Short abstract
Analysis of two genomes of Chlamydia trachomatis using competitive hybridization on DNA microarrays revealed a logarithmic correlation between the signal ratio of the arrays and the 75-99% range of nucleotide identities of the genes.
Abstract
By comparing two fully sequenced genomes of Chlamydia trachomatis using competitive hybridization on DNA microarrays, a logarithmic correlation was demonstrated between the signal ratio of the arrays and the 75-99% range of nucleotide identities of the genes. Variable genes within 14 uncharacterized strains of C. trachomatis were identified by array analysis and verified by DNA sequencing. These genes may be crucial for understanding chlamydial virulence and pathogenesis.
Background
New genomes are continuously being sequenced, offering insight into relationships among a multitude of organisms. Because of the relatively high cost, multiple genomes within a species are rarely sequenced, as it is difficult to justify a full genome effort for the relatively little novel, albeit potentially important, information gained. Fortunately, microarrays can be used to rapidly screen an entire genome for such data. Previously, identification of genomic variability using microarray analysis was limited to those genes that were either absent or highly divergent [1-7]. Only recently has the use of microarrays been expanded to detect differences among closely related strains/isolates at the nucleotide level [8]. This increased resolution offers a greater insight into the level of diversification within a species or population, and this can lead to the characterization of genes linked to unique biological attributes such as pathogenesis.
The power of microarrays for comparative genomic purposes is the ability to discover what may be only a few informative loci among thousands. Additional evolutionary and biological functionality tests can then be pursued on these few genes. Rapid and sensitive assays such as microarrays are important for organisms that are highly conserved and undergo little to no horizontal gene movement (that is, recombination or plasmid acquisition). Traditional genotyping tests, such as pulse-field gel electrophoresis (PFGE) or restriction fragment length polymorphism (RFLP), are relatively insensitive in such circumstances [9]. In these assays, the absence of gene movement results in DNA fragments that differ in size solely due to the loss and/or gain of specific restriction sites, which will be a rare event in very similar genomes. Even if an RFLP assay identifies variability between two samples, it provides no specific information regarding the genes in which these changes are located. It is these nucleotide changes that underlie the amino acid sequence and its corresponding protein function that ultimately influences the fitness of an organism. Our goal was to use microarrays as a comparative genomics tool to identify nucleotide polymorphisms among the many closely related strains of Chlamydia trachomatis.
C. trachomatis is an obligate intracellular bacterium with a worldwide distribution. It has a genome of 1.04 megabases (Mb) consisting of 894 open reading frames (ORFs) between 135 and 5,358 nucleotides long, with a median length of 867 nucleotides. Because of its sequestered lifestyle, acquisition of exogenous DNA is considered to have played a limited part in the subsequent evolution of the species after the organisms moved into their intracellular niche and became environmentally and genetically isolated nearly a billion years ago [10,11]. Consequently, diversity in chlamydial genomes is mostly a result of nucleotide substitutions and gene loss [12]. Over the course of evolutionary history, the accumulation of these differences has led to the present-day biovariants of C. trachomatis, such as those that infect humans or mice. Two biovariants exist within the human-specific strains, which together consist of more than 15 serovariants and occupy one of three distinct biological niches upon infection. Among the trachoma biovar, serovars A, B, Ba and C are associated with ocular infection, and serovars D through K are associated with urogenital infection. Serovars L1, L2 and L3 compose the lymphogranuloma venereum (LGV) biovar and infect lymphatic tissue [13]. Despite these three distinct tissue tropisms among the strains of C. trachomatis, their genomes are all highly similar [14]. In addition to the human strains, there is the closely related C. trachomatis murine biovar, mouse pneumonitis (MoPn), which has been reported to originate from the respiratory epithelial tissue of mice [15].
For C. trachomatis to achieve niche-specific functions without acquiring exogenous DNA, nucleotide changes must have occurred in some genes that could account for diverse biological capabilities. An example of this is the loss of tryptophan synthase function in the strains associated with ocular infection, a change that purportedly facilitates their persistence in this particular biological niche [16-18]. All strains isolated from ocular infections were found to have a defective gene within the trp pathway; no strains of urogenital origin were found to harbor such mutations within this region [16]. Although the loss of function was often a result of a single polymorphism within a gene in the trp pathway, it is evident that such small changes can have a dramatic effect on the resulting phenotype and success of an organism. The genes that possess such critical differences among the serovars of C. trachomatis may be identified through the use of microarrays.
Results
Limiting the effects of bias
Because microarrays are competition-based assays, DNA sequences that are identical for a particular gene in the fluorescently labeled test and reference strains will bind with equal affinity to the corresponding immobilized fragment on the array, thereby yielding equivalent signal ratios. A polymorphism is indicated by an increase in the signal ratio at a particular gene region on the array, due to the preferential hybridization of the most closely related reference DNA fragment to the complementary test sequence.
If there are changes in the signal ratio that do not result from variation in the nucleotide sequence of the test DNA, these regions of the array need to be identified as they will skew the data. To identify the loci in the C. trachomatis microarray that may be intrinsically biased towards a higher signal ratio, the reference/array strain (D/UW-3) was used as both the analyte and reference DNA. In this test, every gene has a 100% match that should result in equal signals for each gene region on the array. Most loci on the array produced equivalent signals, although the results indicated a few anomalous gene regions (data not shown). One such locus, ribE, was found to have a high signal ratio in several of the other serovars as well, which should correlate with a high degree of polymorphism in these strains. However, sequence analysis of the ribE region from all the strains revealed little or no nucleotide variation (99.6-100% identity; GenBank accession nos AY542692-AY542704). It was found that 28 genes from the D/UW-3 versus D/UW-3 array data were above the 95% confidence interval as determined by a one-tailed Z-test, and were therefore removed from subsequent experimental datasets in order to eliminate possible confounding bias in the comparisons of other strains at these regions.
