ABSTRACT
Obligate intracellular bacteria, such as Ehrlichia chaffeensis, perish unless they can enter eukaryotic cells. E. chaffeensis is the etiological agent of human monocytic ehrlichiosis, an emerging infectious disease. To infect cells, Ehrlichia uses the C terminus of the outer membrane invasin entry-triggering protein (EtpE) of Ehrlichia (EtpE-C), which directly binds the mammalian cell surface glycosylphosphatidyl inositol-anchored protein, DNase X. How this binding drives Ehrlichia entry is unknown. Here, using affinity pulldown of host cell lysates with recombinant EtpE-C (rEtpE-C), we identified two new human proteins that interact with EtpE-C: CD147 and heterogeneous nuclear ribonucleoprotein K (hnRNP-K). The interaction of CD147 with rEtpE-C was validated by far-Western blotting and coimmunoprecipitation of native EtpE with endogenous CD147. CD147 was ubiquitous on the cell surface and also present around foci of rEtpE-C-coated-bead entry. Functional neutralization of surface-exposed CD147 with a specific antibody inhibited Ehrlichia internalization and infection but not binding. Downregulation of CD147 by short hairpin RNA (shRNA) impaired E. chaffeensis infection. Functional ablation of cytoplasmic hnRNP-K by a nanoscale intracellular antibody markedly attenuated bacterial entry and infection but not binding. EtpE-C also interacted with neuronal Wiskott-Aldrich syndrome protein (N-WASP), which is activated by hnRNP-K. Wiskostatin, which inhibits N-WASP activation, and cytochalasin D, which inhibits actin polymerization, inhibited Ehrlichia entry. Upon incubation with host cell lysate, EtpE-C but not an EtpE N-terminal fragment stimulated in vitro actin polymerization in an N-WASP- and DNase X-dependent manner. Time-lapse video images revealed N-WASP recruitment at EtpE-C-coated bead entry foci. Thus, EtpE-C binding to DNase X drives Ehrlichia entry by engaging CD147 and hnRNP-K and activating N-WASP-dependent actin polymerization.
IMPORTANCE
Ehrlichia chaffeensis, an obligate intracellular bacterium, causes a blood-borne disease called human monocytic ehrlichiosis, one of the most prevalent life-threatening emerging tick-transmitted infectious diseases in the United States. The survival of Ehrlichia bacteria, and hence, their ability to cause disease, depends on their specific mode of entry into eukaryotic host cells. Understanding the mechanism by which E. chaffeensis enters cells will create new opportunities for developing effective therapies to prevent bacterial entry and disease in humans. Our findings reveal a novel cellular signaling pathway triggered by an ehrlichial surface protein called EtpE to induce its infectious entry. The results are also important from the viewpoint of human cell physiology because three EtpE-interacting human proteins, DNase X, CD147, and hnRNP-K, are hitherto unknown partners that drive the uptake of small particles, including bacteria, into human cells.
INTRODUCTION
Human monocytic ehrlichiosis (HME) is one of the most prevalent life-threatening emerging tick-borne zoonoses in the United States (1). The disease was discovered in 1986 and was designated a nationally notifiable disease in 1998 by the U.S Centers for Disease Control and Prevention. HME is caused by infection with Ehrlichia chaffeensis, an obligate intracellular bacterium in the order Rickettsiales. E. chaffeensis replicates within human monocytes and macrophages and causes severe flulike symptoms accompanied by hematologic abnormalities and signs of hepatitis. No vaccines exist for HME. The broad-spectrum antibiotic doxycycline is the only drug effective for treating HME but is contraindicated for pregnant women and children. Delayed initiation of therapy, the presence of underlying illness, or immunosuppression often lead to severe complications or even death (2). The incidence of tick-borne zoonoses has risen continuously and dramatically in the past 2 decades (3). The Centers for Disease Control and Prevention confirmed 1,404 HME cases in 2014 and 1,509 in 2013 (1), which is more than a 10-fold increase in HME incidence over a 15-year period. The 2011 U.S. Institute of Medicine report “Critical Needs and Gaps in understanding prevention, amelioration, and resolution of Lyme and Other Tick-Borne Diseases” (4) pointed to the urgent need for research on HME.
E. chaffeensis entry into the human acute leukemia cell line THP-1 leads to productive infection and is dependent on host cell surface lipid rafts and glycosylphosphatidyl inositol (GPI)-anchored proteins (5). After entry, E. chaffeensis replicates in an early endosome-like compartment which contains early endosome antigen 1 (EEA1), Rab5, transferrin receptor, and vacuolar-type H+ ATPase (6) but does not contain lysosomal membrane-associated protein 1, CD63, or NADPH oxidase components (6–8). E. chaffeensis cells die if they fail to enter an appropriate host cell. Previous studies using pharmacological inhibitors have suggested that the mechanism of E. chaffeensis entry is distinct from that of classic antimicrobial phagocytosis (5, 9).
E. chaffeensis outer membrane protein ECH1038, with highly strain-conserved N- and C-terminal segments, is highly expressed at the stress-resistant and infectious stage of the E. chaffeensis intracellular developmental cycle (10). We previously named ECH1038 of Ehrlichia entry-triggering protein (EtpE) because we found that EtpE is exposed on the E. chaffeensis surface and serves as an invasin for direct binding of its mammalian receptor DNase X (DNase I-like protein 1), a GPI-anchored ubiquitous cell surface protein, to trigger infectious entry (11).
In patient blood specimens, E. chaffeensis is primarily seen in monocytes and not in neutrophils (12); hence, human monocytic ehrlichiosis is so named to distinguish it from other human ehrlichioses caused by granulocytotropic species. E. chaffeensis has been stably cultivated only in canine macrophage DH82 cells (13) or THP-1 cells (14). To apply transfection and mouse mutagenesis techniques and distinguish ehrlichial infectious entry from classic phagocytosis, we developed an effective E. chaffeensis culture system in nonphagocytic cell lines (human embryonic kidney HEK293 cells and monkey endothelial RF/6A cells), as well as primary phagocytic cells (human and canine peripheral blood monocytes and mouse bone marrow-derived macrophages [BMDMs]) (11). An antibody (Ab) against the E. chaffeensis strain-conserved C-terminal segment of EtpE (EtpE-C) greatly inhibits E. chaffeensis binding, entry, and infection of both phagocytic and nonphagocytic host cells in vitro, and immunization of mice with EtpE-C significantly inhibits infection (11). Although latex beads cannot bind or enter nonphagocytes, EtpE-C-coated beads enter nonphagocytes as well as phagocytes, and the entry is blocked by several compounds that block E. chaffeensis entry (11). An antibody against DNase X or small interfering RNA (siRNA)-mediated knockdown of DNase X significantly reduces E. chaffeensis binding and entry (11). Furthermore, the bacterial loads in the peripheral blood in experimentally infected DNase X−/− mice are significantly reduced compared with those in infected wild-type mice, indicating that DNase X-mediated entry is also relevant in vivo (11).
The actin cytoskeleton has crucial roles in particle uptake into phagocytes and nonphagocytes (15, 16), and a number of intracellular microbial pathogens have evolved ways to manipulate the actin cytoskeleton to facilitate infectious entry into host cells. For example, Salmonella induces plasma membrane ruffles to facilitate host cell entry (17), Chlamydia causes actin remodeling, leading to reorganization and hypertrophy of preexisting cell surface microvilli to induce its entry (18), and Shigella causes retraction of cell surface filopodia via actin remodeling to bring bacteria in close contact with the cell body (19). E. chaffeensis and beads coated with recombinant EtpE-C (rEtpE-C) induce filopodial extensions on the host cell surface at the point of contact (11, 20), suggesting the involvement of actin cytoskeletal remodeling during the entry process. However, the mechanism of EtpE binding to DNase X to trigger infectious entry is unknown, especially considering that the GPI-anchored DNase X does not span the plasma membrane lipid bilayer.
Our present study is the first to investigate the details of the unique molecular interactions triggered by binding of an ehrlichial invasin to host cells, which is required for host cell actin remodeling and the subsequent infectious entry of E. chaffeensis. Our study reveals a previously unknown signaling pathway induced by EtpE for actin cytoskeletal remodeling. EtpE is the first example of a protein that induces DNase X-dependent actin polymerization in vitro.
RESULTS
The EtpE-C–DNase X complex contains CD147 and hnRNP-K.
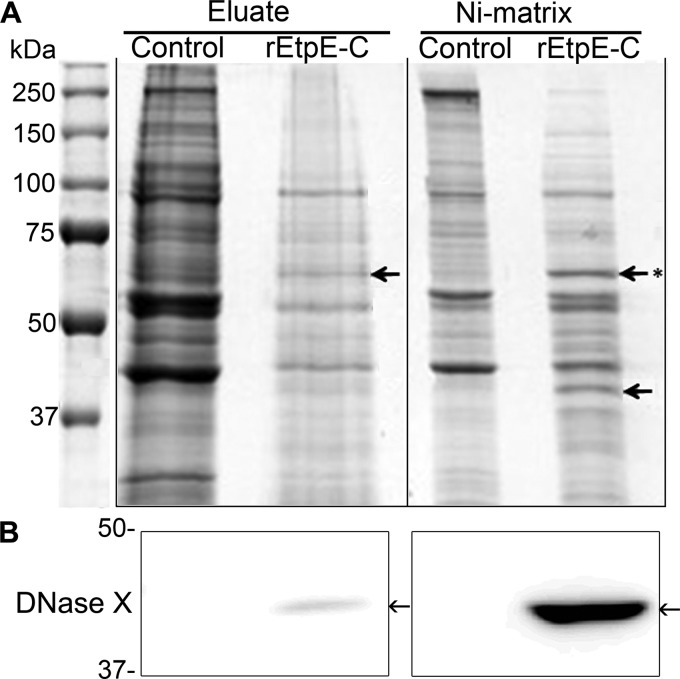
To identify human proteins that may interact with the EtpE-DNase X complex, rEtpE-C (308 amino acid residues) was bound to a Ni-silica column, renatured on the column, and incubated with THP-1 cell lysate. This was followed by washing, elution with imidazole, and finally, separation of eluted proteins and remaining column-bound proteins with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1A). Comparison of separated proteins between the control and rEtpE-C columns revealed two conspicuous bands (Fig. 1A, arrows) stably bound to the rEtpE-C column after imidazole elution; immunoblotting confirmed that one was DNase X (Fig. 1B), but the other was unknown (Fig. 1A, asterisk). Liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS) of this band identified two major proteins, namely, human CD147 (basigin/extracellular matrix metalloproteinase inducer; 26% coverage of the short form of CD147) and human heterogeneous nuclear ribonucleoprotein K (hnRNP-K; 44.3% coverage) (see Table S1 in the supplemental material).
