Abstract
The adult Caenorhabditis elegans hermaphrodite gonad consists of two mirror-symmetric U-shaped arms, with germline nuclei located peripherally in the distal regions of each arm. The nuclei are housed within membrane cubicles that are open to the center, forming a syncytium with a shared cytoplasmic core called the rachis. As the distal germline nuclei progress through meiotic prophase, they move proximally and eventually cellularize as their compartments grow in size. The development and maintenance of this complex and dynamic germline membrane architecture are relatively unexplored, and we have used a forward genetic screen to identify 20 temperature-sensitive mutations in 19 essential genes that cause defects in the germline membrane architecture. Using a combined genome-wide SNP mapping and whole genome sequencing strategy, we have identified the causal mutations in 10 of these mutants. Four of the genes we have identified are conserved, with orthologs known to be involved in membrane biology, and are required for proper development or maintenance of the adult germline membrane architecture. This work provides a starting point for further investigation of the mechanisms that control the dynamics of syncytial membrane architecture during adult oogenesis.
Keywords: C. elegans, whole genome sequencing, gonad development, membrane architecture, membrane dynamics
The adult Caenorhabditis elegans hermaphrodite reproductive system consists of two mirror symmetrical U-shaped tubular gonad arms that each terminate in a spermatheca and share a common central uterus (Hirsh et al. 1976; Kimble and Hirsh 1979) (Figure 1). The gonad arms comprise somatic sheath cells that house a proliferative germline with a complex membrane architecture. At the distal end of each gonad arm, the germline proliferates mitotically. As germline nuclei move proximally and enter meiosis, they adopt peripheral positions, such that the nuclei reside within individual membrane cubicles that are open to and continuous with a shared core cytoplasm called the rachis. As these syncytial meiotic germline nuclei move more proximally, they eventually cellularize and enlarge to become mature oocytes. Although the cellular anatomy of the adult reproductive system and the cell lineages that gives rise to the somatic and germline structures during larval development have been described in detail (Kimble and Hirsh 1979; Hubbard and Greenstein 2000; Kimble and Crittenden 2007; Wong and Schwarzbauer 2012), the membrane dynamics that mediate the movement of syncytial oocyte nuclei and their eventual cellularization during oogenesis in the adult gonad remain poorly understood.
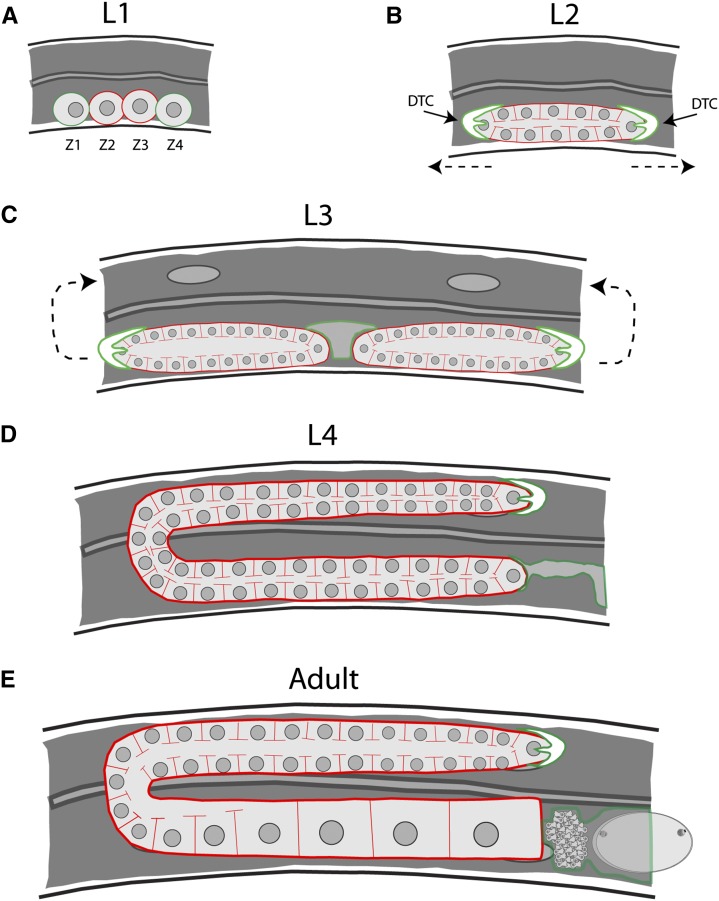
Figure 1.
Germline morphogenesis in C. elegans. In each panel, germ cells are outlined in red and somatic cells/tissues are in green. (A) At the L1 stage, there are four cells that will give rise to the entirety of the gonad. Two of these cells, Z1 and Z4, will give rise to the somatic tissues of the gonad, whereas Z2 and Z3 will generate the germline lineage. (B) Proliferation of the germline begins after the transition to the L2 stage. This is also the stage in which the germline syncytium forms. The direction of distal tip cell (green outline) migration is denoted by the dashed arrows. (C) As proliferation continues during the L3 stage, the single gonad tube is bisected by the developing somatic gonad tissues, giving rise to two mirror symmetrical arms. The distal tip cell migrates first ventrally, then back toward the center of the animal (arrows). (D) By the L4 stage, the gonad has mostly assumed its final shape, and spermatogenesis begins. Only one of the two arms is shown, with the proximal arm connected to the developing spermatheca and uterus (green outline). (E) Oogenesis begins after the transition to adulthood and will happen continuously throughout the lifespan of the animal.
During embryogenesis, four cells called Z1–Z4 are produced such that they align along the anterior–posterior axis and abut the ventral epidermis of the first stage (L1) larva; these four cells then proliferate to form the adult gonad (Figure 1). The two outermost cells, Z1 and Z4, undergo cell divisions during larval development to produce the somatic structures, which include two distal tip cells (DTCs), gonadal sheath cells, two spermathecae, and the uterus (Kimble and Hirsh 1979). The two innermost cells, Z2 and Z3, proliferate during larval and adult development to produce the germline, including approximately 300 sperm that are made late in larval development, and subsequently all of the oocytes produced by an adult hermaphrodite.
Beginning midway through the L1 larval stage, Z1 and Z4 give rise to 12 descendants that, by the early second (L2) larval stage, form a short tube-like structure in which the germline precursor cells reside (Figure 1). These initial descendants include two distal tip cells, one located at each end of the initial tube-like structure. During the third (L3) larval stage, the somatic gonad precursors undergo additional cell division, and the DTCs migrate in opposite directions along the anterior–posterior axis. The DTCs eventually turn to migrate dorsally, and then turn back toward the center of the worm and migrate back toward each other. Concurrent with DTC migration, gonadal sheath cells migrate and elongate to envelop nearly the entire structure. Morphogenesis is mostly complete by the last (L4) larval stage, when the DTCs reach the center of the animal and stop migrating soon after they meet. As a result, the adult gonad has two U-shaped arms joined centrally by the two spermathecae and the uterus.
Proliferation and development of the germline also take place as the somatic gonad develops. The germline precursors Z2 and Z3 begin to proliferate during the L1 larval stage, and proliferation is subsequently maintained locally by Notch signals from the DTCs. The dividing germline cells undergo incomplete cytokinesis as they proliferate, a process that is regulated by two annilins, ANI-1 and ANI-2 (Maddox et al. 2005; Amini et al. 2014). This ultimately creates the shared cytoplasmic germline core (the rachis) that is maintained throughout adulthood. As the germline stem cell nuclei nearest to the DTCs continue to proliferate, descendant nuclei move away and enter meiosis as they leave the influence of Notch signaling. During adult oogenesis, the distal germline continues to proliferate. As the syncytial meiotic nuclei reach the turn of the gonad arm, all meiotic nuclei either undergo programmed cell death or adopt the oocyte fate. The surviving nuclei then begin to cellularize and mature, greatly increasing in size due in part to actomyosin-dependent cytoplasmic streaming (Wolke et al. 2007). The processes of cytoplasmic streaming and oocyte growth and maturation are regulated by at least two signals. One emanates from sperm at the proximal end of the gonad and the other emanates from the DTCs at the distal end (Govindan et al. 2009; Nadarajan et al. 2009). Sperm release the major sperm protein (MSP), a signaling ligand that acts through the somatic gonad sheath cells and cyclic AMP to promote oocyte growth and maturation (Govindan et al. 2009). The DTCs limit oocyte growth through Notch signaling in a process that appears to occur independently of the DTC Notch signaling that promotes mitotic germline proliferation in the distal gonad (Nadarajan et al. 2009). Together, these two signals coordinately regulate oocyte growth. Finally, the most mature oocytes complete cellularization. They close off from the rachis before reaching and ultimately passing through the spermathecae, where they are fertilized before entering the uterus (McCarter et al. 1999; Maddox et al. 2005). In young adult hermaphrodites, ovulation occurs approximately every 23 min, producing in 6.5 hr zygotes approximately equal in volume to the entire body of the worm (Hirsh et al. 1976; McCarter et al. 1999; Wolke et al. 2007). Thus, the hermaphrodite gonad functions like a conveyor belt, with germline nuclei traveling down the length of the arm as they proceed through meiosis, eventually cellularizing, enlarging, and maturing prior to fertilization upon ovulation.
The complex membrane architecture of the adult germline and its ability to produce large numbers of oocytes throughout the adult life span imply a requirement for extensive and dynamic regulation of membrane production and trafficking. Consistent with such a requirement, the v-SNARE SNB-1 localizes to the plasma membrane in the distal mitotic region of the gonad, and loss of function of early secretory pathway components, such as members of the COPII complex, result in a failure of SNB-1 to localize properly, a failure to form new membrane partitions between germline nuclei following division, and severe disruption of the gonad membrane architecture (Green et al. 2011; Hanna et al. 2013). In other contexts, the partitions that arise during cellularization in Drosophila embryos require the trafficking of recycling endosomes for growth (Lecuit 2004), and requirements for endosome trafficking also have been found for the completion of cytokinesis (Schiel and Prekeris 2013). While an extensive RNAi-based screen has identified many essential C. elegans genes required for proper development of the adult gonad (Green et al. 2011), very little is known about the regulation of membrane dynamics during adult oogenesis. As the syncytial nature of the germline is widely conserved (Greenbaum et al. 2015; Haglund et al. 2011), C. elegans provides a useful genetic system for investigating how membrane dynamics are controlled during oogenesis.
