Abstract
Bacterial lipoproteins are lipid-anchored proteins that contain acyl groups covalently attached to the N-terminal cysteine residue of the mature protein. Lipoproteins are synthesized in precursor form with an N-terminal signal sequence (SS) that targets translocation across the cytoplasmic or inner membrane (IM). Lipid modification and SS processing take place at the periplasmic face of the IM. Outer membrane (OM) lipoproteins take the localization of lipoproteins (Lol) export pathway, which ends with the insertion of the N-terminal lipid moiety into the inner leaflet of the OM. For many lipoproteins, the biogenesis pathway ends here. We provide examples of lipoproteins that adopt complex topologies in the OM that include transmembrane and surface-exposed domains. Biogenesis of such lipoproteins requires additional steps beyond the Lol pathway. In at least one case, lipoprotein sequences reach the cell surface by being threaded through the lumen of a beta-barrel protein in an assembly reaction that requires the heteropentomeric Bam complex. The inability to predict surface exposure reinforces the importance of experimental verification of lipoprotein topology and we will discuss some of the methods used to study OM protein topology.
Keywords: localization of lipoproteins pathway, protein topology, surface-exposed protein, RcsF, Bam complex
1. Introduction
Bacterial lipoproteins are important components of the Gram-negative cell envelope. Although the name ‘lipoprotein’ is often used to describe a non-covalent assembly of proteins and lipids, bacterial lipoproteins belong to the class of so-called lipid-anchored proteins. These proteins contain covalently attached acyl groups as a result of a post-translational modification. Eukaryotic lipid-anchored proteins can be modified at different sites and the modifications are sometimes reversible and may be involved in regulation of the protein's activity [1]. By contrast, bacterial lipoproteins are modified at a specific site corresponding to the N-terminal Cys residue of the mature lipoprotein [2–4]. This modification is irreversible and happens on the periplasmic site of inner membrane (IM). The lipid moiety causes membrane association of a protein that usually does not contain a hydrophobic membrane targeting domain. Lipoproteins are then sorted to the IM or the outer membrane (OM) based on the sequences which follow the conserved Cys residue. Lipoproteins which are destined to OM are transported there by the localization of lipoproteins (Lol) pathway and inserted in the inner leaflet of the OM [5]. For several decades, lipoproteins were assumed to be simply soluble periplasmic proteins tethered to the OM by their lipid, partly because methods to study OM topology had not been developed. In this review, we will demonstrate, using Escherichia coli as a model organism, that for many lipoproteins biogenesis does not end with the Lol pathway. Lipoproteins can be further translocated onto the cell surface and they can be assembled into OM protein (OMP) complexes with transmembrane or surface-exposed topologies. We will also discuss some of the commonly used techniques to study the topology of proteins in the OM with its unique barrier properties.
2. Lipoprotein maturation and processing
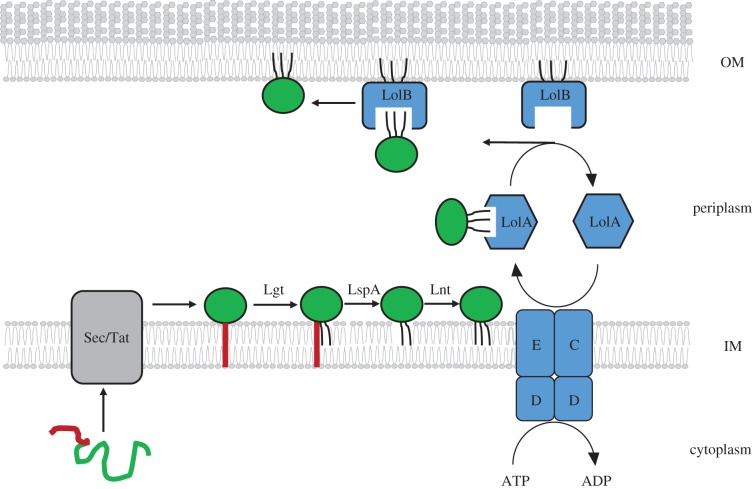
Lipoproteins are synthesized in the cytoplasm as protein precursors with an N-terminal signal sequence (SS) for transport from the cytoplasm. The lipoprotein SS contains a characteristic consensus sequence [LVI][ASTVI][GAS]C known as a lipobox [6,7]. In E. coli, lipoproteins are translocated across the IM by the Sec translocon, but in other organisms there are lipoproteins dependent on Tat translocon [8,9]. Maturation and processing of lipoproteins takes place on the periplasmic side of IM (figure 1). The first step is the addition of diacylglycerol to the sulfhydryl group of the conserved Cys residue by phosphatidylglycerol/prolipoprotein diacylglyceryl transferase (Lgt) [10]. This modification is a prerequisite for the SS cleavage by a dedicated lipoprotein signal peptidase (LspA or signal peptidase II) [2,11,12] and ensures that the apolipoprotein remains anchored in the IM after SS cleavage. Signal peptidase generates diacylated apolipoprotein with a new N-terminus provided by the conserved Cys of the lipobox that is referred to as +1 [3]. Finally, the amino group of the Cys residue is N-acylated by phospholipid/apolipoprotein transacylase (Lnt) generating mature triacylated lipoprotein [13,14] (figure 1). Genes encoding for all three enzymes are essential in E. coli and highly conserved across Gram-negative bacteria [5]. However, lnt was recently found to be dispensable for viability of Francisella tularensis and Neisseria gonorrhoeae [15], suggesting that in some Gram-negative bacteria, mature lipoporteins can exist in a diacylated form similar to what is observed in low GC Gram-positive organisms which do not encode Lnt homologues [16].
Figure 1.
Lipoprotein maturation and export pathway. Lipoprotein (green) is synthesized in the cytoplasm with the N-terminal SS (red) which targets it for translocation across the IM by the Sec or Tat translocon. The lipoprotein remains anchored in the IM by its SS and Lgt adds a diacylglyceryl moiety to the Cys residue. LspA cleaves the SS and Lnt adds another acyl chain to the newly formed N-terminus. The lipoprotein is then recognized by IM LolCDE complex which powers extraction of the lipoprotein from the IM using the energy of ATP. The lipoprotein is released to the periplasm in a complex with the chaperone LolA. LolA delivers lipoprotein to the OM acceptor protein LolB which inserts it in the inner leaflet of the OM. Empty LolA returns to LolCDE and is recycled. (Online version in colour.)
3. Export of lipoproteins to the outer membrane
Most of the mature lipoproteins in E. coli are targeted for translocation to the OM by the Lol pathway unless they contain a so-called Lol avoidance signal. The Lol avoidance signal in E. coli is known as the +2 rule because it is determined by the identity of the amino acid after the conserved Cys [17]. According to this rule, an Asp residue at position +2 causes IM retention of the lipoprotein; it serves as a sorting signal that differentiates IM and OM lipoproteins [17]. Although all native IM lipoproteins in E. coli have Asp at +2 and either Asp, Glu or Gln at position +3, additional combinations of +2 and +3 residues could act as IM retention signals [18–20]. Although the +2 rule is generally conserved in enterobacteria [21], the rule does not always apply for species outside this family. For example, in Pseudomonas aeruginosa amino acids at positions +3 and +4 also play a critical role in lipoprotein sorting [22,23]. OM lipoproteins in Borrelia do not follow the +2 rule either [24].