FIG 1 .
EtpE-C–DNase X complex interacts with host cell downstream signaling proteins. (A) Affinity pulldown of proteins from a THP-1 cell lysate incubated with rEtpE-C bound to a Ni-silica matrix (a control matrix was not bound with rEtpE-C). The eluate of EtpE-C-interacting proteins and postelution Ni-silica matrix were subjected to SDS-PAGE, followed by GelCode blue staining. Arrows indicate protein bands that were distinct in the rEtpE-C eluate/matrix compared with the control. The most conspicuous band, denoted with an asterisk, was further processed for LC-MS/MS (see Table S1 in the supplemental material). (B) Immunoblotting of the eluate and residual matrix from the affinity pulldown assay, probed with anti-DNase X antibody.
Mature human CD147 (i.e., lacking the 21-residue signal sequence) contains 248 residues (short form; ubiquitously expressed isoform of CD147) (21) or 368 residues (long form; retina-specific isoform of CD147) (22). CD147 (short isoform) contains two glycosylated N-terminal extracellular immunoglobulin domains of 185 residues, a 24-residue transmembrane domain, and a 39-residue cytoplasmic region. Glycosylated human CD147 (short isoform) can be detected with apparent molecular masses ranging from 32 to 60 kDa by SDS-PAGE (23).
Human hnRNP-K is a 463-residue protein (51.3 kDa) and a component of the heterogeneous nuclear ribonucleoprotein (hnRNP) complex that localizes both to the nucleus and cytoplasm and can interact with RNA, DNA, and various proteins (24). Two other proteins identified with a MASCOT score above 100 (see Table S1 in the supplemental material) were not investigated further because their interaction was tenuous owing to the relatively low percentage of coverage in our LC-MS/MS analysis.
CD147 interacts with rEtpE-C as well as native EtpE, is recruited around rEtpE-C-coated-bead entry foci, and is required for ehrlichial entry and infection but not for binding.
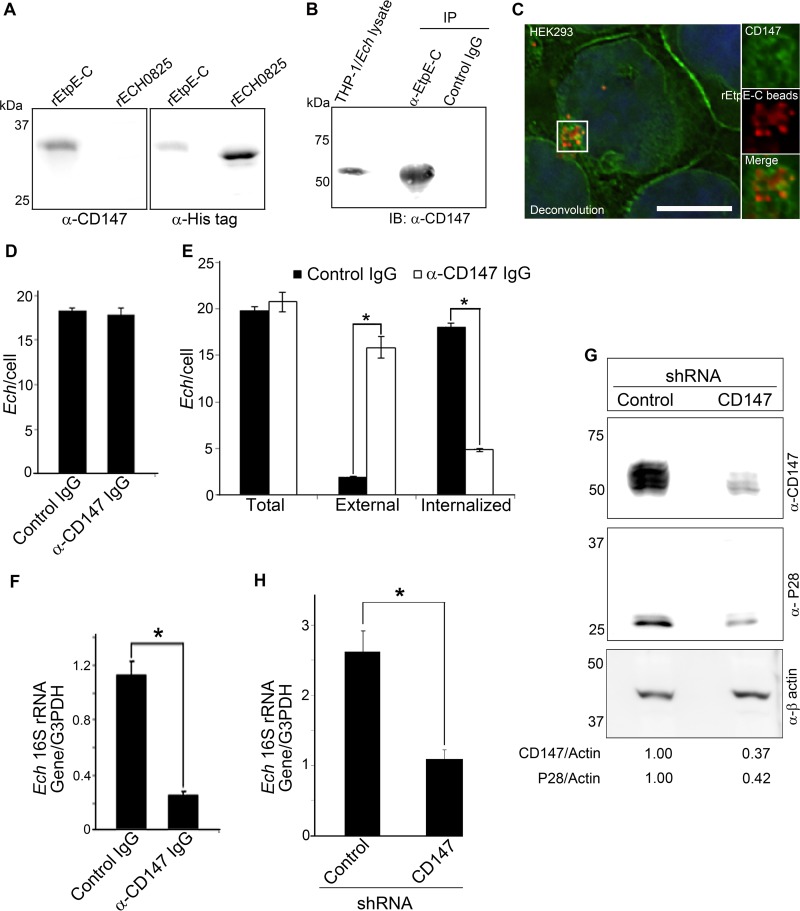
Using yeast two-hybrid analysis, we previously observed that DNase X directly binds EtpE-C and that endogenous DNase X from THP-1 cell lysate binds renatured rEtpE-C on a nitrocellulose membrane, as seen with far-Western blotting (11). In agreement with our pulldown data (Fig. 1), far-Western blotting revealed that endogenous CD147 from the THP-1 cell lysate bound renatured rEtpE-C but not the control recombinant E. chaffeensis protein ECH0825 (rECH0825) that had been treated similarly (Fig. 2A). To validate this interaction with native EtpE, we performed coimmunoprecipitation with lysate of THP-1 cells that had been briefly incubated with E. chaffeensis for 30 min; the lysate was incubated with protein A-agarose beads adsorbed with anti-rEtpE-C antibody or control IgG. The precipitate, which was previously shown to contain DNase X (11), was probed for the presence of CD147. CD147 was present in the complex immunoprecipitated with anti-EtpE-C antibody but not with control IgG (Fig. 2B). CD147 of THP-1 cells is likely glycosylated, because the molecular size of CD147 was >50 kDa by immunoblotting (Fig. 2B), and this is likely the reason why CD147 was found in the same band with hnRNP-K by proteomics analysis (Fig. 1A).
FIG 2 .
CD147 interacts with rEtpE and native EtpE, which is required for E. chaffeensis entry. (A) Far-western blotting of renatured rEtpE-C and rECH0825 on a nitrocellulose membrane that was incubated with THP-1 cell lysate. Native CD147 was detected with anti-CD147 antibody, and recombinant proteins were detected with anti-His antibody. (B) Immunoblotting (IB) of E. chaffeensis-infected THP-1 cell lysate immunoprecipitated (IP) with anti-EtpE-C antibody or control IgG. THP-1 cells were incubated with E. chaffeensis for 30 min, lysed, and immunoprecipitated with anti-EtpE-C antibody- or control IgG-bound protein A-agarose. The precipitates were subjected to immunoblotting with anti-CD147 antibody. (C) Immunofluorescence labeling of rEtpE-C-coated beads (red) incubated (37°C, 1 h) with HEK293 cells, visualized by deconvolution microscopy. Labeling was done with anti-CD147 antibody (green) and DAPI (blue) after saponin permeabilization. The boxed area is enlarged in the smaller panels to the right. Scale bar, 10 µm. (D) Numbers of E. chaffeensis bacteria (Ech) bound to HEK293 cells pretreated with anti-CD147 antibody or isotype-matched control mouse IgG at 30 min p.i. Immunofluorescence labeling with anti-P28 antibody was performed without permeabilization, and the numbers of E. chaffeensis bacteria on 100 HEK293 cells were scored. (E) Numbers of total, external, and internalized E. chaffeensis bacteria after incubation with HEK293 cells pretreated with anti-CD147 antibody (white bar) or control mouse IgG (black bar) at 1 h p.i. Cells were processed for two rounds of immunostaining with anti-P28 antibody as described in Materials and Methods. The numbers of E. chaffeensis bacteria in 100 HEK293 cells were scored. (F) E. chaffeensis load in HEK293 cells pretreated with anti-CD147 antibody or control mouse IgG, determined at 48 h p.i. by qPCR. (G) Immunoblotting of CD147 and P28 in HEK293 cells stably transfected with anti-CD147 lentiviral shRNA or control shRNA. Actin was used as a protein loading control. The values under the bands show the relative ratios of band intensities, with the ratios of those from control shRNA set as 1. (H) E. chaffeensis load in HEK293 cells transfected with CD147 shRNA or control shRNA determined at 76 h p.i. by qPCR. (D, E, F, and H) Data represent the mean results and standard deviations from triplicate samples and are representative of three independent experiments. *, P < 0.05.
To further study whether CD147 is present around the entry foci of rEtpE-C-coated latex beads in the absence of other bacterial proteins, HEK293 cells (which are adherent and nonphagocytic, allowing better visualization of signal localization) were incubated with rEtpE-C-coated red fluorescent beads for 1 h, followed by immunostaining with monoclonal antibody (MAb) MEM-M6/6 directed against the membrane-proximal Ig2 domain of CD147. Fluorescence microscopy with deconvolution showed that CD147 was detected ubiquitously on the plasma membrane and surrounded the entry foci of the coated beads (Fig. 2C). Although pretreatment of HEK293 cells with anti-CD147 antibody did not block E. chaffeensis binding to HEK293 cells at 30 min postinfection (p.i.) (Fig. 2D), it greatly inhibited E. chaffeensis internalization (Fig. 2E). This inhibition led to a significantly reduced bacterial load at 48 h p.i. compared with the bacterial load following control IgG pretreatment (Fig. 2F). This result indicates that CD147 is not a receptor for E. chaffeensis binding but, rather, is specifically required for pathogen entry and for establishing infection.
CD147 gene knockout in mice results in spermatocyte apoptosis, degeneration of germ cells, and infertility (25), and thus, CD147 knockout mice are difficult to produce. Thus, to test the requirement of CD147 for E. chaffeensis entry, we established a stable knockdown of CD147 via lentiviral-based transduction of HEK293 cells with a CD147-specific short hairpin RNA (shRNA). Compared with using siRNA, shRNA provides sustainable knockdown of target genes with fewer off-target effects (26). CD147 expression was markedly reduced in shRNA-transduced cells as observed with immunoblotting (Fig. 2G). Moreover, immunoblotting with an antibody against E. chaffeensis P28 (27) and quantitative PCR (qPCR) of the E. chaffeensis 16S rRNA gene revealed significant reduction of E. chaffeensis infection at 76 h p.i. in CD147 knockdown cells compared with the level in control cells (Fig. 2G and H).
hnRNP-K is required for E. chaffeensis entry and infection.
