Abstract
Clinically, biofilm-associated infections commonly form on intravascular catheters and other hydrogel surfaces. The overuse of antibiotics to treat these infections has led to the spread of antibiotic resistance and underscores the importance of developing alternative strategies that delay the onset of biofilm formation. Previously, it has been reported that during surface contact, bacteria can detect surfaces through subtle changes in the function of their motors. However, how the stiffness of a polymer hydrogel influences the initial attachment of bacteria is unknown. Systematically, we investigated poly(ethylene glycol) dimethacrylate (PEGDMA) and agar hydrogels that were twenty times thicker than the cumulative size of bacterial cell appendages, as a function of Young’s moduli. Soft (44.05 – 308.5 kPa), intermediate (1495 – 2877 kPa), and stiff (5152 – 6489 kPa) hydrogels were synthesized. Escherichia coli and Staphylococcus aureus attachment onto the hydrogels was analyzed using confocal microscopy after 2 and 24 hr incubation periods. Independent of hydrogel chemistry and incubation time, E. coli and S. aureus attachment correlated positively to increasing hydrogel stiffness. For example, after a 24 hr incubation period, there were 52% and 82% less E. coli adhered to soft PEGDMA hydrogels, than to the intermediate and stiff PEGDMA hydrogels, respectively. A 62% and 79% reduction in the area coverage by the Gram-positive microbe S. aureus occurred after 24 hr incubation on the soft versus intermediate and stiff PEGDMA hydrogels. We suggest that hydrogel stiffness is an easily tunable variable that, potentially, could be used synergistically with traditional antimicrobial strategies to reduce early bacterial adhesion, and therefore the occurrence of biofilm-associated infections.
Keywords: Biofilm, Escherichia coli, Staphylococcus aureus, Hydrogel, Poly(ethylene glycol), Young’s moduli
INTRODUCTION
Bacterial colonization and subsequent biofilm formation on polymer surfaces is a pressing challenge. For example, annually in the United States, biofilms formed on intravascular catheters are linked to 250,000 blood stream infections1 with an associated mortality rate of 12–25%.2 To inactivate microbes, antibacterial agents have been released from polymer catheters.3–8 However, this approach yields concerns related to the antibacterial agent release rate, depletion, and toxicity to human cells.9 Furthermore, the continual reports of increasing antibiotic resistance10,11 requires a new, greener strategy12–14 that slows the rate by which bacteria attach to a surface without exerting evolutionary pressure on microorganisms.15,16
Previously, it has been reported that bacteria have the ability to sense and differentiate between surfaces. Surface differentiation occurs through bacterial organelles, specific proteins, or biological complexes that detect signals from the environment and then respond with a transcriptional signal cascade.17,18 While the dynamics of this “swim-or-stick” switch remain unclear, flagella seem central to determining if microbes are going to “stick” and colonize a surface. For motile bacteria species, the flagella drives both their swimming and swarming motility towards a surface; obstructing the motor rotation of motile bacteria induces them to switch to surface-associated behaviors.19 Since biofilms often form on surfaces, the influence of surface chemistry and structure-property relationships (i.e., nanotopographic patterning) on reducing bacterial adhesion have already been investigated and were the topic of several review papers.12–14,20–23 For example, independent of feature dimensions (square, rectangular, or circular posts), Perera-Costa et al.20 reported that organized topography significantly reduced bacterial attachment. Engineered roughness index was proposed as a possible explanation for the reduction of microbial adhesion, however, the general mechanism is poorly understood.24
A different materials approach to reduce bacterial adhesion on hydrogel surfaces could be to utilize stiffness as an easily tunable structure-property relationship. Lichter et al.25 synthesized 50 nm thin polyelectrolyte multilayer (PEM) films from poly(allylamine hydrochloride) and poly(acrylic acid), whose Young’s moduli ranged from 1000 to 100000 kPa. They reported a positive correlation between increasing film stiffness and the adhesion of Escherichia coli and Staphylococcus epidermis. After 1 hr of microbial growth, less bacteria attached to Saha et al.’s26 30 kPa non-crosslinked films than to their 150 kPa crosslinked films, which were comprised of poly(L-lysine) and hyaluronan modified with photoreactive vinylbenzyl groups. However, Saha et al. note that their limited range of elastic modulus ~120 kPa might have caused the relatively small difference in bacterial adhesion observed between their PEM films. Recently, Song and Ren27 found that the stiffness of polydimethylsiloxane (PDMS) substrates, 100 kPa to 2600 kPa, affected the physiology of E. coli RP437 and Pseudomonas aeruginosa PAO1. Attachment and growth was promoted on softer surfaces, but antibiotic susceptibility was enhanced with increasing stiffness. The applicability of correlating bacterial adhesion on ultrathin charge-containing films and PDMS elastomers to biomedically relevant hydrogel coatings is limited. Crosslinked PDMS is a hydrophobic elastomer and polar solvents, such as water, struggle to wet PDMS;28 whereas hydrogels are predominately water and are easily wet by water.29 The unique mechanical properties, elasticity, water content, and mesh size of PEMs, PDMS elastomers, and polymer hydrogels should be well-characterized30,31 in order to gather structure-property relationships. Thus, the effect of a thick hydrogels tunable over a wide range of Young’s moduli will expand our current understanding of how bulk materials properties affect the initial adhesion of bacteria.
To fill this critical gap, here we investigate the attachment of Escherichia coli K12 MG1655, a model Gram-negative bacteria and Staphylococcus aureus SH1000, a model Gram-positive bacteria32,33 to hydrogels that are significantly thicker than the cumulative size of bacterial cell appendages. Model hydrogels were synthesized from the hydrophilic polymer, poly(ethylene glycol) dimethacrylate (PEGDMA), which is known to reduce the nonspecific attachment of proteins and bacterial adsorption/adhesion.34 Chemistry control biopolymer agar hydrogels with mechanical properties analogous to the PEGDMA hydrogels were also investigated. Systematically, as a function of substrate stiffness, E. coli and S. aureus adhesion were assessed after 2 hr and 24 hr to capture the progression of bacterial adhesion. E. coli is a motile microbe that uses its flagella and fimbriae to sense a surface and facilitate adhesion. Whereas the non-motile microbe, S. aureus, relies on surface protein adhesins to facilitate adhesion but lacks a clear mechanism for surface sensing.32,35,36 While beyond the scope of this work, future studies can determine the physiological changes experienced by microbes as a function of hydrogel stiffness, and how this initial adhesion correlates with robust biofilm formation. Innovative catheter design, including hydrogel coatings, are routinely employed to improve the smoothness and lubrication of the catheter exterior while resisting infection.37,38 From our findings, we suggest that improving the performance of hydrogel coatings through a basic design parameter that may not cause evolutionary pressure on pathogens, i.e. stiffness, could be a significant medical contribution.
