Summary
The lysis protein A2, present as a single copy on the surface of Qβ virion particles, was previously shown to inhibit the activity of MurA, an enzyme that catalyzes the first committed step of murein biosynthesis. Here we report experiments with a two-hybrid study that indicates A2 and MurA interact directly. Moreover, experiments with a soluble MBP-A2 fusion indicate that the interaction between MurA and A2 is dependent on a substrate-induced conformational change featured in the UDP-NAG-liganded state of MurA but not the tetrahedral intermediate state. Moreover, based on the location of L138Q, the original dominant A2-resistant mutant that identified MurA as the target, a directed mutagenesis strategy has identified a continuous surface required for A2 binding. This surface spans the catalytic loop/cleft and encompasses both the catalytic and C-terminal domains. These data support a model in which A2 preferentially binds MurA liganded with UDP-NAG, thereby preventing catalysis by occluding PEP from accessing the active site.
Keywords: Qβ, A2, phage, mechanism, MurA, inhibition
Introduction
The Allolevivirus Qβ produces a single protein A2 to effect bacterial lysis, in addition to serving as a structural component of the virion (Fig. 1). Unlike dsDNA phages that produce muralytic enzymes to degrade the cell wall, A2 inhbits MurA, an enzyme that catalyzes the first committed step in cell wall synthesis (Bernhardt et al., 2001b). The key genetic result that led to this conclusion was the isolation of a dominant mutant resistant to Qβ lysis that mapped to the murA locus. This mutant, designated as rat1 (resistant to A-two) was shown to be a Leu to Gln change at position 138 of MurA. In addition, Bernhardt and colleagues showed that MurA expression in trans protected cells from lysis during a Qβ phage infection. Moreover, purified Qβ particles were able to inhibit MurAwt activity, but not MurArat activity, in a crude extract. Qβ mutants that were able to overcome the rat mutant lysis block were isolated and designated as por (plates on rat). These mutants were postulated to be compensatory for the L138Q mutation in the A2-MurA interface (Bernhardt et al., 2001b). However, recent biochemical and genetic evidence has indicated that the suppressor phenotype of these mutants is due to translational up-regulation of A2, deriving from disruption of regulatory RNA stem-loop structures (C.A. Reed, C. Langlais, I. Wang, and R. Young; unpublished).
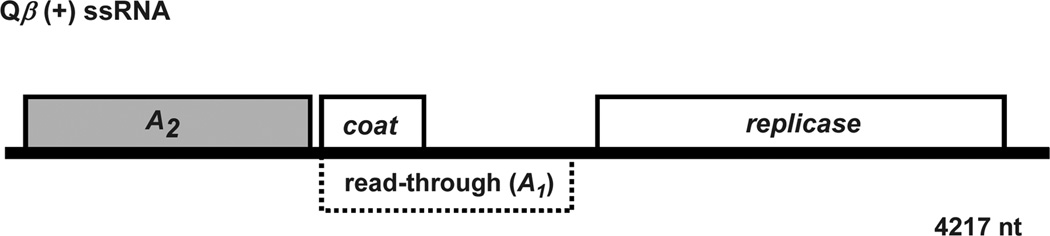
Figure 1. Map of Qβ genome.
Qβ is a (+) ssRNA phage. Replication of the 4.2 kb chromosome is dependent on a viral encoded replicase and four additional host proteins. Qβ capsid is generated from three gene products: Coat, the major virion structural protein, A1 a minor component translated from read-through of the leaky coat UGA stop codon, and a single copy of A2 bound to the RNA.
To address the question of how lysis is regulated in a Qβ infection, a model was proposed in which A2 lysis function was not fully realized until assembled into the capsid of the phage particle (Hatfull, 2001). However, recently we have shown that MurA actually inactivates Qβ particles, indicating that virions are not likely to be involved in lysis (C.A. Reed, C. Langlais, I. Wang, and R. Young; unpublished). Instead, quantitative analysis of the infection cycle indicated that free, unassembled A2 is the inhibitory molecule of MurA. Characterization of the mechanism of inhibition has been hindered due to the insolubility of A2 when it is not assembled onto the virion capsid (Weber et al., 1975).
