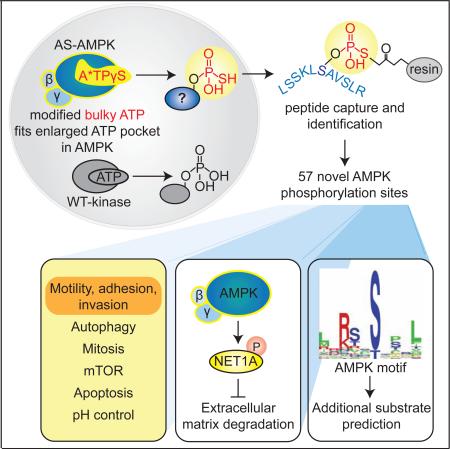
Summary
AMP-activated protein kinase (AMPK) is a central energy gauge that regulates metabolism and has been increasingly involved in non-metabolic processes and diseases. However, AMPK's direct substrates in non-metabolic contexts are largely unknown. To better understand the AMPK network, we use a chemical genetics screen coupled to a peptide capture approach in whole cells, resulting in identification of direct AMPK phosphorylation sites. Interestingly, the high-confidence AMPK substrates contain many proteins involved in cell motility, adhesion, and invasion. AMPK phosphorylation of the RHOA guanine nucleotide exchange factor NET1A inhibits extracellular matrix degradation, an early step in cell invasion. The identification of direct AMPK phosphorylation sites also facilitates large-scale prediction of AMPK substrates. We provide an AMPK motif matrix and a pipeline to predict additional AMPK substrates from quantitative phosphoproteomics datasets. As AMPK is emerging as a critical node in aging and pathological processes, our study identifies potential targets for therapeutic strategies.
Graphical Abstract
Introduction
The ability to adjust to nutrient stress is critical for cellular and organismal functions. Central to this process is AMP-activated protein kinase (AMPK), an energy-sensing protein kinase that regulates metabolic processes (Hardie and Carling, 1997). At the cellular level, AMPK responds to low energy levels by promoting glucose uptake (Kurth-Kraczek et al., 1999), increasing catabolism (Egan et al., 2011; Wang et al., 2001), and inhibiting anabolic processes (Gwinn et al., 2008; Sim and Hardie, 1988) through a repertoire of substrates. Systemically, AMPK integrates hormonal signals to trigger food intake and prevent energy expenditure (Andersson et al., 2004; Hardie and Ashford, 2014). The critical role of AMPK in metabolic regulation has made it an attractive pharmacological target for treatment of metabolic diseases like diabetes (Winder and Hardie, 1999). Indeed, several AMPK activating compounds have recently been identified (Hawley et al., 2012; Zadra et al., 2014; Zhou et al., 2001). As AMPK has been implicated in extending lifespan in several organisms, including potentially humans (Apfeld et al., 2004; Burkewitz et al., 2014; Greer et al., 2007; Mair et al., 2011), it is also an attractive target to delay aspects of aging.
AMPK activity has also been linked with processes that are not directly viewed as metabolic, including mitosis (Banko et al., 2011; Bettencourt-Dias et al., 2004; Vazquez-Martin et al., 2009), development (Lee et al., 2007), and cell polarity (Mirouse et al., 2007; Zhang et al., 2006; Zheng and Cantley, 2007). While many of the substrates that mediate AMPK's effect on metabolism have been well-studied (Chen et al., 2008; Gwinn et al., 2008; Sim and Hardie, 1988), those connecting AMPK to non-metabolic roles are largely unknown. Additionally, aberrant AMPK activity has been associated with diseases like cancer (Kato et al., 2002; Liang and Mills, 2013; Xiang et al., 2004), and the functionally relevant substrates in disease often remain obscure. The emergence of unexpected roles of AMPK and the increasing effort to pharmacologically target this kinase make it critical to fully understand the AMPK substrate network in the context of specific diseases and cellular states.
Central to a kinase-substrate interaction is the exact phosphorylated residue. Previous efforts to characterize the AMPK network in whole cells have focused on protein substrate, but not phosphorylation site, identification (Banko et al., 2011). Additionally, while in vitro phosphorylation motif libraries have helped predict AMPK substrates (Egan et al., 2011; Gwinn et al., 2008), large-scale identification of AMPK phosphorylation sites has never been done. Interestingly, a new approach was developed to identify direct phosphorylation sites of a protein kinase in vitro (Blethrow et al., 2008; Hengeveld et al., 2012). However, this method had not been used in whole cells.
To understand the AMPK substrate network at the resolution of the phosphorylated site, we combined a chemical genetic screen (Banko et al., 2011) and peptide capture approach (Blethrow et al., 2008), allowing us to identify direct AMPKα1 and α2 phosphorylation sites. We provide a comprehensive resource of over 50 AMPK substrates and phosphorylation sites in a human cancer cell line. This screen revealed that AMPK controls different aspects of cellular motility and invasion. We also provide an AMPK motif matrix and pipeline to further predict additional components of the AMPK network, which should have critical implications in pharmacological targeting of AMPK in disease or aging.
Results
A comprehensive physiological screen for AMPKα1 and α2 substrates in a human cancer cell line
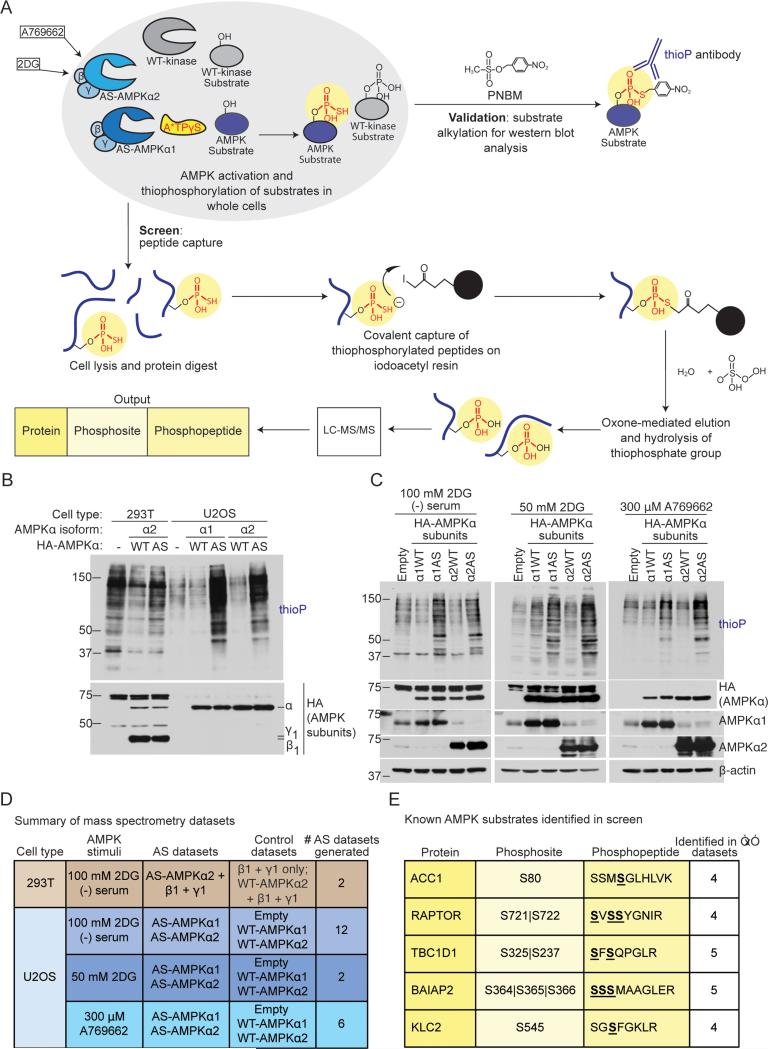
To characterize the AMPK network, we conducted a screen to identify direct AMPK substrates and phosphorylation sites. We used an analog-specific (AS) method of kinase-substrate identification (Banko et al., 2011; Shah et al., 1997), which utilizes a mutant of AMPK that accepts N6-(phenethyl) ATPγS, a bulky form of ATP introduced into cells with gentle digitonin permeabilization, to tag direct substrates with a thiophosphate moiety (Figure 1A). We coupled this approach to a peptide capture system, allowing identification of the exact phosphorylation site by tandem mass spectrometry (Blethrow et al., 2008; Hertz et al., 2010) (Figure 1A).
Figure 1. Screening strategy to identify AMPKα1 andα2 substrates and phosphorylation sites in cells.
(A) Schematic of the peptide-capture technique used to identify analog-specific (AS) AMPKα1 and α2 substrates and phosphorylation sites in whole cells. AS-AMPK uses A*TPγS, a bulky ATP analog, to thiophosphorylate substrates. Upper panel: thiophosphorylated substrates are alkylated by p-nitrobenzyl mesylate (PNBM) and recognized by an antibody to the thiophosphate moiety (thioP). Lower panel: thiophosphorylated peptides are captured on a resin, eluted, and identified using liquid chromatography-tandem mass spectrometry (LC-MS/MS). “2DG”, 2-deoxy-D-glucose.
(B) HA-tagged AS-AMPKα1 and α2 thiophosphorylate endogenous substrates in U2OS cells without over-expression of the β and γ subunits. Cells were serum-starved for 2 hours and stimulated for 5 minutes with 100 mM 2DG, then incubated with A*TPγS. Whole cell lysates were analyzed for the presence of thiophosphorylation (thioP) and exogenous AMPK subunits (HA tag).