EXPERIMENTAL SECTION
Materials
All compounds were used as received. Poly(ethylene glycol) dimethacrylate, (PEGDMA, Mn = 750 Da), 3-(trimethoxysilyl)propyl methacrylate, ampicillin (BioReagent grade), chloramphenicol (BioReagent grade), M9 minimal salts (M9 media), D-(+)-glucose, calcium chloride (anhydrous), phosphate buffered saline (PBS, 1× sterile biograde), tryptic soy broth (TSB), and Luria-Bertani broth (LB) were purchased from Sigma-Aldrich (St. Louis, MO). Irgacure 2959 was obtained from BASF (Ludwigshafen, Germany). Magnesium sulfate anhydrous and molecular grade agar were obtained from Fisher Scientific (Fair Lawn, NJ). Deionized (DI) water was obtained from a Barnstead Nanopure Infinity water purification system (Thermo Fisher Scientific, Waltham, WA).
PEGDMA and Agar Hydrogel Fabrication
PEGDMA hydrogels were prepared using previously established protocols.39 Briefly, PEGDMA solutions (7.5, 10, 15, 25, 40, and 50 vol% in 1 M PBS) were sterile filtered using a 0.2 μm syringe, then degassed using nitrogen gas. For UV-curing the radical photo initiator, 0.8 wt% Irgacure 2959 was added to the polymer precursor solution with induction under a long wave UV light, 365 nm for 10 min. PEGDMA solution (75 μL) was sandwiched between two UV-sterilized coverslips (22-mm, Fisher Scientific) functionalized with 3-(trimethoxysilyl)propyl methacrylate.40 Fabricating the hydrogel between coverslips enabled all hydrogels to have a uniform thickness and limited the oxygen exposure. Following polymerization, the top coverslip was removed with forceps and the hydrogels were swelled for 48 hr in 25 mL of M9 media.
Soft agar hydrogels were prepared by dissolving 3 wt% agar in sterile DI water for 30 min before uniformly heating the solution in a liquid autoclave cycle at 250 °C for 30 min. To achieve a higher weight percent of dissolved agar, hydrothermal preparation was used.41 Here, 9 wt% agar in sterile water was heated for 2 hr in a 95 °C water bath, followed by a liquid autoclave cycle at 250 °C for 30 min. The hot solution was cast into glass petri dishes (Pyrex, Tewksbury MA) and allowed to gel. After the agar gels cooled, a flame sterilized 25.4 mm punch (Spearhead® 130 Power Punch MAXiset, Cincinnati, Ohio) was used to create circular hydrogels that were ~2 mm in height.
Characterization of PEGDMA and Agar Hydrogels
The thickness of PEGDMA and agar hydrogels was determined using a digital micrometer (Mitutoyo Corporation, Kawaski, Japan) by averaging 5 measurements taken on 5 different fully swollen hydrogels. Bulk hydrogel stiffness was measured using a custom-built contact/adhesion test (CAT) test.42 PEGDMA hydrogels were prepared in cylindrical Teflon molds that were 2 mm in diameter and ~2 mm in height, then swollen for 48 hr in PBS before being mounted onto the stage of an inverted microscope to control uniform probe contact. Agar hydrogels were prepared according to the method previously outlined. Care was taken to perform indentation experiments before dehydration occurred. A rigid flat cylindrical steel probe (1.50 mm diameter, High-Speed M2 Tool Steel Hardened Undersized Rod) was brought into contact with the hydrogel and the force (P), displacement (δ), and contact area (A) were recorded via a custom-developed National Instruments LabVIEW software. To minimize any potential viscoelastic contributions, the tests were carried out at a fixed displacement rate of 25 μm/s and a fixed displacement of 250 μm. Force was monitored by a force transducer (Honeywell Sensotec, Columbus, OH) connected in series with a nanoposition manipulator (Burleigh Instruments Inchworm Model IW-820) that controlled the displacement. The interfacial contact area was captured using a CCD camera (Pixelfly, Kelheim Germany) mounted in-line with an inverted optical microscope (bright field, Zeiss Axiovert, Thornwood, NY). To calculate the Young’s modulus, the hydrogel was assumed to reside in “elastic-half space” based on the probe to sample size ratio (Figure S1 of the Supporting Information), which simplified the equation for Young’s modulus with a flat cylindrical probe to:
Where E is the Young’s modulus (N/m2), P is the load (N), a is the measurement depth (m), and R is the radius (m).
Contact angle was determined using HPLC water on a Krüss DSA100 Drop Shape Analysis system (Hamburg, Germany) via a modified dynamic/static test averaged over 5 hydrogels. Hydrogels were fully swollen for 48 hr to determine their wet weight before being lyophilized at 90 °C for 72 hr to determine their dry polymer mass. A modified version of the Flory theory,43 which assumes that the solvent interaction of M9 media with PEGDMA is the same as with PBS was then applied to determine mesh size (ξ):
Where υ2,s is the swollen volume fraction of polymer and is the average end-to-end distance of the crosslinked PEGDMA. Four samples at every polymer concentration were tested.
We quantified protein adsorption to the hydrogels using a fluorescent protein assay. Briefly, hydrogels were polymerized on 15 mm diameter coverslips that were adhered to the bottom of 24-well plates (Fisher Scientific). Samples were then swollen for 48 hr in PBS before being incubated for 48 hr at 23 °C in 1.0 and 10 μg/cm2 of fluorescently tagged Fibronectin. During incubation, samples were gently rotated at 100 RPM. Samples were rinsed 3× with PBS before the adsorption of Fibronectin was assessed using a Zeiss Axiovert Yokogawa Spinning Disk (10× magnification).
