Abstract
Background. An influenza A(H1N1)pdm09 infection was diagnosed in a hematopoietic stem cell transplant recipient during conditioning regimen. He was treated with oral oseltamivir, later combined with intravenous zanamivir. The H275Y neuraminidase (NA) mutation was first detected, and an E119D NA mutation was identified during zanamivir therapy.
Methods. Recombinant wild-type (WT) E119D and E119D/H275Y A(H1N1)pdm09 NA variants were generated by reverse genetics. Susceptibility to NA inhibitors (NAIs) was evaluated with a fluorometric assay using the 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUNANA) substrate. Susceptibility to favipiravir (T-705) was assessed using plaque reduction assays. The NA affinity and velocity values were determined with NA enzymatic studies.
Results. We identified an influenza A(H1N1)pdm09 E119D mutant that exhibited a marked increase in the 50% inhibitory concentrations against all tested NAIs (827-, 25-, 286-, and 702-fold for zanamivir, oseltamivir, peramivir, and laninamivir, respectively). The double E119D/H275Y mutation further increased oseltamivir and peramivir 50% inhibitory concentrations by 790- and >5000-fold, respectively, compared with the WT. The mutant viruses remained susceptible to favipiravir. The NA affinity and velocity values of the E119D variant decreased by 8.1-fold and 4.5-fold, respectively, compared with the WT.
Conclusions. The actual emergence of a single NA mutation conferring pan-NAI resistance in the clinical setting reinforces the pressing need to develop new anti-influenza strategies.
Keywords: H1N1 influenza, zanamivir, oseltamivir, resistance, immunosuppression
Since the 2009 pandemic, influenza A(H1N1)pdm09 has remained the predominant circulating influenza virus in the United States [1, 2] and Europe [1]. Influenza A(H1N1)pdm09 strains are uniformly resistant to adamantanes owing to the S31N matrix M2 mutation [1, 2]. However, these strains remain sensitive to neuraminidase (NA) inhibitors (NAIs), such as oseltamivir and zanamivir, the antiviral drugs most frequently used against influenza A and B viruses, as well as other NAIs (eg, peramivir and laninamivir), which are licensed in a limited number of countries. Laninamivir and zanamivir share a common binding process to the viral NA enzyme that involves the 4-guanidino group, which interacts with the carboxylate of the E119 side chain. By contrast, oseltamivir and peramivir contain an acetamide group at the same position. Consequently, clinical influenza isolates with NA substitutions conferring cross- resistance to zanamivir and oseltamivir have been infrequently reported [3].
In addition to the effect of the NAI structure, the NA subtype of the influenza virus may also influence the nature of the NA substitution that could emerge under drug pressure. For instance, with regard to oseltamivir, the most frequently used NAI, the H275Y substitution predominates in the neuraminidase 1 (N1) subtype, such as in the 2009 pandemic and prepandemic A(H1N1) variants, as well as in avian A(H5N1) strains. By contrast, the E119V and R292K substitutions are molecular markers of oseltamivir resistance in A(H3N2) viruses [4, 5]. Because NA resistance substitutions involve residues in or in close proximity to the NA active site, many are likely to influence viral fitness. This may explain why such mutant viruses have not become generally predominant in the community, or the attenuating effect of these mutations may be neutralized by compensatory or permissive changes similar to those observed in the A/Brisbane/59/2007 background. However, immunosuppression is often associated with protracted viral replication and can promote the selection of resistant variants during prolonged exposure to NAIs [6, 7].
We describe the case of an allogeneic hematopoietic stem cell transplant recipient harboring a multidrug-resistant influenza A(H1N1)pdm09 infection associated with pneumonia and sustained viremia mediated by the H275Y/E119D dual NA substitution. Although the H275Y NA mutation is a well-known marker of N1 resistance to oseltamivir, to our knowledge this is the first report of the actual emergence of zanamivir resistance due to the E119D N1 NA change.
METHODS
Virus Isolation
From nasopharyngeal (NP) swab specimens collected on day 21 after the onset of influenzalike symptoms, 400 µL was inoculated onto duplicate Madin–Darby canine kidney (MDCK) cells transfected with the cDNA of human 2,6-sialyltransferase (SIAT1) (European Collection of Cell Cultures) in 24-well plates. After 1 hour of adsorption at 37°C in a 5% carbon dioxide–containing atmosphere, 1 mL of infection medium (Dulbecco plus 2% fetal calf serum, 2 mmol/L glutamine, 0.5 mg/L trypsin, 20 mmol/L HEPES, 1% dimethyl sulfoxide, and antibiotics) was added before reincubation. Cells were monitored daily with light microscopy for the presence of cytopathic effects. Supernatants were recovered 2 and 7 days after inoculation.
Quantitative Real-Time Reverse-Transcription Polymerase Chain Reaction Analysis
The viral genome was extracted from 400 µL of respiratory or plasma specimen individually using the NucliSENS easyMAG nucleic acid kit (bioMérieux). A quantitative real-time reverse-transcription polymerase chain reaction (RT-PCR) assay [8] was performed using One-Step SuperMix (Invitrogen) in a StepOne Real-Time PCR System (Applied Biosystems). Values below the usual routine positive threshold (<500 copies/mL) were arbitrarily given a value of 250 copies/mL.
