Summary
Photoreceptor proteins enable organisms to sense and respond to light. The newly discovered CarH-type photoreceptors use a vitamin B12 derivative, adenosylcobalamin, as the light-sensing chromophore to mediate light-dependent gene regulation. Here, we present crystal structures of Thermus thermophilus CarH in all three relevant states: in the dark, both free and bound to operator DNA, and after light exposure. These structures provide a visualization of how adenosylcobalamin mediates CarH tetramer formation in the dark, how this tetramer binds to the promoter −35 element to repress transcription, and how light exposure leads to a large-scale conformational change that activates transcription. In addition to the remarkable functional repurposing of adenosylcobalamin from an enzyme cofactor to a light sensor, we find that nature also repurposed two independent protein modules in assembling CarH. These results expand the biological role of vitamin B12 and provide fundamental insight into a new mode of light-dependent gene regulation.
Light allows for photosynthesis and other essential light-dependent chemical reactions. Light also triggers photooxidative stress via generation of reactive oxygen species, which rapidly damage the cell1. Organisms in all domains of life produce proteins capable of sensing light. These biological photoreceptors participate in vision, harness light energy, regulate circadian clocks, and mediate gene expression and major developmental processes2-5. Different classes of photoreceptor proteins with a variety of chromophore cofactors exist to sense light over the visible and ultraviolet spectrum6-7. This group of photoreceptors was recently expanded by a new class that is widespread in bacteria and uses the vitamin B12 derivative adenosylcobalamin (AdoCbl) as the chromophore to orchestrate light perception and response8-9. The prototypes of this class, the CarH-type photoreceptors, have been characterized in Myxococcus xanthus8-9 and Thermus thermophilus10 and typically regulate light-induced expression of carotenoid biosynthetic genes, which results in carotenoid-mediated protection against photo-oxidative damage10-12. This class of photoreceptors thereby allows bacteria to mitigate the detrimental effects of sunlight, a critical determinant of survival in light-exposed conditions, while avoiding unnecessary production of carotenoids in the absence of light.
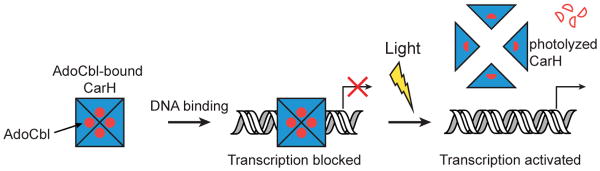
The CarH-type photoreceptors consist of an N-terminal DNA-binding domain and a C-terminal AdoCbl-binding and oligomerization domain to directly sense light and regulate gene expression. In the dark, AdoCbl-bound CarH, a tetramer, binds to the promoter region of target genes to repress transcription. Exposure to blue, green, or ultraviolet light disrupts the photosensitive Co-C bond in AdoCbl, leading to tetramer disassembly, loss of operator-binding, and activation of gene expression (Figure 1)8. Thus, AdoCbl, which is typically used as a cofactor for radical-based enzyme reactions13-14, is now being used for a new biological function as a light sensor. Here, we sought to determine the structural basis for the functional repurposing of one of nature's most complex metallocofactors and for this new mode of light-dependent gene regulation.
Figure 1.
Schematic of CarH-mediated light-dependent gene regulation. Structures of all three relevant states are reported herein. Red circles depict AdoCbl, red half moons photolyzed Cbl, and open red half moons 4′,5′- anhydroadenosine, the recently identified photolysis product of CarH-bound AdoCbl33. See main text for details.
CarH has a modular architecture
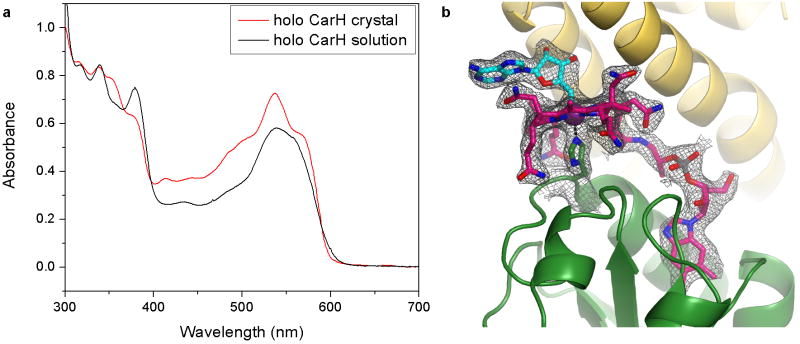
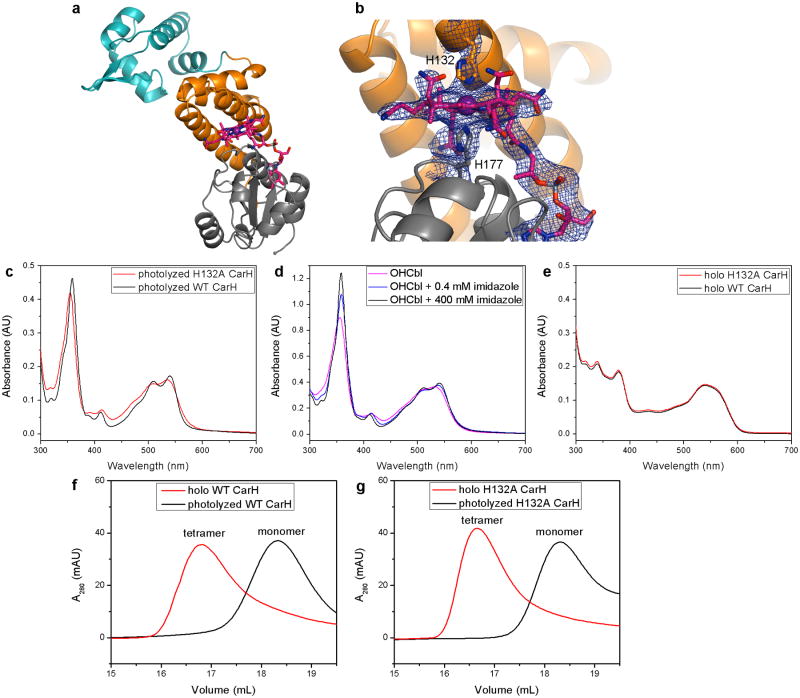
To visualize the CarH domain structure and the architecture of the repressor “dark” state, we first determined two independent structures of AdoCbl-bound CarH from T. thermophilus to 2.15 Å and 2.80 Å resolution (Extended Data Table 1). To prevent cleavage of the light-sensitive AdoCbl Co-C bond, we carried out all crystallization experiments under red light and all diffraction data collection at T=100 K. Both single-crystal UV-Vis spectra and the electron density confirmed that AdoCbl remained intact with a Co-C bond length of 2.0 Å (Extended Data Figure 1), indicating that the structures are in the “dark” state. Each monomer of CarH has a modular three-domain architecture with an N-terminal winged-helix DNA-binding domain followed by the light-sensing domain, composed of a four-helix bundle and a C-terminal Rossmann-fold cobalamin (Cbl)-binding domain (Figure 2a).
Figure 2.
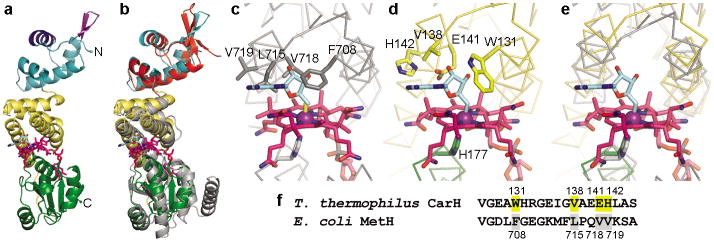
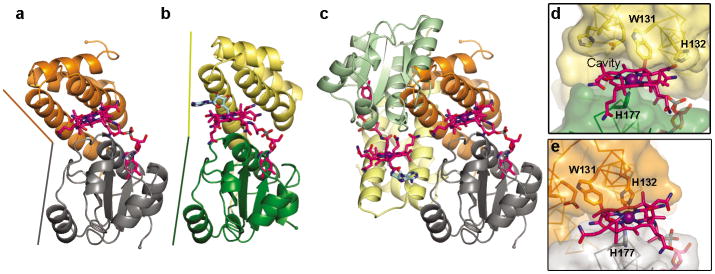
Structure of CarH protomer and comparison to MetH. (a) CarH protomer colored by domain: N-terminal DNA-binding domain (cyan) with recognition helix (dark blue) and β-hairpin wing (purple) highlighted; central four-helix bundle (yellow); C-terminal Cbl-binding domain (green). AdoCbl shown with Cbl carbons in pink, 5′-dAdo group carbons in cyan, cobalt in purple. (b) Overlay of CarH protomer with Cbl-binding module of MetH (gray, PDB ID code 1BMT17) and BmrR DNA-binding domain (red, PDB ID code 1EXJ38). (c) MetH binds MeCbl (methyl group in yellow); modeling 5′-dAdo (cyan) results in steric clashes. (d) CarH accommodates AdoCbl through several substitutions compared to MetH. Cobalt-coordinating His in green. (e) Superposition of MetH and CarH, highlighting shift of the four-helix bundle between MetH (gray) and CarH (yellow). (f) Alignment of CarH and MetH sequences involved in binding the Cbl upper face, highlighting substitutions.
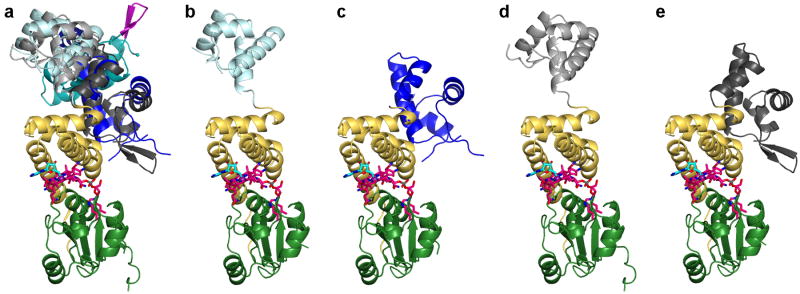
The DNA-binding domain is structurally similar to those of MerR family transcription factors15 (Figure 2b) and to that of the AdoCbl-independent CarH paralog CarA8,16, featuring a canonical recognition helix and a β-hairpin wing for DNA binding (Figure 2a). In our DNA-free structures, the DNA-binding domain samples different orientations for the different CarH protomers in the asymmetric unit, enabled by a flexible linker region and stabilized by crystal lattice contacts (Extended Data Figure 2).
In contrast to the flexible DNA-binding domains, the four-helix bundle and the Rossmann domain are structurally rigid, together forming a module that binds the AdoCbl light sensor. AdoCbl is sandwiched between the four-helix bundle, which interacts with the upper axial 5′-deoxyadenosyl (5′-dAdo) ligand, and the Rossmann-fold domain, which binds the Cbl lower face in the base-off/His-on mode with the side chain of His177 displacing the Cbl dimethylbenzimidazole base (Figure 2a,d, Extended Data Figure 1b). Instead of closely resembling an AdoCbl-dependent enzyme, the CarH light-sensing domain is structurally homologous to the methylcobalamin (MeCbl)-binding module of methionine synthase MetH (Figure 2b)17, even though the AdoCbl 5′-dAdo group is much bulkier than the MeCbl methyl group in MetH. Modeling AdoCbl into MetH leads to several steric clashes (Figure 2c), but in CarH, a small but important shift of 2.5 Å of the four-helix bundle enlarges the cavity on the Cbl upper face (Figure 2d,e). Additionally, four hydrophobic residues at the Cbl upper face in MetH are replaced in CarH, providing the 5′-dAdo group a larger binding pocket (Leu715→Val138), a hydrogen bonding interaction (Val718→Glu141), and more polar environment in general (Val719→His142, Phe708→Trp131) (Figure 2d,f). Although Trp is larger than Phe, the orientation of the Trp side chain on the side of the upper ligand rather than directly above perfectly accommodates the 5′-dAdo group (Figure 2c,d). Notably, Trp131, Glu141, and His142 are highly conserved in CarH homologs, suggesting that this mode of AdoCbl binding is conserved as well (Extended Data Figure 3).
