Abstract
Fifty years ago, a new thymine dimer was discovered as the dominant DNA photolesion in UV irradiated bacterial spores [Donnellan, J. & Setlow R. (1965) Science, 149, 308–310], which was later named the spore photoproduct (SP). Formation of SP is due to the unique environment in the spore core that features low hydration levels favoring an A-DNA conformation, high levels of calcium dipicolinate that acts as a photosensitizer, and DNA saturation with small, acid-soluble proteins that alters DNA structure and reduces side reactions. In vitro studies reveal that any of these factors alone can promote SP formation; however, SP formation is usually accompanied by the production of other DNA photolesions. Therefore, the nearly exclusive SP formation in spores is due to the combined effects of these three factors. SP photoreaction is proved to occur via a unique H-atom transfer mechanism between the two involved thymine residues. Successful incorporation of SP into an oligonucleotide has been achieved via organic synthesis, which enables structural studies that reveal minor conformational changes in the SP-containing DNA. Here, we review the progress on SP photochemistry and photobiology in the past fifty years, which indicates a very rich SP photobiology that may exist beyond endospores.
Graphical abstract

INTRODUCTION
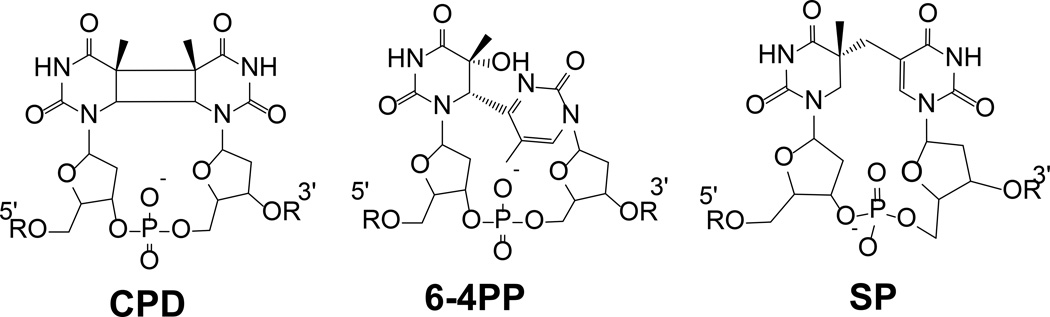
In July 1965, Donnellan and Setlow published a short communication in the journal Science, reporting that in Bacillus megaterium spores irradiated with monochromatic ultraviolet (UV) light at 265 nm, a new thymine lesion was produced as the dominant DNA photo-damage product (1). This lesion is different from the cyclobutane pyridimine dimer (CPD, <Figure 1>), the nature of which was relatively well established at that time. It was also different from the pyrimidine (6-4) pyrimidone photoproduct (6-4PP) discovered by Varghese and Wang two years later in UV irradiated DNA (2). The new spore lesion was soon shown to contribute to spore death (3), and with high UV doses, it may involve up to 28% of total thymine residues in the spore genome (4). Although the spore lesion was also isolated from UV irradiated dry (4, 5) or frozen DNA (6), the yield was never as great as that in irradiated spores. Therefore, this photolesion was commonly referred to as the spore photoproduct (SP). Using DNA irradiated in ice at − 78 °C and then DNA hydrolysis in strong acid at 165 °C, Varghese was able to purify a significant amount of SP base adduct <SPBASE (1), Figure 2> by paper and column chromatography (7). The UV-visible spectrum of the purified SPBASE was also determined and SP was found to possess an extinction coefficient of 8,200 M−1cm−1 at 265 nm, almost identical to that of a single thymine residue. Such an absorption spectrum agrees with the SP structure shown in Figure 1, since the 5′-thymine ring does not absorb at 265 nm due to the loss of this ring’s aromaticity. Using not only UV-visible spectroscopy, but also IR, NMR and mass spectrometry, Varghese then correctly suggested that 5-thyminyl-5,6-dihydrothymine is the most probable structure for SPBASE after both glycosidic bonds were hydrolyzed (7). Later, using NMR spectroscopy coupled with density functional theory (DFT) to examine the dinucleotide SP TpT <SPTIDE (2), Figure 2> obtained from the dinucleotide TpT photoreaction, Mantel et al. determined that the dihydropyrimidine moiety was located at the 5'-end of the dinucleotide and the C5 chiral center adopts an R configuration (8). The SPTIDE 2 was found to be identical to SPs obtained from UV irradiated spores after DNA digestion (9), proving that the SPs formed in spores contain a 5R-chiral center <Figure 1>. The chirality is ascribed to the right-handed DNA helical structure, which prevents any 5S-SP being formed and enables generation of 5R-SP as the dominant DNA photoproduct in UV irradiated spores.
Figure 1.
Chemical structure of the three thymine dimers.
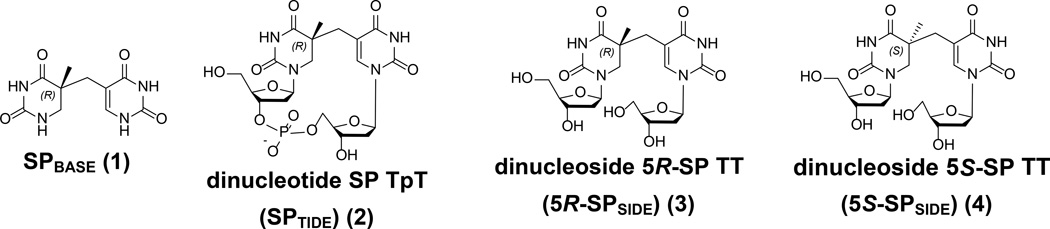
Figure 2.
Structures of SP and SP derivatives discussed in this review. In the literature, dinucleoside SP TT and dinucleotide SP TpT were used respectively for SP dimers with and without the phosphodiester linker; while Desnous et al. used SPSIDE and SPTIDE to refer to these two species (40). Although we used SPSIDE and SPTIDE in our review, we wish to show both nomenclatures.
Shortly after SP was discovered in UV irradiated spores, Donnellan and Stafford established that when spores come back to life in the process of germination, SP was eliminated from genomic DNA by a dark-repair mechanism somewhat different from that responsible for CPD repair in vegetative cells (3). Genetic analysis indicated that at least two genes contribute to the observed high UV resistance of Bacillus subtilis spores (10), with these two genes determining distinct processes for SP removal from DNA during spore germination (11, 12). The first is a spore-specific repair mechanism by which SP is removed rapidly without appearing in an acid-soluble form or released from the germinating spores (13). The second mechanism, now known as the general nucleotide excision repair (NER) pathway, removes SP relatively slowly. In addition to SP, the NER pathway is also responsible for removing CPDs and 6-4PPs and is regarded as the major DNA-repair process involved in the recognition and removal of UV-mediated DNA damage in growing bacteria as well as in eukaryotes (14–17). However, blocking either pathway only slightly increases spore UV sensitivity, while blocking both pathways prevents the efficient removal of SP, resulting in very UV sensitive spores (11, 12).
The spore-specific SP repair mechanism is now known to be mediated by a metalloenzyme – the spore photoproduct lyase (SPL) encoded by the splB gene (18). In the 1970s, it was found that the radioactive tritium label disappearing from SP was recovered stoichiometrically in thymine residues in germinated spores, indicating that SPL directly reverts SP to TpT (3, 13). Bioinformatics studies have established that SPL is a so-called radical S-adenosylmethionine enzyme, which utilizes a [4Fe-4S] cluster coupled with an S-adenosylmethionine cofactor to generate a 5′-deoxyadenosyl radical for catalysis (19). Research from various groups has further established that the 5′-deoxyadenosyl radical initiates the SP repair process by abstracting an H-atom from the C6 position on SP, which triggers a cascade radical transfer process (9, 18–33). The most recent results suggest that SPL uses an unprecedented radical transfer pathway composed of one cysteine and two tyrosine residues to facilitate radical transfer (33). This hypothesis has been fully supported by analysis of the recently-solved SPL structure (34).
Progress in SPL mechanistic studies has been extensively reviewed in the past several years (35–39). In this review, we choose to cover SP photochemistry and photobiology revealed via chemical and photochemical means. The reviews of Desnous et al. (40) and of Yoshimura et al. (41) have covered the SP chemistry and photochemistry till 2010 and 2012 respectively; here we will focus more on progress since 2012. For a number of years, except for SPL-mediated repair, very little was learned about SP photobiology owing to the difficulty in obtaining SP-containing oligonucleotides for biological studies. However, progress in synthetic chemistry in the past several years has largely solved this issue, and opened the door for the understanding of general SP photobiology.
SP FORMATION IN BACTERIAL SPORES
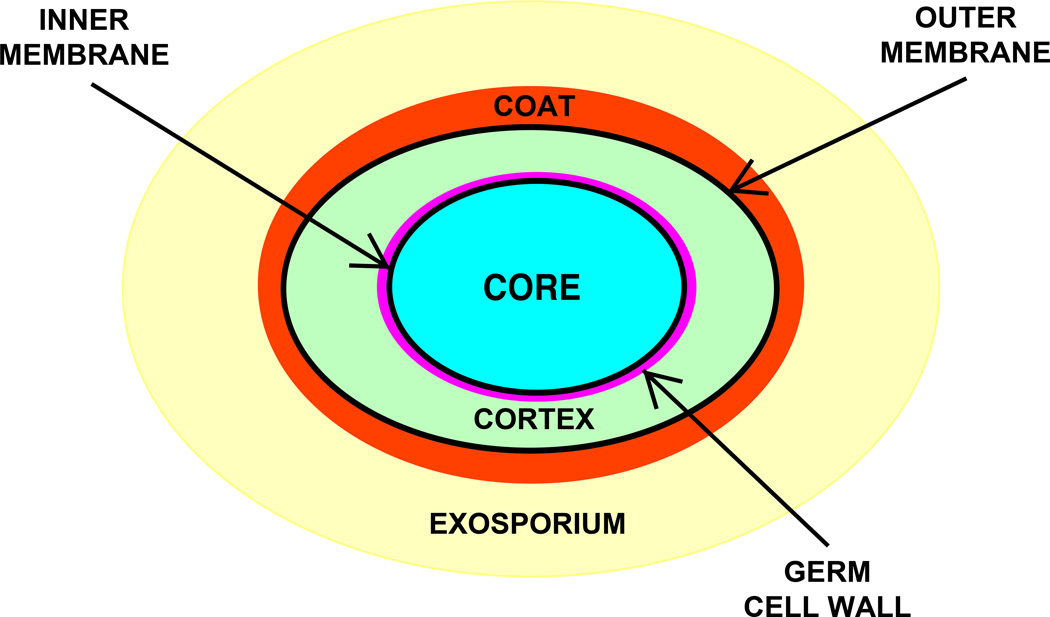
SP is isolated as the dominant DNA photoproduct in UV irradiated spores. This unique photochemistry is largely ascribed to the unique environment inside a spore (40, 42–45). As indicated in <Figure 3>, underneath the outer glycoprotein layer called the exosporium is a proteinaceous coat providing much of spores’ resistance to enzymes and toxic chemicals. Beneath this coat there is an outer membrane, which may not be a functional membrane in spores, and then a thick layer of specialized peptidoglycan called the cortex, which plays an important role in maintaining the low water content in spores’ central core. A germ cell wall composed of peptidoglycan with a structure different from that of the cortex lies under the cortex, followed by the inner membrane, the major permeability barrier against damaging chemicals. The center of the spore, the core, has a relatively low water content, and houses the genomic DNA saturated with spore-specific DNA binding proteins, ribosomes and a large amount (see below) of pyridine-2,6-dicarboxylic acid (dipicolinic acid (DPA)), most of which is chelated to divalent cations, primarily Ca2+ (CaDPA). This unique environment for the spores’ genome is responsible for efficient SP formation in spores. At the same time, formation of other DNA photo-damage as well as DNA damage induced by agents such as heat, desiccation, and genotoxic chemicals is largely suppressed (46).
Figure 3.
Structure of a bacterial endospore (5). The relative sizes of various layers are not drawn to scale, and the large exosporium is not present in spores of all species. Note that there can be several sublayers in individual layers, in particular the coat and probably in the exosporium.
SP formation as a function of UV wavelength
The electromagnetic spectrum of UV irradiation, defined as 100–400 nm in wavelength is generally divided into three categories, UVC (100–280 nm), UVB (280–315 nm) and UVA (315–400 nm). Stratospheric ozone effectively blocks wavelengths shorter than 290 nm from reaching the earth’s surface, filtering out all the UVC and most of the UVB light (47). Consequently, about 95% of the UV radiation from the sun that reaches the earth is UVA, and the remaining 5% is UVB. However, owing to the high energy in UVC photons, UVC has been widely used in germicidal lamps for killing microorganisms in sterilization applications. Indeed, with UVC radiation, SP photochemistry in spores proceeds with slightly higher efficiency than does CPD or 6-4PP formation in growing cells (44, 48). Although formed as the dominant species in spores, SP formation is accompanied by formation of other minor DNA photoproducts, such as CPDs as well as single- and double-strand DNA breaks. The extent of these “side” photoreactions appears to be very dependent on the wavelength of UV light.
Although UVC radiation is unlikely to play a significant role in SP photochemistry in nature, UVC has been used for the vast majority of spore photobiology studies owing to the high photo-efficiency of this radiation. With a dose of 16 kJ/m2 of UVC light predominantly at 254 nm, SP is the principal DNA photoproduct in Bacillus subtilis endospores (~ 4.2 × 105 per chromosome); CPDs and single-/double- strand breaks are also found, but the levels of these lesions are only 0.16 and 0.3% of SP, respectively (49). Therefore, it was concluded that SP generation is the dominant factor determining spore survival after UVC irradiation, and physiological consequences of CPDs and strand breaks may be negligible. Such a conclusion is further supported by a later study, which used high-performance liquid chromatography coupled with tandem mass spectrometry (HPLC-MS/MS) to re-analyze the DNA lesions in UVC irradiated spores, finding that SP was the predominant DNA photoproduct (93 ± 5%); CPDs formed in TpT, TpC, and CpT steps as well as 6-4PPs formed in TpC and TpT steps were also detected, but in small amounts (50).
In contrast to results with UVC, with UVB radiation, SP formation efficiency is reduced by ~ 3 orders of magnitude (51). Moreover, although SP still accounts for the majority of the DNA photo-lesions formed, appreciable quantities of CPDs are generated. As a consequence, the NER and the SPL repair pathways appear to contribute approximately equally to the survival of UVB irradiated spores (49, 52). No DNA strand breaks were detected in UVB irradiated spores; however, they are the major DNA damaging events in UVA irradiated spores, as CPD and SP formation are undetectable in UVA irradiated spore (49, 52). If full spectrum sunlight is used to irradiate spores, the DNA damage pattern resembles the combined effects of UVA and UVB. Therefore, in addition to the NER and SPL pathways to repair DNA photo-dimers, the homologous recombination and nonhomologous-end joining pathways that repair DNA double-strand breaks may also play important roles in DNA repair in spore germination (53, 54). Indeed, proteins involved in these latter repair processes are present in dormant spores to allow for rapid DNA repair when spores germinate (55).
SP formation is the main DNA photochemical event occurring in UVB/UVC irradiated spores. In contrast, in vitro irradiation of spore genomic DNA and/or UV exposure of vegetative B. subtilis cells results in CPDs as the dominant products, with minimal SP formation. Three factors have been suggested to contribute to the unique DNA photochemistry in spores: 1) action of CaDPA as a photosensitizer; 2) the low hydration level in the spore core; and 3) the saturation of spore genomic DNA by a group of DNA-binding proteins termed small, acid-soluble spore proteins (SASPs). As described below, the functions of these factors seem to be intertwined, such that their combined effects enable SP formation as the dominant DNA photochemical event in UV-irradiated spores. However, when spores germinate, CaDPA is excreted, spore core water content rises to that in growing cells, and SASPs are all degraded (56–58). After all these germination events are completed, spore UV resistance and DNA photochemistry revert to that of growing cells.
Function of CaDPA
As noted above, the spore core contains an extremely high level of CaDPA <Figure 4>. This complex comprises 5–15% of the dry weight of spores in both Bacillus and Clostridium species. Given that the core occupies ~ 50% of the overall spore volume, CaDPA accounts for ~ 20% of spore core weight (59). Assuming that the rest of the core is water, the concentration of CaDPA (M.W. = 205 Da) in the core is estimated to be ~ 1.2 M. Using microfluidic Raman tweezers, the CaDPA concentration in the spore core was found to be between 800 mM and 1 M (59), very close to the estimate given above.
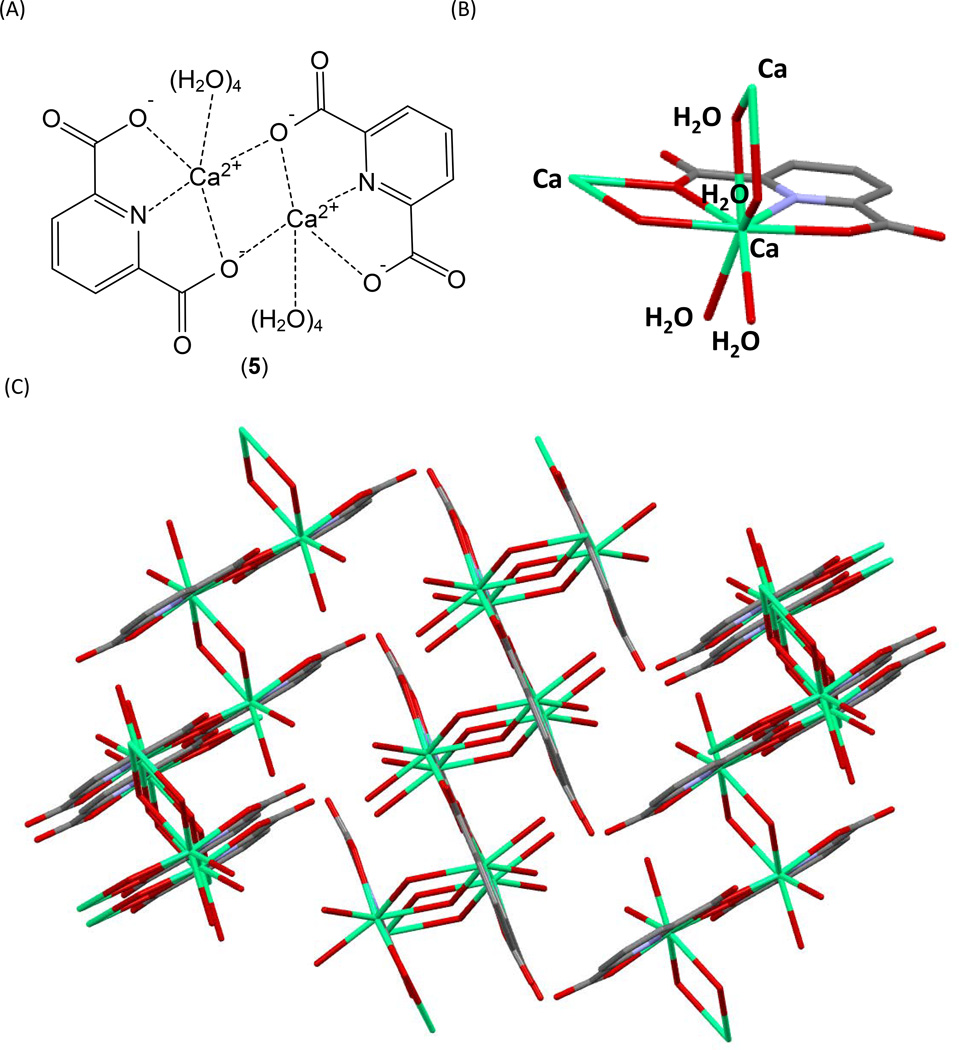
Figure 4.
(A) Structure of the CaDPA dimer as the basic assembly unit for the inorganic polymer (75). The dimer adopts a planar structure. Each Ca2+ is coordinated to eight atoms. Among them, three are from a DPA, one is from a carboxylate moiety of another DPA in the dimer, and the rest are from two pairs of water molecules, with one pair above and the other pair beneath the dimer plane. Both water molecules within the pair coordinate to another Ca2+ from another dimer, thus serving as bridging ligands to link the dimers into an inorganic polymer. (B) The coordination sphere around the calcium ion. The Ca2+ ion is shown in green, nitrogen in blue, and oxygen atoms in red. The four bridging water ligands are also labeled in the structure. (C) The structure of the CaDPA polymer linked by bridging water molecules.
Much work has strongly implicated CaDPA in playing important roles in: 1) spore resistance; 2) spore germination; and 3) spore stability (60, 61). 1) CaDPA accumulation in the spore core decreases the core water content (60), and spores with lower core water content are more resistant to wet heat (60–62). CaDPA is also suggested to interact with DNA in spores, preventing DNA damage from dry heat and desiccation (60, 61). In addition, CaDPA is thought to serve as a photosensitizer, facilitating energy transfer from photo-excited DPA to thymine residues to promote SP formation (63). Hyperresistance to UVC develops upon spore germination following CaDPA release but before SASPs degradation (64–67). This observation is ascribed to the fact that SP is still formed as the dominant DNA photoproduct early in spore germination due to the DNA conformation generated by SASP binding; however, much fewer SPs are produced under the same UV dose after the loss of photosensitizing CaDPA. 2) CaDPA release early in the germination of an individual Bacillus spore is an important part of the signal transduction process in germination (56–58), and spores can even be triggered to germinate by CaDPA (58, 68, 69). As a consequence, it is possible that CaDPA released from one spore may stimulate the germination of neighboring spores, especially if spores are at extremely high concentrations (68). 3) Spores containing no or low levels of CaDPA are unstable and germinate spontaneously (60, 61, 68). Although the mechanism behind this latter behavior is not completely understood, it is likely that the absence of CaDPA results in activation of enzymes that specifically degrade the spore cortex, thus initiating the germination process.
