Natural variants in a gibberelin synthesis gene affect the development of primary dormancy by regulating seed morphogenesis and maturation programs to influence germination capability at harvest.
Abstract
Natural variation in seed dormancy is controlled by multiple genes mapped as quantitative trait loci in major crop or model plants. This research aimed to clone and characterize the Seed Dormancy1-2 (qSD1-2) locus associated with endosperm-imposed dormancy and plant height in rice (Oryza sativa). qSD1-2 was delimited to a 20-kb region, which contains OsGA20ox2 and had an additive effect on germination. Naturally occurring or induced loss-of-function mutations of the gibberellin (GA) synthesis gene enhanced seed dormancy and also reduced plant height. Expression of this gene in seeds (including endospermic cells) during early development increased GA accumulation to promote tissue morphogenesis and maturation programs. The mutant allele prevalent in semidwarf cultivars reduced the seed GA content by up to 2-fold at the early stage, which decelerated tissue morphogenesis including endosperm cell differentiation, delayed abscisic acid accumulation by a shift in the temporal distribution pattern, and postponed dehydration, physiological maturity, and germinability development. As the endosperm of developing seeds dominates the moisture equilibrium and desiccation status of the embryo in cereal crops, qSD1-2 is proposed to control primary dormancy by a GA-regulated dehydration mechanism. Allelic distribution of OsGA20ox2, the rice Green Revolution gene, was associated with the indica and japonica subspeciation. However, this research provided no evidence that the primitive indica- and common japonica-specific alleles at the presumably domestication-related locus functionally differentiate in plant height and seed dormancy. Thus, the evolutionary mechanism of this agriculturally important gene remains open for discussion.
Seed dormancy is primarily developed on the plant before maturation as an adaptive trait to regulate the timing of germination and seasonal fluctuations in plant density. Natural variation of the trait is controlled by multiple genes, which have been mapped as quantitative trait loci (QTLs) in crop (cereal, oil, vegetable, and fruit) or model plants (Anderson et al., 1993; Ullrich et al., 1993; Lin et al., 1998; Fennimore et al., 1999; Lijavetzky et al., 2000; Alonso-Blanco et al., 2003; Argyris et al., 2005; Gandhi et al., 2005; Masojć et al., 2007; Blaker et al., 2013; Schatzki et al., 2013). Allelic variants of a validated QTL for seed dormancy were naturally occurring and may have been selected for the trait itself or for dormancy-interrelated characteristics during evolution or domestication (Gu et al., 2005). Cloning and characterization of the QTLs underlying genes will provide in-depth insights into developmental and evolutionary mechanisms of seed dormancy. This knowledge is also important for an efficient use of the QTL alleles to manipulate germination capability in crop breeding.
Seed development starts with tissue morphogenesis (including double fertilization and cell division, differentiation, or expansion) followed by maturation programs (e.g. nutrient reserve deposition, dehydration, acquisition of desiccation tolerance, and programmed cell death to form protective structures). The GA and abscisic acid (ABA) hormones are involved in regulating the sequential development programs. In cereal crops, such as rice (Oryza sativa) varieties that were selected for weak or no dormancy to obtain uniform germination, the seed GA content started to decline rapidly from the highest level at a few DPA; with the GA declining, the ABA content kept increasing and reached the peak level when the endosperm cell number and grain-filling rate approached the maximum (Zhang et al., 2009). Mutagenesis analysis revealed that developing seeds require a given level of ABA to prevent viviparous germination in maize (Zea mays) and rice (Robertson, 1955; Agrawal et al., 2001; Fang et al., 2008). Introduction of the GA-deficient allele dwarf1 into the maize viviparous5 background resulted in desiccation-tolerant seeds, suggesting that the ABA-to-GA ratio (or balance) could be more important than the level of individual hormones in the induction of maturation programs (White and Rivin, 2000). The hormone balance hypothesis was often used to synthesize germination data, largely from mutagenesis analysis, to infer regulatory mechanisms of seed dormancy (Kucera et al., 2005; Yamaguchi et al., 2007; Finkelstein et al., 2008). However, it is unclear to what extent an ABA-to-GA balance generated by an artificial mutant could represent a natural balance selected for a dormant genotype and if the two hormones synthesized at different stages act antagonistically with each other, or in concert, to influence the development of seed and primary dormancy. In addition, of the genes cloned from the QTLs for primary or conditional dormancy, such as DELAY OF GERMINATION1 and IBO in Arabidopsis (Arabidopsis thaliana; Bentsink et al., 2006; Amiguet-Vercher et al., 2015), Seed dormancy4 (Sdr4) and qSD7-1 in rice (Sugimoto et al., 2010; Gu et al., 2011), QPhs.ocs-3A.1/pseru-3AS in wheat (Triticum aestivum; Nakamura et al., 2011; Liu et al., 2013), and High temperature germination6.1 (Htg6.1) in lettuce (Lactuca sativa; Huo et al., 2013), only qSD7-1 (a transcription factor gene) and Htg6.1 (an ABA synthesis enzyme gene) are known to regulate ABA accumulation in developing or mature seeds, and the other loci are not involved in GA or ABA biosynthesis or signaling. Therefore, the hormone balance hypothesis for the development of seed dormancy has yet to be tested with natural variants.
Bioactive GAs promote stem/cell elongation and seed germination through the GA signaling pathway (Sun and Gubler, 2004; Steber, 2007). During germination, GA synthesized in the embryo is released to the endosperm aleurone cells to trigger GA signaling for the synthesis of hydrolases to soften cell walls of the covering tissues and break down nutrient reserves. For dicot (e.g., Arabidopsis, Lepidium sativum, and tomato [Solanum lycopersicum]) seeds, the embryo is enclosed by one or a few layers of endospermic cells, and the micropylar portion (endosperm cap) is believed to serve as a mechanical constraint to radicle emergence; GA application could induce endosperm cap-specific gene expression to release the constraint (Müller et al., 2006; Martínez-Andújar et al., 2012). For cereal seeds, with a small lateral embryo partially surrounded by the endosperm tissue, the latter may impose dormancy through a different mechanism.
This research aimed to clone and characterize qSD1-2, a QTL that was identified in different mapping populations and associated with endosperm-imposed dormancy and plant height in rice (Ye et al., 2010, 2013; Mispan et al., 2013; Gu et al., 2015). In addition, the qSD1-2 region was reported for the semidwarf1 (sd1) gene, the plant height QTLs CULM LENGTH 1a (qCL1a), qCL1b, ph1, and qPH1, and the QTLs associated with root, seedling, and panicle growth, flowering time, and seed setting (Asano et al., 2011; Kovi et al., 2011; Mispan et al., 2013; Zhang et al., 2013). Thus, the objectives of this research were (1) to isolate the qSD1-2 underlying gene from the multi-QTL region and confirm its functions; (2) to identify the molecular and physiological mechanisms of dormancy development regulated by the cloned and confirmed gene; and (3) to determine the origin, distribution, and functional differentiation of major alleles at the seed dormancy locus.
RESULTS
Fine-Mapping Identified OsGA20ox2 as the Only Candidate Gene for qSD1-2
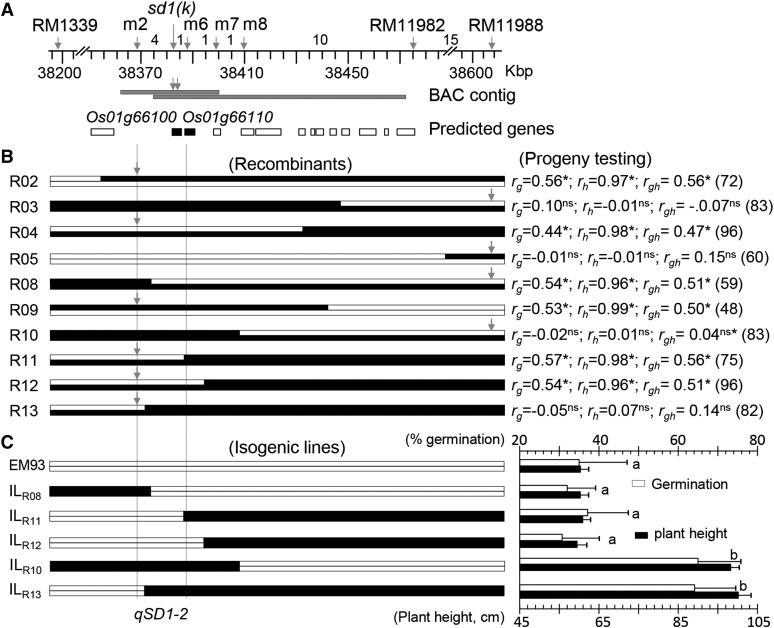
qSD1-2 was colocated with qPH1 on a genomic region of approximately 400 kilo base pairs (Kb), and the QTL-containing region from SS18-2 (weedy rice) was introduced into the EM93-1 (ssp. indica) background to identify recombinants from an F2 population (Supplemental Fig. S1, A and D). A total of 32 recombinants between the m2 and RM11988 markers were identified from 2,592 F2 plants (5,184 gametes) and genotyped with new markers (Supplemental Table S1) to distribute the recombinant events on a partial high-resolution map (Fig. 1A). Ten recombinants (R numbers in Fig. 1B) selected from seven intervals on the map were advanced to the F3 generation to test for marker-trait associations in the progeny lines. Significant correlations between marker genotypes and trait values for germination (rg = 0.44–0.57) and plant height (rh = 0.96–0.99) were detected in six of the 10 F3 lines (Fig. 1B), and the associations were centered on the m2 to m6 interval of approximately 20 Kb. It was estimated, based on data from the six lines, that the 20-Kb interval had an additive effect on germination, which accounted for 19% to 32% of the phenotypic variances, and also had additive and dominance effects on plant height, which together accounted for greater than 90% of the phenotypic variances (Supplemental Table S2). The marker-trait correlations were not significant in the remaining F3 lines (Fig. 1B), as they were fixed for the qSD1-2/qPH1 alleles from EM93-1 (R05) or SS18-2 (R03, R10, and R13; Supplemental Table S1). The absence of correlation in the four progeny lines indicates that there is no seed dormancy or plant height gene, which is differentiated between EM93-1 and SS18-2, in flanking regions of the 20-Kb interval.
