Abstract
Several types of yeasts were isolated from wild flowers around Jangseong Lake in Jeollanam-do, Republic of Korea and identified by comparing the nucleotide sequences of the PCR amplicons for the D1/D2 variable domain of the 26S ribosomal DNA using Basic Local Alignment Search Tool (BLAST) analysis. In total, 60 strains from 18 species were isolated, and Pseudozyma spp. (27 strains), which included Pseudozyma rugulosa (7 strains) and Pseudozyma aphidis (6 strains), was dominant species. Among the 60 strains, Bullera coprosmaensis JS00600 represented a newly recorded yeast strain in Korea, and its microbiological characteristics were investigated. The yeast cell has an oval-shaped morphology measuring 1.4 × 1.7 µm in size. Bullera coprosmaensis JS00600 is an asporous yeast that exhibits no pseudomycelium formation. It grew well in vitamin-free medium as well as in yeast extract-malt extract broth and yeast extract-peptone-dextrose (YPD) broth, and it is halotolerant growing in 10% NaCl-containing YPD broth.
Keywords: Bullera coprosmaensis, Characteristics, Jangseong Lake, Unrecorded yeast, Wild flowers
Generally, yeasts are heterotrophic with relatively simple nutritional needs and live as facultative anaerobe. Therefore, they should be wildly distributed in natural habitats such as in flowers and fruits, cereals, and plant debris in the surface area of soils. However, yeasts are exclusively isolated from various fermentation foods or their raw materials including meju, a traditional Korean fermented soybean [1]. It is necessary to isolate wild yeasts from natural sources and screen for the yeast mycoflora.
We have previously isolated wild yeasts from mountains [2] and fields [3], city gardens [4] and farm villages [5], islands [6], and coastal areas [7] and identified them with molecular biological tools. Furthermore, we have studied the production of bioactive compounds from these wild yeasts. Recently, we screened for new yeast records from among yeasts found in Ulleungdo [8], Yokjido [9], and in orchards and an arboretum [10] and their mycological characteristics were investigated.
In this study, we isolated and identified wild yeast strains from flowers around Jangseong Lake in Jeollanam-do, Republic of Korea. We also found an unrecorded yeast, and their mycological characteristics were investigated.
MATERIALS AND METHODS
Isolation, identification of wild yeasts and screening of unrecorded yeast
Forty-eight wild flowers found lakeside around Jangseong Lake in Jeollanam-do, Republic of Korea were collected in August 2014, and several wild yeasts were isolated and identified as previously described [11].
Unrecorded yeasts in Korea were screened by searching KERIS, PubMed, and other fungal taxonomy databases.
Microbiological characteristics of the unrecorded yeast.
The morphological and cultural characteristics of the unrecorded yeast were investigated as previously described [10,12]. To determine whether the strain forms an ascospore, the unrecorded yeast was cultured in yeast extract-peptone-dextrose (YPD) medium at 30℃ for 24 hr and then, cultured for 5 days in an ascospore-forming medium containing potassium acetate 1%, yeast extract 0.1%, and dextrose 0.05%. The strain was then observed with a microscope for ascospore formation. The unrecorded yeast was successively cultured at 30℃ for 7 days in YPD medium, yeast extract-malt extract medium, potato-dextrose medium, and glucose-peptone-yeast extract agar containing glucose 4%, peptone 0.5%, and yeast extract 0.5%. Pseudomycelium formation was determined by observing the shape of cell in the cultures.
For scanning electron microscopy (SEM), JS00600 strain was cultured in YPD medium and then kept in 20% glycerol stock. The stock was diluted using a 0.05M carcodylate buffer. The diluted solution was centrifuged at 1,300 rpm for 1 min to obtain yeast cell pellet. The pellet was used for fixation. The strain was also cultured in potato-dextrose-broth medium at shaking speed of 150 rpm in darkness at 30℃ for 48 hr. The sample were fixed in 2.5% paraformaldehyde-glutaraldehyde buffer with 0.05 M phosphate (pH 7.2) for 2 hr, washed using the carcodylate buffer, post-fixed in 1% osmium tetroxide in the same buffer for 1 hr, and washed again using the same buffer, dehydrated in graded ethanol followed by isoamyl acetate, and then dried under a fume hood. Finally, the samples were covered in gold in a sputter coater and observed with the Hitachi S4700 field emission scanning electron microscope (Hitachi, Tokyo, Japan).
