Abstract
Hypertension is a prevalent disorder in the world representing one of the major risk factors for heart attack and stroke. These risks are increased in salt sensitive individuals. Hypertension and salt sensitivity are complex phenotypes whose pathophysiology remains poorly understood and, remarkably, salt sensitivity is still laborious to diagnose.
Here we present a urinary proteomic study specifically designed to identify urinary proteins relevant for the pathogenesis of hypertension and salt sensitivity. Despite previous studies that underlined the association of UMOD gene variants with hypertension, this work provides novel evidence showing different uromodulin protein level in the urine of hypertensive patients compared to healthy individuals. Notably, we also show that patients with higher level of uromodulin are homozygous for UMOD risk variant and display a decreased level of salt excretion, highlighting the essential role of UMOD in the regulation of salt reabsorption in hypertension. Additionally, we found that urinary nephrin 1, a marker of glomerular slit diaphragm, may predict a salt sensitive phenotype and positively correlate with increased albuminuria associated with this type of hypertension.
Abbreviations: BP, blood pressure; LC–MS/MS, liquid chromatography coupled to tandem mass spectrometry; BMI, body mass index; SS, salt sensitive; SR, salt resistant; SBP, systolic BP; DBP, diastolic BP; MQ, MaxQuant; GO, Gene Ontology; MBP, mean BP.
Keywords: Salt sensitive hypertension, Urinary biomarker, Quantitative proteomics, Uromodulin, Salt homeostasis, Nephrinuria, Glomerular injury
Highlights
-
•
We identified urinary proteins differently excreted in hypertensive patients.
-
•
Nephrin 1 might predict salt sensitive phenotype and glomerular complications.
-
•
Uromodulin impacts salt homeostasis in hypertension.
-
•
We provide new insights into the pathogenesis of hypertension and salt sensitivity.
1. Introduction
Hypertension is a very common disease, especially in industrialized countries where it is becoming an important issue of public health. Many cases of hypertension are related to incorrect behaviors as a high-fat diet, smoking, and sedentary lifestyle. Hypertension is a very complex disease and even several genetic association studies have been published up to now, these variants explain approximately 20% [1] of the population variation in blood pressure (BP). Clinical and experimental studies have highlighted the implications of abnormal sodium balance in the development of hypertension in both animal models and humans [2], [3], [4], [5]. Generally, the response of BP changes in sodium assumptions is heterogeneous, approximately half of the population with essential hypertension is salt-sensitive as increases BP in response to a salt intake [6]. Salt-sensitive individuals are more prone to develop hypertension which is characterized by glomerular hypertension, microalbuminuria [7] and a higher mortality and morbidity of cardiovascular events [7], [8]. For these individuals, there is no universal consensus of definition or a proper test to assess salt sensitivity. Commonly used tests measure BP response to a change in dietary salt intake. These tests are laborious, expensive and with low patient's compliance [9]. Indeed, the discovery of novel markers of salt sensitivity would be extremely beneficial in focusing treatment with dietary salt restriction only to those patients who really benefit from this therapy. Furthermore, understanding the pathophysiology of hypertension itself might further drive to more specific therapeutic targets for lowering BP and preserving renal function.
Recent advances in clinical research provided by proteomic strategies have accelerated the discovery of urinary biomarkers for kidney diseases [10], [11], [12], [13], [14] as well as coronary artery disease [14], [15] and diabetic nephropathy [16], [17]. Based on these observations, we analyzed the urinary proteome of hypertensive patients [18] in order to discover novel pathways involved in the pathogenesis of hypertension and to identify predictive biomarkers for salt-sensitivity.
2. Experimental procedures
2.1. Patients' selection and study protocol for salt-sensitivity characterization
We enrolled newly discovered, never treated, mild hypertensive male patients in the ‘Outpatient Clinic for Hypertension’ of San Raffaele Hospital, Milan. The Ethics Committee of the Hospital approved the study and a written informed consent was obtained from each individual. The study protocol for the acute salt load test was similar to that reported previously [19]. Briefly, the protocol began with a 2-hour equilibration period during which the subjects were taken to a quiet room and a venous catheter was inserted into an antecubital vein. They remained in the supine position until the end of the salt loading except for voiding. A steady state was considered to be achieved when the volume of urine collection and the values of the BP recordings varied by < 1 mL/min and < 3 mm Hg, respectively. The average equilibration period lasted 2 h.
Acute salt load test: after the equilibration period and achievement of a steady state, a constant-rate i.v. infusion of 2 L of 0.9% NaCl was carried out in 2 h. BP (mean of 3 measurements taken 3 min apart) was measured every 30 min during the 2 h of loading and 3 times at 3-minute intervals at the end of the infusion. These last 3 BP values were averaged and used in the analysis.
Patients were defined salt resistant (SR) or salt sensitive (SS) according to mean BP changes after acute saline infusion: those that displayed a mean BP increase greater than 4 mm Hg were considered salt sensitive. Conversely, patients with a smaller or negative BP change resulted salt resistant. Urines for proteomic analysis were collected during the equilibration period before the acute salt load.
Inclusion criteria for the study were: sex male, age between 18 and 55 years; body mass index (BMI) of < 30 kg/m2; salt intake, evaluated as urinary Na excretion, < 300 mEq/24 h; ambulatory systolic BP (SBP) of > 140 mm Hg and diastolic BP (DBP) of > 90 mm Hg in 3 consecutive visits at their family doctors. Exclusion criteria included history of myocardial infarction, stroke, congestive heart failure, liver disease, secondary cause of hypertension, diabetes, severe hypertension (> 160/110 mm Hg), abuse of drugs or alcohol, and creatinine clearance of < 80 mL/m. Secondary forms of hypertension (e.g., primary aldosteronism) were ruled out with specific investigations when deemed appropriate. Women were excluded from the study as their urinary proteome might be influenced by female hormone cycle.
We added in the study a control of healthy donors, all male, normotensive, with regular BMI, and with no disease assuming that these subjects are salt sensitive or salt resistant subjects according with the frequency of the general population. Collectively, we analyzed urines from 75 subjects: 19 SS, 37 SR and 19 healthy donors. A cohort of 24 subjects was used for the proteomic analysis, a cohort of 52 subjects for the validation phase and 56 hypertensive patients for the microalbuminuria test. The criterion adopted for the selection of all this subsets is that the patients are all consecutive. Personnel blind to the salt test results carried out the proteomic analyses.
2.2. Urine collection and concentration
Urine samples collected from healthy donors and patients were stored immediately at − 80 °C. For each donor, approximately 25 mL of urine was centrifuged at 5000 × g for 15 min at 4 °C to remove cell debris, and were successively concentrated by ultrafiltration (Microcon devices YM-10, Millipore, Billerica, MA, USA). Urine was sequentially passed through a filtration membrane of 10 kDa cut-off at 2500 rpm and at 4 °C. The final concentrate was washed twice with water to remove excess salt occasionally present in urine. Final protein concentration was estimated by the Bradford method and 50 μg of each sample was processed by FASP procedure [20].