Sanger Sequencing Analysis
Extracted viral genome was used for the synthesis of complementary DNA using SuperScript II reverse-transcriptase (Invitrogen) and Uni12W primer updated from previously published assays [9]. The NA gene was amplified by PCR using AmpliTaq polymerase (Applied Biosystems). First-round PCR was done with primers cswN1F1 (5′-AGCAAAAGCAGGAGTTTAAAATG-3′) and cswN1R1440 (5′-TAGTAGAAACAAGGAGTTTTTTTGA-3′), followed by 2 independent heminested PCR assays: one with primers cswN1F1 and cswN1R1099 (5′- CCTATCCAAACACCATTGCCGTAT-3′) and the other with primers cswN1F401 (5′-GGAATGCAGAACCTTCTTCTTGAC-3′) and cswN1R1440 (sequences provided by Rod Daniels, Medical Research Council, London, United Kingdom). Amplicons were sequenced with primers cswN1F1, cswN1R1099, cswN1F401, cswN1R1440, and cswN1R424 (5′-GTCAAGAAGAAGGTTCTGCATTCC-3′), using conventional Sanger sequencing performed with the ABI 3500 × L Genetic Analyzer (Applied Biosystems).
High-Throughput Sequencing Analysis
High-throughput sequencing was performed on 4 selected samples for an in-depth analysis of the viral genome and to investigate the evolution of viral quasispecies populations. The viral nucleic acid extraction and the high-throughput RNA sequencing library preparation procedures were performed as described elsewhere [10]. Specimens were sequenced using the 100-bp paired-end protocol, with indexing on a HiSeq 2500 (Illumina) sequencer. To estimate quasispecies sequence diversity, readings were first mapped [11] to all segments of the reference genomes (A/Brisbane/15/2011 and A/Frankfurt/INS403/2010), and the results were then analyzed [12] to infer the structure of the viral population and the nucleotide sequence composition within each specimen (see Supplementary Tables 1–3).
Generation and Characterization of Recombinant Influenza A(H1N1)pdm09 Viruses
Reverse genetics and site-directed mutagenesis were used to generate the recombinant wild-type (WT), E119D, and E119D/H275Y influenza viruses in the background of A/Quebec/144147/09 (an A(H1N1)pdm09-like virus; GenBank accession Nos. FN434457-FN434464), as described elsewhere [13]. These recombinants were tested by NA inhibition assays using the 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUNANA; Sigma) fluorescent substrate for their susceptibility to laninamivir, zanamivir, oseltamivir and peramivir. Recombinant viruses were standardized to an equivalent dose of 106 plaque-forming units/mL and used in a MUNANA-based assay to determine maximal velocity (Vmax) and Michaelis constant (Km) enzymatic parameters [14]. The phenotype of resistance to favipiravir (T-705) was determined by plaque reduction assays [15] using ST6GalI (MDCK-α2,6) cells. Recombinant WT, E119D, and E119D/H275Y A/Quebec/144147/09 NA proteins were expressed in human embryonic kidney (293T) cells to assess their surface NA activity [16].
NA Protein Structure Analysis
Consensus DNA sequences from clinical specimens and the sequences used for in vitro studies (A/Quebec/144147/09) were translated, modeled, and evaluated using the Modeller software package version 9.13 [17] to confirm that substitutions do not elicit structural changes outside the drug-binding pocket, which could affect functionality and/or stability. Models were built and optimized using reference structures “N1 NA H275Y +oseltamivir” (Protein Data Bank [PDB] code, 3CL0) and “N1 NA H275Y + zanamivir” (PDB code, 3CKZ). These structures were chosen because they contain the 3-dimensional configurations of the particular residues required for coordinating inhibitor binding. In brief, translated consensus NA sequences served as input for structure-guided sequence alignment to reference sequences using the ‘align2d’ command in Modeller. For each NA consensus sequence (including A/Quebec/144147/09), 100 models were built using the automodel class. The best models were selected (based on evaluation with the discrete optimized protein energy “DOPE” score) for subsequent structural analysis. Using the PyMOL molecular graphics system [18], the backbones of the models were structurally aligned to the reference structures to highlight any potential large (>2 angstroms) structural differences. To analyze residues in the binding pocket, side-chain differences (rotamer configurations and displacement between models and references) were calculated using the measurement tools within the PyMOL system; all related images were generated in this system.
Ethical Considerations
After submission of the case to the University of Geneva Hospitals' ethics committee, the requirement for informed consent was waived.