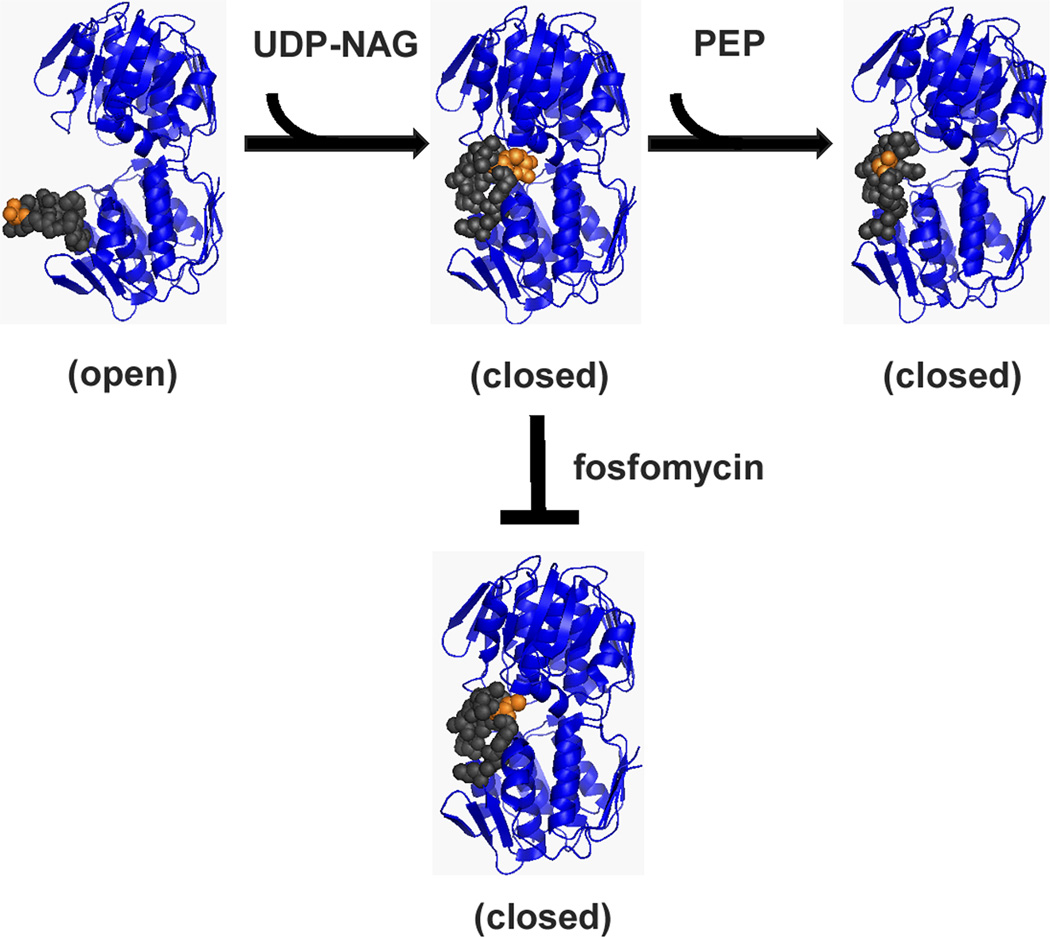
MurA catalyzes the transfer of the enolpyruvyl moiety (EP) from phosphoenolpyruvate (PEP) to uridine 5’-diphosphate-N-acetylglucosamine (UDP-NAG) yielding two products, UDP-NAG-EP and inorganic phosphate (Fig. 2) (Gunetileke & Anwar, 1968). Several catalytic states of MurA have been crystallized: (1) unliganded, “open” conformation (pre-catalysis state; PDB entry 1DLG) (Schönbrunn et al., 2000), (2) UDP-NAG-bound, “closed” conformational state (Han H., unpublished; PDB entry 3KQJ), (3) fosfomycin-inhibited state with UDP-NAG bound (PDB: 1UAE) (Skarzynski et al., 1996), and (4) a tetrahedral intermediate state with both UDP-NAG and PEP liganded (PDB entries 1A2N and 1Q3G) (Skarzynski et al., 1998, Eschenburg et al., 2003). MurA catalysis is an ordered reaction with binding of UDP-NAG, which is responsible for the major conformational change that is required for catalysis prior to PEP association (Schönbrunn, 1998, Zhu et al., 2012). Based on these structures, a major feature of the conformational changes associated with catalysis appears to be the dynamic behavior of the loop (grey spheres in Fig. 2) containing the catalytic Cys residue (orange spheres). An additional structure showed that PEP is excluded from the active site by a covalent bond formation of fosfomycin with residue C115 of MurA and the C-3 hydroxyl of UDP-NAG (Skarzynski et al., 1996). This structure adopts a conformation similar to the UDP-NAG-bound state. Mutation of the C115 residue is the basis for fosfomycin resistance of Mycobacterium tuberculosis MurA (Kim et al., 1996).
Figure 2. Unliganded MurA binds substrates, UDP-NAG and PEP, in an ordered fashion.
Upon binding both substrates, MurA undergoes catalysis. Fosfomycin is a dead-end inhibitor that requires UDP-NAG binding to inactivate MurA. PDB entries: open (1DLG), UDP-NAG bound (3KQJ), tetrahedral intermediate (1A2N), and UDP-NAG/fosfomycin (1UAE). Figures were generated using PyMOL (Schrödinger, 2010).
In this study we address the requirements for an A2-MurA interaction. The results are discussed in terms of the molecular state of MurA when it is bound by A2.
Results
Inhibition of purified MurA by Qβ
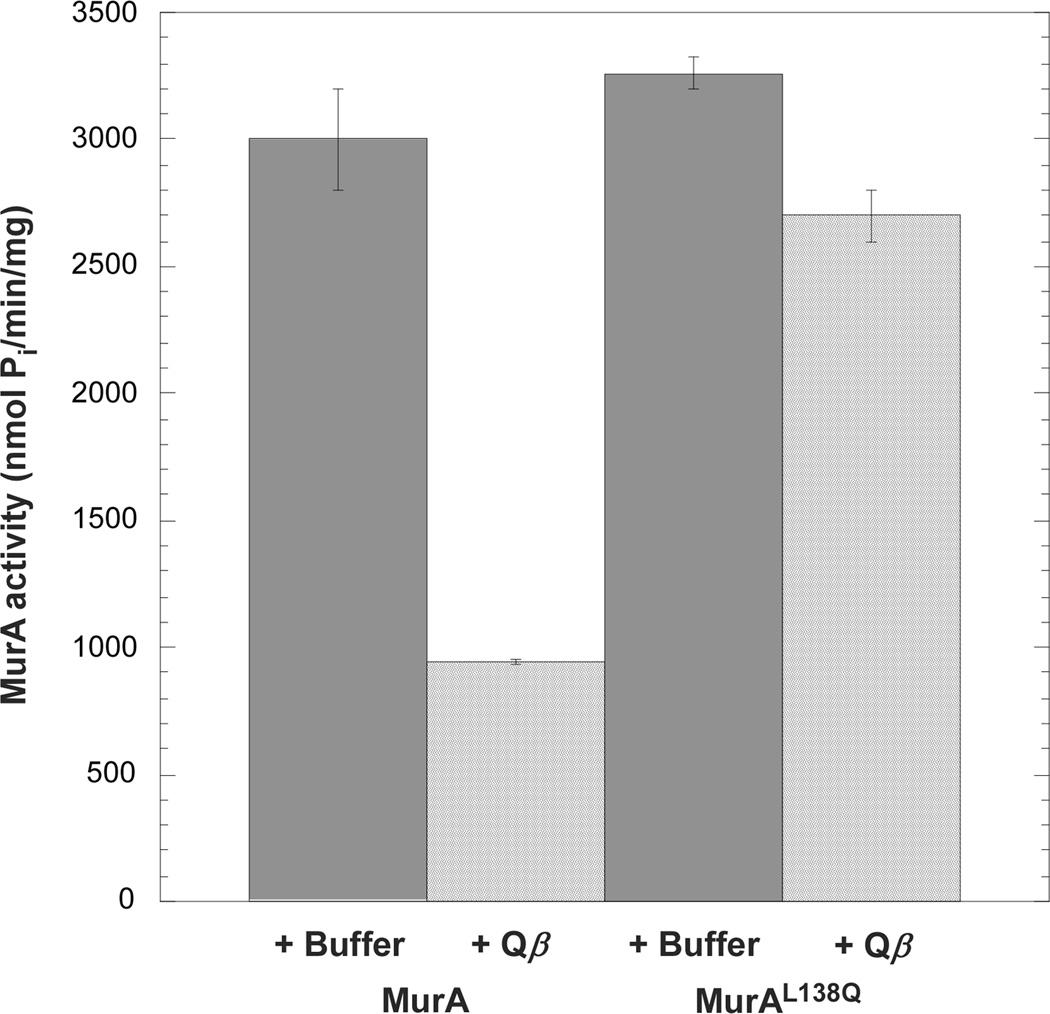
Bernhardt and colleagues (2001) had reported that virion-associated A2 inhibits MurA in a crude lysate. To test whether this inhibition required any other host factor, the in vitro assay was repeated with purified MurA and Qβ particles. We chose a MurA concentration, 400 nM, that reflects the level of MurA in the host cell (C.A. Reed, C. Langlais, I. Wang, and R. Young; unpublished). The results showed that, under these conditions, at the highest possible concentration of virions (~700 nM) Qβ reduced the activity of MurA by 70% when compared to a reaction lacking phage particles (Fig. 3). Under the same conditions, virion-mounted A2 caused only a marginal reduction in MurAL138Q activity (17%). These results rule out the need for other host components in the A2 inhibition of MurA.
