Abstract
The members of the genus Alphavirus are positive-sense RNA viruses, which are predominantly transmitted to vertebrates by a mosquito vector. Alphavirus disease in humans can be severely debilitating, and depending on the particular viral species, infection may result in encephalitis and possibly death. In recent years, alphaviruses have received significant attention from public health authorities as a consequence of the dramatic emergence of chikungunya virus in the Indian Ocean islands and the Caribbean. Currently, no safe, approved or effective vaccine or antiviral intervention exists for human alphavirus infection. The molecular biology of alphavirus RNA synthesis has been well studied in a few species of the genus and represents a general target for antiviral drug development. This review describes what is currently understood about the regulation of alphavirus RNA synthesis, the roles of the viral non-structural proteins in this process and the functions of cis-acting RNA elements in replication, and points to open questions within the field.
Introduction
Alphaviruses are a group of globally distributed arthropod-borne RNA viruses with a broad host range. Although they are most commonly maintained between mosquito vectors and avian hosts, outbreaks of human and livestock infections frequently occur, and are thus of economic and public health concern. Infection of arthropods is persistent, lifelong and asymptomatic, and these differences are recapitulated in cultured cells. In humans, symptoms of alphaviral infections range from fever, rash, nausea and polyarthritis to fatal encephalitis. Whilst mortality is low for many alphaviruses, associated disease can be debilitating, with clinical sequelae lasting from months to years in some patients (Weaver & Lecuit, 2015). Changing vector ranges threaten new populations with emerging or re-emerging disease. Such emergence events are exemplified by the current chikungunya virus (CHIKV) epidemic in the Caribbean (Johansson et al., 2014; Weaver, 2014). At this time, no antiviral therapies or safe, effective vaccines are available. The identification of targets for antiviral intervention and means of rational attenuation for vaccine development require a deep understanding of the mechanisms of virus replication in both the vertebrate host and the vector. Interestingly, whilst a great deal has been elucidated regarding the molecular mechanisms of alphaviral genome replication and RNA synthesis, much of which will be described in this review, the majority of what we know has been derived from work on very few of the 30 recognized alphavirus species, predominantly two: Sindbis virus (SINV) and Semliki Forest virus (SFV). Additionally, the majority of work has been performed in a vertebrate system, leaving many unknowns in terms of the specifics of how different alphaviruses replicate and interact with the vector species.
Taxonomy and distribution
The genus Alphavirus is a member of the family Togaviridae, a group of enveloped positive-sense RNA viruses. The family Togaviridae also includes the genus Rubivirus (Büchen-Osmond, 2006). There are 31 currently recognized alphavirus species that divide into eight phylogenetic groupings with SINV being the type species (Forrester et al., 2012; Nasar et al., 2012). The relatively recent discovery of aquatic alphaviruses that infect fish and Southern Elephant Seal virus, which is likely transmitted by a louse species (La Linn et al., 2001; Villoing et al., 2000; Weston et al., 1999), and the presence of these viruses in basal positions in reported phylogenies suggests an aquatic/oceanic origin for the terrestrial alphaviruses (Forrester et al., 2012). The divergence of the terrestrial alphaviruses is marked by the New World and Old World viruses, which arose after multiple geographical introductions and reintroductions (Forrester et al., 2012; Levinson et al., 1990; Powers et al., 2001). Old World viruses include those of the SFV complex including CHIKV; New World viruses include Venezuelan equine encephalitis virus (VEEV), eastern equine encephalitis virus (EEEV) and western equine encephalitis virus (WEEV). The genus is endemic on six continents with the range of individual species confined to regions by environmental barriers. However, such as in the case of the recent CHIKV outbreak, these ranges are dynamic and new human populations are being exposed to viruses and risk of disease (Weaver, 2014; Weaver & Lecuit, 2015).
The recognition of species that infect fish and aquatic mammals, along with those viruses for which no vertebrate host has been identified, suggests there is an as-yet unrecognized number of alphavirus species inhabiting ecological niches that have not yet been explored. For instance, the recently discovered Eilat virus is apparently restricted to the invertebrate host, and in mammalian cells exhibits defects in both viral protein expression and RNA replication (Nasar et al., 2102, 2014, 2015). Whilst it is likely that there are unifying mechanisms for alphavirus RNA synthesis and replication, the details will vary significantly between species as their evolutionary relatedness diminishes. The following describes what is known about alphavirus non-structural protein functions and the requirements for RNA synthesis based primarily on the work performed using a subset of mosquito-transmitted alphaviruses.
Virion and genome characteristics
The virus particle consists of an RNA genome surrounded by a protein capsid shell within a host-derived lipid envelope decorated with glycoprotein spikes. The capsid shell and glycoprotein spikes are arranged into icosahedral particles of ∼70 nm diameter. There are 240 copies of the capsid protein arranged in a T = 4 lattice, with the surface glycoprotein spikes, consisting of E1 and E2, also forming a T = 4 structure as 80 trimers of heterodimers (Cheng et al., 1995; Zhang et al., 2002). The genome is a single strand of message sense RNA that possesses a type 0 5′ 7-methyl-GpppA cap (Hefti et al., 1975) and a 3′ poly(A) tail (Fig. 1). The genome has two ORFs, the second of which is expressed through production of a subgenomic mRNA from an internal promoter in the minus-strand RNA replication intermediate (Strauss et al., 1984). The second ORF encodes the structural proteins that function in the assembly of new virus particles and the attachment and entry to new cells. The first ORF is translated directly from genomic RNA and encodes the non-structural proteins required for RNA synthesis.
