Complex and distinct regulatory changes contribute to expression differentiation of CYC- and CIN-like TCP genes and morphological divergence of petals in zygomorphic flowers of Petrocosmea spp.
Abstract
CYCLOIDEA (CYC)-like genes, belonging to the plant-specific TCP transcription factor family that is named after TEOSINTE BRANCHED1 (TB1) from maize (Zea mays), CYC from Antirrhinum majus, and the PROLIFERATING CELL FACTORS (PCF) from rice (Oryza sativa), have conserved dorsal identity function in patterning floral zygomorphy mainly through specific expression in dorsal petals of a flower. Their expression changes are usually related to morphological diversity of zygomorphic flowers. However, it is still a challenge to elucidate the molecular mechanism underlying their expression differentiation. It is also unknown whether CINCINNATA (CIN)-like TCP genes, locally controlling cell growth and proliferation, are involved in the evolution of floral zygomorphy. To address these questions, we selected two closely related species, i.e. Petrocosmea glabristoma and Petrocosmea sinensis, with distinct petal morphology to conduct expression, hybridization, mutant, and allele-specific expression analyses. The results show that the size change of the dorsal petals between the two species is mainly mediated by the expression differentiation of CYC1C and CYC1D, while the shape variation of all petals is related to the expression change of CIN1. In reciprocal F1 hybrids, the expression of CYC1C, CYC1D, and CIN1 conforms to an additive inheritance mode, consistent with the petal phenotypes of hybrids. Through allele-specific expression analyses, we find that the expression differentiation of these TCP genes is underlain by distinctly different types of regulatory changes. We suggest that highly redundant paralogs with identical expression patterns and interspecific expression differentiation may be controlled by remarkably different regulatory pathways because natural selection may favor different regulatory modifications rather than coding sequence changes of key developmental genes in generating morphological diversity.
During the evolution of both animals and plants, a common phenomenon is that spatial-temporal expression modifications of key developmental regulators, as a linkage between genotype and phenotype, usually contribute to the origin of morphological novelties (Doebley et al., 1997; Cong et al., 2002; Gompel et al., 2005; Prud’homme et al., 2006, 2007; Werner et al., 2010; Arnoult et al., 2013; Dong et al., 2014). As one key innovation during angiosperm evolution, zygomorphic flowers have arisen independently several times from actinomorphic ancestors and successfully developed, producing several major zygomorphic clades, such as Fabales, Lamiales, Asterales, and Orchidales (Donoghue et al., 1998; Dilcher, 2000). In Antirrhinum majus, a model organism in Lamiales, the establishment of floral zygomorphy depends on the dorsal identity function of CYCLOIDEA (CYC) and its paralog DICHOTOMA (DICH) in determining the fate of dorsal floral organs in the second and third whorls (Luo et al., 1996, 1999). Both CYC and DICH encode proteins belonging to the CYC/TB1 (for TEOSINTE BRANCHED1) or ECE (named after a conserved Glu-Cys-Glu motif) lineage in the plant-specific TCP transcription factor family that is named after TB1 from maize (Zea mays), CYC from A. majus, and the PROLIFERATING CELL FACTORS (PCF) from rice (Oryza sativa; Cubas et al., 1999a; Howarth and Donoghue, 2006). A growing body of evidence has shown that CYC-like TCP genes function in controlling floral zygomorphy widely in eudicots (Feng et al., 2006; Busch and Zachgo, 2007; Broholm et al., 2008; Kim et al., 2008; Wang et al., 2008; Yang et al., 2012). The spatial-temporal expression changes of CYC-like genes usually bring about transformations of floral symmetry and modifications of floral morphology, mainly reflected in size and shape changes of the dorsal petals in the second floral whorl (Cubas et al., 1999b; Hileman et al., 2003; Busch and Zachgo, 2007; Gao et al., 2008; Song et al., 2009; Pang et al., 2010; Zhang et al., 2010; Howarth et al., 2011; Busch et al., 2012; Yang et al., 2012; Zhong and Kellogg, 2015). However, it remains to be elucidated what regulatory changes underlie the expression modifications of these CYC-like genes.
Different from CYC-like genes, CINCINNATA (CIN)-like genes belonging to the CIN lineage in the TCP gene family are mainly implicated in the local morphological control of both leaves and petals in A. majus and Arabidopsis (Arabidopsis thaliana; Nath et al., 2003; Palatnik et al., 2003; Crawford et al., 2004; Koyama et al., 2007, 2010a, 2011; Nag et al., 2009; Das Gupta et al., 2014). Precise control of CIN-like gene activities by microRNA is critical to proper growth and development of leaves and petals, whereas their up- and down-regulation usually results in severe developmental defects, mainly demonstrated by the abnormal serration and curvature of leaves and petals (Nath et al., 2003; Palatnik et al., 2003; Crawford et al., 2004; Koyama et al., 2007; Nag et al., 2009). Therefore, it would be interesting to decipher whether CIN-like gene expression changes bring about morphological transitions in nonmodel organisms. It is also an important issue whether CYC- and CIN-like TCP genes act together to control specific morphologies and generate successive variation forms during the evolution of zygomorphic groups.
Generally, gene expression can be regulated at transcriptional, posttranscriptional, and translational levels. Of these, transcriptional regulation determines when and where a gene is expressed, at what level, and under what environmental conditions (Wray et al., 2003). During the transcriptional regulation process, two types of regulatory units exist: one is cis-regulatory elements or sequences that flank every gene, and the other is trans-acting factors that encode elsewhere and physically interplay with cis-elements to either promote or repress the transcription of the gene (Wray et al., 2003). Correspondingly, gene expression modifications can arise from either cis-regulatory changes that affect transcription in an allele-specific manner or trans-regulatory alterations that modify the expression or activity of related factors (Wray et al., 2003; Wittkopp et al., 2004). Recently, a burgeoning amount of data have shown that distinct portions of cis- and trans-regulatory changes contribute to gene expression divergence in different organisms by quantifying allele-specific expression (ASE) levels in a common, heterozygous diploid individual (Wittkopp et al., 2004, 2008; Kiekens et al., 2006; Stupar et al., 2007; Zhuang and Adams, 2007; Main et al., 2009; von Korff et al., 2009; Bell et al., 2013; Cubillos et al., 2014). In spite of advances regarding cis- and trans-actions, the detailed relationship between regulatory variations of key developmental genes and phenotypic divergences, especially some important morphological novelties, has not been extensively explored yet, particularly in plants (Rosas et al., 2010).
Lamiales is a large clade in angiosperms believed to be ancestrally zygomorphic and further specialized and diversified (Cubas, 2004). As one of the earliest diverging groups in Lamiales, Gesneriaceae has diverse forms of zygomorphic flowers (Cubas, 2004). Recent studies have shown that the spatial-temporal expression alterations of CYC-like genes are highly correlated with the diverse morphologies of zygomorphic flowers in Gesneriaceae (Du and Wang, 2008; Gao et al., 2008; Song et al., 2009; Yang et al., 2012). The genus Petrocosmea, comprising 42 species, is a midsized group in Gesneriaceae with very similar vegetative traits but extremely diverse zygomorphic flowers that are mainly reflected in size and shape variations of the dorsal petals in the second floral whorl (Li and Wang, 2004; Qiu et al., 2011, 2015). Petrocosmea spp. therefore represents an ideal group to address whether different regulatory pathways or their interplay account for the expression differentiation of floral symmetry-related genes and therefore the morphological diversification of the dorsal petals in zygomorphic flowers.
In this study, two Petrocosmea species, i.e. Petrocosmea glabristoma (Pg) and Petrocosmea sinensis (Ps) as the representatives of two clades, were selected to investigate possible regulatory pathways driving the morphological divergence of zygomorphic flowers by employing multiple approaches, including comparative expression analyses of CYC- and CIN-like TCP genes, hybridization, mutant, and ASE assays. The results showed that the expression differentiation of CYC1C, CYC1D, and CIN1 coordinately promoted the morphological divergence of the dorsal petals that is characteristic of different types of zygomorphic flowers in the two species. In addition, both CYC- and CIN-like genes conformed to an additive inheritance mode in reciprocal F1 hybrids, consistent with the phenotypes of the hybrids. We further found that cis-regulatory changes, trans-regulatory changes, and a complex of both changes contributed to the expression differentiation of CYC1C, CYC1D, and CIN1, respectively. This report will shed significant light on how regulatory changes underlie expression differentiation of key developmental genes and therefore morphological variations of the dorsal petals in zygomorphic flowers.