Membrane trafficking also is required for C. elegans eggshell development (Olson et al. 2012; Sato et al. 2006, 2008). The eggshell has a trilaminar structure and is composed of proteins secreted by the zygote following fertilization, with secreted enzymes also participating in eggshell synthesis. Immediately after fertilization, the outermost vitelline layer forms, creating a block to polyspermy. This is closely followed by activation of CHS-1, an enzyme that synthesizes chitin at the cell surface, forming the second layer of the eggshell. Finally, cortical granules carrying the chondroitin proteoglycans CPG-1 and CPG-2 are secreted while CHS-1 is simultaneously internalized, forming the third layer. Failure of cortical granule exocytosis results in embryos that are defective in eggshell formation and are osmotically sensitive. Some membrane trafficking genes are known to be required for cortical granule formation and exocytosis. For example, loss of function of either rab-11.1 or the syntaxin syn-4 results in the aberrant accumulation of caveolin-positive vesicles, normally present at the cortex, around the nuclei of proximal oocytes (Sato et al. 2008). Furthermore, loss of pod-1, a C. elegans coronin homolog, results in osmotic sensitivity due to the misregulation of the timing of cortical granule exocytosis (Rappleye et al. 1999; Olson et al. 2012).
Given the known requirements for membrane trafficking in both germline development and eggshell formation, we have screened a large collection of temperature-sensitive, embryonic lethal C. elegans mutants for defects in both gonad development and eggshell production. These mutants exhibited embryonic osmotic sensitivity when homozygous mutant worms were up-shifted to the restrictive temperature at the L4 larval stage, after the completion of most gonad development. However, when up-shifted to the restrictive temperature at the L1 larval stage, homozygous mutants matured into sterile adults with defects in gonad development. We have identified the causal mutations in a subset of these mutants and present a preliminary analysis of their requirements during gonad development and adult oogenesis.
Materials and Methods
C. elegans strains
All strains were handled and maintained following standard procedures (Brenner 1974). TS alleles were generated by way of a forward genetic screen using chemical mutagenesis, as previously described (Kemphues et al. 1988; Encalada et al. 2000; O’Rourke et al. 2011a). Following isolation, TS mutant strains were outcrossed to wild-type (N2 Bristol) males to remove the lin-2(e1309) mutation present in the parental strain used for screening. TS mutant strains were maintained at the permissive temperature of 15°. Embryonic phenotype characterization was performed by selecting L4 hermaphrodites grown at the permissive temperature and moving them to 26° for a minimum of 6 hr before dissecting embryos into water. The dye permeability assay used to determine permeability barrier defects is described in Carvalho et al. (2011). Postembryonic phenotype characterization was performed by moving synchronized populations of L1 larvae to 26° for several (3–5) days and scoring adult germline morphology. Isolated TS alleles and deletion alleles used are listed here by chromosome: rpl-7(or990ts) I; rpl-7(or1247ts) I; abtm-1(or1400ts) I; abtm-1(tm2721) I; or1184ts I; or1613ts I; or983ts I; or819ts I; ippk-1(or1572ts) II; ippk-1(tm4718) II; vps-15(or1235ts) II; vps-15(ok3132) II; crn-3(or959ts) II; crn-3(ok2269) II; sqv-8(or888ts) II; sqv-8(n2822) II; or1112ts II; atx-2(or821ts) III; atx-2(tm4373) III; ndg-4(or1088ts) III; ndg-4(sa529) III; or1565ts III; or1439ts III; drp-1(or1393ts) IV; drp-1(tm1108) IV; and or1233ts IV. See Supporting Information for a list of mutant strains used for complementation tests to identify causal TS Eos/Ste mutations (Table S2).
Genetic analysis
Embryonic lethality was scored at both 15° and 26° by singling out 10 L4 hermaphrodites onto individual plates and incubating them at the desired temperature overnight. The following day, the parents were removed and the number of eggs and larvae were counted. On the third day, this count was repeated to determine percent hatching rate. To determine heterozygous hatching rate, mutant adult hermaphrodites were mated to N2 males and the F1 progeny were subjected to the above procedure. Complementation tests were scored in a similar manner, with homozygous TS mutant males being crossed into either a heterozygous, balanced strain carrying a lethal deletion allele or a homozygous viable deletion allele. Brood sizes at the permissive and restrictive temperatures were determined by singling out L4 hermaphrodites and culturing them overnight on individual plates. The following day, each parent was transferred to a new plate, and the number of eggs and larvae were counted. This continued until the parents ceased to produce fertilized eggs.
Whole-genome sequencing and data analysis
In preparation for sequencing, mutant hermaphrodites were outcrossed to CB4856 males, and ∼200 F2 worms were singled out from the resultant progeny. These were then assayed for homozygous embryonic lethality by picking 10–12 L4 hermaphrodites of the F3 generation from each F2 plate and incubating them at 26° for 3 d. Gravid adults from each homozygous outcross strain were then pooled together and subjected to hypochlorite treatment to destroy any contaminating microbial DNA, using approximately equally sized populations from each strain. The resulting L1s were then used for genomic DNA extraction, which was performed using a Qiagen DNeasy kit. Libraries were prepared for sequencing using Illumina’s Nextera DNA Library Preparation kit, following the included protocol with no modifications. Sequencing was performed on an Illumina HiSequation 2000 at the University of Oregon’s Genomics Core Facility.
All data analysis tools were run using the Galaxy platform (Giardine et al. 2005; Goecks et al. 2010). The workflow used is the same as that described by Minevich et al. (2013), with some modifications. Read mapping was performed using Bowtie2 (Langmead and Salzberg 2012), alignment quality control was performed using a combination of Picard (http://broadinstitute.github.io/picard/) and GATK (DePristo et al. 2011), and variant detection and analysis were performed using a combination of GATK, CloudMap, snpEff (Cingolani et al. 2012), and Bedtools (Quinlan and Hall 2010). SNP mapping plots were generated using the CloudMap tool.
CRISPR/Cas-9 generation of GFP::DRP-1 transgenic strain
For gfp::drp-1 donor construct, a 2250-bp fragment (IV: 5537396–5539646) amplified from fosmid WRM069aH08 was cloned into the pJET1.2 vector (ThermoScientic cat. no. K1231). The GFP coding region amplified from pSO26 (O’Rourke et al. 2007) was inserted at the N terminus of drp-1 by Gibson assembly (Gibson et al. 2009). A point mutation was then introduced to disrupt the PAM site (CGG to CCG) by inverse PCR. eft-3p::Cas-9 plasmid with drp-1 N terminus targeting sgRNA (GTAGTCGTCGGATCACAGT) was made from pDD162 plasmid (Dickinson et al. 2013). The gfp::drp-1 donor construct, Cas-9-sgRNA plasmid, and selection marker pRF4 were injected into wild-type N2 young adults. The animals with correct knock-in were identified by screening for GFP expression (Kim et al. 2014).
Microscopy
Whole worms were prepared for live imaging by anesthetic treatment with 0.5 mg/mL tetramisole-HCl in M9, and were then mounted on a 4% agarose pad and secured with a cover slip. Imaging was performed on a Leica DMI 4000B inverted compound microscope equipped with a Leica 63X/1.40–0.60 HCX Plan Apo oil objective lens. Image capture was accomplished using a Hamamatsu EM-CCD digital camera. Volocity (PerkinElmer Inc.) software was used to control the system. The entire germline was imaged by acquiring multiple stacks along the length of the gonad; each stack consists of 49 z-planes with a spacing of 0.5 μm. Visualization and analysis of the stacks were performed using ImageJ (http://imagej.nih.gov/ij/). Composite images were prepared using images from each stack corresponding to the same focal plane, and they were then stitched together using Adobe Illustrator.
Data availability
All SNP mapping and whole genome sequence data is available at wormbase.org.
Results
Identifying conditional mutants that are both eggshell-defective and adult sterile
To identify mutants that might have defects in adult germline membrane dynamics, we scored a collection of ∼800 temperature-sensitive, embryonic lethal (TS-EL) mutants, isolated using a previously described procedure (Kemphues et al. 1988; Encalada et al. 2000; O’Rourke et al. 2011a), for eggshell integrity and fertility defects. To identify eggshell-defective mutants (Eos, for embryonic osmotic sensitivity), we up-shifted homozygous mutant worms that had been raised at the permissive temperature of 15° to the restrictive temperature of 26° at the L4 stage. After these L4 mutant larvae matured to adulthood overnight, early embryos were dissected into sterile water for inspection using DIC microscopy. Embryos with eggshell defects either swell or lyse in a hypotonic environment, and we used this phenotype to qualitatively identify 102 eggshell-defective mutants (J. Lowry, unpublished data). We then further examined this group of mutants for defects in fertility by up-shifting populations of homozygous mutant L1 larvae to 26° and scoring them for fertility after they matured to adulthood at the restrictive temperature. While most of the adult mutants were fertile but produced inviable embryos, for 49 of the mutants we found that ∼50% or more of the up-shifted larvae either arrested during larval development or matured into sterile adults (Ste, for adult sterile; J. Lowry, unpublished data). These 49 Eos/Ste mutants were then selected for further characterization based on the relatively high penetrance of their adult sterility.
We next genetically characterized the mutants to determine if they caused recessive and, hence, reduction of function phenotypes. First, we used wild-type (N2) males to backcross each homozygous Eos/Ste mutant strain. The backcross was done to eliminate the recessive mutation lin-2(e1309) from the parental strain used for mutagenesis; e1309 renders adult hermaphrodites egg-laying defective and thus suitable for the screening procedure used to identify embryonic-lethal mutants (Kemphues et al. 1988; Encalada et al. 2000; O’Rourke et al. 2011a). Then, we again crossed each backcrossed and homozygous Eos/Ste mutant strain with wild-type males and scored embryonic viability in the broods produced by self-fertilization after up-shifting heterozygous F1 (ts/+) L4 stage worms to the restrictive temperature. We observed 5% or less embryonic lethality in 20 mutants that we selected for further analysis, reasoning that most of these mutants likely carry recessive loss-of-function mutations, with embryonic lethality at levels similar to that observed when wild-type worms were up-shifted to 26° as L4 larvae. We also scored embryonic lethality for the F2 self-progeny produced by heterozygous F1 ts/+ worms raised at the permissive temperature of 15°, after shifting the F2 self-progeny to the restrictive temperature as L4s. In all 20 cases, we found that ∼25% of the F2 progeny produced dead embryos (unpublished data), consistent with the mutants carrying recessive, loss-of-function mutations in a single locus. We then quantified embryonic lethality and adult sterility in homozygous backcrossed mutants at both the permissive (15°) and restrictive (26°) temperatures, and quantified the eggshell-defective phenotypes in embryos produced at the restrictive temperature using a dye permeability assay (Table 1). We then used whole genome sequencing in an effort to identify the causal mutations, as described below.