Microarray analysis of MoPn
The MoPn biovar of C. trachomatis was selected to establish experimentally that microarray analysis could assess relative levels of nucleotide variation between two related strains. As the genomes of both MoPn and the source strain used to make the microarray (D/UW-3) are known, and were found to be in almost perfect synteny [19,20], the percent nucleotide identity between each DNA fragment on the array and its complementary orthologous sequence in MoPn was determined. Overall, the spectrum of sequence identities among all gene pairs from the array ranged from 43% to 99%, and each pair in an integer continuum of 66-99% identity was represented at least once (Figure 1). This range of identities is broader than that characterized in previous studies [8] and proved to be robust for establishing the sensitivity of the relationship with the array data.
Figure 1.
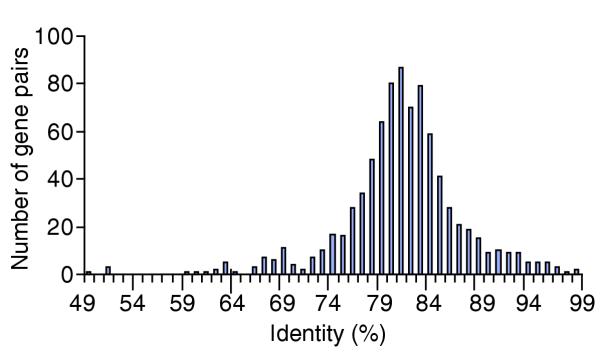
Frequency of gene pair identities between MoPn and D/UW-3. The percent identity for each orthologous gene pair between C. trachomatis serovars D/UW-3 and MoPn was established and rounded to the nearest whole number. The number of times each nucleotide identity occurred was then determined.
When the nucleotide identity for each gene pair between 43% and 99% was compared to its corresponding array signal ratio, the results indicated a logarithmic relationship between the two; as the level of nucleotide identity decreased from 99%, there was an exponential increase in the microarray signal ratio (Figure 2). Below 75% identity, however, the relationship diminished. Because a correlation exists between the signal ratio and nucleotide identities between 75% and 99%, microarrays can be used to assess the relative level of nucleotide differences in genes in otherwise unknown genomes. Those regions below the 75% identity threshold can only be assessed as highly divergent or perhaps absent.
Figure 2.
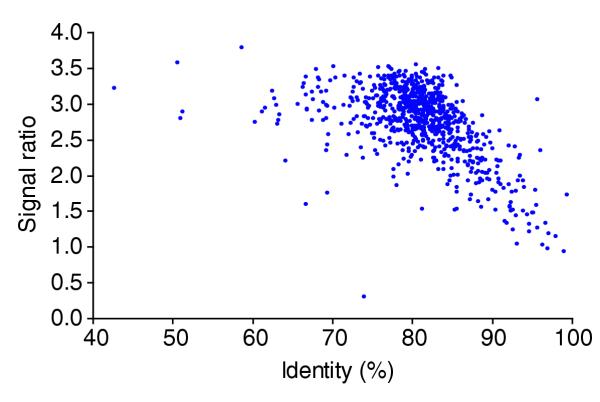
Relationship between the signal ratios and sequence identities for the MoPn vs D/UW-3 DNA microarray. The average signal ratio of each orthologous gene pair between C. trachomatis serovars D/UW-3 and MoPn was log2 transformed. These values were then compared to the corresponding nucleotide identity for each gene pair, yielding a linear association.
Microarray analysis of C. trachomatis
Having demonstrated that nucleotide difference is correlated with the microarray signal ratio, 14 different C. trachomatis strains representing 14 human serovars (A, B, Ba, C, E, F, G, H, I, J, K, L1, L2, L3) were tested to identify gene regions that were polymorphic compared with the reference strain D/UW-3. For each strain, the microarray data were ordered from highest to lowest signal (see Additional data file 1). The highest rank, 1, indicates the highest signal ratio and therefore represents the locus with the most variation between the test and reference strains. Conversely, the low-ranking sites should have little to no sequence polymorphism. As the C. trachomatis genomes are all highly similar, the top-ranking genes within each test strain were of significance, as these should be the regions that contain the most nucleotide differences (Table 1).
Table 1.