Lipoproteins destined for the OM are translocated by the Lol proteins (figure 1). The Lol pathway was discovered and characterized by a series of elegant biochemical experiments in the Tokuda laboratory. In E. coli, five proteins (LolA–E) are involved in lipoprotein transport. In the first step, the OM lipoprotein is extracted from the IM by the LolCDE complex and released to the periplasm in the form of a soluble complex with the chaperone LolA [25,26]. This step is energy-dependent and driven by ATP hydrolysis by the ATPase LolD [26,27]. LolD is a cytoplasmic ABC-type ATPase tethered to the IM by the interaction with the homologous integral IM proteins LolC and LolE [26]. Although homologous LolC and LolE play different functions in lipoprotein extraction from the IM, LolE recognizes and binds lipoprotein substrates [28], whereas LolC recruits the chaperone LolA [29]. ATP hydrolysis powers the transfer of the lipoprotein directly from LolE to LolA [28]. A large hydrophobic cavity within LolA binds the acyl chains of protein substrates shielding this highly hydrophobic region from the aqueous periplasm [30]. LolA delivers lipoprotein substrates to the OM acceptor protein LolB [31]. LolB is an OM lipoprotein itself and is a structural homologue of LolA [30]. It also contains a hydrophobic cavity that has a higher affinity for the acyl chains than LolA [32]. This difference in affinity allows unidirectional ‘mouth-to-mouth’ transfer of the substrate from LolA to LolB [32]. The hydrophobic cavity of LolA undergoes conformational changes between the ‘open’ or substrate-bound and the ‘closed’ or substrate-free form [33]. After lipoprotein release, LolA in its closed form returns to the LolCDE complex and is recycled (figure 1). Lipoprotein release from LolB in the OM is not well understood, but the protruding loop of LolB is somehow important for this final step in lipoprotein transport [34,35].
The lol genes are essential in E. coli and homologues can be found in all Gram-negative bacteria, suggesting that the pathway is conserved. However, conservation of individual genes varies. LolC and LolE are homologues but cannot functionally substitute each other in E. coli [28]. However, some bacterial genomes contain only one copy of a lolC/E gene [15]; interestingly, in such cases, the protein product contains sequence motifs of both LolC and LolE and likely represents a functional hybrid of both proteins. The LolF name was proposed to distinguish such proteins from obvious LolC and LolE homologues [15]. lolB is the other gene that is only conserved in β- and γ-proteobacteria [5]. It is not clear whether other Gram-negative bacteria contain functional analogues which are not related in sequence to LolB or encode a functional hybrid of LolA and LolB since these proteins also have similar structures.
4. Lipoprotein destiny after outer membrane insertion
Until recently, lipoprotein insertion in the OM by LolB was the last known step for lipoprotein export in E. coli. Therefore, the paradigm for OM lipoprotein topology was established that all lipoproteins (at least in E. coli) are simply anchored in the inner leaflet of OM by their lipid moiety with their protein domain facing the periplasm (figure 2). Many surface-exposed lipoproteins have been identified in a number of different organisms [36]. In those organisms, surface-exposed lipoproteins have variety of functions: they participate in iron uptake [37–40]; are enzymes such as phospholipases [41], PPIases [42] or glucanases [43]; they participate in adhesion and binding of host factors [44,45]; and others. In some organisms, like Borrelia, lipoproteins are anchored in the outer leaflet and exposed on the cell surface by default [24].
Figure 2.
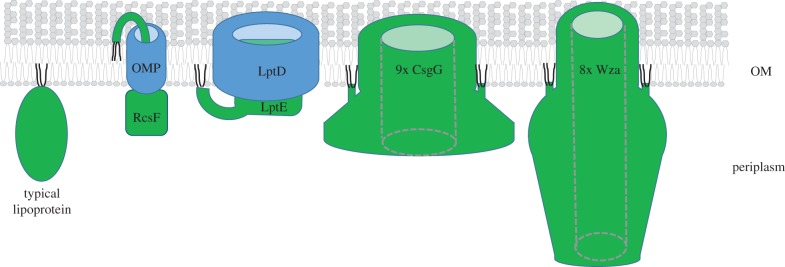
Examples of topologies of OM lipoproteins in E. coli. OM lipoproteins (green) were assumed to adopt a typical topology with the protein domain in the periplasm and lipid anchored in the inner leaflet of the OM. By contrast, the topology of experimentally studied lipoproteins is often more complex. Lipoproteins can be assembled in interlocked complexes with beta-barrel proteins (blue) like RcsF/OMP and LptE/LptD, or they can form transmembrane multimeric channels like CsgG and Wza. Assembly of the lipoproteins at the OM often results in surface exposure of parts of the protein (RcsF, CsgG and Wza). In the case of RcsF, the lipid moiety anchors the protein in the outer leaflet of OM. (Online version in colour.)
In most cases, how lipoproteins get to the cell surface is unclear. Surface-associated lipoprotein pullulanase (PulA) of Klebsiella oxytoca was the first lipoprotein for which the export pathway was identified [43,46]. PulA contains a Lol avoidance signal and is instead exported directly from the IM to the cell surface by a specific transporter Pul, also known as a type 2 secretion system [43,47]. Other known lipoprotein substrates of type 2 secretion systems are surface-exposed lipoprotein cytochromes in Shewanella oneidensis [48] and lipoprotein SslE in enteropathogenic E. coli [49].
Neisseria meningitidis NalP and Bordetella pertussis SphB1 are autotransporters which contain a lipoprotein SS [50–53]. These proteins have a domain architecture typical of autotransporters: a C-terminal translocation domain that is required for the secretion of an N-terminal passenger domain [54]. However, due to the presence of N-terminal lipid on the passenger domain, it must remain associated with the OM, at least for a while, where it functions as a surface protease [50,51].
Many reports of surface-exposed lipoproteins from different organisms are in contrast to the E. coli periplasmic paradigm. However, of the 90 lipoproteins in E. coli fewer than a dozen have been studied experimentally, mainly in connection to peptidoglycan (PG) and envelope biogenesis. Of those lipoproteins that have been examined, many adopt quite complex topologies in the OM: they can exist in complexes with beta-barrel proteins, form transmembrane channels or be surface exposed.
5. Complex topologies of outer membrane lipoproteins
(a). Plug in the barrel
LptE is the OM lipoprotein component of the LPS transport (Lpt) machine; together with its beta-barrel partner LptD, it forms an OM subcomplex that is required for LPS insertion into the outer leaflet of OM [55,56]. N-terminal lipids anchor LptE in the inner leaflet of OM; however, instead of facing the periplasmic space, LptE is almost completely buried inside the large lumen of LptD [57–59] (figure 2).
The interaction between LptE and LptD is highly specific and the complex is very stable [59]. This is achieved by a large number of specific contacts sites between LptE and LptD covering an interaction interface with the surface area of more than 3000 Å2 [57,58]. LptE serves as a scaffold for the folding of the LptD barrel at the Bam complex, and no LptD protein can be assembled without its partner; unassembled LptD is rapidly degraded [60,61]. In addition to its role in LptD folding, LptE functions as a plug that prevents small molecule entry through the LptD barrel and it plays a direct role in the surface assembly of the LPS glycolipid [62,63].
(b). Integral outer membrane lipoproteins
There are two lipoproteins with transmembrane topology in E. coli, CsgG and Wza. Both lipoproteins are OM components of secretion channels. CsgG functions in the transport of curli subunits [64] and Wza transports group 1 (serotype K30) capsular polysaccharides [65]. The lipids of both CsgG and Wza are anchored in the inner leaflet of OM and both proteins form oligomeric complexes with hydrophobic transmembrane domains and large periplasmic domains [66–68]. Each oligomeric complex features a central channel used for the secretion of the corresponding substrates [66–68]. Despite the similarity in topology, CsgG and Wza have completely different structures and interact with the OM in different manners.
The CsgG channel is an oligomer of nine CsgG subunits [66,67] (figure 2). Each subunit has alpha–beta structure, and the transmembrane domain is made by amphipathic beta strands. Each subunits donates four beta strands building up a composite 36-stranded beta-barrel which forms a transmembrane channel with an inner width of 40 Å. Although CsgG does not have long extracellular loops typical of many OMPs, it still can be detected on the cell surface by CsgG antibodies [69]. The large periplasmic domain is made of a mixture of α-helices and β-strands, some of which extend from the transmembrane beta-barrel. The periplasmic domain of the CsgG nonamer also features a large cavity that is open on the periplasmic side but gated in vivo by a partner protein CsgE [67]. Based on the cryo-EM structure of the CsgG/E complex as well as conductance measurements of the CsgG and CsgG/E channels, a model was proposed in which CsgE binding to CsgG entraps curli subunit protein CsgA inside the CsgG chamber, creating conditions that favour diffusion out through CsgG pore [67,70]. After diffusion, CsgA subunits are nucleated into curli amyloid fibres by CsgB [71]. Curli play an important role in adhesion, biofilm formation and colonization of the host [72].