Table 1. Embryonic lethality, adult sterility, and eggshell permeability for temperature-sensitive mutants.
| Allele | Embryonic Lethality (15°C) | Embryonic Lethality (26°C) | Heterozygous Embryonic Lethality (26°C) | Permeability Barrier Defects (26°C) | Adult Sterility (15°C) | Adult Sterility (26°C) |
|---|---|---|---|---|---|---|
| or819ts | 15.8% (336) | 92.7% (55) | 0.52% (191) | 16.3% (92) | 0% (128) | Larval arrest |
| atx-2 (or821ts) | 37.4% (388) | 99.6% (715) | 8.3% (144) | 99.4% (160) | 2.2% (137) | 86% (122) |
| sqv-8 (or888ts) | 1.4% (422) | 99.7% (311) | 2.0% (298) | 92.5% (280) | 2.8% (141) | 100% (127) |
| crn-3 (or959ts) | 4.8% (124) | 74.6% (405) | 2.6% (288) | 68.3% (63) | 1.6% (129) | 100% (107) |
| or983ts | 1.3% (224) | 98.9% (352) | 1.1% (266) | 14.3% (133) | 0% (112) | Larval arrest |
| rpl-7 (or990ts) | 0.5% (551) | 93.8% (130) | 2.9% (450) | 34.2% (73) | 1.4% (140) | 100% (113) |
| ndg-4 (or1088ts) | 16.5% (375) | 84.9% (179) | 4.5% (220) | 4.6% (307) | 1.2% (166) | 100% (102) |
| or1112ts | 0.5% (394) | 100% (837) | 2.7% (296) | 59.0% (122) | 0% (117) | Larval arrest |
| or1184ts | 11.6% (431) | 100% (290) | 33.3% (303) | 0% (169) | 0% (128) | 100% (108) |
| or1188ts | 15.8% (275) | 88.1% (109) | 4.9% (346) | 1.6% (187) | 2.3% (128) | 61% (101) |
| or1233ts | 22.1% (412) | 100% (319) | 5.2% (173) | 89.7% (261) | 14.3% (112) | 50% (121) |
| vps-15 (or1235ts) | 1.5% (275) | 100% (117) | 3.6% (446) | 3.4% (237) | 0.9% (107) | Larval arrest |
| or1247ts | 0.5% (412) | 97.5% (81) | 1.8% (325) | 43.4% (122) | 1.8% (112) | 100% (95) |
| or1353ts | 17.3% (249) | 83.1% (355) | 5.6% (195) | 63.6% (110) | 0% (133) | 55% (98) |
| drp-1 (or1393ts) | 56.6% (571) | 99.7% (294) | 1.1% (363) | 55.2% (145) | 0.6% (160) | 45% (135) |
| abtm-1 (or1400ts) | 3.3% (271) | 100% (181) | 3.9% (442) | 55.2% (375) | 1.2% (167) | 100% (121) |
| or1409ts | 2.2% (543) | 100% (225) | 3.5% (310) | 46.8% (252) | 0% (169) | 100% (113) |
| or1439ts | 2.9% (452) | 91.6% (143) | 1.5% (456) | 23.8% (168) | 0.9% (116) | 83% (105) |
| or1565ts | 45.4% (280) | 90.2% (102) | 10% (289) | 78.7% (389) | 65.2% (125) | 100% (111) |
| ippk-1 (or1572ts) | 9.5% (494) | 100% (465) | 1.1% (366) | 3.8% (366) | 17.8% (185) | Larval arrest |
| or1613ts | 0.03% (324) | 97.2% (253) | 1.1% (190) | 42.8% (180) | 0% (117) | 100% (119) |
| or1621ts | 3.1% (478) | 100% (496) | 4.1% (168) | 97.4% (651) | 0% (116) | 58% (103) |
Embryonic lethality and adult sterility for homozygous mutants were scored at the permissive (15°C) and restrictive (26°C) temperatures. Embryonic lethality for heterozygous mutants was scored at the restrictive temperature. Eggshell permeability barrier defects for homozygous mutants were scored at the restrictive temperature using a dye permeability assay (Carvalho et al. 2011).
Identifying the causal mutations in Eos/Ste mutants using whole genome sequencing
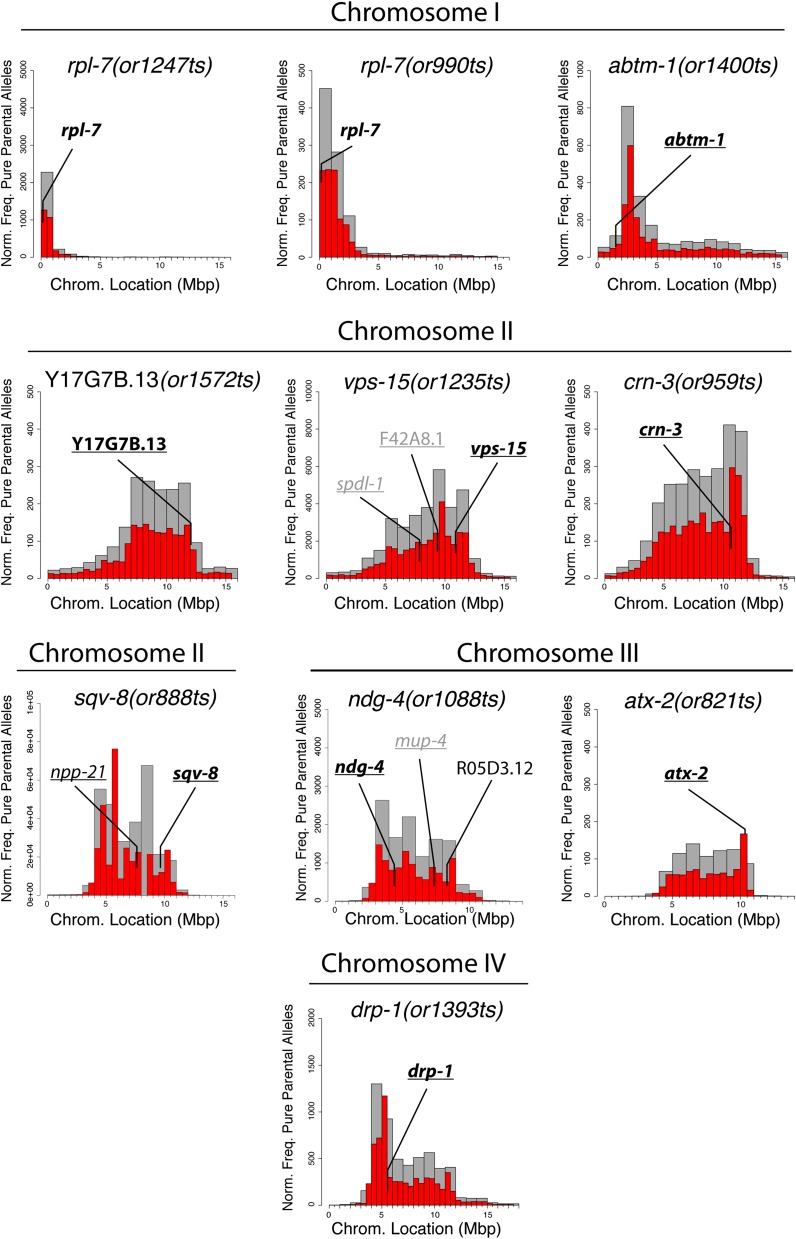
Because mutations in many essential genes required more generally for gene expression or metabolism might indirectly lead to both eggshell defects and larval arrest or sterility, we sought to identify the causal mutations in our set of 20 TS Eos/Ste mutants using a whole genome sequencing strategy before further examining the mutant phenotypes. Chemically mutagenized C. elegans strains have been shown previously to have hundreds of genomic mutations relative to the reference wild-type genome sequence, even after multiple rounds of backcrossing (Sarin et al. 2010). We therefore used an approach that provides both genome-wide SNP mapping and genome sequence data following a single outcross to a polymorphic Hawaiian strain called CB4856 (Doitsidou et al. 2010). In brief, we crossed CB4856 males to homozygous Eos/Ste hermaphrodites at the permissive temperature and then isolated 20–50 homozygous F2 animals scored for embryonic lethality after L4 up-shifts of F3 progeny to the restrictive temperature. We then pooled together for each mutant several individuals from the homozygous (ts/ts) F3 and F4 generations for genomic DNA isolation, and sequenced each pool using paired-end Illumina DNA sequencing (see Materials and Methods). This generated a pool of recombinant chromosomes with a shared lack of introduced CB4856 polymorphisms in the region linked to the causal mutation. This trait can be detected computationally using the CloudMap tool (Minevich et al. 2012), providing genetic mapping data. More specifically, the regions most tightly linked to the causal mutations often have the highest frequency of pure parental alleles (Figure 2). Previous work that applied this approach to viable mutants found that most causal mutations mapped to within 1 Mb of the highest peak in plots of pure parental allele frequency (Doitsidou et al. 2010, Minevich et al. 2012). We therefore used this approach to narrow the genomic regions of interest for these 20 TS Eos/Ste mutations.
Figure 2.
SNP mapping data for temperature-sensitive mutations with identified causal mutations. For each mutant, the frequency of homozygous parental alleles was plotted against chromosomal position in bins of either one megabase (gray bars) or half-megabase (red bars). Gene names on each plot are essential genes in which mis-sense mutations were detected. Underlined gene names are those for which deletion alleles exist for performing complementation tests. For complementation tests in which the mutations failed to complement each other, the gene names are in bold; for tests in which the mutations complemented, the gene names are in gray.
After using pure parental frequency to estimate the location of Eos/Ste mutations in the genome, we then filtered the sequence data for candidate mutations within the region of interest (see Materials and Methods). Our previous work with other TS-EL mutants has shown that most conditional alleles are mis-sense mutations, and less frequently splice-site mutations, in genes that are known to be essential based on genome-wide RNAi screens (O’Rourke et al. 2011a). For each Eos/Ste allele, we generated from the whole genome sequence data short lists of one to five candidate genes found within several megabase intervals derived from the SNP mapping data, focusing on mis-sense and splice site mutations in essential genes. We then used complementation tests, when mutant alleles were available, to identify the causal mutations. For eight of the mutants, the Eos/Ste alleles failed to complement known alleles of candidate genes and complemented available deletion alleles for other candidate genes (Table S1 and Table S2). For two mutants, or990ts and or1247ts, a different mis-sense mutation was found in the same gene in both mutants; this was the only candidate gene shared by both mutants, and the two mutants failed to complement each other (Table S1). Based on these mapping and complementation test data, we have identified the causal mutations in 10 Eos/Ste strains, as described in more detail below. For an additional eight mutants, we have narrowed the candidate genes to one or two loci, but deletion alleles of those genes do not exist, precluding complementation tests that could identify the causal mutation. Finally, in two cases, the TS alleles complemented deletion alleles for all candidate genes (J. Lowry, unpublished data), indicating that in some cases additional analysis will be needed to identify causal mutations (see Discussion).