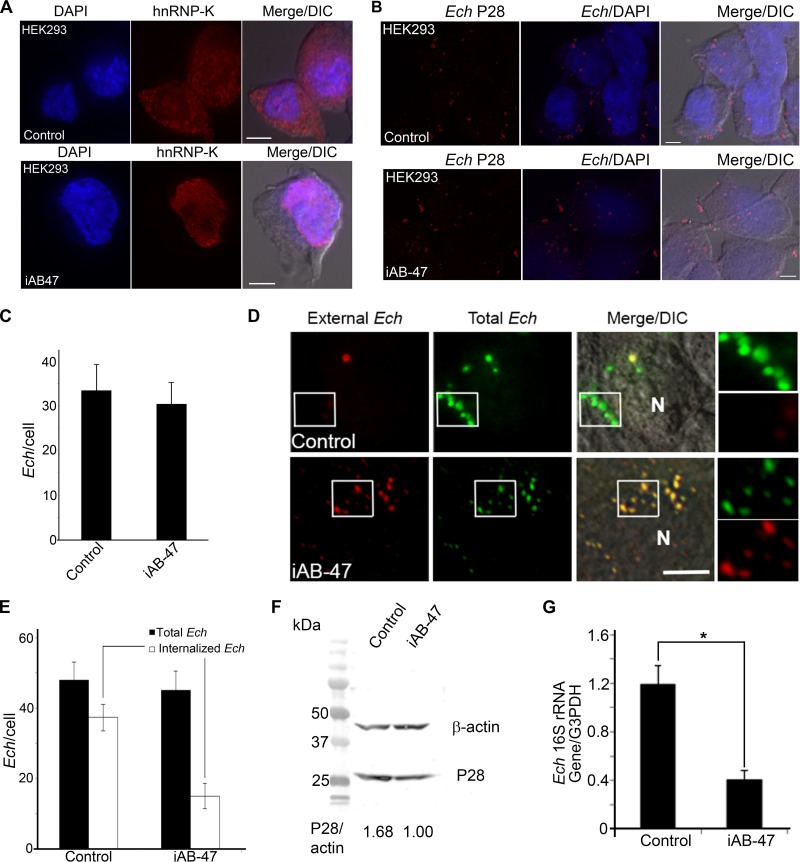
hnRNP-K is present in the nucleus, plasma membrane, and cytoplasm of mammalian cells and acts as a docking platform to integrate signals from various signaling cascades (24). siRNA-mediated depletion of hnRNP-K results in cell death (28), suggesting that it is an indispensable protein. No knockout mouse for hnRNP-K has been reported, probably because hnRNP-K depletion leads to embryonic lethality in mice. Therefore, to study the role of cytoplasmic hnRNP-K in ehrlichial entry and infection, we transfected cells with a plasmid encoding the nanoscale intracellular antibody (iAb) clone number 47 (iAb-47) that contains an hnRNP-K antigen-binding small fragment in the variable domain of the heavy-chain antibodies of camelids that naturally lack light chains (29). iAb-47 has been shown to bind and confine hnRNP-K in the nucleus of HT1080 fibrosarcoma cells without affecting cell viability, and iAb-47 also inhibits the chemotactic migration of HT1080 cells (28). Similar to the results with HT1080 cells, transfection of HEK293 cells with an iAb-47-encoding plasmid resulted in hnRNP-K being confined to the nuclear region, with almost no cytoplasmic localization (Fig. 3A). E. chaffeensis binding was not affected by functional ablation of hnRNP-K by iAb-47 (Fig. 3B and C), whereas internalization was strongly blocked (Fig. 3D and E). Immunoblotting of E. chaffeensis P28 in HEK293 cells transfected with the iAb-47 plasmid revealed a significant reduction in bacterial load compared with the load in cells transfected with the control plasmid pEGFP (Fig. 3F). A similar result was observed with qPCR (Fig. 3G), indicating that cytoplasmic hnRNP-K is required for E. chaffeensis entry and infection of host cells.
FIG 3 .
hnRNP-K is required for Ehrlichia entry and infection. (A) Immunofluorescence labeling of HEK293 cells transfected with iAb-47 plasmid or control plasmid at 48 h, with saponin permeabilization. Labeling was done with anti-hnRNP-K antibody (red) and DAPI (blue). (B) E. chaffeensis (Ech) bound to HEK293 cells transfected with iAb-47 or control plasmid at 30 min p.i. Immunofluorescence labeling with anti-P28 antibody (red) and DAPI (blue) was performed without permeabilization. (C) Quantification of the results from an experiment similar to the one whose results are shown in panel B, done by counting the E. chaffeensis bacteria in 100 HEK293 cells. (D) E. chaffeensis bacteria internalized by HEK293 cells transfected with iAb-47 or control plasmid at 2 h p.i. Cells were processed for two rounds of immunostaining with anti-P28 antibody, the first without permeabilization to detect external E. chaffeensis (red), and the second with saponin permeabilization to detect total E. chaffeensis bacteria (green). Boxed areas are enlarged to the right. N, nucleus. (E) Quantification of the results from an experiment similar to the one whose results are shown in panel D, done by counting E. chaffeensis in 100 HEK293 cells. (F) Immunoblotting of P28 in HEK293 cells transfected with iAb-47 plasmid or the control plasmid at 56 h p.i. Host actin was used as a protein loading control. The values under the bands show the relative ratios of band intensities, with the ratios of those from control plasmid set as 1. (G) Quantification of the results from an experiment similar to the one whose results are shown in panel F, done by qPCR. (A, B, and D) Deconvolution microscopy. Scale bar, 5 µm; Merge/DIC, image merged with differential interference contrast. (C, E, and G) Data represent the mean results and standard deviations from triplicate samples and are representative of three independent experiments. *, P < 0.05.
Activation of N-WASP and actin polymerization are required for E. chaffeensis entry.
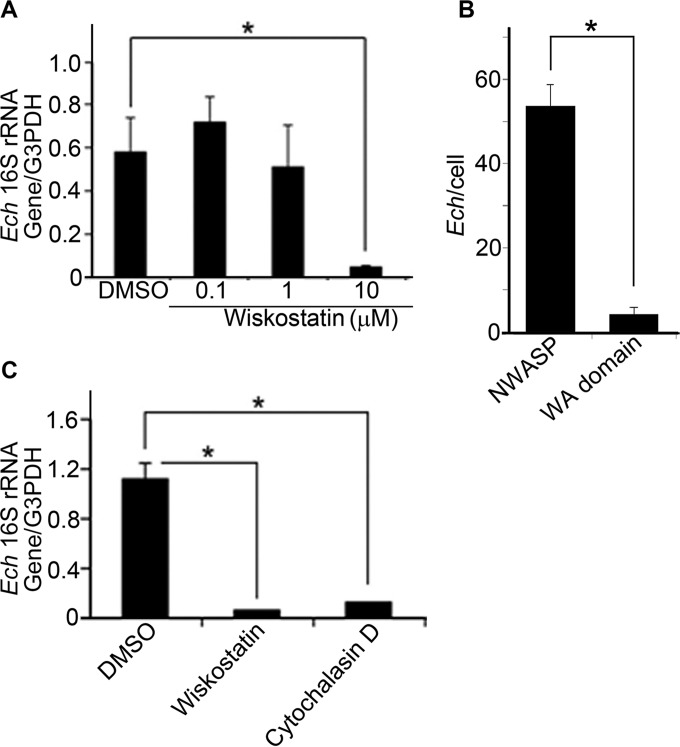
Efficient nucleation of filamentous actin (F-actin) requires actin-nucleating factors, such as the actin-related protein 2 and3 (Arp2/3) complex, and engagement with nucleation-promoting factors, such as neuronal Wiskott-Aldrich syndrome protein (N-WASP), which exists in an inactive closed conformation unless activated by specific stimuli (30, 31). It has been reported that hnRNP-K binds N-WASP and activates the Arp2/3 complex to nucleate actin polymerization in vitro (32). Therefore, we examined whether N-WASP activation is required for E. chaffeensis entry into host cells. The cell-permeable chemical inhibitor wiskostatin binds to the GTPase-binding domain of N-WASP and thereby stabilizes its autoinhibited closed conformation (33). After pretreating DH82 cells with wiskostatin for 30 min and then removing the wiskostatin, E. chaffeensis was added, and the bacterial load was determined at 50 h p.i. Although pretreatment with 100 nM or 1 µM wiskostatin had little effect, a concentration of 10 µM resulted in nearly complete inhibition of infection compared with that in the dimethyl sulfoxide (DMSO) solvent control, suggesting that N-WASP activation is required for E. chaffeensis entry (Fig. 4A). Although wiskostatin at higher concentrations has been reported to nonselectively perturb membrane transport by decreasing the cellular ATP level, wiskostatin at 10 µM does not seem to have this effect (34).
FIG 4 .
N-WASP and actin are required for E. chaffeensis entry and infection. (A, C) E. chaffeensis (Ech) load in DH82 cells pretreated for 30 min with wiskostatin (0.1, 1, or 10 µM) (A) or with wiskostatin (10 µM) or cytochalasin D (10 µM) (C); in each case, cells were then incubated with E. chaffeensis for 50 h and the bacterial load was determined by qPCR. (B) Numbers of internalized E. chaffeensis bacteria at 4 h p.i. in GFP–N-WASP- or GFP-WA-transfected RF/6A cells at 48 h posttransfection. Quantification of E. chaffeensis in 100 cells. (A to C) Data represent the mean results and standard deviations from triplicate samples and are representative of three independent experiments. *, P < 0.05.
N-WASP is composed of an N-terminal WH (WASP homology) 1 domain, the basic region, a Cdc42/Rac-interactive binding domain, a proline-rich domain, and a WA domain (WH2, cofilin binding, acidic domain; also called VCA, verprolin homology, cofilin homology, acidic domain) at the C terminus. The WA domain binds to G-actin and the Arp2/3 complex (30). Because the WA domain sequesters the free Arp2/3 complex in the cytoplasm and prevents it from binding to activated N-WASP, the expression of green fluorescent protein (GFP)-WA is often used to elicit a dominant-negative effect on N-WASP function (31). To validate the role of N-WASP in ehrlichial entry, we transfected RF/6A cells with GFP–N-WASP or GFP-WA. At 48 h posttransfection, the cells were incubated with E. chaffeensis for 4 h, followed by trypsinization to remove noninternalized bacteria, and cells were subjected to immunofluorescence staining with anti-P28 antibody. The internalization of E. chaffeensis was reduced by nearly 90% in WA-transfected cells (Fig. 4B).