Figure 3. Purified Qβ inhibits MurA in vitro.
Activity of purified MurA was measured in the presence of buffer or purified Qβ particles. The rat mutant, MurAL138Q, was tested in parallel. Values are averages of three samples.
Yeast-two-hybrid analysis of the A2-MurA interaction
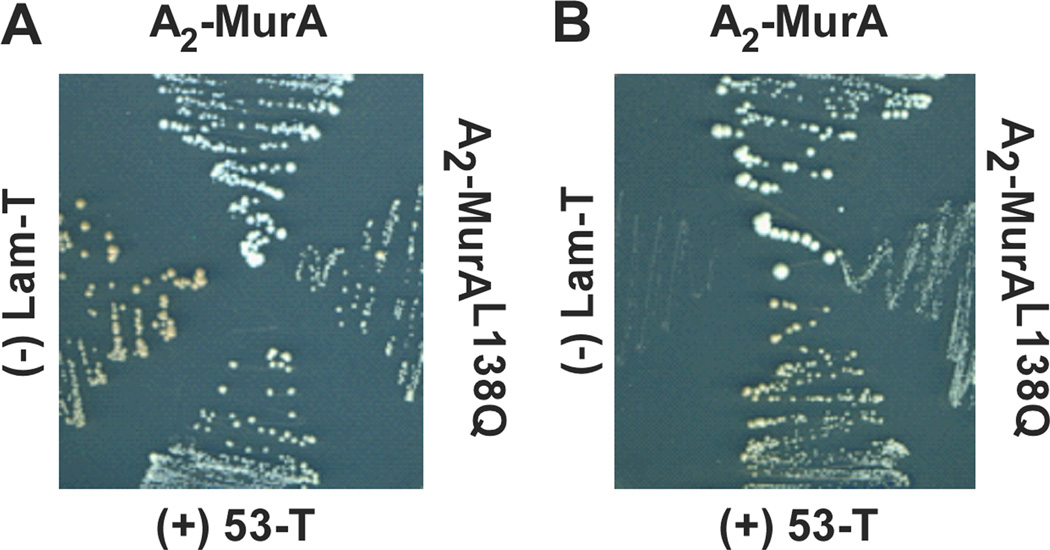
To determine whether there is a direct protein-protein interaction between MurA and A2 apart from other viral components, a yeast-two-hybrid system was utilized. Interaction was assessed with an A2-Gal4 DNA-binding domain fusion (pGBKT7-A2) and a MurA-Gal4 activating domain fusion (pGADT7-murA). AH109 yeast cells harboring both of these vectors were selected for growth on nutrient-deficient medium. Cellular growth was obtained when colonies were transferred from medium lacking leucine and tryptophan (Fig. 4A) to medium deficient of leucine, tryptophan, histidine and adenine (Fig. 4B). Interaction between the fusion proteins is required for supported growth on the latter medium as seen by p53 and T, two proteins known to interact. No growth was seen with Lam and T, which do not interact. Yeast colony growth was not only supported with pGBKT7-A2 and pGADT7-murA but also with a pGBKT7-A2 and pGADT7-murAL138Q combination (Fig. 4B). When cells expressing A2-MurAL138Q were compared to the A2-MurA pair, a difference in colony morphology was observed under conditions where the interaction is required for growth. This difference may reflect a weaker interaction between A2 and MurAL138Q.
Figure 4. The A2-MurA interaction is detectable by yeast-two-hybrid assay.
Yeast cells carrying plasmids expressing Gal4 fusions with A2 and either MurA or MurAL138Q were plated on non-stringent medium (A) or stringent medium, requiring the A2-MurA interaction (B). Negative and positive controls are the Lam-T pair and 53-T pair, respectively. See Experimental procedures for details.
The A2 interaction with MurA is conformation-dependent
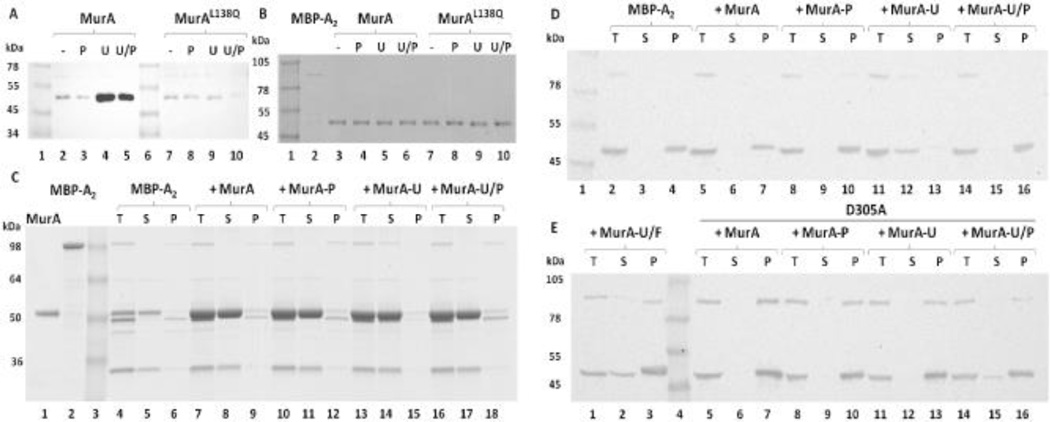
Batch affinity fractionation experiments using a purified fusion protein, MBP-A2, provide in vitro evidence for a direct interaction between A2 and MurA (Fig. 5A–B). Information about the conformational state of MurA required for MBP-A2 association was obtained by assaying binding in the presence of various substrates. MBP-A2 preferentially bound to both the UDP-NAG-liganded and the tetrahedral intermediate states of MurA (Fig. 5A, lanes 2–5), suggesting that A2 associates with MurA in a closed conformational state (Fig. 2). Binding of A2 to MurAL138Q was not observed under these conditions (Fig. 5A, lanes 7–10), suggesting that the mutation reduces the affinity of A2 for MurA.
Figure 5. A2 binds MurA in a substrate-dependent manner.