Urinary albumin and creatinine were measured by immunoturbidimetric technique on a Cobas Mira autoanalyzer (Roche, Basel, Switzerland).
2.3. Protein digestion and peptides preparation
The urinary proteome was digested in-solution with trypsin using the FASP protocol [20] with spin ultrafiltration units of nominal molecular weight cut off of 30,000 Da. Briefly, 200 μL of urinary proteins containing 50 μg of protein concentrates was reduced by adding 50 μL of 0.5 M DTT and 1 M Tris/HCl pH 8.5 and incubating the samples for 1 min at 95 °C, samples were centrifuged at 14,000 × g at 4 °C for 15 min. 200 μL of 8 M urea in 0.1 M Tris/HCl, pH 8.5 (UA) was added to the supernatant and the samples were transferred to YM-30 Microcon filters (Cat No. MRCF0R030, Millipore). Samples were centrifuged at 14,000 × g for 15 min. Then, 50 μL of 0.05 M iodoacetamide in 8 M urea was added to the filters and the samples were incubated in the dark for 5 min. Filters were washed twice with 100 μL of 8 M UA followed by two washes with 100 μL of 40 mM NH4HCO3. Finally, 0.8 μg of trypsin was added in 40 μL of 40 mM NH4HCO3 to each filter. Samples were incubated overnight at 37 °C and the released peptides were collected by centrifugation. The resulting peptides were purified on a C18 StageTip (Proxeon Biosystems, Denmark) [21]. For the analysis of technical replicates, the peptides' concentrate was divided into three independent samples.
2.4. Mass spectrometry analysis and proteins quantitation
Mass spectrometry analysis was performed by LC–MS/MS using an LTQ-Orbitrap mass spectrometer (ThermoScientific, Bremen, Germany). Purified tryptic digests were injected in a capillary chromatographic system (EasyLC, Proxeon Biosystems); peptide mixtures were separated on a homemade 20 cm long fused silica capillary (75-μm inner diameter, Proxeon Biosystems) filled with Reprosil-Pur C18 3 μm resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). A gradient of eluents A (distilled water with 2% (v/v) acetonitrile, 0.5% (v/v) formic acid) and B (80% acetonitrile in distilled water with 0.1% (v/v) formic acid) was used to achieve separation from 4% buffer B to 50% buffer B in 320 min (0.2 μL/min flow rate). MS analysis was performed as reported previously [22]. We used exclusion list for predicted albumin peptides for all the MS runs. Raw MS files were processed with MaxQuant software (v 1.2.2.5) [21] making use of the Andromeda search engine [23], [24]. MS/MS peak lists were searched against the UniProtKB/Swiss-Prot protein sequence complete proteome database (release 2011_12 2010_10 of 07-October-2010) in which trypsin specificity was used with up to two missed cleavages allowed. Searches were performed selecting alkylation of cysteine by carbamidomethylation as fixed modification, and oxidation of methionine and N-terminal acetylation as variable modifications. Mass tolerance was set to 5 ppm and 0.6 Da for parent and fragment ions, respectively. The false discovery rate for both peptides and proteins were set at 0.01. Additionally, we required at least two peptides identifications per protein, of which at least one peptide had to be unique to the protein group. Measurement time for the triplicate analysis was slightly more than 15 h, and total sample consumption was 4 μg (urinary sample yields 0.02 mg/mL of proteins).
Label-free analysis was performed using MaxQuant software for protein quantitation. This included “match between run” option (time window of 5 min) for the quantification of the peptides recognized on the basis of mass and retention time but identified in other LC–MS/MS runs. For label-free proteins quantification a minimum ratio counts of 2 was considered and the “LFQ intensities” which are the intensity values normalized across the entire dataset were used. All the proteomic data as raw files, total proteins and peptides identified with relative intensities and search parameters were loaded on Peptide Atlas repository (accession number http://www.peptideatlas.org/PASS/PASS00383).
2.5. Bioinformatics and statistical analysis
Biological Networks Gene Ontology (BiNGO) program package (v 2.44) [25] with the Cytoscape plugin (v 2.8.0) were used to find statistically over-represented Gene Ontology (GO) categories [26]. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed with the KEGG pathway mapper tool (http://www.genome.jp/kegg/tool/map_pathway2.html) using the Uniprot ID of the modulated proteins.
For patients' study, data are expressed as means ± SEMs. Several hypotheses were tested with ANOVA using a sequence of least squares models.
MultiExperiment Viewer software (MeV) (v 4.6.2) was used for hierarchical clustering. Hierarchical trees were constructed for proteins quantified (y axis) and samples analyzed (x axis). By using MEV, T-tests and ANOVA test were applied on each of the intensities of the quantified proteins.
2.6. Western Blot analysis and antibodies
20 μg of proteins was diluted in loading buffer (10 g/L SDS, 10% glycerol, 1% 2-mercaptoethanol, bromophenol blue 20 g/L, 5 mM Tris HCl, pH 6.8), boiled, separated on a 4–12% gradient SDS-PAGE (NUPAGE from Invitrogen) and transferred onto nitrocellulose membranes (GE Healthcare/Amersham Bioscience, United Kingdom) by electroblotting. Proteins and β-actin abundances in the urine of control group and naïve patients were quantified by densitometric analysis. WB analysis was performed on randomly selected samples.
The following antibodies were used: polyclonal anti-nephrin 1 (H300) antibody produced in rabbit (Santa Cruz Biotechnology, USA), anti-uromodulin polyclonal antibody produced in sheep (Abcam, Cambridge, MA), monoclonal anti-ephrin-B2 antibody produced in mouse and polyclonal anti-IST1 antibody produced in rabbit (both from Sigma-Aldrich, St. Louis, MO), and monoclonal anti-actin produced in mouse (Sigma-Aldrich, St. Louis, MO).
2.7. Genotyping
Genomic DNA was extracted from peripheral blood with standard method [10]. All of the subjects were genotyped for rs4293393 UMOD polymorphism (Assay ID C_27865986_10) by a 5′ nuclease allelic discrimination assay, with allele-specific MGB probes (TaqMan SNP Genotyping Assay — Applied Biosystems, Foster City, CA, USA) according to manufacturer's instructions.
3. Results
3.1. Patients
Clinical characteristics of all naïve hypertensive patients enrolled in the study who underwent acute salt load are summarized in Table 1.
Table 1.
