Abstract
Channelrhodopsin (ChR) is a light-gated cation channel derived from green algae. Since the inward flow of cations triggers the neuron firing, neurons expressing ChRs can be optically controlled even within freely moving mammals. Although ChR has been broadly applied to neuro-science research, little is known about its molecular mechanisms. We determined the crystal structure of chimeric ChR at 2.3 Å resolution and revealed its molecular architecture. The integration of structural, electrophysio-logical, and computational analyses provided insight into the molecular basis for the channel function of ChR, and paved the way for the principled design of ChR variants with novel properties.
Keywords: membrane protein, channelrhodopsin, optogenetics, X-ray crystallography, electrophysiology
Almost all organisms, ranging from human to bacteria, perceive light information by using rhodopsin family proteins, which are covalently bound to the retinal chromo-phore. For example, in our human eyes, animal rhodopsins (type II rhodopsins) catch light and activate the heterotri-meric G protein in the cytoplasmic side, and in some archaea and bacteria, microbial rhodopsins (type I rhodopsins), such as bacteriorhodopsin (BR) and proteorhodopsin, receive light, transfer the proton from the intracellular side to the extracellular side, and sensory rhodopsin transfers the light signal via its transducer, like animal rhodopsins.
Rhodopsin family proteins form a very large superfamily, and during evolution, they have acquired divergent functions, including signal transduction, proton pumping, and anion pumping. However, nobody had found the channel-type rhodopsin family protein until recently.
In 1990’s, Dr. Hegemann’s and Dr. Keszthelyi’s groups found that the fast electrical current was involved in the rhodopsin-mediated phototactic responses in tiny green algae1,2. Since the currents occurred only within 30 ms after light illumination, it was suggested that the photoreceptor and the channel formed a protein complex or were the same protein. In 2002–03, Dr. Hegemann’s, Dr. Spudich’s and Dr. Takahashi’s groups isolated a new microbial rhodopsin from the algae3–5, and Dr. Hegemann’s group identified it as a light-gated cation channel by electrophysiological experiments3. Since this protein was the first channel-type rhodopsin family protein, they named this protein channelrhodop-sins (ChR).
ChR is about 750 amino acids long and is composed of N-terminal seven-transmembrane (7-TM) domain, which is sufficient for the channel function, and C-terminal intracel-lular domain. ChR changes its structure in response to blue light (∼480 nm) and allows various monovalent and divalent cations to pass through its ion-conducting pore. The currents generated by ChRs mediate the phototaxis and photo-phobic responses in native algal cells4,6. However, since the inward flow of cations causes the depolarization of the cell, ChR has attracted more attention as a useful tool for the analysis of specific neuron and/or neural circuit even within freely moving mammals7. ChRs have now been used to control neuronal activity in a wide range of animals, but little is known about the molecular mechanism of ChR. Although a rough helical arrangement was visible in the recently published electron microscopic (EM) structure of ChR at 6 Å resolution8, amino acid positioning and insights into channel function remained completely lacking. A high-resolution structure would be of enormous value, not only to enhance understanding of the mechanism of this new class of rhodopsin family protein, but also to guide the way to design ChR variants with novel function related to absorption spectrum, selectivity, and kinetics.
Expression, purification, and crystallization of ChR
Generally speaking, membrane proteins, especially eukaryotic membrane proteins are notoriously difficult to express, purify, and crystallize. They are often flexible, unstable and easily losses their functions when removed from the lipid bilayer. Thus, to find the promising candidate for structural studies, we tried to express various ChRs from their natural sources and their chimeric ChRs in every expression systems, including E. coli, yeast, mammalian, and insect cells. All ChRs were fused with C-terminal EGFP, and their expression levels and stabilities were evaluated by fluorescence-detection size-exclusion chromatography (FSEC)9. We found that one chimeric ChR (TMs 1–5 are derived from Chlamydomonas ChR1 and TMs 6–7 are from Chlamydomonas ChR2) showed excellent expression level and stability, so we purified this chimeric ChR and crystallize it by vapor diffusion method. However, despite extensive screening of crystallization conditions, we could not obtain this ChR’s crystals of sufficient quality for structural determination.