(C) HA-tagged AS-AMPKα1 and α2 thiophosphorylate endogenous substrates under different AMPK-activating conditions. Whole cell lysates were analyzed for the presence of thiophosphorylation (thioP) and AMPKα (HA tag, AMPKα1, AMPKα2). First panel: 2 hours of serum-starvation with 5 minutes of 100 mM 2DG; second panel: 15 minutes of 50 mM 2DG; third panel: 30 minutes of 300 μM A769662. Representative of 6, 1, and 3 independent experiments for 2DG (−) serum, 2DG, and A769662, respectively. “Empty”, Empty vector; “α1WT”, WT-AMPKα1; “α1AS”, AS-AMPKα1, “α2WT”, WT-AMPKα2, “α2AS”, AS-AMPKα2.
(D) Summary of mass spectrometry datasets. AMPK-activating conditions as in Figure 1C. See Figure S1G and Supplementary List 1 for more information. “Empty”, Empty vector.
(E) Known AMPK substrates identified in multiple AS-AMPK datasets. Underlined and bold residues, phosphorylated sites on the identified phosphopeptide (more than one is shown if the phosphopeptide had multiple or ambiguous phosphorylation site identification). “Phosphosite” column, identified phosphorylation site corresponding to the known AMPK site. “|”, ambiguous site identification. S722, known AMPK site on RAPTOR; S237, known site on TBC1D1; S366, known site on BAIAP2.
AMPK is a heterotrimeric protein kinase composed of a catalytic α subunit (α1 or α2) as well as regulatory β and γ subunits. We had previously screened for AMPKα2 substrates by over-expressing an analog-specific (AS) version of AMPKα2 together with β and γ subunits in 293T cells (Banko et al., 2011). To extend our screen to AMPKα1 and better approximate physiological kinase activity, we generated stable U2OS cell lines that inducibly express AS or wild type (WT) AMPKα1 or α2 upon doxycycline addition (Figures S1A and S1B). While AMPKα1 or α2 subunits were overexpressed in these U2OS cell lines, this did not lead to substantial dysregulation of endogenous AMPK substrates (Figure S1D), probably because the endogenous β and γ subunits keep the activity of the exogenous α subunit in check. Thus, this system provides screening conditions that better approximate physiological AMPK activity.
Both AS-AMPKα1 and α2 were able to use N6-(phenethyl) ATPγS to thiophosphorylate endogenous substrates in U2OS cells upon activation by serum starvation and 2-deoxy-D-glucose (2DG) (Figure 1B). They did so with less background thiophosphorylation than in 293T cells over-expressing all AMPK subunits (Figure 1B) (Banko et al., 2011). AS-AMPKα1 and α2 also thiophosphorylated endogenous substrates when activated by 2DG alone or by the specific AMPK activator A769662 (Cool et al., 2006) (Figure 1C). They also thiophosphorylated the known AMPK substrate PPP1R12C in U2OS cells with less background than in 293T cells (Figure S1C) (Banko et al., 2011).
To use the analog-specific approach to identify not only AMPK substrates, but also the exact AMPK phosphorylation sites, we coupled it to a peptide capture method developed for concurrent AS-substrate and phosphosite identification (Blethrow et al., 2008; Hertz et al., 2010) (Figure 1A). Since this peptide capture method had not yet been used in whole cells, we first determined its efficacy in 239T cells over-expressing AS-AMPKα2, β1, and γ1 subunits or their respective controls (Figure 1D, top row). We successfully identified phosphopeptides unique to AS-AMPKα2 (Figure S1G). We then performed ten biological experiments in U2OS cells expressing AS-AMPKα1 and ASAMPKα2 or their respective controls (Figure 1D, bottom rows; Figure S1G), using several methods of AMPK activation (Figure 1D). Importantly, the known phosphorylation sites on five established AMPK substrates (ACC1 (S80) (Davies et al., 1990), RAPTOR (S722) (Gwinn et al., 2008), TBC1D1 (S237) (Chen et al., 2008), BAIAP2 (S366) (Banko et al., 2011), and KLC2 (S545) (Amato et al., 2011; Johnson et al., 2011)) were identified in multiple independent datasets (Figure 1E). Thus, this whole-cell peptide capture approach can be used to identify AMPK phosphorylation sites. However, the presence of background phosphopeptides (Figures S1E and S1G) and lack of saturation across replicates (Figures S1F and S1G) indicate that this method is not yet sensitive enough to compare AMPK activity between different conditions.
Identification of over 50 potential AMPK substrates and their phosphorylation sites, including 21 previously unknown high confidence substrates
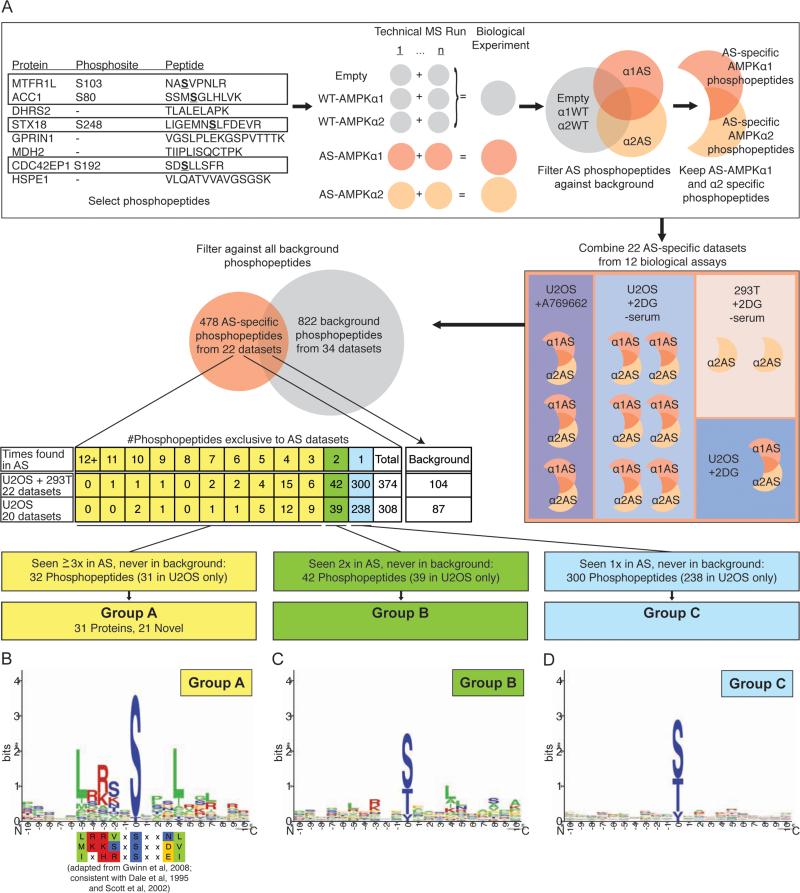
To identify high confidence AMPK substrates and phosphorylation sites, we developed a stringent pipeline to analyze the tandem mass spectrometry data (Figure 2A). Phosphopeptides that were found in experimental datasets, but never in control datasets (see Supplemental Experimental Procedures), were further considered. All AS-AMPK specific phosphopeptides are presented in Supplemental List 2. The motifs surrounding phosphorylation sites on phosphopeptides seen in 3 or more of the 22 experimental datasets (Group A) strongly adhered to the known AMPK motif (Dale et al., 1995; Gwinn et al., 2008; Scott et al., 2002) (Figures 2B and S2B), while those seen less frequently adhered less well to the AMPK motif (Figures 2C, 2D, and S2C). In addition, many known AMPK substrates were identified in Group A (Figure 4A). Thus, there is high confidence that the 21 previously unknown substrates identified in Group A are AMPK targets. We will first focus on the high confidence Group A substrates, but will return to all identified phosphopeptides in Figure 6.
Figure 2. Identification of high confidence AS-AMPK substrates with a tailored pipeline.
(A) Schematic of the pipeline used to identify AS-AMPK substrates in LC-MS/MS datasets. See Supplemental Experimental Procedures. Phosphopeptides only found in ASAMPK datasets were classified as Group A, B, or C based on the number of biological samples in which they were identified. “α1WT”, “α2WT”, “α1AS”, “α2AS”, WT or ASAMPKα1 or α2.
(B-D) Logo motif of the most common phosphorylation sites on each phosphopeptide from Group A (B), Group B (C) and Group C (D). The established in vitro AMPK phosphorylation motif displayed below Group A is modified with permission from (Gwinn et al., 2008), and was generated in that study using a positional scanning peptide library. Green, hydrophobic residues; red, basic; yellow, acidic; blue, neutral polar. See Figure S2A and Supplemental Experimental Procedures for selection of the most common phosphorylation sites.
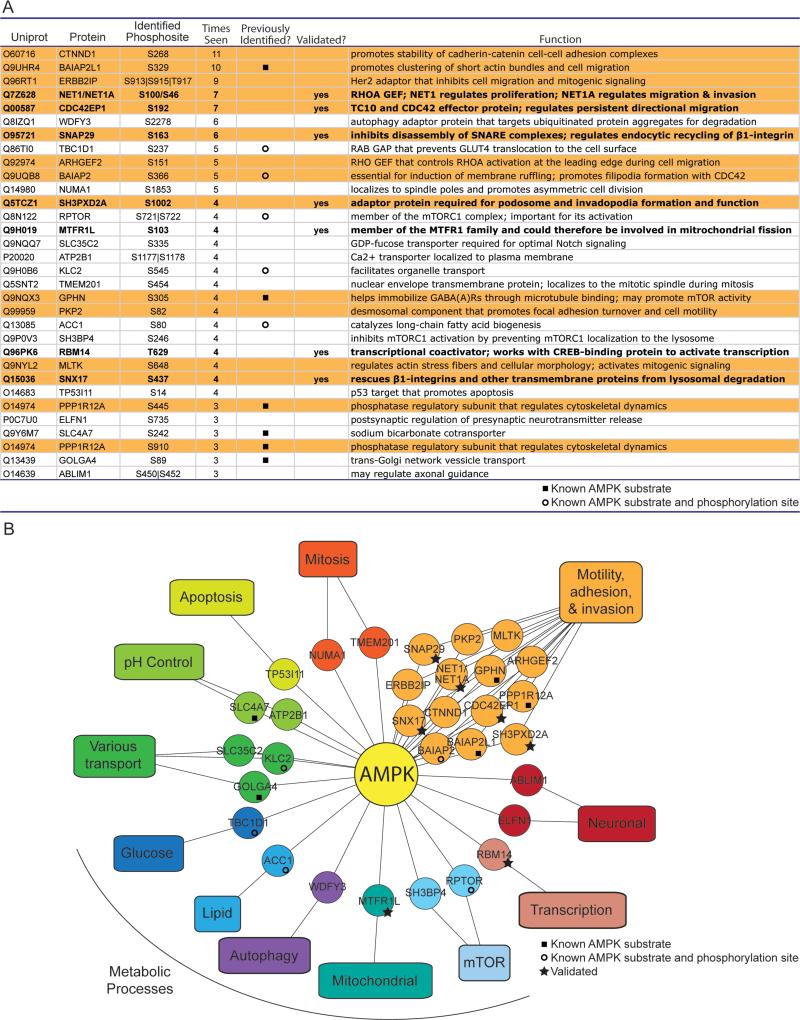
Figure 4. Many high confidence AMPK substrates have known roles in cell motility, adhesion, and invasion.
