Abstract
Scientific activities take place within the structured sets of ideas and assumptions that define a field and its practices. The conceptual framework of evolutionary biology emerged with the Modern Synthesis in the early twentieth century and has since expanded into a highly successful research program to explore the processes of diversification and adaptation. Nonetheless, the ability of that framework satisfactorily to accommodate the rapid advances in developmental biology, genomics and ecology has been questioned. We review some of these arguments, focusing on literatures (evo-devo, developmental plasticity, inclusive inheritance and niche construction) whose implications for evolution can be interpreted in two ways—one that preserves the internal structure of contemporary evolutionary theory and one that points towards an alternative conceptual framework. The latter, which we label the ‘extended evolutionary synthesis' (EES), retains the fundaments of evolutionary theory, but differs in its emphasis on the role of constructive processes in development and evolution, and reciprocal portrayals of causation. In the EES, developmental processes, operating through developmental bias, inclusive inheritance and niche construction, share responsibility for the direction and rate of evolution, the origin of character variation and organism–environment complementarity. We spell out the structure, core assumptions and novel predictions of the EES, and show how it can be deployed to stimulate and advance research in those fields that study or use evolutionary biology.
Keywords: extended evolutionary synthesis, evolutionary developmental biology, developmental plasticity, inclusive inheritance, niche construction, reciprocal causation
1. Introduction
To make progress, scientists must specify phenomena that require explanation, identify causes and decide on what methods, data and analyses are explanatorily sufficient. In doing so, they may inadvertently create a ‘conceptual framework’—a way of thinking for their field, with associated assumptions, concepts, rules and practice, that allows them to get on with their work [1–3]. Conceptual frameworks are necessary in science, but they, and their associated practices, inevitably encourage some lines of research more readily than others. Hence, it is vital that the conceptual frameworks themselves evolve in response to new data, theories and methodologies. This is not always straightforward, as habits of thought and practice are often deeply entrenched. In this regard, alternative conceptual frameworks can be valuable because they draw attention to constructive new ways of thinking, additional causal influences, alternative predictions or new lines of enquiry.
The Modern Synthesis (MS) emerged in the first half of the twentieth century, with the integration of Darwinian natural selection, population-level thinking and Mendelian inheritance, and has provided the dominant conceptual framework for evolutionary biology [4,5]. It is rightly regarded as one of the major achievements of biology and led to the widespread adoption of several core assumptions [6] (table 1). These include: (i) evolutionarily significant phenotypic variation arises from genetic mutations that occur at a low rate independently of the strength and direction of natural selection; (ii) most favourable mutations have small phenotypic effects, which results in gradual phenotypic change; (iii) inheritance is genetic; (iv) natural selection is the sole explanation for adaptation; and (v) macro-evolution is the result of accumulation of differences that arise through micro-evolutionary processes.
Table 1.
A comparison of the core assumptions of the classical MS and the EES.
| classical MS core assumptions | EES core assumptions |
|---|---|
| (i) The pre-eminence of natural selection. The major directing or creative influence in evolution is natural selection, which alone explains why the properties of organisms match the properties of their environments (adaptation) | (i) Reciprocal causation (organisms shape, and are shaped by, selective and developmental environments). Developmental processes, operating through developmental bias and niche construction, share with natural selection some responsibility for the direction and rate of evolution and contribute to organism–environment complementarity |
| (ii) Genetic inheritance. Genes constitute the only general inheritance system. Acquired characters are not inherited | (ii) Inclusive inheritance. Inheritance extends beyond genes to encompass (transgenerational) epigenetic inheritance, physiological inheritance, ecological inheritance, social (behavioural) transmission and cultural inheritance. Acquired characters can play evolutionary roles by biasing phenotypic variants subject to selection, modifying environments and contributing to heritability |
| (iii) Random genetic variation. There is no relationship between the direction in which mutations occur—and hence the supply of phenotypic variants—and the direction that would lead to enhanced fitness | (iii) Non-random phenotypic variation. Developmental bias, resulting from non-random mutation or phenotypic accommodation, means that some phenotypic variants are more likely than others. Developmental systems facilitate well-integrated, functional phenotypic responses to mutation or environmental induction |
| (iv) Gradualism. Evolution via mutations of large effects is unlikely because such mutations have disruptive pleiotropic effects. Phenotypic transitions typically occur through multiple small steps, leading to gradual evolutionary change | (iv) Variable rates of change. Variants of large effect are possible, allowing for rapid evolutionary change. Saltation can occur either through mutations in major regulatory control genes expressed in tissue-, module- or compartment-specific manner, or when developmental processes respond to environmental challenges with change in coordinated suites of traits, or through nonlinear threshold effects |
| (v) Gene-centred perspective. Evolution requires, and is often defined as, change in gene frequencies. Populations evolve through changes in gene frequencies brought about through natural selection, drift, mutation and gene flow | (v) Organism-centred perspective. Developmental systems can facilitate adaptive variation and modify selective environments. Evolution redefined as a transgenerational change in the distribution of heritable traits of a population. There is a broadened notion of evolutionary process and inheritance |
| (vi) Macro-evolution. Macro-evolutionary patterns are explained by micro-evolutionary processes of selection, drift, mutation and gene flow | (vi) Macro-evolution. Additional evolutionary processes, including developmental bias and ecological inheritance, help explain macro-evolutionary patterns and contribute to evolvability |
Following the advent of the MS, the field of evolutionary biology has continued to evolve [7], allowing incorporation of new theoretical and empirical findings (e.g. neutral theory, inclusive fitness theory). As a result, today's evolutionary theory is vastly more sophisticated than the original synthesis and covers a broader range of phenomena. However, while such progress is undeniable, it does not imply that the underlying conceptual framework allows evolutionary biologists to make the most out of progress in biology and other fields. For instance, some more implicit features of contemporary evolutionary thought, such as the assignment of causal and informational privileges to genes in development, or the treating of development and heredity as separate phenomena, remain prevalent in spite of new data that appear to challenge these assumptions [8,9].
In this regard, insights derived from research on: (i) evolutionary developmental biology (‘evo-devo’), (ii) developmental plasticity, (iii) inclusive inheritance, and (iv) niche construction are particularly instructive. Recent findings from these literatures are open to at least two distinct interpretations, which may be regarded as manifestations of a ‘fault line’ in interpretative understanding. These alternatives include a perspective that is in broad agreement with the assumptions of the original MS, and an interpretation that views the same findings as challenging important assumptions of the MS. The latter view is distinctive for its emphasis on organismal causes of development, inheritance and differential fitness, the role of constructive processes in development and evolution, and reciprocal representations of causation. We label this interpretation the ‘extended evolutionary synthesis’ (EES) but emphasize, contrary to some recent claims (e.g. [10]), that the EES is a developing line of contemporary evolutionary thought that exists within the field, and not a denial of the value of past frameworks or of progress in evolutionary biology. To some readers, the use of the EES label might appear grandiloquent but, as we will show, the significance of the proposed changes in evolutionary thinking varies substantially with the researcher's perspective.
While the suggestion that an EES is emerging has been made repeatedly [11–15], its nature and role have remained unclear. As a result, the various lobbies for change within evolutionary biology have been regarded as largely unconnected, or idiosyncratic. Our objective here is to add substance to these debates by providing a clear statement of the structure, assumptions and predictions of the EES that is useful to both enthusiasts and skeptics, allowing its status as an alternative conceptual framework to be evaluated.
We begin by providing brief overviews of evo-devo, developmental plasticity, inclusive inheritance and niche construction, highlighting divergent interpretations. We illustrate how each is subject to two readings, one that accommodates the phenomena without changing the traditional structure of evolutionary explanations, and an alternative interpretation (i.e. the EES) that appears inconsistent with the prevailing framework. We go on to elaborate on the latter, by specifying how the various components of the EES fit together, drawing out its core assumptions and illustrating its ability to generate and test novel hypotheses. We conclude that the EES is not just an extension of the MS but a distinctively different framework for understanding evolution, which, alongside more traditional perspectives, can be put to service constructively within the field.
2. Biological background to the extended evolutionary synthesis
The impetus for an EES is undoubtedly complex and multifaceted. Here, we focus on insights derived from four research areas that, as we describe below, have been subject to alternative interpretations in recent literature, yet nonetheless reveal convergent themes. This section merely presents the relevant findings, while how these findings are interpreted is discussed in later sections. Readers familiar with these literatures may prefer to jump directly to §3.
(a). Evolutionary developmental biology
Evo-devo provides a causal-mechanistic understanding of evolution by using comparative and experimental biology to identify the developmental principles that underlie phenotypic differences within and between populations, species and higher taxa. Among the key empirical insights are that phenotypic variation often involves changes in the gene regulatory machinery that alters the timing, location, amount or type of gene product. This modification of pre-existing developmental processes can bring about coordinated changes in suites of characters, effectively enabling diversification through the differential coupling and decoupling of phenotypic modules [16–19]. As a consequence, developmental properties can affect the rates and patterns of phenotypic evolution [20,21] and contribute to evolvability, the potential of biological lineages for adaptive evolution [19,22–24].
While much evo-devo research is compatible with standard assumptions in evolutionary biology, some findings have generated debate. Of particular interest is the observation that phenotypic variation can be biased by the processes of development, with some forms more probable than others [12,17,25–28]. Bias is manifest, for example, in the non-random numbers of limbs, digits, segments and vertebrae across a variety of taxa [25,26,29,30], correlated responses to artificial selection resulting from shared developmental regulation [31], and in the repeated, differential re-use of developmental modules, which enables novel phenotypes to arise by developmental rearrangements of ancestral elements, as in the parallel evolution of animal eyes [32].