Figure 1.
Fine-mapping of qSD1-2. A, Partial high-resolution map. Arrows indicate marker positions on the reference genome (Kawahara et al., 2013). Figures between markers are numbers of recombinant events identified from 5,184 gametes. Horizontal bars below the map are a bacterial artificial chromosome (BAC) contig screened using the probes (arrows). Boxes below the contig are predicted genes on the reference genome, and genes in the narrowed QTL region are depicted by black color. B, Recombinants and progeny testing. Recombinants (R numbers) for the QTL-containing region are represented by two alleles (chromosomal segments) from SS18-2 (black bars) and/or EM93-1 (white bars). Arrows indicate markers used to genotype the progeny lines. At right, correlation coefficients between marker genotypes and trait values for germination (rg) or plant height (rh) in the lines of n plants are shown. Superscripts indicate the correlations significant at P < 0.001 (*) or not significant (ns). Vertical dotted lines delimit the narrowed qSD1-2 region. Additional statistical and genetic parameters for the lines are listed in Supplemental Table S2. C, Genotypic differences in seed dormancy and plant height among isogenic lines. The isogenic lines were selected from the progeny lines. At right, genotypic means ± sd of 20 to 24 plants for germination (%) and plant height (cm) are shown. The letters a and b indicate results from Duncan’s multiple comparison test for not significant (same letter) and significant (different letter) at P < 0.0001.
Five homozygotes were selected from the F3 lines as a set of isogenic lines for single subsegments from SS18-2 in the EM93-1 background. The five isogenic lines fell into two groups based on phenotypes evaluated under the same conditions (Fig. 1C). Group I lines (ILR08, ILR11, and ILR12) were similar to EM93-1 for germination percentage and plant height and displayed stronger dormancy and shorter stature than group II lines (ILR10 and ILR13). Results from these isogenic lines confirmed the pleiotropic effects of the 20-Kb interval and that the allele from EM93-1 inhibits germination and stem elongation.
The 20-Kb interval encompasses the locus Os01g66100, its flanking intergenic sequences, and part (less than 1 Kb of the 3′ end) of the locus Os01g66110 (Fig. 1A), as predicted based on the genome sequence for the japonica cv Nipponbare (Kawahara et al., 2013). We sequenced the 3′ end of Os01g66110 from EM93-1 and SS18-2 and found no sequence variation between the parental lines. Therefore, Os01g66100, which was annotated as the GA synthesis gene OsGA20ox2, was identified as the only candidate gene for qSD1-2.
Loss-of-Function Mutations of OsGA20ox2 Enhanced Seed Dormancy
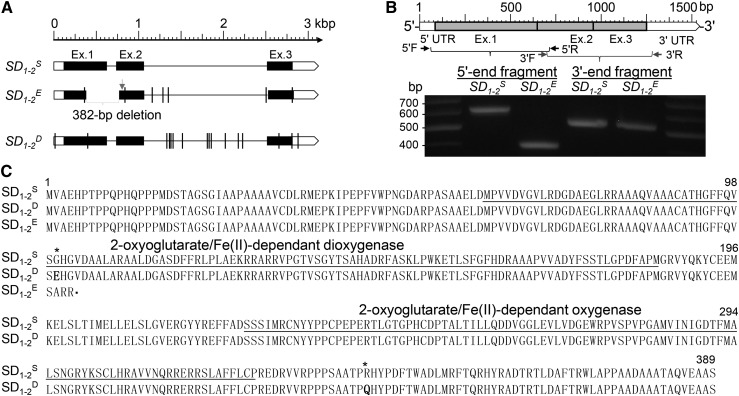
The Os01g66100 allele from SS18-2 (SD1-2S) was isolated from a BAC library (Gu et al., 2010). A BAC contig was aligned across the 20-Kb interval (Fig. 1A) and used to sequence SD1-2S. The sequence was predicted to be 3,183 bp in length and consist of three exons, with untranslated regions (UTRs) in exons 1 (5′ UTR) and 3 (3′ UTR; Fig. 2A). To confirm the gene model, we cloned and sequenced two overlapped complementary DNA (cDNA) fragments (Fig. 2B) from ILR10 (Fig. 1C), an isogenic line for SD1-2S (ILSD1-2S). An assembly of the cDNA fragments is identical to the predicted mRNA sequence for the three exons. The protein sequence deduced from the coding sequence consists of 389 amino acid residues and contains two domains for dioxygenese or oxygenese functions (Fig. 2C). The sequence data suggest that SD1-2S could be a functional GA 20-oxidase.
Figure 2.
Allelic variants of qSD1-2. A, Gene structure. SD1-2S, SD1-2E, and SD1-2D are alleles from SS18-2, EM93-1, and cv Dongjin, respectively. Boxes indicate 5′/3′ UTRs (white) or exons (Ex.; black), and line segments indicate introns. Vertical lines indicate positions of point mutations as compared with SD1-2S. The gap in SD1-2E represents a deletion resulting in a premature stop codon at the arrow-pointed position. B, cDNAs. The scale indicates lengths of UTRs (white boxes) and exons (gray boxes). Arrows indicate positions of PCR primers used to amplify the overlapped fragments on the gel image. C, Protein sequences. Amino acid sequences were deduced based on the cDNA sequences for SD1-2S and SD1-2E or on the genomic DNA sequence for SD1-2D. Underlined sequences are predicted functional domains, asterisks indicate residues different between SD1-2S and SD1-2D, and SD1-2E is a truncated protein.
The Os01g66100 allele of 2,803 bp from EM93-1 (SD1-2E) was cloned by PCR and direct sequencing. Alignment of the SD1-2E with the SD1-2S sequence detected eight point mutations, including a 382-bp deletion (Fig. 2A). This deletion removed intron 1 (102 bp) and its flanking sequences of exons 1 (262 bp) and 2 (18 bp; Fig. 2A), reduced the 5′ end cDNA fragment size (Fig. 2B), and generated a premature stop codon in exon 2, resulting in loss of the two predicted domains (Fig. 2C). The sequence variation suggests that the dormancy-enhancing allele SD1-2E is a loss-of-function allele.
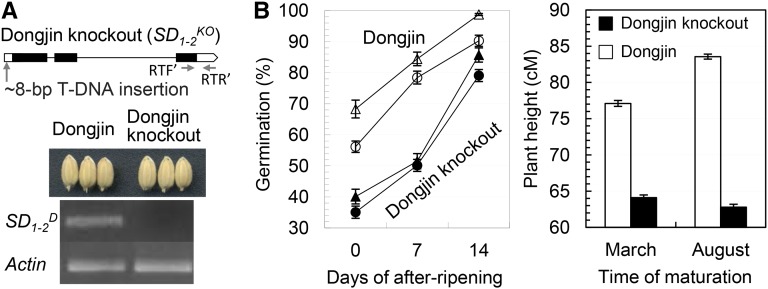
To prove that the loss-of-function mutation could result in enhanced seed dormancy, we analyzed a transfer DNA (T-DNA) insertion line, which has an insertion fragment (approximately 8 Kb) in the 5′ UTR of Os01g66100 (Fig. 3A) in the japonica cv Dongjin (Jeong et al., 2006). Sequence analysis revealed that the cv Dongjin allele SD1-2D is 3,196 bp in length and is different from SD1-2S at 17 sites, including three single-nucleotide polymorphisms (SNPs) in the exons (Fig. 2A). However, the SNPs do not alter the deduced protein length and predicted domains (Fig. 2C), suggesting that SD1-2D could also be functional. Phenotypic identification revealed that both cv Dongjin and the mutant line are similar in seed morphology, but OsGA20ox2 was not transcribed in the mutant (Fig. 3A), suggesting that the T-DNA insertion knocked out the gene function. As expected, the knockout line displayed reduced germinability and plant height, as compared with cv Dongjin, in two independent experiments (Fig. 3B). Thus, the natural (SD1-2E) and induced (SD1-2KO) mutants have a similar effect on delay of germination.
Figure 3.
Mutant analysis of SD1-2KO. A, Seed morphology and transcription. The knockout allele SD1-2KO was generated by a T-DNA insertion at the 5′ UTR of SD1-2D (Fig. 2A) in the cv Dongjin background. Images show the morphology of seeds and semiquantitative reverse transcription PCR products for the segment (horizontal arrows) and ACTIN control from Dongjin and Dongjin knockout M2 plants. Horizontal arrows indicate positions of forward (RTF') and reverse (RTR') primers for the semiquantitative and real-time PCR. B, Genotypic differences in seed dormancy and plant height. Seed dormancy was evaluated by germination of seed samples after-ripened for different days. Data shown are means ± se of 20 to 24 plants harvested in early March or late August.
