A protein from grapevine that transports sodium, potassium, and chloride ions across endomembranes is important for normal growth and salt tolerance.
Abstract
Plant cation-chloride cotransporters (CCCs) have been implicated in conferring salt tolerance. They are predicted to improve shoot salt exclusion by directly catalyzing the retrieval of sodium (Na+) and chloride (Cl−) ions from the root xylem. We investigated whether grapevine (Vitis vinifera [Vvi]) CCC has a role in salt tolerance by cloning and functionally characterizing the gene from the cultivar Cabernet Sauvignon. Amino acid sequence analysis revealed that VviCCC shares a high degree of similarity with other plant CCCs. A VviCCC-yellow fluorescent protein translational fusion protein localized to the Golgi and the trans-Golgi network and not the plasma membrane when expressed transiently in tobacco (Nicotiana benthamiana) leaves and Arabidopsis (Arabidopsis thaliana) mesophyll protoplasts. AtCCC-green fluorescent protein from Arabidopsis also localized to the Golgi and the trans-Golgi network. In Xenopus laevis oocytes, VviCCC targeted to the plasma membrane, where it catalyzed bumetanide-sensitive 36Cl–, 22Na+, and 86Rb+ uptake, suggesting that VviCCC (like AtCCC) belongs to the Na+-K+-2Cl– cotransporter class of CCCs. Expression of VviCCC in an Arabidopsis ccc knockout mutant abolished the mutant’s stunted growth phenotypes and reduced shoot Cl– and Na+ content to wild-type levels after growing plants in 50 mm NaCl. In grapevine roots, VviCCC transcript abundance was not regulated by Cl– treatment and was present at similar levels in both the root stele and cortex of three Vitis spp. genotypes that exhibit differential shoot salt exclusion. Our findings indicate that CCC function is conserved between grapevine and Arabidopsis, but neither protein is likely to directly mediate ion transfer with the xylem or have a direct role in salt tolerance.
Plant salinity stress impacts negatively on crop growth and is a significant limiting factor for agriculture, particularly in arid and semiarid regions, with an estimated cost of U.S. $27 billion because of lost crop production per year (Qadir et al., 2014; Munns and Gilliham, 2015). It has been estimated that irrigated agriculture, which produces 40% of the world’s calories, has one-fifth of its soils salt affected (FAO, 2002). The extent of this salt-affected irrigated agricultural land has been estimated to increase by 4% every year (FAO, 2002; Pimentel et al., 2004). Significant groundwater depletion of the world’s major aquifers through drought, climate change, and mismanagement concentrates dissolved solutes within the groundwater, compounding potential salinity issues for agriculture (Famiglietti, 2014). It is generally recognized that sodium chloride (NaCl) imposes an initial osmotic effect of slowing growth followed by a secondary ionic effect, where sodium (Na+) and chloride (Cl–) accumulate to levels that inhibit essential cellular processes (Munns and Tester, 2008).
Grapevine (Vitis vinifera [Vvi]; Grimplet et al., 2014) is irrigated in many parts of the world and considered moderately sensitive to salinity stress (Maas and Hoffman, 1977). Effects of salt stress on grapevine include a reduced rate of shoot growth and CO2 fixation (Downton, 1977) and reduced yield (Walker et al., 2002). Furthermore, fermentation of grapes that accumulate high concentrations of salt can be prolonged or is prone to stalling before reaching completion (Donkin et al., 2010). The resulting wines from such ferments retain high salt concentrations, can be unpalatable (Walker et al., 2003), and can exceed limits for trade or domestic markets (e.g. NaCl at 1,000 mg L−1 in Australia and 60 mg L−1 free Na+ as recommended by the International Organization of Vine and Wine [Leske et al., 1997]).
Salt tolerance in grapevine as in other woody perennials, such as Citrus spp. and avocado (Persea americana), is correlated with Cl– exclusion from the shoot (Ehlig, 1960; Storey and Walker, 1999; Teakle and Tyerman, 2010). This contrasts with other important crops, such as rice (Oryza sativa), wheat (Triticum aestivum), and the model plant Arabidopsis (Arabidopsis thaliana), where salt tolerance predominantly correlates with shoot Na+ exclusion (Munns et al., 2006, 2012; Jha et al., 2010; Shavrukov et al., 2010). A limited number of studies has highlighted the negative impact of Na+ on grapevine physiology (Shani and Ben-Gal, 2005; Stevens et al., 2011), and although its impact should not be discounted in this species, the major research effort for maintaining viticultural productivity under saline conditions has concentrated on improving shoot Cl– exclusion (Tregeagle et al., 2006; Fort et al., 2015). A key step in accelerating the selection of grapevine germplasm with improved salt tolerance would be to identify the proteins involved in shoot Cl– exclusion (Henderson et al., 2014).
To overcome excessive Cl– accumulation in the field, grapevine is typically grown on Cl– excluding rootstocks. These are usually a hybrid with a complex background of various Vitis spp. For instance, rootstock 140 Ruggeri (Vitis berlandieri ‘Resseguir #2’ × Vitis rupestris ‘St. George’) is known for its ability to limit shoot Cl– accumulation (Gong et al., 2010), whereas rootstock K51-40 (Vitis champini ‘Dogridge’ × Vitis riparia ‘Gloire’) is a poor shoot Cl– excluder, even compared with own rooted grapevines (Tregeagle et al., 2010; Abbaspour et al., 2013). However, the molecular mechanisms that underlie differences in shoot Cl– accumulation between grapevine rootstocks—or between plants of any species–have yet to be determined. Numerous studies have suggested that net shoot Cl– accumulation is regulated by membrane-localized anion transporters that mediate Cl– uptake into roots or control the rate of root to shoot Cl– transport at the root xylem interface (Gilliham and Tester, 2005; Brumós et al., 2009, 2010; Gong et al., 2011; Henderson et al., 2014). Grapevine, with its well-characterized differences in shoot Cl– accumulation, therefore serves as a good model from which to characterize candidate transporters for these processes in plants.
Cation-chloride cotransporters (CCCs) have been proposed to be prime candidates for directly regulating ion concentration in the root xylem (Colmenero-Flores et al., 2007; Flowers and Colmer, 2008; Brumós et al., 2009; Barbier-Brygoo et al., 2011; Shabala, 2013; Wegner, 2014; Fricke, 2015). CCCs from mammals and fish have been relatively well characterized; they transport the cations K+ and/or Na+ with the anion Cl– in a 1:1 ratio, and hence, facilitate electroneutral transport (Gamba, 2005). One of the main roles of mammalian CCCs is the reabsorption of electrolytes and water by the kidney. An example is an Na+-K+-2Cl– cotransporter (NKCC) from rat (Rattus norvegicus) that functions in the thick ascending limb of the loop of Henle of the nephron (Gamba et al., 1994). Given this role of some NKCCs, compounds that show inhibitory activity against CCC proteins are named loop diuretics. There are three types of CCCs classified based on their transport properties. NKCCs are typically inhibited by the loop diuretic bumetanide (Gamba et al., 1993) along with some K+-Cl– cotransporters (Mercado et al., 2000), whereas Na+-Cl– cotransporters are not (Gamba et al., 1993).
The first plant CCC identified was auxin-independent growth protein1 (AXI1) from Nicotiana tabacum, which was originally thought to trigger auxin-independent growth of protoplasts (Harling et al., 1997), but these findings were not reproducible (Schell et al., 1999). Later, Colmenero-Flores et al. (2007) identified the Arabidopsis AtCCC as a bumetanide-sensitive NKCC cotransporter when expressed in Xenopus laevis oocytes. Knockout of Atccc led to greater Na+ and Cl– accumulation in Arabidopsis shoots compared with wild-type plants when treated with high concentrations of K+, Na+, and Cl–, and promoter GUS fusions of AtCCC in planta displayed strong expression in the vasculature at the xylem/symplast boundary (Colmenero-Flores et al., 2007). GFP fusions of OsCCC from rice (ssp. japonica) were reported to localize to the plasma membrane (PM) of onion (Allium cepa) epidermal cells and rice root cells (Kong et al., 2011). Although these data are consistent with the proposed role for plant CCCs—of net xylem loading and/or retrieval of Na+, K+, and Cl– through the PM of root stelar cells (Colmenero-Flores et al., 2007)—this hypothesis was questioned by Teakle and Tyerman (2010). Teakle and Tyerman (2010) argued that the calculated Cl– gradients required for CCCs to function in ion retrieval from the xylem into xylem parenchyma cells would be atypical for plant root cells; instead, Teakle and Tyerman (2010) suggested a potential role of AtCCC in compartmentation of Na+ and Cl– in roots if it functioned on the vacuole, which would also reduce net xylem loading of these ions. Knockout of Atccc also led to a major growth phenotype, even under standard conditions (Colmenero-Flores et al., 2007), which is unlikely to result from the observed small elevation in shoot salt concentration under these conditions. Therefore, further investigations are needed to clarify the putative role of CCCs in conferring shoot Cl– (and Na+) exclusion and salt tolerance in plants.
We identified and functionally characterized the grapevine CCC. We selected Cabernet Sauvignon as the cultivar from which to isolate VviCCC, because it has been used extensively for previous molecular investigations (Cramer et al., 2007; Tattersall et al., 2007; Vincent et al., 2007; Shelden et al., 2009).
RESULTS
Identification of VviCCC
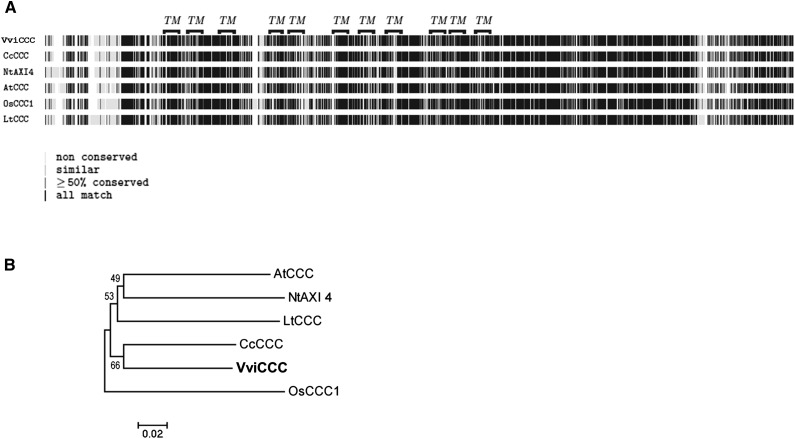
Multiple putative grapevine CCC mRNA sequences exist in the National Center for Biotechnology Information (NCBI) GenBank Database, including truncated versions from Cabernet Sauvignon (accession no. GQ161924.1) and another cultivar, Merlot (accession no. GQ161925.1). Using the amino acid sequence of AtCCC as a query in a BLASTP analysis against the 12× International Grape Genome Program grapevine genome at EnsemblPlants, we identified a single predicted full-length grapevine homolog of the previously characterized Arabidopsis CCC protein. The VviCCC protein was predicted to contain 982 amino acids and encoded by a single-gene locus on chromosome 10 (VIT10s0003g04530). This annotated CCC isoform in Cabernet Sauvignon roots was subsequently confirmed as correct by isolating and sequencing the corresponding full-length coding sequence. Multiple protein sequence alignment showed that VviCCC displays a high degree of similarity with other previously characterized or identified plant CCCs (Fig. 1A). Phylogenetic analysis showed that the most closely related known protein to VviCCC is the Citrus clementina CCC identified by Brumós et al. (2010), which is another woody perennial crop species (Fig. 1B). The VviCCC protein sequence was analyzed by five different transmembrane topology prediction programs (for list, see Supplemental Table S1). The identification of putative transmembrane domains suggested that VviCCC shared membrane transporter function with AtCCC, and therefore, we characterized the protein accordingly.
Figure 1.