(A) MBP-A2 associates with MurA in a closed conformational state. Eluates from amylose magnetic bead fractionation experiments were Western blotted and probed with the α-MurA antibody. MBP-A2 was incubated with MurA under various substrate conditions: No substrate (−), PEP (P), UDP-NAG (U), and both substrates (U/P). MurAL138Q rat mutant was tested in parallel. (B) Unbound fractions of panel A experiments. (C) Fusion cleavage analysis of A2-MurA binding. MBP-A2 was cleaved with TEV protease in the absence or presence of MurA under various substrate conditions (same as in panel A). Binding was assessed as A2 solubility after centrifugation: Total fraction (T), supernatant after centrifugation (S), and pellet fraction (P). Samples were resolved on SDS-PAGE. MBP-A2 has an apparent MW of 100 kDa. Cleaved A2 runs at 50 kDa with the MBP running at the same apparent MW as MurA (~52 kDa). (D) A2 binds MurA liganded to UDP-NAG. Western blot analysis of fusion cleavage assays. Blots were probed with the α-A2 antibody. Binding was tested with substrate conditions as in panel C. (E) A2 does not bind the tetrahedral intermediate state of MurA. UDP-NAG and Fosfomycin (U/F) liganded MurA and MurAD305A binding and immunoblotting was also performed in parallel as in panel D.
Binding was also analyzed by another method in which cleavage of the fusion protein from A2 was performed in the presence of various substrate-induced MurA conformations. A2 cleaved from the MBP fusion protein was insoluble both in the absence and presence of unliganded MurA (Fig. 5C, lanes 4–9; Fig. 5D, lanes 2–7). Addition of PEP to the reaction with MurA did not increase A2 solubility (Fig. 5C, lanes 10–12; Fig. 5D, lanes 8–10), but when UDP-NAG was included in the reaction, A2 remained soluble (Fig. 5C, lanes 13–15; Fig. 5D, lanes 11–13), indicating the formation of an A2-MurA complex. However, in the presence of both substrates, A2 was insoluble (Fig. 5C, lanes 16–18; Fig. 5D, lanes 14–16), indicating a preference for binding to the UDP-NAG liganded form and not the tetrahedral intermediate state. This result is contradictory to the batch affinity data in which MurA association was observed in the presence of both substrates (Fig. 5A, lane 5). Comparison of the two fractions (Fig. 5A, lanes 4–5) revealed a reduction in the amount of MurA bound in the presence of both substrates suggesting that the fraction of MurA bound to A2 could be in the UDP-NAG liganded state, rather than tetrahedral intermediate state. Alternatively, the addition of fosfomycin with UDP-NAG, which locks the enzyme into a closed conformational state (Fig. 2), in the fusion cleavage experiment reduced the amount of soluble A2 but did not eliminate binding altogether (Fig. 5E, lanes 1–3).
Dynamics of the catalytic loop upon addition of both substrates might create unfavorable conditions for A2 association. To address this possibility, MurAD305A was purified and fusion cleavage of MBP-A2 was repeated. MurAD305A is unable to eliminate the product after substrate addition and locks MurA in the tetrahedral intermediate state (Fig. 2) (Eschenburg et al., 2003, Samland et al., 2001). A2 was insoluble with the addition of only UDP-NAG to the D305A mutant MurA which is not the case for the WT protein (compare Fig. 5E, lanes 11–13 to Fig. 5D, lanes 11–13). The purified D305A protein has an increase in absorbance at 260 nm, where UDP-NAG absorbs, compared to WT (data not shown) suggesting that the substrates do co-purify with the protein. Therefore, addition of UDP-NAG would have no effect on A2 solubility since the protein already has both substrates bound and suggests that A2 does not bind the tetrahedral intermediate state. Similarly to the fosfomycin with UDP-NAG bound MurA, a portion of A2 remained soluble in the presence of both UDP-NAG and PEP (Fig. 5E, lanes 14–16). The partial solubility of A2 in the fraction that included both substrates (Fig. 5E, lane 15) was reproducible, possibly indicating that the mutant retains a low level of activity, such that A2 is able to bind in the preferred UDP-NAG-liganded state. When WT MurA is added to a reaction with both substrates without prior incubation on ice, a fraction of A2 appears in the soluble fraction (data not shown) suggesting that A2 is able to bind the UDP-NAG state rather than the tetrahedral intermediate if the enzyme is not permitted to equilibrate with both substrates prior to the A2 fusion addition.
A catalytically inactive state of MurA exists in vivo in which the enzyme is liganded to PEP and uridine 5’-diphosphate-N-acetylmuramic acid (UDP-NAM), the product of the enzyme MurB that immediately follows MurA in the cell wall synthesis pathway (Zhu et al., 2012). This dormant form of MurA could be a potential cellular target for A2 since this liganded state adopts a closed conformation (Zhu et al., 2012). Fusion cleavage of MBP-A2 in the presence of MurA purified under conditions that contain a fraction of protein bound with UDP-NAM and PEP ((Mizyed et al., 2005, Zhu et al., 2012), see Supplemental information) resulted in insoluble A2 protein (Fig. S1, lanes 8–10), suggesting that A2 does not associate with the dormant MurA complex bound to UDP-NAM and PEP. Addition of UDP-NAG to the fusion cleavage reaction only resulted in partial solubility of A2 (Fig. S1, lanes 5–7) unlike cleavage reactions that have MurA bound to UDP-NAG alone (Fig. 5D, lanes 11–13). At 1 mM of UDP-NAG, UDP-NAM is displaced (Zhu et al., 2012); therefore, inclusion of UDP-NAG in the reaction should produce two species: a UDP-NAG bound state and a UDP-NAG/PEP bound state of MurA. The partial binding of A2 under conditions in which UDP-NAG is included in the reaction suggests that A2 is binding the UDP-NAG bound state and not the UDP-NAG/PEP bound state further supporting the notion that the purified MurA was bound to UDP-NAM and PEP.
When conformations of the catalytic loop of various MurA-liganded states were viewed, no apparent conformational difference is seen between the UDP-NAG and the UDP-NAG/fosfomycin-liganded states (Fig. S2A and B); however, a difference between the conformations of the catalytic loop between the UDP-NAG and tetrahedral intermediate states is visible (Fig. S2A and B). Perhaps this slight change in the conformation of the catalytic loop with the addition of another substrate besides UDP-NAG can explain the differences in the solubility of A2 above.
Interestingly, A2 was soluble when cleaved from MBP in the presence of MurAL138Q liganded with UDP-NAG (Fig. S3). This was surprising since the batch affinity experiment in which MBP-A2 did not bind MurAL138Q had higher concentrations of protein; however, the bound fractions were washed several times prior to elution from the magnet beads, unlike the fusion cleavage experiment in which the MurA concentration remains constant. This could explain the lack of the L138Q mutant sequestering if the mutation reduces the affinity that MurA has for A2. The notion that reduction in affinity between A2 and the MurAL138Q mutant is supported by the yeast-two-hybrid data which had reduced colony growth compared to WT (Fig.4B).