Clinical characteristics of all naïve hypertensive patients enrolled in the present study who underwent acute salt load.
| Phenotype | Salt resistant n = 37 | Salt sensitive n = 19 | P value |
|---|---|---|---|
| Age (years) | 42 ± 1.6 | 43.1 ± 2.1 | 0.67 |
| BMI (kg/m2) | 25 ± 0.4 | 26.8 ± 0.8 | 0.071 |
| eGFR (mL/m/1.72 m2) | 99.8 ± 0.6 | 99.6 ± 0.9 | 0.856 |
| 24 h urinary Na (mEq/24 h) | 140.8 ± 7.6 | 167 ± 17.3 | 0.112 |
| SBP baseline (mm Hg) | 142.5 ± 2.1 | 134.5 ± 2.4 | 0.032 |
| DBP baseline (mm Hg) | 89.6 ± 1.6 | 84.2 ± 2.2 | 0.05 |
| SBP T120 (mm Hg) | 140.3 ± 2.1 | 146.2 ± 2.3 | 0.09 |
| DBP T120 (mm Hg) | 86 ± 1.7 | 91.9 ± 1.8 | 0.036 |
| ΔSBP (T120 − T0) (mm Hg) | − 1.8 ± 1 | 11.7 ± 1 | < 0.001 |
| ΔDBP (T120 − T0) (mm Hg) | − 3.7 ± 0.6 | 7.8 ± 1.03 | < 0.001 |
| ΔMBP (T120 − T0) (mm Hg) | − 2.9 ± 0.61 | 8.77 ± 0.92 | < 0.001 |
In particular, SS displayed a large increase in BPs after saline load, however in SR a fall in systolic, diastolic and mean BP was observed. All the other parameters considered were similar at baseline, indicating a good homogeneity of the study population. Notably, we added in the study a control of healthy donors, all male, normotensive, with regular BMI, and with no disease.
3.2. Identification of urinary proteome in hypertensive patients
We characterized urines of 24 subjects by quantitative proteomics: 10 SR and 7 SS hypertensive subjects, plus a control of 7 healthy normotensive donors. Briefly, urinary proteins were concentrated, quantified and trypsin digested. The peptides resulting from trypsin digestion were desalted and analyzed by LC–MS/MS. All of the samples were analyzed in triplicate in order to make the data reliable for statistical analysis and to increase protein coverage using stringent protein identification criteria. The false discovery rate for all the proteomic data was set at 0.01. The total number of identifications increased from 419 in single run to 577 in triplicate runs of the same donor (Supplementary Fig. 1A). This number represents the expected complexity of proteome coverage of a single sample analyzed in triplicate [18], [27].
We performed two sets of experiments using different cohorts of patients in order to validate protein identification and quantitation across independent analyses (Supplementary Fig. 1B). Technical reproducibility of label free quantification was proven to be excellent and label free quantification showed a very high homogeneity between patients (Supplementary Fig. 1C). Collectively, we identified 812 protein groups (Supplementary Table 1). The overlapping proteins identified both in the first and in the second sets were about 80% (Supplementary Fig. 2A).
Cellular component Gene Ontology (GO) terms associated to these proteins are related to extracellular space, cell surface, vacuole, vesicles, plasma membrane, extracellular matrix, collagen and lysosomal proteins (Supplementary Fig. 2B). These ontologies are consistent with urine composition.
3.3. Identification of urinary markers of hypertension
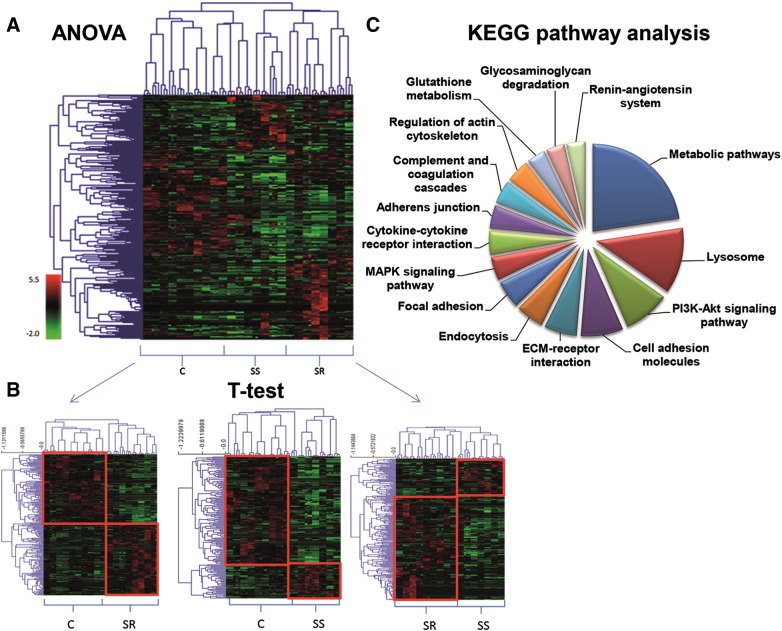
We quantified urinary proteins using protein intensity levels normalized on the total amount of all the identified proteins (Supplementary Fig. 3A). The majority of the proteins resulted up-regulated in both SS and SR patients vs. controls and vice versa (Supplementary Fig. 3C), meaning that most of these proteins reflect the effect of hypertension in common to all patients. Each of the quantified proteins was further subjected to ANOVA-test allowing the identification of 263 proteins undergoing significant changes (P value < 0.01) among SR or SS patients and controls (Supplementary Table 2). As reported in Fig. 1A, hierarchical trees constructed for proteins quantified (y axis) and samples analyzed (x axis) clearly showed the grouping of SS and SR patients with respect to controls. From ANOVA test, we retrieved information on which proteins were differently represented among SR, SS patients and healthy donors, but we did not discriminate if the variation of abundance of these proteins with respect to controls occurred in both groups of patients or if it was specific for one of them. To this aim, proteins significantly affected were further subjected to T-tests analysis as SR vs. C, SS vs. C and SR vs. SS. Fifty six proteins were found up- or down-regulated both in SS and SR patients with respect to the controls, suggesting that these proteins could be associated to hypertension in general (Supplementary Table 3). However, fifty-one proteins were differently regulated in SS patients as compared to both healthy and SR subjects (Supplementary Table 4), while 61 proteins were differently regulated in SR patients (Supplementary Table 5). Hierarchical trees clearly showed the proper clustering of the three groups (Fig. 1B). By using the KEGG pathway analysis tool, we found that these proteins were associated to pathways already known to be involved in hypertension and salt sensitivity, such as those belonging to the PI3K–Akt signaling, MAPK signaling, complement and coagulation factors [28], [29] and renin–angiotensin system [30], plus others involved in the regulation of endocytosis [31], actin cytoskeleton and focal adhesion that might be novel pathways potentially implicated in this pathology (Fig. 1C, Supplementary Fig. 4).
Fig. 1.
Statistical analysis of the quantified proteins. (A) Hierarchical clustering (HCL) of the proteins differentially expressed (ANOVA P value < 0.01) in the urine of SS and SR patients and healthy controls C. (B) HCL of the differentially expressed proteins (T test P value < 0.05) in the urine of SS patients vs. Controls (right), SR patients vs. Controls (left) and SS patients vs. SR patients (middle). Hypertensive patients' specimens co-cluster as well as control ones. Technical replicates of the same sample are always coupled. (C) KEGG pathway analysis of the significant proteins resulting from the ANOVA test: pie chart in terms of numbers of proteins mapped per pathway.