RESULTS
Patient Description
A 63-year-old white male patient with relapsing acute myeloid leukemia was admitted to the hospital in February 2014 for mismatched-donor allogeneic hematopoietic stem cell transplantation (HSCT). At day 1 after admission, during HSCT reduced intensity conditioning with antilymphocytic serum, fludarabine, and melphalan and while he was profoundly neutropenic (leukocytes, <0.5 giga/L), the patient developed fever, sore throat, and a cough. Empiric treatment was immediately started with intravenous piperacillin-tazobactam (4.5 g/8 h) and vancomycin (1 g; single dose) and oral oseltamivir. Oseltamivir was stopped after 24 hours because of a negative real-time RT-PCR screening assay result for influenza A/B viruses in nasopharyngeal secretions. At day 9 after admission and symptom onset, the patient received the peripheral hematopoietic stem cell infusion (unrelated donor; HLA compatibility, 9/10) without any complications; graft-vs-host disease (GVHD) prophylaxis consisted of cyclosporine and mycophenolate mofetil.
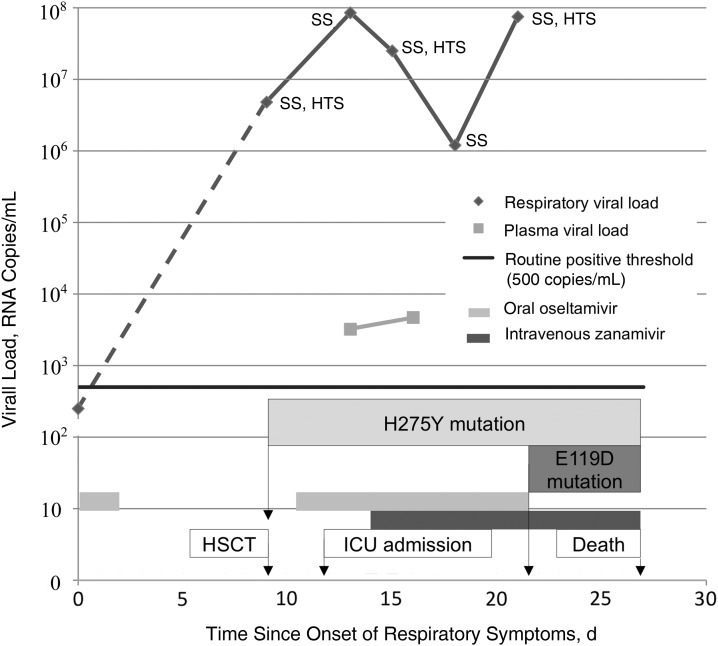
Persistence of fever and worsening of respiratory symptoms at day 1 after HSCT (day 10 after symptom onset) prompted thoracic computed tomographic imaging, which showed new ground-glass pulmonary infiltrates and condensations in all pulmonary lobes, associated with small bilateral pleural effusions. The same day, a new nasopharyngeal swab specimen was positive for influenza A RNA (4.8 ×106 copies/mL), suggesting pneumonia due to the influenza virus infection. Oral oseltamivir therapy (150 mg twice daily) was then resumed. The nasopharyngeal specimen collected at admission was reanalyzed with multiple different PCR assays. One specific assay targeting H1 revealed a slightly positive signal (cycle threshold, 37), suggesting an infection in its very early phase. This signal was below the positive threshold (<500 copies/mL) of the influenza A screening assay used routinely and may have explained why the initial assay result was negative. Subsequently, all respiratory samples yielded positive results for influenza A for the next 12 days, with a respiratory viral load peaking at 8.5 ×107 copies/mL (Figure 1).
Figure 1.
Evolution of upper respiratory and plasma viral load according to neuraminidase inhibitor treatment. Abbreviations: HSCT, hematopoietic stem cell transplantation; HTS, high-throughput sequencing; ICU, intensive care unit; SS, Sanger sequencing.
Despite oseltamivir therapy, the patient's respiratory viral load continued to increase (Figure 1). On day 3 after HSCT (day 12 after symptom onset), the patient presented with hemoptysis, suggesting an alveolar hemorrhage as well as multiorgan failure, requiring intensive care unit admission and intubation. The next day, intravenous zanamivir (600 mg twice daily) was added to oseltamivir on a compassionate-use basis owing to the severity of the clinical presentation. The patient's condition initially improved, with resolution of fever and a decrease in ventilation parameters, but subsequently worsened again. Intravenous methylprednisolone (125 mg once daily) was introduced at day 10 after HSCT (day 19 after symptom onset) for suspected acute digestive GVHD presenting with diarrhea, which was later confirmed by duodenal biopsy. Engraftment occurred at day 12 after HSCT (day 21 after symptom onset), and oseltamivir treatment was stopped because of lack of improvement. Despite zanamivir treatment, the patient's general and respiratory evolution was thereafter unfavorable. At day 18 after HSCT (day 27 after symptom onset), a left middle cerebral artery ischemic stroke associated with cerebral herniation led to the patient's death.
