Abstract
This paper describes patient characteristics, including Ebola viral load, associated with mortality in a Médecins Sans Frontières Ebola case management centre (CMC).
Out of 780 admissions between June and October 2014, 525 (67%) were positive for Ebola with a known outcome. The crude mortality rate was 51% (270/525). Ebola viral load (whole-blood sample) data were available on 76% (397/525) of patients. Univariate analysis indicated viral load at admission, age, symptom duration prior to admission, and distance traveled to the CMC were associated with mortality (P < .05). The multivariable model predicted mortality in those with a viral load at admission greater than 10 million copies per milliliter (P < .05, odds ratio >10), aged ≥50 years (P = .08, odds ratio = 2) and symptom duration prior to admission less than 5 days (P = .14). The presence of confusion, diarrhea, and conjunctivitis were significantly higher (P < .05) in Ebola patients who died.
These findings highlight the importance viral load at admission has on mortality outcomes and could be used to cohort cases with viral loads greater than 10 million copies into dedicated wards with more intensive medical support to further reduce mortality.
Keywords: Ebola virus, outbreak, Sierra Leone, viral load, mortality
The current Ebola virus disease (EVD) outbreak in West Africa [1] is now the largest yet documented, with the countries of Guinea, Liberia, and Sierra Leone most affected [2]. The World Health Organization (WHO) declared the outbreak a public health emergency of international concern on 8 August 2014 [3]. The transmission of EVD via infected body fluids occurs predominantly in 3 settings: (1) family/community through contact with an infected individual or contaminated fomites; (2) funerals, due to touching deceased infected bodies; and (3) nosocomial, via the absence of appropriate infection control measures in healthcare settings [4–7].
Sierra Leone has a population of just under 6 million, a life expectancy of 45 years, and an expenditure on health per capita of 205 international dollars [8]. The district of Kailahun is located in the east of the country, and in response to a number of cases of EVD, Médecins Sans Frontières (MSF) constructed a 50-bed Ebola Case Management Centre (CMC) near Kailahun city (capital of the district), which opened on 26 June 2014. The capacity of this center was further increased to 80 beds in August 2014 due to the increased number of EVD cases in the district.
MSF has been involved in the control of many past EVD outbreaks in central Africa and has developed established guidelines that are regularly referenced by other nongovernmental organizations [9, 10]. The Kailahun CMC (KCMC) [11] is constructed according to these guidelines and is divided into low- and high-risk zones. The low-risk zone comprises medical and nursing administrative tents, laundry area, storage tents, water chlorination points, and other facilities to support the high-risk zone. Within the high-risk zone are located the triage tent; suspect, probable, and confirmed cases tents; morgue; and high-risk waste area. Personal protective equipment must be worn at all times in the high-risk zone.
Patients are initially assessed in the triage area, and if they fulfill the WHO case definition criteria [12] are relocated to the suspect or probable tents as appropriate to await an EVD polymerase chain reaction (PCR) test on a whole-blood sample. When this test is positive, the patient is transferred to the confirmed tents, while a negative result allows the patient to be discharged if the duration of symptoms has been 72 hours or greater. When a suspect/probable patient has a negative PCR result but symptom duration of less than 72 hours, a repeat PCR test is performed at 72 hours or more of symptoms to rule out a false-negative result [10, 13]. A proportion of patients assessed in the triage area were referred from government health centers. This proportion is unknown, as only the home address is recorded on the patient register.
This paper describes the characteristics associated with mortality among EVD-positive cases admitted to the KCMC between June and October 2014.
METHODS
Data Collection
A retrospective cohort study of all patients admitted to the KCMC between 26 June and 12 October 2014 was carried out. A patient register, constructed using Microsoft Excel 2010, was on site at the KCMC. Each patient was designated a unique MSF identification number (MSF ID). Clinical, laboratory, and epidemiological data were collected on all admitted cases and stored in a secure format on the register. Epidemiological information recorded included age, gender, address, distance traveled to KCMC, occupation, symptom duration prior to admission, length of stay, contact history, and patient outcome (classified as cured, death due to EVD, death not due to EVD, defaulter, and not a case). Clinical and laboratory data were restricted to the constellation of symptoms present on admission and the EVD viral load (from whole-blood and blood-swab samples, respectively).