The ten highest-ranked signal ratios and their corresponding genes for each serovar
| Rank | Gene | SR* | SEM† | Gene | SR | SEM | Gene | SR | SEM | |||
| A/Har-1 | B/TW-5 | Ba/Apache-2 | ||||||||||
| 1 | 166 | 4.04 | ± | 0.63 | 163 | 8.21 | ± | 2.78 | 870 (pmpF) | 3.39 | ± | 0.39 |
| 2 | 681 (ompA) | 3.91 | ± | 0.34 | 166 | 6.65 | ± | 0.87 | 861 | 2.77 | ± | 0.16 |
| 3 | 679 (tsf) | 3.56 | ± | 0.15 | 164 | 4.29 | ± | 0.46 | 851 (map) | 2.73 | ± | 0.25 |
| 4 | 622 | 2.93 | ± | 0.21 | 167 | 4.09 | ± | 0.50 | 166 | 2.67 | ± | 0.19 |
| 5 | 51 | 2.84 | ± | 0.06 | 171 (trpA) | 4.02 | ± | 0.73 | 860 | 2.63 | ± | 0.23 |
| 6 | 688 (parB) | 2.81 | ± | 0.10 | 165 | 3.89 | ± | 0.56 | 874 (pmpI) | 2.61 | ± | 0.16 |
| 7 | 870 (pmpF) | 2.76 | ± | 0.16 | 170 (trpB) | 3.81 | ± | 0.41 | 688 (parB) | 2.60 | ± | 0.25 |
| 8 | 161 | 2.72 | ± | 0.11 | 162 | 3.80 | ± | 0.28 | 792 (mutS) | 2.60 | ± | 0.32 |
| 9 | 49 | 2.55 | ± | 0.18 | 168 | 2.94 | ± | 0.19 | 852 (yhgN) | 2.58 | ± | 0.32 |
| 10 | 672 (fliN) | 2.53 | ± | 0.19 | 207 (pfkA) | 2.90 | ± | 1.21 | 855 (fumC) | 2.58 | ± | 0.20 |
| C/TW-3 | E/Bour | F/IC-CAL3 | ||||||||||
| 1 | 166 | 3.66 | ± | 0.29 | 675 (karG) | 3.60 | ± | 0.25 | 680 (rs2) | 6.28 | ± | 1.69 |
| 2 | 681 (ompA) | 2.59 | ± | 0.31 | 161 | 3.40 | ± | 0.33 | 679 (tsf) | 4.48 | ± | 0.17 |
| 3 | 672 (fliN) | 2.40 | ± | 0.15 | 870 (pmpF) | 3.37 | ± | 0.20 | 681 (ompA) | 3.45 | ± | 0.17 |
| 4 | 864 (xerC/D) | 2.39 | ± | 0.11 | 688 (parB) | 3.33 | ± | 0.21 | 677 (frr) | 3.17 | ± | 0.34 |
| 5 | 161 | 2.38 | ± | 0.14 | 792 (mutS) | 3.32 | ± | 0.31 | 873 | 2.87 | ± | 0.09 |
| 6 | 860 | 2.37 | ± | 0.16 | 836 (pheS) | 3.20 | ± | 0.10 | 649 (ygfA) | 2.53 | ± | 0.08 |
| 7 | 696 | 2.35 | ± | 0.10 | 839 | 3.11 | ± | 0.16 | 622 | 2.24 | ± | 0.17 |
| 8 | 675 (karG) | 2.30 | ± | 0.11 | 694 | 3.11 | ± | 0.34 | 696 | 2.19 | ± | 0.12 |
| 9 | 694 | 2.28 | ± | 0.07 | 686 | 3.10 | ± | 0.16 | 49 | 2.04 | ± | 0.22 |
| 10 | 688 (parB) | 2.28 | ± | 0.06 | 761 (murG) | 3.07 | ± | 0.08 | 429 | 1.94 | ± | 0.20 |
| G/UW-57 | H/UW-4 | I/UW-12 | ||||||||||
| 1 | 681 (ompA) | 5.12 | ± | 0.69 | 161 | 2.77 | ± | 0.16 | 870 (pmpF) | 3.80 | ± | 0.08 |
| 2 | 696 | 2.30 | ± | 0.03 | 622 | 2.65 | ± | 0.12 | 792 (mutS) | 3.15 | ± | 0.18 |
| 3 | 291 (ptsN) | 2.23 | ± | 0.22 | 761 (murG) | 2.58 | ± | 0.16 | 161 | 3.12 | ± | 0.09 |
| 4 | 674 (yscC) | 2.10 | ± | 0.13 | 839 | 2.53 | ± | 0.18 | 360 | 3.09 | ± | 0.03 |
| 5 | 175 (oppA) | 2.10 | ± | 0.08 | 427 | 2.40 | ± | 0.11 | 686 | 2.97 | ± | 0.18 |
| 6 | 84 | 2.07 | ± | 0.01 | 870 (pmpF) | 2.38 | ± | 0.09 | 840 (mesJ) | 2.94 | ± | 0.17 |
| 7 | 539 (trxA) | 2.03 | ± | 0.21 | 675 (karG) | 2.38 | ± | 0.08 | 812 (pmpD) | 2.93 | ± | 0.32 |
| 8 | 475 (pheT) | 2.00 | ± | 0.83 | 688 (parB) | 2.30 | ± | 0.09 | 688 (parB) | 2.91 | ± | 0.11 |
| 9 | 672 (fliN) | 1.99 | ± | 0.08 | 216 (xasA) | 2.29 | ± | 0.08 | 49 | 2.90 | ± | 0.14 |
| 10 | 385 (ycfF) | 1.97 | ± | 0.36 | 783 | 2.29 | ± | 0.22 | 598 | 2.89 | ± | 0.34 |
| J/UW-36 | K/UW-31 | L1/440 | ||||||||||
| 1 | 369 (aroB) | 3.83 | ± | 0.30 | 681 (ompA) | 8.45 | ± | 1.42 | 166 | 3.45 | ± | 0.38 |
| 2 | 427 | 3.76 | ± | 0.23 | 84 | 4.07 | ± | 0.48 | 870 (pmpF) | 3.01 | ± | 0.34 |
