Abstract
Objective
Circadian clocks are functional in all light-sensitive organisms, allowing an adaptation to the external world in anticipation of daily environmental changes. In view of the potential role of the skeletal muscle clock in the regulation of glucose metabolism, we aimed to characterize circadian rhythms in primary human skeletal myotubes and investigate their roles in myokine secretion.
Methods
We established a system for long-term bioluminescence recording in differentiated human myotubes, employing lentivector gene delivery of the Bmal1-luciferase and Per2-luciferase core clock reporters. Furthermore, we disrupted the circadian clock in skeletal muscle cells by transfecting siRNA targeting CLOCK. Next, we assessed the basal secretion of a large panel of myokines in a circadian manner in the presence or absence of a functional clock.
Results
Bioluminescence reporter assays revealed that human skeletal myotubes, synchronized in vitro, exhibit a self-sustained circadian rhythm, which was further confirmed by endogenous core clock transcript expression. Moreover, we demonstrate that the basal secretion of IL-6, IL-8 and MCP-1 by synchronized skeletal myotubes has a circadian profile. Importantly, the secretion of IL-6 and several additional myokines was strongly downregulated upon siClock-mediated clock disruption.
Conclusions
Our study provides for the first time evidence that primary human skeletal myotubes possess a high-amplitude cell-autonomous circadian clock, which could be attenuated. Furthermore, this oscillator plays an important role in the regulation of basal myokine secretion by skeletal myotubes.
Keywords: Circadian clock, Human skeletal myotube, Myokine, Interleukin-6, Circadian bioluminescence recording
Highlights
-
•
Human skeletal myotubes possess a self-sustained circadian clock.
-
•
IL-6 secretion is strongly downregulated upon muscle clock disruption.
-
•
The basal secretion of several myokines displays a circadian profile.
-
•
Disruption of the circadian clock affects basal myokine secretion.
Abbreviations
- BMAL1
brain and muscle ARNT-like 1
- CLOCK
circadian locomotor output cycles kaput
- CRY
cryptochrome
- DBP
D-albumin binding protein
- GLP-1
glucagon-like peptide 1
- HPRT
hypoxanthine-guanine phosphoribosyltransferase
- IL-6
interleukin-6
- IL-8
interleukin-8
- Luc
luciferase
- MOI
multiplicity of infection
- MCP-1
monocyte chemotactic protein 1
- M-CSF
macrophage colony-stimulating factor
- PER
period
- RORα
retinoid-related orphan receptor alpha
- REV-ERBα
reverse-erb alpha
- SCN
suprachiasmatic nucleus
- T2D
type 2 diabetes mellitus
- VEGF
vascular endothelial growth factor
- ZT
zeitgeber time
1. Introduction
Circadian oscillations are daily cycles in behavior and physiology that have been described from photosynthetic bacteria to vertebrates. They are reflected by the existence of underlying intrinsic biological clocks with near 24 h periods that generate self-sustained rhythms, influenced by environmental stimuli, such as light and feeding [1]. Under homeostatic conditions, the clock acts as a driver of metabolic processes with remarkable tissue specificity that reflects the unique demand of each tissue [2]. In peripheral organs, a large number of key metabolic functions are subject to daily oscillations, such as carbohydrate and lipid metabolism by the liver, and xenobiotic detoxification by the liver, kidney and small intestine [2], [3]. In rodents, the presence of peripheral circadian oscillators and their impact on gene expression and organ function has been demonstrated in liver, whole pancreas, pancreatic islets, and in adipose tissue (reviewed in [4]). Of note, feeding rhythms represent a potent synchronizing cue for peripheral oscillators. Elegant studies involving inverted or restricted feeding schedules convincingly demonstrate that feeding rhythms are powerful enough to uncouple liver and other organ clocks from the SCN [5]. Moreover, rhythmicity in a number of clock-deficient mouse models could be restored by feeding rhythms [6], [7], [8].
There is growing evidence for a tight reciprocal link between a number of metabolic diseases, including obesity and type 2 diabetes mellitus (T2D), and the circadian clockwork [3], [4]. Mice with circadian clock ablation develop hyperphagia, obesity, hyperglycemia and hypoinsulinemia [9]. The adipocyte-specific Bmal1 knockout leads to obesity development [10], while the islet-specific Bmal1 ablation directly triggers the onset of T2D in mice [11]. In humans, genetic analyses have shown a close association between glucose levels and variants of the CRY2 and melatonin receptor 1B (MTNR1B) genes [2].
Skeletal muscle is the largest insulin sensitive organ in the body, playing an essential role in whole body glucose homeostasis. It is responsible for 70–80% of insulin-stimulated glucose uptake and therefore representing the most important site of insulin resistance in T2D patients [12]. The primary and best-described function of skeletal muscles is their mechanical activity. During the last decade, however, skeletal muscle has been characterized also as a secretory organ, producing and releasing myokines that act on the muscle itself and display endocrine effects on distant organs [13], [14]. Recently, it has been proposed that fully differentiated primary human skeletal muscle cells secrete over 300 potential myokines [15]. Interleukin-6 (IL-6) is one of the first identified myokines, which is produced by the contracting skeletal muscle [16]. Acute high plasma levels of IL-6 are associated with exercise, while chronically elevated IL-6 is observed upon obesity and T2D. Moreover, IL-6 promotes pancreatic alpha cell mass expansion [17] and stimulates GLP-1 production and secretion by alpha and L-cells upon metabolic syndrome, thus exerting beneficial effects in T2D mouse models [18]. Besides IL-6, skeletal muscle produces a number of additional myokines, such as MCP-1 and IL-8, which play a role in skeletal muscle inflammation, recruitment of macrophages and insulin sensitivity [19], [20].
In rodents, about 7% of the skeletal muscle transcriptome is expressed in a circadian manner [21]. Moreover, clock ablation (Bmal1−/−) leads to skeletal muscle pathologies [22]. MyoD, a master regulator of myogenesis, exhibits a robust circadian rhythm and was identified as a direct target of CLOCK and BMAL1. In addition, disruption of myofilament organization was detected in Bmal1−/−, ClockΔ19, and in MyoD−/− mice, suggesting a direct link between the circadian clock and skeletal muscle function in rodents [22]. Furthermore, a recent study suggests that in mice muscle insulin sensitivity might be clock-dependent [23].
In view of the accumulating evidence in rodent models, it has been accepted that the skeletal muscle clock plays an essential role in maintaining proper metabolic functioning (reviewed in [24]), although the mechanism of this important regulation is not entirely clear, particularly in human subjects. Given that, the circadian clock in human skeletal muscle has remained largely unexplored, we aimed to characterize the circadian oscillator in primary human myotubes and explore its impact on the regulation of human skeletal muscle myokine secretion.
2. Material and methods
2.1. Study participants, skeletal muscle tissue sampling and primary cell culture
Muscle biopsies were derived from non-obese or obese donors with the informed consent obtained from all participants (see Table 1 for the donor characteristics). The experimental protocol (‘DIOMEDE’) was approved by the Ethical Committee SUD EST IV (Agreement 12/111) and performed according to the French legislation (Huriet's law). All donors had HbAc1 levels inferior to 6.0% and fasting glycemia inferior to 7 mmol/L, were not diagnosed for T2D, neoplasia or chronic inflammatory diseases, and not doing shift work. Biopsies were taken from the Gluteus maximus (n = 18) or the Rectus abdominus (n = 6) muscles during the planned surgeries. Primary skeletal myoblasts were purified and differentiated into myotubes according to the previously described procedure [25]. Briefly, after removal of apparent connective and fat tissue contaminants, the muscle biopsy was minced with scissors and incubated successively at least 4 times for 20 min in Trypsin-EDTA (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) at 37 °C under agitation. Trypsin-EDTA digested extracts were pooled, and centrifuged (150 g). The pellet was rinsed several times with PBS and cells were filtered on a 70-μm filter before being plated in a Primaria flask (Falcon; Becton Dickinson, Bedford, MA) containing growth medium composed of HAM F-10 supplemented with 2% Ultroser G (BioSepra SA, Cergy-Saint-Christophe, France), 2% fetal bovine serum (FBS, Invitrogen), and 1% antibiotics (Invitrogen). After 4 days, the myoblasts were immuno-selected using a monoclonal human CD56 antibody combined with paramagnetic beads (CD56 MicroBeads, Miltenyi Biotec, Germany), according to the manufacturer's instructions. The selected cells were plated on Primaria plastic ware at 4500 cells/cm2 and cultured in growth medium at 37 °C. After reaching confluence, myoblasts were differentiated into myotubes during 7–10 days in DMEM supplemented with 2% FBS. Muscle differentiation was characterized by the fusion of myoblasts into polynucleated myotubes (Supplementary Figure 1).