Given the importance of N-WASP activation in E. chaffeensis internalization and infection, we examined the requirement for actin mobilization during internalization. DH82 cells were pretreated for 30 min with 10 µM cytochalasin D, which inhibits G-actin polymerization and promotes F-actin depolymerization (35), or with 10 µM wiskostatin followed by incubation with E. chaffeensis in the absence of these inhibitors; qPCR revealed almost complete inhibition of bacterial infection at 50 h p.i. (Fig. 4C). Similar results were obtained with nonphagocytic HEK293 cells incubated with wiskostatin and cytochalasin D that inhibited E. chaffeensis internalization and infection (see Fig. S1 in the supplemental material). Thus, these results demonstrated that N-WASP activation and actin polymerization are necessary for the infectious entry of E. chaffeensis in both phagocytes and nonphagocytes.
N-WASP is recruited to EtpE entry foci in live cells, and N-WASP interacts with rEtpE-C and native EtpE.
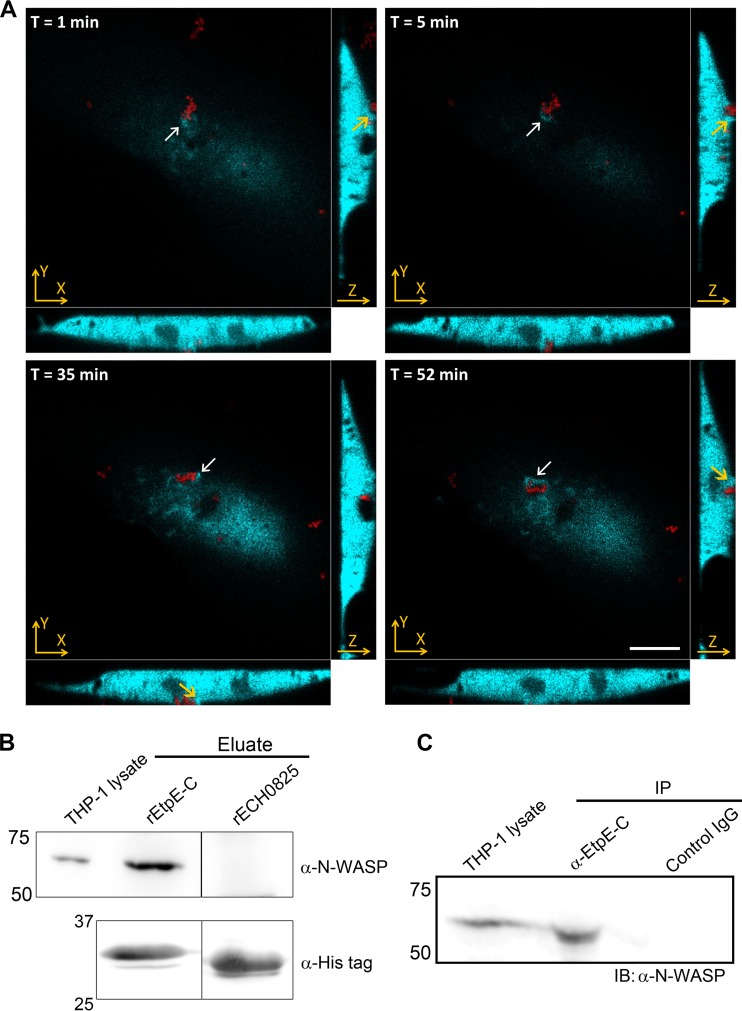
rEtpE-C-coated beads enter nonphagocytic and phagocytic cells in a DNase X-dependent manner (11). To further study whether N-WASP is specifically recruited to entry foci in the absence of other bacterial proteins, we incubated GFP–N-WASP-transfected RF/6A cells with rEtpE-C-coated beads. Live-cell imaging with spinning-disk confocal microscopy revealed that GFP–N-WASP was recruited to entry foci within 1 min of incubation (Fig. 5A, cluster of beads denoted by white arrows; see also Movie S1 in the supplemental material); it appeared to be concentrated at the base of a filopodiumlike cell surface extension embracing the beads and was associated with the beads until the beads were ultimately internalized (Fig. 5A, yellow arrowheads in the xz and yz side views).
FIG 5 .
N-WASP is recruited to rEtpE-C-coated-bead entry foci, and it interacts with rEtpE-C and native EtpE. (A) Spinning-disk confocal microscopy images at four different time points from the three-dimensional time-lapse video of RF/6A cells transfected with GFP–N-WASP (cyan) and then incubated with rEtpE-C-coated beads (red). xy planes show the upper surface of an RF/6A cell that had internalized rEtpE-C-coated beads. Each white arrow indicates a cluster of N-WASP. Cell surface extensions surrounding the cluster of beads are highlighted with yellow arrows in the side views (xz and yz planes). Scale bar, 10 µm. (B) Affinity pulldown of THP-1 cell lysate incubated with rEtpE-C or rECH0825 bound to Ni-silica matrix. Bound proteins were eluted with imidazole and subjected to immunoblot analysis with anti-N-WASP and anti-His antibodies. One lane between rEtpE-C and rECH0825 was deleted to save the space without altering molecular mass or exposure time. (C) THP-1 cells were incubated with E. chaffeensis for 30 min, lysed, and immunoprecipitated (IP) with anti-EtpE-C antibody- or control IgG-bound protein A-agarose. The immunoprecipitates were subjected to immunoblotting with anti-N-WASP antibody. (B and C) Numbers to the left indicate kDa.
It has been reported that the K interaction domain of hnRNP-K binds the WH1 domain of N-WASP and activates N-WASP in cotransfected cells (32). Thus, we next examined whether EtpE-C, which we found to interact with hnRNP-K (Fig. 1; see Table S1 in the supplemental material), could interact with N-WASP. THP-1 cell lysate was incubated with rEtpE-C or control rECH0825 bound to a Ni-silica matrix. N-WASP was present in the eluate from the rEtpE-C-bound column but not the rECH0825 column (Fig. 5B). This interaction was further validated by immunoprecipitation of the lysate of THP-1 cells incubated briefly with E. chaffeensis for 30 min with anti-EtpE-C antibody- or control IgG-adsorbed protein A-agarose beads. N-WASP was immunoprecipitated with anti-rEtpE-C antibody but not with the control antibody (Fig. 5C). These results indicated that N-WASP was recruited to rEtpE-C-coated latex beads or native E. chaffeensis entry foci via protein-protein interaction with EtpE-C.
EtpE-C induces actin polymerization in vitro in a DNase X- and N-WASP-dependent manner.
The recruitment of N-WASP to rEtpE-C-coated-bead entry foci and the requirement of N-WASP activation for E. chaffeensis entry and infection suggest that N-WASP activation upon EtpE-C binding to DNase X receptor drives spatiotemporal actin polymerization. To test this possibility, we examined whether (i) rEtpE-C can induce actin polymerization in the absence of any other bacterial protein and, (ii) if so, whether this is dependent on DNase X and N-WASP.
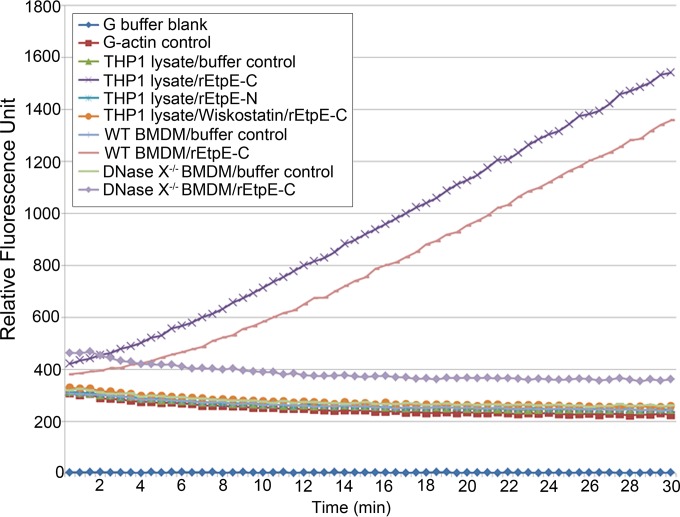
For this, we used an in vitro pyrenyl-actin polymerization time course assay in the presence of the host protein-containing lysate that contains DNase X, CD147, hnRNP-K, N-WASP, and the Arp2/3 complex. The pyrenyl-actin assay is a well-established method to determine the effects of various compounds on actin polymerization based on the enhanced fluorescence of pyrene-conjugated G-actin that occurs during its polymerization (36). Incubation of soluble rEtpE-C with THP-1 lysate indeed induced polymerization of pyrenyl-actin (Fig. 6). This actin polymerization specifically required the C terminus of EtpE, because recombinant N-terminal EtpE (rEtpE-N), which does not bind or induce the entry of coated beads into nonphagocytes (11), did not induce actin polymerization (Fig. 6). The effect of rEtpE-C on actin polymerization was abrogated by the addition of wiskostatin, indicating the requirement for N-WASP activation in EtpE-C-induced actin polymerization in vitro (Fig. 6). To determine the DNase X dependency of actin polymerization, we used the cell lysate of wild-type or DNase X−/− mouse BMDMs rather than THP-1 lysate. rEtpE-C-induced actin polymerization required DNase X, because the lysate of DNase X−/− BMDMs, which lacks DNase X, failed to elicit actin polymerization, whereas the lysate of congenic wild-type BMDMs elicited a polymerization profile similar to that of the THP-1 lysate (Fig. 6). Thus, EtpE-C induced actin polymerization in the absence of any other bacterial proteins and this is dependent on DNase X and N-WASP.
FIG 6 .
EtpE-C induces actin polymerization in vitro in an N-WASP- and DNase X-dependent manner. The results of a fluorometric assay of an in vitro pyrenyl-actin polymerization time course are shown. The assay was performed in the presence of pyrenyl G-actin and the THP-1 lysate by adding rEtpE-C, rEtpE-N, or rEtpE-C plus 10 µM wiskostatin at 0 h or in the presence of pyrenyl G-actin and mouse wild-type or DNase X−/− BMDM lysate by adding rEtpE-C at 0 h. The relative fluorescence of pyrenyl F-actin in each reaction mixture was plotted versus time.