Evaluation of Bacterial Growth on PEGDMA and Agar Hydrogels
Escherichia coli K12 MG1655 (E. coli, DSMZ, Leibniz-Institut, Germany) was transformed using the high copy green florescence plasmid pMF230 (509 nm emission), a generous donation (Dr. Michael Franklin, Montana State University) and an ampicillin resistance marker to select for viable E. coli. Staphylococcus aureus SH1000 (S. aureus) and the high-efficiency pCM29 sGFP plasmid,44 containing a chloramphenicol antibiotic was a generous donation (Dr. Alexander Horswill, University of Iowa). PEGDMA and agar hydrogels (25 mm diameter) were placed at the base of 6-well plates (Fisher Scientific) to which 5 mL of M9 media with 100 μg/mL ampicillin or 10 μg/mL chloramphenicol were added for E. coli or S. aureus, respectively. The growth media in each well was inoculated with an overnight culture of 1.00 × 108 cells/mL of E. coli or S. aureus, which were washed and resuspended in M9 media,45 and then placed in an incubator at 37 °C for 2 or 24 hr. Hydrogels with attached bacteria were removed from the 6-well plates, dipped in M9 media to remove loosely adhered bacteria before being fixed on glass microscope slides using 4% paraformaldehyde for 15 min. E. coli attachment was evaluated using a modified attachment assay46 via confocal microscopy (Nikon microscope D-Eclipse C1 80i, Nikon Corporation, Melville, NY, USA) using a 63× objective, wherein 10–15 randomly acquired images having an area of 3894 μm2 were taken with at least 3 parallel replicates for each hydrogel. E. coli adhesion over the entire captured area of 3894 μm2, was quantified using Image J1.45 software (National Institutes of Health, Bethesda, MD) through direct cell counting.47 S. aureus was imaged using a 50× objective and because S. aureus forms grape-like colonies, the particle analysis function in ImageJ was used to calculate the colony area coverage over the acquired 58716 μm2 area.48,49 Adhered E. coli and S. aureus were confirmed to be viable through propidium iodide staining of GFP bacteria50 after 2 and 24 hr (Figure S2 of the Supporting Information). Significant differences between samples were determined with an unpaired student t-test. Significance (p ≤ 0.05) is denoted in graphs.
RESULTS AND DISCUSSION
Characteristics of PEGDMA and Agar Hydrogels
Six thick PEGDMA and two thick agar hydrogels were successfully synthesized.39 Table 1 summarizes the polymer concentration, thickness, Young’s modulus, mesh size, and contact angle of all hydrogels used in this study. Our PEGDMA and agar hydrogels were all at least 100 μm thick, which is larger than the diameter of the microbes used in this study. Unlike the thin polymer films used in previous studies,25,26 since all of the hydrogels used in this work were thick, the probability that the bacteria are able to “feel” the underlying “hard” glass coverslip mount is reduced. The Young’s moduli of 7.5, 10, 15, 25, 40, and 50% PEGDMA hydrogels spanned nearly 3 orders of magnitude, Figure 1. Based on this wide range, we categorized the PEGDMA hydrogels into three regimes: soft (44.05–308.5 kPa), intermediate (1495–2877 kPa), and stiff (5152–6489 kPa). Prepared 3% and 9% agar hydrogels had soft (44.8 kPa) and intermediate (1336 kPa) Young’s moduli, respectively. Our values are similar to those previously reported for agar hydrogels, however, the measurements vary slightly due to different hydrogel preparation and characterization techniques.51 Agar solubility limited the synthesis of stiff agar hydrogels; thus, agar hydrogels cannot mimic the wide range of mechanical properties that can be achieved using PEGDMA.52 For reader ease, we have rounded the Young’s moduli data throughout the remainder of the results and discussion section.
Table 1.
Properties of hydrogels as a function of polymer concentration. Samples were tested in triplicate with 3–5 measurements per sample. The standard error of each set is displayed.
| Polymer | Concentration (%) | Young’s Modulus (kPa) | Mesh Size (Å) | Thickness (μm) | Contact Angle (°) |
|---|---|---|---|---|---|
| PEGDMA | 7.5 | 44.1 ± 5.6 | 34.3 ± 1.5 | 122.5 ± 5.9 | 72.3 ± 2.3 |
| 10 | 308.5 ± 31.1 | 27.2 ± 0.8 | 153.3 ± 13.9 | 72.3 ± 3.4 | |
| 15 | 1,495.3 ± 80.1 | 25.1 ± 0.7 | 147.3 ± 22.9 | 69.9 ± 3.0 | |
| 25 | 2,877.1 ± 904.8 | 19.3 ± 0.4 | 164.9 ± 20.3 | 75.7 ± 2.5 | |
| 40 | 5,152.5 ± 806.0 | 10.7 ± 0.5 | 184.5 ± 15.1 | 66.6 ± 3.5 | |
| 50 | 6,489.2 ± 116.5 | 10.0 ± 1.0 | 219.7 ± 8.8 | 61.2 ± 4.0 | |
| Agar | 3 | 44.8 ± 1.63 | – | 1,574 ± 29 | 19.1 ± 4.0 |
| 9 | 1,336 ± 589.0 | – | 1,479 ± 39 | 15.8 ± 3.8 |
Figure 1.
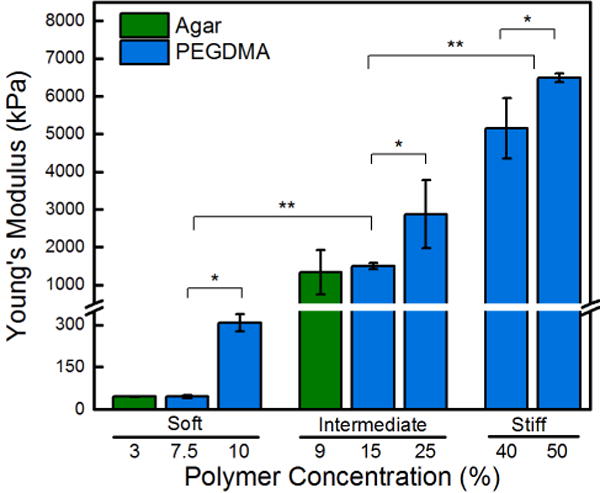
Young’s moduli increases with increasing polymer concentration. Stiffness of PEGDMA hydrogels matched with agar hydrogels form three regimes of statistically different hydrogels: soft (44 – 308 kPa), intermediate (1340 – 2880 kPa), and stiff (5150 – 6500 kPa). An asterisk denotes 95% significance between samples, whereas two asterisks denotes significance between stiffness regimes. Error bars denote standard error.