Table 1.
Characteristics of donors for skeletal muscle biopsies.
| Donor | Sex | Age (years) | BMI (kg/m2) | Biopsy source |
|---|---|---|---|---|
| Non-obese | M = 14, F = 5 | 58 ± 18 | 24.88 ± 3.20 | |
| 1a | M | 48 | 21.7 | Rectus abdominus |
| 2a,f | M | 45 | 21 | Rectus abdominus |
| 3a,f | F | 58 | 20.5 | Rectus abdominus |
| 4a,f | F | 42 | 19.5 | Rectus abdominus |
| 5b,d,k | M | 23 | 29.34 | Gluteus maximus |
| 6a,b,d,e,g,h,i,j | M | 62 | 24.3 | Gluteus maximus |
| 7a,b,f | F | 77 | 25.6 | Gluteus maximus |
| 8b,d,g,i,j | M | 57 | 26 | Gluteus maximus |
| 9b,d,g,h | M | 60 | 24 | Gluteus maximus |
| 10b | F | 88 | 29.64 | Gluteus maximus |
| 11a,b,c,d | F | 65 | 25.8 | Gluteus maximus |
| 12b,d,k | M | 25 | 19.27 | Gluteus maximus |
| 13b,d,g,h | M | 64 | 28 | Gluteus maximus |
| 14b,d | M | 87 | 25.51 | Gluteus maximus |
| 15b,d | M | 85 | 25.54 | Gluteus maximus |
| 16b,d | M | 72 | 26.5 | Gluteus maximus |
| 17b,e,f,h | M | 48 | 24.3 | Gluteus maximus |
| 18b,d,h | M | 45 | 28.1 | Gluteus maximus |
| 19a,b,c,d,g,i,j | M | 57 | 28.1 | Gluteus maximus |
| Obese | M = 1, F = 4 | 53 ± 15 | 39.21 ± 7.16 | |
| 20b | M | 70 | 30.1 | Gluteus maximus |
| 21e | F | 70 | 32.9 | Gluteus maximus |
| 22b,e,g,h,i | F | 43 | 43 | Gluteus maximus |
| 23b,d,g,i | F | 39 | 45.48 | Rectus abdominus |
| 24b,d,g,i | F | 45 | 44.58 | Rectus abdominus |
| All donors | M = 15, F = 9 | 57 ± 17 | 27.87 ± 7.23 |
M, male; F, female.
Non-obese, data are mean ± SD, n = 19.
Obese, data are mean ± SD, n = 5.
All donors, data are mean ± SD, n = 24.
donor cells used for the recording of Bmal1-luc bioluminescence of dexamethasone-synchronized samples.
donor cells used for the recording of Bmal1-luc bioluminescence of forskolin-synchronized samples.
donor cells used for the recording of Bmal1-luc bioluminescence of dexamethasone vs. forskolin-synchronized samples.
donor cells used for the recording of Per2-luc bioluminescence of forskolin-synchronized samples.
donor cells used for the around-the-clock experiment with dexamethasone synchronization.
donor cells used to quantify the silencing of CLOCK in siControl / siClock samples synchronized with dexamethasone.
donor cells used to quantify the silencing of CLOCK in siControl / siClock samples synchronized with forskolin.
donor cells used for the around-the-clock experiment in forskolin-synchronized siControl / siClock samples.
donor cells used for the IL-6 perifusion experiments with forskolin synchronization.
donor cells used for the multiplex assay analysis of perifusion samples synchronized with forskolin.
donor cells used for the IL-6 perifusion experiments with forskolin vs. dexamethasone synchronization.
2.2. siRNA transfection and lentiviral transduction
Human primary myoblasts were differentiated into myotubes as described above. Cells were transfected with 20 nM siRNA targeting CLOCK (siClock), or with non-targeting siControl (Dharmacon, GE Healthcare, Little Chalfont, UK), using HiPerFect transfection reagent (Qiagen, Hombrechtikon, Switzerland) following the manufacturer's protocol, 24 h prior to synchronization. To produce lentiviral particles, Bmal1-luc [26] or Per2-luc [27] lentivectors were transfected into 293T cells using the polyethylenimine method (for detailed procedure see [28]). Myoblasts were transduced with the indicated lentiviral particles with a multiplicity of infection (MOI) = 3 for each, grown to confluence, and subsequently differentiated into myotubes.
2.3. In vitro skeletal myotube synchronization and real-time bioluminescence recording
To synchronize primary myotubes, 10 μM forskolin (Sigma, Saint-Louis, MO, USA) or 100 nM dexamethasone (Alfa Aesar, Johnson Matthey, London, UK) were added to the culture medium, respectively. Following 60 min (forskolin) or 30 min (dexamethasone) incubation at 37 °C in a cell culture incubator, the medium was changed to a phenol red-free recording medium containing 100 μM luciferin and cells were transferred to a 37 °C light-tight incubator (Prolume LTD, Pinetop, AZ, USA), as previously described by us [28]. Bioluminescence from each dish was continuously monitored using a Hamamatsu photomultiplier tube (PMT) detector assembly. Photon counts were integrated over 1 min intervals. Luminescence traces are either shown as raw or detrended data. For detrended time series, luminescence signals were smoothened by a moving average with a window of 144 data points and detrended by an additional moving average with a window of 24 h [29]. For quantification of the circadian amplitude and period the first cycle was not taken into consideration.
2.4. mRNA extraction and quantitative PCR analysis
Differentiated myotubes were synchronized by forskolin or dexamethasone, collected every 4 h during 48 h (0 h–48 h), or during 24 h (12 h–36 h), deep-frozen in liquid nitrogen and kept at −80 °C. Total RNA was prepared using RNeasy Plus Micro kit (Qiagen). 0.5 μg of total RNA was reverse-transcribed using Superscript III reverse transcriptase (Invitrogen) and random hexamers and PCR-amplified on a LightCycler 480 (Roche Diagnostics AG, Rotkreuz, Switzerland). Mean values for each sample were calculated from the technical duplicates of each qRT-PCR analysis, and normalized to the mean of two housekeeping genes (HPRT and 9S or GAPDH and 9S), which served as internal controls. Primers used for this study are listed in Supplementary Table 1.
2.5. Circadian analysis of basal myokine secretion by ELISA and multiplex assay
In vitro synchronized differentiated myotubes, transduced with Bmal1-luc reporter, were placed into an in-house developed two-well horizontal perifusion chamber, connected to the LumiCycle. Cells were continuously perifused for 48 h with culture medium containing 100 μM luciferin. Bioluminescence recordings were performed in parallel to the automated collection of outflow medium in 4 h intervals. Basal IL-6 levels were quantified in the outflow medium using the Human IL-6 Instant ELISA kit (eBioscience, Affymetrix, Santa Clara, CA, USA) following the manufacturer's instructions. Data were normalized to the genomic DNA content, extracted using the QIAamp DNA Blood Mini Kit (Qiagen). Perifusion medium samples were further concentrated using Amicon Ultra 2 ml centrifugal filters (Ultracel-3K, Merck Millipore, Darmstadt, Germany). The evaluation of myokine release from human primary skeletal muscle cells was carried out using a multiplex bead array assay system (R&D Systems, Minneapolis, MN, USA). Custom-made luminex screening plates (CD44, CHI3L1, FABP3, galectin-3, GRO-alpha, IGFBP-3, IL-7, IL-13, IL-17A, M-CSF, MCP-1, MMP-2, Serpin C1, Serpin E1, TIMP-1) and high sensitivity performance plates (IL-1 beta, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12 p70, IFN-gamma, TNF-alpha and VEGF) were analyzed according to the manufacturer's instructions. Plate analysis was performed on a Bio-Plex 200 array reader (Bio-Rad Laboratories, Hercules, CA, USA), with the Bio-Plex software (Bio-Rad) used for data analysis.