Taken together, these results imply that E. chaffeensis infectious entry is induced by EtpE binding to DNase X, with subsequent recruitment of CD147 and hnRNP-K to the entry foci, followed by spatiotemporal mobilization and activation of N-WASP (most likely mediated by hnRNP-K) at the entry foci and spatiotemporal actin polymerization promoted by the activated N-WASP.
DISCUSSION
The present study reveals that EtpE (more specifically EtpE-C), an E. chaffeensis surface invasin that directly binds to the nontransmembrane GPI-anchored protein DNase X (11), can induce actin polymerization in an N-WASP activation-dependent manner. In intact cells, the cell surface EtpE-DNase X complex and cytoplasmic hnRNP-K and N-WASP are topologically separated by the plasma membrane, and thus, it is likely that the transmembrane glycoprotein CD147, which is recruited to entry foci, is responsible for transducing the extracellular EtpE-DNase X binding signals across the plasma membrane to engage hnRNP-K and activate N-WASP-dependent actin polymerization to drive Ehrlichia entry. This is a previously unknown signaling pathway for host cell uptake/infection of obligate intracellular bacteria.
The only known physiologic function of DNase X is the uptake and degradation of exogenous DNA (37). The present study demonstrates for the first time the molecular mechanism by which DNase X mediates endocytosis by mammalian cells. This new information suggests that interaction between CD147 (38) and DNase X (37), both of which are present in plasma membrane lipid rafts, may facilitate ehrlichial uptake. This CD147-mediated mechanism of E. chaffeensis entry may be related to other events of actin dynamics, as CD147 has been reported to colocalize with F-actin and promote cytoskeletal rearrangements (39, 40) and the malarial ligand PfRh5 directly binds to CD147, which leads to parasite entry into erythrocytes (41).
The present work is the first to demonstrate the involvement of hnRNP-K in the host cell entry of any pathogen or molecule. Recombinant glutathione S-transferase (GST)–hnRNP-K was previously shown to activate actin polymerization in an in vitro pyrenyl-actin polymerization assay in the presence of the bovine Arp2/3 complex and GST–N-WASP (32), suggesting that hnRNP-K links the EtpE-C–DNase X signal to N-WASP activation and actin dynamics. hnRNP-K is involved in tumor metastasis and wound healing (28, 42), but its relevant signaling pathways are insufficiently defined. Therefore, our finding may help the discovery of upstream regulators of hnRNP-K that elicit actin-based cytoskeletal reorganization, potentially unifying our understanding of intracellular bacterial infection, tumor metastasis, and wound healing.
The involvement of hnRNP-K in ehrlichial entry is distinct from the classic mechanism of facultative intracellular bacterial entry, which involves the activation of Rho family GTPases and downstream factors to promote actin polymerization, either through the secretion of type III effectors (e.g., Salmonella SopE, SopE2, and SptP) or direct binding of a bacterial ligand to its cognate host cell receptor with a known role in endogenous substrate adhesion and signaling (e.g., Listeria InlB/hepatocyte growth factor and internalin A/E-cadherin and Yersinia invasin/β1 integrin) (43). It remains to be determined, however, whether the activation of Rho family GTPases has any role in ehrlichial entry.
The mammalian WASP/WAVE family contains five members, namely, WASP, N-WASP, WAVE1, WAVE2, and WAVE3, which integrate signaling cascades that lead to Arp2/3-dependent actin polymerization (44). N-WASP and WAVE2 are ubiquitously expressed, whereas WASP is expressed exclusively in leukocytes and WAVE1 and WAVE3 are enriched in the brain (45). The entry of Listeria, Yersinia, Salmonella, and Chlamydia bacteria into nonphagocytes depends on the Rac/WAVE complex pathway (46–49). On the other hand, phagocytosis of an avirulent strain of Yersinia pseudotuberculosis occurs through the activation of N-WASP (50); moreover, a type III secretion effector, Yop2b, which is inoculated into target cells, is critical for evading phagocytosis (51). N-WASP is involved in the internalization of Candia into endothelial cells (52). hnRNP-K binds to the WH1 domain of N-WASP (32), and ehrlichial hnRNP-K-dependent infectious entry likely has coopted N-WASP rather than WAVE2, as the latter lacks a WH1 domain (53).
EtpE-C induced actin nucleation in vitro in a DNase X- and N-WASP-dependent manner. This is distinct from several bacterial proteins that are known to promote actin nucleation for actin-based motility by mimicking Cdc42, N-WASP, or Arp2/3 functions, such as Shigella autotransporter IcsA (31, 54), Listeria ActA (55), the spotted fever group Rickettsia WASP-like protein RickA and Sca2 autotransporter (56), SipC of Salmonella (57), and chlamydial Tarp, a type III secretion effector (58).
E. chaffeensis tandem-repeat protein 120 (TRP120) (glycoprotein 120 [gp120]) was the first protein proposed to mediate E. chaffeensis invasion of human cells, because Escherichia coli transformed with a plasmid encoding TRP120 can invade HeLa cells (59). Antibodies to TRP120 can reduce the E. chaffeensis load in vitro and in mice (60). In addition, an antibody against the E. chaffeensis surface-exposed lipoprotein OmpA inhibits E. chaffeensis binding to and infection of THP-1 cells (10). Immunization of mice with recombinant P28, which is the E. chaffeensis major outer membrane β-barrel protein (27) and functions as a porin (61), protects mice from E. chaffeensis challenge (27). Furthermore, immunization of mice with Ehrlichia muris P28 confers protection from E. muris challenge (62). TRP120 is one of the proteins on the surface of isolated E. chaffeensis cells, the other two proteins being OmpA and VirB6-2, which are selectively degraded by E. chaffeensis HtrA protease upon treatment with the cell-permeable functional antagonist of cyclic-di-GMP; the E. chaffeensis transmembrane outer membrane proteins P28/Omp-1F, VirB9, and heat shock protein 60 (HSP60) were not degraded by the treatment in the same sample (63). Because treatment of E. chaffeensis with the antagonist of cyclic-di-GMP blocks E. chaffeensis entry but not binding (63), this result suggests that TRP120, OmpA, and VirB6-2 are not adhesins. Although some of these Ehrlichia surface proteins may mediate bacterial entry into cells, clear evidence for this has not been reported, and their respective human cell receptors and signaling pathways have not been identified.
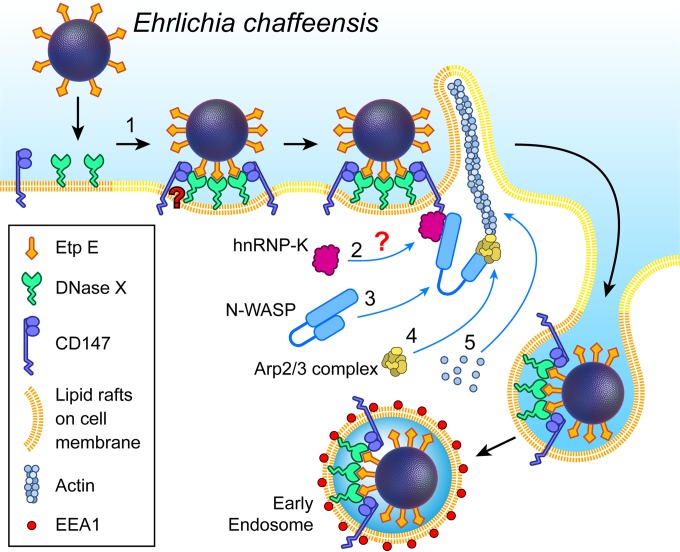
Figure 7 presents our model for the mechanism underlying the infectious entry of Ehrlichia. Upon EtpE binding to DNase X, the lateral redistribution of DNase X within lipid rafts brings CD147, which is enriched in lipid rafts, into association with the EtpE-DNase X complex. Because CD147 is a single-pass transmembrane protein, it can relay a signal to the cytoplasmic side of the cell membrane and recruit hnRNP-K, which may culminate in N-WASP activation, actin polymerization, and filopodial extension that surrounds the bacterium, leading to its uptake. Although further studies are necessary to determine detailed mechanisms of associations between these host cell proteins involved in facilitating Ehrlichia entry, the present study provides insight into a unique signaling pathway for EtpE-mediated infectious entry of an obligate intracellular bacterium.
FIG 7 .
Model for the mechanism of DNase X-mediated E. chaffeensis entry via interaction with CD147 and hnRNP-K and the subsequent N-WASP-dependent actin polymerization. (1) Extracellular E. chaffeensis uses the C-terminal region of its surface protein EtpE to bind to DNase X on the host cell surface. (2) Upon EtpE binding to DNase X, the lateral redistribution of DNase X within dynamic lipid rafts brings CD147 into association with the EtpE-DNase X complex. (3) CD147 potentially relays the signal to the cytoplasmic side of the cell membrane and recruits hnRNP-K to bind N-WASP, leading to activation of N-WASP (conformational change). (4) Activated N-WASP binds Arp2/3 actin nucleation complex. (5) This leads to spatiotemporal actin polymerization and filopodium formation to internalize E. chaffeensis into endosomes.
MATERIALS AND METHODS
Ethics statement.
BMDMs were established from wild-type and DNase X−/− C57BL/6 mice as described previously (11) in accordance with The Ohio State University Institutional Animal Care and Use Committee guidelines under approved e-protocol number 2009A0186. The University program has Full Continued Accreditation by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC-I), number 000028, dated 6 February 2015, and has Public Health Services assurance renewal number A3261-01 through 28 February 2019. The program is licensed by the USDA, 31-R-014, and is in full compliance with Animal Welfare Regulations until 1 August 2017.
Bacterial and host cell culture.
E. chaffeensis type strain Arkansas was propagated in DH82 cells, and host cell-free E. chaffeensis was obtained by controlled sonication as described previously (11). HEK293, RF/6A, THP-1 cells, and BMDMs were cultured as described previously (11).
Plasmids, recombinant proteins, and antibodies.