Identification of a MurA binding surface for A2
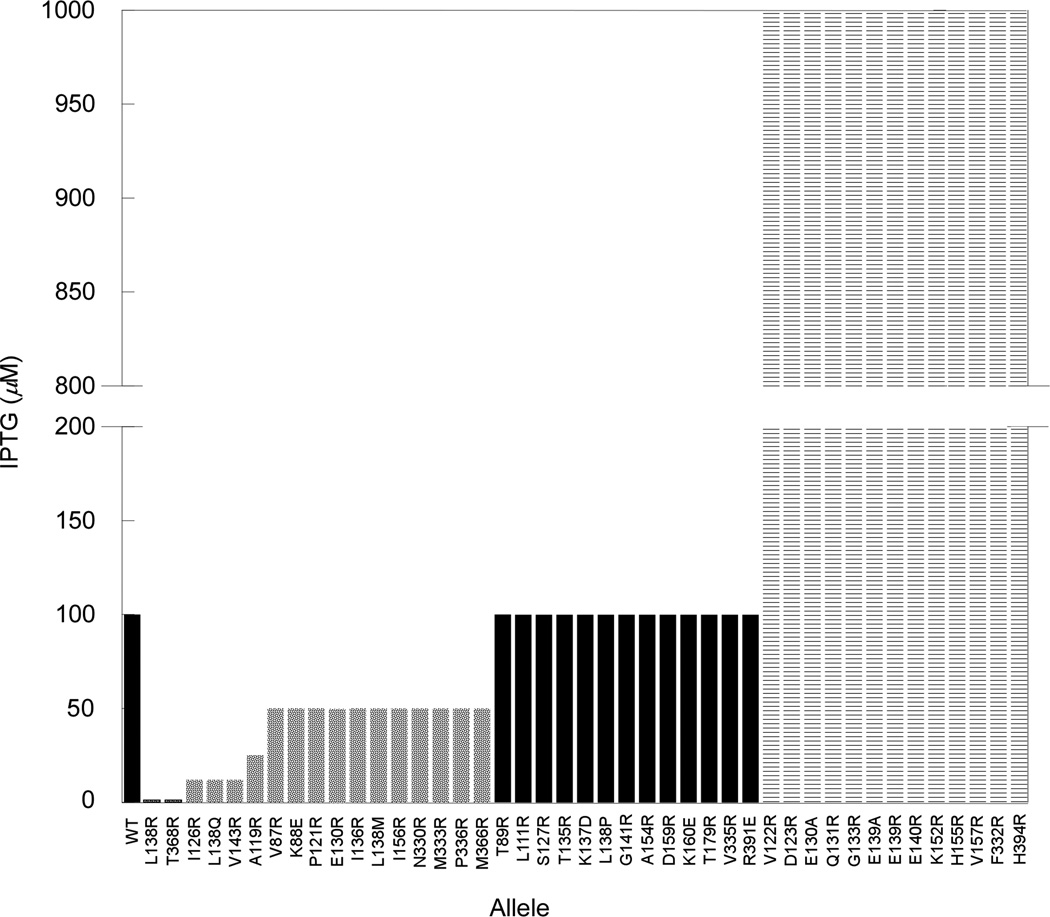
To determine residues involved in the A2 interaction with MurA, arginine-scanning mutagenesis of residues spatially surrounding the position of the original rat mutation, L138Q, was performed, with each allele assessed for its ability to confer resistance to Qβ. Each murA* allele was expressed from a medium-copy plasmid over a range of inducer concentrations, and the level of inducer required to prevent Qβ plaque-formation was determined. Under these conditions, the WT murA allele provides protection at 100 µM IPTG (Fig. 6 and Fig. S4), whereas the L138Q rat allele required only 12.5 µM IPTG. Using these two benchmarks, the new Arg-substituted alleles, and a few alleles where parental Arg and Lys residues were changed to Glu or Asp, were assayed (Fig. 6).
Figure 6. MurArat alleles protect against A2 at lower inducer concentrations.
IPTG induction levels that afford cells expressing MurA variants protection from Qβ infection are depicted by bars. Rats were considered any allele providing protection below the WT level of inducer concentration (100 µM) (solid grey bars). Alleles that protected at inducer concentration levels equivalent to WT are shown as black bars or higher than WT (striped bars).
The results showed that the mutant collection fell into several distinct classes. One class was indistinguishable from WT murA, in providing protection at 100 µM IPTG (black bars in Fig. 6) and were thus considered irrelevant to the A2-MurA interaction. A second class required much higher levels of inducer than the parental allele (striped bars in Fig. 6). These variants accumulated normally in vivo (Fig. S5) but exhibited little to no enzymatic activity (data not shown), suggesting improper folding. A third group of alleles (Y84R, A134R, E337R and R340E) showed no protection at low inducer concentrations and were inviable when inducer was present at 1 mM IPTG (data not shown), presumably due to the accumulation of insoluble material to toxic levels.
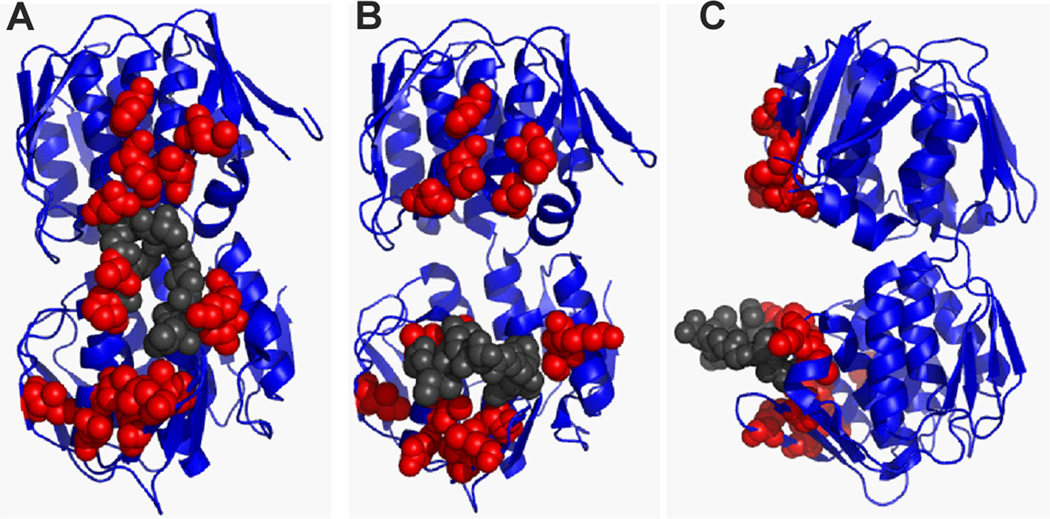
A final class of variants was identified that provided resistance at inducer concentrations less than 100 µM were designated as rat alleles; this class was comprised of 17 allelic substitutions at 15 codons (solid grey bars in Fig. 6). When these mutations were mapped onto the UDP-NAG-liganded MurA crystal structure, an apparent A2-interaction surface could be visualized (Fig. 7, Fig. S6A). Residues important for A2 inhibitory activity localize at sites surrounding the catalytic loop surface of the enzyme, including the catalytic domain, catalytic loop and C-terminal domain (Fig. 7A). Disruption of this interaction surface by conformational changes of the catalytic loop can be observed when the mutations are mapped on the unliganded MurA crystal structure (Fig. 7B–C). This genetic evidence supports the notion that a closed conformation of MurA is required for inhibition by A2.