When it was not possible for medical staff to obtain a whole-blood sample through venesection, a blood-swab sample from a punctured skin site was taken as an alternative sample in consultation with the onsite laboratory. The resulting viral load values from positive blood swabs may not be directly comparable to the viral load values from positive whole-blood venous specimens, as the 2 forms of sampling have not previously been validated against one another. Therefore, viral load blood-swab values were excluded from the primary analysis.
Data Matching
Laboratory data were initially maintained on a separate register to the patient register, and these data were subsequently merged using the MSF ID. The cycle threshold (CT) result from the first EVD PCR whole-blood test carried out on or after admission was used as the measure of viral load at admission. Of note, if the first whole blood EVD PCR test was performed greater than 5 days postadmission, then the result was not considered to represent the viral load at admission. Similar matching was performed for blood-swab results.
Laboratory Analysis
Samples were received by the onsite laboratory from the KCMC medical staff and inactivated using a recognized method [14] inside a flexible film negative-pressure isolator followed by RNA extraction and purification. Quantitative reverse-transcription PCR (qRT-PCR) assays targeting the L and NP genes were used to detect the presence of Zaire Ebola virus using Lightcycler RNA Master Hydrolysis reagents (Roche, Laval, PQ) according to manufacturer's instructions. Assays were performed on the Lightcycler Nano platform (Roche). Primary testing focused on EVD diagnosis using cut-off cycle threshold (CT) values for positive, equivocal, and negative of <36, 36.1–40, and >40, respectively. The lower the CT value, the higher the Ebola viral load and vice versa. The assays detected EVD at 10 genome equivalents (g.e.)/reaction. For differential diagnostics when requested, the laboratory offered qRT-PCR tests for Lassa virus [15] and Plasmodium species [16]. As an internal control for extraction and amplification procedures, MS2 phage was added to each sample as previously described [17].
Statistical Analysis
We used Welch t test, Fisher exact test, or the Wilcoxon rank-sum test, as appropriate, to determine the association between mortality and the variables age, gender, occupation, the presence of gastrointestinal hemorrhage, the number of days between symptom onset and admission, whole-blood CT value at admission, and distance traveled to the KCMC. All variables with a P value <.1 in the univariate analysis were included in the binary (survival vs death) multivariable logistic regression model. Nonsignificant variables were eliminated in a stepwise fashion. Kaplan–Meier methods were used to determine survival curves for various groups of patients, using the interval from admission to death, with data censored at 36 days for survivors.
Survival according to age was examined using a scatter plot and 5-year age categories to determine the most appropriate age comparisons. Additionally, box-plots of whole-blood EVD PCR CT results among survivors and nonsurvivors were constructed to determine the most appropriate dichotomous CT comparisons. All statistical tests were 2-tailed, with a P value of less than .05 considered to indicate statistical significance. All analyses were performed using R software, version 3.0.2.
RESULTS
A total of 780 admissions to the KCMC occurred between 26 June and 12 October 2014. Of these, 525 (67%) had PCR-confirmed EVD with a known outcome (survived or death due to EVD). The remaining 255 (33%) admissions were classified as not a case (232, 91%), current admission awaiting outcome (13, 5%), non-EVD death (7, 3%), and defaulter (3, 1%).