Virologic Investigations
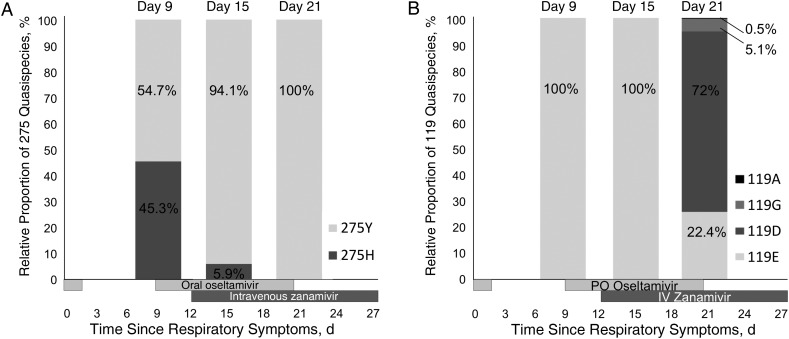
In the first quantifiable swab specimen (day 9 after symptom onset), Sanger sequencing allowed the identification of the H275Y mutation in the NA gene. Importantly, the patient had received 24 hours of oseltamivir treatment 8 days earlier. High-throughput sequencing investigations revealed a mixed population of WT (275H; 45.3%) and mutated (275Y; 54.7%) viruses (Figure 2A). The mutated virus then became dominant with oseltamivir and zanamivir treatment as high-throughput sequencing of the nasopharyngeal samples performed on days 15 and 21 after symptom onset showed proportions of mutated (275Y) viruses of 94.1% and 100%, respectively (Figure 2A).
Figure 2.
Proportion of quasi-species at codons 275 (A) and 119 (B) of the neuraminidase by high-throughput sequencing.
Nasopharyngeal specimens collected on days 9 and 15 after symptom onset showed a 100% population of E119. In the specimen collected pm day 21 after symptom onset and after 8 days of zanamivir treatment, an E119D substitution appeared in a mixed population (119E, 22.4%; 119D, 72%), the remaining readings being intermediary 119G (5.1%) and 119A (0.5%) (Figure 2B). The respiratory specimen displaying the H275Y change and a mixed 119E and 119D population was cultivated in the absence of zanamivir in duplicate. The H275Y and E119D substitutions persisted in the culture stopped after 2 days (moderate cytopathic effect), whereas the culture stopped 7 days after inoculation (stronger cytopathic effect) had the WT genotype for both 119 and 275 codons.
Reverse genetics were used to assess the impact of the E119D mutation in vitro. In NA inhibition assays, the single E119D variant exhibited highly-reduced susceptibility to zanamivir, peramivir, and lanimivir and reduced susceptibility to oseltamivir (50% inhibitory concentration increments of 827-, 286-, 702-, and 25-fold vs WT, respectively) (Table 1). Oseltamivir and peramivir resistance levels were further enhanced in the double E119D/H275Y mutant. All viruses remained susceptible to favipiravir. The E119D substitution caused a significant decrease in the NA affinity (Km, 471.29 vs 58.31 µmol/L) and velocity (Vmax, 3.22 vs 14.40 U/s) compared with the WT (Table 2). This change also significantly reduced surface NA activity (69.4% of the WT) when testing was performed using recombinant NA proteins.
Table 1.
Phenotypic Susceptibility to NAIs and Favipiravir in Recombinant Viruses Containing NA Mutations Identified in a Clinical A(H1N1)pdm09 Isolate
| Recombinant A(H1N1)pdm09 Virus | IC50, Mean (SD), nmol/L [Fold Increase]a |
IC50 for Favipiravir, µg/mLb | |||
|---|---|---|---|---|---|
| Zanamivir | Oseltamivir | Peramivir | Laninamivir | ||
| WT | 0.94 (0.03) [1] | 2.03 (0.01) [1] | 0.14 (0.02) [1] | 0.29 (0.03) [1] | <1 |
| E119D | 778 (64.5) [827] | 50.82 (7.8) [25] | 40.08 (2.6) [286] | 203.63 (9) [702] | <1 |
| E119D/H275Y | 849.13 (31.44) [903] | 1605.26 (31.44) [790] | 834.19 (54.9) [5958] | 106.31 (7.69) [366] | <1 |
Abbreviations: IC50, 50% inhibitory concentration; NA, neuraminidase; NAIs, NA inhibitors; SD, standard deviation; WT, wild type.
a The phenotype of susceptibility to NAIs was determined by NA inhibition assays using the fluorescent 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUNANA)CSITALICSTART CSITALICENDsubstrate; IC50 values are means from 3 independent experiments. Fold increases are in comparison with the recombinant WT virus.
b The phenotype of susceptibility to favipiravir (T-705) was determined by plaque reduction assays using an agarose overlay containing favipiravir at final concentrations ranging from 0.1 to 100 µg/mL. In 2 independent experiments, the 1-µg/mL concentration completely inhibited plaque formation for all tested recombinants.
Table 2.
Enzymatic Properties of the NA Protein of Recombinant Influenza A(H1N1)pdm09 Viruses
| Recombinant A(H1N1)pdm09 Virus | Km, Mean (SD) µmol/La | Vmax, Mean (SD), U/sa | Vmax Ratioa | Surface NA Activity, Mean, (SD) %b |
|---|---|---|---|---|
| WT | 58.31 (4.29) | 14.40 (0.62) | 1 | 100 |
| E119D | 471.29 (46.5)c | 3.22 (0.58)d | 0.22 | 69.4 (0.75)c |
| E119D/H275Y | 252.29 (27.14)c | 1 (0.22)e | 0.07 | 10 (1.03)d |
Abbreviations: Km, Michaelis constant; NA, neuraminidase; SD, standard deviation; Vmax, maximum velocity; WT, wild type.
a Recombinant viruses were used at a standardized amount of 106 plaque-forming units to determine NA enzyme kinetic parameters. Values represent mean Km and Vmax from a representative experiment performed in triplicate.
b Recombinant NA proteins were expressed in 293T cells for the determination of surface NA activity using the fluorescent 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUNANA) substrate. Values represent mean NA activity relative to the recombinant WT protein from triplicate experiments.
c P < .05 (vs recombinant WT).
d P < .01 (vs recombinant WT).
e P < .001 (vs recombinant WT).