Developmental bias may also contribute to the many examples of convergence across the tree of life. For example, cichlid fishes from Lakes Malawi and Tanganyika exhibit striking similarities in body shape, despite being more closely related to species from their own lake than to those from the other lake [17,33]. Such repeated parallel evolution is generally attributed to convergent selection. However, inherent features of development may have channelled morphology along specific pathways, thereby facilitating the evolution of parallel forms in the two lakes [17,33]. If so, then the diversity of organismal form is only partly a consequence of natural selection—the particular evolutionary trajectories taken also depend on features of development.
Some work on developmental bias suggests that phenotypic variation can be channelled and directed towards functional types by the processes of development [27,28]. The rationale is that development relies on highly robust ‘core processes’, from microtubule formation and signal transduction pathways to organogenesis, which at the same time exhibit ‘exploratory behaviour’ [28], allowing them to stabilize and select certain states over others. Exploratory behaviour followed by somatic selection enables core processes to be responsive to changes in genetic and environmental input, while their robustness and conservation maintain their ability to generate functional (i.e. well integrated) outcomes in the face of perturbations. This phenomenon, known as facilitated variation [28,34], provides a mechanistic explanation for how small, genetic changes can sometimes elicit substantial, non-random, well-integrated and apparently adaptive innovations in the phenotype.
(b). Developmental plasticity
Developmental, or phenotypic, plasticity is the capacity of an organism to change its phenotype in response to the environment. Plasticity is ubiquitous across all levels of biological organization, and although it is closely linked to evo-devo, we treat it separately here because it is typically studied in a population context that is rarely central to evo-devo.
While the evolution of plasticity has been studied for decades (e.g. [35–39]), there is renewed interest in plasticity as a cause, and not just a consequence, of phenotypic evolution. For example, plasticity facilitates colonization of novel environments [40,41], affects population connectivity and gene flow [42], contributes to temporal and spatial variation in selection [43–45] and may increase the chance of adaptive peak shifts, radiations and speciation events [27,46–48].
Particularly contentious is the contribution of plasticity to evolution through phenotypic and genetic accommodation [27,48,49]. Phenotypic accommodation refers to the mutual and often functional adjustment of parts of an organism during development that typically does not involve genetic mutation [27]. It has long been argued that phenotypic accommodation could promote genetic accommodation if environmentally induced phenotypes are subsequently stabilized and fine-tuned across generations by selection of standing genetic variation, previously cryptic genetic variation or newly arising mutations [27,47,50,51]. From this viewpoint, developmental processes play a critical role in determining which genetic variants will produce selectable phenotypic differences, and which will not. Genetic accommodation may provide a mechanism for rapid adaptation to novel environments, as those environments simultaneously induce and select for alternative phenotypes [47,52,53]. Consistent with these arguments, plasticity within species has been shown to generate parallel phenotypic differences to those exhibited by closely related species, and ancestral plasticity has been linked to evolutionary divergence among descendant lineages (e.g. [54–57]; reviewed in [58]).
(c). Inclusive inheritance
Biological inheritance is typically defined as the transmission of genes from parents to offspring. However, it is increasingly recognized that there are multiple mechanisms that contribute to heredity [59–61]. Parent–offspring similarity occurs not only because of transmission of DNA, but because parents transfer a variety of developmental resources that enable reconstruction of developmental niches [60,62–65]. These include components of the egg and post-fertilization resources (e.g. hormones), behavioural interactions between parents and offspring (e.g. maternal care), parental modification of other components of the biotic and abiotic environment (e.g. host choice) and inheritance of symbionts directly through the mother's germ cells or by infection. In addition, recent research reveals that vertical and horizontal social transmission is widespread in both vertebrates and invertebrates, and can both initiate population divergence and trigger speciation [66]. Under this broader notion of heredity, inheritance can occur from germ cell to germ cell, from soma to germ cell, from soma to soma, and from soma to soma via the external environment [63], which may provide opportunities for some acquired characteristics to be inherited.
The pathways of inheritance that derive from a parental phenotype (‘parental effects’) have a number of evolutionary consequences similar to those of plasticity, cultural inheritance and niche construction [67]. For example, non-genetic inheritance can bias the expression and retention of environmentally induced phenotypes, thereby influencing the rate and direction of evolution [68]. There is also increasing evidence for more stable transgenerational epigenetic inheritance, or the transmission across generations of cellular states without modification of the DNA sequence, which demonstrates that adaptive evolution may proceed by selection on epigenetic variants as well as variation in DNA sequence [60,69,70].
(d). Niche construction theory
‘Niche construction’ refers to the process whereby the metabolism, activities and choices of organisms modify or stabilize environmental states, and thereby affect selection acting on themselves and other species [71–73]. For example, many species of animals manufacture nests, burrows, webs and pupal cases; algae and plants change atmospheric redox states and modify nutrient cycles; fungi and bacteria decompose organic matter and may fix nutrients and excrete compounds that alter environments. Niche construction frequently scales up, across individuals in a population, and over time, to generate stable and directional changes in environmental conditions [73,74].
Niche construction also influences ontogeny and constitutes an important way in which environmental factors are incorporated into normal development, sometimes to become as dependable as genomic factors [63,73,75]. Ecological inheritance refers to the accumulation of environmental changes, such as altered soil, atmosphere or ocean states that previous generations have brought about through their niche-constructing activity, and that influence the development of descendant organisms [73,76]. Through their activities, organisms may also change the niches of other species in an ecosystem and in so doing lead to direct or diffuse coevolution, including via intermediate abiota, with potentially profound impacts on the stability and dynamics of ecosystems on both micro- and macro-evolutionary timescales [73,76–78].
A body of formal evolutionary theory has shown that niche construction can affect evolutionary dynamics in a variety of ways [79–86], even when it is not an ‘extended phenotype’ [87]; that is, not an adaptation. The evolutionary significance of niche construction stems from: (i) organisms modify environmental states in non-random ways, thereby imposing a systematic bias on the selection pressures they generate; (ii) ecological inheritance affects the evolutionary dynamics of descendants and contributes to the cross-generational stability of environmental conditions; (iii) acquired characters become evolutionarily significant by modifying selective environments; and (iv) the complementarity of organisms and their environments can be enhanced through niche construction (modifying environments to suit organisms), not just through natural selection [73]. These findings have led to the claim that niche construction should be recognized as an evolutionary process through its guiding influence on selection [73], a position that is contested by some evolutionary biologists [88].
3. A traditional interpretation
For many evolutionary biologists, the research described above is not viewed as a challenge to the traditional explanatory framework, but rather developmental bias, plasticity, non-genetic inheritance, and niche construction are considered proximate, but not evolutionary, causes [88–90]. Thus, while these phenomena demand evolutionary explanations, they do not themselves constitute valid, even partial, evolutionary explanations for organismal diversity and adaptation. For example, developmental bias is generally understood as imposing constraints on adaptive evolution (table 2), such as the limit on the absolute size of terrestrial arthropods imposed by breathing via a tracheal system. Constraints, so conceived, are causes of the absence of evolution; they might explain why adaptation has not occurred in a given circumstance, or why phenotypes are not globally optimal, but it is selection that gives directionality in evolution and explains adaptation.
Table 2.
Two alternative interpretations of developmental bias, developmental plasticity, inclusive inheritance and niche construction.
| a traditional interpretation | the EES interpretation | |
|---|---|---|
| developmental bias | sources of bias in phenotypic variation treated as phylogenetic or developmental constraints. Such constraints are important components of optimality models and in analyses of contemporary evolution (e.g. in attempts to quantify the G matrix in quantitative genetics), which may explain why populations are poorly adapted | sources of bias in phenotypic variation considered an important evolutionary process, which does not only constrain but also facilitate and direct evolution. Developmental bias is a major source of evolvability and explanation of its mechanisms, prevalence and direction are crucial to understand evolutionary diversification |
| developmental plasticity | conceptualized as a genetically specified feature of individuals (i.e. a reaction norm) that can evolve under selection and drift. Focus is on the conditions that promote adaptive evolution of plastic versus non-plastic phenotypes. The primary evolutionary role of plasticity is to adjust phenotypes adaptively to variable environments. Plastic responses regarded as pre-filtered by past selection | considers reducing plasticity to a genetic feature to be explanatorily insufficient. Retains an interest in adaptive evolution of plasticity, but also focuses on how plasticity contributes to the origin of functional variation under genetic or environmental change, and how the mechanisms of plasticity limit or enhance evolvability, and initiate evolutionary responses. Many plastic responses viewed as reliant on open-ended (e.g. exploratory) developmental processes, and hence capable of introducing phenotypic novelty |
| inclusive inheritance | heredity defined to exclude non-genetic inheritance. Cultural inheritance treated as a special case. Transmission genetics considered explanatorily sufficient for the evolution of adaptations. Causal effects of parents on offspring are referred to as parental (maternal) effects, which are shown to have a variety of consequences for evolutionary trajectories and may be adaptations | heredity defined to include all causal mechanisms by which offspring come to resemble their parents. Phenotypes are not inherited, they are reconstructed in development. Non-genetic mechanisms of inheritance contribute to heritability and facilitate the origin and spread of environmentally induced novelties |
| niche construction | aspects of niche construction studied under different labels (e.g. extended phenotypes). Environmental states modified by organisms viewed as no different from independent environmental states and treated as a background condition. Niche construction typically reduced to genetically controlled aspects of phenotypes, or adaptations | views evolutionary causation as reciprocal and hence that organisms co-evolve with their environments. Environments modified by organisms viewed as qualitatively different from independent environmental states. Niche construction treated as a process that directs evolution by non-random modification of selective environments. Niche construction may result from acquired characters, by-products and the accumulated outputs of multiple species |
Similarly, the standard view is that phenotypic plasticity and inclusive inheritance are either inconsequential, proximate, causes of variation or outcomes of selection (i.e. adaptations; table 2). Plasticity is typically considered to be a genetically specified, and hence evolvable, trait that allows individuals to match phenotypes to local conditions [91,92], and the same logic is used to accommodate non-genetic inheritance and niche construction in evolutionary theory (e.g. [88,93]).