Next, we tried to crystallize this ChR by lipidic cubic phase (LCP) method. LCP method was originally developed by Dr. Landau and Dr. Rosenbusch in 1996, in which the membrane protein is reconstituted into a continuous curved lipid bilayer, LCP10. We screened about 500 crystallization conditions with commercial crystallization kits using this LCP method, and obtained tiny crystals of ChR. After the refinement of the condition, we got the good-quality crystals that diffracted to 2.8 Å resolution.
In order to solve the crystal structure of ChR, we tried to determine the phase using the SeMet-derivatized ChR protein, since the phase could not be determined by molecular replacement. However, an incorporation rate of SeMet into ChR was too low to solve the phase problem (i.e. about 40∼50% incorporation at maximum), and despite the numerous trials of more than six months, we could not improve the ratio. We are therefore left with the use of heavy-atom derivatized ChR, prepared by soaking or co-crystallization. Since the native ChR crystals exhibited the poor reproducibility under the previous crystallization condition, and citrate in the condition may inhibit the divalent metal ions, especially Hg2+, from binding to ChR by chelating them, we screened another crystallization conditions to find those were adequate for heavy atom soaking or co-crystallization.
Because the commercial screening kits are optimized for the vapor diffusion method, and a number of conditions in these kits (e.g. conditions with too high or too low pH) would break the LCP, we designed a novel crystallization kit optimized for LCP crystallization method, and searched the appropriate conditions for ChR using this homemade kit. We found several new conditions without citrate, refined those, and finally determined the phase by multiple anomalous dispersion (MAD) method, using mercury-derivatized crystals. As far as we know, this was the first example of the phase determination by MAD for the crystal obtained in the lipidic cubic phase. In addition, we could improve the resolution of the native dataset from 2.8 Å to 2.3 Å as a byproduct. The data sets were collected at SPring-8 BL32XU and Swiss Light Source X06SA beamlines.
Overall structure of ChR, and the structural comparison with BR
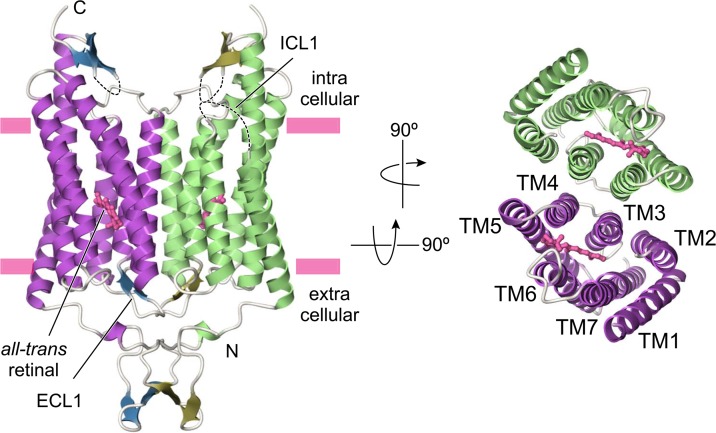
ChR is composed of an N-terminal extracellular domain (24–48), the 7-TM domains connected by three cytoplasmic and extracellular loops (84–317), and the C-terminal intra-cellular domain (318–356) (Fig. 1). Of particular note is that, as previously predicted from EM8, ChR is tightly associated into a dimer via interfacial interactions between the N-terminal domain, ECL1, TMs 3 and 4 of each molecule. This result is a little bit surprising for us because, as far as we know, all other known microbial rhodopsins form homotrimer or heterotetramer with the transducers. This is the first example of microbial rhodopsin to form dimer conformation.
Figure 1.
Overall structure of ChR. Dashed lines represent the disordered regions.