The plasmids used in the study, including those encoding GFP–N-WASP, GFP-WA, and iAb-47 (28), were purified with an EndoFree kit (Qiagen) and transfected using FuGene HD (Promega). The recombinant proteins rEtpE-C, rEtpE-N, and rECH0825 were prepared as described previously (11). The primary antibodies used in the study were mouse anti-EtpE-C antibody (11), rabbit anti-DNase X antibody (Abcam), anti-CD147 MAb (MEM-M6/6; Santa Cruz Biotechnology and Abcam), rabbit anti-N-WASP antibody (Santa Cruz Biotechnology), anti-hnRNP-K MAb (Santa Cruz Biotechnology), rabbit anti-actin antibody (Sigma), and rabbit anti-E. chaffeensis P28 antibody (27). The secondary antibodies were peroxidase-conjugated goat anti-mouse or anti-rabbit antibody (KPL) and goat anti-mouse or anti-rabbit IgG (Invitrogen) conjugated with Alexa Fluor 488 (AF488) or AF555 (Invitrogen). DAPI (4′,6-diamidino-2-phenylindole) (Sigma) was used to stain E. chaffeensis DNA and/or host cell DNA (nuclei).
Affinity pulldown and LC-MS/MS.
For protein pulldown, histidine (His)-tagged rEtpE-C was bound to and renatured on a Ni-silica matrix (Promega). THP-1 cell lysate in NP-40 lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.4, 1% [wt/vol] NP-40, supplemented with 1% protease inhibitor cocktail set III [Calbiochem]) was applied to rEtpE-C-bound or control matrix and incubated for 8 h at 4°C. Unbound proteins were washed from the matrix with 50 mM sodium phosphate buffer (pH 7.4) containing 0.3 M NaCl and 1% (wt/vol) NP-40, followed by 15 ml phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1 mM KH2PO4, pH 7.4) containing 10 mM imidazole and 1% NP-40. rEtpE-C and the bound protein complex were eluted with sodium phosphate buffer containing 0.3 M NaCl and 250 mM imidazole. The eluate and the postelution Ni-silica matrix were resuspended in 2× SDS sample buffer and subjected to SDS-PAGE. The gel was fixed and stained with GelCode blue (Thermo Fisher Scientific), and conspicuously different bands were cut out, trypsin digested, and subjected to LC-MS/MS at the Mass Spectrometry and Proteomics Core Facility at The Ohio State University. Sequence information from the MS/MS data was processed with Mascot Distiller to form a peak list and with the MASCOT MS/MS search engine and Turbo SEQUEST algorithm in BioWorks 3.1 software. Aliquots of samples were also subjected to immunoblotting with anti-DNase X antibody.
Far-Western blotting and coimmunoprecipitation.
rEtpE-C and rECH0825 (5 µg) were separated with SDS-PAGE, transferred to a nitrocellulose membrane, and renatured with serial guanidine-HCl treatment, followed by incubation with THP-1 cell lysate in NP-40 lysis buffer (11). After stringent washing, the membrane was incubated with anti-CD147 antibody, followed by peroxidase-conjugated goat anti-mouse IgG. The membrane was stripped with Restore Western blot stripping buffer (Thermo Fisher Scientific) and reprobed with peroxidase-conjugated anti-His MAb (Sigma). THP-1 cells were incubated with E. chaffeensis for 30 min and lysed in NP-40 lysis buffer. The lysate was immunoprecipitated with anti-EtpE-C antibody (2 µg)- or control mouse IgG (2 µg)-bound protein A-agarose beads. The precipitate was resuspended in 2× SDS sample buffer and subjected to immunoblotting with anti-CD147 and anti-N-WASP antibodies.
CD147 neutralization and RNA interference and iAb-47 functional ablation.
E. chaffeensis was added to HEK293 cells preincubated with 10 µg/ml anti-CD147 MEM-M6/6 MAb (low endotoxin, azide free; Abcam) or control mouse MAb for 30 min at 25°C in serum-free Dulbecco’s modified Eagle medium (DMEM). Binding, internalization, and infection were determined at 30 min, 1 h, and 48 h p.i., respectively. HEK293 cells in 24-well plates were transduced with 1 × 106 transducing units of CD147 shRNA lentiviral transduction particles (Sigma) using 8 µg/ml Polybrene, and resistant cells were selected with three rounds of puromycin selection (2 µg/ml). The resistant clones were replated, lysed with NP-40 lysis buffer, and used to estimate the level of CD147 with immunoblotting. The CD147 levels were normalized to the actin levels. Cells were incubated with E. chaffeensis for 30 min, 1 h, or 76 h to study binding, internalization, and infection, respectively. Coverslip cultures of HEK293 cells were transfected with 0.5 µg iAb-47 or control pEGFP-N1 plasmid. At 48 h posttransfection, cells were harvested, washed, fixed, and immunostained to study the expression pattern of hnRNP-K. Cells were incubated with host cell-free E. chaffeensis for 30 min, 2 h, or 56 h to study binding, internalization, and infection, respectively.
Inhibitors of actin dynamics.
DH82 or HEK293 cells in DMEM were incubated with wiskostatin (Sigma) at 0.1 to 10 µM and cytochalasin D (Calbiochem) at 10 µM for 30 min. The cells were washed and replenished with complete DMEM prior to incubation with freshly isolated host cell-free E. chaffeensis. At 2 h p.i., the cells were washed to remove unbound or loosely attached bacteria and further incubated for 48 h without chemicals. For the internalization and infection study in HEK293 cells, cells were pretreated for 30 min with 2.5 µM cytochalasin D or 5 µM wiskostatin, followed by incubation with host cell-free E. chaffeensis. After removal of chemicals by washing, the cells were cultured for an additional 1 and 48 h, respectively, for internalization and infection. To assess the role of N-WASP in bacterial internalization, RF/6A cells were transfected with GFP–N-WASP or GFP-WA. At 48 h posttransfection, cells were incubated with host cell-free E. chaffeensis at a multiplicity of infection of 50 for 4 h.
Binding, internalization, and infection assays.
For the binding assay, samples were fixed with 3.5% paraformaldehyde and subjected to one round of immunolabeling with anti-P28 antibody, whereas for internalization assays, two steps of labeling of fixed cells with anti-P28 antibody were carried out: the first labeling step was performed without saponin permeabilization to detect bound but not internalized E. chaffeensis using anti-P28 antibody and AF488-conjugated anti-rabbit IgG, and the second labeling step was performed after permeabilization with saponin to detect total E. chaffeensis bacteria using AF555-conjugated anti-rabbit IgG (11). Fluorescence images were acquired using a DeltaVision deconvolution microscope system (GE Healthcare). The number of bacteria on 100 cells was scored. Overall infection was determined at 48 to 76 h p.i. by immunoblotting with anti-P28 antibody or qPCR, using the E. chaffeensis 16S rRNA gene normalized to the host cell glycerol-3-phosphate dehydrogenase (G3PDH) gene (10).
Cellular localization analysis.
Sulfate-modified fluorescent red polystyrene beads (0.5-µm diameter; Sigma) were coated with rEtpE-C as described previously (11). Freshly prepared protein-coated beads were added to HEK293 cells at a ratio of approximately 50 beads per cell and incubated for 1 h at 37°C. Cells were fixed, permeabilized, and labeled with mouse anti-CD147 MAb and AF488-conjugated goat anti-mouse IgG.
Live-cell imaging by spinning-disk confocal microscopy.
RF/6A cells were transfected with GFP–N-WASP, and at 23 h posttransfection, fresh phenol red-free Leibovitz’s L-15 medium supplemented with 10% fetal bovine serum was added, and cells were incubated with flash red beads (0.51-µm diameter; Bangs Laboratories) coated with rEtpE-C (11). Samples were moved to a live-cell imaging chamber at 37°C connected to a TI-E inverted research microscope (Nikon Instruments) controlled by Nikon Elements software and equipped with a spinning-disk confocal unit (Yokogawa Electric), a 100× objective lens (Plan Apochromat Lambda, NA 1.45; Nikon), and an electron multiplying charge-coupled device (EMCCD) camera (iXon DU897 Ultra; Andor Technology). Three-dimensional confocal image stacks were acquired at 1-min intervals for 2 h, with a step size of 0.1 µm along the z axis. The bead internalization movie (see Movie S1 in the supplemental material) was prepared with ImageJ software by creating a stack of confocal images acquired at the upper surface of an RF/6A cell.
Pyrenyl-actin polymerization assay.
The actin polymerization assay was performed according to the manufacturer’s protocol (Cytoskeleton, Inc.). Purified rEtpE-C and rEtpE-N at 20 µg were used for the assay. The baseline fluorescence emitted by pyrenyl-actin in G-actin buffer (Cytoskeleton, Inc.) or G buffer blank was measured at 37°C for 3 min. Control buffer, rEtpE-N, or rEtpE-C was added to the wells in the presence of THP-1 cell lysate or wild-type or DNase X−/− BMDM lysate with or without 10 µM wiskostatin in a total volume of 200 µl. Fluorescence was measured every 30 s for 30 min using the medium photomultiplier setting with an excitation wavelength of 355 nm, an emission wavelength of 430 nm, and a cutoff of 420 nm. At the end of the reading, 10× actin polymerization buffer (Cytoskeleton, Inc.) was added to all wells, and the fluorescence was read again to verify that the assay conditions were appropriate to detect polymerization. The raw data obtained were used to generate a polymerization time course plot.
Statistical analysis.
Statistical analysis was performed with an unpaired, two-tailed Student’s t test. A P value of <0.05 was considered significant.