For biologists schooled in population genetic or quantitative genetic thinking, the starting point for evolutionary analyses is the selection pressures [94]. Leaving aside cases where the source of selection is another organism, environmental change has been treated as a ‘background condition’ (e.g. [88]; table 2). On this perspective, termites evolve to become adapted to the mounds they construct in a manner no different from how organisms adapt to frequent volcanic eruptions. Because niche-constructing activities are seen as proximate sources of variation, they are typically treated as ‘extended phenotypes' [87] that evolve because they enhance inclusive fitness.
We suggest that structuring evolutionary explanations around processes that directly change genotype frequencies is responsible for these interpretations. A widely accepted definition of evolution is change in the genetic composition of populations, which, to many evolutionary biologists, restricts evolutionary processes to those that directly change gene frequencies—natural selection, drift, gene flow and mutation. Phenomena such as developmental bias or niche construction do not directly change gene frequencies, and hence are not viewed as causes of evolutionary processes.
Contemporary evolutionary biology textbooks support this interpretation (see the electronic supplementary material, table S1). Only selection, drift, gene flow and mutation are consistently described as evolutionary processes and coverage of developmental bias, plasticity, inclusive inheritance and niche construction is at best modest (e.g. [95]) and, more commonly, absent [96,97]. What coverage does occur is typically given the traditional interpretation outlined above.
4. The extended evolutionary synthesis perspective
The incorporation of new data into the existing conceptual framework of evolutionary biology may explain why calls for an EES are often met with skepticism; even if the topics discussed above were historically neglected, there is now a substantial amount of research dedicated to them. However, for a second group of evolutionary researchers, the interpretation given in the preceding section underestimates the evolutionary implications of these phenomena (table 2). From this standpoint, too much causal significance is afforded to genes and selection, and not enough to the developmental processes that create novel variants, contribute to heredity, generate adaptive fit, and thereby direct the course of evolution. Under this perspective, the sharp distinction between the proximate and the ultimate is undermined by the fact that proximate causes are themselves often also evolutionary causes [90]. Hence, the EES entails not only new research directions but also new ways to think about, and interpret, new and familiar problems in evolutionary biology.
In this section, we endeavour to draw out the defining themes and structure of the EES. We show how, while the lines of research discussed above arose largely independently, there is considerable coherence across topics. Developmental processes play important evolutionary roles as causes of novel, potentially beneficial, phenotypic variants, the differential fitness of those variants, and/or their inheritance (i.e. all three of Lewontin's [98] conditions for evolution by natural selection). Thus, the burden of creativity in evolution (i.e. the generation of adaptation) does not rest on selection alone [12,19,25,27,60,64,73,99–101].
We see two key unifying themes to these interpretations—constructive development and reciprocal causation.
(a). Constructive development
Constructive development refers to the ability of an organism to shape its own developmental trajectory by constantly responding to, and altering, internal and external states [34,71,102–105]. Constructive development goes beyond the quantitative-genetic concept of gene–environment interaction by attending to the mechanisms of development, and emphasizing how gene (expression) and environment are interdependent. As a consequence, the developing organism cannot be reduced to separable components, one of which (e.g. the genome) exerts exclusive control over the other (e.g. the phenotype). Rather, causation also flows back from ‘higher’ (i.e. more complex) levels of organismal organization to the genes (e.g. tissue-specific regulation of gene expression) (figure 1). Constructive development does not assume a relatively simple mapping between genotype and phenotype, nor does it assign causal privilege to genes in individual development. Instead, the developmental system responds flexibly to internal and external inputs, most obviously through condition-dependent gene expression, but also through physical properties of cells and tissues and ‘exploratory behaviour’ among microtubular, neural, muscular and vascular systems. For example, there is no predetermined map for the distribution of blood vessels in the body; rather, the vascular system expands to regions with insufficient oxygen supply. Such exploratory processes, commonplace throughout development, are powerful agents of phenotype construction, as they enable highly diverse functional responses that need not have been pre-screened by earlier selection [28,34,106,107].
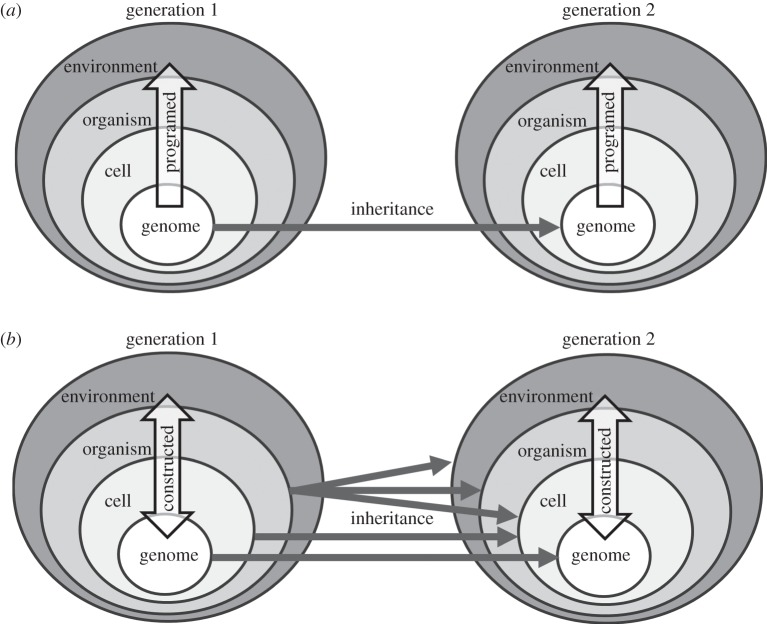
Figure 1.
Contrasting views of development. (a) Programed development. Traditionally, development has been conceptualized as programed, unfolding according to rules and instructions specified within the genome. DNA is ascribed a special causal significance, and all other parts of the developing organism serve as ‘substrate’, or ‘interpretative machinery’ for the expression of genetic information. Evolutionarily relevant phenotypic novelty results solely from genetic mutations, which alter components of the genetic program. Under this perspective, organisms are built from the genome outwards and upwards, with each generation receiving the instruction on how to build a phenotype through the transmission of DNA. (b) Constructive development. By contrast, in the EES, genes and genomes represent one of many resources that contribute to the developing phenotype. Causation flows both upwards from lower levels of biological organization, such as DNA, and from higher levels downwards, such as through tissue- and environment-specific gene regulation. Exploratory and selective processes are important sources of novel and evolutionarily significant phenotypic variation. Rather than containing a ‘program’, the genome represents a component of the developmental system, shaped by evolution to sense and respond to relevant signals and to provide materials upon which cells can draw.
Within evolutionary biology, development has been traditionally viewed as under the direction of a genetic program (e.g. ‘all of the directions, controls and constraints of the developmental machinery are laid down in the blueprint of the DNA genotype as instructions or potentialities' [108, p. 126]). While the terminology of contemporary biologists is typically more nuanced, Moczek [109] shows that genetic ‘blueprint’, ‘program’ or ‘instructions' metaphors remain widespread in evolutionary biology texts. By contrast, the EES regards the genome as a sub-system of the cell designed by evolution to sense and respond to the signals that impinge on it [8]. Organisms are not built from genetic ‘instructions’ alone, but rather self-assemble using a broad variety of inter-dependent resources. Even where there is a history of selection for plasticity, the constructive development perspective entails that prior selection underdetermines the phenotypic response to the environment.
This difference in how development is conceived strongly affects evolutionary interpretations. Readers that view developmental plasticity as programed by genetically specified switches or reaction norms, pre-screened by prior selection, would find it hard to envisage how responses to the environment can be the starting point for evolutionary change as plasticity-led evolution then reduces to selection on genetic variation. Conversely, if, for instance, as a result of exploratory processes, development is constructive and open-ended, entirely new functional phenotypes may be able to emerge with little or no initial genetic modification, yet nonetheless generate critical new raw material for subsequent bouts of selection (e.g. [30]). In such cases, the genetically specified reaction-norm approach is limited, because phenotypic variation results from ontogenetic selective processes, rather than genes, responding to environmental variation.
(b). Reciprocal causation
‘Reciprocal causation’ captures the idea that developing organisms are not solely products, but are also causes, of evolution [90,110,111]. The term ‘reciprocal causation’ simply means that process A is a cause of process B and, subsequently, process B is a cause of process A, with this feedback potentially repeated in causal chains. Reciprocal causation is a common feature of both evolving systems (e.g. when the activities of organisms modify selective environments) and developing systems (where development proceeds through modification of internal and external environments) [102,103,112].
Reciprocal causation can be contrasted with ‘unidirectional’ causation. Consider the example of avian migration: the act of migration does not change the timing or duration of the seasons, and hence migratory behaviour could be portrayed as evolving in response to pre-existing and independent features of the external environment [89]. If correct, this form of evolutionary causation is unidirectional: it starts in the external environment (i.e. with selection) and ends with an adaptive change in the organism (i.e. with modified migratory behaviour). Unidirectional causation has historically been the default assumption within evolutionary biology [71,73,113], and some treatments aligned with traditional perspectives, such as the characterization of niche construction as ‘extended phenotypes’ [87], effectively reduce reciprocally caused phenomena to unidirectional causation.