3.4. Biomarkers verification
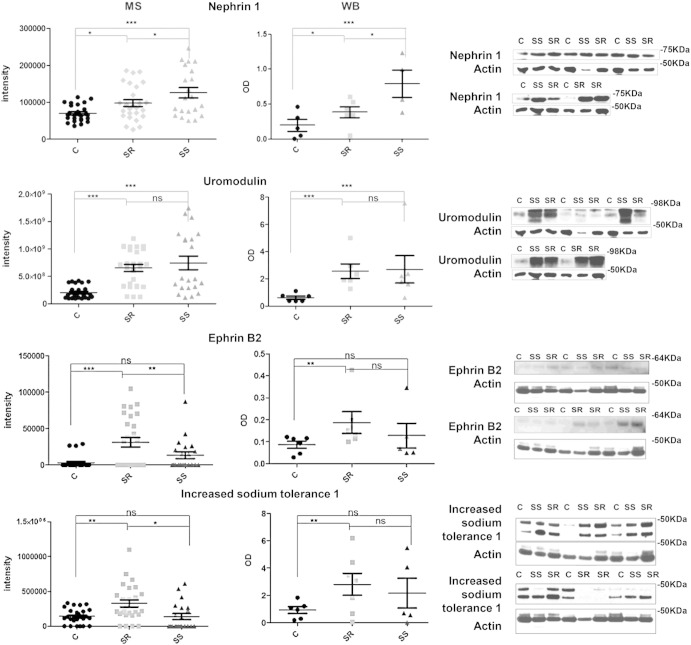
Proteomic data provide a snapshot of the entire processes behind the disease under study. In order to unravel the cause/effect events defining this picture we focused on the candidates known to be involved in the classical BP–kidney relationship. In particular, we validated nephrin 1 as it resides specifically in the glomerulus and its presence in the urine is an indication of podocyte damage [32]. Second, we validated uromodulin as genome-wide association studies (GWAS) have identified susceptibility variants for hypertension in the UMOD gene [33], and therefore it would be interesting to evaluate its behavior at the protein level. Third, we validated IST1 as this protein belongs to the endocytosis pathway and it is involved in the traffic of urinary exosomes and regulates the ESCORTIII complex [31]. An alteration of this protein might modify the trafficking of the numerous ionic channels along the apical membrane, thus influencing salt homeostasis. Last, we validated ephrin B2 as it was published that it may modulate tubular structures in medullary kidney epithelial cells through membrane retraction and rearrangement of the sites of adhesion to the underlying basal lamina [34]. The MS and Western Blot (WB) analysis for nephrin 1, uromodulin, ephrin B2 and IST-1 proteins are reported in Fig. 2 and Table 2. Consistent with the MS data, nephrin 1 was significantly up-regulated in SS vs. both SR and controls, and slightly up-regulated in SR vs. controls; uromodulin was up-regulated in SS and SR vs. controls while IST-1 and ephrin B2 were up-regulated in SR vs. control. As actin intensities from MS data did not change among the three groups of samples, we decided to use actin staining to normalize all the urine samples in the WB analysis (Supplementary Fig. 5).
Fig. 2.
Left panels: scatter plots of the related intensity values from MS analysis. Right panels: scatter plots of optical density values normalized on actin values and relative Western Blot images using antibodies against nephrin 1, uromodulin, ephrin B2 and IST1 (20 μg of concentrated urinary proteins from different subjects per lane was used for the analysis).*0.01 < P value < 0.05; **0.001 < P value < 0.01; ***P < 0.001.
Table 2.
Statistical analysis of the urinary markers for small-scale verification. Proteins significantly modulated either by ANOVA test (P value < 0.01) or T test analysis are reported. For each protein, protein names, gene names, Uniprot ID and averaged intensities values for each group analyzed are reported in the table.
| Protein names | Gene names | Uniprot | P value | Average intensity C | Average intensity SR | Average intensity SS | P value T test C-SR | P value T test C-SS | P value T test SR-SS |
|---|---|---|---|---|---|---|---|---|---|
| Nephrin 1 | NEPH1 | Q96J84 | 4.97E-04 | 69,860.6 | 101,682 | 126,063 | 5.04E-03 | 8.58E-04 | 4.54E-02 |
| Ephrin type-B receptor 2 | EPHB2 | P29323 | 2.59E-04 | 3038.15 | 31,176.4 | 13,283 | 3.15E-04 | ns | 3.31E-02 |
| Uromodulin | UMOD | E9PEA4 | 1.71E-06 | 2.03E + 08 | 6.54E + 08 | 7.45E + 08 | 2.00E-07 | 2.71E-04 | ns |
| Increased sodium tolerance 1 | IST1 | A8KAH5 | 8.83E-04 | 142,782 | 326,882 | 138,294 | 1.61E-03 | ns | 5.89E-03 |
3.5. Biomarkers validation
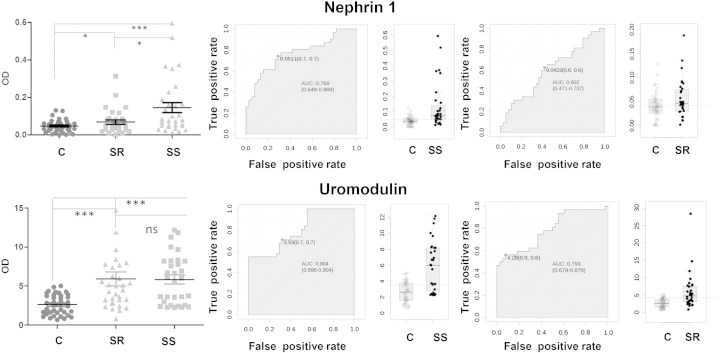
Since the validation step is a laborious and time consuming process, we focused only on the proteins that were potentially more related to hypertension. We queried the Urinary Proteins Biomarker Database [35] and found some diseases sharing the same biomarkers as the ones identified (Supplementary Fig. 6). Therefore, we assumed that these diseases might share the same injury sites or pathophysiological processes similar to those involved in hypertension. Among them, nephrin 1 was found as a relevant biomarker in advanced hypertension due to preeclampsia [36] while uromodulin has been reported as marker of microalbuminuria progression in type 1 diabetes [37]. Both conditions are known to be linked to progressive renal failure often associated with hypertension. Thus, we decided to further validate these two proteins on a larger cohort of patients (19 controls, 16 SR and 15 SS) (Supplementary Fig. 7). As reported in Fig. 3, nephrin 1 was significantly over-represented in SS patients vs. controls with an AUC of 0.77, while the AUC for SR patients was clearly lower. Conversely, uromodulin resulted significantly over-represented in all hypertensive patients compared to controls and no statistical difference between SS and SR patients was observed. The AUCs for the prediction of SS and SR hypertension were 0.80 and 0.79 respectively.
Fig. 3.
Nephrin 1 and uromodulin levels are predictive of salt sensitivity and hypertension. Left panels: scatter plots of optical density values from WB analysis using urines from 19 controls, 16 SR and 15 SS hypertensive patients. Right panels: ROC curves were depicted with associated AUC and sensitivity and specificity at the best cut-off point. As optimal cut off point we used the one closest to top-left corner. In the scatter plots, y-axis indicates the OD of the target proteins normalized against actin values; the lines indicate the optimal cutoff value reported in the ROC curves. *0.01 < P value < 0.05; **0.001 < P value < 0.01; ***P < 0.001.