VviCCC is closely related to CCCs from other plant species. A, Multiple protein sequence alignment of plant CCC. Putative transmembrane (TM) domains of VviCCC and amino acid similarity scores are shown as indicated. Protein accession numbers are reported in Supplemental Table S2, and the full alignment is detailed in Supplemental Figure S7. B, Neighbor-joining phylogenetic tree of plant CCC proteins used in A showing bootstrap values from 5,000 iterations.
VviCCC and AtCCC Localize to the Golgi and Trans-Golgi Network
Although AtCCC has been functionally characterized as a membrane transporter (Colmenero-Flores et al., 2007), its subcellular localization has not been shown previously. We therefore aimed to ascertain the subcellular localizations of both AtCCC and VviCCC in planta. We queried the Subcellular Localization of Proteins in Arabidopsis3 (SUBA3) Database (Tanz et al., 2013) as a starting point to determine AtCCC localization. SUBA3 showed various predicted localizations of AtCCC, including the PM, cytosol, and mitochondria. SUBA3 also summarized four proteomic studies that identified AtCCC peptides in the Golgi, the trans-Golgi network (TGN; Drakakaki et al., 2012; Nikolovski et al., 2012), PM (Benschop et al., 2007), and tonoplast (Whiteman et al., 2008b). Two additional studies have identified AtCCC peptides in the TGN (Sadowski et al., 2008; Groen et al., 2014). Collectively, these studies and the methods that they used to identify AtCCC are summarized in Table I.
Table I. Summary of organellar proteomic studies identifying the subcellular localization of AtCCC by mass spectrometry.
| Subcellular Localization of AtCCC | Tissue Type | Ecotype | Method Used | Reference |
|---|---|---|---|---|
| TGN | Roots | Col-0 | (1) Immunopurification of VHAa1-GFP fraction and (2) LOPIT proteomics | Groen et al. (2014) |
| TGN | Callus | LOPIT proteomics | Nikolovski et al. (2012) | |
| Golgi/TGN | Leaves | Col-0 | Immunopurification of SYP61-CFP vesicles, nanoliquid chromatography/mass spectrometry/mass spectrometry | Drakakaki et al. (2012) |
| Golgi | Callus | Col-0 | LOPIT proteomics | Sadowski et al. (2008) |
| Tonoplast | Leaves | Col-0 | Liquid chromatography/mass spectrometry/mass spectrometry analysis of tonoplast-enriched fraction | Whiteman et al. (2008b) |
| PM | Suspension cells | Col-0 | Nanoscale HPLC-mass spectrometry/mass spectrometry of PM fraction isolated by two-phase partitioning | Benschop et al. (2007) |
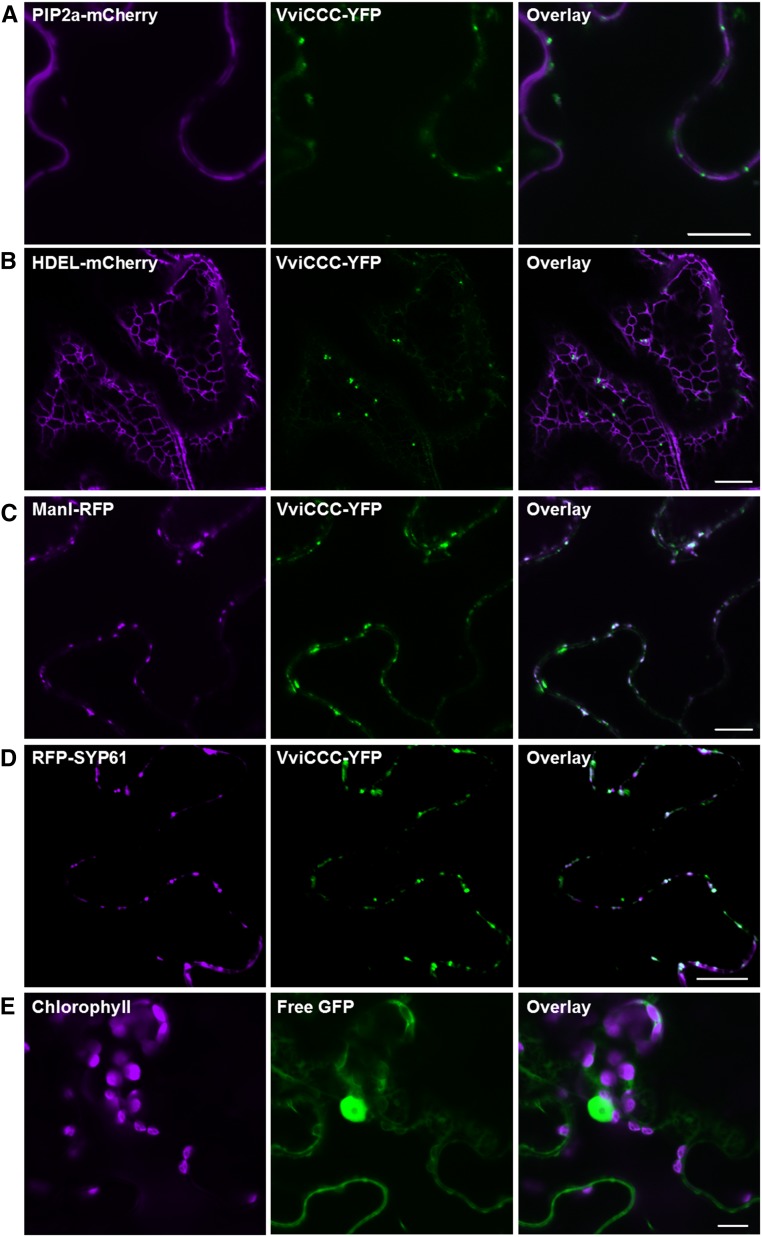
Because of the wide range of putative subcellular localizations of AtCCC, we used fluorescent tagging to establish AtCCC and VviCCC localizations. We constructed a yellow fluorescent protein (YFP) fusion of VviCCC at the carboxy terminus and transiently expressed this protein in tobacco (Nicotiana benthamiana) leaves combined with various subcellular markers fused to red fluorescent protein (RFP) or mCherry by agroinfiltration. Confocal laser-scanning microscopy detected a mobile punctate VviCCC-YFP signal (Supplemental Video S1) that did not overlap with the plasma membrane intrinsic protein 2a (PIP2a) PM or HDEL endoplasmic reticulum (ER) markers (Nelson et al., 2007; Fig. 2, A and B). Partial colocalization was observed between the signals of VviCCC-YFP and either the mannosidase I (Man1)-RFP Golgi marker (Fig. 2C) or the RFP-syntaxin of plants61 (SYP61) TGN marker (Fig. 2D). As a control, we infiltrated free GFP. The pattern of the free GFP signal did not resemble the VviCCC-YFP signal in tobacco leaves (Fig. 2E). Similarly, when a construct encoding AtCCC-GFP was expressed in tobacco leaves, a punctate mobile signal (Supplemental Video S2) was observed that did not overlap with the PM marker (Supplemental Fig. S1A) or the ER marker (Supplemental Fig. S1B). Instead, like VviCCC, the AtCCC-GFP signal overlapped partially with the Golgi marker (Supplemental Fig. S1C) and the TGN marker (Supplemental Fig. S1D).
Figure 2.
VviCCC-YFP localizes to the Golgi and TGN. Transient coexpression of VviCCC-YFP with various subcellular markers in epidermal cells of tobacco. Tobacco leaves were coinfiltrated with A. tumefaciens strains harboring VviCCC-YFP and PM marker AtPIP2a-mCherry (A), ER marker HDEL-mCherry (B), Golgi marker ManI-RFP (C), or TGN marker RFP-SYP61 (D). E, A. tumefaciens harboring free GFP without VviCCC-YFP was infiltrated as a control. Leaf sections were imaged by confocal laser-scanning microscopy. Chlorophyll autofluorescence and mCherry and RFP signals are shown in magenta in left. GFP and YFP signals are shown in green in center. Colocalization of green and magenta signals appears in white in right. Bar = 10 μm.
To further assess the subcellular localization of VviCCC and AtCCC, YFP fusions of these CCC proteins were transiently expressed in Arabidopsis mesophyll protoplasts and imaged by confocal laser-scanning microscopy. YFP signals did not overlap with the PM marker Rho of plants11 (ROP11) fused to enhanced cyan fluorescent protein (eCFP; Molendijk et al., 2008; Supplemental Fig. S2, A and B). However, when colocalization was performed using the teal fluorescent Wave 13T marker (Geldner et al., 2009) encoding the TGN protein vesicle transport v-SNARE12 (AtVTI12), overlap between the VviCCC-YFP or AtCCC-YFP and the TGN-TFP signals was observed (Supplemental Fig. S2, C and D). These data in tobacco epidermal cells and Arabidopsis mesophyll protoplasts suggest that VviCCC and AtCCC are localized to the Golgi and TGN in plants.
VviCCC Transcript Analysis
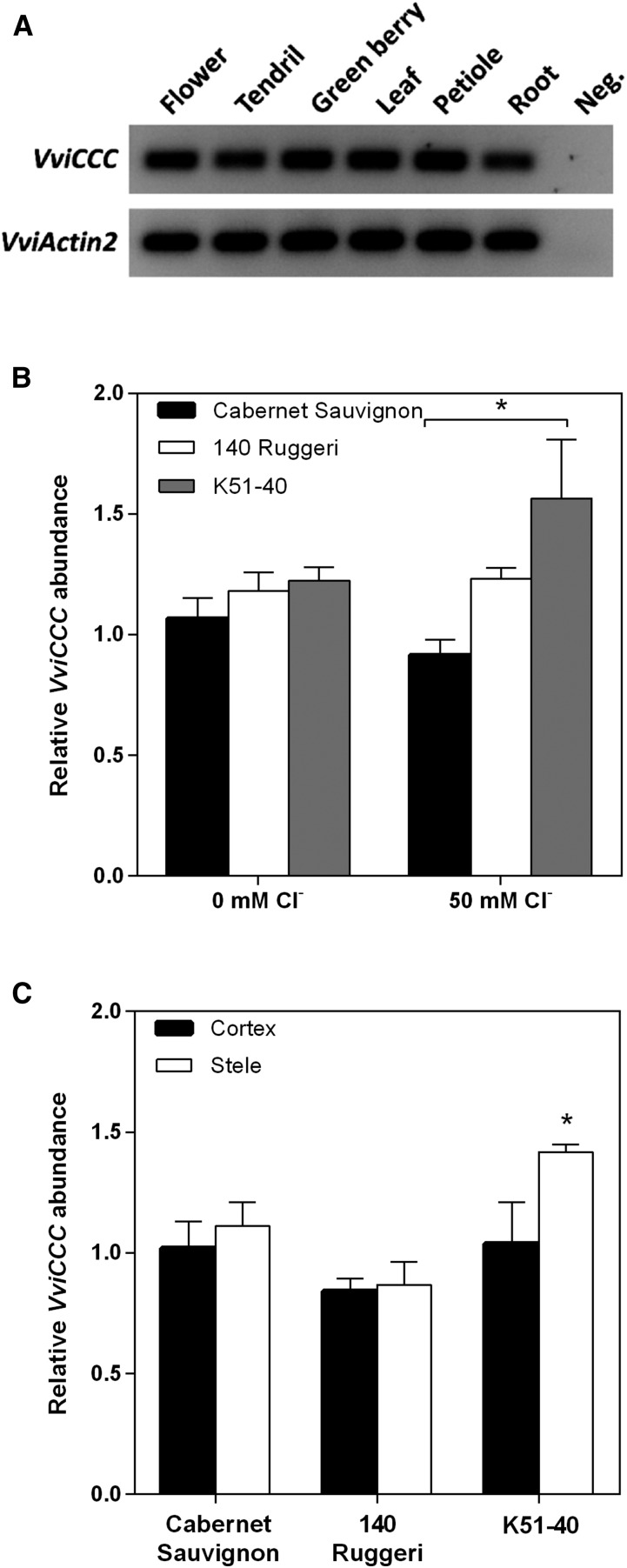
To evaluate the possible function of VviCCC, the tissue specificity of VviCCC transcript was investigated in Cabernet Sauvignon. The flower, tendril, green berry, petiole, leaf, and root were harvested from pot-grown vines and, reverse transcription-PCR was used to detect transcript in these organs. VviCCC was detected in all tissue types analyzed (Fig. 3A).