Contemporary evolutionary biology does recognize reciprocal causation in some cases, such as sexual selection, coevolution, habitat selection and frequency-dependent selection. The peacock's tail (or ‘train’), for instance, evolves through mating preferences in peahens that, in turn, coevolve with the male trait. However, reciprocal causation has generally been restricted to certain domains (largely to direct interactions between organisms), while, many existing analyses of coevolution, habitat- or frequency-dependent selection, are conducted at a level (e.g. genetic, demographic) that removes any consideration of ontogeny. Such studies do capture a core structural feature of reciprocal causation in evolution—namely, selective feedback—but typically fail to recognize that developmental processes can both initiate and co-direct evolutionary outcomes.
By contrast, the EES views reciprocal causation to be a typical, perhaps even universal, feature of evolving and developing systems, characterizing both the developmental origin of phenotypic variation and its evolution in response to changeable features of its environment [27,71,73]. This clearly differs from Mayr's [89] strict separation of proximate and ultimate causation, and his corollary that ontogenetic processes are relevant only to proximate causation [90].
(c). The structure of the extended evolutionary synthesis
This emphasis on constructive development and reciprocal causation leads the EES to recognize several additional classes of evolutionary process (an extension anticipated by Endler [114]), including processes that generate novel variation, bias selection and contribute to inheritance (figure 2).
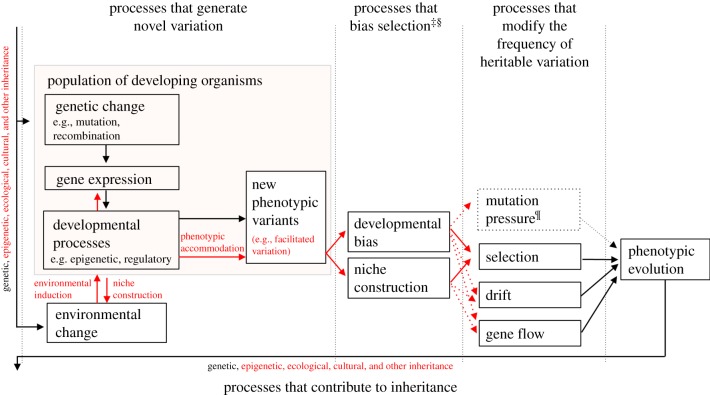
Figure 2.
The structure of the EES. The EES includes as evolutionary causes processes that generate novel variants, bias selection, modify the frequency of heritable variation (including, but not restricted to, genes) and contribute to inheritance. A variety of developmental processes (e.g. epigenetic effects, regulation of gene expression, construction of internal and external developmental environments) contribute to the origin of novel phenotypic variation, which may be viable and adaptive (i.e. ‘facilitated variation’). In addition to accepted evolutionary processes that directly change gene frequencies, the EES recognizes processes that bias the outcome of natural selection, specifically developmental bias and niche construction. All processes that generate phenotypic variation, including developmental plasticity and some forms of inclusive inheritance, are potential sources of bias. A broadened conception of inheritance encompasses genetic, epigenetic and ecological (including cultural) inheritance. Arrows represent causal influences. Processes shown in red are those emphasized by the EES, but not a more traditional perspective. ¶Mutation pressure refers to the population-level consequences of repeated mutation, here depicted as dashed because mutation is also represented in ‘processes that generate novel variation’. ‡In the EES, this category of processes will often need to be broadened to encompass processes that modify the frequencies of other heritable resources. §Developmental bias and niche construction can also affect other evolutionary processes, such as mutation, drift and gene flow. (Online version in colour.)
In agreement with the traditional Darwinian perspective, the EES views variation, differential reproduction and heredity as necessary for adaptive evolution. It differs, however, in how it conceptualizes each of these components and their connections [115]. Explaining the origin of adaptations requires understanding how pre-existing developmental processes generate heritable phenotypic variants from genetic, epigenetic and environmental inputs. Developmental bias and plasticity therefore play central roles in the EES as generators of novel, yet potentially functional and coordinated, phenotypic variation. This conception of bias is different from the traditional characterization of developmental constraints: rather than accounting for the absence of evolution or adaptation, developmental bias is also a source of adaptive variation. Developmental bias and niche construction are, in turn, recognized as evolutionary processes that can initiate and impose direction on selection. Lastly, extra-genetic inheritance mechanisms interact with genetics and environmental inputs to construct the developing organism, thereby contributing to the similarity between ‘transmitting’ and affected individuals.
The EES is thus characterized by the central role of the organism in the evolutionary process, and by the view that the direction of evolution does not depend on selection alone, and need not start with mutation. The causal description of an evolutionary change may, for instance, begin with developmental plasticity or niche construction, with genetic change following [27,73]. The resulting network of processes provides a considerably more complex account of evolutionary mechanisms than traditionally recognized (figure 2).
The most striking and contentious difference from the original MS concerns the relative significance of natural selection versus generative variation in evolution, one of the oldest controversies in evolutionary biology (e.g. [116,117]). In the EES, developmental processes, operating through developmental bias, inclusive inheritance and niche construction, share responsibility for the direction and rate of evolution, the origin of character variation and organism–environment complementarity.
The observations that developmental bias can lead to phenotypic variants that are internally coherent (i.e. well integrated) and can promote functionality in novel environments [25,27,28], result in bias being, at least potentially, probabilistically predictable. The same holds for niche construction, which predictably generates environmental states that are coherent and integrated with the organism's phenotype and its developmental needs, as well as environmental states that are adaptive for the constructor, or its descendants, at least in the short-term [63,73]. Both developmental bias and niche construction impose directionality on evolution, partly because developmental mechanisms have been shaped by prior selection [73], but also because, like other exploratory behaviour within the organism, learning allows organisms to generate and refine novel behavioural variants that are coherent and adaptive [73,118]. Other types of bias may also affect variation and selection, such as systematic biases in mutation [25,116,117,119–121], or other historical contingencies, such as learned traditions [66,73].
As a consequence, the EES predicts that organisms will sometimes have the potential to develop well-integrated, functional variants when they encounter new conditions, which contrasts with the traditional assumption of no relationship between adaptive demand and the supply of phenotypic variation [5,122]. For example, phenotypic plasticity and non-genetic inheritance contributed to the adaptation of the house finch to cold climates during its North American range expansion ([68]; see [27,28,49,101,105,107] for further examples). The EES also anticipates that variants with large phenotypic effect can occur, for example, through mutations in major regulatory control genes (although most such mutations will still be neutral or deleterious) that can be expressed in a tissue- or module-specific manner (e.g. deletion of Pitx1 enhancer that produces pelvis loss and is favoured in sticklebacks) [123]. Other large phenotypic effects occur when developmental processes respond phenotypically to environmental challenges with developmental threshold effects [124], coordinated responses in suites of traits [63] or multiple, stress-induced epigenetic changes [60]. This contrasts with the classical emphasis on gradualism [125,126], which followed from the assumption that, to be adaptive, mutations must have small effects. What the historical rejection of saltationism overlooked was that mechanisms of developmental adjustment allow novel structures to be effectively integrated.
Another distinctive feature of the EES is its recognition that adaptation can arise through both natural selection and internal and external constructive processes. For instance, organisms can respond plastically to novel conditions to generate functional variation. While plasticity is well recognized within the field, what is less well appreciated is that the specific adaptive phenotypes generated need not be the direct targets of past selection, but may be the expression of the more general ability of developmental processes to accommodate novel inputs adaptively, thereby enabling functionally integrated responses to a broad range of conditions [27,34]. Moreover, through niche construction, environments can be changed by organisms to benefit themselves. For instance, Turner [127] notes that, despite living on land for millions of years, earthworms have retained the physiology that is typical of the freshwater oligochaetes from which they evolved. The worms process the soil in ways that allow them to draw water into their bodies more effectively, thereby constructing a simulated aquatic environment on land. The adaptive complementarity of earthworms and soils results to a large extent from the worms changing the soil through niche construction, rather than natural selection changing the worms to a typical terrestrial physiology. Attributing all causal significance to natural selection, by treating earthworm soil-processing as solely proximate causes, linearizes causation, and thereby fails to capture the reciprocal nature of causation in evolution.
This recognition of a variety of distinct routes to phenotype–environment fit furnishes the EES with explanatory resources that traditional perspectives lack. For instance, the well-adapted character of small populations, traditionally regarded as puzzling as selection is weak [128], can potentially be accounted for by the flexible forms of plasticity and niche construction that result from constructive development.
More generally, the EES recognizes that the evolutionary process has a capacity for ‘bootstrapping’ such that prior evolution can generate supplementary information-supplying and adaptation-generating evolutionary processes, expressed in plasticity, learning, non-genetic inheritance, niche construction and culture. In fact, the conceptual change associated with the EES is largely a change in the perceived relationship between genes and development: a shift from a programed to a constructive view of development. Although genes are fundamental to development and heredity, they are not causally privileged in either of these processes [9,129,130]. In the EES, the special evolutionary role of genes (and other components of development) is to be found in a mechanistic description of how DNA affects evolution of life cycles, and not by metaphors such as control, program or blueprint.