(A) All Group A substrates and the most frequently identified phosphorylation site(s) on the phosphopeptide. “|”, ambiguity in the mass spectrometry placement of the phosphorylation site; “Times seen”, number of biological samples the phosphopeptide was identified in; “Previously identified” indicates whether the phosphorylation site (open circle) or the protein (but not phosphorylation site) (closed square) was a previously known AMPK substrate; “Validated?” indicates whether the protein was validated as a substrate of AS-AMPK (Figures 3 and S3). Orange background, proteins with known roles in cell motility, adhesion, or invasion (see Figure S4B); note that 2 different sites were identified on PPP1R12A. Bold type, validated substrates.
(B) Many Group A substrates are proteins involved in cell motility, adhesion, and invasion. Twelve substrates involved in motility, adhesion and invasion were classified by mining the literature (Figure S4B), while an additional 2 were identified using a curated list of GO Terms (Figures S4A and S4B).
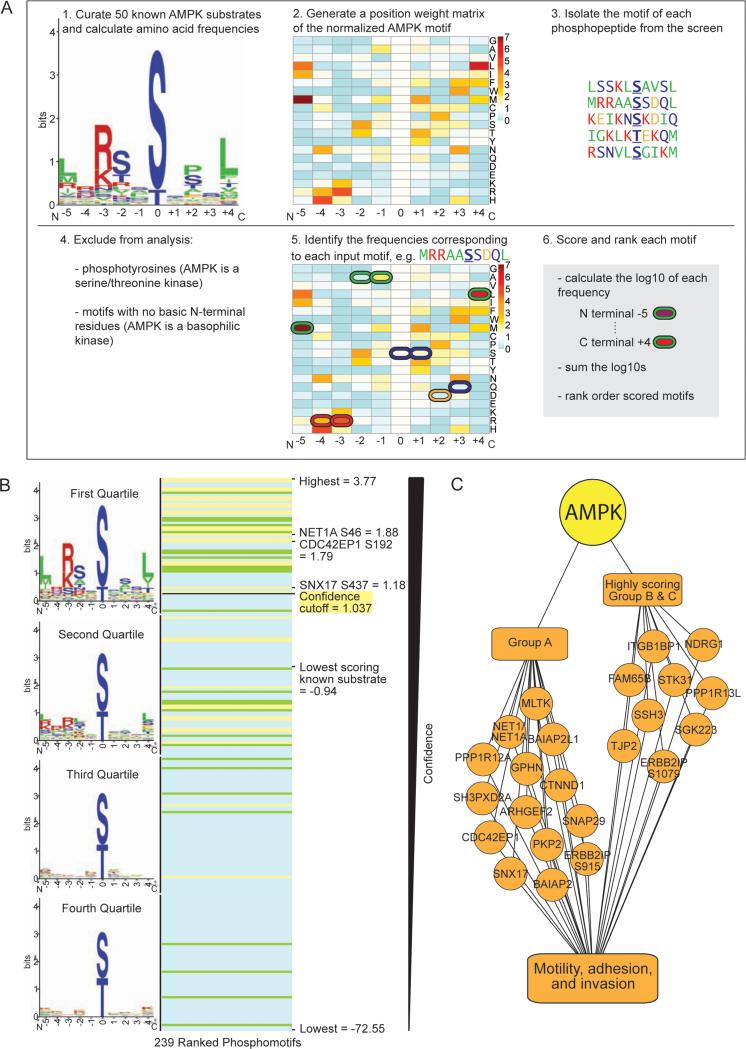
Figure 6. Use of the AMPK phosphorylation motif to rank phosphorylation sites identified at low frequency in the screen.
(A) Schematic of the pipeline used to score resemblance to the AMPK phosphorylation motif. The logo motif (1.) represents the phosphorylation motif of 50 known in vivo AMPK substrates. See Figure 2B for amino acid color-coding. The heat map (2. and 5.) represents the standardized frequencies of the 50 known AMPK substrates, with red indicating enrichment and blue indicating depletion in the AMPK motif compared with background (see Figure S6A).
(B) Ranked list of the scored motifs corresponding to the Group A (yellow lines), B (green lines), and C (blue lines) phosphorylation sites. The logo motif for each quartile of ranked motifs is shown. Scores of interest are noted.
(C) Nine highly scoring Group B and C phosphorylation sites are on proteins with known roles in cell motility, adhesion, and invasion. When combined with Group A, this totals 24 phosphorylation sites on 22 proteins involved in these processes (2 sites are on PPP1R12C and ERBB2IP).
Validation of AMPK substrates and phosphorylation sites
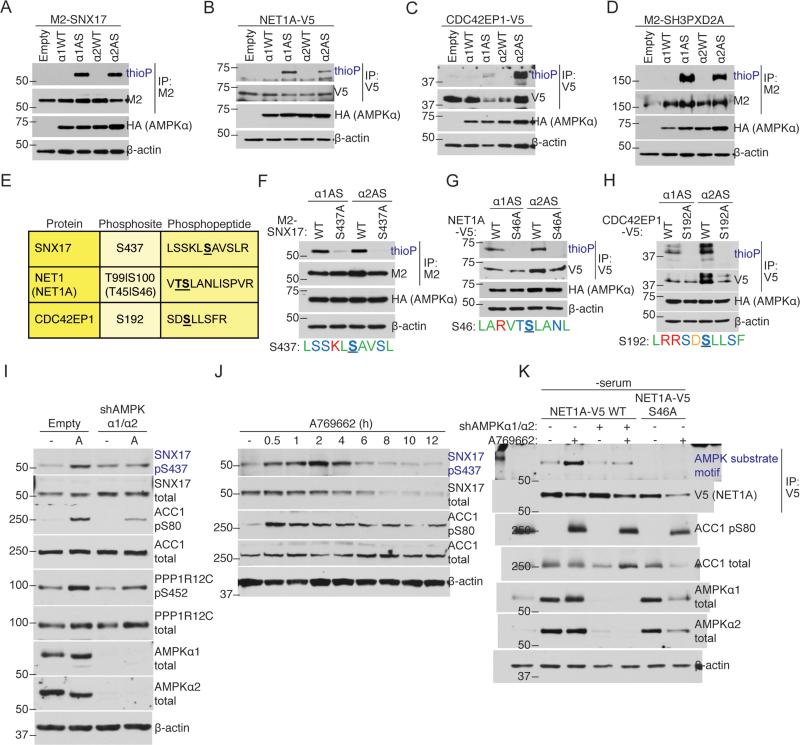
We next validated several AMPK substrates from Group A. AS-AMPKα1 and α2 strongly thiophosphorylated all of the substrates we tested: SNX17 (Figure 3A), NET1A (a short isoform of NET1) (Figure 3B), CDC42EP1 (Figure 3C), SH3PXD2A (Figure 3D), SNAP29 (Figure S3A), MTFR1L (Figure S3B), and RBM14 (Figure S3C). In most cases, AS-AMPKα1 and α2 thiophosphorylated these proteins equally well, although SNAP29 was a better substrate of AS-AMPKα2 (Figure S3A, Figure S3D), while MTFR1L was a better substrate of AS-AMPKα1 (Figure S3B, Figure S3E). Thus, several proteins from Group A validated as good substrates for both AMPK isoforms, though there may be some isoform-preferred substrates.
Figure 3. AS-AMPK directly phosphorylates several high confidence substrates, including SNX17 and NET1A.
(A-D) AS-AMPK thiophosphorylates SNX17, NET1A, CDC42EP1, and SH3PXD2A. Tagged proteins were over-expressed in empty vector or AMPKα-expressing (HA-tagged WT or AS-AMPKα1 or α2) U2OS cell lines, immunoprecipitated, and analyzed by western blot for the presence of thiophosphorylation. AMPK was activated in all conditions with 15 minutes of 50 mM 2DG. Representative of 3, 1, 1, and 2 independent experiments, respectively.
(E) Mass spectrometry-predicted AMPK phosphorylation sites and corresponding phosphopeptides for SNX17, NET1A, and CDC42EP1. Labeled as in Figure 1E. S100 on NET1 (S46 on the short isoform NET1A) was used as the NET1/NET1A site as its surrounding motif resembled the AMPK motif better than that of T99.
(F-H) AS-AMPK thiophosphorylates SNX17, NET1A, and CDC42EP1 at the identified residues. Tagged WT and predicted phosphorylation site mutants of the indicated substrates were over-expressed in U2OS AS-AMPKα2 and α1 cell lines, immunoprecipitated, and analyzed as in Figures 3A-D. The phosphorylation motif for the predicted residue is shown. The phosphorylated residue is underlined and bold. Color-coding as in Figure 2B. Each panel representative of 2 independent experiments.