The ~ 1 M CaDPA concentration found in the spore core is roughly 10 times above its known solubility in water (59). Therefore, the complex is likely to adopt a 3-dimensional structure as a solid matrix rather than existing as a mononuclear complex dissolved in the central protoplast. Indeed, it has been suggested that the spore core adopts a glass-like state, with CaDPA being a major contributor to the matrix structure (70, 71), although this idea has been disputed (71–74). CaDPA has been crystallized as the trihydrate in both H2O (75) and D2O (76), and exhibits the same structure, where the essential unit is a CaDPA dimer 5 <Figure 4A>. Around each Ca2+ cation, eight ligands, seven oxygen-based and one nitrogen-based, can be found. Three of these ligands come from the two carboxylates and the pyridine nitrogen from one DPA, the fourth ligand is from the carboxylate of the other DPA in the dimer unit <Figure 4B>. The dimer unit adopts a planar structure in which the Ca2+ cations in the dimer are coordinated by two pairs of water molecules, with a pair on each side of the plane, to finish the eight coordinated states. Each pair of water molecules also coordinates to the same Ca2+ center in another dimer. Consequently, the dimer units are linked together by these di-water bridges into a polymer-like chain structure <Figure 4C>.
It was suggested more than 40 years ago that a crystalline matrix may exist in the spore core (75). By comparing the signature resonance Raman signals from DPA, recent spectroscopic studies revealed that Raman signals from CaDPA are similar to those from crystalline CaDPA, and these signals clearly differ from those due to DPA or CaDPA dissolved in water (77, 78). This result supports the idea that a CaDPA matrix may contribute to a glass-like state in spore core. However, although somewhat dehydrated, the water content in the spore core is still more than in a CaDPA crystal. Also, the presence of DNA, ribosomes, RNA and proteins could interfere with CaDPA packing, making the structure of the CaDPA matrix in spores likely to be more complicated than that shown in Figure 4 (77).
Resonance Raman data also indicated that CaDPA may intercalate into the DNA bases (78), and in vitro spectroscopic studies found that CaDPA directly interacts with DNA (79, 80). The yield of SP as a function of UV fluence was 10 to 20-fold lower in spores of mutants which do not make DPA than in wild-type spores (81). In dry calf thymus DNA films exposed to UVC radiation, the yield of SP was greatly increased by the presence of CaDPA in the DNA film (63). Moreover, it was suggested that the thymine triplet excited state supports SP formation (82), and CaDPA was therefore suggested to function as a photosensitizer, facilitating energy transfer from the triplet excited state of CaDPA to thymine residues in DNA (63). Thus the involvement of CaDPA in SP photochemistry has been supported by both in vivo and in vitro observations.
Among the four nucleobases, thymine possesses the lowest triplet excited state while cytosine has the highest energy (83). If the energy of the lowest triplet excited state of CaDPA is lower than that of the nucleobase, energy transfer to the nucleobase is not favored resulting in the quenching of certain photoreactions. Given the large amount of CaDPA in the spore core, if its triplet excited state is only higher than that of a thymine residue, photoreactions involving other nucleobases will largely be suppressed. Since in spores thymine dimerization into SP is the dominant reaction and reactions associated with cytosine are negligible, this reaction pattern may be due to the excited state energy of CaDPA, and this is certainly a matter for future experiments.
Low water content in the spore core
Another function of the high CaDPA concentration in the spore is to lower the water content in the core, and this has been strongly implicated as playing a major role in spores’ resistance to wet heat (62). While water comprises 75–80% of the wet weight of the protoplast of a growing cell, water makes up only 27–55% of the spore core wet weight depending on the species (44), and this low water content may result in unique physical properties of the core. Indeed, ions in the spore core were shown long ago to be immobile (84, 85), lipids in the inner membrane of dormant spores of Bacillus species are also largely immobile (86), and at least one normally soluble core protein is immobile in B. subtilis spores (87). These findings further suggested that core water itself may also be immobile.
However, recent results challenge the assumption that the immobility of core macromolecules and ions is directly related to the fixation of water molecules. Two theories have been proposed to describe the physical property of the spore core: a glass-like state where the entire core including the water molecules are immobilized (70), and a gel-like state where the macromolecules are immobilized in the matrix while the water molecules remain mobile (72). Measurements of the 2H and 17O spin relaxation rates in solid-state NMR spectroscopy in D2O or H2O17-exchanged Bacillus subtilis spores indicated that there was high water mobility throughout the spore, which is comparable with binary protein water systems at similar hydration levels (73, 88), supporting the idea that the spore core has a gel-like structure. However, another study using deuterium solid-state NMR found both bound and unbound water in spore core, and the core water exchanged only slowly with water in the medium (76). While the latter observation can be better explained by the glass-like state of the spore core, the low permeability of the spores’ inner membrane may well contribute to slow water exchange between the core and the medium.
While the exact state of water in the spore core is still not completely clear, it is most likely that the low core water content contributes to SP formation. In UVC irradiated wild-type B. subtilis spores, SP is the major (> 98%) DNA photolesion formed by UVC radiation (88). However, while SP was still a major product in UVC irradiated spores lacking either CaDPA or SASPs, significant amounts of CPDs and 6-4PPs were also observed (44, 63, 88). Most notably, in spores lacking both CaDPA and SASPs, only ~ 2% of the pyrimidine dimers formed were SP (88). However, compared with UVC-irradiated vegetative cells where no detectable SP was formed, the 2% yield implies that these spores’ relatively low water content may still be important in facilitating SP photochemistry. Indeed, while the core of B. subtilis spores lacking CaDPA has ~ 30% higher core water content than the wild-type spore core (45% vs 35%), the CaDPA-less spore core still has a much lower water content than a growing cell (50). As described below, an A-like DNA conformation, which is favored at low water content, is suggested to be critical for SP photochemistry. Therefore, the reduced SP formation in DPA-less, SASP-less spores could be due at least in part to the increased water content in these spores’ core.
The immobility of core ions and protein noted above indicates that DNA is also likely to be immobilized in the spore core, and such DNA immobilization may also contribute to SP photochemistry. In aqueous solution, DNA can freely undergo conformational changes; these conformations largely belong to the B-form and support CPD formation upon photo-excitation. However, due to the glass- or gel-like state in the spore core and the binding of SASPs (see below), the conformational changes in the immobile genomic DNA are expected to be greatly slowed. The A-like genomic DNA in spores favored by the low hydration level is further stiffened by the DNA immobility and binding of SASPs (88–90), supporting SP formation as the nearly exclusive photoreaction upon UV irradiation and largely suppressing all other dimerization reactions.
DNA binding by SASPs
In comparison with the presence of CaDPA and low water content in the spore core, the binding of SASPs to spore DNA is generally considered as the most important factor in the occurrence of SP photochemistry (44, 88, 91). Such a conclusion is ascribed to the fact that SASP binding alone is effective in promoting SP photochemistry and quenching the formation of other dimers, while the presence of CaDPA non-selectively increases the yields of both SP and CPDs (63, 92–94). While dormant spores are ~10-fold more UVC resistant than growing cells, spore UVC resistance actually rises dramatically after CaDPA release and before SASPs degradation that takes place minutes after CaDPA release (64–67). SP is still the major UVC photoproduct in these highly UV resistant spores early in germination, but the efficiency of SP formation drops significantly, likely because of the loss of the photosensitizing action of CaDPA on SP photochemistry. However, as α/β-type SASP are degraded, spore UV resistance decreases dramatically to that in growing cells, and SP formation ends. As expected, significant slowing of SASP degradation by loss of the germination protease specific for SASPs greatly prolongs the length of the high UV-resistant state following spore germination as well as continued SP formation. In addition, spores lacking the majority of their α/β-type SASP do not exhibit this period of elevated UV resistance soon after spore germination. These latter results are consistent with the importance of α/β-type SASP in causing SP photochemistry in spores.
The SASPs are synthesized only in the developing spore late in sporulation, and account for 8 to 20% of total spore core protein depending on the species analyzed (91). The SASPs in spores can be divided into two types: the α/β type and the -type. There are multiple α/βtype SASPs in spores of Bacillus and Clostridium species, and one -type SASP in Bacillus spores although γ-type SASP is not present in spores of Clostridium species (91). The α/βtype SASPs have molecular weights of 5–9 kDa, and account for 5–10% of total protein in the spore core. They have a significant percentage of hydrophobic amino acids, share remarkable sequence conservation both within and across species and genera, and their functions are largely interchangeable in spores (91, 95, 96). Therefore, one B. subtilis α/βtype SASP, SspC, has been used in most in vitro studies of SASPs’ properties, even though this protein is not the most abundant α/β-type SASP in spores (91). The -type SASP has a very different amino acid sequence than the α/β-type and has an extremely low content of large hydrophobic amino acids. There is much evidence that α/βtype SASPs bind to and saturate the spore genomic DNA (88). However, -type SASP does not bind to DNA, even though this protein is often the most abundant SASP in spores. All SASPs are digested into amino acids in a process initiated by a protease that recognizes a highly conserved amino acid sequence in SASPs, and the amino acids generated in this process are used for energy generation and protein synthesis (91).
The binding of SASPs to spore DNA is suggested to modify the DNA conformation favoring SP formation upon photoexcitation, and quenching other “side” reactions such as CPD formation (93, 94). In wild-type B. subtilis spores, UVC irradiation generates almost exclusively SP with little if any CPD and 6-4PP (88). In contrast, in spores lacking ~ 85% of their α/β type SASPs, while SP is still the major (~ 70%) DNA photoproduct, cis-syn CPDs formed in TpT, TpC, CpT and CpC steps are also observed (94). Decreased levels of α/βtype SASPs also decrease the resistance of Clostridium perfringens spores to UV radiation, possibly due to the increased formation of CPDs (97, 98). Moreover, UV irradiation of Escherichia coli engineered to have high levels of an α/βtype SASP also yields less CPDs and some SPs, in contrast to the undetectable SP formation in E. coli lacking such a protein (99).
The impact of binding of α/βtype SASPs to spore DNA was examined by immunofluorescence microscopy in developing spores (100, 101). The results indicate that the chromatin adopts a tightly packed ring-shaped morphology as a consequence of SASP binding. To reveal more details of this protein-DNA interaction, a variety of studies were conducted in vitro. Circular dichroism and Fourier-transform infrared spectroscopy showed that binding of SspC to DNA alters the DNA conformation from the B-form to an A-like form (89). The relative affinity of SspC to various DNA sequences was determined to be poly(dG)•poly(dC) > poly(dG-dC)•poly(dG-dC) > plasmid pUC19 > poly(dA-dT)•poly(dA-dT) >> poly(dA) •poly(dT); such an order parallels the ease with which these DNAs adopt an A-like conformation (102). Measured by both electron microscopic and cyclization assays, it was found that SspC binding greatly increases DNA stiffness, but does not alter the rise per base pair (bp) (90). This latter finding is inconsistent with the DNA-SspC complex having the classical structure of A-DNA found in dry DNA fibers. Using cryoelectron microscopy techniques, the SspC binding to DNA was indicated to induce only minor conformational changes, and the DNA pitch (3.18 nm) remains close to that of canonical B-form DNA (103). A tight packing of the DNA-protein filaments was also implied, suggesting that the spore genome may possess a toroidal conformation as a result of SASP-DNA interaction (103).
Although α/β-type SASPs are designed to bind and protect spore DNA, they are not intended to bind permanently. Indeed, if they do not dissociate quickly from DNA soon after spore germination is initiated, then subsequent transcription and spore outgrowth will be dramatically affected leading to spore death (104). Thus, their intrinsic binding affinity to DNA is rather low and this low affinity becomes a major obstacle for in vitro DNA-protein interaction studies. To increase SASP binding affinity for DNA, an SspC mutant, SspCΔN11-D13K-C3, was constructed, which has a ~ 20-fold higher affinity for DNA than wild-type SspC (105). This SspC variant promotes a similar DNA photochemistry as the wild-type protein, suggesting that both wild-type and variant proteins bind to DNA in a similar manner (105).
The SspC variant noted above was used to solve the crystal structure of an oligo(dG)•oligo(dC) 10-mer complexed with SspCΔN11-D13K-C3 (106, 107). The structure suggests that the α/βtype SASPs adopt a helix–turn–helix motif on DNA <Figure 5A>, although the SASPs are largely random coil alone in solution. SASPs interact with DNA through minor groove contacts and bind to ~ 6 bp of DNA as a dimer, agreeing with biochemical data obtained previously (108). More importantly, the DNA in the nucleoprotein complex adopts an A-B type conformation, where the sugar puckering adopts a C3′-endo conformation, a feature of A-form DNA, while the base planes are essentially parallel to each other and normal to the helix axis, which is characteristic of B-DNA <Figure 5B>. In addition, molecular simulation studies replacing the 6th and 7th GC pairs with AT pairs in the 10-mer indicated that the C5 of one T is only 3.4 Å away from the –CH3 of another T (106, 107). This distance is shorter than the 3.9 Å between the two C5 positions which is required to connect in CPD formation (106). The corresponding moieties involved in 6-4PP formation are even further apart. These results offer a rationale for how the DNA conformation in the SASP-DNA complex promotes SP photochemistry and quenches other “side” photoreactions. This structure is also consistent with spectroscopic studies, indicating that the SP photochemistry in spores cannot be simply ascribed to a pure A-like DNA conformation, but at least partially to DNA conformational changes induced by dehydration (109). Although the A-like DNA conformation favors SP formation, the B-A transitional conformations as a consequence of reduced DNA hydration level already support the formation of SP, although such a process is likely accompanied by the formation of other thymine dimers.
Figure 5.
Crystal structure of the α/β-type SASP (SspC)–oligo(dG)•oligo(dC)10-mer complex (pdb entry 2Z3X) (106). (A) Stereo diagram of the complex indicating that SspC adopts a helix–turn–helix motif when binding to DNA. The positions of the N and C termini of each protomer are labeled: orange, SspC1; magenta, SspC2; cyan, SspC3. (B) DNA conformation in the SspC-DNA complex. This conformation is a combination of A- and B-conformations, and is different from the typical A- or B-form DNA. Analyses by molecular simulation indicated that such a DNA conformation favors formation of SP over CPD and 6-4PP.
SP FORMATION IN VITRO
Besides investigations in spores, SP photochemistry has also been extensively studied in vitro. By irradiating single- or double-strand oligonucleotides in aqueous solution, CPDs and 6-4PPs can be readily formed at a given dipyrimidine site (110), and further studies have allowed elucidation of CPD and 6-4PP photochemistry in detail. For instance, in vitro studies have established that the formation of CPDs at a dipyrimidine site is reversible; the “equilibrium” of the reaction appears to be controlled by the wavelength employed (111). In contrast, formation of 6-4PP and SP are irreversible (112). Because of the latter property, with 254 nm UVC irradiation, formation of CPDs quickly reaches a maximum yield of ~ 20% while yields of 6-4PP keep increasing. In vitro studies of CPDs and 6-4PPs in aqueous solution also make the elucidation of their formation mechanism, i.e. the excited states involved and the reaction kinetics, possible (113–120). UV irradiation of DNA in aqueous solution results in no SP, which can be ascribed to the normal DNA hydration level preventing DNA from adopting appropriate reactive conformations to form SP. Consequently, generation of SP in in vitro DNA photochemistry is very inefficient, since reactions have to be conducted under unique environmental conditions.
SP formation in ice
It was discovered by Rahn et al. that DNA conformation at low temperatures favors SP photochemistry and quenches formation of other dimers (6, 121). DNA is less hydrated at low temperatures than at room temperature (122), which may cause a DNA conformational change toward the A-form, providing a rationale for the observed SP formation in ice. In contrast, no SP formation was observed at 25 °C and CPDs were found as the major DNA lesions. However, the percentage of CPDs in all thymine photo-lesions gradually decreased as temperature decreased to −196 °C while the percentage of SP gradually increased at the very beginning of the temperature decrease, reached a maximum at – 100 °C, and then decreased as the temperature was lowered further (121). At a given temperature, the SP yield increased with increasing UV doses. Indeed, as noted earlier, Varghese utilized DNA irradiation in ice to generate a relatively large amount of SP and prove that SP is a thymine dimer, with a structure of 5-thyminyl-5,6-dihydrothymine (7). This chemical structure also allowed Varghese to propose that SP is formed via formation and combination of 5-α-thyminyl and 5,6-dihydrothymin-5-yl radicals, which is now known to be largely correct (see discussion in Section III).
Besides isolated genomic DNA, irradiation of thymidine in ice also produces dinucleoside SP (123). The yield of SPSIDE was not quantified; however, it is likely to have been very low (likely < 1%). Because of the right-handed helical structure of duplex DNA and of TpT, the SP formed can only adopt an R configuration at the C5 chiral center <Figure 1 and Figure 2>. Such a restriction no longer exists in monomeric thymidine residues. As a consequence, a pair of SPSIDE diastereoisomers <Figure 2> adopting 5R (3) and 5S (4) configurations, respectively, are produced from the thymidine photoreaction (124) (see discussion below).
Although only the 5R-SP was predicted to form in spores and be subsequently repaired by the SP repair enzyme SPL during spore germination, some earlier studies suggested that SPL repairs the 5S-isomer (125, 126). The discrepancy was corrected by later SPL enzymology studies (127). In a recent SPL structure solved by Benjdia et al., the enzyme contains a 5R-SPSIDE, further confirming that the 5R-isomer is the SPL substrate (34). Moreover, NMR spectroscopy confirmed that the SPTIDE contains a 5R chiral center (8); this chirality was further established by a dinucleotide SP structure containing a formacetal linker, which is also an SPL substrate (128). After incorporating 5R-SPSIDE and 5S-SPSIDE into a dodecamer sequence, respectively, via solid phase DNA synthesis, subsequent structural studies found that the 5R-SPSIDE fits in the topology of the right-handed helix well, while the 5S-SPSIDE results in a severe strand distortion (129). Therefore, although the thymidine photoreaction in ice produces both 5R- and 5S-SPSIDE, only the 5R-isomer can be formed in the spore genomic DNA.
SP formation in DNA or thymidine films
Besides photoreaction in ice, SP may also be formed in dry DNA films, where DNA molecules exist as fibers. The A ⇔ B conformational change in DNA fibers as a function of DNA hydration level is well documented (130, 131). Thus in the presence of sodium ions, DNA fibers adopt the A-form at 40−88% relative humidity (RH) and the B-form at >88% RH (130). As shown by Rahn et al., when irradiating an E. coli genomic DNA film prepared via evaporation of an aqueous DNA solution on quartz plates under high RH, CPDs were the dominant DNA photoproduct (132). As RH decreased, the DNA underwent conformational changes to a potential A-form, formation of CPDs was suppressed and formation of SP increased dramatically (132). It was also found that at a very low RH level, DNA likely adopts a denatured conformation leading to suppression of formation of both thymine photoproducts, although compared with SP, the reduction in CPD formation was much larger (132).
SP formation in dry films was also observed with short oligonucleotides, such as a 35-mer duplex (133). Air-drying of the aqueous solution of the 35-mer on Saran wrap resulted in a single DNA layer. UV irradiation of this layer at 10% RH produced SP-containing strands; formation of other thymine dimers was not investigated. By preparing the film via freeze-drying an aqueous buffer containing calf thymus DNA, Douki et al. found that irradiation of the dry film produced CPDs at a yield 5 – 10-fold higher than that of SP (63, 134). The SP yield is much lower than that found by Rahn et al. in the earlier work (132), suggesting that the method for DNA film preparation may play a major role in determining the thymine stacking pattern and subsequent SP formation upon UV irradiation. Inclusion of CaDPA in the dry film increased the SP yield by ~ 15-fold, but only increased the CPD yield by ~ 2-fold. As a consequence, comparable amounts of CPDs and SPs were generated.
