Abstract
Sonic hedgehog (Shh) signaling patterns the vertebrate spinal cord by activating a group of transcriptional repressors in distinct neural progenitors of somatic motor neuron and interneuron subtypes. To identify the action of this network, we performed a genome-wide analysis of the regulatory actions of three key ventral determinants in mammalian neural tube patterning: Nkx2.2, Nkx6.1 and Olig2. Previous studies have demonstrated that each factor acts predominantly as a transcriptional repressor, at least in part, to inhibit alternative progenitor fate choices. Here, we reveal broad and direct repression of multiple alternative fates as a general mechanism of repressor action. Additionally, the repressor network targets multiple Shh signaling components providing negative feedback to ongoing Shh signaling. Analysis of chromatin organization around Nkx2.2-, Nkx6.1- and Olig2-bound regions, together with co-analysis of engagement of the transcriptional activator Sox2, indicate that repressors bind to, and probably modulate the action of, neural enhancers. Together, the data suggest a model for neural progenitor specification downstream of Shh signaling, in which Nkx2.2 and Olig2 direct repression of alternative neural progenitor fate determinants, an action augmented by the overlapping activity of Nkx6.1 in each cell type. Integration of repressor and activator inputs, notably activator inputs mediated by Sox2, is probably a key mechanism in achieving cell type-specific transcriptional outcomes in mammalian neural progenitor fate specification.
KEY WORDS: Transcriptional regulation, Fate specification, Neural development, Mouse
Summary: Unbiased functional genomics approaches reveal that Nkx2.2, Nkx6.1 and Olig2 directly repress multiple alternative fates during mammalian neural progenitor fate specification.
INTRODUCTION
Sonic hedgehog (Shh) signaling is crucial for the specification of ventral neural progenitor types that give rise to molecularly and functionally distinct classes of ventral neurons in the developing vertebrate central nervous system (Dessaud et al., 2008). Several lines of evidence, including the direct binding of Gli transcription factors to their cis-regulatory modules, have identified Nkx2.2, Nkx6.1 and Olig2 as direct transcriptional targets of the ventral neural patterning activity of Shh (Lei et al., 2006; Oosterveen et al., 2012; Peterson et al., 2012; Wang et al., 2011). Each of these factors has been shown to function as a transcriptional repressor in neural patterning: Olig2 is required for the specification of somatic motor neuron progenitors, Nkx2.2 for the specification of V3 interneuron progenitors, whereas Nkx6.1 expression overlaps V2 and V3 interneurons and somatic motor neuron progenitors and is essential for the normal specification of both populations (Briscoe et al., 2000, 1999; Lu et al., 2002; Mizuguchi et al., 2001; Muhr et al., 2001; Novitch et al., 2001; Sander et al., 2000; Vallstedt et al., 2001; Zhou and Anderson, 2002; Zhou et al., 2001) (Fig. 1A). Although their general roles in specifying respective neural progenitor types downstream of the Shh pathway have been documented through mis-expression studies (Briscoe et al., 2000; Mizuguchi et al., 2001; Novitch et al., 2001), their direct DNA-associated regulatory actions and target specificity are currently not understood.
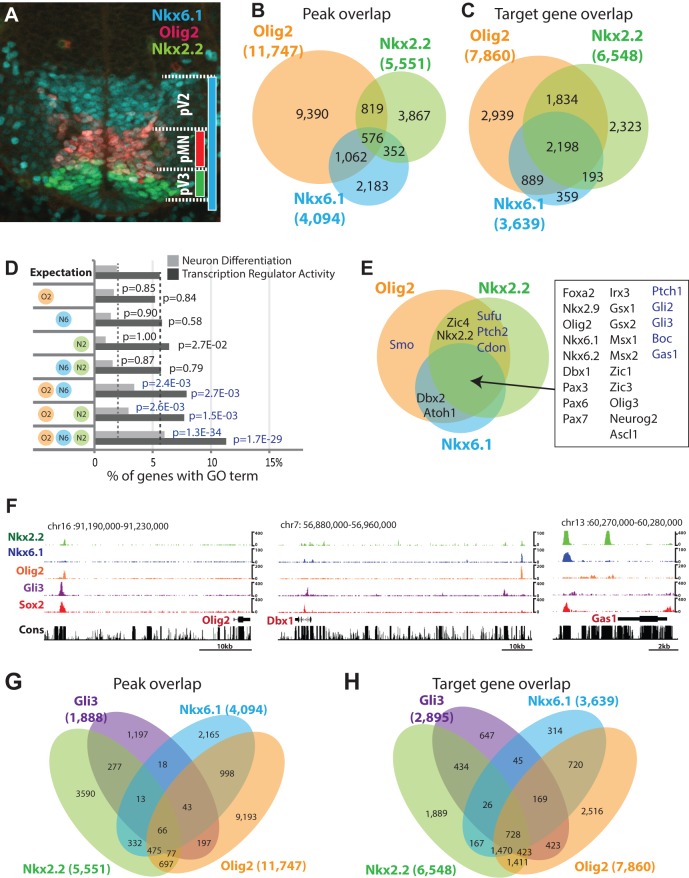
Fig. 1.