The Km and Vmax values for the double E119D/H275Y mutant virus were 252.29 µmol/L and 1 U/s, respectively, whereas the double E119D/H275Y mutant protein demonstrated only 10% of surface NA activity compared with the WT (Table 2). The single E119D genotype (codon GAC) was maintained after 4 passages in MDCKα2,6 cells without zanamivir. By contrast, after only 2 passages, this codon was replaced by a mixture of GAA and GAG codons, with both coding for glutamic acid (E) in the passage of the double E119D/H275Y mutant (data not shown). This D119E reversion was accompanied by the return to the susceptible phenotype against zanamivir and laninamivir and conserved resistance to oseltamivir and peramivir (data not shown). As expected, the double (E119D/H275Y) mutation was maintained when cell culture passages were performed in the presence of zanamivir (final concentration, 0.1 or 1 µmol/L).
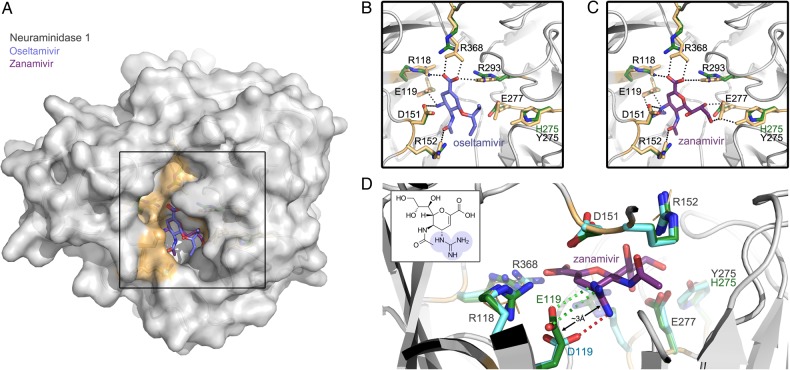
Because amino acid substitutions can alter protein backbone conformation (eg, NA epitopes) and influence functionality, we built models to assess the degree of structural similarity of the recombinant NA proteins used for in vitro studies to WT structures bound to either oseltamivir or zanamivir (PDB code, 3CL0 and 3CKZ) (Figure 3). Structural alignments confirm that our recombinant NA proteins, and those found in clinical specimens, are almost identical to known structures and have similar drug-binding pockets (Figure 3B and 3C).
Figure 3.
Neuraminidase 1 (N1) molecular detail. A, N1 protein highlighting residues (shaded orange) that bind oseltamivir (blue molecule) and zanamivir (purple molecule) inhibitors. B, C, Details of the binding pocket from N1 used for in vitro studies (green side chains) aligned to structure Protein Data Bank codes 3CL0 (B) and 3CKZ (C) (gray backbones, orange side chains). D, Overlay of known structure (orange and gray), in vitro model (green side chains), and model from day 21 specimen (cyan side chains). The positively charged guanidino group of zanamivir (highlighted in blue on the 2D chemical structure inlay) forms 2 salt bridges with E119 (green dotted lines) that stabilize zanamivir binding. The shorter side chain of 119D (cyan) allows only 1 salt bridge to this guanidino group (red dotted line). This reduced ability to form salt bridges with zanamivir probably destabilizes inhibitor binding.
Plasma specimens collected at days 13 and 16 were positive for influenza by quantitative real-time RT-PCR, with a viral load of 3.2 ×103 and 4.7 ×103 copies/mL, respectively (Figure 1). Other plasma specimens collected were positive between days 11 and 24 after symptom onset, but quantification was not possible due to inappropriate storage (data not shown). Owing to a low viral load in blood, it was not possible to culture the virus and confirm the presence of infectious virus in blood.
DISCUSSION
The resistance pattern for N1 viruses observed so far has followed mostly a dichotomous pattern, either spreading in the community without drug pressure [19–21] or limited to highly immunosuppressed hosts treated with antivirals [22, 23]. In the first situation, viral fitness is often preserved, possibly because of other mutations in the NA, whereas fitness is often decreased in the second instance. NA substitutions do not generally confer cross-resistance to zanamivir and oseltamivir owing to the differences between these agents in chemical structure and binding affinity to the active site of the NA enzyme. One notable exception is the R292K substitution, which confers a high level of resistance to oseltamivir and reduced inhibition to zanamivir in H3N2 viruses [3]. The H275Y mutation described elsewhere in H5N1- and H1N1-infected patients [24, 25] impairs the full rotation of E277 (Figure 3B–D), which is necessary for oseltamivir but not for zanamivir high-affinity binding [26].