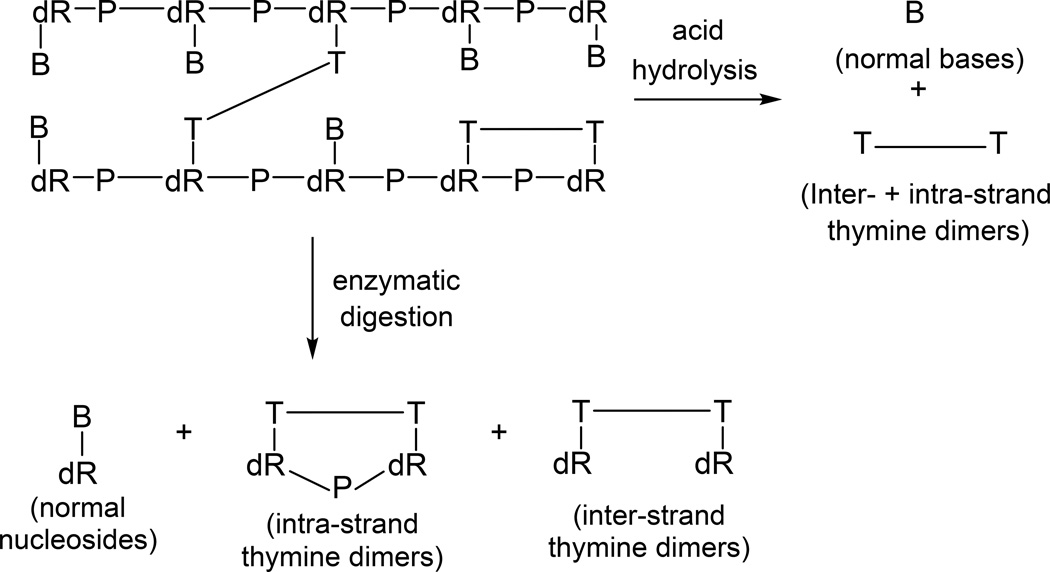
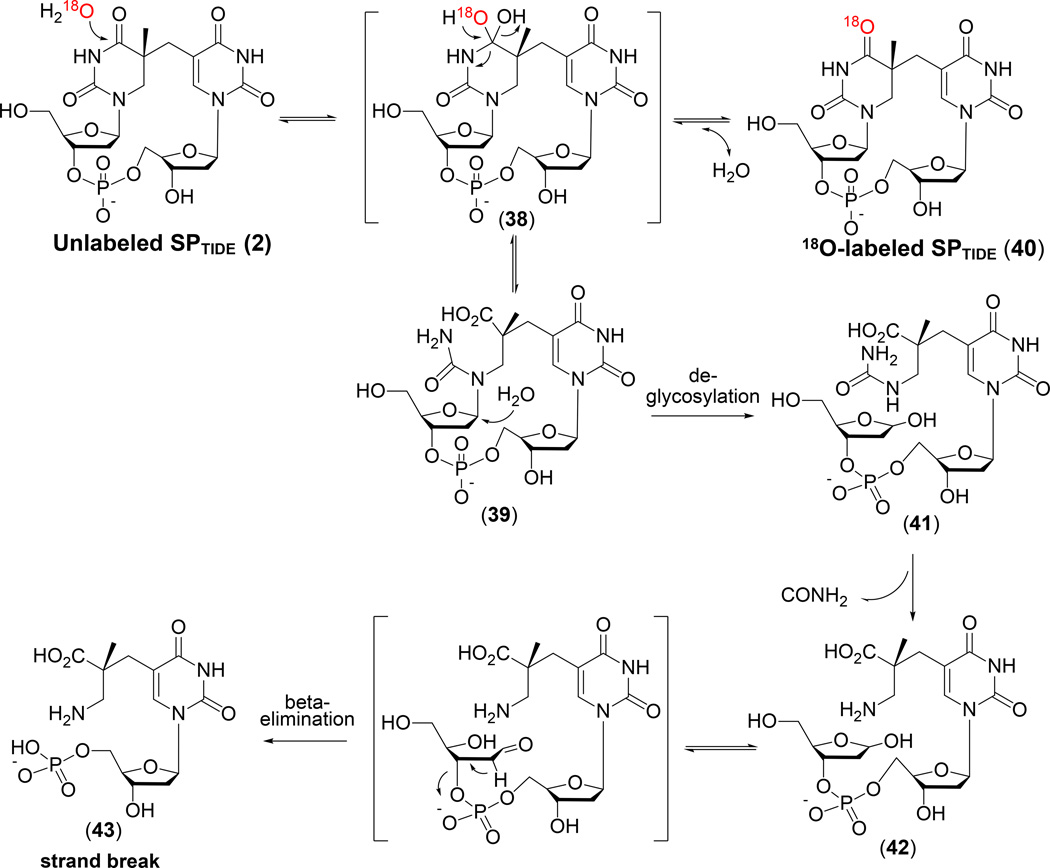
Interestingly, a careful analysis of UVC irradiated calf thymus DNA as a dry film indicated that 30% of thymine dimers (mainly CPD and SP) are formed between thymine residues located on different DNA strands, with the major inter-strand photoproduct being SP (135). The presence of inter-strand dimers was revealed after DNA digestion using a combination of nuclease P1, alkaline phosphatase and phosphodiesterases I and II, with product analysis via HPLC-MS/MS. As shown in Figure 6, the exonucleases do not cleave the phosphodiester bonds in the intra-strand dimer, but readily cleave these bonds in inter-strand dimers. However, if the DNA is hydrolyzed with strong acids at elevated temperatures, such as was used in previous photoproduct analysis on UV irradiated spores, both inter- and intra-strand dimers are converted to the same SPBASE and become indistinguishable. Some DNA in dry films may also exist in denatured states, especially at low RH (130, 131), and the formation of inter-strand SPs may come from denatured DNA. However, DNA in spores is saturated with SASPs, which likely stiffen the duplex DNA and prevent the occurrence of photoreactions other than the formation of intra-strand SP. Notably, yields of inter-strand SP were < 1% of intra-strand SP in UVC-irradiated hydrated spores or a dry or hydrated SspC-DNA complex (5).
Figure 6.
Enzymatic and acid hydrolysis of duplex DNA containing inter- and intra-strand photoproducts (135). B, normal base; dR, 2′-deoxyribose; P, phosphate. The phosphate in intra-strand dimers cannot be cleaved by enzymatic digestion while all phosphates are hydrolyzed by acid, providing an assay to distinguish these two types of DNA photoproducts.
Besides thymidines in duplex DNA, dry films of monomeric thymidine can also give rise to SP photochemistry (5). The thymidine can be dissolved in methanol or ethanol (135), since solvent evaporation gives much better films than are prepared via lyophilization, perhaps because better thymidine stacking interactions are obtained with solvent evaporated films, resulting in much cleaner SP formation. In photoreactions conducted in frozen aqueous solution (135), SP accounts for ~38% of the thymidine dimers formed, with CPDs being the dominant products. In photoreactions with a dry film prepared via lyophilization, the percentage of SP in all thymine dimers was lower at ~ 27%. In contrast, with a thymidine film prepared via ethanol evaporation, SP is the dominant product, accounting for ~ 80% of the dimers formed (135).
As described above, the thymidine photoreaction generates a pair of SPSIDE diastereoisomers, with the C5 chiral center adopting an R or S configuration. With thymidine in ice or in films prepared via lyophilization, roughly equal amounts of the 5R- and 5S- SPSIDE isomers are produced (124). In contrast, using films prepared via methanol or ethanol evaporation, the 5S-SPSIDE isomer becomes the dominant product and the 5R-isomer is a very minor species (~ 20:1 ratio) (135). Such a product distribution pattern indicates that the thymidine stacking conformation is different in these two types of films, resulting in different SPSIDE diastereoisomers upon UV irradiation.
SP formation in aqueous solutions
Successful SP photochemistry requires DNA to possess a reduced hydration level, and such a condition is difficult to fulfill in aqueous solution. Consequently, UV irradiation of DNA in common buffers generates no detectable SP. However, SP photochemistry in aqueous solution can be achieved by binding α/β-type SASP to DNA to achieve the DNA conformation required for SP photochemistry. At moderate or low concentrations of salt and with an ~ 3:1 weight ratio of SASPs/DNA, a DNA/SASP complex can be generated that mimics the DNA environment in a germinated spore when the spore core has been rehydrated and CaDPA has been released (92). In this nucleoprotein complex, the DNA is still in an A-like conformation (89). Consequently, UVC irradiation of this SASP-DNA complex at pH 7.0 in buffer produces SP as the dominant product, with a ratio between SP and CPD formed that is close to 5:1. Such a reaction pattern is close to that found in dormant spores, although the yield of SP as a function of UV fluence is more than an order of magnitude lower, probably due to the absence of CaDPA. When the nucleoprotein complex was irradiated with UVC in the dry state, CPD formation was further suppressed and a SP/CPD ratio of > 40/1 was observed (92).
Another means to obtain A-like DNA conformation supporting SP photochemistry is to add ethanol to an aqueous solution, since DNA adopts an A-form in 80% ethanol (136, 137). Early studies by Patrick and Gray indicated that even in 80% ethanol, only a tiny amount of SP was produced with CPDs being the major thymine dimer formed (109). More recent analysis of DNA’s photoreaction in 80% ethanol by Douki et al. using the HPLC-MS/MS assay found that the intra-strand SP accounted for ~30% of all thymine dimers formed, as well as other dimers including three CPD isomers adopting cis-syn, cis-anti and trans-anti configurations (135). Inter-strand SP was also obtained at a yield of ~ 10% of that for the intra-strand SP. These observations indicate the presence of a wide-range of DNA conformations in the ethanol solution, highlighting the importance of the precise state of the spore core and/or binding interactions between SASPs and DNA to ensure that intra-strand SP photochemistry is the dominant DNA photoreaction in spores.
REACTION MECHANISM FOR SP FORMATION
The progress made on in vivo and in vitro SP studies has paved the way for detailed mechanistic investigations of SP photochemistry. SP formation only involves two thymine residues, and it may not be practical to employ large genomic or plasmid DNAs which contain thousands of nucleotides for SP mechanistic studies in a reaction that features atom movements and C-C/C-H bond formation. Rather, the ideal system for such studies seems likely to be thymidine monomers or the dinucleotide TpT.
Earlier deuterium labeling studies
After determining the structure of SP by NMR spectroscopy, Varghese et al. rationalized that SP is likely formed through a consecutive mechanism via combination of a 5-α-thyminyl and a 5,6-dihydrothymin-5-yl radical (7, 123). To verify this intriguing hypothesis, it is important to know how these radicals are generated. Therefore, Cadet and co-workers utilized a clever approach employing a labeled d3-thymidine residue containing a −CD3 moiety and followed the deuterium during the SP photoreaction. After conducting the thymidine photoreaction on ice to generate SP, they purified the SPSIDE product probably as a mixture of the 5S and 5R diastereoisomers. Analyses of the SPSIDE structure via NMR spectroscopy found that only two thirds of the generated SPs possessed a deuterium on the C6 carbon. Although this finding indicated that a deuterium from the −CD3 moiety migrated to the C6 position during SP formation, such a conclusion was undermined by the incomplete deuterium incorporation observed, leading to a conclusion that the origin of the two H6 atoms was unclear (138). Based on these data, a concerted reaction mechanism was proposed, where formation of the C6-H and C5-C bonds in SP occur simultaneously, possibly via a four-member ring transition state <Figure 7A>, and the 5-α-thyminyl and 5,6-dihydrothymin-5-yl radicals are not involved as reaction intermediates (138, 40).
Figure 7.
(A) The concerted mechanism for SP formation (40, 138). The breakage of the C-H bond in the methyl moiety and the formation of the two new bonds in SP are suggested to occur simultaneously, potentially via a four member-ring transition state. (B) The consecutive mechanism for SP formation (7, 139). The C6 radical abstracts an H-atom from the methyl group, generating a 5-α-thyminyl radical, which combines with the 5,6-dihydrothymin-5-yl radical to generate SP. The right-handed helical structure of duplex DNA ensures that only route (i) is possible in spores, resulting in the 5R-SP. In a solid state photoreaction using monomeric thymidine, route (ii) also occurs, resulting in dinucleoside 5S-SP.
In the dinucleotide TpT
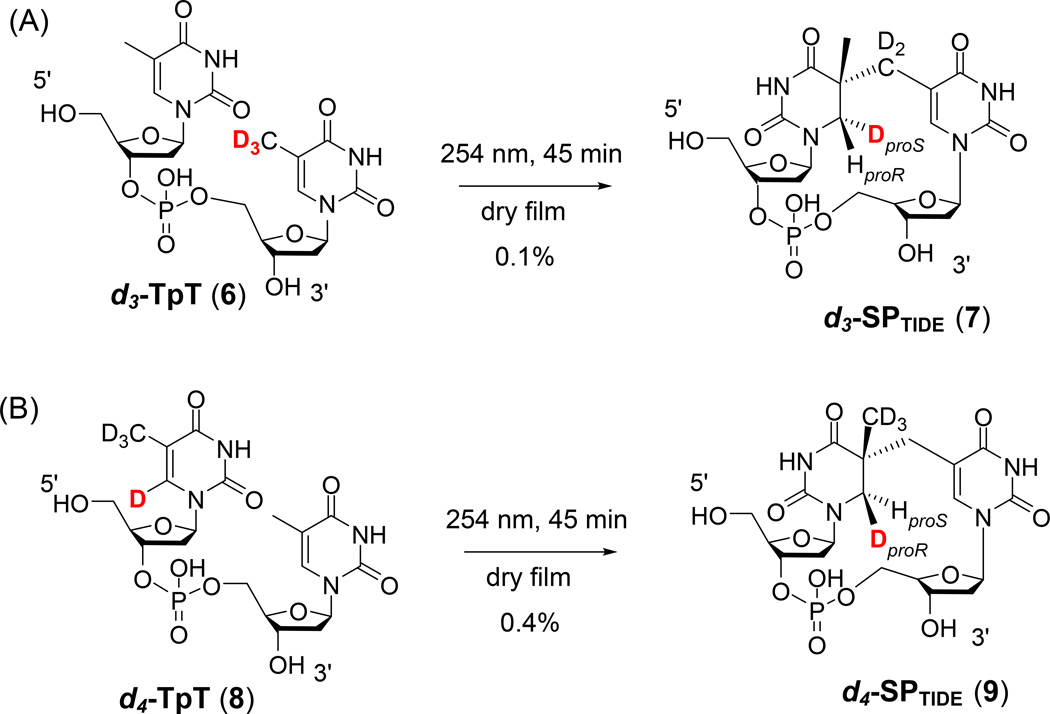
To further investigate the mechanism of SP formation, Lin et al. repeated the deuterium labeling experiments in a dinucleotide context (139). Two dinucleotides were used in their studies: a d3-TpT (6) containing a –CD3 moiety at the 3′-thymine and a d4-TpT (8) with all four carbon-based hydrogen atoms on the 5′-thymine base being replaced by deuteriums <Figure 8>. UVC irradiation of dry films prepared by methanol evaporation led to deuterium-labeled SPTIDEs 7 and 9, which were purified by HPLC and analyzed by NMR spectroscopy. Unlike the previous thymidine labeling studies, LC-MS analysis of the resulting SPTIDEs did not reveal any deuterium loss in either reaction. The NMR data reveal that in the d3-TpT photoreaction, a deuterium is stoichiometrically transferred from the 3′-CD3 group of TpT to the H6proS position in the d3-SPTIDE 7. In the d4-TpT photoreaction, a protium transfer to the H6proS was observed, with the D6 at the 5′-thymine occupying the H6proR position of the resulting d4-SPTIDE 9. Such observations indicate that UV irradiation likely excites the 5'-C5=C6 bond into a diradical, which then abstracts an H atom from the methyl moiety of the adjacent 3'-thymine residue, resulting in 5-α-thyminyl and 5,6-dihydrothymin-5-yl radicals which then combine to yield SP <route (i), Figure 7B>. Moreover, the protium transfer in the d4-TpT photoreaction was found to be 3.5 times faster than the deuterium transfer in the d3-TpT-mediated SP formation. Such a primary deuterium isotope effect indicates that the H-atom abstraction step before the two radical intermediates are formed is involved in the rate-determining process.
Figure 8.
H-atom transfer during SP formation as revealed by the formation of deuterated dinucleotide SPs via solid phase photoreaction using selectively labeled dinucleotide TpTs (139).
Re-examination of earlier deuterium labeling studies
Why does the same labeling strategy result in the two different outcomes described above? To determine the reasons for this discrepancy, the Li and the Cadet groups re-investigated the thymidine photoreaction in both ice and a dry film (124). Their data exclude the possibility that deuterium was washed away by exchanging with protium from the environment. Instead, they found that deuterium is largely discriminated against during SPSIDE formation in ice, as in the formation of 5R-SPSIDE, a 17-fold deuterium discrimination over protium was observed. Generally, the commercially available deuterium reagents are ~ 98% pure and the ~2% protiated impurities usually do not cause significant problems. However, with the 17-fold discrimination against deuterium exhibited in the SP photochemistry, the 2% protiated thymidine residues in d3-thymidine would become 34% in the SPSIDE generated. Such an unusually large protium enrichment offers a reasonable explanation of the previous observation that only two thirds of SPs formed contained a deuterium at the C6 position.
In addition, when SP photochemistry is examined in dry films prepared by methanol evaporation, the deuterium discrimination still exists, albeit to a much smaller extent. Analysis of the resulting 5R- and 5S-SPSIDEs formed in d3-thymidine photoreaction by NMR spectroscopy confirms that a deuterium is transferred exclusively from the methyl group to the H6proR position in 5R-SPSIDE, but to the H6proS position in 5S-SPSIDE. Therefore, observations from both dinucleotide TpT and thymidine photoreactions support the same reaction mechanism <Figure 7B>. It is also worth mentioning that to the best of our knowledge, the SP photoreaction is the only example known to date in which a photo-excited nucleotide residue exhibits a diradical feature.
Although the reaction mechanism for SP formation is largely established as noted above, some questions still remain. One of them concerns the kinetic isotope effects (KIEs) observed in SP formation. It was shown that the 5R-SPTIDE formation in the dinucleotide TpT photoreaction in a dry film exhibits a KIE of 3.5, the KIE becomes 17 ± 1.5 in thymidine photoreaction in ice and 6.5 ± 1.5 in thymidine photoreaction in a dry film (124). Moreover, although the 5R- and 5S-SPSIDEs are generated by the same mechanism, the KIEs exhibited by 5S-SPSIDE photochemistry are different from those found in 5R-SPSIDE formation (124). Currently, these KIEs are tentatively ascribed to the primary isotope effect during the H-atom abstraction process, with the large effects exhibited in the thymidine photoreactions indicating that hydrogen atom tunneling effects may be involved in SP photochemistry. As the tunneling effect is affected by the distance between the two reacting moieties (140), the different KIEs observed may be ascribed to the various distances resulting from the different stacking interactions in free thymidine or in the dinucleotide TpT (124). However, this hypothesis needs further experimental examination.
Reaction in thymidine microcrystals
The remaining KIE issue highlights the importance of DNA conformation controlled by nucleobase stacking interactions on the outcome of DNA photoreaction. As pointed out by Schreier et al., the timescale for excited thymine residues to dimerize is in the range of picoseconds (113), and it has been generally accepted that the original DNA conformation determines the DNA photoproducts (113, 141–143). In genomic DNA, the millions of nucleobases may adopt an equally large number of stacking conformations at any given time, with the vast majority of these conformations being unreactive. Such a problem is even more significant in SP photoreaction in vitro which often occurs in the solid state where conformational changes due to thermal motion are restricted. As a consequence, most in vitro SP photoreactions exhibit extremely low yields, typically lower than 1% after a prolonged UVC irradiation. On the other hand, in some dry film reactions, in particular with film prepared by alcohol evaporation, 5S-SPSIDE is the dominant product produced, suggesting that thymidines adopt a nearly homogeneous conformation in films prepared by alcohol evaporation. However, the yield of SPSIDE is still lower than 1% (124, 135), likely because UVC light can only penetrate several nanometers into the film. Therefore, only the thymidine residues close to the surface can be excited sufficiently to react.
To circumvent the problem of UVC penetration of dry films, Jian et al. examined SPSIDE formation in thymidine microcrystals, where the thymidine residues adopt a homogeneous stacking conformation across the whole crystal lattice <Figure 9A>. To increase the reactive surface area, they suspended the thymidine microcrystals in an organic solvent such as methyl t-butyl ether and irradiated the suspension by unfiltered UVB light centered at 302 nm under vigorous stirring (144). The homogeneous thymidine stacking conformation in the crystal lattice inhibits the formation of other thymine dimers, resulting in 5S-SPSIDE as the dominant photoproduct. After a 32-hr reaction, the yield of 5S-SPSIDE was found to be ~ 85%. Because thymidine residues likely take a random order to dimerize, the reaction yield relies on the dimerization order. If every adjacent thymidine pair takes turns to dimerize, a 100% conversion to SP is expected. In contrast, if dimerization occurs at the first two of every three thymidine residues, the third thymidine will be left between two formed SPSIDEs and remain intact, resulting in the lowest possible yield at 67%. The actual yield of 5S-SPSIDE is expected to fall in between the maximum and minimum yields, with 83%, the average, the most probable choice. Therefore, the ~85% 5S-SPSIDE formation with 302 nm UV irradiation represents the optimal yield in the di-molecular dimerization reaction.
Figure 9.
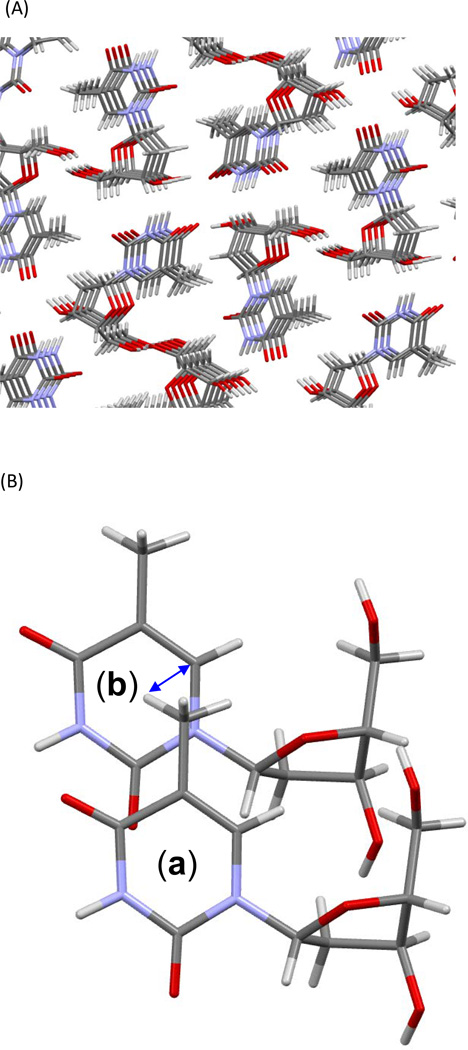
(A) Molecular packing in the thymidine single crystal (145). The distance between two thymidine rings is ~ 3.1 Å, close to the 3.36 Å average rise found in the SASP-oligo(dG)•oligo(dC) nucleoprotein (106). (B) The shortest distance between an H atom in –CH3 and the C6 of another thymine is ~ 3.2 Å (blue arrow), which is close to the 3.4 Å found in the molecular simulation (106), and supports the role of the key H-abstraction step in initiating SP formation (124, 139).
Analysis of the thymidine crystal structure solved by Young et al. (145) also reveals that the methyl group of one thymine residue <a, Figure 9B> is located right above the other thymine ring <b, Figure 9B>, and the shortest distance between an H-atom of the –CH3 moiety and the C6 of the adjacent thymidine is only 3.178Å. As indicated by the SP mechanism in Figure 7B, the C6 radical formed after photo-excitation of the C5=C6 bond is projected to abstract an H-atom from the methyl moiety. Such an abstraction is highly feasible in the thymidine crystal as shown by the short distance between these moieties. In addition, the C5=C6 bonds on adjacent thymidines are 4.86 Å away from each other in the crystal lattice, making it very difficult for these bonds to move closer to allow for CPD formation. The C5=C6 bond is even further from the C4=O moiety of the adjacent thymidine, and thus 6-4PP formation should also be very unfavorable.
Triplet state photochemistry
The experimental data to date strongly support a reaction mechanism for SP formation in which H-atom abstraction by the excited C5=C6 bond initiates SP photochemistry. As both the singlet and the triplet excited states can be involved in thymine photoreaction, the remaining question is which excited state plays a major role in SP photochemistry.
As mentioned above, a major role of CaDPA in spores is to serve as a photosensitizer. Analysis of SP formation in the dry state at the presence of CaDPA reveals that this compound increases the yields of SP and cis-syn CPDs formed between two thymine residues or between a cytosine and a thymine, while generation of 6-4PP and CPD between two cytosine residues is relatively unaffected (63). These observations indicate that formation of both CPD and SP involves the thymine triplet excited state, which likely possesses an energy level lower than the triplet excited state of CaDPA so that the energy transfer from CaDPA to thymine can readily occur. Moreover, cytosine possesses the highest triplet excited state energy among the four nucleobases (83). This energy state may be higher than that of CaDPA, resulting in the suppression of cytosine dimer formation in UV radiated spores.