Characteristics of Nkx2.2, Nkx6.1 and Olig2 target genes. (A) Immunofluorescence assay on transverse E10.5 neural tube section at forelimb level with indicated antibodies. (B) Venn diagram intersection of Nkx2.2-, Nkx6.1- and Olig2-binding regions. (C) Venn diagram intersection of Nkx2.2, Nkx6.1 and Olig2 target genes. (D) Gene Ontology analysis summary for genes targeted by different combinations of factors. (E) Target gene Venn diagram highlighting neural progenitor fate determinants and Sonic hedgehog pathway components. (F) Genome browser snapshots showing indicated ChIP-seq signal. Cons, Phastcon 30 conservation score. (G,H) Venn diagrams for binding region overlap (G) and target gene overlap (H) between Nkx2.2, Nkx6.1, Olig2 and Gli3.
In this report, we undertook an integrative, functional genomic approach to identify genomic binding regions and target genes of Nkx2.2, Nkx6.1 and Olig2 in embryonic stem cell (ESC)-derived neural progenitors. Our data highlight a previously unappreciated breadth of direct fate exclusion, modulation of ongoing upstream Shh signaling input through multiple signaling nodes (Lek et al., 2010) and Sox2 input into available enhancers, with a resulting cell type-specific output directing a specific neural progenitor type (Oosterveen et al., 2012; Peterson et al., 2012).
RESULTS AND DISCUSSION
To examine the direct regulatory actions of the Shh-initiated transcriptional network (Lei et al., 2006; Oosterveen et al., 2012; Peterson et al., 2012), we performed ChIP-seq for Nkx2.2, Nkx6.1 and Olig2 on neural progenitors derived in vitro from mouse embryonic stem cells (mESCs); a model system that recapitulates in vivo patterning processes (Peterson et al., 2012; Wichterle et al., 2002) (supplementary material Table S1). The binding events were reproducibly detected in biological replicates (supplementary material Fig. S1A); moreover, binding was confirmed in neural tube preparations from embryonic day (E)10.5 embryos at 28 out of 36 loci tested (Nkx2.2: 7/11, Nkx6.1: 11/11, Olig2: 10/14) (supplementary material Fig. S1B). DNA regions bound by each factor showed considerable overlap (Fig. 1B); an even greater overlap was observed in the potential target genes: assigned as the nearest 5′ and 3′ neighboring genes to the bound regions (Fig. 1C). These data suggest that the three factors engage a common set of target genes through cis-regulatory elements, many of which bind all three factors, as well as discrete regulatory elements engaging specific members of the regulatory trio.
To assess the significance of the predicted target gene overlap, we performed Gene Ontology (GO) term analysis. ‘Neuron Differentiation’ and ‘Transcription Regulator Activity’ GO terms were strongly enriched in the gene sets targeted by all three repressors (3.1-fold and 2.0-fold, respectively) when compared with single or pairwise targeted gene sets. These data suggest that co-targeting defines the most relevant neural targets within the repressor network in neural fate specification. Detailed analyses showed that a number of known neural fate determinants as well as components of the Hedgehog pathway were co-targeted (Fig. 1E,F; supplementary material Fig. S1C, Figs S2-S4). Targeted neural fate regulators included both progenitor-expressed transcription factors (e.g. Pax6, Irx3) as well as transcriptional regulators active in post-mitotic neurons (e.g. Isl1, En1) (supplementary material Fig. S1C). These results are consistent with at least two regulatory strategies for the ventral repressor program: the repression of alternative neural subtype fates at both progenitor and post-mitotic levels and feedback modulation of the Hedgehog pathway. Interestingly, although Nkx2.2, Nkx6.1 and Olig2 mediate tissue patterning as Shh primary targets (Lei et al., 2006; Oosterveen et al., 2012; Peterson et al., 2012; Wang et al., 2011), intersectional analysis showed a limited overlap with Gli1- and Gli3-bound regions overall (Fig. 1G; supplementary material Fig. S6D). Exceptions were putative cis-regulatory regions around ventral neural progenitor sub-type specifiers, including Nkx2.2, Olig2 and Nkx6.1 (supplementary material Fig. S2) (Lei et al., 2006; Oosterveen et al., 2012; Peterson et al., 2012; Wang et al., 2011). At the target gene-level, Gli factors showed extensive overlap, particularly with genes targeted by all three repressors (Fig. 1H; supplementary material Fig. S6E, Table S1). Thus, the initial Shh/Gli input, and downstream Nkx2.2, Nkx6.1 and Olig2 repressor programs, share common target genes but act through distinct cis-regulatory modules.