SUPPLEMENTAL MATERIAL
(Related to Fig. 5A.) Internalization of rEtpE-C-coated beads (red) at the upper surface of an RF/6A cell transfected with GFP–N-WASP (cyan) captured by spinning-disk confocal microscopy. The movie was prepared with ImageJ software by creating a stack of confocal images acquired at the upper surface of an RF/6A cell at 1-min intervals during the 2-h time span. The beads disappear from the two-dimensional image section as they are fully internalized by the cell and move away from the upper surface. Download
Wiskostatin and cytochalasin D inhibit E. chaffeensis entry into HEK293 cells (related to Fig. 4). HEK293 cells were pretreated for 30 min with 5 µM of wiskostatin or 2.5 µM cytochalasin D and subsequently incubated with E. chaffeensis in the absence of these two inhibitors for an additional 1 and 48 h for internalization and infection, respectively. (A) Internalization analysis. The cells were fixed and subjected to two rounds of immunostaining with anti-P28 antibody, the first without permeabilization to detect bound but not internalized E. chaffeensis bacteria (AF555-conjugated anti-rabbit IgG), and the second with saponin permeabilization to detect total E. chaffeensis bacteria, i.e., bound plus internalized (AF488-conjugated anti-rabbit IgG). The numbers of internalized E. chaffeensis bacteria (Ech, total minus bound) per host cell were determined by counting 100 HEK293 cells. Data represent the mean results and standard deviations from triplicate samples and are representative of three independent experiments. **, P < 0.01. (B, C) Infection analysis. (B) E. chaffeensis infection in DMSO solvent control, wiskostatin, or cytochalasin D-treated cells. The arrowheads indicate E. chaffeensis inclusions. Diff-Quik staining. Scale bar, 5 µm. (C) Quantification of data from the experiment whose results are shown in panel B. Data represent the mean results and standard deviations from triplicate samples and are representative of three independent experiments. *, P < 0.05. Download
LC-MS/MS and protein identification of the band shown by an asterisk in Fig. 1A
ACKNOWLEDGMENTS
We thank Renu Wadhwa (AIST, Tsukuba, Japan) for the iAb-47 plasmid and Michael Way (Cancer Research, United Kingdom) for providing GFP–N-WASP and GFP-WA plasmids. We thank Tim Vojt for assistance in preparing Fig. 7.
Footnotes
Citation Mohan Kumar D, Lin M, Xiong Q, Webber MJ, Kural C, Rikihisa Y. 2015. EtpE binding to DNase X induces ehrlichial entry via CD147 and hnRNP-K recruitment, followed by mobilization of N-WASP and actin. mBio 6(6):e01541-15. doi:10.1128/mBio.01541-15.
REFERENCES
- 1.CDC 2015. Table II. Provisional cases of selected notifiable diseases, United States, weeks ending January 3, 2015 and December 28, 2013 (53rd week). MMWR Morb Mortal Wkly Rep 63(53):ND-733-ND-746. http://www.cdc.gov/mmwr/preview/mmwrhtml/mm6353md.htm?s_cid=mm6353md_w#tab2. [Google Scholar]
- 2.Paddock CD, Childs JE. 2003. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin Microbiol Rev 16:37–64. doi: 10.1128/CMR.16.1.37-64.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paddock CD, Yabsley MJ. 2007. Ecological havoc, the rise of white-tailed deer, and the emergence of Amblyomma americanum-associated zoonoses in the United States. Curr Top Microbiol Immunol 315:289–324. doi: 10.1007/978-3-540-70962-6_12. [DOI] [PubMed] [Google Scholar]
- 4.Committee on Lyme Disease and Other Tick-Borne Diseases: The State of the Science; Institute of Medicine. 2011. Critical needs and gaps in understanding prevention, amelioration, and resolution of Lyme and other tick-borne diseases: the short-term and long-term outcomes. Workshop report. National Academies Press, Washington, DC: http://www.nap.edu/read/13134/chapter/1. [Google Scholar]
- 5.Lin M, Rikihisa Y. 2003. Obligatory intracellular parasitism by Ehrlichia chaffeensis and Anaplasma phagocytophilum involves caveolae and glycosylphosphatidylinositol-anchored proteins. Cell Microbiol 5:809–820. doi: 10.1046/j.1462-5822.2003.00322.x. [DOI] [PubMed] [Google Scholar]
- 6.Mott J, Barnewall RE, Rikihisa Y. 1999. Human granulocytic ehrlichiosis agent and Ehrlichia chaffeensis reside in different cytoplasmic compartments in HL-60 cells. Infect Immun 67:1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin M, Rikihisa Y. 2007. Degradation of p22phox and inhibition of superoxide generation by Ehrlichia chaffeensis in human monocytes. Cell Microbiol 9:861–874. doi: 10.1111/j.1462-5822.2006.00835.x. [DOI] [PubMed] [Google Scholar]
- 8.Barnewall RE, Rikihisa Y, Lee EH. 1997. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect Immun 65:1455–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin M, Zhu MX, Rikihisa Y. 2002. Rapid activation of protein tyrosine kinase and phospholipase C-gamma2 and increase in cytosolic free calcium are required by Ehrlichia chaffeensis for internalization and growth in THP-1 cells. Infect Immun 70:889–898. doi: 10.1128/IAI.70.2.889-898.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng Z, Miura K, Popov VL, Kumagai Y, Rikihisa Y. 2011. Insights into the CtrA regulon in development of stress resistance in obligatory intracellular pathogen Ehrlichia chaffeensis. Mol Microbiol 82:1217–1234. doi: 10.1111/j.1365-2958.2011.07885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohan Kumar D, Yamaguchi M, Miura K, Lin M, Los M, Coy JF, Rikihisa Y. 2013. Ehrlichia chaffeensis uses its surface protein EtpE to bind GPI-anchored protein DNase X and trigger entry into mammalian cells. PLoS Pathog 9:e1003666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maeda K, Markowitz N, Hawley RC, Ristic M, Cox D, McDade JE. 1987. Human infection with Ehrlichia canis, a leukocytic rickettsia. N Engl J Med 316:853–856. doi: 10.1056/NEJM198704023161406. [DOI] [PubMed] [Google Scholar]
- 13.Dawson JE, Anderson BE, Fishbein DB, Sanchez JL, Goldsmith CS, Wilson KH, Duntley CW. 1991. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J Clin Microbiol 29:2741–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barnewall RE, Rikihisa Y. 1994. Abrogation of gamma interferon-induced inhibition of Ehrlichia chaffeensis infection in human monocytes with iron-transferrin. Infect Immun 62:4804–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allison AC, Davies P, De Petris S. 1971. Role of contractile microfilaments in macrophage movement and endocytosis. Nat New Biol 232:153–155. doi: 10.1038/newbio232153a0. [DOI] [PubMed] [Google Scholar]
- 16.Gottlieb TA, Ivanov IE, Adesnik M, Sabatini DD. 1993. Actin microfilaments play a critical role in endocytosis at the apical but not the basolateral surface of polarized epithelial cells. J Cell Biol 120:695–710. doi: 10.1083/jcb.120.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francis CL, Ryan TA, Jones BD, Smith SJ, Falkow S. 1993. Ruffles induced by Salmonella and other stimuli direct macropinocytosis of bacteria. Nature 364:639–642. doi: 10.1038/364639a0. [DOI] [PubMed] [Google Scholar]
- 18.Carabeo RA, Grieshaber SS, Fischer E, Hackstadt T. 2002. Chlamydia trachomatis induces remodeling of the actin cytoskeleton during attachment and entry into HeLa cells. Infect Immun 70:3793–3803. doi: 10.1128/IAI.70.7.3793-3803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romero S, Grompone G, Carayol N, Mounier J, Guadagnini S, Prevost MC, Sansonetti PJ, Van Nhieu GT. 2011. ATP-mediated Erk1/2 activation stimulates bacterial capture by filopodia, which precedes Shigella invasion of epithelial cells. Cell Host Microbe 9:508–519. doi: 10.1016/j.chom.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang JZ, Popov VL, Gao S, Walker DH, Yu XJ. 2007. The developmental cycle of Ehrlichia chaffeensis in vertebrate cells. Cell Microbiol 9:610–618. doi: 10.1111/j.1462-5822.2006.00812.x. [DOI] [PubMed] [Google Scholar]
- 21.Iacono KT, Brown AL, Greene MI, Saouaf SJ. 2007. CD147 immunoglobulin superfamily receptor function and role in pathology. Exp Mol Pathol 83:283–295. doi: 10.1016/j.yexmp.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanna SM, Kirk P, Holt OJ, Puklavec MJ, Brown MH, Barclay AN. 2003. A novel form of the membrane protein CD147 that contains an extra Ig-like domain and interacts homophilically. BMC Biochem 4:17. doi: 10.1186/1471-2091-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia L, Zhou H, Wang S, Cao J, Wei W, Zhang J. 2006. Deglycosylation of CD147 down-regulates matrix metalloproteinase-11 expression and the adhesive capability of murine hepatocarcinoma cell HcaF in vitro. IUBMB Life 58:209–216. doi: 10.1080/15216540600719580. [DOI] [PubMed] [Google Scholar]
- 24.Mikula M, Dzwonek A, Karczmarski J, Rubel T, Dadlez M, Wyrwicz LS, Bomsztyk K, Ostrowski J. 2006. Landscape of the hnRNP K protein-protein interactome. Proteomics 6:2395–2406. doi: 10.1002/pmic.200500632. [DOI] [PubMed] [Google Scholar]
- 25.Chen H, Fok KL, Jiang X, Jiang J, Chen Z, Gui Y, Chan HC, Cai Z. 2012. CD147 regulates apoptosis in mouse spermatocytes but not spermatogonia. Hum Reprod 27:1568–1576. doi: 10.1093/humrep/des050. [DOI] [PubMed] [Google Scholar]
- 26.Rao DD, Vorhies JS, Senzer N, Nemunaitis J. 2009. siRNA vs. shRNA: similarities and differences. Adv Drug Deliv Rev 61:746–759. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Ohashi N, Zhi N, Zhang Y, Rikihisa Y. 1998. Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect Immun 66:132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue A, Sawata SY, Taira K, Wadhwa R. 2007. Loss-of-function screening by randomized intracellular antibodies: identification of hnRNP-K as a potential target for metastasis. Proc Natl Acad Sci U S A 104:8983–8988. doi: 10.1073/pnas.0607595104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jobling SA, Jarman C, Teh MM, Holmberg N, Blake C, Verhoeyen ME. 2003. Immunomodulation of enzyme function in plants by single-domain antibody fragments. Nat Biotechnol 21:77–80. doi: 10.1038/nbt772. [DOI] [PubMed] [Google Scholar]