The physiological functionalities of the supernatants and cell-free extracts from the unrecorded yeast were determined as follows: the unrecorded yeast was cultured in YPD medium at 30℃ for 2 days. After centrifugation at 10,000 ×g for 15 min, the supernatants and cells were separated. The cells were disrupted by vortexing with sonication and centrifuged at 12,000 ×g for 20 min. The mixture was filtered to obtain cell-free extracts and supernatants. The physiological functionalities of the cell-free extracts and supernatants were determined as previously described [6,11,12].
Statistical analysis
Each experiment was performed at least three times, and all quantitative data are expressed as the mean ± SD values.
RESULTS AND DISCUSSION
Isolation and identification of yeasts from wild flowers around Jangseong Lake in Jeollanam-do, Republic of Korea
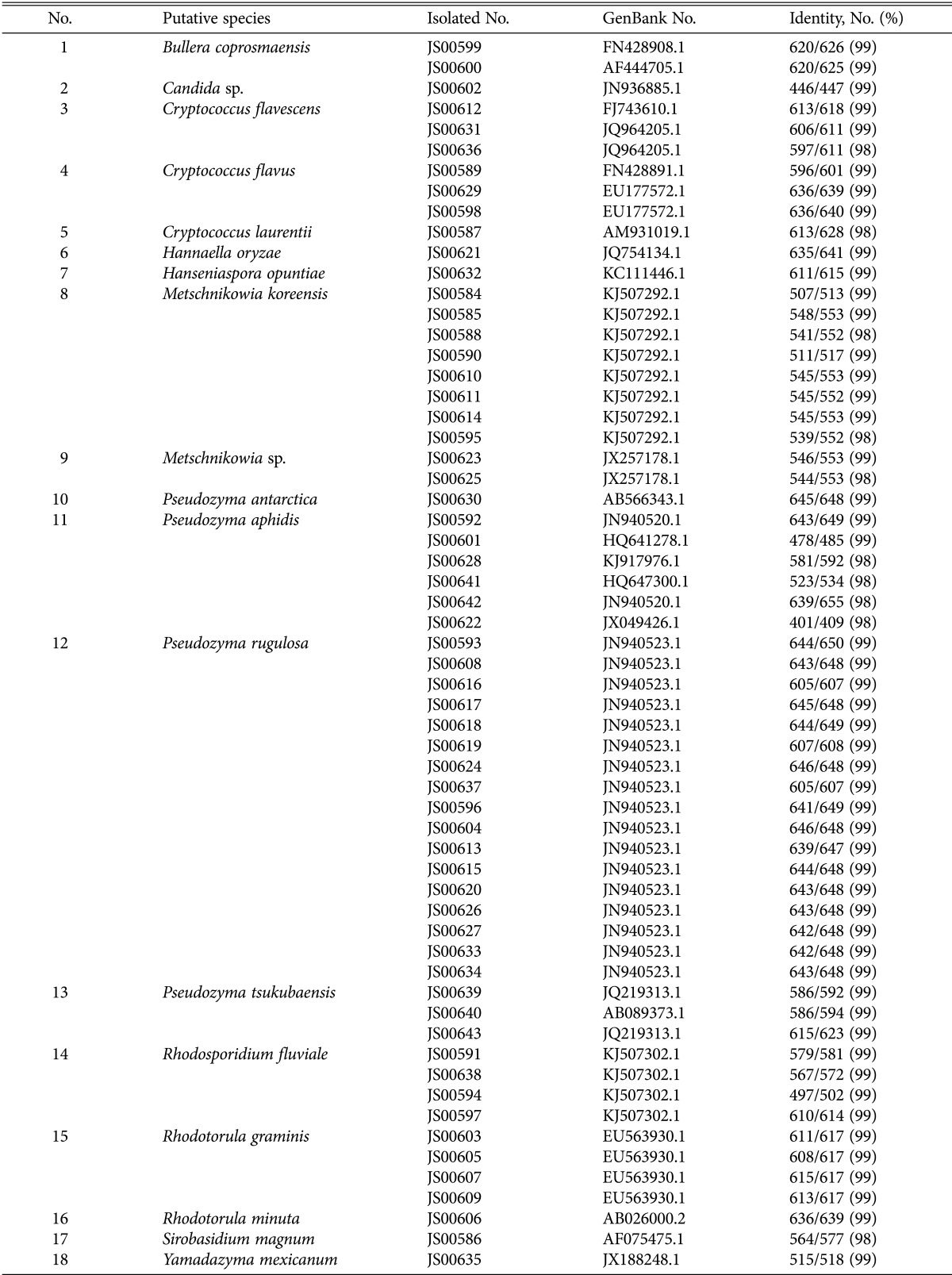
Sixty yeast strains belonging to 18 species were isolated from 48 kinds of blossoms found lakeside around Jangseong Lake during early August in 2014 (Table 1). Among them, the Pseudozyma spp. including 17 strains of Pseudozyma rugulosa, 6 strains of Pseudozyma aphidis, and 3 strains of Pseudozyma tsukubaensis were dominant species.