To address the predicted repressor program functionally, we used a doxycycline-inducible transgenic mESC system (Iacovino et al., 2011; Mazzoni et al., 2011) to express ectopically Nkx2.2, Nkx6.1 and Olig2 individually, or in pairwise combinations: Nkx2.2 and Nkx6.1 (Nkx6.1-2A-Nkx2.2), and Olig2 and Nkx6.1 (Nkx6.1-2A-Olig2). Samples were subjected to global analysis of transcriptional activity by RNA-seq 12 h post-Dox induction, and targeted analysis of a subset of genes by microfluidic-based RT-qPCR (Fluidigm) (Fig. 2A,B). At the global level, we observed an extensive set of targets displaying reduced mRNA levels on individual activation of Nkx2.2, Nkx6.1 and Olig2, as well as their pairwise combinations (Fig. 2C; supplementary material Table S2), which was in good agreement with predictions of the direct DNA interaction data (Fig. 1B-E; supplementary material Fig. S1C). Factor-specific effects were only evident for a small set of genes (clustered toward the left in Fig. 2C). Overall, 76-96 genes were downregulated by individual factor expression and 172-192 in pairwise combinations, setting a twofold cutoff in target gene repression (supplementary material Table S2). Of these downregulated genes, 57-71% were associated with binding of the respective factors, a 1.9- to 4.7-fold enrichment over random expectation (P<0.01) (supplementary material Table S2). Consistent with the ChIP-seq GO profile, the RNA-seq GO profile showed enrichment for neural differentiation and transcription factor terms (supplementary material Table S2). Importantly, alternative fate determinants and Shh pathway components were strongly represented in the highly downregulated gene set (Fig. 2C; supplementary material Table S2).
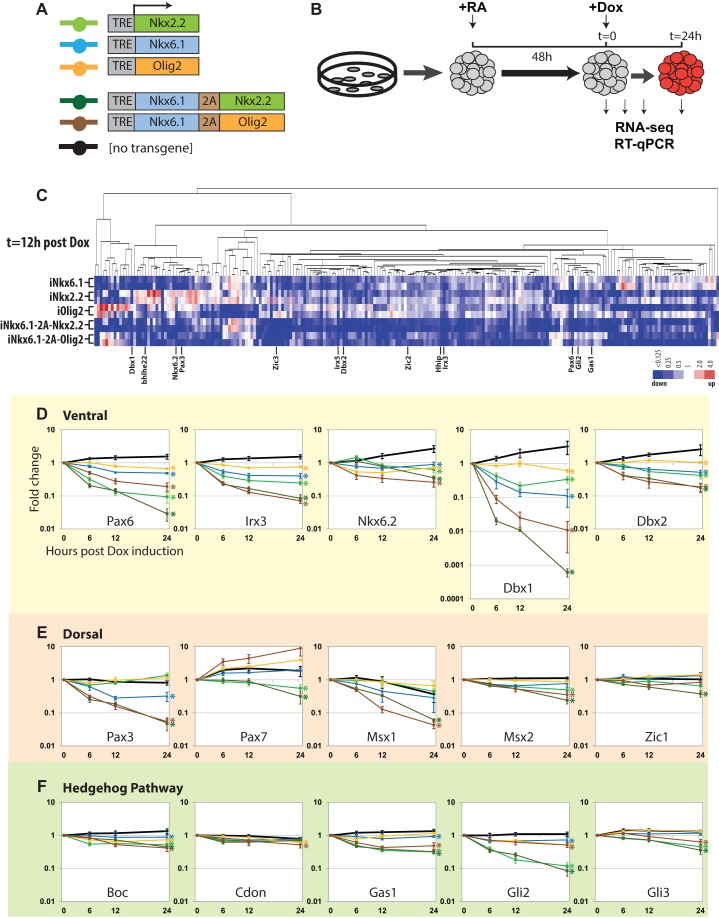
Fig. 2.
Nkx2.2, Nkx6.1 and Olig2 overexpression assay in neural progenitors. (A) Schematic describing transgene structures. (B) Schematic of overexpresssion experiment design. Cell aggregates were generated from mESCs and subjected to neural differentiation and transgene activation. RA, all-trans retinoic acid; Dox, doxycycline. (C) Hierarchical clustering of genes displaying a twofold or greater change in mRNA-seq data relative to the parental cell line 12 h after Dox-mediated activation of transcriptional repressors. Fold change to the parental cell line is shown. (D-F) RT-qPCR time course repression assay. See A for color designations. x-axis, hours post-Dox induction; y-axis, fold change from Dox induction (t=0). Error bars indicate s.e.m. based on three biological replicates. Asterisks indicate significant difference from non-transgenic control based on the s.e.m.
Select genes representing ventral and dorsal neural progenitor fate determinants and Shh pathway components were subjected to a more extensive temporal analysis of regulation by RT-qPCR (Fluidigm). Consistent with the RNA-seq data, Nkx2.2, Nkx6.1 and Olig2 reduced mRNA levels for all tested ventral fate determinants individually (Fig. 2D); their effects were evident within 6 h of Dox-mediated induction. Interestingly, Nkx6.1 enhanced the repressive phenotype observed with Olig2 and Nkx2.2 (Fig. 2D). Together, the results suggest a direct repressive action of the ventral patterning factors on the expression of other transcriptional determinants of neural patterning that is probably through independent regulatory mechanisms, given the additive effects observed in the co-expression analysis. However, genes encoding dorsal neural progenitor fate determinants showed a marked weighting in their specific responses to individual factors: some genes showed little response (e.g. Msx2 and Zic1), whereas expression of others was reduced on activation of a specific factor or combination of factors (e.g. Pax3, Pax7 and Msx1) (Moore et al., 2013) (Fig. 2E; supplementary material Table S2). Such differential sensitivities to repressor input suggest that the regulatory systems restricting dorsal and ventral progenitor fates are largely distinct programs (Briscoe et al., 2000). The targets of the repressor network revealed here are notably broader than previously appreciated (Briscoe et al., 2000; Moore et al., 2013; Oosterveen et al., 2012). This probably reflects redundancies in the regulatory circuitry that obscure de-repression effects in mutant analysis and an emphasis on a restricted set of local cross-repressive interactions in previous ectopic expression studies.