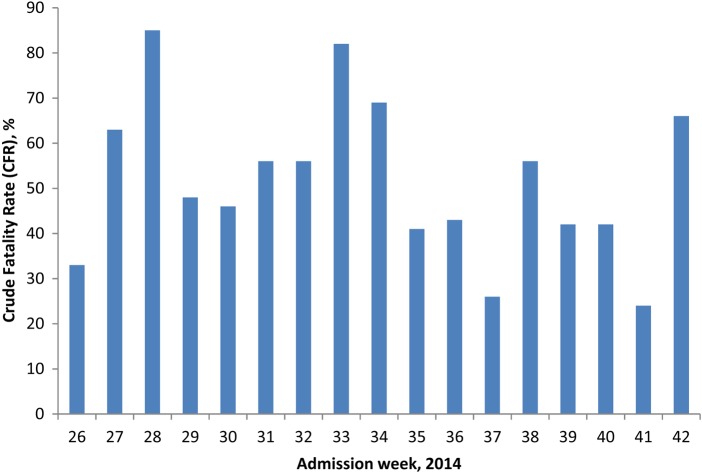
Regarding EVD cases with a known outcome (n = 525), the overall median age was 27 years (interquartile range [IQR], 16–40) and for males and females it was 28 (IQR, 18–40) and 27 (IQR, 15–38) years, respectively. Males represented 272 (52%) cases. The median time intervals from symptom onset to admission, admission to death, and admission to survival were 5 days (IQR, 3–7), 4 days (IQR, 2–6), and 13 days (IQR, 10–18), respectively. The overall case fatality rate of 51% for the KCMC had a weekly fluctuation (by week of admission), ranging from 24% to 85% during the study period (Figure 1). The frequency of symptoms between EVD-positive patients who died and survived were compared, and diarrhea, confusion, and conjunctivitis were found to be statistically different (P < .05) between the 2 outcomes (Supplementary Table 1).
Figure 1.
Fluctuation of crude fatality rate by week of admission (26–42), Kailahun Ebola Case Management Centre, Sierra Leone, 2014.
Among the 525 EVD admissions with a known outcome, 128 (24%) did not have a CT value from a whole-blood sample recorded on the laboratory register. A further 51 (10%) did not have whole-blood EVD PCR test results recorded within 5 days of admission, and later tests were not considered relevant as a measure of viral load at admission. The remaining 346 (66%) admissions had CT values available from whole-blood samples taken within 5 days of admission, of which 319 (61%) were taken within 1 day of admission (Supplementary Figure 1). The median time from admission to death for those with an admission CT < 25 and ≥25 was 4 days (IQR, 2–6) and 4 days (IQR, 3–5), respectively. An EVD CT value of 25 is equivalent to a viral load of 1.28 × 10⁷ copies per milliliter of blood.
A readmission to the KCMC occurs when a patient is initially discharged as not having EVD and then subsequently readmitted to the KCMC after a variable period of time to be reassessed for EVD. The 525 EVD-positive admissions with known outcome comprised 511 (97%) primary admissions and 14 (3%) readmissions, resulting in 270 (51%) deaths (262 primary admissions and 8 readmissions) and 255 survivors (244 primary admissions and 11 readmissions) (Supplementary Figure 2).
A total of 31 healthcare workers were admitted to the KCMC with positive EVD PCR of which 20 died, 9 survived, and 2 were transferred to another health facility. The outcome of those transferred was not known. The overall mortality rate for healthcare workers with a known outcome was 69% (20/69).
Kailahun district is divided into 14 chiefdoms, and the Kailahun MSF CMC is located in the chiefdom of Luawa. Of the 525 confirmed cases with known outcome, 326 (62%) came from the district of Kailahun, with the remaining originating from districts across the rest of the country (Supplementary Figure 3 and 4). Address information was available for 505 (96%) cases, and 181 (36%) had traveled greater than 100 km to reach the KCMC. The overall crude mortality for those who traveled less than and greater than 100 km was 58% (187/324) and 40% (73/181), respectively (P < .001). Additionally, Figure 2 presents the relationship between distances traveled, interval between symptom onset and admission, and overall crude mortality rate.
Figure 2.