RESULTS
Floral Morphology of Parents and F1 Hybrids
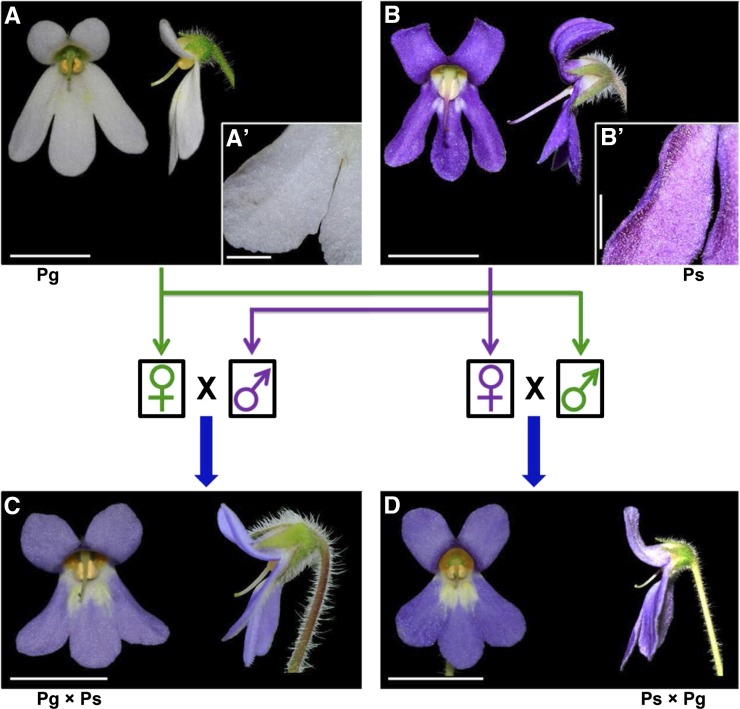
Pg and Ps belong to clade D and clade E in the genus Petrocosmea, respectively (Supplemental Fig. S1; Qiu et al., 2015). They have very similar vegetative traits but exhibit distinctly different zygomorphic flowers that are mainly manifested in the second whorl of floral organs (Fig. 1, A and B). First, the dorsal petals of Pg flowers are significantly short compared with those of Ps. The dorsal petal length of Pg flowers is 4.83 ± 0.21 mm on average, and the length ratio of dorsal to ventral petals is 0.38 ± 0.01. In Ps flowers, the dorsal petal length is 8.18 ± 0.33 mm, and the length ratio of dorsal to ventral petals is 0.84 ± 0.03 (Table I). Second, the curvature degree of their dorsal petals is also distinctly different from each other. As shown in Figure 1, the dorsal petals of Pg are extended upward and almost perpendicular to the corolla tube, while those of Ps are strongly reflexed backward (Fig. 1, A and B, side view). In addition to dorsal petals, their lateral and ventral petals are also clearly different from each other in shape. For example, both lateral and ventral petals of Ps are strongly curved, with lateral margins reflexed backward, while those of Pg are explanate (Fig. 1, A′ and B,′ dorsal view).
Figure 1.
Floral morphology of Pg, Ps, and F1 hybrids. Pg (A) and Ps (B) have distinct zygomorphic flowers, mainly displayed in the different size and shape of dorsal petals, as well as the different shape of lateral and ventral petals (A′ and B′, dorsal view). Both cross (C) and reciprocal cross (D) F1 hybrids have intermediate zygomorphic flowers compared with the two parents. For each flower, both the front (left) and side (right) views are shown. Bars = 1 cm (A–D) and 0.2 cm (A′ and B′).
Table I. The length ratio of dorsal to ventral petals of parents and F1 hybrids.
Data were compared using Fisher’s lsd test (P < 0.05), with values labeled with the same letters being not significantly different. Data are presented as mean ± sd collected from at least 30 flowers. DP, Dorsal petals; VP, ventral petals.
Reciprocal F1 hybrids were constructed using Pg as the maternal parent and Ps as the paternal parent, or vice versa, to see how the petal phenotypes in the two parents are inherited and to rule out parent-of-origin effects. Cross Pg × Ps and reciprocal cross Ps × Pg have very similar floral phenotypes (Fig. 1, C and D). Their flowers exhibit an intermediate phenotype between Pg and Ps that is mainly manifested in the length of the dorsal petals (Fig. 1). First, the dorsal petal length of Pg × Ps and Ps × Pg hybrids is 6.22 ± 0.47 and 5.74 ± 0.35 mm, respectively, both falling somewhere between the two parents (Table I). Second, the length ratio of dorsal to ventral petals is, respectively, 0.58 ± 0.04 and 0.56 ± 0.03 for Pg × Ps and Ps × Pg, also intermediate between Pg and Ps (Table I). The curvature degree of dorsal, lateral, and ventral petals in both Pg × Ps and Ps × Pg hybrids is more similar to Pg, but still located between the two parents (Fig. 1). The above results show that the inheritance of the dorsal petal size and all petal shape in the F1 hybrids generally conforms to an additive model.
Sequence and Expression Analyses of CYC- and CIN-Like Genes in Parents and F1 Hybrids
To know whether the morphological differences of the dorsal petals between Pg and Ps are related to CYC-like gene activity, we isolated four CYC-like genes, i.e. CYC1C, CYC1D, CYC2A, and CYC2B, from both species. Three CIN-like genes, CIN1, CIN2A, and CIN2B, were also isolated due to their potential roles in locally controlling cell growth and proliferation of petals. Phylogenetic analyses showed that CYC- and CIN-like genes isolated here were clustered with A. majus CYC/DICH and CIN, respectively (Supplemental Fig. S2). Sequence analyses showed that Petrocosmea spp. CYC-like genes are characterized by the presence of both TCP and R (an Arg-rich motif of 18–20 residues) domains, while CIN-like genes only contain the TCP domain (Supplemental Fig. S3). In addition, all CIN-like genes isolated herein contain the miR319 (a kind of microRNA) recognition sequence (Supplemental Fig. S3), a typical feature shared by CIN-like genes (Martín-Trillo and Cubas, 2010). Furthermore, there is a very high amino acid sequence similarity between each pair of orthologous genes (Supplemental Figs. S4 and S5), hinting that these CYC- and CIN-like genes are functionally conserved between the two species.
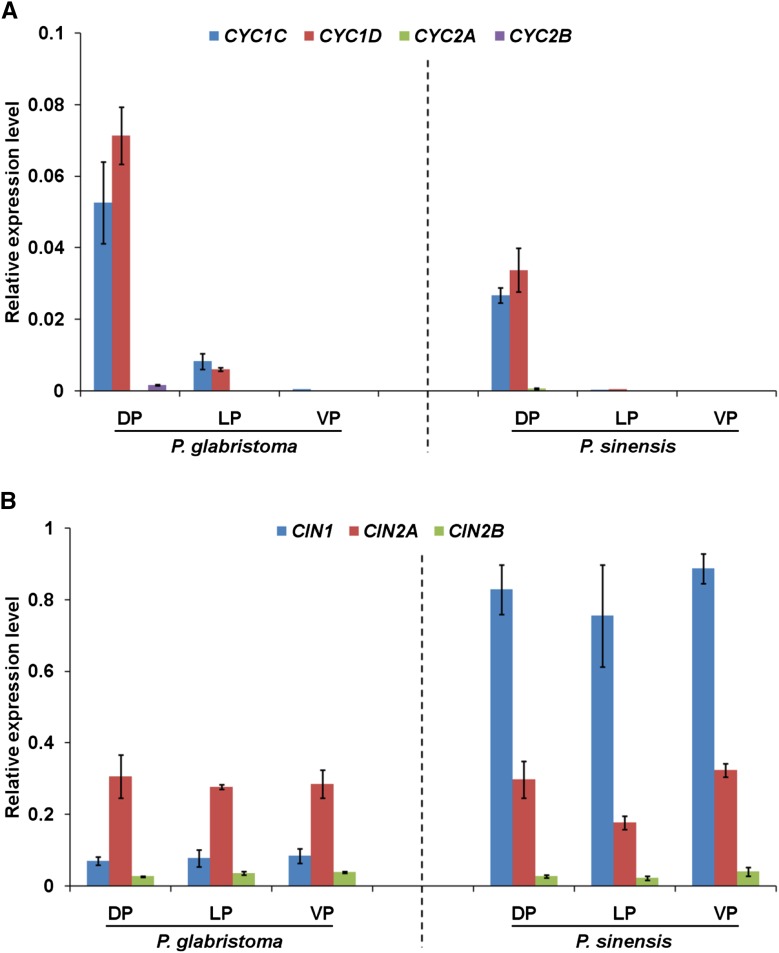
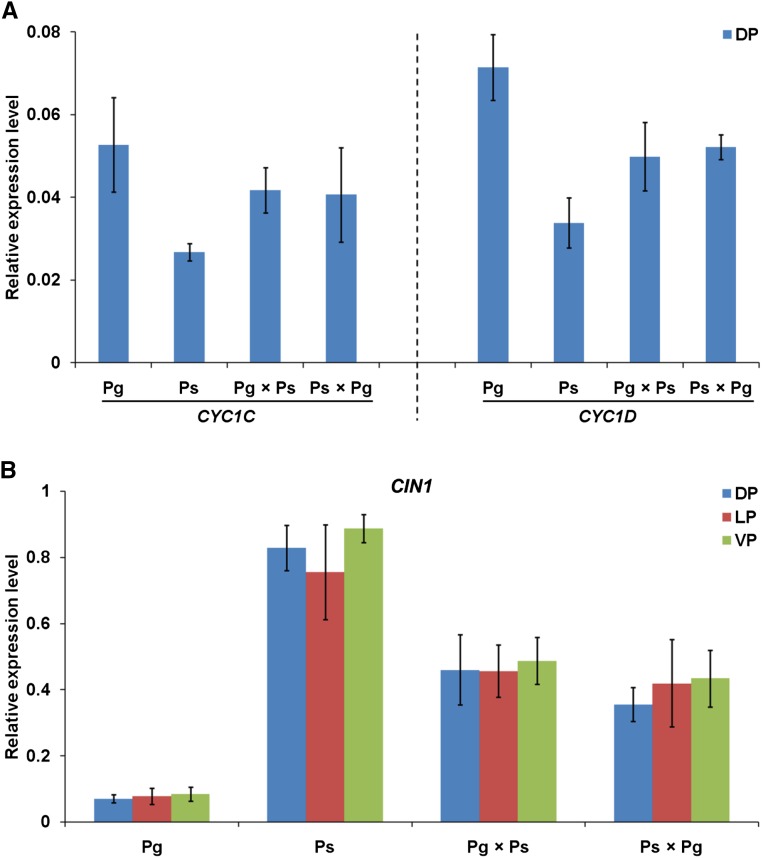
Real-time PCR was conducted to survey the expression patterns of all CYC-like genes in the second whorl of floral organs in Pg, Ps, and F1 hybrids. As shown in Figure 2, both CYC1C and CYC1D were specifically expressed in the dorsal petals of Pg and Ps, with very low expression signal in the lateral and ventral petals (Fig. 2A). By contrast, transcription signals of CYC2A and CYC2B were almost undetectable in all petals of Pg and Ps flowers (Fig. 2A), consistent with a previous report (Gao et al., 2008). Therefore, we only conducted further expression analyses of CYC1C and CYC1D in the dorsal petals. For CYC1C, PgCYC1C was strongly expressed in the dorsal petals of Pg, while PsCYC1C had a much lower expression level in the corresponding regions of Ps (Fig. 3A). The expression ratio of PgCYC1C and PsCYC1C was 1.97 ± 0.20 (Table II), consistent with the morphological variations of the dorsal petals between Pg and Ps. In the Pg × Ps hybrid, the expression signal of CYC1C was obviously weaker than in Pg but stronger than in Ps. Similarly, its expression level in the Ps × Pg hybrid also fell between the two parents (Fig. 3A). Similar to CYC1C, CYC1D also showed a significant expression difference, with a ratio of 2.11 ± 0.27 between PgCYC1D and PsCYC1D (Fig. 3A; Table II), in concert with the morphological changes of the dorsal petals between the two species. In Pg × Ps and Ps × Pg hybrids, CYC1D had very similar expression levels that fell between the two parents (Fig. 3A). As outlined above, CYC1C and CYC1D had very similar dorsal-specific expression patterns in both Pg and Ps and also had almost identical expression differentiation patterns between the two species, with intermediate expression levels in the reciprocal F1 hybrids. Their expression is wholly consonant with the morphology and morphological differentiation of the dorsal petals in parents and hybrids.