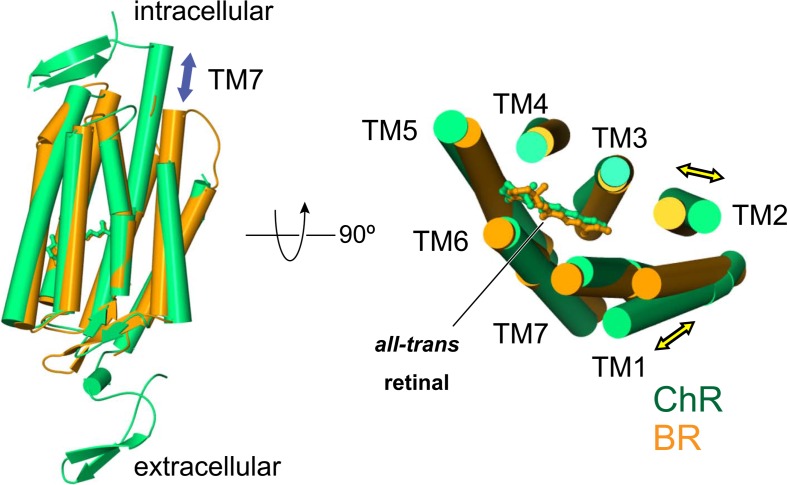
To further understand the ChR structure, we compared our ChR with most well studied microbial rhodopsin, BR11. Although the primary sequence identity between ChR and BR is as low as 15%, the overall structure of ChR is well superimposed on that of BR (Fig. 2). TMs 3–6 are very similar, and the position of the retinal is well conserved, whereas there are two distinct features between ChR and BR. First, ChR has additional N-terminal and C-terminal domains, and more importantly, the extracellular side of TMs 1 and 2 are tilted compared to those of BR.
Figure 2.
Structural comparison between ChR and BR.
Cation-conducting pathway
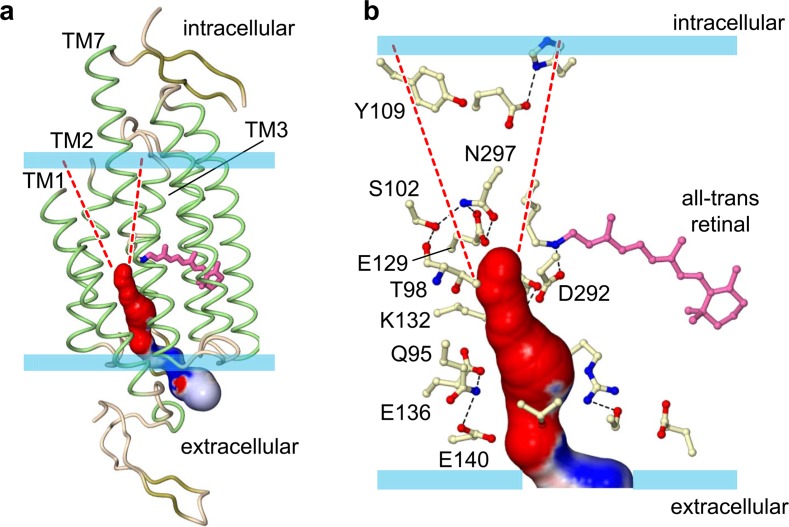
To understand the molecular mechanism of channel function in ChR, it is essential to identify the position of its ion-conducting pathway. Although almost ten years have passed since the discovery of ChR, several hypotheses existed for the molecular nature of its ion-conducting pathway. For example, some scientists have believed that ChR forms dimer or trimer, and cations pass through the interface of the oligomer8, while others have assumed that ChR has its ion-conducting pathway within the monomer. Even among the scientists supporting the latter hypothesis, there is no consensus as to where the pathway exactly is within the monomer12.