- 30.Campellone KG, Welch MD. 2010. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol 11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreau V, Frischknecht F, Reckmann I, Vincentelli R, Rabut G, Stewart D, Way M. 2000. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat Cell Biol 2:441–448. doi: 10.1038/35017080. [DOI] [PubMed] [Google Scholar]
- 32.Yoo Y, Wu X, Egile C, Li R, Guan JL. 2006. Interaction of N-WASP with hnRNPK and its role in filopodia formation and cell spreading. J Biol Chem 281:15352–15360. doi: 10.1074/jbc.M511825200. [DOI] [PubMed] [Google Scholar]
- 33.Peterson JR, Bickford LC, Morgan D, Kim AS, Ouerfelli O, Kirschner MW, Rosen MK. 2004. Chemical inhibition of N-WASP by stabilization of a native autoinhibited conformation. Nat Struct Mol Biol 11:747–755. doi: 10.1038/nsmb796. [DOI] [PubMed] [Google Scholar]
- 34.Guerriero CJ, Weisz OA. 2007. N-WASP inhibitor wiskostatin nonselectively perturbs membrane transport by decreasing cellular ATP levels. Am J Physiol Cell Physiol 292:C1562–C1566. doi: 10.1152/ajpcell.00426.2006. [DOI] [PubMed] [Google Scholar]
- 35.Casella JF, Flanagan MD, Lin S. 1981. Cytochalasin D inhibits actin polymerization and induces depolymerization of actin filaments formed during platelet shape change. Nature 293:302–305. doi: 10.1038/293302a0. [DOI] [PubMed] [Google Scholar]
- 36.Cooper JA, Walker SB, Pollard TD. 1983. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J Muscle Res Cell Motil 4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- 37.Shiokawa D, Matsushita T, Shika Y, Shimizu M, Maeda M, Tanuma S. 2007. DNase X is a glycosylphosphatidylinositol-anchored membrane enzyme that provides a barrier to endocytosis-mediated transfer of a foreign gene. J Biol Chem 282:17132–17140. doi: 10.1074/jbc.M610428200. [DOI] [PubMed] [Google Scholar]
- 38.Tang W, Chang SB, Hemler ME. 2004. Links between CD147 function, glycosylation, and caveolin-1. Mol Biol Cell 15:4043–4050. doi: 10.1091/mbc.E04-05-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlosshauer B, Bauch H, Frank R. 1995. Neurothelin: amino acid sequence, cell surface dynamics and actin colocalization. Eur J Cell Biol 68:159–166. [PubMed] [Google Scholar]
- 40.Qian A, Zhang W, Cao J, Yang P, Gao X, Wang Z, Xu H, Weng Y, Shang P. 2008. Downregulation of CD147 expression alters cytoskeleton architecture and inhibits gelatinase production and SAPK pathway in human hepatocellular carcinoma cells. J Exp Clin Cancer Res 27:50. doi: 10.1186/1756-9966-27-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crosnier C, Bustamante LY, Bartholdson SJ, Bei AK, Theron M, Uchikawa M, Mboup S, Ndir O, Kwiatkowski DP, Duraisingh MT, Rayner JC, Wright GJ. 2011. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature 480:534–537. doi: 10.1038/nature10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao R, Yu Y, Inoue A, Widodo N, Kaul SC, Wadhwa R. 2013. Heterogeneous nuclear ribonucleoprotein K (hnRNP-K) promotes tumor metastasis by induction of genes involved in extracellular matrix, cell movement, and angiogenesis. J Biol Chem 288:15046–15056. doi: 10.1074/jbc.M113.466136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pizarro-Cerdá J, Cossart P. 2006. Bacterial adhesion and entry into host cells. Cell 124:715–727. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 44.Higgs HN, Pollard TD. 2001. Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu Rev Biochem 70:649–676. doi: 10.1146/annurev.biochem.70.1.649. [DOI] [PubMed] [Google Scholar]
- 45.Takenawa T, Miki H. 2001. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J Cell Sci 114:1801–1809. [DOI] [PubMed] [Google Scholar]
- 46.Bosse T, Ehinger J, Czuchra A, Benesch S, Steffen A, Wu X, Schloen K, Niemann HH, Scita G, Stradal TE, Brakebusch C, Rottner K. 2007. Cdc42 and phosphoinositide 3-kinase drive Rac-mediated actin polymerization downstream of c-Met in distinct and common pathways. Mol Cell Biol 27:6615–6628. doi: 10.1128/MCB.00367-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alrutz MA, Srivastava A, Wong KW, D’Souza-Schorey C, Tang M, Ch’Ng L, Snapper SB, Isberg RR. 2001. Efficient uptake of Yersinia pseudotuberculosis via integrin receptors involves a Rac1-Arp 2/3 pathway that bypasses N-WASP function. Mol Microbiol 42:689–703. doi: 10.1046/j.1365-2958.2001.02676.x. [DOI] [PubMed] [Google Scholar]
- 48.Carabeo RA, Grieshaber SS, Hasenkrug A, Dooley C, Hackstadt T. 2004. Requirement for the Rac GTPase in Chlamydia trachomatis invasion of non-phagocytic cells. Traffic 5:418–425. doi: 10.1111/j.1398-9219.2004.00184.x. [DOI] [PubMed] [Google Scholar]
- 49.Humphreys D, Davidson AC, Hume PJ, Makin LE, Koronakis V. 2013. Arf6 coordinates actin assembly through the WAVE complex, a mechanism usurped by salmonella to invade host cells. Proc Natl Acad Sci U S A 110:16880–16885. doi: 10.1073/pnas.1311680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGee K, Zettl M, Way M, Fällman M. 2001. A role for N-WASP in invasin-promoted internalisation. FEBS Lett 509:59–65. doi: 10.1016/S0014-5793(01)03139-8. [DOI] [PubMed] [Google Scholar]
- 51.Rosqvist R, Bolin I, Wolf-Watz H. 1988. Inhibition of phagocytosis in Yersinia pseudotuberculosis: a virulence plasmid-encoded ability involving the Yop2b protein. Infect Immun 56:2139–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shintaku T, Glass KA, Hirakawa MP, Longley SJ, Bennett RJ, Bliss JM, Shaw SK. 2013. Human endothelial cells internalize Candida parapsilosis via N-WASP-mediated endocytosis. Infect Immun 81:2777–2787. doi: 10.1128/IAI.00535-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suetsugu S, Hattori M, Miki H, Tezuka T, Yamamoto T, Mikoshiba K, Takenawa T. 2002. Sustained activation of N-WASP through phosphorylation is essential for neurite extension. Dev Cell 3:645–658. doi: 10.1016/S1534-5807(02)00324-6. [DOI] [PubMed] [Google Scholar]
- 54.Egile C, Loisel TP, Laurent V, Li R, Pantaloni D, Sansonetti PJ, Carlier MF. 1999. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J Cell Biol 146:1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schirmer EC, Yates JR III, Gerace L. 2003. MudPIT: A powerful proteomics tool for discovery. Discov Med 3:38–39. [PubMed] [Google Scholar]
- 56.Harlander RS, Way M, Ren Q, Howe D, Grieshaber SS, Heinzen RA. 2003. Effects of ectopically expressed neuronal Wiskott-Aldrich syndrome protein domains on Rickettsia rickettsii actin-based motility. Infect Immun 71:1551–1556. doi: 10.1128/IAI.71.3.1551-1556.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki K. 2013. Selective autophagy in budding yeast. Cell Death Differ 20:43–48. doi: 10.1038/cdd.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siren J, Sareneva T, Pirhonen J, Strengell M, Veckman V, Julkunen I, Matikainen S. 2004. Cytokine and contact-dependent activation of natural killer cells by influenza A or Sendai virus-infected macrophages. J Gen Virol 85:2357–2364. doi: 10.1099/vir.0.80105-0. [DOI] [PubMed] [Google Scholar]
- 59.Popov VL, Yu X, Walker DH. 2000. The 120-kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb Pathog 28:71–80. doi: 10.1006/mpat.1999.0327. [DOI] [PubMed] [Google Scholar]
- 60.Kuriakose JA, Zhang X, Luo T, McBride JW. 2012. Molecular basis of antibody mediated immunity against Ehrlichia chaffeensis involves species-specific linear epitopes in tandem repeat proteins. Microbes Infect 14:1054–1063. doi: 10.1016/j.micinf.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumagai Y, Huang H, Rikihisa Y. 2008. Expression and porin activity of P28 and OMP-1F during intracellular Ehrlichia chaffeensis development. J Bacteriol 190:3597–3605. doi: 10.1128/JB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas S, Thirumalapura NR, Crocquet-Valdes PA, Luxon BA, Walker DH. 2011. Structure-based vaccines provide protection in a mouse model of ehrlichiosis. PLoS One 6:e27981. doi: 10.1371/journal.pone.0027981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumagai Y, Matsuo J, Hayakawa Y, Rikihisa Y. 2010. Cyclic di-GMP signaling regulates invasion by Ehrlichia chaffeensis of human monocytes. J Bacteriol 192:4122–4133. doi: 10.1128/JB.00132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Related to Fig. 5A.) Internalization of rEtpE-C-coated beads (red) at the upper surface of an RF/6A cell transfected with GFP–N-WASP (cyan) captured by spinning-disk confocal microscopy. The movie was prepared with ImageJ software by creating a stack of confocal images acquired at the upper surface of an RF/6A cell at 1-min intervals during the 2-h time span. The beads disappear from the two-dimensional image section as they are fully internalized by the cell and move away from the upper surface. Download
Wiskostatin and cytochalasin D inhibit E. chaffeensis entry into HEK293 cells (related to Fig. 4). HEK293 cells were pretreated for 30 min with 5 µM of wiskostatin or 2.5 µM cytochalasin D and subsequently incubated with E. chaffeensis in the absence of these two inhibitors for an additional 1 and 48 h for internalization and infection, respectively. (A) Internalization analysis. The cells were fixed and subjected to two rounds of immunostaining with anti-P28 antibody, the first without permeabilization to detect bound but not internalized E. chaffeensis bacteria (AF555-conjugated anti-rabbit IgG), and the second with saponin permeabilization to detect total E. chaffeensis bacteria, i.e., bound plus internalized (AF488-conjugated anti-rabbit IgG). The numbers of internalized E. chaffeensis bacteria (Ech, total minus bound) per host cell were determined by counting 100 HEK293 cells. Data represent the mean results and standard deviations from triplicate samples and are representative of three independent experiments. **, P < 0.01. (B, C) Infection analysis. (B) E. chaffeensis infection in DMSO solvent control, wiskostatin, or cytochalasin D-treated cells. The arrowheads indicate E. chaffeensis inclusions. Diff-Quik staining. Scale bar, 5 µm. (C) Quantification of data from the experiment whose results are shown in panel B. Data represent the mean results and standard deviations from triplicate samples and are representative of three independent experiments. *, P < 0.05. Download
LC-MS/MS and protein identification of the band shown by an asterisk in Fig. 1A