The involvement of the thymine triplet excited state in SP formation is further supported by results of a dry state thymidine photoreaction in the presence of the pyridopsoralen derivatives, 7-methylpyrido[3,4-c]psoralen (MePyPs), pyrido[4,3-c]psoralen (H-PyPs), and 7- methylpyrido[4,3-c]psoralen (2N-MePyPs), under 365 nm UVA irradiation (146). These reactions produce four CPD isomers possessing the cis-syn, trans-syn, cis-anti, and trans-anti configurations and the 5R- and 5S-SPSIDE isomers. Yields of these dimers depend on the nature of the photosensitizer employed, with H-PyPs being the most efficient, followed by MePyPs and 2N-MePyPs. CPD and SPSIDE formation was not observed when photosensitizers, such as 3-carbethoxypsoralen, 5-methoxypsoralen, and 8-methoxypsoralen, which possess a triplet excited state energy lower than that of the thymine, were used.
The involvement of the thymine triplet excited state in SP formation was further supported by a recent DFT calculation by Du et al. (82). This calculation shows that any reaction route involving the singlet excited state S1, the triplet excited state T1, or the singlet ground state S0 alone is energetically unfavorable. The favorable reaction route involves a triplet excited state T1 which enables H-atom abstraction from the thymine methyl group as the rate-limiting process to form the 5-α-thyminyl and 5,6-dihydrothymin-5-yl radicals. The resulting diradical intermediate at the triplet state is very unstable, and interacts with the singlet ground state to undergo triplet-singlet surface crossing to reach the singlet ground state S0 before radical combination occurs, yielding SP. Therefore, the reaction features a consecutive reaction mechanism, as originally proposed by Varghese (7). Moreover, the calculation also shows that with the concerted mechanism shown in Figure 7A, no triplet excited states can be located. Singlet excited states may be involved, but then the reaction has to go over a hefty energy barrier of 70.4 kcal/mol, which is in sharp contrast to the barrier of 14.2 kcal/mol in the consecutive mechanism involving the triplet excited state. Taken together, in the presence of CaDPA, the triplet excited state is likely to be solely responsible for SP formation in bacterial spores.
PROGRESS ON SP SYNTHETIC CHEMISTRY
As discussed above, yields of SP in DNA photoreactions in vitro are generally very low. In most cases, not only is SP always a minor product, but other thymine dimers are also formed. It is also very difficult to separate an SP-containing oligonucleotide from oligonucleotides containing other lesions by HPLC. Therefore, unlike preparation of oligonucleotides containing CPD or 6-4PP via UV irradiation in aqueous solution, it is impractical to utilize photochemistry to prepare large amounts of homogeneous SP-containing oligonucleotides. The latter difficulty may explain the lack of studies to reveal the SP repair efficiency by the NER pathway and the mutagenic effects of SP in germinated spores. Consequently, an effective method for SP incorporation into an oligonucleotide is essential in order to understand intimate details of SP photochemistry and photobiology.
General strategy for SP synthesis
Synthesis of the SPBASE was achieved by Bergstrom et al. shortly after the chemical structure of SP was determined (147). Begley and co-workers later synthesized SPBASE derivatives via different strategies (21, 148). Two routes were adopted in these studies: connection of two pyrimidine base derivatives or building the 5,6-dihydropyrimidine skeleton onto a pre-existing pyrimidine base. The first route developed in these early studies provided the basis for later syntheses of SPSIDE and SPTIDE.
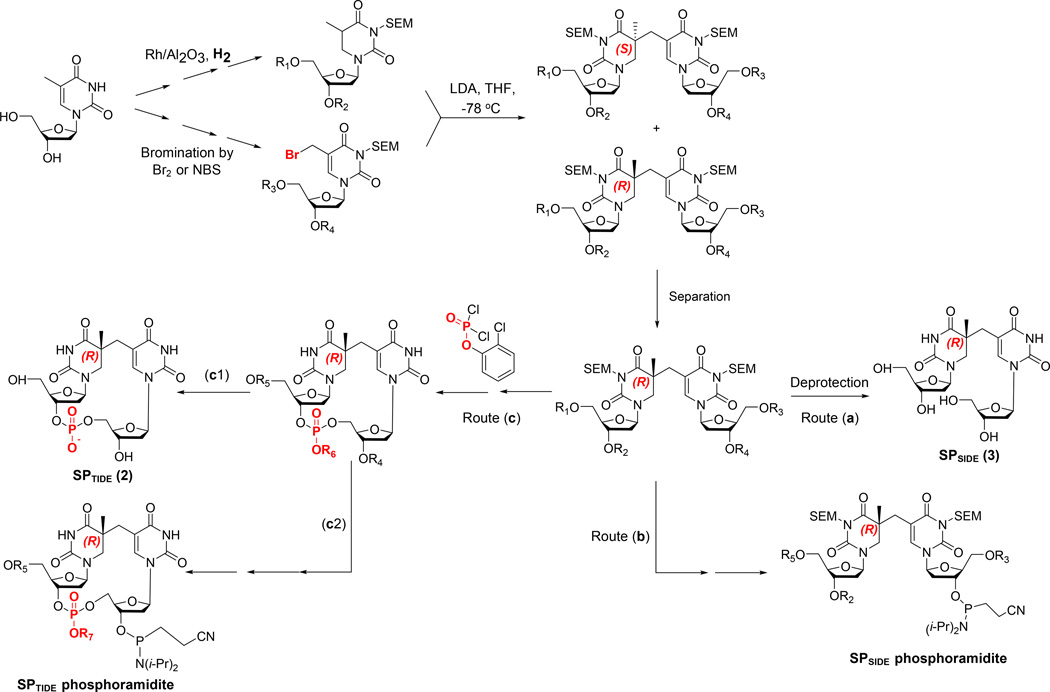
The SPSIDE and SPTIDE as well as their phosphoramidites were prepared via similar strategies. The scheme used by most of these syntheses is summarized in <Figure 10>, which features coupling the enolate of dihydrothymidine with bromomethyl deoxyuridine to give a pair of dinucleoside SP diastereomers with both N3 atoms protected by [2-(trimethylsilyl) ethoxy]methyl acetal (SEM) and the hydroxyl groups on deoxyriboses being protected by other reagents (126, 127, 149, 150). Separation of the protected R and S-diastereomers by flash chromatography followed by de-protection of the resulting 5R-isomer readily yields the 5R-SPSIDE <route (a), Figure 10>, which proved useful in the elucidation of the mechanism of the SP repair enzyme SPL (24, 127). Alternatively, selective deprotection of the hydroxyl groups on the deoxyribose allows the functionalization of the 3'-OH at the 3'-side of the dinucleoside into the 2-cyanoethyl-N,N-diisopropyl phosphoramidite, yielding the SPSIDE phosphoramidite as shown in route (b) (129). Moreover, by using 2-chlorophenyl dichlorophosphate to react with the selectively de-protected dinucleoside SP, the phosphodiester moiety can be inserted producing the protected SPTIDE <route (c), Figure 10>. SPTIDE and SPTIDE phosphoramidites can be subsequently prepared from this protected SPTIDE precursor.
Figure 10.
Summary of a synthetic strategy commonly adopted for the syntheses of dinucleoside and dinucleotide SP as well as their phosphoramidite. Here, both N3 atoms on the thymine ring are protected by [2-(trimethylsilyl) ethoxy]methyl acetal (SEM).
Synthesis of dinucleoside and dinucleotide SP
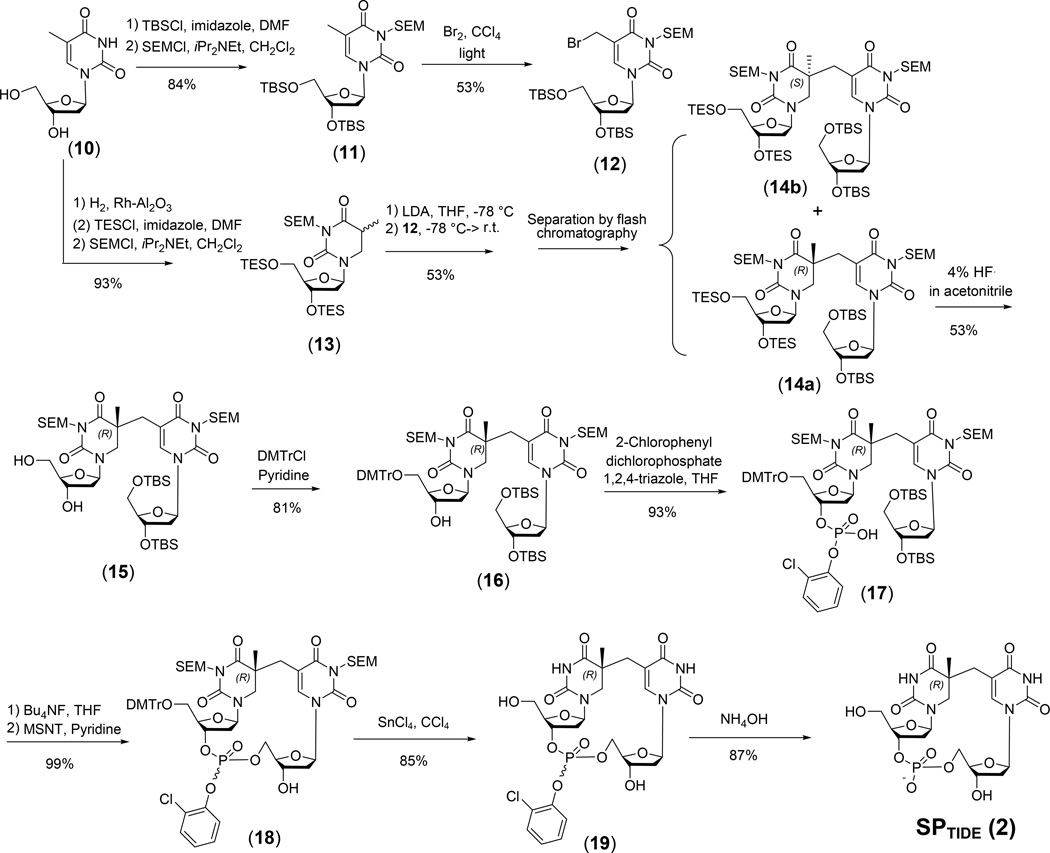
The first SPTIDE was synthesized by Kim et al. (150), which also provided a general strategy for later SPTIDE syntheses. In Kim’s approach, the 5R- and 5S-SPs were not separated until the very last step. However, most recent syntheses separated these isomers via flash chromatography right after the coupling step (127, 139, 149). A reaction scheme for SPTIDE synthesis is shown in Figure 11 which is modified from Kim’s procedure to reflect the latest trend. The N3 atoms on the thymine ring are protected by SEM. Both the 3'-OH and 5'-OH at the bromomethyl deoxyuridine were protected by tert-butyldimethylsilyl ether (TBS), while both hydroxyl groups were protected by triethylsilyl ether (TES) at the dihydrothymidine before they are coupled to form a pair of protected dinucleoside SP diastereomers 14a and 14b. Only the 5R isomer 14a was indicated to be formed in vivo due to the right-handed helical structure of DNA, which prohibits the formation of the 5S-SP (150). After column chromatography to separate these two isomers (127, 149), deprotection of the resulting 14a readily affords the 5R-SPSIDE as indicated by the route (a) in Figure 10 (127). Alternatively, following the route (c1) in Figure 10, both TES groups of 14a was removed before the 5'-OH was protected by dimethoxytrityl (DMTr) (150). The 3'-OH was then allowed to react with 2-chlorophenyl dichlorophosphate to introduce the phosphodiester moiety into the protected dinucleotide SP 17. Deprotection of the resulting compound yields SPTIDE 2.
Figure 11.
Synthesis of the SPTIDE starting from thymidine, which was originally reported by Kim and Begley (150) and later modified by other groups (139, 149). The 5S- and 5R-SP diastereomers were not separated until the last step of Kim’s synthesis, while most recent syntheses separated these isomers via flash chromatography right after the coupling step. The synthesis shown here was thus modified accordingly.
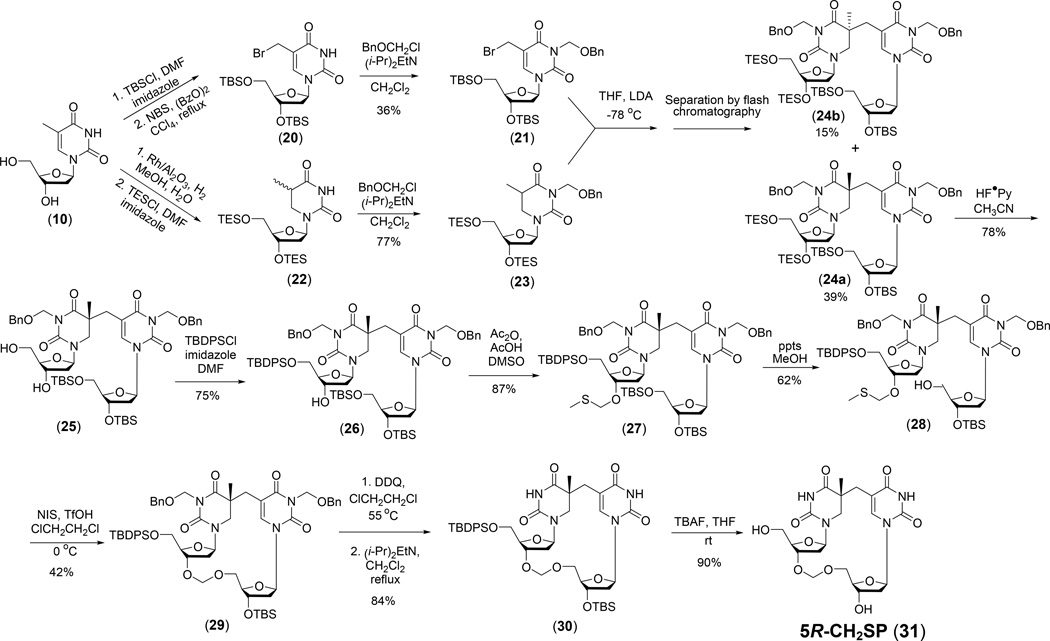
The synthesis of SPTIDE analog 5R-CH2SP (31) containing a formacetal linker adopted by Lin et al. is shown in Figure 12. A formacetal is the simplest and smallest non-chiral isostere for a phosphate moiety and has proved very useful in oligonucleotide biochemical studies (151–156). Although the general strategy used for the 5R-CH2SP synthesis was similar to the SPTIDE synthesis shown in Figure 11, the key protection/de-protection strategies were different due to the instability of the formacetal linkage (128). Consequently, SEM was no longer used as the protecting reagent for the thymine N3 atom as the SnCl4-mediated SEM de-protection also detaches the formacetal linker. The benzyloxymethyl group (CH2OBn) was therefore chosen for N3 protection, which increased the diastereoselectivity of the 5R (24a) over the 5S isomer (24b) to 2.6 : 1, in contrast to the non-selective 1:1 mixture (13a vs 13b) obtained in the SPTIDE synthesis (150). After removing the CH2OBn moiety via oxidation using 2,3-dichloro-5,6-dicyano-1,4-benzoquinone (DDQ) followed by deprotection, the 5R-CH2SP was readily obtained.
Figure 12.
Synthesis of SPTIDE analog 5R-CH2SP (30) containing a formacetal linker (128).
Synthesis of dinucleoside SP phosphoramidite
The synthesis described above provides the foundation for the synthesis of SPSIDE phosphoramidite, making its incorporation into an oligonucleotide possible. As shown in Figure 11, the synthesis of SPTIDE is lengthy and tedious, and preparation of large amounts of the SPTIDE phosphoramidite to enable the preparation of SP-containing oligonucleotides is challenging. Therefore, although 5R-SPSIDE 3 is not considered to be truly biologically relevant, it provides a proof-of-concept example for the preparation of SP phosphoramidite. Moreover, the SPSIDE-containing oligonucleotide is a substrate of SPL, the repair of which releases two smaller oligonucleotide fragments, facilitating the examination of the SPL activity (26, 157).
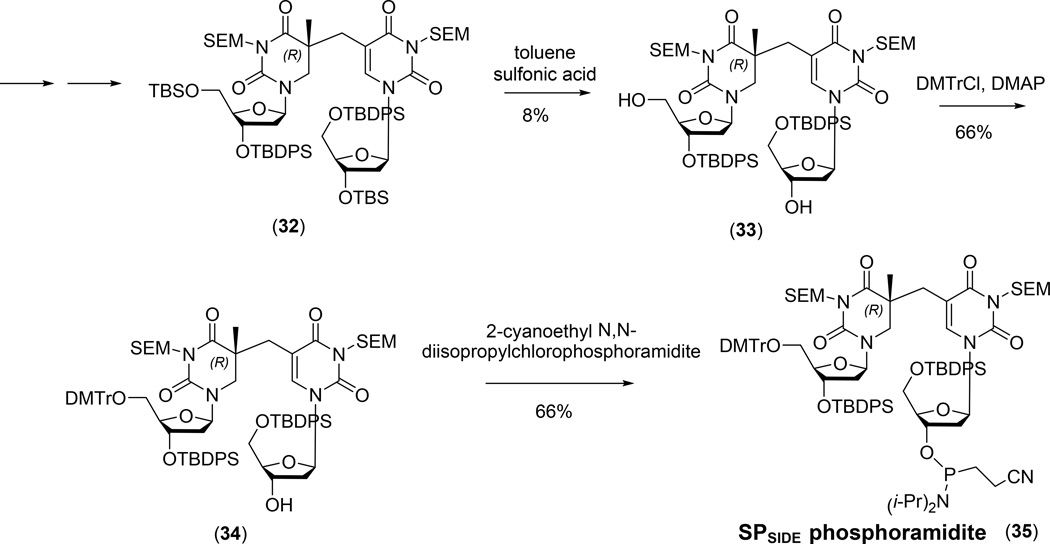
The strategy for the synthesis of SPSIDE phosphoramidite has been summarized in route (b), Figure 10. Compared with SPTIDE synthesis, the major difference in the preparation of SPSIDE phosphoramidite 35 is the selective protection of the hydroxyl groups at the 2'-deoxyriboses (129). The 3'-OH group of the bromomethyl deoxyuridine and the 5'-OH group of the dihydrothymidine were protected by TBS, while the other two hydroxyl groups were protected by tert-butyldiphenylsilyl ether (TBDPS) before the protected dimer 32 was allowed to form <Figure 13>. After removal of the TBS moieties, the 5'-OH group of the dihydrothymidine was protected by DMTr before the phosphoramidite was introduced on the 3'-OH group to give the dinucleoside SP phosphoramidite 35. This phosphoramidite readily supports solid phase DNA synthesis for SPSIDE incorporation, although the yield for the SP coupling step was probably low as reflected by the overall 15% yield in the synthesis of a dodecamer oligonucleotide (129).
Figure 13.
Key steps for the synthesis of SPSIDE phosphoramidite from the protected SPSIDE (129).
Synthesis of dinucleotide SP phosphoramidite
The CPD TpT (158–165) and 6-4PP TpT (166) phosphoramidites are available, enabling their incorporation into oligonucleotides via standard solid phase DNA synthesis. These phosphoramidites were generated via hybrid approaches where the corresponding dimers were first produced by photochemistry using partially protected dinucleotide TpT before the phosphoramidite moiety was introduced on the 3′-OH group at the 3′-end of the dimer via organic synthesis. Such an approach, however, proves futile in preparing the SPTIDE phosphoramidite, since UV irradiation of the unprotected dinucleotide TpT in the solid state generates SP in ~1% yield (139). In addition, once the phosphodiester moiety is protected by esterization with either –CH3 or –CH2CH2CN, the resulting dinucleotide no longer supports SP formation (149), suggesting that the negative charge carried by the phosphodiester linker is essential for the two thymine residues to adopt the “right” stacking conformation to enable SP photochemistry. Therefore, the hybrid strategy cannot be applied to the preparation of SPTIDE phosphoramidite.
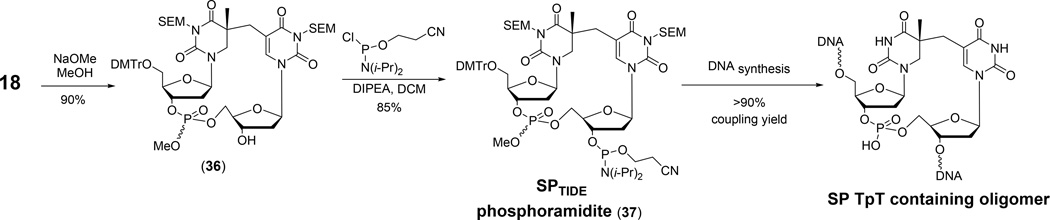
To prepare SPTIDE phosphoramidite in a relatively large scale, Jian et al. built from the synthesis of the dinucleotide SP (149), as shown in route (c2), Figure 10. Importantly, they found that the bulky 2-chlorophenyl moiety as the phosphate protecting group used in the SPTIDEsynthesis may create steric hindrance, preventing an efficient coupling reaction from occurring during the solid phase DNA synthesis. By exchanging the 2-chlorophenyl for a much small methyl moiety, the resulting phosphoramidite 37 improved the coupling efficiency from ~15% to > 90% <Figure 14>. This high coupling efficiency enables the large scale preparation of SPTIDE-containing oligonucleotides.
Figure 14.
Synthesis of dinucleotide SP phosphoramidite from the penultimate intermediate during the synthesis of SPTIDE shown in Figure 11 (149).
SP STRUCTURAL STUDIES
As noted above, progress on SP synthetic chemistry and photochemistry has now made SP photobiology studies possible. These studies have led to breakthroughs in analyses of SP structure, offering much-needed insight into the impact of SP on genomic DNA.