Table 1. Yeast species isolated from wild flowers of Jangseong lakeside in Jeollanam-do, Korea.
Screening of the unrecorded yeast, Bullera coprosmaensis JS00600
The unrecorded yeast Bullera coprosmaensis JS00600 (NIBR No. KOSPFGC000129189) was found among the sixty yeast strains isolated in this study. B. coprosmaensis JS00600 was isolated from Plantago asiatica which was in full bloom on August 4, 2014.
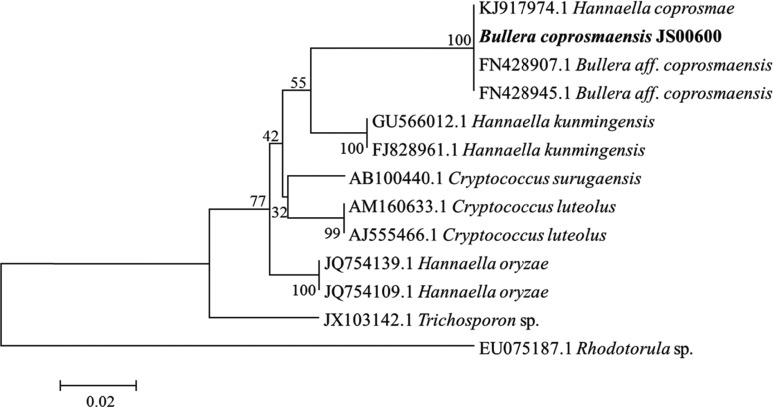
A phylogenetic tree for the unrecorded yeast B. coprosmaensis JS00600 was constructed based on the largesubunit rDNA D1/D2 domain sequence using the MEGA 5.1 program. B. coprosmaensis JS00600 was closely grouped to B. aff. coprosmaensis FN428907.1 (Fig. 1). Therefore, we reconfirmed the newly recorded yeast as B. coprosmaensis JS00600 and submitted its sequence to the GenBank database with the accession number KT277522.
Fig. 1. Phylogenetic tree of the unrecorded yeast Bullera coprosmaensis JS00600 (NIBR No. KOSPFGC000129189) from this study based on the nucleotide sequences of large subunit 26S ribosomal DNA. The tree was generated by the neighbor-joining method, using MEGA v5.1.
B. coprosmaensis was the first reported from the surface of plants materials collected in New Zealand [13]. Few studies on B. coprosmaensis have been done.