Figure 2.
Real-time PCR expression analyses of CYC- and CIN-like genes in all petals of Pg and Ps. A, Relative expression levels of CYC1C, CYC1D, CYC2A, and CYC2B in dorsal (DP), lateral (LP), and ventral petals (VP) of Pg and Ps. B, Relative expression levels of CIN1, CIN2A, and CIN2B in dorsal, lateral, and ventral petals of the two species. ACTIN was amplified as an internal reference. Data are mean ± sd of three biological replicates, with each including three technical replicates.
Figure 3.
Real-time PCR expression analyses of CYC1C, CYC1D, and CIN1 in petals of parents and F1 hybrids. A, Relative expression levels of CYC1C and CYC1D in dorsal petals (DP) of parents and hybrids. B, Relative expression levels of CIN1 in dorsal, lateral (LP), and ventral petals (VP) of parents and hybrids. ACTIN was amplified as an internal reference. Data represent the mean ± sd of three biological replicates, with each including three technical replicates.
Table II. Expression ratios of CYC- and CIN-like genes between two parents and allele expression biases in F1 hybrids.
Data are mean ± sd of three independent biological replicates, with each including three technical replicates. The asterisk represents the ratio being significantly different (Fisher’s lsd test; H0: Pg/Ps = 1, PgF1/PsF1 = 1, P < 0.01). DP, Dorsal petals; LP, lateral petals; VP, ventral petals.
Real-time PCR was also performed to investigate the expression patterns of all CIN-like genes in all floral organs of the second whorl in both parents and F1 hybrids. The results showed that CIN1 expression levels were similar in the dorsal, lateral, and ventral petals in both Pg and Ps but were much different between the two species (Fig. 2B). Different from CIN1, CIN2A and CIN2B had similar expression levels between Pg and Ps in all petals (Fig. 2B), ruling out the possibility of their involvement in the petal shape variation between the two species. Therefore, we further carried out expression analyses of only CIN1 in both parents and hybrids. The results showed that PgCIN1 and PsCIN1 exhibited an extreme expression divergence in the dorsal petals between Pg and Ps, with a ratio of 0.09 ± 0.01 (Fig. 3B; Table II), in line with the distinct curvature degree of the dorsal petals that is upward extended in Pg while extremely backward reflexed in Ps. In addition, CIN1 also showed an extreme expression difference in both lateral and ventral petals between Pg and Ps (Fig. 3B), consistent with the different curvature status of these petals between the two species. In Pg × Ps hybrid, CIN1 expression levels were higher than in Pg but lower than in Ps in all petals, thus showing an intermediate expression level between the two parents. Similarly, its expression level in the Ps × Pg hybrid fell between the two parents.
Analysis of the Dorsalized Mutant Flowers in F1 Hybrids
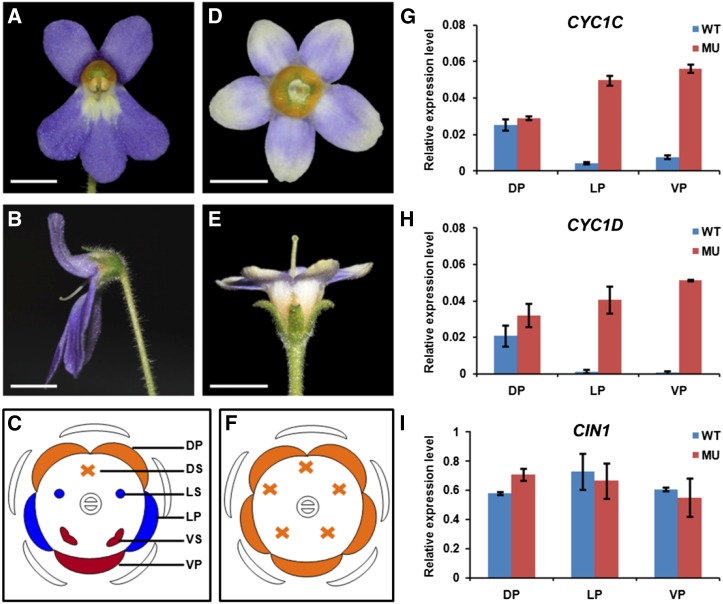
The vast majority of F1 hybrids have normal zygomorphic flowers featuring two smaller dorsal petals and two fertile ventral stamens with both dorsal and lateral stamens aborted (Fig. 4, A–C). Nevertheless, a few Ps × Pg F1 hybrids produce dorsalized actinomorphic flowers with all petals resembling the dorsal petals of wild-type flowers and all stamens aborted (Fig. 4, D–F). These mutant flowers provide an opportunity to investigate the function of CYC-like genes. Real-time PCR results showed that both CYC1C and CYC1D expression expanded from the dorsal to lateral and ventral petals in the mutant flowers, in contrast with their expression specifically restricted to the dorsal petals in the wild-type flowers (Fig. 4, G and H). Meanwhile, the expression level of CIN1 was not affected, consistent with the unchanged curvature status of the five petals in the mutant flowers compared with the wild-type flowers (Fig. 4I). The close correlation between gene expression and petal identity indicates that these dorsalized mutant flowers are formed due to an ectopic expression of the dorsal identity genes CYC1C and CYC1D in lateral and ventral regions.
Figure 4.
Expanded expression of CYC1C and CYC1D accounts for the formation of peloric flowers in F1 hybrid offspring. A, Normal Ps × Pg offspring have typical zygomorphic flowers, with two smaller dorsal petals (DP) compared with lateral (LP) and ventral petals (VP) and only two fertile ventral stamens (VS). B, The side view of wild-type (WT) flowers. C, The floral diagram of wild-type flowers. D, Peloric flowers have five petals that adopt the dorsal petal identity of wild-type flowers, as well as five staminodes. E, The side view of mutant (MU) flowers. F, The floral diagram of mutant flowers. The expression domains of both CYC1C (G) and CYC1D (H) expanded from dorsal to lateral and ventral petals in mutant flowers. I, The expression pattern of CIN1 was unaffected in mutant flowers. ACTIN was used as an internal reference. Data are mean ± sd of three biological replicates, with each including three technical replicates. DS, Dorsal stamens; LS, lateral stamens. Bars = 1 cm.
ASE Analyses
Given that CYC1C, CYC1D, and CIN1 were differentially expressed in petals of Pg and Ps, we further conducted ASE analyses in reciprocal F1 hybrids to determine what regulatory changes account for their expression differentiation between the two parents. First, we identified the single nucleotide polymorphism (SNP) sites for each orthologous pair by sequencing the PCR products of CYC1C, CYC1D, and CIN1 amplified from about 30 different genomic DNA samples of Pg and Ps, respectively, which were further confirmed by genotyping the reciprocal F1 hybrids (Supplemental Fig. S6). For each gene, all sequences from 30 individuals of each species were identical, indicating the homozygosity of the two parents.