Repression was also highly selective for Hedgehog signaling components, with Nkx2.2 displaying a stronger inhibitory activity on gene expression than Olig2. Although genes enhancing (e.g. Gli2, Gas1 and Boc) and inhibiting (e.g. Gli3) Shh signaling showed reduced expression, the strongest effect was observed on key genes that promote Shh signaling, including Gli2, the predominant transcriptional activator in the Hedgehog pathway, and Gas1, a co-receptor in Shh signaling (Fig. 2F). These results are consistent with the notion that Nkx2.2 exerts its patterning action in part by negative-feedback regulation of Hedgehog pathway components (Lek et al., 2010).
To determine whether the observed repressive effects were dependent on DNA binding, point mutations predicted to abolish direct DNA binding (see Materials and Methods) were introduced into the DNA-binding domains of the Nkx2.2, Nkx6.1 and Olig2 transgenes. Each mutant form showed a loss of repression in the RT-qPCR assay (supplementary material Fig. S5). Thus, direct DNA binding is probably the primary mode by which each regulatory factor controls gene activity. This conclusion is supported by motif analysis that recovered centrally positioned DNA recognition motifs for each of the factors in factor specific ChIP-seq (Fig. 3A-C).
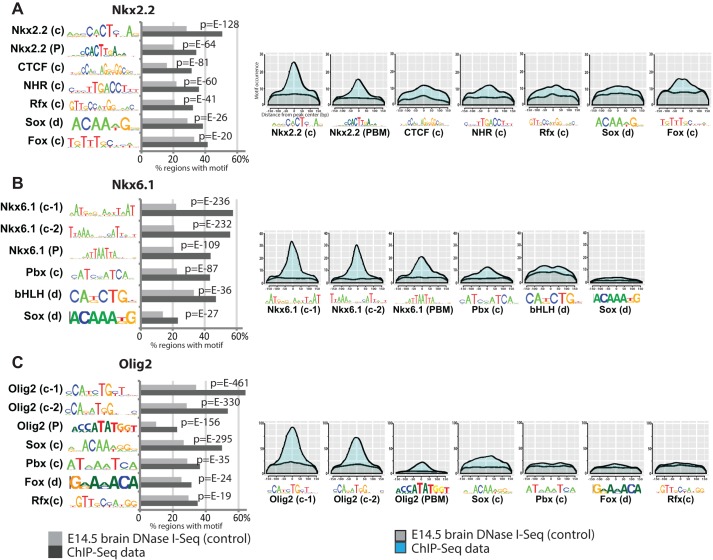
Fig. 3.
Analysis of enriched binding motifs. (A-C) Occurrence of ChIP-recovered and in vitro-determined motifs. c, CisGenome motif recovery; d, DREME motif recovery; P, protein binding microarray. Gray, E14.5 brain DNaseI-seq control dataset; black, ChIPseq data. (Left) Motif distribution histogram relative to binding peak center. x-axis, bp from peak center; y-axis, cumulative motif occurrence. Gray, E14.5 brain DNaseI-seq control; light blue, ChIPseq data.
To gain additional mechanistic insights into regulatory processes by Nkx2.2, Nkx6.1 and Olig2, we analyzed bound regions for overrepresented motifs. The clear similarity of the primary ChIP motif to motifs determined by in vitro binding of factors supports the argument of direct DNA engagement by each factor (Fig. 3A-C). Moreover, the data revealed additional features of DNA engagement modes: the Nkx6.1 primary motifs appear to contain the Nkx6.1 in vitro binding motif and an additional motif separated by a spacer, consistent with complex formation, possibly with Pbx [Fig. 3B, compare Nkx6.1 (c-2), Nkx6.1 (P) and Pbx (c)]. bHLH factors such as Olig2 bind an E-box motif (CAXXTG). Comparison between the unambiguous in vitro Olig2 homodimer motif (CATATG) and the more flexible in vivo motifs (CA T/G A/G TG), as well as inspection of E-box sequences at ChIP peaks (data not shown) suggest that Olig2 binds as both homo- and heterodimers (Fig. 3C). Interestingly, centered Fox and nuclear hormone receptor (NHR) motif predictions in Nkx2.2-bound regions, and a Pbx motif recovered from Nkx6.1-bound regions, suggest a direct regulatory interplay (Fig. 3A,B). SoxB1 transcription factors (Sox1, 2 and 3) play key roles in the active maintenance and fate determination of neural progenitors (Bergsland et al., 2011; Bylund et al., 2003; Graham et al., 2003; Oosterveen et al., 2012; Peterson et al., 2012). Examination of the Nkx2.2, Nkx6.1 and Olig2 datasets showed a consistent enrichment of a Sox motif in bound regions (Fig. 3A-C).