| 3 | 476 | 3.72 | ± | 0.37 | 175 (oppA) | 2.86 | ± | 0.31 | 161 | 2.72 | ± | 0.28 |
| 4 | 870 (pmpF) | 3.64 | ± | 0.25 | 291 (ptsN) | 2.37 | ± | 0.10 | 760 (ftsW) | 2.69 | ± | 0.20 |
| 5 | 792 (mutS) | 3.53 | ± | 0.12 | 696 | 2.29 | ± | 0.09 | 868 | 2.66 | ± | 0.16 |
| 6 | 360 | 3.48 | ± | 0.15 | 557 (lpdA) | 2.26 | ± | 0.09 | 167 | 2.59 | ± | 0.08 |
| 7 | 836 (pheS) | 3.40 | ± | 0.18 | 298 (sms) | 2.17 | ± | 0.11 | 144 | 2.59 | ± | 0.12 |
| 8 | 797 (pgsA) | 3.38 | ± | 0.29 | 577 | 2.14 | ± | 0.19 | 872 (pmpH) | 2.53 | ± | 0.21 |
| 9 | 839 | 3.35 | ± | 0.08 | 656 | 2.12 | ± | 0.11 | 761 (murG) | 2.47 | ± | 0.13 |
| 10 | 507 (rpoA) | 3.33 | ± | 0.14 | 461 (yaeI) | 2.07 | ± | 0.11 | 839 | 2.46 | ± | 0.09 |
| L2/434 | L3/404 | |||||||||||
| 1 | 166 | 9.90 | ± | 1.26 | 166 | 11.61 | ± | 0.29 | ||||
| 2 | 167 | 5.72 | ± | 1.23 | 681 (ompA) | 6.46 | ± | 0.39 | ||||
| 3 | 84 | 5.68 | ± | 1.12 | 167 | 6.15 | ± | 0.46 | ||||
| 4 | 165 | 5.27 | ± | 1.60 | 165 | 4.93 | ± | 0.55 | ||||
| 5 | 173 | 3.82 | ± | 0.50 | 622 | 3.85 | ± | 0.23 | ||||
| 6 | 681 (ompA) | 3.25 | ± | 0.37 | 173 | 3.69 | ± | 0.23 | ||||
| 7 | 144 | 3.23 | ± | 0.21 | 144 | 3.67 | ± | 0.32 | ||||
| 8 | 622 | 3.09 | ± | 0.12 | 619 | 3.23 | ± | 0.05 | ||||
| 9 | 619 | 2.95 | ± | 0.29 | 870 (pmpF) | 2.71 | ± | 0.19 | ||||
| 10 | 870 (pmpF) | 2.57 | ± | 0.05 | 293 (accD) | 2.65 | ± | 0.03 |
*Average signal ratio; †Standard error of the mean.
Verification of array data
To confirm that the microarray analysis had identified nucleotide variability among the various genomes, several high-ranking genes were chosen, on the basis of biological interest, to be sequenced and compared to the reference DNA sequence. These genes, which were predicted to be polymorphic from the array data, were found to contain nucleotide differences that were evenly distributed throughout the sequences (Table 2a). Several of the same genes from strains in which the locus was not predicted to be variable were then sequenced to verify the discriminatory powers of the assay; as expected, these regions did not contain sequence polymorphisms (Table 2b). In addition, those genes that had previously been described to harbor few to no polymorphisms were not found to have any significant signal ratios (that is, gseA, trpB, 16S) [17,21]. The well-characterized and sequence-variable ompA gene [22] was among the highest in rank and nucleotide diversity among many of the strains. B/TW-5 was the only strain used in this study with known gene deletions as it is lacking the trp operon and several neighboring genes (CT162-171) [18]. Congruent with these findings, the array data from B/TW-5 indicated that these genes are absent or otherwise highly divergent (Table 1). As this region was not highly ranked in any of the other strains, the results indicated that the trp operon was present in the remainder of the strains. The CT868 gene region of strain L1/440 was interesting because of the fact that its high signal ratio and high rank were not entirely due to nucleotide variation (2.2% difference); there was also a 33 base-pair (bp) deletion in the L1/440 sequence, indicating that the array is sensitive to insertions/deletions as well as nucleotide variations and gene loss.
Table 2.