Figure 3.
VviCCC is expressed across multiple organs and not strongly regulated by salt stress in roots. A, Semi-qRT-PCR of different tissues of pot-grown Cabernet Sauvignon. Results for 30 cycles are shown. Negative control uses a water template. B, Quantitative real-time PCR of CCC in total root tissue of hydroponically grown Cabernet Sauvignon, 140 Ruggeri, and K51-40 rooted leaves under 0 or 50 mm Cl− stress for 4 d. Data are the means ± sem of four biological replicates, each of four pooled individuals. Data are relative to the untreated Cabernet Sauvignon replicate with the greatest transcript abundance. Significant differences were measured by one-way ANOVA with Bonferroni post hoc test (P < 0.05). *, Significant difference between genotypes. C, qRT-PCR analysis of CCC in root cortical- and root stelar-enriched fractions derived from rooted leaves of Cabernet Sauvignon, 140 Ruggeri, and K51-40 exposed to 50 mm Cl− for 4 d. Data are the means ± sem of three biological replicates of pooled tissue from multiple plants. Data are relative to the Cabernet Sauvignon cortical replicate with the greatest transcript abundance. *, Significant difference between cortex and stele (P < 0.05; Student’s t test).
Next, we aimed to elucidate changes in VviCCC transcript abundance in grapevine root tissue in response to salt stress and between Vitis spp. rootstocks of contrasting Cl– exclusion capacity. Grapevine cultivars were established as rooted leaves and grown hydroponically using the method by Schachtman and Thomas (2003). CCC expression was compared between the roots of Cabernet Sauvignon, the salt-tolerant rootstock 140 Ruggeri, and the salt-sensitive rootstock K51-40 by real-time quantitative reverse transcription (qRT)-PCR. No significant difference in the relative transcript abundance between varieties was observed under nonstressed conditions (Fig. 3B). In addition, salt stress applied as 50 mm Cl– did not significantly alter the transcript abundance within varieties (Fig. 3B). Between varieties, salt stress conditions induced an approximate 50% increase in CCC transcript levels in K51-40 compared with Cabernet Sauvignon (Fig. 3B), but there was no significant difference between the rootstocks in CCC expression under salt treatment. The Cl– content of the roots, leaves, and petioles of this material was measured and described previously (see fig. 1 in Henderson et al., 2014). The expected Cl– compartmentation pattern was found, and K51-40 had greater Cl– in the leaf but lower Cl– in the root than in the corresponding tissues of 140 Ruggeri.
To further characterize CCC expression, the stele and epidermis/cortex of grapevine roots were separated by hand, resulting in samples that were enriched in cell types from these compartments (Henderson et al., 2014). RNA was extracted, and expression levels were determined by qRT-PCR. The poor Cl– excluder K51-40 had significantly greater expression of CCC in the root stele under 50 mm Cl– stress (Fig. 3C). The other varieties displayed no significant difference between these two tissue types (Fig. 3C).
Sequence Analysis of CCC between Contrasting Vitis spp.
Although related, Cabernet Sauvignon, 140 Ruggeri, and K51-40 are different Vitis spp. Therefore, it would be possible that nucleotide sequence differences and subsequent amino acid changes in CCC proteins between species could have functional implications that might contribute to differential Cl– exclusion capacity. To address this, we amplified the coding sequences of Vitis spp. CCCs from all three grapevine varieties and sequenced them. Some heterozygosity could be observed as overlapping peaks on the electrophoretogram. Only one of these heterozygous regions had the potential for an amino acid substitution (N123I). The heterozygosity that could bring about this potential substitution was identical in the CCC complementary DNA (cDNA) from both 140 Ruggeri and K51-40, and therefore, we concluded that there was no difference in the CCC gene sequence that could contribute to differential Cl– exclusion between the grapevine varieties examined in this study. The cDNA sequences are shown in Supplemental Figure S3.
VviCCC Transports Sodium, Potassium, and Chloride
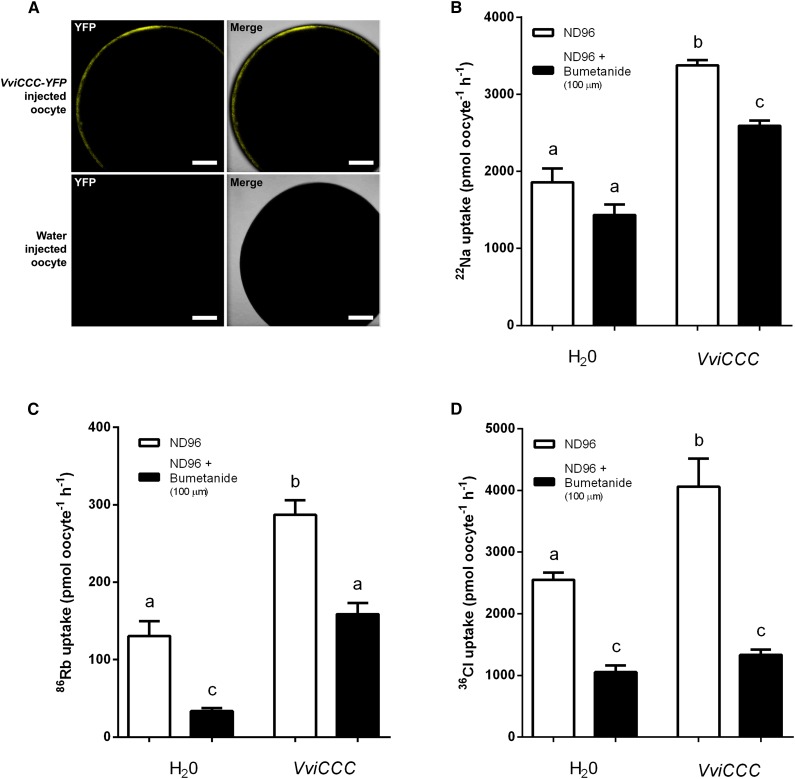
AtCCC is a member of the NKCC family (Colmenero-Flores et al., 2007). We therefore investigated the transport properties of VviCCC in X. laevis oocytes. Because VviCCC is localized to the Golgi and TGN in plant cells, we first examined whether VviCCC targeted to the PM of oocytes, which is a common end point in oocytes for proteins that are targeted to plant endomembranes (Maurel et al., 1993). Injection of capped RNA (cRNA) encoding VviCCC-YFP into oocytes resulted in detectable fluorescence at the PM, and this fluorescence was not detectable in water-injected control oocytes (Fig. 4A). Given that VviCCC-YFP was PM localized in X. laevis oocytes, we assayed the uptake in oocytes of the radiotracers 22Na+, 86Rb+, and 36Cl– after injection with untagged VviCCC cRNA to determine its transport properties. Compared with water-injected control oocytes, VviCCC mediated significant uptake of 22Na+ (Fig. 4B), 86Rb+ (a K+ tracer; Fig. 4C; Supplemental Fig. S4), and 36Cl– (Fig. 4D), suggesting that VviCCC is able to transport Na+, K+, and Cl−. Furthermore, radiotracer uptake by VviCCC-injected oocytes was significantly inhibited by 100 µm NKCC-specific loop diuretic bumetanide (Fig. 4, B–D). The reduced uptake of 86Rb+ and 36Cl– after bumetanide treatment in water-injected oocytes (Fig. 4, C and D) is likely caused by the activity of endogenous NKCC of X. laevis oocytes (Suvitayavat et al., 1994). This suggests that VviCCC functions as a bumetanide-sensitive NKCC transporter.
Figure 4.
VviCCC displays NKCC function in X. laevis oocytes. A, Localization of VviCCC in X. laevis oocytes. Oocytes were injected with cRNA encoding VviCCC-YFP (upper) or water (lower) and analyzed by confocal laser-scanning microscopy. YFP signal is shown in left, and YFP signal plus the bright-field image are overlaid in right. Bar = 200 µm. B, Oocytes (injected with water or untagged VviCCC cRNA as indicated) were bathed in radioactive uptake buffer (ND96) containing 22Na (white bars) or uptake buffer containing 22Na and 100 µm bumetanide (black bars) for 1 h. Data are the means ± sem of at least 17 oocytes and representative of multiple experiments. C, Oocytes were bathed in radioactive uptake buffer (ND96) containing 86Rb (white bars) or uptake buffer containing 86Rb and 100 µm bumetanide (black bars) for 1 h. Data are the means ± sem of at least 16 oocytes and representative of multiple experiments. D, Oocytes were bathed in radioactive uptake buffer (ND96) containing 36Cl (white bars) or uptake buffer containing 36Cl and 100 µm bumetanide (black bars) for 1 h. Data are the means ± sem of 10 oocytes. All oocyte data are representative of multiple experiments using oocytes from different frogs. In B to D, different letters indicate significant differences (ANOVA with Tukey’s post hoc test; P < 0.05).
Expression of VviCCC Complements the Arabidopsis ccc Mutant
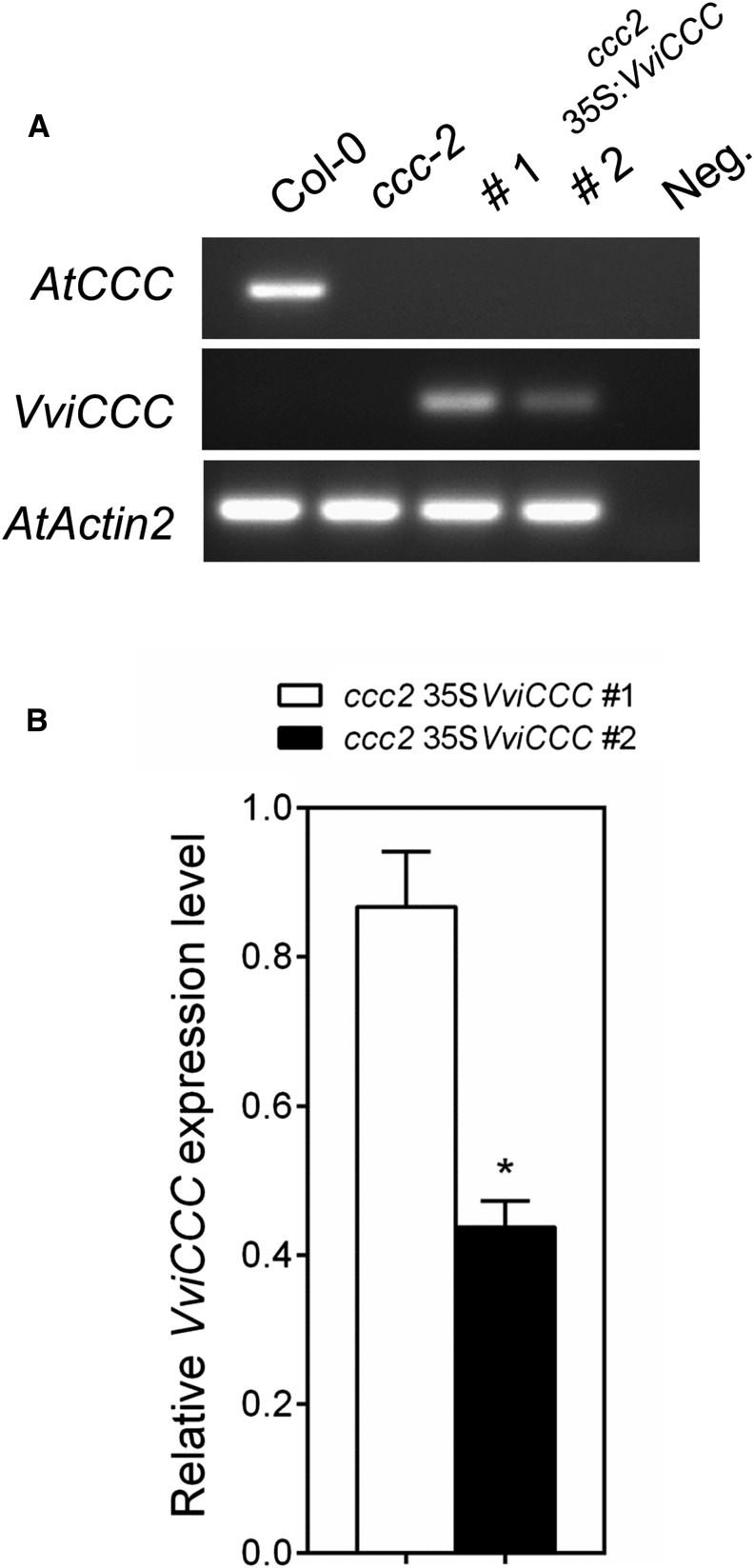
Two previously characterized Arabidopsis transfer DNA (T-DNA) insertion mutants with reduced expression of AtCCC (ccc1 and ccc2) display a stunted growth phenotype with late flowering and bushy inflorescences (Colmenero-Flores et al., 2007). We used the ccc2 mutant (Salk_145300) for complementation analysis. Salk_145300 is not part of the homozygous collection, and therefore, it was self-fertilized; the presence of a homozygous T-DNA insertion was confirmed by PCR in subsequent generations (Supplemental Fig. S5). Salk_145300 was transformed with VviCCC under the constitutive Cauliflower mosaic virus 35S promoter, and two independent nonsegregating T3 individuals were obtained. Semi-qRT-PCR showed that the AtCCC transcript was only detectable in the Columbia-0 (Col-0) wild-type plant and absent from ccc2 and both transgenic lines made in this study (Fig. 5A). The presence of VviCCC transcript was seen in the two independent VviCCC-expressing lines (Fig. 5A). Using real-time quantitative PCR performed on whole-plant samples, we determined that the VviCCC-expressing line number 1 had approximately 2-fold greater expression of VviCCC than line number 2 (Fig. 5B).