We explored a potential Sox factor association at repressor-bound regions by intersecting Sox2 binding data in neural progenitors (Peterson et al., 2012). Sox2 is best known in the neural lineage for its role in progenitor state maintenance, a general property shared by all progenitors independent of progenitor specificity (Bylund et al., 2003; Graham et al., 2003). In our data, we uncovered extensive overlap of Sox2 binding and DNA regions targeted by all three repressors: 57% of Sox2-associated regions intersected with DNA regions bound by Nkx2.2, Nkx6.1 or Olig2 (supplementary material Fig. S6A). Collectively, these data suggest that both repressor and activator inputs governing progenitor programs are mediated through a common set of regulatory elements (Fig. 4A). Similarly, Sox3 DNA target interactions in neural progenitors (Bergsland et al., 2011) showed an extensive overlap with the repressor trio (46%) (supplementary material Fig. S6B). As only a small percentage of these Sox2/repressor trio-bound regions can be identified in the ESC regulatory profile of Sox2 (9%; see supplementary material Fig. S6C; and Marson et al., 2008), the data reveal a distinct Sox2 engagement with the neural regulatory genome.
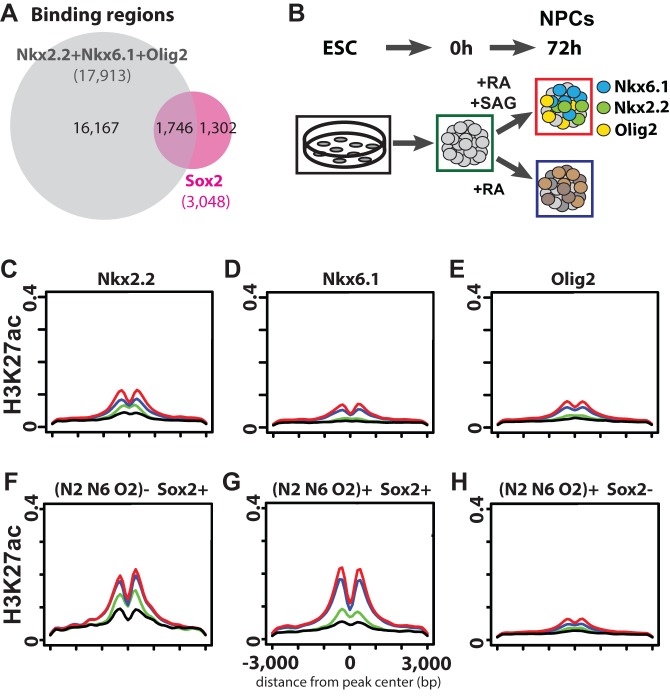
Fig. 4.
Analysis of Sox2 inputs into ventral repressor-bound regions. (A) Venn diagram intersection between Sox2-binding regions and the union of Nkx2.2-, Nkx6.1- and Olig2-binding regions. (B) Schematic showing neural progenitor differentiation protocol. Each condition analyzed in C-H is annotated with a solid box of the corresponding color. (C-H) Aggregate analysis of H3K27ac modification status along neural progenitor differentiation paths. Black, ESCs; green, pre-neural induction; blue, dorsal neural progenitors; red, ventral neural progenitors. Also see B for color coding. (C-E) Individual plot for Nkx2.2-, Nkx6.1- and Olig2-binding regions. (F) Sox2-binding regions that do not overlap with Nkx2.2, Nkx6.1 or Olig2. (G) Sox2-binding regions that overlap with Nkx2.2, Nkx6.1 or Olig2. (H) Nkx2.2-, Nkx6.1- or Olig2-binding regions that do not overlap with Sox2 binding.
To address the regulatory role of the repressor- and activator-bound regions, we analyzed the presence of histone modifications associated with active cis-regulatory elements accompanying neural progenitor specification in vitro (Fig. 4B) (Creyghton et al., 2010; Heintzman et al., 2007; Rada-Iglesias et al., 2011). Regions bound by Nkx2.2, Nkx6.1 and Olig2 associated overall with acetylation of lysine 27 on histone 3 (H3K27ac) in the mixed populations of dorsal and ventral neural progenitors (Fig. 4C-E; supplementary material Fig. S7G), suggesting that repressors probably engage at active transcriptional enhancers (Creyghton et al., 2010; Rada-Iglesias et al., 2011). Interestingly, Nkx2.2-, Nkx6.1- or Olig2-binding regions that do not overlap with Sox2 binding showed only low levels of H3K27ac modification in neural progenitors, or in ESC-derivatives prior to neural specification (Fig. 4H), whereas those overlapping with Sox2 showed markedly elevated H3K27ac levels in a primarily neural progenitor-specific manner (Fig. 4G). Thus, Sox2 engagement correlates with an active enhancer signature at this subset of the repressor targeted genome. Importantly, Sox2-binding regions that do not overlap with Nkx2.2-, Nkx6.1- or Olig2-bound regions showed similar enrichment of H3K27ac, suggesting that these are also active enhancers (Fig. 4F). Nkx2.2-, Nkx6.1- and Olig2-bound regions that do not show significant H3K27ac signal could act as transcriptional silencer domains, a possibility that requires further study. Overall, we observed similar observations and correlations to those with H3K27ac analyzing H3K4me2, a second chromatin modification linked to enhancer signatures (He et al., 2010) (supplementary material Fig. S7A-F,H). Whether the set of enhancers identified here is engaged by distinct repressor networks in more dorsally located neural progenitors or acts independently of repressor networks remains to be determined.