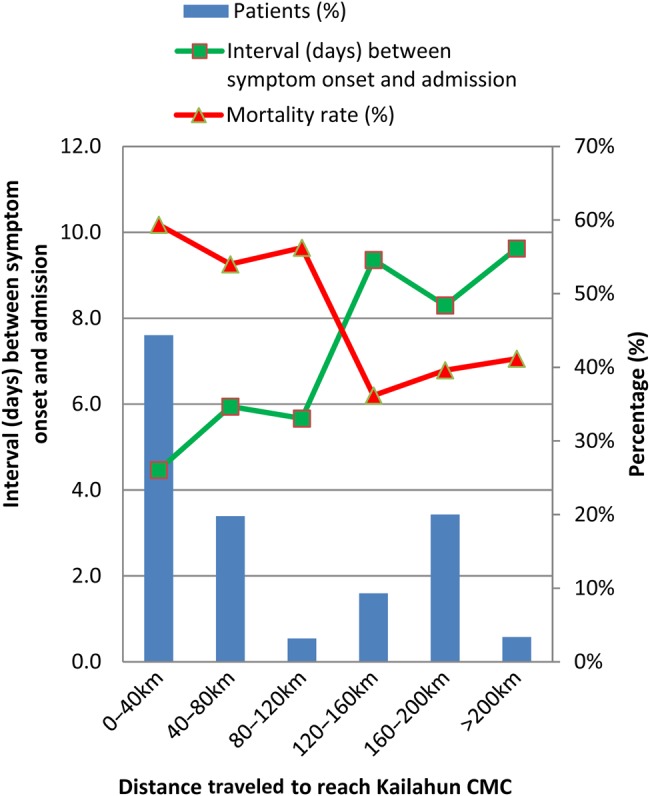
Mortality rate and interval (days) between symptom onset and admission by distance traveled to Kailahun CMC. Abbreviation: CMC, case management centre.
A number of variables between survivors and nonsurvivors were shown to be significantly (P < .05) different among the 525 EVD-positive patients with known outcomes, including age, whole-blood CT value at admission, interval between symptom onset and admission, length of stay, and distance traveled to KCMC (Table 1). Of note, median interval between symptom onset and admission was longer for survivors (6 days) than nonsurvivors (4 days).
Table 1.
Differences in Characteristics Between EVD-Positive Patients Who Died and Survived, Kailahun Ebola Case Management Centre, Sierra Leone, 2014
| Characteristic | Survivor (n = 255) | Death (n = 270) | P Value |
|---|---|---|---|
| Median CT value at admission (IQR) | 31 (25–34) | 22 (20–25) | 2.20 × 10−16 |
| Median length of stay, days (IQR) | 13 (10–18) | 4 (2–6) | 2.20 × 10−16 |
| Median distance traveled to KCMC, km (IQR) | 49 (28–160) | 38 (27–113) | 8.00 × 10−05 |
| Median interval between symptom onset and admission, days (IQR) | 6 (3–9) | 4 (3–7) | .0001 |
| Median age, years (IQR) | 25 (14–35.5) | 30 (19–42.25) | .0007 |
| Healthcare worker, no. (%) | 9 (3.5) | 20 (7.4) | .0808 |
| Hemorrhage, no. (%) | 1 | 10 | .1703 |
| Male : female ratio | 131:124 | 141:129 | .9145 |
Abbreviations: CT, cycle threshold; EVD, Ebola virus disease; IQR, interquartile range; KCMC, Kailahun CMC.
Multivariable Logistic Regression Model
The selected logistic regression model adjusted for age, whole-blood CT value at admission, and time from symptom onset to KCMC admission. The parameter estimates, P values, and odds ratios are shown in Table 2. All other variables (both clinical and epidemiological), including healthcare worker status and distanced traveled to the KCMC, did not contribute to the multivariable model and thus were removed. The chosen model indicates that whole-blood CT value on admission was the most significant predictor of mortality in the dataset with age (P = .08) and duration of symptoms prior to admission (P = .14) being less so. The receiver operating characteristic value for the preferred model was 0.82. The effect of admission whole-blood CT value on survival is represented by a Kaplan–Meier curve in Figure 3. The survival curve of EVD-positive infants approximates that of adults aged >50 years (Figure 4), while cases aged between 1 and 50 years demonstrate better survival outcomes when compared with the prior 2 age groups.