Characteristics of the unrecorded yeast Bullera coprosmaensis JS00600
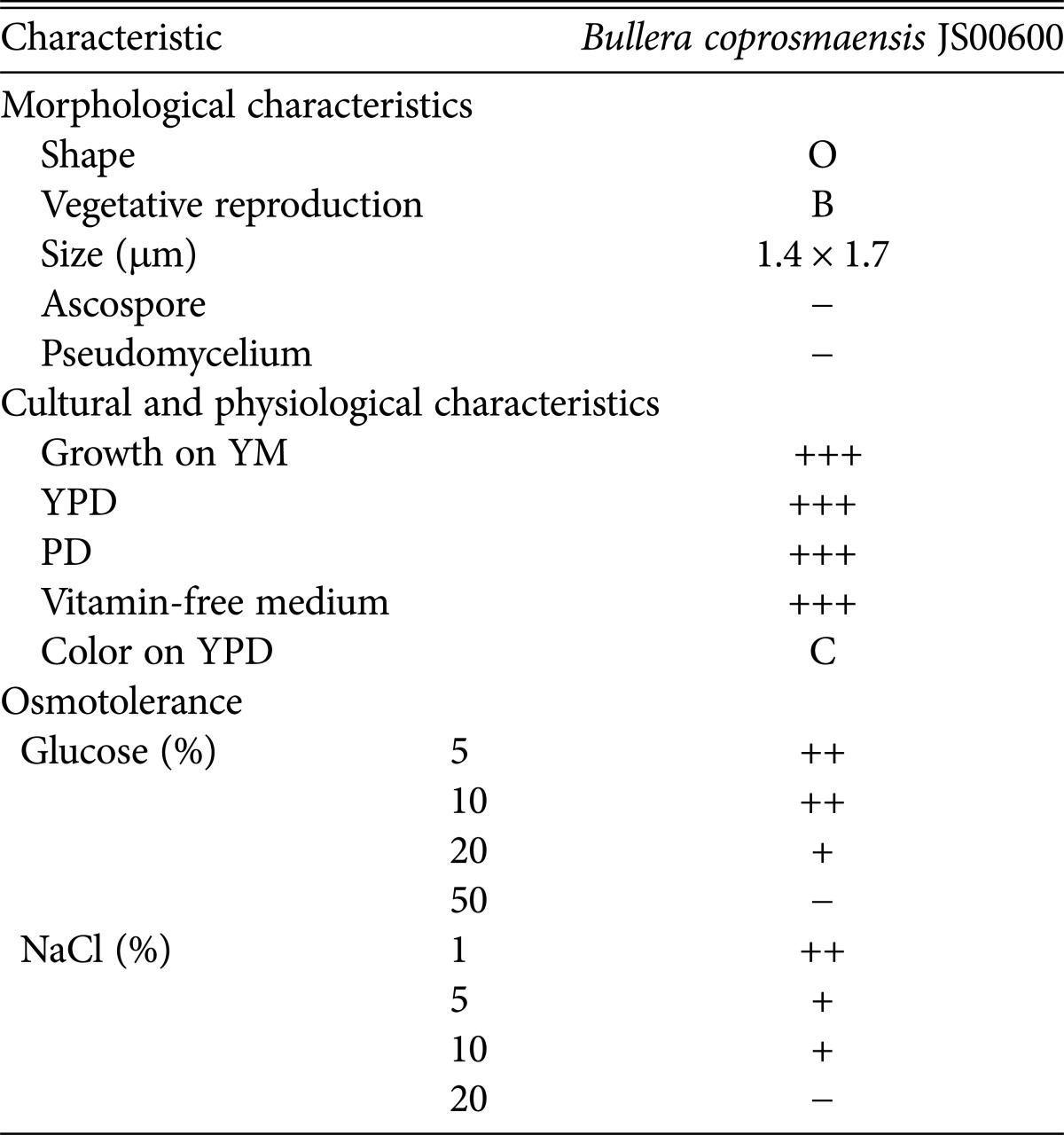
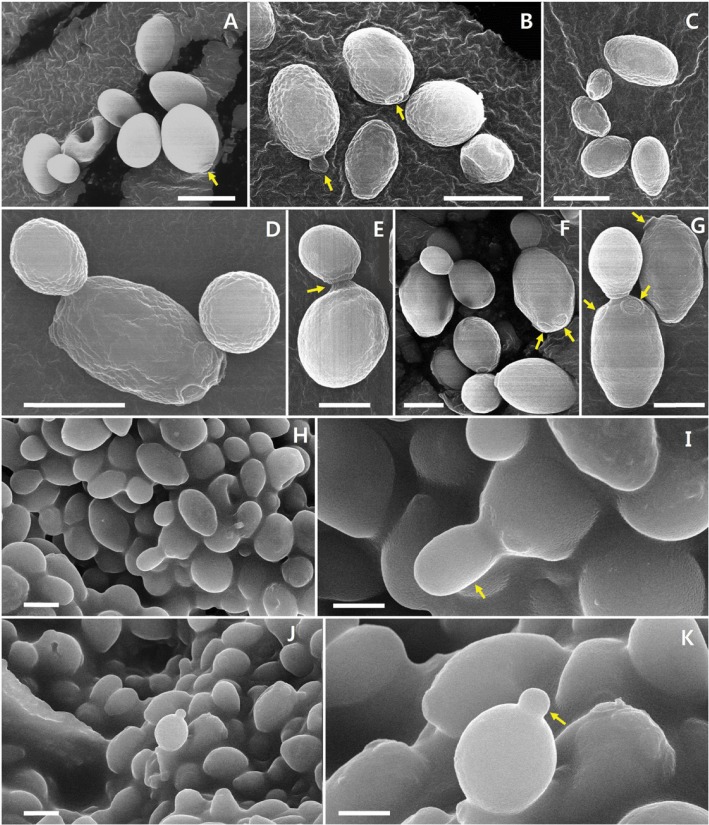
The morphological and cultural characteristics of B. coprosmaensis JS00600 are presented in Table 2 and Fig. 2. B. coprosmaensis JS00600 has an oval shaped morphology and a budding system for vegetative reproduction. Fig. 1 shows the SEM of JS00600 strain in different media and conditions. The typical shapes of vegetative cells of JS00600 were ellipsoidal to oval, showing commonly single cell forms and budding system. The strain did not form an ascospore and pseudomycelium. B. coprosmaensis JS00600 grew well in YPD medium, yeast extract-malt extract medium, and vitamin-free medium.
Table 2. Morphological and cultural characteristics of the unrecorded yeast Bullera coprosmaensis JS00600.
O, oval-shaped; B, budding; +++, very good growth; ++ or +, good growth; -, no growth; C, cream color; YM, yeast extractmalt extract medium; YPD, yeast extract-peptone-dextrose broth; PD, potato-dextrose medium.
Fig. 2. Scanning electron microscopy (SEM) of vegetative cells of Bullera coprosmaensis JS00600 strain in different media and conditions (A~G and H~K, photos from two samples cultured in potato-dextrose-broth and yeast extract-peptone-dextrose [diluted from glycerol stock stored at -80℃], respectively). A~C, Typical cell shapes of JS00600; D, E, Cell shapes and budding forms with age barrier (arrow); A, B, F, G, Budding form and cells budding off daughter cells, and scars (arrows); H, J, Cell shapes and budding forms; I, K, Their magnified forms and cells budding off daughter cell (arrows) (scale bars = 2 µm).
The osmotolerance of B. coprosmaensis JS00600 against glucose and NaCl were investigated. B. coprosmaensis JS00600 grew well in YPD medium containing 20% glucose and 10% NaCl, respectively. Few studies on halophilic yeasts have been done except for Zygosaccharomyces rouxii from soybeans [14] and halotolerant protease-producing Saccharomyces lipolytica [15] and Hansenula polymorpha S-9 from traditional Meju in a previous study [14,16]. It is generally known that halophilic microorganisms produce halotolerant enzymes with some advantages such as preventing microbial contamination in the enzyme industry and enhancing the flavor of salted foods during aging. Therefore, B. coprosmaensis JS00600 in this study should be very useful in preparing halotolerant enzymes and bioactive compounds for the food and medical industry.