Between 2007 and 2009, most seasonal A/Brisbane/59/2007 (H1N1) strains acquired this mutation, thus highlighting a preserved viral fitness despite the absence of antiviral pressure [27]. This phenomenon was attributed to permissive NA mutations, whose presence in that background may have facilitated the emergence and worldwide spread of the H275Y variant [16, 28]. Fortunately, global prevalence of the H275Y NA mutation in the A(H1N1)pdm09 background has remained sporadic worldwide since 2009 [29–32]. This risk may eventually increase because most A(H1N1)pdm09 circulating strains carry V241I and N369K NA mutations that have a potential permissive effect [33, 34]. Moreover, these 2 mutations have been recently reported in oseltamivir-resistant community clusters with no previous NAI exposure [35, 36]. Immunosuppression and previous oseltamivir treatment are known risk factors for resistance selection [6, 7, 22]. The rapid emergence of the H275Y mutation in our patient suggests it was already present within the viral population and selected during oseltamivir treatment.
Glutamic acid at position 119 is highly conserved in influenza A and B NAs. This residue is known to stabilize the NA catalytic site and therefore enhances cleavage of sialic acid residues [37]. Besides its interaction with the sialic acid substrate, E119 plays a crucial role in the binding of the NA enzyme to the guanidine group present in zanamivir, laninamivir and peramivir (Figure 3D). E119 is also involved in the interaction between the NA enzyme and the C4 hydroxyl group present in oseltamivir. The replacement of glutamic acid (E) by aspartic acid (D), which has a shorter side chain, may alter these interactions and result in the cross-resistance phenotype. Indeed, our 3-dimensional structure analysis revealed that although the overall integrity of the binding pocket is maintained, the shorter side chain of 119D removes 1 of 2 salt bridges that coordinate the guanidine group of zanamivir (Figure 3D).
Other substitutions of residue E119 may result also in altered binding processes and NAI resistance in influenza A or B variants. A change to glycine or alanine (as observed on day 21), or even valine (nonpolar side chain) at position 119 would remove both interactions, thus further destabilizing inhibitor binding. E119 V is a known marker of oseltamivir resistance in H3N2 viruses. The same substitution conferred cross-resistance to zanamivir, peramivir, oseltamivir, and A-315675 when introduced in recombinant influenza A/WSN/33 (H1N1) variants [38]. A similar resistant phenotype was obtained in the A(H1N1)pdm09 E119V recombinant [13]. More recently, we reported the selection of the E119K NA substitution in influenza A(H1N1)pdm09 viruses submitted to laninamivir pressure in vitro [14]. Therefore, it is plausible that the E119D mutation recovered in our patient was due to zanamivir pressure. Nevertheless, the simultaneous presence of the E119D and H275Y mutations in the A(H1N1)pdm09 background is worrisome because of the resulting multidrug-resistant phenotype.
The E119D mutation was initially absent in our patient and appeared after 8 days of zanamivir treatment in a mixed population, with >70% harboring the 119D codon. As shown in Figure 1, there was a slight transient decrease in viral load after zanamivir introduction, followed by a secondary increment. This could be explained by the induction of in vivo resistance (E119D) but may also be explained by an increase of immunosuppression secondary to GVHD. Owing to this secondary 5–10-fold increase in respiratory viral load, it can be hypothesized that the fitness of the viral mutant was relatively preserved under drug pressure. Although it has still to be proved, one may speculate that the viral mutant detected in our immunosuppressed patient would have been cleared in an immunocompetent patient.
The double mutant E119D/H275Y virus demonstrated extensive resistance to all NAIs. A few reports have described in vivo resistance to oseltamivir, zanamivir, and peramivir [23, 39] in patients carrying a double mutation (ie, H275Y and I223R). Nevertheless, the level of resistance to zanamivir was relatively low in these cases. Recently, Baek et al [40] showed that recombinant influenza A/California/04/2009 [A(H1N1)pdm09] viruses harboring E119D and E119D/H275Y mutations also displayed reduced sensitivity to oseltamivir, zanamivir and peramivir with a synergistic effect being observed for the double E119D/H275Y mutant compared with the single E119D mutant. However, in that report, both the single (E119D) and double (E119D/H275Y) variants were found to be genetically stable after 4 passages in MDCK cells, as well as in mice. By contrast, in our double (E119D/H275Y) recombinant mutant, the E119D mutation reverted to the WT genotype after 2 passages, whereas the E119D genotype was not altered after cell-culture passaging of the single mutant.
Thus, our findings suggest a compromising effect for the double mutation that could be attributed to the more detrimental impact on the NA velocity (Vmax) and cell surface NA activity observed in our experiments (Table 2). The fact that the E119D NA mutation was present in a mixed population in our clinical specimen impaired the ability to accurately determine the impact of this change on the A(H1N1)pdm09 NA properties. On the other hand, this information could be obtained by the use of reverse genetics. It should be noted that the NA proteins from the clinical isolates described in this study share approximately 98% amino acid identity with our recombinant (A/Quebec/144147/09) virus and exhibit almost identical structures (Figure 3). In addition, there was a 98.4% amino acid identity between the HA of A/QC/144147/09 and that of the clinical isolate, suggesting a close genetic and antigenic relationship among these strains.