Table 2.
Multivariable Logistic Regression Model Parameters
| Estimate | P Value | Odds Ratio | 95% CI | |
|---|---|---|---|---|
| Intercept | −0.9759 | .0004 | 0.3769 | .2167–.6358 |
| Age ≥50 y | 0.7644 | .0791 | 2.1477 | .9278–5.1602 |
| CT <25 at admission | 2.3810 | 9.68e−16 | 10.8156 | 6.1322–19.6558 |
| Duration of symptoms prior to admission ≥5 d | −0.4354 | .1404 | 0.6470 | .3632–1.1602 |
Abbreviations: CI, confidence interval; CT, cycle threshold.
Figure 3.
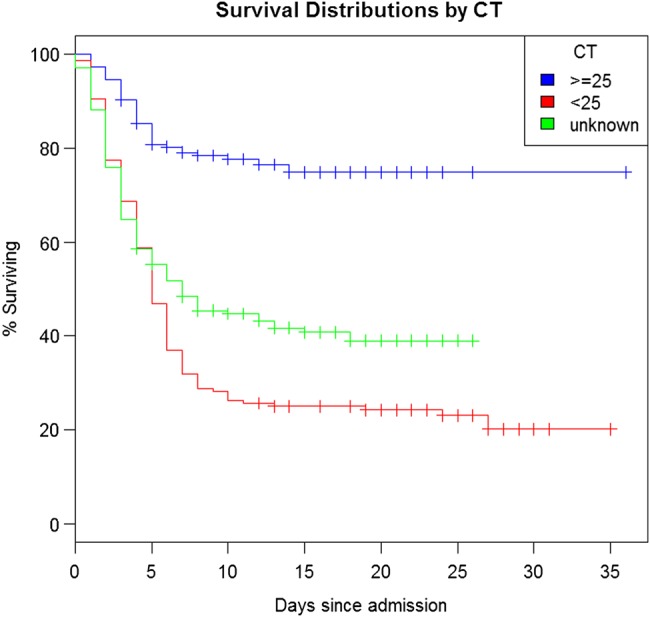
Kaplan–Meier curve of the probability of survival among patients with EVD according to CT value. Abbreviations: CT, cycle threshold; EVD, Ebola virus disease.
Figure 4.
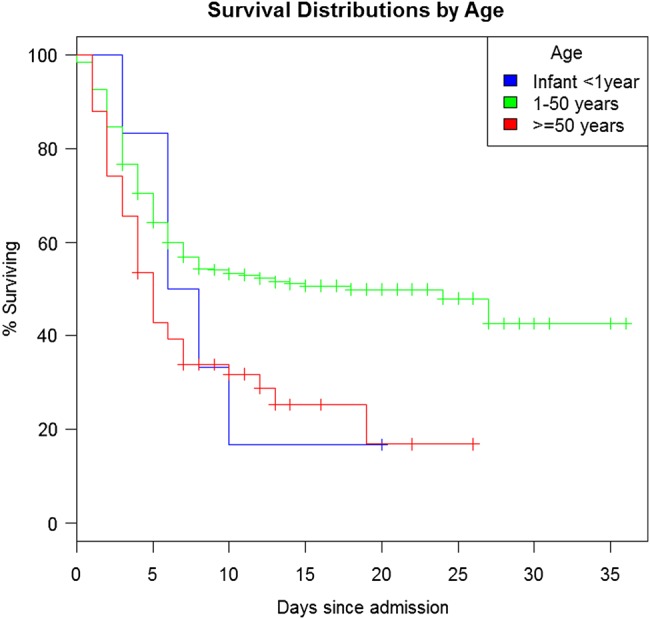
Kaplan–Meier curve of the probability of survival among patients with EVD according to age. Abbreviation: EVD, Ebola virus disease.
