Abstract
The transition from ductal carcinoma in situ (DCIS) to invasive breast cancer (IBC) is a crucial step in breast cancer progression. The specific alterations that govern this transition have not been elucidated. HER2/neu is frequently overexpressed in DCIS but is less common in IBC, thereby suggesting additional requirements for transformation. To identify genes capable of cooperating with HER2/neu to fully transform mammary epithelial cells, we used an insertional mutagenesis screen on cells isolated from wild-type neu expressing mice and identified the E3 ligase HACE1 as HER2 cooperative tumor suppressor gene. Loss of HACE1 expression is commonly seen in clinical breast cancer data sets. HACE1 downregulation in normal human mammary epithelial cells (HMECs) results in the accumulation of the activated GTP-bound Rac1 partially transforming these cells. Overexpression of HER2 activates Rac1, which further accumulates upon HACE1 loss resulting in Rac1 hyperactivation. Although the knockdown of HACE1 or overexpression of HER2 alone in HMECs is not sufficient for tumorigenesis, HER2 overexpression combined with HACE1 downregulation fully transforms HMECs resulting in robust tumor formation. The pharmaceutical interference of Rac function abrogates the effects of HACE1 loss both in vitro and in vivo, resulting in marked reduction in tumor burden. Our work supports a critical role for HACE1 in breast cancer progression and identifies patients that may benefit from Rac-targeted therapies.
Introduction
Breast cancer develops through a multistep process driven by genetic and epigenetic changes that gradually transform normal breast epithelium into pre-invasive (pre-cancerous) lesions such as ductal carcinoma in situ (DCIS) and finally culminate in invasive breast cancer. Although the transition from DCIS to invasive disease has been implicated as the key transition in breast cancer progression, it is a non-obligate step.1, 2 Therefore, the elucidation of alterations that regulate cellular transformation are required to further understand disease progression.
Previous clinical studies identified HER2 (also know as neu) overexpression in DCIS lesions as a predictive indicator for the transition of in situ to invasive breast cancer.3 However, while HER2 overexpression is seen in up to 50% of all DCIS lesions, <25% of invasive breast carcinomas overexpress HER2,4 suggesting that HER2 alone is not sufficient for full malignant transformation.
Genetically engineered mice that express the wild-type neu proto-oncogene under the control of the mouse mammary tumor virus promoter spontaneously develop focal mammary tumors after a prolonged latency period, suggesting that additional alterations are required for tumorigenesis.5 Interbreeding of mouse mammary tumor virus-neu mice with other genetically engineered mice models has identified numerous neu cooperating genes.6 However, this process is both costly and time consuming.
The use of unbiased forward screening approaches has identified novel cancer-associated genes as well as validating known cancer genes. One selection system: validation-based insertional mutagenesis (VBIM), uses a lentiviral-based random insertion of a strong promoter into the genome.7 VBIM was used to identify genes capable of cooperating with HER2/neu enabling malignant transformation. We now provide evidence that the loss of HECT and Ankyrin domain containing E3 ubiquitin ligase 1 (HACE1), an E3 ligase that tags activated Rac1 for proteosomal degradation, leads to breast cancer transformation. Molecular characterization of HACE1 in breast cancer shows that HACE1 attenuates Rac signaling in mammary epithelial cells, and that loss of HACE1 results in enhanced Rac signaling resulting in tumorigenicity. The pharmaceutical inhibition of Rac diminished the effects of HACE1 loss-induced transformation. The clinical significance of loss of HACE1 expression was validated in multiple clinical data sets. Therefore, the ramifications of HACE1 loss further substantiate the importance of Rac signaling in breast cancer and its role in tumor progression.
Results
Insertional mutagenesis screen identifies HACE1 loss that leads to anchorage-independent growth
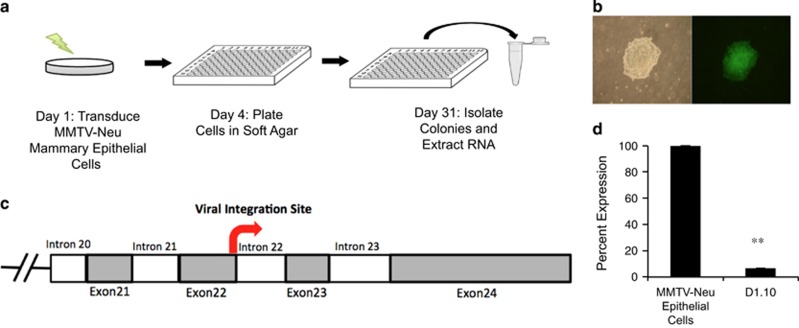
To identify novel contributing factors to breast cancer development and progression, we performed an insertional mutagenesis screen on primary mouse mammary epithelial cells (MMECs) that are partially transformed by expression of the neu proto-oncogene under the control of the mouse mammary tumor virus promoter.5 At 7 weeks of age, these mice show hyperplasia of their ductal trees indicative of neu overexpression. However, isolated MMECs at this time point are unable to form colonies under anchorage-independent conditions. To generate additional genetic events, cells from post-pubescent mice were isolated, transduced with the VBIM lentivirus to generate a library of random transformants7 and then screened for anchorage-independent growth (Figure 1a). Colonies that were formed in soft agar were isolated and 3′ RACE was conducted on isolated RNA using gene-specific primers against components of the virus to identify insertion sites. Sequencing results revealed a candidate list of putative oncogenes and tumor suppressor genes, as well as canonical tumor suppressor genes such as caspase-9 and genes shown to be associated with HER2+ breast cancer such as the mitochondrial transporter protein SLC25A438 (Supplementary Table 1). Sequencing of clone 10 (Figure 1b) identified an out-of-frame insertion in Exon 22 of the murine HACE1, which has previously been identified as an E3 ubiquitin ligase lost in sporadic Wilms' tumor patients (Figure 1c).9 The out-of-frame insertion in clone 10 resulted in a decrease in HACE1 mRNA expression compared with parental cells (Figure 1d). These data support the notion of HACE1 as a breast cancer tumor suppressor gene that is capable of cooperating with HER2/neu.
Figure 1.
Identification of HACE1 as a HER2/neu cooperating tumor suppressor gene. (a) VBIM soft agar screen design. (b) Typical colony formed in soft agar. Bright-field and green fluorescent protein (GFP) filter images taken at × 10 magnification. (c) Mapping of viral integration site of clone 1.10 to murine HACE1 Exon22. (d) RT–PCR for HACE1 on cDNA from parental mouse mammary tumor virus-neu mouse mammary epithelial cells and clone SD1.10. (**P< 0.001 between groups, Student's t-test).
HACE1 expression is reduced in human breast tumors
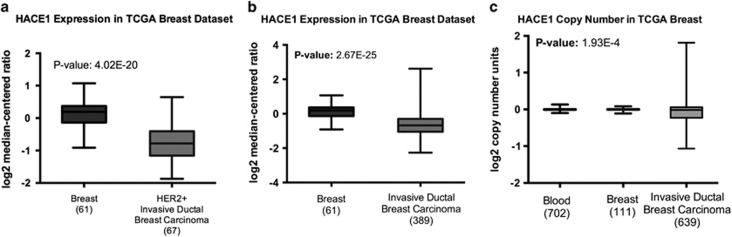
Previous studies have provided evidence that HACE1 is lost in multiple cancer types due to allelic loss or promoter methylation.10, 11, 12, 13 In addition, HACE1 knockout mice have been shown to develop spontaneous tumors at multiple locations, including the breast, after a prolonged latency period.13 HACE1 expression was analyzed in multiple clinical patient data sets including The Cancer Genome Atlas (TCGA) breast data set. HACE1 was found to be highly significantly underexpressed in HER2+ invasive ductal breast carcinoma compared with normal breast epithelium (Figure 2a). Interestingly, HACE1 expression was also highly significantly underexpressed in all invasive ductal breast carcinoma compared with normal breast, suggesting that HACE1 loss is not confined to HER2 positivity (Figure 2b and Supplementary Figure 1). Moreover, allelic loss of HACE1 in invasive ductal breast carcinoma was also observed in the TCGA breast data set (Figure 2b). Thus, underexpression of HACE1 in breast cancer can be attributed to allelic loss of the HACE1 locus.