Mutational inactivation of genes with general roles in gene expression or metabolism can cause the Eos/Ste phenotype
For all but one of the 10 Eos/Ste mutants in which we have identified causal mutations, the affected genes are conserved across vertebrate species (Figure 3), with published data providing some insight regarding their possible roles in C. elegans (see Discussion). While our goal has been to identify genes that are important for membrane dynamics in the adult gonad during oogenesis, we anticipated that mutations in genes required more generally for metabolism and gene expression, without being directly involved in the regulation of membrane dynamics, might also cause both eggshell defects and adult sterility after L1 and L4 up-shifts, respectively. For example, RNAi knockdown screens have found that genes encoding ribosomal proteins are required both for eggshell integrity and for adult fertility (Green et al. 2011; Sönnischen et al. 2005; Kamath et al. 2003) .
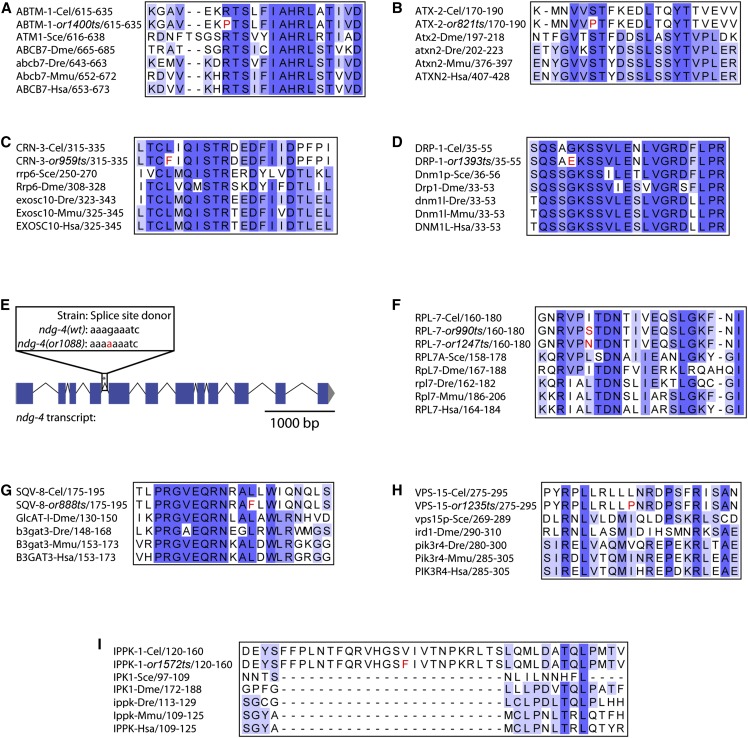
Figure 3.
Conservation of amino acids for identified causal mutations. (A–D, F–I) For each conserved gene, both the wild-type and mutant C. elegans protein sequences were aligned against homologs from other species (Cel = C. elegans; Sce = S. cerevisiae; Dme = D. melanogaster; Dre = D. rerio; Mmu = M. musculus; Hsa = H. sapiens). The level of sequence conservation has been highlighted using Jalview’s Color by Percentage Identity function (Waterhouse et al. 2009). (E) Schematic of the ndg-4 transcript, with the mutated splice donor site highlighted with an asterisk (*).
Consistent with our expectation that our screening procedure would identify more generally required genes, we have identified two alleles of rpl-7, or990ts and or1247ts, which encode a ribosomal protein subunit required for rRNA processing (Jakovljevic et al. 2012). We also have identified one allele of abtm-1, or1400ts, which encodes an ABC family iron transporter active in the mitochondrial inner membrane (Zutz et al. 2009); one allele of crn-3, or959ts, which encodes an RNA endonuclease subunit of the exosome complex (Parrish and Xue, 2003; Chlebowski et al. 2013); and one allele of atx-2, or821ts, which encodes an RNA-binding protein that regulates translation in the C. elegans germline and is known to be required for eggshell formation (Maine et al. 2004; Ciosk et al. 2004; Sönnischen et al. 2005). We suspect that these genes only indirectly influence membrane dynamics during gonad development and adult oogenesis.
Some Eos/Ste genes are more directly implicated in membrane biology
Five genes we have identified may more directly impact membrane architecture and dynamics. We identified or888ts as an allele of sqv-8, which encodes an enzyme required for glycosaminoglycan synthesis (Herman et al. 1999). In C. elegans, loss of chondroitin synthesis, which requires sqv-8 function, leads to cytokinesis defects in the early embryo, possibly due to a loss of extracellular matrix integrity during furrow ingression (Mizuguchi et al., 2003; Izumikawa et al. 2004; Olson et al. 2012). Thus, it is possible that sqv-8 similarly influences membrane architecture or dynamics during gonad development. Two genes we have identified appear most likely to more directly influence membrane dynamics. One, or1235ts, is an allele of vps-15, which encodes a regulator of the lipid kinase VPS-34 that, based on studies in yeast and vertebrates, is required for both endocytosis and autophagy (Backer 2008). Another, or1393ts, is an allele of drp-1, which encodes a dynamin-related protein required for mitochondrial membrane fission (Otera et al. 2013). Finally, two of the identified genes are involved in lipid metabolism. One, or1572ts, is an allele of Y17G7B.13, which is homologous to human IPPK, a kinase required for the production of inositol 6-polyphosphate (Tsui and York 2010). Based on this homology, we have named the C. elegans gene ippk-1. The second, or1088ts, is an allele of ndg-4, which encodes a transmembrane protein that is involved in the transport of dietary lipids into the germline. While homologs of ndg-4 have not been described in vertebrates, an ortholog in Drosophila is important for longevity (Brejning et al. 2014).
Germline membrane architecture defects in adult Eos/Ste mutants after temperature up-shifts during larval development
To investigate whether the genes we identified influence gonad membrane morphology, we next examined adult gonad membrane architecture in live mutants expressing a PH domain fused to mCherry to mark the plasma membrane and Histone2B fused to GFP to mark chromosomes after shifting TS Eos/Ste mutants from the permissive to the restrictive temperature during larval development. We focused most of our analysis on four widely conserved genes that appeared likely to most directly influence membrane dynamics: sqv-8, vps-15, drp-1, and ippk-1 (see Figure S2 for a more limited analysis of abtm-1, atx-2, crn-3, and rpl-7 mutants). Homozygous mutant worms were grown at the permissive temperature (15°) and then shifted to the restrictive temperature (26°) at either the L1 or the L4 stage until they matured into young adults that were then mounted for imaging using spinning disc confocal microscopy (see Materials and Methods).
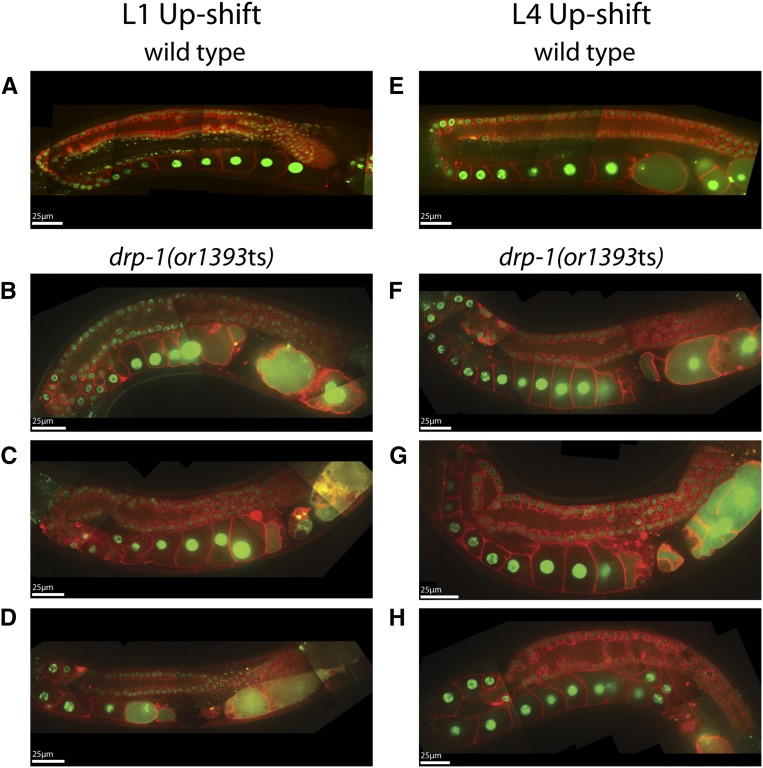
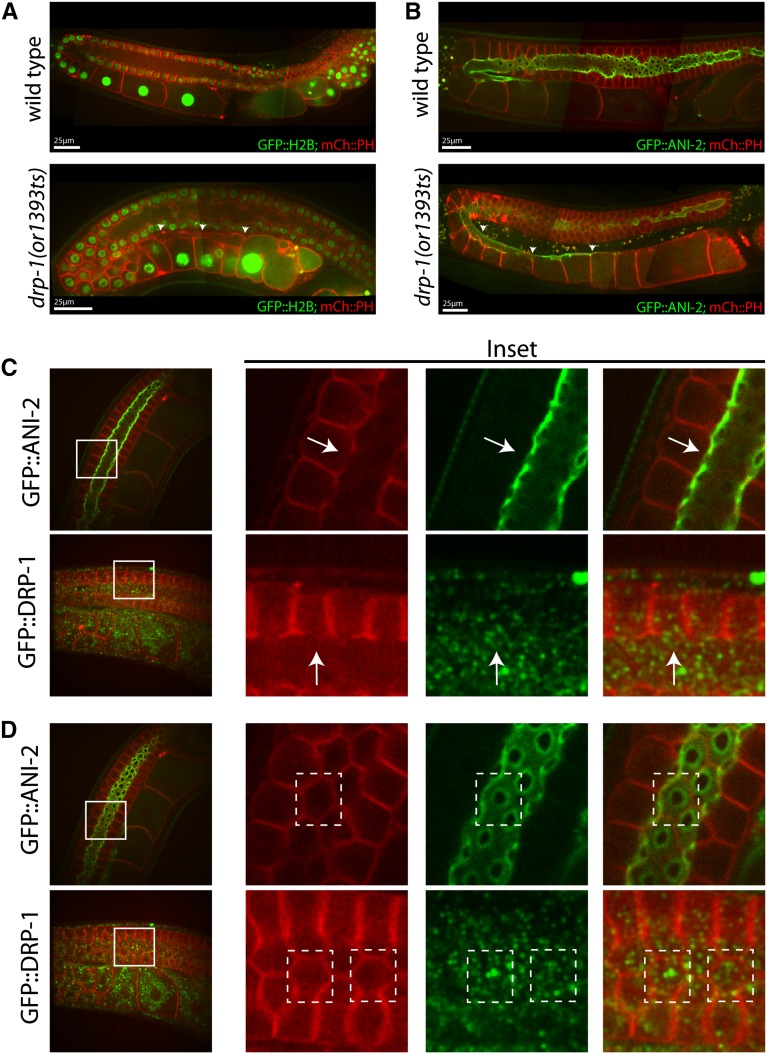
The dynamin-related protein mutant, drp-1(or1393ts), was only weakly penetrant for adult sterility after L1 up-shifts (Table 1), and the adult germline in these mutants was relatively normal after both L1 and L4 up-shifts (Figure 4). We observed the most severe defects in the proximal gonad, with multiple small, membrane-bound compartments often visible near the most mature oocytes (Figure 4, B–D, F–H). These membrane structures may result from defective ovulation, as we observed frequent failures in ovulation in live animals using DIC optics (J. Lowry, unpublished data). In some mutant gonads, the rachis appeared to extend abnormally far into the proximal arm, with the most mature oocyte still connected (Figures 4, F and G, Figure 5, A and B; Figure S3A). We considered the possibility that DRP-1 might be involved in narrowing the rachis during oocyte cellularization, or perhaps in maintenance of the openings of the membrane cubicles to the rachis in the distal gonad. However, because mitochondria have been shown to be abnormally long in drp-1(tm1108) mutants (Labrousse et al. 1999; Breckenridge et al. 2009), and because we have found that a CRISPR/Cas-9 generated GFP fusion to the wild-type drp-1 locus was expressed only in mitochondria and was not detectable elsewhere in the germline (Figure 5, C and D), we suspect the gonad morphology and eggshell defects may be indirect consequences of abnormal mitochondrial function or morphology.