Wza has different structure and domain organization [68]. Wza is an octamer with four rings (R1–R4) of individual domains (figure 2). The Wza octamer is described as an amphora without handles, in which the R1–R3 domains make up the periplasmic ‘vessel’ domain and R4 makes the integral transmembrane ‘neck’ domain. The transmembrane domains of each subunit are made by one long amphiphatic helix, with the C-terminus exposed on the cell surface. Eight amphiphatic helices create an alpha-barrel with a hydrophobic exterior for membrane insertion and a hydrophilic interior for polysaccharide export. Thus, Wza represents a new class of integral OMPs with a transmembrane domain in the form of a barrel made of α-helices. At the centre of the octamer, there is long hydrophilic cavity. Although the structure shows the periplasmic side of the Wza octamer is closed, it can probably undergo a conformation change to open a translocation channel for polysaccharide chain extrusion [73].
Assembly of CsgG and Wza oligomers at the OM does not require the Bam complex, but the assembly pathways remain known [74]. In both cases, the lipid moiety is required for formation of functional multimers. When lipid is absent, as a result of SS replacement, CsgG proteins accumulate as soluble monomers in the periplasm and the few multimers that can be observed have the incorrect symmetry. By contrast, lipid-less Wza can be still targeted to the OM to some extent, but no stable, functional multimers are formed [75]. Interestingly, when both the transmembrane helix and the lipid are removed, soluble multimers of the R1–R3 domains are formed similar to what are observed in the full-length Wza structure [76,77]. Hence, the lipid moiety of both proteins is specifically required to direct multimer formation into the OM.
(c). Surface-exposed lipoproteins
Perhaps another reason that contributed to the periplasmic paradigm for lipoproteins is that the two best-studied lipoproteins in E. coli, Lpp and Pal, have clearly defined periplasmic functions where they interact with the cell wall. Lpp, also known as Braun's lipoprotein, is the major lipoprotein in E. coli. It is a small 6 kDa protein that exists in two forms: one known as the ‘bound form’ represents Lpp molecules that are covalently cross-linked to PG [78,79]; another fraction is not attached to the PG and is known as the ‘free form’ [80]. The function of the free form is not understood. In contrast to Lpp, Pal interacts with the PG by non-covalent binding [81]. Both lipoproteins contribute to general envelope stability by attaching the OM to the cell wall and null mutations in either gene causes increased OM vesiculation, sensitivity to detergents and other envelope-related defects [82–84]. Both Lpp and Pal were used as model lipoproteins in studies of the Lol pathway [25,26,31]. Because of their clear function in the attachment of the OM to the cell wall, periplasmic localization of these proteins had never been questioned until we discovered that the ‘free form’ of Lpp adopts a cell surface-exposed orientation [85]. We uncovered this topology of Lpp in our effort to characterize the surface proteome of E. coli by selective surface biotinylation: Lpp was one of the major proteins that are labelled in E. coli. Although we do not know the precise topology of surface-exposed Lpp, studies of biotinylation sites demonstrate that at least the C-terminus of Lpp is exposed on the cell surface [85]. Since then, the topology of Pal has also been challenged and new data demonstrate that Pal, like Lpp, may exist in this dual orientation with a fraction of the protein being surface exposed [86]. Additional surface-exposed lipoproteins reported in E. coli are TraT [87], BamC [88] and YaiW [89].
The surface exposure of Lpp posed the question as to whether there is a specific transporter to localize lipoproteins to the cell surface. However, because the function of free form Lpp is not known, we decided to look for a more experimentally tractable system to probe the mechanism(s) that catalyse lipoprotein surface exposure.
The lipoprotein RcsF stood out because of its function as a sensory component of Rcs envelope stress response [90,91]. The Rcs stress response monitors envelope integrity and can be activated by a number of cues such as LPS stress [92,93], PG stress [94,95], osmotic stress [96,97] and defects in lipoprotein biogenesis [98,99]. Our hypothesis that RcsF may localize to the outer leaflet to sense LPS defects led to the discovery of yet another surface-exposed lipoprotein in E. coli [100]. Later, it was shown that RcsF activation to the PG stress does not require surface localization RcsF [101], suggesting distinct mechanisms for monitoring integrity of different envelope compartments.
Not only is RcsF a surface-exposed lipoprotein, it displays a novel OM topology: the N-terminal domain and the lipids of RcsF are surface exposed, while the C-terminal domain resides in the periplasm (figure 2). How could RcsF span the OM if it does not contain a transmembrane domain? The solution to this topological problem proved to rely on RcsF interaction with several OMPs via a short amino acid sequence which exists in an extended conformation that is threaded through the OMP lumen where it is shielded from the hydrophobic membrane interior. Therefore, like LptE, RcsF is also found in an interlocked complex with a beta-barrel protein. However, there are number of important differences between RcsF/OMP and LptE/D complexes. Unlike LptE, RcsF interacts with the beta-barrel via a short unstructured region only, without extensive site-specific contacts. This lack of barrel-specificity is further evidenced by the fact that RcsF is capable of interacting with several different OMPs such as OmpA, OmpC and OmpF. It is likely that the presence of an unstructured region, rather than sequence specificity, is the requirement for formation RcsF/OMP complexes. Strikingly, a similar type of interaction was observed for the unstructured linker region of a colicin during its entry through the OmpF lumen [102,103]. Finally, LptE is absolutely required for the folding and assembly of LptD. RcsF is not required for the folding and assembly of any OMP.
Having established the surface exposure and topology of RcsF, we wanted to examine the assembly mechanism responsible. How can the unstructured region of RcsF be threaded into the lumen of an OMP? RcsF cannot enter the lumen from N-terminus because even though it is unstructured [104,105], it is tethered to the membrane by a lipid moiety that cannot be extracted from the membrane without an energy source, and even if it could be, the hydrophilic lumen of a folded OMP would not allow the passage of the hydrophobic lipid. Likewise, the folded C-terminal domain of RcsF cannot pass through the lumen because it is simply too big and is folded at an early step of RcsF biogenesis with the help of periplasmic disulfide oxidase DsbA, and the disulfide isomerase DsbC, which catalyse the formation of the two non-consecutive disulfide bonds necessary to stabilize the structure [104,105]. Therefore, to explain the transmembrane topology, it seemed necessary to suggest that the OMP beta-barrel is folded around the RcsF during the process of OMP assembly.
We have provided two lines of evidence to support the hypothesis that the RcsF/OMP interaction is formed during OMP assembly. First, we showed that RcsF cannot form a complex with folded OmpA in vitro; however, the complex can be reconstituted when urea-denatured OmpA is refolded in the presence of RcsF. Second, all OMPs are folded in vivo by the heteropentomeric Bam complex [106], and we showed that RcsF also interacts with BamA, an essential component of the Bam complex. Although BamA is itself a beta-barrel, site-specific cross-linking revealed that the RcsF/BamA interaction is different from the RcsF/OMP interaction, suggesting that the RcsF/BamA cross-linking observed could represent a step in the assembly pathway of RcsF/OMP at the Bam complex. Indeed, when the function of the Bam complex is compromised, fewer RcsF/OMP complexes are assembled at the OM. These results argue that Bam complex catalyses RscF surface exposure by assembling the RcsF/OMP complexes at the OM.