The most striking feature of our study is that the single E119D mutation alone led to increased resistance to all NAIs. To our knowledge, this is the first clinical report of a single mutation conferring pan-NAI resistance and highlights the plasticity of influenza viruses and their endless ability to evolve [41]. Due to intrinsic resistance to the adamantanes, there is no effective antiviral treatment against an influenza A(H1N1)pdm09 virus harboring the E119D mutation, except favipiravir, an inhibitor of the viral RNA polymerase [15], which is currently not licensed. These results confirm the importance of monitoring drug resistance and the continuous need for antiviral research and development.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We acknowledge Caroline Tapparel for her constructive criticism, Bruno Lina for his work, and Rosemary Sudan for editorial assistance.
Disclaimer. The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Financial support. This work was supported by the Laboratory of Virology in Geneva (research grant), the Swiss Federal Office of Public Health (grant 11.004805/301.0001-705), the Swiss National Foundation (grant 32003B-146993), and the Canadian Institutes of Health Research (grant 229733 to G. B.).
Potential conflicts of interest. T. J. P. has received a research grant from the Louis-Jeantet Foundation. Y. C. has served as a consultant for Novartis, Bristol-Myers Squibb, Pfizer, and Ariad, and Merck Sharp & Dohme and Celgene have contributed to his meeting expenses. G. B. has received research grants from GlaxoSmithKline, Canadian Institutes of Health Research, and Biota. L. K. has received research grants from the Swiss Federal Office of Public Health, the Swiss National Science Foundation, and the University of Geneva Hospitals. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. Review of the 2013–2014 winter influenza season, northern hemisphere. Wkly Epidemiol Rec 2014; 89:245–56. [PubMed] [Google Scholar]
- 2.Epperson S, Blanton L, Kniss K et al. Influenza activity - United States, 2013–14 season and composition of the 2014–15 influenza vaccines. MMWR Morb Mortal Wkly Rep 2014; 63:483–90. [PMC free article] [PubMed] [Google Scholar]
- 3.McKimm-Breschkin JL. Influenza neuraminidase inhibitors: antiviral action and mechanisms of resistance. Influenza Other Respir Viruses 2013; 7(Suppl 1):25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zurcher T, Yates PJ, Daly J et al. Mutations conferring zanamivir resistance in human influenza virus N2 neuraminidases compromise virus fitness and are not stably maintained in vitro. J Antimicrob Chemother 2006; 58:723–32. [DOI] [PubMed] [Google Scholar]
- 5.Baz M, Abed Y, Simon P, Hamelin ME, Boivin G. Effect of the neuraminidase mutation H274Y conferring resistance to oseltamivir on the replicative capacity and virulence of old and recent human influenza A(H1N1) viruses. J Infect Dis 2010; 201:740–5. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization. Oseltamivir resistance in immunocompromised hospital patients. Pandemic (H1N1) briefing note 18. 2009. http://www.who.int/csr/disease/swineflu/notes/briefing_20091202/en/. Accessed 16 December 2014.
- 7.Graitcer SB, Gubareva L, Kamimoto L et al. Characteristics of patients with oseltamivir-resistant pandemic (H1N1) 2009, United States. Emerg Infect Dis 2011; 17:255–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piralla A, Daleno C, Pariani E et al. Virtual quantification of influenza A virus load by real-time RT-PCR. J Clin Virol 2013; 56:65–8. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 2001; 146:2275–89. [DOI] [PubMed] [Google Scholar]
- 10.Petty TJ, Cordey S, Padioleau I et al. Comprehensive human virus screening using high-throughput sequencing with a user-friendly representation of bioinformatics analysis: a pilot study. J Clin Microbiol 2014; 52:3351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zagordi O, Bhattacharya A, Eriksson N, Beerenwinkel N. ShoRAH: estimating the genetic diversity of a mixed sample from next-generation sequencing data. BMC Bioinformatics 2011; 12:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pizzorno A, Bouhy X, Abed Y, Boivin G. Generation and characterization of recombinant pandemic influenza A(H1N1) viruses resistant to neuraminidase inhibitors. J Infect Dis 2011; 203:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samson M, Abed Y, Desrochers FM et al. Characterization of drug-resistant influenza virus A(H1N1) and A(H3N2) variants selected in vitro with laninamivir. Antimicrob Agents Chemother 2014; 58:5220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sleeman K, Mishin VP, Deyde VM, Furuta Y, Klimov AI, Gubareva LV. In vitro antiviral activity of favipiravir (T-705) against drug-resistant influenza and 2009 A(H1N1) viruses. Antimicrob Agents Chemother 2010; 54:2517–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abed Y, Pizzorno A, Bouhy X, Boivin G. Role of permissive neuraminidase mutations in influenza A/Brisbane/59/2007-like (H1N1) viruses. PLoS Pathog 2011; 7:e1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 1993; 234:779–815. [DOI] [PubMed] [Google Scholar]
- 18.PyMOL. The PyMOL molecular graphics system. Version 1.7.4. http://www.pymol.org Accessed 7 May 2015.