Figure 4.
Adult germline defects in drp-1(or1393ts) mutants following temperature up-shifts to the restrictive temperature (26°C) at the L1 and L4 larval stages. Spinning disk fluorescent confocal microscopy was used to make composite images of the adult hermaphrodite gonads in both wild-type (A and E) and drp-1(or1393ts) (B–D and F–H) worms. In this and subsequent figures, plasma membranes were marked with an mCherry::PH domain fusion, chromosomes were marked with a histone2B::GFP fusion (see Materials and Methods), and scale bars are shown in white.
Figure 5.
Rachis extension in drp-1(or1393ts) mutants and GFP::DRP-1 localization. (A) The rachis appears to extend to the most mature oocyte (arrowheads) in an adult drp-1(or1393ts) mutant gonad, as detected using mCherry::PH to mark the membranes. (B) ANI-2::GFP localizes to the surface of the rachis (Maddox et al. 2005), which terminates just after the turn in wild-type hermaphrodites. The rachis again extends proximally in an adult drp-1(or1393ts) mutant gonad (arrowheads). (C and D) GFP::DRP-1 (see Materials and Methods) does not localize to openings to the rachis in the distal gonad or to the rachis more proximally, arguing against a direct role in oocyte cellularization. (C) An orthogonal view of the meiotic germ cell openings to the rachis of wild-type strains carrying either ANI-2::GFP or GFP::DRP-1. GFP::DRP-1 does not localize to the germ cell opening (arrows). (D) A view in the plane of the meiotic germ cell openings from the rachis surface. Solid white lines in (C) and (D) mark insets, and dashed white lines indicate individual germ cells with openings to rachis.
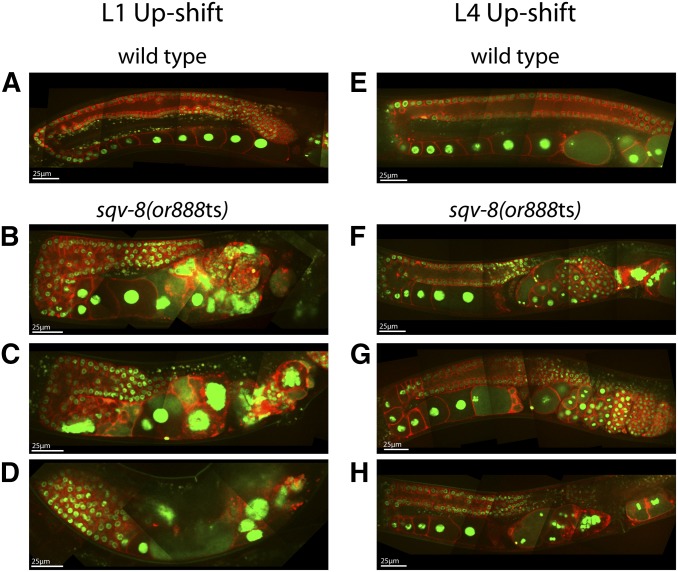
We observed severe adult gonad morphology defects following L1 up-shifts of sqv-8(or888ts) worms. The distal and proximal regions of each arm appeared to be wider than in wild-type, and the arms were shorter in length from the distal end to the spermathecum (Figure 6, B and C). In some cases, the arms consisted of a single wide region with no turn (Figure 6D), and in some animals one gonad arm was small or absent (Figure S3B). The rachis structure in the distal region was severely disrupted, with tightly packed and disorganized germ cell precursors throughout (Figure 6, B–D). The uteri were filled with disorganized membrane and DNA (Figure 6, B–D), perhaps due to the accumulation of burst eggshell-defective embryos within the worms, as sqv-8 is required for proper vulval development and egg-laying (Herman et al. 1999; Herman and Horvitz 1999). The gonad morphology was much more normal after L4 up-shifts, but the adults were still egg-laying defective (Figure 6, F–H).
Figure 6.
Adult germline defects in sqv-8(or888ts) mutants following temperature up-shifts to the restrictive temperature (26°C) at the L1 and L4 larval stages. Composite images were prepared as previously described (Figure 3) for the adult hermaphrodite gonads in wild-type (A and E) and sqv-8(or888ts) (B–D and F–H) worms.
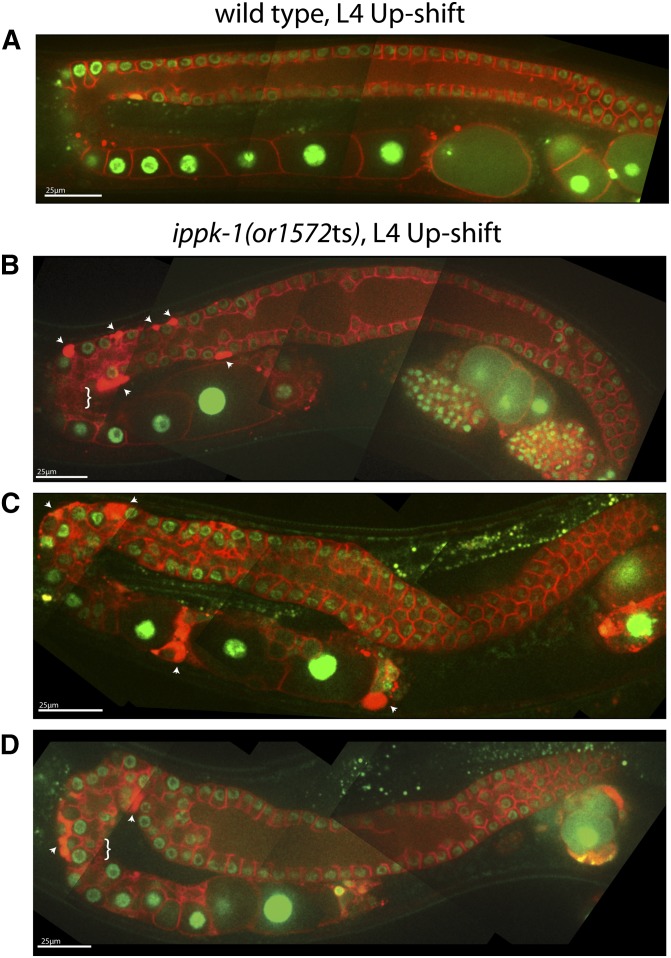
Because ippk-1(or1572ts) mutants arrest during larval development after L1 up-shifts, we examined adult gonad morphology only after L4 up-shifts and observed severe abnormalities. The width of the distal region varied along its length, as did the rachis (Figure 7, B–D). We also observed numerous small membrane vesicles near the turn and areas with densely concentrated mCherry::PH signal (arrowheads in Figure 7, B–D). Finally, the distal regions of the gonad arms were in some cases displaced ventrally, rather than lying against the overlying dorsal surface (Figure 7, C and D). This displacement could result from abnormal migration of the distal tip cell during larval development (see Introduction), or could be due to a later displacement.
Figure 7.
Adult germline defects in ippk-1(or1572ts) mutants following temperature up-shifts to the restrictive temperature (26°C) at the L4 larval stage. Composite images were prepared as previously described (Figure 3) for the adult hermaphrodite gonad in wild-type (A) and ippk-1(or1572ts) (B–D) worms. Arrowheads point to membrane accumulations in the germline; braces highlight regions of rachis vesiculation.
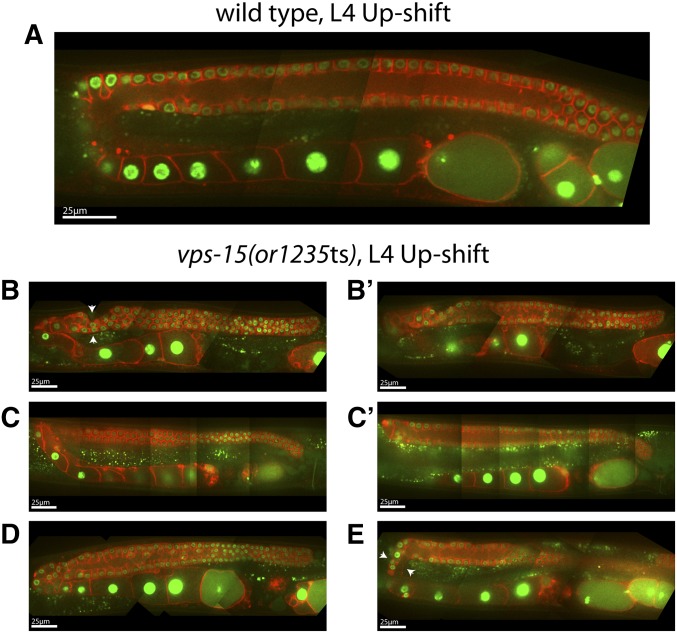
We also observed larval arrest after L1 up-shifts of vps-15(or1235ts) mutants and thus examined adult gonads for morphology only after L4 up-shifts. The most prominent defect we observed was in the proximal part of the gonad arms, with fewer and often highly elongated oocytes present (Figure 8, B, C, E). The proximal portion of the gonad arms also was abnormally narrow near the turn, with oocytes becoming much wider near the spermathecum (Figure 8, B, D, E). In the distal arm, the rachis appeared abnormally narrow, with a membrane/rachis interface that was less uniform than in wild-type (Figure 8, B′, C′, D, E). The turns of the gonad arms were also abnormally narrow, and we observed abnormal constrictions near the turns (arrowheads in Figure 8, B and E).