Mutation of OsGA20ox2 Altered Transcription Patterns of GA Metabolic and Signaling Genes in Seeds at Early Development
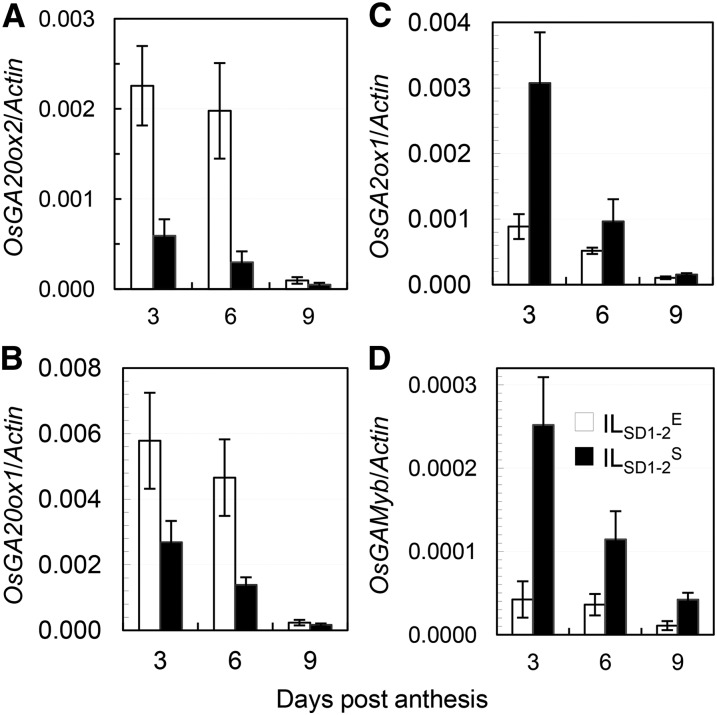
To determine the regulatory role of qSD1-2 in seed development, genes known for GA synthesis (OsGA20ox2 and its paralog OsGA20ox1), catabolism (OsGA2ox1), or signal transduction (OsGAMyb; Gubler et al., 1997; Sakamoto et al., 2004; Tsuji et al., 2006) were quantified for transcription levels in developing caryopses from the isogenic lines. A caryopsis is a true seed united with the covering tissue pericarp. The four selected genes were all transcribed in the caryopses, with the abundance decreased from 3 to 9 DPA (Fig. 4). However, the transcription level was higher for the GA synthesis genes but lower for the catabolism and signaling genes in ILSD1-2E than in ILSD1-2S (Fig. 4). The contrasting expression levels between the synthesis and catabolism genes in the isogenic lines strongly suggest that an OsGA20ox feedback mechanism (Middleton et al., 2012) existed in the developing seeds, presumably to regulate GA homeostasis, signal transduction, and responses.
Figure 4.
Gene expression in developing seeds from ILSD1-2E and ILSD1-2S. A and B, GA biosynthesis enzyme genes (OsGA20ox2 and OsGA20ox1). C, GA catabolic enzyme gene (OsGA2ox1). D, GA signaling gene (OsGAMyb). RNA samples were prepared from caryopses at 3, 6, and 9 DPA and quantified by qRT-PCR. Data shown are mean transcription levels ± sd, relative to the ACTIN control, for three biological replicates.
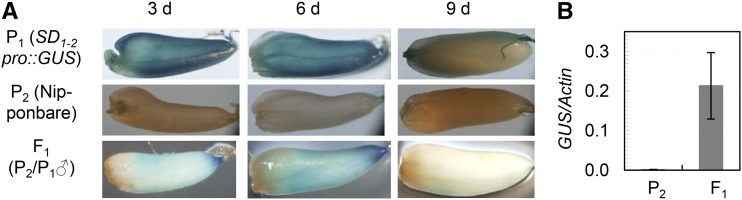
The OsGA20ox2 transcripts in the developing caryopses (Fig. 4A) may include those from the endospermic cells, as qSD1-2 was associated with endosperm-imposed dormancy. To prove this hypothesis, a transgenic line with the SD1-2promoter::GUS reporter system was used to pollinate cv Nipponbare (the recipient of the transgene) to produce hybrid F1 seeds. GUS activity was visualized on caryopses self-pollinated by the transgenic plants and also on the F1 caryopses at 3 to 9 DPA, and the staining intensity decreased dramatically after 6 DPA (Fig. 5A). Quantitative real-time (qRT)-PCR analysis detected transcripts of the reporter gene in the embryo-removed F1 caryopses (Fig. 5B), which consist of the endosperm and maternal (testa and pericarp) tissues. Because the maternal tissues did not receive the reporter gene from the pollen parent, the transcripts detected in the embryoless F1 caryopses must be expressed in the endosperm. These transcriptional data demonstrated that OsGA20ox2 was expressed in developing seeds, including the endosperm tissue, and is likely involved in the regulation of dormancy induction through the GA signaling pathway.
Figure 5.
Histochemical localization of GUS reporter activity in developing seeds. A, GUS-stained caryopses. Caryopses were produced by self-pollinating the SD1-2pro::GUS transgenic (P1) and cv Nipponbare (P2) plants, or by crossing P2 with the pollen parent P1 (F1), and sampled at 3, 6, and 9 DPA. B, Expression levels of the GUS reporter gene in embryoless P2 and F1 caryopses. RNA samples were prepared from 6-d caryopses after the embryo portion was removed and quantified by qRT-PCR. Data shown are means ± sd, relative to the ACTIN control, of two biological replicates.
Loss-of-Function Mutation of OsGA20ox2 Reduced the Seed GA Level, Resulting in Delay of Tissue Morphogenesis, ABA Accumulation, and Subsequent Maturation Programs
To gain deep insight into qSD1-2-regulated mechanisms for dormancy development, a series of experiments were conducted to compare seed hormone, morphological, histological, and physiological profiles between the isogenic lines. In the first experiment, caryopses at 5 DPA were quantified for the GA, ABA, auxin, and cytokinin metabolites. Several GA forms on the early 13-hydroxylation (GA53, GA44, GA19, and GA8) and nonhydroxylation (GA24, GA9, GA51, and GA34) pathways were detected in both lines, and trivial amounts of bioactive GAs were present in ILSD1-2S (GA3 and GA7) or ILSD1-2E (GA1; Supplemental Table S3). The presence of the GA 20-oxidase-catalized metabolites in ILSD1-2E indicates that OsGA20ox2’s paralogs, such as OsGA20ox1 (Fig. 4B), were functional in the genetic background. Genotypic differences were also observed for the other three hormones or their metabolites. For example, the ABA and indole acetic acid contents were relatively higher, while the cytokinin metabolites were relatively lower, in ILSD1-2E than in ILSD1-2S (Supplemental Table S3). Thus, the functional mutation of OsGA20ox2 may affect seed development at the early stage, as the indole acetic acid, cytokinin, and ABA quantities were correlated with endosperm cell-division and grain-filling rates in rice (Zhang et al., 2009).
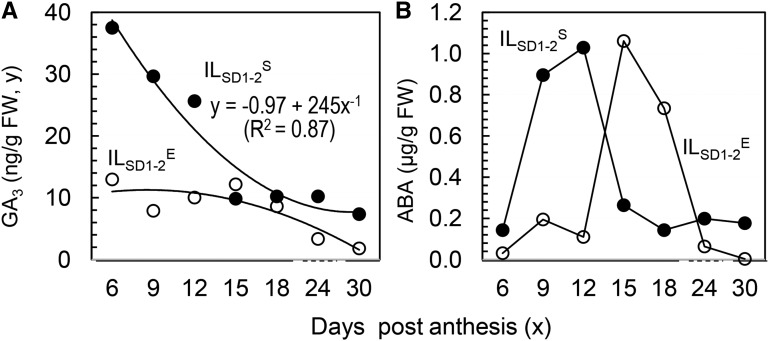
In the second experiment, caryopses sampled at 3-d intervals from 6 to 30 DPA were quantified for GA3 and ABA contents to model temporal distribution patterns. The GA distribution fits to an inverse first-order polynomial, with the highest level at approximately 6 DPA in ILSD1-2S, while the GA level was reduced by up to approximately 2-fold in ILSD1-2E during the 30-d period (Fig. 6A). The ABA distribution was unimodal, with the peak value at approximately 10 DPA in ILSD1-2S or at approximately 15 DPA in ILSD1-2E (Fig. 6B). Interestingly, the isogenic lines were similar in the peak concentration, indicating that the ABA synthesis/accumulation was delayed, but not repressed or promoted, in ILSD1-2E.
Figure 6.
Temporal distributions of seed GA (A) and ABA (B) contents in ILSD1-2S and ILSD1-2E. The nonlinear regression equation was developed to model the GA distribution in ILSD1-2S. FW, Fresh weight.