3.6. Nephrin and albumin positively correlate in salt sensitive hypertensive patients
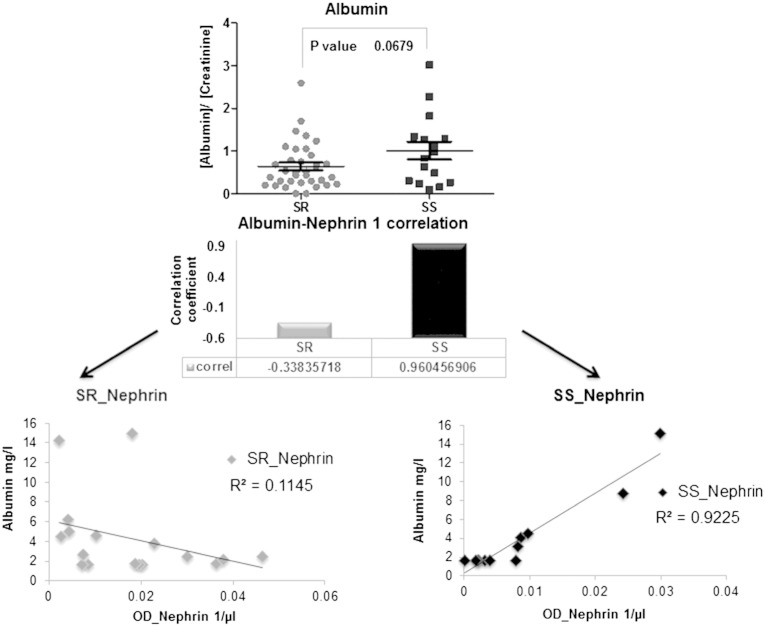
As nephrinuria might be associated to a putative crumbling of the glomerular filtration barrier, we explored the urinary albumin excretion in 56 hypertensive subjects including also the patients previously analyzed. As expected, no subject presented microalbuminuria (albumin:creatinine ratio < 3 mg/mmol) while albumin excretion normalized to creatinine values was higher in SS than in SR patients (Fig. 4). Interestingly, albumin and nephrin 1 positively correlated exclusively in SS patients (correlation coefficient = 0.96) suggesting the presence of an early glomerular damage that might alter the glomerular filtration barrier.
Fig. 4.
Albumin excretion was analyzed from urine of 56 patients, 19 SS and 37 SR. Upper panel represents scatter plot of albumin excretion values (mg/L) normalized on creatinine (mmol/L) concentration for SR and SS patients (albumin:creatinine ratio were all < 3 mg/mmol). Lower panels show Pearson correlation of albumin and nephrin 1 values both normalized on volume for SR and SS patients (albumin is expressed as mg/L and nephrin as optical density (OD)/μL).
3.7. Uromodulin secretion in hypertension correlates with homozygous UMOD risk variant and with decreased level of salt excretion
As previously discussed, we found uromodulin significantly more abundant in the urine of both salt sensitive and salt resistant hypertensive patients with respect to normotensive individuals, suggesting that this alteration is independent from the salt sensitivity. In fact, no significant differences in UMOD amounts have been observed between salt-sensitive and salt-resistant urine samples. GWAS studies identified common variants in the promoter of the UMOD gene associated to hypertension [38], and, in particular, the UMOD risk variant rs4293393 TT was recently found associated with higher expression of uromodulin both at transcript and protein level [33]. Therefore, we analyzed the frequency of this last variant in the patients enrolled in the study.
We divided the patients in two groups according to urinary uromodulin levels, using as cutoff the value derived by the ROC curve. Interestingly, we found that patients secreting higher level of uromodulin were carriers, with a high frequency (0.75), of the UMOD risk variant rs4293393 TT suggesting that higher uromodulin excretion might be linked to a major protein expression. This finding might be extremely relevant for this disease given the high frequency of UMOD risk variant in the general population [33]; obviously, further prospective studies using larger cohort of patients will be necessary to confirm this data in hypertensive population.
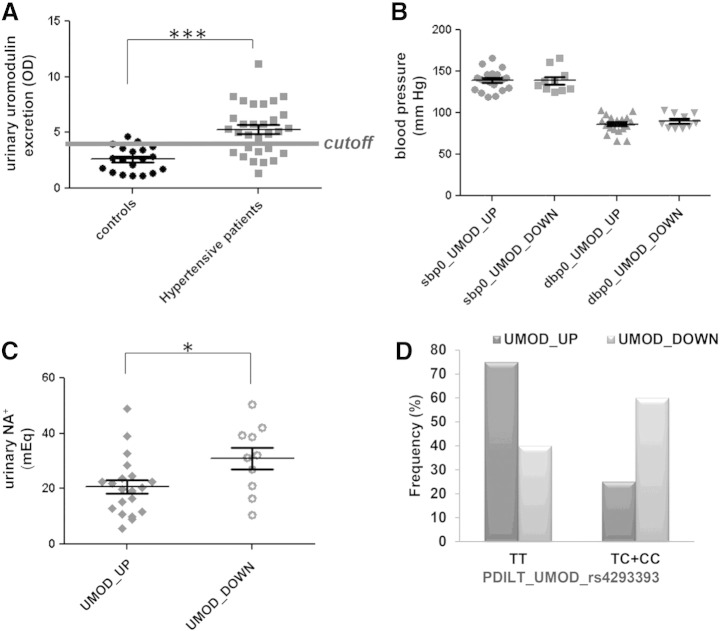
Finally, we evaluated the clinical significance of our findings by testing whether the level of BP and sodium excretion associated to uromodulin level in the urine. Patients with higher level of uromodulin in the urine displayed lower sodium excretion, indicating an increased sodium reabsorption along the nephron (Fig. 5).
Fig. 5.
Association of uromodulin excretion level with blood pressure, salt excretion and UMOD promoter risk variant: (A) scatter plots of optical density values from WB analysis using urines from 19 controls and 31 hypertensive patients, the cutoff threshold derived from the ROC curve is plotted on the graph (optimal cut off point was the one closest to top-left corner). (B, C) scatter plots of values for systolic (SBP) and diastolic (DBP) blood pressure and changes in sodium excretion in hypertensive patients divided in high and low uromodulin excretion levels based on the cutoff filter. (D) Frequency of the protective (C) and risk (T) alleles of rs4293393 SNP in hypertensive cohort divided in high and low uromodulin excretion levels.
These data reflect for the first time a direct link of uromodulin urinary concentration with an altered salt homeostasis in hypertensive patients.