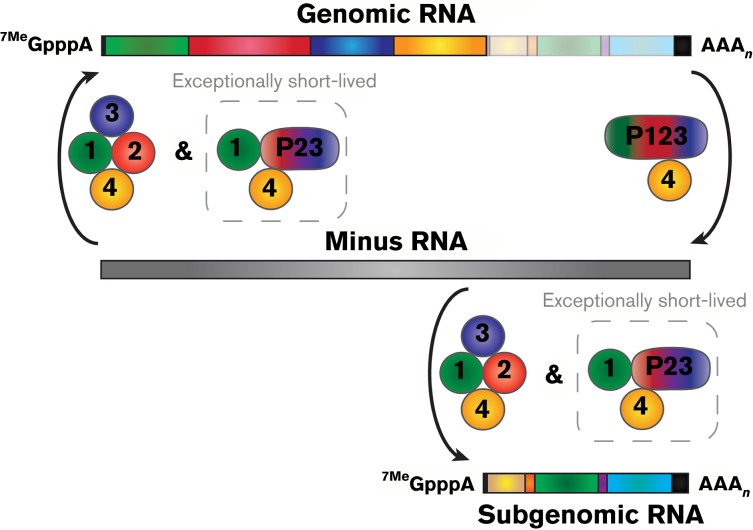
Fig. 1. Alphavirus RNA synthesis scheme. After infection the alphaviral genomic RNA is translated to yield the P1234 polyprotein, which following proteolytic cleavage via nsP2 forms the minus-strand replicase complex consisting of P123 and nsP4. Together, P123 and nsP4 initiate the synthesis of the minus-strand RNA, which serves as a template for synthesis of the genomic and subgenomic RNAs. Further processing of the P123 polyprotein, to nsP1 and P23, results in a switch from minus-strand synthesis to positive-strand synthesis, resulting in the production of genomic and subgenomic RNAs. The P23 polyprotein is exceptionally short-lived, with a presumed half-life of a few seconds. Proteolytic cleavage of P23 into nsP2 and nsP3 individually results in the fully cleaved viral replicase complex which, whilst capable of genomic and subgenomic RNA synthesis, strongly favours production of the subgenomic RNA.
Alphavirus replication
Alphavirus RNA synthesis requires all four viral non-structural proteins, individually and in the context of non-structural polyprotein precursors. The requirements for each of the individual non-structural proteins, and their respective functions, in RNA synthesis were initially identified by way of temperature-sensitive mutations (Hahn et al., 1989a, b; Sawicki & Sawicki, 1985). As described later in this review, subsequent characterizations have definitively assigned specific enzymic functions to the individual non-structural proteins through genetic and biochemical analyses.
Non-structural protein regulation of RNA synthesis
Translation of the alphaviral genomic RNA to yield the membrane-associated viral RNA synthetic complex results in the production of the non-structural polyprotein. The majority of the translation events, as much as ∼90 %, produce the P123 polyprotein; readthrough of the opal stop codon at the junction of nsP3 and nsP4 results in the production of the P1234 polyprotein (Li & Rice, 1993; Strauss et al., 1983). Interestingly, some isolates of SFV and O'nyong-nyong virus (ONNV) have an arginine codon in lieu of the opal stop codon, resulting in the constitutive production of P1234. The precise importance of this phenomenon is unclear, but it should be noted that the avirulent SFV isolate A7(74) possesses the opal codon, suggesting a potential role in pathogenesis (Tuittila et al., 2000). Regardless, the RNA synthetic properties of the alphaviral replicase complex are highly regulated in a sequential fashion at the level of polyprotein processing – a molecular event that leads to the individualization of the polyprotein domains into their mature protein species through the protease activity of nsP2. The P123 polyprotein, in itself, lacks any intrinsic RNA synthetic activity as the RNA-dependent RNA polymerase (RdRp; the nsP4 protein) is absent. Similarly, the P1234 form of the polyprotein is incapable of RNA synthesis until proteolytic processing releases the nsP4 component of the polyprotein (Shirako & Strauss, 1994). In complex, the P123 polyprotein and nsP4 exhibit RNA synthetic activity resulting in the synthesis of the minus-strand RNA (Fig. 1, right side). The P123/nsP4 RNA synthetic complex can be, depending on the time post-infection, short-lived; existing data indicate that an individual P123/nsP4 complex may synthesize as few as a single minus-strand RNA molecule before being further processed (Lemm & Rice, 1993; Lemm et al., 1994, 1998; Sawicki & Sawicki, 1980, 1994; Shirako & Strauss, 1994). Processing of the P123 polyprotein results in the liberation of the nsP1 component forming the nsP1/P23/nsP4 replicase complex. Cleavage of the P123 polyprotein into nsP1 and P23 effectively marks the functional transition between the synthesis of negative-sense to positive-sense RNAs (Lemm et al., 1994; Shirako & Strauss, 1994) (Fig. 1, left side and bottom). The inhibition of minus-strand RNA synthesis is a direct result of shutoff of viral non-structural protein expression and rapid processing of the non-structural polyprotein. Whilst polyprotein processing is clearly an impetus regulating viral RNA synthesis, it is unclear if this is a strict result of polyprotein cleavage or structural rearrangement of the non-structural proteins themselves leading to the switch from minus-strand to positive-sense RNA synthesis. Work by Gorchakov et al. (2008a) indicated that the P123/nsP4 complex was capable of synthesizing positive-sense RNAs, indicating that cleavage of the P123 polyprotein was not an absolute requirement for the transition from minus to positive-sense RNA synthesis.
Positive-sense RNA synthetic complexes must regulate the synthesis of genome and subgenome from the same minus-strand template. Mutations in nsP2 have implied nsP2 may act as a transcription factor associating with the subgenomic promoter to recruit the RNA synthetic complex (Sawicki et al., 1978; Suopanki et al., 1998). The template for positive-sense RNA synthesis is the minus-strand; however, evidence suggests that the template remains double-stranded (Kääriäinen & Ahola, 2002; Simmons & Strauss, 1972a, b). This mode of templating results in different dsRNA species depending on which RNA is being synthesized. When these dsRNAs were isolated by RNase treatment, three forms were released dubbed replicative forms (RFs). RFI, RFII and RFIII correspond to minus-strand RNA base-paired with full-length genome, the non-structural protein ORF and the subgenome, respectively (Simmons & Strauss, 1972a, b). As RFII does not correspond to an RNA with function outside of the intermediate, this is likely a paused genomic ternary complex which can, with some frequency, reactivate and finish synthesis (Wielgosz & Huang, 1997). The genome and subgenome forms both rapidly incorporate labelled nucleotides, whilst RFII does not, suggesting that complexes synthesizing genome and those synthesizing subgenome are both stable (Simmons & Strauss, 1972b). The regulation of synthesis of the two positive-sense RNAs has also been shown to depend on nsP4 itself, as distinct sites of nsP4 bind the two promoters (Li & Stollar, 2004, 2007; Li et al., 2010). Also, as stated above, nsP2 may be involved in regulating template recognition, leading to differential association of the template RNA with specific individual binding sites on nsP4. The nsP1/P23/nsP4 replication complex is capable of synthesizing both the genomic and subgenomic RNA species from the nascent minus-strand RNAs (Lemm et al., 1994; Shirako & Strauss, 1994). However, the P23 intermediate is exceptionally short lived and can only be detected following mutation of the 2/3 cleavage site (Hardy et al., 1990). Further proteolytic cleavage to yield the individual nsP1/nsP2/nsP3/nsP4 non-structural proteins, which together represent the main complex responsible for the synthesis of positive-sense viral RNAs, results in the synthesis of both the genomic and subgenomic RNAs, of which the subgenomic RNA is produced in excess of the viral genome (Lemm et al., 1994; Shirako & Strauss, 1994) (Fig. 1, bottom).