2.6. Data analysis
Actimetrics LumiCycle analysis software (Actimetrics LTD) and the JTK_CYCLE algorithm [30] were used for bioluminescence and myokine secretion profile data analyses, respectively. For the ELISA and multiplex data analysis, 2 technical duplicates from 3 biological samples were analyzed for each myokine. From these values the average ± the SEM was calculated for each time point. For JTK_CYCLE analysis the period width was set at 20–24 h. Statistical analyses were performed using a paired Student's t-test. Differences were considered significant for p < 0.05 (*), p < 0.01 (**) and p < 0.001 (***).
3. Results
3.1. High-amplitude self-sustained clocks are functional in primary human skeletal myotubes
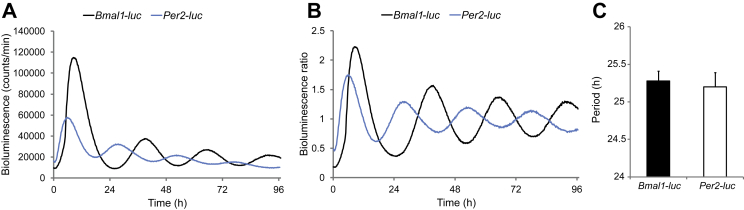
Circadian bioluminescence recordings in living cells allow for the study of molecular clocks in human peripheral tissues, as previously demonstrated by us [28], [31] and others [32]. We applied this powerful methodology to assess clock properties in human primary skeletal myotubes established from human donor biopsies and differentiated in vitro (see Supplementary Figure 1 and Table 1 for donor characteristics). Multiple in vitro stimuli have previously been demonstrated to efficiently synchronize cultured cells, among them forskolin and dexamethasone [28], [33]. As shown in Supplementary Figure 2, short pulses of dexamethasone or forskolin were able to strongly synchronize human myotubes bearing the Bmal1-luc lentivector. Both forskolin and dexamethasone induced oscillations with comparable period length: 25.29 ± 0.13 h, n = 19 for forskolin (Table 2), and 24.48 ± 0.24 h, n = 8 for dexamethasone (data not shown). However, the forskolin pulse induced more sustained cycles with higher circadian amplitude compared to dexamethasone-synchronized cells (Supplementary Figure 2) and was therefore mainly used in this study as the in vitro synchronization stimulus. The here established experimental settings allowed for continuous recording of oscillation profiles in human primary myotubes for several days with high resolution (Figure 1A, B). As expected, the profiles of the Bmal1-luc and Per2-luc reporters were antiphasic (Figure 1A, B; [3]).
Table 2.
Circadian period length of forskolin-synchronized human myotubes assessed by circadian bioluminescent reporters.
|
Bmal1-luc period (h) |
Per2-luc period (h) |
Mean period (h) |
||||
|---|---|---|---|---|---|---|
| n | Mean ± SEM | n | Mean ± SEM | n* | Mean ± SEM | |
| Non-obese | 15 | 25.35 ± 0.14 | 12 | 25.25 ± 0.22 | 27 | 25.31 ± 0.12 |
| Obese | 4 | 25.04 ± 0.35 | 2 | 24.85 ± 0.30 | 6 | 24.98 ± 0.24 |
| All donors | 19 | 25.29 ± 0.13 | 14 | 25.20 ± 0.19 | 33 | 25.25 ± 0.11 |
*n represents the number of experimental repetitions.
Figure 1.
High-amplitude cell autonomous oscillators are functional in differentiated human primary myotubes. Human primary myoblasts were transduced with lentiviral particles expressing Bmal1-luc (black line) or Per2-luc (blue line). Cells were differentiated into myotubes, synchronized with forskolin, and transferred to the Actimetrics LumiCycle for bioluminescence recording. Raw (A) and detrended (B) oscillation profiles are representative of 19 and 14 independent experiments, respectively (one donor per experiment). (C) The period length of Bmal1-luc or Per2-luc was on average 25.29 ± 0.13 h, (n = 19) or 25.20 ± 0.19 h, (n = 14), respectively. Data represent the mean ± SEM.
High-amplitude self-sustained oscillations were reproducibly recorded from human primary myotubes for Bmal1-luc and Per2-luc reporters with an average period length of 25.29 ± 0.13 h and 25.20 ± 0.19 h, respectively (Figure 1C). Of note, no significant difference in period length of the Bmal1-luc or Per2-luc reporter was observed between myotubes established from non-obese and obese donors (Table 2), which might reflect the resistance of the core clock to metabolic changes.
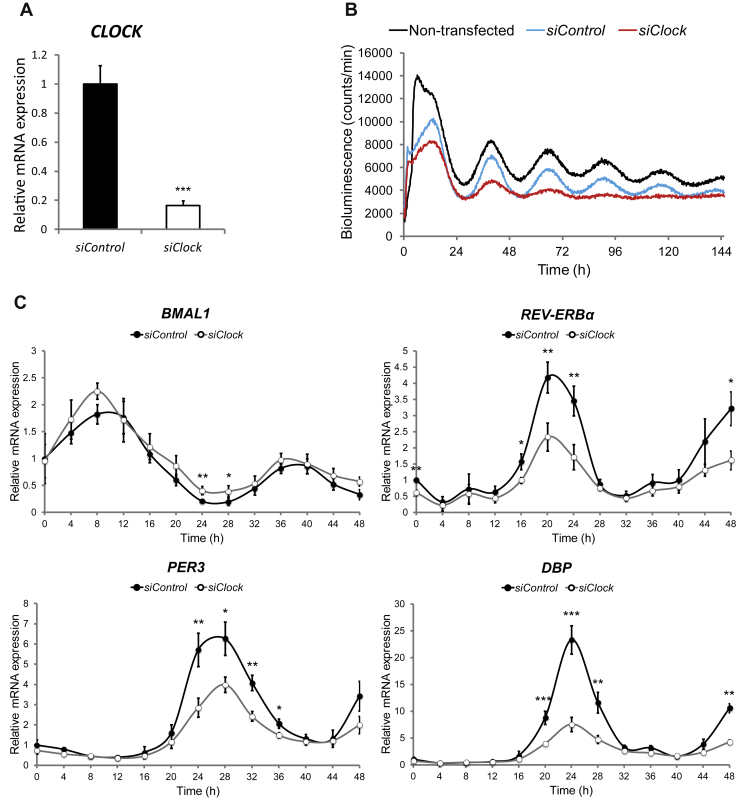
To further validate the results obtained by circadian bioluminescence reporter studies, we examined endogenous core clock gene expression profiles in forskolin-synchronized myotubes (Figure 2C, closed circles). mRNA accumulation patterns from synchronized skeletal myotubes were monitored every 4 h during 48 h by quantitative RT-PCR, using amplicons for BMAL1, REV-ERBα, PER3 and DBP. The values obtained were normalized to the mean of the housekeeping genes HPRT and 9S. In agreement with our Bmal1-luc reporter experiments, endogenous BMAL1 transcript levels exhibited circadian oscillations over 48 h in synchronized myotubes (compare Figure 2C BMAL1 panel to Figure 1A), clearly antiphasic to the profiles of the endogenous REV-ERBα, PER3, and DBP transcripts (Figure 2C). Similar experiments were conducted in dexamethasone-synchronized myotubes (Supplementary Figure 3). BMAL1 and CRY1 mRNAs exhibited oscillatory profiles antiphasic to those of REV-ERBα and PER2-3, consistent with the dexamethasone-induced oscillations of the Bmal1-luc reporter (Supplementary Figure 2).
Figure 2.
Silencing of CLOCK expression attenuates circadian oscillations in human skeletal myotubes. (A) CLOCK mRNA was measured in human myotubes transfected with siControl or siClock by RT-qPCR and normalized to the mean of 9S and HPRT. CLOCK expression was reduced by 83.5 ± 3.4% (mean ± SEM, n = 8; ***p < 0.001) in siClock-transfected cells. (B) Amplitude of the Bmal1-luc reporter is strongly reduced in siClock-transfected myotubes. Representative Bmal1-luc oscillation profiles are shown for non-transfected (black line), siControl (blue line), and siClock (red line) transfected myotubes. Bmal1-luc oscillation profiles were recorded in duplicates (3 experiments, one donor per experiment). (C) RT-qPCR was performed for BMAL1, REV-ERBα, PER3 and DBP on RNA samples extracted from forskolin-synchronized human myotubes, transfected with siClock (open circles) or siControl (closed circles). Samples were collected every 4 h and normalized to the mean of 9S/HPRT. Profiles (mean ± SEM) are representative of 3 experiments (2 donors for time points 0 h–48 h and 3 donors for time points 12 h–36 h) with duplicates per time point.