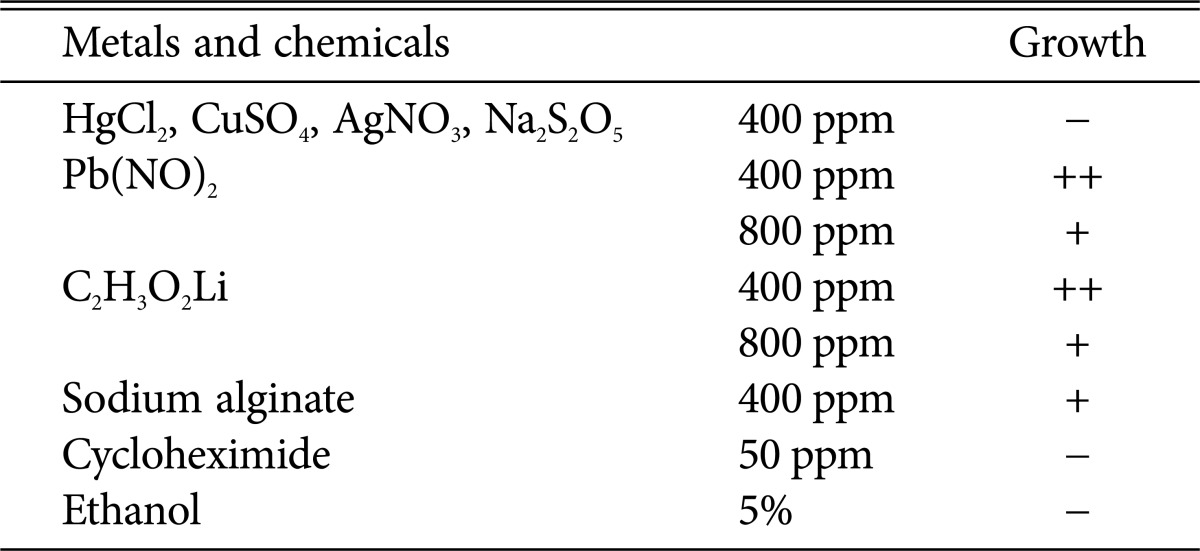
Table 3 shows the resistance of the unrecorded yeast B. coprosmaensis JS00600 to heavy metals and cycloheximide. B. coprosmaensis JS00600 exhibited tolerance to 800 ppm Pb2+ and Li+, whereas it showed growth inhibition for 400 ppm Hg2+, Cu2+, Ag+, Na2S2O5, and 50 ppm cycloheximide. Lee et al. [17] reported some Saccharomyces sp. and Zygosaccharomyces sp. from traditional Meju were resistant to 800 ppm Pb2+.
Table 3. Resistance to heavy metals and chemicals of the unrecorded yeast, Bullera coprosmaensis JS00600.
++, very good growth; +, good growth; and -, no growth on yeast extract-peptone-dextrose medium.
Assimilation and fermentation on carbon sources
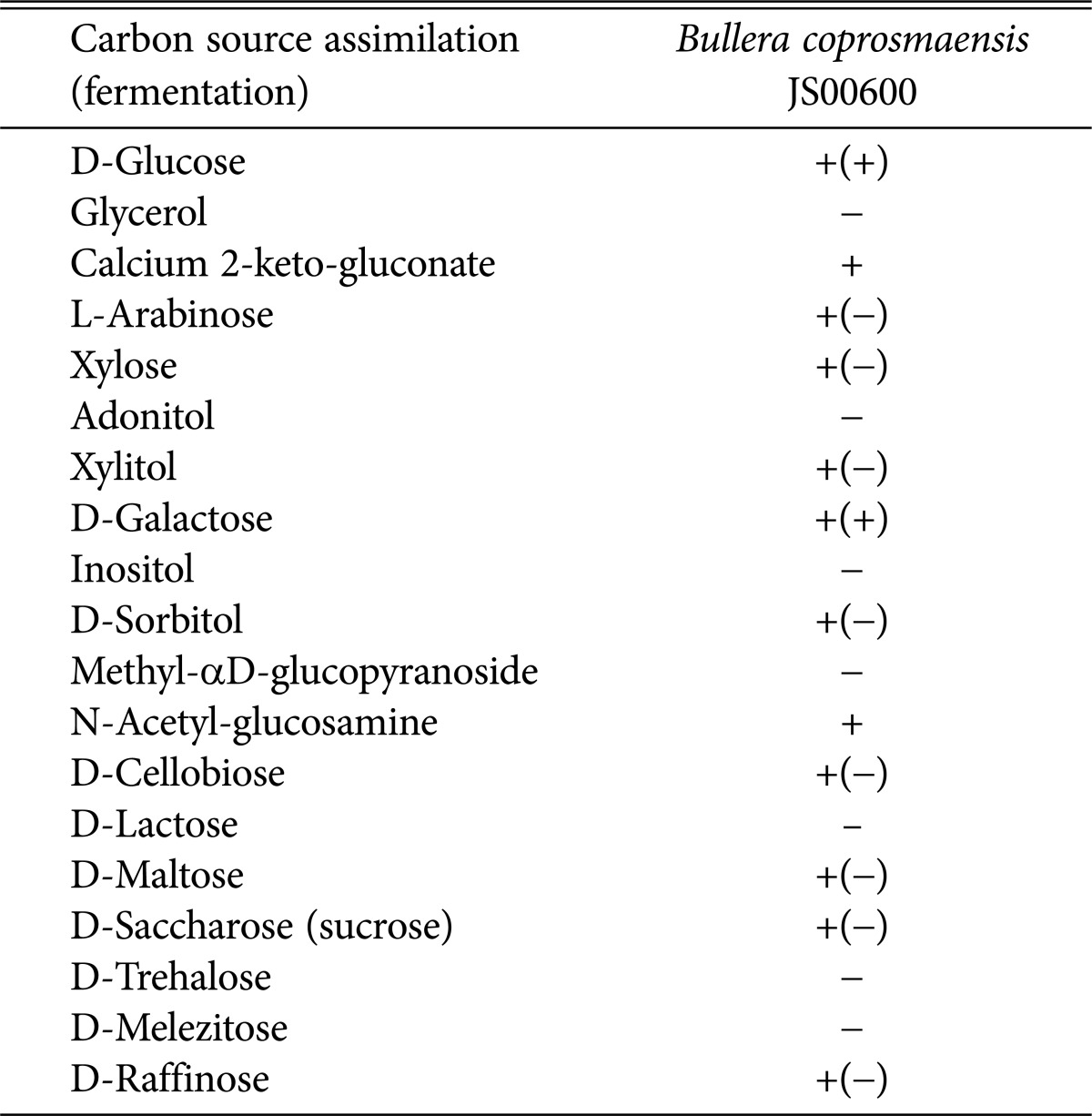
Assimilation and fermentation of B. coprosmaensis JS00600 were investigated with several kinds of sugars and sugar alcohol (Table 4). B. coprosmaensis JS00600 assimilated glucose, maltose, sucrose, raffinose and cellobiose in hexose. Additionally, arabinose, xylose in pentose and sorbitol, xylitol N-acetyl glucosamine and calcium 2-keto-gluconate were also assimilated. However, only glucose and galactose in sugars used in this study were fermented by B. coprosmaensis JS00600.