4. Discussion
This study uses a label free quantitative proteomic approach to address salt sensitivity and hypertension in humans. We used a robust approach with high sensitivity and reproducibility that allowed the identification of 812 urinary proteins. Among them, we found several excreted proteins differentially present in both salt sensitive and salt resistant hypertensive patients compared to controls and other proteins excreted differently in SS compared to SR patients. This finding supports the view that high BP and the abnormal response to salt are multi-factorial and polygenic disorders that are under the control of multiple mechanisms and complex signaling activities. Using bioinformatics tools we found pathways potentially involved in salt dependent hypertension. In particular, in SS patients we found down-regulated the proteins glutamyl aminopeptidase A (ENPEP) and alanyl (membrane) aminopeptidase (ANPEP), which are both enzymes involved in angiotensin conversion and belong to the renin–angiotensin system. Interestingly, also in the SS animal model, the renin–angiotensin system in the kidney has been suggested to play significant roles and, consistent with our data, the SS rat is considered a low-renin model of hypertension [39], [40]. Moreover, in SR patients we found over-represented proteins related to endocytosis, lysosome and glycolysis; the involvement of this last pathway is completely new in the hypertension scenario. Further, by querying the Urinary Proteins Biomarker Database we found that some of the proteins differently excreted in this study were also identified as urinary markers of diabetic nephropathy, ureteropelvic junction obstruction and IgA nephropathy therefore we speculated that they might indicate the same injury sites or pathophysiological processes. Moreover, we found nephrin 1 present in an advanced hypertension disease as preeclampsia and uromodulin as marker associated with progressive renal failure, as a risk factor for kidney disease, and as a biomarker for the development of chronic kidney disease. We have confirmed four interesting candidates (nephrin 1, uromodulin, IST1 and ephrin b2) as potential markers that might justify previously described key pathophysiological aspects of these diseases. In particular, we proceeded with the validation of nephrin 1 and uromodulin on a larger cohort of patients. Nephrin 1 was validated as a biomarker of salt sensitivity. Nephrin 1 is an essential component of the glomerular podocyte junction and participates in the formation of the glomerular filtration barrier via interactions with associated proteins. Among others, the Src family protein kinase Fyn favors nephrin 1 phosphorylation and the recruitment of adaptor proteins involved in the regulation of podocyte junction formation and actin cytoskeletal dynamics. Mutations of podocyte proteins, such as podocin, adducin, laminin-β-2 and knocking out of Fyn or nephrin 1 in mice, induce actin cytoskeleton rearrangement and disruption of the filtration barrier leading to renal disease. This evidence supports the hypothesis that in hypertension, and in particular in salt sensitive subjects, the podocyte integrity might be compromised [41], [42], [43], [44]. As mentioned above, nephrinuria was also found altered in preeclampsia, a pregnancy-specific, multi-systemic disorder characterized by the new-onset of hypertension [45].
Here, we found that SS patients, besides an increased nephrinuria compared to SR, show a higher (although within normal values) albumin excretion, which positively correlates with nephrin 1 excretion. This finding is in line with the view indicating nephrin 1 as a predictive biomarker of glomerular damage earlier than microalbuminuria. Such hypothesis is also in agreement with a recent study where nephrinuria was detected in normoalbuminuric compared to microalbuminuric patients with diabetic nephropathy [46]. Altogether, these findings streamline the possibility of using nephrin 1 to predict glomerular complications. Obviously, to definitively associate nephrinuria with disease progression, other prospective studies will be necessary.
On the other hand, uromodulin was validated as a marker of hypertension. The exclusive expression of uromodulin in the thick portion of the ascending limb, where physiologically crucial mechanisms of sodium handling are located, and its potential to act as modulator of other salt transport systems located downstream, suggests that alterations of this protein might affect hypertension and renal sodium handling through an effect on sodium homeostasis. In agreement with our finding, Torffvit et al. [47] showed that urinary excretion of uromodulin is dependent on sodium intake; while Genome Wide Association Study of BP extremes identified variants near UMOD associated with hypertension [38] and increased urinary uromodulin levels [48]. Here we found that uromodulin is more secreted in hypertensive patients with respect to healthy individuals, and that the level of this protein correlates with lower sodium excretion. These findings point out the hypothesis that uromodulin increases salt reabsorption favoring high blood pressure by impairing sodium excretion. Indeed our group has already demonstrated that, in an animal model of kidney related hypertension, the Na–K–2Cl (NKCC2) and the Na–Cl (NCC) transporters, two potential targets of uromodulin action, are up-regulated in the development and maintenance of the hypertensive disease [49], [50].
We also showed that patients secreting increased level of uromodulin carry the UMOD risk variant PDILT_UMOD_rs4293393 with high frequency. Interestingly, this variant was recently found associated to increased level of uromodulin both at transcript and at protein level [33]. In the same study, the relationship of uromodulin and hypertension was proved in animal models where uromodulin overexpression leads to hypertension and renal damage.
Even though we know that hypertension is a complex disease, implying the existence of other factors that influence blood pressure, our work highlights key markers of hypertension and salt sensitivity. Both nephrin and uromodulin are associated to renal damage, therefore these findings are open to novel therapeutic targets aimed to preserve renal function and to lower blood pressure, thus personalizing the pharmacological treatment of patients.
Disclosure statement
The authors declare that they have no conflict of interest.
Acknowledgements
We thank the Biomolecular Mass Spectrometry group and Dr. Luca Rampoldi for precious suggestions and scientific discussion. This work was partially supported by the Italian Ministry of Research PRIN grant 2008W5AZEC003.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbacli.2014.10.001.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Cowley A.W., Jr. The genetic dissection of essential hypertension. Nature Reviews Genetics. 2006;7:829–840. doi: 10.1038/nrg1967. [DOI] [PubMed] [Google Scholar]
