Abstract
Arterial tortuosity syndrome (ATS) is an autosomal recessive connective tissue disorder caused by loss-of-function mutations in SLC2A10, which encodes facilitative glucose transporter 10 (GLUT10). The role of GLUT10 in ATS pathogenesis remains an enigma, and the transported metabolite(s), i.e. glucose and/or dehydroascorbic acid, have not been clearly elucidated. To discern the molecular mechanisms underlying the ATS aetiology, we performed gene expression profiling and biochemical studies on skin fibroblasts. Transcriptome analyses revealed the dysregulation of several genes involved in TGFβ signalling and extracellular matrix (ECM) homeostasis as well as the perturbation of specific pathways that control both the cell energy balance and the oxidative stress response. Biochemical and functional studies showed a marked increase in ROS-induced lipid peroxidation sustained by altered PPARγ function, which contributes to the redox imbalance and the compensatory antioxidant activity of ALDH1A1. ATS fibroblasts also showed activation of a non-canonical TGFβ signalling due to TGFBRI disorganization, the upregulation of TGFBRII and connective tissue growth factor, and the activation of the αvβ3 integrin transduction pathway, which involves p125FAK, p60Src and p38 MAPK. Stable GLUT10 expression in patients' fibroblasts normalized redox homeostasis and PPARγ activity, rescued canonical TGFβ signalling and induced partial ECM re-organization. These data add new insights into the ATS dysregulated biological pathways and definition of the pathomechanisms involved in this disorder.
Introduction
Arterial tortuosity syndrome (ATS; OMIM #208050) is a rare autosomal recessive connective tissue disorder (CTD) characterized by the tortuosity and elongation of large- and medium-sized arteries. The main cardiovascular features include arterial and pulmonary valve stenosis, a propensity towards aneurysm formation and vascular dissection, aberrant origin of aortic branches, and vasomotor instability. ATS also shows additional signs shared with other CTDs, including soft/velvety/hyperextensible skin, mild dysmorphic facial features, i.e. elongated face, hypertelorism, cleft palate and/or bifid uvula, and micro/retrognathia, abdominal hernias, joint hypermobility and instability (1–8).
ATS is caused by mutations in the SLC2A10 gene, which encodes the 541-amino acid facilitative glucose transporter 10 (GLUT10) (3). GLUT10 belongs to the SLC2A transporter family, the members of which facilitate uptake of several substrates, including glucose, galactose, fructose, mannose, glucosamine, urate and dehydroascorbic acid (DAA) (9,10). In vitro-grown ATS skin fibroblasts and aortic vascular smooth muscle cells (AVSMCs) do not organize the GLUT10 in plasma and intracellular membranes (3).
Twenty-three SLC2A10 gene mutations have been reported (8). Although the pathogenic mechanism underlying the SLC2A10 mutations is clearly a loss of function, the specific role of GLUT10 in the ATS aetiology remains an enigma. Mouse models failed to recapitulate the ATS vascular phenotype because vascular abnormalities were absent in mice with Slc2a10 homozygous missense substitutions (11,12). The zebrafish model better highlighted the essential role of GLUT10 in cardiovascular development because knockdown of the slc2a10 orthologue, which is broadly expressed during embryogenesis (13), was associated with several cardiovascular defects (14).
Loss of function of SLC2A10 activates TGFβ signalling (3,15,16). Induction of the TGFβ pathway was indicated by the enhanced expression of the connective tissue growth factor (CTGF) in ATS arterial tissue and by the reduced expression of decorin (DCN), which is a TGFβ signalling inhibitor proteoglycan, in in vitro cultured ATS cells (3). ATS is associated with abnormal collagens (COLLs) and elastin (ELN) synthesis (3), due to the key role of TGFβ signalling in extracellular matrix (ECM) fibrillogenesis and homeostasis. The importance of GLUT10 in facilitating TGFβ signalling was also indicated by comparing slc2a10 zebrafish morphants with TGFBR1 receptor-inhibited embryos, which are phenocopies that exhibit the same cardiovascular abnormalities, such as a wavy notochord as well as incomplete and irregular vascular patterning (14).
In the last years, increasing evidence indicates that GLUT10 might act as an intracellular transporter of DAA, which is the oxidized form of ascorbic acid (AA) (10,17,18). As an antioxidant, AA protects against oxidative stress-induced cellular damage by scavenging free radicals and reactive oxygen species (ROS), neutralizing by a vitamin E-dependent way lipid hydroperoxyl radicals, and protecting proteins from alkylation using electrophilic lipid peroxidation (LPO) products (19). Furthermore, AA is an essential cofactor for α-ketoglutarate-dependent dioxygenases (10). Segade used rat aortic smooth muscle cells to show GLUT10 colocalization with the rough endoplasmic reticulum (ER), wherein proline and lysine residues are hydroxylated, and COLLs and ELN are prepared for secretion by the Golgi apparatus. Thus, the author hypothesized that the loss of GLUT10 in ATS decreases DAA uptake by the ER, which yields an inadequate level of available AA for prolyl and lysyl hydroxylases inside the ER as well as synthesis and extracellular deposition of abnormal COLL and ELN (17). AA regulates the activity of many AA-dependent dioxygenases also in the nucleus, such as ten-eleven translocation demethylases and the Jumonji protein family, which catalyse epigenetically relevant reactions, including nucleic acid demethylation and histone demethylation/hydroxylation (10). Based on these data, Bánhegyi and coworkers hypothesized that DAA could enter the nucleus from a pool of DAA in the nuclear envelope (NE), which is a subdomain of the ER, through GLUT10, which is located either in the ER membrane or the inner membrane of the NE (10). In ATS, abnormal AA nuclear homeostasis, due to the lack of GLUT10, should lead to altered gene expression.
Lee and colleagues showed that GLUT10 localized to mitochondria in mouse AVSMCs, where it enhanced DAA uptake, which was accompanied by a decrease in ROS levels in H2O2-treated cells, and suggested that GLUT10 protects cells from ROS-induced vascular damage (18). Despite these findings, a recent study suggested that GLUT10 is not likely to be present in mitochondria because this transporter got far the lowest mitochondrial localization scores using in silico prediction tools (20). Moreover, mitochondrial AA transport should not depend on GLUT10 because it is mainly mediated by a low-affinity form of the sodium-coupled ascorbic acid transporter-2 (21). Overall, the dependence of AA availability in the ER, in the nucleus, and in the mitochondria on GLUT10 activity and its relevance to ATS remains to be demonstrated.
For insight into the ATS pathomechanisms, we used skin fibroblasts from healthy donors and ATS patients, grown in medium containing fetal bovine serum (FBS) as the only source of AA, in a combined approach using transcriptome, biochemical and functional studies. Our results revealed that the GLUT10 deficiency alters energetic/redox homeostasis and activates non-canonical TGFβ signalling, which involves TGFBRII, and the αvβ3 integrin transduction pathway, in association with the ECM disarray.
Results
Lack of GLUT10 expression is associated with the disarray of several structural ECM components in ATS fibroblasts
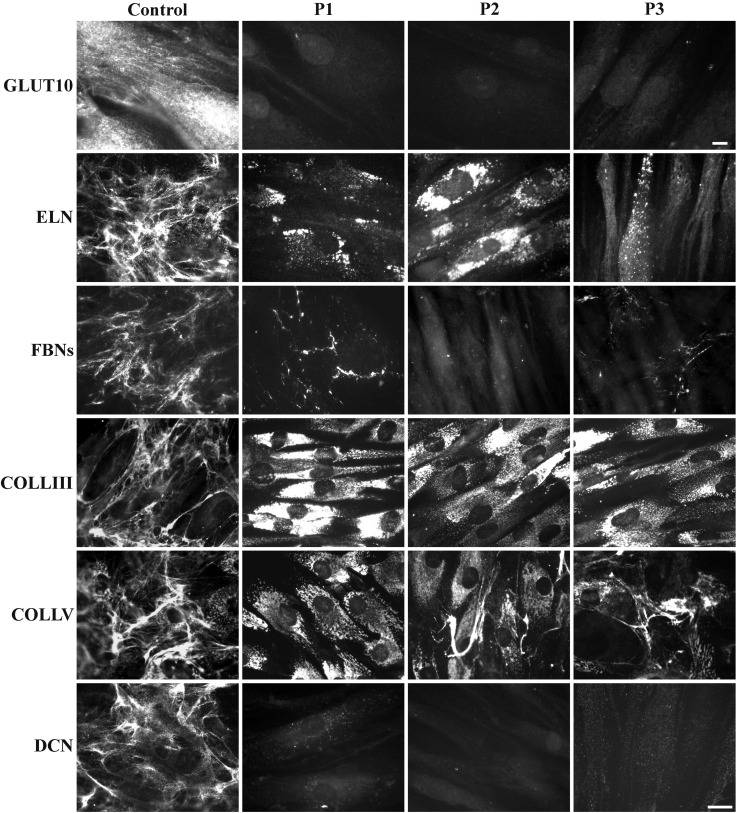
Three ATS fibroblast strains derived from patients with different SLC2A10 mutations were analysed for GLUT10 expression. Indirect immunofluorescence (IF) analysis showed that the protein was abundantly expressed by control skin fibroblasts, whereas it was undetectable in the ATS cells regardless of the SLC2A10 mutation type (Fig. 1). Western blotting (WB) analysis confirmed these results (data not shown). To validate the association of GLUT10 deficiency with ECM disarray, we used IF to investigate the organization of different structural proteins, type III COLL (COLLIII), type V COLL (COLLV), ELN, all of the fibrillin isoforms (FBNs) and DCN. As shown in Figure 1, ELN was organized in a fibrillar network covering 7-day-grown control fibroblasts, whereas it was not assembled into the ECM of ATS cells, which retained this protein at different levels in the cytoplasm also after 15 days of culture (data not shown). ATS fibroblasts did not organize FBNs, COLLIII, COLLV and DCN in the ECM. In particular, only a few FBN fibrils were localized in the intercellular spaces; all of the COLLs investigated were retained in the cytoplasm, and DCN was almost undetectable in all of the ATS patients' cells. These results demonstrate that GLUT10 deficiency is associated with the disassembly of several ECM components, which highlights the key role of this transporter in ECM homeostasis.
Figure 1.
Expression of GLUT10 and ECM organization in control and ATS fibroblasts. IF of GLUT10 in control and ATS fibroblasts grown for 48 h and immunoreacted with anti-GLUT10 Ab. Scale bar, 4 µm. IF of ELN, FBNs, COLLIII, COLLV and the core protein of DCN organized in the ECM was performed for the control and ATS fibroblasts 48 h after seeding or 7 days after seeding for ELN using the specific Abs. Scale bar: 10.8 µm. The experiments were repeated three times. The images are representative of two control (C1 and C2) and the three ATS cell strains (P1, P2, and P3).
Transcriptome-wide expression profiling of ATS skin fibroblasts shows differentially expressed genes that influence lipid metabolism, intracellular redox homeostasis and ECM maintenance
To screen for candidate genes that may contribute to ATS pathogenesis, we performed a transcriptome-wide expression analysis, wherein we compared the gene expression patterns of three ATS patients' skin fibroblasts with those of fibroblasts from three healthy individuals. We identified 217 differentially expressed genes (DEGs) in the patients by applying a fold-change threshold greater than or equal to ±1.5 with an uncorrected P < 0.05. In particular, 112 genes were significantly upregulated, and 105 were significantly downregulated (Supplementary Material, Table S1). Table 1 shows a selection of the up- and downregulated transcripts. A scatter plot for all of the microarray data and a heatmap for the DEGs are shown in Supplementary Material, Figure S1. Although fibroblasts from only three control subjects and three ATS patients were investigated, this analysis showed the presence of two distinct clusters of transcripts that clearly distinguish the mutant from control cells, which indicates relative low variability within each group and demonstrates that our findings are statistically significant.
Table 1.
Selection of DEGs in ATS fibroblasts.