3.2. Human myotube clock disruption by siRNA-mediated CLOCK knockdown
In order to disrupt the circadian clock in cultured human myotubes, we set up an efficient siClock transfection protocol, resulting in more than 80% knockdown of CLOCK transcript levels (Figure 2A, Supplementary Figure 4A). Circadian expression of the Bmal1-luc reporter was blunted in siClock-transfected myotubes upon forskolin or dexamethasone synchronization, if compared to cells transfected with non-targeting sequences (siControl) or to non-transfected counterparts (Figure 2B, Supplementary Figure 4B), thus validating circadian clock disruption. Moreover, the amplitudes of endogenous REV-ERBα, PER3 and DBP transcript profiles were strongly blunted in siClock-transfected cells, in comparison to siControl cells (Figure 2C, Supplementary Table 2). By contrast, BMAL1 transcript levels were slightly upregulated (Figure 2C, Supplementary Table 2).
3.3. Regulation of basal IL-6 secretion by the circadian clock in human primary myotubes
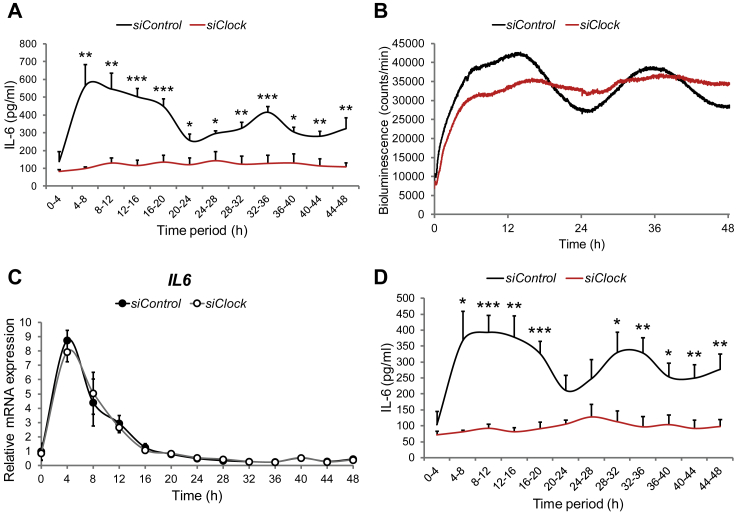
Given the accumulating evidence on the essential role of IL-6 secretion by skeletal muscle under physiological conditions and upon metabolic diseases [14], [17], [18], we next monitored basal circadian IL-6 secretion by human primary myotubes established from non-obese and obese donors. To this end, we developed an in-house perifusion system connected to the LumiCycle chamber that allows for parallel cell perifusion and bioluminescence profile recordings. Basal IL-6 secretion from in vitro synchronized skeletal myotubes was monitored in “around-the-clock” experimental settings, with a continuous flow of culture medium (see Material and methods for details). The perifusion experiments suggested that forskolin-synchronized myotubes exhibited a circadian profile of basal IL-6 secretion over 48 h, with a Zenith around 8 h–12 h and 32 h–36 h and a Nadir of 20 h–24 h (Figure 3A). JTK_CYCLE analysis [30] revealed that average profile of basal IL-6 secretion did not reach significance to be qualified as circadian over the entire time span of 48 h. However it was significantly circadian over the first 36 h of perifusion following in vitro synchronization (**p < 0.01, n = 6). Similar basal IL-6 secretion profiles were observed from dexamethasone-synchronized myotubes (Supplementary Figure 5).
Figure 3.
Basal IL-6 secretion by human skeletal myotubes is strongly inhibited in the absence of a functional circadian clock. Myoblasts were transduced with the Bmal1-luc lentivector, differentiated into myotubes and transfected with either siControl or siClock siRNA. 24 h following transfection, myotubes were synchronized with forskolin and subjected to continuous perifusion with parallel bioluminescence recording. (A) Basal IL-6 secretion profile (mean + SEM) in the presence or absence of a functional clock. The perifusion outflow medium was collected continuously in an automated manner in 4 h intervals until 48 h (0–4 corresponds to the accumulation of IL-6 between 0 h and 4 h). IL-6 levels in the perifusion outflow medium were assessed by ELISA. The results represent basal IL-6 levels normalized to the total DNA content. 2 technical duplicates from 3 independent experiments (3 non-obese donors, see Table 3) were analyzed for each time point. (B) Bmal1-luc bioluminescence profiles of siControl-transfected myotubes (black line) and siClock-transfected myotubes (red line), representative of 3 experiments, with one donor cell line used per experiment. (C) RT-qPCR was performed for IL6 on RNA samples extracted from forskolin-synchronized human myotubes, transfected with siClock (open circles) or siControl (closed circles). Samples were collected every 4 h and normalized to the mean of 9S/HPRT. Profiles (mean ± SEM) are representative of 3 experiments (2 donors for time points 0 h–48 h and 3 donors for time points 12 h–36 h) with duplicates per time point. (D) Basal IL-6 secretion profiles in the presence or absence of a functional clock obtained from concentrated perifusion samples, assessed by multiplex analysis. Data shown as mean + SEM of 2 technical duplicates from 3 biological samples for each time point, normalized to the total DNA content.
We next tested the effect of CLOCK depletion on IL-6 secretion. Similar to previous experiments (Figure 2), CLOCK transcript expression was at least 80% downregulated in siClock-transfected myotubes (***p < 0.001, paired t-test, Table 3) compared to siControl-transfected cells. The achieved clock disruption was also confirmed by parallel Bmal1-luc bioluminescence recording in perifused cells (Figure 3B). Importantly, IL-6 secretion decreased on average 64% in siClock-transfected myotubes compared to siControl-transfected counterparts (*p < 0.05, Table 3). Furthermore, the profile of basal IL-6 secretion became flat upon clock disruption if compared to the siControl-transfected profile (see red line vs. black line, Figure 3A). Of note, overall basal IL-6 secretion was on average higher in obese siControl-transfected subjects (n = 3) compared to non-obese siControl-expressing counterparts (n = 3), although values did not reach statistical significance (Table 3).
Table 3.
Overall basal IL-6 secretion after forskolin synchronization in human myotubes.
| Non-obese (n = 3) |
Obese (n = 3) |
All donors (n = 6) |
|
|---|---|---|---|
| Mean ± SEM | Mean ± SEM | Mean ± SEM | |
| IL-6/48h in siControl samples (pg) | 4386.54 ± 411.71 | 5888.25 ± 1547.86 | 5137.40 ± 796.43 |
| IL-6/48h in siClock samples (pg) | 1424.12 ± 408.83 | 2395.52 ± 625.60 | 1909.82 ± 385.20 |
| Inhibition of secretion (%) | 69.30 ± 10.61 | 59.66 ± 7.04 | 64.48 ± 6.09 |
| Clock mRNA silencing (%) | 91.32 ± 2.67 | 78.57 ± 8.15 | 84.94 ± 4.57 |
To get an insight into the regulation of basal IL-6 secretion by the circadian clock, we assessed IL6 transcript levels following in vitro synchronization with forskolin. No clear circadian pattern was observed (Figure 3C). The strong immediate early peak of IL6 transcription induced by the forskolin pulse might be attributed to the presence of a cAMP response element previously identified in IL-6 promoter region [34]. Moreover, CLOCK depletion by siClock had no effect on basal IL6 transcription (Figure 3C).
3.4. Multiplex screen identifies additional clock-regulated myokines
Since we obtained convincing evidence on the requirement of a functional circadian clock for basal IL-6 secretion by skeletal myotubes (Figure 3A,B), we selected an additional panel of myokines for analysis by multiplex assay. The around-the-clock secretory profiles of IL-6 and 24 other myokines were assessed in perifusion samples obtained from primary human myotubes as described in Figure 3, and further concentrated to allow for the detection of basal myokine secretion in siControl and siClock-transfected myotubes. Selected myokines were chosen based on their presence in the secretome of differentiated primary skeletal muscle cells [15], their function and implication in metabolic diseases, and their availability and compatibility for multiplex array analysis. Out of the selected panel of 25 myokines, 15 myokines were detected in the concentrated perifusion samples, while 10 myokines remained undetectable, due to their low levels of basal secretion (Table 4).