Table 4. Carbon source assimilation (fermentation) by unrecorded yeast, Bullera coprosmaensis JS00600.
+, good growth; and -, no growth on yeast extract-peptone-dextrose medium.
Physiological functionalities
To obtain data for application of B. coprosmaensis JS00600 in functional foods or the medicinal industry, we prepared supernatants and cell-free extracts of B. coprosmaensis JS00600, and then, some of their physiological functionalities were determined (Table 5). The cell-free extracts exhibited a very high hypoglycemic α-glucosidase inhibitory activity of 94.7% and an antihypertensive angiotensin I-converting enzyme inhibitory activity of 75.0%. This α-glucosidase inhibitory activity was similar to that of Pichia burtonii (90.9%) in our previous paper [18]. B. coprosmaensis JS00600 newly recorded in this study should be useful in the production of bioactive anti-diabetic and anti-hypertensive agents.
Table 5. Physiological functionalities of unrecorded yeast Bullera coprosmaensis JS00600.
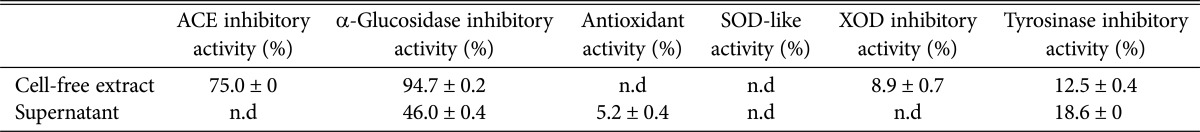
ACE, angiotensin I-converting enzyme; SOD, superoxide dismutase; XOD, xanthine oxidase; n.d, not detected or < 5%.
ACKNOWLEDGEMENTS
This work was supported by a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea.