(I) AMPK phosphorylates S437 on SNX17 endogenously. AMPK was activated in U2OS cells stably expressing an shRNA against AMPKα1 and α2 or empty vector control. Phosphorylation of the known substrates ACC1 S80 and PPP1R12C S452 are shown as controls for AMPK activation. Note that there is still some degree of AMPK substrate phosphorylation in cells with stable knockdown of AMPKα1 and α2, probably due to residual AMPK expression in these cells. “-“, no drug (DMSO vehicle control); A, A769662, 300 μM for 30 minutes. Representative of 2 independent experiments.
(J) Specific activation of AMPK decreases SNX17 protein levels. AMPK was activated in U2OS cells with 300 μM of A769662 for the indicated amount of time. Representative of 2 independent experiments.
(K) Over-expressed NET1A is phosphorylated at S46 in response to endogenous AMPK activation. NET1A-V5 WT or S46A was expressed in a doxycycline-inducible manner in U2OS cell lines. NET1A-V5 WT was also expressed in U2OS cell lines with shRNA-knockdown of both AMPKα1 and α2. Cells were serum-starved overnight, which was important to decrease basal NET1A phosphorylation, and NET1A-V5 expression was induced by 2 hours of doxycycline exposure (see Supplemental Experimental Procedures). AMPK was activated with 300 μM A769662 for 30 minutes. Following NET1A-V5 immunoprecipitation, samples were immunoblotted with an AMPK substrate motif antibody. Representative of 3 independent experiments.
To confirm the identified AMPK phosphorylation sites on three validated substrates, SNX17, NET1A, and CDC42EP1, we mutated the identified site on these proteins (Figure 3E). These mutations reduced (SNX17 S437A; Figure 3F) or completely removed (NET1A S46A, CDC42EP1 S192A; Figures 3G and 3H) thiophosphorylation by AS-AMPK. Mutation of the identified site on another validated substrate, SH3PXD2A, did not decrease thiophosphorylation (Figure S3F), though this may be due to the presence of many potential AMPK phosphorylation sites on this protein (Figures S3F and S3G). Thus, the identified sites are likely bona fide AMPK phosphorylation sites.
To validate phosphorylation sites independently of the analog-specific approach, we focused on SNX17 pS437 and NET1A pS46. For SNX17, we generated a phosphospecific antibody against pS437. We verified that this antibody recognized the WT, but not the S437A phosphorylation mutant, form of SNX17 (Figure S3H). Activation of endogenous AMPK by the specific AMPK activator A769662 increased the phosphorylation of endogenous SNX17 at S437 in U2OS cells (Figure 3I). Furthermore, shRNA knockdown of both AMPKα1 and α2 diminished the phosphorylation of endogenous SNX17 at S437 in response to A769662 (Figure 3I). Long-term activation of AMPK with either A769662 or nutrient deprivation correlated with decreased total levels of SNX17 (Figures 3J and S3I), raising the possibility that phosphorylation at this site leads to degradation of SNX17. Together, these results indicate that SNX17 pS437 is an endogenous AMPK target. For NET1A, we generated a phosphospecific antibody against pS46, but it was not potent enough to recognize NET1A pS46 in cells (data not shown). We thus used an antibody that recognizes the general AMPK phosphorylation motif (Ducommun et al., 2015; Gwinn et al., 2008; Zhang et al., 2002). Activation of endogenous AMPK by the specific AMPK activator A769662 increased the phosphorylation of the exogenously expressed WT, but not the S46A phosphorylation mutant, form of NET1A in U2OS cells (Figure 3K). In addition, knockdown of both AMPKα1 and α2 diminished phosphorylation of NET1A in response to A769662 (Figure 3K). These results indicate that endogenous AMPK is important for exogenous NET1A phosphorylation at S46, although other kinases could be contributing to NET1A basal phosphorylation. Collectively, these results confirm that the phosphorylation sites in Group A are likely bona fide endogenous AMPK targets.
Many high confidence AMPK substrates have known roles in cell motility, adhesion, and invasion
We mined the literature and used GO Terms to determine the functions of the AMPK substrates in Group A (Figure 4A, Supplemental List 3). As expected, some of the substrates further link AMPK with metabolic signaling pathways (Figures 4A & 4B). For example, WDFY3, a phosphatidylinositol 3⊠phosphate binding protein, promotes recycling of protein aggregates by autophagy (Simonsen et al., 2004). Interestingly, 14 AMPK substrates are involved in cellular motility, adhesion, or invasion as defined by literature mining and GO Terms (Figure 4A orange rows, Figures 4B, S4A, and S4B). For example, NET1A is a RHOA guanine nucleotide exchange factor (GEF) that can promote cell migration and invasion (Carr et al., 2013), and ERBB2IP is an adaptor protein that can inhibit cell migration (Liu et al., 2013). While unbiased GO analysis did not reveal any significant enrichment, probably due to the small size of the list, 11 of the 31 substrates were identified in a compiled list of GO Terms encompassing aspects of cell motility, adhesion, and invasion (Figures S4A and S4B). Consistently, studies have recently implicated AMPK in cell motility, adhesion and invasion, in part via the known substrates ACC1 and CLIP-170 (Nakano et al., 2010; Scott et al., 2012; Zhang et al., 2006). Thus, our screen provides additional substrates and phosphorylation sites that could help in functionally analyzing the role of AMPK in cell motility, adhesion, and invasion, processes that are key for wound healing and metastasis.
AMPK phosphorylation of NET1A inhibits extracellular matrix degradation
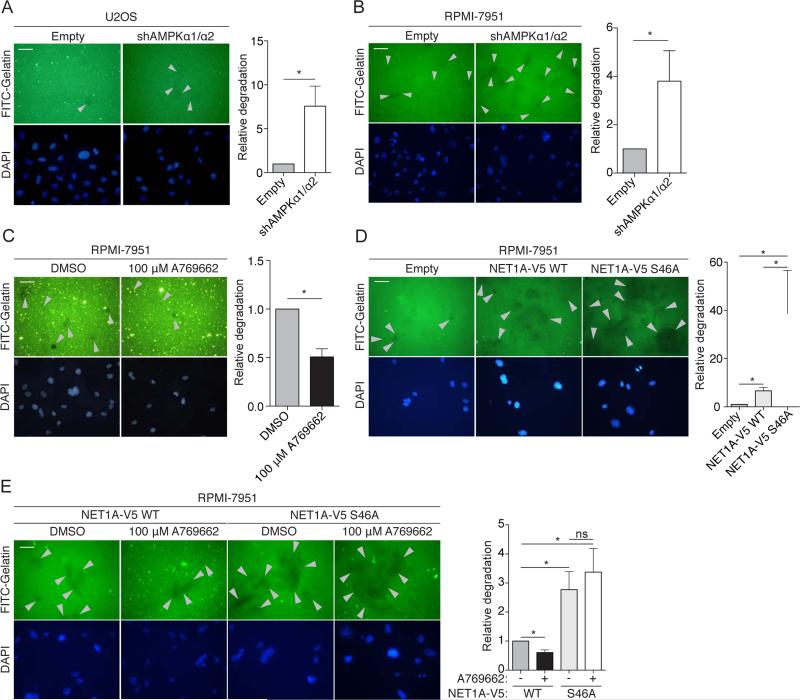
Cell invasion was recently found to be regulated by ACC1, a well-known AMPK substrate (Scott et al., 2012), but the exact roles of AMPK in cell invasion and the other potential substrates involved are largely unknown. We first tested whether manipulating AMPK activity indeed impacted a cell's ability to degrade the extracellular matrix (ECM), an early step in cell invasion, by using a gelatin-degradation assay (Bowden et al., 2001). U2OS cells stably expressing an shRNA against both AMPKα1 and α2 (Figure S5A and (Banko et al., 2011)) displayed a slight but significant increase in gelatin degradation (Figure 5A). Because U2OS cells are not a highly invasive cell line (Yuan et al., 2009) (Figure 5A, see basal levels), we also used RPMI-7951 cells, a metastatic melanoma cell line that extends invadopodia – protrusions that degrade the ECM (Seals et al., 2005). RPMI-7951 cells stably expressing an shRNA against AMPKα1 and α2 (Figure S5B) also displayed a significant increase in ECM degradation (Figure 5B). Conversely, activation of AMPK by A769662 in RPMI-7951 cells inhibited ECM degradation (Figure 5C). Together, these results suggest that AMPK impedes this early step of cell invasion.
Figure 5. AMPK phosphorylation of NET1A inhibits extracellular matrix (ECM) degradation.
(A) Knockdown of AMPK increases ECM degradation in U2OS cells. Cells stably expressing an shRNA against both AMPKα1 and α2 or empty vector control (Empty) cells were cultured on Fluorescein isothyocyanite (FITC)-conjugated gelatin-coated coverslips for 3 days. Fixed cells were stained for DAPI and analyzed. Grey arrowheads indicate points of gelatin degradation. Within each experiment, approximately 15-20 20× fields per sample were quantified and averaged; displayed images are 40x. Error bars represent mean +/− SEM of the averaged values from 6 independent experiments; the control samples in 4 of the experiments were the same used in 4 of the experiments in Figure S5G. *p < 0.05 by two-tailed Wilcoxon matched-pairs signed ranked test; scale bar = 50 μM.
(B) Knockdown of AMPK increases ECM degradation in RPMI-7951 cells. Cells stably expressing an shRNA against both AMPKα1 and α2 or empty vector control cells were analyzed and results are represented as in Figure 5A. Seven independent experiments were quantified.