Sequence differences in those regions predicted to contain polymorphisms on the basis of microarray data
| Serovar | Gene | SR* | Rank† | Difference (%)‡ | GenBank ID§ |
| (a) High-ranking genes | |||||
| A/Har-1 | CT679 (tsf) | 3.6 | 3 | 7.9 | AY539791 |
| A/Har-1 | CT622 | 2.9 | 4 | 2.2 | AY539765 |
| A/Har-1 | CT870 (pmpF) | 2.8 | 7 | 7.5 | AY539793 |
| A/Har-1 | CT681 (ompA) | 3.9 | 2 | 21.6 | J03813 |
| B/TW-5 | CT870 (pmpF) | 2.1 | 23 | 7.5 | AY539794 |
| B/TW-5 | CT681 (ompA) | 2.2 | 16 | 6.4 | M17342 |
| Ba/AP-2 | CT870 (pmpF) | 3.4 | 3 | 7.5 | AY539795 |
| Ba/AP-2 | CT675 (karG) | 2.6 | 25 | 2.9 | AY539779 |
| C/TW-3 | CT870 (pmpF) | 2.3 | 13 | 7.5 | AY539796 |
| C/TW-3 | CT681 (ompA) | 2.6 | 2 | 22.0 | M17343 |
| E/Bour | CT622 | 2.8 | 30 | 9.2 | AY539768 |
| E/Bour | CT675 (karG) | 3.6 | 2 | 4.2 | AY539781 |
| F/IC-CAL3 | CT622 | 2.2 | 7 | 9.1 | AY539769 |
| F/IC-CAL3 | CT679 (tsf) | 4.5 | 2 | 7.6 | AY539790 |
| F/IC-CAL3 | CT681 (ompA) | 3.5 | 3 | 15.8 | X52080 |
| G/UW-57 | CT681 (ompA) | 5.1 | 1 | 19.1 | AF063199 |
| H/UW-4 | CT622 | 2.7 | 4 | 1.2 | AY539771 |
| J/UW-36 | CT870 (pmpF) | 3.6 | 14 | 7.5 | AY539798 |
| K/UW-31 | CT681 (ompA) | 8.5 | 1 | 21.2 | AF063204 |
| L1/440 | CT622 | 2.5 | 17 | 13.3 | AY539775 |
| L1/440 | CT868 | 2.7 | 9 | 2.2¶ | AY539792 |
| L1/440 | CT870 (pmpF) | 3.0 | 4 | 14.6 | AY539803 |
| L2/434 | CT144 | 3.2 | 7 | 10.8 | AY539751 |
| L2/434 | CT293 (accD) | 2.5 | 11 | 3.4 | AY539763 |
| L2/434 | CT622 | 3.1 | 8 | 13.3 | AY539776 |
| L2/434 | CT870 (pmpF) | 2.6 | 10 | 14.6 | AY539804 |
| L2/434 | CT681 (ompA) | 3.2 | 6 | 10.5 | M14738 |
| L3/404 | CT293 (accD) | 2.7 | 10 | 3.4 | AY539764 |
| L3/404 | CT681 (ompA) | 6.5 | 2 | 21.1 | X55700 |
| L3/404 | CT622 | 3.8 | 5 | 13.3 | AY539777 |
| L3/404 | CT870 (pmpF) | 2.7 | 9 | 14.6 | AY539805 |
| (b) Intermediate- to low-ranking genes | |||||
| A/Har-1 | CT293 (accD) | 1.9 | 307 | 0.0 | AY539752 |
| B/TW-5 | CT293 (accD) | 1.5 | 406 | 0.0 | AY539753 |
| Ba/AP-2 | CT293 (accD) | 1.7 | 330 | 0.0 | AY539754 |
| C/TW-3 | CT293 (accD) | 1.6 | 403 | 0.0 | AY539755 |
| E/Bour | CT293 (accD) | 1.8 | 372 | 0.4 | AY539756 |
| F/IC-CAL3 | CT293 (accD) | 1.2 | 521 | 0.4 | AY539757 |
| G/UW-57 | CT293 (accD) | 1.3 | 365 | 0.0 | AY539758 |
| H/UW-4 | CT293 (accD) | 1.7 | 305 | 0.0 | AY539759 |
| J/UW-36 | CT293 (accD) | 2.0 | 299 | 0.0 | AY539761 |
| K/UW-31 | CT622 | 1.3 | 375 | 0.0 | AY539774 |
| G/UW-57 | CT675 (karG) | 1.2 | 469 | 0.0 | AY539783 |
| K/UW-31 | CT675 (karG) | 1.3 | 475 | 0.0 | AY539787 |
| F/IC-CAL3 | CT870 (pmpF) | 1.2 | 489 | 0.0 | AY539798 |
| G/UW-57 | CT870 (pmpF) | 1.1 | 673 | 0.0 | AY539799 |
| K/UW-31 | CT870 (pmpF) | 1.2 | 626 | 0.0 | AY539802 |
*Average signal ratio; †rank based on signal ratio within that strain; ‡percent nucleotide difference of region between test strain and reference strain; §GenBank accession number; ¶contained 33-bp deletion.
Differences between biological groups
An organism with a specific tissue tropism will have evolved differences in its genome as a result of the selection of mutations that promote its survival in that particular biological niche. Therefore, those genes that are different within all the strains of one of the three pathobiological groups of C. trachomatis (ocular, urogenital or lymphogranuloma) may have been selected as a result of niche-specific pressures (Table 3). For example, a gene identified as variable in serovars A, B, Ba and C may confer an advantage in the ocular environment. Such group-specific changes may be directly associated with differences in phenotypes, and would be important for future functional experiments. However, there were very few genes overall that were classified as different within one biological niche from all the strains tested. Of the 31 niche-specific genes identified, 16 coded for hypothetical genes (5.3% of all hypothetical ORFs) and 15 were from known genes (2.6% of all named genes).
Table 3.
Predicted niche-specific genes for each of the three different biological tropisms
| Ocular* | Urogenital† | LGV‡ |
| CT158 | CT161 | CT116 (incE) |
| CT210 (hemL) | CT166 | CT144 |
| CT216 (xasA) | CT622 | CT167 |
| CT360 | CT672 (fliN) | CT223 (inc) |
| CT398 | CT870 (pmpF) | CT288 |
| CT470 | CT872 (pmpH) | CT293 (accD) |
| CT675 (karG) | CT312 (fer) | |
| CT686 | CT618 | |
| CT688 (parB) | CT664 | |
| CT690 (dppD) | CT696 | |
| CT694 | CT760 (ftsW) | |
| CT792 (mutS) | ||
| CT860 | ||
| CT874 (pmpI) |
*Ocular strains A/Har1, B/TW-5, Ba/Apache-2, C/TW-3; †urogenital strains D/UW-3, E/Bour, F/IC-Cal 3, G/UW-57, H/UW-4, I/UW-12, J/UW-36, K/UW-31; ‡LGV strains L1/440, L2/434, L3/404.