NMR Structure of dinucleotide SP
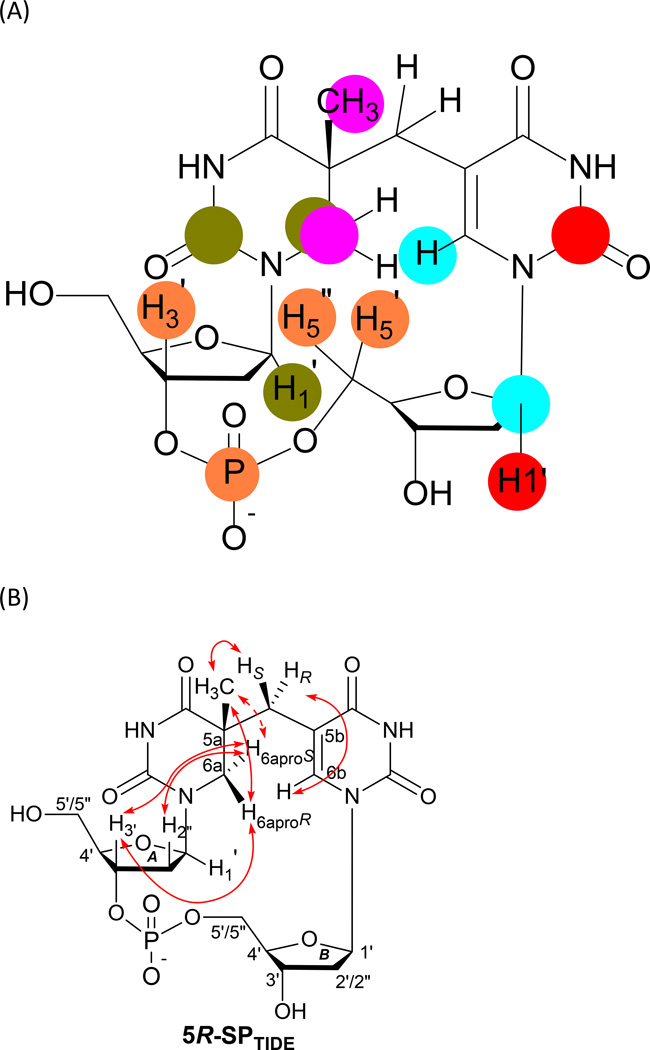
Starting from the dinucleotide TpT and conducting DNA photochemistry in dry films formed via lyophilization, Mantel et al. purified sufficient SPTIDE to elucidate its solution structure via 2D-NMR spectroscopy coupled with DFT calculations (8). As shown in Figure 15A, using 1H-13C and 1H-31P Heteronuclear Multiple-Bond Correlation (HMBC) spectroscopy, the through-bond correlations in SP TpT were determined, revealing the enchainment of different chemical groups (thymine, deoxyribose, and phosphate). The results unambiguously demonstrate that SP is formed via addition of the 3-methyl moiety to the C5=C6 bond at the 5-thymine residue, excluding the involvement of the 5-methyl group in SP photochemistry. In addition, after analyzing the through-space 1H-1H interaction via Rotating frame Overhauser Effect SpectroscopY (ROESY) experiments, it was found that H6aproS displays a weaker coupling to the methyl group at the 5′-dihydrothymine moiety than does H6aproR, indicating that the C6a position is located above the sugar ring <Figure 15B>. In addition, the strong interaction between H6aproR and the methyl group unequivocally proves that the configuration of the C5a carbon is R because if an S configuration is encountered, the proton H6aproR could not display strong coupling signals both to H3′A and to protons of the methyl group. Such an observation is further supported by DFT calculations that found ten favorable correlations for the R isomer but only seven for the S isomer. Taken together, these studies strongly indicate that the C5a chiral center in SP adopts an R configuration, agreeing with the assumption proposed by Kim et al. based on the right-handed DNA helical structure (150).
Figure 15.
(A) Structure of SPTIDE determined from 1H-13C and 1H-31P Heteronuclear Multiple-Bond Correlation (HMBC) spectroscopic studies (8). From the through-bond correlations revealed by HMBC, the enchainment of different chemical groups (thymine, sugar, and phosphate) in SP was determined. The groups exhibiting strong HMBC signals are indicated by the same color. (B) Summary of key NOESY cross-peaks determined from the NMR spectra of SPTIDE (8, 128).
Structure of the formacetal analog of the dinucleotide SP
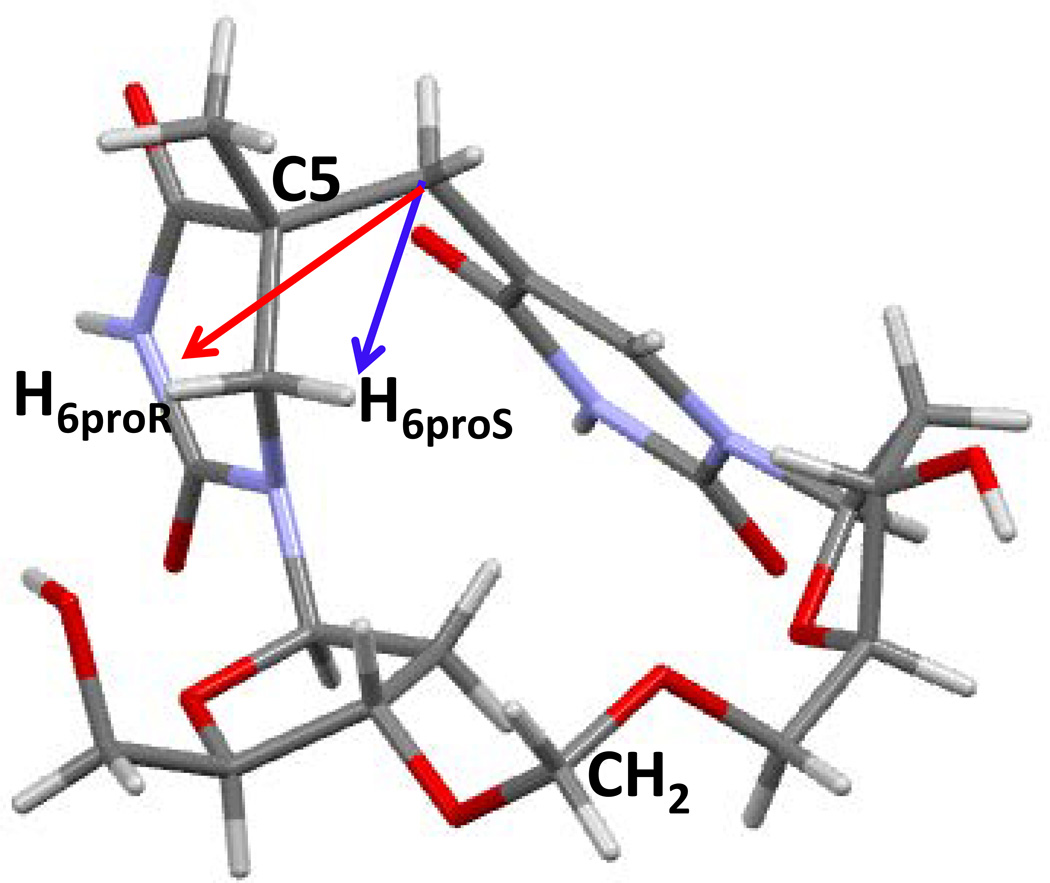
Although the NMR spectroscopic studies indicated that 5R-SP is the biologically relevant species, a crystal structure allowing direct observation of the atom arrangements in SP would clearly be significant. Given that the dinucleotide SP possesses the size of a small organic molecule, it may be crystalized via solvent evaporation, a method widely used for small molecule crystallization. However, the negative charge associated with the phosphate moiety may prevent crystal growth as reflected by the fact that all CPD dimers crystallized to date contain either no or a neutral linker between the two T residues (155, 167). Formacetal is a non-chiral isostere for a phosphate moiety which was used to allow for crystallization of a dinucleotide CPD formed between two uracil residues (155). After synthesizing the 5R-SP analog containing a formacetal linker with its steric configuration indicated by 2D-NMR spectroscopy, Lin et al. dissolved the resulting CH2SP in water, and obtained a single crystal via solvent evaporation (128). The solved structure of 5R-CH2SP <Figure 16> reveals that the two thymine rings are connected by a methylene bridge. The C5 carbon at the 5'-thymine moiety clearly adopts an R configuration, confirming the assignment by previous NMR spectroscopic studies (8). The 3′-thymine ring in SP is planar, and the 5′-dihydrothymine is distorted owing to the loss of C5=C6 bond and ring aromaticity. Moreover, the distance between the H6proS and the bridging methylene carbon was found to be 2.63 Ǻ in the SP structure, which is 0.7 Ǻ longer than that between the H6proR and the methylene carbon. This observation supports the conclusion in previous labeling studies that a H atom from the methyl group on the 3'-thymine migrates to the H6proS position during 5R-SP formation and that SP is formed via an intramolecular H-atom transfer process (139).
Figure 16.
X-ray structure of the 5R-CH2SP with the nitrogen and oxygen atoms labeled (128). The structure clearly shows that the C5 chiral center adopts an R configuration. The distance between the H6proS and the methylene carbon (blue arrow) is 2.63 Ǻ and that between the H6proR and the methylene carbon (red arrow) is 3.36 Ǻ, which supports the reaction mechanism in which the H6proS of SP originates from the methyl group in 3'-thymine of the TpT step before SP formation.
Structure of a dinucleoside SP in a duplex strand-protein complex
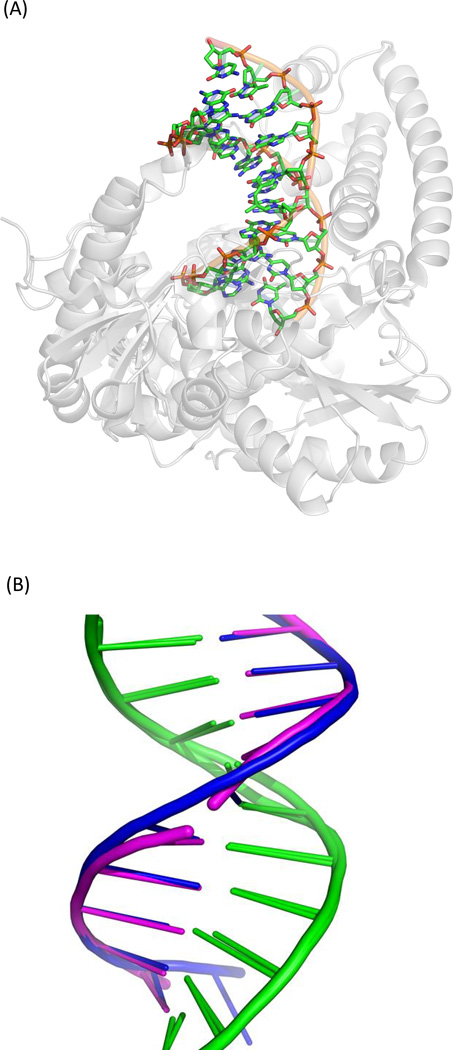
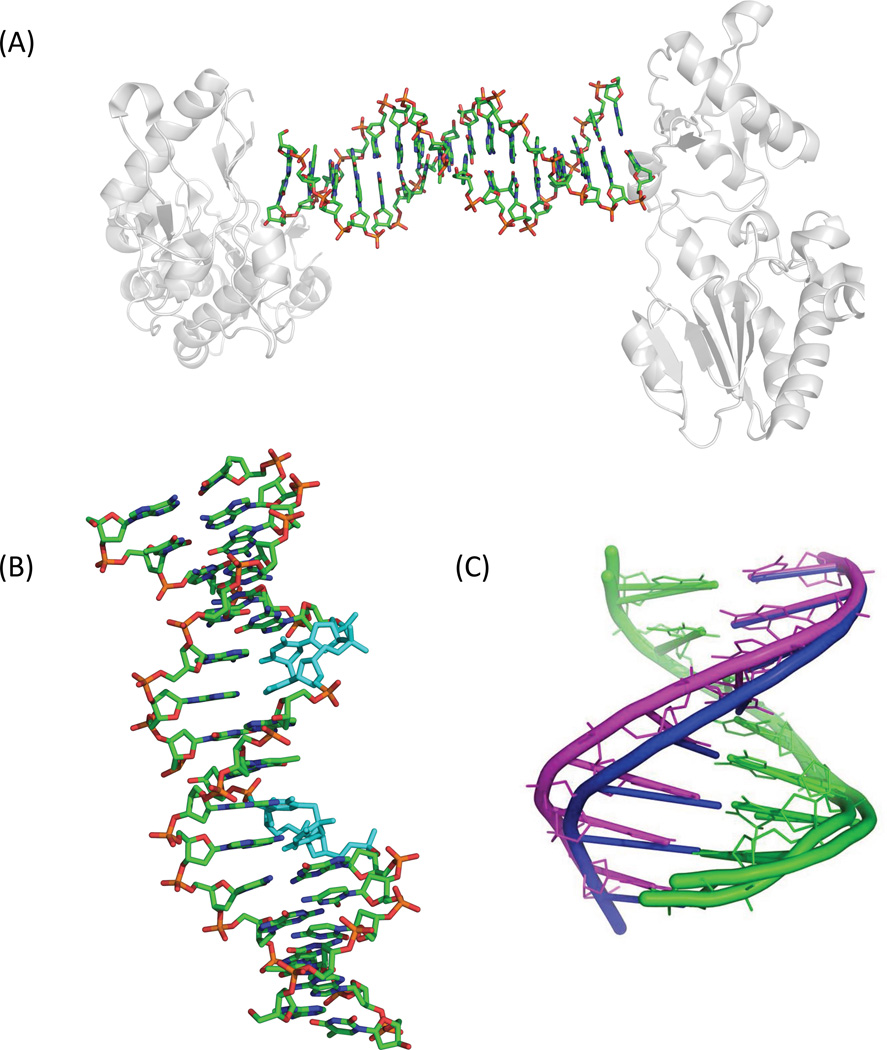
The synthesis of the dinucleoside SP phosphoramidite enables its incorporation into an oligonucleotide. Using a duplex dodecamer with SP at positions 6 and 7 of one strand, Heil et al. obtained its structure in complex with the Bacillus stearothermophilus (now Geobacillus stearothermophilus) DNA polymerase I (B. st. Pol I, <Figure 17>) (129). The structure shows that the 5R-SPSIDE 3 fits quite well into the DNA duplex framework, forming two nearly perfect Watson-Crick base pairs with the apposed adenine residues. As a control, they also obtained the structure of the strand with identical sequence except replacing SP by undamaged TpT. These two structures can be almost perfectly superimposed, suggesting that the 5R-SPSIDE does not induce much local distortion. Such an observation may partially reflect the property of SPTIDE in duplex DNA. However, due to the lack of phosphodiester bonds in the dinucleoside, the two thymine residues may freely rotate, releasing any distortion resulting from the presence of SP. Also, much of the oligonucleotide was embedded in the B. st. Pol I protein. As indicated by the analysis using a software package named 3DNA (168, 169), the duplexes exhibit an A-conformation near the polymerase active site. The unusual groove width and helical forms indicate that the conformation may be controlled by the protein framework, but not by the nucleic acid sequence. Therefore, the structure revealed by the SPSIDE-containing DNA may differ somewhat from that in the SPTIDE-containing genomic DNA in germinated spores.
Figure 17.
(A) Crystal structure of the dinucleoside 5R-SP-containing duplex complexed with B. st. Pol I (pdb entry 2YLJ) (129). (B) Superimposed structures of the undamaged DNA (pdb entry 1NJY) and the dinucleoside SP-containing DNA bound to B. st. Pol I (129). The DNA backbone in the latter case is broken due to the lack of phosphodiester group in the dinucleoside SP. Except for the broken backbone, the two DNA structures overlay nearly perfectly.
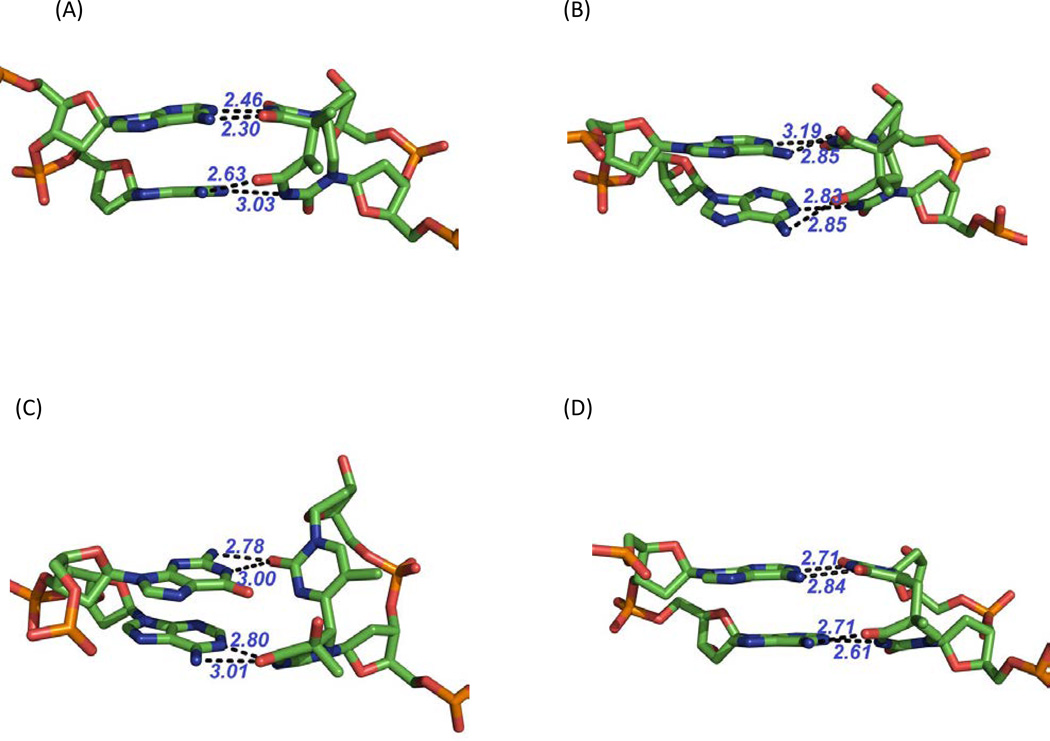
Structure of the dinucleotide SP in a duplex strand-protein complex
Employing a 16-mer oligonucleotide prepared via solid phase DNA synthesis using SPTIDE phosphoramidite, the duplex was crystalized as a complex with the N-terminal fragment of Moloney murine leukemia virus reverse transcriptase (MMLV RT) (170). In the 16-mer sequence used, the 3-mers at the ends are recognized by the MMLV RT fragment and the internal 10-mer sequence can be variable. The central 10 bps in this 16-mer are thus free of interactions with the protein, allowing the nucleic acid to adopt a structure dictated by the sequence. The sequence used also needs to be symmetric against the middle point of the 16-mer sequence, as the 8 bps are a part of the asymmetric component, the unique repeating unit within the crystal (171). Therefore, two SPTIDEs, one in each strand, were incorporated into the 16-mer duplex oligonucleotide. As a control, the 16-mer duplex with SPs replaced by undamaged TpTs was also crystalized to facilitate structural comparisons.
As shown in Figure 18, the SPTIDE-containing 16-mer duplex adopts a right-handed helical conformation with continuous base stacking (170). However, significant unwinding at the SP step followed by overwinding at the next three nucleotides was observed, preventing the duplex from adopting a perfect B-conformation. In contrast, in the undamaged oligonucleotide 16-mer duplex complexed with the MMLV RT crystallized as a control, the DNA adopts a perfect B-conformation throughout the entire sequence (170). Surprisingly, all hydrogen bonding interactions between SP and apposed adenines are maintained; the bonding distances are even ~ 0.5 Å shorter than those with undamaged TpT. This result is in contrast to the drastically weakened hydrogen bonds between the 5'-thymine residue in CPD and its complementary adenine as revealed by an X-ray crystallographic study (172). On the other hand, the unwinding at the 5'-T of SP followed by overwinding results in the widening of the minor groove by ~ 3 Å, from 9.7 Å in the undamaged DNA strand to 12.5 Å in the SP-containing strand.
Figure 18.
(A) Crystal structure of the nucleoprotein complex including two MMLV RT molecules and one 16-base-pair oligonucleotide duplex (170). The asymmetric unit of the crystal includes one protein molecule and eight base pairs of the duplex oligomer. (B) Structure of the 16-mer duplex strand containing two SP dimers (shown in cyan) (170), where all H-bonding interactions in the duplex are maintained. The two 8-mer asymmetric units are disconnected in the figure although an integral 16-mer strand was used for crystallization. Such a fragmentation is ascribed to the artificial effect of the crystallographic data; it does not imply cleavage of the 16-mer strand. (C) Superimposed structures of the 8-base-pair asymmetric unit of the undamaged oligomer (in blue, pdb entry 4M95) and the SPTIDE-containing oligomer (in magenta, pdb entry 4M94) bound to MMLV RT. The minor groove is widened from 9.7 Å in the undamaged strand to 12.5 Å in the SP-containing strand.
These local structural changes may explain the thermal destabilization induced by the presence of SP as observed by a DNA denaturation study (149). By comparing the thermal denaturation curves of SPTIDE-containing oligonucleotide duplexes with those of the corresponding undamaged duplexes, Jian et al. found that SPTIDE destabilizes the duplex by 10–20 kJ/mole, with the precise extent of destabilization dependent on the oligonucleotide sequence examined (149). CPD with a cis-syn conformation is known to destabilize an oligonucleotide duplex by 5.7 – 8.4 kJ/mole (173, 174). Therefore, barring effects of the sequence difference between oligonucleotides used in these different studies, the larger Gibbs free energy change induced by SP indicates that even though SPTIDE maintains all hydrogen bonding interactions with opposite adenine residues, it collectively disturbs the duplex local structure more than cis-syn CPD.
IMPACTS OF SP ON THE GENOMIC DNA
Mutagenesis potential indicated by the thymine dimer structures
The X-ray crystallographic structure of a cis-syn CPD-containing oligonucleotide has been solved in a duplex decamer sequence (172); the solution-state NMR structure of a dodecamer duplex with a cis-syn CPD is also available (174). Although 6-4PP-containing oligonucleotides have been crystallized when complexed with a 6-4PP antibody (175), the 6-4PP photolyase (176), and the nucleotide-excision repair protein DDB1–DDB2 (177) respectively, the 6-4PP lesion was flipped out of the duplex context into the protein binding pocket in all structures. Therefore, how 6-4PP alters the local structure of the duplex DNA was not revealed. Luckily, the solution-state NMR structures of two 6-4PP-containing decamer duplexes with ApA and GpA steps opposite to the 6-4PP respectively are available (178, 179). Together with the SP-containing 16-mer duplex structure solved by Singh,(170) a side-by-side comparison of the structural changes induced by these three thymine dimers becomes possible <Figure 19>.