The study of RcsF uncovered novel functions for Bam complex in the biogenesis of surface-exposed lipoproteins. BamA can recognize lipoproteins and translocate the lipidated N-terminus into the outer leaflet and then assemble the OMP beta-barrel around the short, unstructured linker between the N- and C-terminus. We think it is likely that other lipoproteins exist in similar OMP complexes in E. coli as well as other bacteria and that the Bam complex is involved in their assembly. Clearly, as is demonstrated by PulA, the Bam complex is not required for the surface localization of every lipoprotein. We think it is possible that other structural groups (e.g. α-helical proteins) have their own dedicated assembly pathway.
6. Common methods to study topology of outer membrane lipoproteins
The examples above demonstrate that lipoproteins vary in their structures and topologies, and the latter has to be experimentally determined. Generally, any method commonly used to study membrane protein topology can be employed to study OM lipoproteins [107,108] but the unique features of the OM have to be taken into account when designing the experiment. All methods can be divided into three groups: those based on protein labelling; based on antibody binding; and accessibility for proteolysis.
Techniques based on selective protein labelling use compounds that chemically modify certain amino acids in the protein and allow the detection of the modified protein. The two most commonly used chemistries are N-hydroxysuccinimide (NHS) esters which react with primary amines (e.g. protein lysines), or maleimides which react with sulfhydryls (e.g. non-oxidized cysteines) [109]. These functional groups can be coupled chemically to biotin and this allows detection of the modifications using anti-biotin antibodies or streptavidin conjugates. PEG linkers of different length, which allow detection of the modification based on a protein size shift during immunoblot analysis, can also be used. There are a huge variety of labels that vary in their size and hydrophobicity, and choosing the right one requires understanding the two important properties of the OM that differentiate it from other biological membranes [110]. First, the OM is not permeable to hydrophobic compounds because of the presence of charged, highly interactive lipopolysaccharide molecules that cover the cell surface. Second, the OM is permeable to the small soluble compounds due to the presence of porins which allow water and nutrients to freely enter the cell [110]. Therefore, selection of compounds for labelling OM proteins is somewhat counterintuitive compared to other membrane proteins: hydrophobic compounds will selectively label the cell surface [85], while small hydrophilic compounds can enter the periplasm and can label protein on both sides of the OM. The size exclusion limit of porins in E. coli was determined to be around 600 Da [111], so using water-soluble labels of much larger molecular weight will result in selective surface labelling as well. In our laboratory, we used a hydrophobic biotin label to study Lpp surface exposure [85] as well as for the initial discovery of surface-exposed RcsF [100]. We also used a hydrophilic maleimide with high molecular weight to study the conformation change in extracellular loop 6 of BamA [112]. It is worth mentioning that perhaps not all Gram-negative bacteria have such an asymmetric OM and the natural resistance to hydrophobic OM compounds that E. coli does, and protein labels should be evaluated in each case.
Surface proteolysis is another commonly used method to test protein surface exposure [46,113]. Extracellularly added proteases cannot enter the cell and hence periplasmic proteins are protected from cleavage while surface-exposed proteins are degraded. Cleavage or complete degradation can be analysed by immunoblotting. However, it should be noted that many surface-exposed proteins in bacteria are inherently protease resistant, so negative results cannot be interpreted with confidence, even if proper controls have been done.
Using extracellularly added antibodies follows the same logic as surface proteolysis. Antibodies can bind only surface-exposed epitopes and this binding can be detected either by using antibodies labelled with fluorescent dyes or enzyme conjugates. Whole-cell antibody-based assays include dot blots, whole cell ELISA, immunofluorescence and FACS analysis [114–116]. However, cell fixation techniques can disrupt OM integrity as well as producing other artefacts and it is best for antibody labelling to be performed with live cells [117].
Labelling techniques, especially biotinylation, are widely used for proteomic discovery of surface-exposed proteins because they do not require the generation of protein-specific antibodies: labelled proteins can be easily affinity purified and analysed by mass spectrometry [118,119]. Quantitative proteomics can be also employed to study surface-exposed proteins by comparing relative abundance of the protein before and after the protease treatment [120]. Employing a combination of different methods provides more reliable identification of surface-exposed proteins [113].
All of the methods described can be used to study native proteins and do not require making protein fusions or mutants for site-specific labelling. However, the limitation is that ability to detect a protein with any of the methods depends not only on protein localization to the cell surface but also on the presence of accessible sites for labelling, proteolysis or antibody binding. Lack of such sites can generate false-negative results. One example of such false-negative results that we have encountered is BamC. BamC is an OM lipoprotein component of the Bam complex [106]. We have highly specific polyclonal anti-BamC antibodies [121] which do not bind to whole cells [100] and we could not detect BamC biotinylation [85]. Therefore, we concluded that BamC is a periplasmic lipoprotein, and we have used BamC as a negative-control for lipoprotein surface exposure [85,100]. However, independently raised polyclonal antibodies against BamC can recognize the protein on the cell surface, indicating that at least part of BamC is surface exposed [88]. The apparent discrepancy between these results can be explained by differences in antibody epitope recognition. Identifying these epitopes would help in understanding the topology of BamC. It is likely that only a portion of the protein is exposed on the cell surface; the structure of BamC/BamD complex shows that the N-terminal domain of BamC is involved in the interaction with periplasmically localized lipoprotein BamD [122].
One of the other disadvantages of using these methods to study native proteins is that they seldom give information about precise OM topology. Therefore, a number of approaches were developed to introduce labelling sites (e.g. Cys accessibility assay), proteolytic sites for specific proteases (e.g. TEV protease, enterokinase) or epitopes for antibody-binding (His, HA, FLAG, Strep epitopes) at specific positions of the protein. This method was successfully used to study OMPs. For example, labelling of native and engineered Cys was used in studies of the ferrichrome-binding extracellular loop of FhuA [123], and epitope insertions were used to study the topology of several OMPs in E. coli [124–127]. Insertion of proteolytic sites was used to determine that surface-exposed lipoproteins in Borrelia are anchored to the outer leaflet of OM [128]. These approaches are recommended only when assays to test protein function and/or folding are available to demonstrate that epitope insertions do not interfere with native protein topology. In our study of RcsF, we used biotinylation, proteolysis and polyclonal RcsF antibodies to show that RcsF is surface exposed [100]. However, FLAG epitope walking across the entire protein sequence allowed us to uncover the fact that RcsF has an N-out C-in topology.
7. Concluding remarks
The lipoprotein biogenesis pathway, including lipoprotein maturation and export to the OM by the Lol machine, has been characterized in great molecular detail. Because lipoprotein insertion into the inner leaflet of OM is the last step in the Lol pathway, the periplasmic paradigm for lipoproteins, in which all lipoproteins are viewed as soluble periplasmic proteins simply anchored to the inner leaflet of the OM by N-terminal lipid moieties, was established. This assumption delayed the discovery of many lipoproteins that interact with the OM differently: some lipoproteins are anchored in the outer leaflet of the OM, some are transmembrane proteins and some exist in protein complexes shielded from the periplasm. Identification of such proteins made us acknowledge that our long-held view on lipoprotein topology was oversimplified and our understanding of lipoprotein biogenesis was far from complete. We suggest that a periplasmic topology should no longer be assumed for OM lipoproteins but instead a predicted topology should be experimentally tested. We believe that this would lead to the discovery of even more examples of OM lipoproteins with unusual structures and topologies, presenting us with even more challenges for understanding OM biogenesis.
Acknowledgements
We thank Marcin Grabowicz for critical reading of the manuscript and members of Silhavy Lab for helpful discussion.
Authors' contributions
A.K. and T.J.S. contributed to conception, design and writing of the article. Both authors gave final approval for publication.
Competing interests
We have no competing interests.
Funding
The research in the laboratory of T.J.S. is supported by National Institute of General Medical Sciences grant GM34821.