Figure 8.
Adult germline defects in vps-15(or1235ts) mutants following temperature up-shifts to the restrictive temperature of 26°C at the L4 larval stage. Composite images were prepared as previously described (Figure 3) for the adult hermaphrodite gonad in wild-type (A) and vps-15(or1235ts) (B–E) worms. (B and B′, C and C′) Images from the same animal at two different focal planes. Arrows point to regions of constriction at or near the turn of the gonad.
Adult up-shifts reveal genetic requirements for the maintenance of adult germline membrane architecture
To determine if the genes we have identified are important for maintenance of the gonad membrane architecture during oogenesis in adults, we next examined mutants up-shifted as young adults from the permissive to the restrictive temperature for 18 hr, again using a PH domain fused to mCherry to mark the plasma membrane and GFP fused to Histone2B to mark chromosomes (Figure 9). As a control, we first examined wild-type adults 18 hr after up-shift and found no significant perturbations in gonad membrane morphology (Figure 9, A and B). Similarly, the membrane morphology in adult mutants grown for 18 hr at 15° largely resembled wild-type (Figures 9, C and D; Figure S4C and D). We did not detect significant defects in drp-1(or1393ts) mutants after adults were fed for 18 hr at the restrictive temperature (Figure S4E), which was not surprising given the minor defects we observed following both L1 and L4 up-shifts. We also failed to detect significant defects in sqv-8(or888ts) mutants (Figure S4F), even though we had detected severe defects in gonad morphogenesis after L1 and L4 up-shifts (Figure 6). Thus, although sqv-8 function is important for gonad development, we did not find any sqv-8 requirements for maintenance of adult gonad membrane morphology.
Figure 9.
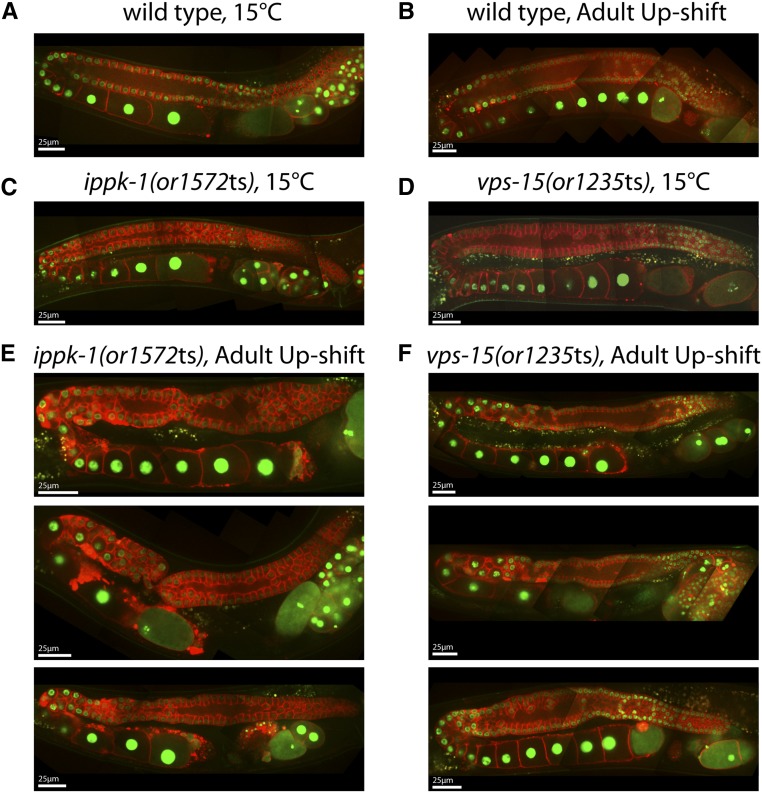
Germline defects in ippk-1(or1572ts) and vps-15(or1235ts) mutants after young adult up-shifts to the restrictive temperature (26°C). Composite images of the germline were prepared as previously described for wild-type (A, B), ippk-1(or1572ts) (C, E), and vps-15(or1235ts) (D, F) worms. These animals were either grown entirely at 15°C or shifted to 26°C as young adults for 18 hr.
We did detect membrane architecture defects in both ippk-1(or1572ts) and vps-15(or1235ts) mutants after adult up-shifts (Figure 9, E and F), and in both cases the defects resembled those observed after L4 up-shifts. In ippk-1(or1572ts) mutants, the distal arm varied in width along its length, the mCherry::GFP signal accumulated to high levels in foci near the turns of the gonad arms, and in some animals severe constrictions were formed near the turn (Figure 9E). In vps-15(or1235ts) mutants, proximal oocyte precursors were disorganized near the turns, and in some animals the oocytes near the turns were narrow and elongated compared to wild-type (Figure 9F). We conclude that vps-15 and ippk-1 are required to maintain gonad membrane architecture during adulthood and therefore may be important for regulating adult gonad membrane dynamics during oogenesis.
Discussion
Although C. elegans gonad development has been studied comprehensively, the extensive membrane dynamics that must accompany adult oogenesis have been largely ignored. As the syncytial nature of the germline is widely conserved (Greenbaum et al. 2011; Haglund et al. 2011), studies of this phenomenon in C. elegans are likely to be of general significance. We have therefore attempted to identify genes that mediate membrane dynamics during adult oogenesis by identifying temperature-sensitive, embryonic-lethal (TS-EL) C. elegans mutants that are both eggshell-defective (osmotically sensitive) after late larval up-shifts and adult sterile after early larval up-shifts (Eos/Ste mutants), reasoning that both processes involve membrane trafficking. Because general metabolic and gene expression functions might also affect both of these processes, we applied a high-throughput whole genome sequencing strategy to identify the causal mutations in a collection of TS Eos/Ste mutants that all share these two defects. Although we identified alleles of several genes that likely indirectly affect membrane trafficking due to more general roles in gene expression and metabolism, we also identified genes that appear likely to more directly influence membrane dynamics, including two that are required to maintain proper gonad membrane architecture during oogenesis in adult worms. As discussed below, further analysis of these genes may improve our understanding of membrane dynamics during oogenesis. In addition, our identification of TS alleles of other genes that indirectly influence oogenesis provides useful genetic tools for the study of widely conserved gene functions.
High-throughput identification of temperature-sensitive loss of function alleles of essential C. elegans genes
Temperature-sensitive alleles of essential genes are powerful genetic tools, as many essential genes have multiple requirements throughout the life of an organism: with TS alleles, one can bypass early requirements at the permissive temperature and study later requirements after up-shifts to the restrictive temperature (O’Rourke et al. 2011a). Moreover, fast-acting TS alleles (that rapidly inactivate and reactivate upon temperature up-shifts and down-shifts) also can be used to define temperature-sensitive periods when their functions are required (Severson et al. 2000; Schmidt et al. 2005; Davies et al. 2014), and TS mutants grown at intermediate temperatures provide sensitized genetic backgrounds for modifier screens that can identify other genes involved in the same processes (Kemp et al. 2007; Dorfman et al. 2009). The C. elegans genome is estimated to contain ∼20,170 protein-coding genes (Shaye and Greenwald 2011), and at least 2500 have essential functions based on genome-wide screens that have used RNA interference to knockdown their functions (Sonnichsen et al. 2005; Kamath et al. 2003). However, TS alleles have been identified for only a small fraction of these essential genes (we estimate no more than ∼200); therefore, we have explored high-throughput methods for identifying the causal mutations in a large collection of TS-EL mutants (O’Rourke et al. 2011b; data reported here).
To identify the causal mutations in the recessive Eos/Ste mutants we isolated, we used a previously developed approach that combines genome-wide single nucleotide polymorphism (SNP) mapping with whole genome sequencing (Jaramillo-Lambert et al. 2015; Doitsidou et al. 2010; Minevich et al. 2013). Because the genome of a single mutant strain isolated after chemical mutagenesis can contain hundreds of mis-sense mutations (Sarin et al. 2010), SNP mapping is needed to narrow the interval in which candidate mutations can be found and then further analyzed to identify the causal mutations. Initially developed for homozygous viable mutants, we have modified this procedure for use with TS-EL mutants (see Materials and Methods). Using this approach, we were able to identify one to a few candidate mutations (mis-sense mutations, splice site mutations, and mutations in 3′ and 5′ UTRs) within megabase intervals for each mutant (see Materials and Methods). To determine if candidate mutations were responsible for embryonic lethality, we used complementation tests with previously isolated mutant alleles, often by taking advantage of deletion alleles that have been isolated by genome-wide knockout consortiums. Of the 20 Eos/Ste mutants we processed for whole genome sequencing, we were able to identify the causal mutations in 10 mutants, based on complementation test results. For eight of the remaining Eos/Ste mutants, deletion alleles were not available for all candidate genes; nevertheless, we were able to narrow down the candidates to one or two loci. For these mutants, we anticipate that CRISPR/Cas-9 technology will make it possible to isolate deletion alleles of the candidate genes and that the causal mutations can then be determined with additional complementation tests. However, for two of the Eos/Ste mutants, deletion alleles were available for all candidate genes, but all of them complemented the TS mutations in genetic crosses. We suspect that these strains harbor mutations that were not captured in our filtering process. For example, they may have mutations in currently uncharacterized genes for which loss of function data are not available, or they may have small deletions that would have been missed with our filtering strategy. Nevertheless, the approach we have used should be able to identify the causal mutations in most TS-EL mutants processed. We plan to systematically apply these methods for high-throughput analysis of large numbers of TS-EL C. elegans mutants in an effort to provide a resource of temperature-sensitive mutations for the analysis of essential C. elegans gene requirements.