Whilst the proteolytic processing of the non-structural polyprotein is readily observable, the functional arrangement within the replicase complex remains poorly understood. Attempts have been made to elucidate the structural and functional arrangement of the non-structural proteins in RNA synthetic complexes. Pairwise interactions between each of the non-structural proteins has been tested by yeast two-hybrid screening by several groups without yielding conclusive results (Salonen et al., 2003). Co-immunoprecipitation experiments have found evidence for interactions between nsP1 and nsP4, and between nsP1 and nsP3, although the significance of these interactions for complex activities is not known (Lulla et al., 2008; Salonen et al., 2003; Zusinaite et al., 2007). Thus, despite large amounts of genetic data for functional interactions and structural data for several domains in the non-structural proteins, a model for the arrangements within the functioning complexes is lacking. The study of alphavirus RNA synthesis would benefit from a model for how and when the non-structural proteins interact, and what functions these interactions facilitate.
Host factor involvement in RNA synthesis and gene expression
Viral RNA synthesis has long been thought to involve host proteins, as host proteins were found to co-isolate with viral factors (Barton et al., 1991; Pardigon & Strauss, 1992). A number of host factors have been implicated to be involved; however, it is likely that many have yet to be identified. In most cases, host proteins have been demonstrated to interact with viral RNA or non-structural proteins; however, very few have been clearly shown to impact viral RNA synthesis. One such example is the La antigen which, whilst clearly shown to bind to SINV minus-strand RNA, has not yet been demonstrated to function during viral RNA synthesis (Pardigon & Strauss, 1996). In contrast, a study investigating the role of host proteins known to recognize dsRNA has demonstrated a role for RNase L in minus-strand synthesis shutoff facilitating the transition from early to late replication complexes (Sawicki et al., 2003).
Modern proteomic techniques have recently increased the identification of host factors associated with viral RNA synthetic complexes. Several groups have engineered viruses with tagged non-structural proteins for affinity isolation, and they have identified numerous host proteins that co-isolate with non-structural proteins by MS (Cristea et al., 2006; Cristea et al., 2010; Frolova et al., 2006). The capacity to interact with cellular factors represents a mechanism by which alphaviruses may exhibit different activities in the vertebrate and invertebrate host. Any roles these proteins play in RNA synthesis have only just begun to be determined. RNA interference (RNAi) co-knockdown of two of the proteins identified, G3BP1 and G3BP2, resulted in an increase in non-structural protein production; however, viral RNA synthesis was only minimally affected (Cristea et al., 2010). These interactions were found to be due to the nsP3 protein, which has been demonstrated to influence vector specificity in ONNV (Saxton-Shaw et al., 2013). Another study identified heterogeneous nuclear ribonucleoprotein (hnRNP) K as enriched in the membrane fraction of infected cells containing RNA synthetic complexes (Burnham et al., 2007). RNAi knockdowns of hnRNP K led to a modest reduction in viral infectivity, and immunoprecipitation experiments showed hnRNP K to interact with non-structural proteins and subgenomic RNA, but not genomic RNA. The role of hnRNP A1 has also been investigated and found to play a significant role in SINV RNA synthesis (Gui et al., 2010; Lin et al., 2009). Lin et al. (2009) found that RNAi knockdown of hnRNP A1 resulted in a significant decrease in viral RNA and protein production. RNA binding assays demonstrated direct binding of hnRNP A1 to genomic and subgenomic promoter probes, and in vitro RNA synthesis assays showed hnRNP A1 to be required for RNA synthesis. More recently, RNA-binding proteins were isolated from lysosomes within SFV-infected cells; these lysosomes contained intact RNA synthetic complexes in close proximity to a number of host proteins including PCBP 1, hnRNP M, hnRNP C and hnRNP K. RNAi knockdowns of all four proteins had an effect on alphavirus RNA replication and/or protein expression (Varjak et al., 2013).
These studies have examined a handful of the dozens of host proteins that co-isolate with alphavirus RNA synthetic complexes; thus, we are only just beginning to characterize the host factors involved in alphavirus RNA synthesis.
Site of replication
Alphaviral RNA synthesis is a membrane-associated process. Early studies found that RNA synthetic activity co-fractionated with membranes (Friedman et al., 1972; Gomatos et al., 1980). Alphaviruses and other RNA viruses induce rearrangement of host membranes into cytoplasmic structures known as type-1 cytopathic vacuoles (CPVs), which have long been suspected to be the sites of RNA synthesis (Grimley et al., 1968; Salonen et al., 2005). RNA synthesis occurs on the cytoplasmic side of these modified membrane structures, but is at least partially sequestered within invaginations dubbed spherules (Froshauer et al., 1988; Kujala et al., 2001). The formation of these spherules has been found to depend not only on non-structural proteins, but also active RNA synthesis (Frolova et al., 2010; Spuul et al., 2011). The specific protein determinants of spherule formation remain unknown and similarities of viral proteins to known curvature-inducing proteins (e.g. BAR superfamily proteins) have not been observed. Nonetheless, nsP3 has been shown to bind to cellular ampiphysin-1 and -2, indicating a role for nsP3 in the induction of membrane curvature (Neuvonen et al., 2011). Interestingly, the length of the RNA template appears to influence the spherule size during SFV replication, indicating that protein–protein interactions are not the only determinants of spherule assembly and formation (Kallio et al., 2013). Recent studies using fluorescence microscopy, immunolabelling and electron microscopy have further shown that the RNA synthetic complexes are assembled and form spherules at the host plasma membrane, and that these structures are then incorporated into CPVs through endocytosis (Frolova et al., 2010; Kujala et al., 2001; Spuul et al., 2010). These studies also found that the transport of endocytosed spherules and assembly of CPVs has discrete steps dependent on distinct cytoskeletal elements that could be disrupted with microtubule and actin/myosin inhibitors (Spuul et al., 2010). Nevertheless, pharmacological disruption of CPV assembly had limited or no effects on viral RNA synthesis (Frolova et al., 2010; Spuul et al., 2010). Thus, the mechanism of formation and movement of higher-order membrane structures containing viral replication complexes remains unclear. However, the cytoskeletal association of nsP3 suggests it plays a role in the formation of CPVs (Frolova et al., 2006; Gorchakov et al., 2008b).