Number of Whole-Blood EVD PCR Tests Per Patient
Of the 525 EVD-positive admissions with known outcome, 150 (29%) had 2 or more whole-blood PCR tests performed (a minimum of several days apart) during their admission. Interestingly, the overall mortality rate for this group of patients was 9% (13/150) as opposed to 69% (257/375) for the remaining admissions within the group. Furthermore, for those patients with 2 or more whole-blood EVD PCR tests performed during the same admission, the mortality rate when the second PCR test had a lower CT (higher viral load) than the first was 58% (7/12), while the mortality rate for those with a second PCR test with the same or higher CT (lower viral load) than the first was 4% (6/138). This difference in mortality rate associated with the CT value of the second whole-blood sample was statistically significant (P < .05).
Nonwhole-Blood Sample Types (Blood Swabs)
A total of 71 admissions had blood swabs performed by medical staff of which 31 (44%) were EVD positive, had a CT value, age, interval between symptom onset and admission, and known outcome recorded and were performed within 5 days of admission to the KCMC (Supplementary Figure 5). The whole-blood multivariable logistic regression model was used to calculate the expected number of deaths for blood-swab cohorts with a CT <25 and ≥25. A χ2 test was applied. The null hypothesis stated that the parameters from the whole-blood multivariable logistic regression model could be applied to blood swab results to predict mortality. The test failed to reject the null hypothesis (Table 3), which may indicate that the whole-blood model could be used for blood swabs to predict mortality (although the result should be treated with caution as the sample size was small). Blood swabs with a CT value <25 were a closer fit to the whole-blood model than those ≥25 (z² of 0.16 and 3.39, respectively).
Table 3.
Blood Swabs, Stratified by CT Value, Actual Versus Expected Deaths
| Group | CT | No. Patients | Actual Deaths | Expected Deaths | Estimated Risk | z2 |
|---|---|---|---|---|---|---|
| 1 | ≥25 | 21 | 10 | 6 | 0.27 | 3.39 |
| 2 | <25 | 10 | 9 | 8 | 0.79 | 0.16 |
| Total | 31 | 19 | 14 | 0.44 | 3.54 |
N = 31; crude mortality rate = 61% (19/31).
Critical value for χ2 test = 3.84.
Abbreviation: CT, cycle threshold.
DISCUSSION
Various EVD case fatality rates (CFRs) have been reported during this outbreak [18–20] and our study has shown a favorable rate of 51% among confirmed cases. There are many reasons why certain centers have lower EVD CFRs than others, including: the sickest EVD patients dying before they reach the CMC; well-established health promotion activities in the community, allowing patients to avail of medical support as early as possible; and the availability of professional care according to designated protocols. It is also notable that within the KCMC, the weekly CFR varied between 24% and 85% (by admission week) during the study period. The reasons for this variation are unclear as all patients received standardized medical care during their admission according to MSF guidelines. Additionally, inexperienced staff are always paired with more senior staff to ensure that the level of care provided is consistent throughout the life of the KCMC. These measures are of particular importance in an environment where there is a high turnover of staff due to the physical and emotional burden of the workload.
It is noteworthy that the interval between symptom onset and admission was longer (6 days) for survivors than nonsurvivors (4 days). This is a counterintuitive finding and is potentially explained by those more likely to die, due to the longer interval between symptom onset and admission, having done so prior to admission (selection bias). However, in the multivariable regression model, the interval between symptom onset and admission was not significant (P = .14) and age was marginally significant (P = .08). The most influential predictor of mortality within the multivariable regression model was whole-blood CT value at admission (P = 9.7 × 10−16) with an odds ratio of 10.8 in favor of mortality when the CT value was <25 (equivalent to 107 viral copies per mL or greater). This finding could be used in the field to cohort patients with CT values of <25 into dedicated wards where they could receive enhanced medical support and palliative care if resources allow due to their much-increased risk of death. These high-risk patients could also be prioritized for experimental treatments under strict ethical protocol. The increased risk of death with higher Ebola viral loads documented here supports the findings in a previous outbreak of Ebola in Uganda in 2000 [21, 22].