| RefSeq | Gene symbol | Gene description | Fold change |
|---|---|---|---|
| Upregulated genes | |||
| NM_000689 | ALDH1A1 | Aldehyde dehydrogenase 1 family, member A1 | 9.83738 |
| NM_005159 | ACTC1 | Actin, alpha, cardiacmuscle 1 | 4.75185 |
| NM_018099 | FAR2 | Fatty-acyl-CoA reductase 2 | 4.50713 |
| NM_000965 | RARB | Retinoic acid receptor, beta | 3.87470 |
| NM_003206 | TCF21 | Transcription factor 21 | 3.84177 |
| NM_000597 | IGFBP2 | Insulin-like growth factor-binding protein 2 | 3.69562 |
| NM_006547 | IGF2BP3 | Insulin-like growth factor 2 mRNA-binding protein 3 | 3.64178 |
| NM_024420 | PLA2G4A | Phospholipase A2, group IVA (cytosolic, calcium-dependent) | 3.56846 |
| NM_138712 | PPARG | Peroxisome proliferator-activated receptor gamma | 3.43780 |
| NM_032961 | PCDH10 | Protocadherin 10 | 3.32056 |
| NM_000921 | PDE3A | Phosphodiesterase 3A, cGMP-inhibited | 3.03603 |
| NM_181458 | PAX3 | Paired box 3 | 3.02732 |
| NM_133459 | CCBE1 | Collagen and calcium-binding EGF domains 1 | 2.95478 |
| NM_001957 | EDNRA | Endothelin receptor type A | 2.93793 |
| NM_021110 | COL14A1 | Collagen, type XIV, alpha 1 | 2.50414 |
| NM_001144996 | ITGA7 | Integrin, alpha 7 | 2.30934 |
| NM_021219 | JAM2 | Junctional adhesion molecule 2 | 2.21089 |
| NM_206943 | LTBP1 | Latent transforming growth factor beta-binding protein 1 | 2.18537 |
| NM_004265 | FADS2 | Fatty acid desaturase 2 | 2.06592 |
| NM_145176 | SLC2A12 | Solute carrier family 2 (facilitated glucose transporter), member 12 | 1.88461 |
| NM_005328 | HAS2 | Hyaluronan synthase 2 | 1.80415 |
| NM_000669 | ADH1C | Alcohol dehydrogenase 1C (class I), gamma polypeptide | 1.79118 |
| NM_001080508 | TBX18 | T-box 18 | 1.78918 |
| NM_002207 | ITGA9 | Integrin, alpha 9 | 1.70987 |
| Downregulated genes | |||
| NM_181486 | TBX5 | T-box 5 | −5.38536 |
| NM_003326 | TNFSF4 | Tumour necrosis factor (ligand) superfamily, member 4 | −4.52814 |
| NM_002016 | FLG | Filaggrin | −3.53054 |
| NM_000561 | GSTM1 | Glutathione S-transferase mu 1 | −3.42929 |
| NM_000095 | COMP | Cartilage oligomeric matrix protein | −3.13002 |
| NM_000600 | IL6 | Interleukin 6 (interferon, beta 2) | −2.98091 |
| NM_003266.3 | TLR4 | Toll-like receptor 4 | −2.80100 |
| NM_198148 | CPXM2 | Carboxypeptidase X (M14 family), member 2 | −2.74518 |
| NM_000782 | CYP24A1 | Cytochrome P450, family 24, subfamily A, polypeptide 1 | −2.68277 |
| NM_016352 | CPA4 | Carboxypeptidase A4 | −2.67307 |
| NM_004613 | TGM2 | Transglutaminase 2 | −2.62830 |
| NM_002214 | ITGB8 | Integrin, beta 8 | −2.47052 |
| NM_032211 | LOXL4 | Lysyl oxidase-like 4 | −2.10516 |
| NM_017993 | ENOX1 | Ecto-NOX disulfide-thiol exchanger 1 | −1.80740 |
| NM_173354 | SIK1 | Salt-inducible kinase 1 | −1.75467 |
| NM_032470 | TNXB | Tenascin XB | −1.75391 |
| NM_003236 | TGFA | Transforming growth factor, alpha | −1.71934 |
| NM_005904 | SMAD7 | SMAD family member 7 | −1.55245 |
To identify the biological processes that are over- or underrepresented in GLUT10-deficient fibroblasts, we grouped the DEGs according to the Gene Ontology (GO) categories using DAVID functional annotation clustering and selecting only terms with P ≤ 0.05 and ≥3 genes in each annotation term. An enrichment analysis for the 112 upregulated genes generated 21 distinct clusters (Supplementary Material, Table S2A). Among the different GO clusters, the most enriched genes involved in “lipid metabolic process” and “response to organic substance” were PPARG, PLA2G4A, IGFBP2, ALPL, ADCY4 and ACTC1 and in “oxidation reduction process” and “response to ROS” were ALDH1A1, CYP2U1, FAR2, ADH1C, FADS2, SOD3 and PLA2G4A. Among the upregulated genes, ALDH1A1 encodes the aldehyde dehydrogenase (ALDH) family 1 member A1 (ALDH1A1) involved in retinoic acid metabolism and LPO-derived reactive aldehyde detoxification (22,23). PPARG, FAR2, FADS2, CYP2U1 and PLA2G4A are involved in the PPAR signalling pathway and fatty acid metabolism. In particular, FAR2 encodes a fatty-acyl-CoA reductase 2, which is a peroxisomal enzyme involved in the long-chain fatty-acyl-CoA metabolic process (24). The fatty acid desaturase FADS2 is involved in alpha-linolenic (ω-3) and linoleic (ω-6) acid metabolism (25). The phospholipase PLA2G4A catalyses membrane phospholipid hydrolysis to release arachidonic acid, which is metabolized into eicosanoids (26). PPARG is a member of the nuclear hormone receptor superfamily that acts as a ligand-activated transcription factor and is a master regulator in a range of physiological and pathophysiological events, including vascular remodelling, adipogenesis, insulin action stimulation, lipid metabolism regulation, mitochondrial and redox homeostasis maintenance, and inflammatory processes (27).
A functional analysis of 105 downregulated genes yielded 20 different clusters (Supplementary Material, Table S2B). The significantly downregulated processes include genes related to ECM homeostasis, i.e. LOXL4, IL6, TNXB, COMP, MMP7, TGFA and PDGFC; genes with transcription regulator activity, i.e. EGR3, TSHZ2, TSHZ1, TBX5, HOXA11, MSX2, ACVR2A and HOXA7; genes related to regulating cell proliferation, i.e. IL6, TNFSF4, TBX5, MSX2, TGFA and PDGFC; and genes related to blood vessel morphogenesis and heart development, i.e. MSX2, SMAD7, HAND2, TBX5, EMCN, TGM2 and TGFA. Processes related to regulating apoptosis, signal transduction and phosphate metabolic processes were also underrepresented. Among the downregulated genes, LOXL4 encodes an extracellular copper-dependent amine oxidase that catalyses the first step in cross-link formation in COLL and ELN, and TGM2 encodes a member of the transglutaminase enzyme family that plays a role in ECM remodelling (28,29). A pathway enrichment analysis performed using all of the DEGs confirmed that the GLUT10 deficiency perturbs several functions that are mainly involved in ECM homeostasis (Supplementary Material, Table S2C).
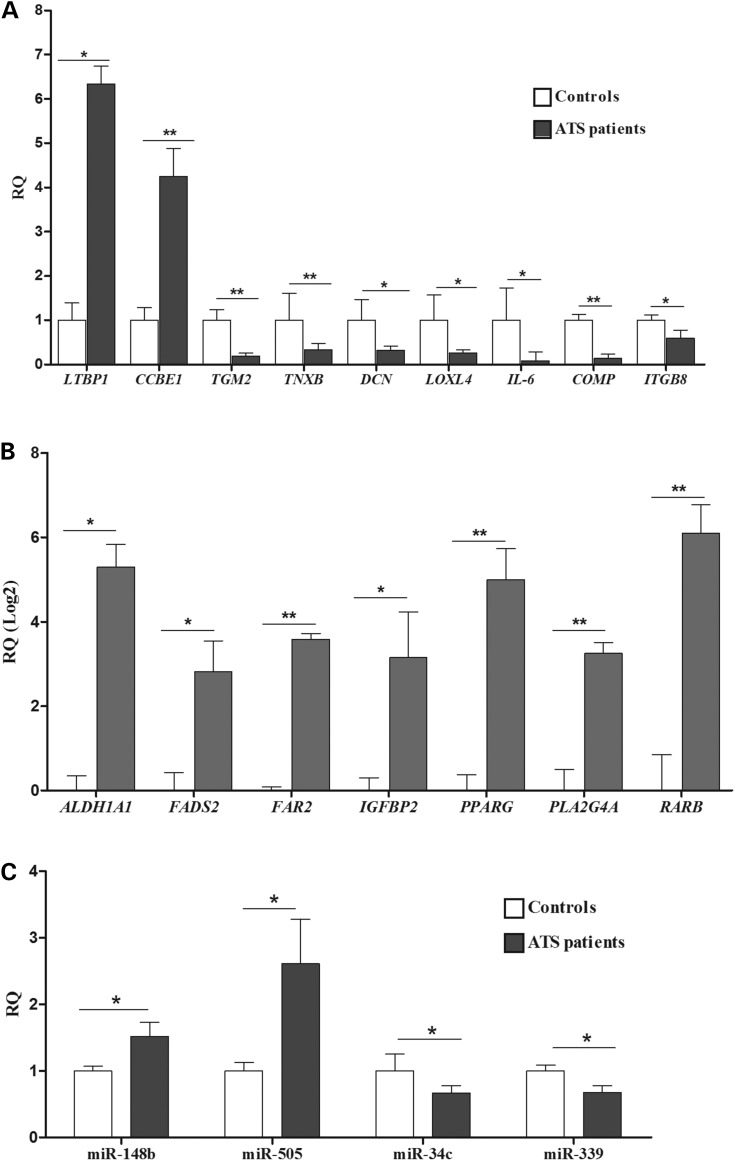
Quantitative real-time PCR (qPCR) was performed on a selection of genes involved in ECM homeostasis and TGFβ signalling, i.e. LTBP1, TGM2, CCBE1, TNXB, DCN, LOXL4, IL-6, ITGB8 and COMP, and in different metabolic processes, i.e. ALDH1A1, FADS2, FAR2, IGFBP2, PPARG, PLA2G4A and RARB. The data confirmed the reliability of the microarray data because the above-reported genes were differentially expressed (Fig. 2A and B). Although the array did not indicate the differential expression of DCN, which is a proteoglycan involved in COLL fibrils formation and the regulation of TGFβ bioavailability, we analysed it using qPCR and demonstrated that it was significantly downregulated (Fig. 2A). Other dysregulated genes that enter the TGFβ pathway include LTBP1, which encodes a member of the latent-binding protein family, and ITGB8, which encodes the β8 integrin subunit involved in integrin-mediated activation of TGFβ. TNXB encodes a member of the tenascins family (TN-X) that regulates elastic fibre structure and stability, COLL synthesis and deposition in the ECM, and TGFβ bioavailability (30). Similarly, COMP, which encodes a non-collagenous ECM glycoprotein that binds several COLLs, fibronectin (FN), and aggregan, regulates TGFβ bioavailability (31). Together, the transcriptome-wide expression profiling results indicate that SLC2A10 mutations perturb many physiological processes related to energetic/redox homoeostasis, TGFβ signalling maintenance, and ECM architecture.
Figure 2.
Validation of microarray expression data by qPCR. Relative quantification (RQ) of selected genes related to ECM organization and TGFβ signalling (A) and involved in different metabolic processes (B) was generated using the 2−(ΔΔCt) method and normalized using the geometric mean of the HPRT, GAPDH, CYC1, ATP5B and RPLP0 reference genes. The expression levels of selected miRNAs (C) was calculated using RNU66 as an internal normalization transcript. The bars represent the mean ratio of the target gene/miRNA expression in patients’ fibroblasts compared with three unrelated healthy individuals. qPCR was performed in triplicate, and the results are expressed as the mean ± SEM. In (A) and (C), the relative expression levels of the target genes/miRNA in patients versus controls were expressed as the non-transformed relative fold change. In (B), the log 2 transformed values are shown. Statistical significance was determined using an unpaired Student's t-test or the one-way ANOVA followed by Tukey's post hoc test (*P < 0.05 and **P < 0.01).
We also analysed the global expression levels of miRNAs in the three ATS patients' skin fibroblasts. By selecting a ≥1.5-fold-change threshold relative to the controls, 30 miRNAs showed a differential expression patterns: 15 were upregulated and 15 were downregulated (Supplementary Material, Table S3). The most upregulated miRNA was hsa-miR-505. Interestingly, this miRNA was upregulated in dissected aortic tissues, which suggests that its altered expression plays an essential role in aortic dissection pathogenesis (32). We compared the DEG list with the differentially expressed miRNAs to investigate a possible correlation between the expressed miRNA and mRNA in ATS cells (see Supplementary Material, Table S4 for a full list of the miRNA-mRNA interactions). This analysis showed that the upregulated hsa-miR-505, hsa-miR-148b and hsa-miR-4708-5p miRNA might regulate the level of transcription for many downregulated ECM genes, i.e. TGM2, ITGB8, LOXL4 and TNXB. On the other hand, the downregulated hsa-miR-34c-3p, hsa-miR-339-3p and hsa-miR-200c miRNAs have as potential targets several upregulated DEGs related to metabolic processes and ECM remodelling, i.e. ALDH1A1, PLA2G4A, LTBP1 and CCBE1 (Supplementary Material, Table S4). qPCR validation confirmed the differential expression pattern of a selection of miRNAs in ATS cells, i.e. hsa-miR-505, hsa-miR-148b, hsa-miR-34c-3p and hsa-miR-339-3p (Fig. 2C). The modulation of the expression levels of several miRNAs and the compelling correlation between miRNA-DEG levels suggests that epigenetics may be involved in the altered gene expression observed in GLUT10-deficient cells, which merits future investigation.
ATS fibroblasts show increased synthesis of ALDH1A1 and PPARγ as well as ROS-induced oxidative stress
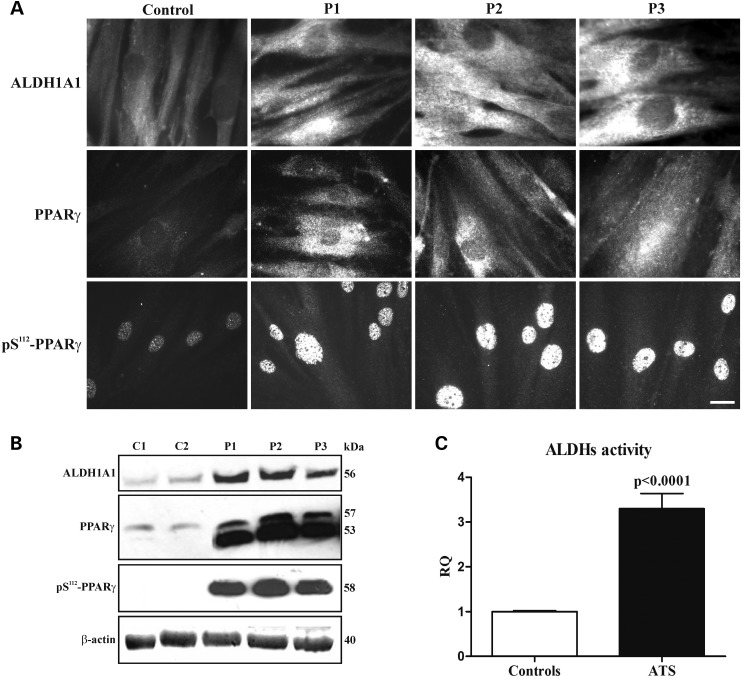
The expression levels and activity of ALDH1A1, which is encoded by the most upregulated gene in the ATS cells versus the control fibroblasts, were investigated by IF, WB and flow cytometry (Fig. 3). Using IF, cytosolic ALDH1A1 was not well detected in control fibroblasts, but it was highly expressed in ATS cells (Fig. 3A). These results were confirmed by WB (Fig. 3B) and by the Aldefluor assay, which showed ∼3-fold higher ALDH activity in the ATS cells versus the controls (Fig. 3C and Supplementary Material, Fig. S2). Because the other two ALDH isoforms, i.e. ALDH1A2 and ALDH1A3, were expressed in the ATS cells and control fibroblasts at comparable levels (data not shown), the enhanced ALDH activity in ATS cells is mainly attributed to ALDH1A1.
Figure 3.