Table 4.
Myokines with clock-regulated basal secretion.
| Circadian analysis |
Fold change of secretion |
||||
|---|---|---|---|---|---|
| (p-values calculated by JTK_CYCLE, n = 3) |
(mean ± SEM, n = 3) |
||||
| siControl | siClock | siControl | siClock | Paired t-test | |
| CD44a | 0.777 | 0.692 | 1.00 | 0.67 ± 0.13 | 0.025* |
| CHI3L1/YKL40a | 1.000 | 1.000 | 1.00 | 2.12 ± 1.22 | 0.378 |
| FABP3/H-FABPa | 1.000 | 0.231 | 1.00 | 0.53 ± 0.11 | 0.002** |
| Galectin-3a | 1.000 | 0.096 | 1.00 | 0.41 ± 0.07 | 5.19E-6*** |
| GRO-alpha/CXCL1a | 0.096 | 1.000 | 1.00 | 0.55 ± 0.23 | 0.083 |
| IGFBP-3a | 1.000 | 1.000 | 1.00 | 0.70 ± 0.23 | 0.226 |
| IL-6b | 0.01* | 1.000 | 1.00 | 0.44 ± 0.16 | 0.006** |
| IL-8b | 0.020* | 1.000 | 1.00 | 0.47 ± 0.20 | 0.036* |
| MCP-1/CCL2a | 0.020* | 0.096 | 1.00 | 0.59 ± 0.27 | 0.157 |
| M-CSF/CSF1a | 0.059 | 0.777 | 1.00 | 0.64 ± 0.16 | 0.053 |
| MMP-2a | 1.000 | 1.000 | 1.00 | 0.78 ± 0.19 | 0.268 |
| Serpin E1/PAI-1a | 1.000 | 1.000 | 1.00 | 0.69 ± 0.16 | 0.070 |
| Serpin C1a | 0.492 | 1.000 | 1.00 | 0.64 ± 0.19 | 0.123 |
| TIMP-1a | 1.000 | 1.000 | 1.00 | 0.68 ± 0.14 | 0.043* |
| VEGFb | 0.949 | 0.231 | 1.00 | 0.60 ± 0.10 | 0.002** |
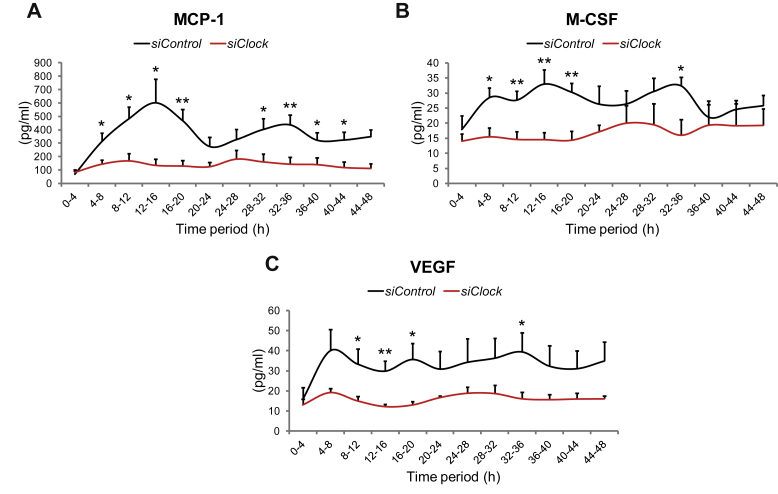
Importantly, the profile and concentration levels of around-the-clock IL-6 secretion measured by multiplex analysis were very similar to those obtained by ELISA (compare Figure 3D–A). JTK_CYCLE analysis [30] confirmed that the average profile of secreted IL-6 measured by multiplex analysis was significantly circadian within 48 h (Figure 3D, Table 4). In addition to IL-6, MCP-1 and IL-8 were secreted in a circadian manner according to JTK_CYCLE analysis (Figure 4A, Table 4). M-CSF and GRO-alpha exhibited a secretion profile that might be clock-controlled; however, the adjusted minimal p-value did not reach significance according to JTK_CYCLE analysis (Figure 4B, Table 4). Furthermore, although the temporal secretion patterns of VEGF, CD44, FABP3, Galectin-3 and TIMP-1 were not identified as circadian, the overall secretion levels of these myokines were significantly reduced upon CLOCK depletion (Figure 4C, Table 4).
Figure 4.
Basal myokine secretion by human skeletal myotubes is affected by circadian clock disruption. Myoblasts were transduced with the Bmal1-luc lentivector, differentiated into myotubes, transfected with either siControl or siClock siRNA, and subjected to continuous perifusion with parallel bioluminescence recording. Concentrated perifusion samples were assessed by multiplex analysis. 2 technical duplicates from 3 biological samples were analyzed for each time point, and normalized to the total DNA content. Basal secretion profiles (mean + SEM) in the presence or absence of a functional clock are shown for (A) MCP-1, (B) M-CSF, and (C) VEGF.
4. Discussion
4.1. Molecular makeup of the circadian oscillator operative in human skeletal muscle
Our study provides for the first time evidence for cell-autonomous self-sustained circadian oscillators, operative in human primary skeletal myotubes. Molecular characteristics of the human myotube clock were assessed by two complementary approaches. Pronounced circadian oscillations were recorded with high temporal resolution for at least 4–5 consecutive days from in vitro synchronized primary skeletal myotubes, transduced with Bmal1-luciferase or Per2-luciferase lentivectors (Figure 1A,B). The circadian characteristics of human skeletal myotubes were in accordance with those reported for human primary thyrocytes, pancreatic islets, skin fibroblasts [28], [31], [35], and for mouse skeletal muscle assessed in vivo [21], [22]. Sustained circadian oscillations were efficiently induced in our system by both forskolin (Figure 1) and dexamethasone (Supplementary Figure 2) pulses, suggesting that these oscillators are functional irrespective of the synchronization stimulus or entrainment pathway. It might be of interest to further explore whether other physiologically relevant stimuli like glucose, insulin, or myokine-induced signaling pathways play a role in human myotube synchronization.
In line with the outcome of our reporter experiments, endogenous around-the-clock gene expression analyses suggested that the core clock genes BMAL1, REV-ERBα, and PER3, as well as the clock output gene DBP, exhibit circadian oscillatory patterns in forskolin and dexamethasone-synchronized myotubes (Figure 2C closed circles, Supplementary Figure 3). Of note, the circadian amplitude of oscillations induced in vitro is typically lower if compared to in vivo oscillations from the same tissue, as demonstrated, for instance, for mouse islets synchronized in vitro or collected in vivo [11]. Thus, identifying clock-controlled genes in vitro by RT-qPCR represents a significant challenge due to rather low amplitudes [21], [28]. More accurate methods like RNA sequencing of samples collected with higher temporal resolution might represent a solution to this problem.
4.2. Experimental model for circadian clock disruption in human primary myotubes
Here, we have established experimental settings for a highly reproducible siRNA-mediated CLOCK transcript knockdown of more than 80% in human primary muscle cells (Figure 2A, Supplementary Figure 4A). Upon such CLOCK silencing, significant flattening of the circadian amplitude was observed for the Bmal1-luc reporter (Figure 2B, Figure 3B, Supplementary Figure 4B) and for endogenous REV-ERBα, PER3 and DBP transcripts (Figure 2C), confirming circadian core clock disruption. The discrepancy between Bmal1-luc reporter data and endogenous BMAL1 expression upon siClock might be related to the fact that the knockdown effect of CLOCK on the Bmal1-luc reporter is evident primarily after 48 h, and that the promoter length of the Bmal1 reporter is different from the endogenous gene. While REV-ERBα, PER3 and DBP transcript profiles only exhibited residual circadian oscillations, which might be explained by incomplete clock ablation, the amplitude of the endogenous BMAL1 oscillatory profile was not reduced and even slightly increased (Figure 2C). Similarly, siRNA-mediated depletion of CLOCK in U2OS cells was reported to increase BMAL1 expression levels [36]. Moreover, Clock-deficient mice continue to exhibit circadian patterns of behavioral and molecular rhythms. In the SCN and liver of Clock-deficient mice, Bmal1 mRNA was elevated, as well as in pancreatic islets of Clock-mutant mice, which was attributed to reduced REV-ERBα expression and a compensatory effect of NPAS2 [11], [37].