In the third experiment, caryopses sampled at 3 to 30 DPA were examined for morphological and histological changes to visualize the mutant effect on seed development. An intact developing caryopsis has a green color because of the presence of chlorophyll in the pericarp; as the development proceeds, the green color disappears gradually due to chlorophyll decomposition in the maternal tissue. In this experiment, the green pigment disappeared at approximately 12 d in ILSD1-2S and at approximately 18 d in ILSD1-2E (Fig. 7A). A similar genotypic difference in the green pigment was also observed in leaves of flowering plants, as shown by higher chlorophyll meter readings in ILSD1-2E than in ILSD1-2S during seed development (Fig. 7B). Thus, OsGA20ox2 was involved in the regulation of chlorophyll metabolism, and the functional mutation delayed the pigment decomposition in green tissues. Histological analysis demonstrated that cell development and differentiation were slower in ILSD1-2E than in ILSD1-2S (Fig. 7C). For example, the aleurone cell layer and the aleurone cell protein bodies became visible at approximately 7 and 10 d, respectively, in ILSD1-2S, and at approximately 10 and 15 d, respectively, in ILSD1-2E. The developmental delay of ILSD1-2E seeds from early to middle stages did not affect the fresh weight of caryopses at approximately 24 DPA (Fig. 7D), but it postponed the physiological maturity. Physiological maturity is the time point when the seed dry mass reaches the maximum weight and is followed by a rapid decline in seed moisture (or dehydration) to gain germinability and longevity (Bewley et al., 2013). The above results demonstrated that the mutation of qSD1-2 slowed down the sequential steps of seed development from tissue morphogenesis to maturation.
Figure 7.
Genotypic differences in seed development and chlorophyll content between ILSD1-2S and ILSD1-2E. A, Caryopsis morphology at 3 to 30 DPA. The green color shows the presence of chlorophyll in the pericarp tissue. B, Temporal distributions of leaf chlorophyll meter readings. The measurement was made for the flag leaves of flowering plants. C, Histological profiles of developing caryopses. Images show the tissues pericarp (pe), testa (te), and endosperm (en) cells. Arrows point to protein bodies (pb) in aleuronic (al) cells. Bars = 50 μm. D, Temporal distributions of caryopsis fresh weight. Arrows indicate estimated time points of physiological maturity.
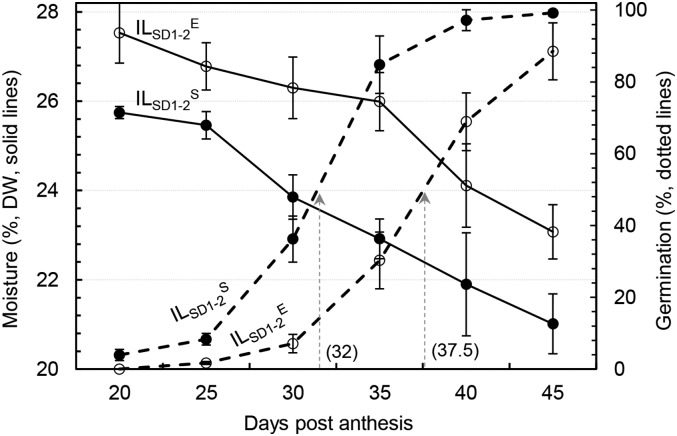
In the fourth experiment, seeds sampled at 5-d intervals from 20 to 45 DPA were measured for moisture content immediately after sample collection and germination percentage after 7 d of air drying to model temporal distribution patterns for dehydration and germinability development. The seed moisture kept declining in an approximately linear manner from approximately 26% at 20 d to 21% at 45 d for ILSD1-2S and was approximately 2% higher for ILSD1-2E during this period (Fig. 8). The ILSD1-2S seeds started to gain germinability at approximately 20 d (approximately 4%); then, the capability increased in an approximately normal distribution pattern, with 50% germination at approximately 32 d (Fig. 8). In contrast, for ILSD1-2E seeds, it took 25 d of development to gain germinability (less than 2%) and approximately 38 d to reach 50% germination. The germinability was negatively correlated with the moisture content in each of the two lines (r > 0.97). Similar distribution patterns for and high correlations (r = −0.98) between germinability and moisture content were also observed in a preliminary experiment, in which seeds from the isogenic lines were sampled at 3-d intervals from 24 to 45 DPA (Supplemental Fig. S2). These two independent observations demonstrated that qSD1-2 acted as a regulator of dehydration and desiccation (maturation drying), which is critical for orthodox seeds to switch from the development to the germination program (Angelovici et al., 2010).
Figure 8.
Temporal distribution patterns of seed moisture content and germination capability in ILSD1-2S and ILSD1-2E. Arrows indicate the time of development required to reach 50% germination. Data shown are means ± sd of three biological replicates. DW, Dry weight.
OsGA20ox2 Was Expressed Only in the Embryo of Germinating Seeds, and GA Application Accelerated the Release of Primary Dormancy Associated with qSD1-2
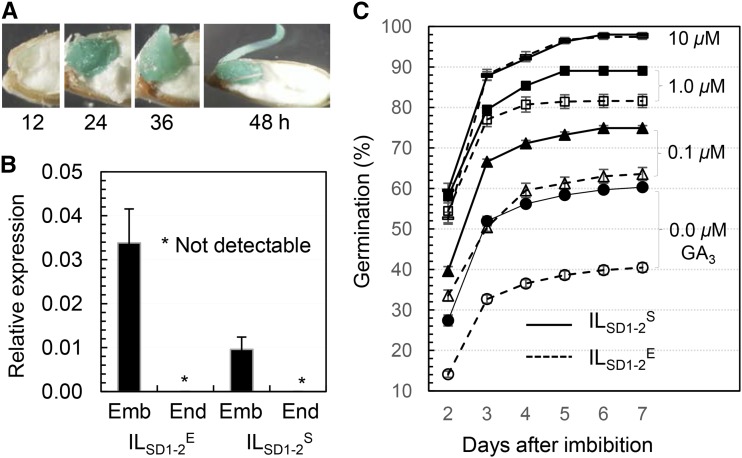
Both GUS activity and OsGA20ox2’s transcripts were detected in the embryo, but not the endosperm, of germinating seeds. The SD1-2promoter::GUS reporter system was activated in the embryo tissue after imbibition, and the activity became visible after 12 h and was extended to the rapidly growing radicle after the completion of germination (Fig. 8). The transcription level of OsGA20ox2 in the embryo tissue of seeds imbibed for 24 h was higher in ILSD1-2E than in ILSD1-2S (Fig. 8). This genotypic difference implies that OsGA20ox2 does not express in the aleuronic cells where the GA signaling pathway is triggered to promote germination.
The loss-of-function mutation of OsGA20ox2 lowered the GA content in the developing seeds (Fig. 6A) and is also expected to reduce the hormone in the embryo of germinating seeds. Thus, the mutant effect on delayed germination can be caused by the primary dormancy and/or by the reduced GA production in the embryo after imbibition. The effect due to the GA reduction in the embryo could be compensated by a GA treatment with the concentration equivalent to the endogenous GA level. To test this hypothesis, non-after-ripened seeds from ILSD1-2S and ILSD1-2E were germinated in 0.0, 0.1, 1, and 10 μm GA3. Genotypic differences in germination distribution pattern and germination percentage at day 7 were observed at 0.1 and 1 μm; with the increase in GA concentration, the difference diminished and finally disappeared at 10 μm (Fig. 9C). Provided that the GA concentration of 0.1 or 1 μm was higher than, or equivalent to, the embryonic GA level in ILSD1-2S, the observed genotypic difference in germinability could be an effect of the primary dormancy imposed by the tissue(s), including the endosperm. The requirement for a high GA concentration (approximately 10 μm) to completely offset the effect of qSD1-2 on germination was similar to the GA response of the GA-deficient mutant ga-1 in Arabidopsis (Koornneef and van der Veen, 1980). It is noted that the germination response of qSD1-2 to the 10 μm GA application occurred only in the genetic background where the qSD7-2 locus was fixed for the dormancy-reducing allele (Ye et al., 2013).
Figure 9.
Gene expression in germinating seeds and germination responses to GA applications. A, Histochemical localization of GUS reporter activity. Seeds from SD1-2promoter::GUS transgenic plants were imbibed for 12 to 48 h. B, Relative expression levels of OsGA20ox2 in embryo and endosperm tissues. Seeds from ILSD1-2E and ILSD1-2S plants were imbibed for 24 h; RNA samples prepared from the embryo (Emb) and endosperm (End) tissues were quantified for transcription levels by qRT-PCR. Data shown are means ± sd, relative to the ACTIN control, of three biological replicates. C, Genotypic differences in the GA response of germination. Seeds from ILSD1-2E and ILSD1-2S were germinated in the indicated GA3 solutions. Data shown are means ± se of 10 plants.
Origin, Distribution, and Functional Differentiation of SD1-2E, SD1-2S, and SD1-2N
The dormancy-enhancing allele SD1-2E was traced back to the semidwarf line IR1545339-2-2 through the pedigree of the donor parent EM93-1 (Supplemental Fig. S1A). Like the other semidwarf varieties developed by the International Rice Research Institute, IR1545339-2-2 carries the Green Revolution gene sd1 initially introduced from the traditional cv Dee-Geo-Woo-Gen (IRRI, 1981). Because sd1 has been used to develop high-yield semidwarf varieties worldwide since the 1960s, SD1-2E should be common in modern cultivars.