Figure 5.
Two independent transformants of the Arabidopsis ccc mutant show variation in the expression level of VviCCC. A, Semi-qRT-PCR of the Arabidopsis ccc (Salk_145300) wild type (Col-0) and two independently transformed lines of ccc complemented with VviCCC. Thirty cycles of PCR were performed. Negative control is water template. B, Quantitative real-time PCR analysis of the expression level of VviCCC in whole ccc plants of two independent transformants. Data are the means of four biological replicates ± sem. Data are relative to the biological replicate with the greatest transcript abundance in line number 1. *, Significant difference (P < 0.05; Student’s t test).
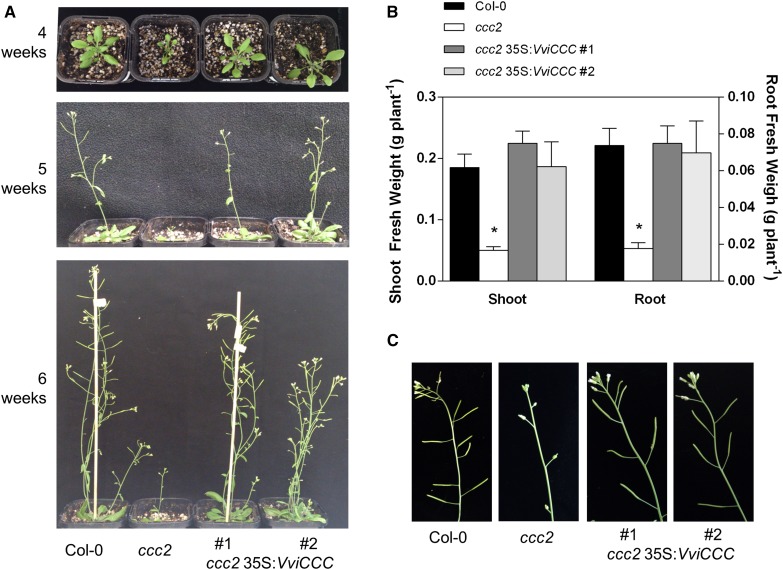
The stunted growth phenotype of ccc was fully reversed in VviCCC-expressing line number 1 and partially ameliorated in line number 2. The rosette size of both lines more closely resembled that of wild-type Col-0 (Fig. 6A). Line 2 displayed bushy and slightly shorter inflorescences compared with the wild type and line 1, and this was more evident after 6 weeks of growth in long-day conditions (Fig. 6A; Supplemental Fig. S6). After 5 weeks in hydroponics under short-day conditions, the two independent VviCCC-expressing lines showed full complementation of a root and shoot fresh weight phenotype of the ccc mutant (Fig. 6B). Silique development was restored to normal in both VviCCC-expressing lines (Fig. 6C).
Figure 6.
Expression of VviCCC complements a growth phenotype of the ccc mutant. A, Dwarf phenotype of the Arabidopsis ccc mutant is rescued by constitutive expression of VviCCC in two independent lines; 4-, 5-, and 6-week-old soil-grown plants under nonstress conditions are shown. B, Root and shoot fresh weights of 6-week-old hydroponically grown wild-type, mutant, and VviCCC-expressing lines. Bars are means ± sem of at least 17 individuals from two independent experiments. *, Significant difference from the wild type (ANOVA with Tukey’s post hoc test; P < 0.05). C, Although the ccc mutant line shows undeveloped siliques at 6 weeks of age, expression of VviCCC in both independent complemented lines results in normal silique development, reminiscent of wild-type Col-0.
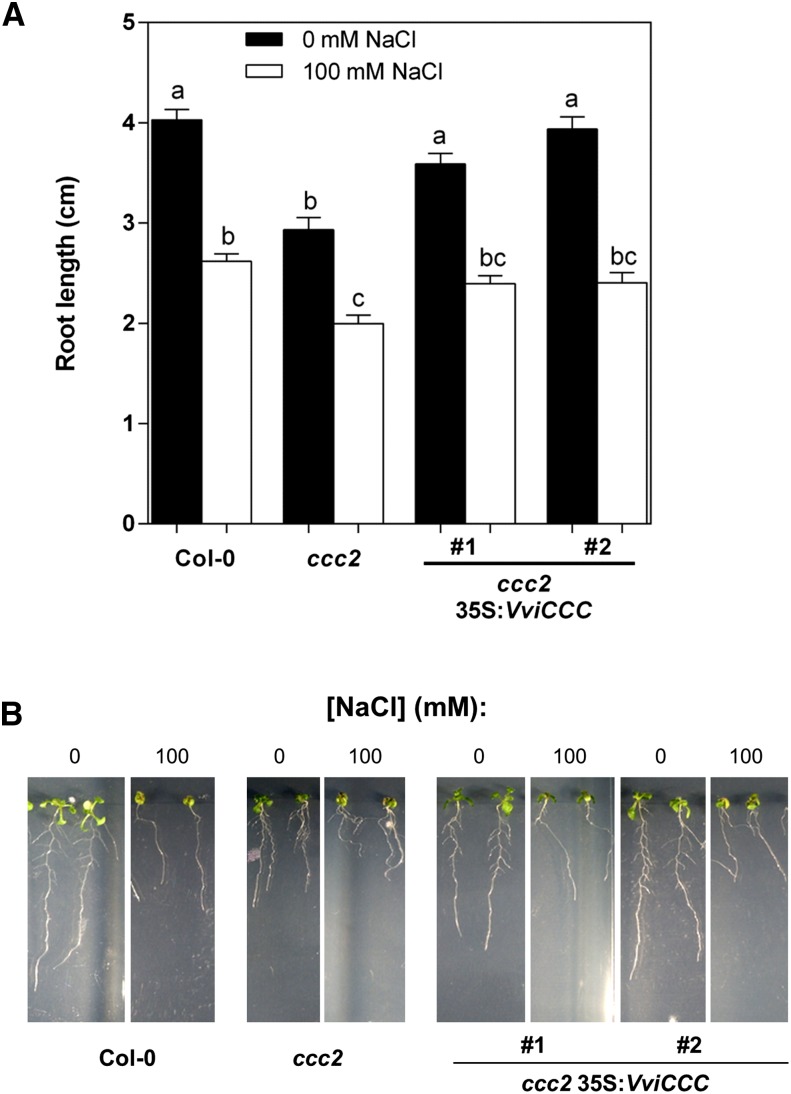
Compared with wild-type Col-0, the ccc mutant showed a reduced root length phenotype during the first 2 weeks of growth in vitro (Fig. 7A). Expression of VviCCC in the mutant background could complement this phenotype (Fig. 7A). We also investigated whether this phenotype was exacerbated by salt stress. When seedlings were transferred to one-half-strength Murashige and Skoog medium (MS) plates supplemented with 100 mm NaCl for 10 d, a significant reduction in root length was observed in all lines compared with control conditions (Fig. 7A). There was also a significant difference between the root length of the wild type and ccc when grown on 100 mm NaCl, and this was complemented by VviCCC expression in two independent transformants (Fig. 7). VviCCC expression in the mutant ccc did not improve growth above wild-type levels in any conditions tested.
Figure 7.
Reduced root length of the ccc mutant is abolished in two 35S:VviCCC lines under normal conditions and 100 mm NaCl stress. A, Root length of wild-type, mutant, and VviCCC-expressing lines 10 d after transfer to one-half-strength MS plates containing 0 (black bars) or 100 mm NaCl (white bars). Bars are means ± sem of at least 16 individuals from two independent experiments. Different letters indicate statistically significant differences (ANOVA with Bonferroni’s post hoc test; P < 0.05). C, Physical representation of plants used to collect data for A.
VviCCC Mediates Sodium and Chloride Homeostasis in the Arabidopsis ccc Mutant
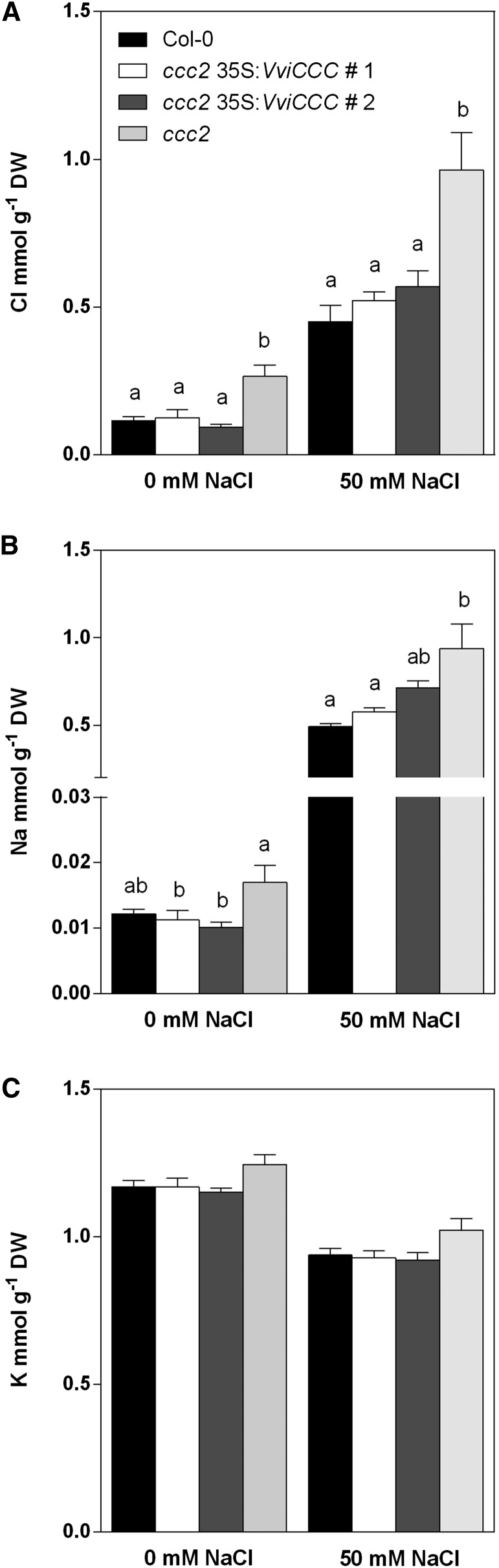
Arabidopsis CCC is implicated in controlling long-distance Cl– transport from the root to shoot. Two Atccc mutant lines accumulated more Cl– in aerial tissues and less in root tissue, and these differences were exacerbated by both the duration and strength of Cl– treatment (Colmenero-Flores et al., 2007). To investigate whether VviCCC has a similar role in Cl– homeostasis in planta, we used inductively coupled plasma optical emission spectrometry (ICP-OES) to compare the shoot ion concentration of hydroponically grown wild-type, mutant, and VviCCC-expressing Arabidopsis lines exposed to 0 and 50 mm NaCl. The Cl– concentration of the ccc mutant was significantly greater under control conditions compared with wild-type and VviCCC-expressing lines (Fig. 8A). This was exacerbated by salt treatment, with all lines accumulating more shoot Cl– (Fig. 8A). The Arabidopsis ccc mutant also accumulated more shoot Na+ than the wild type, and this mutant phenotype was reversed by expression of VviCCC in the two independent ccc lines (Fig. 8B). Salt treatment led to greater shoot Na+ accumulation, and this was reversed in VviCCC-expressing line 1, whereas the weaker complemented line number 2 shoot Na+ concentration was slightly higher (Fig. 8B). K+ concentration was unchanged between mutant, wild-type, and VviCCC -expressing lines under control and salt-stress conditions (Fig. 8C).