Despite these advances, the specific role membranes play in RNA synthesis itself remains somewhat speculative. It is likely that the spherules provide protection of dsRNA replication intermediates from host cell detection and disruption. It has also been proposed that the membrane structures act as scaffolds and effectively increase the concentration of replication factors at the sites of synthesis. It may be that the spherule plays a role in stabilizing the complex once the polyprotein is cleaved and it is known that assembly of the complexes requires the polyprotein stage (Salonen et al., 2003). For enzymic functions, as discussed above, nsP1 was thought to require host lipids for capping activity, but these activities have since been seen in membrane-free nsP1 preparations (Tomar et al., 2011).
Functions of the individual viral non-structural proteins
nsP1
The ∼60 kDa alphavirus nsP1 protein primarily serves two functions during alphavirus replication. As depicted in Fig. 2, the N-terminal domain of nsP1 contains Rossman-like methyltransferase (MTase) motifs that direct the alphaviral capping reaction (Martin & McMillan, 2002; Rozanov et al., 1992; Schluckebier et al., 1995). Recent bioinformatic analysis of the alphaviral nsP1 protein and other viral capping proteins indicated that homologous MTase and guanylyltransferase (GTase) domains are present in members of the family Nodaviridae (Ahola & Karlin, 2015). Additionally, these analyses have revealed a number of residues common amongst the alphavirus nsP1 proteins which have yet to be functionally characterized. Following the N-terminal domain are tandem features that confer association of the nsP1 protein to host membranes. An amphipathic helix and palmitoylation both act to anchor the nsP1 protein, and nsP1-containing non-structural polyproteins, to the host membrane (Ahola et al., 1999, 2000; Laakkonen et al., 1996; Lampio et al., 2000; Peränen et al., 1995; Spuul et al., 2007), although palmitoylation is not essential for the enzymic activities of the nsP1 protein (Laakkonen et al., 1994; Mi & Stollar, 1991). Indeed, purified SINV nsP1 has been shown to be functional in the absence of lipids, indicating that membrane association is not necessary for nsP1 enzymic function (Tomar et al., 2011). In contrast, the enzymic activities of purified SFV nsP1 were found to require lipids (Ahola et al., 1999). Therefore, the requirement for lipid association in regard to nsP1 enzymic activities seems to vary amongst members of the genus. Nevertheless, it should be noted that depalmitoylation mutants exhibit diminished pathogenesis in mice despite lacking a phenotype in tissue culture models of infection (Ahola et al., 2000). This may be in part due to the alternative functions of nsP1 during alphavirus infection including membrane and cytoskeletal rearrangement, the development of cell filopodia, and cell-to-cell transmission of alphaviruses (Karo-Astover et al., 2010; Laakkonen et al., 1998; Martinez et al., 2014). These additional functions of nsP1, whilst clearly important to viral infection, are beyond the scope of this particular review.
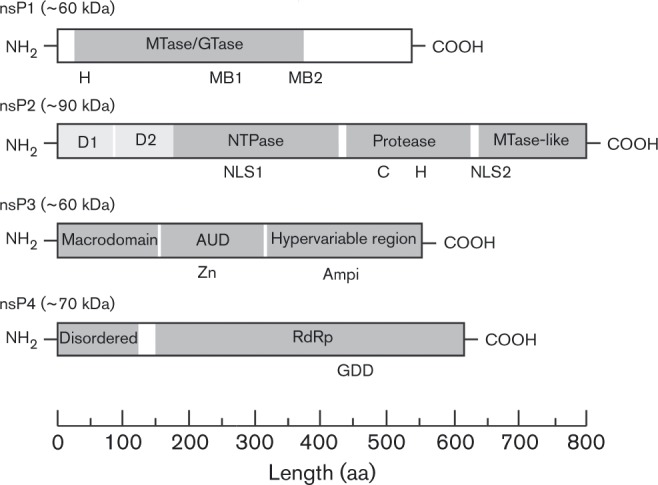
Fig. 2. Alphavirus non-structural proteins. Shown are domain organization of the individual alphaviral non-structural proteins nsP1, nsP2, nsP3 and nsP4. Recognized domains, in terms of either structural or genetic evidence, are indicated as grey boxes in their relative position in their respective proteins; specific features, as indicated on their respective proteins, are briefly listed below. nsP1: H designates the histidine residue determined to bind covalently to the 7MeGMP moiety; MB1 and MB2 indicate the sites of the membrane-binding amphipathic helix and the palmitoylation site of nsP1, respectively. MTase, methyltransferase; GTase, guanylyltransferase. nsP2: NLS1 and NLS2 indicate the sites of canonical nuclear localization sites; the cysteine and histidine residues of the protease active site are indicated with a C and H, respectively. NTPase, nucleoside triphosphatase. nsP3: the location of the zinc ion coordination site is denoted with Zn; the presence of ampiphysin interaction sites is denoted with Ampi. AUD, alphavirus unique domain. nsP4: the functional GDD catalytic triad of RdRp is shown.
During replication nsP1 is responsible for the addition of the 5′ cap to viral genomic and subgenomic RNAs after the preparation of the nascent RNA by the triphosphatase activity of nsP2 (Vasiljeva et al., 2000). The capping of the positive-sense viral RNAs have been ascribed to nsP1, namely by way of the MTase and GTase-like activities of nsP1. The MTase and GTase activities were first identified by biochemical assays and genetic work (Cross, 1983; Mi & Stollar, 1990; Scheidel & Stollar, 1991). Using infected cell lysates, or lysates from cells with individually expressed nsP1, Ahola & Kääriäinen (1995) found the GTase reaction catalysed by nsP1 to be distinct from the typical eukaryotic GTase reaction. nsP1 was found to require S-adenosylmethionine to link to GTP and acid hydrolysis of nsP1-GMP yielded 7MeGMP and not GMP. The GT activity of nsP1 is dependent on successful MTase activity, as GTase activity is not observed in the absence of a methyl donor or in the presence of pre-methylated GMP (Ahola & Kääriäinen, 1995). Together these findings along with mutational evidence indicate that the MTase activity of nsP1 occurs prior to the transfer to the 5′ end of the substrate RNA (Ahola & Kääriäinen, 1995). This phenomenon is in stark contrast to the eukaryotic capping mechanism in which methylation occurs after the transfer of the guanylate moiety to the substrate RNA. Whilst nsP1 mutants lacking MTase activity or GTase activity are non-viable, recent evidence has indicated that the alphavirus capping reaction is not absolute and non-capped positive-sense viral RNAs are generated during infection (Ahola et al., 1997; Sokoloski et al., 2015; Wang et al., 1996). The precise importance of the non-capped viral RNAs during infection is unclear; however, the capping activities of nsP1 are clearly important at the organismal level during infection (Cruz et al., 2010; Sokoloski et al., 2015; Stoermer Burrack et al., 2014).