References
- 1.Resh MD. 2013. Covalent lipid modifications of proteins. Curr. Biol. 23, R431–R435. ( 10.1016/j.cub.2013.04.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tokunaga M, Tokunaga H, Wu HC. 1982. Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc. Natl Acad. Sci. USA 79, 2255–2259. ( 10.1073/pnas.79.7.2255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inouye S, Franceschini T, Sato M, Itakura K, Inouye M. 1983. Prolipoprotein signal peptidase of Escherichia coli requires a cysteine residue at the cleavage site. EMBO J. 2, 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hantke K, Braun V. 1973. Covalent binding of lipid to protein: diglyceride and amide-linked fatty acid at the N-terminal end of the murein-lipoprotein of the Escherichia coli outer membrane. Eur. J. Biochem. 34, 284–296. ( 10.1111/j.1432-1033.1973.tb02757.x) [DOI] [PubMed] [Google Scholar]
- 5.Okuda S, Tokuda H. 2011. Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 65, 239–259. ( 10.1146/annurev-micro-090110-102859) [DOI] [PubMed] [Google Scholar]
- 6.von Heijne G. 1989. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 2, 531–534. ( 10.1093/protein/2.7.531) [DOI] [PubMed] [Google Scholar]
- 7.Hayashi S, Wu HC. 1990. Lipoproteins in bacteria. J. Bioenerg. Biomembr. 22, 451–471. ( 10.1007/BF00763177) [DOI] [PubMed] [Google Scholar]
- 8.Shruthi H, Babu MM, Sankaran K. 2010. TAT-pathway-dependent lipoproteins as a niche-based adaptation in prokaryotes. J. Mol. Evol. 70, 359–370. ( 10.1007/s00239-010-9334-2) [DOI] [PubMed] [Google Scholar]
- 9.Kudva R, Denks K, Kuhn P, Vogt A, Muller M, Koch HG. 2013. Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res. Microbiol. 164, 505–534. ( 10.1016/j.resmic.2013.03.016) [DOI] [PubMed] [Google Scholar]
- 10.Sankaran K, Wu HC. 1994. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269, 19 701–19 706. [PubMed] [Google Scholar]
- 11.Dev IK, Ray PH. 1984. Rapid assay and purification of a unique signal peptidase that processes the prolipoprotein from Escherichia coli B. J. Biol. Chem. 259, 11 114–11 120. [PubMed] [Google Scholar]
- 12.Yamagata H, Taguchi N, Daishima K, Mizushima S. 1983. Genetic characterization of a gene for prolipoprotein signal peptidase in Escherichia coli. Mol. Gen. Genet. 192, 10–14. ( 10.1007/BF00327640) [DOI] [PubMed] [Google Scholar]
- 13.Gupta SD, Gan K, Schmid MB, Wu HC. 1993. Characterization of a temperature-sensitive mutant of Salmonella typhimurium defective in apolipoprotein N-acyltransferase. J. Biol. Chem. 268, 16 551–16 556. [PubMed] [Google Scholar]
- 14.Gupta SD, Wu HC. 1991. Identification and subcellular localization of apolipoprotein N-acyltransferase in Escherichia coli. FEMS Microbiol. Lett. 62, 37–41. ( 10.1111/j.1574-6968.1991.tb04413.x) [DOI] [PubMed] [Google Scholar]
- 15.LoVullo ED, Wright LF, Isabella V, Huntley JF, Pavelka MS Jr. 2015. Revisiting the Gram-negative lipoprotein paradigm. J. Bacteriol. 197, 1705–1715. ( 10.1128/JB.02414-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchings MI, Palmer T, Harrington DJ, Sutcliffe IC. 2009. Lipoprotein biogenesis in Gram-positive bacteria: knowing when to hold 'em, knowing when to fold 'em. Trends Microbiol. 17, 13–21. ( 10.1016/j.tim.2008.10.001) [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi K, Yu F, Inouye M. 1988. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53, 423–432. ( 10.1016/0092-8674(88)90162-6) [DOI] [PubMed] [Google Scholar]
- 18.Gennity JM, Inouye M. 1991. The protein sequence responsible for lipoprotein membrane localization in Escherichia coli exhibits remarkable specificity. J. Biol. Chem. 266, 16 458–16 464. [PubMed] [Google Scholar]
- 19.Seydel A, Gounon P, Pugsley AP. 1999. Testing the '+2 rule' for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34, 810–821. ( 10.1046/j.1365-2958.1999.01647.x) [DOI] [PubMed] [Google Scholar]
- 20.Terada M, Kuroda T, Matsuyama SI, Tokuda H. 2001. Lipoprotein sorting signals evaluated as the LolA-dependent release of lipoproteins from the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 276, 47 690–47 694. ( 10.1074/jbc.M109307200) [DOI] [PubMed] [Google Scholar]
- 21.Lewenza S, Vidal-Ingigliardi D, Pugsley AP. 2006. Direct visualization of red fluorescent lipoproteins indicates conservation of the membrane sorting rules in the family Enterobacteriaceae. J. Bacteriol. 188, 3516–3524. ( 10.1128/JB.188.10.3516-3524.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narita S, Tokuda H. 2007. Amino acids at positions 3 and 4 determine the membrane specificity of Pseudomonas aeruginosa lipoproteins. J. Biol. Chem. 282, 13 372–13 378. ( 10.1074/jbc.M611839200) [DOI] [PubMed] [Google Scholar]
- 23.Lewenza S, Mhlanga MM, Pugsley AP. 2008. Novel inner membrane retention signals in Pseudomonas aeruginosa lipoproteins. J. Bacteriol. 190, 6119–6125. ( 10.1128/JB.00603-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulze RJ, Zuckert WR. 2006. Borrelia burgdorferi lipoproteins are secreted to the outer surface by default. Mol. Microbiol. 59, 1473–1484. ( 10.1111/j.1365-2958.2006.05039.x) [DOI] [PubMed] [Google Scholar]
- 25.Matsuyama S, Tajima T, Tokuda H. 1995. A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane. EMBO J. 14, 3365–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yakushi T, Masuda K, Narita S, Matsuyama S, Tokuda H. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2, 212–218. ( 10.1038/35008635) [DOI] [PubMed] [Google Scholar]
- 27.Yakushi T, Yokota N, Matsuyama S, Tokuda H. 1998. LolA-dependent release of a lipid-modified protein from the inner membrane of Escherichia coli requires nucleoside triphosphate. J. Biol. Chem. 273, 32 576–32 581. ( 10.1074/jbc.273.49.32576) [DOI] [PubMed] [Google Scholar]
- 28.Mizutani M, Mukaiyama K, Xiao J, Mori M, Satou R, Narita S, Okuda S, Tokuda H. 2013. Functional differentiation of structurally similar membrane subunits of the ABC transporter LolCDE complex. FEBS Lett. 587, 23–29. ( 10.1016/j.febslet.2012.11.009) [DOI] [PubMed] [Google Scholar]
- 29.Okuda S, Tokuda H. 2009. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc. Natl Acad. Sci. USA 106, 5877–5882. ( 10.1073/pnas.0900896106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeda K, Miyatake H, Yokota N, Matsuyama S, Tokuda H, Miki K. 2003. Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J. 22, 3199–3209. ( 10.1093/emboj/cdg324). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuyama S, Yokota N, Tokuda H. 1997. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 16, 6947–6955. ( 10.1093/emboj/16.23.6947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taniguchi N, Matsuyama S, Tokuda H. 2005. Mechanisms underlying energy-independent transfer of lipoproteins from LolA to LolB, which have similar unclosed β-barrel structures. J. Biol. Chem. 280, 34 481–34 488. ( 10.1074/jbc.M507388200) [DOI] [PubMed] [Google Scholar]
- 33.Oguchi Y, Takeda K, Watanabe S, Yokota N, Miki K, Tokuda H. 2008. Opening and closing of the hydrophobic cavity of LolA coupled to lipoprotein binding and release. J. Biol. Chem. 283, 25 414–25 420. ( 10.1074/jbc.M804736200) [DOI] [PubMed] [Google Scholar]
- 34.Tsukahara J, Mukaiyama K, Okuda S, Narita S, Tokuda H. 2009. Dissection of LolB function: lipoprotein binding, membrane targeting and incorporation of lipoproteins into lipid bilayers. FEBS J. 276, 4496–4504. ( 10.1111/j.1742-4658.2009.07156.x) [DOI] [PubMed] [Google Scholar]