Figure 8.
VviCCC is involved in Na+ and Cl− homeostasis in planta but does not enhance shoot Cl– exclusion in ccc under saline conditions. Shoot ion concentration of hydroponically grown Arabidopsis wild type, ccc mutant, and two independent ccc lines expressing VviCCC under normal and 50 mm NaCl conditions. A, Shoot Cl concentration. B, Shoot Na concentration. C, Shoot K concentration. One-way ANOVA with Fisher’s lsd post hoc test (P < 0.01) was used to compare means within treatments. DW, Dry weight.
DISCUSSION
CCCs have been extensively studied in mammalian systems because of their vital role in renal function (Gamba, 2005). Comparatively few studies have been conducted in plants, and the role of plant CCCs has yet to be fully established. We have isolated and functionally characterized VviCCC—a gene encoding a CCC from grapevine. Radioactive tracer uptake studies in X. laevis oocytes revealed that VviCCC mediates Na+, Rb+ (a K+ tracer), and Cl– uptake. Radioisotope uptake was inhibited by bumetanide, a loop diuretic that targets NKCCs, indicating that VviCCC belongs to the NKCC class of CCC. This is supported by the observation that VviCCC, when expressed in an Arabidopsis ccc mutant devoid of its native NKCC, complemented phenotypes relating to both growth and ion accumulation. Phenotypic complementation included reduced levels of Na+ and Cl– in shoots. This provides evidence that VviCCC is involved in Na+ and Cl– transport in planta.
Suggestions have been made that AtCCC may actively retrieve Cl– from the root xylem (Colmenero-Flores et al., 2007) and that orthologous proteins might therefore mediate shoot Cl– exclusion and salt tolerance in Cl–-sensitive plant species, such as grapevine and citrus (Brumós et al., 2010). For VviCCC to directly mediate Cl– (and Na+ and K+) efflux to or retrieval from the root xylem, it must be embedded within the PM. In our study, both GFP-tagged AtCCC and YFP-tagged VviCCC were absent from the PM when expressed in tobacco leaves and Arabidopsis mesophyll protoplasts. Instead, CCC fluorescence was detected in the Golgi and TGN. This contrasts with the described localization of OsCCC, where PM localization was reported to be present in onion epidermis and rice root cells (Kong et al., 2011). Yet, in agreement with our findings, AtCCC peptides have been identified in the Golgi and TGN in four independent proteomic studies (Sadowski et al., 2008; Drakakaki et al., 2012; Nikolovski et al., 2012; Groen et al., 2014; Table I). This shows that plant CCCs localize to these subcellular compartments in planta. In particular, one of these studies identified AtCCC in TGN vesicles that were immunopurified using antibodies against SYP61 (Drakakaki et al., 2012). In our study, RFP-SYP61 colocalized with VviCCC-YFP (Fig. 2D) and AtCCC-GFP in tobacco cells (Supplemental Fig. S1D). AtCCC was previously identified in a tonoplast-enriched fraction of Arabidopsis (Whiteman et al., 2008b). However, when the same technique was applied to rice, OsCCC was not identified in the tonoplast or PM (Whiteman et al., 2008a). In another study, AtCCC was found in PM-enriched preparations, but a small number of potential contaminants was also observed (Benschop et al., 2007). Using localization of organelle proteins by isotope tagging (LOPIT), Groen et al. (2014) were able to determine the steady-state positions of TGN proteins in Arabidopsis roots and discriminate between contaminants and cargo proteins in transit from full-time residents of this organelle. The steady-state location of AtCCC was determined to be the TGN.
Based on our data and the salt-related phenotypes of other plants misexpressing proteins localized to Golgi and TGN membranes, it is increasingly clear that these compartments have an important role in plant salt tolerance (Munns and Gilliham, 2015). For instance, double knockouts of TGN-localized Na+-H+ antiporters, nhx5/nhx6, are hypersensitive to moderate salinity and disrupt vesicle trafficking to the vacuole (Bassil et al., 2011). Furthermore, both a phosphate transporter in Arabidopsis (AtPHT4;6) and a monosaccharide transporter from rice (OsGMST1) are Golgi localized, and reduced expression of either transporter reduces salt tolerance of plants (Cubero et al., 2009; Cao et al., 2011). Many vesicle trafficking-related proteins, such as RAB GTPases, and soluble N-ethyl-maleimide sensitive factor attachment protein receptors (SNARE) also localize to the Golgi and TGN, and knockout of these factors also results in altered salt sensitivity (Kim and Bassham, 2011; Asaoka et al., 2013). The role of endosomal-localized transporters in the control of shoot salt exclusion is unclear but might involve sensing cytoplasmic salt concentrations or regulating luminal pH or ion homeostasis, which is required for normal Golgi and TGN function. Disrupted endosomal function could result in the misprocessing of other transport proteins important in shoot salt exclusion. Alternatively, vesicle trafficking, which may improve salt tolerance by removal of salts from the cytoplasm to the apoplast through exocytosis or the secretion of salts into the vacuole, could be disrupted in plants lacking the full complement of Golgi/TGN transporters. This is clearly an area of growing research interest that warrants further attention.
We localized AtCCC and VviCCC to the Golgi and TGN; however, it is impossible to fully rule out alternative localizations for these proteins under different conditions considering the role of the Golgi in delivering proteins to different compartments. Another membrane protein, Arabidopsis thaliana Phosphate1 (AtPHO1), and also, its mammalian homolog, Mus musculus Xenotropic Polytropic Virus Receptor1 (MmXPR1), localize to the plant Golgi and TGN under normal conditions but somehow mediate inorganic phosphate release to the extracellular space (Arpat et al., 2012; Wege and Poirier, 2014). In these studies, it was hypothesized that phosphate export proteins might be present in too low abundance at the PM for detection by fluorophore tagging or alternatively, that phosphate might be loaded into vesicles for exocytosis followed by rapid recycling away from the PM (Arpat et al., 2012; Wege and Poirier, 2014). Iron-regulated transporter1 (IRT1), which mediates iron and divalent metal uptake from the soil by root hairs, was also localized to the TGN and early endosome in Arabidopsis (Barberon et al., 2011). IRT1 could also be observed at the PM but only when endocytosis was chemically disrupted or monoubiquitination was prevented by mutation of key amino acids, and this resulted in toxicity (Barberon et al., 2011). It could be envisaged that the localization of plant CCCs and how they affect shoot ion concentration might be regulated by similar mechanisms. However, examination of concentration gradients that determine the direction of net flux through NKCC indicated that influx from xylem vessels to xylem parenchyma cells would only occur through a PM-localized CCC if the xylem parenchyma had very low cytosolic concentrations of Na+ and Cl–, below the normal physiological range of plant root cells (Teakle and Tyerman, 2010). These observations and our data showing that VviCCC and AtCCC are localized at the TGN and not other membranes cast serious doubt on the hypothesis that plant CCCs are directly involved in ion reabsorption from the xylem across the PM of xylem-associated cells.
Additionally, our analyses of VviCCC transcript did not support a direct role of this gene in grapevine shoot chloride exclusion. Promoter GUS studies in Arabidopsis have shown that AtCCC is expressed in the root and shoot vasculature, suggesting a possible role in long-distance ion transport (Colmenero-Flores et al., 2007). In our study, quantitative PCR on RNA isolated from stele and cortex root fractions revealed that Vitis spp. CCC transcripts were equally abundant in the root stele and cortex of three grapevine genotypes. Expression of CCC in roots was also not regulated by a physiologically relevant (50 mm) Cl– stress in Cabernet Sauvignon or two grapevine rootstocks of contrasting Cl– exclusion capacity. Similarly, AtCCC transcript in Arabidopsis roots was not responsive to applications of 50 or 100 mm NaCl (Supplemental Fig. S7). Although transcript levels might not correspond to protein levels, expression data indicate no primary role of VviCCC or AtCCC in chloride exclusion.
Overexpression or ectopic expression of genes that are important for salt tolerance in plants can often improve plant growth under salt stress (for example, high affinity potassium transporters [HKTs; Plett et al., 2010; Munns et al., 2012], glutathione S-transferase [GST; Roxas et al., 1997], and chloride channels [CLCs; Wei et al., 2013]). Expression of VviCCC restored growth of the Atccc mutant to wild-type levels under salt stress but did not enhance growth beyond that of wild-type Col-0. The severe growth phenotype of Atccc occurs in the absence of salt, and it is possible that the salt sensitivity of this mutant is an exacerbation of the existing phenotype. Insertional mutagenesis of the AtCCC gene has been linked with abnormal pollen grain development and reduced fertility (Johnson et al., 2004), highlighting other roles of plant CCCs that are unrelated to salt tolerance. It is clear, however, that plant CCCs are crucial for maintaining the balance of inorganic ions in shoot tissues. Interestingly, Zeuthen and MacAulay (2012) observed bumetanide-sensitive water and Rb fluxes in X. laevis oocytes expressing mouse NKCC1 but only Rb fluxes when human NKCC2 was expressed. Based on this finding, it could be speculated that plant CCC transports Na+, K+, and Cl– to generate osmotic gradients for water to follow through aquaporins or the CCC pore itself, thereby contributing to compartmental volume regulation.
The data presented here indicate that VviCCC and AtCCC are TGN-localized NKCCs and that plant CCC function is conserved across these two species. Our data do not support the hypothesis that AtCCC and VviCCC directly control the flux of Na+, Cl–, and K+ from the xylem through the PM of xylem parenchyma cells. Furthermore, we suggest that CCC proteins perform a function in the Golgi and TGN that heavily impacts plant performance, which indirectly influences the ability of plants to tolerate high salinity. Future structure-function investigations of plant CCC proteins and further studies of their ion transport profiles, such as ion dependencies, will help identify the mechanisms of CCC regulation and further clarify their function in planta.
MATERIALS AND METHODS
Gene Identification
The Arabidopsis (Arabidopsis thaliana) CCC protein sequence (NCBI accession no. NP_849732.1) was used as the basis for BLASTP searches of the 12× IGGP grapevine (Vitis vinifera) genome at EnsemblPlants (http://plants.ensembl.org/). Multiple sequence alignments were performed using ClustalW2 (Larkin et al., 2007) and edited by visual inspection using Geneious, version 5.1.7. Shading and final alignment were generated with TEXshade (Beitz, 2000). The full alignment is shown in Supplemental Figure S8. A phylogenetic tree of previously identified plant CCC family members and VviCCC (Supplemental Table S2) was generated using MEGA5 (Tamura et al., 2011) by the neighbor-joining method with 5,000 bootstrap iterations and default settings. The resulting branch lengths were set to be proportional to the amount of inferred evolutionary change. Putative transmembrane domains were identified by running the VviCCC amino acid sequence through various topology prediction algorithms (Supplemental Table S1).