CarH is a dimer-of-dimers-type tetramer
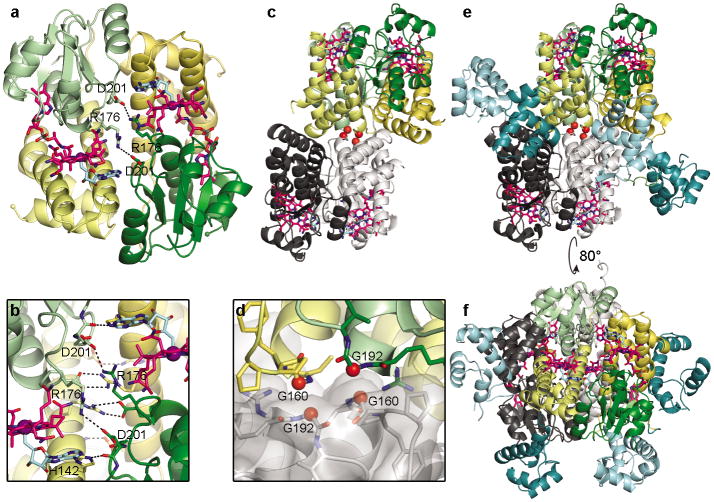
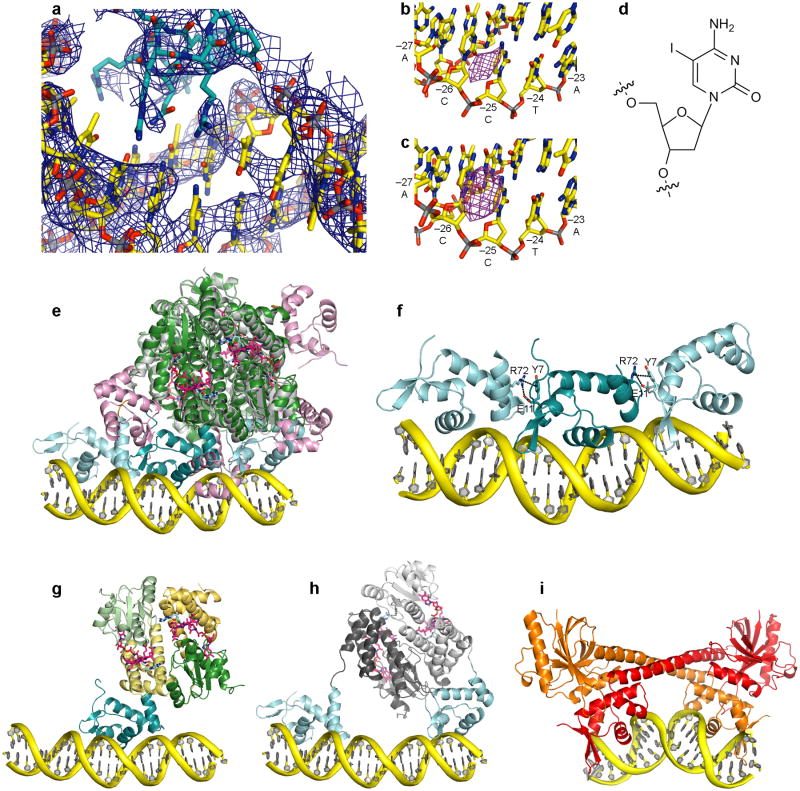
AdoCbl-bound CarH is a tetramer in the crystal structure (Figure 3), consistent with results from size exclusion chromatography (SEC) and analytical ultracentrifugation8,18. Four light-sensing domains comprise the core of the tetramer (Figure 3a-d) with the DNA-binding domains extending outward (Figure 3e-f). The core has a dimer-of-dimers architecture, with each constituent dimer composed of two CarH light-sensing domains in a head-to-tail orientation (Figure 3a). The extensive head-to-tail dimer interface is formed by the four-helix bundles and the Cbl-binding domains and features a solvent-buried area of 1430 Å2 on each protomer as well as numerous hydrogen bonds and ionic interactions involving various side chains and the 5′-dAdo group of AdoCbl (Figures 3a,b). Two such head-to-tail dimers assemble to a tetramer in a staggered fashion (Figures 3c-f). This dimer-dimer interface is again formed by the light-sensing domains, creating a buried surface area of 1590 Å2 on each of the two head-to-tail dimers, whereas the four DNA-binding domains are positioned on the surface of the tetramer and only make minor contributions to the interface (Figures 3e,f). It was previously demonstrated by SEC that CarH lacking the DNA-binding domain still forms tetramers8, and here we find that CarH adopts the same tetramer architecture when the DNA-binding domains are proteolytically removed during crystallization (Figures 3c,e). Thus, the light-sensing domains appear to mediate tetramerization, positioning the DNA-binding domains on the surface to engage DNA.
Figure 3.
CarH oligomerization. (a) CarH protomers arranged in a head-to-tail dimer, colored by domain (helix bundle: yellow; Cbl-binding domain: green) with left protomer shown in lighter colors. AdoCbl shown as in Fig. 2a. DNA-binding domains hidden for clarity. (b) Close-up of selected residues at dimer interface. (c) Core of CarH tetramer, assembled from two head-to-tail dimers. Top dimer is colored as in (a) and bottom dimer is colored in black and gray. Gly160 and Gly192 at the dimer-dimer interface are shown as red spheres. Structure is from a sample of CarH that degraded during crystallization and lacks the DNA-binding domains (crystal form 2, see methods). (d) Close-up of Gly160 and Gly192 (red spheres) from two protomers at the dimer-dimer interface. (e) Tetramer of full-length CarH including DNA-binding domains (crystal form 3), colored as in (c). DNA-binding domains of colored dimer are shown in dark cyan, those of gray dimer are shown in light cyan. (f) Additional view of CarH tetramer, revealing how DNA-binding domains are positioned on the protein surface.
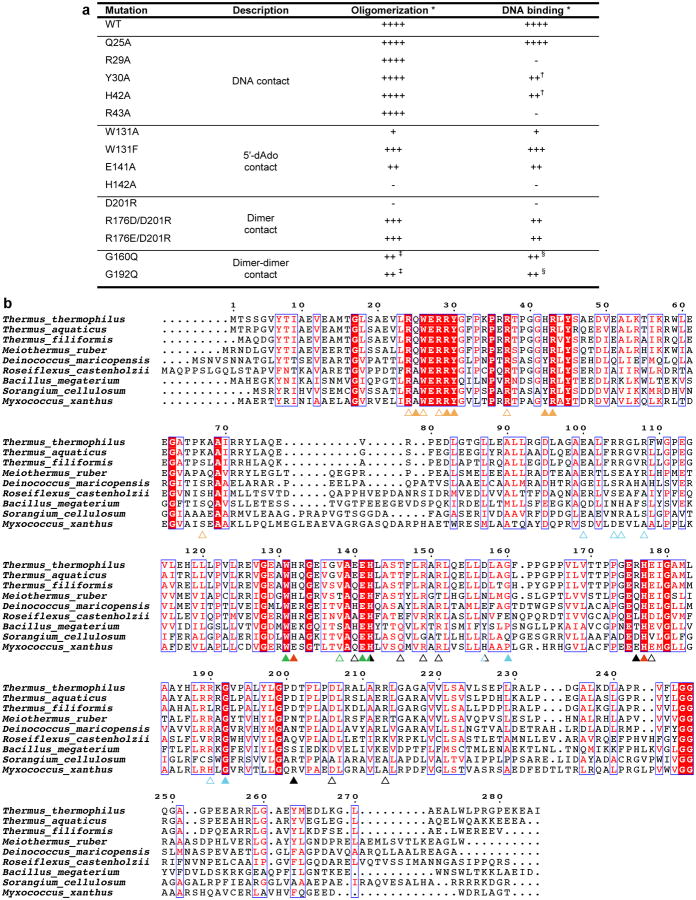
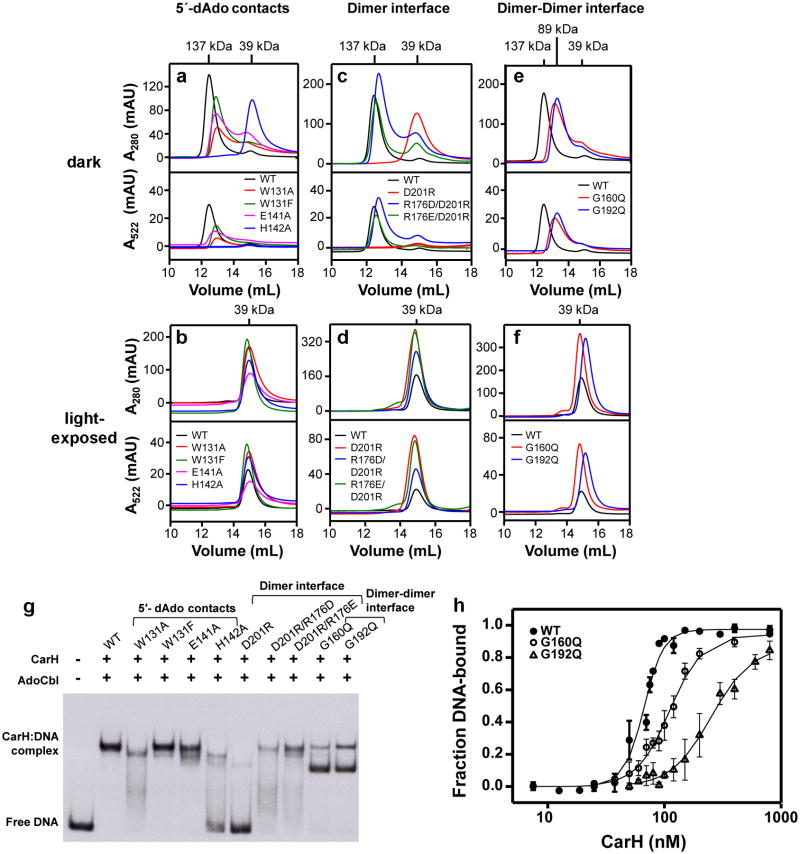
To confirm the CarH tetramer architecture and the mode of AdoCbl binding, we mutated residues in the AdoCbl binding site, at the head-to-tail dimer interface, and at the dimer-dimer interface and analyzed these mutants using SEC and electrophoretic mobility shift assays (EMSAs) (Extended Data Figures 3,4). Non-conservative mutations in the binding site for the 5′-dAdo group impaired tetramer formation (W131A, E141A, Figure 2d) and mutations near the head-to-tail dimer interface completely abolished it (H142A, D201R, Figure 3b). Remarkably, the most drastically adverse H142A and D201R mutations also appeared to impair AdoCbl binding (Extended Data Figure 4a,c, absorbance traces at 522 nm). Moreover, DNA binding affinity weakened with decreased ability to form tetramers in the W131A, E141A, H142A, and D201R mutants (Extended Data Figures 3a, 4g). For comparison, we also introduced the conservative W131F mutation, which behaved like wild-type (WT) CarH in its oligomerization and DNA binding properties (Extended Data Figure 4a,g). The inability of the D201R mutant protein to oligomerize is consistent with the observed Asp201/Arg176 interaction playing an important role in stabilizing the head-to-tail dimer interface (Figure 3a). We expected a second compensatory mutation, R176D or R176E, to restore the interaction and, indeed, the D201R/R176D and D201R/R176E double mutants could form tetramers (Extended Data Figure 4c) and bind to DNA, albeit less efficiently than WT CarH (Extended Data Figure 4g). Finally, replacing Gly160 and Gly192 at the CarH dimer-dimer interface (Figure 3d) by the bulkier Gln resulted in dimers in the presence of AdoCbl, a form previously never observed for WT CarH (Extended Data Figure 4e). Although both these mutants bound to the DNA probe in the dark, they formed a smaller size (higher mobility) complex than WT CarH, suggesting that their binding mode is distinct (Extended Data Figure 4g). Both mutants furthermore bound DNA with reduced affinity and cooperativity compared to WT CarH (Extended Data Figure 4h). Together, these results are consistent with the observed CarH tetramer architecture and indicate that this architecture is critical for DNA binding.
