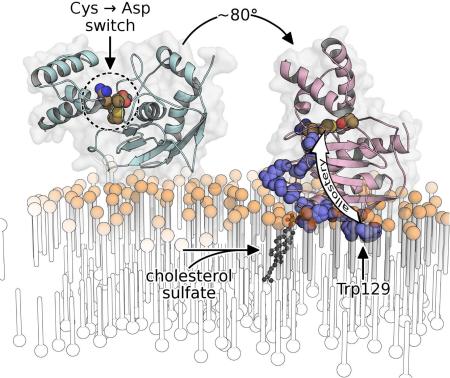
SUMMARY
Matrix metalloproteinase-7 (MMP-7) sheds signaling proteins from cell surfaces to activate bacterial killing, wound healing, and tumorigenesis. The mechanism targeting soluble MMP-7 to membranes has been investigated. NMR structures of the zymogen, free and bound to membrane mimics without and with anionic lipid, reveal peripheral binding to bilayers through paramagnetic relaxation enhancements. Addition of cholesterol sulfate partially embeds the protease in the bilayer, restricts its diffusion, and tips the active site away from the bilayer. Its insertion of hydrophobic residues organizes the lipids, pushing the head groups and sterol sulfate outward towards the enzyme's positive charge on the periphery of the enlarged interface. Fluorescence probing demonstrates a similar mode of binding to plasma membranes and internalized vesicles of colon cancer cells. Binding of bilayered micelles induces allosteric activation and conformational change in the auto-inhibitory peptide and the adjacent scissile site, illustrating a potential intermediate in the activation of the zymogen.
Graphical Abstract
INTRODUCTION
The organization and transient features of plasma membranes support signaling across them (Kusumi et al., 2012). Responses to membrane components by proteins that bind membranes peripherally can regulate and localize signaling and trafficking (Moravcevic et al., 2012). Pericellular proteolysis by metalloproteinases contributes to shaping of transmembrane signaling, such as in inflammation, tissue remodeling, and cancer via crosstalk with growth factor, receptor tyrosine kinase, and inflammatory signaling pathways (Bellac et al., 2014; Dean et al., 2008; Lu et al., 2009; Murphy, 2008; Yu et al., 2002). Pericellular proteolytic activities of matrix metalloproteinase-7 (MMP-7 or matrilysin) govern the health and inflammation of mucosal epithelia by triggering an antibacterial response in the small intestine through activation of α-defensins (Wilson et al., 1999) and by promoting wound closure in the lung by shedding of syndecan-1 (Chen et al., 2009). MMP-7 regulates the remodeling of female reproductive organs by proteolytic activation of heparin-binding epidermal growth factor precursor (pro-HB-EGF) to activate its ErbB4 receptor in uterine and mammary epithelia and tumor cells (Yu et al., 2002).
MMP-7 has been associated with tumorigenesis and aggressiveness of cancers of the breast (Chaturvedi and Hass, 2011; Hulboy et al., 2004; Lynch et al., 2007), colon (Wilson et al., 1997), and pancreas (Fukuda et al., 2011; Sawey et al., 2007). Several instances of its proteolytic release (shedding) of the soluble ectodomain of transmembrane proteins from cell surfaces promote tumorigenesis (Ii et al., 2006) or other key tumor cell behaviors. For example, MMP-7 shedding of HB-EGF stimulates EGF receptor signaling and the proliferation of squamous cell carcinoma cells (Kivisaari et al., 2010). Its shedding of a domain from the ErbB4 receptor boosts the proliferation of mammary tumor cells (Lynch et al., 2007). Its release of the ectodomain of E-cadherin increases tumor cell invasiveness (Noe et al., 2001). MMP-7 shedding of Fas ligand (FasL) and syndecan-1 are each associated with resistance to chemotherapy (Mitsiades et al., 2001; Wang et al., 2014). Understanding how MMP-7 can catalyze these pivotal shedding events will benefit from insight into the structural basis of its pericellular positioning.
Two major strategies of anchorage of MMP-7 to cells have been reported: binding of heparan sulfate proteoglycan (HSPG)-containing complexes (Ryu et al., 2009; Yu and Woessner, 2000; Yu et al., 2002) and binding of cholesterol 3-sulfate (CS) extracted from colon cancer cells (Berton et al., 2007; Yamamoto et al., 2006). The cholesterol sulfate cycle among layers of the epidermis shifts CS content between 1 and 5% of lipids there and potently regulates differentiation, barrier function, cohesion, and scaling (Elias et al., 2014). CS is a marker of precancerous and cancerous lesions of the cervix and prostate (Cooks et al., 2011; Eberlin et al., 2010; Kiguchi et al., 1998). It anchors MMP-7 to membranes in some colon cancer cell lines, causing them to cluster and increasing their metastatic potential (Yamamoto et al., 2006). MMP-7 interactions with CS boost and alter its proteolytic activities upon a number of proteins on cell surfaces and in the extracellular matrix, which are likely to affect tumor cell behaviors (Yamamoto et al., 2006; Yamamoto et al., 2010, 2014). Liposomes, particularly those containing anionic lipids, bind MMP-7 (Ganguly et al., 2007). The anionic lipids CS, sulfatide and cardiolipin (CL) from cell surfaces bind and modulate the activity of MMP-7 in distinct and allosteric ways (Yamamoto et al., 2014). Four residues distant from the active site, plus the C-terminus, were found by mutagenesis to promote binding of this protease to CS and cultured colon cancer cells (Higashi et al., 2008).
Bilayer binding by MMP-7 has been scrutinized here by determining NMR structures of the proMMP-7 zymogen free in solution and bound to membrane mimics, as well as by probing with a membrane-responsive fluor. Structural studies of proteins in fluid, membranous environments have historically been challenging. Paramagnetic structural NMR methods using micelles containing a spin label (Hilty et al., 2004; Kutateladze et al., 2004) have been enhanced and extended to the bilayer phase of disk-like, bilayered micelles known as bicelles (Koppisetti et al., 2014; Lenoir et al., 2015). This approach found MMP-12 to bind bilayer mimics and cellular membranes on opposite sides of the catalytic domain nearer either the β-sheet or α-helices (Koppisetti et al., 2014). ProMMP-7 is reported herein to bind lipid bilayers and mimics near the edges of its β-sheet, but with rather different angles of approach. It binds zwitterionic bicelles by peripherally skimming the head groups using three loops. Addition of anionic lipids like CS to bicelles or vesicles draws proMMP-7 into the bilayer with a more than three-fold larger interface and an altered angle of approach that tips the active site away from the membrane. The dramatic, electrostatically-driven partial insertion and reorientation appears also to be triggered by other bioactive, anionic lipids. Bicelle binding triggers activation, preceded by allosteric changes that propagate into the active site to rearrange the auto-inhibitory peptide (the “cysteine switch”), replacing inhibitory Cys binding of the zinc by Asp in a prospective intermediate in zymogen activation.
RESULTS
For investigating the structural basis of interactions of MMP-7 with membranes, the first NMR structure of an MMP zymogen (247 residues) was determined (PDB: 2mze). 2981 NOEs joined by assignments of aromatic and stereospecific methyl groups defined the structure of proMMP-7 (stabilized by E195A replacement of its general base) with an RMSD of 0.55 Å across the backbone (Figure 1A; Table 1). It is similar to the crystal structures of the catalytic domain (1.7 Å RMSD) and zymogens of other MMPs (2.4 Å RMSD). However, the exposed II-III loop relevant to this study is shifted ~6.5 Å in solution. Disorder is noticeable at the termini, the scissile interdomain linker (Ala73— Pro82, numbered according to (Fulcher et al., 2014)), and the adjoining “bait” region (which recruits proteolytic activation) connecting the first two helices in the pro-domain.
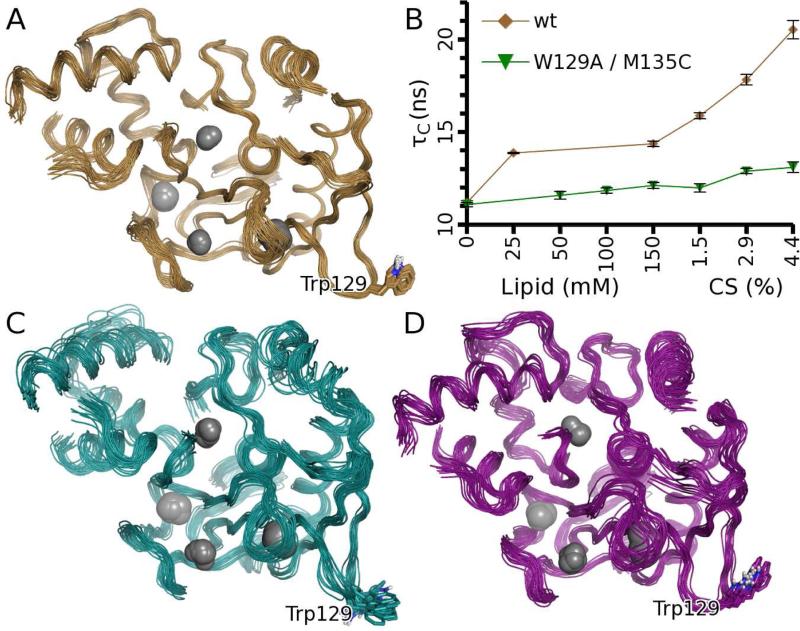
Figure 1. Solution structures and interactions of proMMP-7 with bicelles lacking or containing cholesterol sulfate.
(A) NMR structure of free proMMP-7(E195A) (gold). Flexible loops (specified in Table 1) have been removed for clarity.