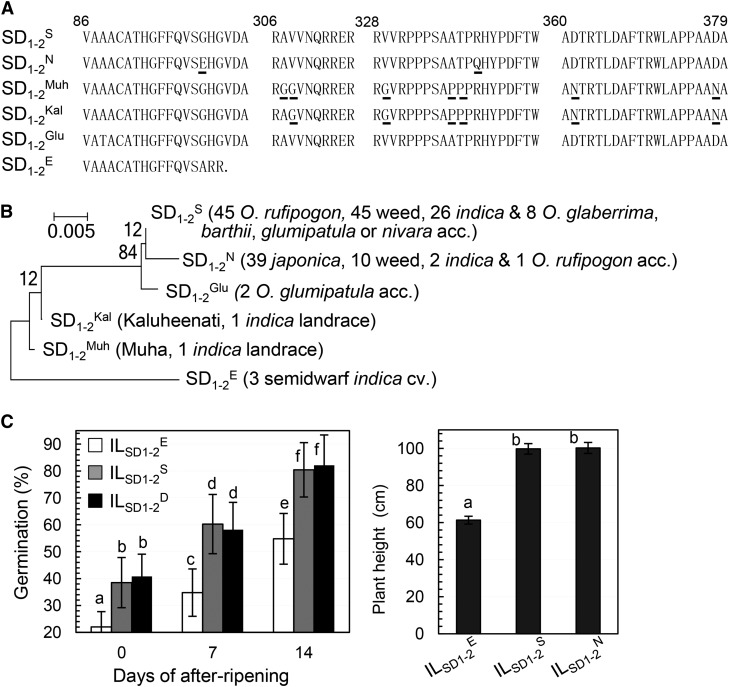
Phylogenetic analysis based on OsGA20ox2 genomic DNA sequences from 183 accessions (Supplemental Table S4) revealed that the allele SD1-2S from SS18-2 represents a primitive one in wild (Oryza rofipogon), indica-type cultivated, and indica-like weedy rice (Supplemental Fig. S3). The allele SD1-2D from cv Dongjin is identical to the allele from cv Nipponbare (SD1-2N) and belongs to a clade consisting of japonica cultivars and japonica-like weedy rice (Supplemental Fig. S3). Based on deduced protein sequences, the 183 accessions consist of six alleles, and the primitive (SD1-2S) and japonica specific (SD1-2N) alleles are present in 68% and 28% of the accessions, respectively (Fig. 10A).
Figure 10.
Allelic variants of OsGA20ox2 in protein sequences and phenotypes. A, Partial protein sequences. The six sequences represent deduced proteins in 183 accessions of wild, weedy, or cultivated rice (Supplemental Table S4; Supplemental Fig. S3) and are represented by the alleles from SS18-2 (SD1-2S), cv Nipponbare (SD1-2N), cv Muha (SD1-2Muh), cv Kaluheenati (SD1-2Kal), Oryza glumipatula (SD1-2Glu), and EM93-1 (SD1-2E). Underlined residues are different from the primitive allele SD1-2S. B, Protein dendrogram. Shown in parentheses are numbers of accessions (acc.) from Oryza spp., rice ssp. indica and japonica, or weed type. C, Genotypic differences in germinability and plant height. ILSD1-2E, ILSD1-2S, and ILSD1-2N are isogenic lines for the SD1-2E, SD1-2S, and SD1-2N alleles, respectively (Supplemental Fig. S1D). Data shown are means ± sd of 20 plants. The letters (a–f) indicate results of Duncan’s multiple comparison test for nonsignificance (same letter) or significance (different letter) at P < 0.0001.
Five of the six protein sequences (excluding SD1-2E) differentiated at 10 residues. The 100th (G) and 340th (R) residues in SD1-2S were substituted with E and Q, respectively, in SD1-2N (Fig. 10A). To determine if there is a functional differentiation between the two common alleles, we introduced SD1-2N into the EM93-1 background (Supplemental Fig. S1, C and D). The isogenic lines ILSD1-2N, ILSD1-2S, and ILSD1-2E were evaluated for seed dormancy and plant height under controlled conditions. Genotypic differences in both traits were present between ILSD1-2E and ILSD1-2S or ILSD1-2N but absent between ILSD1-2S and ILSD1-2N. The data from the isogenic background indicate that the GR-EQ differentiation between SD1-2S and SD1-2N has no effect on seed dormancy and plant height.
DISCUSSION
Natural Variants of GA Synthesis Genes Contribute to Genetic Variation in Seed Dormancy
The qSD1-2 QTL was identified as the OsGA20ox2 locus by map-based cloning and mutant analysis. There are four paralogs encoding GA 20-oxidase catalyzing the next to last steps of bioactive GA synthesis in rice (Sakamoto et al., 2004). Of these loci, OsGA20ox2 was reported for natural variants based on the mutant effect on reduced plant height, such as the allele sd1 widespread in semidwarf cultivars. This research demonstrated that a loss-of-function mutation of the GA synthesis gene, naturally occurring or induced, also enhances seed dormancy.
Discovery of the dormancy function explained the perpetuation of sd1 in populations of wild and weedy rice and also revealed that the Green Revolution gene has contributed to the resistance to preharvest sprouting (PHS) in crop production. The sd1 mutant originally occurred in an O. rufipogon population and may have been hidden in the genetic background of tall plants before introgression into traditional varieties (e.g. cv Dee-Geo-Woo-Gen), because short-statured genotypes are less competitive in natural populations (Nagano et al., 2005). However, the selective disadvantage that sd1 carries in plant height could be compromised by a gain in adaptability from the enhanced seed dormancy in a wild population. Similarly, because of the gained adaptability, if sd1 escapes from semidwarf cultivars to accompanying plants of weedy rice by cross-pollination, it would promote the persistence of sd1-containing weeds in the local ecosystems. As expected, sd1 has been found in newly established populations of weedy rice in the United States (Reagon et al., 2011). It is noticed that sd1 is also present in male-sterile lines of hybrid rice, and these lines usually have panicles partially enclosed in the leaf sheath (Virmani et al., 1997). Seed producers have to apply GA3 to promote the peduncle elongation, which could offset sd1’s dormancy effect and increase the susceptibility to PHS. Thus, additional dormancy genes are required to improve male-sterile lines for resistance to PHS.
Some other GA synthesis or signaling genes may also influence seed dormancy. Dozens of dwarf mutants assigned to the GA biosynthesis pathways in rice (Sakamoto et al., 2004) have not been determined for an effect on seed dormancy. Reasons may include that these mutants usually have detrimental effects on gametophyte development or seed setting, some may not express in developing seeds, and research was focused on postgermination events (Kaneko et al., 2003). Natural mutants used to develop semidwarf varieties in the barley (Hordeum vulgare), sorghum (Sorghum bicolor), and wheat crops usually have a mild negative effect on plant development, and some of them are GA sensitive (Gale and Marshall, 1975; Franckowiak and Pecio, 1992; Rodríguez et al., 2012). Some seed dormancy QTLs in barley and wheat have been associated with GA metabolism genes by comparative genomics research using rice as the reference (Li et al., 2004; Cabral et al., 2014). Thus, the information from this research would encourage exploring new functions of GA mutants to enrich knowledge on seed development and to test if orthologous genes are involved in the regulation of seed dormancy in different species.
A Developmental Mechanism of Seed Dormancy
qSD1-2 controls the variation of primary dormancy through a GA-regulated dehydration mechanism outlined in the working model (Fig. 11). The expression of OsGA20ox2 in seeds at early development increases the GA content to stimulate cell development and tissue morphogenesis, which in turn accelerates ABA accumulation to promote dehydration and the likely acquisition of desiccation tolerance. The loss-of-function mutation reduces the GA level, which slows down tissue morphogenesis, delays ABA accumulation and subsequent maturation programs, and eventually results in increased dormancy at harvest. The GA and ABA molecules signal and induce the early (cell elongation of the maternal tissues and endospermic cell differentiation) and late development events, respectively, rather than act antagonistically with each other, to influence moisture content and germinability. Given that dormancy is an adaptive mechanism for the embryo to survive in a dehydrated environment after physiological maturity to prevent vivipary or PHS, seed moisture would be critical for the acquisition of desiccation tolerance and the onset of after-ripening to release primary dormancy. However, in previous research, we detected no difference in the seed moisture content at 40 DPA between the isogenic lines for the qSD7-1 dormancy QTL in the EM93-1 background (Gu et al., 2011). qSD7-1 encodes a basic helix-loop-helix family transcription factor, which promotes ABA accumulation and activates flavonoid biosynthesis in the lower epidermal cell layer of the pericarp tissue to control maternal tissue-imposed dormancy (Gu et al., 2011). Therefore, the model (Fig. 11) represents an additional mechanism for the development of primary dormancy.
Figure 11.
A working model for the development of primary dormancy regulated by qSD1-2.
The association of qSD1-2 with endosperm-imposed dormancy was demonstrated by four distinct germination distribution patterns, each for one of the four triploid genotypes of seeds (F2) from the same plant (F1) that is heterozygous (SD1-2SSD1-2E) for the QTL (Gu et al., 2015). From a genetic point of view, the dormancy gene must express in the endosperm and have an additive effect on germination to display the four distribution patterns. As expected, SD1-2S was expressed in the triploid tissue (Fig. 5), promoted endosperm cell differentiation (Fig. 8C), and had only an additive effect on germination (Supplemental Table S2). From a physiological point of view, the endosperm tissue at the middle to late development stages accounts for a vast majority of the seed fresh weight and, accordingly, dominates the dehydration and desiccation programs to influence germination potential at harvest. Thus, the GA-regulated dehydration mechanism (Fig. 11) may apply only to endosperm-imposed dormancy.