Despite a lack of specific molecular understanding, nsP1 is known to be important for minus-strand RNA synthesis (Hahn et al., 1989b; Wang et al., 1991). Numerous studies have identified suppressor mutations in nsP1 for minus-strand deficient mutants of nsP4 (Fata et al., 2002b; Rupp et al., 2011; Shirako et al., 2003; Shirako & Strauss, 1998). Indeed, mutations have been shown to negatively affect minus-strand synthesis without negatively impacting the enzymic properties of nsP1 (Lulla et al., 2008). The mechanism of nsP1 function in this regard remains elusive, although interactions within replicative complexes are likely critical. Studies attempting to determine these interactions using co-immunoprecipitation have found a strong interaction between nsP1 and nsP4 as well as between nsP1 and nsP3 (Salonen et al., 2003; Zusinaite et al., 2007), and it is possible that more fragile interactions with nsP2 also occur.
nsP2
The ∼90 kDa alphavirus nsP2 protein serves multiple important functions during virus infection. nsP2 was initially described as consisting of two domains, an N-terminal helicase domain, which also exhibits nucleoside triphosphatase (NTPase) activity and a C-terminal protease domain (Fig. 2). The C-terminal protease domain shows similarity to known cathepsins; however, the structure is a distinct and novel cysteine protease fold (Russo et al., 2006). Crystallographic analyses of the C-terminal protease domain have revealed a third domain resembling a MTase fold (Russo et al., 2006). The MTase-like domain shows structural similarity to the FtsJ MTase; however, the nsP2 MT-like domain lacks the active-site residues responsible for enzymic activity, indicating that the domain likely does not exhibit enzymic activity. Mutational analysis has further indicated the putative presence of at least two additional domains in the amino-proximal region of nsP2 (Atasheva et al., 2007; Frolov et al., 1999). The first putative N-terminal domain has exhibited cofactor-like properties regarding the activity of the nsP2 protease domain. The second putative domain appears to function in promoter selection, as this domain has been the site of suppressor mutations developed in response to promoter site mutations. Further characterization is warranted for each of these putative domains.
In the context of viral replication, nsP2 exhibits three important functions, acting as a helicase, a triphosphatase and a protease. In addition to these roles, nsP2 is intimately involved in the shutoff of host macromolecular synthesis; this particular topic, whilst fascinating and a vital component of alphavirus infection, is beyond the scope of this review (Frolov et al., 1999; Garmashova et al., 2006, 2007; Gorchakov et al., 2005; Kim et al., 2004).
NTPase/RNA triphosphatase (RTPase) domain functions
nsP2 functions as a helicase to unwind RNA secondary structures formed during viral RNA replication. The helicase/NTPase motifs responsible for this activity were first identified by bioinformatic analyses and later confirmed by mutational and recombinant studies of nsP2 (Gomatos et al., 1980; Gomez de Cedrón et al., 1999; Gorbalenya et al., 1988). The helicase activity of nsP2 is dependent on the NTPase activity of the N-terminal domain, as mutations in the Walker A motif ablated helicase activity in both recombinant and tissue culture models (Rikkonen et al., 1994). Recent data has indicated that the helicase activity of nsP2 likely acts in coordination with the polymerase activity of nsP4 and is dependent on the full-length protein, and that a severable helicase domain is not present (Das et al., 2014). Collectively, these data indicate that the helicase activity of nsP2 is essential for viability, presumably due to its function during viral replication (Rikkonen, 1996).
A second activity of nsP2 that specifically pertains to viral replication is the RTPase activity associated with the N-terminal domain. The RTPase activity of nsP2 is responsible for the removal of the γ-phosphate from the 5′ end of nascent positive-sense RNAs to yield a diphosphate moiety at the 5′ terminus, enabling the RNA to act as a substrate for the nsP1-mediated capping reaction (Vasiljeva et al., 2000). The RTPase activity appears to be dependent on the same active site as the NTPase activity, as the inhibition of NTPase activity by mutation or chemical inhibition results in the loss of RTPase activity.
Protease domain functions
The C-terminal domain of nsP2 was genetically identified as the protease responsible for the processing of the non-structural polyprotein (Hahn et al., 1989b; Hardy & Strauss, 1989). This function is absolutely essential for virus replication and has been shown to be functionally discrete from the nsP2 functions described above (Lulla et al., 2006). Nevertheless, the activities of the protease are modulated by other domains of nsP2 and nsP2 containing polyproteins, so the domain is not entirely functionally independent, but is essential for processing of the non-structural polyprotein species (Vasiljeva et al., 2003). Several mechanisms combine to regulate the processing of the non-structural polyproteins. In SINV, each junction has been found to be cleavable in trans and under some conditions the 1/2 and 3/4 junctions are also cleavable in cis (de Groot et al., 1990; Hardy & Strauss, 1989). The cleavage of the 1/2 junction in cis is readily observed in P12 polyproteins and this observation was also made in SFV (Vasiljeva et al., 2003). In P123, however, cis cleavage of 1/2 appeared to be inefficient, at least in SINV (de Groot et al., 1990). The efficiency of cleavage in cis or in trans gives rise to different activities over the infectious cycle. Early in infection the concentration of non-structural proteins is low and cis cleavage is thus favoured, whilst later when the concentration of protease is high trans cleavage increases. In addition to cis/trans differences the cleavage efficiency of the protease is different for each of the three junctions and these efficiencies are altered by the polyprotein context of nsP2. Specifically, within a polyprotein containing nsP1 the ability of the protease to cleave the 2/3 junction is poor and within a polyprotein that lacks nsP3 the ability to cleave the 3/4 junction is reduced (de Groot et al., 1990; Shirako & Strauss, 1990). The result of these activity shifts is a preference for cleaving the 3/4 junction early in infection and a preference for cleaving the 2/3 junction later in infection.