The mesh size of the PEGDMA hydrogels was inversely correlated to their Young’s moduli, the softest hydrogel had a mesh size of 34.3 ± 1.5 Å, whereas the stiffest had a mesh size of 10.0 ± 1.0 Å. This is consistent with similar hydrogel systems found in literature,43 is an order of magnitude smaller than E. coli, and is small enough to avoid adsorption of most proteins (1–100 nm) and molecules. Water contact angle measurements were statistically equivalent across all PEGDMA hydrogels. Agar hydrogels were superhydrophilic with a contact angle below 20°. Thus, surface energy can be ruled out as a confounding variable. Confocal micrographs in Figure S3 of the Supporting Information display that no detectible Fibronectin protein adhered to the PEGDMA hydrogels after 24 hr, consistent with previous reports for poly(ethylene glycol)-based materials.34,39 Agar hydrogels adsorbed significant Fibronectin, exceeding glass controls, which adsorbed significantly more protein than the PEGDMA. Since most mechanisms of bacterial adhesion are protein-mediated,53,54 the presence of protein on the agar hydrogel surface serves as a positive control for the PEGDMA hydrogels that resist protein adsorption over the timescale of this study, 24 hr.
Attachment of E. coli and S. aureus after 2 hr Incubation on PEGDMA and Agar Hydrogels
PEGDMA hydrogels, agar hydrogels, and internal control glass coverslips were placed at the base of 6-well polystyrene plates to which E. coli in M9 media were incubated for 2 hr, Figure 2. Confocal microscopy paired with ImageJ software enabled the total cell attachment to be determined. The average attachment of E. coli was 21 ± 7 cells and 315 ± 67 cells for the soft 7.5% PEGDMA and stiff 50% PEGDMA hydrogels, respectively. Attachment of E. coli onto the intermediate and stiff hydrogels was significantly greater than onto the soft hydrogels for both PEGDMA and agar hydrogels. There was a strong linear correlation between PEGDMA moduli and bacterial attachment, 0.90 R2, which was calculated through linear least squares regression. Soft agar hydrogels with statistically similar moduli to PEGDMA (45 kPa versus 44 kPa) had significantly more bacteria attached, 42 ± 8 cells. This may be an effect of the surface chemistry or the larger mesh size of the agar hydrogels.
Figure 2.
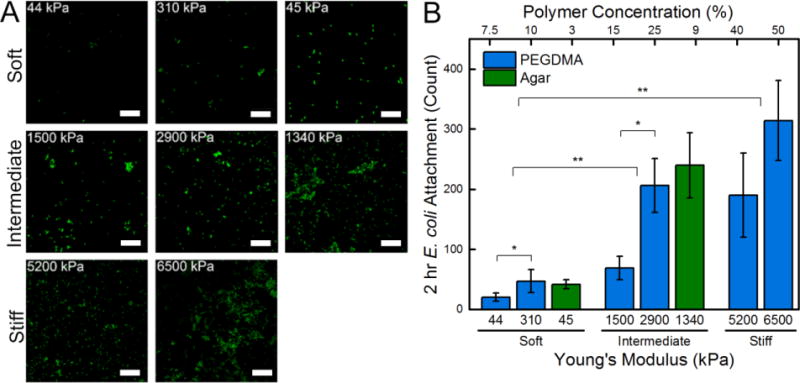
(A) Representative confocal micrographs (3894 μm2) of E. coli attached after a 2 hr incubation period on soft (44 – 308 kPa), intermediate (1340 – 2880 kPa), and stiff (5150 – 6500 kPa) PEGDMA and agar hydrogels. A 10 μm scale bar is displayed. (B) Total cell count quantified that there was a significant increase in E. coli attachment with increasing hydrogel stiffness. An asterisk denotes 95% significance between samples, whereas two asterisks denotes significance between stiffness regimes. Error bars denote standard error.
Further evaluation was conducted by challenging a subset of soft, intermediate, and stiff PEGDMA and agar hydrogels with the Gram-positive bacteria, S. aureus, Figure S4 of the Supporting Information. Since this microbe has a different mechanism of attachment, we can gain insight into whether stiffness sensing is an adhesion mechanism-independent process. After a 2 hr incubation time, a small quantity of S. aureus adhered to the soft and intermediate PEGDMA hydrogels. There was a 0.16% S. aureus area colony coverage on the soft 45 kPa PEGDMA hydrogels, a 0.27% area colony coverage on the intermediate 1500 kPa PEGDMA hydrogels, and a sharp increase to 1.8% coverage on the stiff 6500 kPa PEGDMA hydrogels. Substantially more S. aureus adhered to agar hydrogels; the soft agar hydrogel had a 2.4% area colony coverage and the intermediate agar hydrogel had a 3.7% area colony coverage. This indicates that independent of hydrogel chemistry, and independent of adhesion mechanism, increasing the hydrogel stiffness increases the amount of E. coli or S. aureus that adheres to a stiffer hydrogel after a 2 hr incubation period.
Attachment of E. coli and S. aureus after 24 hr Incubation on PEGDMA and Agar Hydrogels
After a 24 hr incubation period, E. coli displayed early development into 3D microstructures, Figure 3A. The presence of microcolonies suggests that the signaling involved in the early stages of biofilm formation (limited quorum sensing, twitching motility) are active. Qualitatively, colony formation and proliferation was observed to be more robust on 9% agar hydrogels and all stiff regime PEGDMA hydrogels. Stiff regime PEGDMA hydrogels had significantly more attachment and growth than all other PEGDMA hydrogels, Figure 3B. Attachment to the 3% and 9% agar hydrogels was more than twice that of PEGDMA hydrogels that had a statistically equivalent Young’s moduli, while attachment to glass substrates experienced a 4.5 fold increase, or an increase of ~600 E. coli cells. This data further suggests that the larger mesh size of agar hydrogels or their different surface chemistry may be promoting the attachment of E. coli.