Figure 2.
Reduced HACE1 expression in breast cancer data sets. (a) mRNA expression analysis for human HACE1 on normal breast and HER2+ invasive ductal breast carcinoma in TCGA breast data set graphed as log2 median-centered ratio. (b) mRNA expression analysis for human HACE1 on normal breast and invasive ductal breast carcinoma in TCGA breast data set graphed as log2 median-centered ratio. (c) HACE1 DNA copy number for blood, normal breast and invasive ductal breast carcinoma in TCGA breast data set graphed as log2 copy number units. Statistical analyses between normal breast and invasive ductal breast carcinoma were conducted using a Student's t-test between groups.
To determine whether HACE1 loss also occurs in other cancer types, we queried HACE1 expression and/or copy number in multiple clinical data sets. Notably, HACE1 was underexpressed or underwent allelic loss in cancer compared with respective normal tissues in glioblastoma, melanoma, lymphoma, lung and pancreatic cancers (Supplementary Figure 1). Taken together, these data show that HACE1 is significantly decreased during the transformation from the normal to malignant state in breast cancer as well as other many other types of cancer.
HACE1 knockdown in human mammary epithelial cells leads to transformation
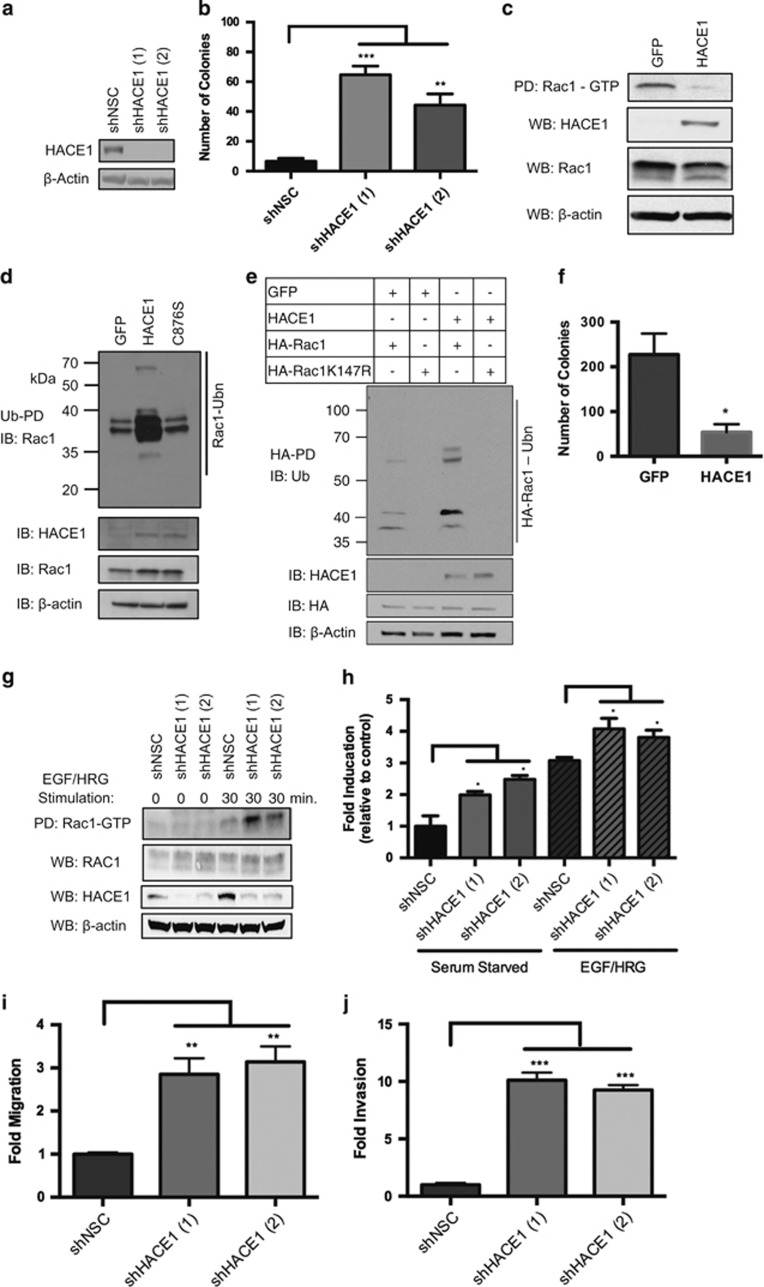
To further validate HACE1 as a human tumor suppressor gene, we determined whether HACE1 ablation in the normal human mammary epithelial cell line MCF12A increased malignant potential. Two independent short hairpin RNAs (shRNAs) targeting HACE1 (shHACE1, 1 and 2) and a non-silencing control (shNSC) were introduced into the MCF12A cells (Figure 3a). Transformed cells were plated in soft agar and colony numbers were determined. Although control MCF12A cells were unable to form colonies, cells expressing both shRNAs against HACE1 showed robust colony formation (Figure 3b). Analysis of HACE1 ablation in another human mammary epithelial cell line, HME3, recapitulated the results found using the MCF12A cells (Supplementary Figures 2A and B). Thus, HACE1 knockdown in normal untransformed mammary epithelial cells is sufficient to drive anchorage-independent growth.
Figure 3.
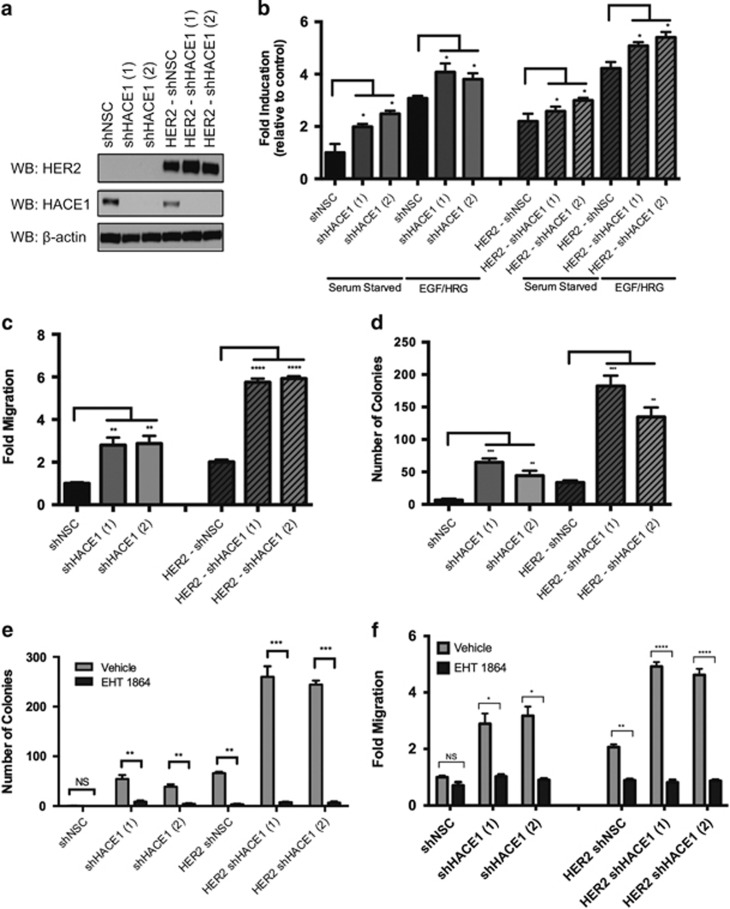
HACE1 controls transformation through regulation of Rac1. (a) HACE1 expression in MCF12A cells after treatment with two independent HACE1-specific shRNAs (shHACE1, 1 and 2) as determined by western blotting. Non-silencing control (NSC) shRNA is shown as control. (b) Colony formation of MCF12A shHACE1 (1 and 3) and shNSC cells. Data are expressed as mean±s.e.m. of three separate experiments. (c) Rac-GTP levels in MCF7 HACE1 overexpressing and GFP control cells stimulated with EGF and HRG as determined by Rac-GTP pull-down. (d) Immunoblots show Rac1 ubiquitylation in HRG- and EGF-stimulated MCF7 cells expressing GFP control cells, HACE1 and HACE1 C876S cells. Ub cross-linked forms of Rac1 were purified (Ub-enrichment), resolved on 10% SDS–PAGE and detected by immunoblot for Rac1. (e) Immunoblots shows Rac1 ubiquitylation in HRG- and EGF-stimulated MCF7 cells expressing GFP control, HACE1, HA-Rac1 and HA-Rac1K147R. Ubiquitylated HA-Rac was enriched by pull-down, resolved by SDS–PAGE and detected by immunoblot for Ubiquitin. (f) Colony formation of MCF7 GFP control cells and MCF7 HACE1 cells. Data are expressed as mean±s.e.m. of three separate experiments. (g) Rac1-GTP levels in MCF12A shHACE1 (1 and 2) knockdown and shNSC cells as determined by Rac1-GTP pull-down. Cells were stimulated for 30 min with 100 ng/ml EGF and 10 ng/ml HRG after overnight starvation. (h) Rac1 fold activation of MCF12A shHACE1 (1 and 2) and shNSC cells as determined by Rac1 G-LISA. Cells were stimulated for 30 min with 100 ng/ml EGF and 10 ng/ml HRG after overnight starvation. Data from triplicates (fold increase relative to NSC in the absence of stimuli) are presented as mean±s.e.m. of three independent experiments. (i) Migration (Boyden chamber) and (j) invasion (modified Boyden chamber) of MCF12A shHACE1 (1 and 2) and shNSC cells (20 h). A quantity of 100 ng/ml EGF and 10 ng/ml HRG was used as chemoattractant between groups, Student's t-test). Data are expressed as mean±s.e.m. of three separate experiments. (*P<0.01, **P<0.001, ***P<0.0001 between groups, Student's t-test).