CarH binds the promoter −35 element
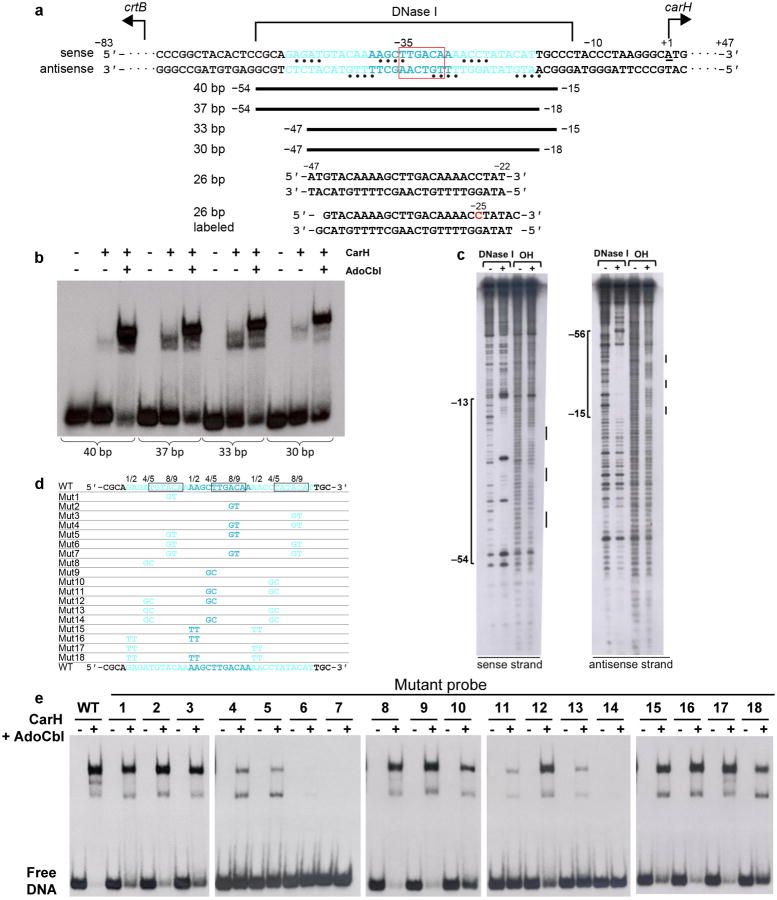
To reveal the mode of transcriptional repression, we next sought to visualize CarH bound to its cognate DNA operator. The CarH operator lies within a 110-bp segment of the intergenic region between carH and the carotenogenic crtB8,10. Using systematically truncated DNA probes in EMSAs (Extended Data Figure 5a,b), this operator sequence was narrowed down to a 30-bp segment. Diffraction quality crystals of AdoCbl-bound CarH were obtained in complex with a blunt-ended 26-bp DNA segment (−47 to −22 relative to carH transcription start site, Extended Data Figure 5a), allowing us to determine the crystal structure of DNA-bound CarH to 3.89 Å resolution (Figure 4, Extended Data Figure 6a-h, Extended Data Table 1).
Figure 4.
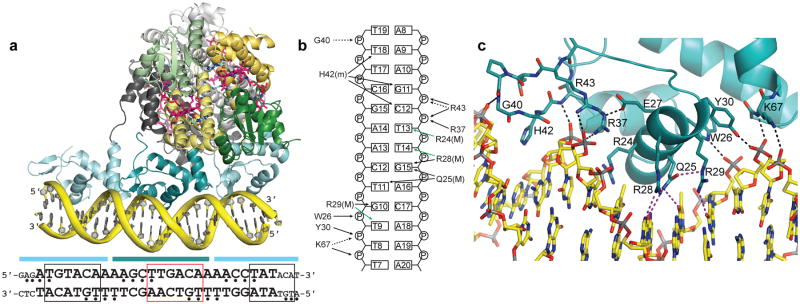
CarH DNA binding. (a) CarH tetramer bound to a 26-bp DNA segment (yellow). CarH is shown in ribbons with one head-to-tail dimer in green (Cbl-binding domains) and yellow (helix bundles) and the other dimer in gray. AdoCbl shown as in Fig. 2a. DNA-binding domains are shown in cyan. Sequence of DNA segment used for crystallization (larger font) as well as flanking sequences in the operator (smaller font) are shown below. Cyan bars indicate base pairs covered by each DNA-binding domain. Base pairs covered by the recognition helix are boxed; red box highlights the promoter −35 element. Nucleotides protected from hydroxyl radical cleavage are indicated by bullets. The orientation of the DNA in the structure was confirmed by heavy atom labeling (Extended Data Figure 6b-d). (b) Schematic of CarH:DNA interactions for the central DNA-binding domain, denoted as follows: black arrows: hydrogen bonds/electrostatic interactions; green arrows: van der Waals interactions; solid lines: contacts from protein side chains; dashed lines: contacts from main chain; (M): contacts in the DNA major groove; (m): contacts in the DNA minor groove. (c) Close-up of interactions between CarH (cyan) and DNA (yellow). Hydrogen bonds and ionic interactions to the phosphate backbone are shown as black dashed lines. Contacts to DNA bases are shown in purple. Side chain orientation is not unambiguous due to the modest resolution of this structure, but many of the contacts shown are supported by mutagenesis (Extended Data Figure 7).
The structure revealed a unique mode of DNA binding involving three of the four DNA-binding domains of tetrameric CarH (Figure 4a). The fourth DNA-binding domain is disordered and not visible in the electron density. The overall architecture of the tetramer is the same before and after DNA binding except for a reorientation of the DNA-binding domains (Extended Data Figure 6e). All three DNA-binding domains face the same way on the DNA segment and bind to a set of three 11-bp repeats with a consensus sequence (A/G)A(G/C)(A/C)T(A/G/T)(T/G)ACA(A/T) (Figure 4a). This parallel orientation is stabilized by specific interactions between adjacent DNA-binding domains (Extended Data Figure 6f). The central DNA-binding domain comes from one head-to-tail dimer, whereas the two flanking DNA-binding domains come from the second head-to-tail dimer (Extended Data Figure 6g,h). These structures suggest that the two individual head-to-tail dimers would bind to DNA less tightly, consistent with the reduced affinity and cooperativity for the dimeric G160Q and G192Q CarH mutants (Extended Data Figure 4g,h). CarH rendered monomeric by light exposure8,18 or mutagenesis (H142A, D201R) binds DNA with even further reduced affinity (Extended Data Figures 3a, 4g).
To obtain support for this unusual DNA binding mode, hydroxyl radical and DNase I footprinting were used to examine the regions of DNA protected by CarH. The DNase I footprint, which was obtained using a longer DNA segment (130-bp) than the one used in the crystal structure (26-bp), still matches the crystal structure footprint (Extended Data Figure 5a,c). Additionally, three evenly spaced 4-bp hydroxyl radical footprints are observed on both the sense and the antisense strand (Extended Data Figure 5c) that correlate with where the “wings” of the three DNA-binding domains contact the minor groove (Figure 4a). Taken together, the size of the DNase I footprint, the presence of three hydroxyl radical footprints, and the observation that the DNA sequence contains three repeats (see above), suggest that CarH binds to DNA using three of its four DNA-binding domains.
To determine whether all three repeats are important for high affinity binding of CarH to DNA, we tested the effect of mutating DNA bases in the CarH operator (Extended Data Figure 5d,e). Mutating dinucleotides in any single repeat only led to a small decrease in affinity as evidenced by the intense retarded bands for the CarH:DNA complex and the small amounts of free DNA in EMSAs (Mutants 1-3 or 8-10 in Extended Data Figure 5e). In contrast, simultaneously mutating dinucleotides in any two of the three repeats or in all three repeats almost completely abolished DNA binding (Mutants 4-7 and 11-14 in Extended Data Figure 5e). As a control, we also mutated DNA bases in the operator that CarH does not contact directly, and as expected, WT CarH bound to these mutants with similar affinity (Mutants 15-18 in Extended Data Figure 5e). Given that the results of mutations were similar for each of the three repeats, it appears that all three repeats are important in determining CarH:DNA affinity.
Each DNA-binding domain forms hydrogen bonds and electrostatic interactions to the phosphate backbone, contributed from both the peptide backbone and the side chains of Trp26, Tyr30, Arg37, Arg43, and Lys67 (Figure 4b,c), and each domain also inserts His42 of its β-hairpin “wing” into the DNA minor groove. Finally, each DNA-binding domain places its recognition helix in the DNA major groove, where it recognizes a 6-bp stretch (Figure 4a) using specific hydrogen bonds from the side chains of Gln25, Arg28, and Arg29 (Figure 4b,c). Strikingly, the major groove sequence occupied by the central DNA-binding domain spans the promoter −35 element (TTGACA, red box in Figure 4a) for the major σA/σ70-associated bacterial RNA polymerase (RNAP). Thus, these structures reveal the mechanism of transcriptional repression: CarH occupies the −35 element, blocks access by the RNAP-σA holoenzyme, and thereby prevents transcription initiation.
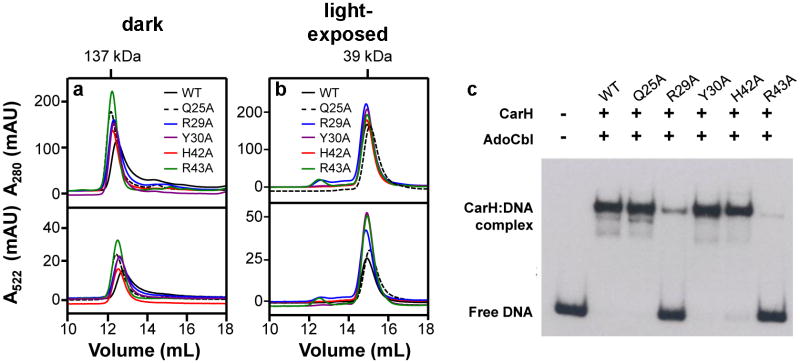
To verify the observed mode of DNA binding, we generated the Q25A, R29A, Y30A, H42A, and R43A CarH mutants and tested their DNA binding capacity using EMSAs. Mutating the conserved Arg29 or Arg43 to Ala abolished DNA binding but did not affect AdoCbl-dependent tetramerization (Extended Data Figures 3a, 7), consistent with a role of these residues in DNA binding. Notably, the Q25A mutant retained DNA-binding capacity, indicating that Gln25 could be more important for mediating DNA specificity than affinity (Extended Data Figure 7c). Finally, the H42A and Y30A mutants only showed mildly reduced affinity, suggesting that their interactions are not essential for DNA binding (Extended Data Figure 3a, 7c).
Light triggers tetramer disassembly through helix bundle movement
Finally, to examine how light exposure causes tetramer disassembly, we determined the crystal structure of light-exposed CarH to 2.65 Å resolution (Figure 5, Extended Data Figure 8a,b, Extended Data Table 1). The structure contains monomeric CarH with bound Cbl but without the 5′-dAdo group, which dissociated as a consequence of light exposure (Figure 5a). The four-helix bundle and the Cbl-binding domain individually do not exhibit major conformational changes compared to the dark AdoCbl-bound structure. However, the orientation of the helical bundle relative to the Cbl-binding domain has changed drastically with a >8 Å displacement (Figure 5a,b). This helix bundle movement would disrupt the head-to-tail dimer interface (Figure 5c), leading to tetramer disassembly, dissociation from DNA, and transcriptional activation.
Figure 5.
Light-induced conformational changes in CarH. (a) Structure of light-exposed CarH, with the helix bundle in orange and Cbl-binding domain in gray. DNA-binding domain is hidden for clarity (see Extended Data Figure 8a). Cbl shown with carbons in pink and cobalt in purple. (b) Structure of a CarH protomer in the dark state, shown with helix bundle in yellow, Cbl-binding domain in green, and 5′-dAdo group in cyan. Colored lines highlight domain orientations. (c) Light-induced helix bundle movement causes tetramer disassembly. Shown is a head-to-tail dimer of CarH in the dark state, but the right protomer is replaced by the structure of light-exposed CarH to show the steric clash. (d) Departure of the 5′-dAdo group after light exposure leaves a large cavity on the Cbl upper face. The helix bundle (yellow) and the Cbl-binding domain (green) are shown in surface representation with selected residues shown as sticks. (e) Helix bundle movement fills the cavity at the Cbl upper face and brings His132 to the cobalt, where it occupies the open coordination site. Coloring as in (a).
Tetramer disassembly is triggered by loss of the AdoCbl 5′-dAdo group: its presence in the AdoCbl-bound structure blocks movement of the helix bundle, due in large part to the positioning of W131 against the upper Cbl ligand (Figure 2d), keeping the CarH protomers in the extended “upright” conformation required for tetramerization. Loss of the 5′-dAdo group leaves a large cavity on the Cbl upper face (Figure 5d), prompting movement of the helix bundle to occupy this void and cover the Cbl (Figure 5e).
Strikingly, the helix bundle motion brings His132 from the protein surface to the Cbl upper face, where it binds to the cobalt to form bis-His ligated Cbl (Figures 5d,e, Extended Data Figure 8b). Such bis-His ligation, common for hemes, has not been reported for Cbl, although bis-His Cbl ligation was recently proposed based on mass spectrometry for the Cbl-dependent transcription factor AerR19. We therefore validated formation of bis-His ligated Cbl using UV-Vis spectroscopy. Spectra of light-exposed WT CarH and a H132A mutant, which is unable to form the bis-His ligation, resemble those of free Cbl with two or one nitrogen-based ligands20, respectively (Extended Data Figure 8c,d). In contrast, the spectra of the AdoCbl-bound proteins are identical (Extended Data Figure 8e). These results provide unambiguous evidence for a bis-His-ligated Cbl in light-exposed CarH and suggest that this mode of coordination might be employed more frequently in non-heme proteins.