Discussion
The ability to use the C. trachomatis array as a screening tool for DNA polymorphisms was first demonstrated by determining the nucleotide identities for each of 830 gene regions between two known genomes - C. trachomatis D/UW-3 and C. trachomatis MoPn [19,20] - and then comparing these identities to their corresponding signal ratios. As a locus became more variable on the nucleotide level, its relative signal ratio concomitantly increased as a logarithmic function. Previous studies using Helicobacter pylori microarrays concluded that the relationship between gene identity and the array signal was valid only for those regions above 81% [8]. However, the strains used in that analysis lacked sufficient regions below 81% identity to assess the prospect of a lower cut-off. As the two C. trachomatis genomes had a strong representation of orthologous gene regions with a continuous range of identities from 66% to 99%, it was determined that the logarithmic relationship of the signal ratios diminished in regions below 75% identity, thus delineating this as the lower limit of the association.
One factor that may skew the logarithmic relationship between signal ratios and the corresponding nucleotide identities for some of the gene regions is insertion and deletion. Insertions and deletions can affect hybridization in two ways depending on whether they are present in the test- or in the reference-strain gene. An insertion in a test-strain gene will cause the region to be longer than the complementary sequence on the array, forcing the test DNA at that point to fold during hybridization. A deletion in part of a test-strain gene will prevent proper alignment with the target region, as the DNA sequence on the microarray has a novel segment for which the test strain lacks a complement for hybridization. In addition, the test-strain DNA will not span the unique sequence on the array to align with the nucleotides on both sides. Either an insertion or a deletion will result in a signal ratio higher than expected from the overall nucleotide identity of the gene region, as was seen with the 33 bp deletion in gene CT868 of the L1/440 strain.
Another factor that could affect the correlation of the signal ratio with nucleotide identity is the presence of multiple homologous sequences within a genome. Even if a gene is identical between a test and reference strain, regions of nucleotide similarity found elsewhere in the genome would compete for hybridization to the target region on the array, thereby skewing the signal ratio of the intended gene pair. This may have a profound effect when one is studying genes that have paralogs due to recent gene duplication events, as they may confound the array data because of their regions of similarity. In C. trachomatis this is not an issue, as its paralogs [12] lack significant nucleotide similarity as a result of their ancient duplication events pre-dating chlamydial diversification. Therefore, enough changes have occurred to avoid such bias.
Differences among C. trachomatis strains were identified by microarray analysis and confirmed by subsequent DNA sequencing. Specific genes that were found to vary in one or more genomes may have become fixed either by chance or by selection. If these genes were selected because they offered an advantage in fitness, then they may contribute to phenotypic differences between the serovars. A possible example of this is the tsf (elongation factor TS (EF-TS)) gene, which is a GDP-dissociation protein that plays an important role in protein biosynthesis and may have a direct role in the chlamydial developmental cycle [23]. This region was found to be polymorphic in strains A/Har1 and F/IC-Cal3, and the respective nucleotide differences resulted in 12 and 13 amino-acid substitutions over a portion of the coding region when compared to strain D/UW-3. Although none of the predicted binding sites of EF-TS for EF-Tu was variable, conformational changes in a protein involved in the regulation of other proteins, especially those associated with the developmental cycle, may have a direct effect on the overall fitness or phenotype of an organism.
Genes that were found to be polymorphic within all serovars of a particular biological or tissue tropism group may represent the selection of mutations due to niche-specific pressures for survival within that environment. The fliN (flagellar motor switch domain) gene, which is thought to serve a role in the type III secretion system in C. trachomatis, was found to be variable in serovars A-C and L1-L3. In other organisms with type III secretion systems, this gene encodes a protein involved in the switch complex, and amino-acid changes in this region have been shown to have an effect on levels of secretion [24]. By analogy, changes in the fliN gene of C. trachomatis may have resulted in altered phenotypes, leading to an increase in fitness for a particular biological niche and its subsequent selection.
It appears that the three distinct tissue tropisms for strains of C. trachomatis (that is, ocular, urogenital and lymph node) are due to relatively few changes within the coding regions, as there were only 31 genes that were classified as being variable within all strains of a biological niche; interestingly, the majority of these genes code for hypothetical proteins of unknown function. With such globally low differences between all the genomes of C. trachomatis, those few substitutions that have become fixed within the population would have conferred a benefit; otherwise they would have been lost by chance. These genes could be essential in the goal of establishing the genetic basis for the different tissue tropisms of C. trachomatis, as well as providing a basis for future functionality tests.
Conclusions
DNA microarray technology can serve as a rapid tool for identifying regions of polymorphisms in otherwise unknown isolates of C. trachomatis as it has the potential to quickly reduce a genome of a thousand genes to a handful of meaningful sites. Without a genetic model for C. trachomatis, the nucleotide differences identified in this study may offer the best insights into assessing gene function among phenotypically distinct strains.