Figure 3.
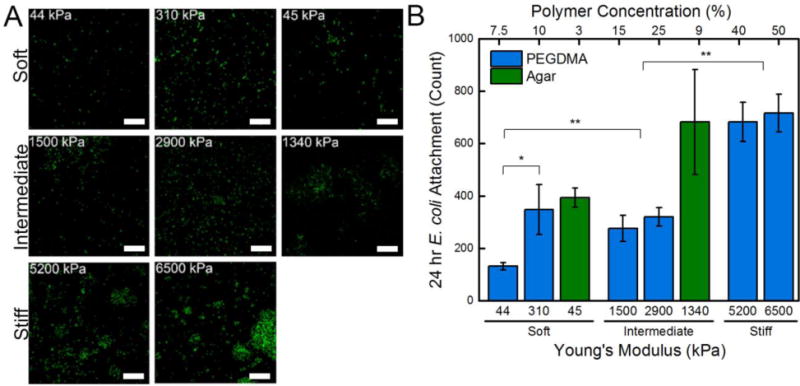
(A) Representative confocal micrographs (3894 μm2) of E. coli attached after a 24 hr incubation period on soft (44 – 308 kPa), intermediate (1340 – 2880 kPa), and stiff (5150 – 6500 kPa) PEGDMA and agar hydrogels. A 10 μm scale bar is displayed. (B) Total cell count quantified that there was a significant increase in E. coli attachment with increasing stiffness. An asterisk denotes 95% significance between samples, whereas two asterisks denotes significance between stiffness regimes. Error bars denote standard error.
After a 24 hr incubation period, S. aureus displayed characteristic grape-like colony formation on intermediate and stiff PEGDMA hydrogels, as well as on soft and intermediate agar hydrogels, Figure 4A. Additionally, the same strong correlation observed for E. coli held true: bacterial adhesion increased with increasing hydrogel stiffness. Less than 1.0% of the soft 7.5% PEGDMA hydrogels exhibited area colony coverage by S. aureus, whereas the stiff 50% PEGDMA hydrogels had a statistically significant greater area colony coverage of 4.7%, Figure 4B. As reported with E. coli, substantially more S. aureus adhered to the agar hydrogels than to the PEGDMA hydrogels. More than double the area colony coverage of S. aureus was present on agar hydrogels that had a Young’s moduli that was statistically equivalent to the PEGDMA hydrogels.
Figure 4.
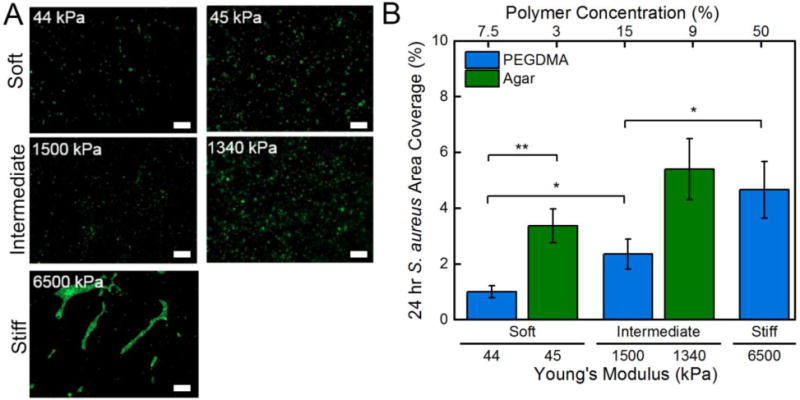
(A) Representative micrographs (58716 μm2) of S. aureus attached after a 24 hr incubation period on soft (44, 45 kPa), intermediate (1500, 1340 kPa), and stiff (6500 kPa) PEGDMA and agar hydrogels. A 10 μm scale bar is displayed. (B) Area coverage quantified that there was a significant increase in S. aureus area coverage with increasing stiffness. An asterisk denotes 95% significance between samples, whereas two asterisks denotes significance between stiffness regimes. Error bars denote standard error.
The colonization of medical devices by bacteria is a problem of increasing concern. Poly(ethylene glycol)-based coatings are frequently used on medical implants to enhance their biocompatibility while mitigating fouling. If the hydrogel coatings were softer, potentially, less bacteria would initially adhere, thus delaying the onset of biofilms that are hard to combat using commercial antibiotics. After a 2 hr incubation period, a 15 fold decrease or ~295 less E. coli attached to the softest PEGDMA hydrogels as compared to stiffest PEGDMA hydrogels. Our results suggest that this trend is independent of chemistry, and in our study, microbial adhesion mechanism. After a 24 hr incubation period 42% less E. coli attached to soft (45 kPa) versus intermediate (1336 kPa) agar hydrogels, whereas on PEGDMA hydrogels, there was a 52% reduction of E. coli attachment over the same stiffness regimes, 44 kPa versus 1500 kPa. S. aureus displayed a strikingly similar trend; after the 24 hr incubation period, between the soft and intermediate hydrogels there was a 38% and 58% reduction in area colony coverage for the PEGDMA and agar hydrogels, respectively. While we report the same observed trend for the Gram-negative and Gram-positive microbes, the full mechanism is beyond the scope of this study. Receptor specific binding through E. coli organelle, specifically type 1 fimbriae, auto transporter proteins, and aggregative fimbriae, can permanently bind bacteria to a surface,55 but the ability of poly(ethylene glycol) to resist protein adhesion suggests that another mechanism might be responsible. S. aureus lack these extracellular organelle and instead rely on protein adhesions through microbial surface components recognizing adhesive matrix molecules (MSCRAMM).56,57 As an inexpensive synergistic mode of modulating bacterial attachment, the stiffness of hydrogels can be tuned in combination with traditional nanoparticle or antibiotic loading to further delay the onset of biofilm formation in health care applications.