5. Novel predictions made, and new research stimulated, by the extended evolutionary synthesis
Conceptual frameworks should be evaluated on their ability to stimulate useful research [1,131]. The EES does make novel predictions, several of which are summarized in table 3, together with an account of the equivalent expectation deriving from a more traditional standpoint. For example, the EES predicts that stress-induced phenotypic variation can initiate adaptive divergence in morphology, physiology and behaviour because of the ability of developmental mechanisms to accommodate new environments (consistent with predictions 1–3 and 7 in table 3). This is supported by research on colonizing populations of house finches [68], water fleas [132] and sticklebacks [55,133] and, from a more macro-evolutionary perspective, by studies of the vertebrate limb [57]. The predictions in table 3 are a small subset of those that characterize the EES, but suffice to illustrate its novelty, can be tested empirically, and should encourage deriving and testing further predictions. Naturally, perspectives encompass a range of views on evolutionary dynamics, and we fully recognize that contemporary evolutionary biologists are represented in this continuum. Nonetheless, table 3 should prove useful because, if we are correct that adoption of an EES requires conceptual change and not just a shift in focus, researchers will tend to favour one set of predictions over another and, ultimately, one set may prove to be more useful. Research in evolutionary biology already provides sufficient data to validate several EES expectations [27,56,73].
Table 3.
A comparison of predictions made by a traditional interpretation and the EES.
| traditional predictions | proposed EES predictions |
|---|---|
| (i) genetic change causes, and logically precedes, phenotypic change, in adaptive evolution | (i) phenotypic accommodation can precede, rather than follow, genetic change, in adaptive evolution |
| (ii) genetic mutations, and hence novel phenotypes, will be random in direction and typically neutral or slightly disadvantageous | (ii) novel phenotypic variants will frequently be directional and functional |
| (iii) isolated mutations generating novel phenotypes will occur in a single individual | (iii) novel, evolutionarily consequential, phenotypic variants will frequently be environmentally induced in multiple individuals |
| (iv) adaptive evolution typically proceeds through selection of mutations with small effects | (iv) strikingly different novel phenotypes can occur, either through mutation of a major regulatory control gene expressed in a tissue-specific manner, or through facilitated variation |
| (v) repeated evolution in isolated populations is due to convergent selection | (v) repeated evolution in isolated populations may be due to convergent selection and/or developmental bias |
| (vi) adaptive variants are propagated through selection | (vi) in addition to selection, adaptive variants are propagated through repeated environmental induction, non-genetic inheritance, learning and cultural transmission |
| (vii) rapid phenotypic evolution requires strong selection on abundant genetic variation | (vii) rapid phenotypic evolution can be frequent and can result from the simultaneous induction and selection of functional variants |
| (viii) taxonomic diversity is explained by diversity in the selective environments | (viii) taxonomic diversity will sometimes be better explained by features of developmental systems (evolvability, constraints) than features of environments |
| (ix) heritable variation will be unbiased | (ix) heritable variation will be systematically biased towards variants that are adaptive and well-integrated with existing aspects of the phenotype |
| (x) environmental states modified by organisms are not systematically different from environments that change through processes independent of organismal activity | (x) niche construction will be systematically biased towards environmental changes that are well suited to the constructor's phenotype, or that of its descendants, and enhance the constructor's, or its descendant's, fitness |
The predictions given in table 3 are all short-term. The EES opens up the possibility of more informed longer term forecasting, by drawing on insights from developmental biology, ecology and computer science to make probabilistic predictions concerning how organisms will respond developmentally to future environmental conditions, and how organisms will modify environments (and hence what selection pressures they will encounter). The EES proposes that variation is more predictable and selection pressures less exogenous than hitherto thought. While it will probably remain difficult to make predictions about how specific populations will evolve, it may be feasible to make and test stochastic predictions concerning future trends, or patterns, across multiple populations. This point relates to Sober's [134] distinction between ‘source laws' (concerned with the properties of processes) and ‘consequence laws' (concerned with their outcomes): a deeper understanding of ecology and developmental biology can potentially provide source laws for natural selection, which will complement those consequence laws currently studied through population genetics [114], enhancing the predictive power of evolutionary analyses.
The EES also raises new questions, informs established lines of inquiry and helps to provide more complete explanations for evolutionary phenomena. EES-style thinking has already contributed constructively to several research questions, including: how do complex novel traits originate? [27,49,57,131,135–137]; how does inclusive inheritance affect the evolutionary process? [60–63,67,68,79,80,138–140]; and how do macro-evolutionary patterns arise? [16,22,56,60,76,141]. In addition, the EES points to some novel lines of inquiry that hitherto have received little attention. Documenting the extent of developmental bias and niche construction becomes of far greater interest to evolutionary biologists once they are recognized as sources of adaptation and diversification. Likewise, questions about the role of plasticity in evolutionary innovation become far more fundamental with a constructive rather than a programed conception of development. Exactly how constructive development can be incorporated into formal evolutionary models is a central issue for the future.
6. The value of an extended evolutionary synthesis
Evolutionary biology has never been more vibrant, and it would be a distortion to characterize it as in a (Kuhnian) state of ‘crisis’. In the EES, all processes central to contemporary evolutionary theory (e.g. natural selection, genetic drift, Mendelian inheritance), and its empirical findings, remain important; in this respect, the EES requires no ‘revolution’. In fact, modern thinking in philosophy of science challenges the hypothesis that scientific change occurs through a single kind of revolution [1,142].
Nevertheless, our analysis suggests that the EES is more than simply an extension of ‘business as usual’ science: it requires conceptual change [15]. The additional evolutionary processes that the EES highlights are more than just non-essential ‘add-ons’ [10] and may be as important in shaping evolution as those recognized within the field over the past century. Consequently, the requisite changes are non-trivial. Irrespective of how this debate unfolds, researchers will continue to make use of the existing quantitative machinery of evolutionary theory; indeed, formal models that incorporate aspects of developmental plasticity, inclusive inheritance and niche construction are already being developed.
Our analysis is motivated by the belief that there is heuristic value in specifying its conceptual structure in sufficient detail for the EES to serve as an alternative ‘ecological-developmental perspective’, to be deployed alongside more traditional standpoints to stimulate useful work. We believe that a plurality of perspectives in science is healthy, as it encourages consideration of a greater diversity of hypotheses, and instigates empirical research, including the investigation of new phenomena. This stance is shared by Arnold [7], who writes: ‘to synthesize, we need diverse perspectives and bridges between them’. By highlighting differences in perspective, we hope to encourage research that distinguishes between alternative expectations and resolves contention. By drawing attention to the need for source laws, we believe that the EES offers the prospect of greater predictive power within the field. By encouraging greater reflection on the plurality of the underlying causes of evolution, the EES should deepen understanding of the mechanisms of evolution.
A further benefit potentially comes through strengthening ties to adjacent disciplines, such as ecology [76,143,144], or the human sciences, including archaeology, biological anthropology, developmental psychology, epidemiology and economics [145–150], where some of these ideas are already starting to have an impact. Moreover, other advances in biology potentially take on new significance within the EES. For instance, the emphasis on inclusive inheritance potentially gives multi-level selection even greater significance, as selection can operate on all forms of heritable variation. Another case is genome evolution, where horizontal gene transfer in prokaryotes, and genetic transfer from endosymbionts in eukaryotes [151], can be understood as part of a broader suite of phenomena with the propensity to propagate horizontally (e.g. social transmission, ecological inheritance). The recognition that genome change is an active cell-mediated physiological process that responds to challenging life-history events [152] fits neatly with the EES's treatment of plasticity. The EES perspective may also facilitate implementation of approaches from computer science that enable mathematical representation of complex dynamic systems, such as connectionist models of memory and learning applied to model genotype to phenotype relations [153]. The EES will be of value in bringing together researchers from diverse fields who share its ecological-developmental perspective.
We expect that evolutionary biology will now enter a phase in which the merits of the EES will be evaluated through empirical and theoretical research, and anticipate that it will contribute constructively to the further evolution of evolutionary theory.
Acknowledgements
We are grateful to Wallace Arthur, Patrick Bateson, Gillian Brown, Doug Erwin, Doug Futuyma, Scott Gilbert, Marc Kirschner, Thomas Morgan and Mary Jane West-Eberhard for helpful comments on earlier drafts.
Authors' contributions
K.L., T.U. and J.O.-S. conceived of the project and took the lead in design, discussion, synthesis, coordination and writing. All authors brought distinctive expertise to the collaboration and contributed importantly to the ideas represented, as well as through the drafting and revising of the article.
Competing interests
We declare we have no competing interests.
Funding
Research supported in part by an ERC Advanced Grant to K.N.L., a Royal Society URF and a Wallenberg Academy Fellowship to T.U., and by the Morrison Institute for Population and Resource Studies at Stanford University (M.W.F.).