- 35.Hayashi Y, Tsurumizu R, Tsukahara J, Takeda K, Narita S, Mori M, Miki K, Tokuda H. 2014. Roles of the protruding loop of factor B essential for the localization of lipoproteins (LolB) in the anchoring of bacterial triacylated proteins to the outer membrane. J. Biol. Chem. 289, 10 530–10 539. ( 10.1074/jbc.M113.539270) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuckert WR. 2014. Secretion of bacterial lipoproteins: through the cytoplasmic membrane, the periplasm and beyond. Biochim. Biophys. Acta 1843, 1509–1516. ( 10.1016/j.bbamcr.2014.04.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson JE, Sparling PF, Cornelissen CN. 1994. Gonococcal transferrin-binding protein 2 facilitates but is not essential for transferrin utilization. J. Bacteriol. 176, 3162–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ostberg KL, DeRocco AJ, Mistry SD, Dickinson MK, Cornelissen CN. 2013. Conserved regions of gonococcal TbpB are critical for surface exposure and transferrin iron utilization. Infect. Immunol. 81, 3442–3450. ( 10.1128/IAI.00280-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersson A, Prinz T, Umar A, van der Biezen J, Tommassen J. 1998. Molecular characterization of LbpB, the second lactoferrin-binding protein of Neisseria meningitidis. Mol. Microbiol. 27, 599–610. ( 10.1046/j.1365-2958.1998.00707.x) [DOI] [PubMed] [Google Scholar]
- 40.Dashper SG, Hendtlass A, Slakeski N, Jackson C, Cross KJ, Brownfield L, Hamilton R, Barr I, Reynolds EC. 2000. Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J. Bacteriol. 182, 6456–6462. ( 10.1128/JB.182.22.6456-6462.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pride AC, Guan Z, Trent MS. 2014. Characterization of the Vibrio cholerae VolA surface-exposed lipoprotein lysophospholipase. J. Bacteriol. 196, 1619–1626. ( 10.1128/JB.01281-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leuzzi R, et al. 2005. Ng-MIP, a surface-exposed lipoprotein of Neisseria gonorrhoeae, has a peptidyl-prolyl cis/trans isomerase (PPIase) activity and is involved in persistence in macrophages. Mol. Microbiol. 58, 669–681. ( 10.1111/j.1365-2958.2005.04859.x) [DOI] [PubMed] [Google Scholar]
- 43.d'Enfert C, Ryter A, Pugsley AP. 1987. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 6, 3531–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin S, Joe A, Lynett J, Hani EK, Sherman P, Chan VL. 2001. JlpA, a novel surface-exposed lipoprotein specific to Campylobacter jejuni, mediates adherence to host epithelial cells. Mol. Microbiol. 39, 1225–1236. ( 10.1111/j.1365-2958.2001.02294.x) [DOI] [PubMed] [Google Scholar]
- 45.Fuchs H, Wallich R, Simon MM, Kramer MD. 1994. The outer surface protein A of the spirochete Borrelia burgdorferi is a plasmin(ogen) receptor. Proc. Natl Acad. Sci. USA 91, 12 594–12 598. ( 10.1073/pnas.91.26.12594) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pugsley AP, Kornacker MG, Ryter A. 1990. Analysis of the subcellular location of pullulanase produced by Escherichia coli carrying the pulA gene from Klebsiella pneumoniae strain UNF5023. Mol. Microbiol. 4, 59–72. ( 10.1111/j.1365-2958.1990.tb02015.x) [DOI] [PubMed] [Google Scholar]
- 47.Korotkov KV, Sandkvist M, Hol WG. 2012. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 10, 336–351. ( 10.1038/nrmicro2762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi L, et al. 2008. Direct involvement of type II secretion system in extracellular translocation of Shewanella oneidensis outer membrane cytochromes MtrC and OmcA. J. Bacteriol. 190, 5512–5516. ( 10.1128/JB.00514-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baldi DL, et al. 2012. The type II secretion system and its ubiquitous lipoprotein substrate, SslE, are required for biofilm formation and virulence of enteropathogenic Escherichia coli. Infect. Immunol. 80, 2042–2052. ( 10.1128/IAI.06160-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roussel-Jazede V, Grijpstra J, van Dam V, Tommassen J, van Ulsen P. 2013. Lipidation of the autotransporter NalP of Neisseria meningitidis is required for its function in the release of cell-surface-exposed proteins. Microbiology 159, 286–295. ( 10.1099/mic.0.063982-0) [DOI] [PubMed] [Google Scholar]
- 51.van Ulsen P, van Alphen L, ten Hove J, Fransen F, van der Ley P, Tommassen J. 2003. A neisserial autotransporter NalP modulating the processing of other autotransporters. Mol. Microbiol. 50, 1017–1030. ( 10.1046/j.1365-2958.2003.03773.x) [DOI] [PubMed] [Google Scholar]
- 52.Coutte L, Willery E, Antoine R, Drobecq H, Locht C, Jacob-Dubuisson F. 2003. Surface anchoring of bacterial subtilisin important for maturation function. Mol. Microbiol. 49, 529–539. ( 10.1046/j.1365-2958.2003.03573.x) [DOI] [PubMed] [Google Scholar]
- 53.Coutte L, Antoine R, Drobecq H, Locht C, Jacob-Dubuisson F. 2001. Subtilisin-like autotransporter serves as maturation protease in a bacterial secretion pathway. EMBO J. 20, 5040–5048. ( 10.1093/emboj/20.18.5040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leyton DL, Rossiter AE, Henderson IR. 2012. From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat. Rev. Microbiol. 10, 213–225. ( 10.1038/nrmicro2733) [DOI] [PubMed] [Google Scholar]
- 55.Chng SS, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. 2010. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc. Natl Acad. Sci. USA 107, 5363–5368. ( 10.1073/pnas.0912872107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl Acad. Sci. USA 103, 11 754–11 759. ( 10.1073/pnas.0604744103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. 2014. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111. ( 10.1038/nature13484) [DOI] [PubMed] [Google Scholar]
- 58.Dong H, et al. 2014. Structural basis for outer membrane lipopolysaccharide insertion. Nature 511, 52–56. ( 10.1038/nature13464) [DOI] [PubMed] [Google Scholar]
- 59.Freinkman E, Chng SS, Kahne D. 2011. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc. Natl Acad. Sci. USA 108, 2486–2491. ( 10.1073/pnas.1015617108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chimalakonda G, Ruiz N, Chng SS, Garner RA, Kahne D, Silhavy TJ. 2011. Lipoprotein LptE is required for the assembly of LptD by the β-barrel assembly machine in the outer membrane of Escherichia coli. Proc. Natl Acad. Sci. USA 108, 2492–2497. ( 10.1073/pnas.1019089108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruiz N, Chng SS, Hiniker A, Kahne D, Silhavy TJ. 2010. Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc. Natl Acad. Sci. USA 107, 12 245–12 250. ( 10.1073/pnas.1007319107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Malojcic G, Andres D, Grabowicz M, George AH, Ruiz N, Silhavy TJ, Kahne D. 2014. LptE binds to and alters the physical state of LPS to catalyze its assembly at the cell surface. Proc. Natl Acad. Sci. USA 111, 9467–9472. ( 10.1073/pnas.1402746111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grabowicz M, Yeh J, Silhavy TJ. 2013. Dominant negative lptE mutation that supports a role for LptE as a plug in the LptD barrel. J. Bacteriol. 195, 1327–1334. ( 10.1128/JB.02142-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. 2006. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 59, 870–881. ( 10.1111/j.1365-2958.2005.04997.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Drummelsmith J, Whitfield C. 2000. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J. 19, 57–66. ( 10.1093/emboj/19.1.57) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao B, Zhao Y, Kou Y, Ni D, Zhang XC, Huang Y. 2014. Structure of the nonameric bacterial amyloid secretion channel. Proc. Natl Acad. Sci. USA 111, E5439–E5444. ( 10.1073/pnas.1411942111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goyal P, et al. 2014. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 516, 250–253. ( 10.1038/nature13768) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dong C, Beis K, Nesper J, Brunkan-Lamontagne AL, Clarke BR, Whitfield C, Naismith JH. 2006. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature 444, 226–229. ( 10.1038/nature05267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Epstein EA, Reizian MA, Chapman MR. 2009. Spatial clustering of the curlin secretion lipoprotein requires curli fiber assembly. J. Bacteriol. 191, 608–615. ( 10.1128/JB.01244-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Den Broeck I, Goyal P, Remaut H. 2015. Insights in peptide diffusion channels from the bacterial amyloid secretor CsgG. Channels. 9, 65–67. ( 10.1080/19336950.2015.1017172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bian Z, Normark S. 1997. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J. 16, 5827–5836. ( 10.1093/emboj/16.19.5827) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. 2012. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 20, 66–73. ( 10.1016/j.tim.2011.11.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nickerson NN, Mainprize IL, Hampton L, Jones ML, Naismith JH, Whitfield C. 2014. Trapped translocation intermediates establish the route for export of capsular polysaccharides across Escherichia coli outer membranes. Proc. Natl Acad. Sci. USA 111, 8203–8208. ( 10.1073/pnas.1400341111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dunstan RA, et al. In press. Assembly of the secretion pores GspD, Wza and CsgG into bacterial outer membranes does not require the Omp85 proteins BamA or TamA. Mol. Microbiol. ( 10.1111/mmi.13055) [DOI] [PubMed] [Google Scholar]
- 75.Nesper J, Hill CM, Paiment A, Harauz G, Beis K, Naismith JH, Whitfield C. 2003. Translocation of group 1 capsular polysaccharide in Escherichia coli serotype K30. Structural and functional analysis of the outer membrane lipoprotein Wza. J. Biol. Chem. 278, 49 763–49 772. ( 10.1074/jbc.M308775200) [DOI] [PubMed] [Google Scholar]
- 76.Ford RC, Brunkan-LaMontagne AL, Collins RF, Clarke BR, Harris R, Naismith JH, Whitfield C. 2009. Structure-function relationships of the outer membrane translocon Wza investigated by cryo-electron microscopy and mutagenesis. J. Struct. Biol. 166, 172–182. ( 10.1016/j.jsb.2009.02.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hagelueken G, Ingledew WJ, Huang H, Petrovic-Stojanovska B, Whitfield C, ElMkami H, Schiemann O, Naismith JH. 2009. PELDOR spectroscopy distance fingerprinting of the octameric outer-membrane protein Wza from Escherichia coli. Angew. Chem. Int. Ed. Engl. 48, 2904–2906. ( 10.1002/anie.200805758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Braun V, Wolff H. 1970. The murein-lipoprotein linkage in the cell wall of Escherichia coli. Eur. J. Biochem. 14, 387–391. ( 10.1111/j.1432-1033.1970.tb00301.x) [DOI] [PubMed] [Google Scholar]
- 79.Braun V, Rehn K. 1969. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur. J. Biochem. 10, 426–438. ( 10.1111/j.1432-1033.1969.tb00707.x) [DOI] [PubMed] [Google Scholar]
- 80.Braun V, Hantke K. 1975. Characterization of the free form of murein-lipoprotein from the outer membrane of Escherichia coli B/r. FEBS Lett. 60, 26–28. ( 10.1016/0014-5793(75)80410-8) [DOI] [PubMed] [Google Scholar]
- 81.Bouveret E, Benedetti H, Rigal A, Loret E, Lazdunski C. 1999. In vitro characterization of peptidoglycan-associated lipoprotein (PAL)-peptidoglycan and PAL-TolB interactions. J. Bacteriol. 181, 6306–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hirota Y, Suzuki H, Nishimura Y, Yasuda S. 1977. On the process of cellular division in Escherichia coli: a mutant of E. coli lacking a murein-lipoprotein. Proc. Natl Acad. Sci. USA 74, 1417–1420. ( 10.1073/pnas.74.4.1417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suzuki H, Nishimura Y, Yasuda S, Nishimura A, Yamada M, Hirota Y. 1978. Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Mol. Gen. Genet. 167, 1–9. [DOI] [PubMed] [Google Scholar]
- 84.Lazzaroni JC, Portalier R. 1992. The excC gene of Escherichia coli K-12 required for cell envelope integrity encodes the peptidoglycan-associated lipoprotein (PAL). Mol. Microbiol. 6, 735–742. ( 10.1111/j.1365-2958.1992.tb01523.x) [DOI] [PubMed] [Google Scholar]
- 85.Cowles CE, Li Y, Semmelhack MF, Cristea IM, Silhavy TJ. 2011. The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol. Microbiol. 79, 1168–1181. ( 10.1111/j.1365-2958.2011.07539.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Michel LV, Shaw J, MacPherson V, Barnard D, Bettinger J, D'Arcy B, Surendran N, Hellman J, Pichichero ME. 2015. Dual Orientation of the outer membrane lipoprotein Pal in Escherichia coli. Microbiology 161, 1251–1259. ( 10.1099/mic.0.000084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Manning PA, Beutin L, Achtman M. 1980. Outer membrane of Escherichia coli: properties of the F sex factor traT protein which is involved in surface exclusion. J. Bacteriol. 142, 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Webb CT, Selkrig J, Perry AJ, Noinaj N, Buchanan SK, Lithgow T. 2012. Dynamic association of BAM complex modules includes surface exposure of the lipoprotein BamC. J. Mol. Biol. 422, 545–555. ( 10.1016/j.jmb.2012.05.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arnold MF, Caro-Hernandez P, Tan K, Runti G, Wehmeier S, Scocchi M, Doerrler WT, Walker GC, Ferguson GP. 2014. Enteric YaiW is a surface-exposed outer membrane lipoprotein that affects sensitivity to an antimicrobial peptide. J. Bacteriol. 196, 436–444. ( 10.1128/JB.01179-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Majdalani N, Heck M, Stout V, Gottesman S. 2005. Role of RcsF in signaling to the Rcs phosphorelay pathway in Escherichia coli. J. Bacteriol. 187, 6770–6778. ( 10.1128/JB.187.19.6770-6778.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Castanie-Cornet MP, Cam K, Jacq A. 2006. RcsF is an outer membrane lipoprotein involved in the RcsCDB phosphorelay signaling pathway in Escherichia coli. J. Bacteriol. 188, 4264–4270. ( 10.1128/JB.00004-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Parker CT, Kloser AW, Schnaitman CA, Stein MA, Gottesman S, Gibson BW. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 174, 2525–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Farris C, Sanowar S, Bader MW, Pfuetzner R, Miller SI. 2010. Antimicrobial peptides activate the Rcs regulon through the outer membrane lipoprotein RcsF. J. Bacteriol. 192, 4894–4903. ( 10.1128/JB.00505-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Callewaert L, Vanoirbeek KG, Lurquin I, Michiels CW, Aertsen A. 2009. The Rcs two-component system regulates expression of lysozyme inhibitors and is induced by exposure to lysozyme. J. Bacteriol. 191, 1979–1981. ( 10.1128/JB.01549-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Laubacher ME, Ades SE. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J. Bacteriol. 190, 2065–2074. ( 10.1128/JB.01740-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ophir T, Gutnick DL. 1994. A role for exopolysaccharides in the protection of microorganisms from desiccation. Appl. Environ. Microbiol. 60, 740–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sledjeski DD, Gottesman S. 1996. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178, 1204–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shiba Y, Miyagawa H, Nagahama H, Matsumoto K, Kondo D, Matsuoka S, Hara H. 2012. Exploring the relationship between lipoprotein mislocalization and activation of the Rcs signal transduction system in Escherichia coli. Microbiology 158, 1238–1248. ( 10.1099/mic.0.056945-0) [DOI] [PubMed] [Google Scholar]