HACE1 attenuates anchorage-independent growth of human breast cancer cells by reducing levels of activated Rac1
Previous studies have identified the role of ubiquitin-mediated proteosomal degradation of GTP-bound Rac1 in mitigating downstream signaling.14, 15 Recent studies identified HACE1 as the E3 ubiquitin ligase that binds preferentially to GTP-bound Rac1 and catalyzes its polyubiquitylation and subsequent degradation via the proteasome.16, 17 Consequently, the loss of HACE1 resulted in the accumulation of GTP-bound Rac1 resulting in hyperactive Rac signaling. In breast cancer, Rac1 has been reported to be overexpressed and hyperactive.18 Upon activation by guanine nucleotide exchange factors (GEFs), active (GTP-bound) Rac1 has been shown to increase cancer cell migration, proliferation, transcription and survival by activating a multitude of downstream effectors.19 To determine whether HACE1 controlled Rac activation in breast cancer cells, HACE1 was overexpressed in MCF7 breast cancer cell line (Figure 3c). Rac was activated by epidermal growth factor (EGF) and heregulin (HRG), ligands known to potently activate Rac1 in breast cancer cell lines.20 Upon activation of Rac1, control MCF7 cells showed potent activation of Rac1 as determined by Rac1-GTP pull-down. Consistent with previous studies, the overexpression of HACE1 resulted in decreased levels of GTP-bound Rac1 (Figure 3c).9 To confirm that the decrease in activated Rac1 levels was due to the polyubiquitation of Rac1 by HACE1, we performed ubiquitin precipitation experiments in Rac-stimulated MCF7 cells that overexpress HACE1. The overexpression of HACE1 enhanced Rac1 polyubiquitination compared with control cells, whereas the overexpression of catalytically inactive HACE1 (C876S) showed no change in levels of Rac1 polyubiquitination (Figure 3d)16 Moreover, to verify that HACE1 ubiquitylation of Rac1 occurs at lysine 147 (citation), we ectopically expressed HA-tagged wild-type Rac1 as well as a K147R mutant. Consistent with previous groups, we observed that HACE1 overexpression enhances polyubiquitination of wild-type Rac1 but not the K147R mutant (Figure 3d). Thus, the decrease in levels of activated Rac1 in HACE1 overexpressing MCF7 cells is due to the proteosomal degradation of polyubiquitination of Rac1 at lysine 147 by HACE1. Although breast cancer cell lines have reduced levels of HACE1 compared with their normal counterparts (Supplementary Figure 3), we wished to see whether the reduction of HACE1 contributes to the their initial transformation. To do so, MCF7 cells that express HACE1 or control cells were plated in soft agar. Overexpression of HACE1 in MCF7 breast cancer cells reduced colony formation in soft agar compared with control cells (Figure 3d). These results were recapitulated in another breast cancer cell line, ZR-75-1 (Supplementary Figure 4). Together, these data indicate that breast cancer cells have lost HACE1 during their transformative process allowing cells to accumulate activated Rac1. HACE1 expression in breast cancer cell lines can attenuate the levels of activated Rac1 resulting in diminishing their clonogenic potential.
Loss of HACE1 leads to accumulation of active Rac signaling in normal mammary epithelial cells
To determine whether HACE1 loss leads to enhanced Rac1 activity in mammary epithelial cells, MCF12A cells that have HACE1 knocked down were tested for Rac1 activation. Control and shHACE1 knockdown MCF12A cells were serum starved and stimulated with EGF and HRG that induced Rac1 activation (Figure 3e). Knockdown of HACE1 resulted in enhanced Rac1 activation levels over control cells as determined by Rac1-GTP pull-down. Rac1-GTP levels in HACE1 knockdown cells were enhanced >2-fold over control cells as determined by Rac1-GTP enzyme-linked immunosorbent assay21 corroborating the results of the Rac1-GTP pull-down assays (Figure 3f). Together, these results show that HACE1 loss in normal mammary epithelial cells results in enhanced levels of Rac1 signaling.
Growth factor-induced Rac activation has long been associated with actin-cytoskeletal rearrangements resulting in enhanced cell motility.22 We reasoned that the accumulation of active Rac1 due to HACE1 loss would result in an increased migratory ability of normal mammary epithelial cells. To investigate whether HACE1 loss can enhance the migratory ability of normal breast epithelial cells, HACE1-depleted MCF12A cells were placed in the upper portion of a Boyden chamber and allowed to migrate through the semi-permeable membrane toward a chemoattractant. HACE1 knockdown cells showed significant increase in migratory ability (>3-fold) over control cells (Figure 3g and Supplementary Figure 3C). To determine whether HACE1 loss increases invasiveness (a critical element of the transition from DCIS to invasive breast cancer), HACE1-depleted MCF12A cells were plated in the upper portion of a modified Boyden chamber coated with Matrigel. Loss of HACE1 stimulated the ability of the noninvasive MCF12A cells to invade through Matrigel (Figure 3h). Together, HACE1 loss in normal mammary epithelial cells leads to elevated levels of Rac activation resulting in enhanced migratory and invasive capabilities.