(B) DMPC/DH6PC (1:2 or q=0.5) bicelles increased the rotational correlation time τC of wt proMMP-7(E195A) (gold), especially upon additions of CS up to ~4.4% (w/w), but not with the W129A lesion present (green). (M135C is inconsequential to membrane binding; see Figure 4F). Error bars represent RMS of curve fitting.
(C) NMR structure of proMMP-7(E195A) when bound to DMPC/DH6PC (q=0.5) bicelles (cyan). Lipids are omitted for clarity.
(D) NMR structure of proMMP-7(E195A) when bound to such bicelles containing ~4.4% CS (magenta). Lipids are omitted for clarity. See also Figure S1 and Table S1.
Table 1.
NMR Structural Statistics.
| Free | DMPC-bound | DMPC,CS-bound | |
|---|---|---|---|
| PDB ID | 2MZE | 2MZH | 2MZI |
| BMRB code | 25485 | 25488 | 25489 |
| Avg. backbone RMSD (Å)* | 0.55 ± 0.08 | 0.92 ± 0.35 | 0.85 ± 0.11 |
| Avg. heavy RMSD (Å)* | 1.01 ± 0.09 | 1.17 ± 0.36 | 1.12 ± 0.11 |
| Restraints | |||
| Total intraprotein NOEs | 2981 | 827 | 838 |
| Intraresidue NOEs | 896 | 171 | 169 |
| Sequential NOEs | 834 | 361 | 338 |
| Medium-range NOEs | 421 | 145 | 151 |
| Long-range NOEs | 859 | 150 | 180 |
| Interdomain NOEs | 66 | 13 | 14 |
| Dihedrals† | 358 | 358 | 358 |
| Protein-lipid PREs | n/a | 66 | 107 |
| Protein-lipid NOE | n/a | 3 | 13 |
| Restraint violations | |||
| Protein NOE. > 0.5 Å | 0 | 4‡,§ | 4‡ |
| Dihedral > 5° | 0 | 29† | 22† |
| Protein-lipid PRE > 1.0 Å | n/a | 0‡ | 0‡ |
| Protein-lipid NOE > 0.5 Å | n/a | 0‡ | 1‡ |
| Structural quality | |||
| Ramachandran analysis | |||
| Most favored (%) | 87.4 | 91.0 | 86.9 |
| Allowed (%) | 9.5 | 6.2 | 9.1 |
| Dihedral G-factor, overall | −0.32 | −0.60 | −0.58 |
| Bad contacts/1000 residues | 11.92 | 1.07 | 1.08 |
| Verify3D | 0.40 | 0.43 | 0.47 |
Residue ranges used: 12-27, 31-72, 83-216, and 224-238. Uncertainties are SD.
Dihedral restraints of the free form were used in all calculations, but were relaxed in MD simulations with bilayers to a tolerance of at least ± 15° and by 10-fold weaker force constant.
Time-averaged over 12 ps.
Interdomain linker adopted at least two mutually incompatible structures during restrained MD trajectories. Its violations are not included.
Association of Bilayers with proMMP-7
Potential interactions of the II-III loop and interdomain linker with bilayers are suggested by (i) NMR chemical shift perturbations (CSPs) by bicelles prepared in the proportions of one dimyristoyl phosphatidylcholine (DMPC) per two dihexanoyl phosphatidylcholine (DH6PC) and (ii) structure-based predictions (Figure S1A). DMPC/DH6PC bicelles (q=0.5) slowed the rotational diffusion of proMMP-7 by 28%, i.e. the correlation time τc from 15N NMR relaxation increased from 11.2 to 14.4 ns (Figure 1B), suggestive of a transient complex. Additions of the anionic sterol partner CS up to six per bicelle slowed τc 45% further (Figure 1B). The translational diffusion coefficients of the bicelles are insensitive to the addition of CS and are consistently near 1.02×10−10 m2/s (Table S1). This suggests that the CS-slowed rotational diffusion of proMMP-7 results primarily from increased association with the bicelles.
Doping of bicelles with spin-labeled phospholipids at ~1 per leaflet was used to measure proximity of the enzyme to the spin label placed at two different depths (14- or 5-positions) within the fatty acyl chains of the labeled dipalmitoyl phosphatidylcholine (DPPC). The resulting paramagnetic NMR relaxation enhancements (PREs) were measured as differences of single exponential fits from the diamagnetic decays without spin label (Figure S2A). Four scenarios of proximity to the bicelles are illustrated in Figure S2A. Spin labels in the zwitterionic bicelles introduced PREs to three loops of the catalytic domain, with the largest being in the II-III loop and smaller PREs in the I-A and III-IV loops (Figure S2B). The absence of PREs to accompany the CSPs in the interdomain linker ruled out direct interaction of this linker with bilayers and suggests this segment to respond indirectly to bicelles. The three loops nearest the spin-labeled lipids form a roughly planar surface where the catalytic domain may meet the bilayer surface. In one of these loops, Trp129 was found to contribute to association with anionic lipids (Higashi et al., 2008). The W129A mutation nearly abrogates the slowing of rotational diffusion (τc) by DMPC/DH6PC bicelles, with or without CS present (Figure 1B), suggesting the importance of Trp129 to association with bilayers, both without and with net negative charge.
Structures of ProMMP-7 Bound to Bicelles
With saturating DMPC/DH6PC (1:2) bicelles present, ~800 methyl and amide NOEs within proMMP-7(E195A, 15N/12C/2H/ ILV 13CH3-labeled) were used to revise the structure bound to bicelles to an average backbone RMSD of the protein of 0.92Å (Figure 1C and Table 1). The coordinates are quite similar to the unbound structure, with backbone RMSD averaging 1.9 Å across the catalytic domain and 2.3 Å across the pro-domain where some localized adjustment occurs (see below). When saturated by CS-containing DMPC/DH6PC bicelles (1:2, ~6 CS/bicelle), ~850 methyl and amide NOEs in the ILV-labeled pro-enzyme component provided its NMR structure with average backbone RMSD of 0.85 Å (Figure 1D). These coordinates differ from the free form with backbone RMSD of 1.6 Å across the catalytic domain and 2.1 Å across the pro-domain (omitting the disordered segments listed in Table 1).
Bicelles containing spin-labeled DPPC introduce the largest PREs to the II-III loop of proMMP-7 (Figure S2). Especially large PREs at Val128 and Trp129 (Figures S2B,C and S3) suggest that the II-III loop dominates in molecular recognition of the DMPC bilayer. NOEs between the indole NH of Trp129 and each moiety of choline head groups of the zwitterionic bicelles (Figure S4) suggest these to be significant peripheral contacts. Protein-bilayer depth restraints estimated from the PREs were used to dock proMMP-7 and a simulated DMPC bilayer, initially as rigid bodies. These served as starting coordinates for molecular dynamics (MD) trajectories restrained by the PRE-based protein-lipid depths, protein-lipid NOEs, and NOEs among protein amide and methyl groups. The proMMP-7 component of the ensemble of 20 structures sampled from the three trajectories is shown in Figures 1C and 2A. The peripheral association with the bilayer (PDB: 2mzh) is shallow with a modest 670 ± 70 Å2 of buried surface area (BSA; Table 2), consistent with transiency in the association. The indole ring of Trp129 from the II-III loop and Ala142 from the III-IV loop display hydrophobic interactions with choline head groups (Table S2). The side chain of Tyr96 brushes head groups. Arg92 (from β-strand I near the II-III loop) approaches phosphoryl groups. Neighbors Arg98 (I-A loop) and Arg140 (III-IV loop) consistently form apparent salt bridges with phosphoryl moieties of the head groups (Table S2).
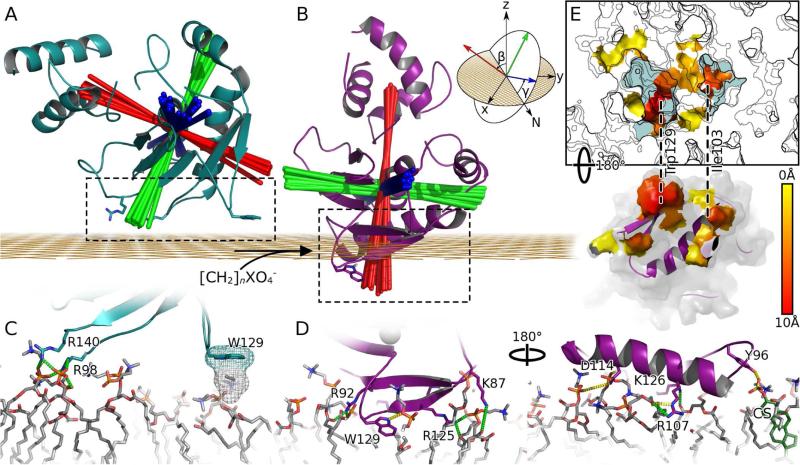
Figure 2. The orientation, penetration, and interface of proMMP-7 for the DMPC bilayer within bicelles in NMR structures are changed by cholesterol sulfate.
(A) The complex with the zwitterionic DMPC bilayer is depicted by the cyan ribbon of the lowest energy NMR structure (model 1). Red lines indicate the longitudinal axis of each of the 20 members of the ensemble. Blue lines mark the lateral axis parallel to the helix at the active site, and green lines the orthogonal axis. The axes intersect at the center-of-mass of the catalytic domain. The grid in orange represents the average depth of lipid phosphoryl groups.
(B) The complex with DMPC bilayer made anionic by CS is represented by the magenta ribbon of its lowest energy structure (model 1). A cartoon (inset) shows the nautical coordinate system and Euler angles β (pitch) and γ (roll); the cartoon is modified from Wikipedia under Creative Commons 2.5 license.
(C) The interfacial contacts with the zwitterionic DMPC head groups mark apparent salt bridges with green dashes and hydrophobic contacts with mesh.