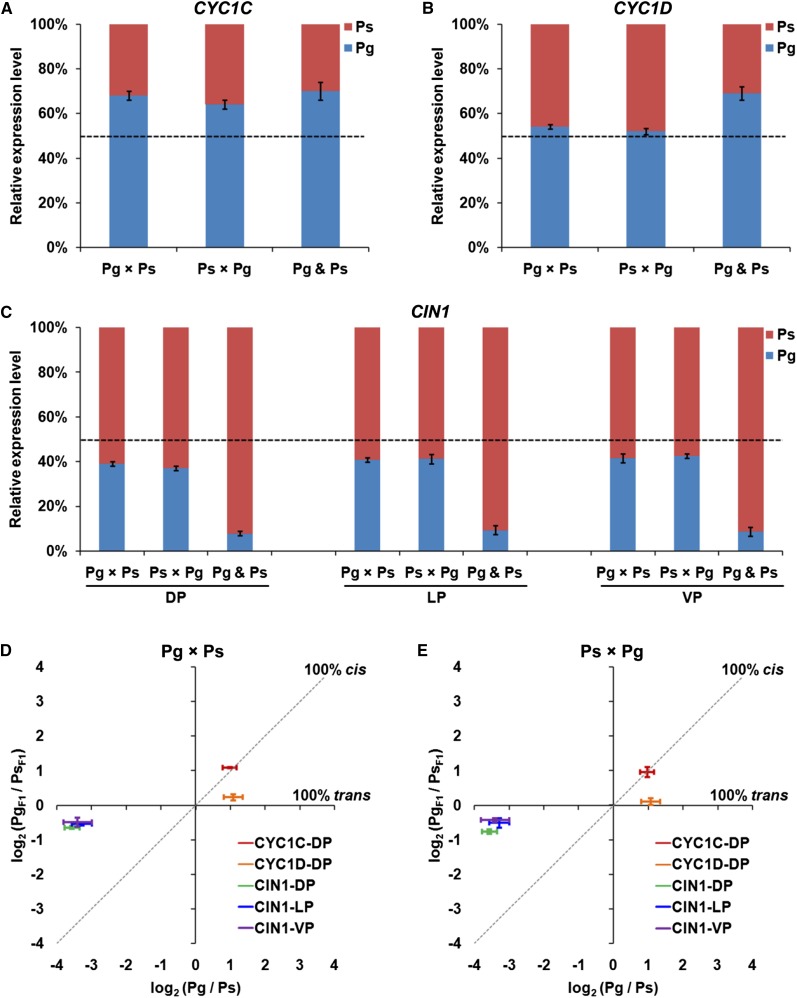
In ASE assays, allele expression ratios in the reciprocal F1 hybrids for each gene were normalized by genomic DNA in which allelic ratios were assumed to be present in equal amounts. For CYC1C, the expression levels of PgCYC1C and PsCYC1C alleles in both Pg × Ps and Ps × Pg hybrids were significantly different from each other, with the ratios of 2.13 ± 0.15 and 1.98 ± 0.14, respectively (Fig. 5A; Table II). Because CYC1C showed the same expression bias between parents and hybrids, cis-regulatory changes were inferred to account for, at least predominantly, the observed expression changes between the two parents. For CYC1D, PgCYC1D and PsCYC1D alleles showed no significant expression difference in both Pg × Ps and Ps × Pg hybrids, with the ratios of 1.17 ± 0.06 and 1.07 ± 0.04, respectively (Fig. 5B; Table II). Because CYC1D showed biased expression in parents but equal expression level in hybrids, the expression difference of PgCYC1D and PsCYC1D genes between the two parents could be mainly explained by trans-regulatory changes.
Figure 5.
Distinct regulatory changes account for expression differences of CYC- and CIN-like genes in Petrocosmea spp. A, Relative expression levels of two CYC1C alleles in dorsal petals (DP) of F1 hybrids. B, Relative expression levels of two CYC1D alleles in dorsal petals of F1 hybrids. C, Relative expression levels of two CIN1 alleles in dorsal, lateral (LP), and ventral petals (VP) of F1 hybrids. For comparison, relative expression levels of each gene in two parents are also shown on the right column of each histogram. D and E, Determination of regulatory changes for expression differences of CYC- and CIN-like genes in Pg × Ps (D) and Ps × Pg (E) hybrids. For each gene, the expression ratio of two orthologs between two parents was plotted against the expression ratio of two alleles in F1 hybrids. Genes for which cis-regulatory changes can explain all (100% cis) of the expression differences between parents fall on the diagonal y = x line. Genes for which trans-regulatory differences can explain all (100% trans) of the expression modifications between parents fall along the horizontal y = 0 line. Error bars are ses calculated from at least three biological replicates, with each including three technical replicates.
PgCIN1 and PsCIN1 alleles showed an ASE imbalance in the dorsal petals of the reciprocal F1 hybrids, with expression ratios of 0.64 ± 0.01 and 0.59 ± 0.01 in Pg × Ps and Ps × Pg hybrids, respectively (Fig. 5C; Table II). They also showed the extreme ASE imbalance in both lateral and ventral petals of the reciprocal F1 hybrids (Fig. 5C; Table II). Therefore, cis-regulatory changes should be involved in the expression difference of PgCIN1 and PsCIN1 genes in the dorsal, lateral, and ventral petals between the two parents. Nevertheless, the allele expression differences in all petals of the reciprocal F1 hybrids were much lower than the expression bias in the corresponding regions of the two parents (Fig. 5C), indicating that the expression differentiation of CIN1 between the two parents might also be attributed to trans-regulatory changes. The above results also indicate that there is little parent-of-origin effect on the expression of CYC1C, CYC1D, and CIN1 in the reciprocal F1 hybrids.
To understand whether cis- or trans-regulatory changes are the primary causes of parental expression differences, or if they contribute concertedly to expression changes, we compared the expression differences of CYC1C, CYC1D, and CIN1 between parents (Pg/Ps) with the relative abundance of allele-specific transcripts in F1 hybrids (PgF1/PsF1; Fig. 5) according to Wittkopp et al. (2004, 2008). Expression ratio for each gene was log2 transformed at first. Then, the expression ratio between parents (log2Pg/Ps) was plotted against the allelic expression ratio in F1 hybrids (log2PgF1/PsF1). The relative contribution of cis- and trans-regulatory changes was determined based on the position of the point, as a plot of the relative allelic expression bias against the relative parental expression differentiation (Fig. 5). For CYC1C, the allelic expression ratio in the Pg × Ps hybrid and the expression ratio between parents were almost identical (log2Pg/Ps = log2PgF1/PsF1), with the point fallen on the diagonal (y = x; Fig. 5D). Similarly, the point for the allelic expression ratio in the Ps × Pg hybrid and the parental expression ratio fell on the y = x line (Fig. 5E). Therefore, cis-regulatory changes can completely explain the expression difference of CYC1C between the two parents. For CYC1D, PgCYC1D and PsCYC1D alleles were equally expressed in both Pg × Ps and Ps × Pg hybrids (PgF1/PsF1 = 1), and the points fell along the horizontal axis (y = 0; Fig. 5, D and E). Therefore, trans-regulatory changes are solely responsible for the expression difference of CYC1D between the two parents. For CIN1, the allelic expression ratios in the dorsal, lateral, and ventral petals of both Pg × Ps and Ps × Pg hybrids strongly deviated from the parental expression ratios in the corresponding regions, with the points fallen outside of both lines but much closer to the y = 0 line (Fig. 5, D and E). Therefore, the expression differences of CIN1 in all petals between the two parents are mainly affected by trans-regulatory divergences, with cis-regulatory changes only accounting for a small portion of the expression differentiation.
DISCUSSION
The Petrocosmea genus is divided into five clades according to molecular phylogenetic analyses (Supplemental Fig. S1; Qiu et al., 2015). In clade A, the two dorsal petals of the upper lip are slightly shorter than the three petals of the lower lip. During the successive morphological evolution and diversification process, the two dorsal petals tend to be shortened and specialized in size and shape from clade A through B and C to clade D, with coordinated variations of correlative characters. However, the dorsal petal size of the upper lip demonstrates a reversal in clade E in which the dorsal petals are almost equal to the ventral petals in length, in contrast with the dorsal petals in clade D that are about one-half of the ventral petals. Meanwhile, a unique morphological feature occurs in the dorsal petals that are highly reflexed backward in clade E versus extended upward in clade D (Fig. 1; Qiu et al., 2015). The issue we focus on here is what molecular mechanisms underlie the size and shape variation of the dorsal petals characterizing the rich diversity of zygomorphic flowers in Petrocosmea spp., exemplified by two representative species, Pg in clade D and Ps in clade E.
Differential Expression of CYC- and CIN-Like Genes Relating to the Morphological Divergence of Zygomorphic Flowers
How new morphological traits originate is a central question for evolutionary developmental biologists. A growing body of evidence has shown that expression modifications of key developmental genes are crucially important for the origin of phenotypic novelties in both animals and plants. For example, expression changes of the yellow gene account for the frequent gain or loss of a male wing pigmentation pattern in different lineages of Drosophila spp. (Gompel et al., 2005; Prud’homme et al., 2006, 2007), and the doubled expression level of TB1 resulting from a distant upstream enhancer plays an important role in the domestication process of modern maize from its wild ancestor teosinte (Doebley et al., 1997; Clark et al., 2006; Studer et al., 2011).