(C) Activation of AMPK inhibits ECM degradation. RPMI-7951 cells were plated on FITC-conjugated gelatin-coated coverslips for 3-4 hours prior to administration of 100 μM A769662 or DMSO vehicle control for 16 hours. Analysis and results are represented as in Figure 5A. Six independent experiments were quantified.
(D) Loss of the AMPK phosphorylation site on NET1A increases ECM degradation. RPMI-7951 cells expressing similar levels of doxycycline-inducible NET1A-V5 WT or S46A (Figures S5C and S5D) were plated on FITC-conjugated gelatin coated coverslips and allowed to adhere overnight. 2 μg/ml doxycycline was added, and cells were cultured for an additional 2 days. Analysis and results are as in Figure 5A. Six independent experiments were quantified.
(E) Activation of AMPK inhibits ECM degradation in the presence of WT, but not S46A, NET1A. RPMI-7951 cells expressing similar levels of doxycycline-inducible NET1A-V5 WT or S46A (Figures S5C and S5D) were plated on FITC-conjugated gelatin coated coverslips for 3 hours prior to addition of 2 μg/ml doxycycline and either 100 μM A769662 or DMSO vehicle control for 20 hours. Media and drugs were replaced with fresh stocks after 10 hours. Analysis and results are represented as in Figure 5A. Six independent experiments were quantified; ns, not significant.
As NET1A can promote cell invasion (Carr et al., 2013), we next asked if the AMPK phosphorylation site on NET1A (S46, Figures 3G and 3K) mediates ECM degradation. To this end, we generated RPMI-7951 and U2OS cell lines that express either WT or phosphomutant (S46A) NET1A in a doxycycline inducible manner (Figures S5C-F). We verified that doxycycline induces similar levels of WT and S46A NET1A in these cell lines at the doses used in these assays (Figures S5C-F). Over-expression of NET1A S46A resulted in a strong increase in gelatin degradation in RPMI-7951 cells compared to both WT NET1A and empty vector control (Figure 5D). This trend was also observed in U2OS cells (Figure S5G). These observations suggest that phosphorylation of S46 on NET1A inhibits ECM degradation. Activation of AMPK by A769662 reduced gelatin degradation in the presence of WT, but not S46A, NET1A in RPMI-7951 cells (Figure 5E). AMPK phosphorylation of NET1A did not seem to affect NET1A localization or the ability to activate RHOA (data not shown). Collectively, these results suggest that AMPK inhibits ECM degradation in part by phosphorylating NET1A at S46, although how phosphorylation affects NET1A function is still unclear. Given that other AMPK substrates are involved in cell motility, adhesion, and invasion, AMPK likely modulates a network of proteins to affect these processes, perhaps dependent on cellular or environmental contexts.
Using the AMPK phosphorylation motif to computationally rank and identify low-frequency substrates
Given the successful validation of Group A substrates from our screen, we surmised that some of the phosphopeptides identified only once (Group C) or twice (Group B) could also be AMPK substrates instead of background. Group A substrates resemble the in vitro AMPK motif (Dale et al., 1995; Gwinn et al., 2008; Scott et al., 2002) (Figure 2B). They also closely match a curated motif that we built from 50 published AMPK substrates that were validated in cells (Figures 6A, top left panel, Supplemental Experimental Procedures). This similarity suggests that the AMPK phosphorylation motif could be used to identify additional likely AMPK substrates identified at low frequency in the screen.
To rank the phosphorylation sites from our screen based on their similarity to the AMPK motif, we built a ‘position-weight matrix’ (PWM) algorithm using the 50 published AMPK substrates (Figures 6A and S6A, Supplemental List 4, Supplemental Experimental Procedures). PWM algorithms are frequently used to score the likelihood of a motif being targeted by a specific kinase (e.g. Scansite (Obenauer et al., 2003)). Constructing our own PWM algorithm allowed us to base it on well-validated AMPK phosphorylation sites. We then used our algorithm to score and rank each phosphorylation site from the screen (Figures 6A and 6B, Supplemental List 5). The motifs of highly ranked sites matched the validated AMPK motif, whereas the lower ranked ones did not (Figure 6B). Consistently, Group A sites (in yellow) ranked higher than Group B sites (in green), which themselves ranked higher than Group C sites (in blue) (Figures 6B and S6B). We noted that there was also a cluster of lower ranked Group A sites (Figures 6B and S6B): those sites mostly resembled the AMPK motif (Figure S6C), but contained one amino acid that was not present in the validated AMPK motif, resulting in a scoring penalty. Therefore, our PWM algorithm selectively and stringently identifies phosphorylation motifs that closely resemble the AMPK motif, although it can also miss some substrates.
To predict which Group B and C phosphorylation sites could be real AMPK substrates, we applied a stringent cutoff score (1.037, see Figure S6D) to the ranked list (Figure 6B). Fifty phosphorylation sites scored above this cutoff, including most sites from Group A as well as 31 sites from Groups B and C (Figure S6E). These 31 additional sites, while found infrequently in our screen, likely represent real AMPK substrates. Indeed, two known substrates, CDC27 and TP53BP2 (Banko et al., 2011), are present in this group. Similar to Group A substrates, the 31 highly scoring Group B and C proteins are involved in a variety of cellular processes, including aspects of cell motility, adhesion, and invasion (Figures 6C and S6E, second tab of Supplemental List 5), further suggesting that these processes likely represent an important aspect of the AMPK network in U2OS cancer cells. However, because the phosphorylation motif of other AMPK family members is highly similar to that of AMPK (Goodwin et al., 2014), some of these sites may also be targeted by other kinases, and indeed S227 of RAB11FIP2 is a known target of the AMPK-related kinase, MARK2 (Ducharme et al., 2006). Thus, this AMPK motif matrix algorithm helped maximize our screen, revealing a total of 57 previously unknown AMPK phosphorylation sites. These sites serve as a resource for future studies seeking to functionally understand the AMPK-substrate network.
In silico analysis of AMPK network dynamics and prediction of AMPK phosphorylation sites
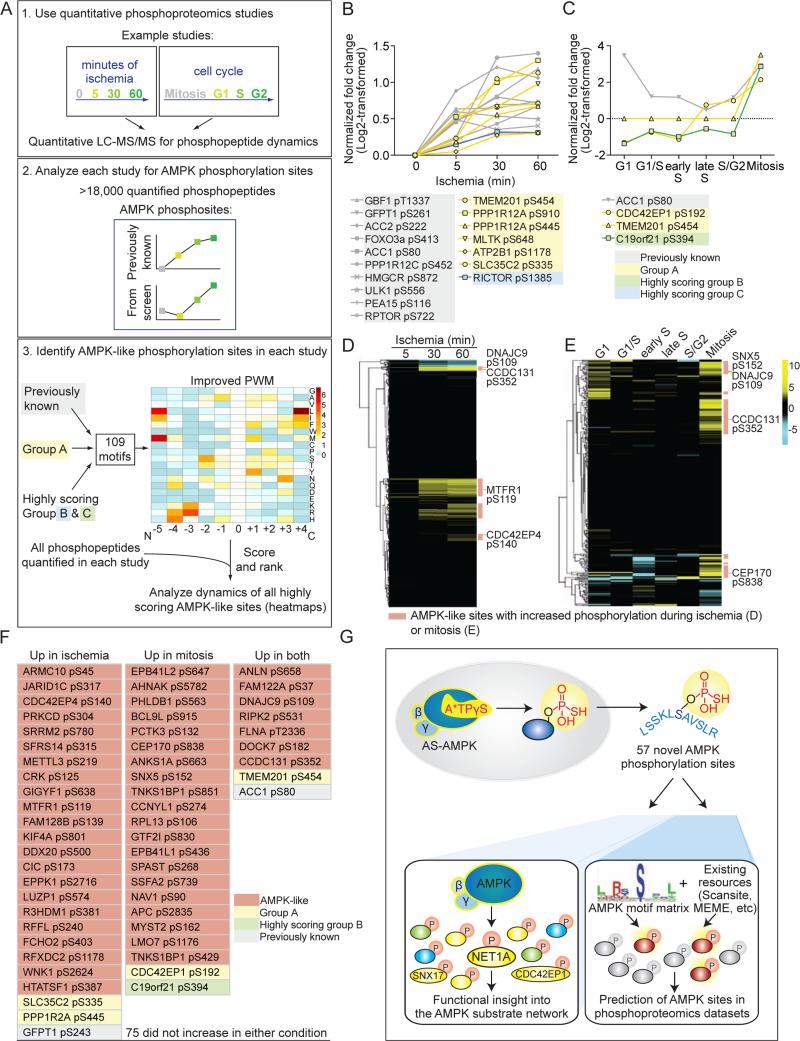
We next sought to extend our analysis of the AMPK network to other contexts (e.g. cell type, stimuli) and use our algorithm to facilitate large-scale substrate prediction. We first examined the dynamics of AMPK phosphorylation sites in publicly available quantitative phosphoproteomic datasets (Figure 7A, top panel). We selected large-scale datasets generated under conditions where AMPK is likely active. One dataset was generated from human luminal breast cancer xenografts, where ischemia – a condition that can lead to AMPK activation (Kudo et al., 1995; Russell et al., 2004) – was mimicked by delaying sample processing (Mertins et al., 2014) (Figure S7A). In the other dataset (Olsen et al., 2010), HeLa S3 cells were synchronized in different phases of the cell cycle, including mitosis – a context that can result in AMPK activation (Banko et al., 2011; Vazquez-Martin et al., 2009) (Figure S7B). Searching these datasets for AMPK phosphorylation sites (Figure 7A, middle panel) revealed that phosphorylation of ACC1 at S80 increased during both ischemia (Figure 7B) and mitosis (Figure 7C), consistent with the notion that AMPK is likely active under these conditions. Not all previously known AMPK sites were dynamically regulated during these processes (Figures S7C and S7D), which suggests that AMPK activity may not be optimal and/or is directed towards other substrates under these conditions. Several of the AMPK phosphorylation sites we identified in our screen, including a number of high confidence sites, also increased during ischemia (Figures 7B and S7C) or mitosis (Figures 7C and S7D). Thus, this type of analysis may help generate hypotheses for the relevance of AMPK substrates in specific contexts.