Expression and organization of ALDH1A1 and PPARγ and evaluation of ALDH activity in the control and ATS fibroblasts. (A) IF of ALDH1A1, PPARγ and phospho-serine 112 PPARγ (pS112-PPARγ) in the control and ATS fibroblasts. Scale bar: 10.8 µm. The experiments were repeated three times. The images are representative of two control (C1 and C2) and the three ATS cell strains (P1, P2 and P3). (B) WB of ALDH1A1, PPARγ and pS112-PPARγ in two controls (C1 and C2) and in three ATS fibroblasts (P1, P2 and P3). ALDH1A1 and pS112-PPARγ are shown as a 56 and 58 kDa band, respectively, whereas the anti-PPARγ mAb that recognizes the two PPARγ isoforms identified a 53 and 57 kDa band corresponding to PPARγ1 and PPARγ2, respectively. Loading control: β-actin. (C) FACScan analysis of ALDHs activity in the two control and three ATS fibroblasts; the Bodipy-conjugated aminoacetate developed in BAAA-treated versus untreated cells was used for the measurements. The results are expressed as a relative quantification (RQ) of ALDHs activity in ATS versus the controls; the experiments were repeated three times. Statistical analyses were performed using the GraphPad Prism software (***P < 0.0001).
We further studied PPARγ using IF and WB. As shown in Figure 3A, PPARγ, which is not well detected in control cells, was highly expressed in the cytoplasm and in the perinuclear region of ATS fibroblasts. The increased PPARγ expression in ATS fibroblasts was confirmed by WB, which showed high levels of a 53 kDa band corresponding to PPARγ1, which was almost undetectable in the controls, and a 57 kDa band corresponding to PPARγ2, which was not well detected in the control fibroblasts (Fig. 3B). PPARγ undergoes several post-translational modifications (PTMs) in response to exogenous signals, such as growth factors and adipokines, which modulate its functions. The most well-described PTM is PPARγ2 phosphorylation at Ser112 (pS112-PPARγ) (corresponding to Ser82 in PPARγ1) (33), which we investigated by IF and WB (Fig. 3A and B) using a specific polyclonal antibody (Ab) that recognizes both phosphorylated isoforms. The ATS cells showed higher levels of pS112-PPARγ than the control cells, and elevated PPARγ signalling should be activated in ATS fibroblasts because the protein is mainly translocated to the nucleus.
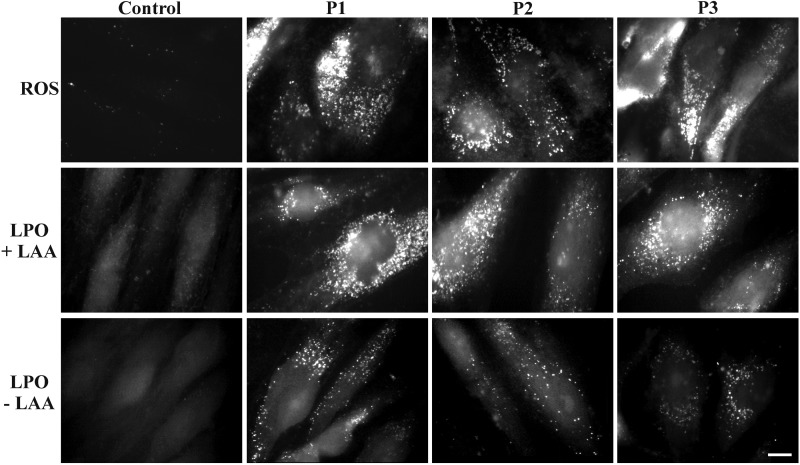
To validate that the GLUT10 deficiency is associated with the alteration of the intracellular redox homeostasis, we analysed both in control and ATS fibroblasts the ROS levels using the CellROX® Green assay. As shown in Figure 4, high ROS levels were detected in ATS cells, but ROS activity was not observed in the control fibroblasts without adding the ROS-specific pro-oxidant agent tert-butyl hydroperoxidase (TBHP) (Supplementary Material, Fig. S3). To investigate the possible involvement of PPARγ in the ROS overproduction observed in ATS cells, we inhibited PPARγ activity using the selective antagonist T0070907 at sublethal concentrations, 1.0–2.5 µM. After inhibiting either control or ATS fibroblasts, we evaluated the pS112-PPARγ distribution and the ROS levels. As shown in Supplementary Material, Figure S4, the antagonist significantly reduced the levels of the nuclear pS112-PPARγ in a dose-dependent manner; in particular, the nuclear receptor was undetectable in both cell types at 2.5 μm. At this optimal inhibition dose, the control fibroblasts showed a marked increase in ROS production, which confirmed that PPARγ attenuates oxidative stress under physiological conditions by inducing antioxidant pathways (27). In ATS cells, PPARγ inhibition elicited the opposite effect because it was associated with a marked decrease in ROS levels. This result suggests that aberrant activity of PPARγ in ATS cells, likely linked to Ser112 phosphorylation, contributes to oxidative stress-induced injury, which merits future investigation.
Figure 4.
Evaluation of the ROS activity and LPO process in the control and ATS fibroblasts. The intracellular ROS activity was evaluated, wherein the control and ATS fibroblasts were exposed to the non-fluorescent cell-permeable CellROX® reduced reagent, and its conversion was detected in a fluorescent molecule upon oxidation by ROS. To analyse the LPO process, the control and ATS fibroblasts were grown for 48 h in complete MEM and treated (+LAA) or not (−LAA) with 50 µm LAA, and the LPO-derived adducts were detected by fluorescence microscopy, wherein the cells reacted with the Click-iT® reaction cocktail containing the Alexa Fluor 488 azide. Scale bar: 6 µm. The experiments were repeated three times. The images are representative of the two control (C1 and C2) and three ATS cell strains (P1, P2 and P3).
The elevated ROS levels identified in ATS fibroblasts indicate that these cells are under oxidative stress and suggest an enhanced ROS-induced LPO process, which may explain the induction of ALDH1A1. To validate this hypothesis, LPO was investigated both in the control and ATS cells using the Click-iT® LPO detection kit with linoleamide alkyne (LAA). ATS fibroblasts showed high levels of LPO-derived adducts (Fig. 4, +LAA), whereas these modifications were undetectable in the control cells without adding the LPO-specific pro-oxidant compound cumene hydroperoxide (Supplementary Material, Fig. S3). Furthermore, in ATS cells, the LPO-derived modifications were measurable even without adding the specific substrate LAA (Fig. 4, −LAA), and the levels of these adducts were higher than in cumene hydroperoxide-treated control cells (Supplementary Material, Fig. S3). These results suggest that a GLUT10 deficiency is associated with the hyperactivation of the ROS-induced LPO process in ATS cells, which is likely sustained by the enhanced lipid metabolism that upregulates ALDH1A1 as a compensatory mechanism to attenuate oxidative stress-mediated injury.
In ATS fibroblasts a non-canonical TGFβ signalling is activated that exhibits cross-talk with the αvβ3 integrin transduction pathway
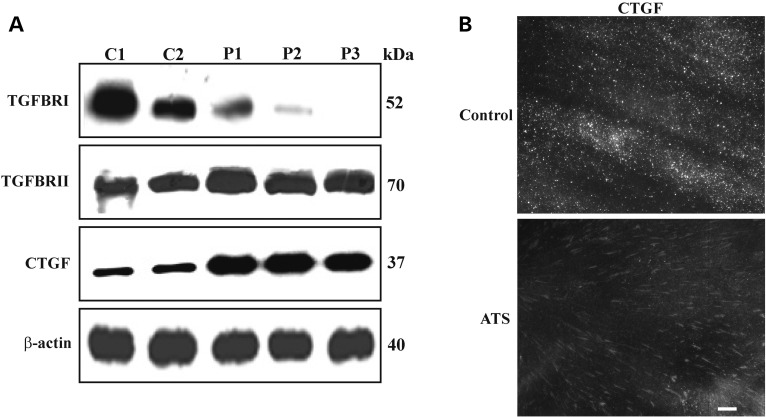
To better define the involvement of TGFβ signalling in ATS aetiology, we used WB to analyse the expression of plasma membrane-bound TGFBRI and TGFBRII receptor subunits as well the total CTGF, which is a downstream modulator of the TGFβ pathway. ATS fibroblasts showed markedly lower levels of TGFBRI compared with the control cells, which was particularly evident in patient 3 (P3), wherein the protein was undetectable (Fig. 5A). In contrast, in the ATS cells, TGFBRII was expressed at higher levels than in the control fibroblasts. These data indicate that GLUT10 deficiency leads to differential regulation of TGFBRI and TGFBRII. Despite this clear dysregulation of the canonical TGFβ receptor subunit organization, the ATS fibroblasts synthesized higher levels of CTGF than the control cells (Fig. 5A), which confirmed activation of paradoxical TGFβ signalling (15). Next, we used IF to analyse distribution of the cell membrane-associated CTGF, which was only organized as elongated patches in ATS cells (Fig. 5B). Given the known co-distribution and interaction between CTGF and αv-dependent integrins (35), such as αvβ3 and αvβ8, we used IF to analyse the organization of the αvβ3 integrin and β8 integrin subunit. As shown in Figure 6A, αvβ3 was almost undetectable in the control fibroblasts, whereas, in ATS cells, it was organized in linear patches on the cell surface with a pattern resembling the CTGF distribution (Fig. 5B). Furthermore, we observed much lower expression of the β8 integrin subunit in the ATS fibroblasts compared with the control cells (Fig. 6A), which is consistent with the lower ITGB8 gene expression observed using microarray (Table 1). We investigated αvβ3 integrin activation through IF by analysing phosphorylation of Tyr 773 in the β3 integrin subunit (pY773β3 integrin). In the ATS cells, but not the controls, the phosphorylated β3 integrin subunit was abundantly organized with a pattern similar to that of αvβ3 (Fig. 6A) and CTGF (Fig. 5B).
Figure 5.
TGFBRI, TGFBRII and CTGF expression in ATS fibroblasts. (A) WB of 50 µg cell membrane-bound proteins extracted from control (C1 and C2) and ATS (P1, P2 and P3) fibroblasts immunoreacted with the anti-human TGFBRI and TGFBRII Abs, detecting a 52 and 70 kDa band, respectively. To analyse the expression of CTGF, which was indicated through a 37 kDa band, the total extract from both cell types was used. Loading control: β-actin. (B) To detect the distribution of the membrane-associated insoluble fraction of CTGF, the control and ATS fibroblasts were fixed and immunoreacted as previously described (34). Scale bar: 4 µm. The experiments were repeated three times. A representative image of two control (C1 and C2) and the three ATS fibroblasts (P1, P2 and P3) is shown.
Figure 6.
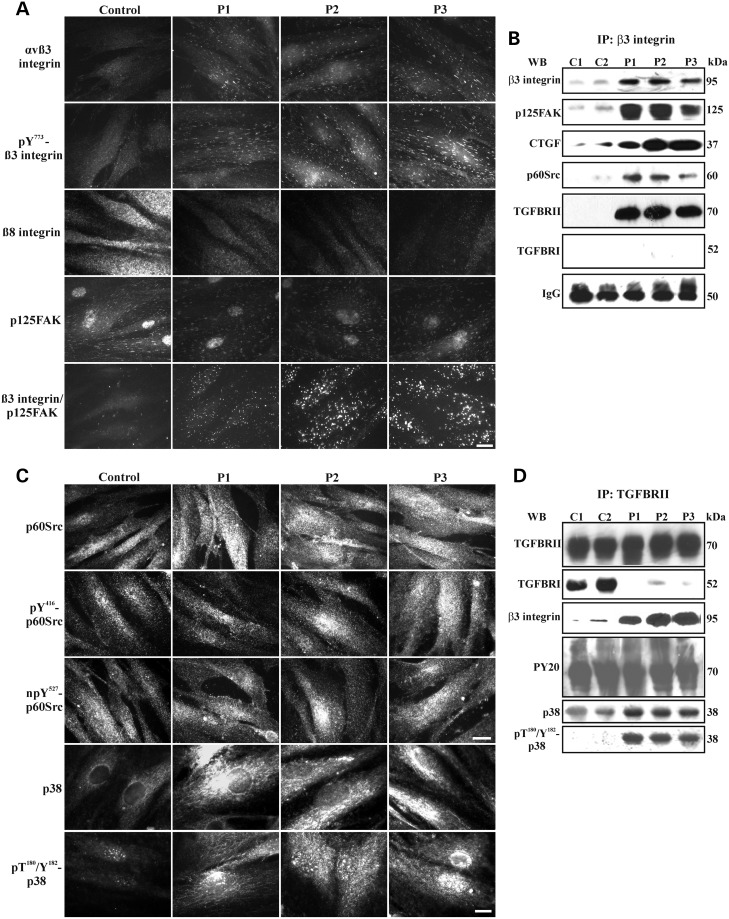
Activation of αvβ3 integrin signalling that cross-talks with TGFBRII in ATS fibroblasts. (A) IF of αvβ3 integrin, pY773β3, β8 integrin subunit and p125FAK in the control and ATS fibroblasts. The co-localization of the β3 integrin subunit with p125FAK was evaluated using the Duolink in situ PLA probe kit. Scale bar: 4.5 µm. The experiments were repeated three times. The images are representative of two control (C1 and C2) and the three ATS cell strains (P1, P2 and P3). (B) WB of the β3 integrin subunit immunoprecipitated from the control (C1 and C2) and ATS (P1, P2 and P3) fibroblasts, which were immunoreacted with the anti-β3 integrin subunit mAb, anti-p125FAK, anti-CTGF, anti-p60Src, anti-TGFBRII and anti-TGFBRI Abs to detect bands at 95, 125, 37, 60, 70 and 52 kDa, respectively. Loading control: IgG. (C) IF of p60Src and p38 MAPK in the control and ATS fibroblasts. p60Src activation was monitored using two Abs that recognize the phospho-Tyr416 (pY416-p60Src) and non-phospho-Tyr527 (npY527-p60Src) residues of the protein; detection of both signals indicates the active form of p60Src. p38 MAPK activation was analysed using an Ab that recognizes the phospho-Threo180 and Tyr182 (pT180/Y182-p38). Scale bar: 8.3 µm for p60Src and 4.5 µm for p38. The experiments were repeated three times. The images are representative of two control (C1 and C2) and the three ATS cell strains (P1, P2 and P3). (D) WB of the cell membrane-bound TGFBRII immunoprecipitated from the control and ATS fibroblasts and immunoreacted with the anti-TGFBRII, anti-TGFBRI and anti-pT180/Y182-p38 MAPK Abs as well as with the anti-β3 integrin subunit, anti-phospho-Tyr PY20 and anti-p38 MAPK mAbs to detect bands at 70, 52, 38, 95, 70 and 38 kDa, respectively.