These cis/trans and preference shifts of the protease result in specific cleavage timing. When P123 and P1234 are translated at the start of infection cleavage in cis of the 3/4 junction produces the initial non-structural protein species, P123 and nsP4. In this context the 1/2 junction must be cleaved for processing to continue, and thus the rate of this step provides key temporal regulation of processing. Strauss and Strauss argue that as the concentration of P123 rises trans cleavage becomes favourable (Strauss & Strauss, 1994). Vasiljeva et al. (2003) argue that slow 1/2 cleavage in cis, with ∼30 min half-life, produces the delay in processing. Nevertheless, whilst cleavage of the 1/2 junction in cis has been observed for P12 in both SINV and SFV there is evidence to suggest this cleavage is inhibited in the context of P123 (Hardy & Strauss, 1989). Taken together these data indicate that processing at the 1/2 junction is temporally regulated, and that the half-life of P123 varies during infection depending on whether cis or trans cleavage is favoured.
Once the 1/2 junction is cleaved the protease is activated for trans cleavage of the 2/3 junction, both in SFV and SINV (Merits et al., 2001; Shirako & Strauss, 1990). As the concentration of 2/3 competent protease rises, the cleavage of the 2/3 junction becomes more rapid than that of the other junctions. Structural analysis of a nsP2/nsP3 fragment has indicated that the 2/3 junction is distant to the protease active site of nsP2 indicating that cleavage of the 2/3 junction must occur in trans (Shin et al., 2012). Thus, newly translated non-structural proteins are cleaved into P12 and P34. At this later stage of infection the concentration of protease competent for cleavage of the 1/2 and 2/3 junctions is high, and thus protease competent for cleavage of the 3/4 junction (P23, P123, P1234) is rapidly processed away. This was found to result in P34 becoming a stable polyprotein species later in SINV infection (de Groot et al., 1990; Hardy & Strauss, 1989). However, the stability of P34 is largely a phenomenon unique to SINV as P34 of SFV is always actively processed.
A study regarding the cleavage of artificial substrates by the purified protease domains of nsP2 from SFV and VEEV did not indicate significant differences in substrate specificity, and hence differences in protease activities between the alphaviruses was examined (Zhang et al., 2009). For both SFV and VEEV the cleavage of the 1/2 and 3/4 junctions was found to be highly efficient; whilst the cleavage of the 2/3 junction was poor. The poor cleavage of the 2/3 junction observed with the purified C-terminal protease domain of nsP2 is consistent with previous results indicating the entire nsP2 domain is required for cleavage of the 2/3 junction (Vasiljeva et al., 2003). Further examination of the requirements for cleavage at this junction have shown the need for precise assembly of the cleavage complex (Lulla et al., 2012). The macrodomain of nsP3 and the N terminus of nsP2 are necessary for substrate positioning in order for cleavage of the 2/3 junction to occur. (Lulla et al., 2013)
Examination of alphaviral polyprotein cleavage sites reveals commonalities across the genus. These amino acid positions of the polyprotein cleavage sites are described below using the Schechter and Berger nomenclature for protease site residues (Schechter & Berger, 1967). Using this nomenclature, amino acid residues surrounding the peptide bond cleaved are designate P1 on the N-terminal side and P1′ on the C-terminal side. It should be noted that this nomenclature has similarities to that used for the naming of alphavirus polyproteins and to avoid confusion italics have been used for designation of residues around protease cleavage sites. Characterizations of the plasticity of nsP2 cleavage sites indicated that the context in which the cleavage site is presented influences the capacity of the particular site to be cleaved (Lulla et al., 2013). The P2 position of the recognition motif of all three sites (i.e. the 1/2, 2/3 and 3/4 junctions) is a glycine and this is conserved across the genus. Based on amino acid conservation or differences, Strauss and Strauss argue that positions P1, P2 and P3 are the most important for protease recognition of a site, and P1′, P2′ and P3′ are tolerant of different residues (Strauss & Strauss, 1994). The cleavage site requirements were tested biochemically using SFV nsP2pro, and P1, P2 and P3 were found to be important, as well as P4 and P1′ (Lulla et al., 2006). Tests using synthetic substrates and purified protease domains from SINV, SFV and VEEV found that VEEV nsP2pro recognized and cleaved SFV's 1/2 junction, but no other cross-species recognition was identified in that study (Zhang et al., 2009)
nsP2 may also perform functions in RNA synthesis beyond its roles as protease, RTPase, and helicase. It has been proposed that nsP2 acts as a transcription factor for subgenome synthesis by binding to the subgenomic promoter (Hahn et al., 1989a; Sawicki et al., 1978; Suopanki et al., 1998). This was suggested by the temperature-sensitive mutation ts4, which mapped to nsP2 and was found to decrease subgenomic RNA synthetic activity specifically. Direct binding of nsP2 to the subgenomic promoter on the minus-strand template has not been shown and ts4 also alters the protease activity of nsP2. However, the shift to non-permissive temperature was found to dissociate ts4–nsP2 from replicative complexes, possibly supporting its role as a transcription factor (Suopanki et al., 1998). Additional temperature-sensitive mutations were characterized in nsP2 that also affect subgenome synthesis. Whilst these mutations again decreased polyprotein processing, subgenome synthesis was still inhibited upon shift to the non-permissive temperature, even once complexes of cleaved non-structural proteins had formed (Sawicki & Sawicki, 1993). Other mutations in nsP2 have been characterized that alter switching from minus-strand to positive-sense RNA synthesis; however, these likely are due to alterations in nsP2’s protease activity or perhaps in some other conformational conversion within the non-structural protein complex (Dé et al., 1996) or its alteration of host effects including RNase L activation (Sawicki et al., 2006).
nsP3
Historically, the functional importance of ∼60 kDa nsP3 has been less clear and has been the subject of extensive examination. The nsP3 protein is clearly necessary for RNA synthesis; mutations in nsP3 have been shown to exhibit defects in the initiation of minus-strand synthesis or subgenomic RNA synthesis (LaStarza et al., 1994b; Rupp et al., 2011; Wang et al., 1994). Nonetheless, despite the functions and activities of nsP3 described below, the precise role(s) of nsP3 during replication is unknown.