Floral zygomorphy as a key novelty contributes significantly to the explosive radiation of angiosperms (Dilcher, 2000). The modified expression of CYC-like TCP genes usually plays a decisive role in generating unique morphologies in floral symmetry (Hileman and Cubas, 2009). For example, in A. majus, the dorsal-specific expression of CYC and DICH produces typical zygomorphic flowers, while their expansion or loss of expression gives rise to backpetals mutants or ventralized peloric flowers (Luo et al., 1996, 1999). Similar phenomena have been widely observed in other angiosperms (Cubas et al., 1999b; Citerne et al., 2006; Busch and Zachgo, 2007; Pang et al., 2010; Yang et al., 2012). Furthermore, morphological transitions in zygomorphic flowers and between zygomorphy and actinomorphy in closely related species are usually related to the expression changes of CYC-like genes (Zhang et al., 2010; Howarth et al., 2011; Busch et al., 2012; Zhong and Kellogg, 2015). As representatives of two different clades in Petrocosmea spp., Pg and Ps are morphologically different from each other, with the former featuring two short dorsal petals about one-half of the ventral petal and the latter characterized by two long dorsal petals almost equal to the ventral petal. Real-time PCR results show that the dorsal-specific expression level of both CYC1C and CYC1D is negatively correlated with the dorsal petal size, indicative of their implication in the morphological changes of the dorsal petals. In addition, the identical expression levels between CYC1C and CYC1D in both species demonstrate their high redundancy in patterning the dorsal petals in Petrocosmea spp. Considering their constitutive expression in the second whorl of the dorsalized mutant flowers herein (reminiscent of the snapdragon backpetals mutant due to ectopic CYC expression; Luo et al., 1999), we suggest that CYC-like genes in Petrocosmea spp. might repress the dorsal petal growth as previously demonstrated in other Gesneriaceae plants (Yang et al., 2012; Liu et al., 2014). Therefore, we conclude that the expression differentiation of CYC1C and CYC1D, specifically controlling the dorsal petals, brings about, at least partially, the morphological divergence of the dorsal petals between Pg and Ps.
Different from CYC-like genes, CIN-like genes mainly control the local morphology of leaves and petals, with curvature produced upon their up- or down-regulation (Nath et al., 2003; Palatnik et al., 2003; Crawford et al., 2004; Koyama et al., 2007, 2010a, 2011; Nag et al., 2009; Das Gupta et al., 2014). In this study, Pg has upward extended dorsal petals and explanate lateral/ventral petals, while Ps has highly backward reflexed dorsal petals and obviously outward curved lateral/ventral petals, sharply different from each other in all petals. Accordingly, PgCIN1 and PsCIN1 show extreme expression differences in all petals, tightly correlated with the petal shape changes between the two species. Even though there is no functional confirmation so far, the strong correlation between gene expression and petal morphology suggests that CIN-like genes may be involved in petal shape variations in Petrocosmea spp.
The TCP transcription factor family is divided into the PCF subfamily and the CYC/TB1 subfamily, with the latter further classified into CIN lineage and ECE lineage (Cubas et al., 1999a; Howarth and Donoghue, 2006). While CYC-like genes belong to the CYC2 clade in the ECE lineage, CIN-like genes are grouped into the CIN lineage. To our knowledge, this is the first report regarding CYC- and CIN-like genes acting together in patterning a specific zygomorphic morphology with phenotypic variation upon their coordinate expression differentiation. However, CYC- and CIN-like genes may have no direct regulatory relationship in specifying floral zygomorphy in Petrocosmea spp., at least at the transcriptional level, evident in the unaffected expression of CIN1 in the dorsalized mutant flowers and its independent actions in lateral/ventral petals.
Expression Modes of CYC- and CIN-Like Genes Consistent with Petal Phenotypes of F1 Hybrids
Hybridization usually results in phenotypic variations that, at least in part, attribute to modifications of gene expression (Springer and Stupar, 2007; Chen, 2010). A large number of studies have shown that the majority of genes differentially expressed between two parents are often expressed at additive levels in the F1 hybrid offspring (Guo et al., 2004, 2006; Stupar and Springer, 2006; Swanson-Wagner et al., 2006; Stupar et al., 2007). In this study, expression levels of both CYC1C and CYC1D in the reciprocal F1 hybrids fall somewhere between the two parents, thus exhibiting an additive pattern, consistent with the petal phenotype of F1 hybrids. The additive expression patterns of these CYC-like TCP genes might contribute to the intermediate phenotype of the dorsal petals in F1 hybrids. CIN1 expression in both Pg × Ps and Ps × Pg hybrids also conforms to an additive mode, consonant with the petal morphology in F1 hybrids. Therefore, the results herein demonstrate a close correlation between TCP gene expression patterns and petal phenotypes in F1 hybrids. The genotype-phenotype correlation further implies that CYC-like genes may actually control the size of the dorsal petals, while CIN-like genes might be related to the shape of all petals in zygomorphic flowers of Petrocosmea spp. However, their roles in floral development are hypothesized only based on limited data herein. Further expression, genetic, and functional studies will be necessary to establish a causal relationship between TCP gene activities and petal morphologies in Petrocosmea spp. flowers.
Distinct Regulatory Changes Underlying Expression Differentiation of CYC- and CIN-Like TCP Genes
Differences in gene expression can be caused by either cis-regulatory changes that result from changes in enhancer or promoter sequences, changes in the transcribed region, or changes in chromatin structure or trans-regulatory changes that are caused by genetic and epigenetic changes affecting the activity or availability of proteins and RNAs that mediate gene expression (Wittkopp et al., 2004, 2008). While cis-regulatory changes alter gene expression in an allele-specific manner, trans-regulatory variations influence expression of both alleles in a diploid cell. It has been hypothesized that natural selection may favor one type of change over the other (Wittkopp et al., 2008).
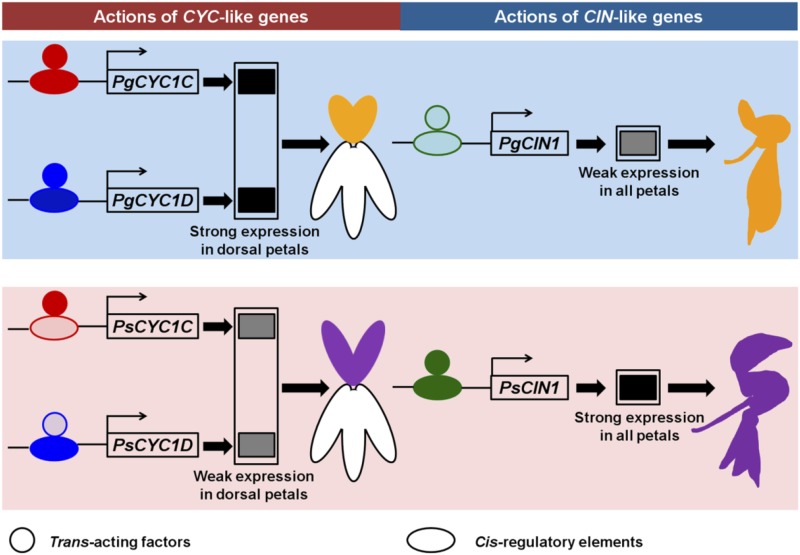
In this study, we examined ASE patterns of CYC- and CIN-like genes in F1 hybrids to elucidate the regulatory mechanisms underlying their expression modifications between Pg and Ps. ASE analyses preclude the possibility of a parent-of-origin (or genome imprinting) effect accounting for allele expression differences because of identical ASE patterns of both CYC- and CIN-like genes in the reciprocal F1 hybrids. The results further demonstrate that the expression differentiation of these TCP genes is underlain by distinctly different types of regulatory changes. The expression differentiation of CYC1C is only (or at least predominantly) caused by cis-regulatory changes, while that of CYC1D is predominantly underlain by trans-acting variations. In angiosperms, especially in eudicots, the CYC2 clade genes have undergone frequent duplications that gave rise to multiple copies of CYC2 genes in most members of zygomorphic clades (Howarth and Donoghue, 2006). Of these, a pair of CYC2 genes are usually involved in controlling the dorsal petals redundantly in zygomorphic flowers, with others having no or transient expression signals (Luo et al., 1996, 1999; Citerne et al., 2006; Feng et al., 2006; Gao et al., 2008; Wang et al., 2008; Song et al., 2009; Zhang et al., 2010; Howarth et al., 2011; Yang et al., 2012). A recent study shows that a double positive autoregulatory feedback loop has evolved repeatedly in different zygomorphic clades of angiosperms to maintain the expression of these gene pairs (Yang et al., 2012). However, it is still an open question as to what regulatory changes have led to their expression divergence. This report indicates that a pair of CYC-like paralogs might have similar expression levels in each species and identical differentiation patterns among related species. However, their underlying regulatory mechanisms would be wholly different from each other as mentioned here (Fig. 6).
Figure 6.