Rooted Leaf Preparation and Salt Treatment
Rooted leaves of 140 Ruggeri, K51-40, and Cabernet Sauvignon were established as outlined by Gong et al. (2011). After pretreatment, the rooted leaves were subjected to 50 mm Cl– (Na+:Ca2+:Mg2+ [6:1:1]) in nutrient solution for 4 d. A nutrient solution control without 50 mm Cl– (Na+:Ca2+:Mg2+ [6:1:1]) was included for comparison. There were four biological replicates, each consisting of four rooted leaves, for each cultivar in each treatment. Roots were removed from rooted leaves and rinsed free of nutrient solution with the surface water removed, and then, total roots were used for RNA extraction. When stated, the stele and cortex were first separated before RNA extraction; lateral roots were removed from main roots by hand, and then, the cortex was stripped from stele using fine tweezers. All samples were immediately frozen in liquid nitrogen and stored at −80°C.
RNA Extraction
RNA was isolated from either total roots or root fractions enriched in stelar or cortical tissue. Root tissue was ground to a fine powder under liquid nitrogen in a mortar and pestle. RNA was isolated using the Spectrum Plant Total RNA Kit (Sigma) following the manufacturer’s procedures. RNA was treated with DNaseI using Turbo DNA-Free (Ambion) for 1 h at 37°C; then, it was ethanol precipitated and resuspended in water. An RNA quality threshold was set for 260:280 and 260:230 absorbance ratios at >1.8.
cDNA Synthesis
One microgram of total RNA was reverse transcribed in a 20-µL reaction using the iScript cDNA Synthesis Kit (Bio-Rad) following the manufacturer’s procedures with the following modifications: before synthesis, RNA was heated at 65°C for 5 min and then, placed on ice for 1 min to prevent secondary structure formation; cDNA synthesis was carried out at 42°C for 1 h.
Quantitative Real-Time PCR
Each biological replicate consisted of roots from four pooled individuals. For each primer pair, a fragment was amplified from cDNA, recombined into pCR8/GW/TOPO (Life Technologies), and sequenced to confirm amplification of the correct target. The subsequent plasmid was linearized by restriction digest, and a 10-fold dilution series was prepared over seven orders of magnitude and used as a template for qRT-PCR in duplicate. iCycler iQ Optical System software, version 3.1 (Bio-Rad) was used to generate standard curves. The reaction efficiency (E) of each primer pair was determined using the formula E = 10−1/slope. The primers used for qRT-PCR are detailed in Supplemental Table S3. Real-time PCR reactions (20 µL) were performed in 96-well plates with an iCycler iQ Thermocycler (Bio-Rad). Reactions consisted of 250 nm forward and reverse primer, 1× KAPA SYBR Fast qRT-PCR Master Mix (KAPA Biosystems), and 2 µL of cDNA template (diluted 1:5). Reactions were performed in triplicate with 40 cycles of 95°C for 15 s and 56°C for 20 s (plus data acquisition). To ensure single-product amplification, melt curve analysis was performed by heating the PCR products for 40 cycles starting at 52°C and increasing by 0.5°C per cycle with continuous fluorescence detection. To facilitate comparison between plates, an interrun control was loaded on each plate in duplicate. Relative transcript abundances were quantified using the E−ΔΔCt method (Pfaffl, 2001) and normalized to the geometric mean of Actin, Ubiquitin, and Elongation Factor 1α (Vandesompele et al., 2002). Statistical analyses were performed using Prism, version 5.01 (GraphPad Software Inc.).
For measuring AtCCC expression levels, Arabidopsis ecotype Col-0 was grown hydroponically as described previously (Conn et al., 2013). After 5 weeks, plants were transferred to a nutrient solution containing 50 mm NaCl and treated for 7 d. Whole roots from three biological replicates were used for RNA extraction. cDNA synthesis and qRT- PCR were carried out as described previously (Conn et al., 2011).
Cloning
The coding sequence of VviCCC was obtained by PCR amplified from root cDNA of grapevine ‘Cabernet Sauvignon’ and the corresponding gene from grapevine rootstocks 140 Ruggeri and K51-40. Phusion High-Fidelity Polymerase (Finnzymes) was used in a 20-µL final volume. PCR products were purified using the ISOLATE PCR and Gel Kit (Bioline) and sequenced for comparison and to ensure consistency with the available sequence at NCBI (accession no. GQ161924.1). Purified PCR products were A tailed for 1 h at 70°C with Taq Polymerase (New England Biolabs) and recombined into pCR8/GW/TOPO (Life Technologies) following the manufacturer’s procedures.
Subcellular Localization
For transient expression of fluorescent fusion proteins in tobacco (Nicotiana benthamiana), the VviCCC-YFP coding sequence was recombined into pMDC32 using LR recombination (Life Technologies). AtCCC without a stop codon was recombined into pMDC83 to generate AtCCC-GFP. For colocalization studies, the following subcellular markers were used: PM AtPIP2a-mCherry, ER HDEL-mCherry, Golgi ManI-RFP, and TGN AtSYP61-mCherry. All constructs were transformed into Agrobacterium tumefaciens strain Agl-1 using the freeze-thaw method. Tobacco was maintained in a controlled growth chamber at 23°C with a 16-h photoperiod. Agroinfiltration was performed on fully expanded leaves of 4- to 6-week-old plants. Overnight cultures of A. tumefaciens harboring the fluorescently tagged CCC, the subcellular marker of interest, and the P19 viral suppressor of gene silencing (Voinnet et al., 2003) were resuspended in buffer containing 10 mm MgCl2, 150 µm acetosyringone, and 10 mm MES (pH 5.6). Cultures were mixed together to achieve a final OD600 of 0.1 (P19) to 0.5 (VviCCC-YFP) and infiltrated into the abaxial side of tobacco leaves with a 1-mL syringe. After 2 to 3 d, leaf sections were imaged using a Nikon A1R Confocal Laser-Scanning Microscope equipped with a 63× water objective lens and NIS-Elements C software (Nikon Corporation). Excitation/emission conditions were YFP and GFP (488 nm/500–550 nm) and RFP and mCherry (561 nm/570–620 nm).
For localization in Arabidopsis protoplasts, VviCCC and AtCCC coding sequences without stop codons were recombined into pBS AttR-YFP (Subramanian et al., 2006) by LR recombination (Life Technologies) to create C-terminal YFP translational fusions. Plasmids were isolated from Escherichia coli using the GenElute HP Plasmid Maxiprep Kit (Sigma) and ethanol precipitated to increase the concentration to 1 µg µL−1. Mesophyll protoplasts were isolated from 5-week-old hydroponically grown Arabidopsis following the method by Wu et al. (2009). Protoplasts were transfected with 10 µg of plasmid DNA by the polyethylene glycol method. After overnight incubation, transgenic protoplasts were visualized using a Leica SP5 Spectral Scanning Confocal Microscope. Fluorescence images were captured sequentially and merged using LEICA SPS software (Leica Microsystems). Excitation/emission conditions were YFP (514 nm/527–552 nm) and chlorophyll autofluorescence (488 nm/664–696 nm).
Xenopus laevis Expression Studies
The VviCCC coding region was recombined into the Gateway-enabled X. laevis expression vector pGEMHE-DEST (Shelden et al., 2009) by LR recombination (Life Technologies). For localization of VviCCC in X. laevis oocytes, the VviCCC-YFP translational fusion from pBS AttR-YFP was amplified by PCR and recombined into pCR8/GW/TOPO before LR recombination. Plasmids were linearized with restriction endonuclease NheI and used as templates to synthesize cRNA in vitro with the mMessage mMachine T7 Kit (Ambion). After synthesis, cRNA was purified by phenol/chloroform extraction followed by ethanol precipitation and elution in water.
Stages IV and V oocytes were injected with 25 to 32 ng of cRNA in a final volume of 42 nL or with sterile water using a Nanoject II Injector (Drummond Scientific Company). Oocytes were analyzed 2 to 3 d post injection. For YFP imaging, oocytes were incubated in calcium Ringers solution (96 mm NaCl, 2 mm KCl, 5 mm MgCl, 0.6 mm CaCl2, 5 mm HEPES, 5% [v/v] horse serum, and 500 µg mL−1 tetracycline) and visualized with a Zeiss LSM 5 Pascal Confocal Laser-Scanning Microscope equipped with an argon laser. YFP excitation/emission conditions were 514 nm/523 to 600 nm.
For radiotracer studies, oocytes were incubated in an isotonic (220 mOsm kg−1) Cl– free ND96 solution (96 mm Na-gluconate, 2 mm K-gluconate, 1.8 mm Ca-gluconate, 1 mm Mg-gluconate, and 5 mm HEPES [pH 7.4] adjusted with 1 m Tris) for 2 to 16 h before the experiment. This is known to increase the driving force for radiotracer uptake (Gamba et al., 1994). Oocytes were then transferred to 500 µL of one of three possible uptake buffers: (1) standard ND96 (96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, and 5 mm HEPES adjusted to pH 7.4 with 1 m Tris) supplemented with 1 mm ouabain and H36Cl (1 µCi mL−1; Amersham); (2) same as buffer 1 but without H36Cl (replaced with 86RbCl [PerkinElmer] at 8 µCi mL−1 from a 16.16 mCi mg−1 stock); or (3) Na uptake buffer (40 mm NaCl, 56 mm N-methyl-d-glucamine-Cl, 2 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, and 5 mm HEPES/Tris [pH 7.4] plus 1 mm ouabain and 2 μCi mL−1 22NaCl from a 1 mCi mL−1 stock). Ouabain was added to prevent tracer uptake by the Na+-K+-ATPase. Each buffer was prepared with and without the addition of 100 µm bumetanide. Uptake was allowed to proceed for 1 h. Oocytes were then washed three times with ice cold uptake buffer without isotope and incubated overnight in 200 µL of 10% (w/v) SDS to dissolve the membrane and release radionuclides. Oocytes were transferred to 4 mL of scintillation cocktail (IRGA SAFE PL; PerkinElmer). Tracer uptake was measured by β-scintillation counting on a Tri-Carb 2500 TR Liquid Scintillation Counter (PerkinElmer), and total uptake per oocyte was calculated using the concentration of unlabeled Na+, Cl–, or K+ (for Rb+) in the uptake buffer; 200 µL of 10% (w/v) SDS was used to measure the background, and 20 µL of uptake buffer was measured to calculate the specific activity of the uptake buffer.
Arabidopsis Complementation Studies
Seed was obtained from the Arabidopsis Biological Resource Centre, and all lines used in this study were from a Col-0 background. An Arabidopsis (Salk_145300) line homozygous for a T-DNA insertion in AtCCC (At1g30450) was used for complementation studies, because this has been published previously (Colmenero-Flores et al., 2007). Confirmation of a homozygous T-DNA insertion was performed by PCR using gene-specific primers and the pROK2 T-DNA-specific primer LBa1. Col-0 was used as the wild type.
VviCCC in pCR8/GW/TOPO was recombined into the constitutive expression vector pMDC32 (Curtis and Grossniklaus, 2003) by LR recombination (Life Technologies). A. tumefaciens strain Agl-1 was transformed with 35S:VvCCC by the freeze-thaw method and then used to transform Arabidopsis by floral dipping (Clough and Bent, 1998). Transformed (T1) seed were selected on hygromycin plates (15 µg mL−1) following the rapid method reported previously (Harrison et al., 2006). T-DNA copy number was determined by selecting T2 seed on hygromycin plates and measuring the segregation ratios (Supplemental Fig. S9).