Clockwork perturbations may develop in humans with ageing and upon a number of disorders [3], [4]. In this respect, our experimental model, which allows for reproducible clock disruption mediated by CLOCK depletion in differentiated skeletal myotubes, represents a valuable tool for getting significant insights into the roles of clock perturbations in different aspects of human skeletal muscle function. In order to detect oscillatory alterations caused by metabolic changes it might be necessary to develop further readouts involving clock output genes, as the here assessed Bmal1-luc and Per2-luc reporter profiles, although definitely informative regarding the core clock, might be of limited value as readout for metabolic perturbations.
4.3. Basal IL-6 secretion by human skeletal muscle is regulated by the circadian clock
Importantly, our study demonstrates that human skeletal myotubes, synchronized in vitro by forskolin or dexamethasone, secrete basal IL-6 in a circadian manner (Figure 3A,D, Supplementary Figure 5). Furthermore, this circadian pattern is flattened under siClock-mediated clockwork disruption, with overall basal IL-6 secretion being strongly downregulated by oscillator perturbation (Figure 3A,D, Table 3). The experimental settings we have developed, combining continuous perifusion and bioluminescence recording, allow for the direct assessment of IL-6 secretion by cultured human myotubes. Studies in healthy adults have suggested that IL-6 levels in the cerebrospinal fluid exhibit a 24 h oscillatory profile, and plasma levels of IL-6 have a biphasic 12 h component [38], [39]. However, it has to be taken into account that plasma levels are originating from different sources of IL-6 and are also influenced by absorption in effector organs.
Our finding that basal IL-6 secretion is regulated locally by the skeletal muscle clock is in line with accumulating evidence that endocrine body rhythms are tightly regulated in humans by the circadian system at several levels (reviewed in [40]). Insulin secretion by beta cells in rodents was suggested to be a subject for clock regulation [11]. Moreover, a number of proinflammatory cytokines exhibit pronounced circadian alterations in the magnitude of their response to an endotoxin challenge at different times of the day [41], among them IL-6. Such circadian gating of the inflammatory response was lost for IL-6 in REV-ERBα knockout mice. Moreover, attenuation of REV-ERBα levels in human macrophages implied a direct link between REV-ERBα and IL-6 secretion [41].
Given a general controversy around the roles of IL-6 level alterations in the etiology of metabolic diseases [42], such downregulation might have a positive or negative impact on skeletal muscle function and body metabolism. A recent in vivo study in humans suggested that injection of IL-6 to T2D patients did not affect insulin-stimulated glucose uptake [43], while it was reported to have beneficiary effects on the glucose uptake in young, healthy subjects [44]. Indeed, acute high levels of IL-6 found after exercise improve insulin sensitivity, glucose metabolism [18], [45], and fat metabolism [46], whereas chronic exposure to IL-6 causes insulin resistance in mice [47]. At the same time, IL6 knockout mice develop mature-onset obesity and glucose intolerance [48]. We speculate that similar to cortisol, thyroid hormones, insulin, and other key hormones, which exhibit circadian rhythmicity of their levels [40], [49], the oscillatory pattern of IL-6 secretion by the skeletal muscle might give an advantage for the temporal coordination of this myokine with the rest–activity cycle, thus ensuring optimal IL-6 response to external cues.
Our results presented in Figure 3C suggest that the circadian regulation of basal IL-6 secretion may not occur at the transcriptional level. As it has been reported, the clock is quite insensitive to large fluctuations of transcription rate [50]. Moreover, in mouse livers almost 50% of the cycling proteins do not have rhythmic steady-state mRNA levels [51], [52]. Similarly, the IL6 transcriptional profile was not circadian in cultured mHypoE-37 neurons [53], while a low amplitude oscillatory profile of IL6 transcription was detected in human monocyte-derived macrophages [41] and in mouse astrocytes, where Il6 transcription was suggested to be directly regulated by RORα [41], [54]. Application of more sensitive transcription analysis approaches (RNA sequencing in samples with a higher temporal resolution), as well as addressing the potential regulation of IL6 transcription in human skeletal muscle in particular by REV-ERBα and RORα, might shed additional light on this conjunction. As IL-6 is extensively modified at the post-translational level, one plausible hypothesis beyond transcription could be that the circadian profile of IL-6 secretion is controlled by post-translational modification [55]. Visualizing IL-6 secretion at the single cell level in synchronized human myotubes would be necessary to provide an insight into the mechanism of this phenomenon.
4.4. Functional circadian oscillator in human skeletal myotubes is required for proper basal secretion of a broader range of myokines
Of note, our screening for additional clock-controlled myokines, employing a human cytokine array, suggests that the regulatory role of the skeletal muscle clock is not restricted solely to basal IL-6 secretion but might represent a more general mechanism involved in the fine-tuning of a panel of myokines (Figure 4, Table 4). Specifically, the temporal profiles and/or overall levels of basal secretion of IL-8, MCP-1, M-CSF, GRO-alpha, VEGF, CD44, Galectin-3, FABP3 and TIMP-1 by human skeletal myotubes imply that these myokines might be regulated by the circadian clock (Figure 4, Table 4).
The following limitations of this part of the study should be taken into account: the low number of around-the-clock perifusion experiments analyzed by multiplex, due to the high experimental complexity of the automated perifusion system, and the low basal concentration levels of many myokines. Therefore, although the technical reproducibility for the duplicates in each around-the-clock experiment was high, in view of the large variability among the human donors with respect to their myokine secretion levels, these experiments must be interpreted with caution. More experimental repetitions will therefore be required to claim the circadian regulation of the myokines identified in our screen. Furthermore, in order to allow for the detection and quantification of myokines with low concentration levels in a circadian manner, more sensitive tools need to be developed.
Of note, myokines identified in our multiplex screen as potentially clock-regulated for their basal secretion (Table 4, Figure 4) have been previously linked to obesity or T2D. For instance, serum IL-8 levels are increased in T2D patients, with a significant diurnal variation observed for IL-8 in blood upon LPS-stimulation [56], [57]. Furthermore, insulin-resistant human myotubes secrete higher levels of IL-8 [19]. MCP-1, which mediates skeletal muscle macrophage recruitment, was also suggested to play a role in the etiology of T2D [20]. Interestingly, this proinflammatory cytokine has been previously shown to exhibit pronounced circadian alterations in the magnitude of its response to endotoxin challenge at different times of the day [41]. M-CSF has been shown to play an important role in inflammatory diseases including obesity [58]. VEGF, known for its angiogenic properties, is transcriptionally regulated by the circadian clock [59], and has also been implicated in T2D and insulin resistance [60], [61]. Plasma levels of CD44 were positively correlated with insulin resistance in humans [62], [63]. Furthermore, genetic association studies have linked CD44 with T2D [62], [64]. Taken together, our data suggest that disruption of the circadian clock might affect the level and temporal profile of basal secretion for a number of myokines that play a role in the etiology of T2D and obesity.
Inevitably, our findings raise the question on the physiological relevance of basal myokine secretion. One plausible argument is that while induced myokine secretion is regulated acutely (for instance by exercise), circadian regulation of basal myokine secretion might represent a fine-tuning mechanism, allowing adaptation of the skeletal muscle function to the rest–activity cycle. Given the major role of the circadian clock in allowing organisms to anticipate daily environmental changes rather than react to them, circadian regulation of basal myokine secretion might represent such an anticipatory mechanism that coordinates skeletal muscle “availability”. Furthermore, this newly discovered link between the functional skeletal muscle clock and basal secretion of a number of myokines might bear potential consequences for the development of chronic diseases, such as obesity and T2D.
5. Conclusion
Skeletal muscle represents the most important site of insulin resistance in T2D patients [12]. Moreover, the emerging critical role of inflammation in the etiology of T2D makes inflammatory cytokines plausible candidates for developing new therapeutic approaches for the treatment of this disease [65]. It is therefore of highest scientific and clinical importance to provide further insight into the emerging connection between circadian oscillator function, metabolic regulation, and T2D. Given that human primary myotubes, established from T2D patient biopsies and cultured in vitro, have been demonstrated to maintain their in vivo phenotypes of inflammation and insulin resistance [66], our model of synchronized cultured primary myotubes represents a valuable experimental tool that allows for studying the role of the circadian oscillator in human skeletal muscle function upon metabolic diseases. This work is the first detailed characterization of the human skeletal myotube circadian oscillator and its critical impact on basal myokine secretion. It opens the way for future studies that may link defects in these pathways with insulin resistance, obesity, and T2D. Given obvious obstacles for studying the human circadian oscillator in vivo, our experimental approach, using human primary cell cultures established from biopsies that express a luciferase reporter driven from a circadian promoter, constitutes a powerful model system for human skeletal muscle clock studies.