Materials and methods
Bacterial strains, growth conditions, and preparation of genomic DNA
C. trachomatis strains (A/Har1, B/TW-5, Ba/Apache-2, E/Bour, F/IC-Cal 3, G/UW-57, H/UW-4, I/UW-12, J/UW-36, K/UW-31) were kindly provided by J. Schachter, University of California, San Francisco. C. trachomatis strains (A/Har1, B/TW-5, Ba/Apache-2, C/TW-3, D/UW-3, E/Bour, F/IC-Cal 3, G/UW-57, H/UW-4, I/UW-12, J/UW-36, K/UW-31, L1/440, L2/434, L3/404, MoPn) were propagated in HeLa229 cell monolayers in T-150 flasks containing RPMI medium (Invitrogen) supplemented with 10% fetal bovine serum and 50 μg/ml vancomycin. Chlamydial elementary bodies were isolated by sonic treatments of cell suspensions and purified by ultracentrifugation over 30% and 30/44% discontinuous Renografin gradients (E.R. Squibb and Sons, Princeton, NJ) as previously described [25]. Aliquots were frozen at -80°C in sucrose-phosphate-glutamate buffer. Before hybridization, chlamydial elementary bodies were washed and genomic DNA from each strain was prepared by proteinase-K digestion, phenol/chloroform extraction, and ethanol precipitation [26].
Hybridizations and data analysis
PCR fragments representing an average of 60% of each ORF from the genome of C. trachomatis strain D/UW-3 were spotted in duplicate per microarray slide [27]. The gene region represented by each array probe was chosen on the basis of the ability to create primer pairs specific for amplification of the longest possible target region, thereby preventing any bias in the selection of particular regions. For each slide, hybridization with the immobilized microarray DNA was measured between the reference strain DNA (D/UW-3) and one test strain DNA. Using random primers as stated in the BioPrime DNA Labeling System Kit (Invitrogen), D/UW-3 genomic DNA (0.2 μg) was labeled with Cy5 dye-labeled nucleotides, whereas all test DNA (0.2 μg) were labeled with Cy3 dye-labeled nucleotides (Invitrogen). Buffer exchange, purification, and concentration of the labeled-DNA products were accomplished as previously described [27]. The two labeled-DNA samples to be compared were mixed, heat denatured (95°C for 3 min), and applied to a chlamydial-DNA microarray in a hybridization mixture containing 3.5 × SSC, 0.3% SDS, and 10 μg yeast tRNA [27]. All hybridizations took place under a glass coverslip in a 75°C water bath overnight, except for the MoPn versus D/UW-3 comparison, which was conducted in a 65°C water bath overnight. The slides were washed, dried, and scanned using a GenePix Scanner 4000A and the resulting 16-bit TIFF images were analyzed using GenePix Pro 4.0 software (Axon Instruments). Only spots with greater than 60% of all pixels having intensities greater than average background intensities were used for analysis. To reduce the effects of variation in array quality, each hybridization was performed at least twice, giving a minimum of four data points for each gene region of a strain as the genome is printed twice per slide. Data for duplicate readings and each hybridization experiment were normalized on the basis of the overall median percent intensity to eliminate slide-to-slide variation.
Percent identity between strains D/UW-3 and MoPn
Each gene region from the D/UW-3 array was aligned with the corresponding orthologous sequence from MoPn using ClustalX [28], and Mega2 was used to assess the number of nucleotide differences [29]. The percent identity for each region was determined by dividing the number of identical sites between two sequences by the total number of sites, and then multiplying by 100.
Sequence analysis
For sequence analysis, the gene regions corresponding to the array probe of interest were amplified by PCR and sequenced in both the 5' and 3' direction on an ABI PRISM 377 DNA Sequencer (Applied Biosystems) and were deposited in GenBank (accession numbers AY539751-AY539805; AY542692-AY542704). Sequences for the ompA gene were taken from Stothard et al. [30]. The percent nucleotide identity between each test region and the reference sequence represented on the array was calculated as described above.
Additional data files
A complete table (Additional data file 1) containing the signal ratios for each gene (about 900) and their corresponding rank within each serovar (14 each) is available with the online version of this article. Table 1 of the text is a subset of these data.
Supplementary Material
A complete table containing the signal ratios for each gene (about 900) and their corresponding rank within each serovar (14 each)
Acknowledgments
Acknowledgements
We thank Gary K. Schoolnik and Kevin Visconti for assistance in printing the microarray slides and Jeanne Moncada for help in the propagation of several of the chlamydial strains. We also thank George F. Sensabaugh for input and suggestions and P. Scott Hefty for laboratory assistance and critical review of the manuscript. This research was supported by the National Institutes of Health grant AI042156.