Figure 7. In silico analysis of AMPK network dynamics and prediction of AMPK phosphorylation sites.
(A) 1. General schematic of quantitative phosphoproteomic studies. 2. Datasets were queried for the presence of known AMPK phosphorylation sites and their dynamics during the biological processes analyzed. 3. A PWM constructed from 109 AMPK phosphorylation sites from both the literature and this study (Supplemental List 6) was generated to score and rank the motifs surrounding each quantified site in 1. The dynamics of sites with motifs scoring above the cutoff (Figure S7F) were analyzed.
(B) Seventeen AMPK phosphorylation sites present in the ischemia study (Mertins et al., 2014) increased significantly during ischemia (significance determined in (Mertins et al., 2014)). Sites in yellow are from Group A; blue, highly scoring Group C substrates; grey, previously known. Phosphorylation site location was standardized to isoform 1 in Uniprot. The 5, 30, and 60 minute time-points were standardized to the 0 minute time-point and log2-transformed in (Mertins et al., 2014).
(C) Four AMPK phosphorylation sites present in the cell cycle dataset (Olsen et al., 2010) increased at least 2-fold (following log2-transformation) during mitosis. Sites with yellow lines are from Group A; green, highly scoring Group B substrates; grey, previously known. Site location was standardized to isoform 1 in Uniprot. All timepoints were standardized to an asynchronously cycling population, normalized to protein level, and log2-transformed in (Olsen et al., 2010).
(D) 630 AMPK-like phosphorylation sites were present in the ischemia dataset, and a subset are dynamically phosphorylated (Supplemental List 7). Yellow, relative increase; blue, relative decrease compared to the 0 minute time-point. The time-points were standardized as in Figure 7B and (Mertins et al., 2014).
(E) 266 AMPK-like phosphorylation sites were present in the cell cycle dataset, and a subset are dynamically phosphorylated (Supplemental List 7). Yellow, relative increase; blue, relative decrease compared to an asynchronously cycling population. The relative changes were standardized as in Figure 7C and (Olsen et al., 2010).
(F) Highly scoring AMPK-like sites quantified in both the ischemia and cell cycle datasets and whose phosphorylation increased during ischemia and/or mitosis. A total of 131 quantified AMPK-like sites were present in both datasets (Supplemental List 7). Phosphorylation sites are as reported in the ischemia study (based on Protein GI Accessions).
(G) Summary of the proteomic and in silico approaches used here to identify AMPK phosphorylation sites and understand the AMPK functional network.
As these quantitative phosphoproteomic datasets contain thousands of phosphopeptides, they could also be analyzed more globally to predict additional AMPK-like sites and their dynamics (Figure 7A, bottom panel). We first increased the robustness of the PWM algorithm by combining the phosphorylation sites identified in our screen with the well-validated AMPK sites, generating a matrix from 109 AMPK phosphorylation sites (Figure S7E, Supplemental List 6). We then scored and ranked the motifs of sites in each phosphoproteomic dataset (Figures S7E and S7F, see Experimental Procedures). This analysis identified 630 AMPK-like phosphorylation sites in the ischemia dataset and 266 in the cell cycle dataset (Figures 7D, 7E and S7F; Supplemental List 7). These AMPK-like sites could be AMPK substrates, although they could also be targets of other related kinases. During ischemia, phosphorylation increased on 150 of the 630 AMPK-like sites and decreased on 28 (Figure 7D, Supplemental List 7). During mitosis, phosphorylation increased on 74 of the 266 AMPK-like sites and decreased on 13 (Figure 7E; Supplemental List 7). Of the 131 AMPK-like sites that were present in both studies (Supplemental List 7), 47 increased specifically in either ischemia (25) (e.g. CDC42EP4 S140) or mitosis (22) (e.g. SNX5 S152), while 9 increased in both (e.g. CCDC131 S352 and the Group A substrate TMEM201 S454) (Figures 7F; Supplemental List 7). This predictive approach allows rapid identification of potential AMPK substrates and their regulation in different contexts, although this could also reflect experimental conditions and/or contributions of other kinases.
As high quality phosphoproteomics datasets become available, analyzing AMPK site dynamics and predicting new substrates in context-specific scenarios will likely yield important hypotheses to test experimentally. To facilitate this, we have made both the AMPK motif matrix and our algorithm available to the community at https://github.com/BrunetLabAMPK/AMPK_motif_analyzer (see Supplemental Experimental Procedures). These tools complement existing resources that scan proteins for specific motifs, such as Scansite (Obenauer et al., 2003) and MEME (FIMO) (Bailey et al., 2009; Grant et al., 2011)) (Figure S7G). The comprehensive AMPK motif matrix we generated (Supplemental List 6) can also be uploaded to Scansite and MEME to query protein sequences for the presence of AMPK-like motifs (Figure S7G and Supplemental Experimental Procedures).
In summary, using a biochemical screen for direct AMPK substrate identification, we identified 57 previously unknown AMPK phosphorylation sites (Figure 7G), highlighting a role for this energy sensor in cell motility, adhesion, and invasion. We also developed an in silico approach to predict AMPK substrates and analyze their phosphorylation dynamics (Figure 7G), providing resources for future studies on AMPK.
Discussion
Identification of direct AMPK substrates and phosphorylation sites
Our screen is the first large-scale identification of direct kinase substrates and phosphorylation sites in cells and identified 57 previously unknown AMPK phosphorylation sites. Seven substrates and three phosphorylation sites validated as direct targets of AS-AMPK. It is possible that the AS system, which involves digitonin permeabilization and over-expression of a mutated form of the alpha subunit of AMPK, alters the specificity of AMPK. However, our screen identified known substrates of AMPK (e.g. ACC1 S80 and RAPTOR S722) and two of the previously uncharacterized substrates (SNX17 and NET1A) were confirmed to be substrates of endogenous AMPK, suggesting that this screen identified bona fide AMPK targets.
While successful in identifying direct AMPK phosphorylation sites, the screening conditions were subject to background and low saturation. In addition, not all known AMPK substrates were found. Several factors may help explain this. For example, not all tryptic peptides are detectable by mass spectrometry, and the peptide capture approach results in the loss of cysteine-containing peptides from the sample, as they irreversibly bind to the iodoacetyl groups (Blethrow et al., 2008). Furthermore, some endogenous kinases may also use the bulky ATPγS. Developing methods to decrease background phosphopeptides and increase retention of bona fide thiophosphopeptides will help make this method more widely applicable to other kinases.
AMPK phosphorylates many substrates involved in cell motility, adhesion, and invasion
An important finding of this screen is the number of substrates involved in different aspects of cell motility, adhesion, and invasion. We found that AMPK inhibits ECM degradation, a key early step in cell invasion, in part through NET1A. NET1A was recently implicated in cell invasion (Carr et al., 2013), but its importance during ECM degradation and regulation by AMPK were completely unknown. NET1A does not appear to localize to invadopodia (B.E.S. and A.B., unpublished data), but it does localize to focal adhesions and binds focal adhesion kinase (FAK) (Carr et al., 2013). FAK helps regulate the balance between invadopodia and focal adhesions (Chan et al., 2009), so AMPK phosphorylation of NET1A could alter the ability of FAK to modulate this balance. Alternatively, phosphorylation of NET1A by AMPK could affect the function of specific invadopodia components. Importantly, AMPK may regulate additional targets to inhibit ECM degradation, including other previously unknown substrates, like the invadopodia component SH3PXD2A (Seals et al., 2005), or well-known ones such as ACC1. Indeed, ACC1 mediates AMPK-dependent inhibition of ECM degradation (Scott et al., 2012). As ECM degradation is an initial step in metastasis (Eckert et al., 2011), these results raise the possibility that AMPK could inhibit early stages of metastasis.
In silico approach to predict AMPK phosphorylation sites in phosphoproteomics datasets
Exploring AMPK substrates in different contexts is an important but difficult experimental task, and a well-defined AMPK motif can be used to more accurately predict substrates in other contexts. Previous approaches have successfully used an AMPK motif defined by in vitro peptide scanning (Gwinn et al., 2008) or by mutational analyses of AMPK substrates (Marin et al., 2015; Towler and Hardie, 2007). Here, we develop an in silico approach based on in vivo AMPK phosphorylation sites to predict likely AMPK sites in phosphoproteomics datasets, notably quantitative ones. As quantitative datasets already contain information about the dynamics of phosphorylation sites, data are immediately available regarding the regulation of an AMPK-like site in that context. Caveats of this approach are that phosphoproteomic datasets are often generated using different experimental procedures and do not capture all phosphorylated peptides. In addition, the experimental conditions used (e.g. pharmacological cell synchronization) may result in aberrant AMPK activity, and AMPK-like sites may be phosphorylated by other kinases. Nevertheless, comparison of the predicted AMPK phosphorylation sites between different phosphoproteomic datasets could generate hypotheses concerning context-specific phosphorylation.
Our work provides the first large-scale identification of direct AMPK phosphorylation sites, extending the AMPK network and facilitating the generation of a platform to help predict additional AMPK targets. As promising therapeutic roles of AMPK continue to emerge for diseases like cancer (Kato et al., 2002; Liang and Mills, 2013; Xiang et al., 2004) and diabetes (Winder and Hardie, 1999), as well as for aging (Apfeld et al., 2004; Greer et al., 2007; Mair et al., 2011), the substrates we identified and the platform we created could help properly harness AMPK's full therapeutic potential.