The finding that a follow-up improving CT value resulted in a mortality rate of 4% versus 58% for a deteriorating one is noteworthy. The very fact that an EVD patient has survived long enough to undergo a follow-up phlebotomy procedure indicates they have an overall improved chance of survival regardless of the viral load at admission.
Sometimes, in the field it is difficult for medical staff to obtain a whole-blood sample from patients with poor venous access. In such cases, the skin is punctured under aseptic technique and a blood swab is taken. This procedure is easier and safer for staff to perform; however, it was unknown how well a CT value from a blood swab correlates with that from a whole-blood sample. Although the sample size was small, our results indicate that a blood swab, particularly with a CT value <25, may be used to predict mortality, as we found no evidence that the results do not approximate that of whole-blood samples. We propose that further monitoring of blood swab results be undertaken to substantiate these early findings. This is of potential importance, as it would provide staff with a safe, alternative procedure for obtaining a blood sample if they fail to attain a whole-blood sample through phlebotomy.
The mapping of confirmed cases indicates that a large proportion stem from districts outside Kailahun, as far away as the capital, Freetown. This has important implications for control of the EVD outbreak at a national level, as the transfer of large numbers of cases across the country by various modes of transport could potentially facilitate the unintentional spread of the virus. The large number of admissions originating outside Kailahun was associated with the lack of operational CMCs in other districts of Sierra Leone at that time. Notably, patients who traveled greater than 100 km to reach the KCMC had a lower mortality rate than those who journeyed less than 100 km. This paradoxical result could be explained by the possibility that for the cohort who traveled greater than 100 km, cases within it who had a low chance of survival already died before reaching the KCMC, therefore causing an artificially low crude mortality rate for this group within the KCMC.
There are many challenges regarding the accurate collection of epidemiological, clinical, and laboratory data in the field setting in West Africa, including heavy workload, difficulty obtaining information from patients due to language barriers, and lack of an information technology infrastructure to allow healthcare professionals to record data electronically at point of patient contact. These factors contributed to the incomplete data presented in our results, which is an obvious limitation of the study. Another drawback was that the study included no clinical data while cases were inpatients in the KCMC. This information was collected on individual patient paper records but was not transcribed electronically to the patient register.
This study documents the importance of viral load at admission on predicting mortality in a large MSF Ebola case management center in a remote jungle region of Eastern Sierra Leone.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. To all healthcare staff who lost their lives fighting Ebola, their bravery endures.
Gabriel Fitzpatrick wrote the first draft of the paper. All other authors reviewed the draft and suggested amendments. Theses amendments were then incorporated by Gabriel Fitzpatrick into the final draft.