A diagram showing putative molecular regulatory mechanisms for the formation of distinct zygomorphic flowers in Petrocosmea spp. Distinct regulatory changes for CYC1C (cis), CYC1D (trans), and CIN1 (cis + trans) contribute to their expression differences between Pg and Ps, and their coordinating roles give rise to distinctly different zygomorphic flowers, displayed in both dorsal and lateral/ventral petals. Ellipses and circles (red for CYC1C, blue for CYC1D, and green for CIN1) represent cis-regulatory sequences and trans-acting factors, respectively, with dark and light colors indicating strong and weak actions.
Natural selection driving morphological diversification has received much attention at the level of protein evolution but very little for gene expression evolution (Fraser, 2011). In fact, adaptive gene expression is the primary fuel, or at least an important source, for morphological diversification (Prud’homme et al., 2007; Fraser et al., 2010). Because natural selection acts on the phenotype of organisms, molecular changes that increase fitness or at least minimize fitness penalties would be much more subject to natural selection than those causing deleterious effects or less tolerance under natural selection. As a kind of highly pleiotropic body plan patterning gene in controlling both reproductive and vegetative growth of plants, CYC-like genes’ overexpression or knockout usually results in seriously abnormal phenotypes (Costa et al., 2005; Busch and Zachgo, 2007; Guo et al., 2010; Koyama et al., 2010b; Yang et al., 2012). By contrast, the regulatory changes affecting their expression usually have a specific effect on plant growth, with others unaffected. The regulatory changes can occur in the cis-regulatory elements themselves, such as with CYC1C, or in the related trans-acting factors interacting with them, such as with CYC1D (Fig. 6). These results provide, to our knowledge, the first insight into diverse regulatory pathways of paralogous CYC-like TCP genes in promoting morphological diversification of zygomorphic flowers. Natural selection might not favor the changes in the proteins (or coding sequences) themselves but the modifications in their regulatory pathways that shape their expression patterns associated with evolution of floral zygomorphy.
In addition, genome duplication initially generates redundant gene copies followed by evolution and functional divergence. Many duplicate genes can retain a considerable degree of expressional or functional similarity over very long periods, which is usually assumed due to purifying selection maintaining the ancestral function in both duplicates (Lynch and Conery, 2000; Kondrashov and Kondrashov, 2006; Dean et al., 2008; Vavouri et al., 2008). This may be true with respect to the preservation of the CYC-like TCP genes in a genome for a long period of time in zygomorphic clades. However, the high redundancy of CYC1C and CYC1D with identical expression patterns and interspecific expression differentiation in Petrocosmea species is merely shaped by the regulatory context rather than their own coding sequences. This similarity is maintained by remarkably different regulatory mechanisms (Fig. 6). Therefore, our findings herein pose a challenge to the traditional way of evaluating and understanding the role of selection in fixation of gene duplications and their evolutionary fates.
Unlike CYC-like genes, CIN1 expression divergence between Pg and Ps is mediated coordinately by cis- and trans-regulatory changes, with the latter interpreting a large portion of expression changes. It has been reported that miR319 is involved in preventing aberrant activity of CIN-like genes expressed from its native promoter in both leaves and petals of Arabidopsis by direct mRNA cleavage (Palatnik et al., 2003; Nag et al., 2009). In the TCP gene family, only CIN lineage genes contain sequences matching the miR319 target sequence (Palatnik et al., 2003; Martín-Trillo and Cubas, 2010). Therefore, trans-regulatory changes of CIN1 might be related to the modification of miR319 activity and function. This scenario might be sound because both PgCIN1 and PsCIN1 contain potential target sequences of miR319. It is also possible that the interaction of modified miR319 activity and changed cis-elements might have finally given rise to the expression divergence of CIN1 between Pg and Ps (Fig. 6). It would be important to further decipher how the miR319 activity is coopted to the CIN1 regulatory pathway, whether there is a complex combinatorial mechanism underlying a possible dynamic regulatory interaction or network in and between CYC- and CIN-like genes, and if and how they are modified coordinately in creating the expression differentiation of these genes responsible for related morphological evolution in zygomorphic flowers.
CONCLUSION
We here report that CYC- and CIN-like TCP genes have undergone expression differentiation, strongly correlated with the morphological divergence between Pg and Ps. It is, to our knowledge, the first report that CYC- and CIN-like TCP genes act together in patterning a specific morphology of floral zygomorphy, with their coordinate expression differentiation giving rise to phenotypic variations (Fig. 6). In F1 hybrids, their expression conforms to an additive pattern, consistent with the petal phenotypes. We further reveal a new phenomenon that significantly distinct regulatory pathways underlie the identical expression patterns and interspecific expression differentiation of highly redundant paralogous genes in which cis-regulatory changes contribute to the CYC1C expression differentiation while trans-acting variations account for the CYC1D expression divergence. The remarkably differential expression of CIN1 between the two species is mediated coordinately by cis- and trans-regulatory changes. Given that the role of selection in fixation of gene duplications has been usually evaluated based on coding sequences, our findings herein offer new insight into understanding the evolutionary fate of duplicate genes. Further identification and functional analyses of related cis-elements and trans-regulators would be important to decipher how cis- and trans-regulatory changes underlie the expression differentiation of CYC- and CIN-like genes and subsequent morphological evolution and diversification of floral zygomorphy in angiosperms.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Petrocosmea glabristoma and Petrocosmea sinensis, two diploid species belonging to the genus Petrocosmea (Gesneriaceae), were sampled from Menglun Town, Yunnan Province, and Leshan Giant Buddha Scenic Area in Leshan City, Sichuan Province, respectively. The plants were transplanted to 8-cm pots containing a mixture of vermiculite, perlite, and Pindstrup substrate (1:1:1) in the greenhouse at the Institute of Botany, Chinese Academy of Sciences, under a natural photoperiod, except for a high relative humidity (50%–70%) and a certain degree of shading. The hybrid Pg × Ps (the first plant denotes the maternal parent) was repeatedly constructed during the flowering seasons from 2008 to 2012. The reciprocal cross Ps × Pg was also generated to detect parent-of-origin effects.
The F1 hybrid seeds were surface sterilized in 70% (v/v) ethanol for 1 min, rinsed with sterile water once, disinfected with 2.5% (v/v) sodium hypochlorite for 3 to 5 min, and finally rinsed five times with sterile water. The sterilized seeds were germinated on Murashige and Skoog medium in a growth chamber at 26°C under 10-h-light/14-h-dark photoperiod (100 µmol m–2 s–1). The plantlets were transferred to 8-cm pots and grown under the same conditions as the parents.
DNA Extraction and Gene Isolation
Genomic DNA of Pg and Ps was extracted from fresh leaves following a modified cetyl-trimethyl-ammonium bromide procedure of Doyle and Doyle (1987). A forward primer F1 and a reverse primer R (Gao et al., 2008; Pang et al., 2010) were used to amplify partial coding sequences of CYC-like genes. To isolate CIN-like genes, a pair of primers, PCIN-F and PCIN-R, were designed according to the sequences of Antirrhinum majus CIN and Arabidopsis (Arabidopsis thaliana) CIN-like genes. The full-length coding sequences of CYC- and CIN-like genes were further isolated using Genome Walking Kit (TaKaRa). All primers are listed in Supplemental Table S1. The PCR products were cloned into the pEASY-T1 Simple Cloning Vector (TransGen Biotech) and sequenced (Beijing Genomics Institute Tech).
Phylogenetic Analyses of CYC- and CIN-Like Genes
DNAMAN software (Lynnon Biosoft) was used to translate the isolated Petrocosmea spp. CYC- and CIN-like genes. Neighbor-joining analyses were performed using the amino acid sequences of Petrocosmea spp. CYC- and CIN-like genes, CYC/DICH/CIN genes from A. majus (Luo et al., 1996, 1999; Nath et al., 2003), one CYC-like gene (AtTCP1) and eight CIN-like genes (AtTCP2/AtTCP3/AtTCP4/AtTCP5/AtTCP10/AtTCP13/AtTCP17/AtTCP24) from Arabidopsis (Martín-Trillo and Cubas, 2010), and four CYC-like genes (PhCYC1C/PhCYC1D/PhCYC2A/PhCYC2B) from Primulina heterotricha (Yang et al., 2012). The rice (Oryza sativa) PCF1 and PCF2 (Kosugi and Ohashi, 1997) were used as outgroups. The sequences were first aligned using Clustal X (Thompson et al., 1997) and further adjusted manually with BioEdit (Hall, 1999). MEGA6 software (Sudhir Kumar) was used to construct neighbor-joining trees using the following settings: p-distance and pairwise deletion, with the bootstrap values calculated for 1,000 replicates.