When we see the cross-section of ChR from the extra-cellular side, we found that although there is no space to allow the cations to pass through the dimer interface, there is a pore between TMs 1, 2, 3, and 7 within the monomer. Since the pore is formed by the tilts of TMs 1 and 2 that we mentioned before, this pore is the unique to ChR. Moreover, the calculated electrostatic surface potential reveals that this pore is strongly electronegative, so we assume that this pore would work as the cation-conducting pathway in ChR (Fig. 3). To verify this hypothesis, we expressed the mutants of the pore-lining residues in HEK293 cells and recorded photocurrents in response to blue light pulses. Most mutants showed the altered properties, including photo-conductance, kinetics, and the ion preference. Therefore, we suggest that this pore actually works as the cation-conducting pathway. Our recent molecular dynamics (MD) simulation showing that water molecules penetrate into this pore, also support our hypothesis (Koyama M, Kato E. H., Ishitani R, and Nureki, O. unpublished data).
Figure 3.
The putative ion-conducting pathway. Dashed lines are putative intracellular vestibule of the pathway. (a) Overall view. (b) Magnified view of the pathway.
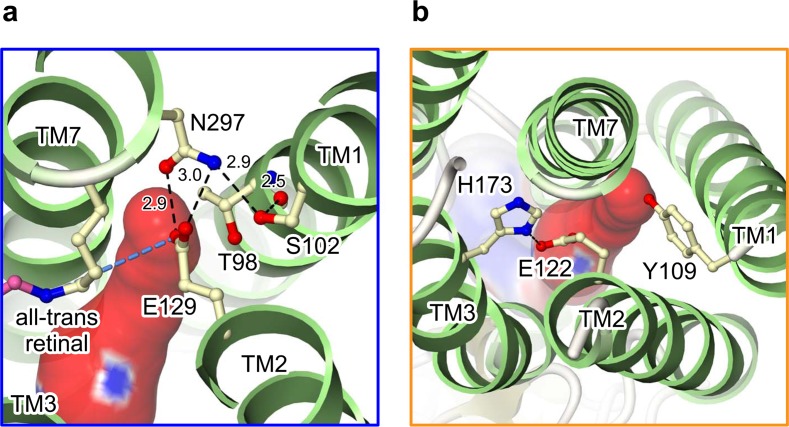
Two constrictions in the pathway
In this study, since we incubated the ChR crystals in the dark, and harvested them under dim red light, our crystal structure represents the closed/dark state. Actually, while this putative ion-conducting pathway is opened toward the extracellular side, its cytoplasmic side is occluded by two constriction sites (Fig. 4). The first constriction is formed by three highly conserved polar residues: Ser102, Glu129 and Asn297 (Fig. 4a). In this constriction site, Ser102, with a b-OH group that hydrogen bonds to the main-chain carbo-nyl oxygen of Thr98, fixes the position of Asn297 and, in turn, Asn297 fixes Glu129 by hydrogen bonds. Glu129 protrudes into and occludes the pore. To further analyze the importance of Glu129, we substituted Glu129 with several residues and measured their photocurrents. We found that although E129A showed the similar current to that of wild-type, E129Q mutant showed the decreased photocurrent. Since the calculated pKa of Glu129 suggests that this residue is protonated in the closed/dark state, we assume that Glu129 is once deprotonated during the photocycle and the change of the protonation state is important to the channel activity. Recent FT-IR study performed by Dr. Gerwert group, showing that Glu90 in ChR2 (Glu129 in our chimera) is protonated in the closed/dark state and deproto-nated at the early stage of photocycle, supports our idea13. His group also performed MD simulation using ChR2 homology model made from the crystal structure of BR and showed that when Glu90 is deprotonated, the diameter of our pathway would be wider13. Because there are some differences between their ChR homology model and our crystal structure, we recently performed MD simulation in similar conditions using the actual crystal structure. We verified that the deprotonated Glu129 lost interaction with Asn297, the side chain of Glu129 changes its orientation, the diameter of our pathway is wider, and the water molecules penetrate deeper into the pore. Based on these results, we assume that Glu129 would work as a putative channel gate in ChR and it is regulated by its protonation change (Koyama M., Kato E. H., Ishitani R., and Nureki, O. unpublished data).