To identify the signalling proteins that interact with the activated αvβ3 integrin in ATS fibroblasts, p125FAK and p60Src were investigated using IF and WB. As shown in Figure 6A, p125FAK was abundantly organized in the control and ATS cells, but it only co-localized with the β3 integrin subunit in ATS fibroblasts, as demonstrated by the Duolink analysis. Co-localization of the αvβ3 integrin with p125FAK was confirmed by WB of the β3 integrin subunit immunoprecipitates that were immunoreacted with the anti-p125FAK Ab (Fig. 6B). This analysis showed higher levels of the β3 integrin subunit that co-precipitated with higher levels of p125FAK in ATS cells compared with controls.
As shown in Figure 6C, both the controls and ATS fibroblasts abundantly expressed p60Src, which was in its active and transducing form, as demonstrated by the phosphorylation at Tyr 416 (pY416-p60Src) and the concomitant non-phosphorylation at Tyr 527 (npY527-p60Src). Because, in ATS cells, the β3 integrin subunit immunoprecipitates showed high levels of p60Src or CTGF (Fig. 6B), the αvβ3 integrin-CTGF complex should transduce to p125FAK and p60Src in the GLUT10-deficient fibroblasts.
To verify a possible correlation between the αvβ3 integrin-mediated signal transduction and the dysregulated expression of TGFBRI and TGFBRII observed in the ATS fibroblasts, co-immunoprecipitation of these receptor subunits with β3 integrin was investigated using WB. In the control cells, the poorly expressed β3 integrin subunit did not co-immunoprecipitate at detectable levels with either TGFBRII or TGFBRI (Fig. 6B), whereas the β3 integrin subunit co-immunoprecipitated with high levels of TGFBRII, but not TGFBRI, in ATS cells (Fig. 6B). These results were confirmed through immunoprecipitation of TGFBRII and analysis of the complexes using the anti-TGFBRI and anti-β3 monoclonal antibodies (mAbs). As shown in Figure 6D, high and comparable levels of TGFBRII were recovered in the control and ATS extracts. In the control cells, TGFBRII immunoprecipitated with high levels of TGFBRI, whereas, in the ATS cells, TGFBRII co-immunoprecipitated with significant levels of the β3 integrin subunit without TGFBRI (Fig. 6D). Together, these data suggest an activated cross-talk between αvβ3 integrin-CTGF and TGFBRII, but not TGFBRI, in ATS cells, which likely involves p125FAK and p60Src.
To investigate TGFBRII phosphorylation, the global tyrosine phosphorylation pattern on the TGFBRII immunoprecipitates was analysed using the PY20 mAb, which recognizes all of the phosphorylated tyrosine residues (Fig. 6D). Among the numerous bands in the control and ATS complexes, we detected a 70 kDa band, which should include the TGFBRII and indicates that the receptor is in the active form in both cell types. This result with the presence of p60Src in the immunocomplexes containing the β3 integrin subunit and TGFBRII, but not TGFBRI, suggests a role for p60Src in tyrosine phosphorylation of TGFBRII and in activation of non-canonical TGFβ signalling involving the CTGF-αvβ3 integrin transduction pathway.
To define whether canonical downstream signalling transducers of the TGFβ pathway are recruited in ATS fibroblasts, we analysed the expression, distribution and phosphorylation of SMAD2 using IF and either an anti-SMAD2 mAb that detects the total protein or a specific anti-phospho-SMAD2 Ab that only recognizes the phosphorylated 465/467 serine residues in active SMAD2 (pSMAD2). In the control and ATS cells, SMAD2 was present either in the cytoplasm or nucleus, and SMAD2 was detected at much lower levels in ATS fibroblasts (Supplementary Material, Fig. S5). pSMAD2 was detected in control cell nuclei and, at low levels, in ATS cell nuclei. IF also showed increased levels of total and phosphorylated p38 MAPK in GLUT10-deficient cells compared with the controls (Fig. 6C). As shown in Figure 6D, the TGFBRII recovered from the ATS cell extracts immunoprecipitated with increased levels of p38 MAPK compared with the controls, which was in the active form, as demonstrated by the phosphorylation at Threo180 or Tyr182 (pT180/Y182-p38). In conclusion, these data suggest that, in ATS cells, TGFβ signalling should be activated by an SMAD2-independent TGFBRII-mediated non-canonical pathway that exhibits cross-talk with αvβ3 integrin signalling, which involves CTGF, p125FAK, p60Src and p38 MAPK. This MAPK seems to play a pivotal role in ATS aetiopathogenesis because it might also be activated by stimuli other than TGFβ signalling, including an imbalance of redox homeostasis due to ROS-induced LPO-derived aldehydes.
Stable expression of GLUT10 in ATS fibroblasts normalizes ALDH1A1 and PPARγ activity, ROS production and the LPO process, rescues canonical αvβ3-independent TGFβ signalling and partially restores the ECM
To demonstrate that the alteration of gene expression, protein synthesis and organization observed in the ATS fibroblasts are downstream effects of the SLC2A10 loss-of-function mutations, we generated a stable GLUT10 expression vector (pG10), which was transfected into the ATS patient 1 (P1) cells, and analysed a selection of dysregulated genes/proteins by qPCR, IF and WB. As shown in Figure 7A, pG10-transfected fibroblasts stably expressed and abundantly organized GLUT10 similarly to control fibroblasts (Fig. 1). We used qPCR analyses for a small selection of upregulated genes, i.e. ALDH1A1 and PPARG, or downregulated genes, i.e. LOXL4, TNXB and DCN, in the ATS versus the control cells to compare the mRNA levels of the pG10 versus the empty vector (mock)-transfected ATS P1 cells. In particular, the ALDH1A1 and PPARG mRNA levels showed an approximately 2-fold decrease in the pG10 versus mock cells, and LOXL4 and TNXB were upregulated ∼2.5-fold after transfection (Supplementary Material, Fig. S6A).
Figure 7.
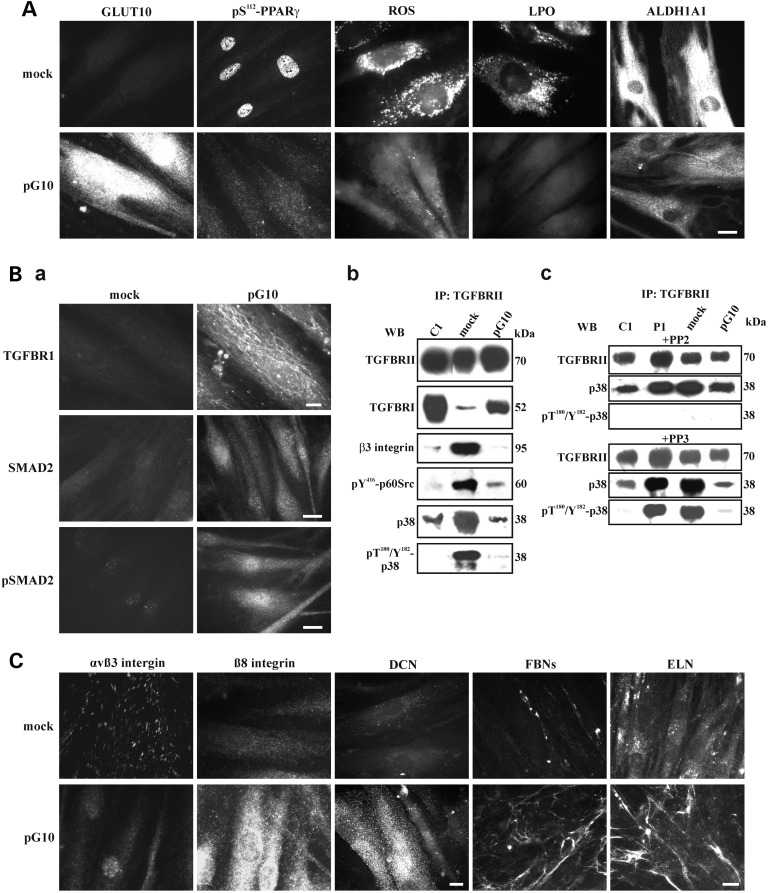
Phenotypic rescue of ATS fibroblasts after stable GLUT10 expression. (A) IF of GLUT10, pS112-PPARγ and ALDH1A1 in mock- and pG10-transfected ATS P1 fibroblasts using the anti-human GLUT10, anti-pS112-PPARγ Ab and anti-ALDH1A1 mAb. Intracellular ROS and LPO were evaluated using the CellROX® Green assay and the Click-iT® LAA kit, respectively. Scale bar: 5 µm. The experiments were repeated three times. Representative images are shown. (B-a) IF of TGFBRI, SMAD2 and its phosphorylation (pSMAD2) in mock- and pG10-transfected ATS P1 fibroblasts using the anti-TGFBRI and anti-phospho-SMAD2 Abs as well as the anti-SMAD2 mAb. Scale bar: 4 µm for TGFBRI, 8.3 µm for SMAD2 and pSMAD2. The experiments were repeated three times. Representative images are shown. (B-b) WB of the cell membrane-bound TGFBRII immunoprecipitated from the control (C1), mock- and pG10-transfected P1 fibroblasts, which were immunoreacted with the anti-TGFBRII, anti-TGFBRI, anti- pT180/Y182-p38 MAPK and anti-pY416-p60Src Abs to detect bands at 70, 52, 38 and 60 kDa, respectively, as well as with the anti-β3 integrin subunit and anti-p38 MAPK mAbs to detect bands at 95 and 38 kDa, respectively. (B-c) The involvement of p60Src in p38 MAPK phosphorylation was investigated through treating the untransfected control (C1) and ATS (P1) fibroblasts as well as the mock- and pG10-transfected ATS cells with 50 µm of PP2 and PP3. The TGFBRII immunoprecipitates were reacted with the anti-TGFBRII and anti-pT180/Y182-p38 Abs as well as the anti-p38 MAPK mAb. (C) IF of the αvβ3 integrin, β8 integrin subunit, DCN, FBNs and ELN in mock- and pG10-transfected ATS P1 fibroblasts. The cells were grown for 48 h before immunodetection with the anti-αvβ3, β8 and DCN mAbs and for 15 days before reacting with the anti-FBN and anti-ELN mAbs. Scale bar: 3.5 µm for αvβ3 integrin, β8 integrin subunit and DCN; 7 µm for FBN and ELN. The experiments were repeated three times. Representative images are shown.
In the transfected ATS fibroblasts, IF and WB showed that GLUT10 expression significantly decreased the pS112-PPARγ levels (Fig. 7A and Supplementary Material, Fig. S6B), which were undetectable in the nuclei (Fig. 7A), in association with normalized intracellular ROS production, LPO levels (Fig. 7A) and ALDH1A1 synthesis and activity (Fig. 7A and Supplementary Material, Fig. S6B,C). Overall, these data demonstrate that GLUT10 plays a key role in maintaining redox homeostasis and corroborates the hypothesis that a PPARγ-dependent mechanism contributes to ROS overproduction and is associated with ATS aetiopathogenesis.
For TGFβ signalling, TGFBRI, which was undetected by IF in the mock-transfected cells, was upregulated and organized in the plasma membrane in the GLUT10-expressing ATS cells (Fig. 7B-a). Consistent with this result, for these transfected cells, WB showed higher levels of the TGFBRI isoforms compared with the untransfected and mock-transfected cells (Supplementary Material, Fig. S6B). Furthermore, we only detected SMAD2, total and phosphorylated, in the cytoplasm and nucleus of pG10-transfected cells (Fig. 7B-a). These data suggest that GLUT10 expression rescues canonical TGFβ signalling in ATS cells. To corroborate these findings, TGFBRII was immunoprecipitated from the control and mock- and pG10-transfected fibroblast extracts, and the immunocomplexes were analysed for the presence of the TGFBRI isoforms and β3 integrin subunit. As reported in Figure 7B-b, similar levels of TGFBRII were recovered from the control and both transfected ATS fibroblast cell types. In the control cells and in ATS fibroblasts expressing GLUT10, TGFBRII immunoprecipitated higher levels of TGFBRI than in mock-transfected counterparts. The β3 integrin subunit was recovered in abundance by TGFBRII in the mock-transfected cells, but was almost undetected in the GLUT10-expressing counterpart cells. These results are supported by IF, which showed that the GLUT10 expression was associated with αvβ3 integrin disorganization and β8 integrin subunit re-organization (Fig. 7C). Furthermore, we examined the co-immunoprecipitation of both the p60Src and p38 MAPK active forms with TGFBRII using the anti-pY416-p60Src and anti-pT180/pY182-p38 Abs, respectively. The two phosphorylated proteins were abundantly recovered in the immunocomplexes from the mock-transfected ATS cells but were recovered at significantly lower levels in the GLUT10-expressing fibroblast and control cell immunoprecipitates (Fig. 7B-b), which is consistent with lower levels of p38 MAPK in pG10-transfected fibroblasts.
To confirm that the p38 MAPK phosphorylation was due to the presence of the active form of p60Src, the control, untransfected and mock- and pG10-transfected ATS fibroblasts were treated with subapoptotic concentrations of the p60Src inhibitor PP2 and its inactive analogue PP3. TGFBRII was then immunoprecipitated and tested for binding to p38 and its phosphorylated form. As shown in Figure 7B-c, the PP3 treatment did not affect p38 phosphorylation in the untransfected and mock-transfected ATS cells, whereas the PP2 treatment completely removed phosphorylation in these cells, even though the total protein levels were maintained. This result suggests that, in ATS cells, p38 phosphorylation is due to non-canonical TGFβ signalling and occurs downstream of the p60Src kinase activity, which should be involved in phosphorylating TGFBRII. Together, these data confirm that loss of GLUT10 expression affects canonical TGFβ signalling and recruits an alternative pathway that involves TGFBRII cross-talk with the αvβ3 integrin through p60Src activation, which is involved in TGFBRII phosphorylation, activating p38 MAPK. Supporting this novel disease-associated pathway, ATS fibroblasts expressing GLUT10 rescue canonical TGFBRI/TGFBRII signalling, which is αvβ3 integrin, p60Src and p38 independent.
Finally, to verify whether the recovery of normal redox homeostasis and canonical TGFβ signalling is associated with fibrillar ECM organization in ATS fibroblasts, the pG10-transfected cells were analysed using IF for COLLIII-, COLLV-, DCN-, FBN- and ELN-ECM organization (Fig. 7C). COLLIII and COLLV organization in the ECM was not rescued by GLUT10 expression at up to 15 days of culture (data not shown). However, DCN, which was absent in the untransfected (Fig. 1) and mock-transfected ATS fibroblasts, was abundantly organized in GLUT10-expressing cells grown for 48 h (Fig. 7C), even without modulated gene expression (Supplementary Material, Fig. S6A). At this period during culture, the FBNs- and ELN-ECMs were not organized, but 15 days after seeding, the GLUT10-expressing fibroblasts secreted and organized thin fibrils of FBNs and ELN (Fig. 7C). These data are consistent with the upregulated LOXL4 expression observed using qPCR in the GLUT10-expressing ATS cells (Supplementary Material, Fig. S6A) and with restored canonical TGFβ signalling. These data suggest that GLUT10 rescue is associated with organization of several ECM components and yields control-like ECM homeostasis.