In sum, our data support a model in which neural progenitor diversity in the developing mammalian central nervous system follows from the suppression of alternative neural pathway choices by the action of Shh-dependent transcriptional repressors coupled with Sox-family-mediated transcriptional activation of available cis-regulatory modules within a given progenitor type. The core dorso-ventral neural patterning network is ancient: the spatial arrangements and actions of several key transcriptional components, including vnd/Nkx2, ind/Gsh and msh/Msx are conserved from insects to mammals (Cornell and Ohlen, 2000). In the mammalian pancreas, transcriptional networks involving Nkx2.2 and Nkx6.1 also play central roles in islet cell specification, together with a number of other factors linked to neural fate determination, including Foxa2, Mnx1 and Isl1 (Arda et al., 2013). Exploring the mechanisms at play in neural systems in invertebrate organisms and patterning in mammalian pancreatic development might prove useful for defining further the underlying operating principles of these repressor networks in cell fate specification in animal development.
MATERIALS AND METHODS
Chromatin immunoprecipitation (ChIP) analysis
Neural progenitors were derived from mESCs in serum-free suspension culture in all-trans retinoic acid (RA, Sigma; 500 nM) and SAG (Calbiochem; 100-800 nM) as described previously (Peterson et al., 2012; Wichterle et al., 2002). ChIP-seq was performed as described in Peterson et al. (2012) and analyzed using the CisGenome 2-sample conditional binomial algorithm in an mm9 genome assembly; data are accessible through GEO (accession number GSE65462). In bioinformatics studies, the nearest genes 5′ and 3′ of transcription factor binding regions were considered potential transcriptional targets. Conformational and potential isoform bias in ChIP-seq was minimized by employing a cocktail of factor-specific antibodies: Millipore (AB9610) for Olig2, the cocktail of [DSHB (74.5A5), Sigma (HPA003468) and custom rabbit polyclonal (a gift from T. Jessell)] for Nkx2.2, and the cocktail of [DSHB (F55A10, F55A12, F64A6B4 and F65A2), RD Systems (AF5857) and custom rabbit polyclonal (a gift from T. Jessell)] for Nkx6.1. Gli3-FLAG ChIP was performed with anti-FLAG (M2, Sigma) on a mESC line with a 3× FLAG-Avi tag inserted immediately upstream of the Gli3 start codon. Motif analysis was performed on the top 2000 peaks with CisGenome or DREME combined with TOMTOM (Bailey, 2011). Aggregate plot and heatmap clustering were performed with HOMER (Heinz et al., 2010). Gene ontology annotation was performed through the DAVID program (https://david.ncifcrf.gov/). Sox2 (ESC; accession number GSE11724), Gli1, Sox2, H3K4me2 and H3K27ac (all NPC; accession number GSE42132), H3K4me2 (ESC; accession number GSE11172), H3K27ac (ESC; accession number GSE24164) and DNaseI-seq (E14.5 brain; accession number GSM1014197) were used in this study. A crude neural tube preparation for ChIP-qPCR was performed on limbless, decapitated E10.5 mouse trunk samples, in biological and technical duplicates. See supplementary material Table S3 for primer sequences.
Ectopic expression study
Open reading frames (ORF) were targeted to the engineered Hprt locus in the mA2.lox.Cre mESC line to express the ORFs under the control of a tetracycline response element (Iacovino et al., 2011). Neural progenitors were induced with 500 nM RA as described in the previous section, and the transgene was activated with 1 µg/ml doxycycline (Dox) after two days of RA treatment. RNA samples were collected up to 24 h post-Dox induction and subjected to RNA-seq or BioMark (Fluidigm) RT-qPCR assay. RNA-seq was performed at 12 h post-induction in biological duplicates, and analyzed with STAR aligner and DEseq2 (GEO accession number: GSE65462). The following residues highlighted in bold were mutated to alanine to generate DNA binding-deficient mutants: WFQNHRY [Nkx2.2; Pradhan et al. (2012)], WFQNRRT [Nkx6.1; Lee et al. (2008)], NSRERKR [Olig2; Longo et al. (2008)]. See supplementary material Table S3 for RT-qPCR primer sequences.