(D) The more extensive contacts with the anionic bilayer are viewed from opposite sides, with H-bonds marked with yellow dashes. The carbon atoms of the CS are green and the sulfur yellow at far right.
(E) Consistent hydrophobic contacts between CS-doped bicelles and proMMP-7 are colored. Membrane surface is contoured at 1 A intervals with light blue representing a depth of > 4 A. Areas of protein and membrane surface involved in hydrophobic contacts with one another are shown as surface colored by distance from lipid phosphate groups from yellow (0 A) to red (10 A). Trp129 and IIe103 insert among the lipid acyl chains; dashed lines connect them with the cavities where they lie in the proximal leaflet. See also Figures S2 – S4 and Table S2.
Table 2.
Features of proMMP-7 binding to bicelles in the absence and presence of cholesterol 3-sulfate*.
| Property | Zwitterionic DMPC/D6PC | Anionic CSa+DMPC/D6PC |
|---|---|---|
| Tc of rotational diffusion | 14.4 ns | 20.9 ns |
| Vertical displacement† | 39.6 ± 0.5 Å | 28.4 ± 0.4 Å |
| Pitch (β)‡ | 62.1 ± 2.7° | 7.5 ± 1.8° |
| Roll (γ)§ | 12.7 ± 3.8° | 4.6 ± 1.2° |
| Buried surface area (BSA) | 670 ± 70 Å2 | 2395 ± 116 Å2 |
| Polar BSA∥ | 448 ± 45 Å2 | 1485 ± 99 Å2 |
| Hydrophobic BSA | 223 ± 39 Å2 | 910 ± 47 Å2 |
| Convergent hydrophobic BSA∥ | 70 ± 23 Å2 | 745 ± 42 Å2 |
at ~3 per leaflet per bicelle on average. Uncertainties are SD.
measured between the centers of mass of the bilayer and catalytic domain
This nautical angle is the Euler angle β between the membrane normal and the principal (longitudinal) axis of the zymogen (red in Fig. 2A,B).
This nautical angle is the Euler angle γ, defined as the rotation about the longitudinal axis, measured between the lateral axis along the active site helix and the bilayer surface.
A detailed list of consistent, apparent contacts is provided in Table S1.
Cholesterol Sulfate Reorients and Inserts proMMP-7 into Bicelles
Anionic lipids are an important component of membranes and CS potently stimulates MMP-7 interactions with tumor cells (Yamamoto et al., 2006) and substrates (Yamamoto et al., 2010, 2014). The sulfate of CS should be attracted to the nominal +9 charge of proMMP-7. Titration of CS into the bicelles up to three per leaflet slows the rotational diffusion of proMMP-7 significantly, increasing its τC from 14.4 to 20.9 ns (Figure 1B), strongly suggesting a longer-lived complex. PRE NMR provided insight into CS-altered proMMP-7 – bicelle complexes. Examples of sites induced by CS to hug bicelles are Ile103 and Lys126 (Figure S2A). With CS present (at ~3 per leaflet of the bicelles), PREs to 14-doxyl PC are systematically higher throughout the II-III loop (Figure S2C), suggesting the insertion of this loop among the acyl chains. CS also introduced significant PREs throughout the N-terminal turn of helix A from Ile103 through Asp106 (Figure S2C), suggesting it to approach the bicelles closely. Not far away, CS resulted in significant PREs in the V-B loop (Gly181) and helix B (Ala192 especially). These changes contrast Asp145 in the III-IV loop which steadily retains Γ2 values around 15 s−1, regardless of CS (Figure S2). Overall, CS brings about closer hugging of the bicelles (consistent with the 45% increase in τC), accompanied by changes in the angle of approach.
Addition of CS introduces NOEs indicating penetration into the hydrophobic interior of the bilayer, e.g., acyl CH2 groups of PC to the Trp129 indole NH and CS groups to Leu108 (Figure S4). The broader membrane interface with CS present is evident from its introduction of NOEs between choline CH3 groups and the amide groups of Tyr91 and His123 in β-strands I and II (Figure S4). Addition of CS to the bicelles induced CSPs extending from the interfacial II-III loop to one end of helix A and across the I-A, V-B and III-IV loops skirting the edge of the β-sheet to reach into the active site (Figure S1B and C).
Addition of CS resulted in 41 more protein-bilayer distance restraints from significant PREs and 10 from additional NOEs to phospholipids, which were used in calculating coordinates of the main complex formed by proMMP-7 and bilayer models containing CS (Table 1; PDB: 2mzi). This structural ensemble reveals that CS switches proMMP-7 from its superficial, reclining posture on the bicelles essentially to “sit up” and dip more deeply into the DMPC bilayer (Figure 2A,B). The deepening due to CS is apparent from the shift of the center of mass of the catalytic domain around 11 Å closer to the bilayer (Table 2). The CS-induced reorientation of the pitch of the longitudinal axis exceeds 55°, moving to within 8° from the membrane normal on average (Table 2). This and the roll angle near 5° indicate that the catalytic cleft is pointing almost directly away from the bilayer, with the length of the cleft orthogonal to the membrane normal (Figure 2B). The CS-induced reorganization buries an additional ~1725 Å2 of surface area in the interface (Table 2) and brings a number of additional contacts into play. Lost are the salt bridges between Arg98 and Arg140 and the head groups. Yet they are replaced by new apparent salt bridges formed by Lys87, Arg92, Arg107, Arg125 and Lys126 (Figure 2D, Table S2) which form a positively charged swath across β-strands I and II, the II-III loop, and helix A. Arg92, Tyr96, Arg107, Asn114, Phe124, Arg125, Lys126, and Trp129 form new H-bonds to lipid head groups and ester groups (Table S2). The hydrophobic surface area buried in the interface is increased nearly 10-fold by the presence of CS (Table 2). The Trp129 indole shifts from merely tickling the lipid head groups to penetrating the acyl chains of the bilayer (Figure 2E). The II-III loop adjusts as Trp129 satisfies both the hydrophobic and hydrophilic proclivities of its side chain. Its indole ring shifts in the presence of CS to insert in a hydrophobic cavity among lipid alkyl and glyceroyl moieties and position the indole Hε1 in an apparent H-bond with a phosphoryl moiety of a head group. Collectively these changes explain the slowing of rotational diffusion of proMMP-7 upon addition of CS.
Variability in Positioning on Bicelles and Restriction by CS
Variability exists in the lateral, vertical, and rotational positioning of the enzyme and its II-III loop relative to the surface of the fluid zwitterionic bilayer (Figures 3A, 2A, Movie S1, and Table 2). The extreme broadening of the Trp129 indole NH peak and the Val128 amide peak by PREs from 5-doxyl PC, while the Val127 NH remains largely unaffected (Figures S2B and S3), is not compatible with a static depth of the spin label below these positions. Transiently closer approaches between the spin-labeled phospholipid and these sites offer an explanation for these PRE patterns. For example, if close approaches of the 5-doxyl label to the Val128 NH of ~16 Å suggested by the NMR ensemble are halved to 8 Å or less, fast exchange with a 2% population of the closer approach would average in enough additional PRE (≥ 54 s−1 at τc of 14.4 ns) to broaden away the NH of Val128 with much less effect on the NH of Val127 merely 3.5 Å more distant. Transient insertion of the Trp129 indole and Val128 is one possible means of such close approach.
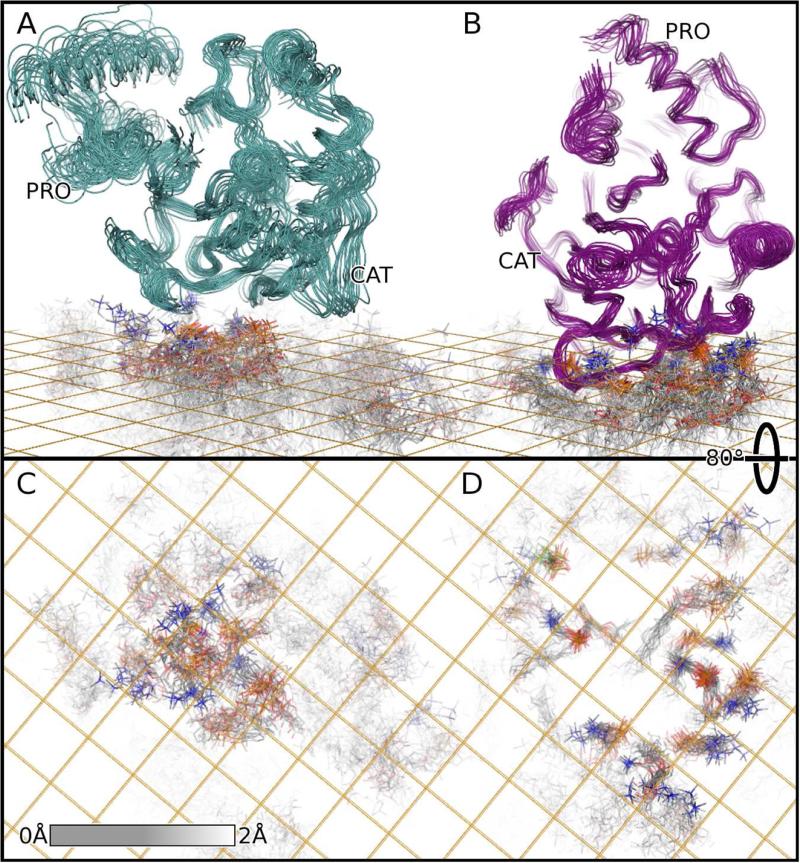
Figure 3. NMR ensembles of complexes of proMMP-7 with DMPC bilayer models showing ordering of some lipids within the fluid bilayers.