Some progress has been made in computationally predicting amino acid changes that make a protein temperature-sensitive in function (Chakshusmathi et al. 2004; Poultney et al. 2011; Lockwood et al. 2011), and the identification of causal TS-EL mutations from chemical mutagenesis screens may help further guide such efforts. As expected, most (9 of 10) of the mutations we identified are mis-sense mutations; the one exception is ndg-4(or1088ts), which alters a splice donor nucleotide (Figure 3 and Table 2). In a previous survey of 24 TS-EL mutations affecting 13 C. elegans genes, we similarly found that 20 were mis-sense mutations, two were intronic splice site mutations, one was an in-frame deletion, and one caused a premature stop codon (O’Rourke et al. 2011a). The mis-sense mutations we report here most often affected hydrophobic amino acids, changing them to charged or polar amino acids, and five of the nine involve highly conserved residues. Three others affect hydrophobic amino acids that are not fully conserved but are replaced by other hydrophobic residues in different species. Only one, ippk-1(or1572ts), affects an amino acid that is not conserved to any extent. The TS-EL mutations that affect conserved amino acids may be useful for engineering conditional mutations in other species (e.g., Schmidt et al. 2005).
Table 2. Causal mutations for 10 Osm/Ste mutants identified by whole genome sequencing and complementation tests.
| Allele | Transcript Name | Codon Mutated | Amino Acid Change | Nucleotide Change | Transcript Nucleotide (Spliced) | Average Depth of Coverage |
|---|---|---|---|---|---|---|
| atx-2 (or821ts) | D2045.1a | 175 | S > P | T > C | 523 | 75.3X |
| sqv-8 (or888ts) | ZK1307.5 | 187 | L > F | C > T | 559 | 24.2X |
| crn-3 (or959ts) | C14A4.4 | 318 | L > F | C > T | 954 | 25.6X |
| rpl-7 (or990ts) | F53G12.10 | 165 | I > S | T > G | 494 | 30.4X |
| ndg-4 (or1088ts) | F56F3.2 | —a | — | C > T | — | 25.6X |
| vps-15 (or1235ts) | ZK930.1 | 284 | L > P | T > C | 851 | 17.1X |
| rpl-7 (or1247ts) | F53G12.10 | 165 | I > N | T > A | 494 | 32.7X |
| drp-1 (or1393ts) | T12E12.4 | 39 | G > E | G > A | 116 | 28.3X |
| abtm-1 (or1400ts) | Y74C10AR.3 | 621 | R > P | C > G | 1862 | 35.0X |
| ippk-1 (or1572ts) | Y17G7B.13 | 137 | V > F | G > T | 409 | 32.3X |
Nature and location of each mutation and the average depth of sequencing coverage are shown for each mutant.
The mutation in ndg-4(or1088) affects a splice site donor at the end of exon IV.
Identification of genes required for oogenesis
We pursued phenotypic analysis of Eos/Ste TS-EL mutants in an effort to identify genes that are required for the membrane dynamics that mediate oogenesis as oocyte precursors travel from the distal to the proximal end of the gonad and eventually undergo cellularization and enlarge to produce mature oocytes (see Introduction). Of the nine genes in which we identified causal mutations, all of them are required for proper gonad development after larval up-shifts, and five of them (drp-1, ippk-1, ndg-4, sqv-8, and vps-15) have been shown in other studies to be relevant to membrane biology (see Results). We did not detect any significant requirements for drp-1 or sqv-8 in adults, and we did not investigate ndg-4 due to a lack of known vertebrate orthologs. However, vps-15 and ippk-1 did appear to be required for maintenance of the syncytial gonad membrane architecture in adult worms and thus may be required for membrane dynamics during oogenesis.
The vps-15 gene was identified in budding yeast due to its requirement for vacuolar (lysosomal) protein sorting and acts in multiple membrane trafficking processes (Backer 2008). It encodes a regulator of vps-34, a class 3 phosphatidylinositol-3 kinase (PI3K), and also is required for autophagy (Backer 2008). More recently, it has been shown that a core PI3K complex consisting of Vps15-Vps34-Atg6/Beclin-1 is required for endocytosis. During endosome maturation, membrane lipid composition is altered through the action of lipid kinases. Rab5 localizes to early endosomes and interacts directly with Vps15, which in turn recruits the rest of the PI3K complex. The PI3K complex can then generate phosphatidylinositol-3 phosphate (PI(3)P) in the outer leaflet of the endosome, aiding in the recruitment of PI(3)P-binding proteins that continue the early endosome maturation process (Huotari and Helenius 2011). Interactions between the PI3K complex and other effector proteins such as Atg14L, UVRAG, and Rubicon serve to regulate the formation of autophagosomes in a similar manner (Barth and Köhler 2014).
The ippk-1 gene has not been studied in C. elegans but is homologous to the human inositol 1,3,4,5,6-pentakisphosphate 2-kinase (IPPK). Conserved from yeast to humans, IPPK is an enzyme responsible for the production of inositol-6-phosphate (IP6) (Tsui and York 2010). IP6 is generated from the sequential addition of phosphate groups to IP3, which itself is formed from the cleavage of the lipid phosphatidylinositol 4,5-bisphosphate, a membrane lipid. IP6 is found at high intercellular concentrations, in some systems reaching as high as 50 μM (Shears et al. 2012). In yeast, IP6 is required for mRNA export from the nucleus, whereas in higher eukaryotes it functions in the nonhomologous end joining DNA repair pathway (Monserrate and York 2010). In both cases, IP6 acts as a cofactor for enzymes required for these processes. IP6 also acts as a building block for the more highly phosphorylated inositol pyrophosphates, which themselves have been implicated in a number of cellular roles, including the control of vesicular trafficking (Wilson et al. 2013), and thus IPPK-1 might be important for membrane dynamics in the C. elegans germline. The enzyme itself is essential in vertebrates, as homozygous Ippk knockout mice are embryonic lethal (Verbsky et al. 2005). Furthermore, the zebrafish homolog, ippk, has been shown to play a role in generating left–right asymmetry in zebrafish (Tsui and York 2010). Given its diverse roles in other organisms, the or1572ts allele may be a useful tool for analyzing its requirements in C. elegans.
A large-scale RNAi-based screen that classified sterile phenotypes after individually knocking down more than 600 gene functions in L4 larvae did not report defects for ippk-1, sqv-8, or vps-15, but we have compared their mutant phenotypes to the reported categories (Green et al. 2011). While sqv-8(or888ts) mutants resemble the phenotype category that includes RNAi knockdown of sqv-4, which functions in the same glycosaminoglycan synthesis pathway as sqv-8, the mutant phenotypes we have found for vps-15(or1235ts) and ippk-1(or1577ts) do not match any of the reported categories in Green et al. (2011). The lack of any match to the reported phenotype categories may reflect difference degrees in loss of gene function due to RNAi vs. our mutant alleles, or indicate that additional gonad membrane defect categories remain to be discovered.
Our results suggest that vps-15 and ippk-1 are important during adult oogenesis and may contribute to the dynamic remodeling of membranes that must accompany the movement of oocyte precursors in the syncytial hermaphrodite gonad, and their eventual cellularization and enlargement prior to maturation and fertilization. Although the development of the adult membrane architecture of the syncytial hermaphrodite gonad has been the subject of some investigation (Green et al. 2011; Amini et al. 2014), very little is known about the processes that control membrane dynamics in adult worms during oogenesis. Our identification of TS alleles of essential genes required for maintenance of adult gonad membrane architecture may make the investigation of membrane dynamics during adult oogenesis more tractable.
To understand how vps-15 and ippk-1 influence membrane dynamics in the adult gonad will require further investigation. Examining the expression of these two proteins over time in live animals using fluorescent protein fusions to the endogenous loci may provide insights. It also will be interesting to examine membrane dynamics over time in immobilized live adults using fluorescent protein fusions that mark different membrane compartments and spinning disk or light sheet confocal microscopy in wild-type and in TS-EL mutants after temperature up-shifts.
Finally, although some of the genes we have identified (rpl-7, crn-3, abtm-1, drp-1, and atx-2) likely affect oogenesis only indirectly, conditional alleles of these essential loci should prove useful for investigating their different requirements, and the 10 causal TS mutations we identified are the first conditional alleles reported for all nine genes. Furthermore, the high-throughput identification of TS-EL mutations made possible by next-generation DNA sequencing methods now make it possible to more efficiently isolate conditional alleles of many more of the ∼2500 C. elegans genes known to be essential. A systematic effort to isolate large numbers of TS-EL mutations in C. elegans should provide a valuable resource for the detailed investigation of many different biological processes in this model organism.
Supplementary Material
Acknowledgments
We thank the C. elegans Genetic Center (funded by the National Institutes of Health National Center for Research Resources) and the Shohei Mitani lab (National Bio-Resource Project of the MEXT, Japan) for strains, and Diana Libuda, David Greenstein, and members of the Bowerman lab for comments on the manuscript. This work was supported by National Institutes of Health Grants R01GM049869 and R21HG006540 (J. L., J. Y., K. S., C.-H. C., A.C., and B.B.) and the Human Frontiers Science Program (K.S.).