Figure 7. Rat residues define a Qβ A2 interaction surface important for inhibition of MurA.
(A) Rat residues highlighted on the MurA UDP-NAG-bound state (closed conformation, front view) (Han H., unpublished; PDB entry 3KQJ) defines a continuous surface for A2 interaction. The structure of (B) MurA unliganded (front view) (Schönbrunn et al., 2000) and (C) MurA unliganded (side view) disrupts the A2 interaction surface with the conformational change in the catalytic loop backbone. Rat residues depicted as red spheres. Residues of the catalytic loop are shown as grey spheres. Figures were generated using PyMOL (Schrödinger, 2010).
To determine whether the MurA* variants are not simply complementing the loss of the endogenous MurA activity, an additional D305A mutation was introduced to inactivate the enzyme (Samland et al., 2001). Under these conditions, protection from Qβ requires that each D305A variant titrates out the inhibitory A2 produced during the infection cycle, thus sparing the endogenous parental MurA activity. However, if A2 is unable to associate with the mutant protein because of disruption of the A2-MurA binding interface, no protection should be observed. In this experiment, the D305A version of the parental MurA provides protection, whereas the D305A version of the canonical rat allele does not (Table S1). In this assay, all of the rat alleles failed to provide protection against Qβ lysis, supporting the notion that these alleles define an A2- binding surface.
When the rat residues are differentiated based upon the level of inducer needed to provide protection, (Fig. 6, Fig. S6B) there appears to be a correlation with the position of the residue. The residues with the greatest effect are located at the base of the catalytic loop as well as the point where the loop contacts the C-terminal domain. This could suggest that dynamics of the catalytic loop play an important role in the inhibition process. However, when the catalytic activity of MurAL138Q was assayed, no difference was found between the mutant and WT (Fig. 3). From this perspective, it is more likely that these residues are important contacts for A2 in preventing movement of the catalytic loop and blocking substrate accessibility once bound to MurA.
Discussion
A2-MurA interaction: Model for inhibition
The combined results of the enzymatic assays with purified components, binding experiments using the MBP-A2 fusion, and yeast-two-hybrid analysis demonstrate that the Qβ A2 maturation protein inhibits MurA by forming a complex that requires no other host or viral protein. This finding is important for several reasons: (1) it demonstrates that cellular lysis is not dependent on the participation of host components, (2) it militates against the involvement of viral particles in lysis, and (3) suggests that structural analysis of the A2-MurA complex would be informative for identification of a potential peptide-based inhibitor of MurA.
Regarding the structure, the in vivo protection assays allowed us to characterize a potential A2-MurA binding interface. These genetic results identified residues of importance that cover the catalytic loop/cleft and include residues that encompass both the catalytic and C-terminal domains of MurA. The fact that the surface spans the catalytic loop/cleft suggests that A2 binds MurA in a closed conformation. This is supported by binding studies where we showed that A2 associates with MurA in the UDP-NAG singly-liganded state. Moreover, addition of a second substrate besides UDP-NAG either severely reduces the ability of A2 to complex with MurA or abrogates it altogether, suggesting that the catalytic cleft is in fact an important contact site for A2. This binding interface of MurA is dominated by regions of negatively charged residues surrounding the rather hydrophobic catalytic loop (Fig. S7). A2 seems to have hydrophobic character, based on its strong tendency to aggregate even when fused to highly soluble protein domains, and, in its role as the maturation protein, must be able to associate with the viral RNA. Thus the putative MurA binding surface we have described by genetic means has features consistent with binding properties of A2. We hypothesize that the reason A2 was able to acquire the role as lysis protein in the Qβ system, in addition to binding the F-pilus, protecting the viral RNA, and assembly onto the virion capsid, was due to the hydrophobic and negatively charged nature of the MurA surface.
Proteins as inhibitors of enzymes
Protein inhibitors of enzymes involved in biosynthetic pathways are rare. One inhibitor, E, the lysis protein of ssDNA Microviridae phage ϕX174, targets MraY, an enzyme in the cell wall synthesis pathway (Bernhardt et al., 2001a). It was proposed that upon E binding to MraY a conformation change occurs in two transmembrane domains of MraY that in turn inactivates the enzyme irrespective of substrate binding (Zheng et al., 2009). A second inhibitor, T7 Lysozyme, binds to the T7 RNA polymerase palm and finger sub-domains and locks the protein in a non-processive conformation (Jeruzalmi & Steitz, 1998, Zhang & Studier, 1997). This association is independent of substrate binding and does not occlude the active site thereby, inhibiting the protein indirectly by preventing a conformation change (Zhang & Studier, 1997). A2-MurA complex formation on the other hand, requires a conformational change of the catalytic loop produced from UDP-NAG binding to MurA prior to A2 association. The presence of an additional substrate molecule, such as PEP, prevents this A2-MurA interaction (Fig. 5D, lanes 11–16). Therefore, A2 must bind UDP-NAG-liganded MurA over the catalytic loop/cleft and occlude PEP from the active site. Neither T7 lysozyme nor A2 bind their targets directly in the active site. Perhaps evolving the ability to become a competitive inhibitor requires a protein to adopt a highly specific tertiary structure which limits the number of roles a protein can fill. Thus, a form of mixed inhibition is better suited for these proteins to maintain additional functions apart from inhibition.
Experimental procedures
Bacterial strains and growth conditions
Standard molecular biology techniques were performed as described elsewhere (Sambrook et al., 1989). Escherichia coli strains and plasmids used in this study are listed in Table 1. Plasmid construction is described in supplementary information. XL-1 Blue was used for all plasmid constructions, HfrH served as lawns for bacteriophage plaque assays, and ER2738 was used for phage propagation and protein expression. E. coli strains were grown with aeration at 37°C in standard LB medium supplemented with 100 µg ml−1 ampicillin, 40 µg ml−1 kanamycin, or 10 µg ml−1 tetracycline when appropriate.
Table 1.