Allelic Differentiation of OsGA20ox2 between the indica and japonica Subspecies
This research provided no evidence that the indica- and japonica-specific alleles at OsGA20ox2 are functionally differentiated. The allelic distribution was associated with the subspeciation of indica and japonica rice (Supplemental Fig. S3; Nagano et al., 2005; Asano et al., 2011). A discussion was focused on whether the nonrandom distribution arose from an artificial selection for reduced plant height early during the domestication of japonica rice (Asano et al., 2011; Paterson and Li, 2011). SD1-2S and SD1-2N are prevalent in the indica and japonica subspecies, respectively (Supplemental Fig. S3), and SD1-2S (GR) differs from SD1-2N (EQ) in two residues (Fig. 2, A and C). The similarity between the SD1-2S and SD1-2N isogenic lines in plant height and seed dormancy (Fig. 10C) suggests that both alleles are functional, and the two-residue substitution may not cause a discernible change in the gene function in the EM93-1 background. Considering the additive effect of qSD1-2 on plant height (Supplemental Table S2), it was surprising that the addition of an SD1-2N as a transgene to the cv Nipponbare background did not change the plant height (Asano et al., 2011). In any event, SD1-2S is identical to the allele (GR) from the indica landrace Kasalath, based on the genomic DNA sequence for SD1-2K reported by Reagon et al. (2011). The reported association between plant height and the sd1-containing segment of approximately 300 kb in the advanced generation of the cv Nipponbare/Kasalath cross (Asano et al., 2011) may arise from a functional differentiation of the OsGA20ox2 locus (including the noncoding sequences) between SD1-2K and SD1-2N or from a genetic differentiation outside the locus through an uncharacterized molecular mechanism. It is noticed that SD1-2S represents a more primitive/frequent allele than SD1-2K in the clade that consists of wild and weedy rice and indica cultivars (Supplemental Fig. S3). It is likely that SD1-2K, together with the rare alleles SD1-2Kal and SD1-2Mhl in the clade (Fig. 10, A and B), differentiated from SD1-2S after the divergence between SD1-2S and SD1-2N. Thus, Kasalath is not an ideal genotype to be used as the wild-type control to elucidate the evolutionary mechanism of the putatively domestication-related gene in japonica rice. Consequently, additional research is needed to address driving forces for the fixation of the japonica-specific allele and to determine if the rare alleles are functionally differentiated from SD1-2S and SD1-2N.
MATERIALS AND METHODS
Plant Materials and Cultivation
The rice (Oryza sativa) BC5F3 line used to develop the F2 population was selected from a recombinant that has only one chromosomal segment from SS18-2 in the EM93-1 background (Ye et al., 2013). The pedigree of EM93-1 (Supplemental Fig. S1A) was made based on information from Gu et al. (2004) and from Hanfu Qian, who developed the parent line Wujinxianxian at the Jiangsu Wujin Rice Research Institute. ILSD1-2N was developed by recurrent backcrossing with marker-assisted selection (Supplemental Fig. S1C) and the backcross initiated to introduce the Rc/SD7-1 transgene from cv Nipponbare into the EM93-1 background (Gu et al., 2011).
To develop plant populations, after-ripened seeds were germinated at 30°C for 5 d before being cultured with nutrition solution (Yoshida et al., 1976) in 200-cell plug trays for 2 to 3 weeks to sample the leaf tissue for marker genotyping. Seedlings with known genotypes for qSD1-2 or its flanking regions were transplanted into pots in a greenhouse to harvest seeds.
Progeny Testing and Seed Dormancy Assessment
Seedlings (F3) from selected recombinants (F2) were sorted based on marker genotypes and grown under temperature (29°C/21°C for day/night)-controlled conditions. Plants were tagged for flowering date when the first panicle of a plant emerged from the leaf sheath and measured for plant height at harvest. Seeds were harvested at 40 d after flowering and dried in the greenhouse for 3 d before being stored at −20°C in a freezer to maintain the dormant status.
Seed dormancy was evaluated by germination testing. Prior to a test, seed samples from the freezer were partially after-ripened at room temperature (approximately 24°C) for several hours to 30 d, depending on the experiment. A sample of approximately 50 well-developed seeds was distributed on filter paper in a 9-cm petri dish, soaked with 8 mL of water, and incubated at 30°C and 100% relative humidity in the dark. Germination was evaluated visually by protrusion of the radicle from the hull by more than 3 mm and counted daily from day 2 to day 7 or at day 7. A test for a plant at a time point was replicated three times, and germination percentages were averaged for genetic analysis.
Marker Development and Genotyping
The high-resolution map (Fig. 1A) was developed with simple sequence repeat (SSR) and SNP markers (Supplemental Table S1). Nine new SSRs were selected from the reference genome and screened for polymorphism between EM93-1 and SS18-2. Six genomic DNA fragments were sequenced from EM93-1 and SS18-2 and their sequences aligned to identify SNPs. Genomic DNA preparation, sequencing and sequence alignment, and genotyping with SSR markers were performed using previously described methods (Gu et al., 2011). Recombinant genotypes for the SNP loci were determined by PCR and direct sequencing.
Marker-Trait Association Analysis and Estimation of Genic Effects
Linear correlation analysis was used to determine the marker-trait associations in progeny lines. Genotypes for a marker locus were coded as i (i = 1, 2, and 3 for EM93-1-like homozygote, heterozygote, and SS18-2-like homozygote, respectively) for the correlation analysis. Additive and dominance effects of the locus on germination or plant height were estimated using the linear regression model:
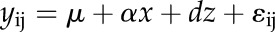 |
where yij is the trait value for the jth plant of the ith marker genotype; μ is the model mean; x is the dummy variable for the additive component and was coded as −1, 0, and 1 for i = 1, 2, and 3, respectively; z is the dummy variable for the dominance component and was coded as 0.5 for i = 2 or −0.5 for i = 1 or 3; a and d are regression coefficients and estimates of the additive and dominance effects, respectively; and εij is the error term of the model. Correlation and regression analyses were implemented using SAS (SAS Institute, 2011).
T-DNA Insertion Mutant Analysis
The mutant 3A-14927 (M1) and its recipient cv Dongjin were introduced from the Crop Biotech Institute, Kyung Hee University, in Korea. The M1 seedlings and thermal asymmetric interlaced-PCR were used to confirm the T-DNA insertion site at the 5′ UTR of Os01g66100 (Jeong et al., 2006). The knockout effect on Os01g66100’s transcription was determined by semiquantitative PCR. The PCR primers are listed in Supplemental Table S1. The M2 and M3 generations of plants were grown in the greenhouse and evaluated for seed dormancy and plant height.
BAC Library Screening, DNA and cDNA Cloning, Sequencing, and Sequence Analysis
Southern blotting was used to screen the BAC library constructed specifically for SS18-2 (Gu et al., 2010). Two 32P-labeled Overgo sequences designed to target the narrowed qSD1-2 region (Supplemental Table S2) were used to probe the library membranes. Positive clones were end sequenced for approximately 500 bp and sequences aligned against the reference genome to develop the contig. A positive clone covering the 20-Kb region was sequenced for the entire Os01g66100 and partial Os01g66110 loci with the listed primers (Supplemental Table S1). These primers were also used to clone Os01g66100 from EM93-1 and cv Dongjin by PCR to assemble SD1-2E and SD1-2D, respectively. The sequence assemblies were annotated with the RiceGAAS annotation system (Sakata et al., 2002) to predict the gene structures and also aligned by the SeqMan program (DNASTAR) to identify the allelic variation among SD1-2S, SD1-2E, and SD1-2D.
To isolate full-length cDNAs for the SD1-2S and SD1-2E alleles, total RNA samples were prepared from the ILSD1-2E and ILSD1-2S seedlings using TriReagent (Sigma-Aldrich) and reverse transcribed using SuperScript III First-Strand (Invitrogen). Two pairs of primers (Supplemental Table S2) were designed to clone the overlapped cDNA fragments (Fig. 2B), the fragments were sequenced, and the sequences were assembled to obtain full-length cDNAs. The cDNAs were aligned against their genomic DNA sequences to confirm the gene structures and also used to predict functional domains in deduced proteins using the Conserved Domain Database (Marchler-Bauer et al., 2013).
Transcriptional Analysis by qRT-PCR
To compare transcriptional profiles of the GA metabolism and signaling genes between the isogenic lines, three biological replicates of the ILSD1-2E and ILSD1-2S plants were grown in the greenhouse, and spikelets were labeled for anthesis dates to collect fresh caryopses at 3, 6, and 9 DPA in liquid N2. A sample of approximately 50 caryopses was used to prepare total RNA, and 1 µg of the RNA was reverse transcribed into cDNAs. PCR primers were designed based on the cv Nipponbare genome annotation release 7 (Kawahara et al., 2013). The primers and Power SYBR Green PCR Master Mix (Applied Biosystems) were used to amplify the cDNA templates in the ABI 7900HT Fast Real-Time PCR System. The ACTIN gene was used as the control.
Development of the SD1-2promoter::GUS System and Detection of the Reporter Gene in Endosperm
A sequence of 1.6 Kb covering the promoter and 5′ UTR of SD1-2S was ligated into pCXGUS-P vector (Chen et al., 2009), and the construct was delivered to the cv Nipponbare background using the Agrobacterium tumefaciens-mediated transformation system (http://agron-www.agron.iastate.edu/ptf/protocol/rice.pdf). To identify the reporter gene expression in the endosperm tissue, T1 plants as the pollen parent were crossed with cv Nipponbare to obtain hybrid F1 seeds. Developing seeds at 3, 6, and 9 DPA and mature seeds imbibed for 12, 24, 36, and 48 h were stained and destained following the protocol of Jefferson et al. (1987) and photographed using a stereomicroscope.