Implications for the Future Design of Antifouling Materials
This work explored bacterial adhesion as a function of hydrogel stiffness by comparing the attachment of two different microbes onto PEGDMA and agar hydrogels. The adhesion of both E. coli K12 MG1655 and S. aureus SH1000 was reduced for 24 hr on the softest surfaces we tested. This was surprising given the different modes of adhesion possessed by these two microbes, as well as the very different surface chemistries of our two hydrogel platforms. We acknowledge that our findings cannot necessarily be extrapolated to form a sweeping statement about all microbial adhesion mechanisms, as other bacteria strains, changes to bacterial physiology, and various growth conditions should be tested. While it has yet to be determined if equivalent biofilms will eventually form on soft and stiff hydrogels, providing a clinician a longer time to identify and combat bacteria at the catheter implant site has implications on decreasing the amount of infections associated with mortality. We suggest that such fundamental insights could be used to define design principles for bacterial resistant surfaces because, surfaces that delay the onset of microbial attachment could transform a variety of industries, including, medical, marine, water treatment, and food processing.
CONCLUSION
We have fabricated PEGDMA and agar hydrogels over a wide range of Young’s moduli. The adhesion of E. coli correlated positively with hydrogel stiffness over the range of Young’s moduli that was investigated, 44 – 6500 kPa. This range exceeds previous stiffness ranges investigated and represents the first time that thick polymer hydrogels were used as a testing substrate. After a 24 hr incubation period, the soft 44 kPa PEGDMA hydrogels had ~52% less E. coli adhered to them than the intermediate 1500 kPa PEGDMA hydrogels, and 82% less E. coli than the stiff 6500 kPa PEGDMA hydrogels. Similarly, the adhesion of E. coli on soft 45 kPa agar hydrogels was reduced by 42% when compared to intermediate 1340 kPa agar hydrogels. After 24 hrs, the adhesion of S. aureus was reduced by 62% on soft 44 kPa PEGDMA compared to intermediate 1500 kPa PEGDMA, and by 79% when compared to the stiff 6500 kPa PEGDMA hydrogels. The attachment of S. aureus onto soft 45 kPa agar hydrogels was reduced by 38% when compared to intermediate 1340 kPa agar hydrogels. For the first time, we have determined that more E. coli and S. aureus adhere to stiffer hydrogels and that this relationship occurs independent of hydrogel chemistry. We suggest that stiffness, a structure-property relationship could potentially reduce the initial adhesion of bacteria on both synthetic and biopolymer hydrogels.
Supplementary Material
Acknowledgments
This investigation was supported by National Research Service Award T32 GM008515 from the National Institutes of Health. This work was partially supported by the Prof. James M. Douglas Career Development Faculty Award and the Armstrong Fund for Science. S.R.P was supported by a career development award from Barry and Afsaneh Siadat and by a New Innovator Award from the NIH (DP2 CA186573-01). S.R.P. is a Pew Biomedical Scholar supported by the Pew Charitable Trusts. Thanks to Dr. Michael Franklin (Montana State University) for kindly providing the florescence plasmid pMF 230 and to Dr. Alexander Horswill (University of Iowa) for the high-efficiency pCM29 sGFP plasmid. Thanks to Drs. Julian McClements (UMass, Food Science) and Alfred Crosby (UMass, Polymer Science and Engineering) for access to equipment. We acknowledge the use of facilities at the W.M. Keck Center for Electron Microscopy and the MRSEC at UMass Amherst.
Footnotes
Supporting Information: Figure S1 provides force displacement curves for PEGDMA and agar hydrogels obtained through contact-adhesion testing. Figure S2 shows that E. coli and S. aureus have high viability on PEGDMA hydrogels. Figure S3 shows the adsorption of fluorescently labeled Fibronectin onto PEGDMA and agar hydrogels. Figure S4 displays that after a 2 hr incubation period, statistically less S. aureus attached to soft hydrogels than to intermediate and stiff PEGDMA hydrogels or to intermediate agar hydrogels. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.O’Grady NP, Alexander M, Burns LA, Dellinger EP, Garland J, Heard SO, Lipsett PA, Masur H, Mermel LA, Pearson ML, Raad II, Randolph A, Rupp ME, Saint S. Guidelines for the Prevention of Intravascular Catheter-Related Infections. Clin Infect Dis. 2011:1–83. doi: 10.1093/cid/cir257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goede MR, Coopersmith CM. Catheter-Related Bloodstream Infections. Surg Clin North Am. 2009;89:463–474. doi: 10.1016/j.suc.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Jansen B, Schumacher-Perdreau F, Peters G, Pulverer G. New Aspects in the Pathogenesis Prevention of Polymer-Associated Foreign-Body Infections Caused by Coagulase-Negative Staphylococci. J Investig Surg. 1989;2:361–380. doi: 10.3109/08941938909018262. [DOI] [PubMed] [Google Scholar]
- 4.Dizman B, Elasri MO, Mathias LJ. Synthesis, Characterization, and Antibacterial Activities of Novel Methacrylate Polymers Containing Norfloxacin. Biomacromolecules. 2005;6:514–520. doi: 10.1021/bm049383+. [DOI] [PubMed] [Google Scholar]
- 5.Kim H-W, Knowles JC, Kim H-E. Hydroxyapatite Porous Scaffold Engineered with Biological Polymer Hybrid Coating for Antibiotic Vancomycin Release. J Mater Sci ater Med. 2005;16:189–195. doi: 10.1007/s10856-005-6679-y. [DOI] [PubMed] [Google Scholar]