Strains and plasmids used in this study.
| Genotypes and relevant features | Sources and references |
|
|---|---|---|
| E. coli strains | ||
| XL-1 Blue | recA endA1 gyrA96 thi1 hsdR17 supE44 relA1 lac (F’ proAB lacZΔM15::Tn10) | Stratagene |
| HfrH | λ− relA1 spoT1 thi-1 lacIq1 lacZ::Tn5 | Laboratory stock |
| ER2738 | (F´proA+B+ lacIq Δ(lacZ)M15 zzf::Tn10) fhuA2 glnV Δ(lac-proAB) thi-1 Δ(hsdS-mcrB)5 | New England Biolabs |
| Yeast strain | ||
| AH109 | MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ | Clontech |
| Plasmids | ||
| pGADT7-murA | E. coli murA cloned into pGADT7 (Clontech) | This study |
| pGADT7-murAL138Q | Substitution of Leu138Gln in E. coli murA | This study |
| pGADT7-T | Encodes the Gal4 activation domain fused with SV40 large T-antigen | Clontech |
| pGBKT7-53 | Encodes the Gal4 DNA-binding domain fused with murine p53 | Clontech |
| pGBKT7-A2 | Qβ A2 cloned into pGBKT7 (Clontech) | This study |
| pGBKT7-Lam | Encodes the Gal4 DNA-binding domain fused with lamin | Clontech |
| pETMBP-A2 | Qβ A2 cloned into pET28b-MBP, contains a TEV protease cleavage site in the linker between MBP and A2 | This study |
| pZE12-murA | E. coli murA cloned under the PLlacO-1 promoter which is IPTG inducible | Bernhardt et al. (2001) |
| pZE12-murAHis | an oligo-histidine tag (G2H6G2) was adjoined to the C-terminus of murA | This study |
| pZA31-murABs | Bacillus subtilis murAA cloned under the PLtet0-1 promoter which is constitutively on in a tetR− background | This study |
Protein expression and purification
MurA was expressed for 3 hrs in ER2738 cells harboring pZE12-murAHis grown at 37°C with aeration and induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG). Cells were collected by centrifugation at 4,000 ×g for 15 min at 4°C and resuspended in MurA buffer (0.1 M Tris-HCl [pH 8]) with 20 µg ml−1 DNase and 10 µg ml−1 RNase. Cells were disrupted by passage through a SLM-Aminco French pressure cell (Spectronic Instruments) at 16,000 psi, and lysate was clarified by centrifugation at 5,800 ×g for 15 min at 4°C. The clarified lysate was stored as an ammonium sulfate precipitant (70%). The precipitate was collected by centrifugation at 5,800 ×g for 10 min at 4°C, and resuspended with equivalent volume of MurA buffer. The resuspended sample was then applied to a Talon metal affinity resin (Clontech), washed with buffer, and eluted with a 0 to 0.5 M imidazole gradient in MurA buffer. Fractions were collected with a micro-fractionator (Gilson) and assessed for protein concentration (A280). Activity of each fraction was assayed as previously described (Marquardt et al., 1992), and the fractions with the highest protein concentration and activity were pooled and stored as an ammonium sulfate precipitant at 4°C until further use.
The plasmid pETMBP-A2, a construct encoding A2 fused to the E. coli maltose-binding protein (MBP), was generated to assist in purification of a soluble form of A2. To prevent lysis, this plasmid was induced in cells constitutively expressing the Bacillus subtilis protein MurAA from the plasmid pZA31-murABs. Cells were grown at 37°C with aeration until chilling in an ice bath prior to induction with 1 mM IPTG and expression at 16°C for 18 hrs. Cells were harvested by centrifugation at 16,000 ×g for 15 min at 4°C and resuspended in amylose buffer (20 mM Tris-HCl [pH 8], 150 mM NaCl, 1% glycerol, and 1 mM EDTA) with 20 µg ml−1 DNase, 10 µg ml−1 RNase, 5 mM MgCl2, and protease inhibitor cocktail (Sigma). Cells were disrupted as described above. The lysate was clarified by centrifugation at 12,000 ×g for 15 min at 4°C and filtered through a 0.45 µm filter prior to loading onto an amylose column (Amylose High Flow resin, NEB). Protein was eluted with 10 mM maltose in amylose buffer (above) lacking glycerol. Typical yield for the protein was 8–11 mg/L with ~95 % purity.
MurA activity assay
Qβ inhibition of MurA was performed as previously reported (Bernhardt et al., 2001b) but with 2 µg of purified MurA (0.4 µM) instead of cell lysate and 60 µl of Qβ (4 × 1013 physical particles) (~0.7 µM A2) in a 100 µl reaction. Qβ was purified as previously described (Strauss & Sinsheimer, 1963).
Yeast-two-hybrid analyses
Interaction between A2 and MurA was assessed with the Matchmaker Gal4 Two-Hybrid System 3 (Clontech) according to manufacturer’s standard protocols. Yeast strain AH109 (See Table 1) was co-transformed with pGBKT7-A2 and pGADT7-murA or pGADT7-murAL138Q by the lithium acetate method. Strains were selected on solid media deficient of tryptophan and leucine. After incubation at 30°C for four days, emergent colonies were individually streaked on more stringent media deficient of leucine, tryptophan, histidine and adenine. Yeast were co-transformed and tested with pGBKT7-53 and pGADT7-T or pGBKT7-Lam and pGADT7-T as positive and negative controls, respectively.
Batch affinity fractionation experiments
Amylose magnetic beads were chosen for batch affinity fractionation experiments since MBP binds specifically to amylose resin. An aliquot (40 µl, NEB) of beads was washed with of amylose buffer. A magnet was applied for 1 minute and supernatant was removed. MBP-A2 (90 µl of 6 µM) was incubated for 30 min with rolling at 4°C. Beads were washed three times, as above, to remove unbound MBP-A2, vortexing briefly before application of magnet and removal of supernatant. Beads with MBP-A2 bound were resuspended in buffer containing 100 µl of 10 µM of MurA with the following substrate conditions: none, 1 mM PEP (Sigma), 1 mM UDP-NAG (Sigma), and 1mM PEP/UDP-NAG. Reactions were incubated at 4°C for 1 hr with rolling. Samples were washed three times and eluted by resuspending beads in 2× 40 µl of amylose elution buffer. Samples were analyzed by SDS-PAGE and immunoblotting as described previously (Gründling et al., 2000). Unbound fractions (supernatant of first wash) were analyzed by SDS-PAGE and bound fractions were immunoblotted with a 1:3,000 dilution of the α-MurA antibody raised against the synthetic peptide: CHGKRPKAVNVRTAP (GenScript) and a 1:3,000 dilution of goat-anti-rabbit 2° antibody (Pierce). Chemiluminescent detection was achieved with the SuperSignal West Femto developer kit (Pierce).
Fusion cleavage assay
Reactions containing 2.5 µM of MBP-A2, 5 µM MurA under different substrate conditions: none, 1 mM PEP, 1 mM UDP-NAG, 1mM PEP/UDP-NAG, and 1 mM UDP-NAG/Fosfomycin (Sigma), and 10 µg of MBP-TEV protease in buffer (10 mM sodium phosphate [pH 7.4], 100 mM NaCl, and 1 mM DTT) were incubated overnight at 4°C. MurA was incubated with substrates on ice for 30 min prior to preparing reactions. An aliquot of samples was removed prior to centrifugation at 18,000 ×g, 15 min at 4°C. Supernatant was removed and the pellet was resuspended in equivalent volume of buffer. Fractions were analyzed with SDS-PAGE and immunoblotting, probed with a 1:5,000 dilution of the α-A2 antibody raised against a synthetic peptide: PKLPRGLRFGA (Bethyl Laboratories, Inc.) and a 1:3,000 dilution of goat-anti-rabbit 2° antibody.