Rac1 is required for transformation in HACE1-depleted cells
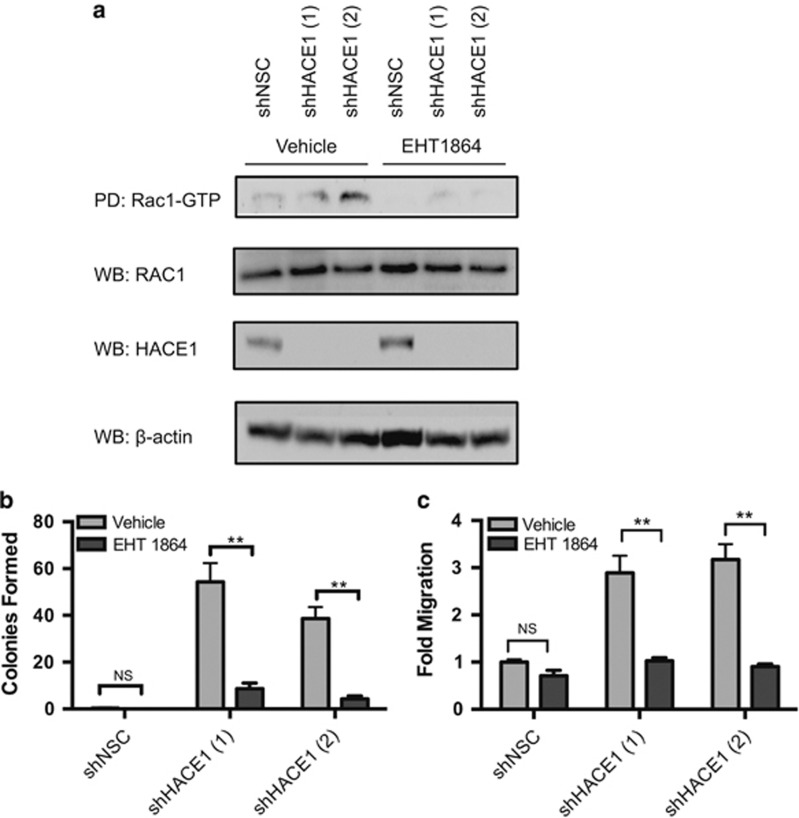
Owing to the exquisite spatial, temporal and substrate specificity requirements for E3 ligases to properly ubiquitylate their target substrates, the number of target substrates is limited. However, E3 ubiquitin ligases have been shown to have multiple target substrates.23 Although HACE1 has been shown to ubiquitylate activated Rac1 for proteosomal degradation, HACE1 has also been reported to be involved in ubiquitylating a Rab GTPase.24, 25 However, Rac1 knockdown in MCF12A cells that had HACE1 knocked down was capable of reverting their migratory, invasive and clonogenic potential, indicating that the phenotypic effects of HACE1 loss is driven by the accumulation of activated Rac1 signaling (Supplementary Figure 5). After establishing Rac1 as the major signaling component resulting from HACE1 loss, we used a small molecule inhibitor EHT1864 known to inhibit Rac by promoting the loss of the nucleotide-bound Rac1 in an inert and inactive state, thus inhibiting downstream signal transduction.26
MCF12A cells were treated with EHT1864 and then stimulated with EGF and HRG to induce Rac1 activation. Pretreatment with the Rac inhibitor EHT1864 at a concentration of 25 μM inhibited Rac signaling in the control MCF12A cells as well as MCF12A cells with HACE1 knocked down, while the vehicle control had no suppression of Rac1 activation (Figure 4a). These results support previous studies that EHT1864 is a potent RAC inhibitor and indicate that EHT1864 can be used to inhibit Rac1 signaling of human mammary epithelial cells. To rule out nonspecific toxicity, we performed a proliferation and cell viability assay on MCF12A cells. Although EHT1864 caused a marked decrease in cellular proliferation in both control and HACE1 knockdown cells, cellular viability was unaffected (Supplementary Figure 6).
Figure 4.
Rac inhibition reverses phenotypic effects of HACE1 loss. (a) Rac1-GTP levels in MCF12A shHACE1 (1 and 2) knockdown and MCF12A shNSC cells as determined by Rac1-GTP pull-down. Cells were stimulated for 30 min with 100 ng/ml EGF and 10 ng/ml HRG in the presence of 25 μM EHT1864 or vehicle after overnight serum starvation. (b) Soft agar colony formation of MCF12A shHACE1 (1 and 2) and MCF12A shNSC cells in the presence of 50 μM EHT1864 or vehicle (**P<0.001 between groups, Student's t-test). Data are expressed as mean±s.e.m. of three separate experiments. (c) In vitro migration (20 h) of MCF12A shHACE1 (1 and 2) and MCF12A shNSC cells as determined by Boyden chamber in the presence of 25 μM EHT1864 or vehicle. A quantity of 100 ng/ml EGF and 10 ng/ml HRG was used chemoattractant (*P<0.01 between groups, Student's t-test). Data are expressed as mean±s.e.m. of three separate experiments.
Just as the knockdown of Rac1 halted anchorage-independent growth (Supplementary Figure 5B), the inhibition of Rac by EHT1864 eliminated the ability of the normal MCF12A cells with HACE1 knockdown to grow colonies in agar (Figure 4b). These results indicate that EHT1864 blockade of Rac1 signaling is as efficacious as Rac1 ablation by RNA interference in inhibiting the growth of HER2 driven cells as well as the GTP-Rac1 accumulated cell lines.
In order to determine the efficacy of EHT1864 on migration, cells were treated with the Rac inhibitor or vehicle control and plated in a Boyden chamber. Treatment of cells with the Rac inhibitor recapitulated the results of Rac1 knockdown resulting in a twofold reduction in migratory ability (Figure 4c). These results suggest that the inhibition of Rac signaling has the ability to reduce migratory gains caused by HACE1 loss similar to that of the molecular knockdown of Rac1, and support the mechanism of action of EHT1864 as an inhibitor of Rac signaling.
HER2 cooperates with HACE1 loss resulting in accumulation of activated RAC1
We identified HACE1 as a tumor suppressor gene that cooperates with HER2. The overexpression of HER2 in normal cells has been previously reported to activate Rac1 by activating RAC1 GEFs such as DOCK1, PREX1 and VAV2.27, 28, 29, 30 To determine whether HACE1 loss would further enhance Rac1 activity when HER2 is overexpressed, MCF12A cells were constructed that stably overexpress HER2 (MCF12A-HER2; Figure 5a). HACE1 was then stably knocked down as described above. Upon activation of Rac1 using HRG and EGF after serum starvation, potent Rac1 activation was observed in the MCF12A-HER2 control cells as determined by Rac1-GTP enzyme-linked immunosorbent assay. Moreover, HACE1 knockdown resulted in even greater levels of Rac1 activation than that of the control cells (Figure 5b), suggesting that Rac1 activation by HER2 overexpression cooperates with HACE1 loss to further enhance Rac1 downstream signaling.
Figure 5.
HER2 cooperates with HACE1 to enhance Rac-mediated transformation. (a) HACE1 expression in MCF12A cells and MCF12A-HER2 cells after treatment with two independent HACE1-specific shRNAs (shHACE1 (1 and 2) as determined by western blot analysis. Non-silencing control (NSC) shRNA is shown as a control. (b) Rac1 fold activation of MCF12A shHACE1 (1 and 2), shNSC, HER2 shHACE1 (1 and 2) and HER2 shNSC cells as determined by Rac1 G-LISA. Cells were stimulated for 30 min with 100 ng/ml EGF and 10 ng/ml HRG after overnight starvation. Data from triplicates (fold increase relative to NSC in the absence of stimuli) are presented as mean±s.e.m. of three independent experiments. (c) Migration (20 h) of MCF12A shHACE1 (1 and 2), shNSC, HER2 shHACE1 (1 and 2) and HER2 shNSC cells. A quantity of 100 ng/ml EGF and 10 ng/ml HRG was used as chemoattractant. Data are expressed as mean±s.e.m. of three separate experiments. (d) Soft agar colony formation of MCF12A shHACE1 (1 and 2), shNSC, HER2 shHACE1 (1 and 2) and HER2 shNSC cells. Data are expressed as mean±s.e.m. of three separate experiments. (e) Colony formation of MCF12A shHACE1 (1 and 2), shNSC, HER2 shHACE1 (1 and 2) and HER2 shNSC cells in the presence of 50 μM EHT1864 or vehicle. Data are expressed as mean±s.e.m. of three separate experiments. (f) Migration (20 h) of MCF12A shHACE1 (1 and 2), shNSC, HER2 shHACE1 (1 and 2) and HER2 shNSC cells in the presence of 25 μM EHT1864 or vehicle. A quantity of 100 ng/ml EGF and 10 ng/ml HRG was used as chemoattractant. Data are expressed as mean±s.e.m. of three separate experiments. (*P<0.01, **P<0.001, ***P<0.0001, ****P<0.00001 between groups, Student's t-test).
To show that elevated Rac1 activation in the MCF12A-HER2 shHACE1 cells correlated with an enhanced cellular phenotype, we performed migration and colony formation assays. HER2 has been previously been shown to enhance migratory and invasive ability of mammary cells through Rac1.31 As expected, HER2 overexpression resulted in migratory ability over the control MCF12A cells (Figure 5c). Knockdown of HACE1 in the MCF12A-HER2 cells resulted in a threefold increase in migration compared with control MCF12A-HER2 cells, suggesting additive effects of HER2 overexpression and HACE1 loss.