(A) Bound to the zwitterionic DMPC bilayer, the proMMP-7 ensemble (cyan) was iteratively fitted to the 1000 best-defined atoms of the enzyme and the 1000 best defined among the lipids, both here and for Figure 2. The grid indicates the average depth of the phosphoryl groups.
(B) The proMMP-7 ensemble (magenta) bound to DMPC bilayers harboring CS, also superposed iteratively, is plotted.
(C) After rotating the view 80°, only DMPC molecul es are plotted from the ensemble containing zwitterionic bilayer. The best defined atoms of the fluid bilayer have full intensity. The shading fades to white where the RMSD of the carbon atoms reaches 2 Å. The nitrogen of the choline groups is blue, oxygen of the phosphoryl groups is red, and the phosphorous orange.
(D) The DMPC molecules of the CS-containing ensemble are plotted in like manner.
Rocking of the enzyme on the bicelles is suggested by variation in the pitch angle with σ of 2.7° and in the roll with σ of 3.8° (Table 2). NOEs between Trp129 and phospholipids suggest their persistent interaction in spite of any transient dipping and rocking (Figure S4). Addition of CS restricts the variation in all of the simulated angular and positional parameters (Table 2, Figures 3B, 2B, and Movie S2). In the complex with the CS-containing DMPC bilayer, several lipid molecules are ordered near proMMP-7 (Figure 3). Ordered dimpling in the bilayer surface looks like a pair of footprints under proMMP-7 (Figures 2E and 3D). This shaping of the head groups places multiple phosphoryl anions next to the positive charge of proMMP-7 (Figures 2C,D and 3). Most of the positively charged choline head groups are displaced outward away from the dimples to the periphery of the interface (Figure 3D).
Vesicle Insertion Promoted by Anionic Lipids
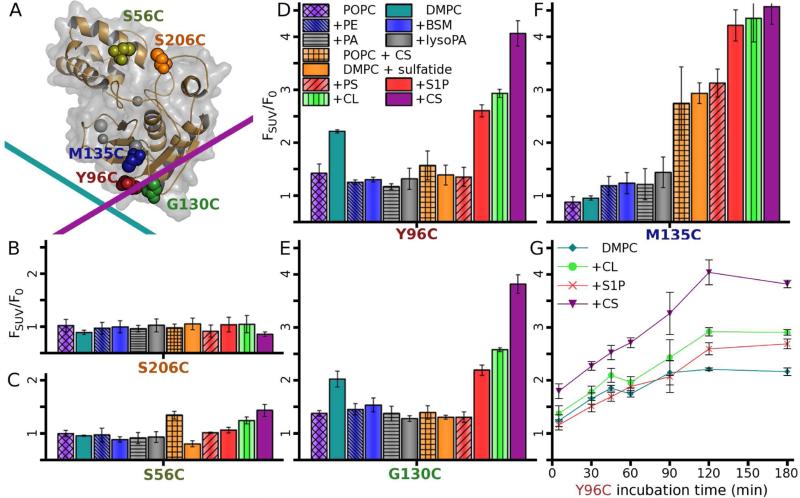
To test the relevance of these modes of binding to vesicles, as well as dependencies on lipid composition, single cysteines were placed in the putative bilayer interface in the I-A loop at Y96C and in the II-III loop at G130C (Figure 4A). M135C was placed near but outside the interface in β-strand III. Negative control sites for labeling were prepared at surfaces distant from the interface in the B-C loop as S206C or at S56C in the pro-domain (Figure 4A). Each Cys was conjugated to the polarity-sensitive fluor IANBD (site-directed fluor labeling; SDFL), preserving the NMR spectra (Figure S5) and protein fold. The fluor at the interfacial Y96C (I-A loop) and G130C (II-III loop) sites were entirely free to dip into both vesicles of DMPC and of POPC (unsaturated 1-palmitoyl-2-oleoyl-sn-phosphatidylcholine), judging from at least 2-fold increases of F/F0 (Figure 4D,E). In contrast, these zwitterionic vesicles failed to interact with the label placed at S206C as negative control, S56C in the pro-domain, or M135C partly recessed on the β-sheet (Figure 4B,C, and F). The behavior of the fluor at M135C agrees well with the NMR structures, which imply that it should be separated from zwitterionic membranes but draw nearer to CS-containing membranes.
Figure 4. Some anionic lipids trigger deeper binding of proMMP-7 to vesicles.
(A) The polarity-sensitive fluor IANBD was conjugated to single Cys substitutions of residues marked with spheres with color shared with labels on the other panels. The interface for zwitterionic DMPC bilayers is represented by the cyan line and the interface for CS-containing anionic bilayers by the magenta line.
(B-G) Interactions of vesicles (SUVs) of various lipid compositions with fluor-tagged proMMP-7 (10 nM) are revealed by elevated emission, FSUV, normalized by the emission F0 without vesicles. The principal lipid, DMPC or POPC, was 300 μM and the supplementary lipid (denoted by a ”+”) added to 10% (w/w), or 15% (w/w) for brain sphingomyelin (BSM). FSUV/F0 was measured after 2 h of incubation with lipids, in quadruplicate with SD given as error bars. Fluorescence responses to the lipids were minimal when monitored by the fluor at negative control site of (B) S206C or small at (C) S56C. FSUV/F0 responses are significant from the fluor placed in the interface at (D) Y96C or (E) G130C, or outside but near the interface with CS present at M135C on the β-sheet (F). See also Figure S5.
(G) Fluorescence increases from the fluor at centrally located Y96C are plotted as a function of time after addition of vesicles containing bioactive anionic lipids.
Incorporation of anionic and bioactive CS, sphingosine 1-phosphate (S1P), or CL into DMPC vesicles dramatically increased the emission from the fluor at M135C about 4-fold, while sulfatides and phosphatidylserine (PS) increased this emission about 3-fold (Figure 4F). These anionic lipids apparently bring the fluor at M135C into intimate contact with the vesicles, in complete accord with the change of orientation observed in the solution structure with CS present. A greater increase of emission from the fluor at M135C results from addition of S1P and PS than from phosphatidate (PA) or lyso-PA. (While the reason is unclear, S1P and PS each have an amino group that could donate an H-bond). The fluor placed at Y96C or G130C responded ~2-fold to the DMPC vesicles and more strongly when doped with S1P, CL, and especially CS, which increased fluorescence > 3-fold (Figure 4D,E). Smaller enhancements by anionic vesicles of IANBD fluorescence at Y96C and G130C than at M135C might suggest that substitutions at the former two key interfacial sites might slightly impair membrane binding. The trends of greater fluorescence changes with CS or other anionic lipids are consistent with the deeper and broader protein-bilayer interface in the NMR structure with CS present. Two hours were required for complete insertion of the interfacial fluor in the presence of anionic lipids (Figure 4G), suggesting a thermal activation barrier to insertion.
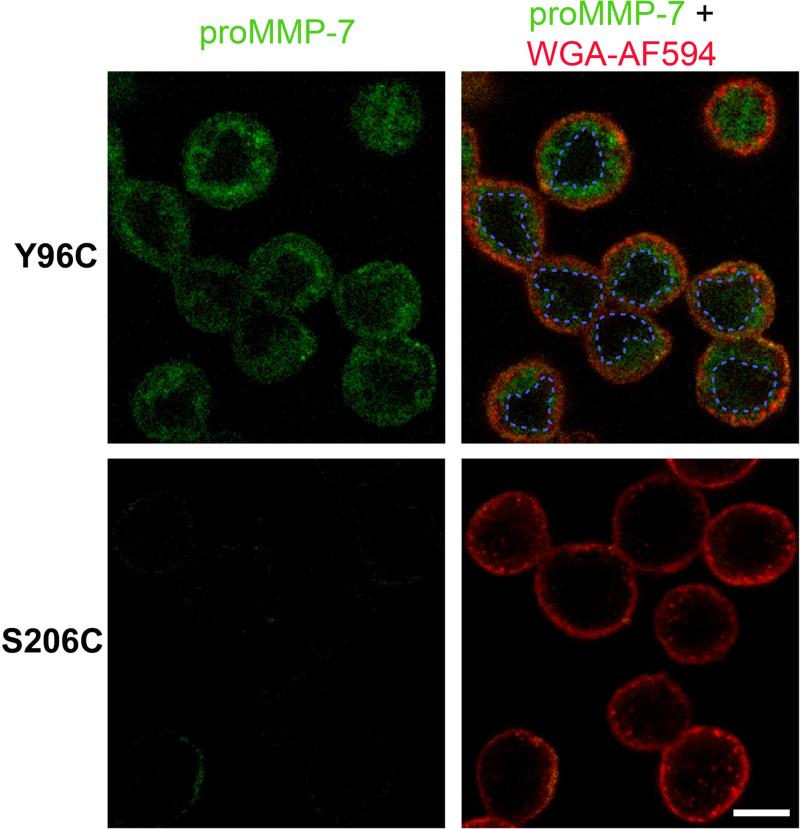
Binding of Plasma Membranes and Internalization
Since MMP-7 binds cells via CS and heparin sulfate partners (Berton et al., 2007; Kioi et al., 2003; Yamamoto et al., 2006; Yu et al., 2002), we tested for direct binding of cell membrane bilayers using the SDFL approach. IANBD-conjugated proMMP-7 was added exogenously to cultured colon cancer cells for confocal imaging of any green fluorescence evidence of IANBD insertion into cell membranes. ProMMP-7 labeled with IANBD at Y96C emitted fluorescence at the plasma membranes, but the zymogen labeled at S206C did not (Figure 5). Within five min. after application, proMMP-7 labeled at Y96C was detected at both plasma membranes and intracellular hydrophobic compartments, presumably vesicles throughout the cytoplasm, while the zymogen labeled at S206C showed virtually no fluorescence (Figure 5). The imaging implies that proMMP-7 readily binds directly to bilayers of plasma membranes and internalized membranes in similar orientations as those characterized with membrane models.