Figure 19.
X-ray crystallographic or solution-state NMR structures of duplex oligonucleotides containing the three thymine dimers, cis-syn CPD, 6-4PP and SP, respectively. A) Crystal structure of a decamer duplex containing a cis-syn CPD lesion (pdb entry 1N4E) (172). B) Solution-state NMR structure of a dodecamer duplex containing a cis-syn CPD lesion (pdb entry 1TTD) (174). C) Solution-state NMR structure of a decamer duplex containing a stable GA•6-4PP mismatch (pdb entry 1CFL)(179). D) Structure of a 16-mer duplex strand containing two SPTIDEs deduced from a nucleoprotein complex (pdb entry 4M94) (170). The crystal structure shown in Panel A was indicated to only represent one of many solution structures that was captured during the crystallization process; the solution-state NMR structure shown in Panel B thus may better reflect the local structure of a cis-syn CPD-containing DNA (180). It is also evident that 6-4PP drastically changes the duplex oligonucleotide structure; while CPD and SP only induce minor changes.
The crystal structure shows that the cis-syn CPD bends the overall helical axis by 30 toward the major groove and unwinds the helix by 9° <Figure 19 A>. However, the solution-state NMR structure reveals only minor structural changes from undamaged duplex DNA, including a slightly narrower minor groove, a small helix bend toward the major groove, and a slight unwinding of the duplex at the lesion site <Figure 19 B> (174). Although these two structures vary significantly, the larger bend observed in the crystal structure was indicated to result from crystal packing forces and the structure likely represents one of many conformational states of the duplex in solution (180). Therefore, it was concluded that cis-syn CPD induces only minor DNA conformational changes in cells. In contrast, the solution-state NMR structure reveals that the two pyrimidine rings in 6-4PP become almost perpendicular, making it impossible for simultaneous base pairing and stacking inside the duplex (178, 179). Such a drastic local structural alteration led to a 44 bending in the overall helix and a 30 helix unwinding at the lesion site (178). Replacing the AA by the GA step opposite to the 6-4PP removes the local helix unwinding; however, an overall helix bend of 27.4 ± 1.5° was still observed <Figure 19C> (179). Although the solution-state NMR structure for SP-containing duplex oligonucleotides is currently unavailable, the crystal structure shown in Figure 19D indicates that SP only induces minor DNA structural changes. The extent of changes is probably comparable to that caused by cis-syn CPD but much smaller than caused by 6-4PP.
Such a hypothesis is further supported by the hydrogen bonding pattern observed within the dimer sites. As indicated by the key distances between non-hydrogen atoms, the H-bonding interactions associated with the 5'-T are largely maintained in the cis-syn CPD crystal structure, while the H-bonds associated with the 3'-T are much weakened <Figure 20A> (172). In contrast, in the solution-state NMR structure, the H-bonds associated with the 5'-T are weakened while those with the 3'-T were maintained. Moreover, examination of the weakened H-bonds reveals that the H-bond vectors deviate from the planes of the T and A bases. The H-bonding changes are even more drastic with the 6-4PP. Although the 5'-residue, i.e. the 5,6-dihydro-5-hydroxythymine, maintains hydrogen bonds with its partner adenine, no hydrogen bond is present with the 3'-residue of the 6-4PP. Interestingly, when inserting a GA step opposite to the 6-4PP lesion, the C2=O of the 3'-T form two H-bonds with the G residue <Figure 20C> (179). Such an H-bonding pattern may explain the A→G mutation resulting from the 6-4PP bypass event during translesion synthesis. In contrast, SP maintains all H-bonding interactions with the opposing adenine residues, and the H-bonds are even shortened relative to those found between TT and AA in undamaged oligonucleotides (170).
Figure 20.
Zoom-in view of the two base pairs containing the thymine dimers in a duplex oligonucleotide. The distances between the non-hydrogen atoms involved in hydrogen bond formation are also shown. From Panel A (from the crystal structure) and Panel B (from the solution-state NMR structure), it is clear that the hydrogen bonding interactions in one of the base pairs associated with the cis-syn CPD are weakened. The 6-4PP maintains weaker hydrogen bonds with its 5'-5,6-dihydro-5-hydroxythymine residue; its 3'-residue does not form any hydrogen bond with the opposite adenine. However, two hydrogen bonds were found if the opposite nucleobase becomes a guanine as shown in Panel C, explaining its tendency to induce an A→G mutation during the translesion synthesis event. In contrast, SP maintains all four hydrogen bonds (Panel D); these bonds may be even stronger than those formed between undamaged nucleobases as indicated by the shortened distances between the involving atoms. Again, due to the restriction of the 8-mer asymmetric units in the crystallographic data, the 16-mer strand was shown as two fragments. This “artificial” fragmentation however does not reflect the integrity of the 16-mer oligonucleotide and does not alter the DNA conformation.
Pyrimidine photo-dimers are mainly recognized and repaired by the NER pathway in prokaryotes and eukaryotes (14–17). NER can be divided into two subpathways: global genomic NER (GG-NER) and transcription coupled NER (TC-NER). Local conformational changes are thought to be the major recognition elements for the GG-NER machinery (14, 15). 6-4PP drastically distorts the local TpT structure, which is responsible for the sharp helix kink and significant local distortions at the 6-4PP site in duplex DNA (180). As a consequence, 6-4PP is readily recognized and repaired by GG-NER in live cells (half-life of 6-4PP = 2.3 hrs) (181). Analysis of CPD and SP suggests that both lesions may only result in minor local structural changes in DNA, as the involved thymine residues still retain the stacking conformation. Repair of CPD by the GG-NER pathway is slow; many CPDs remain unrepaired unless they are located in a transcriptionally active region and are repaired by the TC-NER pathway (15, 16, 182–184). By the same token, the smaller duplex structural changes induced by SP (170) indicate that SP repair by the GG-NER pathway is also likely to be slow.
The slow repair of CPDs indicates that these lesions are more likely to encounter a replicative polymerase and induce mutations when being bypassed during translesion synthesis (184). When a CPD TpT is bypassed by human Y-polymerase Pol η, two “A”s can be inserted in the opposite strand in a nearly error-free manner (<10−2) (185), although another study suggests that the accuracy rate is only 83% for both positions (186). Thus, although CPD is the dominant photo-lesion in DNA, the vast majority of these lesions may be harmless. In contrast, human Pol η is 7-fold more likely to insert a G than an A at the position opposite the 3′-residue of a 6-4PP (187), indicating that 6-4PP is highly mutagenic. As the repair of SP by GG-NER is likely to be slow, SP would have a good chance to encounter error-prone Y-polymerases and induce mutagenesis during translesion synthesis. Therefore, the unrepaired SP may be highly mutagenic in germinating spores, as suggested by a previous competent cell study (188). Confirmation of this hypothesis and the nature of the mutations generated by SP await future investigations.
Possible DNA secondary lesions induced by SP
Besides mutagenesis, the presence of SP may physically destabilize the genomic DNA and induce secondary lesions such as strand breaks. Recently, by treating the SPTIDE 2 with a strong base such as 200 mM KOH, Lin et al. found that a tetrahedral hemiaminal intermediate (38) is generated at the C4=O at the saturated 5'-thymine residue of SP. The hemiaminal species subsequently converts to a ring-opened hydrolysis product 39 through cleavage of the pyrimidine N3−C4 bond <Figure 21>. Alternatively, during the hemiaminal decay process, one of the C4-O bonds can be cleaved, reverting the hemiaminal species back to SP. As a consequence, the original oxygen atom attached with the C4=O moiety only possesses a 50% chance to be retained with the re-generated SP after one reaction cycle, and it can eventually be replaced by an oxygen atom from water if the reaction is allowed to proceed long enough. This possibility is supported by 18O labeling experiments, where almost all remaining SP molecules carried an 18O label after a 24-hr reaction using 18O labeled water at pH 12 (189). Such an oxygen exchange process mediated by the hemiaminal intermediate is facilitated by basic conditions; however, it also occurs at physiological pH albeit at a slower rate (189).
Figure 21.
Formation of the hemiaminal intermediate 37 at the C4=O moiety of the 5'-residue of SP. Either C-O bond associated with the hemiaminal species is cleavable. As a consequence, the oxygen atom at the C4=O moiety becomes exchangeable with the oxygen atom from water. Decomposition of the hemiaminal intermediate produces a SP hydrolysis product 38, whose further decomposition would induce a cascade of elimination reactions, eventually leading to DNA strand break.
Further analysis of the ring-opened SP hydrolysis product 39 reveals that it readily deglycosylates to 41, which further decays via the intermediate 42 to eliminate the 2'-deoxyribose at the 5'-residue of SP, yielding 43 <Figure 21>. Should such a process occur within an SP-containing oligonucleotide, a DNA strand break would be generated (189). Taken together, these data suggest that due to the loss of the ring aromaticity, the C4 position at the 5'-thymine residue of SP becomes a “hot spot” for the formation of a tetrahedral intermediate, the decay of which triggers a cascade of elimination reactions that can convert the SP modification into a DNA strand break. Even though the SP-induced DNA strand cleavage is slow under physiological pH, the process is irreversible. As dormant spores of Bacillus and Clostridium strains can survive for hundreds of years and perhaps longer (190–192), this SP-induced strand cleavage would gradually accumulate in dormant spores during this period and potentially become a serious DNA damage event threating spore’s survival.
SUMMARY AND PERSPECTIVES
The seminal finding by Donnellan and Setlow fifty years ago in their studies of UV irradiated bacterial spores initiated photochemical and photobiological studies of SP. Among the three thymine dimers, significant formation of SP requires DNA to be less hydrated and/or adopt an A-like conformation, while CPD and 6-4PP formation requires B-DNA, the conformation a duplex DNA usually adopts in aqueous solution. As a consequence, although all these dimers were discovered in the 1960s, the photochemistry and photobiology of CPDs and 6-4PPs, which can be readily generated by UV irradiation of oligonucleotides in aqueous solution, have been extensively studied in vivo and in vitro. In contrast, due to the difficulty in maintaining an A-DNA conformation in vitro, the SP photoreaction is always a minor event, and its occurrence is accompanied by many other “side” reactions. Such photochemical behavior makes the preparation of SP-containing oligonucleotides for subsequent biological studies problematic, thus hindering the development of SP photobiology. SP photobiology in spores is relatively well studied, because it is the nearly exclusive photo-lesion formed under UV irradiation. As described in this review, the combination of low water content, the presence of CaDPA, and DNA binding by SASPs in spores ensures that SP photochemistry is the dominant photochemical event in dormant spores. However, SP photobiology beyond dormant spores, such as its repair and mutagenic impacts in germinating spores, is still an area where questions remain.
SP was first discovered in UV irradiated bacterial spores, and the DNA photochemistry and photobiology field generally ascribes SP formation to the unique environment inside spores. However, is it truly just a spore photoproduct? It is now known that in dry or desiccated microorganisms exposed to UVC or UVB photons, SP is formed as a major DNA photo-lesion (193). SP was also formed in UV irradiated dried yeast cells (194, 195), and the yeast cells likely carrying SP exhibit greatly increased UV sensitivity. SP formation has also been suggested to occur in UV-irradiated frozen bacteria, bacteriophage (196), and yeast (197), and the frozen cells appear to be more sensitive to UV irradiation possibly again due to the presence of SP. However, SP formation in these non-spore cells has not been verified and the molecular basis for the enhanced UV sensitivity for these microorganisms has not been studied. Interestingly, very recent work has found that a virus that infects a hyperthermophilic bacterium encapsidates its DNA in an A-form due to association of the virus capsid protein with the DNA (198). It will be most interesting to learn the UVC photochemistry of the DNA in this virus.
For future studies of SP photobiology beyond endospores, several tools are needed. The first is an effective method to incorporate SP into an oligonucleotide in vitro. Although SP DNA photochemistry in vitro has proven to be extremely difficult, SP incorporation into oligonucleotides via chemical synthesis using phosphoramidites and solid phase DNA synthesis has been successful (149). The second tool is an effective means for SP analysis, which can now be achieved by the HPLC-MS/MS assay developed by Douki et al. (63). Using dinucleoside or dinucleotide SP as a standard for instrument calibration, the HPLC-MS/MS assay makes SP global analysis in living cells possible. Ideally, a method enabling similar SP analysis in living cells is also needed, which could well be achieved by immunological techniques using anti-SP antibodies. Monoclonal and polyclonal CPD and 6-4PP antibodies are available (199–202), and it is fair to state that what we know about CPD and 6-4PP photobiology in living cells is largely due to the availability of these antibodies. Certainly, having antibodies specifically recognizing SP would be a major asset to analysis of SP photobiology in living cells.
In summary, in the past half a century, much progress has been made towards a general understanding of SP photochemistry and photobiology. What has been revealed so far predicts a rich SP photobiology in nature, as the impact of SP photochemistry is likely to extend well beyond bacterial spores. It is fair to state that despite the extensive effort in the past fifty years, the progress summarized in this review only represents the starting point for further exciting SP research, as what we now know about SP is probably only the “tip of the iceberg”. With the tools that have been and could readily be developed, we are now positioned to significantly extend chemical and biological studies of SP. In addition, since spore-forming bacteria are responsible for a number of serious human diseases, including botulism (Clostridium botulinum) and anthrax (Bacillus anthracis) (203), two of the six category A bioterrorism agents, understanding SP photobiology may allow one to selectively target this photochemical process in spore-forming bacterial to design better strategies to combat these deadly diseases, improving human health as well as national security.
ACKNOWLEDGEMENTS
We thank the reviewers for their insightful comments. This work has been supported in part by funds from NIH ES017177 and NSF CHE1506598 grants (LL). Work in the PS laboratory over the years was supported by grants from the NIH as well as the Army Research Office. One of us (PS) is grateful to Richard (Dick) Setlow who died while this review article was in preparation. Dick’s contribution to DNA photobiology was immense, and his co-discovery with Ed Donnellan of SP in UV-irradiated Bacillus spores in 1965 initiated a new area of research, a good bit of it in my laboratory, even though I will admit I never paid attention to Dick’s work in my younger years. However, I certainly did as my research career progressed, and the work in my laboratory on SP was instrumental in enabling the funding of the college education of two of Dick’s grandchildren as well as paying the salary of one of Dick’s daughter-in-laws for many years!
REFERENCES
- 1.Donnellan JE, Jr, Setlow RB. Thymine photoproducts but not thymine dimers found in ultraviolet-irradiated bacterial spores. Science. 1965;149:308–310. doi: 10.1126/science.149.3681.308. [DOI] [PubMed] [Google Scholar]
- 2.Varghese AJ, Wang SY. Ultraviolet irradiation of DNA in vitro and in vivo produces a third thymine-derived product. Science. 1967;156:955–957. doi: 10.1126/science.156.3777.955. [DOI] [PubMed] [Google Scholar]
- 3.Donnellan JE, Stafford RS. The ultraviolet photochemistry and photobiology of vegetative cells and spores of Bacillus megaterium. Biophys. J. 1968;8:17–28. doi: 10.1016/S0006-3495(68)86471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith KC, Yoshikawa H. Variation in the photochemical reactivity of thymine in the DNA of B. subtilis spores, vegetative cells and spores germinated in chloramphenicol. Photochem. Photobiol. 1966;5:777–786. doi: 10.1111/j.1751-1097.1966.tb05773.x. [DOI] [PubMed] [Google Scholar]
- 5.Varghese AJ. Photochemistry of thymidine as a thin solid film. Photochem. Photobiol. 1971;13:357–364. [Google Scholar]
- 6.Rahn RO, Setlow JK, Hosszu JL. Ultraviolet inactivation and photoproducts of transforming DNA irradiated at low temperatures. Biophys. J. 1969;9:510–517. doi: 10.1016/S0006-3495(69)86401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varghese AJ. 5-Thyminyl-5,6-dihydrothymine from DNA irradiated with ultraviolet light. Biochem. Biophys. Res. Commun. 1970;38:484–490. doi: 10.1016/0006-291x(70)90739-4. [DOI] [PubMed] [Google Scholar]
- 8.Mantel C, Chandor A, Gasparutto D, Douki T, Atta M, Fontecave M, Bayle PA, Mouesca JM, Bardet M. Combined NMR and DFT studies for the absolute configuration elucidation of the spore photoproduct, a UV-induced DNA lesion. J. Am. Chem. Soc. 2008;130:16978–16984. doi: 10.1021/ja805032r. [DOI] [PubMed] [Google Scholar]
- 9.Chandor A, Berteau O, Douki T, Gasparutto D, Sanakis Y, Ollagnier-De-Choudens S, Atta M, Fontecave M. Dinucleotide spore photoproduct, a minimal substrate of the DNA repair spore photoproduct lyase enzyme from Bacillus subtilis. J. Biol. Chem. 2006;281:26922–26931. doi: 10.1074/jbc.M602297200. [DOI] [PubMed] [Google Scholar]
- 10.Munakata N. Genetic analysis of a mutant of Bacillus subtilis producing ultraviolet-sensitive spores. Mol. Gen. Genet. 1969;104:258–263. doi: 10.1007/BF02539290. [DOI] [PubMed] [Google Scholar]
- 11.Munakata N, Rupert CS. Genetically controlled removal of "spore photoproduct" from deoxyribonucleic acid of ultraviolet-irradiated Bacillus subtilis spores. J. Bacteriol. 1972;111:192–198. doi: 10.1128/jb.111.1.192-198.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munakata N, Rupert CS. Dark repair of DNA containing “spore photoproduct” in Bacillus subtilis. Mol. Gen. Genet. 1974;130:239–250. doi: 10.1007/BF00268802. [DOI] [PubMed] [Google Scholar]
- 13.Wang T-CV, Rupert CS. Evidence for the monomerization of spore photoproduct to two thymines by the light-independent "spore repair" process in Bacillus subtilis. Photochem. Photobiol. 1977;25:123–127. doi: 10.1111/j.1751-1097.1977.tb07432.x. [DOI] [PubMed] [Google Scholar]
- 14.Sancar A. DNA Excision Repair. Ann. Rev. Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 15.Cline SD, Hanawalt PC. Who's on first in the cellular response to DNA damage? Nat. Rev. Mol. Cell Biol. 2003;4:361–373. doi: 10.1038/nrm1101. [DOI] [PubMed] [Google Scholar]
- 16.Shah P, He Y-Y. Molecular regulation of UV-induced DNA repair. Photochem. Photobiol. 2015;91:254–264. doi: 10.1111/php.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kisker C, Kuper J, Van Houten B. Prokaryotic nucleotide excision repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012591. doi: 10.1101/cshperspect.a012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedraza-Reyes M, Gutierrez-Corona F, Nicholson WL. Spore photoproduct lyase operon (splAB) regulation during Bacillus subtilis sporulation: modulation of splB-lacZ fusion expression by P1 promoter mutations and by an in-frame deletion of splA. Curr. Microbiol. 1997;34:133–137. doi: 10.1007/s002849900157. [DOI] [PubMed] [Google Scholar]
- 19.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rebeil R, Sun Y, Chooback L, Pedraza-Reyes M, Kinsland C, Begley TP, Nicholson WL. Spore photoproduct lyase from Bacillus subtilis spores is a novel iron-sulfur DNA repair enzyme which shares features with proteins such as class III anaerobic ribonucleotide reductases and pyruvate-formate lyases. J. Bacteriol. 1998;180:4879–4885. doi: 10.1128/jb.180.18.4879-4885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehl RA, Begley TP. Mechanistic studies on the repair of a novel DNA photolesion: The spore photoproduct. Org. Lett. 1999;1:1065–1066. doi: 10.1021/ol9908676. [DOI] [PubMed] [Google Scholar]