Discussion
Although SLC2A10 was identified as causal gene for ATS in 2006 (3), the specific role of GLUT10 remains largely unknown and controversial. Different hypotheses, i.e. glucose and/or DAA transport into different intracellular compartments (3,10,17,18), have been drawn, but neither the exact localization nor the transported substances have been clarified with certainty. Furthermore, the interaction between GLUT10 and the TGFβ pathway (3,14) was not elucidated.
Transcriptome analysis confirmed the crucial role of GLUT10 in ATS, wherein GLUT10 maintains ECM homeostasis because several ECM-related genes involved in focal adhesion, ECM-receptor interaction and TGFβ signalling were dysregulated. Interestingly, the array results also suggested oxidative stress in ATS fibroblasts, which was validated by increased intracellular levels of ROS- and LPO-derived adducts. Specifically, we observed significant upregulation of several genes associated either with maintaining redox homeostasis, including the cellular response to ROS- and LPO-derived aldehyde detoxification mechanisms or cell energy balance, i.e. polyunsaturated fatty acid (PUFA) metabolism. The LPO process is initiated by interactions between ROS and PUFAs, such as linoleic, γ-linolenic, or arachidonic acids, which trigger chain peroxidation (36). Because LPO was detected in ATS fibroblasts even without the specific LAA substrate, the increased lipid metabolism observed in ATS cells may have sustained this process. The resulting lipid radicals interact with oxygen, which propagates the reaction via peroxyl radical intermediates and generates lipid hydroperoxides and aldehydes, such as 4-hydroxy-2-nonenal (4-HNE), which is considered a major bioactive marker of LPO (37). LPO can be terminated by so-called chain-breaking antioxidants, such as vitamin E. Notably, AA protects against this deleterious process either by acting as an ROS scavenger or by the one-electron reduction of lipid hydroperoxyl radicals via the vitamin E redox cycle (19). LPO-derived aldehydes are increasingly recognized as markers of cellular dysfunction and degeneration in a range of disorders including cardiovascular diseases (38). They interact with lipids, nucleic acids and proteins, which impairs their function (39). Failure to eliminate protein adducts results in proteasome inhibition, altered protein turnover and stress response pathway activation, such as MAPKs and ER stress induction, which compromises the ER redox balance and, ultimately, leads to cell death (40). The main LPO-derived aldehyde detoxification mechanisms include GSH, glutathione-S-transferases, aldo-keto reductases and ALDHs, including ALDH1A1, which is upregulated in ATS fibroblasts. This enzyme converts aldehydes to carboxylic acids, contributes to the antioxidant arsenal because the NAD(P)H produced during this reaction is used to regenerate GSH and acts as direct antioxidant for ROS (41). The increased ALDH1A1 activity can be considered a compensatory mechanism to rescue ATS cells from oxidative stress triggered by increased ROS-induced LPO.
Oxidative stress triggers a series of responses, which results in altered PPARs expression and activity. Redox states may regulate PPARs signalling via transcription regulation and PTMs (42). PPAR expression and activity have been observed in endothelial cells (43) and AVSMCs (44), which suggests that PPARs are redox-sensitive transcription factors in the vasculature and are selectively activated by oxidized fatty acids. Peroxidized PUFAs promote PPARγ-mediated transcription and PPARγ binding to specific target genes, including the PPARG itself (45). PPARγ also regulates lipid and glucidic metabolism (27); therefore, in ATS cells, the increased PPARγ may be involved in upregulating genes related to peroxisomal β-oxidation and PUFA metabolism, as shown in the array. Overall, PPAR expression and activity may be altered by the cellular energy metabolism status, and oxidative stress is attributed to altered PPAR expression and activity as adaptive or maladaptive feedback (42). In ATS cells, increased PPARG expression, enhanced protein synthesis, and preferential distribution of phosphorylated (Ser112) PPARγ at the nuclear level with the evidence of ROS-induced oxidative stress sustained by enhanced lipid, suggest aberrant PPARγ activity. Such aberrant activity may finally produce a vicious cycle that sustains oxidative stress. This hypothesis is supported by the preliminary results of PPARγ inhibition. In control cells, the specific antagonist enhanced ROS synthesis, which is consistent with a pivotal antioxidant role for PPARγ, which, under physiological conditions and via transcriptional regulation, activates scavenger genes (42). In ATS fibroblasts, PPARγ inhibition attenuated oxidative stress, which indicates that, despite its positive benefits to other cells/tissues, PPARγ activation is detrimental under oxidative stress-induced injury. Formal proof that GLUT10 deficiency causes oxidative stress due to enhanced ROS-induced LPO likely sustained by increased lipid metabolism through a PPARγ-mediated mechanism was generated by stable GLUT10 expression in ATS fibroblasts, which normalized redox homeostasis and ALDH1A1 synthesis. One study recently reported that ROS-induced PPARγ alterations, which are associated with PPARγ Ser112 phosphorylation, enhance oxidative stress-mediated renal tubular cell damage (46). Therefore, we supposed that, in ATS cells, modulated PPARγ activity involves Ser112 phosphorylation. Given that this PTM can produce different transcription outcomes depending on the physiological context and kinases involved (33), in ATS cells, the specific protein kinase(s) should be identified in future studies.
ROS involvement in the ATS aetiology is intriguing because ROS are involved in different vascular-related pathological conditions, i.e. atherosclerosis, hypertension, stroke, cardiomyopathies, cardiac hypertrophy and cardiac congestive insufficiency (47,48).
ROS can affect ECM organization through protein fragmentation, which is also mediated by proteolytic enzymes, and sugar structure variation. Recently, oxidized ELN fragments were identified as associated with arterial diseases (49); in AVSMC, ROS induce ECM degradation during vascular ECM remodelling in aneurysms (50); and ROS can damage ECM proteoglycan glycosaminoglycan chains (51). However, ECM disassembly can promote ROS production and induce the LPO process (52). Loss of matrix contacts decreases cell uptake of nutrients, such as glucose and glutamine, which exacerbates fuel deprivation for the citric acid cycle and leads to an ATP shortage. Moreover, diminished activity of the pentose phosphate pathway leads to a critical shortage of NADPH, which is an ROS-reducing/scavenging agent. To compensate for the low levels of ATP and NADPH, peroxisomal fatty acid degradation is turned on with a concomitant release of additional ROS (52). Furthermore, in regulating intracellular homeostasis, a complex interaction between different subcellular compartments occurs. ROS can be generated in mitochondria as a respiration by-product and in ER as a by-product of protein synthesis, PTMs, oxidative protein folding, trafficking and detoxification, which require redox constituents, carbohydrate precursors and lipids for disulphide bond and glycoprotein formation. ROS can perturb ER protein folding as well as induce ER stress, and oxidants, such as peroxides and LPO-derived aldehydes, may activate the unfolded protein response (53,54). In addition to ROS overproduction, in ATS patient fibroblasts, disorganized ELN-, COLLIII-, COLLV-, FBNs- and DCN-ECM were observed concomitant with cytoplasmic retention of certain such proteins, such as ELN, COLLIII and COLLV. Generating high levels of LPO-mediated adducts in ATS fibroblasts may affect the amino acid structure of ELN and COLLs and negatively influence their secretion and organization in the ECM. The intracellular protein retention may lead to defective trafficking and ultimately generate ER stress. Furthermore, the reduced LOXL4 expression that was observed in ATS cells using the array might affect COLL and ELN strength and elasticity and may be involved in generating misfolded or unfolded proteins, which might ultimately increase ROS production. Similarly, expression of the TGM2 transglutaminase involved in PTMs and organization of several ECM proteins was also downregulated in ATS cells, which is consistent with the FN and DCN disorganization observed in these cells (1,3, this work). Thus, although not investigated in this study, ER stress in ATS fibroblasts cannot be excluded. Moreover, based on the cofactor enzyme model of ATS aetiology, the defective synthesis and assembly of COLLs and ELN in ATS cell ECM may be due to impaired DAA uptake in the ER (10,17).
TGFβ is an important determinant of matrix deposition, vessel structure and neointima formation. This growth factor is secreted in a large latent complex (LLC) that consists of TGFβ, an inhibitor, i.e. latency-associated protein (LAP), and one of the latent TGFβ-binding proteins (LTBPs). LTBPs are covalently cross-linked to ECM proteins such as FBNs, DCN, FN, TN-X and COMP by transglutaminases and thus localize latent TGFβ in the ECM. The TGFβ activation process involves LLC release from the ECM followed by LAP proteolysis to release active TGFβ to its receptors. Alternatively, upon mechanical stretch, integrins, such as αvβ3 and αvβ8, activate TGFβ by binding the RGD motif in LAP and inducing TGFβ release from its latent complex (55). Decreased expression of DCN and disarray of DCN- and FBNs-ECMs was demonstrated in ATS fibroblasts, which suggests higher TGFβ bioavailability, which is consistent with the upregulated TGFβ signalling indicated by enhanced CTGF expression (3, this study). Furthermore, because ROS can activate latent TGFβ1 (56), oxidative stress in ATS cells likely contributes to this process. Dysregulated TGFβ signalling associated with arterial aneurysm formation is common in several conditions that are referred to as TGFβ vasculopathies such as Marfan syndrome and Loeys-Dietz syndrome, which involve the constitutive stimulation of canonical and paradoxical signalling, respectively (16).The canonical TGFβ pathway requires ligand binding to a heteromeric complex composed of TGFBRI and TGFBRII serine/threonine kinase receptors. The TGFBRI receptor directly phosphorylates SMAD2/3, which then binds SMAD4, allowing it to accumulate in the nucleus, where the complex transcriptionally activates many target genes. TGFβ also activates non-SMAD-signalling pathways, such as the p38 and JNK MAPK pathway (57).
To discern the molecular interactors involved in TGFβ signalling activated in ATS cells, TGFBRI, TGFBRII and CTGF expression was investigated. The high level of TGFBRII at the ATS cell membrane is not consistent with TGFBRI but is associated with upregulated CTGF and αvβ3 integrin, which co-immunoprecipitate with TGFBRII and are abundantly and similarly organized at the cell membrane. αvβ3 integrin is not organized by control cells, which, in turn, express the β8-dependent integrins that are almost absent in ATS fibroblasts. A cell survival role for αvβ3 integrin was reported in Ehlers-Danlos syndrome skin fibroblasts, wherein this receptor rescues the cells from apoptosis induced by ECM disassembly due to COL5A1 and COL3A1 mutations (58,59).
In ATS cells, the β3 integrin subunit is phosphorylated at tyrosine 773 and co-immunoprecipitates with p125FAK and phosphorylated p60Src, which suggests activation of canonical αvβ3 integrin-mediated signal transduction. Moreover, in ATS cells, this integrin co-immunoprecipitates with TGFBRII but not with TGFBRI, which is downregulated. p125FAK recruitment by the β3 integrin subunit is consistent with its reported role as a key mediator of TGFβ signalling (60,61). Together, these data suggest that GLUT10 loss leads to non-canonical αvβ3 integrin-TGFBRII cross-talk.
A dual role for αvβ3 integrin has been reported, which support its cross-talk with TGFBRII. The first role is in binding to CTGF, which was reported in endothelial cells, where it stimulates angiogenesis and fibrotic mechanisms (62). CTGF-αvβ3 integrin complex signalling through p60Src activation and downstream ERK pathway modulation have been reported (35). Second, the LLC complex binds αv-dependent integrins, including αvβ3, because LAP contains an RGD domain by which TGFβ may be activated (55,63). TGFβ activation induces αvβ3 integrin association with TGFBRII, which initiates cross-talk (55,64,65) and leads to p60Src-mediated tyrosine 284 (Y284) phosphorylation of TGFBRII that, in turn, activates p38 MAPK, which is involved in several functions, such as adhesion, migration and ECM remodelling (66,67). ATS fibroblasts exhibit a similar transduction pathway based on the following. The ATS cells' TGFBRII co-immunoprecipitates with the β3 integrin subunit, which is tyrosine phosphorylated as well as in the control fibroblasts; however, only in the GLUT10-deficient cells, TGFBRII co-immunoprecipitates phosphorylated p60Src and p38 MAPK. PP2-mediated inhibition of p60Src leads to complete elimination of p38 MAPK phosphorylation within TGFβ signalling in the ATS fibroblasts. These data support a p60Src-dependent TGFBRII-mediated activation of p38 MAPK. Furthermore, stable GLUT10 expression in ATS cells abolishes the cross-talk between TGFBRII and αvβ3 and rescues the SMAD2-dependent TGFBRI/TGFBRII canonical pathway, which is associated with partial re-organization of the ECM. These results clearly demonstrate that GLUT10 is involved in maintaining the canonical TGFβ pathway. For p38-mediated TGFβ signalling, currently, we cannot characterize the specific TGFBRII tyrosine residues phosphorylated by p60Src (68), due to a lack of specific Abs.
p38 MAPK is a stress-activated protein and a key enzyme for survival, inflammation and regulation of ER homeostasis. LPO and, particularly, 4-HNE, activate ERK, JNK and p38 MAPK as negative regulators of cellular stress and confer cytoprotection against LPO-mediated cell injury (69). Because ATS cells show an increase in the LPO process, the derived toxic aldehydes might also be involved in p38 MAPK activation, which is indicated by the increased levels of total and phosphorylated p38. Interestingly, an oxidative stress imbalance is associated with aberrant elastogenic activity, which is regulated by TGFβ signalling. Recently, impairment of the TGFβ pathway by oxidative exposure of human skin fibroblasts has been reported (70), and inhibition of TGFβ signalling by 4-HNE, which triggers ELN assembly in the ECM, was reported in oxidative stress-associated aging using human skin fibroblasts (71). Moreover, CTGF production by TGFβ-activated p38 MAPK signalling independent of SMAD action was reported (72,73).