Although control MCF12A cells were unable to form colonies in soft agar, the overexpression of HER2 in these cells resulted in anchorage-independent growth (Figure 5d). The knockdown of HACE1 in these MCF12A-HER2 cells resulted in a threefold increase in the number of colonies that were formed, again showing synergy between HER2 and HACE1 loss. The knockdown of Rac1 using shRNA (Supplementary Figure 7) or pharmaceutical inhibition using the Rac inhibitor EHT1864 reversed the cooperative effects observed by HER2 overexpression and HACE1 loss. Rac inhibition suppressed colony formation (Figure 5e) as well as migration (Figure 5f), thereby supporting the role of Rac signaling as the key component of transformation. HACE1 knockdown in a primary HER2+ breast cancer cell line, DT13,32 also resulted in enhanced anchorage-independent growth in soft agar owing to enhanced levels of activated Rac1 (Supplementary Figure 8). EHT1864 suppressed the ability of the DT13 cells to grow in soft agar as well as MCF7 cells (Supplementary Figure 9), suggesting that breast cancers including those that overexpress HER2 may be susceptible to Rac-targeted therapies. Therefore, we propose a model in which overexpression of HER2 results in elevated activation of Rac1. When HACE1 is lost, activated Rac1 accumulates resulting in hyperactive Rac1 signaling (Figure 6).
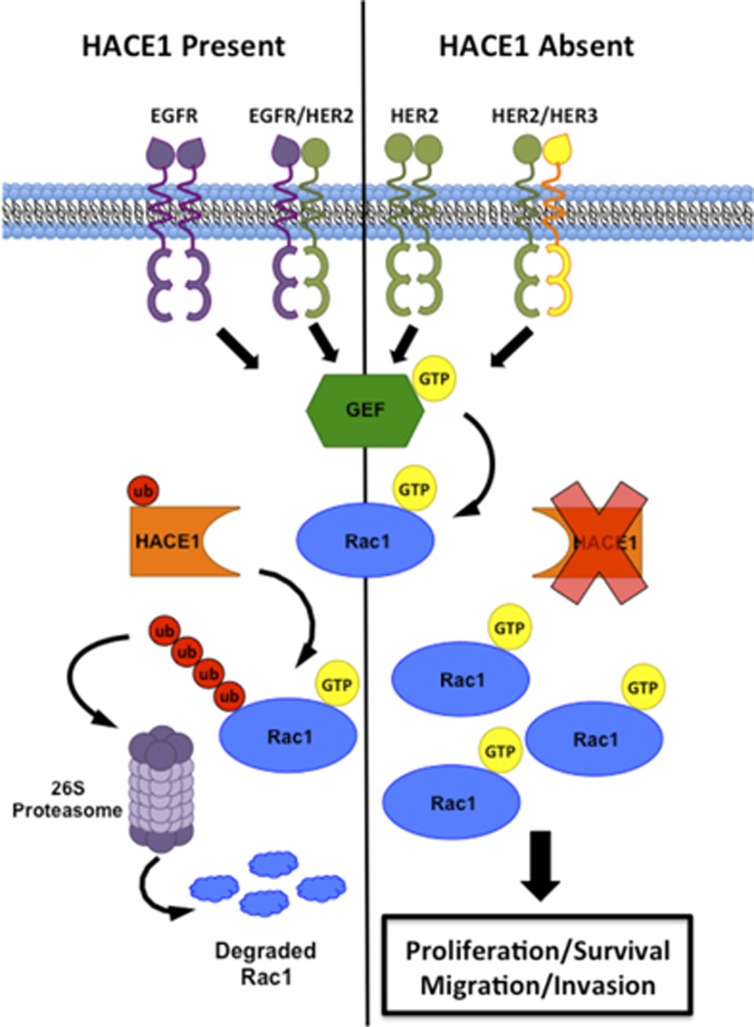
Figure 6.
Schematic overview of HACE1 loss in breast cancer. The overexpression of HER2 results in Rac1 activation by homo- or hetero-dimerization of HER family members. Ubiquitylation of Rac1 occurs upon activation when HACE1 is present and Rac1 is degraded via the 26 S proteasome. When HACE1 is lost, activated Rac1 accumulated in the cell driving proliferation, cell survival, migration and invasion.
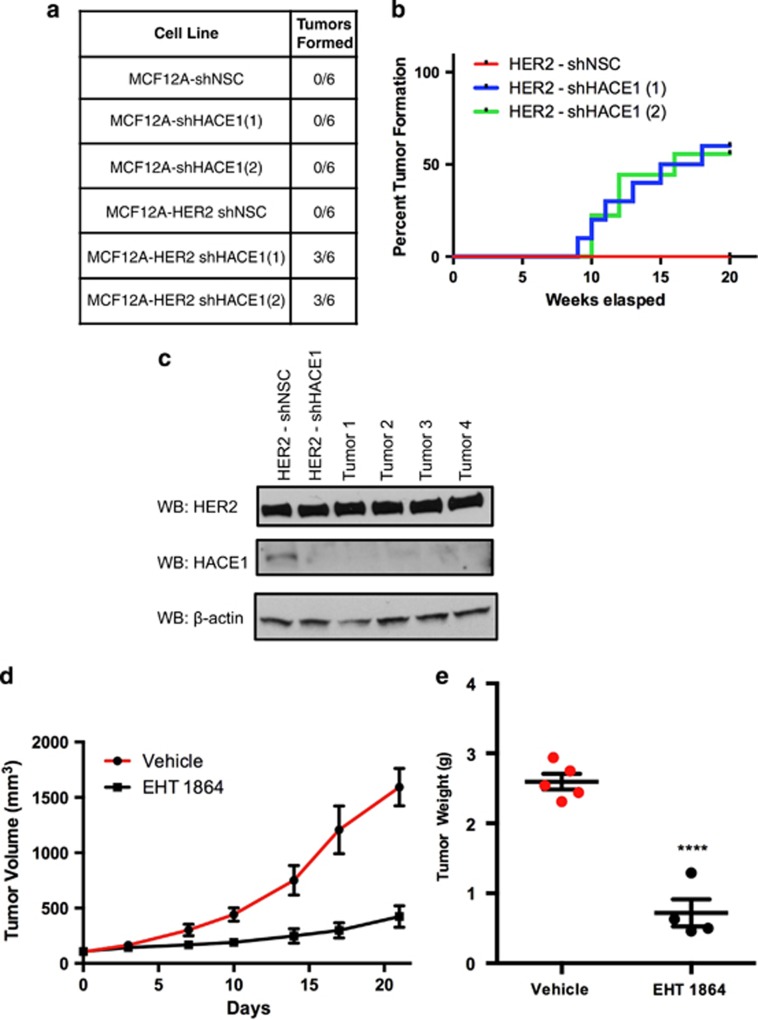
To test whether colony formation translates to tumor formation in vivo, MCF12A and MCF12A-HER2 cell lines with HACE1 knockdown were implanted orthotopically into mammary fat pads of NOD-SCID mice. Concordant with our initial hypothesis, the overexpression of HER2 in the MCF12A cells was not enough to allow full malignant transformation (Figure 7a). The MCF12A cells that had enhanced Rac1 activation due to HACE1 knockdown were also unable to form tumors. Interestingly, only the combination of HER2 and HACE1 loss were able to generate tumors in (6 out of 12) NOD-SCID mice (Figures 7a and b). These results establish HACE1 as a tumor suppressor gene that cooperates with HER2 overexpression to fully transform human mammary epithelial cells. HER2 overexpression results in activation of Rac1 signaling, signaling that is then prolonged when HACE1 is absent. This hyperactivation of Rac1 signaling results in enhanced anchorage-independent growth, migration and tumorigenicity in NOD/SCID mice.
Figure 7.
HER2 cooperates with HACE1 allowing full malignant transformation. (a) In vivo tumor formation of MCF12A shHACE1 (1 and 2), MCF12A shNSC, MCF12A-HER2 shHACE1 (1 and 2) and MCF12A-HER2 shNSC cells. Cells were injected into NOD/SCID mice and allowed to grow for 30 weeks; n=6 per group. (b) Time course of In vivo tumor formation of MCF12A-HER2 shHACE1 (1 and 2) and MCF12A-HER2 shNSC cells. Cells were injected into NOD/SCID mice and allowed to grow for 20 weeks; n=10 per group. (c) Western blot analysis of MCF12A-HER2 shNSC and shHACE1 cell lines and primary tumors (d) MCF12A-HER2 shHACE1 tumor volumes from mice treated with intraperitoneal injections of 30 mg/kg EHT1864 or vehicle. Graphs show mean±s.e.m. of tumor volume at indicated days. n=4 or 5 mice per group. (d) Graph shows tumor weight after 21 days of vehicle or EHT1864 treatment of MCF12A-HER2 shHACE1 tumors (****P<0.00001 between groups, Student's t-test). Graphs show mean±s.e.m. of tumor weight; n=4 or 5 mice per group.