References
- 1.Lakatos I. 1978. The methodology of scientific research programmes. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 2.Fleck L. 1979. The genesis and development of a scientific fact. Chicago, IL: Chicago University Press. [Google Scholar]
- 3.Kuhn TS. 1962. The structure of scientific revolutions. Chicago, IL: Chicago University Press. [Google Scholar]
- 4.Provine WB. 1971. The origins of theoretical population genetics. Chicago, IL: Chicago University Press. [Google Scholar]
- 5.Mayr E. 1982. The growth of biological thought: diversity, evolution and inheritance. Cambridge, MA: Belknap Press. [Google Scholar]
- 6.Futuyma DJ. 1998. Evolutionary biology. Sunderland, MA: Sinauer. [Google Scholar]
- 7.Arnold SJ. 2014. Phenotypic evolution: the ongoing synthesis. Am. Nat. 1836, 729–746. ( 10.1086/675304) [DOI] [PubMed] [Google Scholar]
- 8.Keller EF. 2014. From gene action to reactive genomes. J. Physiol. 592, 2423–2429. ( 10.1113/jphysiol.2014.270991) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths P, Stotz K. 2013. Genetics and philosophy. An introduction. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 10.Wray GA, Hoekstra HE, Futuyma DJ, Lenski RE, Mackay TFC, Schluter D, Strassman JE. 2014. Does evolutionary theory need a rethink? No, all is well. Nature 514, 161–164. ( 10.1038/514161a) [DOI] [PubMed] [Google Scholar]
- 11.Gilbert SF, Opitz J, Raff RA. 1996. Resynthesizing evolutionary and developmental biology. Dev. Biol. 173, 357–372. ( 10.1006/dbio.1996.0032) [DOI] [PubMed] [Google Scholar]
- 12.Gould SJ. 2002. The structure of evolutionary theory. Cambridge, MA: Belknap Press. [Google Scholar]
- 13.Pigliucci M, Müller GB (eds). 2010. Evolution: the extended synthesis. Cambridge, MA: MIT Press. [Google Scholar]
- 14.Noble D, Jablonka E, Joyner M, Müller GB, Omholt S. 2014. The integration of evolutionary biology with physiological science. J. Physiol. 592, 2237–2244. ( 10.1113/jphysiol.2014.273151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laland KN. et al. 2014. Does evolutionary theory need a rethink? Nature 514, 161–164. [DOI] [PubMed] [Google Scholar]
- 16.Davidson EH, Erwin DH. 2006. Gene regulatory networks and the evolution of animal body parts. Science 311, 796–800. ( 10.1126/science.1113832) [DOI] [PubMed] [Google Scholar]
- 17.Brakefield P. 2006. Evo-devo and constraints on selection. Trends Ecol. Evol. 21, 362–368. ( 10.1016/j.tree.2006.05.001) [DOI] [PubMed] [Google Scholar]
- 18.Brakefield P. 2011. Evo-devo and accounting for Darwin's endless forms. Phil. Trans. R. Soc. B 366, 2069–2075. ( 10.1098/rstb.2011.0007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller GB. 2007. Evo-devo: extending the evolutionary synthesis. Nat. Rev. Genet. 8, 943–950. ( 10.1038/nrg2219) [DOI] [PubMed] [Google Scholar]
- 20.Atchley W. 1987. Developmental quantitative genetics and the evolution of ontogenies. Evolution 41, 316–330. ( 10.2307/2409141) [DOI] [PubMed] [Google Scholar]
- 21.Badyaev AV, Walsh JB. 2014. Epigenetic processes and genetic architecture. In Quantitative genetics in the wild (eds Charmantier WA, Garant D, Kruuk LEB), pp. 177–189. Oxford, UK: Oxford University Press. [Google Scholar]
- 22.Wagner GP, Altenberg L. 1996. Perspective: complex adaptations and the evolution of evolvability. Evolution 50, 967–976. ( 10.2307/2410639) [DOI] [PubMed] [Google Scholar]
- 23.Sterelny K. 2007. What is evolvability? In Philosophy of biology (eds Matthen M, Stephens C), pp. 163–178. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 24.Wagner GP, Draghi J. 2010. Evolution of evolvability In Evolution: the extended synthesis (eds Pigliucci M, Müller GB), pp. 218–228. Cambridge, MA: MIT Press. [Google Scholar]
- 25.Arthur W. 2004. Biased embryos and evolution. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 26.Arthur W. 2011. Evolution: a developmental approach. Oxford, UK: Wiley-Blackwell. [Google Scholar]
- 27.West-Eberhard MJ. 2003. Developmental plasticity and evolution. Oxford, UK: Oxford University Press. [Google Scholar]
- 28.Kirschner M, Gerhart J. 2005. The plausibility of life: resolving Darwin's Dilemma. New Haven, CT: Yale University Press. [Google Scholar]
- 29.Galis F, Arntzen JW, Lande R. 2010. Dollo's law and the irreversibility of digit loss in Bachia. Evolution 64, 2466–2476. ( 10.1111/j.1558-5646.2010.01041.x) [DOI] [PubMed] [Google Scholar]
- 30.Lange A, Nemeschkal HL, Müller GB. 2014. Biased polyphenism in polydactylous cats carrying a single point mutation: the Hemingway model for digit novelty. Evol. Biol. 41, 262–275. ( 10.1007/s11692-013-9267-y) [DOI] [Google Scholar]
- 31.Beldade P, Koops K, Brakefield PM. 2002. Developmental constraints versus flexibility in morphological evolution. Nature 416, 844–847. ( 10.1038/416844a) [DOI] [PubMed] [Google Scholar]
- 32.Shubin N, Tabin C, Carroll S. 2009. Deep homology and the origins of evolutionary novelty. Nature 457, 818–823. ( 10.1038/nature07891) [DOI] [PubMed] [Google Scholar]
- 33.Albertson RC, Kocher TD. 2006. Genetic and developmental basis of cichlid trophic diversity. Heredity 97, 211–221. ( 10.1038/sj.hdy.6800864) [DOI] [PubMed] [Google Scholar]
- 34.Gerhart JC, Kirschner MW. 2007. The theory of facilitated variation. Proc. Natl Acad. Sci. USA 104, 8582–8589. ( 10.1073/pnas.0701035104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levins R. 1968. Evolution in changing environments. Princeton, NJ: Princeton University Press. [Google Scholar]
- 36.Via S, Lande R. 1985. Genotype–environment interaction and the evolution of phenotypic plasticity. Evolution 39, 505–522. ( 10.2307/2408649) [DOI] [PubMed] [Google Scholar]
- 37.Schlichting CD. 1986. The evolution of phenotypic plasticity in plants. Annu. Rev. Ecol. Syst. 17, 667–693. ( 10.1146/annurev.es.17.110186.003315) [DOI] [Google Scholar]
- 38.Stearns S. 1989. The evolutionary significance of phenotypic plasticity. Bioscience 39, 436–445. ( 10.2307/1311135) [DOI] [Google Scholar]
- 39.Pigliucci M. 2001. Phenotypic plasticity: beyond nature and nurture. Baltimore, MD: John Hopkins University Press. [Google Scholar]
- 40.Yeh PJ, Price TD. 2004. Adaptive phenotypic plasticity and the successful colonization of a novel environment. Am. Nat. 164, 531–542. ( 10.1086/423825) [DOI] [PubMed] [Google Scholar]
- 41.Sol D, Duncan RP, Blackburn TM, Cassey P, Lefebvre L. 2005. Big brains, enhanced cognition, and response of birds to novel environments. Proc. Natl Acad. Sci. USA 102, 5460–5465. ( 10.1073/pnas.0408145102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crispo E. 2008. Modifying effects of phenotypic plasticity on interactions among natural selection, adaptation and gene flow. J. Evol. Biol. 21, 1460–1469. ( 10.1111/j.1420-9101.2008.01592.x) [DOI] [PubMed] [Google Scholar]
- 43.Huey RB, Hertz PE, Sinervo B. 2003. Behavioral drive versus behavioral inertia in evolution: a null model approach. Am. Nat. 161, 357–366. ( 10.1086/346135) [DOI] [PubMed] [Google Scholar]
- 44.Duckworth RA. 2009. The role of behavior in evolution. Evol. Ecol. 23, 513–531. ( 10.1007/s10682-008-9252-6) [DOI] [Google Scholar]
- 45.Cornwallis CK, Uller T. 2010. Towards an evolutionary ecology of sexual traits. Trends Ecol. Evol. 25, 145–152. ( 10.1016/j.tree.2009.09.008) [DOI] [PubMed] [Google Scholar]
- 46.Price TD, Qvarnström A, Irwin DE. 2003. The role of phenotypic plasticity in driving genetic evolution. Proc. R. Soc. Lond. B 270, 1433–1440. ( 10.1098/rspb.2003.2372) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lande R. 2009. Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. J. Evol. Biol. 22, 1435–1446. ( 10.1111/j.1420-9101.2009.01754.x) [DOI] [PubMed] [Google Scholar]
- 48.Pfennig DW, McGee M. 2010. Resource polyphenism increases species richness: a test of the hypothesis. Phil. Trans. R. Soc. B 365, 577–591. ( 10.1098/rstb.2009.0244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moczek AP, et al. 2011. The role of developmental plasticity in evolutionary innovation. Proc. R. Soc. B 278, 2705–2713. ( 10.1098/rspb.2011.0971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baldwin JM. 1902. Development and evolution. New York, NY: MacMillan. [Google Scholar]
- 51.Schmalhausen I. 1949. Factors of evolution: the theory of stabilizing selection. Toronto, Canada: Blakiston. [Google Scholar]
- 52.Schlichting CD, Pigliucci M. 1998. Phenotypic evolution: a reaction norm perspective. Sunderland, MA: Sinauer. [Google Scholar]
- 53.Chevin LM, Lande R. 2010. When do adaptive plasticity and genetic evolution prevent extinction of a density-regulated population? Evolution 64, 1143–1150. ( 10.1111/j.1558-5646.2009.00875.x) [DOI] [PubMed] [Google Scholar]
- 54.Suzuki Y, Nijhout HF. 2006. Evolution of a polyphenism by genetic accommodation. Science 311, 650–652. ( 10.1126/science.1118888) [DOI] [PubMed] [Google Scholar]