RNA Extraction and Real-Time PCR
Dorsal, lateral, and ventral petals of Pg, Ps, Pg × Ps, and Ps × Pg flowers before anthesis were dissected, immediately frozen in liquid nitrogen, and stored at –80°C until extraction. Total RNA was extracted using an SV Total RNA Isolation System (Promega) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized using a RevertAid H Minus First-Strand cDNA Synthesis Kit (Thermo). Gene-specific primers were used to amplify all genes, and ACTIN was amplified as a reference gene (Supplemental Table S1). It is noteworthy that in real-time PCR, different alleles for each TCP gene in F1 hybrids were indiscriminately amplified using a pair of primers that anneal to the conserved regions. The specificity of all primers was confirmed by sequencing PCR products. The SYBR Premix Ex Taq (TaKaRa) was used to perform real-time PCR using the StepOne Plus Real-Time PCR System (AB Applied Biosystems). The PCR conditions were: initial denaturation at 95°C at 30 s and 40 cycles of 95°C at 10 s and 60°C at 40 s, after which dissociation curves were recorded using one cycle of 95°C at 30 s, 60°C at 30 s, and 95°C at 30 s. The relative expression levels were determined by normalizing the PCR threshold cycle number of each gene with that of ACTIN. The data were the average of three biological replicates, with each including three technical replicates. The expression ratio of each gene between paternal and maternal parents (Pg/Ps) was calculated and compared using the Fisher’s lsd test to identify the parental expression imbalance (null hypothesis, H0: Pg/Ps = 1, P < 0.01). All statistical analyses were performed using SPSS software version 14.0.
SNP Identification and ASE Assays by Single-Base Primer Extension Experiment
For SNP identification, at least 30 individuals from either Pg or Ps were sampled to isolate genomic DNA. DNA fragments for CYC1C, CYC1D, and CIN1 were amplified using real-time PCR primers, and the PCR products were directly sequenced. The homozygosity of two parents was identified as the identity of all sequences from different individuals of a species for each gene. SNP sites were identified as single-base differences between two parents. The SNP sites were further confirmed by genotyping reciprocal F1 hybrids in which two peaks would appear in a position corresponding to two different reads, respectively, from two parents in the same position using the Chromas software (Chromos MFC Application). One fixed SNP site for each gene was selected for ASE analyses.
For ASE assays, the gene-specific segment surrounding the SNP site was first amplified from the cDNA pool of reciprocal F1 hybrids using real-time PCR primers. Single-base extension experiments were conducted using the ABI PRISM SNaPshot Multiplex Kit (AB Applied Biosystems) according to the manufacturer’s instructions. Briefly, following PCR thermal cycling, unincorporated primers and deoxyribonucleoside triphosphates were removed by adding 5 units of shrimp alkaline phosphatase (TaKaRa) and one unit of Exonuclease I (TaKaRa) to each 15-µL PCR product. Reactions were mixed briefly and incubated at 37°C for 60 min, followed by 75°C for 15 min to inactivate the enzyme. The PCR products were then subjected to a primer extension assay using single-base extension primers that were designed to anneal to the amplified DNA fragments adjacent to the SNP sites (Supplemental Table S1). Primer extension reactions were carried out in a total volume of 10 µL containing 5 µL of SNaPshot Multiplex Ready Reaction Mix, 0.4 µL of 5 µm extension primer, 2 µL of PCR product, and 2.6 µL of double-distilled water. Thermal cycling conditions for extension reactions were carried out with the following program: initial denaturation at 94°C for 2 min and 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 30 s. After cycling, the unincorporated fluorescent dideoxyribonucleoside triphosphates were removed by adding one unit of shrimp alkaline phosphatase and incubating for 60 min at 37°C, followed by 15 min at 65°C for enzyme inactivation. The resulting primer extension products were analyzed on an ABI 3730 capillary electrophoresis DNA instrument using the Peak Scanner Software version 1.0 (Applied Biosystems) according to the manufacturer’s protocol. The expression ratio of two alleles was measured by comparing the height of peaks. Allelic ratios of genomic DNA from F1 hybrids assumed to be present in equal amounts (1:1) were used to normalize allelic ratios of cDNA samples to adjust PCR bias (Wittkopp et al., 2004, 2008; Zhuang and Adams, 2007). ses were calculated from three independent biological replicates (including three technical replicates for each). For each gene, the expression ratio of two alleles in F1 hybrids (PgF1/PsF1) was compared with interspecific expression bias of two orthologs in two parents (Pg/Ps) to identify cis-regulatory divergence by using the Fisher’s lsd test (H0: PgF1/PsF1 = 1, P < 0.01). The statistical analyses were performed using SPSS software version 14.0.
Determination of Cis- and Trans-Regulatory Changes
Gene expression modifications can result from either cis- or trans-regulatory changes. To discriminate the two changes, we compared the expression ratio of two alleles in heterozygous F1 hybrids with the expression ratio of two orthologs between two homozygous parents. In the same genetic background of hybrids, two alleles inherited from different parents are exposed to a common set of trans-acting factors. Therefore, any observed allelic expression imbalance in hybrids is characteristic of cis-regulatory divergence. Trans-acting variation can then be deduced if genes show expression differences in two parents but no allelic expression imbalance in hybrids.
To further determine relative contributions of cis- and trans-regulatory changes to gene expression differences between two parents, we compared the expression ratio of two orthologs between parents (Pg/Ps) with the expression ratio of two alleles in F1 hybrids (PgF1/PsF1) according to Wittkopp et al. (2004, 2008). Expression ratios for each gene were log2 transformed at first. Then, the expression ratio between parents (log2Pg/Ps; the horizontal axis) was plotted against the allelic expression bias in F1 hybrids (log2PgF1/PsF1; the vertical axis). If the hybrid and parental expression ratios are identical (log2Pg/Ps = log2PgF1/PsF1) and points fall on the diagonal (y = x), cis-regulatory changes are believed to completely explain the expression differences between parents. By contrast, if the Pg and Ps alleles are equally expressed in F1 hybrids (PgF1/PsF1 = 1) and points fall along the horizontal axis (y = log21 = 0), trans-regulatory changes are the sole causes for the expression differences between parents. If points fall outside of these lines, the expression differences are affected by both cis- and trans-regulatory divergences.
Sequence data from this article can be found in the GenBank data library under accession numbers PgCYC1C, KT596748; PgCYC1D, KT596749; PgCYC2A, KT596750; PgCYC2B, KT596751; PgCIN1, KT596752; PgCIN2A, KT596753; PgCIN2B, KT596754; PsCYC1C, T596755; PsCYC1D, KT596756; PsCYC2A, KT596757; PsCYC2B, KT596758; PsCIN1, KT596759; PsCIN2A, KT596760; and PsCIN2B, KT596761.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The majority rule consensus Bayesian tree generated from analysis of combined chloroplast DNA and nuclear DNA data.
Supplemental Figure S2. The neighbor-joining tree of CYC- and CIN-like proteins.
Supplemental Figure S3. Gene structures of Petrocosmea spp. CYC- and CIN-like TCP genes.
Supplemental Figure S4. Amino acid sequence alignment of Petrocosmea spp. CYC-like TCP genes.
Supplemental Figure S5. Amino acid sequence alignment of Petrocosmea spp. CIN-like TCP genes.
Supplemental Figure S6. SNP identification of CYC1C, CYC1D, and CIN1 genes.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank James F. Smith for constructive comments on and language improvements to this article.
Glossary
- ASE
allele-specific expression
- Pg
Petrocosmea glabristoma
- Ps
Petrocosmea sinensis
- cDNA
complementary DNA
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31170198 to Y.-Z.W. and 31300197 to X.Y.).
Articles can be viewed without a subscription.