Since our tumor model of HER2 overexpression and HACE1 loss is driven by Rac1 hyperactivation, we evaluated the antitumor activity of EHT1864 in orthotopic-implanted MCF12A-HER2 shHACE1 cells. Mice were pre-randomized to either drug or vehicle control group. Once tumors reached 100 mm3 in volume, a bi-weekly treatment regimen began. As shown in Figure 7c, 21 days of EHT1864 treatment significantly reduced tumor size and tumor weight (Figure 7d) relative to vehicle-treated controls while having no obvious toxicity to the mice. These results support the role of Rac in mammary epithelial cell transformation and demonstrate the in vivo utilization of the Rac inhibitor EHT1864 to suppress tumor growth.
Discussion
Over the course of the last few decades, the incidence of DCIS has significantly increased.33 This increase results in the overtreatment of patients in whom DCIS will not progress to IDC,34 indicating a clear need for predictive markers of disease progression.35, 36 HER2/neu occurs in up to 50% of in situ and only 25% of invasive ductal carcinomas,37 suggesting that additional alterations are required to transform DCIS into invasive disease.
Here, using an insertional mutagenesis screen in mammary epithelial cells that express wild-type neu, we identify and establish HACE1 as a breast tumor suppressor gene that attenuates the levels of activated Rac1. We showed that HACE1 deficiency results in the accumulation of activated Rac1 resulting in enhanced clonogenicty, migration/invasion and tumorigenesis in mice.
We observed that HACE1 is underexpressed in not only HER2+ breast cancers but also in all human breast cancer patients compared with normal breast epithelium. HACE1 was found to undergo allelic loss in breast cancer as well as many other cancers while hypermethylation of the HACE1 promoter has also been observed.13, 38, 39, 40 Gene inactivation of Hace1 in mice results in spontaneous cancers after long latency in multiple sites including the breast, further supporting the notion of HACE1 being a breast tumor suppressor gene.13
The tightly regulated balance between active GTP-bound and inactive GDP-bound Rac1 is governed by GEFs and GAPs, respectively. Overexpression of Rac1 GEFs in breast cancer shift the equilibrium toward active GTP-bound Rac1.19 Activation of ErbB family members have also been shown to activate Rac1 and its subsequent downstream effectors leading to enhanced mitogenic and motile effects through the activation of GEFs. HER2, in particular, activates numerous Rac GEFs including P-Rex1, VAV2 and DOCK1 resulting in Rac activation in breast cancer.28, 29, 30 Constitutively, active Rac1 variants have also been identified in breast cancers such as the splice variant Rac1b18, 41 and activating mutational variants,42 suggesting that Rac1 signaling is a major component in breast cancer.
In this study, we found that knockdown of HACE1 in normal breast epithelial cells resulted in enhanced Rac1 activation causing enhanced migratory and invasive ability as well as anchorage-independent growth. Moreover, chronic activation of Rac1 via HER2 overexpression paired with HACE1 knockdown further enhanced the in vitro transformation of these cells. Although knockdown of HACE1 alone was not sufficient for normal mammary epithelial cells to become tumorigenic in mice, HER2 overexpression coupled with knockdown of HACE1 allowed robust tumor formation. The ability to identify the active state of Rho GTPases histochemically makes it difficult to identify tumors driven by these Rho GTPases. Hence, the overexpression of upstream transmembrane receptors or Rac GEFs that can either directly or indirectly activate Rac1 combined with HACE1 loss may identify Rac-driven tumors.
Our findings demonstrate that while sustained Rac1 signaling (due to HER2 overexpression) may be a potent driver of transformation in human breast cancer, HACE1 is capable of tempering its downstream signaling ability. The Rac inhibitor EHT1864 was able to inhibit Rac1 activation nullifying the phenotypic effects of enhanced Rac signaling in vitro.26, 43 Although Rac1 knockout mice are embryonic lethal, tissue-specific Rac1 knockout mice are viable, suggesting that Rac1 inhibition in the adult animal may be tolerable.44 Our findings using EHT1864 in an in vivo setting demonstrate that systemic administration of a Rac inhibitor can be biologically active and may be tolerable to the host, although further studies optimizing dosing and duration is merited. Furthermore, we propose that patients that have both HER2 overexpression combined with HACE1 loss have hyperactivation of the Rac signaling axis and may benefit from Rac-targeted therapies.
In summary, we performed a screen to identify genes capable of cooperating with HER2 allowing malignant transformation and identified HACE1 as a breast cancer tumor suppressor gene. We found that HACE1 loss in mammary epithelial cells and breast cancer leads to enhanced Rac1 signaling resulting in enhanced migration, invasion and anchorage-independent growth. HACE1 loss coupled with the overexpression of a Rac1 activator such as HER2 results in hyperactivation of Rac signaling and is sufficient to transform normal mammary epithelial cells allowing tumor formation in mice. Because aberrant Rac signaling drives these tumors, treatment with the Rac inhibitor diminishes the oncogenic addiction, thus hauling the Rac-driven phenotypes both in vitro and in vivo. HACE1 loss may therefore be utilized as a surrogate for Rac activation, thus identifying patients at risk of disease progression as well as those susceptible to Rac-targeted therapy.
Materials and methods
Cell culture and reagents
MCF12A cells (American Type Culture Collection) were grown in DMEM/F12 supplemented with 10 mM HEPES, 10 μg/ml insulin, 20 ng/ml EGF, 20 ng/ml cholera toxin, 30, mM sodium bicarbonate, 0.5 μg/ml hydrocortisone and 5% normal horse serum in a humidified atmosphere containing 5% CO2 at 37 °C. MCF7 cells (ATCC) were grown in Improved Minimum Essential Medium (IMEM) (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum. HRG was purchased from R&D Systems (Minneapolis, MN, USA). EGF was purchased from Peprotech (Rocky Hill, NJ, USA). EHT1864 was purchased from Tocris (Minneapolis, MN, USA).
Oncomine and TCGA analysis
Microarray data from oncomine and TCGA analysis were downloaded for different studies, and expression analysis is plotted as log2 median-centered ratio on the y axis for gene expression analysis. DNA copy number analysis was plotted as log2 copy number units.
Virus production and infection
VBIM-SD1 was provided by GR Stark (Cleveland Clinic, Cleveland, OH, USA). The GIPZ shRNAs against HACE1 (shRNA 1: V2LHS_2035241 and shRNA 2: V2LHS_203231) as well as GIPZ shNSC (HS4346) were obtained from Thermo Scientific (Waltham, MA, USA). Viral vectors for HACE1 (E2838-Lv105), HACE1 C876S (E2838-C876S-Lv105), HA-Rac1 (EX-A0247-Lv157), control vector (EX-EGFP-Lv105) and HER2 (EX-Z2866-Lv151) were obtained from Genecopoeia (Rockville, MD, USA). HA-Rac1K147R mutant was created using site-directed mutagenesis (QuickChange II, Agilent Technologies, Santa Clara, CA, USA). Lentiviruses were prepared by transfecting 293 T cells with 6 μg plasmid, 4 μg psPAX2 and 2 μg pMD2G using the Lipofectamine 2000 reagent (Invitrogen). Virus-containing supernatants were harvested 48 h after transfection and were used to infect cells.
Real-time–PCR analysis
mRNA expression was detected by real-time PCR using standard procedures. Primers used include the following: murine HACE1, forward 5′-CGTCAACCCTGACTATGCAC-3′ and reverse 5′-CTGCCTGTGGTTCAAAGCTA-3′ and murine GAPDH, forward 5′-ACCCCAGCAAGGACACTGAGCAA-3′ and reverse 5′-TGGGGGTCTGGGATGGAAATTGTG-3′.