- 6.Ng VWL, Ke X, Lee ALZ, Hedrick JL, Yang YY. Synergistic Co-Delivery of Membrane-Disrupting Polymers with Commercial Antibiotics against Highly Opportunistic Bacteria. Adv Mater. 2013;25:6730–6736. doi: 10.1002/adma.201302952. [DOI] [PubMed] [Google Scholar]
- 7.Cook G, Costerton JW, Darouiche RO. Direct Confocal Microscopy Studies of the Bacterial Colonization In Vitro of a Silver-Coated Heart Valve Sewing Cuff. Int J Antimicrob Agents. 2000;13:169–173. doi: 10.1016/s0924-8579(99)00120-x. [DOI] [PubMed] [Google Scholar]
- 8.Ruparelia JP, Chatterjee AK, Duttagupta SP, Mukherji S. Strain Specificity in Antimicrobial Activity of Silver and Copper Nanoparticles. Acta Biomater. 2008;4:707–716. doi: 10.1016/j.actbio.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Ahamed M, Alsalhi MS, Siddiqui MKJ. Silver Nanoparticle Applications and Human Health. Clin him Acta. 2010;411:1841–1848. doi: 10.1016/j.cca.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Tenover FC. Mechanisms of Antimicrobial Resistance in Bacteria. Am J Med. 2006;119:S3–S10. doi: 10.1016/j.amjmed.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Stewart PS, Costerton JW. Antibiotic Resistance of Bacteria in Biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 12.Grozea CM, Walker GC. Approaches in Designing Non-Toxic Polymer Surfaces to Deter Marine Biofouling. Soft Matter. 2009;5:4088–4100. [Google Scholar]
- 13.Gu H, Ren D. Materials and Surface Engineering to Control Bacterial Adhesion and Biofilm Formation: A Review of Recent Advances. Front Chem Sci Eng. 2014;8:20–33. [Google Scholar]
- 14.Dobosz KM, Kolewe KW, Schiffman JD. Green Materials Science and Engineering Reduces Biofouling: Approaches for Medical and Membrane-Based Technologies. Front Microbiol. 2015;6:1–8. doi: 10.3389/fmicb.2015.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costerton JW. Bacterial Biofilms: A Common Cause of Persistent Infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 16.Yang L, Liu Y, Wu H, Song Z, Høiby N, Molin S, Givskov M. Combating Biofilms. FEMS Immunol Med Mic. 2012;65:146–157. doi: 10.1111/j.1574-695X.2011.00858.x. [DOI] [PubMed] [Google Scholar]
- 17.Belas R. Biofilms Flagella and Mechanosensing of Surfaces by Bacteria. Trends Microbiol. 2014:1–11. doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Thomas WE. Mechanochemistry of Receptor-Ligand Bonds. Curr Opin Struct Biol. 2009;19:50–55. doi: 10.1016/j.sbi.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Ellison C, Brun YV. Mechanosensing: A Regulation Sensation. Curr Biol. 2014;25:R113–R115. doi: 10.1016/j.cub.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perera-Costa D, Bruque JM, González-Martín ML, Gómez-García AC, Vadillo-Rodríguez V. Studying the Influence of Surface Topography on Bacterial Adhesion Using Spatially Organized Microtopographic Surface Patterns. Langmuir. 2014;30:4633–4641. doi: 10.1021/la5001057. [DOI] [PubMed] [Google Scholar]
- 21.Scardino AJ, de Nys R. Mini Review: Biomimetic Models and Bioinspired Surfaces for Fouling Control. Biofouling. 2011;27:73–86. doi: 10.1080/08927014.2010.536837. [DOI] [PubMed] [Google Scholar]
- 22.Meel C, Kouzel N, Oldewurtel ER, Maier B. Three-Dimensional Obstacles for Bacterial Surface Motility. Small. 2012;8:530–534. doi: 10.1002/smll.201101362. [DOI] [PubMed] [Google Scholar]
- 23.Bakker DP, Van der Plaats A, Verkerke GJ, Busscher HJ, Van der Mei HC. Comparison of Velocity Profiles for Different Flow Chamber Designs Used in Studies of Microbial Adhesion to Surfaces. Appl Environ Microbiol. 2003;69:6280–6287. doi: 10.1128/AEM.69.10.6280-6287.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bazaka K, Crawford RJ, Ivanova EP. Do Bacteria Differentiate between Degrees of Nanoscale Surface Roughness? Biotechnol J. 2011;6:1103–1114. doi: 10.1002/biot.201100027. [DOI] [PubMed] [Google Scholar]
- 25.Lichter JA, Thompson MT, Delgadillo M, Nishikawa T, Rubner MF, Van Vliet KJ. Substrata Mechanical Stiffness Can Regulate Adhesion of Viable Bacteria. Biomacromolecules. 2008;9:1571–1578. doi: 10.1021/bm701430y. [DOI] [PubMed] [Google Scholar]
- 26.Saha N, Monge C, Dulong V, Picart C, Glinel K. Influence of Polyelectrolyte Film Stiffness on Bacterial Growth. Biomacromolecules. 2013;14:520–528. doi: 10.1021/bm301774a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song F, Ren D. Stiffness of Cross-Linked Poly(dimethylsiloxane) Affects Bacterial Adhesion and Antibiotic Susceptibility of Attached Cells. Langmuir. 2014;30:10354–10362. doi: 10.1021/la502029f. [DOI] [PubMed] [Google Scholar]
- 28.Mata A, Fleischman AJ, Roy S. Characterization of Polydimethylsiloxane (PDMS) Properties for Biomedical Micro/nanosystems. Biomed Microdevices. 2005;7:281–293. doi: 10.1007/s10544-005-6070-2. [DOI] [PubMed] [Google Scholar]
- 29.Lin-Gibson S, Bencherif S, Cooper JA, Wetzel SJ, Antonucci JM, Vogel BM, Horkay F, Washburn NR. Synthesis and Characterization of PEG Dimethacrylates and Their Hydrogels. Biomacromolecules. 2004;5:1280–1287. doi: 10.1021/bm0498777. [DOI] [PubMed] [Google Scholar]
- 30.Trappmann B, Gautrot JE, Connelly JT, Strange DGT, Li Y, Oyen ML, Cohen Stuart MA, Boehm H, Li B, Vogel V, Spatz JP, Watt FM, Huck WTS. Extracellular-Matrix Tethering Regulates Stem-Cell Fate. Nat Mater. 2012;11:742–742. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 31.Oyen ML. Mechanical Characterisation of Hydrogel Materials. Int Mater Rev. 2014;59:44–59. [Google Scholar]