- 99.Chen MH, Takeda S, Yamada H, Ishii Y, Yamashino T, Mizuno T. 2001. Characterization of the RcsC→YojN→RcsB phosphorelay signaling pathway involved in capsular synthesis in Escherichia coli. Biosci. Biotechnol. Biochem. 65, 2364–2367. ( 10.1271/bbb.65.2364) [DOI] [PubMed] [Google Scholar]
- 100.Konovalova A, Perlman DH, Cowles CE, Silhavy TJ. 2014. Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. Proc. Natl Acad. Sci. USA 111, E4350–E4358. ( 10.1073/pnas.1417138111). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cho SH, et al. 2014. Detecting envelope stress by monitoring beta-barrel assembly. Cell 159, 1652–1664. ( 10.1016/j.cell.2014.11.045) [DOI] [PubMed] [Google Scholar]
- 102.Housden NG, Wojdyla JA, Korczynska J, Grishkovskaya I, Kirkpatrick N, Brzozowski AM, Kleanthous C. 2010. Directed epitope delivery across the Escherichia coli outer membrane through the porin OmpF. Proc. Natl Acad. Sci. USA 107, 21 412–21 417. ( 10.1073/pnas.1010780107). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yamashita E, Zhalnina MV, Zakharov SD, Sharma O, Cramer WA. 2008. Crystal structures of the OmpF porin: function in a colicin translocon. EMBO J. 27, 2171–2180. ( 10.1038/emboj.2008.137). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Leverrier P, Declercq JP, Denoncin K, Vertommen D, Hiniker A, Cho SH, Collet JF. 2011. Crystal structure of the outer membrane protein RcsF, a new substrate for the periplasmic protein-disulfide isomerase DsbC. J. Biol. Chem. 286, 16 734–16 742. ( 10.1074/jbc.M111.224865) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rogov VV, Rogova NY, Bernhard F, Lohr F, Dotsch V. 2011. A disulfide bridge network within the soluble periplasmic domain determines structure and function of the outer membrane protein RCSF. J. Biol. Chem. 286, 18 775–18 783. ( 10.1074/jbc.M111.230185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245. ( 10.1016/j.cell.2005.02.015). [DOI] [PubMed] [Google Scholar]
- 107.van Geest M, Lolkema JS. 2000. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol. Mol. Biol. Rev. 64, 13–33. ( 10.1128/MMBR.64.1.13-33.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Green N, Fang H, Kalies K-U, Canfield V. 2001. Determining the topology of an integral membrane protein. Curr. Protoc. Cell Biol. 5.2.1–5.2.27. ( 10.1002/0471143030.cb0502s00). [DOI] [PubMed] [Google Scholar]
- 109.Hermanson GT. 2013. Bioconjugate techniques, 3rd edn San Diego, CA: Academic Press. [Google Scholar]
- 110.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656. ( 10.1128/MMBR.67.4.593-656.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rosenbusch JP. 1990. Structural and functional properties of porin channels in E. coli outer membranes. Experientia 46, 167–173. [PubMed] [Google Scholar]
- 112.Rigel NW, Ricci DP, Silhavy TJ. 2013. Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for beta-barrel assembly in Escherichia coli. Proc. Natl Acad. Sci. USA 110, 5151–5156. ( 10.1073/pnas.1302662110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pinne M, Haake DA. 2009. A comprehensive approach to identification of surface-exposed, outer membrane-spanning proteins of Leptospira interrogans. PLoS ONE 4, e6071 ( 10.1371/journal.pone.0006071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Matsunaga J, Werneid K, Zuerner RL, Frank A, Haake DA. 2006. LipL46 is a novel surface-exposed lipoprotein expressed during leptospiral dissemination in the mammalian host. Microbiology 152, 3777–3786. ( 10.1099/mic.0.29162-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pinne M, Haake D. 2011. Immuno-fluorescence assay of leptospiral surface-exposed proteins. J. Vis. Exp. 53, e2805 ( 10.3791/2805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Blom K, Lundin BS, Bolin I, Svennerholm A. 2001. Flow cytometric analysis of the localization of Helicobacter pylori antigens during different growth phases. FEMS Immunol. Med. Microbiol. 30, 173–179. ( 10.1111/j.1574-695X.2001.tb01567.x) [DOI] [PubMed] [Google Scholar]
- 117.Schnell U, Dijk F, Sjollema KA, Giepmans BN. 2012. Immunolabeling artifacts and the need for live-cell imaging. Nat. Methods 9, 152–158. ( 10.1038/nmeth.1855) [DOI] [PubMed] [Google Scholar]
- 118.Voss BJ, Gaddy JA, McDonald WH, Cover TL. 2014. Analysis of surface-exposed outer membrane proteins in Helicobacter pylori. J. Bacteriol. 196, 2455–2471. ( 10.1128/JB.01768-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cao Y, Bazemore-Walker CR. 2014. Proteomic profiling of the surface-exposed cell envelope proteins of Caulobacter crescentus. J. Proteomics 97, 187–194. ( 10.1016/j.jprot.2013.08.011) [DOI] [PubMed] [Google Scholar]
- 120.Wilson MM, Anderson DE, Bernstein HD. 2015. Analysis of the outer membrane proteome and secretome of Bacteroides fragilis reveals a multiplicity of secretion mechanisms. PLoS ONE 10, e0117732 ( 10.1371/journal.pone.0117732). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. 2007. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317, 961–964. ( 10.1126/science.1143993) [DOI] [PubMed] [Google Scholar]
- 122.Kim KH, Aulakh S, Paetzel M. 2011. Crystal structure of beta-barrel assembly machinery BamCD protein complex. J. Biol. Chem. 286, 39 116–39 121. ( 10.1074/jbc.M111.298166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bos C, Lorenzen D, Braun V. 1998. Specific in vivo labeling of cell surface-exposed protein loops: reactive cysteines in the predicted gating loop mark a ferrichrome binding site and a ligand-induced conformational change of the Escherichia coli FhuA protein. J. Bacteriol. 180, 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Harms N, Oudhuis WC, Eppens EA, Valent QA, Koster M, Luirink J, Oudega B. 1999. Epitope tagging analysis of the outer membrane folding of the molecular usher FaeD involved in K88 fimbriae biosynthesis in Escherichia coli. J. Mol. Microbiol. Biotechnol. 1, 319–325. [PubMed] [Google Scholar]
- 125.Newton SM, Klebba PE, Michel V, Hofnung M, Charbit A. 1996. Topology of the membrane protein LamB by epitope tagging and a comparison with the X-ray model. J. Bacteriol. 178, 3447–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moeck GS, Bazzaz BS, Gras MF, Ravi TS, Ratcliffe MJ, Coulton JW. 1994. Genetic insertion and exposure of a reporter epitope in the ferrichrome-iron receptor of Escherichia coli K-12. J. Bacteriol. 176, 4250–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ehrmann M, Bolek P, Mondigler M, Boyd D, Lange R. 1997. TnTIN and TnTAP: mini-transposons for site-specific proteolysis in vivo. Proc. Natl Acad. Sci. USA 94, 13 111–13 115. ( 10.1073/pnas.94.24.13111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen S, Kumru OS, Zuckert WR. 2011. Determination of Borrelia surface lipoprotein anchor topology by surface proteolysis. J. Bacteriol. 193, 6379–6383. ( 10.1128/JB.05849-11) [DOI] [PMC free article] [PubMed] [Google Scholar]