- 55.Wund MA, Baker JA, Clancy B, Golub JL, Foster SA. 2008. A test of the ‘Flexible stem’ model of evolution: ancestral plasticity, genetic accommodation, and morphological divergence in the threespine stickleback radiation. Am. Nat. 172, 449–462. ( 10.1086/590966) [DOI] [PubMed] [Google Scholar]
- 56.Pfennig DW, et al. 2010. Phenotypic plasticity's impacts on diversification and speciation. Trends Ecol. Evol. 25, 459–467. ( 10.1016/j.tree.2010.05.006) [DOI] [PubMed] [Google Scholar]
- 57.Standen EM, Du TY, Larsson HCE. 2014. Developmental plasticity and the origin of tetrapods. Nature 513, 54–58. ( 10.1038/nature13708) [DOI] [PubMed] [Google Scholar]
- 58.Schlichting CD, Wund MA. 2014. Phenotypic plasticity and epigenetic marking: an assessment of evidence for genetic accommodation. Evolution 68, 656–672. ( 10.1111/evo.12348) [DOI] [PubMed] [Google Scholar]
- 59.Cavalli-Sforza LL, Feldman MW. 1981. Cultural transmission and evolution. Princeton, NJ: University of Princeton Press. [Google Scholar]
- 60.Jablonka E, Lamb MJ. 2014. Evolution in four dimensions, 2nd edn Cambridge, MA: MIT Press. [Google Scholar]
- 61.Danchin E, et al. 2011. Beyond DNA: integrating inclusive inheritance into an extended theory of evolution. Nat. Rev. Genet. 12, 475–486. ( 10.1038/nrg3028) [DOI] [PubMed] [Google Scholar]
- 62.West MJ, King AP. 1987. Settling nature and nurture into an ontogenetic niche. Dev. Psychobiol. 20, 549–562. ( 10.1002/dev.420200508) [DOI] [PubMed] [Google Scholar]
- 63.Badyaev AV, Uller T. 2009. Parental effects in ecology and evolution: mechanisms, processes and implications. Phil. Trans. R. Soc. B 364, 1169–1177. ( 10.1098/rstb.2008.0302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gilbert SF, Epel D. 2009. Ecological developmental biology: integrating epigenetics, medicine, and evolution. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 65.Uller T, Helanterä H. In press. Heredity and evolution. In Challenges to evolutionary theory: development and heredity (eds Walsh K, Hunemann O). Oxford, UK: Oxford University Press. [Google Scholar]
- 66.Hoppitt W, Laland KN. 2013. Social learning: an introduction to mechanisms, methods, and models. Princeton, NJ: Princeton University Press. [Google Scholar]
- 67.Uller T. 2012. Parental effects in development and evolution. In Evolution of parental care (eds Royle NJ, Smiseth PT, Kölliker M), p. 376 Oxford, UK: Oxford University Press. [Google Scholar]
- 68.Badyaev AV. 2009. Evolutionary significance of phenotypic accommodation in novel environments: an empirical test of the Baldwin effect. Phil. Trans. R. Soc. B 364, 1125–1141. ( 10.1098/rstb.2008.0285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heard E, Martienssen RA. 2014. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157, 95–109. ( 10.1016/j.cell.2014.02.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van der Graaf A, Wardenaar R, Neumann DA, Taudt A, Shaw RG, Jansen RC, Schmitz RJ, Colome-Tatche M, Johannes F. 2015. Rate, spectrum, and evolutionary dynamics of spontaneous epimutations. Proc. Natl Acad. Sci. USA 112, 6676–6681. ( 10.1073/pnas.1424254112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lewontin RC. 1983. Gene, organism and environment. In Evolution from molecules to men (ed. Bendall DS.), pp. 273–285. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 72.Odling-Smee FJ, Laland KN, Feldman MW. 1996. Niche construction. Am. Nat. 147, 641–648. ( 10.1086/285870) [DOI] [Google Scholar]
- 73.Odling-Smee FJ, Laland KN, Feldman MW. 2003. Niche construction: the neglected process in evolution. Monographs in population biology, 37 Princeton, UK: Princeton University Press. [Google Scholar]
- 74.Odling-Smee J. 2010. Niche inheritance. In Evolution: the extended synthesis (eds Pigliucci M, Müller GB), pp. 175–208. Cambridge, MA: MIT Press. [Google Scholar]
- 75.Donohue K. 2014. Why ontogeny matters during adaption: developmental niche construction and pleiotropy across the life cycle in Arabidopsis thaliana. Evolution 68, 32–47. ( 10.1111/evo.12284) [DOI] [PubMed] [Google Scholar]
- 76.Erwin DH. 2008. Macroevolution of ecosystem engineering, niche construction and diversity. Trends Ecol. Evol. 23, 304–310. ( 10.1016/j.tree.2008.01.013) [DOI] [PubMed] [Google Scholar]
- 77.Odling-Smee FJ, Erwin D, Palkovacs E, Feldman M, Laland KN. 2013. Niche construction theory: a practical guide for ecologists. Q. Rev. Biol. 88, 3–28. ( 10.1086/669266) [DOI] [PubMed] [Google Scholar]
- 78.Krakauer DC, Page KM, Erwin DH. 2009. Diversity, dilemmas, and monopolies of niche construction. Am. Nat. 173, 26–40. ( 10.1086/593707) [DOI] [PubMed] [Google Scholar]
- 79.Laland KN, Odling-Smee FJ, Feldman MW. 1996. On the evolutionary consequences of niche construction. J. Evol. Biol. 9, 293–316. ( 10.1046/j.1420-9101.1996.9030293.x) [DOI] [Google Scholar]
- 80.Laland KN, Odling-Smee FJ, Feldman MW. 1999. Evolutionary consequences of niche construction and their implications for ecology. Proc. Natl Acad. Sci. USA 96, 10 242–10 247. ( 10.1073/pnas.96.18.10242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kerr B, Schwilk DW, Bergman A, Feldman MW. 1999. Rekindling an old flame: a haploid model for the evolution and impact of flammability in resprouting plants. Evol. Ecol. Res. 1, 807–833. [Google Scholar]
- 82.Creanza N, Fogarty L, Feldman MW. 2012. Models of cultural niche construction with selection and assertive mating. PLoS ONE 7, e42744 ( 10.1371/journal.pone.0042744) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Silver M, Di Paolo EA. 2006. Spatial effects favour the evolution of niche construction. Theor. Pop. Biol. 70, 387–400. ( 10.1016/j.tpb.2006.08.003) [DOI] [PubMed] [Google Scholar]
- 84.Lehmann L. 2008. The adaptive dynamics of niche constructing traits in spatially subdivided populations: evolving posthumous extended phenotypes. Evolution 62, 549–566. ( 10.1111/j.1558-5646.2007.00291.x) [DOI] [PubMed] [Google Scholar]
- 85.van Dyken JD, Wade M. 2012. Origins of altruism diversity II: runaway coevolution of altruistic strategies via ‘reciprocal niche construction’. Evolution 66, 2498–2513. ( 10.1111/j.1558-5646.2012.01629.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kylafis G, Loreau M. 2008. Ecological and evolutionary consequences of niche construction for its agent. Ecol. Lett. 11, 1072–1081. ( 10.1111/j.1461-0248.2008.01220.x) [DOI] [PubMed] [Google Scholar]
- 87.Dawkins R. 1982. The extended phenotype. Oxford, UK: Oxford University Press. [Google Scholar]
- 88.Scott-Phillips TC, Laland KN, Shuker DM, Dickins TE, West SA. 2013. The niche construction perspective. A critical appraisal. Evolution 68, 1231–1243. ( 10.1111/evo.12332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mayr E. 1961. Cause and effect in biology. Science 134, 1501–1506. ( 10.1126/science.134.3489.1501) [DOI] [PubMed] [Google Scholar]
- 90.Laland KN, Sterelny K, Odling-Smee FJ, Hoppitt W, Uller T. 2011. Cause and effect in biology revisited: is Mayr's proximate–ultimate dichotomy still useful? Science 334, 1512–1516. ( 10.1126/science.1210879) [DOI] [PubMed] [Google Scholar]
- 91.DeWitt TJ, Scheiner SM. 2004. Phenotypic plasticity: functional and conceptual approaches. Oxford, UK: Oxford University Press. [Google Scholar]
- 92.De Jong G. 2005. Evolution of phenotypic plasticity: patterns of plasticity and the emergence of ecotypes. New Phytol. 166, 101–117. ( 10.1111/j.1469-8137.2005.01322.x) [DOI] [PubMed] [Google Scholar]
- 93.Dickins T, Rahman Q. 2012. The extended evolutionary synthesis and the role of soft inheritance in evolution. Proc. R. Soc. B 278, 1721–1727. ( 10.1098/rspb.2010.1726) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Endler JA. 1986. Natural selection in the wild. Princeton, NJ: Princeton University Press. [Google Scholar]
- 95.Futuyma DJ. 2013. Evolution. Sunderland, MA: Sinauer. [Google Scholar]
- 96.Barton N, et al. 2007. Evolution. Cold Spring Harbor, NY: Cold Spring Harbor. [Google Scholar]
- 97.Ridley M. 2004. Evolution, 3rd edn Cambridge, MA: Blackwell. [Google Scholar]
- 98.Lewontin RC. 1970. The units of selection. Annu. Rev. Ecol. Syst. 1, 1–18. ( 10.1146/annurev.es.01.110170.000245) [DOI] [Google Scholar]
- 99.Darwin C. 1859. The origin of species. London, UK: John Murray Press. [Google Scholar]
- 100.Bateson P. 2013. New thinking about biological evolution. Biol. J. Linn. Soc. 112, 268–275. ( 10.1111/bij.12125) [DOI] [Google Scholar]
- 101.Bateson P, Gluckman P. 2011. Plasticity, robustness, development and evolution. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 102.Waddington CH. 1969. Paradigm for an evolutionary process. In Towards a theoretical biology (ed. Waddington CH.), pp. 106–124. Edinburgh, UK: Edinburgh University Press. [Google Scholar]