- 2.Barba G., Cappuccio F.P., Russo L., Stinga F., Iacone R., Strazzullo P. Renal function and blood pressure response to dietary salt restriction in normotensive men. Hypertension. 1996;27:1160–1164. doi: 10.1161/01.hyp.27.5.1160. [DOI] [PubMed] [Google Scholar]
- 3.Campese V.M., Parise M., Karubian F., Bigazzi R. Abnormal renal hemodynamics in black salt-sensitive patients with hypertension. Hypertension. 1991;18:805–812. doi: 10.1161/01.hyp.18.6.805. [DOI] [PubMed] [Google Scholar]
- 4.Hall J.E., Brands M.W., Shek E.W. Central role of the kidney and abnormal fluid volume control in hypertension. J. Hum. Hypertens. 1996;10:633–639. [PubMed] [Google Scholar]
- 5.Roman R.J. Abnormal renal hemodynamics and pressure–natriuresis relationship in Dahl salt-sensitive rats. Am. J. Physiol. 1986;251:F57–F65. doi: 10.1152/ajprenal.1986.251.1.F57. [DOI] [PubMed] [Google Scholar]
- 6.Williams G.H., Hollenberg N.K. Sodium-sensitive essential hypertension: emerging insights into an old entity. J. Am. Coll. Nutr. 1989;8:490–494. doi: 10.1080/07315724.1989.10720318. [DOI] [PubMed] [Google Scholar]
- 7.Cubeddu L.X., Hoffmann I.S., Aponte L.M., Nunez-Bogesits R., Medina-Suniaga H., Roa M., Garcia R.S. Role of salt sensitivity, blood pressure, and hyperinsulinemia in determining high upper normal levels of urinary albumin excretion in a healthy adult population. Am. J. Hypertens. 2003;16:343–349. doi: 10.1016/s0895-7061(03)00057-8. [DOI] [PubMed] [Google Scholar]
- 8.Weinberger M.H., Fineberg N.S., Fineberg S.E., Weinberger M. Salt sensitivity, pulse pressure, and death in normal and hypertensive humans. Hypertension. 2001;37:429–432. doi: 10.1161/01.hyp.37.2.429. [DOI] [PubMed] [Google Scholar]
- 9.Felder R.A., White M.J., Williams S.M., Jose P.A. Diagnostic tools for hypertension and salt sensitivity testing. Curr. Opin. Nephrol. Hypertens. 2013;22:65–76. doi: 10.1097/MNH.0b013e32835b3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Citterio L., Simonini M., Zagato L., Salvi E., Delli Carpini S., Lanzani C., Messaggio E., Casamassima N., Frau F., D'Avila F., Cusi D., Barlassina C., Manunta P. Genes involved in vasoconstriction and vasodilation system affect salt-sensitive hypertension. PLoS One. 2011;6:e19620. doi: 10.1371/journal.pone.0019620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuccurullo M., Beneduci A., Anand S., Mignani R., Cianciaruso B., Bachi A., Capasso G. Fabry disease: perspectives of urinary proteomics. J. Nephrol. 2010;23(Suppl. 16):S199–S212. [PubMed] [Google Scholar]
- 12.Matafora V., Bachi A., Capasso G. Genomics and proteomics: how long do we need to reach clinical results? Blood Purif. 2013;36:7–11. doi: 10.1159/000350578. [DOI] [PubMed] [Google Scholar]
- 13.Varghese S.A., Powell T.B., Budisavljevic M.N., Oates J.C., Raymond J.R., Almeida J.S., Arthur J.M. Urine biomarkers predict the cause of glomerular disease. J. Am. Soc. Nephrol. JASN. 2007;18:913–922. doi: 10.1681/ASN.2006070767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Zur Muhlen C., Schiffer E., Zuerbig P., Kellmann M., Brasse M., Meert N., Vanholder R.C., Dominiczak A.F., Chen Y.C., Mischak H., Bode C., Peter K. Evaluation of urine proteome pattern analysis for its potential to reflect coronary artery atherosclerosis in symptomatic patients. J. Proteome Res. 2009;8:335–345. doi: 10.1021/pr800615t. [DOI] [PubMed] [Google Scholar]
- 15.Zimmerli L.U., Schiffer E., Zurbig P., Good D.M., Kellmann M., Mouls L., Pitt A.R., Coon J.J., Schmieder R.E., Peter K.H., Mischak H., Kolch W., Delles C., Dominiczak A.F. Urinary proteomic biomarkers in coronary artery disease. Mol. Cell Proteomics MCP. 2008;7:290–298. doi: 10.1074/mcp.M700394-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Rossing K., Mischak H., Dakna M., Zurbig P., Novak J., Julian B.A., Good D.M., Coon J.J., Tarnow L., Rossing P., Network P. Urinary proteomics in diabetes and CKD. J. Am. Soc. Nephrol. JASN. 2008;19:1283–1290. doi: 10.1681/ASN.2007091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snell-Bergeon J.K., Maahs D.M., Ogden L.G., Kinney G.L., Hokanson J.E., Schiffer E., Rewers M., Mischak H. Evaluation of urinary biomarkers for coronary artery disease, diabetes, and diabetic kidney disease. Diabetes Technol. Ther. 2009;11:1–9. doi: 10.1089/dia.2008.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagaraj N., Mann M. Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J. Proteome Res. 2011;10:637–645. doi: 10.1021/pr100835s. [DOI] [PubMed] [Google Scholar]
- 19.Manunta P., Cusi D., Barlassina C., Righetti M., Lanzani C., D'Amico M., Buzzi L., Citterio L., Stella P., Rivera R., Bianchi G. Alpha-adducin polymorphisms and renal sodium handling in essential hypertensive patients. Kidney Int. 1998;53:1471–1478. doi: 10.1046/j.1523-1755.1998.00931.x. [DOI] [PubMed] [Google Scholar]
- 20.Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 21.Rappsilber J., Ishihama Y., Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 22.Matafora V., D'Amato A., Mori S., Blasi F., Bachi A. Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol. Cell Proteomics MCP. 2009;8:2243–2255. doi: 10.1074/mcp.M900079-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 24.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 25.Maere S., Heymans K., Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 26.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kentsis A., Monigatti F., Dorff K., Campagne F., Bachur R., Steen H. Urine proteomics for profiling of human disease using high accuracy mass spectrometry. Proteomics Clin. Appl. 2009;3:1052–1061. doi: 10.1002/prca.200900008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ocaranza M.P., Jalil J.E. Mitogen-activated protein kinases as biomarkers of hypertension or cardiac pressure overload. Hypertension. 2010;55:23–25. doi: 10.1161/HYPERTENSIONAHA.109.141960. [DOI] [PubMed] [Google Scholar]
- 29.Sata M., Nagai R. Phosphatidylinositol 3-kinase: a key regulator of vascular tone? Circ. Res. 2002;91:273–275. doi: 10.1161/01.res.0000031956.29928.62. [DOI] [PubMed] [Google Scholar]
- 30.Kobori H., Nangaku M., Navar L.G., Nishiyama A. The intrarenal renin–angiotensin system: from physiology to the pathobiology of hypertension and kidney disease. Pharmacol. Rev. 2007;59:251–287. doi: 10.1124/pr.59.3.3. [DOI] [PubMed] [Google Scholar]
- 31.Shestakova A., Hanono A., Drosner S., Curtiss M., Davies B.A., Katzmann D.J., Babst M. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol. Biol. Cell. 2010;21:1059–1071. doi: 10.1091/mbc.E09-07-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrandi M., Molinari I., Matafora V., Zerbini G., Trevisani F., Rastaldi M.P., Simonini M., Giardino L., Ferrari P., Manunta P. SIK1 localizes with nephrin in glomerular podocytes and its polymorphism predicts kidney injury. Hum. Mol. Genet. 2014;23:4371–4382. doi: 10.1093/hmg/ddu154. [DOI] [PubMed] [Google Scholar]