Figure 5. ProMMP-7 binds plasma membranes and internalized vesicles of live colon cancer cells by the interface present in both NMR structures.
Confocal images of human colon adenocarcinoma COLO 205 cells were acquired after incubating 5 min. at 37 °C with proMMP-7 labeled wi th IANBD at Y96C (top row) or the non-interacting S206C site (bottom row). The external surface of the plasma membrane emits red from wheat germ agglutinin conjugated to Alexa Fluor 594 (WGA-AF594). The dashed blue lines delimit the cell nuclei stained by Hoechst 33342 (not shown). Scale bar, 10 μm.
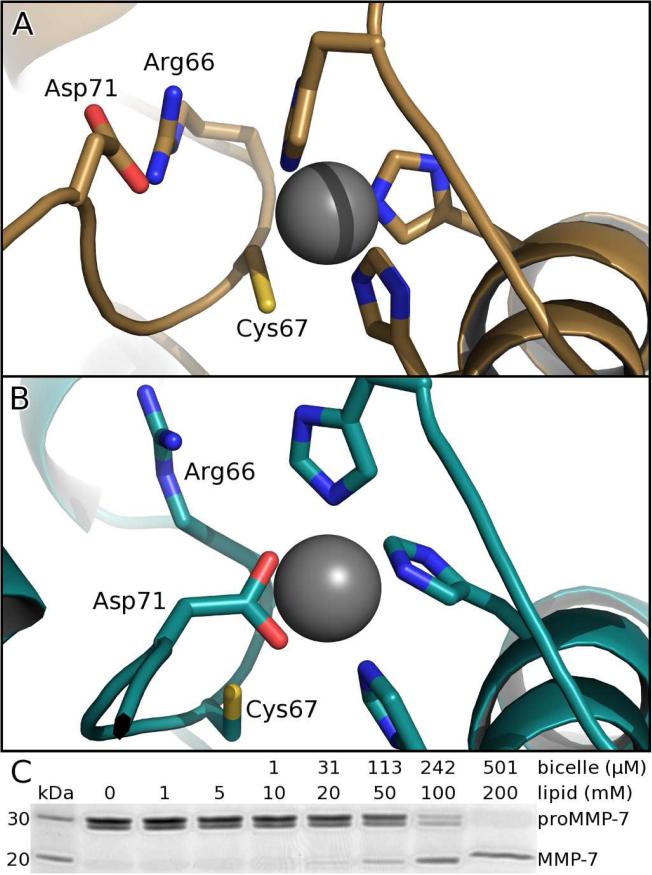
Bicelle-Induced Removal of Auto-inhibitory Conformation
In the conserved auto-inhibitory peptide (known as the “Cys switch”) running through the catalytic cleft between the domains of free proMMP-7, NOEs position Cys67 close enough to coordinate the catalytic zinc, while Arg66 and Asp71 form a salt bridge (Figure 6A) which is stabilized by a Π-cation interaction with Phe22. Addition of bicelles (without or with CS), completely changes the NOE patterns of the auto-inhibitory sequence (Figure S6), revealing the displacement of Cys67 from the zinc and disruption of the Arg66-Asp71 salt bridge (Figure 6B). The changed NOEs and structural refinement suggest ~5 Å of movement of Asp71 to position both carboxyl oxygens to bind the zinc (Figure 6B). The side chain of Arg66 remains stationary as its interaction with Phe22 remains unaffected. In concert with removal of auto-inhibition, bicelles introduce large peaks shifts (Figure S1A) and NOEs to the Glu74 – Asn80 region that imply its change from disorder in the free state to formation of an ordered hairpin (Figure S6). This makes the scissile Glu74-Tyr75 bond accessible for the proteolysis that completes activation. The inactivating E195A lesion used prevented this proteolysis and facilitated the capture of these structural changes. Addition of DMPC/DH6PC bicelles, without or with CS, activates wt proMMP-7 similarly well (Figure 6C), but DMPC vesicles failed to activate the zymogen. Thus, the bicelle-induced conformational change that removes the auto-inhibitory zinc coordination and orders the subsequent scissile sequence may facilitate the activation of the wt zymogen.
Figure 6. Bicelles alter auto-inhibitory conformation and stimulate activation.
(A) Free proMMP-7 features normal proximity of the catalytic zinc to the conserved cysteine of the auto-inhibitory segment of the pro-domain.
(B) When bound to DMPC/DH6PC bicelles, the conserved auto-inhibitory peptide switches to place the conserved aspartate at the zinc, according to the altered NOE patterns highlighted in Figure S6.
(C) DMPC/DH6PC bicelles (q=0.5) of sufficient concentration accelerate activation (2 h at 37°C) of proMMP-7 (7 μM) to mature MMP-7 (migrating faster by SDS-PAGE).
DISCUSSION
Significance of Modes of Binding to Bilayers
This is the third MMP where the II-III loop jutting from the catalytic domain has been found important for localization to cellular membranes. The corresponding but longer MT-loop of MT1-MMP is required for its localization to integrin-containing cell adhesion complexes in plasma membranes where it is enabled to digest collagen fibrils for tumor cell invasion (Woskowicz et al., 2013). The corresponding II-III loop of MMP-12 is the only sequence element that its membrane interfaces (lacking a Trp)(Koppisetti et al., 2014) share with MMP-7. Might other MMPs employ the II-III loop to bind cell membranes as well?
MMP-7 association with cell surfaces is likely to include direct but nonspecific interactions with phospholipids. Nonspecific bilayer interactions, i.e. superficial and mobile MMP-7 binding to zwitterionic bilayers (Figures 2 and 3; Table 2), potentially expedite its search for specific partners such as proteoglycan assemblies, e.g. CD44/HB-EGF/ErbB4 complexes (Yu et al., 2002) and syndecans (Ryu et al., 2009; Wang et al., 2014), as well as for anionic patches of bilayers. Diffusion reduced to a 2D surface, along with repeated nonspecific association with the surface, tends to broaden the effective radius and accessibility of the functional sites (Berg, 1985).
Once recruited to its partners and substrates, MMP-7 can carry out its proteolytic shedding and maturation activities on complexes that influence epithelial health and tumor cell behavior. Binding of mature MMP-7 to zwitterionic vesicles and bicelles leaves the catalytic channel open and facing laterally (leftward in views of Figures 2A and 3A). The CS content of bilayers turns the catalytic cleft to face out and away from the membrane (Figs. 2B and 3B). Fluorescence results (Figure 4F) imply that bioactive S1P and CL probably also point the active site away from the membrane, and that PS and sulfatide SM4 very possibly do as well. Anionic lipids positioning of MMP-7 to face outward probably maximizes and prolongs the accessibility of the active site to pericellular protein substrates.
Punctate patterns of proMMP-7 binding to plasma membranes (Figure 5) suggest preferred binding sites with partners to recruit MMP-7 there. Internalization of the inactivated proMMP-7 into live cells is rapid and associated with membranes (Figure 5). This agrees with and complements the report of internalization of MMP-7 into cultured neurons (to cleave a SNARE protein for neurotransmitter release), which proceeds with a similar distribution across the cytoplasm and sensitivity to inhibitors of clathrin-dependent endocytosis (Szklarczyk et al., 2007).
Acidic Lipids, Broader Interface, and Partial Insertion
Striking CS stabilization of MMP-7 association with vesicles and bicelles has been elucidated (Figures 2 – 4). CS is attracted to the vicinity of the N-terminal end of helix A according to its NOEs to Leu108 (Figure S4) and interference by mutations of IIe103 or Arg107 (Higashi et al., 2008). CS freely migrated to this vicinity into an apparent H-bond with the Tyr96 –OH group in two of three restrained MD trajectories (which omitted the NOEs to CS). Arg125 and Trp129, implicated by mutagenesis in association with CS (Higashi et al., 2008), also reside in the bilayer interface (Figure 2D). Several additional salt bridges and H-bonds to phospholipid head groups (Figure 2D and Table S2) should be borne in mind. C-terminal Arg245 - Lys247 (disordered in solution) were important for interaction with highly acidic lipid mixtures of CS with CL (Higashi et al., 2008). Deletion of the seven C-terminal residues perturb the NMR peaks of a distinct patch on β-strands I and II, suggesting the C-terminus could reside there part-time and be close enough for contact with anionic membranes.
Anionic lipids shift an equilibrium from superficial binding of proMMP-7 on bilayers without net charge towards the partly inserted state predominating where the negative charge of CS, CL, S1P, or PS is present (Figures 2 and 3). SDFL measurements find the negative charge-induced insertion to be slow (Figure 4G), suggesting an activation barrier to insertion. Part of the thermal barrier must be for the displacement of lipids by the insertion of segments containing Val128, Trp129, and Ile103 (Figures 2E and 3). The attraction of anionic lipids for the positive charge of MMP-7 probably pays the energetic costs for partial insertion of the enzyme, the ordering of lipids around it, and displacement of positively charged choline head groups away from the interface (Figure 3). Trp129 might first penetrate among acyl chains of the bilayer ahead of other residues forming non-covalent interactions that broaden the interface (Figure 2).