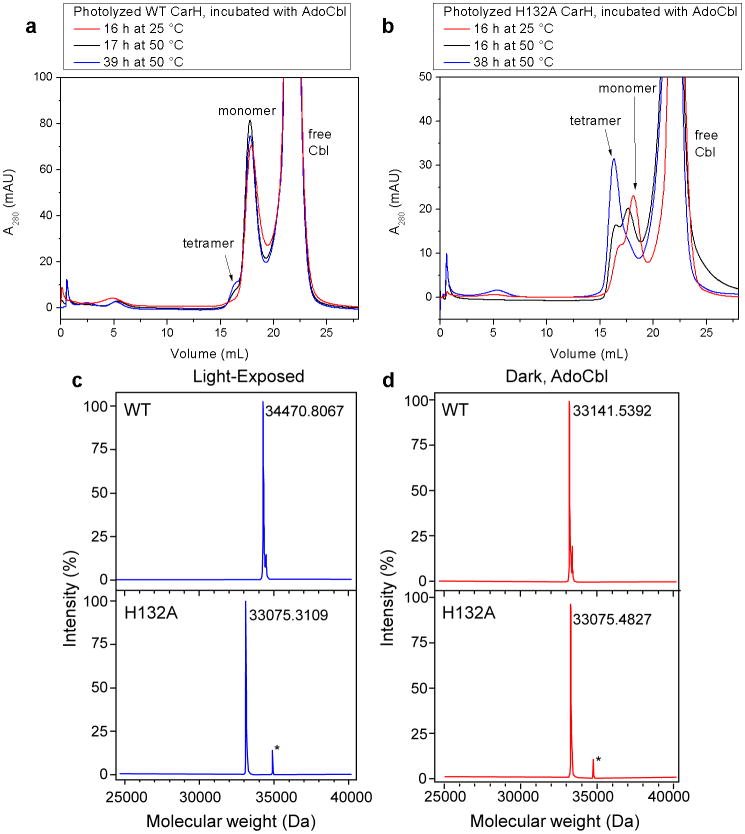
Notably, both WT and H132A CarH undergo light-dependent tetramer disassembly, indicating that bis-His ligation is not required for disassembly (Extended Data Figure 8f,g). However, Cbl dissociation after light exposure is faster for H132A CarH than for WT CarH, which forms a very tight and stable complex with the photolyzed Cbl, as indicated by the relative abilities of the protein to be reconstituted with fresh AdoCbl (Extended Data Figure 9a,b) and by the observation of a CarH:Cbl adduct in mass spectrometry for WT CarH but not for the H132A mutant (Extended Data Figure 9c,d). Thus, the bis-His ligation could be important to retain the Cbl cofactor after photolysis.
Discussion
Impressively, CarH-type photoreceptors are found in hundreds of bacterial genomes, including bacteria that uptake rather than biosynthesize AdoCbl. CarH is distinguished from most known classes of photoreceptors (with the exception of some LOV-type photoreceptors21-23) in that it can bind to DNA directly, instead of requiring additional proteins for gene regulation. Beyond gene regulation, the CarH light-sensing domains can be found fused to effector domains such as histidine kinases and in stand-alone modules that could undergo light-dependent protein:protein interactions. This versatility likely explains the broad distribution of CarH-like proteins in bacteria.
CarH's use of AdoCbl as a light-sensing chromophore is biologically unprecedented. AdoCbl is structurally and photochemically distinct from known photoreceptor chromophores such as bilin24, flavins25-27, retinal4, or Trp side chains28: light exposure leads to breakage of a covalent Co-C bond, whereas other chromophores undergo less drastic changes such as light-induced electron transfer or cis-trans isomerizations. In all cases, however, light energy is ultimately harnessed to drive a large-scale conformational change, highlighting the convergence of different light sensing mechanisms. AdoCbl was previously best-known as a cofactor for radical-based enzyme reactions, in which reversible homolytic cleavage of the Co-C bond provides access to the 5′-dAdo radical for catalysis13, and as a modulator of gene expression via riboswitches29-32. Our structures now allow us to visualize how AdoCbl is repurposed as a light sensor in CarH: in the dark, the AdoCbl 5′-dAdo group acts as a molecular doorstop that keeps CarH protomers in an extended “upright” conformation for tetramerization, and light exposure triggers collapse into a kinked conformation. Whereas AdoCbl photolysis is an unwanted side reaction in enzyme catalysis because it leads to cofactor inactivation, in CarH this light sensitivity is harnessed to drive a light-dependent gene expression switch and a change in physiology. Remarkably, use of AdoCbl in this alternative function appears to come with a safeguard: the product of CarH-bound AdoCbl photolysis is not a 5′-dAdo radical, but rather 4′,5′- anhydroadenosine33, which differs by one proton and one electron, and cannot cause radical damage. Thus, AdoCbl now joins the list of enzyme cofactors that have been repurposed as sensors; a list that already includes flavins (as light sensors in LOV, BLUF, and cryptochrome photoreceptors25-27) and hemes (as sensors of oxygen and other small molecules34).
Our CarH structures additionally reveal the functional repurposing of two different protein modules. The CarH light-sensing domain mirrors the Cbl-binding module of methionine synthase MetH, an enzyme that uses a MeCbl intermediate in the transfer of a methyl group from methyltetrahydrofolate to homocysteine, generating tetrahydrofolate and methionine35. But whereas MetH employs its helix bundle to position Phe708 over the MeCbl methyl group and protect it from photolysis36, CarH, enabled by specific substitutions at the Cbl upper face, uses this fold as an AdoCbl-binding light-sensing domain, in which the corresponding Trp131 senses the presence of the 5′-dAdo group and transmits the signal of AdoCbl photolysis by leading a conformational change of the helix bundle. Furthermore, whereas the Cbl-binding module of MetH is embedded in a 136 kDa multi-domain protein and juxtaposed to different substrate-binding domains via transient domain-domain interactions during a catalytic cycle37, the light-sensing domain of CarH is used to assemble a tetramer that is stable enough to occlude the −35 element from RNAP. Thus, this module has been repurposed from a methyl group carrier in primary metabolism to a light-sensing modulator of oligomerization state.
Similarly, the CarH DNA-binding domain resembles those of MerR-type transcription factors such as MerR, BmrR, and SoxR, whose role as transcriptional activators in the presence of heavy metals or other stresses has been established and whose DNA-bound structures have been reported15,38-39. Whereas MerR proteins bind as dimers to a (pseudo-)palindromic DNA sequence and distort the DNA, which brings promoter elements into alignment for transcriptional activation (Extended Data Figure 6i), the CarH tetramer uses its DNA-binding domains to bind to three contiguous repeat sequences in a parallel mode, which occludes the −35 element and represses transcription. This unique DNA binding mode rationalizes CarH's tetramer architecture of two-head-to-tail dimers, which is unusual for transcription factors but here enables the DNA-binding domains to arrange in a parallel fashion and cooperatively engage the repeat sequences.
AdoCbl is a biologically expensive molecule, requiring arduous pathways for biosynthesis or specialized machinery for uptake. For CarH-using organisms, it would not be surprising if there were a recovery mechanism for Cbl following tetramer disassembly, and it is tempting to suggest that formation of bis-His ligated Cbl in some CarHs might be the first step of such a recovery pathway. In this regard, it is interesting to note that His132 is strictly conserved in thermophilic bacteria (Extended Data Figure 3b), where perhaps the bis-His ligated Cbl is as an adaption to elevated temperatures. Although use of AdoCbl as a light sensor comes at a price, it appears that the physiological benefits make this repurposing worthwhile.
Altogether, our results provide fundamental insight into a new mode of light-dependent gene regulation and reveal an exquisite example of cofactor and protein domain repurposing. The structures furthermore provide a basis for deployment of the modular CarH photoreceptors, in which the light-sensing and DNA-binding activities rest on different domains, for engineering light-modulated transcriptional control or protein-protein interactions.
Methods
Protein Constructs
Cloning of the pET15b-CarH construct encoding for Thermus thermophilus CarH with an N-terminal His6-tag was described previously8. The H132A mutation was introduced into pET15b-CarH using QuikChange PCR mutagenesis (Stratagene) with Pfu Turbo DNA polymerase. All other mutants were obtained by gene synthesis (Genscript) with appropriate 5′ and 3′ restriction sites for cloning into the pET15b expression vector.
Protein purification
WT CarH and mutants were purified as described previously8. A slightly modified protocol was employed for His6-tagged CarH for crystallization. After expression and affinity chromatography, performed as described previously8, a 3-fold molar excess of AdoCbl (Sigma) was added and the mixture was incubated on ice for 1 h. All subsequent handling was performed in a dark room under red light. The protein solution was applied to a HiLoad 26/60 Superdex 200 size exclusion column (GE Healthcare) pre-equilibrated with CarH buffer (0.1 M NaCl, 0.05 M Tris•HCl, pH 8). Under these conditions, tetrameric AdoCbl-bound CarH eluted as a single peak, separate from residual amounts of monomeric CarH. Fractions containing AdoCbl-bound CarH were combined and concentrated to about 8 mg/mL, as judged by the absorbance at 280 nm using the combined ε280 for AdoCbl (22.5 mM-1cm-1, determined spectroscopically based on published extinction coefficients at 260 nm, 288 nm, and 522 nm40-42) and for CarH (37.9 mM-1cm-1, calculated from the protein sequence using ProtParam at http://web.expasy.org/protparam).
Purified native and mutant protein identities were verified prior to use by HPLC-ESI TOF or Trap mass spectrometry using an Agilent 1100 Series HPLC equipped with a μ-well plate autosampler and a capillary pump and connected to an Agilent Ion Trap XCT Plus Mass Spectrometer with an electrospray (ESI) interface (Agilent Technologies). Samples were injected into an Zorbax Poroshell 300 SB-C18 HPLC column (Agilent Technologies) that was coupled online to the mass spectrometer using an electrospray interface. Samples were separated at 60 °C at a flow rate of 0.2 mL/min using a linear gradient of buffer A (water/acetonitrile/formic acid, 95:4.9:0.1) to 90% buffer B (water/acetonitrile/formic acid, 10:89.9:0.1) over 30 min and protein elution was monitored at 210 nm and 280 nm. Mass spectra were acquired in the positive ion mode in an m/z range from 100–2200.
The integrity of the AdoCbl Co-C bond was assessed by UV-Vis spectroscopy (described below). Protein containing intact AdoCbl was flash-frozen in liquid nitrogen until further use. CarH containing photolyzed AdoCbl was generated by exposing the protein solution to ambient light for 30 min at 4 °C. Complete photolysis was assessed by UV-Vis spectroscopy (described below). Light-exposed CarH was used for crystallization experiments immediately. CarH:DNA complexes for crystallization were generated by mixing protein and DNA at the desired ratio and incubating the mixture for 1 h on ice in the dark prior to crystallization experiments.
Preparation of DNA segments for crystallization
HPLC-purified single-stranded DNA oligonucleotides without heavy atom labels (Integrated DNA technologies) or containing a single 5-iodo-deoxycytidine (Jena Bioscience) were dissolved to 1 mM in CarH buffer. Equimolar amounts of complementary oligonucleotides were mixed, heated to 95 °C for 10 min, and then slowly left to cool down to 4 °C in a thermocycler over the course of 1 h for annealing. Final double-stranded DNA concentrations were assessed by the absorbance at 260 nm using the calculated sequence-specific ε260 (http://biophysics.idtdna.com/UVSpectrum.html).
Crystallization
Purified AdoCbl-bound CarH was crystallized in three different crystal forms. All crystallization procedures for AdoCbl-bound CarH were carried out in a dark room under red light. Crystals of AdoCbl-bound CarH in crystal form 1 were obtained by the hanging drop vapor diffusion technique at 25 °C. An aliquot (1 μL) of a protein solution (7 mg/mL AdoCbl-bound CarH in CarH buffer) was mixed with 1 μL of a precipitant solution (10 % (w/v) PEG 8000, 10 % (v/v) glycerol, 0.04 M KH2PO4) on a glass cover slip. The cover slip was sealed with grease over a reservoir containing 500 μL of the precipitant solution. Octahedral crystals appeared within 3 d and grew to maximum size within 7 d. Under these conditions, the protein underwent proteolysis at the linker region between the DNA-binding domain and the four-helix bundle, as judged by SDS-PAGE. The crystals consisted only of the C-terminal light-sensing domains. Crystals were transferred in 2 steps of increasing glycerol concentration into a cryogenic solution containing 10 % (w/v) PEG 8000, 20 % (v/v) glycerol, 0.04 M KH2PO4, 0.05 M Tris•HCl pH 8, and 0.1 M NaCl, soaked in that solution for 20 s, and then flash-frozen in liquid nitrogen.
A second crystal form of AdoCbl-bound CarH was obtained by the sitting drop vapor diffusion technique at 25 °C. An aliquot (0.15 μL) of a protein solution (5.9 mg/mL AdoCbl-bound CarH in CarH buffer, supplemented with 70 μM of a 31-bp DNA oligonucleotide) was mixed with 0.15 μL of a precipitant solution (20 % (w/v) PEG 3350, 0.2 M KCl) using a Phoenix liquid handling robot (Art Robbins Instruments). The drop was equilibrated against 70 μL of the precipitant solution. Rectangular crystals appeared within 6 months. Again, the protein underwent proteolysis and the crystals only consisted of the C-terminal light-sensing domains. Crystals were transferred in 2 steps of increasing glycerol concentration into a cryogenic solution containing the precipitant supplemented with 20 % (v/v) glycerol, soaked in that solution for 5 s, and then flash-frozen in liquid nitrogen.