- 19.Chen H, Cheung CL, Tai H et al. Oseltamivir-resistant influenza A pandemic (H1N1) 2009 virus, Hong Kong, China. Emerg Infect Dis 2009; 15:1970–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le QM, Wertheim HF, Tran ND et al. A community cluster of oseltamivir-resistant cases of 2009 H1N1 influenza. New Engl J Med 2010; 362:86–7. [DOI] [PubMed] [Google Scholar]
- 21.Hauge SH, Dudman S, Borgen K, Lackenby A, Hungnes O. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007–08. Emerg Infect Dis 2009; 15:155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention. Oseltamivir-resistant novel influenza A (H1N1) virus infection in two immunosuppressed patients - Seattle, Washington, 2009. MMWR Morbid Mortal Wkly Rep 2009; 58:893–6. [PubMed] [Google Scholar]
- 23.Nguyen HT, Fry AM, Loveless PA, Klimov AI, Gubareva LV. Recovery of a multidrug-resistant strain of pandemic influenza A 2009 (H1N1) virus carrying a dual H275Y/I223R mutation from a child after prolonged treatment with oseltamivir. Clin Infect Dis 2010; 51:983–4. [DOI] [PubMed] [Google Scholar]
- 24.Le QM, Kiso M, Someya K et al. Avian flu: isolation of drug-resistant H5N1 virus. Nature 2005; 437:1108. [DOI] [PubMed] [Google Scholar]
- 25.Ives JA, Carr JA, Mendel DB et al. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res 2002; 55:307–17. [DOI] [PubMed] [Google Scholar]
- 26.Collins PJ, Haire LF, Lin YP et al. Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature 2008; 453:1258–61. [DOI] [PubMed] [Google Scholar]
- 27.Moscona A. Global transmission of oseltamivir-resistant influenza. New Engl J Med 2009; 360:953–6. [DOI] [PubMed] [Google Scholar]
- 28.Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 2010; 328:1272–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurt AC, Chotpitayasunondh T, Cox NJ et al. Antiviral resistance during the 2009 influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect Dis 2012; 12:240–8. [DOI] [PubMed] [Google Scholar]
- 30.Hurt AC, Deng YM, Ernest J et al. Oseltamivir-resistant influenza viruses circulating during the first year of the influenza A(H1N1) 2009 pandemic in the Asia-Pacific region, March 2009 to March 2010. Euro Surveill 2011; 16:pii:19770. [PubMed] [Google Scholar]
- 31.Leang SK, Deng YM, Shaw R et al. Influenza antiviral resistance in the Asia-Pacific region during 2011. Antiviral Res 2013; 97:206–10. [DOI] [PubMed] [Google Scholar]
- 32.Okomo-Adhiambo M, Nguyen HT, Abd Elal A, Sleeman K, Fry AM, Gubareva LV. Drug susceptibility surveillance of influenza viruses circulating in the United States in 2011–2012: application of the WHO antiviral working group criteria. Influenza Other Respir Viruses 2014; 8:258–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler J, Hooper KA, Petrie S et al. Estimating the fitness advantage conferred by permissive neuraminidase mutations in recent oseltamivir-resistant A(H1N1)pdm09 influenza viruses. PLoS Pathog 2014; 10:e1004065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abed Y, Pizzorno A, Bouhy X, Rheaume C, Boivin G. Impact of potential permissive neuraminidase mutations on viral fitness of the H275Y oseltamivir-resistant influenza A(H1N1)pdm09 virus in vitro, in mice and in ferrets. J Virol 2014; 88:1652–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hurt AC, Hardie K, Wilson NJ et al. Characteristics of a widespread community cluster of H275Y oseltamivir-resistant A(H1N1)pdm09 influenza in Australia. J Infect Dis 2012; 206:148–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takashita E, Ejima M, Itoh R et al. A community cluster of influenza A(H1N1)pdm09 virus exhibiting cross-resistance to oseltamivir and peramivir in Japan, November to December 2013. Euro Surveill 2014; 19:pii:20666. [DOI] [PubMed] [Google Scholar]
- 37.Yang Z, Yang G, Zhou L. Mutation effects of neuraminidases and their docking with ligands: a molecular dynamics and free energy calculation study. J Comput Aided Mol Des 2013; 27:935–50. [DOI] [PubMed] [Google Scholar]
- 38.Abed Y, Boivin G. Treatment of respiratory virus infections. Antiviral Res 2006; 70:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Vries E, Stelma FF, Boucher CA. Emergence of a multidrug-resistant pandemic influenza A (H1N1) virus. New Engl J Med 2010; 363:1381–2. [DOI] [PubMed] [Google Scholar]
- 40.Baek YH, Song MS, Lee EY et al. Profiling and characterization of potentially multidrug-resistant influenza neuraminidase 1 (N1) strains against neuraminidase inhibitors. J Virol 2015; 89:287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaiser L. Influenza will not miss opportunities. Clin Infect Dis 2006; 43:1562–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.