Measurement of Plant Hormone Metabolites in Developing Seeds
To have a snapshot of metabolic profiles of the plant hormones involved in early seed development, 200 spikelets sampled from ILSD1-2E and ILSD1-2S plants at 5 DPA had their hulls removed and fresh caryopses freeze dried to measure the ABA, auxin, cytokinin, and GA metabolites. The measurement was conducted using an HPLC-electrospray ionization-tandem mass spectrometry system following published procedures (Ross et al., 2004; Lulsdorf et al., 2013) at the National Research Council Plant Biotechnology Institute, Canada.
To identify temporal distribution patterns of the GA and ABA hormones, spikelets from ILSD1-2E and ILSD1-2S plants were sampled at the indicated DPA to collect caryopses at −80°C. A sample of 50 caryopses was used to prepare and purify extracts using methods modified from Chen and Yang (2005). The hormones were quantified using HPLC-mass spectrometry with an external standard. The chromatographic analysis was performed in a Dubhe C18 column (4.6 mm × 250 mm, 5 μm) at 30°C with detection wavelength at 254 nm. A solution containing 5% (v/v) acetonitrile, 50% (v/v) methanol, and 0.27% (v/v) acetic acid was used as the mobile phase in the analytical system. The washing procedure was 0.6% (v/v) acetic acid at 0 min and 0.3% (v/v) acetic acid in 50% (v/v) methanol at 10 and 30 min. For a test, 10 μL of the extract was injected, and the recovery rate was 96.5% ± 2.6%. The test was repeated four times.
Histochemical Analysis
The histological experiment was conducted basically following the protocol of Luna (1968). Intact seeds from ILSD1-2E and ILSD1-2S plants were sampled at the indicated DPA, and fresh caryopses were fixed with 4% (w/v) paraformaldehyde solution before embedding in Paraplast Plus (Sigma-Aldrich). Longitudinal microtome sections of 8 to 10 µm thick were stained with hematoxylin and eosin (Sigma-Aldrich) and imaged using an Olympus AX70 upright compound microscope.
Tests of Moisture Content and Germinability of Developing Seeds
To identify the genotypic differences in dehydration and germinability acquisition during seed development, three biological replicates of the ILSD1-2E and ILSD1-2S plants, 24 plants per replicate, were grown in the greenhouse, spikelets on the main tillers were marked for anthesis dates, and 200 to 250 seeds were sampled from each replicate at the indicated DPA and allocated into two bags. One bag of seeds was weighed immediately after harvesting and then dried at 105°C for 72 h to measure dry weight to calculate the moisture content. The other bag of seeds was air dried for 7 d and germinated at 30°C and 100% relative humidity in the dark.
Phylogenetic Analysis
The phylogenetic tree (Supplemental Fig. S3) was generated based on genomic DNA sequences of OsGA20ox2 from 183 accessions of wild (Oryza spp.), weedy, or cultivated rice. Information about geographical origins and types of these accessions and sequence identifiers are listed in Supplemental Table S4. Protein sequences were deduced from coding sequences of the gene model (Fig. 2C). Dendrograms for genomic or protein sequences were generated using the maximum likelihood method with partial deletion after ClustalW alignment with MEGA 6.06 software (Tamura et al., 2013). Bootstrap values (%) were obtained by 500 replicates.
Sequence data used for this article were submitted to GenBank under accession numbers KT354861 to KT354865.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Pedigrees and development of plant genotypes for this research.
Supplemental Figure S2. Temporal distribution patterns for seed moisture content and germination capability in ILSD1-2S and ILSD1-2E.
Supplemental Figure S3. Dendrogram of 183 rice accessions for the OsGA20ox2 locus.
Supplemental Table S1. List of PCR primers used in this research.
Supplemental Table S2. Summary of genic effects of qSD1-2 on germination and plant height in recombinant-derived progeny lines.
Supplemental Table S3. Summary of hormone metabolites in 5-d developing caryopses from ILSD1-2E and ILSD1-2S.
Supplemental Table S4. List of accessions of rice germplasm for phylogenetic analysis of OsGA20ox2.
Supplementary Material
Acknowledgments
We thank Bradley Carsrud, Matthew Josephson, and Zhiqin Wang for technical assistance and Dr. Sharon Clay for help in analysis of the GA distribution pattern.
Glossary
- QTL
quantitative trait locus
- ABA
abscisic acid
- Kb
kg bp
- BAC
bacterial artificial chromosome
- UTR
untranslated region
- cDNA
complementary DNA
- T-DNA
transfer DNA
- SNP
single-nucleotide polymorphism
- qRT
quantitative real-time
- PHS
preharvest sprouting
- SSR
simple sequence repeat
Footnotes
This work was supported by the National Science Foundation (grant nos. IOS 1021382 and IOS 0641376), the U.S. Department of Agriculture National Research Initiative (grant no. 2008–35301–19058) and National Institute of Food and Agriculture (grant no. 2013–03572), and the South Dakota Agricultural Extension Station.
References
- Agrawal GK, Yamazaki M, Kobayashi M, Hirochika R, Miyao A, Hirochika H (2001) Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion: tagging of a zeaxanthin epoxidase gene and a novel OSTATC gene. Plant Physiol 125: 1248–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M (2003) Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164: 711–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiguet-Vercher A, Santuari L, Gonzalez-Guzman M, Depuydt S, Rodriguez PL, Hardtke CS (2015) The IBO germination quantitative trait locus encodes a phosphatase 2C-related variant with a nonsynonymous amino acid change that interferes with abscisic acid signaling. New Phytol 205: 1076–1082 [DOI] [PubMed] [Google Scholar]
- Anderson JA, Sorrells ME, Tanksley SD (1993) RFLP analysis of genomic regions associated with resistance to preharvest sprouting in wheat. Crop Sci 33: 453–459 [Google Scholar]
- Angelovici R, Galili G, Fernie AR, Fait A (2010) Seed desiccation: a bridge between maturation and germination. Trends Plant Sci 15: 211–218 [DOI] [PubMed] [Google Scholar]
- Argyris J, Truco MJ, Ochoa O, Knapp SJ, Still DW, Lenssen GM, Schut JW, Michelmore RW, Bradford KJ (2005) Quantitative trait loci associated with seed and seedling traits in Lactuca. Theor Appl Genet 111: 1365–1376 [DOI] [PubMed] [Google Scholar]
- Asano K, Yamasaki M, Takuno S, Miura K, Katagiri S, Ito T, Doi K, Wu J, Ebana K, Matsumoto T, et al. (2011) Artificial selection for a Green Revolution gene during japonica rice domestication. Proc Natl Acad Sci USA 108: 11034–11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103: 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Bradford K, Hilhorst H, Nonogaki H (2013) Seeds: Physiology of Development, Germination and Dormancy, Ed 3 Springer, New York [Google Scholar]
- Blaker KM, Chaparro JX, Beckman TG (2013) Identification of QTLs controlling seed dormancy in peach (Prunus persica). Tree Genet Genomes 9: 659–668 [Google Scholar]
- Cabral AL, Jordan MC, McCartney CA, You FM, Humphreys DG, MacLachlan R, Pozniak CJ (2014) Identification of candidate genes, regions and markers for pre-harvest sprouting resistance in wheat (Triticum aestivum L.). BMC Plant Biol 14: 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Songkumarn P, Liu J, Wang GL (2009) A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol 150: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YP, Yang WY (2005) Determination of GA3, IAA, ABA and ZT in dormant buds of Allium ovalifolium by HPLC. J Sichuan Agric Univ 23: 498–500 [Google Scholar]
- Fang J, Chai C, Qian Q, Li C, Tang J, Sun L, Huang Z, Guo X, Sun C, Liu M, et al. (2008) Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J 54: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennimore SA, Nyquist WE, Shaner GE, Doerge RW, Foley ME (1999) A genetic model and molecular markers for wild oat (Avena fatua L.) seed dormancy. Theor Appl Genet 99: 711–718 [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Franckowiak JD, Pecio A (1992) Coordinator’s report. Semi-dwarf gene: a listing of genetic stocks. Barley Genet Newsl 21: 116–126 [Google Scholar]
- Gale MD, Marshall GA (1975) The nature and genetic control of gibberellin insensitivity in dwarf wheat grain. Heredity 35: 55–65 [Google Scholar]
- Gandhi SD, Heesacker AF, Freeman CA, Argyris J, Bradford K, Knapp SJ (2005) The self-incompatibility locus (S) and quantitative trait loci for self-pollination and seed dormancy in sunflower. Theor Appl Genet 111: 619–629 [DOI] [PubMed] [Google Scholar]
- Gu XY, Foley ME, Chen ZX (2004) A set of three genes regulates photoperiodic responses of flowering in rice (Oryza sativa). Genetica 122: 127–140 [DOI] [PubMed] [Google Scholar]
- Gu XY, Foley ME, Horvath DP, Anderson JV, Feng J, Zhang L, Mowry CR, Ye H, Suttle JC, Kadowaki K, et al. (2011) Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice. Genetics 189: 1515–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu XY, Kianian SF, Hareland GA, Hoffer BL, Foley ME (2005) Genetic analysis of adaptive syndromes interrelated with seed dormancy in weedy rice (Oryza sativa). Theor Appl Genet 110: 1108–1118 [DOI] [PubMed] [Google Scholar]