Protein binding microarray (PBM) experiments
Full-length mouse Olig2 was purified from E. coli as a GST fusion. PBM experiments were conducted using 200 nM Olig2 in the PBM binding reactions essentially as described previously, with the addition of 0.023% Triton X-100 to the binding buffer, using two custom ‘all-10mer’ array designs (AMADID #015681 and #016060) (Berger and Bulyk, 2009). PBM data were quantified and normalized, and data from the two arrays were combined as described previously (Berger and Bulyk, 2009) to determine the in vitro DNA binding specificity of Olig2. The resulting PWMs were trimmed as described previously (Gordân et al., 2011) to remove flanking sequences of low information content.
Acknowledgements
We would like to thank Michael Kyba (University of Minnesota) for the mA2.lox.Cre ESC line, Esteban Mazzoni (NYU) for iOlig2 ESCs and expression vectors for mA2.lox.Cre targeting, Tom Jessell (Columbia) for Nkx2.2 and Nkx6.1 antibodies, Ben Novitch (UCLA) for plasmids, Eva Kutejova and James Briscoe (MRC, London) for sharing unpublished results.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Y.N. performed the ChIPseq and ectopic expression experiments, K.A.P. purified Olig2 protein, A.V. performed the Olig2 protein binding microarray experiments and analysis, and Y.N., X.Z., J.J. and K.A.P. performed the computational analysis. Y.N. and A.P.M. conceived and designed the experiments and wrote the manuscript. M.L.B., W.A.H. and A.P.M. supervised research. All authors contributed to and approved the final manuscript.
Funding
This work was supported by grants from the National Institutes of Health (NIH) [R37 NS033642 to A.P.M.; R01 HG003985 to M.L.B.; T32 HL087735 to K.A.P.]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.124636/-/DC1
References
- Arda H. E., Benitez C. M. and Kim S. K. (2013). Gene regulatory networks governing pancreas development. Dev. Cell 25, 5-13. 10.1016/j.devcel.2013.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. (2011). DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27, 1653-1659. 10.1093/bioinformatics/btr261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M. F. and Bulyk M. L. (2009). Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat. Protoc. 4, 393-411. 10.1038/nprot.2008.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsland M., Ramsköld D., Zaouter C., Klum S., Sandberg R. and Muhr J. (2011). Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 25, 2453-2464. 10.1101/gad.176008.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J., Sussel L., Serup P., Hartigan-O'Connor D., Jessell T. M., Rubenstein J. L. and Ericson J. (1999). Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature 398, 622-627. 10.1038/19315 [DOI] [PubMed] [Google Scholar]
- Briscoe J., Pierani A., Jessell T. M. and Ericson J. (2000). A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435-445. 10.1016/S0092-8674(00)80853-3 [DOI] [PubMed] [Google Scholar]
- Bylund M., Andersson E., Novitch B. G. and Muhr J. (2003). Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nat. Neurosci. 6, 1162-1168. 10.1038/nn1131 [DOI] [PubMed] [Google Scholar]
- Cornell R. A. and Von Ohlen T. (2000). Vnd/nkx, ind/gsh, and msh/msx: conserved regulators of dorsoventral neural patterning? Curr. Opin. Neurobiol. 10, 63-71. 10.1016/S0959-4388(99)00049-5 [DOI] [PubMed] [Google Scholar]
- Creyghton M. P., Cheng A. W., Welstead G. G., Kooistra T., Carey B. W., Steine E. J., Hanna J., Lodato M. A., Frampton G. M., Sharp P. A et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107, 21931-21936. 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaud E., McMahon A. P. and Briscoe J. (2008). Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development 135, 2489-2503. 10.1242/dev.009324 [DOI] [PubMed] [Google Scholar]
- Gordân R., Murphy K. F., McCord R. P., Zhu C., Vedenko A. and Bulyk M. L. (2011). Curated collection of yeast transcription factor DNA binding specificity data reveals novel structural and gene regulatory insights. Genome Biol. 12, R125 10.1186/gb-2011-12-12-r125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V., Khudyakov J., Ellis P. and Pevny L. (2003). SOX2 functions to maintain neural progenitor identity. Neuron 39, 749-765. 10.1016/S0896-6273(03)00497-5 [DOI] [PubMed] [Google Scholar]
- He H. H., Meyer C. A., Shin H., Bailey S. T., Wei G., Wang Q., Zhang Y., Xu K., Ni M., Lupien M. et al. (2010). Nucleosome dynamics define transcriptional enhancers. Nat. Genet. 42, 343-347. 10.1038/ng.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A. et al. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311-318. 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H. and Glass C. K. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576-589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovino M., Bosnakovski D., Fey H., Rux D., Bajwa G., Mahen E., Mitanoska A., Xu Z. and Kyba M. (2011). Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells 29, 1580-1588. 10.1002/stem.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee B., Joshi K., Pfaff S. L., Lee J. W. and Lee S.-K. (2008). A regulatory network to segregate the identity of neuronal subtypes. Dev. Cell 14, 877-889. 10.1016/j.devcel.2008.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Q., Jeong Y., Misra K., Li S., Zelman A. K., Epstein D. J. and Matise M. P. (2006). Wnt signaling inhibitors regulate the transcriptional response to morphogenetic Shh-Gli signaling in the neural tube. Dev. Cell 11, 325-337. 10.1016/j.devcel.2006.06.013 [DOI] [PubMed] [Google Scholar]