New means of structural elucidation of bilayer penetration by peripheral membrane proteins have been needed (Moravcevic et al., 2012), but detailed examples have only begun to emerge using NMR, i.e. for the FAPP1 PH domain (Lenoir et al., 2015) and MMP-7. Just as the FAPP1 PH domain inserts a central hydrophobic “wedge” flanked by basic surfaces on the periphery for recognizing phospholipid head groups (Lenoir et al., 2015), so MMP-7 in the presence of CS (or S1P or CL) also inserts its hydrophobic II-III loop surrounded by basic residues that make favorable contacts with the phosphoryl groups, ester linkages, and acidic lipids (Figures. 2D,E and 3B). MMP-7 differs in multiple ways, however. It inserts a second hydrophobic feature of the Ile103 side chain from the end of helix A. In contrast to the norm in phosphoinositide recognition of polar groups fitting into a pocket (Moravcevic et al., 2012), the sulfate of CS instead abuts a convex, positively charged region of MMP-7. The 2400 Å2 interface between MMP-7 and CS-containing DMPC bilayers joins MMP-12 (Koppisetti et al., 2014) among the broadest interfaces for membrane surfaces characterized. Though membrane bilayers cross different sides of the MMP-7 and MMP-12 catalytic domains, both enzymes exploit multiple basic side chains to form salt bridges and donate H-bonds to phosphoryl moieties and ester linkages of the head groups.
Allosteric Coupling to Active Site
Allosteric effects of anionic lipids (especially sulfated CS and sulfatide) on MMP-7 binding and digestion of substrates, and conversely an inhibitor diminishing affinity for the anionic lipids, were proposed (Yamamoto et al., 2010, 2014). NMR results support the hypothesis of allostery elicited by bicelles and CS added to them being transmitted to the active site. NMR peak shifts induced by bicelles or the subsequent addition of CS suggest an arc of residues connecting the catalytic cleft to the shared interface for bilayers (Figure S1). Within this arc, the structural ensembles suggest a site of bicelle-induced switching of connectivity and a site of CS-induced structural switching (Figure S1C,D). In crystal and solution structures, the carboxyl group of Asp99 is H-bonded to the backbone of Trp178, while Arg177 is disordered in solution. However, in the bicelle complexes with proMMP-7, Asp99 instead forms a salt bridge with the guanidinium group of Arg177 (Figure S1C). This appears to contort Trp178 into hydrophobic contact with Leu156, which is in direct contact with the backbone of Arg66 of the autoinhibitory sequence. Addition of CS to the bicelles appears to modify another connection within the potential path of transmission and to adjust the active site. The Thr104 withdraws from its hydrophobic packing with Leu100 and the Thr104 γ –OH then donates an H-bond to the O of Pro101 (Figure S1D). The breadth of the catalytic cleft of the free proMMP-7 NMR structure widens in the complex with CS-containing bilayer by 2.1 ± 0.3 Å at the Cα backbone at β-strand IV. Perhaps the subtle changes in binding and activities (Yamamoto et al., 2010, 2014) could be related to these changes in the proposed arc of allosteric transmission and active site.
Switch of Zinc from Cys to Asp during Activation
Among the conserved residues in the “Cys switch” sequence of MMPs, emboldened in PRCGΦPD, the Arg and Asp form a salt bridge (Figure 6A) in each structure of a latent zymogen available. This salt bridge now appears to prevent this Asp from coordinating the zinc. Just as mutations of this conserved Arg auto-activated MMP-3 (Park et al., 1991), R66A-substituted proMMP-7 also auto-activates during purification. These results are consistent with a need for the salt bridge to maintain latency. The bicelle-associated disruption of this Arg-Asp salt bridge, movement of Asp71 to supplant Cys67 in binding the zinc (Figure 6B), and ordering of the scissile sequence could represent an important intermediate en route to activation. The conformational changes raise the question of the cysteine switch better being regarded as switching from Cys to Asp binding of the zinc, or equivalently as switching of the Asp between the Arg and zinc during the activation process. This change and disruption of autoinhibition is possibly transmitted from the bilayer interface by the allosteric pathway proposed above (Figure S1). Perhaps altered contacts between Leu156 and Arg66 help trigger the disruption of the Arg66-Asp71 salt bridge (Figure 6A,B), as well as transient pentacoordination of the zinc during activation.
The displacement of the conserved Cys to 4.8 ± 0.3 Å from the zinc (Figure 6B) has prior precedent. Within 1 sec of mixing kallikrein with proMMP-9, the S coordination of the zinc was replaced by an O or N ligand (Rosenblum et al., 2007). Simulations suggested this to be the carboxylate group from glutamate in the active site on 10% of occasions. Carboxylate binding of the zinc instead is provided by the conserved Asp in the pro-domain in the NMR structures of proMMP-7 bound to bilayers from bicelles (Figure 6B). If these structures (captured with inactivated proMMP-7) portray an intermediate on the pathway to activation (Figure 6B), then the wt zymogen should be more susceptible to self-proteolysis. Indeed, bicelles at high enough concentration do activate wt proMMP-7 (Figure 6C).
In summary, novel structure refinement of a protein-lipid assembly has captured peripheral bilayer binding by the protease MMP-7 and its partial insertion and reorientation in a bilayer by its partner cholesterol sulfate. A key membrane-binding loop has distinctive counterparts recently implicated in the cell membrane targeting of two other MMPs. Bicelle binding results in allosteric structural changes faraway in the auto-inhibitory and scissile sequences that could represent an intermediate in the process of activation of MMPs. MMP-7 binds plasma membranes and internalized membranes of a colon cancer cell line with the interface identified by NMR. Such binding is likely to support its influential proteolytic activities at epithelial and tumor cells.
EXPERIMENTAL PROCEDURES
Bicelle – Enzyme Distance Measurements by NMR
15N/12C/2H/ILV 13CH3 labeling was used for intermolecular PRE and NOE measurements of complexes with bicelles. NMR samples were typically 400 μM in 20 mM imidazole (pH 6.6), 10 mM CaCl2, 20 μM ZnCl2, 10 mM 2-mercaptoethanol, 0.02% NaN and 10% 2H2O. DMPC/DH6PC bicelles were prepared at 1:2 molar ratio (q=0.5) (Koppisetti et al., 2014). Spectra were acquired at 37°C using a Bruker Avance III 800 MHz spectrometer with TCI cryoprobe. NOEs were obtained from simultaneously 13C- and 15N-edited NOESY-HSQC (Pascal et al., 1994).
Bicelle-enzyme PREs were recorded in the presence of spin-labeled DPPC, absence or presence of ~4.4% (w/w) CS. A CPMG train that suppresses 1H-1H J couplings (Aguilar et al., 2012) was inserted in 15N HSQC, TROSY, and 13C HMQC pulse sequences for measuring relaxation accurately as exponential decays for both paramagnetic samples with spin-labeled DPPC and diamagnetic controls without spin label (Koppisetti et al., 2014). The difference of these decay rates represents the highly distance-dependent PRE, Γ2, from which each depth restraint was derived as described (Koppisetti et al., 2014). Six-point series measured on a protonated complex highlight the inherently exponential decays (Figure S2A) resulting from successful suppression of cosine modulation by 3JHnHα couplings using the PROJECT-CPMG train of the pulse sequence. Relaxation from deuterated samples measured with four points was chosen for estimating Γ2 because deuteration slows diamagnetic relaxation, thereby enhancing dynamic range for estimating Γ2 (Figure S2A).
Distances were estimated from Γ2 values with calibration using τc and expressions available (Battiste and Wagner, 2000; Koppisetti et al., 2014).To obtain τc, 15N NMR transverse cross-correlated relaxation was measured (Liu and Prestegard, 2008) and compared against spectral density expressions available (Lee et al., 2006).
Structural Calculations, Free and Bound to Bilayer Coordinates
Iterative calculations of the NMR structure of the free form of proMMP-7 were carried out using Cyana 2.1 (Güntert and Buchner, 2015). DMPC bilayer coordinates (Poger and Mark, 2010) and the coordinates of free proMMP-7 were docked as rigid bodies using the PRE- and NOE-based distance restraints between protein and phospholipids and HADDOCK 2.1 (Dominguez et al., 2003) with the electrostatic and van der Waals terms turned off and extensions to accommodate bilayers (Koppisetti et al., 2014). From both of these independent sets of rigid body docking calculations, the three structures with smallest violations of protein-bilayer distance restraints were drawn. Martini 2.0 (Marrink and Tieleman, 2013) was used to insert CS into coordinates of the DMPC bilayer. Each of the three low energy structures from each of the rigid body dockings (using restraint sets measured without and with CS) were inserted into the DMPC bilayer coordinates without or with CS by the procedure of (Kandt et al., 2007). The sets of NOE restraints measured on ILV-labeled proMMP-7 bound to bicelles without or with CS were critically evaluated and violations removed by iterative refinement starting from the coordinates of free proMMP-7; this was performed in vacuo using GROMACS 4.6 (Hess et al., 2008). The validated, bicelle-compatibile NOE lists were used for restrained MD refinement of each threesome of rigid body-docked starting structures packed with a bilayer and immersed in TIP3 water. The refinement of the protein-bilayer assemblies with the intraprotein NOE restraints and the protein – phospholipid distance restraints from NOEs and PREs proceeded in GROMACS 4.6 using the OPLS force field modified for lipids (Berger et al., 1997) and periodic boundary conditions. A unique set of initial velocities was assigned to each starting structure based on a Maxwell distribution at 310 K and the structures were equilibrated without distance restraints for 1.5 ns. Distance restraints were then applied with disre_tau set to 12 ps to avoid overly constraining the protein in the xy plane and to allow local fluctuations within the protein, and the system equilibrated until stabilization of potential energy for at least 5 ns. The trajectories were continued to a total restrained simulation time of 20 ns at 310 K. Each threesome of restrained MD trajectories with DMPC bilayers, without or with CS incorporated, was inspected for reproducibility among the independent trajectories. The structures with lowest average restraint violations (and spaced by > 100 ps) were collected from a trajectory to serve as the final ensemble.