References
- Arnoult L, Su KFY, Manoel D, Minervino C, Magriña J, Gompel N, Prud’homme B (2013) Emergence and diversification of fly pigmentation through evolution of a gene regulatory module. Science 339: 1423–1426 [DOI] [PubMed] [Google Scholar]
- Bell GDM, Kane NC, Rieseberg LH, Adams KL (2013) RNA-seq analysis of allele-specific expression, hybrid effects, and regulatory divergence in hybrids compared with their parents from natural populations. Genome Biol Evol 5: 1309–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broholm SK, Tähtiharju S, Laitinen RAE, Albert VA, Teeri TH, Elomaa P (2008) A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc Natl Acad Sci USA 105: 9117–9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Horn S, Mühlhausen A, Mummenhoff K, Zachgo S (2012) Corolla monosymmetry: evolution of a morphological novelty in the Brassicaceae family. Mol Biol Evol 29: 1241–1254 [DOI] [PubMed] [Google Scholar]
- Busch A, Zachgo S (2007) Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proc Natl Acad Sci USA 104: 16714–16719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. (2010) Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci 15: 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citerne HL, Pennington RT, Cronk QCB (2006) An apparent reversal in floral symmetry in the legume Cadia is a homeotic transformation. Proc Natl Acad Sci USA 103: 12017–12020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, Wagler TN, Quijada P, Doebley J (2006) A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat Genet 38: 594–597 [DOI] [PubMed] [Google Scholar]
- Cong B, Liu J, Tanksley SD (2002) Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc Natl Acad Sci USA 99: 13606–13611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MMR, Fox S, Hanna AI, Baxter C, Coen E (2005) Evolution of regulatory interactions controlling floral asymmetry. Development 132: 5093–510116236768 [Google Scholar]
- Crawford BCW, Nath U, Carpenter R, Coen ES (2004) CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol 135: 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P. (2004) Floral zygomorphy, the recurring evolution of a successful trait. BioEssays 26: 1175–1184 [DOI] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E (1999a) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Cubas P, Vincent C, Coen E (1999b) An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401: 157–161 [DOI] [PubMed] [Google Scholar]
- Cubillos FA, Stegle O, Grondin C, Canut M, Tisné S, Gy I, Loudet O (2014) Extensive cis-regulatory variation robust to environmental perturbation in Arabidopsis. Plant Cell 26: 4298–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Gupta M, Aggarwal P, Nath U (2014) CINCINNATA in Antirrhinum majus directly modulates genes involved in cytokinin and auxin signaling. New Phytol 204: 901–912 [DOI] [PubMed] [Google Scholar]
- Dean EJ, Davis JC, Davis RW, Petrov DA (2008) Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet 4: e1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilcher D. (2000) Toward a new synthesis: major evolutionary trends in the angiosperm fossil record. Proc Natl Acad Sci USA 97: 7030–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Dong Y, Yang X, Liu J, Wang BH, Liu BL, Wang YZ (2014) Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat Commun 5: 3352. [DOI] [PubMed] [Google Scholar]
- Donoghue MJ, Ree RH, Baum DA (1998) Phylogeny and the evolution of flower symmetry in the Asteridae. Trends Plant Sci 3: 311–317 [Google Scholar]
- Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15 [Google Scholar]
- Du ZY, Wang YZ (2008) Significance of RT-PCR expression patterns of CYC-like genes in Oreocharis benthamii (Gesneriaceae). J Syst Evol 46: 23–31 [Google Scholar]
- Feng X, Zhao Z, Tian Z, Xu S, Luo Y, Cai Z, Wang Y, Yang J, Wang Z, Weng L, et al. (2006) Control of petal shape and floral zygomorphy in Lotus japonicus. Proc Natl Acad Sci USA 103: 4970–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB. (2011) Genome-wide approaches to the study of adaptive gene expression evolution: Systematic studies of evolutionary adaptations involving gene expression will allow many fundamental questions in evolutionary biology to be addressed. BioEssays 33: 469–477 [DOI] [PubMed] [Google Scholar]
- Fraser HB, Moses AM, Schadt EE (2010) Evidence for widespread adaptive evolution of gene expression in budding yeast. Proc Natl Acad Sci USA 107: 2977–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Tao JH, Yan D, Wang YZ, Li ZY (2008) Expression differentiation of CYC-like floral symmetry genes correlated with their protein sequence divergence in Chirita heterotricha (Gesneriaceae). Dev Genes Evol 218: 341–351 [DOI] [PubMed] [Google Scholar]
- Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB (2005) Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433: 481–487 [DOI] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Yang X, Crasta O, Zinselmeier C, Smith OS, Bowen B (2006) Genome-wide transcript analysis of maize hybrids: allelic additive gene expression and yield heterosis. Theor Appl Genet 113: 831–845 [DOI] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Zinselmeier C, Habben J, Bowen BA, Smith OS (2004) Allelic variation of gene expression in maize hybrids. Plant Cell 16: 1707–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Fujioka S, Blancaflor EB, Miao S, Gou X, Li J (2010) TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Hileman LC, Cubas P (2009) An expanded evolutionary role for flower symmetry genes. J Biol 8: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman LC, Kramer EM, Baum DA (2003) Differential regulation of symmetry genes and the evolution of floral morphologies. Proc Natl Acad Sci USA 100: 12814–12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth DG, Donoghue MJ (2006) Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc Natl Acad Sci USA 103: 9101–9106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth DG, Martins T, Chimney E, Donoghue MJ (2011) Diversification of CYCLOIDEA expression in the evolution of bilateral flower symmetry in Caprifoliaceae and Lonicera (Dipsacales). Ann Bot (Lond) 107: 1521–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiekens R, Vercauteren A, Moerkerke B, Goetghebeur E, Van Den Daele H, Sterken R, Kuiper M, van Eeuwijk F, Vuylsteke M (2006) Genome-wide screening for cis-regulatory variation using a classical diallel crossing scheme. Nucleic Acids Res 34: 3677–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Cui ML, Cubas P, Gillies A, Lee K, Chapman MA, Abbott RJ, Coen E (2008) Regulatory genes control a key morphological and ecological trait transferred between species. Science 322: 1116–1119 [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Kondrashov AS (2006) Role of selection in fixation of gene duplications. J Theor Biol 239: 141–151 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (1997) PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9: 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2010a) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Ohme-Takagi M, Sato F (2011) Generation of serrated and wavy petals by inhibition of the activity of TCP transcription factors in Arabidopsis thaliana. Plant Signal Behav 6: 697–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Sato F, Ohme-Takagi M (2010b) A role of TCP1 in the longitudinal elongation of leaves in Arabidopsis. Biosci Biotechnol Biochem 74: 2145–2147 [DOI] [PubMed] [Google Scholar]
- Li ZY, Wang YZ (2004) Petrocosmea Oliv. In Li ZY, Wang YZ, eds, Plants of Gesneriaceae in China. Henan Science and Technology Publishing House, Zhengzhou, China, pp 154–166 [Google Scholar]
- Liu BL, Pang HB, Yang X, Wang YZ (2014) Functional and evolutionary analyses of Primulina heterotricha CYC1C gene in tobacco and Arabidopsis transformation systems. J Syst Evol 52: 112–123 [Google Scholar]
- Luo D, Carpenter R, Copsey L, Vincent C, Clark J, Coen E (1999) Control of organ asymmetry in flowers of Antirrhinum. Cell 99: 367–376 [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E (1996) Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Main BJ, Bickel RD, McIntyre LM, Graze RM, Calabrese PP, Nuzhdin SV (2009) Allele-specific expression assays using Solexa. BMC Genomics 10: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Nag A, King S, Jack T (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106: 22534–22539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U, Crawford BCW, Carpenter R, Coen E (2003) Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Pang HB, Sun QW, He SZ, Wang YZ (2010) Expression pattern of CYC-like genes relating to a dorsalized actinomorphic flower in Tengia (Gesneriaceae). J Syst Evol 48: 309–317 [Google Scholar]
- Prud’homme B, Gompel N, Carroll SB (2007) Emerging principles of regulatory evolution. Proc Natl Acad Sci USA (Suppl 1) 104: 8605–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh SD, True JR, Carroll SB (2006) Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440: 1050–1053 [DOI] [PubMed] [Google Scholar]
- Qiu ZJ, Lu YX, Li CQ, Dong Y, Smith JF, Wang YZ (2015) Origin and evolution of Petrocosmea (Gesneriaceae) inferred from both DNA sequence and novel findings in morphology with a test of morphology-based hypotheses. BMC Plant Biol 15: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu ZJ, Yuan ZL, Li ZY, Wang YZ (2011) Confirmation of a natural hybrid species in Petrocosmea (Gesneriaceae) based on molecular and morphological evidence. J Syst Evol 49: 449–463 [Google Scholar]
- Rosas U, Barton NH, Copsey L, Barbier de Reuille P, Coen E (2010) Cryptic variation between species and the basis of hybrid performance. PLoS Biol 8: e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CF, Lin QB, Liang RH, Wang YZ (2009) Expressions of ECE-CYC2 clade genes relating to abortion of both dorsal and ventral stamens in Opithandra (Gesneriaceae). BMC Evol Biol 9: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Stupar RM (2007) Allelic variation and heterosis in maize: how do two halves make more than a whole? Genome Res 17: 264–275 [DOI] [PubMed] [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J (2011) Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet 43: 1160–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Hermanson PJ, Springer NM (2007) Nonadditive expression and parent-of-origin effects identified by microarray and allele-specific expression profiling of maize endosperm. Plant Physiol 145: 411–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Springer NM (2006) Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173: 2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS (2006) All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci USA 103: 6805–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri T, Semple JI, Lehner B (2008) Widespread conservation of genetic redundancy during a billion years of eukaryotic evolution. Trends Genet 24: 485–488 [DOI] [PubMed] [Google Scholar]
- von Korff M, Radovic S, Choumane W, Stamati K, Udupa SM, Grando S, Ceccarelli S, Mackay I, Powell W, Baum M, et al. (2009) Asymmetric allele-specific expression in relation to developmental variation and drought stress in barley hybrids. Plant J 59: 14–26 [DOI] [PubMed] [Google Scholar]
- Wang Z, Luo Y, Li X, Wang L, Xu S, Yang J, Weng L, Sato S, Tabata S, Ambrose M, et al. (2008) Genetic control of floral zygomorphy in pea (Pisum sativum L.). Proc Natl Acad Sci USA 105: 10414–10419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Koshikawa S, Williams TM, Carroll SB (2010) Generation of a novel wing colour pattern by the Wingless morphogen. Nature 464: 1143–1148 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG (2004) Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG (2008) Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet 40: 346–350 [DOI] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA (2003) The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 20: 1377–1419 [DOI] [PubMed] [Google Scholar]
- Yang X, Pang HB, Liu BL, Qiu ZJ, Gao Q, Wei L, Dong Y, Wang YZ (2012) Evolution of double positive autoregulatory feedback loops in CYCLOIDEA2 clade genes is associated with the origin of floral zygomorphy. Plant Cell 24: 1834–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Kramer EM, Davis CC (2010) Floral symmetry genes and the origin and maintenance of zygomorphy in a plant-pollinator mutualism. Proc Natl Acad Sci USA 107: 6388–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Kellogg EA (2015) Duplication and expression of CYC2-like genes in the origin and maintenance of corolla zygomorphy in Lamiales. New Phytol 205: 852–868 [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Adams KL (2007) Extensive allelic variation in gene expression in Populus F1 hybrids. Genetics 177: 1987–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]