References
- Murray AE, Lies D, Li G, Nealson K, Zhou J, Tiedje JM. DNA/DNA hybridization to microarrays reveals gene-specific differences between closely related microbial genomes. Proc Natl Acad Sci USA. 2001;98:9853–9858. doi: 10.1073/pnas.171178898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama N, Guillemin K, McDaniel TK, Sherlock G, Tompkins L, Falkow S. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc Natl Acad Sci USA. 2000;97:14668–14673. doi: 10.1073/pnas.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel DA, Salama N, Krishna U, Rieger UM, Atherton JC, Falkow S, Peek RM., Jr Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc Natl Acad Sci USA. 2001;98:14625–14630. doi: 10.1073/pnas.251551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell N, Mangan JA, Laing KG, Hinds J, Linton D, Al-Ghusein H, Barrell BG, Parkhill J, Stoker NG, Karlyshev AV, et al. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 2001;11:1706–1715. doi: 10.1101/gr.185801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Baker S, Kim CC, Detweiler CS, Dougan G, Falkow S. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar typhimurium DNA microarray. J Bacteriol. 2003;185:553–563. doi: 10.1128/JB.185.2.553-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkholm B, Lundin A, Sillen A, Guillemin K, Salama N, Rubio C, Gordon JI, Falk P, Engstrand L. Comparison of genetic divergence and fitness between two subclones of Helicobacter pylori. Infect Immun. 2001;69:7832–7838. doi: 10.1128/IAI.69.12.7832-7838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, Small PM. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- Kim CC, Joyce EA, Chan K, Falkow S. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 2002;3:research0065.1–0065.17. doi: 10.1186/gb-2002-3-11-research0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez P, Allardet-Servent A, de Barbeyrac B, Ramuz M, Bebear C. Genetic variability among Chlamydia trachomatis reference and clinical strains analyzed by pulsed-field gel electrophoresis. J Clin Microbiol. 1994;32:2921–2928. doi: 10.1128/jcm.32.12.2921-2928.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G, Raoult D. History of the ADP/ATP-translocase-encoding gene, a parasitism gene transferred from a Chlamydiales ancestor to plants 1 billion years ago. Appl Environ Microbiol. 2003;69:5530–5535. doi: 10.1128/AEM.69.9.5530-5535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS. Chlamydial evolution: a billion years and counting. In: Schacter J, Chrstiansen G, Clarke IN, Hammerschlag MR, Kaltenboek B, Kuo C-C, Rank GL, Ridgeway GL, Saikku P, Stamm WE, editor. In Chlamydial Infections, Proceedings of the Tenth International Symposium on Human Chlamydial Infections:16-21 June 2002 Antalya, Turkey. International Chlamydia Symposium; 2002. pp. 3–16. [Google Scholar]
- Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman RW, Olinger L, Grimwood J, Davis RW, Stephens RS. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet. 1999;21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- Wang SP, Grayston JT. Micro-immunofluorescence antibody responses to trachoma vaccines. Int Ophthalmol. 1988;12:73–80. doi: 10.1007/BF00133785. [DOI] [PubMed] [Google Scholar]
- Stephens RS. Genomic autobiographies of Chlamydiae. In: Stephens RS, editor. In Chlamydia: Intracellular Biology, Pathogenesis, and Immunity. Washington, DC: American Society for Microbiology; 1999. pp. 6–26. [Google Scholar]
- Nigg C. Unidentified virus which produces pneumonia and systemic infection in mice. Science. 1942;95:49–50. doi: 10.1126/science.95.2454.49-a. [DOI] [PubMed] [Google Scholar]
- Caldwell HD, Wood H, Crane D, Bailey R, Jones RB, Mabey D, Maclean I, Mohammed Z, Peeling R, Roshick C, et al. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J Clin Invest. 2003;111:1757–1769. doi: 10.1172/JCI200317993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehlner-Gardiner C, Roshick C, Carlson JH, Hughes S, Belland RJ, Caldwell HD, McClarty G. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J Biol Chem. 2002;277:26893–26903. doi: 10.1074/jbc.M203937200. [DOI] [PubMed] [Google Scholar]
- Shaw AC, Christiansen G, Roepstorff P, Birkelund S. Genetic differences in the Chlamydia trachomatis tryptophan synthase alpha-subunit can explain variations in serovar pathogenesis. Microbes Infect. 2000;2:581–592. doi: 10.1016/S1286-4579(00)00368-3. [DOI] [PubMed] [Google Scholar]
- Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Mamat U, Lobau S, Persson K, Brade H. Nucleotide sequence variations within the lipopolysaccharide biosynthesis gene gseA (Kdo transferase) among the Chlamydia trachomatis serovars. Microb Pathog. 1994;17:87–97. doi: 10.1006/mpat.1994.1055. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Zhang YX, Watkins NG, Caldwell HD. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect Immun. 1989;57:1040–1049. doi: 10.1128/iai.57.4.1040-1049.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tao J, Zhou M, Meng Q, Zhang L, Shen L, Klein R, Miller DL. Elongation factor Ts of Chlamydia trachomatis: structure of the gene and properties of the protein. Arch Biochem Biophys. 1997;344:43–52. doi: 10.1006/abbi.1997.0178. [DOI] [PubMed] [Google Scholar]
- Kubori T, Yamaguchi S, Aizawa S. Assembly of the switch complex onto the MS ring complex of Salmonella typhimurium does not require any other flagellar proteins. J Bacteriol. 1997;179:813–817. doi: 10.1128/jb.179.3.813-817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler JE, Burgess RR, Thompson NE, Stephens RS. Chlamydia trachomatis RNA polymerase major sigma subunit. Sequence and structural comparison of conserved and unique regions with Escherichia coli sigma 70 and Bacillus subtilis sigma 43. J Biol Chem. 1990;265:13206–13214. [PubMed] [Google Scholar]
- Davis LG, Kuehl WM, Battey JF. In: In Basic Methods in Molecular Biology. 2. Davis LG, Kuehl WM, Battey JF, editor. Norwalk, CT: Appleton and Lange; 1994. pp. 16–21. [Google Scholar]
- Nicholson TL, Olinger L, Chong K, Schoolnik G, Stephens RS. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J Bacteriol. 2003;185:3179–3189. doi: 10.1128/JB.185.10.3179-3189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Stothard DR, Boguslawski G, Jones RB. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect Immun. 1998;66:3618–3625. doi: 10.1128/iai.66.8.3618-3625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A complete table containing the signal ratios for each gene (about 900) and their corresponding rank within each serovar (14 each)