Site-Directed Fluor Labeling and Emission Promoted by Vesicles
Sites for linking the membrane-sensitive fluor IANBD (Invitrogen) were introduced to proMMP-7(E195A) as single Cys substitutions via QuikChange™ with PCR master mix (Agilent Technologies). The mutant zymogens were folded and purified as described (Fulcher et al., 2014). The conjugation reactions followed Invitrogen's protocol using 1 μM of each Cys mutant with 10 μM IANBD in the dark overnight at 4°C in a vacuum chamber with excess 2-mercaptoethanol in fluorescence buffer of 20 mM Tris, pH 7.2, 5 mM CaCl2, 0.1 mM ZnCl2. Desalting on Sephadex G-25 (Sigma) was used to eliminate unreacted IANBD. Full completion of each conjugation reaction was monitored by absorbance.
SUVs of DMPC or POPC (Avanti Polar Lipids) were prepared as described (Sommer et al., 2012) and used at 400 μM monomer concentrations in fluoresence assays with the IANBD conjugates of proMMP-7 (Koppisetti et al., 2014). Additions of the lipids CL, CS, PA, lyso PA, PE, PS, and S1P were at 10% (w/w). Zwitterionic BSM was added to 15% (w/w). A BioTek Synergy MX plate reader was used to excite the fluorochrome at 478 nm and to detect emission at 541 nm from each IANBD conjugate of proMMP-7 (10 nM) in the fluorescence buffer before and after mixing with liposomes. Emission was measured after incubations of 10, 30, 45, 60, 90, 120, and 180 min. with the liposomes (FSUV) and is normalized by the emission without liposomes (F0). Errors, propagated through this ration, report σ from quadruplicate samples.
Lipid Activation of ProMMP-7
Wt proMMP-7 (5 M) was incubated with DMPC/DH6PC bicelles (q=0.5), or 0.01 mg/ml CS with the DMPC/DH6PC bicelles. The reaction mixtures were incubated for 2 h at 37°C, stopped using lithium dodecyl sulfate sample bu ffer (Novex), separated by Bis-Tris 4–12% SDS-PAGE (Novex), and stained with Coomassie Blue. The protein bands were scanned and quantified using Quantity One software (Biorad).
Live cell imaging
COLO 205 cells were imaged using a Leica TCP SP8 laser scanning confocal microscope equipped with 405 nm diode laser and a tunable supercontinuum white light laser. The excitation/emission band-pass wavelengths were set as follows: 405/415-470 nm (Hoechst 33342), 472/485-525 nm (IANBD - proMMP-7 conjugates) and 594/610-680 nm (Alexa Fluor 594 – WGA). The microscope was equipped with an environmental chamber and all imaging experiments were conducted at 37°C. After acquiring the first multichannel image, 1 μM of each IANBD-labeled proMMP-7(E195A + Cys) variant was added to the cell culture dish and images captured at 5 min intervals.
Supplementary Material
Highlights.
ProMMP-7 forms a transient complex with bilayer mimics lacking net charge.
Cholesterol sulfate causes it to reorient, partly insert, and slow in diffusion.
Binding of bilayered micelles changes the auto-inhibitory region by allostery.
MMP-7 binds directly to bilayers of cells by the interface measured.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01 GM57289. The 800 MHz spectrometer was purchased with funds from the University of Missouri and NIH grant S10 RR022341. We thank Y. Jeong for confirmatory backbone peak assignments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
S.H.P., S.R.V., and Y.G.F. designed research; S.H.P., Y.G.F., R.K.K., and A.J. performed research; S.H.P. and S.R.V. wrote the paper.
ACCESSION NUMBERS
Atomic coordinates, restraints, and topology files were deposited in the Protein Data Bank accession codes 2mze for proMMP-7, 2mzh bound to DMPC bilayer, and 2mzi bound to DMPC bilayer containing CS. NMR chemical shift tables were deposited in BioMagResBank under accession codes 25485 for free proMMP-7, 25488 bound to bicelles, and 25489 bound to bicelles containing CS.
SUPPLEMENTAL INFORMATION
Supplemental Information comprising six figures, two tables, Supplemental Experimental Methods, two movies, and three 3D molecular models can be found online with this article.
REFERENCES
- Aguilar JA, Nilsson M, Bodenhausen G, Morris GA. Spin echo NMR spectra without J modulation. Chem. Commun. 2012;48:811–813. doi: 10.1039/c1cc16699a. [DOI] [PubMed] [Google Scholar]
- Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- Bellac Caroline L., Dufour A, Krisinger Michael J., Loonchanta A, Starr Amanda E., auf dem Keller U, Lange Philipp F., Goebeler V, Kappelhoff R, Butler Georgina S., et al. Macrophage Matrix Metalloproteinase-12 Dampens Inflammation and Neutrophil Influx in Arthritis. Cell Reports. 2014;9:618–632. doi: 10.1016/j.celrep.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Berg OG. Orientation constraints in diffusion-limited macromolecular association. The role of surface diffusion as a rate-enhancing mechanism. Biophys. J. 1985;47:1–14. doi: 10.1016/S0006-3495(85)83870-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger O, Edholm O, Jähnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berton A, Selvais C, Lemoine P, Henriet P, Courtoy PJ, Marbaix E, Emonard H. Binding of matrilysin-1 to human epithelial cells promotes its activity. Cell. Mol. Life Sci. 2007;64:610–620. doi: 10.1007/s00018-007-6415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi S, Hass R. Extracellular signals in young and aging breast epithelial cells and possible connections to age-associated breast cancer development. Mech. Ageing Dev. 2011;132:213–219. doi: 10.1016/j.mad.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Chen P, Abacherli LE, Nadler ST, Wang Y, Li Q, Parks WC. MMP7 Shedding of Syndecan-1 Facilitates Re-Epithelialization by Affecting α2β1 Integrin Activation. PLoS ONE. 2009;4:e6565. doi: 10.1371/journal.pone.0006565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooks RG, Dill AL, Eberlin LS, Ifa DR. Methods for diagnosing or monitoring for recurrence of prostate cancer. (Google Patents) 2011 [Google Scholar]
- Dean RA, Cox JH, Bellac CL, Doucet A, Starr AE, Overall CM. Macrophage-specific metalloelastase (MMP-12) truncates and inactivates ELR+ CXC chemokines and generates CCL2, -7, -8, and -13 antagonists: potential role of the macrophage in terminating polymorphonuclear leukocyte influx. Blood. 2008;112:3455–3464. doi: 10.1182/blood-2007-12-129080. [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Eberlin LS, Dill AL, Costa AB, Ifa DR, Cheng L, Masterson T, Koch M, Ratliff TL, Cooks RG. Cholesterol Sulfate Imaging in Human Prostate Cancer Tissue by Desorption Electrospray Ionization Mass Spectrometry. Anal. Chem. 2010;82:3430–3434. doi: 10.1021/ac9029482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias PM, Williams ML, Choi E-H, Feingold KR. Role of cholesterol sulfate in epidermal structure and function: Lessons from X-linked ichthyosis. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2014;1841:353–361. doi: 10.1016/j.bbalip.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A, Wang Sam C., Morris John P.I.V., Folias Alexandra E., Liou A, Kim Grace E., Akira S, Boucher Kenneth M., Firpo Matthew A., Mulvihill Sean J., et al. Stat3 and MMP7 Contribute to Pancreatic Ductal Adenocarcinoma Initiation and Progression. Cancer Cell. 2011;19:441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher YG, Sanganna Gari RR, Frey NC, Zhang F, Linhardt RJ, King GM, Van Doren SR. Heparinoids activate a protease, secreted by mucosa and tumors, via tethering supplemented by allostery. ACS Chem Biol. 2014;9:957–966. doi: 10.1021/cb400898t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly B, Banerjee J, Elegbede AI, Klocke DJ, Mallik S, Srivastava DK. Intrinsic selectivity in binding of matrix metalloproteinase-7 to differently charged lipid membranes. FEBS Lett. 2007;581:5723–5726. doi: 10.1016/j.febslet.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güntert P, Buchner L. Combined automated NOE assignment and structure calculation with CYANA. J. Biomol. NMR. 2015:1–19. doi: 10.1007/s10858-015-9924-9. [DOI] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. Journal of Chemical Theory and Computation. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Higashi S, Oeda M, Yamamoto K, Miyazaki K. Identification of amino acid residues of matrix metalloproteinase-7 essential for binding to cholesterol sulfate. J. Biol. Chem. 2008;283:35735–35744. doi: 10.1074/jbc.M806285200. [DOI] [PubMed] [Google Scholar]
- Hilty C, Wider G, Fernandez C, Wuthrich K. Membrane protein-lipid interactions in mixed micelles studied by NMR spectroscopy with the use of paramagnetic reagents. ChemBioChem. 2004;5:467–473. doi: 10.1002/cbic.200300815. [DOI] [PubMed] [Google Scholar]
- Hulboy DL, Gautam S, Fingleton B, Matrisian LM. The influence of matrix metalloproteinase-7 on early mammary tumorigenesis in the multiple intestinal neoplasia mouse. Oncol. Rep. 2004;12:13–17. [PubMed] [Google Scholar]
- Ii M, Yamamoto H, Adachi Y, Maruyama Y, Shinomura Y. Role of Matrix Metalloproteinase-7 (Matrilysin) in Human Cancer Invasion, Apoptosis, Growth, and Angiogenesis. Exp. Biol. Med. 2006;231:20–27. doi: 10.1177/153537020623100103. [DOI] [PubMed] [Google Scholar]