- 32.Beloin C, Roux A, Ghigo J. Escherichia coli Biofilms. Curr Top Microbiol Immunol. 2008;322:249–289. doi: 10.1007/978-3-540-75418-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flemming H-C, Wingender J. The Biofilm Matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 34.Krishnan S, Weinman CJ, Ober CK. Advances in Polymers for Anti-Biofouling Surfaces. J Mater Chem. 2008;18:3405–3413. [Google Scholar]
- 35.Götz F. Staphylococcus and Biofilms. Mol Microbiol. 2002;43:1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- 36.Beloin C, Houry A, Froment M, Ghigo JM, Henry N. A Short-Time Scale Colloidal System Reveals Early Bacterial Adhesion Dynamics. PLoS Biol. 2008;6:1549–1558. doi: 10.1371/journal.pbio.0060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence EL, Turner IG. Materials for Urinary Catheters: A Review of Their History and Development in the UK. Med Eng Phys. 2005;27:443–453. doi: 10.1016/j.medengphy.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 38.Hetrick EM, Schoenfisch MH. Reducing Implant-Related Infections: Active Release Strategies. Chem Soc Rev. 2006;35:780–789. doi: 10.1039/b515219b. [DOI] [PubMed] [Google Scholar]
- 39.Herrick WG, Nguyen TV, Sleiman M, McRae S, Emrick TS, Peyton SR. PEG-Phosphorylcholine Hydrogels as Tunable and Versatile Platforms for Mechanobiology. Biomacromolecules. 2013;14:2294–2304. doi: 10.1021/bm400418g. [DOI] [PubMed] [Google Scholar]
- 40.Liu V, Bhatia S. Three-Dimensional Photopatterning of Hydrogels Containing Living Cells. Biomed Microdevices. 2002;4:257–266. [Google Scholar]
- 41.Ayyad O, Muñoz-Rojas D, Agulló N, Borrós S, Gómez-Romero P. High-Concentration Compact Agar Gels from Hydrothermal Synthesis. Soft Matter. 2010;6:2389. [Google Scholar]
- 42.Shull KR, Ahn D, Chen W-L, Flanigan CM, Crosby AJ. Axisymmetric Adhesion Tests of Soft Materials. Macromol Chem Phys. 1998;199:489–511. [Google Scholar]
- 43.Canal T, Peppas N. Correlation between Mesh Size and Equilibrium Degree of Swelling of Polymeric Networks. J Biomed Mater Res. 1989;23:1183–1193. doi: 10.1002/jbm.820231007. [DOI] [PubMed] [Google Scholar]
- 44.Pang YY, Schwartz J, Thoendel M, Ackermann LW, Horswill AR, Nauseef WM. Agr-Dependent Interactions of Staphylococcus Aureus USA300 with Human Polymorphonuclear Neutrophils. J Innate Immun. 2010;2:546–559. doi: 10.1159/000319855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zodrow K, Schiffman J, Elimelech M. Biodegradable Polymer (PLGA) Coatings Featuring Cinnamaldehyde and Carvacrol Mitigate Biofilm Formation. Langmuir. 2012;28:13993–13999. doi: 10.1021/la303286v. [DOI] [PubMed] [Google Scholar]
- 46.Fletcher M, Loeb GI. Influence of Substratum Characteristics on the Attachment of a Marine Pseudomonad to Solid Surfaces Influence of Substratum Characteristics on the Attachment of a Marine Pseudomonad to Solid Surfaces. Appl Enviromental Microbiol. 1979;37:67–72. doi: 10.1128/aem.37.1.67-72.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schiffman JD, Wang Y, Giannelis EP, Elimelech M. Biocidal Activity of Plasma Modified Electrospun Polysulfone Mats Functionalized with Polyethyleneimine-Capped Silver Nanoparticles. Langmuir. 2011;27:13159–13164. doi: 10.1021/la202605z. [DOI] [PubMed] [Google Scholar]
- 48.Fux CA, Wilson S, Stoodley P. Detachment Characteristics and Oxacillin Resistance of Staphoccocus aureus Biofilm Emboli in an In Vitro Catheter Infection Model. J Bacteriol. 2004;186:4486–4491. doi: 10.1128/JB.186.14.4486-4491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung KK, Schumacher JF, Sampson EM, Burne Ra, Antonelli PJ, Brennan AB. Impact of Engineered Surface Microtopography on Biofilm Formation of Staphylococcus aureus. Biointerphases. 2007;2:89–94. doi: 10.1116/1.2751405. [DOI] [PubMed] [Google Scholar]
- 50.Lehtinen J, Nuutila J, Lilius E-M. Green Fluorescent Protein-Propidium Iodide (GFP-PI) Based Assay for Flow Cytometric Measurement of Bacterial Viability. Cytometry A. 2004;60:165–172. doi: 10.1002/cyto.a.20026. [DOI] [PubMed] [Google Scholar]
- 51.Nayar VT, Weiland JD, Nelson CS, Hodge AM. Elastic and Viscoelastic Characterization of Agar. J Mech Behav Biomed Mater. 2012;7:60–68. doi: 10.1016/j.jmbbm.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 52.Peyton SR, Kalcioglu ZI, Cohen JC, Runkle AP, Van Vliet KJ, Lauffenburger DA, Griffith LG. Marrow-Derived Stem Cell Motility in 3D Synthetic Scaffold Is Governed by Geometry along with Adhesivity and Stiffness. Biotechnol Bioeng. 2011;108:1181–1193. doi: 10.1002/bit.23027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moriarty TF, Poulsson AHC, Rochford ETJ, Richards RG. Bacterial Adhesion and Biomaterial Surfaces. Comprehensive Biomaterials. 2011;4:75–100. [Google Scholar]
- 54.Tuson HH, Weibel DB. Bacteria-Surface Interactions. Soft Matter. 2013;9:4368–4380. doi: 10.1039/C3SM27705D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Dunne WM. Bacterial Adhesion: Seen Any Good Biofilms Lately? Clin Microbiol Rev. 2002;15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foster TJ, Höök M. Surface Protein Adhesins of Staphylococcus aureus. Trends Microbiol. 1998;6:484–488. doi: 10.1016/s0966-842x(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 57.Clarke SR, Foster SJ. Surface Adhesins of Staphylococcus aureus. Ad Microb Physiol. 2006;51:187–224. doi: 10.1016/S0065-2911(06)51004-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