A third crystal form of AdoCbl-bound CarH containing full-length protein was obtained by the sitting drop vapor diffusion technique at 25 °C. An aliquot (0.15 μL) of a protein solution (6 mg/mL AdoCbl-bound CarH in CarH buffer, supplemented with 70 μM of a 28-bp DNA segment) was mixed with 0.15 μL of a precipitant solution (20 % (w/v) PEG 3350, 0.1 M ammonium citrate tribasic pH 7) using a Phoenix liquid handling robot (Art Robbins Instruments). The drop was equilibrated against 70 μL of the precipitant solution. Rod crystals appeared within 1 month. These crystals contained full-length AdoCbl-bound CarH but no DNA. Crystals were transferred in 3 steps of increasing glycerol concentration into a cryogenic solution containing the precipitant supplemented with 20 % (v/v) glycerol, soaked in that solution for 10 s, and then flash-frozen in liquid nitrogen.
Light-exposed CarH was crystallized by the hanging drop vapor diffusion technique at 25 °C. An aliquot (1 μL) of a protein solution (4.5 mg/mL light-exposed CarH in CarH buffer) was mixed with 1 μL of a precipitant solution (3.4 M NaCl, 0.1 M Bis-Tris pH 6) on a glass cover slip. The cover slip was sealed with grease over a reservoir containing 500 μL of the precipitant solution. Octahedral crystals appeared within 8 months. Crystals were transferred in 3 steps of increasing glycerol concentration into a cryogenic solution containing the precipitant supplemented with 18 % (v/v) glycerol, incubated in that solution for 10 s, and then flash-frozen in liquid nitrogen.
CarH bound to both AdoCbl and a 26-bp DNA segment was crystallized by the hanging drop vapor diffusion technique at 25 °C. An aliquot (1 μL) of a protein solution (6 mg/mL AdoCbl-bound CarH in CarH buffer, supplemented with 67.5 μM of a 26-bp DNA segment, 1.5-fold molar excess) was mixed with 1 μL of a precipitant solution (16% PEG 3350, 0.2 M l-proline, 0.1 M HEPES pH 7.5) on a glass cover slip. The cover slip was sealed with grease over a reservoir containing 500 μL of the precipitant solution. Tetragonal bipyramidal crystals appeared within 3 weeks. Crystals were transferred in 3 steps of increasing PEG 400 concentration into a cryogenic solution containing the precipitant supplemented with 15 % (w/v) PEG 400, incubated in that solution for 20 s, and then flash-frozen in liquid nitrogen.
CarH bound to both AdoCbl and a 26-bp DNA segment containing 5-iodo-deoxycytidine (Extended Data Figure 6b-d) in position −25 of the sense strand (Extended Data Figure 5a) was crystallized by the hanging drop vapor diffusion technique at 25 °C. An aliquot (1 μL) of a protein solution (5 mg/mL AdoCbl-bound CarH in CarH buffer, supplemented with 94 μM of the iodine-labeled 26-bp DNA segment, 2.5-fold molar excess) was mixed with 1 μL of a precipitant solution (11.5% PEG 3350, 0.28 M l-proline, 0.1 M Tris pH 8.5) on a glass cover slip. The cover slip was sealed with grease over a reservoir containing 500 μL of the precipitant solution. Crystals appeared within 4 months. Crystals were transferred in 5 steps of increasing xylitol concentration into a cryogenic solution containing the precipitant supplemented with 25 % (w/v) xylitol, incubated in that solution for 30 s, and then flash-frozen in liquid nitrogen.
Data collection and processing
All data were collected at the Advanced Photon Source (Argonne, IL) at beamline 24ID-C using a Pilatus 6M pixel detector at a temperature of 100 K. Crystals of AdoCbl-bound CarH crystal form 1 belong to space group P43212. An initial AdoCbl-bound CarH crystal was used for a fluorescence scan to determine the Co peak wavelength for anomalous data collection. Another crystal was then used for collection of both native data and anomalous peak data. Native data were collected in a single wedge of 75° at a wavelength of 0.9792 Å (12662 eV). The crystal was displaced continuously along its major macroscopic axis during data collection. Anomalous peak data were collected in a single wedge of 345° at a wavelength of 1.6039 Å (7730 eV). The crystal was aligned using a mini-κ goniometer such that Bijvoet mates were recorded on the same frame.
All other data except for iodine anomalous data and native data of light-exposed CarH (see below) were collected at a wavelength of 0.9795 Å (12658 eV). Crystals of AdoCbl-bound CarH crystal form 2 belong to space group P212121. Data were collected in a single wedge of 100°. Crystals of AdoCbl-bound CarH crystal form 3 belong to space group P1. Data were collected in a single wedge of 270° and the crystal was displaced continuously along its major macroscopic axis during data collection. Crystals of light-exposed CarH belong to space group I4122 and data were collected at a wavelength of 0.9791 Å (12663 eV) in a single wedge of 150°. Crystals of DNA-bound CarH both with and without the iodine label belong to space group P21212. Data for crystals with unlabeled DNA were collected in a single wedge of 180°. Data for crystals of CarH in complex with iodine-labeled DNA were collected at a wavelength of 1.7365 Å (7140 eV) in a single wedge of 200° and the crystal was displaced continuously along its major macroscopic axis during data collection.
Data for the AdoCbl-bound CarH (crystal form 1) Co peak data set were integrated in HKL2000 and scaled in Scalepack43. Data for all other data sets were integrated in XDS and scaled in XSCALE44. Data collection statistics are summarized in Extended Data Table 1.
Structure building and refinement
The structure of AdoCbl-bound CarH in crystal form 1 (space group P43212) was determined to 2.80 Å resolution using single-wavelength anomalous diffraction. Positions of two cobalt sites, corresponding to two CarH protomers in the asymmetric unit, were located using ShelxD45 in the HKL2MAP shell46 and refined using SHARP/autoSHARP47. The initial overall figure of merit (acentric) was calculated by SHARP to be 0.43 to 5.1 Å resolution. Experimental maps from the SHARP output, solvent flattened using SOLOMON48 and extended to 3.3 Å resolution, were of sufficient quality to place two copies of the Cbl-binding domain of MetH17 (PDB ID code 1BMT, residues 745-868), eight additional helices, and AdoCbl in the electron density. This initial model was used to better define solvent boundaries in another round of solvent flattening of SOLOMON. Using the resulting electron density, loop regions were modified and side chains with visible electron density were added. A near-complete model of AdoCbl-bound CarH (containing 374 amino acid residues and bound AdoCbl) was then used for rigid body refinement in Phenix49 against the native AdoCbl-bound CarH data set (crystal form 1) using data from 100 to 2.80 Å resolution. The resulting R-factors were 42.0% and 44.1% for the working and the free R-factor, respectively. The model was refined by manual adjustment in Coot50 until rigid body refinement in Phenix yielded R-factors of 30.8% and 34.7% for the working and the free R-factor, respectively. Subsequent cycles of refinement included positional refinement with non-crystallographic symmetry (NCS) restraints and individual B-factor refinement in Phenix until the R-factors were 20.9% and 24.2% for the working and the free R-factor, respectively. This model was not refined to completion. The near-complete model was used to determine the structures of AdoCbl-bound CarH in crystal form 2 (space group P212121) and crystal form 3 (space group P1), which are of higher resolution (crystal form 2) or contain the full-length protein (crystal form 3).
The structure of AdoCbl-bound CarH in crystal form 2 was determined to 2.15 Å resolution by molecular replacement in Phaser51. The structure in crystal form 2 contains four CarH protomers in the asymmetric unit, corresponding to a tetramer. After molecular replacement, ten cycles of simulated annealing refinement were carried out in Phenix to remove model bias. The model was then refined by iterative cycles of manual adjustment in Coot and refinement in Phenix. Initially, strict NCS restraints were applied for the two head-to-tail dimers in the asymmetric unit. Subsequently, these restraints were loosened for residues that are in unique environments either because of the asymmetric tetramer architecture or because of crystal contacts. In advanced stages of refinement, water molecules were added manually in Coot and refined in Phenix, with placement of additional water molecules until their number was stable. Final cycles of refinement included TLS parametrization52 with one TLS group per CarH protomer.
The structure of AdoCbl-bound CarH in crystal form 3 was determined to 2.80 Å resolution using molecular replacement. First, two CarH tetramers were placed in the asymmetric unit using Phaser, accounting for all eight protomers in the asymmetric unit. Subsequently, four CarH DNA-binding domains from the structure of light-exposed CarH (see below) were placed using Phaser. After refinement in Phenix, there was clear electron density for an additional DNA-binding domain as well as fragments of the three remaining DNA-binding domains, accounting for all eight DNA-binding domains in the asymmetric unit. The model was refined by iterative cycles of manual adjustment in Coot and refinement in Phenix. Strict NCS restraints were applied for all CarH protomers in the asymmetric unit and loosened in later stages of refinement as described above. No water molecules were added to this structure. Final cycles of refinement included TLS parametrization52. For each CarH protomer, the light-sensing domain was defined as a single TLS group and, if fully present, the DNA-binding domain was defined as an additional TLS group.
The structure of light-exposed CarH was determined to 2.65 Å resolution by molecular replacement in Phaser using consecutive searches for the CarH Cbl-binding domain, the four-helix bundle, and the first conformation of the NMR structure of the CarA DNA-binding domain (PDB ID code 2JML16). The structure contains one protomer in the asymmetric unit and all three domains could be placed unambiguously. Ten cycles of simulated annealing refinement were carried out in Phenix. The model was then refined by iterative cycles of manual adjustment in Coot and refinement in Phenix. In advanced stages of refinement, water molecules were added manually in Coot and refined in Phenix, with placement of additional water molecules until their number was stable. Final cycles of refinement included TLS parametrization52 with one TLS group.
The structure of CarH bound to AdoCbl and a 26-bp DNA segment was determined to 3.89 Å resolution by molecular replacement in Phaser using consecutive searches for two CarH tetramers without the DNA-binding domains and for two 26-bp DNA segments (models generated by the 3D- DART server53; http://haddock.science.uu.nl/services/3DDART/). After molecular replacement, there was clear electron density for six DNA-binding domains in the asymmetric unit, which were positioned manually in the electron density from the structure of light-exposed CarH. The model was refined by iterative cycles of manual adjustment in Coot and refinement in Phenix. B-factors were refined grouped by residue and positions of individual atoms were restrained using NCS restraints. Planarity and hydrogen bonding restraints were applied to DNA base pairs. Final cycles of refinement included TLS parametrization using one TLS group for each CarH protomer and each DNA segment. Anomalous difference maps, calculated from data collected on crystals that contained a DNA segment with an iodine label at position −25 (Extended Data Figure 5a) were used to unambiguously determine the orientation of the DNA segment in the crystal structure and thus validate the sequence assignments. Maps, calculated using FFT54 in the CCP4 software suite55, revealed a strong anomalous difference density peak at one position for each of the two CarH:DNA complexes in the asymmetric unit, allowing for position −25 of the sense strand to be assigned in the structure (Extended Data Figure 6b-d). Note that the iodine-labeled DNA segment differed slightly from the DNA segment used in the structure determination (Extended Data Figure 5a), but both crystallize in the same space group and with the same crystal packing.
Parameter files for cobalamin were generously provided by Oliver Smart at Global Phasing (Cambridge, UK). Refinement restrains for the 5′-dAdo group were generated using the Grade Web Server (Global Phasing, Cambridge, UK).
Crystallographic refinement of all CarH structures yielded models that possess low free R-factors, excellent stereochemistry, and small root mean square deviations from ideal values for bond lengths and angles. In all models, side chains without visible electron density were truncated to the last atom with electron density and amino acids without visible electron density were not included in the model. All refinement statistics are summarized in Extended Data Table 1. The models were validated using simulated annealing composite omit maps (AdoCbl-bound CarH, light-exposed CarH) or regular refinement composite omit maps (DNA-bound CarH) calculated in CNS56 and Phenix. Model geometry was analyzed using MolProbity57 and ProCheck58. Analysis of the Ramachandran statistics using MolProbity indicated that for AdoCbl-bound CarH (crystal form 2), 98.1%, 1.9%, and 0.0% of residues are in the favored, allowed, and disallowed regions, respectively, for AdoCbl-bound CarH (crystal form 3), 97.7%, 2.3%, and 0.0% of residues are in the favored, allowed, and disallowed regions, respectively, for light-exposed CarH, 97.8%, 2.2%, and 0.0% of residues are in the favored, allowed, and disallowed regions, respectively, and for AdoCbl- and DNA-bound CarH, 97.1%, 2.7%, and 0.2% of residues are in the favored, allowed, and disallowed regions, respectively. The larger number of residues in the disallowed region of the Ramachandran plot of DNA-bound CarH is due to the mediocre resolution of the structure. Figures were generated using PyMoL59. Interfaces between subunits were analyzed using the ‘Protein interfaces, surfaces and assemblies’ service PISA at the European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html)60. Crystallography software packages were compiled by SBGrid61.
