOI] [PubMed] [Google Scholar]
- 69.Lim JH, Lee YM, Chun YS, Chen JJ, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1α. Mol Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 70.Ueda H, Nakajima H, Hori Y, Fujita T, Nishimura M, Goto T. FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum No. 968 I. Taxonomy, fermentation, isolation, physico-chemical and biological properties, and antitumor activity. J Antibiot. 1994;47:301–310. doi: 10.7164/antibiotics.47.301. [DOI] [PubMed] [Google Scholar]
- 71.Furumai R, Matsuyama A, Kobashi N, Lee KH, Nishiyama M, Nakajima H. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 2002;62:4916–4921. [PubMed] [Google Scholar]
- 72.Kong XG, Lin Z, Liang DM, Fath D, Sang NL, Caro J. Histone deacetylase inhibitors induce VHL and ubiquitin-independent proteasomal degradation of hypoxia-inducible factor 1α. Mol Cell Biol. 2006;26:2019–2028. doi: 10.1128/MCB.26.6.2019-2028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee K, Kang JE, Park SK, Jin YL, Chung KS, Kim HM. LW6, a novel HIF-1 inhibitor, promotes proteasomal degradation of HIF-1α via upregulation of VHL in a colon cancer cell line. Biochem Pharmacol. 2010;80:982–989. doi: 10.1016/j.bcp.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 74.Lee K, Zhang HF, Qian DZ, Rey S, Liu JO, Semenza GL. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc Natl Acad Sci USA. 2009;106:17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Lee K, Qian DZ, Rey S, Wei H, Liu JO, Semenza GL. Anthracycline chemotherapy inhibits HIF-1 transcriptional activity and tumor-induced mobilization of circulating angiogenic cells. Proc Natl Acad Sci USA. 2009;106:2353–2358. doi: 10.1073/pnas.0812801106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Vergis R, Corbishley CM, Norman AR, Bartlett J, Jhavar S, Borre M. Intrinsic markers of tumour hypoxia and angiogenesis in localised prostate cancer and outcome of radical treatment: a retrospective analysis of two randomised radiotherapy trials and one surgical cohort study. Lancet Oncol. 2008;9:342–351. doi: 10.1016/S1470-2045(08)70076-7. [DOI] [PubMed] [Google Scholar]
- 77.Staab A, Loeffler J, Said HM, Diehlmann D, Katzer A, Beyer M. Effects of HIF-1 inhibition by chetomin on hypoxia-related transcription and radiosensitivity in HT 1080 human fibrosarcoma cells. BMC Cancer. 2007;7:213. doi: 10.1186/1471-2407-7-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Richardson PG, Hideshima T, Anderson KC. Bortezomib (PS-341): a novel, first-in-class proteasome inhibitor for the treatment of multiple myeloma and other cancers. Cancer Control. 2003;10:361–369. doi: 10.1177/107327480301000502. [DOI] [PubMed] [Google Scholar]
- 79.Befani CD, Vlachostergios PJ, Hatzidaki E, Patrikidou A, Bonanou S, Simos G. Bortezomib represses HIF-1α protein expression and nuclear accumulation by inhibiting both PI3K/Akt/TOR and MAPK pathways in prostate cancer cells. J Mol Med. 2012;90:45–54. doi: 10.1007/s00109-011-0805-8. [DOI] [PubMed] [Google Scholar]
- 80.Goda AE, Erikson RL, Sakai T, Ahn JS, Kim BY. Preclinical evaluation of bortezomib/dipyridamole novel combination as a potential therapeutic modality for hematologic malignancies. Mol Oncol. 2015;9:309–322. doi: 10.1016/j.molonc.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang BL, Gu Y. Bortezomib inhibits gastric carcinoma HGC-27 cells through the phospho-Jun N-terminal kinase (p-JNK) pathway in vitro. Gene. 2015;559:164–171. doi: 10.1016/j.gene.2015.01.035. [DOI] [PubMed] [Google Scholar]
- 82.Huang B, Li J, Xu X, Zheng D, Zhou Z, Liu J. Successful treatment of renal light chain (AL) amyloidosis with bortezomib and dexamethasone (VD) Pathol Biol. 2015;63:17–20. doi: 10.1016/j.patbio.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 83.Ko FN, Wu CC, Kuo SC, Lee FY, Teng CM. YC-1, a novel activator of platelet guanylate cyclase. Blood. 1994;84:4226–4233. [PubMed] [Google Scholar]
- 84.Li SH, Shin DH, Chun YS, Lee MK, Kim MS, Park JW. A novel mode of action of YC-1 in HIF inhibition: stimulation of FIH-dependent p300 dissociation from HIF-1α. Mol Cancer Ther. 2008;7:3729–3738. doi: 10.1158/1535-7163.MCT-08-0074. [DOI] [PubMed] [Google Scholar]
- 85.Chun YS, Yeo EJ, Choi E, Teng CM, Bae JM, Kim MS. Inhibitory effect of YC-1 on the hypoxic induction of erythropoietin and vascular endothelial growth factor in Hep3B cells. Biochem Pharmacol. 2001;61:947–954. doi: 10.1016/s0006-2952(01)00564-0. [DOI] [PubMed] [Google Scholar]
- 86.Park JW, Chun Y-G, Bair K, Cho HS, inventors; HIF Bio, Inc., assignee. Compound for treating angiogenesis. US patent 7,226,941. 2007 Jun 5.
- 87.Koh MY, Spivak-Kroizman T, Venturini S, Welsh S, Williams RR, Kirkpatrick DL. Molecular mechanisms for the activity of PX-478, an antitumor inhibitor of the hypoxia-inducible factor-1α. Mol Cancer Ther. 2008;7:90–100. doi: 10.1158/1535-7163.MCT-07-0463. [DOI] [PubMed] [Google Scholar]