As shown in Fig. 2, the alphavirus nsP3 protein has three recognized domains: the macrodomain, the alphavirus unique domain (AUD) and the hypervariable region. The N-terminal portion of nsP3 is conserved amongst the alphaviruses and contains a macrodomain with detectable homologues across the domains of life (Allen et al., 2003; Park & Griffin, 2009; Pehrson & Fuji, 1998). The alphavirus macrodomain exhibits both nucleic acid binding and phosphatase capabilities (Malet et al., 2009). The macrodomains of CHIKV and VEEV were found to bind to DNA, RNA and polyADP-ribose in addition to exhibiting adenosine diphosphoribose 1′-phosphate phosphatase activity. Interestingly, this finding is in contrast to the severe acute respiratory syndrome coronavirus macrodomain which, whilst capable of binding ADP-containing molecules, exhibited low phosphatase activity indicating that all viral macrodomains may not be functionally identical (Egloff et al., 2006). The precise role of polyADP-ribose binding in the context of viral replication remains unclear. Moreover, mutational analysis has indicated that polyADP-ribose binding is not the only function of the alphaviral macrodomain and evidence exists that the macrodomain is involved in one, or more, host protein interactions (Park & Griffin, 2009).
The AUD is located within the central portion of nsP3 – a region that shares a strong sequence homology across the alphaviruses. To date, the most well-defined structural data for the alphavirus replication complexes is at the level of the nsP2/nsP3 junction. The crystal structure of the nsP2/nsP3 fragment revealed the presence of a previously unknown zinc coordination site within the AUD (Shin et al., 2012). Genetic manipulations within the AUD have resulted in defects in minus-strand and subgenomic RNA synthesis, polyprotein processing, and neurovirulence (Dé et al., 2003; LaStarza et al., 1994a; Tuittila & Hinkkanen, 2003), but the mechanism of these defects has yet to be determined.
The C-terminal domain of nsP3 is characterized as being hypervariable, exhibiting poor conservation between alphaviruses in terms of length and sequence composition. Comparisons of the hypervariable regions across isolates of individual alphavirus species have indicated conservation and selection for certain elements of the hypervariable domain indicating an, as of yet unknown, advantage to their evolutionary retention (Aaskov et al., 2011; Oberste et al., 1996). Mutational analyses, including deletion and duplication studies, have indicated that the hypervariable region is largely tolerant of significant changes in this domain in tissue culture models; however, these mutations often result in the attenuation of virulence in mice (Davis et al., 1989; Galbraith et al., 2006; LaStarza et al., 1994a; Tuittila & Hinkkanen, 2003). nsP3 is known to be multiply phosphorylated and has been observed to exist in various phosphorylated states during infection (Li et al., 1990; Peränen et al., 1988). The lack of sequence conservation ultimately results in the formation of grossly different phosphorylation states between alphavirus species (Davis et al., 1989; LaStarza et al., 1994a; Vihinen et al., 2001). In SFV, mutation of the phosphorylation sites exhibited mild effects on viral growth kinetics and RNA synthesis in culture, and demonstrated attenuation of pathogenicity in mouse models (Vihinen et al., 2001). Work by Foy et al. (2013b) demonstrated that the phosphorylation state of the hypervariable domain of VEEV nsP3 did not affect virus replication in vertebrate cells, but was important for replication in mosquito cells. Additionally, whilst the virus could tolerate mutations in this domain when replicating in BHK-21 cells (a highly permissive cell line), these same mutations were not tolerated in other cell lines. Additionally it has been shown that the hypervariable domain is responsible for the formation of virus-species specific complexes in infected cells (Foy et al., 2013a). Together these data suggest that the hypervariable domain plays an important role in the virus–host interaction and may be a significant determinant of pathogenesis through interactions with cell-type-specific factors.
Several studies have sought directly to address the functional relationships of nsP3 with host factors during viral infection. Immunoprecipitation of SINV nsP3 from infected cells has revealed numerous interactions with host factors, most notably perhaps with the cellular G3BP proteins (Cristea et al., 2006, 2010; Frolova et al., 2006; Scholte et al., 2015). The association of G3BP1 and G3BP2 reduces the translation of viral RNAs early during infection indicating a potential role for the G3BPs in enhancing replication indirectly. At this time it is not clear that the identified interactions of nsP3 with host factors influence viral RNA synthesis or whether they are indicative of another nsP3 function that regulates the host cell environment. Work by Fros et al. (2012), demonstrated that the interactions of CHIKV nsP3 with the host G3BP proteins prevents further assembly of stress granules during viral infection.
nsP4
Whilst all of the non-structural proteins are involved in alphavirus RNA synthesis, the ∼70 kDa nsP4 protein is solely responsible for the RNA synthetic properties of the viral replicase complex. The nsP4 protein contains the core RdRp domain and motifs. The ∼100 N-terminal residues are unique the alphaviral RdRp, whilst the remaining ∼500 residues exhibit, as predicted by alignment, the typical RdRp structure with fingers, palm containing the GDD active site and thumb domains (O‘Reilly & Kao, 1998; Rubach et al., 2009; Tomar et al., 2006) (Fig. 2). As described earlier, many alphavirus species produce significantly less nsP4 than the other non-structural proteins. Additionally, nsP4 is targeted for degradation by the N-end rule pathway and is presumably stabilized via incorporation into the replicase complex (de Groot et al., 1991).
Due to the presence of the GDD motif and mapping of RNA synthesis-defective mutants, the nsP4 protein was proposed as the alphavirus RdRp (Hahn et al., 1989a). Recombinant expression of full-length nsP4 has historically proven exceptionally difficult. N-terminal truncation mutants of nsP4 have demonstrated terminal adenylyltransferase (TATase) activity, indicating a potential role in polyadenylation, but were ultimately found to lack de novo copying activity (Tomar et al., 2006). The TATase activity observed with the N-terminal truncated nsP4 was independent of other viral factors; however, a predilection towards viral RNA substrates was observed. Whether or not this is the genuine mechanism for viral polyadenylation is still unknown; evidence indicating the presence of a 5′ poly(U) tract on the minus-strand RNA and the ability of truncated alphavirus genomic RNAs to be polyadenylated during infection highlight the complexity of viral polyadenylation (Hill et al., 1997; Raju et al., 1999; Sawicki & Gomatos, 1976). Purification of full-length nsP4 has been accomplished using an N-terminal SUMO tag method. Full-length recombinant nsP4 exhibited TATase activity and was capable of de novo RNA synthetic activity only after the addition of the other viral non-structural proteins supplied from mammalian cell membrane fractions (Rubach et al., 2009). Thus, despite nsP4 being the sole viral protein with RdRp activity, viral replication is the sum of coordinated non-structural protein activity.