References
- 1.Kim JH, Kim NM, Lee JS. Physiological characteristics and ethanol fermentation of thermotolerant yeast Saccharomyces cerevisiae OE-16 from traditional Meju. Korean J Food Nutr. 1999;12:490–495. [Google Scholar]
- 2.Min JH, Ryu JJ, Kim HK, Lee JS. Isolataion and identification of yeasts from wild flowers in Gyejoksan, Oseosan and Beakamsan of Korea. Korean J Mycol. 2013;41:47–51. [Google Scholar]
- 3.Han SM, Park WJ, Lee JS. Isolation and diversity of wild yeasts from some cereals. Korean J Mycol. 2015;43:64–67. [Google Scholar]
- 4.Hyun SH, Min JH, Kim SA, Lee JS, Kim HK. Yeasts associated with fruits and blossoms collected from Hanbat arboretum, Daejeon, Korea. Korean J Mycol. 2014;42:178–182. [Google Scholar]
- 5.Hyun SH, Lee JG, Park WJ, Kim HK, Lee JS. Isolation and diversity of yeasts from fruits and flowers of orchard in Sinam-myeon of Yesan-gun, Chungcheongnam-do, Korea. Korean J Mycol. 2014;42:21–27. [Google Scholar]
- 6.Hyun SH, Han SM, Lee JS. Isolation and physiological functionality of yeasts from wild flowers in Seonyudo of Gogunsanyeoldo, Jeollabuk-do, Korea. Korean J Mycol. 2014;42:201–206. [Google Scholar]
- 7.Min JH, Lee HB, Lee JS, Kim HK. Identification of yeasts isolated from wild flowers collected in coast areas of Korea based on the 26S rDNA sequences. Korean J Mycol. 2013;41:185–191. [Google Scholar]
- 8.Hyun SH, Min JH, Lee HB, Kim HK, Lee JS. Characteristics of two unrecorded yeasts from wild flowers in Ulleungdo, Korea. Korean J Mycol. 2014;42:170–173. [Google Scholar]
- 9.Hyun SH, Lee JS. Microbiological characteristics and physiological functionality of new records of yeasts from wild flowers in Yokjido, Korea. Mycobiology. 2014;42:198–202. doi: 10.5941/MYCO.2014.42.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han SM, Hyun SH, Shin JW, Kim HK, Lee JS. Mycological characteristics of nine unrecorded yeasts from flowers in the orchard of Yesan-gun, Chungcheongnam-do and Hanbat arboretum in Daejeon City, Korea. Korean J Mycol. 2014;42:231–234. [Google Scholar]
- 11.Han SM, Hyun SH, Lee JS. Isolation, identification of yeasts from wild flowers in Deogyu mountain in Korea and their physiological funtionalities. Korean J Mycol. 2015;43:47–52. [Google Scholar]
- 12.Hyun SH, Han SM, Lee JS. Characteristics and physiological functionalities of unrecorded yeasts from wild flowers in Seonyudo of Jeollabuk-do, Korea. Korean J Microbiol Biotechnol. 2014;42:402–406. [Google Scholar]
- 13.Hamamoto M, Nakase T. Ballistosporous yeasts found on the surface of plant materials collected in New Zealand. The genera Bensingtonia and Bullera with descriptions of five new species. Antonie Van Leeuwenhoek. 1996;69:279–291. doi: 10.1007/BF00399617. [DOI] [PubMed] [Google Scholar]
- 14.Jeong SC, Hyun KW, Kim JH, Lee JS. Isolation of halotolerant yeast and the production of extracellular protease. Korean J Biotechnol Bioeng. 2001;16:158–162. [Google Scholar]
- 15.Kang KH, Bae IH, Lee CH. Studies on extracellular protease from Saccharomyces lipolytica: conditions of enzyme production. Korean J Appl Microbiol Biotechnol. 1987;15:279–285. [Google Scholar]
- 16.Lee JS, Yi SH, Kwon SJ, Ahn C, Yoo JY. Enzyme activities and physiological functionality of yeasts from traditional Meju. Korean J Appl Microbiol Bioeng. 1997;25:448–453. [Google Scholar]
- 17.Lee JS, Choi YJ, Kwon SJ, Yoo JY, Chung DH. Screening and characterization of osmotolerant and gas-producing yeasts from traditional Doenjang and Kochujang. Food Sci Biotechnol. 1996;5:54–58. [Google Scholar]
- 18.Kim YH, Shin JW, Lee JS. Production and anti-hyperglycemic effects of α-glucosidase inhibitor from yeast, Pichia burtonii Y257-7. Korean J Microbiol Biotechnol. 2014;42:219–224. [Google Scholar]