Experimental Procedures
Full protocols and additional information are provided in Supplemental Experimental Procedures
Thiophosphorylation of AS-AMPK substrates
In-cell thiophosphorylation of AS-AMPK substrates was performed as in (Banko et al., 2011), but without addition of AMP to the thiophosphorylation labeling buffer (see Supplemental Experimental Procedures). Samples processed for western blot analysis were alkylated using p-nitrobenzyl mesylate. Samples generated for peptide capture were processed as outlined below.
Peptide capture of thiophosphorylated peptides and identification via liquid chromatography-tandem mass spectrometry (LC-MS/MS)
Peptide capture of thiophosphorylated peptides was performed according to (Blethrow et al., 2008; Hertz et al., 2010). Experiment-specific modifications to the full protocol detailed under Supplemental Experimental Procedures are in Supplemental List 1. LC-MS/MS and processing was performed as in (Hengeveld et al., 2012).
Analysis of mass spectrometry data
Mass spectrometry peaklists were generated with an in-house (UCSF) software named PAVA and searched against the SwissProt Homo Sapiens database (downloaded on March 21, 2012) using Protein Prospector (version 5.10.10). Phosphopeptides identified in the AS-AMPK samples were compared to those identified in control samples across all experiments (phosphopeptides identified in control samples: Supplemental List 2, second tab), and overlapping phosphopeptides were removed as background. The peptide, irrespective of phosphosite placement, was used for this filtering step, accounting for possible missed trypsinization events or other peptide overlap. See Supplemental Experimental Procedures for more details.
Scoring phosphorylation motifs based on similarity to the AMPK motif
To determine the similarity of phosphorylation motifs to the AMPK motif, a position weight matrix (PWM) was constructed using the frequencies of amino acids in the motifs surrounding 50 well-validated in vivo AMPK phosphorylation sites (see “Curation of known in vivo AMPK substrates” in Supplemental Experimental Procedures and Supplemental List 4) (Figure 6) or 109 AMPK phosphorylation sites that were previously validated or were discovered in our screen (Figure 7). Each amino acid frequency in the AMPK motif was standardized to a background frequency generated by averaging the amino acid occurrences in 10,000 randomly sampled datasets of matched number and phosphoS:T ratio from a compendium of human phosphorylation sites. Query motifs were scored by summing the log10 of each location's amino acid standardized frequency from the N terminal 5 to the C terminal 4 position. Motifs lacking basic residues within 5 N terminal amino acids or surrounding phosphotyrosines were discarded. If a phosphopeptide contained more than one possible phosphorylation site, the site with the highest scoring motif was used (Figure 6). Scored motifs were rank ordered. The standardized frequencies of the 50 well-validated AMPK sites are in the second tab of Supplemental List 4; those of the 109 AMPK sites are in Supplemental List 6 (matrix form) and on GitHub (tab-delimited list form). The code to score and rank input motifs is available on GitHub (https://github.com/BrunetLabAMPK/AMPK_motif_analyzer) (see Supplemental Experimental Procedures).
Logo motif generation
To visualize phosphorylation motifs, logo motifs were generated using Berkeley's Weblogo generator (Crooks et al., 2004; Schneider and Stephens, 1990).
Antibody information, primer sequences and construct generation, cell culture methods, generation of stable cell lines, cell lysis and immunoprecipitation in non-AS-based assays, immunoprecipitation of AS-AMPK substrates, preparation of Fluorescein isothyocyanite (FITC)-conjugated Gelatin coated coverslips, analysis of gelatin degradation, using the AMPK motif matrix and GitHub access, and general data plotting and statistical analyses are detailed in Supplemental Experimental Procedures
Supplementary Material
Highlights.
A screen identifies AMPK substrates and phosphorylation sites in human cancer cells
Many novel AMPK substrates have roles in cell motility, adhesion, and invasion
AMPK phosphorylation of one new substrate, NET1A, inhibits cell invasion
A computational pipeline using the AMPK motif can predict additional substrates
Acknowledgements
We thank Lauren Booth, Salah Mahmoudi, Itamar Harel, Elizabeth Pollina, Aaron Daugherty, Parag Mallick, Elizabeth Schroeder, Josh Elias, and Monte Winslow for helpful discussion and carefully reading the manuscript. We thank Param Priya Singh for establishing the GitHub repository and verifying the Perl codes, Aaron Daugherty and Katja Hebestreit for verifying R and Perl codes, and Carman Li and Tyler Jacks for the protocol on FITC-gelatin coated coverslip preparation. This work was supported by grants CIRM RB4-06087 and NIH R01 AG031198 (A.B.), NSF GRFP (B.E.S.), the Robert M. and Anne T. Bass Stanford Graduate Fellowship (B.E.S.), NIH T32 CA09302 (B.E.S), Howard Hughes Medical Institute (K.M.S.), and NIH R01 DK080425 and P01 CA120964 (R.J.S.). Mass Spectrometry was provided by the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (A.L. Burlingame, Director) supported by the Biomedical Technology Research Centers program of the NIH National Institute of General Medical Sciences, NIH NIGMS 8P41GM103481 (the Thermo Scientifi LTQOrbitrap Velos is specifically supported by P41GM103481 and Howard Hughes Medical Institute) and by the Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University Mass Spectrometry, with specific thanks to Chris Adams and NIH S10RR027425 (National Center For Research Resources).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
B.E.S. and A.B. designed the study. B.E.S. conducted all the experiments unless otherwise indicated and constructed the pipelines for data analysis. N.T.H. and R.S.L. contributed equally: they helped develop the peptide capture approach and conducted mass spectrometry experiments under the supervision of K.M.S.. T.J.M. and R.J.S. provided intellectual contributions and mentorship to B.E.S.. M.L.S. performed most of the extracellular matrix degradation assays and some western-blots. P.E.H. designed and conducted assays to detect NET1A phosphorylation under the guidance of R.J.S.. B.A.B. provided guidance in computational analyses. M.R.B. cloned several AMPKα1 constructs. K.M.S. provided intellectual contributions for the analog-specific system. R.J.S. and K.M.S. had equal contributions. B.E.S. and A.B. wrote the paper, and other authors commented on the manuscript.
References
- Amato S, Liu X, Zheng B, Cantley L, Rakic P, Man HY. AMP-activated protein kinase regulates neuronal polarization by interfering with PI 3-kinase localization. Science. 2011;332:247–251. doi: 10.1126/science.1201678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson U, Filipsson K, Abbott CR, Woods A, Smith K, Bloom SR, Carling D, Small CJ. AMP-activated protein kinase plays a role in the control of food intake. J Biol Chem. 2004;279:12005–12008. doi: 10.1074/jbc.C300557200. [DOI] [PubMed] [Google Scholar]
- Apfeld J, O'Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banko MR, Allen JJ, Schaffer BE, Wilker EW, Tsou P, White JL, Villen J, Wang B, Kim SR, Sakamoto K, et al. Chemical genetic screen for AMPKalpha2 substrates uncovers a network of proteins involved in mitosis. Mol Cell. 2011;44:878–892. doi: 10.1016/j.molcel.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW, et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc Natl Acad Sci U S A. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden ET, Coopman PJ, Mueller SC. Invadopodia: unique methods for measurement of extracellular matrix degradation in vitro. Methods Cell Biol. 2001;63:613–627. doi: 10.1016/s0091-679x(01)63033-4. [DOI] [PubMed] [Google Scholar]
- Burkewitz K, Zhang Y, Mair WB. AMPK at the nexus of energetics and aging. Cell Metab. 2014;20:10–25. doi: 10.1016/j.cmet.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr HS, Zuo Y, Oh W, Frost JA. Regulation of focal adhesion kinase activation, breast cancer cell motility, and amoeboid invasion by the RhoA guanine nucleotide exchange factor Net1. Mol Cell Biol. 2013;33:2773–2786. doi: 10.1128/MCB.00175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KT, Cortesio CL, Huttenlocher A. FAK alters invadopodia and focal adhesion composition and dynamics to regulate breast cancer invasion. J Cell Biol. 2009;185:357–370. doi: 10.1083/jcb.200809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Murphy J, Toth R, Campbell DG, Morrice NA, Mackintosh C. Complementary regulation of TBC1D1 and AS160 by growth factors, insulin and AMPK activators. Biochem J. 2008;409:449–459. doi: 10.1042/BJ20071114. [DOI] [PubMed] [Google Scholar]
- Cool B, Zinker B, Chiou W, Kifle L, Cao N, Perham M, Dickinson R, Adler A, Gagne G, Iyengar R, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab. 2006;3:403–416. doi: 10.1016/j.cmet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale S, Wilson WA, Edelman AM, Hardie DG. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 1995;361:191–195. doi: 10.1016/0014-5793(95)00172-6. [DOI] [PubMed] [Google Scholar]
- Davies SP, Sim AT, Hardie DG. Location and function of three sites phosphorylated on rat acetyl-CoA carboxylase by the AMP-activated protein kinase. Eur J Biochem. 1990;187:183–190. doi: 10.1111/j.1432-1033.1990.tb15293.x. [DOI] [PubMed] [Google Scholar]