Funding
This work was funded by the Swiss National Science Foundation Grant No. 31003A_146475/1, Fondation Romande pour la Recherche sur Diabète, Fondation Ernst et Lucie Schmidheiny, Société Académique de Genève (CD), and the Sinergia Swiss National Science Foundation Grant No. CRSII3-154405 (CD, EL).
Acknowledgments
We are grateful to our colleagues from University Hospital of Geneva: Jacques Philippe for constructive comments on this work and Thierry Berney for generously providing human samples; and to our colleagues from the University of Geneva: Ueli Schibler for invaluable help with the perifusion system development and for scientific inspiration; Andre Liani and George Severi for assistance with the perifusion experiments; Anne-Marie Makhlouf and Camille Saini for lentivirus preparations; Christelle Barraclough and Mylene Docquier (iGE3 Genomics Platform) for assistance in performing qPCR experiments; Nicolas Hulo (iGE3 BioSC) for assistance with the statistical analysis.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2015.07.009.
Conflict of interest
The authors have no conflict of interest to declare.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Albrecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron. 2012;74:246–260. doi: 10.1016/j.neuron.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 3.Dibner C., Schibler U. Circadian timing of metabolism in animal models and humans. Journal of Internal Medicine. 2015 May;277(5):513–527. doi: 10.1111/joim.12347. [DOI] [PubMed] [Google Scholar]
- 4.Marcheva B., Ramsey K.M., Peek C.B., Affinati A., Maury E., Bass J. Circadian clocks and metabolism. Handbook of Experimental Pharmacology. 2013:127–155. doi: 10.1007/978-3-642-25950-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damiola F., Le Minh N., Preitner N., Kornmann B., Fleury-Olela F., Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes & Development. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adamovich Y., Rousso-Noori L., Zwighaft Z., Neufeld-Cohen A., Golik M., Kraut-Cohen J. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metabolism. 2014;19:319–330. doi: 10.1016/j.cmet.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feillet C.A., Albrecht U., Challet E. “Feeding time” for the brain: a matter of clocks. Journal of Physiology Paris. 2006;100:252–260. doi: 10.1016/j.jphysparis.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Iijima M., Yamaguchi S., van der Horst G.T., Bonnefont X., Okamura H., Shibata S. Altered food-anticipatory activity rhythm in Cryptochrome-deficient mice. Neuroscience Research. 2005;52:166–173. doi: 10.1016/j.neures.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Turek F.W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paschos G.K., Ibrahim S., Song W.L., Kunieda T., Grant G., Reyes T.M. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nature Medicine. 2012;18:1768–1777. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcheva B., Ramsey K.M., Buhr E.D., Kobayashi Y., Su H., Ko C.H. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeFronzo R.A., Tripathy D. Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care. 2009;32(Suppl. 2):S157–S163. doi: 10.2337/dc09-S302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bostrom P., Wu J., Jedrychowski M.P., Korde A., Ye L., Lo J.C. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedersen B.K., Febbraio M.A. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nature Reviews. Endocrinology. 2012;8:457–465. doi: 10.1038/nrendo.2012.49. [DOI] [PubMed] [Google Scholar]
- 15.Hartwig S., Raschke S., Knebel B., Scheler M., Irmler M., Passlack W. Secretome profiling of primary human skeletal muscle cells. Biochimica et Biophysica Acta. 2014;1844:1011–1017. doi: 10.1016/j.bbapap.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Ostrowski K., Rohde T., Zacho M., Asp S., Pedersen B.K. Evidence that interleukin-6 is produced in human skeletal muscle during prolonged running. Journal of Physiology. 1998;508(Pt 3):949–953. doi: 10.1111/j.1469-7793.1998.949bp.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellingsgaard H., Ehses J.A., Hammar E.B., Van Lommel L., Quintens R., Martens G. Interleukin-6 regulates pancreatic alpha-cell mass expansion. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13163–13168. doi: 10.1073/pnas.0801059105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellingsgaard H., Hauselmann I., Schuler B., Habib A.M., Baggio L.L., Meier D.T. Interleukin-6 enhances insulin secretion by increasing glucagon-like peptide-1 secretion from L cells and alpha cells. Nature Medicine. 2011;17:1481–1489. doi: 10.1038/nm.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouzakri K., Plomgaard P., Berney T., Donath M.Y., Pedersen B.K., Halban P.A. Bimodal effect on pancreatic beta-cells of secretory products from normal or insulin-resistant human skeletal muscle. Diabetes. 2011;60:1111–1121. doi: 10.2337/db10-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patsouris D., Cao J.J., Vial G., Bravard A., Lefai E., Durand A. Insulin resistance is associated with MCP1-mediated macrophage accumulation in skeletal muscle in mice and humans. PLoS One. 2014;9:e110653. doi: 10.1371/journal.pone.0110653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy J.J., Andrews J.L., McDearmon E.L., Campbell K.S., Barber B.K., Miller B.H. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiological Genomics. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrews J.L., Zhang X., McCarthy J.J., McDearmon E.L., Hornberger T.A., Russell B. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dyar K.A., Ciciliot S., Wright L.E., Bienso R.S., Tagliazucchi G.M., Patel V.R. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Molecular Metabolism. 2014;3:29–41. doi: 10.1016/j.molmet.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harfmann B.D., Schroder E.A., Esser K.A. Circadian rhythms, the molecular clock, and skeletal muscle. Journal of Biological Rhythms. 2014 doi: 10.1177/0748730414561638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agley C.C., Rowlerson A.M., Velloso C.P., Lazarus N.L., Harridge S.D. Isolation and quantitative immunocytochemical characterization of primary myogenic cells and fibroblasts from human skeletal muscle. Journal of Visualized Experiments. 2015 Jan 12;(95):52049. doi: 10.3791/52049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu A.C., Tran H.G., Zhang E.E., Priest A.A., Welsh D.K., Kay S.A. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genetics. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujioka A., Takashima N., Shigeyoshi Y. Circadian rhythm generation in a glioma cell line. Biochemical and Biophysical Research Communications. 2006;346:169–174. doi: 10.1016/j.bbrc.2006.05.094. [DOI] [PubMed] [Google Scholar]
- 28.Pulimeno P., Mannic T., Sage D., Giovannoni L., Salmon P., Lemeille S. Autonomous and self-sustained circadian oscillators displayed in human islet cells. Diabetologia. 2013;56:497–507. doi: 10.1007/s00125-012-2779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saini C., Morf J., Stratmann M., Gos P., Schibler U. Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes & Development. 2012;26:567–580. doi: 10.1101/gad.183251.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes M.E., Hogenesch J.B., Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. Journal of Biological Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mannic T., Meyer P., Triponez F., Pusztaszeri M., Le Martelot G., Mariani O. Circadian clock characteristics are altered in human thyroid malignant nodules. The Journal of Clinical Endocrinology and Metabolism. 2013;98:4446–4456. doi: 10.1210/jc.2013-2568. [DOI] [PubMed] [Google Scholar]
- 32.Gaspar L., van de Werken M., Johansson A.S., Moriggi E., Owe-Larsson B., Kocks J.W. Human cellular differences in cAMP–CREB signaling correlate with light-dependent melatonin suppression and bipolar disorder. European Journal of Neuroscience. 2014;40:2206–2215. doi: 10.1111/ejn.12602. [DOI] [PubMed] [Google Scholar]