DNA-binding assays
All DNA binding assays were repeated three to five times for each experimental condition. Electrophoretic mobility shift assays (EMSAs) were carried out in the dark as described previously8. A 177-bp DNA probe PCR-amplified using primers with one 5′-end 32P-labeled with T4 polynucleotide kinase (T4PK; Takara) prior to the PCR or shorter HPLC-purified synthetic probes (Biolegio) were used in the EMSAs. With the latter, one strand was 32P-labeled at the 5′-end with T4PK and then mixed with a two-fold excess of the unlabeled complementary strand to ensure that all of the labeled strand was present as double-stranded probe. The strand mixture was incubated at 100 °C for 2 min and then slowly left to cool down for hybridization. For EMSAs, a 20 μL reaction volume containing the DNA probe (1.2 nM, approximately 13,000 cpm) and protein with a five-fold excess of AdoCbl in 0.1 M KCl, 0.025 M Tris•HCl, pH 8, 1 mM DTT, 10% (v/v) glycerol, 200 ng/μL BSA, and 1 μg of sheared salmon sperm DNA as nonspecific competitor was incubated for 30 min at 65 °C (177-bp probe) or 30 °C (shorter probes). They were then loaded onto 6% native polyacrylamide gels (37.5:1 acrylamide:bisacrylamide) pre-run for 30 min in 0.5 × TBE buffer (0.045 M Tris base, 0.045 M boric acid, 1 mM EDTA) and subjected to electrophoresis for 1.5 h at 200 V, 10 °C. Gels were vacuum-dried and analyzed by autoradiography. Autoradiograms were scanned using an Image Scanner II imager with LabScan 5.0 software (GE Healthcare). Band intensities were quantified using ImageJ (NIH) with those of free DNA used to estimate the fraction bound, which was fit to the 3-parameter Hill equation using SigmaPlot (Systat Software) to estimate KD, the apparent equilibrium dissociation constant equivalent to the protein concentration for half-maximal binding, and n, the Hill coefficient. The latter, for example expected to be 2 for dimer or 4 for tetramer DNA-binding models, can vary due to cooperativity effects, contributions from monomer-tetramer equilibria and/or deviations from true equilibrium.
DNase I and hydroxyl radical footprinting
DNase I and hydroxyl radical footprinting analyses were carried out under solution conditions similar to EMSA using previously described protocols8,62. A 130-bp CarH operator-promoter DNA probe (Extended Data Figure 5a) was 32P-radiolabeled at the 5′ end of its sense or anti-sense strand by PCR using appropriately labeled primers, as described above. For DNase I footprinting, 20 μL of 32P-radiolabeled DNA probe (∼20,000 cpm) with 800 nM CarH and five-fold excess of AdoCbl in EMSA buffer lacking glycerol and with 0.01 M MgCl2 were incubated for 30 min at 37 °C, then treated with 0.07 units of DNase I for 2 min and finally quenched with 0.025 M EDTA. For hydroxyl radical footprinting, samples (as for DNase I footprints but without MgCl2) were treated with 2 μL each of freshly prepared Fe(II)-EDTA solution (1 mM ammonium iron (II) sulphate, 2 mM EDTA), 0.01 M sodium ascorbate and 0.6% hydrogen peroxide for 4 min at 25 °C. The reaction was stopped with 2 μL each of 0.1 M thiourea and 0.5 M EDTA (pH 8). Footprinting reactions were done under dim light and, after quenching, under normal light. DNA from each sample was ethanol precipitated, washed twice with 70% ethanol, dried, and resuspended in formamide loading buffer. The 5 μL samples were heated at 95 °C for 3 min and loaded onto a 6% polyacrylamide-8 M urea sequencing gel together with G+A chemical sequencing ladders. Gels were vacuum-dried and analyzed by autoradiography, and the bands quantitated using GelAnalyzer 2010a (http://www.gelanalyzer.com). Each experiment was repeated at least three times.
Analytical size exclusion chromatography (SEC)
Analytical SEC for all CarH mutants except for H132A CarH was carried out using an ÄKTAbasic unit and a Superdex 200 analytical SEC column (GE Healthcare)8. The calibration curve was: log Mr = 7.885 – 0.221Ve, where Mr is the apparent molecular weight and Ve is the elution volume. Pure protein (100 μL, 50–100 μM) was incubated with a five-fold molar excess of AdoCbl for at least 15 min and analyzed by SEC in the dark or after light irradiation for 5 min with white light from fluorescent lamps at 10 W/m2. Elution at 0.4 mL/min flow rate was tracked by absorbance at 280 nm and 522 nm and Mr was estimated from Ve. Each SEC experiment was performed at least three times.
Analytical SEC for H132A CarH was carried out using an ÄKTA FPLC unit and a Superose 6 10/300 GL column (GE Healthcare) equilibrated with CarH buffer. The calibration curve was: log Mr = 9.74 – 0.30Ve. WT or H132A CarH (300 μL, 20 – 50 μM) with stoichiometric AdoCbl with or without exposure to white light for 1 h were injected onto the column and elution was tracked by absorbance at 280 nm. For AdoCbl exchange studies, WT or H132A CarH samples were exposed to light as described and then incubated with a 10-fold excess of free AdoCbl for the given time periods and temperatures and analyzed by SEC.
Solution UV-Vis spectroscopy
Solution UV-Vis spectra were recorded at 25 °C on a SpectraMax Plus 384 (Molecular Devices) using SoftMax Pro 5 software (Molecular Devices) and a 1 cm path length quartz cuvette (Starna). WT or H132A CarH in CarH buffer were transferred to the cuvette under red light or after exposure to white light for 20 min and UV-Vis spectra were recorded from 250–800 nm. The spectrum of pure CarH buffer was used for background subtraction. No photolysis occurred on the timescale of spectrum acquisition, as repeated acquisition did not lead to spectral changes.
Spectra of Cbl with increasing imidazole concentrations, similar to spectra reported previously20, were obtained with the same experimental parameters. Cbl solutions contained 50 μM OHCbl•HCl (Sigma) in 50 mM Tris with 0 mM, 0.4 mM, or 400 mM imidazole, adjusted to a final pH of 8 to match the protein solutions. All solutions were incubated for 16 h at 25 °C to allow for complete ligand exchange to take place.
Single crystal UV-Vis spectroscopy
Single-crystal UV-Vis spectra were recorded at a temperature of 100 K at Stanford Synchrotron Radiation Laboratory beamline 11-1 (Menlo Park, CA) using a UV-Vis microspectrophotometer. The setup used a Hamamatsu light source (50 μm light spot) with deuterium and halogen lamps, UV solarization-resistant optical fibers, reflective Newport Schwardchild objectives, and an Ocean Optics QE65000 Spectrum Analyzer. Spectra were acquired as 50 averages with an integration time of 0.03 s and a boxcar width of 3. A crystal of AdoCbl-bound CarH was cryoprotected, transferred to a nylon fiber loop, and frozen in liquid nitrogen as described above. A background spectrum was acquired on a region of the fiber loop containing just cryoprotectant. A sample spectrum was then acquired on the crystal.
Extended Data
Extended Data Figure 1.
CarH crystals contain intact AdoCbl. (a) UV-Vis spectra obtained from AdoCbl-bound CarH crystals at T=100 K (red trace) or AdoCbl-bound CarH in solution at T=298 K (black trace) exhibit good qualitative agreement and similar features, including a peak centered around 540 nm with a shoulder around 560 nm. Because many band intensities are orientation-dependent and the crystal spectrum changes with orientation but molecules are rotationally averaged in solution, quantitative comparison of the spectra is difficult. Note also that individual bands appear sharper in the crystal spectrum because the molecules have fewer rotational degrees of freedom and because fewer vibrational states are populated at T=100 K. (b) Simulated annealing composite omit electron density contoured around AdoCbl at 1.0 σ (gray). The electron density covers the entire AdoCbl molecule including the Co-C bond, indicating that the Co-C bond remained intact during crystallization and data collection. AdoCbl is shown in stick representation with Cbl carbons in pink and 5′-dAdo group carbons in cyan. Co is shown as a purple sphere. The Co-coordinating His177 is shown in sticks with carbons in green. CarH shown in ribbons with helix bundle in yellow and Cbl-binding domain in green.
Extended Data Figure 2.
The CarH DNA-binding domain is flexible in the absence of DNA. (a) Overlay of five CarH protomers, including the protomer shown in Figure 2a, highlighting flexibility of DNA-binding domains. Structures are aligned by the Cbl-binding domains (green) and helix bundles (yellow) and shown in the same orientation as Figure 2a. DNA-binding domains are colored in dark cyan, light cyan, dark blue, black, and gray. AdoCbl is shown with Cbl carbons in pink, 5′-dAdo group carbons in cyan, and cobalt in purple. (b-e) Individual CarH protomers shown side by side. Orientation and coloring as in (a).
Extended Data Figure 3.
CarH mutant analysis and multiple sequence alignment. (a) Results summary for in vitro CarH mutant analysis. (b) Alignment of CarH sequences from different bacterial species. Sequence identity is shown in white font with red background, sequence similarity in red font. Colored triangles highlight functionally important positions, with filled triangles indicating residues analyzed by mutagenesis in this study and empty triangles indicating residues not analyzed by mutagenesis. Mutating the highly conserved His177 of the Cbl-binding motif, the lower axial ligand of bound AdoCbl, has previously been shown to impair AdoCbl binding and tetramerization8. Coloring as follows: hydrogen bonds/ionic interactions to DNA: orange; contact to 5′-dAdo: green; histidines coordinating Cbl (His132 only coordinates after light exposure): red; hydrogen bonds/ionic interactions at dimer interface: black; hydrogen bonds/ionic interactions as well as Gly160 and Gly192 at dimer-dimer interface: cyan. Residues involved in more than one type of interaction are colored half/half. Residues at protein interfaces are less well conserved than other functionally important residues, likely because compensatory mutations and local structural deformations are possible. Note, however, that the T. thermophilus Arg176-Asp201 pair observed in our structure is inverted in Myxococcus xanthus, suggesting that the interaction is conserved. Alignment generated using ESPript63.
Extended Data Figure 4.
Characterization of CarH mutants affecting oligomerization state. (a-f) SEC traces (Superdex 200 analytical SEC column) of CarH carrying mutations (a,b) near the 5′-dAdo group, (c,d) at the head-to-tail dimer interface, and (e,f) at the dimer-dimer interface. Shown are traces of mutants incubated with AdoCbl in the dark (top panels) and after light exposure (bottom panels). In all panels, both A280 (tracking protein) and A522 (tracking Cbl) traces are shown. Molecular weights are calculated from the observed elution volumes as described in the methods, and are consistent with a tetrameric species (137 kDa), a dimeric species (89 kDa), and a monomeric species (39 kDa). Notably, mutant CarH proteins that do not tetramerize in the presence of AdoCbl (D201R, H142A) also do not appear to bind AdoCbl (see 522 nm traces of dark samples). This finding is consistent with previous studies that show cooperativity of AdoCbl binding and tetramerization, a feature that does not hold for other forms of Cbl (methylcobalamin, CNCbl and Cbl)8. Both of these mutant proteins can still bind Cbl (see 522 nm traces of light-exposed samples), which further suggests that these mutants are properly folded and that the lack of AdoCbl binding stems from inability to oligomerize. Although the degree of tetramerization of CarH mutant proteins in the dark varied, all of these mutant proteins form Cbl-bound monomers after light exposure (g) DNA-binding capacity of WT and mutant proteins (800 nM) as determined by EMSAs after incubation with AdoCbl (4 μM) in the dark. (h) EMSA data for WT CarH and the G160Q and G192Q mutants fit to the Hill equation, as described in the methods. KD (in nM) and Hill coefficients from the fits are, respectively, (67±2) and (5.1±0.7) for WT CarH, (111±18) and (3.0±0.2) for G160Q CarH, and (253±17) and (2.5±0.3) for G192Q CarH. The data shown are the mean values and standard errors of three to five repeat experiments.
Extended Data Figure 5.