- Ducharme NA, Hales CM, Lapierre LA, Ham AJ, Oztan A, Apodaca G, Goldenring JR. MARK2/EMK1/Par-1Balpha phosphorylation of Rab11-family interacting protein 2 is necessary for the timely establishment of polarity in Madin-Darby canine kidney cells. Mol Biol Cell. 2006;17:3625–3637. doi: 10.1091/mbc.E05-08-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducommun S, Deak M, Sumpton D, Ford RJ, Nunez Galindo A, Kussmann M, Viollet B, Steinberg GR, Foretz M, Dayon L, et al. Motif affinity and mass spectrometry proteomic approach for the discovery of cellular AMPK targets: Identification of mitochondrial fission factor as a new AMPK substrate. Cell Signal. 2015;27:978–988. doi: 10.1016/j.cellsig.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Eckert MA, Lwin TM, Chang AT, Kim J, Danis E, Ohno-Machado L, Yang J. Twist1-induced invadopodia formation promotes tumor metastasis. Cancer Cell. 2011;19:372–386. doi: 10.1016/j.ccr.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin JM, Svensson RU, Lou HJ, Winslow MM, Turk BE, Shaw RJ. An AMPK-independent signaling pathway downstream of the LKB1 tumor suppressor controls Snail1 and metastatic potential. Mol Cell. 2014;55:436–450. doi: 10.1016/j.molcel.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ashford ML. AMPK: regulating energy balance at the cellular and whole body levels. Physiology (Bethesda) 2014;29:99–107. doi: 10.1152/physiol.00050.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Carling D. The AMP-activated protein kinase--fuel gauge of the mammalian cell? Eur J Biochem. 1997;246:259–273. doi: 10.1111/j.1432-1033.1997.00259.x. [DOI] [PubMed] [Google Scholar]
- Hawley SA, Fullerton MD, Ross FA, Schertzer JD, Chevtzoff C, Walker KJ, Peggie MW, Zibrova D, Green KA, Mustard KJ, et al. The ancient drug salicylate directly activates AMP-activated protein kinase. Science. 2012;336:918–922. doi: 10.1126/science.1215327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengeveld RC, Hertz NT, Vromans MJ, Zhang C, Burlingame AL, Shokat KM, Lens SM. Development of a chemical genetic approach for human aurora B kinase identifies novel substrates of the chromosomal passenger complex. Mol Cell Proteomics. 2012;11:47–59. doi: 10.1074/mcp.M111.013912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz NT, Wang BT, Allen JJ, Zhang C, Dar AC, Burlingame AL, Shokat KM. Chemical genetic approach for kinase-substrate mapping by covalent capture of thiophosphopeptides and analysis by mass spectrometry. Curr Protoc Chem Biol. 2010;2:15–36. doi: 10.1002/9780470559277.ch090201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Tinti M, Wood NT, Campbell DG, Toth R, Dubois F, Geraghty KM, Wong BH, Brown LJ, Tyler J, et al. Visualization and biochemical analyses of the emerging mammalian 14-3-3-phosphoproteome. Mol Cell Proteomics. 2011;10:M110 005751. doi: 10.1074/mcp.M110.005751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Ogura T, Kishimoto A, Minegishi Y, Nakajima N, Miyazaki M, Esumi H. Critical roles of AMP-activated protein kinase in constitutive tolerance of cancer cells to nutrient deprivation and tumor formation. Oncogene. 2002;21:6082–6090. doi: 10.1038/sj.onc.1205737. [DOI] [PubMed] [Google Scholar]
- Kudo N, Barr AJ, Barr RL, Desai S, Lopaschuk GD. High rates of fatty acid oxidation during reperfusion of ischemic hearts are associated with a decrease in malonyl-CoA levels due to an increase in 5'-AMP-activated protein kinase inhibition of acetyl-CoA carboxylase. J Biol Chem. 1995;270:17513–17520. doi: 10.1074/jbc.270.29.17513. [DOI] [PubMed] [Google Scholar]
- Kurth-Kraczek EJ, Hirshman MF, Goodyear LJ, Winder WW. 5' AMP-activated protein kinase activation causes GLUT4 translocation in skeletal muscle. Diabetes. 1999;48:1667–1671. doi: 10.2337/diabetes.48.8.1667. [DOI] [PubMed] [Google Scholar]
- Lee JH, Koh H, Kim M, Kim Y, Lee SY, Karess RE, Lee SH, Shong M, Kim JM, Kim J, et al. Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature. 2007;447:1017–1020. doi: 10.1038/nature05828. [DOI] [PubMed] [Google Scholar]
- Liang J, Mills GB. AMPK: a contextual oncogene or tumor suppressor? Cancer Res. 2013;73:2929–2935. doi: 10.1158/0008-5472.CAN-12-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Shi M, Duan C, Chen H, Hu Y, Yang Z, Duan H, Guo N. Downregulation of Erbin in Her2-overexpressing breast cancer cells promotes cell migration and induces trastuzumab resistance. Mol Immunol. 2013;56:104–112. doi: 10.1016/j.molimm.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Mair W, Morantte I, Rodrigues AP, Manning G, Montminy M, Shaw RJ, Dillin A. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470:404–408. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin TL, Gongol B, Martin M, King SJ, Smith L, Johnson DA, Subramaniam S, Chien S, Shyy JY. Identification of AMP-activated protein kinase targets by a consensus sequence search of the proteome. BMC Syst Biol. 2015;9:13. doi: 10.1186/s12918-015-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Yang F, Liu T, Mani DR, Petyuk VA, Gillette MA, Clauser KR, Qiao JW, Gritsenko MA, Moore RJ, et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol Cell Proteomics. 2014;13:1690–1704. doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouse V, Swick LL, Kazgan N, St Johnston D, Brenman JE. LKB1 and AMPK maintain epithelial cell polarity under energetic stress. J Cell Biol. 2007;177:387–392. doi: 10.1083/jcb.200702053. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nakano A, Kato H, Watanabe T, Min KD, Yamazaki S, Asano Y, Seguchi O, Higo S, Shintani Y, Asanuma H, et al. AMPK controls the speed of microtubule polymerization and directional cell migration through CLIP-170 phosphorylation. Nat Cell Biol. 2010;12:583–590. doi: 10.1038/ncb2060. [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Russell RR, 3rd, Li J, Coven DL, Pypaert M, Zechner C, Palmeri M, Giordano FJ, Mu J, Birnbaum MJ, Young LH. AMP-activated protein kinase mediates ischemic glucose uptake and prevents postischemic cardiac dysfunction, apoptosis, and injury. J Clin Invest. 2004;114:495–503. doi: 10.1172/JCI19297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Norman DG, Hawley SA, Kontogiannis L, Hardie DG. Protein kinase substrate recognition studied using the recombinant catalytic domain of AMP-activated protein kinase and a model substrate. J Mol Biol. 2002;317:309–323. doi: 10.1006/jmbi.2001.5316. [DOI] [PubMed] [Google Scholar]
- Scott KE, Wheeler FB, Davis AL, Thomas MJ, Ntambi JM, Seals DF, Kridel SJ. Metabolic regulation of invadopodia and invasion by acetyl-CoA carboxylase 1 and de novo lipogenesis. PLoS One. 2012;7:e29761. doi: 10.1371/journal.pone.0029761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals DF, Azucena EF, Jr., Pass I, Tesfay L, Gordon R, Woodrow M, Resau JH, Courtneidge SA. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell. 2005;7:155–165. doi: 10.1016/j.ccr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Shah K, Liu Y, Deirmengian C, Shokat KM. Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc Natl Acad Sci U S A. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim AT, Hardie DG. The low activity of acetyl-CoA carboxylase in basal and glucagon-stimulated hepatocytes is due to phosphorylation by the AMP-activated protein kinase and not cyclic AMP-dependent protein kinase. FEBS Lett. 1988;233:294–298. doi: 10.1016/0014-5793(88)80445-9. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- Towler MC, Hardie DG. AMP-activated protein kinase in metabolic control and insulin signaling. Circ Res. 2007;100:328–341. doi: 10.1161/01.RES.0000256090.42690.05. [DOI] [PubMed] [Google Scholar]
- Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. The active form of the metabolic sensor: AMP-activated protein kinase (AMPK) directly binds the mitotic apparatus and travels from centrosomes to the spindle midzone during mitosis and cytokinesis. Cell Cycle. 2009;8:2385–2398. doi: 10.4161/cc.8.15.9082. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wilson WA, Fujino MA, Roach PJ. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol Cell Biol. 2001;21:5742–5752. doi: 10.1128/MCB.21.17.5742-5752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder WW, Hardie DG. AMP-activated protein kinase, a metabolic master switch: possible roles in type 2 diabetes. Am J Physiol. 1999;277:E1–10. doi: 10.1152/ajpendo.1999.277.1.E1. [DOI] [PubMed] [Google Scholar]
- Xiang X, Saha AK, Wen R, Ruderman NB, Luo Z. AMP-activated protein kinase activators can inhibit the growth of prostate cancer cells by multiple mechanisms. Biochem Biophys Res Commun. 2004;321:161–167. doi: 10.1016/j.bbrc.2004.06.133. [DOI] [PubMed] [Google Scholar]
- Yuan J, Ossendorf C, Szatkowski JP, Bronk JT, Maran A, Yaszemski M, Bolander ME, Sarkar G, Fuchs B. Osteoblastic and osteolytic human osteosarcomas can be studied with a new xenograft mouse model producing spontaneous metastases. Cancer Invest. 2009;27:435–442. doi: 10.1080/07357900802491477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadra G, Photopoulos C, Tyekucheva S, Heidari P, Weng QP, Fedele G, Liu H, Scaglia N, Priolo C, Sicinska E, et al. A novel direct activator of AMPK inhibits prostate cancer growth by blocking lipogenesis. EMBO Mol Med. 2014;6:519–538. doi: 10.1002/emmm.201302734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, Polakiewicz RD, Comb MJ. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J Biol Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li J, Young LH, Caplan MJ. AMP-activated protein kinase regulates the assembly of epithelial tight junctions. Proc Natl Acad Sci U S A. 2006;103:17272–17277. doi: 10.1073/pnas.0608531103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Cantley LC. Regulation of epithelial tight junction assembly and disassembly by AMP-activated protein kinase. Proc Natl Acad Sci U S A. 2007;104:819–822. doi: 10.1073/pnas.0610157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.