Rac activity assays
Rac1 pull-downs were conducted essentially as previously described (Yang et al.20). In brief, Rac1 pull-down assays were performed on MCF12A and MCF7 cells. Cells were starved in serum-free media when indicated. Activation was stimulated with 100 ng/ml EGF (Peprotech) and 10 ng/ml HRG (R&D Systems). Activation of Rac1 GTPase was also determined by G-LISA Rac1 colorimetric-based kit (Cytoskeleton, Denver, CO, USA). In brief, MCF12A cells were treated as previously described. When indicated, cells were lysed in the provided lysis buffer and were centrifuged to obtain the soluble extract. After correction of total protein concentration according to the manufacturer's instructions, half of the extract volume (40 μl) was mixed with the provided binding buffer, then added to the enzyme-linked immunosorbent assay plates and incubated on ice for 30 min. Wells were washed and active GTPase was detected by specific antibodies according to the manufacturer's instructions in a microplate spectrophotometer (Bio-Rad, Hercules, CA, USA). An aliquot was also analyzed by western blot analysis to confirm shRNA-mediated HACE1 depletion. In our hands, the GTPase activation measured in the G-LISA assays was comparable to that obtained in classical Rac1-PBD pull-down assay but with higher sensitivity and thus requiring considerably less cell lysate.
Western blot analysis
Western blots were conducted as previously described in Yang et al.20 In brief, cells were lysed with RIPA buffer containing protease inhibitors (Thermo Scientific). Lysates (10–50 μg per lane) were separated by SDS–PAGE, and protein was transferred onto a nitrocellulose membrane (Protran, GE Healthcare Life Sciences, Pittsburgh, PA, USA). The membranes were blocked in 5% milk and were incubated overnight with primary antibodies in 5% bovine serum albumin or milk. Specific antibodies to HACE1 (1:1000; ab133637, Abcam, Cambridge, MA, USA), HER2/ErbB2 (1:1000; 29D8, Cell Signaling, Danvers, MA, USA), Rac1 (1:500; 05–389, Millipore, Billerica, MA, USA) and actin (1:5,000; ab49900, Abcam) were detected using the appropriate secondary horseradish peroxidase-conjugated antibodies (Bio-Rad) and were visualized by an enhanced chemiluminescence detection system (Pierce, Rockford, IL, USA).
Ubiquitylation assays
For the enrichment of any ubiquitinated proteins, polyubiquitinated proteins were isolated using high-binding affinity resin (Ubiquitinated Protein Enrichment kit). Cells were serum starved for 16 h and Rac1 activation was achieved using with EGF and HRG while simultaneously being treated with the proteasome inhibitor MG132 (10 μm) for 2 h before cell harvest. Ubiquitin resin was incubated with samples in lysis buffer for 2 h. Resin was then washed four times in the same buffer and bound protein was eluted for SDS–PAGE. The ubiquitinated protein-enriched samples were subjected to immunoblotting for Rac1. For experiments using HA-Rac1, cells were serum starved for 16 h and treated as previously described. Anti-HA agarose (Pierce) was incubated with sample in lysis buffer at 4 °C overnight. Agarose was then washed four times in tris-buffered saline and bound protein was eluted for SDS–PAGE.
Soft agar assay
Soft agar assays were performed as previously described (Yang et al.20). In brief, 5 × 105 cells were suspended in 0.6% granulated agar (Sigma, St Louis, MO, USA) diluted in complete medium (× 2) and poured onto a 0.8% layer of agar. Fresh medium was added every 4 days, and 28 days later colonies were stained with crystal violet and counted.
Transwell migration and invasion assays
Cells were serum starved and 20 000 cells were plated in triplicate into the upper portion of migration or invasion (migration chambers coated with Matrigel) chambers with 8 μM pores (BD Biosciences, San Jose, CA, USA) in 200 μl of serum-free media. A volume of 750 μl of full cell culture medium supplemented with HRG (10 ng/ml) was added to lower chambers as chemoattractant. In experiments using EHT1864, 25 μM EHT1864 or dimethyl sulfoxide was added to media for top and bottom of transwells. After 20 h after plating, media and cells from the upper chambers were removed by aspiration and scrubbing of membranes with cotton swabs. Chambers were fixed in 4% paraformaldehyde for 30 min, washed with phosphate-buffered saline and stained with crystal violet. Five images of the three filters were imaged using a light microscope (× 10 objective) and cells were counted. Data were normalized to migratory counts of control cells and fold migration was calculated. Data were plotted as a fold migration and Student's t-tests were used to determine significance.
Tumor xenograft growth
Experiments involving animals were approved by the Institutional Animal Care and Use Committee of the University of Miami. MCF12A cells were harvested by trypsin digestion and resuspended in phosphate-buffered saline. The MCF12A shHACE1 (1 and 2), MCF12A shNSC, MCF12A-HER2 shHACE1 (1 and 2) and MCF12A-HER2 shNSC cells were mixed with an equal volume of Matrigel (BD Biosciences). Cell/Matrigel mixture (2 × 106 cells in 100 μl) was inoculated orthotopically into the mammary fat pads of 5-week-old NOD/SCID mice (Jackson Laboratories, Bar Harbor, ME, USA). Tumor size was measured as indicated with a caliper. Tumor size was calculated using the following formula: 0.5 × length × width2.
In vivo EHT1864 experiments
Tumors were inoculated into NOD/SCID mice as described above. When tumors reached ~100 mm3 in volume, EHT1864 was administered subcutaneously twice a week at a dose of 30 mg/kg body weight (control animals received equal volumes of vehicle, dimethyl sulfoxide). Tumor volume measurements were recorded every 3–4 days until the end point was reached. Tumor size was calculated using the following formula: 0.5 × length × width2.
Statistical analysis
All of the results shown are expressed as means. Standard errors are depicted by error bars in the graphs. All P-values were calculated between two groups using Student's t-test. P<0.05 was considered statistically significant, unless otherwise indicated.