In summary, our approach demonstrates that GLUT10 deficiency alters redox/energetic homeostasis, which leads to oxidative stress and is likely to be sustained by altered lipid metabolism and PPARγ maladaptive feedback that contribute to the enhanced ROS-induced LPO process, which, in turn, elicits ALDH1A1 antioxidant activity. Furthermore, our data show that GLUT10 is crucial for both TGFβ signalling and ECM homeostasis because, in ATS fibroblasts, non-canonical αvβ3 integrin-TGFBRII cross-talk is activated, which is ineffective at supporting physiological ECM organization. Our findings support both “antioxidant-” and “enzyme cofactor-” models of a vitamin C-related pathology, but a new hypothesis of ATS aetiology can also be envisaged. In particular, since the GLUT10 re-expressing fibroblasts rescue the redox homeostasis, canonical TGFβ signalling and ECM organization under low non-physiological AA concentrations (74), a transport-independent function for GLUT10 cannot be excluded. Several transporters or channels are known to possess multiple functions (75), for instance the glucose-6-phosphate transporter in the ER was recently shown to be a key regulator of autophagy in a transport-independent manner (76). Further studies are necessary to verify the existence of such a function for GLUT10.
In conclusion, our data add new insights into the ATS dysregulated biological pathways and definition of the pathomechanisms involved in this disorder and may provide a basis for identifying potential therapeutic options.
Materials and Methods
Patients and ethics statement
Skin fibroblasts cultures from three ATS patients and three unrelated age-matched healthy donors were established in our lab from skin biopsies by standard protocols. All of the three ATS patients were previously characterized for three different mutations in the SLC2A10 gene (3,4,7). In particular, ATS patient 1 (P1) was homozygous for the c.1334del microdeletion (p.Gly445Glufs*40) that leads to the activation of the nonsense-mediated mRNA decay (3); patient 2 (P2) was homozygous for the c.1411+1G>A splice donor mutation, which causes in frame exon 3 skipping (p.Val430_Ile470del) (7); patient 3 (P3) was compound heterozygous for the c.1309G>A (p.Glu437Lys) and the c.1330C>T mutations (p.Arg444*) (4). Written informed consent was obtained from each patient for skin biopsy. This study was approved by the medical ethical committee of the University Hospital Spedali Civili of Brescia and was performed in accordance with the Declaration of Helsinki Principles.
Cell cultures and antibodies
Dermal fibroblasts were grown in vitro at 37°C in a 5% CO2 atmosphere in Earle's modified Eagle's medium (MEM) supplemented with 2 mm l-glutamine, 10% FBS, 100 µg/ml penicillin and streptomycin (Life Technologies, Carlsbad, CA, USA) (complete MEM). Fibroblasts were analysed at the same in vitro passage number (from 3rd to 7th).
Polyclonal rabbit Ab against the human GLUT10 was from Alpha Diagnostic Int. Inc. (San Antonio, TX, USA), the mAb against β-actin (clone AC-74) and the horseradish peroxidase (HRP)-conjugated anti-rabbit, anti-goat and anti-mouse secondary Abs were from Sigma Chemicals (St. Louis, MO, USA). The goat anti-COLLV and anti-COLLIII Abs were purchased from LifeSpan BioSciences, Inc. (Seattle, WA, USA). The anti-DCN mAb (clone 115402) and the goat anti-human TGFBRI/ALK5 Ab were from R&D Systems, Inc. (Minneapolis, MN, USA). The rabbit anti-TGFBRI, which recognizes all of the receptor isotypes, the anti-phospho-Tyr416 p60Src (pY416-p60Src), the anti-non-phospho-Tyr527 p60Src (npY527-p60Src), the anti-phospho-SMAD2 (pSMAD2) (Ser465/467) Abs, the rabbit anti-PPARγ (clone 81B8), the anti-SMAD2 (clone 86F7) and the rabbit anti-p60Src (clone 36D10) mAbs were purchased from Cell Signalling Technology, Inc. (Boston, MA, USA); the affinity purified anti-phospho-S112 PPARγ (pS112-PPARγ) Ab, the rabbit anti-p38 MAPK phospho-T180/Y182 (pT180/Y182-p38) Ab, the mouse anti-β8 integrin subunit Ab, and the mouse anti-phospho-tyrosine (PY20) and anti-p38 MAPK (M138) mAbs were from Abcam (Cambridge, UK); the anti-FBN (clone 11C1.3) mAb was from NeoMarkers (Fremont, CA); the mouse anti-ELN (clone 10B8), the anti-β3 integrin (clone B3A) and the anti-αvβ3 (clone LM609) integrin mAbs were from Millipore-Chemicon Int. (Billerica, MA, USA). The rabbit anti-human TGFBRII, the anti-human p125FAK, the goat anti-CTGF affinity purified Abs, and the mouse anti-ALDH1A1 mAb (clone B-5) were from Santa Cruz Biotec. Inc. (Heidelberg, Germany). The rabbit anti-integrin β3 pY773phosphospecific Ab was from Biosource Int. Inc. (Camarillo, CA, USA) and the rabbit anti-CTGF was from Thermo Scientific Pierce (Rockford, IL, USA).
Rhodamine-conjugated anti-goat secondary Ab was obtained from Calbiochem-Novabiochem INTL, the Alexa Fluor® 488 anti-rabbit and Alexa Fluor®594 anti-mouse were purchased from Life Technologies.
Microarray procedures
To screen for candidate genes that may contribute to the pathogenesis of ATS, we performed a transcriptome-wide expression profiling using the Affymetrix Gene 1.0 ST platform by comparing the gene expression patterns of skin fibroblasts of the three ATS patients with those of three healthy individuals. The microarray analysis was performed starting from 250 ng of total RNA per sample; labelled targets were prepared using Ambion® Whole Transcript Expression Kit (Life Technologies) and GeneChip® WT Terminal Labelling and Controls Kit (Affymetrix) in accordance with manufacturers' instructions. In brief, total RNA was primed with synthetic primers containing a T7 promoter sequence, reverse transcribed into first-strand cDNA and converted into double-stranded cDNA. Following the in vitro transcription, 10 µg of cRNA was reverse transcribed using random primers. 5.5 µg of second-cycle cDNA was fragmented, biotin labelled and hybridized for 16 h at 45°C onto the arrays. The chips were then washed in the Affymetrix GeneChip Fluidics station FS 450, scanned using the Affymetrix GeneChip scanner 3000 7G system and analysed with the GeneChip® Operating Software. Analysis of the miRNA expression profile was performed on three ATS patients' fibroblasts and controls in accordance with the manufacturer's instructions, starting from 250 ng of total RNA labelled with the Affymetrix Flash Tag Biotin Labelling Kit, followed by the hybridization on the GeneChip miRNA 3.0 array. The resulting CEL files were analysed using Partek® Genomics Suite software version 6.6 (Partek Inc., St. Louis, MO, USA). Analysis of variance (ANOVA) was used to identify the DEGs between ATS patients and controls, using a combination of fold-change value >1.5 and an uncorrected P-value threshold at 0.05 (P < 0.05). To identify miRNAs that showed a significantly different expression in ATS cells, one-way ANOVA was performed and the samples were filtered for fold changes >1.5 with an uncorrected P < 0.05. To identify significantly perturbed biological processes and enriched pathways in ATS patients, the DAVID database was queried to grouped DEGs into the functional clusters of GO annotations. The miRNA target prediction databases miRWalk, TargetScan and miRDB were queried to correlate the differentially expressed miRNAs with the DEGs. All microarray data are MIAME compliant, and the raw data have been deposited in the MIAME compliant GEO database with the accession numbers GSE70683 and GSE71172.
Quantitative real-time PCR
The relative mRNA levels of a series of selected genes/miRNAs identified by array analysis were confirmed by qPCR. In particular, 3 µg of total RNA purified from skin fibroblasts of three ATS patients and unrelated healthy controls were reverse-transcribed with random primers by standard procedure. qPCR reactions were performed with SYBR Green qPCR Master Mix (Life Technologies), 10 ng of cDNA, and with 10 µM of each primers set. Amplifications were performed in triplicate using the ABI PRISM 7500 Real-Time PCR System by standard thermal cycling conditions. The HPRT, GAPDH, ATP5B, CYC1 and RPLP0 reference genes were also amplified for normalization of cDNA loading. Relative mRNA expression levels were normalized to the geometric mean of these housekeeping genes and analysed using the 2−(ΔΔCt) method. Expression of miRNA was assayed using stem-loop RT–PCR starting from 50 ng of total RNA in a final volume of 15 µl followed by TaqMan-based qPCR profiling in accordance with the manufacturer's instructions (Life Technologies). The qPCR reaction contained 1.3 µl of reverse transcriptase product, 10 µl of TaqMan 2× Universal PCR Master Mix and 1 µl of the appropriate TaqMan MicroRNA assay containing primers and probes for the target miRNA. The expression of selected miRNA was based on the 2−(ΔΔCt) method, using RNU66 as internal normalization transcript, and all of the qPCR reactions were run in triplicate. Amplification plots, dissociation curves and threshold cycle values were generated by ABI Sequence detection System software version 1.3.1. Statistical analyses were performed using GraphPad Prism software (San Diego, CA, USA). The results were expressed as the mean value of relative quantification (RQ) ± SEM. Statistical significance between groups was determined using unpaired Student's t-test, or the one-way ANOVA followed by Tukey's post hoc test (*P < 0.05, **P < 0.01 and ***P < 0.001).
Construction of a stable GLUT10 expression vector
The full-length human SLC2A10 cds (refseq: NM_030777.3), from the Kozak consensus sequence to the stop codon, was amplified using the total RNA from normal skin fibroblasts with the SuperScript III One-Step RT-PCR System. The sequence was inserted into the eukaryotic pEF6/V5-His-TOPO™ expression vector (containing the blasticidine selectable marker gene and the strong human elongation factor 1α promoter) using standard procedures (Life Technologies). This expression vector (pG10) and the empty cloning vector (mock) were stably transfected into skin fibroblasts of ATS P1 using the FUGENE 6 liposomal transfection reagent in accordance with the manufacturer's instruction (Roche). The cells were selected by adding selective medium (2 µg/ml of blasticidine) that was changed every 3 days until all sensitive cells were eliminated. Established cell lines with an integrated pG10 construct or empty vector were expanded for subsequent experiments. The total RNA from all untransfected and transfected fibroblasts was extracted using the Qiagen RNeasy kit in accordance with the manufacturer's instructions (Qiagen, Hilden, Germany). RNA quality control was assessed using an Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA).
Immunofluorescence microscopy
To analyse the GLUT10 distribution, control, untransfected and transfected ATS fibroblasts were grown for 48 h and reacted for 2 min with 3% paraformaldehyde (PFA)/0.5% Triton X-100, 20 min with 3% PFA, washed with 100 mm glycine/PBS, blocked for 30 min with 5% BSA and immunoreacted ON at +4°C with 20 µg/ml anti-GLUT10 Ab. To analyse the ELN organization, the cells were grown 7–15 days in complete MEM, fixed in 1% PFA for 20 min, treated 1 h at 37°C with 10 U/ml hyaluronidase diluted in 5% FBS-MEM, and, after washing, immunoreacted with 1:50 diluted anti-ELN mAb for 1 h. The DCN organization was analysed 48 h after seeding fixing cells with 3% PFA/PBS for 20 min. Subsequently, the cells were incubated for 1 h with 25 µg/ml anti-DCN mAb, which recognized the core protein. The COLLIII, COLLV and FBN organization into ECM, was monitored for the control, untransfected, and transfected ATS fibroblasts from 48 h to 15 days, and the TGFBRI, PPARγ, ALDH1A1 and pSer112-PPARγ distribution was analysed in cells treated or not ON with increasing quantities, from 1.0 to 2.5 µM, of the PPARγ antagonist T0070907. The cells were fixed in cold methanol and reacted for 1 h with anti-COLLIII or anti-COLLV Abs; anti-FBN, which recognized all of the FBN isoforms; anti-ALDH1A1 mAbs diluted 1:100 in 1% BSA or 14 µg/ml anti-TGFBRI Ab, 2.5 µg/ml anti-pS112-PPARγ Ab and 1:200 diluted anti-PPARγ mAb. Before the anti-pS112-PPARγ Ab incubation, we permeabilized and blocked the cells with 1% BSA/10% normal goat serum/0.3 m glycine in 0.1% PBS-Tween 20. The analyses for Src and its activation were performed by fixing cells for 10 min in 3% PFA, permeabilizing for 2 min with 0.5% Triton X-100 and immunoreacting with 1:100 anti-pY416-p60Src and anti-npY527-p60Src Abs as well as 2 µg/ml anti-p60Src mAb. To investigate the distribution of the αvβ3 integrin, β8 integrin subunit, pY773β3 integrin subunit, p125FAK, SMAD2 and its phosphorylation, the cells were fixed as previously reported (58,59). The cells were then immunoreacted for 2 h with 1:100 anti-αvβ3 integrin, anti-β8 and anti-pY773β3, and 1:300 anti-SMAD2 mAbs, and with 1:100 anti-pSMAD2 (Ser465/467) and anti-p125FAK Abs. To detect the membrane-associated insoluble fraction of CTGF, 48 h grown control and ATS fibroblasts were incubated in prechilled 1% Triton X-100 on ice for 20 min, washed in PBS/0.15 M glycine, fixed in 3% PFA for 7 min, blocked in 1% BSA and immunoreacted for 1 h at room temperature (RT) with 1:50 anti-CTGF Ab (34). The analysis of p38 and its T180/Y182 phosphorylation was performed on cells fixed in cold methanol, permeabilized for 1 h and immunoreacted with 2.5 µg/ml anti-p38 mAb or 5.0 µg/ml anti-pT180/Y182-p38 Ab. After washing in PBS, the cells were incubated for 1 h with the Alexa Fluor® 488 anti-rabbit and Alexa Fluor® 594 anti-mouse secondary Abs or the rhodamine-conjugated anti-goat IgG. The IF signals were collected using a CCD black and white TV camera (SensiCam-PCO Computer Optics GmbH, Germany) mounted on a Zeiss fluorescence-Axiovert microscope and digitalized using the Image Pro Plus program (Media Cybernetics, Silver Spring, MD). The experiments were repeated three times.
To investigate β3 integrin subunit co-distribution with p125FAK, the Duolink in situ PLA probe and reagents (Olink® Bioscience, Sigma Chemicals) were used in accordance with the manufacturers' instructions. In brief, control and ATS fibroblasts were grown on glass slides for 48 h, fixed and immunoreacted ON at +4°C as previously reported (58,59). The cells were then washed and incubated for 1 h at 37°C with the two PLA probes, the anti-mouse MINUS probe and the anti-rabbit PLUS probe directed against the anti-β3 mAb and anti-p125FAK Ab, respectively. The two probes are secondary Abs that are labelled with an oligonucleotide generating a fluorescent signal only when they are bound to the sample in close proximity. After removing the probes and washing, the slides were incubated in a pre-heated humidity chamber for 30 min at 37°C with ligase diluted 1:40 in the ligation buffer containing the oligonucleotides that hybridize to PLA probes. The slides were washed, reacted in the dark for 100 min at 37°C with 1:80 diluted polymerase, the rolling circle amplification was performed in the presence of fluorophore-labelled oligonucleotide probes, and the slides mounted for fluorescent spot detection using microscopy.