Semi-qRT-PCR was performed by extracting RNA from Arabidopsis leaves using the Plant RNeasy Mini Kit (Qiagen). This was followed by DNaseI treatment with Turbo DNA Free (Ambion) and reverse transcription with Superscript III following the manufacturer’s procedures (Life Technologies). PCR was carried out in a 20-µL volume using 0.4 units of KAPA Taq Polymerase (KAPA Biosystems), 0.4 µm forward and reverse primers, and 0.5 µL of cDNA. Reactions were carried for 25, 30, 35, and 40 cycles; 10 µL of PCR product was run on 1.3% (w/v) agarose gel stained with SYBR Safe (Life Technologies) and visualized by UV illumination.
Two nonsegregating T3 lines were used for analysis. For salt stress studies, plants were surface sterilized with 30% (v/v) bleach and 0.05% (v/v) Triton X-100 for 10 min, washed five times with sterile water, and plated on one-half-strength MS (1% [w/v] agar) with 1% (w/v) Suc. After a 2-d stratification period at 4°C in the dark, plants were transferred to an MLR-351 Controlled Environmental Chamber (Sanyo) with a 16-h photoperiod and a light intensity of 130 to 150 µE m2 s−1. After the roots had reached 3 mm in length, seedlings were transferred to one-half-strength MS (1% [w/v] agar) with 1% (w/v) Suc and 100 mm NaCl. After 10 d, plates were photographed, and root length measurements were taken using ImageJ software.
For ion concentration analysis, plants were grown in aerated hydroponics in a basal nutrient solution (BNS) buffered at pH 5.6 as described previously (Conn et al., 2013). After 5 weeks, plants were transferred to BNS or BNS containing 50 mm NaCl for 3 d. After treatment, plants were rinsed in deionized water and separated into roots and shoots, and fresh weights were taken. For each biological replicate, three plants were pooled together. Plant material was dried overnight at 60°C, dry weights were measured, and the material was ground to a fine powder directly in the tube with a sterilized metal rod. Measurement of elemental concentrations was performed by Waite Analytical Services. Cation concentration was measured by acid extraction and radial-view ICP-OES (Wheal et al., 2011). Chloride concentration was measured by axial-view ICP-OES of 4% (v/v) nitric acid extracts as described by Wheal and Palmer (2010).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Transient coexpression of AtCCC-GFP with various subcellular markers in epidermal cells of tobacco.
Supplemental Figure S2. Subcellular localization of VviCCC and AtCCC in Arabidopsis mesophyll protoplasts.
Supplemental Figure S3. Comparison of cDNA coding sequences of VviCCC from rootstocks K51-40 and 140-Ruggeri.
Supplemental Figure S4. 86Rb uptake by X. laevis oocytes expressing VviCCC in high-potassium buffer.
Supplemental Figure S5. Confirmation of a homozygous T-DNA insertion in the Arabidopsis mutant line Salk_145300.
Supplemental Figure S6. Complementation of the ccc phenotype with VviCCC.
Supplemental Figure S7. AtCCC transcript is not regulated by salt stress in Arabidopsis roots.
Supplemental Figure S8. Multiple sequence alignment of six plant CCC family members.
Supplemental Figure S9. Identification of T-DNA copy number in complemented Arabidopsis mutants.
Supplemental Table S1. Summary of outputs generated from different trans-membrane prediction algorithms used to analyze the VviCCC full-length amino acid sequence.
Supplemental Table S2. Accession numbers of plant CCC genes used for multiple sequence alignment.
Supplemental Table S3. Primers used in this study.
Supplemental Video S1. VviCCC-YFP forms mobile, punctate structures when transiently expressed in epidermal cells of tobacco.
Supplemental Video S2. AtCCC-GFP forms mobile, punctate structures when transiently expressed in epidermal cells of tobacco.
Supplementary Material
Acknowledgments
We thank Grant Cramer for initiating the study of AtCCC in Adelaide while on sabbatical and providing the Atccc knockout mutant, Sunita Ramesh (University of Adelaide) and Wendy Sullivan (University of Adelaide) for harvesting Xenopus spp. oocytes and advising on oocyte transformation and radiotracer uptake studies, Virginie Masson (University of Adelaide) for assisting with LaTeX, George Dimitroff (University of Adelaide) for donating tobacco seeds and the pEAQ-HT vector, Stuart Roy for discussions and cosupervision of J.Q. with M.G, the Australian Centre for Plant Functional Genomics PCR Service for performing qRT-PCR for Supplemental Figure S6, and UA and Commonwealth Scientific and Industrial Research Organization, which are both members of the Wine Innovation Cluster.
Glossary
- BNS
basal nutrient solution
- cDNA
complementary DNA
- Col-0
Columbia-0
- cRNA
capped RNA
- ER
endoplasmic reticulum
- ICP-OES
inductively coupled plasma optical emission spectrometry
- LOPIT
localization of organelle proteins by isotope tagging
- MS
Murashige and Skoog medium
- NCBI
National Center for Biotechnology Information
- PM
plasma membrane
- qRT
quantitative reverse transcription
- SUBA3
Subcellular Localization of Proteins in Arabidopsis3
- T-DNA
transfer DNA
- TGN
trans-Golgi network
Footnotes
This work was supported by the Australian grape growers and winemakers through their investment body, the Australian Grape and Wine Authority, with matching funds from the Australian Government (Student Scholarship no. GWR PH1001 to S.W.H. and Project Grant no. CSP1002 to A.R.W., R.R.W., and M.G.) and the Australian Research Council (Centre of Excellence Funding Grant no. CE140100008 to S.D.T. and M.G. and Future Fellowship no. FT130100709 to M.G.).
Articles can be viewed without a subscription.
References
- Abbaspour N, Kaiser B, Tyerman S (2013) Chloride transport and compartmentation within main and lateral roots of two grapevine rootstocks differing in salt tolerance. Trees 27: 1317–1325 [Google Scholar]
- Arpat AB, Magliano P, Wege S, Rouached H, Stefanovic A, Poirier Y (2012) Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate. Plant J 71: 479–491 [DOI] [PubMed] [Google Scholar]
- Asaoka R, Uemura T, Ito J, Fujimoto M, Ito E, Ueda T, Nakano A (2013) Arabidopsis RABA1 GTPases are involved in transport between the trans-Golgi network and the plasma membrane, and are required for salinity stress tolerance. Plant J 73: 240–249 [DOI] [PubMed] [Google Scholar]
- Barberon M, Zelazny E, Robert S, Conéjéro G, Curie C, Friml J, Vert G (2011) Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier-Brygoo H, De Angeli A, Filleur S, Frachisse JM, Gambale F, Thomine S, Wege S (2011) Anion channels/transporters in plants: from molecular bases to regulatory networks. Annu Rev Plant Biol 62: 25–51 [DOI] [PubMed] [Google Scholar]
- Bassil E, Ohto MA, Esumi T, Tajima H, Zhu Z, Cagnac O, Belmonte M, Peleg Z, Yamaguchi T, Blumwald E (2011) The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 23: 224–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitz E. (2000) TEXshade: shading and labeling of multiple sequence alignments using LATEX2 epsilon. Bioinformatics 16: 135–139 [DOI] [PubMed] [Google Scholar]
- Benschop JJ, Mohammed S, O’Flaherty M, Heck AJR, Slijper M, Menke FLH (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics 6: 1198–1214 [DOI] [PubMed] [Google Scholar]
- Brumós J, Colmenero-Flores JM, Conesa A, Izquierdo P, Sánchez G, Iglesias DJ, López-Climent MF, Gómez-Cadenas A, Talón M (2009) Membrane transporters and carbon metabolism implicated in chloride homeostasis differentiate salt stress responses in tolerant and sensitive Citrus rootstocks. Funct Integr Genomics 9: 293–309 [DOI] [PubMed] [Google Scholar]
- Brumós J, Talón M, Bouhlal R, Colmenero-Flores JM (2010) Cl− homeostasis in includer and excluder citrus rootstocks: transport mechanisms and identification of candidate genes. Plant Cell Environ 33: 2012–2027 [DOI] [PubMed] [Google Scholar]
- Cao H, Guo S, Xu Y, Jiang K, Jones AM, Chong K (2011) Reduced expression of a gene encoding a Golgi localized monosaccharide transporter (OsGMST1) confers hypersensitivity to salt in rice (Oryza sativa). J Exp Bot 62: 4595–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colmenero-Flores JM, Martínez G, Gamba G, Vázquez N, Iglesias DJ, Brumós J, Talón M (2007) Identification and functional characterization of cation-chloride cotransporters in plants. Plant J 50: 278–292 [DOI] [PubMed] [Google Scholar]
- Conn SJ, Gilliham M, Athman A, Schreiber AW, Baumann U, Moller I, Cheng NH, Stancombe MA, Hirschi KD, Webb AAR, et al. (2011) Cell-specific vacuolar calcium storage mediated by CAX1 regulates apoplastic calcium concentration, gas exchange, and plant productivity in Arabidopsis. Plant Cell 23: 240–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn SJ, Hocking B, Dayod M, Xu B, Athman A, Henderson S, Aukett L, Conn V, Shearer MK, Fuentes S, et al. (2013) Protocol: optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods 9: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer GR, Ergül A, Grimplet J, Tillett RL, Tattersall EAR, Bohlman MC, Vincent D, Sonderegger J, Evans J, Osborne C, et al. (2007) Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Funct Integr Genomics 7: 111–134 [DOI] [PubMed] [Google Scholar]
- Cubero B, Nakagawa Y, Jiang XY, Miura KJ, Li F, Raghothama KG, Bressan RA, Hasegawa PM, Pardo JM (2009) The phosphate transporter PHT4;6 is a determinant of salt tolerance that is localized to the Golgi apparatus of Arabidopsis. Mol Plant 2: 535–552 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkin R, Robinson S, Sumby K, Harris V, McBryde C, Jiranek V (2010) Sodium chloride in Australian grape juice and its effect on alcoholic and malolactic fermentation. Am J Enol Vitic 61: 392–400 [Google Scholar]
- Downton WJS. (1977) Photosynthesis in salt-stressed grapevines. Aust J Plant Physiol 4: 183–192 [Google Scholar]
- Drakakaki G, van de Ven W, Pan S, Miao Y, Wang J, Keinath NF, Weatherly B, Jiang L, Schumacher K, Hicks G, et al. (2012) Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res 22: 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlig CF. (1960) Effects of salinity on four varieties of table grapes grown in sand culture. Proc Am Soc Hortic Sci 76: 323–331 [Google Scholar]
- Famiglietti JS. (2014) The global groundwater crisis. Nat Clim Chang 4: 945–948 [Google Scholar]
- FAO (2002) Extent and causes of salt affected soils in participating countries. FAO. http://www.fao.org/nr/aboutnr/nrl/en/ (April 23, 2015)
- Flowers TJ, Colmer TD (2008) Salinity tolerance in halophytes. New Phytol 179: 945–963 [DOI] [PubMed] [Google Scholar]
- Fort KP, Heinitz CC, Walker MA (2015) Chloride exclusion patterns in six grapevine populations. Aust J Grape Wine Res 21: 147–155 [Google Scholar]
- Fricke W. (2015) The significance of water co-transport for sustaining transpirational water flow in plants: a quantitative approach. J Exp Bot 66: 731–739 [DOI] [PubMed] [Google Scholar]
- Gamba G. (2005) Molecular physiology and pathophysiology of electroneutral cation-chloride cotransporters. Physiol Rev 85: 423–493 [DOI] [PubMed] [Google Scholar]
- Gamba G, Miyanoshita A, Lombardi M, Lytton J, Lee WS, Hediger MA, Hebert SC (1994) Molecular cloning, primary structure, and characterization of two members of the mammalian electroneutral sodium-(potassium)-chloride cotransporter family expressed in kidney. J Biol Chem 269: 17713–17722 [PubMed] [Google Scholar]