Identification and validation of CarH operator sequence by EMSAs and footprinting. (a) Location of CarH operator in the intergenic region between carH and the carotenogenic crtB of the T. thermophilus genome. Structural and biochemical data are mapped onto the sequence. Three 11-bp CarH binding sites are shown in cyan font and the promoter −35 element is highlighted with a red box. Nucleotides protected from hydroxyl radical cleavage are indicated with bullets. The ∼42-nucleotide DNase I footprint on the sense strand is shown above the sequence and that of the antisense strand has been omitted for clarity. Nucleotide numbering on the sense strand is relative to the carH transcription start site (underlined, +1)10. To identify suitable DNA constructs for crystallization, operator sequences were systematically trimmed around a ∼40-bp segment, as indicated by the black bars, and binding was assessed by EMSAs (shown in b). The sequences of two 26-bp DNA segments used for co-crystallization are also shown. The blunt-ended 26-bp segment was used for determination of the CarH:DNA structure. The second 26-bp segment contained 1-nucleotide 3′-overhangs and 5-iodo-deoxycytidine in position −25 (red) and was used to validate the mode of DNA binding. (b) Binding of CarH (800 nM) to DNA segments of different lengths after incubation with AdoCbl (4 μM) in the dark. Substantial DNA-binding was observed for a probe as small as 30-bp. (c) DNase I and hydroxyl radical footprints of CarH on a 130-bp operator DNA segment. Disappearance of bands in the presence of CarH indicates protection from cleavage. Protected regions are marked on the side and were mapped onto the operator sequence using G+A chemical sequencing experiments performed in parallel. (d,e) CarH binding to 40-bp operators carrying mutations. (d) Sequences of tested operator variants. WT operator sequence shown at the top and bottom, with repeat sequences that CarH recognizes shown in cyan. 6-bp stretch contacted by CarH recognition helix is boxed. Mutations are as follows: Mut1-7: single (1-3), pairwise (4-6), and triple (7) mutations of AC to GT (Positions 8/9); Mut8-14: single (8-10), double (11-13), and triple (14) mutations of (A/C)T to GC (Positions 4/5); Mut15-18: pairwise (15-17) and triple (18) mutations of (A/G)A) to TT (Positions 1/2). (e) EMSAs with WT CarH (800 nM) and each of the 40-bp operator variants after incubation with AdoCbl (4 μM) in the dark. Note that two additional lower mobility complexes are observed, most apparent with the WT operator and its variants with comparable binding. The origin of these complexes is unknown, but they likely arise from oligomeric equilibria and residual amounts of light-exposed protein in the sample.
Extended Data Figure 6.
CarH DNA binding, conformational changes upon binding, and comparison to BmrR. (a) 2Fo–Fc omit electron density for DNA-bound CarH, calculated after performing full refinement of the model with DNA omitted and contoured at 1.0 σ. DNA is shown with carbons in yellow and recognition helix of a CarH DNA-binding domain with carbons in cyan. (b,c) Validation of DNA-binding mode using heavy-atom derivatized DNA segments. CarH was crystallized with a DNA segment containing 5-iodo-deoxycytidine in position −25 of the sense strand. Shown is the resulting anomalous difference density (purple mesh), contoured at 6 σ, for both CarH:DNA complexes in the asymmetric unit, with peaks directly adjacent to the C5 atom of deoxycytidine in position −25. (d) Chemical structure of 5-iodo-deoxycytidine. (e) Comparison of CarH before and after DNA binding, revealing rearrangement of DNA-binding domains. CarH before DNA binding is shown with helix bundles and Cbl-binding domains in gray and DNA-binding domains in pink. CarH bound to DNA is shown with helix bundles and Cbl-binding domains in green and DNA-binding domains in cyan. The fourth DNA-binding domain of DNA-bound CarH is disordered and not modelled. DNA is shown in yellow. AdoCbl is shown with Cbl carbons in pink and 5′-dAdo group carbons in cyan. (f) Contacts between residues in neighboring DNA-binding domains, colored by domain. Each interface between two DNA-binding domains buries 280 Å2 of surface from solvent on each DNA-binding domain. Interactions of Arg72 to Tyr7 and Glu11 are indicated by black dashed lines. Coloring as in (e). (g,h) Models of individual CarH head-to-tail dimers bound to DNA. (g) Head-to-tail dimer contributing the middle of the three DNA-binding domains, colored by domain with DNA-binding domain in cyan, helix bundles in yellow, and Cbl-binding domains in green. The DNA-binding domain of the second protomer (right) is disordered and not modeled. DNA and AdoCbl are shown as in (e). (h) Head-to-tail dimer contributing the flanking DNA-binding domains. Helix bundles and Cbl-binding domains are shown in gray, remaining coloring as in (e). (i) BmrR bound to DNA (PDB ID code 1EXJ38). A BmrR dimer is shown in ribbon representation in orange and red. DNA is shown in yellow. BmrR binds as a dimer to a palindromic sequence and distorts the DNA double strand from its ideal conformation.
Extended Data Figure 7.
In vitro characterization of CarH DNA binding mutants. (a,b) SEC traces (Superdex 200 analytical SEC column) of CarH carrying mutations in the DNA-binding domain. Shown are traces of mutants (a) incubated with AdoCbl in the dark and (b) after light exposure. In all panels, both A280 (tracking protein) and A522 (tracking Cbl) traces are shown. Molecular weights are calculated from the observed elution volumes as described in the methods, and are consistent with a tetrameric species (137 kDa) and a monomeric species (39 kDa). (c) DNA-binding capacity of mutants (800 nM) as determined by EMSAs after incubation with AdoCbl (4 μM) in the dark.
Extended Data Figure 8.
Light-exposed CarH has bis-His ligated Cbl. (a) Structure of light-exposed CarH including the DNA-binding domain (cyan) and other domains colored as in Figure 5a. (b) Close-up view of the Cbl in light-exposed CarH, with both coordinating His side chains shown in sticks. Simulated annealing composite omit electron density is shown in blue, contoured at 1.0 σ. (c) UV-Vis spectra of light-exposed WT CarH (black) and H132A CarH (red) exhibit pronounced differences, indicating that the bis-His ligation is also formed in solution. (d) UV-Vis spectra of free OHCbl (50 μM) with increasing imidazole concentration. The spectrum of bis-imidazole ligated Cbl (black, Cbl with 400 mM imidazole contains 60% bis-imidazole ligated Cbl and 40% Cbl with dimethylbenzimidazole and imidazole as ligands20) resembles that of light-exposed WT CarH, whereas the spectrum of free OHCbl (pink) resembles that of light-exposed H132A CarH. Note that the latter two are expected to be slightly different because free OHCbl contains a dimethylbenzimidazole group as the lower axial ligand, whereas light-exposed H132A CarH contains a histidine imidazole as the lower axial ligand. Experimental conditions chosen were similar to those reported elsewhere20. (e) UV-Vis spectra of AdoCbl-bound WT CarH (black) and H132A CarH (red) are virtually identical, suggesting that the mode of AdoCbl binding is unchanged, as is expected from the structure. (f,g) Size exclusion chromatograms (Superose 6 10/300 GL column) of AdoCbl-bound and light-exposed (f) WT CarH and (g) H132A CarH, demonstrating that H132A CarH, like WT CarH, forms a tetramer in the dark and undergoes light-dependent tetramer disassembly.
Extended Data Figure 9.
Disruption of bis-His ligation by H132A mutation facilitates Cbl dissociation after photolysis. (a,b) WT and H132A CarH were exposed to light, rendering them monomeric, and then incubated with free AdoCbl at the indicated temperatures and time periods. For AdoCbl to bind to CarH and induce tetramerization, the photolyzed Cbl has to dissociate from the protein first. Thus, the extent of tetramer formation, as assessed by size exclusion chromatography (Superose 6 10/300 GL column), is indicative of the affinity of the protein for photolyzed Cbl. That is, lack of tetramer formation in the presence of fresh AdoCbl indicates that the photolyzed Cbl is still bound to the protein. The observed differential in tetramer formation between H132A and WT CarH is substantial: WT CarH retains its photolyzed Cbl, showing only a small amount of tetramer formation, whereas H132A CarH loses its photolyzed Cbl, reforming tetramers upon AdoCbl addition. (c) ESI-TOF mass spectra of WT and H132A CarH after light exposure also reveal differential affinity for photolyzed Cbl. Light-exposed WT CarH is 1329 Da larger in molecular weight than light-exposed H132A CarH, a number corresponding to the molecular weight of Cbl. This difference in molecular weight suggests that, even under the harsh conditions of this experiment, photolyzed Cbl remains bound to the WT CarH monomer but not to H132A CarH, indicating that Cbl dissociates more readily without the bis-His ligation. The minor peak next to WT CarH arises from protein bound to a potassium ion (mass shift 39 Da). Species marked with an asterisk (*) correspond to an unidentified impurity in the H132A CarH sample. (d) Control experiment showing ESI-TOF mass spectra of WT and H132A CarH in the AdoCbl-bound dark state. The very similar molecular weights obtained (differing only due to the H132A mutation) indicate that the mutation has no effect on AdoCbl binding, consistent with the fact that His132 is not coordinated to Cbl when the upper 5′-dAdo ligand is present. Both WT and H132A CarH lose their AdoCbl cofactor as the tetramer disassembles into monomeric units.
Extended Data Table 1.
Crystallographic data collection and refinement statistics.
| AdoCbl-bound CarH form 1 Native * | AdoCbl-bound CarH form 1 Co Peak† | AdoCbl-bound CarH form 2 | AdoCbl-bound CarH form 3 | AdoCbl- and DNA-bound CarH | AdoCbl- and DNA-bound CarH (iodine-labeled) *† | light-exposed CarH | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| PDB code | 5C8A | 5C8D | 5C8E | 5C8F | |||
| Data collection | |||||||
| Space group | P43212 | P43212 | P212121 | P1 | P21212 | P21212 | I4122 |
| Cell dimensions | |||||||
| a, b, c (Å) | 94.5, 94.5, 180.5 | 94.5, 94.5, 180.6 | 51.4, 99.7, 144.0 | 78.7, 79.7, 118.4 | 177.9, 141.8, 162.7 | 176.7, 141.7, 162.6 | 126.9, 126.9, 149.5 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.7, 96.6, 117.3 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Wavelength | 0.9792 | 1.6039 | 0.9795 | 0.9795 | 0.9795 | 1.7365 | 0.9791 |
| Resolution (Å) | 200–2.80 (2.87–2.80) | 200–3.30 (3.36–3.30) | 100–2.15 (2.21 –2.15) | 100–2.80 (2.87–2.80) | 100–3.89 (3.99–3.89) | 100–5.00 (5.13–5.00) | 100–2.65 (2.72–2.65) |
| Rsym (%) ‡ | 6.5 (69.0) | 12.4 (38.6) | 5.3 (60.8) | 10.5 (83.2) | 9.8 (126.4) | 7.2 (107.1) | 6.9 (167.8) |
| Rmeas (%) ‡ | 7.2 (76.5) | – § | 6.2 (71.5) | 11.7 (91.9) | 10.2 (131.9) | 8.6 (127.2) | 7.2 (174.4) |
| CCl/2‡ | 99.9 (77.1) | – § | 99.9 (87.7) | 99.7 (84.3) | 99.9 (87.0) | 99.9 (54.3) | 100.0 (68.5) |
| <I/σ(I)> ‡ | 16.4 (2.3) | 15.7 (7.0) | 15.2 (2.1) | 12.0 (2.0) | 15.7 (2.0) | 7.4 (1.1) | 26.8 (1.8) |
| Completeness (%)‡ | 99.5 (99.9) | 99.5 (100.0) | 98.5 (99.4) | 94.9 (95.5) | 99.9 (100.0) | 97.7 (98.2) | 100.0 (99.9) |
| Redundancy‡ | 5.3 (5.4) | 11.9 (11.5) | 3.6 (3.6) | 5.6 (5.6) | 12.2 (12.4) | 3.5 (3.3) | 12.8 (13.4) |
| Refinement | |||||||
| Resolution (Å) ‡ | 100–2.15 (2.21–2.15) | 100–2.80 (2.87–2.80) | 100–3.89 (3.99–3.89) | 100–2.65 (2.72–2.65) | |||
| No. reflections‡ | 40511 (2970) | 59284 (4419) | 38480 (2819) | 18067 (1319) | |||
| Rwork/Rfree | 0.183/0.227 | 0.183/0.230 | 0.250/0.257 | 0.172/0.203 | |||
| No. atoms | |||||||
| protein | 5766 | 14668 | 14500 | 2090 | |||
| Cbl | 364 | 728 | 728 | 91 | |||
| 5′-deoxyadenosine | 72 | 144 | 144 | – | |||
| water | 259 | – | – | 19 | |||
| DNA | – | – | 2120 | – | |||
| glycerol | 6 | – | – | 6 | |||
| chloride | – | – | – | 2 | |||
| B-factors | |||||||
| protein | 48.5 | 76.4 | 164.4 | 85.9 | |||
| Cbl | 45.1 | 65.2 | 153.9 | 100.8 | |||
| 5′-deoxyadenosine | 46.1 | 75.8 | 158.5 | – | |||
| water | 47.3 | – | – | 71.2 | |||
| DNA | – | – | 231.3 | – | |||
| glycerol | 54.3 | – | – | 95.3 | |||
| chloride | – | – | – | 94.7 | |||
| R.m.s deviations | |||||||
| Bond lengths (Å) | 0.004 | 0.005 | 0.005 | 0.004 | |||
| Bond angles (°) | 0.82 | 0.92 | 0.79 | 0.74 | |||
| Rotamer outliers (%) | 5 (0.9%) | 8 (0.6%) | 10(0.8%) | 0 (0.0%) | |||
Structure was not refined to completion. Oligomerization was probed by size exclusion chromatography and DNA binding by gel shift analysis.