ROS and LPO detection and ALDH activity measurement
To analyse the oxidative stress status in ATS versus control fibroblasts, the ROS production was monitored using the CellROX® Green Assay Kit (Molecular Probes® by Life Technologies), in accordance with the manufacturers' instructions. This kit efficiently detects the hydroxyl radical (OH−), superoxide anion () and TBHP. In brief, control, untransfected and transfected ATS fibroblasts, grown for 48 h on a coverslide in complete MEM, were treated or not ON with increasing amounts, from 1.0 to 2.5 µm, of PPARγ antagonist T0070907 (Sigma); the cells were then treated with 500 nm non-fluorescent cell-permeable reduced CellROX® reagent for 1 h at 37°C. Under these conditions, the CellROX® reagent developed a fluorogenic signal upon oxidation, which indicated the presence of ROS in live cells. The fluorescence developed by the ROS-mediated reaction was monitored in 3% PFA-fixed cells using microscopy with a 488 nm excitation filter. Control fibroblasts were treated with TBHP to induce the ROS production as a positive control or with a mix of THBP and antioxidant N-acetylcysteine (NAC) as a negative control.
To analyse the LPO process in ATS versus control fibroblasts, the Click-iT® LPO detection kit with LAA (Molecular Probes® by Life Technologies) was used in accordance with the manufacturers' instructions. In brief, the LAA substrate was incorporated or not into the cells, grown for 48 h on a coverslide in complete MEM. When LAA is oxidized, it produces hydroperoxides that decompose to unsaturated aldehydes, which modify proteins and DNA. These modifications were detected through fixing cells with 3.7% PFA, permeabilizing with 0.5% Tween 20, blocking with 1% BSA, and reacting for 30 min with the Click-iT® reaction cocktail, which contains the Alexa Fluor 488 azide. Control fibroblasts were treated with cumene hydroperoxide to induce LPO. Labelling was monitored by fluorescence microscopy.
To measure the ALDH enzyme activity, we used the ALDEFLUORTM detection kit in accordance with the manufacturers' instructions (STEMCELL Technologies, Vancouver, Canada). In particular, control, untransfected and transfected ATS cells were reacted for 1 h at 37°C with the ALDEFLUORTM Bodipy-conjugated aminoacetaldehyde (BAAA) reagent, which is a non-toxic substrate that is converted into Bodipy-conjugated aminoacetate in the presence of ALDH that is retained inside the cells. The fluorescence was monitored by flow cytometry using the BD FACSCanto™ Flow Cytometer and analysed using BD FACSDiva Software 6.0 (BD Biosciences). In particular, the fold change of fluorescence greater than the background mean fluorescence measured without BAAA was calculated for all of the cell types; the experiments were repeated three times. The results were expressed as RQ of ALDH activity in ATS versus controls and in pG10 versus mock-transfected cells. Statistical analyses were performed using GraphPad Prism software (***P < 0.0001). A specific ALDH inhibitor, the diethylaminobenzaldehyde, was used as a negative control.
Cell extracts and western blotting
To analyse the ALDH1A1 enzyme and CTGF, control, untransfected and transfected ATS fibroblast were grown for 48 h in complete MEM, washed twice in ice-cold PBS and extracted for 1 h at +4°C with the RIPA buffer containing 150 mm NaCl, 20 mm Tris–HCl, pH 7.4, 1% Nonidet P-40, 0.1% SDS, 1 mm EDTA, 1 µg/ml leupeptin, 4 µg/ml pepstatin, 1 mm PMSF, 1 mm Na3VO4, 25 mm NaF and 10 mm Na4P2O7. After scraping, the cell extracts were centrifuged at 14 000 rpm and +4°C for 12 min.
The PPARγ analysis was performed through scraping cells in wash buffer (10 mm PBS, 1 mm NaF and 1 mm Na3VO4), recovering the cells after a 3000 rpm centrifugation and treating the cells at +4°C for 1 h with agitation with a lysis buffer containing 50 mm KCl, 1% Nonidet P-40, 25 mm HEPES, pH 7.8, 10 µg/ml leupeptin, 20 µg/ml aprotinin, 125 µm DTT, 1 mm PMSF, 1 mm Na3VO4 and 1 mm NaF. The cell extracts were sonicated four times for 8 s and centrifuged at 12 000 rpm and +4°C for 10 min.
The membrane-bound TGFBRI and TGFBRII analysis was performed using the cell extracts obtained by lysing the cells with 1% Triton X-100, 0.1% SDS in 50 mm Tris–HCl, pH 7.4, 150 mm NaCl, 1 mm EGTA, 1 mm PMSF, 1 mm NaF, 1 mm Na3VO4, 1 µg/ml leupeptin and pepstatin, and 20 µg/ml aprotinin.
The integrin extraction was performed by lysing the cells with 0.5% Triton X-100, 150 mm NaCl, 1 mm CaCl2, 1 mm MgCl2, 20 mm Tris–HCl, pH 7.4, 1 mm Na3VO4, 10 mm NaF and 10 mm Na4P2O7, 10 µg/ml leupeptin, 4 µg/ml pepstatin and 0.1 U/ml aprotinin.
The protein concentration was evaluated using detergent compatible Bio-Rad DC Protein Assay (Bio-Rad Laboratories, Hercules, CA, USA); 20–50 μg of the total protein was separated by electrophoresis using 8% SDS–PAGE. After nitrocellulose sheet transfer, the membranes were blocked ON at 37°C or at +4°C with 5% non-fat dry milk (w/v) in TBS-0.1% Tween 20 (TBS-T) (blocking buffer) and immunoreacted for 2 h at RT with 0.4 μg/ml anti-ALDH1A1 mAb or anti-CTGF Ab, with 1:1000 anti-TGFBRI and 1:500 anti-TGFBRII Abs and with 1:1000 anti-PPARγ mAb or 2 μg/ml anti-pS112-PPARγ Ab ON at +4°C and diluted in TBS-T. After washing in TBS-T, the membranes were incubated for 2 h at RT with HRP-conjugated anti-rabbit, anti-mouse or anti-goat IgGs, diluted in blocking buffer and developed using the ECL method (Pierce). The filters were stripped, tested for the absence of a residual signal and reprobed with 1 μg/ml anti-β actin mAb diluted in TBS-T for 2 h at RT.
To analyse tyrosine phosphorylation of TGFBRII and its co-precipitation with β3 integrin subunit, 1 mg of TGFBR cell extract was immunoprecipitated with 10 µg of anti-TGFBRII Ab in accordance with the immunoprecipitation Dynabeads protein G kit instructions (Novex by Life Technologies). The immunocomplexes were tested through WB and immunoreacting with anti-TGFBRII (1:500); 1:1000 anti-TGFBRI; anti-pY416-p60Src; anti-pT180/Y182-p38 MAPK; and 1:1000 anti-PY20, anti-p38 MAPK and anti-β3 integrin subunit mAbs.
To investigate the role of p60Src in p38 MAPK phosphorylation, the control, untransfected and transfected ATS P1 cells were treated for 2 h with a subapoptotic dose (50 µM) of the specific Src family kinase inhibitor PP2 or its inactive analogue PP3 (Calbiochem), extracted for TGFBR, immunoprecipitated with anti-TGFBRII Ab and immunoreacted with anti-TGFBRII and anti-pT180/Y182-p38 Abs and anti-PY20 and anti-p38 mAbs, as reported above. Alternatively, 1 mg of integrin cell extract was immunoprecipitated with 10 µg of anti-β3 integrin subunit mAb and tested for co-immunoprecipitation of TGFBRII, TGFBRI and CTGF by WB using the above conditions; of p125FAK through immunoreacting with 0.2 µg/ml anti-p125FAK Ab; and of p60Src using 1:1000 rabbit anti-p60Src mAb. Each experiment was performed two times.
Supplementary Material
Funding
This work was supported by the Telethon Foundation: grant number GGP13167 to M.C. Funding to pay the Open Access publication charges for this article was provided by the Telethon Foundation.
Supplementary Material
Acknowledgments
Conflict of Interest statement. None declared.
References
- 1.Gardella R., Zoppi N., Assanelli D., Muiesan M.L., Barlati S., Colombi M. (2004) Exclusion of candidate genes in a family with arterial tortuosity syndrome. Am. J. Med. Genet. A, 126, 221–228. [DOI] [PubMed] [Google Scholar]
- 2.Wessels M.W., Catsman-Berrevoets C.E., Mancini G.M., Breuning M.H., Hoogeboom J.J., Stroink H., Frohn-Mulder I., Coucke P.J., Paepe A.D., Niermeijer M.F., Willems P.J. (2004) Three new families with arterial tortuosity syndrome. Am. J. Med. Genet. A, 131, 134–143. [DOI] [PubMed] [Google Scholar]
- 3.Coucke P.J., Willaert A., Wessels M.W., Callewaert B., Zoppi N., De Backer J., Fox J.E., Mancini G.M., Kambouris M., Gardella R. et al. (2006) Mutations in the facilitative glucose transporter GLUT10 alter angiogenesis and cause arterial tortuosity syndrome. Nat. Genet., 38, 452–457. [DOI] [PubMed] [Google Scholar]
- 4.Drera B., Guala A., Zoppi N., Franceschini P., Barlati S., Colombi M. (2007) Two novel SLC2A10/GLUT10 mutations in a patient with arterial tortuosity syndrome. Am. J. Med. Genet. A., 143, 216–218. [DOI] [PubMed] [Google Scholar]
- 5.Callewaert B.L., Willaert A., Kerstjens-Frederikse W.S., De Backer J., Devriendt K., Albrecht B., Ramos-Arroyo M.A., Doco-Fenzy M., Hennekam R.C., Pyeritz R.E. et al. (2008) Arterial tortuosity syndrome: clinical and molecular findings in 12 newly identified families. Hum. Mutat., 29, 150–158. [DOI] [PubMed] [Google Scholar]
- 6.Ritelli M., Drera B., Vicchio M., Puppini G., Biban P., Pilati M., Prioli M.A., Barlati S., Colombi M. (2009) Arterial tortuosity syndrome in two Italian paediatric patients. Orphanet J. Rare. Dis., 4, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castori M., Ritelli M., Zoppi N., Molisso L., Chiarelli N., Zaccagna F., Grammatico P., Colombi M. (2012) Adult presentation of arterial tortuosity syndrome in a 51-year-old woman with a novel homozygous c.1411+1G>A mutation in the SLC2A10 gene. Am. J. Med. Genet. A., 58, 1164–1169. [DOI] [PubMed] [Google Scholar]
- 8.Ritelli M., Chiarelli N., Dordoni C., Reffo E., Venturini M., Quinzani S., Monica M.D., Scarano G., Santoro G., Russo M.G. et al. (2014) Arterial tortuosity syndrome: homozygosity for two novel and one recurrent SLC2A10 missense mutations in three families with severe cardiopulmonary complications in infancy and a literature review. BMC Med. Genet., 15, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueckler M., Thorens B. (2103) The SLC2 (GLUT) family of membrane transporters. Mol. Aspects Med., 34, 121–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bánhegyi G., Benedetti A., Margittai E., Marcolongo P., Fulceri R., Németh C.E., Szarka A. (2014) Subcellular compartmentation of ascorbate and its variation in disease states. Biochim. Biophys. Acta, 1843, 1909–1916. [DOI] [PubMed] [Google Scholar]
- 11.Callewaert B.L., Loeys B.L., Casteleyn C., Willaert A., Dewint P., De Backer J., Sedlmeier R., Simoens P., De Paepe A.M., Coucke P.J. (2008) Absence of arterial phenotype in mice with homozygous slc2A10 missense substitutions. Genesis, 46, 385–389. [DOI] [PubMed] [Google Scholar]
- 12.Cheng C.H., Kikuchi T., Chen Y.H., Sabbagha N.G., Lee Y.C., Pan H.J., Chang C., Chen Y.T. (2009) Mutations in the SLC2A10 gene cause arterial abnormalities in mice. Cardiovasc. Res., 81, 381–388. [DOI] [PubMed] [Google Scholar]
- 13.Chiarelli N., Ritelli M., Zoppi N., Benini A., Borsani G., Barlati S., Colombi M. (2011) Characterization and expression pattern analysis of the facilitative glucose transporter 10 gene (slc2a10) in Danio rerio. Int. J. Dev. Biol., 55, 229–236. [DOI] [PubMed] [Google Scholar]
- 14.Willaert A., Khatri S., Callewaert B.L., Coucke P.J., Crosby S.D., Lee J.G., Davis E.C., Shiva S., Tsang M., De Paepe A., Urban Z. (2012) GLUT10 is required for the development of the cardiovascular system and the notochord and connects mitochondrial function to TGFβ signaling. Hum. Mol. Genet., 21, 1248–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akhurst R.J. (2006) A sweet link between TGFbeta and vascular disease? Nat. Genet., 38, 400–401. [DOI] [PubMed] [Google Scholar]
- 16.Akhurst R.J. (2012) The paradoxical TGF-β vasculopathies. Nat. Genet., 44, 838–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segade F. (2010) Glucose transporter 10 and arterial tortuosity syndrome: the vitamin C connection. FEBS Lett., 584, 2990–2994. [DOI] [PubMed] [Google Scholar]
- 18.Lee Y.C., Huang H.Y., Chang C.J., Cheng C.H., Chen Y.T. (2010) Mitochondrial GLUT10 facilitates dehydroascorbic acid import and protects cells against oxidative stress: mechanistic insight into arterial tortuosity syndrome. Hum. Mol. Genet., 19, 3721–3733. [DOI] [PubMed] [Google Scholar]
- 19.Traber M.G., Stevens J.F. (2011) Vitamins C and E: beneficial effects from a mechanistic perspective. Free Radic. Biol. Med., 51, 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szarka A., Balogh T. (2015) In silico aided thoughts on mitochondrial vitamin C transport. J. Theoret. Biol., 365, 181–189. [DOI] [PubMed] [Google Scholar]
- 21.Muñoz-Montesino C., Roa F.J., Peña E., González M., Sotomayor K., Inostroza E., Muñoz C.A., González I., Maldonado M., Soliz C. et al. (2014) Mitochondrial ascorbic acid transport is mediated by a low-affinity form of the sodium-coupled ascorbic acid transporter-2. Free Radic. Biol. Med., 70, 241–254. [DOI] [PubMed] [Google Scholar]