- Gu XY, Liu T, Feng J, Suttle JC, Gibbons J (2010) The qSD12 underlying gene promotes abscisic acid accumulation in early developing seeds to induce primary dormancy in rice. Plant Mol Biol 73: 97–104 [DOI] [PubMed] [Google Scholar]
- Gu XY, Zhang J, Ye H, Zhang L, Feng J (2015) Genotyping of endosperms to determine seed dormancy genes regulating germination through embryonic, endospermic, or maternal tissues in rice. G3 (Bethesda) 5: 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Watts RJ, Kalla R, Matthews P, Keys M, Jacobsen JV (1997) Cloning of a rice cDNA encoding a transcription factor homologous to barley GAMyb. Plant Cell Physiol 38: 362–365 [DOI] [PubMed] [Google Scholar]
- Huo H, Dahal P, Kunusoth K, McCallum CM, Bradford KJ (2013) Expression of 9-cis-EPOXYCAROTENOID DIOXYGENASE4 is essential for thermoinhibition of lettuce seed germination but not for seed development or stress tolerance. Plant Cell 25: 884–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IRRI (1981) Research Highlights for 1980. International Rice Research Institute, Los Banos, Philippines [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, et al. (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45: 123–132 [DOI] [PubMed] [Google Scholar]
- Kaneko M, Itoh H, Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Ashikari M, Matsuoka M (2003) Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J 35: 104–115 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Kovi MR, Zhang Y, Yu S, Yang G, Yan W, Xing Y (2011) Candidacy of a chitin-inducible gibberellin-responsive gene for a major locus affecting plant height in rice that is closely linked to Green Revolution gene sd1. Theor Appl Genet 123: 705–714 [DOI] [PubMed] [Google Scholar]
- Kucera B, Cohn MA, Leubner-Metzger G (2005) Plant hormone interactions during seed dormancy release and germination. Seed Sci Res 15: 281–307 [Google Scholar]
- Li C, Ni P, Francki M, Hunter A, Zhang Y, Schibeci D, Li H, Tarr A, Wang J, Cakir M, et al. (2004) Genes controlling seed dormancy and pre-harvest sprouting in a rice-wheat-barley comparison. Funct Integr Genomics 4: 84–93 [DOI] [PubMed] [Google Scholar]
- Lijavetzky D, Marinez MC, Carrari F, Hopp E (2000) QTL analysis and mapping of pre-harvest sprouting resistance in sorghum. Euphytica 112: 125–135 [Google Scholar]
- Lin SY, Sasaki T, Yano M (1998) Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor Appl Genet 96: 997–1003 [Google Scholar]
- Liu S, Sehgal SK, Li J, Lin M, Trick HN, Yu J, Gill BS, Bai G (2013) Cloning and characterization of a critical regulator for preharvest sprouting in wheat. Genetics 195: 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulsdorf MM, Yuan HY, Slater SMH, Vandenberg A, Han X, Zaharia LI, Abrams SR (2013) Endogenous hormone profiles during early seed development of C. arietinum and C. anatolicum. J Plant Growth Regul 71: 191–198 [Google Scholar]
- Luna LG. (1968) Manual of Histologic Staining Methods of the Armed Forces Institute of Pathology. McGraw-Hill, New York [Google Scholar]
- Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, et al. (2013) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41: D348–D352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Andújar C, Pluskota WE, Bassel GW, Asahina M, Pupel P, Nguyen TT, Takeda-Kamiya N, Toubiana D, Bai B, Górecki RJ, et al. (2012) Mechanisms of hormonal regulation of endosperm cap-specific gene expression in tomato seeds. Plant J 71: 575–586 [DOI] [PubMed] [Google Scholar]
- Masojć P, Banek-Tabor A, Milczarski P, Twardowska M (2007) QTLs for resistance to preharvest sprouting in rye (Secale cereale L.). J Appl Genet 48: 211–217 [DOI] [PubMed] [Google Scholar]
- Middleton AM, Úbeda-Tomás S, Griffiths J, Holman T, Hedden P, Thomas SG, Phillips AL, Holdsworth MJ, Bennett MJ, King JR, et al. (2012) Mathematical modeling elucidates the role of transcriptional feedback in gibberellin signaling. Proc Natl Acad Sci USA 109: 7571–7576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mispan MS, Zhang L, Feng J, Gu XY (2013) Quantitative trait locus and haplotype analyses of wild and crop-mimic traits in U.S. weedy rice. G3 (Bethesda) 3: 1049–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Tintelnot S, Leubner-Metzger G (2006) Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol 47: 864–877 [DOI] [PubMed] [Google Scholar]
- Nagano H, Onishi K, Ogasawara M, Horiuchi Y, Sano Y (2005) Genealogy of the “Green Revolution” gene in rice. Genes Genet Syst 80: 351–356 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, Utsugi S, Ogawa T, Handa H, Ishida H, et al. (2011) A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23: 3215–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Li ZK (2011) Paleo-Green Revolution for rice. Proc Natl Acad Sci USA 108: 10931–10932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagon M, Thurber CS, Olsen KM, Jia Y, Caicedo AL (2011) The long and the short of it: SD1 polymorphism and the evolution of growth trait divergence in U.S. weedy rice. Mol Ecol 20: 3743–3756 [DOI] [PubMed] [Google Scholar]
- Robertson DS. (1955) The genetics of vivipary in maize. Genetics 40: 745–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez MV, Mendiondo GM, Cantoro R, Auge GA, Luna V, Masciarelli O, Benech-Arnold RL (2012) Expression of seed dormancy in grain sorghum lines with contrasting pre-harvest sprouting behavior involves differential regulation of gibberellin metabolism genes. Plant Cell Physiol 53: 64–80 [DOI] [PubMed] [Google Scholar]
- Ross AR, Ambrose SJ, Cutler AJ, Feurtado JA, Kermode AR, Nelson K, Zhou R, Abrams SR (2004) Determination of endogenous and supplied deuterated abscisic acid in plant tissues by high-performance liquid chromatography-electrospray ionization tandem mass spectrometry with multiple reaction monitoring. Anal Biochem 329: 324–333 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al. (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata K, Nagamura Y, Numa H, Antonio BA, Nagasaki H, Idonuma A, Watanabe W, Shimizu Y, Horiuchi I, Matsumoto T, et al. (2002) RiceGAAS: an automated annotation system and database for rice genome sequence. Nucleic Acids Res 30: 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute (2011) Base SAS 9.3 Procedures Guide. SAS Institute, Cary, NC [Google Scholar]
- Schatzki J, Schoo B, Ecke W, Herrfurth C, Feussner I, Becker HC, Möllers C (2013) Mapping of QTL for seed dormancy in a winter oilseed rape doubled haploid population. Theor Appl Genet 126: 2405–2415 [DOI] [PubMed] [Google Scholar]
- Steber CM. (2007) De-repression of seed germination by GA signaling. In Bradford K, Nonogaki H, eds, Seed Development, Dormancy and Germination. Blackwell Publishing, London, pp 248–263 [Google Scholar]
- Sugimoto K, Takeuchi Y, Ebana K, Miyao A, Hirochika H, Hara N, Ishiyama K, Kobayashi M, Ban Y, Hattori T, et al. (2010) Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc Natl Acad Sci USA 107: 5792–5797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Aya K, Ueguchi-Tanaka M, Shimada Y, Nakazono M, Watanabe R, Nishizawa NK, Gomi K, Shimada A, Kitano H, et al. (2006) GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J 47: 427–444 [DOI] [PubMed] [Google Scholar]
- Ullrich SE, Hayes PM, Dyer WE, Blake TK, Clancy JA (1993) Quantitative trait locus analysis of seed dormancy in ‘Steptoe’ barley. In Walker-Simmons MK, Ried JL, eds, Pre-Harvest Sprouting in Cereals. American Association of Cereal Chemists, St Paul, pp 136–145 [Google Scholar]
- Virmani SS, Viraktamath BC, Casal CL, Toledo RS, Lopez MT, Manalo JO (1997) Hybrid Rice Breeding Manual. International Rice Research Institute, Los Banos, Philippines [Google Scholar]
- White CN, Rivin CJ (2000) Gibberellins and seed development in maize: II. Gibberellin synthesis inhibition enhances abscisic acid signaling in cultured embryos. Plant Physiol 122: 1089–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Kamiya Y, Nambara E (2007) Regulation of ABA and GA levels during seed development and maturation in Arabidopsis. In Bradford K, Nonogaki H, eds, Seed Development, Dormancy and Germination. Blackwell Publishing, London, pp 224–247 [Google Scholar]
- Ye H, Beighley DH, Feng J, Gu XY (2013) Genetic and physiological characterization of two clusters of quantitative trait loci associated with seed dormancy and plant height in rice. G3 (Bethesda) 3: 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Foley ME, Gu XY (2010) New seed dormancy loci detected from weedy rice-derived advanced populations with major QTL alleles removed from the background. Plant Sci 179: 612–619 [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory Manual for Physiological Studies of Rice, Ed 3 International Rice Research Institute, Manila, Philippines [Google Scholar]
- Zhang F, Jiang YZ, Yu SB, Ali J, Paterson AH, Khush GS, Xu JL, Gao YM, Fu BY, Lafitte R, et al. (2013) Three genetic systems controlling growth, development and productivity of rice (Oryza sativa L.): a reevaluation of the ‘Green Revolution’. Theor Appl Genet 126: 1011–1024 [DOI] [PubMed] [Google Scholar]
- Zhang H, Tan G, Yang L, Yang J, Zhang J, Zhao B (2009) Hormones in the grains and roots in relation to post-anthesis development of inferior and superior spikelets in japonica/indica hybrid rice. Plant Physiol Biochem 47: 195–204 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.