- Lek M., Dias J. M., Marklund U., Uhde C. W., Kurdija S., Lei Q., Sussel L., Rubenstein J., Matise M. P., Arnold H.-H. et al. (2010). A homeodomain feedback circuit underlies step-function interpretation of a Shh morphogen gradient during ventral neural patterning. Development 137, 4051-4060. 10.1242/dev.054288 [DOI] [PubMed] [Google Scholar]
- Longo A., Guanga G. P. and Rose R. B. (2008). Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: heterodimer selectivity and DNA recognition. Biochemistry 47, 218-229. 10.1021/bi701527r [DOI] [PubMed] [Google Scholar]
- Lu Q. R., Sun T., Zhu Z., Ma N., Garcia M., Stiles C. D. and Rowitch D. H. (2002). Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109, 75-86. 10.1016/S0092-8674(02)00678-5 [DOI] [PubMed] [Google Scholar]
- Marson A., Foreman R., Chevalier B., Bilodeau S., Kahn M., Young R. A. and Jaenisch R. (2008). Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell 3, 132-135. 10.1016/j.stem.2008.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni E. O., Mahony S., Iacovino M., Morrison C. A., Mountoufaris G., Closser M., Whyte W. A., Young R. A., Kyba M., Gifford D. K. et al. (2011). Embryonic stem cell–based mapping of developmental transcriptional programs. Nat. Methods 8, 1056-1058. 10.1038/nmeth.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi R., Sugimori M., Takebayashi H., Kosako H., Nagao M., Yoshida S., Nabeshima Y.-i., Shimamura K. and Nakafuku M. (2001). Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron 31, 757-771. 10.1016/S0896-6273(01)00413-5 [DOI] [PubMed] [Google Scholar]
- Moore S., Ribes V., Terriente J., Wilkinson D., Relaix F. and Briscoe J. (2013). Distinct regulatory mechanisms act to establish and maintain Pax3 expression in the developing neural tube. PLoS Genet. 9, e1003811 10.1371/journal.pgen.1003811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhr J., Andersson E., Persson M., Jessell T. M. and Ericson J. (2001). Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell 104, 861-873. 10.1016/S0092-8674(01)00283-5 [DOI] [PubMed] [Google Scholar]
- Novitch B. G., Chen A. I. and Jessell T. M. (2001). Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 31, 773-789. 10.1016/S0896-6273(01)00407-X [DOI] [PubMed] [Google Scholar]
- Oosterveen T., Kurdija S., Alekseenko Z., Uhde C. W., Bergsland M., Sandberg M., Andersson E., Dias J. M., Muhr J. and Ericson J. (2012). Mechanistic differences in the transcriptional interpretation of local and long-range shh morphogen signaling. Dev. Cell 23, 1006-1019. 10.1016/j.devcel.2012.09.015 [DOI] [PubMed] [Google Scholar]
- Peterson K. A., Nishi Y., Ma W., Vedenko A., Shokri L., Zhang X., McFarlane M., Baizabal J.-M., Junker J., van Oudenaarden A. et al. (2012). Neural-specific Sox2 input and differential Gli-binding affinity provide context and positional information in Shh-directed neural patterning. Genes Dev. 26, 2802-2816. 10.1101/gad.207142.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan L., Genis C., Scone P., Weinberg E. O., Kasahara H. and Nam H.-J. (2012). Crystal structure of the human NKX2.5 homeodomain in complex with DNA target. Biochemistry 51, 6312-6319. 10.1021/bi300849c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A. and Wysocka J. (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279-283. 10.1038/nature09692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M., Paydar S., Ericson J., Briscoe J., Berber E., German M., Jessell T. M. and Rubenstein J. L. R. (2000). Ventral neural patterning by Nkx homeobox genes: Nkx6.1 controls somatic motor neuron and ventral interneuron fates. Genes Dev. 14, 2134-2139. 10.1101/gad.820400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallstedt A., Muhr J., Pattyn A., Pierani A., Mendelsohn M., Sander M., Jessell T. M. and Ericson J. (2001). Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron 31, 743-755. 10.1016/S0896-6273(01)00412-3 [DOI] [PubMed] [Google Scholar]
- Wang H., Lei Q., Oosterveen T., Ericson J. and Matise M. P. (2011). Tcf/Lef repressors differentially regulate Shh-Gli target gene activation thresholds to generate progenitor patterning in the developing CNS. Development 138, 3711-3721. 10.1242/dev.068270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H., Lieberam I., Porter J. A. and Jessell T. M. (2002). Directed differentiation of embryonic stem cells into motor neurons. Cell 110, 385-397. 10.1016/S0092-8674(02)00835-8 [DOI] [PubMed] [Google Scholar]
- Zhou Q. and Anderson D. J. (2002). The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109, 61-73. 10.1016/S0092-8674(02)00677-3 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Choi G. and Anderson D. J. (2001). The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron 31, 791-807. 10.1016/S0896-6273(01)00414-7 [DOI] [PubMed] [Google Scholar]