- Kandt C, Ash WL, Peter Tieleman D. Setting up and running molecular dynamics simulations of membrane proteins. Methods. 2007;41:475–488. doi: 10.1016/j.ymeth.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Kiguchi K, Iwamori M, Yamanouchi S, Ishiwata I, Saga M, Amemiya A. Coexpression of cholesterol sulfate and cytokeratin as tumor markers in well-differentiated squamous cell carcinoma of the human uterine cervix. Clin. Cancer. Res. 1998;4:2985–2990. [PubMed] [Google Scholar]
- Kioi M, Yamamoto K, Higashi S, Koshikawa N, Fujita K, Miyazaki K. Matrilysin (MMP-7) induces homotypic adhesion of human colon cancer cells and enhances their metastatic potential in nude mouse model. Oncogene. 2003;22:8662–8670. doi: 10.1038/sj.onc.1207181. [DOI] [PubMed] [Google Scholar]
- Kivisaari AK, Kallajoki M, Ala-aho R, McGrath JA, Bauer JW, Königová R, Medvecz M, Beckert W, Grénman R, Kähäri VM. Matrix metalloproteinase-7 activates heparin-binding epidermal growth factor-like growth factor in cutaneous squamous cell carcinoma. Br. J. Dermatol. 2010;163:726–735. doi: 10.1111/j.1365-2133.2010.09924.x. [DOI] [PubMed] [Google Scholar]
- Koppisetti RK, Fulcher YG, Jurkevich A, Prior SH, Xu J, Lenoir M, Overduin M, Van Doren SR. Ambidextrous binding of cell and membrane bilayers by soluble matrix metalloproteinase-12. Nature communications. 2014;5:5552. doi: 10.1038/ncomms6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A, Fujiwara TK, Chadda R, Xie M, Tsunoyama TA, Kalay Z, Kasai RS, Suzuki KGN. Dynamic Organizing Principles of the Plasma Membrane that Regulate Signal Transduction: Commemorating the Fortieth Anniversary of Singer and Nicolson's Fluid-Mosaic Model. Annu. Rev. Cell. Dev. Biol. 2012;28:215–250. doi: 10.1146/annurev-cellbio-100809-151736. [DOI] [PubMed] [Google Scholar]
- Kutateladze TG, Capelluto DG, Ferguson CG, Cheever ML, Kutateladze AG, Prestwich GD, Overduin M. Multivalent mechanism of membrane insertion by the FYVE domain. J. Biol. Chem. 2004;279:3050–3057. doi: 10.1074/jbc.M309007200. [DOI] [PubMed] [Google Scholar]
- Lee D, Hilty C, Wider G, Wuthrich K. Effective rotational correlation times of proteins from NMR relaxation interference. J. Magn. Reson. 2006;178:72–76. doi: 10.1016/j.jmr.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Lenoir M, Grzybek M, Majkowski M, Rajesh S, Kaur J, Whittaker SBM, Coskun Ü, Overduin M. Structural Basis of Dynamic Membrane Recognition by trans-Golgi Network Specific FAPP Proteins. J. Mol. Biol. 2015;427:966–981. doi: 10.1016/j.jmb.2014.12.023. [DOI] [PubMed] [Google Scholar]
- Liu Y, Prestegard JH. Direct measurement of dipole-dipole/CSA cross-correlated relaxation by a constant-time experiment. J. Magn. Reson. 2008;193:23–31. doi: 10.1016/j.jmr.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Wang Q, Hu G, Van Poznak C, Fleisher M, Reiss M, Massagué J, Kang Y. ADAMTS1 and MMP1 proteolytically engage EGF-like ligands in an osteolytic signaling cascade for bone metastasis. Genes Dev. 2009;23:1882–1894. doi: 10.1101/gad.1824809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch CC, Vargo-Gogola T, Martin MD, Fingleton B, Crawford HC, Matrisian LM. Matrix metalloproteinase 7 mediates mammary epithelial cell tumorigenesis through the ErbB4 receptor. Cancer Res. 2007;67:6760–6767. doi: 10.1158/0008-5472.CAN-07-0026. [DOI] [PubMed] [Google Scholar]
- Marrink SJ, Tieleman DP. Perspective on the Martini model. Chem Soc Rev. 2013;42:6801–6822. doi: 10.1039/c3cs60093a. [DOI] [PubMed] [Google Scholar]
- Mitsiades N, Yu W.-h., Poulaki V, Tsokos M, Stamenkovic I. Matrix Metalloproteinase-7-mediated Cleavage of Fas Ligand Protects Tumor Cells from Chemotherapeutic Drug Cytotoxicity. Cancer Res. 2001;61:577–581. [PubMed] [Google Scholar]
- Moravcevic K, Oxley Camilla L., Lemmon Mark A. Conditional Peripheral Membrane Proteins: Facing up to Limited Specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer. 2008;8:932–941. doi: 10.1038/nrc2459. [DOI] [PubMed] [Google Scholar]
- Noe V, Fingleton B, Jacobs K, Crawford HC, Vermeulen S, Steelant W, Bruyneel E, Matrisian LM, Mareel M. Release of an invasion promoter E-cadherin fragment by matrilysin and stromelysin-1. J. Cell Sci. 2001;114:111–118. doi: 10.1242/jcs.114.1.111. [DOI] [PubMed] [Google Scholar]
- Park AJ, Matrisian LM, Kells AF, Pearson R, Yuan ZY, Navre M. Mutational analysis of the transin (rat stromelysin) autoinhibitor region demonstrates a role for residues surrounding the “cysteine switch”. J. Biol. Chem. 1991;266:1584–1590. [PubMed] [Google Scholar]
- Pascal SM, Muhandiram D, Yamazaki T, Formankay J, Kay LE. Simultaneous acquisition of 15N-and 13C-edited NOE spectra of proteins dissolved in H2O. Journal of Magnetic Resonance, Series B. 1994;103:197–201. [Google Scholar]
- Poger D, Mark AE. On the validation of molecular dynamics simulations of saturated and cis-monounsaturated phosphatidylcholine lipid bilayers: a comparison with experiment. J. Chem. Theory Comput. 2010;6:11. doi: 10.1021/ct900487a. [DOI] [PubMed] [Google Scholar]
- Rosenblum G, Meroueh S, Toth M, Fisher JF, Fridman R, Mobashery S, Sagi I. Molecular structures and dynamics of the stepwise activation mechanism of a matrix metalloproteinase zymogen: challenging the cysteine switch dogma. J. Am. Chem. Soc. 2007;129:13566–13574. doi: 10.1021/ja073941l. [DOI] [PubMed] [Google Scholar]
- Ryu H-Y, Lee J, Yang S, Park H, Choi S, Jung K-C, Lee S-T, Seong J-K, Han I-O, Oh E-S. Syndecan-2 Functions as a Docking Receptor for Pro-matrix Metalloproteinase-7 in Human Colon Cancer Cells. J. Biol. Chem. 2009;284:35692–35701. doi: 10.1074/jbc.M109.054254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawey ET, Johnson JA, Crawford HC. Matrix metalloproteinase 7 controls pancreatic acinar cell transdifferentiation by activating the Notch signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19327–19332. doi: 10.1073/pnas.0705953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer LAM, Meier MA, Dames SA. A fast and simple method for probing the interaction of peptides and proteins with lipids and membrane-mimetics using GB1 fusion proteins and NMR spectroscopy. Protein Sci. 2012;21:1566–1570. doi: 10.1002/pro.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk A, Oyler G, McKay R, Gerfen C, Conant K. Cleavage of neuronal synaptosomal-associated protein of 25 kDa by exogenous matrix metalloproteinase-7. J. Neurochem. 2007;102:1256–1263. doi: 10.1111/j.1471-4159.2007.04625.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Zuo D, Chen Y, Li W, Liu R, He Y, Ren L, Zhou L, Deng T, Wang X, et al. Shed Syndecan-1 is involved in chemotherapy resistance via the EGFR pathway in colorectal cancer. Br. J. Cancer. 2014;111:1965–1976. doi: 10.1038/bjc.2014.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CL, Heppner KJ, Labosky PA, Hogan BL, Matrisian LM. Intestinal tumorigenesis is suppressed in mice lacking the metalloproteinase matrilysin. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1402–1407. doi: 10.1073/pnas.94.4.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- Woskowicz AM, Weaver SA, Shitomi Y, Ito N, Itoh Y. MT-LOOP-dependent Localization of Membrane Type I Matrix Metalloproteinase (MT1-MMP) to the Cell Adhesion Complexes Promotes Cancer Cell Invasion. J. Biol. Chem. 2013;288:35126–35137. doi: 10.1074/jbc.M113.496067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Higashi S, Kioi M, Tsunezumi J, Honke K, Miyazaki K. Binding of active matrilysin to cell surface cholesterol sulfate is essential for its membrane-associated proteolytic action and induction of homotypic cell adhesion. J. Biol. Chem. 2006;281:9170–9180. doi: 10.1074/jbc.M510377200. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Miyazaki K, Higashi S. Cholesterol sulfate alters substrate preference of matrix metalloproteinase-7 and promotes degradations of pericellular laminin-332 and fibronectin. J. Biol. Chem. 2010;285:28862–28873. doi: 10.1074/jbc.M110.136994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Miyazaki K, Higashi S. Pericellular proteolysis by matrix metalloproteinase-7 is differentially modulated by cholesterol sulfate, sulfatide, and cardiolipin. FEBS J. 2014;281:3346–3356. doi: 10.1111/febs.12865. [DOI] [PubMed] [Google Scholar]
- Yu WH, Woessner JF., Jr. Heparan sulfate proteoglycans as extracellular docking molecules for matrilysin (matrix metalloproteinase 7). J. Biol. Chem. 2000;275:4183–4191. doi: 10.1074/jbc.275.6.4183. [DOI] [PubMed] [Google Scholar]
- Yu WH, Woessner JF, Jr., McNeish JD, Stamenkovic I. CD44 anchors the assembly of matrilysin/MMP-7 with heparin-binding epidermal growth factor precursor and ErbB4 and regulates female reproductive organ remodeling. Genes Dev. 2002;16:307–323. doi: 10.1101/gad.925702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.