- 22.Rebeil R, Nicholson WL. The subunit structure and catalytic mechanism of the Bacillus subtilis DNA repair enzyme spore photoproduct lyase. Proc. Natl. Acad. Sci. 2001;98:9038–9043. doi: 10.1073/pnas.161278998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fajardo-Cavazos P, Rebeil R, Nicholson W. Essential cysteine residues in Bacillus subtilis spore photoproduct lyase identified by alanine scanning mutagenesis. Curr. Microbiol. 2005;51:331–335. doi: 10.1007/s00284-005-0052-8. [DOI] [PubMed] [Google Scholar]
- 24.Pieck J, Hennecke U, Pierik A, Friedel M, Carell T. Characterization of a new thermophilic spore photoproduct lyase from Geobacillus stearothermophilus (SplG) with defined lesion containing DNA substrates. J. Biol. Chem. 2006;281:36317–36326. doi: 10.1074/jbc.M607053200. [DOI] [PubMed] [Google Scholar]
- 25.Kneuttinger AC, Heil K, Kashiwazaki G, Carell T. The radical SAM enzyme spore photoproduct lyase employs a tyrosyl radical for DNA repair. Chem. Commun. 2013;49:722–724. doi: 10.1039/c2cc37735g. [DOI] [PubMed] [Google Scholar]
- 26.Benjdia A, Heil K, Winkler A, Carell T, Schlichting I. Rescuing DNA repair activity by rewiring the H-atom transfer pathway in the radical SAM enzyme, spore photoproduct lyase. Chem. Commun. 2014;50:14201–14204. doi: 10.1039/c4cc05158k. [DOI] [PubMed] [Google Scholar]
- 27.Chandor-Proust A, Berteau O, Douki T, Gasparutto D, Ollagnier-de-Choudens S, Fontecave M, Atta M. DNA repair and free radicals, new insights into the mechanism of spore photoproduct lyase revealed by single amino Acid substitution. J. Biol. Chem. 2008;283:36361–36368. doi: 10.1074/jbc.M806503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheek J, Broderick J. Direct H atom abstraction from spore photoproduct C-6 initiates DNA repair in the reaction catalyzed by spore photoproduct lyase: Evidence for a reversibly generated adenosyl radical intermediate. J. Am. Chem. Soc. 2002;124:2860–2861. doi: 10.1021/ja017784g. [DOI] [PubMed] [Google Scholar]
- 29.Buis J, Cheek J, Kalliri E, Broderick J. Characterization of an active spore photoproduct lyase, a DNA repair enzyme in the radical S-adenosylmethionine superfamily. J. Biol. Chem. 2006;281:25994–26003. doi: 10.1074/jbc.M603931200. [DOI] [PubMed] [Google Scholar]
- 30.Silver S, Chandra T, Zilinskas E, Ghose S, Broderick W, Broderick J. Complete stereospecific repair of a synthetic dinucleotide spore photoproduct by spore photoproduct lyase. J. Biol. Inorg. Chem. 2010;15:943–955. doi: 10.1007/s00775-010-0656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, Lin G, Liu D, Dria KJ, Telser J, Li L. Probing the reaction mechanism of spore photoproduct lyase (SPL) via diastereoselectively labeled dinucleotide SP TpT substrates. J. Am. Chem. Soc. 2011;133:10434–10447. doi: 10.1021/ja110196d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L, Lin G, Nelson RS, Jian Y, Telser J, Li L. Mechanistic studies of the spore photoproduct lyase via a single cysteine mutation. Biochemistry. 2012;51:7173–7188. doi: 10.1021/bi3010945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, Nelson RS, Benjdia A, Lin G, Telser J, Stoll S, Schlichting I, Li L. A radical transfer pathway in spore photoproduct lyase. Biochemistry. 2013;52:3041–3050. doi: 10.1021/bi3016247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjdia A, Heil K, Barends TRM, Carell T, Schlichting I. Structural insights into recognition and repair of UV-DNA damage by spore photoproduct lyase, a radical SAM enzyme. Nucleic Acids Res. 2012;40:9308–9318. doi: 10.1093/nar/gks603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L, Li L. The enzyme-mediated direct reversal of a dithymine photoproduct in germinating endospores. Int. J. Mol. Sci. 2013;14:13137–13153. doi: 10.3390/ijms140713137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang L, Li L. Spore photoproduct lyase: the known, the controversial, and the unknown. J. Biol. Chem. 2015;290:4003–4009. doi: 10.1074/jbc.R114.573675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L. Mechanistic studies of the radical SAM enzyme spore photoproduct lyase (SPL) Biochim. Biophys. Acta, Proteins Proteomics. 2012;1824:1264–1277. doi: 10.1016/j.bbapap.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kneuttinger AC, Kashiwazaki G, Prill S, Heil K, Müller M, Carell T. Formation and direct repair of UV-induced dimeric DNA pyrimidine lesions. Photochem. Photobiol. 2014;90:1–14. doi: 10.1111/php.12197. [DOI] [PubMed] [Google Scholar]
- 39.Benjdia A. DNA photolyases and SP lyase: structure and mechanism of light-dependent and independent DNA lyases. Curr. Opin. Struct. Biol. 2012;22:711–720. doi: 10.1016/j.sbi.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Desnous Cl, Guillaume D, Clivio P. Spore photoproduct: A key to bacterial eternal life. Chem. Rev. 2010;110:1213–1232. doi: 10.1021/cr0781972. [DOI] [PubMed] [Google Scholar]
- 41.Yoshimura Y, Takahata H. Recent advances in cyclonucleosides: C-cyclonucleosides and spore photoproducts in damaged DNA. Molecules. 2012;17:11630–11654. doi: 10.3390/molecules171011630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerhardt P, Marquis RE. Spore thermoresistance mechanisms. In: Smith I, Slepecky RA, Setlow P, editors. Regulation of prokaryotic development. Washington, DC: American Society for Microbiology; 1989. pp. 43–63. [Google Scholar]
- 43.Henriques AO, Moran CP., Jr Structure, assembly, and function of the spore surface layers. Annu. Rev Microbiol. 2007;61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- 44.Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 45.McKenney PT, Driks A, Eichenberger P. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 2013;11:33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Setlow P. Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu. Rev. Microbiol. 1995;49:29–54. doi: 10.1146/annurev.mi.49.100195.000333. [DOI] [PubMed] [Google Scholar]
- 47.Duthie MS, Kimber I, Norval M. The effects of ultraviolet radiation on the human immune system. Br. J. Dermatol. 1999;140:995–1009. doi: 10.1046/j.1365-2133.1999.02898.x. [DOI] [PubMed] [Google Scholar]
- 48.Nicholson W, Schuerger A, Setlow P. The solar UV environment and bacterial spore UV resistance: considerations for Earth-to-Mars transport by natural processes and human spaceflight. Mutat. Res.-Fund. Mol. M. 2005;571:249–264. doi: 10.1016/j.mrfmmm.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 49.Slieman TA, Nicholson WL. Artificial and solar UV radiation induces strand breaks and cyclobutane pyrimidine dimers in Bacillus subtilis spore DNA. Appl. Environ. Microbiol. 2000;66:199–205. doi: 10.1128/aem.66.1.199-205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moeller R, Douki T, Cadet J, Stackebrandt E, Nicholson WL, Rettberg P, Reitz G, Horneck G. UV-radiation-induced formation of DNA bipyrimidine photoproducts in Bacillus subtilis endospores and their repair during germination. Int. Microbiol. 2007;10:39–46. [PubMed] [Google Scholar]
- 51.Tyrrell RM. Solar dosimetry with repair deficient bacterial spores: action spectra, photoproduct measurements and a comparison with other biological systems. Photochem. Photobiol. 1978;27:571–579. doi: 10.1111/j.1751-1097.1978.tb07648.x. [DOI] [PubMed] [Google Scholar]
- 52.Xue Y, Nicholson WL. The two major spore DNA repair pathways, nucleotide excision repair and spore photoproduct lyase, are sufficient for the resistance of Bacillus subtilis spores to artificial UV-C and UV-B but not to solar radiation. Appl. Environ. Microbiol. 1996;62:2221–2227. doi: 10.1128/aem.62.7.2221-2227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moeller R, Stackebrandt E, Reitz G, Berger T, Rettberg P, Doherty AJ, Horneck G, Nicholson WL. Role of DNA repair by nonhomologous-end joining in Bacillus subtilis spore resistance to extreme dryness, mono- and polychromatic UV, and ionizing radiation. J. Bacteriol. 2007;189:3306–3311. doi: 10.1128/JB.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moeller R, Reitz G, Li Z, Klein S, Nicholson WL. Multifactorial resistance of Bacillus subtilis spores to high-energy proton radiation: Role of spore structural components and the homologous recombination and non-homologous end joining DNA repair pathways. Astrobiology. 2012;12:1069–1077. doi: 10.1089/ast.2012.0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, Setlow P, Losick R, Eichenberger P. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- 56.Setlow P. When the sleepers wake: the germination of spores of Bacillus species. J. Appl. Microbiol. 2013;115:1251–1268. doi: 10.1111/jam.12343. [DOI] [PubMed] [Google Scholar]
- 57.Setlow P. Germination of spores of Bacillus species: what we know and do not know. J. Bacteriol. 2014;196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moir A. How do spores germinate? J. Appl. Microbiol. 2006;101:526–530. doi: 10.1111/j.1365-2672.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- 59.Huang SS, Chen D, Pelczar PL, Vepachedu VR, Setlow P, Li YQ. Levels of Ca2+-dipicolinic acid in individual bacillus spores determined using microfluidic Raman tweezers. J. Bacteriol. 2007;189:4681–4687. doi: 10.1128/JB.00282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paidhungat M, Setlow B, Driks A, Setlow P. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J. Bacteriol. 2000;182:5505–5512. doi: 10.1128/jb.182.19.5505-5512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Setlow B, Atluri S, Kitchel R, Koziol-Dube K, Setlow P. Role of dipicolinic acid in resistance and stability of spores of Bacillus subtilis with or without DNA-protective α/β-type small acid-soluble proteins. J. Bacteriol. 2006;188:3740–3747. doi: 10.1128/JB.00212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bender GR, Marquis RE. Spore heat resistance and specific mineralization. Appl. Environ. Microbiol. 1985;50:1414–1421. doi: 10.1128/aem.50.6.1414-1421.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Douki T, Setlow B, Setlow P. Photosensitization of DNA by dipicolinic acid, a major component of spores of Bacillus species. Photochem. Photobiol. Sci. 2005;4:591–597. doi: 10.1039/b503771a. [DOI] [PubMed] [Google Scholar]
- 64.Stafford RS, Donnellan JE., Jr Photochemical evidence for conformation changes in DNA during germination of bacterial spores. Proc. Natl. Acad. Sci. 1968;59:822–828. doi: 10.1073/pnas.59.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Setlow B, Setlow P. Absence of transient elevated UV resistance during germination of Bacillus subtilis spores lacking small, acid-soluble spore proteins alpha and beta. J. Bacteriol. 1988;170:2858–2859. doi: 10.1128/jb.170.6.2858-2859.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanchez-Salas JL, Santiago-Lara ML, Setlow B, Sussman MD, Setlow P. Properties of Bacillus megaterium and Bacillus subtilis mutants which lack the protease that degrades small, acid-soluble proteins during spore germination. J. Bacteriol. 1992;174:807–814. doi: 10.1128/jb.174.3.807-814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Setlow P. Resistance of spores of Bacillus species to ultraviolet light. Environ. Mol. Mutagen. 2001;38:97–104. doi: 10.1002/em.1058. [DOI] [PubMed] [Google Scholar]
- 68.Setlow P. Spore germination. Curr. Opin. Microbiol. 2003;6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 69.Riemann H, Ordal ZJ. Germination of bacterial endospores with calcium and dipicolinic acid. Science. 1961;133:1703–1704. doi: 10.1126/science.133.3465.1703. [DOI] [PubMed] [Google Scholar]
- 70.Ablett S, Darke AH, Lillford PJ, Martin DR. Glass formation and dormancy in bacterial spores. Int. J. Food Sci. Tech. 1999;34:59–69. [Google Scholar]
- 71.Leuschner RGK, Lillford PJ. Thermal properties of bacterial spores and biopolymers. Int. J. Food Microbiol. 2003;80:131–143. doi: 10.1016/s0168-1605(02)00139-3. [DOI] [PubMed] [Google Scholar]
- 72.Black SH, Gerhardt P. Permeability of bacterial spores IV.: Water content, uptake, and distribution. J. Bacteriol. 1962;83:960–967. doi: 10.1128/jb.83.5.960-967.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaieda S, Setlow B, Setlow P, Halle B. Mobility of core water in Bacillus subtilis spores by 2H NMR. Biophys. J. 2013;105:2016–2023. doi: 10.1016/j.bpj.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stecchini ML, Del Torre M, Venir E, Morettin A, Furlan P, Maltini E. Glassy state in Bacillus subtilis spores analyzed by differential scanning calorimetry. Int. J. Food Microbiol. 2006;106:286–290. doi: 10.1016/j.ijfoodmicro.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 75.Strahs G, Dickerson RE. The crystal structure of calcium dipicolinate trihydrate (a bacterial spore metabolite) Acta Crystallogr. B. 1968;24:571–578. doi: 10.1107/s0567740868002852. [DOI] [PubMed] [Google Scholar]
- 76.Friedline AW, Zachariah MM, Johnson K, Thomas KJ, Middaugh AN, Garimella R, Powell DR, Vaishampayan PA, Rice CV. Water behavior in bacterial spores by deuterium NMR spectroscopy. J. Phys. Chem. B. 2014;118:8945–8955. doi: 10.1021/jp5025119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kong L, Setlow P, Li Y-q. Analysis of the Raman spectra of Ca2+-dipicolinic acid alone and in the bacterial spore core in both aqueous and dehydrated environments. Analyst. 2012;137:3683–3689. doi: 10.1039/c2an35468c. [DOI] [PubMed] [Google Scholar]
- 78.Ghiamati E, Manoharan R, Nelson WH, Sperry JF. UV resonance Raman-spectra of Bacillus spores. Appl. Spectrosc. 1992;46:357–364. [Google Scholar]
- 79.Lindsay J, Murrell W. Solution spectroscopy of dipicolinic acid interaction with nucleic acids: Role in spore heat resistance. Curr. Microbiol. 1986;13:255–259. [Google Scholar]
- 80.Lindsay J, Murrell W. Changes in density of DNA after interaction with dipicolinic acid and its possible role in spore heat resistance. Curr. Microbiol. 1985;12:329–333. [Google Scholar]
- 81.Setlow B, Setlow P. Dipicolinic acid greatly enhances production of spore photoproduct in bacterial spores upon UV irradiation. Appl. Environ. Microbiol. 1993;59:640–643. doi: 10.1128/aem.59.2.640-643.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Du Q, Zhao H, Song D, Liu K, Su H. Consecutive reaction mechanism for the formation of spore photoproduct in DNA photolesion. J. Phys. Chem. B. 2012;116:11117–11123. doi: 10.1021/jp305915e. [DOI] [PubMed] [Google Scholar]
- 83.Lamola AA. Triplet photosensitization and the photobiology of thymine dimers in DNA. Pure Appl. Chem. 1970;24:599–610. doi: 10.1351/pac197024030599. [DOI] [PubMed] [Google Scholar]
- 84.Carstensen EL, Marquis RE, Child SZ, Bender GR. Dielectric properties of native and decoated spores of Bacillus megaterium. J. Bacteriol. 1979;140:917–928. doi: 10.1128/jb.140.3.917-928.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carstensen EL, Marquis RE, Gerhardt P. Dielectric study of the physical state of electrolytes and water within Bacillus cereus spores. J. Bacteriol. 1971;107:106–113. doi: 10.1128/jb.107.1.106-113.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cowan AE, Olivastro EM, Koppel DE, Loshon CA, Setlow B, Setlow P. Lipids in the inner membrane of dormant spores of Bacillus species are largely immobile. Proc. Natl. Acad. Sci. 2004;101:7733–7738. doi: 10.1073/pnas.0306859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cowan AE, Koppel DE, Setlow B, Setlow P. A soluble protein is immobile in dormant spores of Bacillus subtilis but is mobile in germinated spores: Implications for spore dormancy. Proc. Natl. Acad. Sci. 2003;100:4209–4214. doi: 10.1073/pnas.0636762100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Setlow P. I will survive: DNA protection in bacterial spores. Trends microbiol. 2007;15:172–180. doi: 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 89.Mohr SC, Sokolov NV, He CM, Setlow P. Binding of small acid-soluble spore proteins from Bacillus subtilis changes the conformation of DNA from B to A. Proc. Natl. Acad. Sci. 1991;88:77–81. doi: 10.1073/pnas.88.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Griffith J, Makhov A, Santiagolara L, Setlow P. Electron-microscopic studies of the interaction between a Bacillus-Subtilis alpha/beta-type small, acid-soluble spore protein with DNA - protein-binding is cooperative, stiffens the DNA, and induces negative supercoiling. Proc. Natl. Acad. Sci. 1994;91:8224–8228. doi: 10.1073/pnas.91.17.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Setlow P. Small, acid-soluble spore proteins of Bacillus species: structure, synthesis, genetics, function, and degradation. Annu. Rev. Microbiol. 1988;42:319–338. doi: 10.1146/annurev.mi.42.100188.001535. [DOI] [PubMed] [Google Scholar]
- 92.Nicholson WL, Setlow B, Setlow P. Ultraviolet irradiation of DNA complexed with alpha/beta-type small, acid-soluble proteins from spores of Bacillus or Clostridium species makes spore photoproduct but not thymine dimers. Proc. Natl. Acad. Sci. 1991;88:8288–8292. doi: 10.1073/pnas.88.19.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fairhead H, Setlow P. Binding of DNA to alpha/beta-type small, acid-soluble proteins from spores of Bacillus or Clostridium species prevents formation of cytosine dimers, cytosine-thymine dimers, and bipyrimidine photoadducts after UV irradiation. J. Bacteriol. 1992;174:2874–2880. doi: 10.1128/jb.174.9.2874-2880.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Douki T, Setlow B, Setlow P. Effects of the binding of alpha/beta-type small, acid-soluble spore proteins on the photochemistry of DNA in spores of Bacillus subtilis and in vitro. Photochem. Photobiol. 2005;81:163–169. doi: 10.1562/2004-08-18-RA-278. [DOI] [PubMed] [Google Scholar]
- 95.Mason JM, Setlow P. Different small, acid-soluble proteins of the alpha/beta type have interchangeable roles in the heat and UV radiation resistance of Bacillus subtilis spores. J. Bacteriol. 1987;169:3633–3637. doi: 10.1128/jb.169.8.3633-3637.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tovar-Rojo F, Setlow P. Effects of mutant small, acid-soluble spore proteins from Bacillus subtilis on DNA in vivo and in vitro. J. Bacteriol. 1991;173:4827–4835. doi: 10.1128/jb.173.15.4827-4835.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Raju D, Setlow P, Sarker MR. Antisense-RNA-mediated decreased synthesis of small, acid-soluble spore proteins leads to decreased resistance of Clostridium perfringens spores to moist heat and UV radiation. Appl. Environ. Microbiol. 2007;73:2048–2053. doi: 10.1128/AEM.02500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Raju D, Waters M, Setlow P, Sarker M. Investigating the role of small, acid-soluble spore proteins (SASPs) in the resistance of Clostridium perfringens spores to heat. BMC. Microbiol. 2006;6:50. doi: 10.1186/1471-2180-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Setlow B, Hand AR, Setlow P. Synthesis of a Bacillus subtilis small, acid-soluble spore protein in Escherichia coli causes cell DNA to assume some characteristics of spore DNA. J. Bacteriol. 1991;173:1642–1653. doi: 10.1128/jb.173.5.1642-1653.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ragkousi K, Cowan AE, Ross MA, Setlow P. Analysis of nucleoid morphology during germination and outgrowth of spores of Bacillus species. J. Bacteriol. 2000;182:5556–5562. doi: 10.1128/jb.182.19.5556-5562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pogliano K, Harry E, Losick R. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol. Microbiol. 1995;18:459–470. doi: 10.1111/j.1365-2958.1995.mmi_18030459.x. [DOI] [PubMed] [Google Scholar]
- 102.Setlow B, Sun D, Setlow P. Interaction between DNA and alpha/beta-type small, acid-soluble spore proteins: a new class of DNA-binding protein. J. Bacteriol. 1992;174:2312–2322. doi: 10.1128/jb.174.7.2312-2322.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Frenkiel-Krispin D, Sack R, Englander J, Shimoni E, Eisenstein M, Bullitt E, Horowitz-Scherer R, Hayes CS, Setlow P, Minsky A, Wolf SG. Structure of the DNA-SspC complex: implications for DNA packaging, protection, and repair in bacterial spores. J. Bacteriol. 2004;186:3525–3530. doi: 10.1128/JB.186.11.3525-3530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hayes CS, Setlow P. An {alpha}/{beta}-type, small, acid-soluble spore protein which has very high affinity for DNA prevents outgrowth of Bacillus subtilis spores. J. Bacteriol. 2001;183:2662–2666. doi: 10.1128/JB.183.8.2662-2666.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kosman J, Setlow P. Effects of carboxy-terminal modifications and pH on binding of a Bacillus subtilis small, acid-soluble spore protein to DNA. J. Bacteriol. 2003;185:6095–6103. doi: 10.1128/JB.185.20.6095-6103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee KS, Bumbaca D, Kosman J, Setlow P, Jedrzejas MJ. Structure of a protein-DNA complex essential for DNA protection in spores of Bacillus species. Proc. Natl. Acad. Sci. 2008;105:2806–2811. doi: 10.1073/pnas.0708244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bumbaca D, Kosman J, Setlow P, Henderson RK, Jedrzejas MJ. Crystallization and preliminary X-ray analysis of the complex between a Bacillus subtilis [alpha]/[beta]-type small acid-soluble spore protein and DNA. Acta Crystallogr. F. 2007;63:503–506. doi: 10.1107/S1744309107022750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nicholson WL, Setlow B, Setlow P. Binding of DNA in vitro by a small, acid-soluble spore protein from Bacillus subtilis and the effect of this binding on DNA topology. J. Bacteriol. 1990;172:6900–6906. doi: 10.1128/jb.172.12.6900-6906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Patrick MH, Gray DM. Independence of photoproduct formation on DNA conformation. Photochem. Photobiol. 1976;24:507–513. doi: 10.1111/j.1751-1097.1976.tb06867.x. [DOI] [PubMed] [Google Scholar]