- 33.Balsalobre A., Brown S.A., Marcacci L., Tronche F., Kellendonk C., Reichardt H.M. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 34.Dendorfer U., Oettgen P., Libermann T.A. Multiple regulatory elements in the interleukin-6 gene mediate induction by prostaglandins, cyclic AMP, and lipopolysaccharide. Molecular and Cellular Biology. 1994;14:4443–4454. doi: 10.1128/mcb.14.7.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown S.A., Fleury-Olela F., Nagoshi E., Hauser C., Juge C., Meier C.A. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biology. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baggs J.E., Price T.S., DiTacchio L., Panda S., Fitzgerald G.A., Hogenesch J.B. Network features of the mammalian circadian clock. PLoS Biology. 2009;7:e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debruyne J.P., Noton E., Lambert C.M., Maywood E.S., Weaver D.R., Reppert S.M. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 38.Agorastos A., Hauger R.L., Barkauskas D.A., Moeller-Bertram T., Clopton P.L., Haji U. Circadian rhythmicity, variability and correlation of interleukin-6 levels in plasma and cerebrospinal fluid of healthy men. Psychoneuroendocrinology. 2014;44:71–82. doi: 10.1016/j.psyneuen.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 39.Vgontzas A.N., Bixler E.O., Lin H.M., Prolo P., Trakada G., Chrousos G.P. IL-6 and its circadian secretion in humans. Neuroimmunomodulation. 2005;12:131–140. doi: 10.1159/000084844. [DOI] [PubMed] [Google Scholar]
- 40.Philippe J., Dibner C. Thyroid circadian timing: roles in physiology and thyroid malignancies. Journal of Biological Rhythms. 2014 doi: 10.1177/0748730414557634. [DOI] [PubMed] [Google Scholar]
- 41.Gibbs J.E., Blaikley J., Beesley S., Matthews L., Simpson K.D., Boyce S.H. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pedersen B.K., Febbraio M.A. Muscle as an endocrine organ: focus on muscle-derived interleukin-6. Physiological Reviews. 2008;88:1379–1406. doi: 10.1152/physrev.90100.2007. [DOI] [PubMed] [Google Scholar]
- 43.Harder-Lauridsen N.M.N., Krogh-Madsen R., Holst J.J., Plomgaard P., Leick L., Pedersen B.K.B. The effect of IL-6 on the insulin sensitivity in patients with type 2 diabetes. American Journal of Physiology Endocrinology and Metabolism. 2014 Apr 1;306(7):E769–E778. doi: 10.1152/ajpendo.00571.2013. [DOI] [PubMed] [Google Scholar]
- 44.Carey A.L., Steinberg G.R., Macaulay S.L., Thomas W.G., Holmes A.G., Ramm G. Interleukin-6 increases insulin-stimulated glucose disposal in humans and glucose uptake and fatty acid oxidation in vitro via AMP-activated protein kinase. Diabetes. 2006;55:2688–2697. doi: 10.2337/db05-1404. [DOI] [PubMed] [Google Scholar]
- 45.Weigert C., Hennige A.M., Lehmann R., Brodbeck K., Baumgartner F., Schauble M. Direct cross-talk of interleukin-6 and insulin signal transduction via insulin receptor substrate-1 in skeletal muscle cells. Journal of Biological Chemistry. 2006;281:7060–7067. doi: 10.1074/jbc.M509782200. [DOI] [PubMed] [Google Scholar]
- 46.Wueest S., Item F., Boyle C.N., Jirkof P., Cesarovic N., Ellingsgaard H. Interleukin-6 contributes to early fasting-induced free fatty acid mobilization in mice. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology. 2014;306:R861–R867. doi: 10.1152/ajpregu.00533.2013. [DOI] [PubMed] [Google Scholar]
- 47.Nieto-Vazquez I., Fernandez-Veledo S., de Alvaro C., Lorenzo M. Dual role of interleukin-6 in regulating insulin sensitivity in murine skeletal muscle. Diabetes. 2008;57:3211–3221. doi: 10.2337/db07-1062. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Wallenius V., Wallenius K., Ahren B., Rudling M., Carlsten H., Dickson S.L. Interleukin-6-deficient mice develop mature-onset obesity. Nature Medicine. 2002;8:75–79. doi: 10.1038/nm0102-75. [DOI] [PubMed] [Google Scholar]
- 49.Kalsbeek A., Fliers E. Daily regulation of hormone profiles. Handbook of Experimental Pharmacology. 2013:185–226. doi: 10.1007/978-3-642-25950-0_8. [DOI] [PubMed] [Google Scholar]
- 50.Dibner C., Sage D., Unser M., Bauer C., d'Eysmond T., Naef F. Circadian gene expression is resilient to large fluctuations in overall transcription rates. The EMBO Journal. 2009;28:123–134. doi: 10.1038/emboj.2008.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mauvoisin D., Wang J., Jouffe C., Martin E., Atger F., Waridel P. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:167–172. doi: 10.1073/pnas.1314066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reddy A.B., Karp N.A., Maywood E.S., Sage E.A., Deery M., O'Neill J.S. Circadian orchestration of the hepatic proteome. Current Biology. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 53.Greco J.A., Oosterman J.E., Belsham D.D. Differential effects of omega-3 fatty acid docosahexaenoic acid and palmitate on the circadian transcriptional profile of clock genes in immortalized hypothalamic neurons. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology. 2014;307:R1049–R1060. doi: 10.1152/ajpregu.00100.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Journiac N., Jolly S., Jarvis C., Gautheron V., Rogard M., Trembleau A. The nuclear receptor ROR(alpha) exerts a bi-directional regulation of IL-6 in resting and reactive astrocytes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21365–21370. doi: 10.1073/pnas.0911782106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simpson R.J., Hammacher A., Smith D.K., Matthews J.M., Ward L.D. Interleukin-6: structure-function relationships. Protein Science. 1997;6:929–955. doi: 10.1002/pro.5560060501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zozulinska D., Majchrzak A., Sobieska M., Wiktorowicz K., Wierusz-Wysocka B. Serum interleukin-8 level is increased in diabetic patients. Diabetologia. 1999;42:117–118. doi: 10.1007/s001250051124. [DOI] [PubMed] [Google Scholar]
- 57.Hermann C., von Aulock S., Dehus O., Keller M., Okigami H., Gantner F. Endogenous cortisol determines the circadian rhythm of lipopolysaccharide–but not lipoteichoic acid–inducible cytokine release. European Journal of Immunology. 2006;36:371–379. doi: 10.1002/eji.200535470. [DOI] [PubMed] [Google Scholar]
- 58.Chitu V., Stanley E.R. Colony-stimulating factor-1 in immunity and inflammation. Current Opinion in Immunology. 2006;18:39–48. doi: 10.1016/j.coi.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 59.Jensen L.D., Cao Y. Clock controls angiogenesis. Cell Cycle (Georgetown, Tex) 2013;12:405–408. doi: 10.4161/cc.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elias I., Franckhauser S., Ferre T., Vila L., Tafuro S., Munoz S. Adipose tissue overexpression of vascular endothelial growth factor protects against diet-induced obesity and insulin resistance. Diabetes. 2012;61:1801–1813. doi: 10.2337/db11-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hagberg C.E., Mehlem A., Falkevall A., Muhl L., Fam B.C., Ortsater H. Targeting VEGF-B as a novel treatment for insulin resistance and type 2 diabetes. Nature. 2012;490:426–430. doi: 10.1038/nature11464. [DOI] [PubMed] [Google Scholar]
- 62.Kodama K., Horikoshi M., Toda K., Yamada S., Hara K., Irie J. Expression-based genome-wide association study links the receptor CD44 in adipose tissue with type 2 diabetes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7049–7054. doi: 10.1073/pnas.1114513109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu L.F., Kodama K., Wei K., Tolentino L.L., Choi O., Engleman E.G. The receptor CD44 is associated with systemic insulin resistance and proinflammatory macrophages in human adipose tissue. Diabetologia. 2015;58:1579–1586. doi: 10.1007/s00125-015-3603-y. [DOI] [PubMed] [Google Scholar]
- 64.Kodama K., Toda K., Morinaga S., Yamada S., Butte A.J. Anti-CD44 antibody treatment lowers hyperglycemia and improves insulin resistance, adipose inflammation, and hepatic steatosis in diet-induced obese mice. Diabetes. 2015;64:867–875. doi: 10.2337/db14-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Donath M.Y. Targeting inflammation in the treatment of type 2 diabetes: time to start. Nature Reviews Drug Discovery. 2014;13:465–476. doi: 10.1038/nrd4275. [DOI] [PubMed] [Google Scholar]
- 66.Green C.J., Pedersen M., Pedersen B.K., Scheele C. Elevated NF-kappaB activation is conserved in human myocytes cultured from obese type 2 diabetic patients and attenuated by AMP-activated protein kinase. Diabetes. 2011;60:2810–2819. doi: 10.2337/db11-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.