- 22.Makia N.L., Bojang P., Falkner K.C., Conklin D.J., Prough R.A. (2011) Murine hepatic aldehyde dehydrogenase 1a1 is a major contributor to oxidation of aldehydes formed by lipid peroxidation. Chem. Biol. Interact., 191, 278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh S., Brocker C., Koppaka V., Chen Y., Jackson B.C., Matsumoto A., Thompson D.C., Vasiliou V. (2013) Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic. Biol. Med., 56, 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng J.B., Russell D.W. (2004) Mammalian wax biosynthesis. I. Identification of two fatty acyl-Coenzyme A reductases with different substrate specificities and tissue distributions. J. Biol. Chem., 279, 37789–37797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burdge G.C. (2006) Metabolism of alpha-linolenic acid in humans. Prostaglandins Leukot. Essent. Fatty Acids, 75, 161–168. [DOI] [PubMed] [Google Scholar]
- 26.Buczynski M.W., Dumlao D.S., Dennis E.A. (2009) Thematic review series: proteomics. An integrated omics analysis of eicosanoid biology. J. Lipid. Res., 50, 1015–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchesi C., Schiffrin E.L. (2008) Peroxisome proliferator-activated receptors and the vascular system: beyond their metabolic effects. J. Am. Soc. Hypertens., 2, 227–238. [DOI] [PubMed] [Google Scholar]
- 28.Busnadiego O., González-Santamaría J., Lagares D., Guinea-Viniegra J., Pichol-Thievend C., Muller L., Rodríguez-Pascual F. (2013) LOXL4 is induced by transforming growth factor β1 through Smad and JunB/Fra2 and contributes to vascular matrix remodeling. Mol. Cell. Biol., 33, 2388–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nurminskaya M.V., Belkin A.M. (2012) Cellular functions of tissue transglutaminase. Int. Rev. Cell. Mol. Biol., 294, 1–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valcourt U., Alcaraz L.B., Exposito J.Y., Lethias C., Bartholin L. (2015) Tenascin-X: beyond the architectural function. Cell. Adh. Migr., 9, 154–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Acharya C., Yik J.H., Kishore A., Van Dinh V., Di Cesare P.E., Haudenschild D.R. (2014) Cartilage oligomeric matrix protein and its binding partners in the cartilage extracellular matrix: interaction, regulation and role in chondrogenesis. Matrix Biol., 37, 102–111. [DOI] [PubMed] [Google Scholar]
- 32.Hu Z.Y., Luo J.F., Zhong S.L., Xue L., Chen Y.F., Fan R.X. (2012) MicroRNAs expression in normal and dissected aortic tissue. Zhonghua Xin Xue Guan Bing Za Zhi, 40, 406–410. [PubMed] [Google Scholar]
- 33.Ahmadian M., Suh J.M., Hah N., Liddle C., Atkins A.R., Downes M., Evans R.M. (2013) PPARγ signaling and metabolism: the good, the bad and the future. Nat. Med., 19, 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tall E.G., Bernstein A.M., Oliver N., Gray J.L., Masur S.K. (2010) TGF-β-stimulated CTGF production enhanced by collagen and associated with biogenesis of a novel 31-kDa CTGF form in human corneal fibroblasts. Invest. Ophthalmol. Vis. Sci., 51, 5002–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakerakanti S.S., Bujor A.M., Trojanowska M. (2011) CCN2 is required for the TGF-β induced activation of Smad1-Erk1/2 signaling network. PLoS ONE, 6, e21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girotti A.W. (1998) Lipid hydroperoxide generation, turnover, and effector action in biological systems. J. Lipid Res., 39, 1529–1542. [PubMed] [Google Scholar]
- 37.Yin H., Xu L., Porter N.A. (2011) Free radical lipid peroxidation: mechanisms and analysis. Chem. Rev., 111, 5944–5972. [DOI] [PubMed] [Google Scholar]
- 38.Leonarduzzi G., Chiarpotto E., Biasi F., Poli G. (2005) 4-Hydroxynonenal and cholesterol oxidation products in atherosclerosis. Mol. Nutr. Food Res., 49, 1044–1049. [DOI] [PubMed] [Google Scholar]
- 39.Poli G., Biasi F., Leonarduzzi G. (2008) 4-Hydroxynonenal-protein adducts: a reliable biomarker of lipid oxidation in liver diseases. Mol. Aspects Med., 29, 67–71. [DOI] [PubMed] [Google Scholar]
- 40.Dalleau S., Baradat M., Guéraud F., Huc L. (2013) Cell death and diseases related to oxidative stress: 4-hydroxynonenal (HNE) in the balance. Cell. Death Differ., 20, 1615–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasiliou V., Thompson D.C., Smith C., Fujita M., Chen Y. (2013) Aldehyde dehydrogenases: from eye crystallins to metabolic disease and cancer stem cells. Chem. Biol. Interact., 202, 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim T., Yang Q. (2013) Peroxisome-proliferator-activated receptors regulate redox signaling in the cardiovascular system . World J. Cardiol., 5, 164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanquicett C., Kang B.Y., Ritzenthaler J.D., Jones D.P., Hart C.M. (2010) Oxidative stress modulates PPAR gamma in vascular endothelial cells. Free Radic. Biol. Med., 48, 1618–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staels B., Koenig W., Habib A., Merval R., Lebret M., Torra I.P., Delerive P., Fadel A., Chinetti G., Fruchart J.C. et al. (1998) Activation of human aortic smooth-muscle cells is inhibited by PPARalpha but not by PPARgamma activators. Nature, 393, 790–793. [DOI] [PubMed] [Google Scholar]
- 45.Almeida M., Ambrogini E., Han L., Manolagas S.C., Jilka R.L. (2009) Increased lipid oxidation causes oxidative stress, increased peroxisome proliferator-activated receptor-gamma expression, and diminished pro-osteogenic Wnt signaling in the skeleton. J. Biol. Chem., 284, 27438–27448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Small D.M., Morais C., Coombes J.S., Bennett N.C., Johnson D.W., Gobe G.C. (2014) Oxidative stress-induced alterations in PPAR-γ and associated mitochondrial destabilization contribute to kidney cell apoptosis. Am. J. Physiol. Renal. Physiol., 307, F814–F822. [DOI] [PubMed] [Google Scholar]
- 47.Valko M., Rhodes C.J., Moncol J., Izakovic M., Mazur M. (2006) Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact., 160, 1–40. [DOI] [PubMed] [Google Scholar]
- 48.Brown D.I., Griendling K.K. (2015) Regulation of signal transduction by reactive oxygen species in the cardiovascular system. Circ. Res., 116, 531–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chuang C.Y., Degendorfer G., Davies M.J. (2014) Oxidation and modification of extracellular matrix and its role in disease. Free Radic. Res., 48, 970–989. [DOI] [PubMed] [Google Scholar]
- 50.Griendling K.K., FitzGerald G.A. (2003) Oxidative stress and cardiovascular injury. Part I. Basic mechanisms and in vivo monitoring of ROS. Circulation., 108, 912–916. [DOI] [PubMed] [Google Scholar]
- 51.Nieuwdorp M., van Haeften T.W., Gouverneur M.C., Mooij H.L., van Lieshout M.H., Levi M., Meijers J.C., Holleman F., Hoekstra J.B., Vink H., Kastelein J.J., Stroes E.S. (2006) Loss of endothelial glycocalyx during acute hyperglycemia coincides with endothelial dysfunction and coagulation activation in vivo. Diabetes, 55, 480–486. [DOI] [PubMed] [Google Scholar]
- 52.Eble J.A., de Rezende F.F. (2014) Redox-relevant aspects of the extracellular matrix and its cellular contacts via integrins. Antioxid. Redox. Signal., 20, 1977–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao S.S., Kaufman R.J. (2014) Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid. Redox. Signal., 21, 396–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haynes C.M., Titus E.A., Cooper A.A. (2004) Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol. Cell, 15, 767–776. [DOI] [PubMed] [Google Scholar]
- 55.Munger J.S., Sheppard D. (2011) Cross talk among TGF-β signaling pathways, integrins, and the extracellular matrix. Cold Spring Harb. Perspect. Biol., 3, a005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Annes J.P., Munger J.S., Rifkin D.B. (2003) Making sense of latent TGFbeta activation. J. Cell. Sci., 116, 217–224. [DOI] [PubMed] [Google Scholar]
- 57.Mu Y., Gudey S.K., Landström M. (2012) Non-Smad signaling pathways. Cell. Tissue Res., 347, 11–20. [DOI] [PubMed] [Google Scholar]
- 58.Zoppi N., Barlati S., Colombi M. (2008) FAK-independent alphavbeta3 integrin-EGFR complexes rescue from anoikis matrix-defective fibroblasts. Biochim. Biophys. Acta, 1783, 1177–1188. [DOI] [PubMed] [Google Scholar]
- 59.Zoppi N., Gardella R., De Paepe A., Barlati S., Colombi M. (2004) Human fibroblasts with mutations in COL5A1 and COL3A1 genes do not organize collagens and fibronectin in the extracellular matrix, down-regulate alpha2beta1 integrin, and recruit alphavbeta3 Instead of alpha5beta1 integrin. J. Biol. Chem., 279, 18157–18168. [DOI] [PubMed] [Google Scholar]
- 60.Liu S., Xu S.W., Kennedy L., Pala D., Chen Y., Eastwood M., Carter D.E., Black C.M., Abraham D.J., Leask A. (2007) FAK is required for TGFbeta-induced JNK phosphorylation in fibroblasts: implications for acquisition of a matrix-remodeling phenotype. Mol. Biol. Cell., 18, 2169–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leask A. (2013) Focal adhesion kinase: a key mediator of transforming growth factor beta signaling in fibroblasts. Adv. Wound. Care., 2, 247–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Babic A.M., Chen C.C., Lau L.F. (1999) Fisp12/mouse connective tissue growth factor mediates endothelial cell adhesion and migration through integrin alphavbeta3, promotes endothelial cell survival, and induces angiogenesis in vivo . Mol. Cell Biol., 19, 2958–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ludbrook S.B., Barry S.T., Delves C.J., Horgan C.M. (2003) The integrin alphavbeta3 is a receptor for the latency-associated peptides of transforming growth factors beta1 and beta3. Biochem. J., 369, 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scaffidi A.K., Petrovic N., Moodley Y.P., Fogel-Petrovic M., Kroeger K.M., Seeber R.M., Eidne K.A., Thompson P.J., Knight D.A. (2004) Alpha(v)beta(3) integrin interacts with the transforming growth factor beta (TGFbeta) type II receptor to potentiate the proliferative effects of TGFbeta1 in living human lung fibroblasts. J. Biol. Chem., 279, 37726–37733. [DOI] [PubMed] [Google Scholar]
- 65.Margadant C., Sonnenberg A. (2010) Integrin-TGF-beta crosstalk in fibrosis, cancer and wound healing. EMBO Rep., 11, 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Galliher A.J., Schiemann W.P. (2007) Src phosphorylates Tyr284 in TGF-beta type II receptor and regulates TGF-beta stimulation of p38 MAPK during breast cancer cell proliferation and invasion. Cancer Res., 67, 3752–3768. [DOI] [PubMed] [Google Scholar]
- 67.Pechkovsky D.V., Scaffidi A.K., Hackett T.L., Ballard J., Shaheen F., Thompson P.J., Thannickal V.J., Knight D.A. (2008) Transforming growth factor beta1 induces alphavbeta3 integrin expression in human lung fibroblasts via a beta3 integrin-, c-Src-, and p38 MAPK-dependent pathway. J. Biol. Chem., 283, 12898–13908. [DOI] [PubMed] [Google Scholar]
- 68.Chen X., Wang H., Liao H.J., Hu W., Gewin L., Mernaugh G., Zhang S., Zhang Z.Y., Vega-Montoto L., Vanacore R.M. et al. (2014) Integrin-mediated type II TGF-β receptor tyrosine dephosphorylation controls SMAD-dependent profibrotic signaling. J. Clin. Invest., 124, 3295–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ayala A., Muñoz M.F., Argüelles S. (2014) Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal . Oxid. Med. Cell. Longev., 2014, 360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He T., Quan T., Shao Y., Voorhees J.J., Fisher G.J. (2014) Oxidative exposure impairs TGF-β pathway via reduction of type II receptor and SMAD3 in human skin fibroblasts. Age, 36, 9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larroque-Cardoso P., Mucher E., Grazide M.H., Josse G., Schmitt A.M., Nadal-Wolbold F., Zarkovic K., Salvayre R., Nègre-Salvayre A. (2014) 4-Hydroxynonenal impairs transforming growth factor-β1-induced elastin synthesis via epidermal growth factor receptor activation in human and murine fibroblasts. Free Radic. Biol. Med., 71, 427–436. [DOI] [PubMed] [Google Scholar]
- 72.Ding Z.Y., Jin G.N., Liang H.F., Wang W., Chen W.X., Datta P.K., Zhang M.Z., Zhang B., Chen X.P. (2013) Transforming growth factor β induces expression of connective tissue growth factor in hepatic progenitor cells through Smad independent signaling. Cell. Signal., 25, 1981–1992. [DOI] [PubMed] [Google Scholar]
- 73.Leask A., Holmes A., Black C.M., Abraham D.J. (2003) Connective tissue growth factor gene regulation. Requirements for its induction by transforming growth factor-beta 2 in fibroblasts. J. Biol. Chem., 278, 13008–13015. [DOI] [PubMed] [Google Scholar]
- 74.Arigony A.L., de Oliveira I.M., Machado M., Bordin D.L., Bergter L., Prá D., Henriques J.A. (2013) The influence of micronutrients in cell culture: a reflection on viability and genomic stability. Biomed. Res. Int., 2013, 597282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salaün C., Leroy C., Rousseau A., Boitez V., Beck L., Friedlander G. (2010) Identification of a novel transport-independent function of PiT1/SLC20A1 in the regulation of TNF-induced apoptosis. J. Biol. Chem., 285, 34408–34418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahn H.H., Oh Y., Lee H., Lee W., Chang J.W., Pyo H.K., Nah do H., Jung Y.K. (2015) Identification of glucose-6-phosphate transporter as a key regulator functioning at the autophagy initiation step. FEBS Lett., 589, 2100–2109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.