Bijvoet pairs were not merged during data processing. Y30A, H42A: weakened binding at 100 nM protein.
Values in parentheses indicate highest resolution bin. G160Q, G192Q: dimer, no tetramer.
Values were not reported in the version of Scalepack used for scaling. G160Q, G192Q: binds with reduced affinity and cooperativity and as a higher mobility (smaller size) complex.
Acknowledgments
We thank Prof. Francisco Murillo for his invaluable contributions and support, José Antonio Madrid for technical assistance (Universidad de Murcia), and Dr. Aina Cohen (SSRL) and Dr. Nozomi Ando (MIT) for help with single crystal UV-Vis spectroscopy. This work was supported in part by the National Institutes of Health (NIH, Grant GM069857 to C.L.D), the Ministerio de Economía y Competitividad, Spain (grants BFU2012-40184-C02-01 co-financed by FEDER funds to M.E.-A.; BFU2012-40184-C02-02 to S.P.; Ph.D. fellowship to J.F.-Z.), a CSIC-JAE-Predoc (Spain) fellowship (to J.M.O.-G.), and an MIT Poitras pre-doctoral fellowship (to M.J.). C.L.D. is a Howard Hughes Medical Institute Investigator. This work used the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source (APS), which are supported by National Institute of General Medical Sciences (NIGMS) grant P41GM103403 and the NIH. APS is a U.S. Department of Energy (DOE) Office of Science User Facility operated under Contract No. DE-AC02-06CH11357. Use of the Stanford Synchrotron Radiation Lightsource is supported by the U.S. DOE, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515, the DOE Office of Biological and Environmental Research, and by the NIH, NIGMS (including P41GM103393).
Footnotes
Author Information: Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank (http://www.rcsb.org) under accession codes 5C8A (AdoCbl-bound CarH crystal form 2), 5C8D (AdoCbl-bound CarH crystal form 3), 5C8E (AdoCbl- and DNA-bound CarH), and 5C8F (light-exposed CarH).
The authors declare no competing financial interests.
Author Contributions: M.J. performed crystallographic experiments, J.F.-Z., M.C.P., and J.M.O.-G. performed in vitro mutant analyses, and G.K. and P.Y.-T.C. assisted with crystal structure refinement. M.J., S.P., M.E.-A., and C.L.D. designed experiments, analyzed the data, and wrote the manuscript.
References
- 1.Ziegelhoffer EC, Donohue TJ. Bacterial responses to photo-oxidative stress. Nat Rev Microbiol. 2009;7:856–863. doi: 10.1038/nrmicro2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eberhard S, Finazzi G, Wollman FA. The Dynamics of Photosynthesis. Annu Rev Genet. 2008;42:463–515. doi: 10.1146/annurev.genet.42.110807.091452. [DOI] [PubMed] [Google Scholar]
- 3.Kami C, Lorrain S, Hornitschek P, Fankhauser C. Light-Regulated Plant Growth and Development. Plant Development. 2010;91:29–66. doi: 10.1016/S0070-2153(10)91002-8. [DOI] [PubMed] [Google Scholar]
- 4.Palczewski K. Chemistry and Biology of Vision. J Biol Chem. 2012;287:1612–1619. doi: 10.1074/jbc.R111.301150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- 6.Purcell EB, Crosson S. Photoregulation in prokaryotes. Curr Opin Microbiol. 2008;11:168–178. doi: 10.1016/j.mib.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Möglich A, Yang XJ, Ayers RA, Moffat K. Structure and Function of Plant Photoreceptors. Annu Rev Plant Biol. 2010;61:21–47. doi: 10.1146/annurev-arplant-042809-112259. [DOI] [PubMed] [Google Scholar]
- 8.Ortiz-Guerrero JM, Polanco MC, Murillo FJ, Padmanabhan S, Elias-Arnanz M. Light-dependent gene regulation by a coenzyme B12-based photoreceptor. Proc Natl Acad Sci U S A. 2011;108:7565–7570. doi: 10.1073/pnas.1018972108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Marin MC, Padmanabhan S, Polanco MC, Murillo FJ, Elias-Arnanz M. Vitamin B12 partners the CarH repressor to downregulate a photoinducible promoter in Myxococcus xanthus. Mol Microbiol. 2008;67:804–819. doi: 10.1111/j.1365-2958.2007.06086.x. [DOI] [PubMed] [Google Scholar]
- 10.Takano H, et al. Involvement of CarA/LitR and CRP/FNR Family Transcriptional Regulators in Light-Induced Carotenoid Production in Thermus thermophilus. J Bacteriol. 2011;193:2451–2459. doi: 10.1128/JB.01125-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elias-Arnanz M, Padmanabhan S, Murillo FJ. Light-dependent gene regulation in nonphototrophic bacteria. Curr Opin Microbiol. 2011;14:128–135. doi: 10.1016/j.mib.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Takano H, et al. LdrP, a cAMP receptor protein/FNR family transcriptional regulator, serves as a positive regulator for the light-inducible gene cluster in the megaplasmid of Thermus thermophilus. Microbiology. 2014;160:2650–2660. doi: 10.1099/mic.0.082263-0. [DOI] [PubMed] [Google Scholar]
- 13.Frey PA. In: Comprehensive Natural Products II Chemistry and Biology. Mander L, Liu HW, editors. Vol. 7. Elsevier; 2010. pp. 501–546. [Google Scholar]
- 14.Banerjee R. Radical carbon skeleton rearrangements: catalysis by coenzyme B12-dependent mutases. Chem Rev. 2003;103:2083–2094. doi: 10.1021/cr0204395. [DOI] [PubMed] [Google Scholar]
- 15.Brown NL, Stoyanov JV, Kidd SP, Hobman JL. The MerR family of transcriptional regulators. FEMS Microbiol Rev. 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 16.Navarro-Aviles G, et al. Structural basis for operator and antirepressor recognition by Myxococcus xanthus CarA repressor. Mol Microbiol. 2007;63:980–994. doi: 10.1111/j.1365-2958.2006.05567.x. [DOI] [PubMed] [Google Scholar]
- 17.Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig ML. How a protein binds B12: A 3.0 Å X-ray structure of B12-binding domains of methionine synthase. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 18.Diez AI, et al. Analytical ultracentrifugation studies of oligomerization and DNA-binding of TtCarH, a Thermus thermophilus coenzyme B12-based photosensory regulator. Eur Biophys J. 2013;42:463–476. doi: 10.1007/s00249-013-0897-x. [DOI] [PubMed] [Google Scholar]
- 19.Cheng Z, Li KR, Hammad LA, Karty JA, Bauer CE. Vitamin B12 regulates photosystem gene expression via the CrtJ antirepressor AerR in Rhodobacter capsulatus. Mol Microbiol. 2014;91:649–664. doi: 10.1111/mmi.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marques HM, Marsh JH, Mellor JR, Munro OQ. The Coordination of Imidazole and Its Derivatives by Aquocobalamin. Inorg Chim Acta. 1990;170:259–269. [Google Scholar]
- 21.Linden H, Macino G. White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J. 1997;16:98–109. doi: 10.1093/emboj/16.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Froehlich AC, Liu Y, Loros JJ, Dunlap JC. White collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- 23.Nash AI, et al. Structural basis of photosensitivity in a bacterial light-oxygen-voltage/helix-turn-helix (LOV-HTH) DNA-binding protein. Proc Natl Acad Sci U S A. 2011;108:9449–9454. doi: 10.1073/pnas.1100262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgie ES, Vierstra RD. Phytochromes: An Atomic Perspective on Photoactivation and Signaling. Plant Cell. 2014;26:4568–4583. doi: 10.1105/tpc.114.131623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Losi A, Gärtner W. The Evolution of Flavin-Binding Photoreceptors: An Ancient Chromophore Serving Trendy Blue-Light Sensors. Annu Rev Plant Biol. 2012;63:49–72. doi: 10.1146/annurev-arplant-042811-105538. [DOI] [PubMed] [Google Scholar]
- 26.Conrad KS, Manahan CC, Crane BR. Photochemistry of flavoprotein light sensors. Nat Chem Biol. 2014;10:801–809. doi: 10.1038/nchembio.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrou J, Crosson S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat Rev Microbiol. 2011;9:713–723. doi: 10.1038/nrmicro2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkins GI. The UV-B Photoreceptor UVR8: From Structure to Physiology. Plant Cell. 2014;26:21–37. doi: 10.1105/tpc.113.119446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nahvi A, Barrick JE, Breaker RR. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;32:143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nahvi A, et al. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 31.Johnson JE, Reyes FE, Polaski JT, Batey RT. B12 cofactors directly stabilize an mRNA regulatory switch. Nature. 2012;492:133–137. doi: 10.1038/nature11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peselis A, Serganov A. Structural insights into ligand binding and gene expression control by an adenosylcobalamin riboswitch. Nat Struct Mol Biol. 2012;19:1182–1184. doi: 10.1038/nsmb.2405. [DOI] [PubMed] [Google Scholar]
- 33.Jost M, Simpson JH, Drennan CL. The Transcription Factor CarH Safeguards Use of Adenosylcobalamin as a Light Sensor by Altering the Photolysis Products. Biochemistry. 2015 doi: 10.1021/acs.biochem.5b00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girvan HM, Munro AW. Heme sensor proteins. J Biol Chem. 2013;288:13194–13203. doi: 10.1074/jbc.R112.422642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matthews RG. Cobalamin-dependent methyltransferases. Acc Chem Res. 2001;34:681–689. doi: 10.1021/ar0000051. [DOI] [PubMed] [Google Scholar]
- 36.Jarrett JT, et al. Protein radical cage slows photolysis of methylcobalamin in methionine synthase from Escherichia coli. Bioorg Med Chem. 1996;4:1237–1246. doi: 10.1016/0968-0896(96)00119-8. [DOI] [PubMed] [Google Scholar]
- 37.Bandarian V, Ludwig ML, Matthews RG. Factors modulating conformational equilibria in large modular proteins: A case study with cobalamin-dependent methionine synthase. Proc Natl Acad Sci U S A. 2003;100:8156–8163. doi: 10.1073/pnas.1133218100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heldwein EEZ, Brennan RG. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature. 2001;409:378–382. doi: 10.1038/35053138. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe S, Kita A, Kobayashi K, Miki K. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc Natl Acad Sci U S A. 2008;105:4121–4126. doi: 10.1073/pnas.0709188105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barker HA, et al. Isolation and Properties of Crystalline Cobamide Coenzymes Containing Benzimidazole or 5,6-Dimethylbenzimidazole. J Biol Chem. 1960;235:480–488. [PubMed] [Google Scholar]
- 41.Firth RA, Hill HAO, Pratt JM, Williams RJP, Jackson WR. The Circular Dichroism and Absorption Spectra of Some Vitamin B12 Derivatives. Biochemistry. 1967;6:2178–2189. doi: 10.1021/bi00859a040. [DOI] [PubMed] [Google Scholar]
- 42.Hill JA, Williams RJP, Pratt JM. Chemistry of Vitamin B12. Part I. The Valency and Spectrum of the Coenzyme. Journal of the Chemical Society. 1964:5149–5153. [Google Scholar]
- 43.Otwinowski Z, Minor W. Methods Enzymol. In: Carter CW Jr, Sweet RM, editors. Macromolecular crystallography, part A. Vol. 276. Academic Press; 1997. pp. 307–326. [Google Scholar]
- 44.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheldrick GM, Schneider TR. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 46.Schneider TR, Pape T. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Crystallogr. 2004;37:843–844. [Google Scholar]
- 47.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 48.Abrahams JP, Leslie AGW. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 53.van Dijk M, Bonvin AMJJ. 3D-DART: a DNA structure modelling server. Nucleic Acids Res. 2009;37:W235–W239. doi: 10.1093/nar/gkp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ten Eyck LF. Fast Fourier transform calculation of electron density maps. Methods Enzymol. 1985;115:324–337. doi: 10.1016/0076-6879(85)15024-x. [DOI] [PubMed] [Google Scholar]
- 55.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 57.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 59.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010 [Google Scholar]
- 60.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 61.Morin A, et al. Collaboration gets the most out of software. Elife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jain SS, Tullius TD. Footprinting protein-DNA complexes using the hydroxyl radical. Nat Protoc. 2008;3:1092–1100. doi: 10.1038/nprot.2008.72. [DOI] [PubMed] [Google Scholar]
- 63.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]