MurA bioassay
Gradient induction of MurA in bacterial lawns of HfrH harboring pZE12-murA, or various alleles (murA*), was generated by plating 200 µl of a mid-log phase culture in a 0.7 % soft agar overlay supplemented with 2 mM CaCl2 onto LB agar plates containing ampicillin and 0 µM, 12.5 µM, 25 µM, 50 µM, 100 µM or 1 mM of IPTG. An aliquot of cell-free Qβ lysate (~650 plaque forming units) was included in the overlay. Plates were incubated at 37°C for 12–16 hrs prior to screening for plaque formation (Fig. S3).
Supplementary Material
Acknowledgments
We thank the Young lab members, past and present, for their helpful criticisms and suggestions. This work was supported by Public Health Service grant GM27099 to R.Y., the Robert A. Welch Foundation, and the Program for Membrane Structure and Function, a Program of Excellence grant from the Office of the Vice President for Research at Texas A&M University.
References
- Bernhardt TG, Struck DK, Young R. The lysis protein E of ϕX174 is a specific inhibitor of the MraY-catalyzed step in peptidoglycan synthesis. J. Biol. Chem. 2001a;276:6093–6097. doi: 10.1074/jbc.M007638200. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, Wang IN, Struck DK, Young R. A protein antibiotic in the phage Qβ virion: Diversity in lysis targets. Science. 2001b;292:2326–2329. doi: 10.1126/science.1058289. [DOI] [PubMed] [Google Scholar]
- Eschenburg S, Kabsch W, Healy ML, Schönbrunn E. A new view of the mechanisms of UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) and 5-enolpyruvylshikimate-3-phosphate synthase (AroA) derived from X-ray structures of their tetrahedral reaction intermediate states. J. Biol. Chem. 2003;278:49215–49222. doi: 10.1074/jbc.M309741200. [DOI] [PubMed] [Google Scholar]
- Gründling A, Bläsi U, Young R. Biochemical and genetic evidence for three transmembrane domains in the class I holin, λ S. J. Biol. Chem. 2000;275:769–776. doi: 10.1074/jbc.275.2.769. [DOI] [PubMed] [Google Scholar]
- Gunetileke KG, Anwar RA. Biosynthesis of Uridine Diphospho-N-acetylmuramic acid. J. Biol. Chem. 1968;243:5770–5778. [PubMed] [Google Scholar]
- Hatfull GF. The great escape. Science. 2001;292:2263–2264. doi: 10.1126/science.1062957. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D, Steitz TA. Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme. EMBO J. 1998;17:4101–4113. doi: 10.1093/emboj/17.14.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Lees WJ, Kempsell KE, Lane WS, Duncan K, Walsh CT. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry. 1996;35:4923–4928. doi: 10.1021/bi952937w. [DOI] [PubMed] [Google Scholar]
- Marquardt JL, Siegele DA, Kolter R, Walsh CT. Cloning and sequencing of Escherichia coli murZ and purification of its product, a UDP-N-acetylglucosamine enolpyruvyl transferase. J. Bacteriol. 1992;174:5748–5752. doi: 10.1128/jb.174.17.5748-5752.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizyed S, Oddone A, Byczynski B, Hughes DW, Berti PJ. UDP-N-acetylmuramic acid (UDP-MurNAc) is a potent inhibitor of MurA (enolpyruvyl-UDP-GlcNAc synthase) Biochemistry. 2005;44:4011–4017. doi: 10.1021/bi047704w. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. pp. 1.1–1.52. [Google Scholar]
- Samland AK, Etezady-Esfarjani T, Amrhein N, Macheroux P. Asparagine 23 and aspartate 305 are essential residues in the active site of UDP-N-acetylglucosamine enolpyruvyl transferase from Enterobacter cloacae . Biochemistry. 2001;40:1550–1559. doi: 10.1021/bi001490a. [DOI] [PubMed] [Google Scholar]
- Schönbrunn E, Eschenburg S, Krekel F, Luger K, Amrhein N. Role of the loop containing residue 115 in the induced-fit mechanism of the bacterial cell wall biosynthetic enzyme MurA. Biochemistry. 2000;39:2164–2173. doi: 10.1021/bi991091j. [DOI] [PubMed] [Google Scholar]
- Schönbrunn E, Svergun DI, Amrhein N, Koch MHJ. Studies on the conformational changes in the bacterial cell wall biosynthetic enzyme UDP-N-acetylglucosamine enolpyruvyltransferase (MurA) Eur. J. Biochem. 1998;253:406–412. doi: 10.1046/j.1432-1327.1998.2530406.x. [DOI] [PubMed] [Google Scholar]
- Schrödinger LLC. The PyMOL Molecular Graphics System. Version 0.99. 2010 [Google Scholar]
- Skarzynski T, Kim DH, Lees W, Walsh CT, Duncan K. Stereochemical course of enzymatic enolpyruvyl transfer and catalytic conformation of the active site revealed by the crystal structure of the fluorinated analogue of the reaction tetrahedral intermediated bound to the active site of the C115A mutant of MurA. Biochemistry. 1998;37:2572–2577. doi: 10.1021/bi9722608. [DOI] [PubMed] [Google Scholar]
- Skarzynski T, Mistry A, Wonacott A, Hutchinson SE, Kelly VA, Duncan K. Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin. Structure. 1996;4:1465–1474. doi: 10.1016/s0969-2126(96)00153-0. [DOI] [PubMed] [Google Scholar]
- Strauss JH, Sinsheimer RL. Purification and properties of bacteriophage MS2 and of its ribonucleic acid. J. Mol. Biol. 1963;7:43–54. doi: 10.1016/s0022-2836(63)80017-0. [DOI] [PubMed] [Google Scholar]
- Weber K, Konigsberg W, Zinder ND. Proteins of the RNA phages. In: Zinder ND, editor. RNA phages. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1975. pp. 51–84. [Google Scholar]
- Zhang X, Studier FW. Mechanism of inhibition of bacteriophage T7 RNA polymerase by T7 lysozyme. J. Mol. Biol. 1997;269:10–27. doi: 10.1006/jmbi.1997.1016. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Struck DK, Young R. Purification and functional characterization of ϕX174 Lysis Protein E. Biochemistry. 2009;48:4999–5006. doi: 10.1021/bi900469g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-Y, Yang Y, Han H, Betzi S, Olesen SH, Marsilio F, Schönbrunn E. Functional consequence of covalent reaction of phosphoenolpyruvate with UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA) J. Biol. Chem. 2012;287:12657–12667. doi: 10.1074/jbc.M112.342725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.