- 110.Cadet J, Courdavault S, Ravanat JL, Douki T. UVB and UVA radiation-mediated damage to isolated and cellular DNA. Pure and Applied Chemistry. 2005;77:947–961. [Google Scholar]
- 111.Johns HE, Pearson ML, Helleiner CW, Leblanc JC. Ultraviolet photochemistry of thymidylyl-(3' to 5') -thymidine. J. Mol. Biol. 1964;9:503–524. doi: 10.1016/s0022-2836(64)80223-0. [DOI] [PubMed] [Google Scholar]
- 112.Sztumpf E, Shugar D. Photochemistry of model oligo- and polynucleotides 6. Photodimerization and its reversal in thymine dinucleotide analogues. Biochim. Biophys. Acta. 1962;61:555–566. [Google Scholar]
- 113.Schreier WJ, Schrader TE, Koller FO, Gilch P, Crespo-Hernandez CE, Swaminathan VN, Carell T, Zinth W, Kohler B. Thymine dimerization in DNA is an ultrafast photoreaction. Science. 2007;315:625–629. doi: 10.1126/science.1135428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kwok W-M, Ma C, Phillips DL. A doorway state leads to photostability or triplet photodamage in thymine DNA. J. Am. Chem. Soc. 2008;130:5131–5139. doi: 10.1021/ja077831q. [DOI] [PubMed] [Google Scholar]
- 115.Cuquerella MC, Lhiaubet-Vallet V, Bosca F, Miranda MA. Photosensitised pyrimidine dimerisation in DNA. Chem. Sci. 2011;2:1219–1232. [Google Scholar]
- 116.Schreier WJ, Kubon J, Regner N, Haiser K, Schrader TE, Zinth W, Clivio P, Gilch P. Thymine dimerization in DNA model systems: cyclobutane photolesion is predominantly formed via the singlet channel. J. Am. Chem. Soc. 2009;131:5038–5039. doi: 10.1021/ja900436t. [DOI] [PubMed] [Google Scholar]
- 117.Kozak CR, Kistler KA, Lu Z, Matsika S. Excited-state energies and electronic couplings of DNA base dimers. J. Phys. Chem. B. 2010;114:1674–1683. doi: 10.1021/jp9072697. [DOI] [PubMed] [Google Scholar]
- 118.Banyasz A, Douki T, Improta R, Gustavsson T, Onidas D, Vaya I, Perron M, Markovitsi D. Electronic excited states responsible for dimer formation upon UV absorption directly by thymine strands: joint experimental and theoretical study. J. Am. Chem. Soc. 2012;134:14834–14845. doi: 10.1021/ja304069f. [DOI] [PubMed] [Google Scholar]
- 119.Giussani A, Serrano-Andrés L, Merchán M, Roca-Sanjuán D, Garavelli M. Photoinduced formation mechanism of the thymine–thymine (6-4) adduct. J. Phys. Chem. B. 2013;117:1999–2004. doi: 10.1021/jp307200g. [DOI] [PubMed] [Google Scholar]
- 120.Pilles BM, Bucher DB, Liu L, Clivio P, Gilch P, Zinth W, Schreier WJ. Mechanism of the decay of thymine triplets in DNA single strands. J. Phys. Chem. Lett. 2014;5:1616–1622. doi: 10.1021/jz500364g. [DOI] [PubMed] [Google Scholar]
- 121.Rahn RO, Hosszu JL. Photochemistry of polynucleotides: a summary of temperature effects. Photochem. Photobiol. 1968;7:637–642. [Google Scholar]
- 122.Eisenstein M, Frolow F, Shakked Z, Rabinovich D. The structure and hydration of the A-DNA fragement d(GGGTACCC) at room temperature and low temperature. Nucleic Acids Res. 1990;18:3185–3194. doi: 10.1093/nar/18.11.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Varghese AJ. Photochemistry of thymidine in ice. Biochemistry. 1970;9:4781–4787. doi: 10.1021/bi00826a023. [DOI] [PubMed] [Google Scholar]
- 124.Ames DM, Lin G, Jian Y, Cadet J, Li L. Unusually large deuterium discrimination during spore photoproduct formation. J. Org. Chem. 2014;79:4843–4851. doi: 10.1021/jo500775b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Friedel M, Berteau O, Pieck J, Atta M, Ollagnier-de-Choudens S, Fontecave M, Carell T. The spore photoproduct lyase repairs the 5S- and not the 5R-configured spore photoproduct DNA lesion. Chem. Commun. 2006:445–447. doi: 10.1039/b514103f. [DOI] [PubMed] [Google Scholar]
- 126.Friedel M, Pieck J, Klages J, Dauth C, Kessler H, Carell T. Synthesis and stereochemical assignment of DNA spore photoproduct analogues. Chem. Eur. J. 2006;12:6081–6094. doi: 10.1002/chem.200600169. [DOI] [PubMed] [Google Scholar]
- 127.Chandra T, Silver SC, Zilinskas E, Shepard EM, Broderick WE, Broderick JB. Spore photoproduct lyase catalyzes specific repair of the 5R but not the 5S spore photoproduct. J. Am. Chem. Soc. 2009;131:2420–2421. doi: 10.1021/ja807375c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lin G, Chen C-H, Pink M, Pu J, Li L. Chemical synthesis, crystal structure and enzymatic evaluation of a dinucleotide spore photoproduct analogue containing a formacetal linker. Chem. Eur. J. 2011;17:9658–9668. doi: 10.1002/chem.201101821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Heil K, Kneuttinger AC, Schneider S, Lischke U, Carell T. Crystal structures and repair studies reveal the identity and the base-pairing properties of the UV-induced spore photoproduct DNA lesion. Chem. Eur. J. 2011;17:9651–9657. doi: 10.1002/chem.201100177. [DOI] [PubMed] [Google Scholar]
- 130.Valle-Orero J, Wildes A, Garden J-L, Peyrard M. Purification of A-form DNA fiber samples by the removal of B-form DNA residues. J. Phys. Chem. B. 2013;117:1849–1856. doi: 10.1021/jp311199f. [DOI] [PubMed] [Google Scholar]
- 131.Falk M, Hartman KA, Lord RC. Hydration of deoxyribonucleic acid. III. A spectroscopic study of the effect of hydration on the structure of deoxyribonucleic acid. J. Am. Chem. Soc. 1963;85:391–394. [Google Scholar]
- 132.Rahn RO, Hosszu HL. Influence of relative humidity on the photochemistry of DNA films. Biochim. Biophys. Acta. 1969;190:126–131. doi: 10.1016/0005-2787(69)90161-0. [DOI] [PubMed] [Google Scholar]
- 133.Slieman TA, Rebeil R, Nicholson WL. Spore photoproduct (SP) lyase from Bacillus subtilis specifically binds to and cleaves SP (5-thyminyl-5,6-dihydrothymine) but not cyclobutane pyrimidine dimers in UV-irradiated DNA. J. Bacteriol. 2000;182:6412–6417. doi: 10.1128/jb.182.22.6412-6417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Douki T, Cadet J. Formation of the spore photoproduct and other dimeric lesions between adjacent pyrimidines in UVC-irradiated dry DNA. Photochem. Photobiol. Sci. 2003;2:433–436. doi: 10.1039/b300173c. [DOI] [PubMed] [Google Scholar]
- 135.Douki T, Laporte G, Cadet J. Inter-strand photoproducts are produced in high yield within A-DNA exposed to UVC radiation. Nucleic Acids Res. 2003;31:3134–3142. doi: 10.1093/nar/gkg408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gray DM, Ratliff RL. Circular-dichroism spectra of poly[d(AC)-d(GT)], poly[r(AC)-r(GU)], and hybrids poly[d(AC)-r(GU)] and poly[r(AC)-d(GT)] in presence of ethanol. Biopolymers. 1975;14:487–498. doi: 10.1002/bip.1975.360140305. [DOI] [PubMed] [Google Scholar]
- 137.Ivanov VI, Minchenk Le, Minyat EE, Frankkam Md, Schyolki Ak. B to A transition of DNA in solution. J. Mol. Biol. 1974;87:817–833. doi: 10.1016/0022-2836(74)90086-2. [DOI] [PubMed] [Google Scholar]
- 138.Cadet J, Vigny P. Photochemistry and the nucleic acids. In: Morrison H, editor. Bioorganic photochemistry v. 1. Vol. 1. Wiley, New York: 1990. pp. 1–272. [Google Scholar]
- 139.Lin G, Li L. Elucidation of spore-photoproduct formation by isotope labeling. Angew. Chem. Int. Ed. 2010;49:9926–9929. doi: 10.1002/anie.201005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Krishtalik LI. The mechanism of the proton transfer: an outline. Biochim. Biophys. Acta Bioenergetics. 2000;1458:6–27. doi: 10.1016/s0005-2728(00)00057-8. [DOI] [PubMed] [Google Scholar]
- 141.Ostrowski T, Maurizot J-C, Adeline M-T, Fourrey J-L, Clivio P. Sugar conformational effects on the photochemistry of thymidylyl(3'-5')thymidine. J. Org. Chem. 2003;68:6502–6510. doi: 10.1021/jo030086p. [DOI] [PubMed] [Google Scholar]
- 142.Moriou C, Thomas M, Adeline M-T, Martin M-T, Chiaroni A, Pochet S, Fourrey J-L, Favre A, Clivio P. Crystal structure and photochemical behavior in solution of the 3'-N-sulfamate analogue of thymidylyl(3'-5')thymidine. J. Org. Chem. 2007;72:43–50. doi: 10.1021/jo061488a. [DOI] [PubMed] [Google Scholar]
- 143.Desnous C, Babu BR, Moriou C, Mayo JUO, Favre A, Wengel J, Clivio P. The sugar conformation governs (6-4) photoproduct formation at the dinucleotide level. J. Am. Chem. Soc. 2008;130:30–31. doi: 10.1021/ja077095q. [DOI] [PubMed] [Google Scholar]
- 144.Jian Y, Ames DM, Ouyang H, Li L. Photochemical reactions of microcrystalline thymidine. Org. Lett. 2015;17:824–827. doi: 10.1021/ol5036276. [DOI] [PubMed] [Google Scholar]
- 145.Young DW, Tollin P, Wilson HR. The crystal and molecular structure of thymidine. Acta Crystallogr. B. 1969;25:1423–1432. [Google Scholar]
- 146.Moysan A, Viari A, Vigny P, Voituriez L, Cadet J, Moustacchi E, Sage E. Formation of cyclobutane thymine dimers photosensitized by pyridopsoralens: quantitative and qualitative distribution within DNA. Biochemistry. 1991;30:7080–7088. doi: 10.1021/bi00243a007. [DOI] [PubMed] [Google Scholar]
- 147.Bergstrom DE, Rash KF. Synthesis of 5,6-dihydro-5-(alpha-thyminyl)thymine. J. Org. Chem. 1979;44:1414–1417. [Google Scholar]
- 148.Nicewonger R, Begley TP. Synthesis of the spore photoproduct. Tetrahedron Lett. 1997;38:935–936. [Google Scholar]
- 149.Jian Y, Li L. Chemical syntheses of oligodeoxyribonucleotides containing spore photoproduct. J. Org. Chem. 2013;78:3021–3029. doi: 10.1021/jo400013n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kim SJ, Lester C, Begley TP. Synthesis of the dinucleotide spore photoproduct. J. Org. Chem. 1995;60:6256–6257. [Google Scholar]
- 151.Matteucci M. Deoxyoligonucleotide analogs based on formacetal linkages. Tetrahedron Lett. 1990;31:2385–2388. [Google Scholar]
- 152.Gao X, Brown FK, Jeffs P, Bischofberger N, Lin KY, Pipe AJ, Noble SA. Probing structural factors stabilizing antisense oligonucleotide duplexes: NMR studies of a DNA•DNA duplex containing a formacetal linkage. Biochemistry. 1992;31:6228–6236. doi: 10.1021/bi00142a009. [DOI] [PubMed] [Google Scholar]
- 153.Jones RJ, Lin KY, Milligan JF, Wadwani S, Matteucci MD. Synthesis and binding properties of pyrimidine oligodeoxynucleoside analogs containing neutral phosphodiester replacements: the formacetal and 3'-thioformacetal internucleoside linkages. J. Org. Chem. 1993;58:2983–2991. [Google Scholar]
- 154.Rozners E, Strömberg R. Synthesis and properties of oligoribonucleotide analogs having formacetal internucleoside linkages. J. Org. Chem. 1997;62:1846–1850. [Google Scholar]
- 155.Butenandt J, Eker APM, Carell T. Synthesis, crystal structure, and enzymatic evaluation of a DNA-photolesion isostere. Chem. Eur. J. 1998;4:642–654. [Google Scholar]
- 156.Varma RS. Synthesis of oligonucleotide analogs with modified backbones. Synlett. 1993:621–637. [Google Scholar]
- 157.Heil K, Pearson D, Carell T. Chemical investigation of light induced DNA bipyrimidine damage and repair. Chem. Soc. Rev. 2011;40:4271–4278. doi: 10.1039/c000407n. [DOI] [PubMed] [Google Scholar]
- 158.Taylor J-S, Brockie IR. Synthesis of a trans-syn thymine dimer building block. Solid phase synthesis of CGTAT[t,s]TATGC. Nucleic Acids Res. 1988;16:5123–5136. doi: 10.1093/nar/16.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Taylor JS, Brockie IR, O'Day CL. A building block for the sequence-specific introduction of cis-syn thymine dimers into oligonucleotides. Solid-phase synthesis of TpT[c,s]pTpT. J. Am. Chem. Soc. 1987;109:6735–6742. [Google Scholar]
- 160.Ortiz Mayo JU, Thomas M, Saintomé C, Clivio P. Facile synthesis of a cis–syn thymine dimer building block and its incorporation into oligodeoxynucleotides. Tetrahedron. 2003;59:7377–7383. [Google Scholar]
- 161.Friedel MG, Gierlich J, Carell T. The Chemistry of Cyclobutanes. John Wiley & Sons, Ltd; 2006. Cyclobutane pyrimidine dimers as UV-induced DNA lesions; pp. 1031–1059. [Google Scholar]
- 162.Murata T, Iwai S, Ohtsuka E. Synthesis and characterization of a substrate for T4 endonuclease V containing a phosphorodithioate linkage at the thymine dimer site. Nucleic Acids Res. 1990;18:7279–7286. doi: 10.1093/nar/18.24.7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tommasi S, Swiderski PM, Tu Y, Kaplan BE, Pfeifer GP. Inhibition of transcription factor binding by ultraviolet-induced pyrimidine dimers. Biochemistry. 1996;35:15693–15703. doi: 10.1021/bi962117z. [DOI] [PubMed] [Google Scholar]
- 164.Kosmoski JV, Smerdon MJ. Synthesis and nucleosome structure of DNA containing a UV photoproduct at a specific site. Biochemistry. 1999;38:9485–9494. doi: 10.1021/bi990297h. [DOI] [PubMed] [Google Scholar]
- 165.Butenandt J, Burgdorf LT, Carell T. Synthesis of DNA lesions and DNA-lesion-containing oligonucleotides for DNA-repair studies. Synthesis. 1999:1085–1105. [Google Scholar]
- 166.Iwai S, Shimizu M, Kamiya H, Ohtsuka E. Synthesis of a phosphoramidite coupling unit of the pyrimidine (6-4) pyrimidone photoproduct and its incorporation into oligodeoxynucleotides. J. Am. Chem. Soc. 1996;118:7642–7643. [Google Scholar]
- 167.Cadet J, Voituriez L, Hruska FE, Grand A. Crystal structure of the cis–syn photodimer of thymidylyl (3′-5′) thymidine cyanoethyl ester. Biopolymers. 1985;24:897–903. doi: 10.1002/bip.360240512. [DOI] [PubMed] [Google Scholar]
- 168.Zheng G, Lu X-J, Olson WK. Web 3DNA—a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009;37:W240–W246. doi: 10.1093/nar/gkp358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lu X-J, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protocols. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Singh I, Jian Y, Li L, Georgiadis MM. The structure of an authentic spore photoproduct lesion in DNA suggests a basis for recognition. Acta Crystallogr. D. 2014;70:752–759. doi: 10.1107/S1399004713032987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Goodwin KD, Long EC, Georgiadis MM. A host-guest approach for determining drug-DNA interactions: an example using netropsin. Nucleic Acids Res. 2005;33:4106–4116. doi: 10.1093/nar/gki717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Park H, Zhang K, Ren Y, Nadji S, Sinha N, Taylor J-S, Kang C. Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc. Natl. Acad. Sci. 2002;99:15965–15970. doi: 10.1073/pnas.242422699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Taylor JS, Garrett DS, Brockie IR, Svoboda DL, Telser J. Proton NMR assignment and melting temperature study of cis-syn and trans-syn thymine dimer containing duplexes of d(CGTATTATGC)•d(GCATAATACG) Biochemistry. 1990;29:8858–8866. doi: 10.1021/bi00489a049. [DOI] [PubMed] [Google Scholar]
- 174.McAteer K, Jing Y, Kao J, Taylor JS, Kennedy MA. Solution-state structure of a DNA dodecamer duplex containing a cis-syn thymine cyclobutane dimer, the major UV photoproduct of DNA. J. Mol. Biol. 1998;282:1013–1032. doi: 10.1006/jmbi.1998.2062. [DOI] [PubMed] [Google Scholar]
- 175.Yokoyama H, Mizutani R, Satow Y. Structure of a double-stranded DNA (6-4) photoproduct in complex with the 64M-5 antibody Fab. Acta Crystallogr. D. 2013;69:504–512. doi: 10.1107/S0907444912050007. [DOI] [PubMed] [Google Scholar]
- 176.Maul M, Barends TM, Glas AF, Cryle M, Domratcheva T, Schneider S, Schlichting I, Carell T. Crystal structure and mechanism of a DNA (6-4) photolyase. Angew. Chem. Int. Ed. 2008;47:10076–10080. doi: 10.1002/anie.200804268. [DOI] [PubMed] [Google Scholar]
- 177.Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jong-Ki Kim R, Byong-Seok C. The solution structure of DNA duplex-decamer containing the (6-4) photoproduct of thymidylyl (3'→5')thymidine by NMR and relaxation matrix refinement. Eur. J. Biochem. 1995;228:849–854. doi: 10.1111/j.1432-1033.1995.tb20331.x. [DOI] [PubMed] [Google Scholar]
- 179.Lee J-H, Hwang G-S, Choi B-S. Solution structure of a DNA decamer duplex containing the stable 3′ T•G base pair of the pyrimidine(6-4)pyrimidone photoproduct [(6-4) adduct]: Implications for the highly specific 3′ T → C transition of the (6-4) adduct. Proc. Natl. Acad. Sci. 1999;96:6632–6636. doi: 10.1073/pnas.96.12.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lukin M, de los Santos C. NMR structures of damaged DNA. Chem. Rev. 2006;106:607–686. doi: 10.1021/cr0404646. [DOI] [PubMed] [Google Scholar]
- 181.Young AR, Chadwick CA, Harrison GI, Hawk JLM, Nikaido O, Potten CS. The In Situ repair kinetics of epidermal thymine dimers and 6-4 photoproducts in human skin types I and II. J. Investig. Dermatol. 1996;106:1307–1313. doi: 10.1111/1523-1747.ep12349031. [DOI] [PubMed] [Google Scholar]
- 182.de Laat WL, Jaspers NGJ, Hoeijmakers JHJ. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 183.de Boer J, Hoeijmakers JHJ. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21:453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- 184.Jans J, Schul W, Sert Y-G, Rijksen Y, Rebel H, Eker APM, Nakajima S, van Steeg H, de Gruijl FR, Yasui A, Hoeijmakers JHJ, van der Horst GTJ. Powerful skin cancer protection by a CPD-photolyase transgene. Curr. Biol. 2005;15:105–115. doi: 10.1016/j.cub.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 185.Yoon J-H, Prakash L, Prakash S. Highly error-free role of DNA polymerase η in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc. Natl. Acad. Sci. 2009;106:18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Taggart DJ, Camerlengo TL, Harrison JK, Sherrer SM, Kshetry AK, Taylor J-S, Huang K, Suo Z. A high-throughput and quantitative method to assess the mutagenic potential of translesion DNA synthesis. Nucleic Acids Res. 2013;41:e96. doi: 10.1093/nar/gkt141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Johnson RE, Haracska L, Prakash S, Prakash L. Role of DNA polymerase η in the bypass of a (6-4) TT photoproduct. Mol. Cell. Biol. 2001;21:3558–3563. doi: 10.1128/MCB.21.10.3558-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Muñoz-Sánchez J, Cabrera-Juárez E. In vitro mutation of Haemophilus influenzae transforming deoxyribonucleic acid by ultraviolet radiation at −70°C. Mutat. Res.-Fund. Mol. M. 1991;251:21–29. doi: 10.1016/0027-5107(91)90212-7. [DOI] [PubMed] [Google Scholar]
- 189.Lin G, Jian Y, Dria KJ, Long EC, Li L. Reactivity of damaged pyrimidines: DNA cleavage via hemiaminal formation at the C4 positions of the saturated thymine of spore photoproduct and dihydrouridine. J. Am. Chem. Soc. 2014;136:12938–12946. doi: 10.1021/ja505407p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cano RJ, Borucki MK. Revival and identification of bacterial spores in 25- to 40-million-year-old Dominican amber. Science. 1995;268:1060–1064. doi: 10.1126/science.7538699. [DOI] [PubMed] [Google Scholar]
- 191.Vreeland RH, Rosenzweig WD, Powers DW. Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal. Nature. 2000;407:897–900. doi: 10.1038/35038060. [DOI] [PubMed] [Google Scholar]
- 192.Kennedy MJ, Reader SL, Swierczynski LM. Preservation records of micro-organisms: evidence of the tenacity of life. Microbiology. 1994;140(Pt 10):2513–2529. doi: 10.1099/00221287-140-10-2513. [DOI] [PubMed] [Google Scholar]
- 193.Cadet J, Grand A, Douki T. Solar UV Radiation-Induced DNA Bipyrimidine Photoproducts: Formation and Mechanistic Insights. In: Barbatti M, Borin AC, Ullrich S, editors. Photoinduced Phenomena in Nucleic Acids II. Vol. 356. Springer International Publishing; 2015. pp. 249–275. [DOI] [PubMed] [Google Scholar]
- 194.Hieda K. Effects of dehydration on the sensitivity of yeast to ultraviolet light. Mutat. Res.-Fund. Mol. M. 1971;12:365–380. [Google Scholar]
- 195.Hieda K. Induction of genetic changes in Saccharomyces cerevisiae by partial drying in air of constant relative humidity and by UV. Mutat. Res. 1981;84:17–27. doi: 10.1016/0027-5107(81)90046-4. [DOI] [PubMed] [Google Scholar]
- 196.Ashwood-Smith MJ, Bridges BA, Munson RJ. Ultraviolet damage to bacteria and bacteriophage at low temperatures. Science. 1965;149:1103–1105. doi: 10.1126/science.149.3688.1103. [DOI] [PubMed] [Google Scholar]
- 197.Hieda K, Ito T. The enhancement of UV sensitivity of yeast in frozen state. Mutat. Res. 1968;6:325–327. doi: 10.1016/0027-5107(68)90049-3. [DOI] [PubMed] [Google Scholar]
- 198.DiMaio F, Yu X, Rensen E, Krupovic M, Prangishvili D, Egelman EH. Virology. A virus that infects a hyperthermophile encapsidates A-form DNA. Science. 2015;348:914–917. doi: 10.1126/science.aaa4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mori T, Matsunaga T, Hirose T, Nikaido O. Establishment of a monoclonal antibody recognizing ultraviolet light-induced (6-4) photoproducts. Mutat. Res. 1988;194:263–270. doi: 10.1016/0167-8817(88)90028-4. [DOI] [PubMed] [Google Scholar]
- 200.Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6-4) photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem. Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 201.Strickland PT, Boyle JM. Characterisation of two monoclonal antibodies specific for dimerised and non-dimerised adjacent thymidines in single stranded DNA. Photochem. Photobiol. 1981;34:595–601. [PubMed] [Google Scholar]
- 202.Strickland PT, Nikaido O, Matsunaga T, Boyle JM. Further characterization of monoclonal antibody indicates specificity for (6-4)-dipyrimidine photoproducts. Photochem. Photobiol. 1992;55:723–727. [PubMed] [Google Scholar]
- 203.Prescott LM, Harley JP, Klein DA. Microbiology. Dubuque, IA: McGraw-Hill Higher Education; 2005. [Google Scholar]