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.021451/-/DC1
Communicating editor: D. G. Moerman
Literature Cited
- Amini R., Goupil E., Labella S., Zetka M., Maddox A. S., et al. , 2014. C. elegans Anillin proteins regulate intercellular bridge stability and germline syncytial organization. J. Cell Biol. 206: 129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer J. M., 2008. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem. J. 410: 1–17. [DOI] [PubMed] [Google Scholar]
- Barth J. M. I., Köhler K., 2014. How to take autophagy and endocytosis up a notch. BioMed Res. Int. 2014: 960803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckenridge D. G., Kang B. H., Xue D., 2009. Bcl-2 Proteins EGL-1 and CED-9 do not regulate mitochondrial fission or fusion in Caenorhabditis elegans. Curr. Biol. 19: 768–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brejning J., Nørgaard S., Schøler L., Morthorst T. H., Jakobsen H., et al. , 2014. Loss of NDG-4 extends lifespan and stress resistance in Caenorhabditis elegans. Aging Cell 13: 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A., Olson S. K., Gutierrez E., Zhang K., Noble L. B., et al. , 2011. Acute drug treatment in the early C. elegans embryo. PLoS One 6: e24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakshusmathi G., Mondal K., Lakshmi G. S., Singh G., Roy A., et al. , 2004. Design of temperature-sensitive mutants solely from amino acid sequence. Proc. Natl. Acad. Sci. USA 101: 7925–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski A., Lubas M., Jensen T. H., Dziembowski A., 2013. RNA decay machines: The exosome. Biochim. Biophys. Acta. Gene Regul. Mech. 1829: 552–560. [DOI] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w 1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R., DePalma M., Priess J. R., 2004. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development 131: 4831–4841. [DOI] [PubMed] [Google Scholar]
- Davies T., Jordan S. N., Chand V., Sees J. A., Laband K., et al. , 2014. High-resolution temporal analysis reveals a functional timeline for the molecular regulation of cytokinesis. Dev. Cell 30: 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M., Poole R. J., Sarin S., Bigelow H., Hobert O., 2010. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS One 5: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman M., Gomes J. E., O’Rourke S., Bowerman B., 2009. Using RNA interference to identify specific modifiers of a temperature-sensitive, embryonic-lethal mutation in the Caenorhabditis elegans ubiquitin-like Nedd8 protein modification pathway E1-activating gene rfl-1. Genetics 182: 1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encalada S. E., Martin P. R., Phillips J. B., Lyczak R., Hamill D. R., et al. , 2000. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev. Biol. 228: 225–238. [DOI] [PubMed] [Google Scholar]
- Giardine B., Riemer C., Hardison R. C., Burhans R., Elnitski L., et al. , 2005. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 15: 1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A., 3rd, et al. , 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Goecks J., Nekrutenko A., Taylor J., 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan J. A., Nadarajan S., Kim S., Starich T. A., Greenstein D., 2009. Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans. Development 136: 2211–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. A., Kao H., Audhya A., Arur S., Mayers J. R., et al. , 2011. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell 145: 470–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum M. P., Iwamori T., Buchold G. M., Matzuk M. M., 2011. Germ cell intercellular bridges. Cold Spring Harb. Perspect. Biol. 3: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K., Nezis I. P., Stenmark H., 2011. Structure and functions of stable intercellular bridges formed by incomplete cytokinesis during development. Commun. Integr. Biol. 4: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna M., Wang L., Audhya A., 2013. Worming our way in and out of the Caenorhabditis elegans germline and developing embryo. Traffic 14: 471–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman T., Horvitz H. R., 1999. Three proteins involved in Caenorhabditis elegans vulval invagination are similar to components of a glycosylation pathway. Proc. Natl. Acad. Sci. USA 96: 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman T., Hartwieg E., Horvitz H. R., 1999. sqv mutants of Caenorhabditis elegans are defective in vulval epithelial invagination. Proc. Natl. Acad. Sci. USA 96: 968–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D., Oppenheim D., Klass M., 1976. Development of the reproductive system of Caenorhabditis elegans. Dev. Biol. 49: 200–219. [DOI] [PubMed] [Google Scholar]
- Hubbard E. J., Greenstein D., 2000. The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev. Dyn. 218: 2–22. [DOI] [PubMed] [Google Scholar]
- Huotari J., Helenius A., 2011. Endosome maturation. EMBO J. 30: 3481–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa T., Kitagawa H., Mizuguchi S., Nomura K. H., Nomura K., et al. , 2004. Nematode chondroitin polymerizing factor showing cell-/organ-specific expression is indispensable for chondroitin synthesis and embryonic cell division. J. Biol. Chem. 279: 53755–53761. [DOI] [PubMed] [Google Scholar]
- Jakovljevic J., Ohmayer U., Gamalinda M., Talkish J., Alexander L., et al. , 2012. Ribosomal proteins L7 and L8 function in concert with six A3 assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits. RNA 18: 1805–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Fuchsman A. S., Fabritius A. S., Smith H. E., Golden A., 2015. Rapid and efficient identification of Caenorhabditis elegans legacy mutations using Hawaiian SNP-based mapping and whole genome sequencing. G3 (Bethesda) 5: 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kemp C. A., Mi H. S., Addepalli M. K., Hunter G., O’Connell K., 2007. Suppressors of zyg-1 define regulators of centrosome duplication and nuclear association in Caenorhabditis elegans. Genetics 176: 95–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues K. J., Priess J. R., Morton D. G., Cheng N. S., 1988. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52: 311–320. [DOI] [PubMed] [Google Scholar]
- Kim H., Ishidate T., Ghanta K. S., Seth M., Conte D., Jr, et al. , 2014. A Co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., Hirsh D., 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70: 396–417. [DOI] [PubMed] [Google Scholar]
- Kimble J., Crittenden S. L., 2007. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 23: 405–433. [DOI] [PubMed] [Google Scholar]
- Labrousse A. M., Zappaterra M. D., Rube D. A., van der Bliek A. M., 1999. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4: 815–826. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T., 2004. Junctions and vesicular trafficking during Drosophila cellularization. J. Cell Sci. 117: 3427–3433. [DOI] [PubMed] [Google Scholar]
- Lockwood S., Krishnamoorthy B., Ye P., 2011. Neighborhood properties are important determinants of temperature sensitive mutations. PLoS One 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox A. S., Habermann B., Desai A., Oegema K., 2005. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development 132: 2837–2848. [DOI] [PubMed] [Google Scholar]
- Maine E. M., Hansen D., Springer D., Vought V. E., 2004. Caenorhabditis elegans atx-2 promotes germline proliferation and the oocyte fate. Genetics 168: 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T., 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205: 111–128. [DOI] [PubMed] [Google Scholar]
- Minevich G., Park D. S., Blankenberg D., Poole R. J., Hobert O., 2012. CloudMap: A cloud-based pipeline for analysis of mutant genome sequences. Genetics 192: 1249–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi S., Uyama T., Kitagawa H., Nomura K. H., Dejima K., et al. , 2003. Chondroitin proteoglycans are involved in cell division of Caenorhabditis elegans. Nature 423: 443–448. [DOI] [PubMed] [Google Scholar]
- Monserrate J. P., York J. D., 2010. Inositol phosphate synthesis and the nuclear processes they affect. Curr. Opin. Cell Biol. 22: 365–373. [DOI] [PubMed] [Google Scholar]
- Nadarajan S., Govindan J. A., McGovern M., Hubbard E. J. A., Greenstein D., 2009. MSP and GLP-1/Notch signaling coordinately regulate actomyosin-dependent cytoplasmic streaming and oocyte growth in C. elegans. Development 136: 2223–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S. M., Dorfman M. D., Carter J. C., Bowerman B., 2007. Dynein modifiers in C. elegans: light chains suppress conditional heavy chain mutants. PLoS Genet. 3: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S. M., Carter C., Carter L., Christensen S. N., Jones M. P., et al. , 2011a A survey of new temperature-sensitive, embryonic-lethal mutations in C. elegans: 24 alleles of thirteen genes. PLoS One 6: e16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S. M., Yochem J., Connolly A. A., Price M. H., Carter L., et al. , 2011b Rapid mapping and identification of mutations in Caenorhabditis elegans by restriction site-associated DNA mapping and genomic interval pull-down sequencing. Genetics 189: 767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson S. K., Greenan G., Desai A., Müller-Reichert T., Oegema K., 2012. Hierarchical assembly of the eggshell and permeability barrier in C. Elegans. J. Cell Biol. 198: 731–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H., Ishihara N., Mihara K., 2013. New insights into the function and regulation of mitochondrial fission. Biochim. Biophys. Acta 1833: 1256–1268. [DOI] [PubMed] [Google Scholar]
- Parrish J. Z., Xue D., 2003. Functional genomic analysis of apoptotic DNA degradation in C. elegans. Mol. Cell 11: 987–996. [DOI] [PubMed] [Google Scholar]
- Poultney C. S., Butterfoss G. L., Gutwein M. R., Drew K., Gresham D., et al. , 2011. Rational design of temperature-sensitive alleles using computational structure prediction. PLoS One 6: e24937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye C. A., Paredez A. R., Smith C. W., McDonald K. L., Aroian R. V., 1999. The coronin-like protein POD-1 is required for anterior-posterior axis formation and cellular architecture in the nematode Caenorhabditis elegans. Genes Dev. 13: 2838–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S., Bertrand V., Bigelow H., Boyanov A., Doitsidou M., et al. , 2010. Analysis of multiple ethyl methanesulfonate-mutagenized Caenorhabditis elegans strains by whole-genome sequencing. Genetics 185: 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Sato M., Audhya A., Oegema K., Schweinsberg P., et al. , 2006. Dynamic regulation of caveolin-1 trafficking in the germ line and embryo of Caenorhabditis elegans. Mol. Biol. Cell 17: 3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Grant B. D., Harada A., Sato K., 2008. Rab11 is required for synchronous secretion of chondroitin proteoglycans after fertilization in Caenorhabditis elegans. J. Cell Sci. 121: 3177–3186. [DOI] [PubMed] [Google Scholar]
- Severson A. F., Hamill D. R., Carter J. C., Bowerman B., 2000. The Aurora/Ipl1p-related kinase AIR-2 is required for cytokinesis and functions at metaphase to recruit the kinesin-like protein ZEN-4/MKLP1 to the mitotic spindle midzone. Curr. Biol. 10: 1162–1171. [DOI] [PubMed] [Google Scholar]
- Schiel J. A., Prekeris R., 2013. Membrane dynamics during cytokinesis. Curr. Opin. Cell Biol. 25: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D. J., Rose D. J., Saxton W. M., Strome S., 2005. Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol. Biol. Cell 16: 1200–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye D. D., Greenwald I., 2011. Ortholist: A compendium of C. elegans genes with human orthologs. PLoS One 6: e20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears S. B., Ganapathi S. B., Gokhale N. A., Schenk T. M. H., Wang H., et al. , 2012. Defining Signal Transduction by Inositol Phosphates, pp. 389–412 Phosphoinositides II: The Diverse Biological Functions, edited by T. Balla, M. Wyman, and J. D. York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., et al. , 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469. [DOI] [PubMed] [Google Scholar]
- Tsui M. M., York J. D., 2010. Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv. Enzyme Regul. 50: 324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbsky J., Lavine K., Majerus P. W., 2005. Disruption of the mouse inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene, associated lethality, and tissue distribution of 2-kinase expression. Proc. Natl. Acad. Sci. USA 102: 8448–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J., 2009. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinfomratics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. S. C., Livermore T. M., Saiardi A., 2013. Inositol pyrophosphates: between signalling and metabolism. Biochem. J. 452: 369–379. [DOI] [PubMed] [Google Scholar]
- Wolke U., Jezuit E. A., Priess J. R., 2007. Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development 134: 2227–2236. [DOI] [PubMed] [Google Scholar]
- Wong M. C., Schwarzbauer J. E., 2012. Gonad morphogenesis and distal tip cell migration in the Caenorhabditis elegans hermaphrodite. Wiley Interdiscip. Rev. Dev. Biol. 1: 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zutz A., Gompf S., Schägger H., Tampé R., 2009. Mitochondrial ABC proteins in health and disease. Biochim. Biophys. Acta. Bioenerg. 1787: 681–690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All SNP mapping and whole genome sequence data is available at wormbase.org.