Figure 4.
Two constriction sites located in the putative ion-conducting pathway. (a) The first constriction mainly formed by conserved Glu129. (b) The second constriction mainly formed by Tyr109 and Glu122. Black dashed lines indicate the hydrogen bonds.
The second constriction is formed by Tyr109, Glu122, and His173 (Fig. 4b). Although we do not exactly know how the constriction is regulated, we recently found that Glu122 was protonated in the closed/dark state and E122Q mutant showed the decreased current like E129Q. We assume therefore that Glu122 plays important role in channel function by changing its protonation state during photo-cyle (Koyama M., Kato E. H., Ishitani R., and Nureki, O. unpublished data).
Concluding remarks
In this review, we overviewed the crystal structure of chimeric ChR at 2.3 Å resolution, revealing its essential molecular architecture, including the retinal-binding pocket, the ion-conducting pathway, and the putative channel gate. Although the complete understanding of the molecular mechanism of ChR awaits the high-resolution structure of ChR in the open state, the integration of structural, electro-physiological, and computational analyses, mentioned in this review, provided important insight into the molecular basis for the function of ChRs, and paved the way for the precise and principled design of ChR variants with novel properties.
References
- 1.Sineshchekov OA, Litvin FF, Keszthelyi L. Two components of photoreceptor potential in phototaxis of the flagellated green alga Haematococcus pluvialis. Biophys J. 1990;57:33–39. doi: 10.1016/S0006-3495(90)82504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deininger W, Kröger P, Hegemann U, Lottspeich F, Hegemann P. Chlamyrhodopsin represents a new type of sensory photoreceptor. EMBO J. 1995;14:5849–5858. doi: 10.1002/j.1460-2075.1995.tb00273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 4.Sineshchekov OA, Jung KH, Spudich JL. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2002;99:8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki T, Yamasaki K, Fujita S, Oda K, Iseki M, Yoshida K, Watanabe M, Daiyasu H, Toh H, Asamizu E, Tabata S, Miura K, Fukuzawa H, Nakamura S, Takahashi T. Archaeal-type rhodopsins in Chlamydomonas: model structure and intracellular localization. Biochem Biophys Res Commun. 2003;301:711–717. doi: 10.1016/s0006-291x(02)03079-6. [DOI] [PubMed] [Google Scholar]
- 6.Govorunova EG, Jung KH, Sineshchekov OA, Spudich JL. Chlamydomonas sensory rhodopsins A and B: cellular content and role in photophobic responses. Biophys J. 2004;86:2342–2349. doi: 10.1016/S0006-3495(04)74291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Müller M, Bamann C, Bamberg E, Kuhlbrandt W. Projection structure of channelrhodopsin-2 at 6 A resolution by electron crystallography. J Mol Biol. 2011;414:86–95. doi: 10.1016/j.jmb.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 9.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Landau EM, Rosenbusch JP. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc Natl Acad Sci USA. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsui Y, Sakai K, Murakami M, Shiro Y, Adachi Si, Okumura H, Kouyama T. Specific damage induced by X-ray radiation and structural changes in the primary photoreaction of bacteriorhodopsin. J Mol Biol. 2002;324:469–481. doi: 10.1016/s0022-2836(02)01110-5. [DOI] [PubMed] [Google Scholar]
- 12.Bamann C, Gueta R, Kleinlogel S, Nagel G, Bamberg E. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry (Mosc) 2010;49:267–278. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- 13.Eisenhauer K, Kuhne J, Ritter E, Berndt A, Wolf S, Freier E, Bartl F, Hegemann P, Gerwert K. In channelrhodopsin-2 Glu-90 is crucial for ion selectivity and is deprotonated during the photocycle. JBiol Chem. 2012;287:6904–6911. doi: 10.1074/jbc.M111.327700. [DOI] [PMC free article] [PubMed] [Google Scholar]