Acknowledgments
We thank George R Stark for providing us the VBIM plasmids; Tan Ince for providing us the HME3 cells; Dorraya El-Ashry for providing us DT13 cells; and Katie Baker, Nanette Bishopric, Enrique Mesri, Stefan Gluck and Diana Lopez for discussions and technical support.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- 1Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ et al. The genomic landscapes of human breast and colorectal cancers. Science 2007; 318: 1108–1113. [DOI] [PubMed] [Google Scholar]
- 2Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007; 128: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Roses RE, Paulson EC, Sharma A, Schueller JE, Nisenbaum H, Weinstein S et al. HER-2/neu overexpression as a predictor for the transition from in situ to invasive breast cancer. Cancer Epidemiol Biomarkers Prev 2009; 18: 1386–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Latta EK, Tjan S, Parkes RK, O'Malley FP. The role of HER2/neu overexpression/amplification in the progression of ductal carcinoma in situ to invasive carcinoma of the breast. Mod Pathol 2002; 15: 1318–1325. [DOI] [PubMed] [Google Scholar]
- 5Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA 1992; 89: 10578–10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Marcotte R, Muller WJ. Signal transduction in transgenic mouse models of human breast cancer—implications for human breast cancer. J Mammary Gland Biol Neoplasia. 2008; 13: 323–335. [DOI] [PubMed] [Google Scholar]
- 7Lu T, Jackson MW, Singhi AD, Kandel ES, Yang M, Zhang Y et al. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFkappaB. Proc Natl Acad Sci USA 2009; 106: 16339–16344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Tina E, Lindqvist BM, Gabrielson M, Lubovac Z, Wegman P, Wingren S. The mitochondrial transporter SLC25A43 is frequently deleted and may influence cell proliferation in HER2-positive breast tumors. BMC Cancer 2012; 12: 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Anglesio MS, Evdokimova V, Melnyk N, Zhang L, Fernandez CV, Grundy PE et al. Differential expression of a novel ankyrin containing E3 ubiquitin-protein ligase, Hace1, in sporadic Wilms' tumor versus normal kidney. Hum Mol Genet. 2004; 13: 2061–2074. [DOI] [PubMed] [Google Scholar]
- 10Hibi K, Sakata M, Sakuraba K, Shirahata A, Goto T, Mizukami H et al. Aberrant methylation of the HACE1 gene is frequently detected in advanced colorectal cancer. Anticancer Res 2008; 28: 1581–1584. [PubMed] [Google Scholar]
- 11Hyytinen ER, Saadut R, Chen C, Paull L, Koivisto PA, Vessella RL et al. Defining the region(s) of deletion at 6q16-q22 in human prostate cancer. Genes Chromosomes Cancer 2002; 34: 306–312. [DOI] [PubMed] [Google Scholar]
- 12Sakata M, Kitamura YH, Sakuraba K, Goto T, Mizukami H, Saito M et al. Methylation of HACE1 in gastric carcinoma. Anticancer Res 2009; 29: 2231–2233. [PubMed] [Google Scholar]
- 13Zhang L, Anglesio MS, O'Sullivan M, Zhang F, Yang G, Sarao R et al. The E3 ligase HACE1 is a critical chromosome 6q21 tumor suppressor involved in multiple cancers. Nat Med 2007; 13: 1060–1069. [DOI] [PubMed] [Google Scholar]
- 14Nethe M, Hordijk PL. The role of ubiquitylation and degradation in RhoGTPase signalling. J Cell Sci 2010; 123: 4011–4018. [DOI] [PubMed] [Google Scholar]
- 15Visvikis O, Maddugoda MP, Lemichez E. Direct modifications of Rho proteins: deconstructing GTPase regulation. Biol Cell 2010; 102: 377–389. [DOI] [PubMed] [Google Scholar]
- 16Torrino S, Visvikis O, Doye A, Boyer L, Stefani C, Munro P et al. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev Cell 2011; 21: 959–965. [DOI] [PubMed] [Google Scholar]
- 17Castillo-Lluva S, Tan CT, Daugaard M, Sorensen PH, Malliri A. The tumour suppressor HACE1 controls cell migration by regulating Rac1 degradation. Oncogene 2012. [DOI] [PubMed]
- 18Schnelzer A, Prechtel D, Knaus U, Dehne K, Gerhard M, Graeff H et al. Rac1 in human breast cancer: overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene 2000; 19: 3013–3020. [DOI] [PubMed] [Google Scholar]
- 19Wertheimer E, Gutierrez-Uzquiza A, Rosemblit C, Lopez-Haber C, Sosa MS, Kazanietz MG. Rac signaling in breast cancer: a tale of GEFs and GAPs. Cell Signal 2012; 24: 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Yang C, Liu Y, Lemmon MA, Kazanietz MG. Essential role for Rac in heregulin beta1 mitogenic signaling: a mechanism that involves epidermal growth factor receptor and is independent of ErbB4. Mol Cell Biol 2006; 26: 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Hernandez E, De La Mota-Peynado A, Dharmawardhane S, Vlaar CP. Novel inhibitors of Rac1 in metastatic breast cancer. P R Health Sci J 2010; 29: 348–356. [PubMed] [Google Scholar]
- 22Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 1992; 70: 401–410. [DOI] [PubMed] [Google Scholar]
- 23Melino G, Gallagher E, Aqeilan RI, Knight R, Peschiaroli A, Rossi M et al. Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ 2008; 15: 1103–1112. [DOI] [PubMed] [Google Scholar]
- 24Lachance V, Degrandmaison J, Marois S, Robitaille M, Genier S, Nadeau S et al. Ubiquitylation and activation of a Rab GTPase is promoted by a beta2AR-HACE1 complex. J Cell Sci 2014; 127(Pt 1): 111–123. [DOI] [PubMed] [Google Scholar]
- 25Tang D, Xiang Y, De Renzis S, Rink J, Zheng G, Zerial M et al. The ubiquitin ligase HACE1 regulates Golgi membrane dynamics during the cell cycle. Nat Commun 2011; 2: 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Shutes A, Onesto C, Picard V, Leblond B, Schweighoffer F, Der CJ. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J Biol Chem 2007; 282: 35666–35678. [DOI] [PubMed] [Google Scholar]
- 27Ueda Y, Wang S, Dumont N, Yi JY, Koh Y, Arteaga CL. Overexpression of HER2 (erbB2) in human breast epithelial cells unmasks transforming growth factor beta-induced cell motility. J Biol Chem 2004; 279: 24505–24513. [DOI] [PubMed] [Google Scholar]
- 28Sosa MS, Lopez-Haber C, Yang C, Wang H, Lemmon MA, Busillo JM et al. Identification of the Rac-GEF P-Rex1 as an essential mediator of ErbB signaling in breast cancer. Mol Cell 2010; 40: 877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Wang SE, Shin I, Wu FY, Friedman DB, Arteaga CL. HER2/Neu (ErbB2) signaling to Rac1-Pak1 is temporally and spatially modulated by transforming growth factor beta. Cancer Res 2006; 66: 9591–9600. [DOI] [PubMed] [Google Scholar]
- 30Laurin M, Huber J, Pelletier A, Houalla T, Park M, Fukui Y et al. Rac-specific guanine nucleotide exchange factor DOCK1 is a critical regulator of HER2-mediated breast cancer metastasis. Proc Natl Acad Sci USA 2013; 110: 7434–7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Kim IY, Yong HY, Kang KW, Moon A. Overexpression of ErbB2 induces invasion of MCF10A human breast epithelial cells via MMP-9. Cancer Lett 2009; 275: 227–233. [DOI] [PubMed] [Google Scholar]
- 32Drews-Elger K, Brinkman JA, Miller P, Shah SH, Harrell JC, da Silva TG et al. Primary breast tumor-derived cellular models: characterization of tumorigenic, metastatic, and cancer-associated fibroblasts in dissociated tumor (DT) cultures. Breast Cancer Res Treat 2014; 144: 503–517. [DOI] [PubMed] [Google Scholar]
- 33Ernster VL, Barclay J. Increases in ductal carcinoma in situ (DCIS) of the breast in relation to mammography: a dilemma. J Natl Cancer Inst Monogr 1997. 151–156. [DOI] [PubMed]
- 34Baxter NN, Virnig BA, Durham SB, Tuttle TM. Trends in the treatment of ductal carcinoma in situ of the breast. J Natl Cancer Inst 2004; 96: 443–448. [DOI] [PubMed] [Google Scholar]
- 35Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J et al. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res 2003; 1: 362–375. [PubMed] [Google Scholar]
- 36Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003; 100: 5974–5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Barnes DM, Bartkova J, Camplejohn RS, Gullick WJ, Smith PJ, Millis RR. Overexpression of the c-erbB-2 oncoprotein: why does this occur more frequently in ductal carcinoma in situ than in invasive mammary carcinoma and is this of prognostic significance? Eur J Cancer 1992; 28: 644–648. [DOI] [PubMed] [Google Scholar]
- 38Sakata M, Kitamura YH, Sakuraba K, Goto T, Mizukami H, Saito M et al. Methylation of HACE1 in gastric carcinoma. Anticancer Res 2009; 29: 2231–2233. [PubMed] [Google Scholar]
- 39Hibi K, Sakata M, Sakuraba K, Shirahata A, Goto T, Mizukami H et al. Aberrant methylation of the HACE1 gene is frequently detected in advanced colorectal cancer. Anticancer Res 2008; 28: 1581–1584. [PubMed] [Google Scholar]
- 40Sakata M, Yokomizo K, Kitamura Y, Sakuraba K, Shirahata A, Goto T et al. Methylation of the HACE1 gene is frequently detected in hepatocellular carcinoma. Hepatogastroenterology 2013; 60: 781–783. [DOI] [PubMed] [Google Scholar]
- 41Singh A, Karnoub AE, Palmby TR, Lengyel E, Sondek J, Der CJ. Rac1b, a tumor associated, constitutively active Rac1 splice variant, promotes cellular transformation. Oncogene 2004; 23: 9369–9380. [DOI] [PubMed] [Google Scholar]
- 42Kawazu M, Ueno T, Kontani K, Ogita Y, Ando M, Fukumura K et al. Transforming mutations of RAC guanosine triphosphatases in human cancers. Proc Natl Acad Sci USA 2013; 110: 3029–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43Onesto C, Shutes A, Picard V, Schweighoffer F, Der CJ. Characterization of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. Methods Enzymol 2008; 439: 111–129. [DOI] [PubMed] [Google Scholar]
- 44Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol 2008; 9: 690–701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.