- 103.Oyama S, Griffiths PE, Gray RD (eds). 2001. Cycles of contingency: developmental systems and evolution. Cambridge, MA: MIT Press. [Google Scholar]
- 104.Noble D. 2006. The music of life. Oxford, UK: Oxford University Press. [Google Scholar]
- 105.Hallgrímsson B, Hall BK. 2011. Epigenetics. Linking genotype and phenotype in development and evolution. Berkeley, CA: University of California Press. [Google Scholar]
- 106.Newman SA. 2012. Physico-genetic determinants in the evolution of development. Science 338, 217–219. ( 10.1126/science.1222003) [DOI] [PubMed] [Google Scholar]
- 107.Snell-Rood EC. 2012. Selective processes in development: implications for the costs and benefits of phenotypic plasticity. Integr. Comp. Biol. 52, 31–42. ( 10.1093/icb/ics067) [DOI] [PubMed] [Google Scholar]
- 108.Mayr E. 1984. The triumph of the evolutionary synthesis. Times Lit. 2, 1261–1262. [Google Scholar]
- 109.Moczek AP. 2012. The nature of nurture and the future of evodevo: toward a comprehensive theory of developmental evolution. Integrative Comp. Biol. 52, 108–119. ( 10.1093/icb/ics048) [DOI] [PubMed] [Google Scholar]
- 110.Laland KN, Odling-Smee J, Hoppitt W, Uller T. 2013. More on how and why: cause and effect in biology revisited. Biol. Phil. 28, 719–745. ( 10.1007/s10539-102-9335-1) [DOI] [Google Scholar]
- 111.Laland KN, Odling-Smee J, Hoppitt W, Uller T. 2013. More on how and why: a response to commentaries. Biol. Phil. 28, 793–810. ( 10.1007/s10539-013-9380-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gilbert SF. 2003. The morphogenesis of evolutionary developmental biology. Int. J. Dev. Biol. 47, 467–477. [PubMed] [Google Scholar]
- 113.Godfrey-Smith P. 1996. Complexity and the function of mind in nature. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 114.Endler JA. 1986. The newer synthesis? Some conceptual problems in evolutionary biology. Oxford Surv. Evol. Biol. 3, 224–243. [Google Scholar]
- 115.Badyaev AV. 2011. Origin of the fittest: link between emergent variation and evolutionary change as a critical question in evolutionary biology. Proc. R. Soc. B 278, 1921–1929. ( 10.1098/rspb.2011.0548) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mivart SGJ. 1871. On the genesis of species. London, UK: Macmillan and Company. [Google Scholar]
- 117.Bateson W. 1894. Materials for the study of variation: treated with especial regard to discontinuity in the origin of species. London, UK: Macmillan and Company. [Google Scholar]
- 118.Plotkin HC, Odling-Smee FJ. 1981. A multiple-level model of evolution and its implications for sociobiology. Behav. Brain Sci. 4, 225–268. ( 10.1017/S0140525X00008566) [DOI] [Google Scholar]
- 119.Nei M. 2013. Mutation driven evolution. Oxford, UK: Oxford University Press. [Google Scholar]
- 120.Stoltzfus A. 1999. On the possibility of constructive neutral evolution. J. Mol. Evol. 49, 169–181. ( 10.1007/PL00006540) [DOI] [PubMed] [Google Scholar]
- 121.Stoltzfus A. 2012. Constructive neutral evolution: exploring evolutionary theory's curious disconnect. Biol. Direct 7, 35 ( 10.1186/1745-6150-7-35) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fisher RA. 1930. The genetical theory of natural selection. Oxford, UK: Clarendon Press. [Google Scholar]
- 123.Chan YF, et al. 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx 1 enhancer. Science 327, 302–305. ( 10.1126/science.1182213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Müller GB. 2010. Epigenetic innovation. In Evolution: the extended synthesis (eds Pigliucci M, Müller GB), pp. 307–332. Cambridge, MA: MIT Press. [Google Scholar]
- 125.Charlesworth B, Lande R, Slatkin M. 1982. A neo-Darwinian commentary on macroevolution. Evolution 36, 474–498. ( 10.2307/2408095) [DOI] [PubMed] [Google Scholar]
- 126.Dawkins R. 1986. The blind watchmaker. New York, NY: Norton. [Google Scholar]
- 127.Turner JS. 2000. The extended organism: the physiology of animal-built structures. Cambridge, MA: Harvard University Press. [Google Scholar]
- 128.Charlesworth B, Charlesworth D. 2010. Elements of evolutionary genetics. Greenwood Village, CO: Roberts and Company Publishers. [Google Scholar]
- 129.Griffiths PE, Gray RD. 1994. Developmental systems and evolutionary explanation. J. Phil. 91, 277–304. ( 10.2307/2940982) [DOI] [Google Scholar]
- 130.Oyama S. 1985. The ontology of information: developmental systems and evolution. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 131.Wagner GP. 2014. Homology, genes, and evolutionary innovation. Princeton, NJ: Princeton University Press. [Google Scholar]
- 132.Scoville AG, Pfrender ME. 2010. Phenotypic plasticity facilitates recurrent rapid adaptation to introduced predators. Proc. Natl Acad. Sci. USA 107, 4260–4263. ( 10.1073/pnas.0912748107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Foster SA, Wund MA. 2011. Epigenetic contributions to adaptive radiation. Insights from threespine stickleback. In Epigenetics: linking genotype and phenotype in development and evolution (eds Hallgrimsson B, Hall BK), pp. 317–336. Berkeley and Los Angeles, CA: University of California Press. [Google Scholar]
- 134.Sober E. 1984. The nature of selection. Cambridge, MA: MIT Press. [Google Scholar]
- 135.Müller GB, Newman SA. 2005. The innovation triad: an EvoDevo agenda. J. Exp. Zool. 304, 487–503. ( 10.1002/jez.b.21081) [DOI] [PubMed] [Google Scholar]
- 136.Moczek AP. 2008. On the origins of novelty in development and evolution. Bioessays 30, 432–447. ( 10.1002/bies.20754) [DOI] [PubMed] [Google Scholar]
- 137.Hallgrimsson B, Jamniczky HA, Young NM, Rolian C, Schmidt-Ott U, Marcucio RS. 2012. The generation of variation and the developmental basis for evolutionary novelty. J. Exp. Zool. B Mol. Dev. Evol. 318, 501–517. ( 10.1002/jez.b.22448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Boyd R, Richerson P. 1985. Culture and the evolutionary process. Chicago, IL: University of Chicago Press. [Google Scholar]
- 139.Bonduriansky R, Day T. 2009. Nongenetic inheritance and its evolutionary implications. Annu. Rev. Ecol. Evol. Syst. 40, 103–125. ( 10.1146/annurev.ecolsys.39.110707.173441) [DOI] [Google Scholar]
- 140.Uller T. 2014. Evolutionary perspectives on transgenerational epigenetics. In Transgenerational epigenetics: evidence and debate (ed. Tollefsbol T.), pp. 175–185. London, UK: Elsevier. [Google Scholar]
- 141.Erwin DH, Valentine JW. 2013. The Cambrian explosion: the construction of animal biodiversity. Greenwood Village, CO: Roberts and Company. [Google Scholar]
- 142.Andersen H, Barker P, Chen X. 2006. The cognitive structure of scientific revolutions. New York, NY: Cambridge University Press. [Google Scholar]
- 143.Sultan SE. 2007. Development in context: the timely emergence of eco-devo. Trends Ecol. Evol. 22, 575–582. ( 10.1016/j.tree.2007.06.014) [DOI] [PubMed] [Google Scholar]
- 144.Matthews B, De Meester L, Jones CG, Iberlings BW, Bouma TJ, Nuutinen V, van der Koppel J, Odling-Smee FJ. 2014. Under niche construction: an operational bridge between ecology, evolution, and ecosystem science. Ecol. Monogr. 84, 245–263. ( 10.1890/13-0953.1) [DOI] [Google Scholar]
- 145.Smith B. 2007. Human niche construction and the behavioral context of plant and animal domestication. Evol. Anthropol. 16, 188–199. ( 10.1002/evan.20135) [DOI] [Google Scholar]
- 146.Bickerton D. 2009. Adam's Tongue. How humans made language, how language made humans. New York, NY: Hill and Wang. [Google Scholar]
- 147.Kendal J, Tehrani JJ, Odling-Smee FJ. 2011. Human niche construction in interdisciplinary focus. Phil. Trans. R. Soc. B 366, 785–792. ( 10.1098/rstb.2010.0306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.O'Brien M, Laland KN. 2012. Genes, culture and agriculture: an example of human niche construction. Curr. Anthropol. 53, 434–470. ( 10.1086/666585) [DOI] [Google Scholar]
- 149.O'Brien M, Laland KN (eds). 2011. Cultural niche construction. Special edition of biological theory, vol. 6 Berlin, Germany: Springer Science and Business Media. [Google Scholar]
- 150.Flynn E, Laland KN, Kendal R, Kendal J. 2013. Developmental niche construction. Dev. Sci. 16, 296–313. ( 10.1111/desc.12030) [DOI] [PubMed] [Google Scholar]
- 151.Koonin EV. 2009. Darwinian evolution in the light of genomics. Nucleic Acids Res. 37, 1011–1034. ( 10.1093/nar/gkp089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shapiro JA. 2011. Evolution: a view from the 21st century. Upper Saddle River, NJ: FT Press Science. [Google Scholar]
- 153.Watson RA, Wagner GP, Pavlicev M, Weinreich DM, Mills R. 2014. The evolution of phenotypic correlations and ‘developmental memory’. Evolution 68, 1124–1138. ( 10.1111/evo.12337) [DOI] [PMC free article] [PubMed] [Google Scholar]