This work fulfilled the Médecins Sans Frontières (MSF) Ethics Review Board (Geneva, Switzerland) approved criteria for analysis of routinely collected anonymous program data. All activities conducted by MSF were approved by the national authorities of Sierra Leone.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Baize S, Pannetier D, Oestereich L et al. Emergence of Zaire Ebola virus disease in guinea—preliminary report. N Engl J Med 2014; 371:1418–25. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Situation report, Ebola response roadmap. Geneva, Switzerland: World Health Organisation, 2014. http://www.who.int/csr/disease/ebola/situation-reports/en/. Accessed 28 December 2014. [Google Scholar]
- 3.WHO. Ebola response roadmap. Geneva, Switzerland: World Health Organisation, 2014. http://www.who.int/csr/resources/publications/ebola/response-roadmap/en/. Accessed 28 December 2014. [Google Scholar]
- 4.Ftika L, Maltezou HC. Viral haemorrhagic fevers in healthcare settings. J Hosp Infect 2013; 83:185–92. [DOI] [PubMed] [Google Scholar]
- 5.Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet 2011; 377:849–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacNeil A, Rollin PE. Ebola and Marburg hemorrhagic fevers: neglected tropical diseases? PLOS Negl Trop Dis 2012; 6:e1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowell SF, Mukunu R, Ksiazek TG, Khan AS, Rollin PE, Peters CJ. Transmission of Ebola hemorrhagic fever: a study of risk factors in family members, Kikwit, Democratic Republic of the Congo, 1995. Commission de Lutte contre les Epidémies à Kikwit. J Infect Dis 1999; 179(suppl 1):S87–91. [DOI] [PubMed] [Google Scholar]
- 8.WHO. Sierra Leone country profile. Geneva, Switzerland: World Health Organization, 2014. http://www.who.int/countries/sle/en/. Accessed 28 December 2014. [Google Scholar]
- 9.Kerstiëns B, Matthys F. Interventions to control virus transmission during an outbreak of Ebola hemorrhagic fever: experience from Kikwit, Democratic Republic of the Congo, 1995. J Infect Dis 1999; 179(suppl 1):S263–7. [DOI] [PubMed] [Google Scholar]
- 10.Sterk E. Medecins Sans Frontieres Filovirus haemorrhagic fever guidelines. 2008. www.medbox.org/ebola-guidelines/filovirus-haemorrhagic-fever-guideline/preview. Accessed 28 December 2014.
- 11.Wolz A. Face to face with Ebola—an emergency care center in Sierra Leone. N Engl J Med 2014; 371:1081–3. [DOI] [PubMed] [Google Scholar]
- 12.WHO. Case definition recommendations for Ebola or Marburg virus diseases. Geneva, Switzerland: World Health Organisation, 2014. www.who.int/csr/resources/publications/ebola/ebola-case-definition-contact-en.pdf?ua=1. Accessed 28 December 2014. [Google Scholar]
- 13.Fitzpatrick G, Vogt F, Moi Gbabai O et al. Describing readmissions to an Ebola case management centre (CMC), Sierra Leone, 2014. Euro Surveill 2014; 19:pii:20924. [PubMed] [Google Scholar]
- 14.Grolla A, Mehedi M, Lindsay R, Bosio C, Duse A, Feldmann H. Enhanced detection of Rift Valley fever virus using molecular assays on whole blood samples. J Clin Virol 2012; 54:313–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demby AH, Chamberlain J, Brown DW, Clegg CS. Early diagnosis of Lassa fever by reverse transcription-PCR. J Clin Microbiol 1994; 32:2898–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee MA, Tan CH, Aw LT et al. Real-time fluorescence-based PCR for detection of malaria parasites. J Clin Microbiol 2002; 40:4343–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreier J, Störmer M, Kleesiek K. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. J Clin Microbiol 2005; 43:4551–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bah EI, Lamah MC, Fletcher T et al. Clinical presentation of patients with Ebola virus disease in Conakry, Guinea. N Engl J Med 2014; 372:40–7. [DOI] [PubMed] [Google Scholar]
- 19.Schieffelin JS, Shaffer JG, Goba A et al. Clinical illness and outcomes in patients with Ebola in Sierra Leone. N Engl J Med 2014; 371:2092–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Team WER. Ebola virus disease in West Africa—the first 9 months of the epidemic and forward projections. N Engl J Med 2014; 371:1481–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Towner JS, Rollin PE, Bausch DG et al. Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome. J Virol 2004; 78:4330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez A, Lukwiya M, Bausch D et al. Analysis of human peripheral blood samples from fatal and nonfatal cases of Ebola (Sudan) hemorrhagic fever: cellular responses, virus load, and nitric oxide levels. J Virol 2004; 78:10370–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.