- Gamba G, Saltzberg SN, Lombardi M, Miyanoshita A, Lytton J, Hediger MA, Brenner BM, Hebert SC (1993) Primary structure and functional expression of a cDNA encoding the thiazide-sensitive, electroneutral sodium-chloride cotransporter. Proc Natl Acad Sci USA 90: 2749–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliham M, Tester M (2005) The regulation of anion loading to the maize root xylem. Plant Physiol 137: 819–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, Blackmore D, Clingeleffer P, Sykes S, Jha D, Tester M, Walker R (2011) Contrast in chloride exclusion between two grapevine genotypes and its variation in their hybrid progeny. J Exp Bot 62: 989–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, Blackmore DH, Walker RR (2010) Organic and inorganic anions in Shiraz and Chardonnay grape berries and wine as affected by rootstock under saline conditions. Aust J Grape Wine Res 16: 227–236 [Google Scholar]
- Grimplet J, Adam-Blondon AF, Bert PF, Bitz O, Cantu D, Davies C, Delrot S, Pezzotti M, Rombauts S, Cramer GR (2014) The grapevine gene nomenclature system. BMC Genomics 15: 1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen AJ, Sancho-Andrés G, Breckels LM, Gatto L, Aniento F, Lilley KS (2014) Identification of trans-golgi network proteins in Arabidopsis thaliana root tissue. J Proteome Res 13: 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harling H, Czaja I, Schell J, Walden R (1997) A plant cation-chloride co-transporter promoting auxin-independent tobacco protoplast division. EMBO J 16: 5855–5866 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Harrison SJ, Mott EK, Parsley K, Aspinall S, Gray JC, Cottage A (2006) A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson SW, Baumann U, Blackmore DH, Walker AR, Walker RR, Gilliham M (2014) Shoot chloride exclusion and salt tolerance in grapevine is associated with differential ion transporter expression in roots. BMC Plant Biol 14: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha D, Shirley N, Tester M, Roy SJ (2010) Variation in salinity tolerance and shoot sodium accumulation in Arabidopsis ecotypes linked to differences in the natural expression levels of transporters involved in sodium transport. Plant Cell Environ 33: 793–804 [DOI] [PubMed] [Google Scholar]
- Johnson MA, von Besser K, Zhou Q, Smith E, Aux G, Patton D, Levin JZ, Preuss D (2004) Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168: 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Bassham DC (2011) TNO1 is involved in salt tolerance and vacuolar trafficking in Arabidopsis. Plant Physiol 156: 514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong XQ, Gao XH, Sun W, An J, Zhao YX, Zhang H (2011) Cloning and functional characterization of a cation-chloride cotransporter gene OsCCC1. Plant Mol Biol 75: 567–578 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Leske PA, Sas AN, Coulter AD, Stockley CS, Lee TH (1997) The composition of Australian grape juice: chloride, sodium and sulfate ions. Aust J Grape Wine Res 3: 26–30 [Google Scholar]
- Maas EV, Hoffman GJ (1977) Crop salt tolerance: current assessment. J Irrig Drain Div 103: 115–134 [Google Scholar]
- Maurel C, Reizer J, Schroeder JI, Chrispeels MJ (1993) The vacuolar membrane protein gamma-TIP creates water specific channels in Xenopus oocytes. EMBO J 12: 2241–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado A, Song L, Vázquez N, Mount DB, Gamba G (2000) Functional comparison of the K+-Cl- cotransporters KCC1 and KCC4. J Biol Chem 275: 30326–30334 [DOI] [PubMed] [Google Scholar]
- Molendijk AJ, Ruperti B, Singh MK, Dovzhenko A, Ditengou FA, Milia M, Westphal L, Rosahl S, Soellick TR, Uhrig J, et al. (2008) A cysteine-rich receptor-like kinase NCRK and a pathogen-induced protein kinase RBK1 are Rop GTPase interactors. Plant J 53: 909–923 [DOI] [PubMed] [Google Scholar]
- Munns R, Gilliham M (June 24, 2015) Salinity tolerance of crops: what is the cost? New Phytol 10.1111/nph.13519 [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Läuchli A (2006) Approaches to increasing the salt tolerance of wheat and other cereals. J Exp Bot 57: 1025–1043 [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, et al. (2012) Wheat grain yield on saline soils is improved by an ancestral Na⁺ transporter gene. Nat Biotechnol 30: 360–364 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nikolovski N, Rubtsov D, Segura MP, Miles GP, Stevens TJ, Dunkley TPJ, Munro S, Lilley KS, Dupree P (2012) Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol 160: 1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel D, Berger B, Filiberto D, Newton M, Wolfe B, Karabinakis E, Clark S, Poon E, Abbett E, Nandagopal S (2004) Water resources: agricultural and environmental issues. Bioscience 54: 909–918 [Google Scholar]
- Plett D, Safwat G, Gilliham M, Skrumsager Møller I, Roy S, Shirley N, Jacobs A, Johnson A, Tester M (2010) Improved salinity tolerance of rice through cell type-specific expression of AtHKT1;1. PLoS One 5: e12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadir M, Quillérou E, Nangia V, Murtaza G, Singh M, Thomas RJ, Drechsel P, Noble AD (2014) Economics of salt‐induced land degradation and restoration. Nat Resour Forum 38: 282–295 [Google Scholar]
- Roxas VP, Smith RK Jr., Allen ER, Allen RD (1997) Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat Biotechnol 15: 988–991 [DOI] [PubMed] [Google Scholar]
- Sadowski PG, Groen AJ, Dupree P, Lilley KS (2008) Sub-cellular localization of membrane proteins. Proteomics 8: 3991–4011 [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Thomas MR (2003) A rapid method for generating sufficient amounts of uniform genotype-specific material from the woody perennial grapevine for ion transport studies. Plant Soil 253: 195–199 [Google Scholar]
- Schell J, Bisseling T, Dülz M, Franssen H, Fritze K, John M, Kleinow T, Leßnick A, Miklashevichs E, Pawlowski K, et al. (1999) Re-evaluation of phytohormone-independent division of tobacco protoplast-derived cells. Plant J 17: 461–466 [Google Scholar]
- Shabala S. (2013) Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Ann Bot (Lond) 112: 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani U, Ben-Gal A (2005) Long-term response of grapevines to salinity: osmotic effects and ion toxicity. Am J Enol Vitic 56: 148–154 [Google Scholar]
- Shavrukov Y, Gupta NK, Miyazaki J, Baho MN, Chalmers KJ, Tester M, Langridge P, Collins NC (2010) HvNax3--a locus controlling shoot sodium exclusion derived from wild barley (Hordeum vulgare ssp. spontaneum). Funct Integr Genomics 10: 277–291 [DOI] [PubMed] [Google Scholar]
- Shelden MC, Howitt SM, Kaiser BN, Tyerman SD (2009) Identification and functional characterisation of aquaporins in the grapevine, Vitis vinifera. Funct Plant Biol 36: 1065–1078 [DOI] [PubMed] [Google Scholar]
- Stevens RM, Harvey G, Partington DL (2011) Irrigation of grapevines with saline water at different growth stages: effects on leaf, wood and juice composition. Aust J Grape Wine Res 17: 239–248 [Google Scholar]
- Storey R, Walker RR (1999) Citrus and salinity. Sci Hortic 78: 39–81 [Google Scholar]
- Subramanian C, Woo J, Cai X, Xu X, Servick S, Johnson CH, Nebenführ A, von Arnim AG (2006) A suite of tools and application notes for in vivo protein interaction assays using bioluminescence resonance energy transfer (BRET). Plant J 48: 138–152 [DOI] [PubMed] [Google Scholar]
- Suvitayavat W, Palfrey HC, Haas M, Dunham PB, Kalmar F, Rao MC (1994) Characterization of the endogenous Na(+)-K(+)-2Cl- cotransporter in Xenopus oocytes. Am J Physiol 266: C284–C292 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanz SK, Castleden I, Hooper CM, Vacher M, Small I, Millar HA (2013) SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res 41: D1185–D1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall EAR, Grimplet J, DeLuc L, Wheatley MD, Vincent D, Osborne C, Ergül A, Lomen E, Blank RR, Schlauch KA, et al. (2007) Transcript abundance profiles reveal larger and more complex responses of grapevine to chilling compared to osmotic and salinity stress. Funct Integr Genomics 7: 317–333 [DOI] [PubMed] [Google Scholar]
- Teakle NL, Tyerman SD (2010) Mechanisms of Cl(-) transport contributing to salt tolerance. Plant Cell Environ 33: 566–589 [DOI] [PubMed] [Google Scholar]
- Tregeagle JM, Tisdall JM, Blackmore DH, Walker RR (2006) A diminished capacity for chloride exclusion by grapevine rootstocks following long-term saline irrigation in an inland versus a coastal region of Australia. Aust J Grape Wine Res 12: 178–191 [Google Scholar]
- Tregeagle JM, Tisdall JM, Tester M, Walker RR (2010) Cl- uptake, transport and accumulation in grapevine rootstocks of differing capacity for Cl- exclusion. Funct Plant Biol 37: 665–673 [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent D, Ergül A, Bohlman MC, Tattersall EAR, Tillett RL, Wheatley MD, Woolsey R, Quilici DR, Joets J, Schlauch K, et al. (2007) Proteomic analysis reveals differences between Vitis vinifera L. cv. Chardonnay and cv. Cabernet Sauvignon and their responses to water deficit and salinity. J Exp Bot 58: 1873–1892 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Walker RR, Blackmore DH, Clingeleffer PR, Correll RL (2002) Rootstock effects on salt tolerance of irrigated field-grown grapevines (Vitis vinifera L. cv. Sultana). I. Yield and vigour inter-relationships. Aust J Grape Wine Res 8: 3–14 [Google Scholar]
- Walker RR, Blackmore DH, Clingeleffer PR, Godden P, Francis L, Valente P, Robinson E (2003) Salinity effects on vines and wines. Bull Office Int Vigne Vin 76: 200–227 [Google Scholar]
- Wege S, Poirier Y (2014) Expression of the mammalian Xenotropic Polytropic Virus Receptor 1 (XPR1) in tobacco leaves leads to phosphate export. FEBS Lett 588: 482–489 [DOI] [PubMed] [Google Scholar]
- Wegner LH. (2014) Root pressure and beyond: energetically uphill water transport into xylem vessels? J Exp Bot 65: 381–393 [DOI] [PubMed] [Google Scholar]
- Wei Q, Liu Y, Zhou G, Li Q, Yang C, Peng SA (2013) Overexpression of CsCLCc, a chloride channel gene from Poncirus trifoliata, enhances salt tolerance in Arabidopsis. Plant Mol Biol Rep 31: 1548–1557 [Google Scholar]
- Wheal MS, Fowles TO, Palmer LT (2011) A cost-effective acid digestion method using closed polypropylene tubes for inductively coupled plasma optical emission spectrometry (ICP-OES) analysis of plant essential elements. Anal Methods 3: 2854–2863 [Google Scholar]
- Wheal MS, Palmer LT (2010) Chloride analysis of botanical samples by ICP-OES. J Anal At Spectrom 25: 1946–1952 [Google Scholar]
- Whiteman SA, Nühse TS, Ashford DA, Sanders D, Maathuis FJM (2008a) A proteomic and phosphoproteomic analysis of Oryza sativa plasma membrane and vacuolar membrane. Plant J 56: 146–156 [DOI] [PubMed] [Google Scholar]
- Whiteman SA, Serazetdinova L, Jones AME, Sanders D, Rathjen J, Peck SC, Maathuis FJM (2008b) Identification of novel proteins and phosphorylation sites in a tonoplast enriched membrane fraction of Arabidopsis thaliana. Proteomics 8: 3536–3547 [DOI] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS (2009) Tape-Arabidopsis Sandwich: a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuthen T, Macaulay N (2012) Cotransport of water by Na⁺-K⁺-2Cl⁻ cotransporters expressed in Xenopus oocytes: NKCC1 versus NKCC2. J Physiol 590: 1139–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.