- 33.Trudu M., Janas S., Lanzani C., Debaix H., Schaeffer C., Ikehata M., Citterio L., Demaretz S., Trevisani F., Ristagno G., Glaudemans B., Laghmani K., Dell'Antonio G., Swiss Kidney Project on Genes in Hypertension, Loffing J., Rastaldi M.P., Manunta P., Devuyst O., Rampoldi L. Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat. Med. 2013;19:1655–1660. doi: 10.1038/nm.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogawa K., Wada H., Okada N., Harada I., Nakajima T., Pasquale E.B., Tsuyama S. EphB2 and ephrin-B1 expressed in the adult kidney regulate the cytoarchitecture of medullary tubule cells through Rho family GTPases. J. Cell Sci. 2006;119:559–570. doi: 10.1242/jcs.02777. [DOI] [PubMed] [Google Scholar]
- 35.Shao C., Li M., Li X., Wei L., Zhu L., Yang F., Jia L., Mu Y., Wang J., Guo Z., Zhang D., Yin J., Wang Z., Sun W., Zhang Z., Gao Y. A tool for biomarker discovery in the urinary proteome: a manually curated human and animal urine protein biomarker database. Mol. Cell Proteomics MCP. 2011;10(M111):010975. doi: 10.1074/mcp.M111.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y., Zhao S., Loyd S., Groome L.J. Increased urinary excretion of nephrin, podocalyxin, and betaig-h3 in women with preeclampsia. Am. J. Physiol. Ren. Physiol. 2012;302:F1084–F1089. doi: 10.1152/ajprenal.00597.2011. [DOI] [PubMed] [Google Scholar]
- 37.Schlatzer D., Maahs D.M., Chance M.R., Dazard J.E., Li X., Hazlett F., Rewers M., Snell-Bergeon J.K. Novel urinary protein biomarkers predicting the development of microalbuminuria and renal function decline in type 1 diabetes. Diabetes Care. 2012;35:549–555. doi: 10.2337/dc11-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padmanabhan S., Melander O., Johnson T., Di Blasio A.M., Lee W.K., Gentilini D., Hastie C.E., Menni C., Monti M.C., Delles C., Laing S., Corso B., Navis G., Kwakernaak A.J., van der Harst P., Bochud M., Maillard M., Burnier M., Hedner T., Kjeldsen S., Wahlstrand B., Sjogren M., Fava C., Montagnana M., Danese E., Torffvit O., Hedblad B., Snieder H., Connell J.M., Brown M., Samani N.J., Farrall M., Cesana G., Mancia G., Signorini S., Grassi G., Eyheramendy S., Wichmann H.E., Laan M., Strachan D.P., Sever P., Shields D.C., Stanton A., Vollenweider P., Teumer A., Volzke H., Rettig R., Newton-Cheh C., Arora P., Zhang F., Soranzo N., Spector T.D., Lucas G., Kathiresan S., Siscovick D.S., Luan J., Loos R.J., Wareham N.J., Penninx B.W., Nolte I.M., McBride M., Miller W.H., Nicklin S.A., Baker A.H., Graham D., McDonald R.A., Pell J.P., Sattar N., Welsh P., Global B.C., Munroe P., Caulfield M.J., Zanchetti A., Dominiczak A.F. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010;6:e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang P.X., Sanders P.W. Mechanism of hypertensive nephropathy in the Dahl/Rapp rat: a primary disorder of vascular smooth muscle. Am. J. Physiol. Ren. Physiol. 2005;288:F236–F242. doi: 10.1152/ajprenal.00213.2004. [DOI] [PubMed] [Google Scholar]
- 40.Trepiccione F., Zacchia M., Capasso G. The role of the kidney in salt-sensitive hypertension. Clin. Exp. Nephrol. 2012;16:68–72. doi: 10.1007/s10157-011-0489-y. [DOI] [PubMed] [Google Scholar]
- 41.Ferrandi M., Cusi D., Molinari I., Del Vecchio L., Barlassina C., Rastaldi M.P., Schena F.P., Macciardi F., Marcantoni C., Roccatello D., Peters L.L., Armelloni S., Min L., Giardino L., Mattinzoli D., Camisasca C., Palazzo F., Manunta P., Ferrari P., Bianchi G. Alpha- and beta-adducin polymorphisms affect podocyte proteins and proteinuria in rodents and decline of renal function in human IgA nephropathy. J. Mol. Med. 2010;88:203–217. doi: 10.1007/s00109-009-0549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagase M., Shibata S., Yoshida S., Nagase T., Gotoda T., Fujita T. Podocyte injury underlies the glomerulopathy of Dahl salt-hypertensive rats and is reversed by aldosterone blocker. Hypertension. 2006;47:1084–1093. doi: 10.1161/01.HYP.0000222003.28517.99. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura T., Kataoka K., Tokutomi Y., Nako H., Toyama K., Dong Y.F., Koibuchi N., Yamamoto E., Yasuda O., Ogawa H., Kim-Mitsuyama S. Novel mechanism of salt-induced glomerular injury: critical role of eNOS and angiotensin II. J. Hypertens. 2011;29:1528–1535. doi: 10.1097/HJH.0b013e328348ca95. [DOI] [PubMed] [Google Scholar]
- 44.Ponnuchamy B., Khalil R.A. Cellular mediators of renal vascular dysfunction in hypertension. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009;296:R1001–R1018. doi: 10.1152/ajpregu.90960.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carty D.M., Siwy J., Brennand J.E., Zurbig P., Mullen W., Franke J., McCulloch J.W., Roberts C.T., North R.A., Chappell L.C., Mischak H., Poston L., Dominiczak A.F., Delles C. Urinary proteomics for prediction of preeclampsia. Hypertension. 2011;57:561–569. doi: 10.1161/HYPERTENSIONAHA.110.164285. [DOI] [PubMed] [Google Scholar]
- 46.Jim B., Ghanta M., Qipo A., Fan Y., Chuang P.Y., Cohen H.W., Abadi M., Thomas D.B., He J.C. Dysregulated nephrin in diabetic nephropathy of type 2 diabetes: a cross sectional study. PLoS One. 2012;7:e36041. doi: 10.1371/journal.pone.0036041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torffvit O., Melander O., Hulten U.L. Urinary excretion rate of Tamm-Horsfall protein is related to salt intake in humans. Nephron. Physiology. 2004;97:31–36. doi: 10.1159/000077600. [DOI] [PubMed] [Google Scholar]
- 48.Olden M., Corre T., Hayward C., Toniolo D., Ulivi S., Gasparini P., Pistis G., Hwang S.J., Bergmann S., Campbell H., Cocca M., Gandin I., Girotto G., Glaudemans B., Hastie N.D., Loffing J., Polasek O., Rampoldi L., Rudan I., Sala C., Traglia M., Vollenweider P., Vuckovic D., Youhanna S., Weber J., Wright A.F., Kutalik Z., Bochud M., Fox C.S., Devuyst O. Common variants in UMOD associate with urinary uromodulin levels: a meta-analysis. J. Am. Soc. Nephrol. JASN. 2014;25:1869–1882. doi: 10.1681/ASN.2013070781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Capasso G., Rizzo M., Evangelista C., Ferrari P., Geelen G., Lang F., Bianchi G. Altered expression of renal apical plasma membrane Na + transporters in the early phase of genetic hypertension. Am. J. Physiol. Ren. Physiol. 2005;288:F1173–F1182. doi: 10.1152/ajprenal.00228.2004. [DOI] [PubMed] [Google Scholar]
- 50.Capasso G., Rizzo M., Garavaglia M.L., Trepiccione F., Zacchia M., Mugione A., Ferrari P., Paulmichl M., Lang F., Loffing J., Carrel M., Damiano S., Wagner C.A., Bianchi G., Meyer G. Upregulation of apical sodium-chloride cotransporter and basolateral chloride channels is responsible for the maintenance of salt-sensitive hypertension. Am. J. Physiol. Ren. Physiol. 2008;295:F556–F567. doi: 10.1152/ajprenal.00340.2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.