Whilst functional viral RNA synthesis is dependent on the formation of the viral replicase complex consisting of, in one form or another, the viral non-structural proteins, mutational analyses have indicated that determinants for RNA synthesis exist within nsP4. For instance, mutation of a conserved arginine at 183 resulted in a specific minus-strand defect in chicken cells (Fata et al., 2002a). Genetic evidence suggests nsP4’s absolutely conserved N-terminal tyrosine interacts with nsP1 for minus-strand synthesis (Shirako & Strauss, 1998) and different mutations in the predicted disordered N-terminal domain resulted in either minus-strand or positive-sense RNA defects, suggesting roles in each activity (Rupp et al., 2011). Stollar and colleagues have determined several determinants of promoter binding in nsP4 as residues of nsP4 were found to contact the subgenomic and genomic promoters as determined by RNA cross-linking experiments (Li & Stollar, 2004, 2007). Mutations of the regions identified as binding specifically to either the genomic or subgenomic promoters were found to abrogate the corresponding RNA synthetic activity (Li & Stollar, 2004, 2007; Li et al., 2010).
Cis-acting elements
Viral RNA synthesis requires the appropriate recognition of sequence/structure elements in the template RNAs by the viral RNA synthetic complex. For alphaviruses these cis-acting elements predominantly correspond to UTRs in the viral genome. There are three UTRs in the alphavirus genome: one at the 5′ end, one at the 3′ end, and one at the junction region between the non-structural and structural ORFs. Additionally, elements exist in coding regions of the genome and subgenome that function in the synthesis of viral RNA, viral protein expression and viral genome packaging (Fig. 3). These elements are conserved to varying degrees across the genus, and were first identified by sequence comparisons of the three UTRs and adjacent coding sequence (Ou et al., 1981, 1982a); their role(s) in alphavirus replication continues to be clarified and refined.
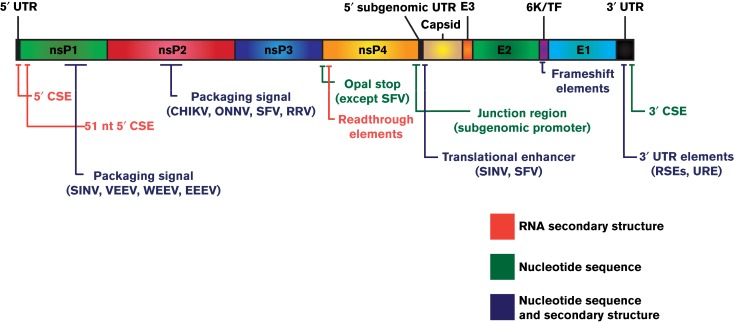
Fig. 3. Alphavirus conserved RNA sequence elements (CSEs): schematic of the alphaviral genomic RNA with conserved RNA elements indicated in their relative positions on the genomic RNA. The nature of the conserved elements is colour coded to indicate whether RNA secondary structure, primary nucleotide sequence or both is conserved. Exceptions to either element location or presence are indicated. RRV, Ross river virus; RSE, repeat sequence element; URE, U-rich element.
5′ Elements
The 5′ end of the genome, or its complement in the 3′ end of the minus-strand, contains two conserved sequence elements (CSEs): there is one in the 60 nt 5′ UTR and the 51 nt CSE within nsP1 coding region (Fig. 3). In the genomic strand these elements also possess defined RNA secondary structures. The 5′ UTR contains a conserved stem–loop critical for its function in RNA synthesis (Niesters & Strauss, 1990a). The modelled structure of this stem–loop has been confirmed and further refined by chemical analyses (Nickens & Hardy, 2008). The complementary sequence at the 3′ end of the minus-strand is also predicted to be structured and this structure functions in genomic RNA synthesis (Frolov et al., 2001; Niesters & Strauss, 1990a). The stability of this structure and the ability of the polymerase to access the promoter is thought to be important for the regulation and initiation of RNA synthesis (Shirako & Strauss, 1998). In close proximity to, but distinct from the 5′ CSE, is the 51 nt CSE in the nsP1-coding region that forms two stem–loops. Mutational analyses of the 51 nt CSE suggested that both the sequence and structure of the loop may be important for the CSE's function as a transcriptional enhancer (Niesters & Strauss, 1990b).
Whilst it has been known for some time that RNA synthesis, templated by the genomic RNA, initiates at its 3′ end, 5′ elements must be involved as the subgenome, which contains the same 3′ elements, is not copied into a minus-strand copy. Circularization of template RNAs for initiation of copying has been identified as a strategy by which other positive-sense RNA viruses, such as poliovirus and dengue virus, regulate their RNA synthesis and translation (Herold & Andino, 2001; You & Padmanabhan, 1999). Circularization of the alphavirus genome has been investigated and observed in vitro; however, those studies did not define the sequences involved or the functional consequence of circularization (Frey et al., 1979). Studies using chimeric SINV/SFV RNA templates found that the 51 nt CSE is likely an enhancer element for RNA synthesis, whilst the 5′ UTR and the 3′ elements from the same virus species are necessary for RNA replication. Characterizations of the 5′ UTR of VEEV, and in particular the 51 nt CSE of VEEV, have similarly revealed a vital role in the regulation of viral RNA synthesis (Kulasegaran-Shylini et al., 2009a, b; Michel et al., 2007). This shows that the 5′ UTR is essential for minus-strand RNA synthesis initiating at the 3′ end of the genome, suggesting circularization mediated in part by the 5′ UTR element is necessary for minus-strand synthesis (Frolov et al., 2001).
Several studies have found that changes in the cis elements result in host species-specific phenotypes, suggestive of host factor interactions with the elements (Fayzulin & Frolov, 2004; Gorchakov et al., 2004; Niesters & Strauss, 1990a, b). Studies comparing replication between vertebrate and arthropod systems found that mutations to the 51 nt CSE had a greater effect on replication in the mosquito host than the mammalian host (Fayzulin & Frolov, 2004). Additionally, passage of the 51 nt CSE mutants resulted in second site suppressor mutations in nsP2, nsP3 and the 5′ UTR, suggestive of functional interactions between the 51 nt CSE and these factors. Further study of chimeras of 5′ UTR and 51 nt CSE from SINV and SFV found adaptive mutations that relieved inhibition RNA synthesis through AU additions at the 5′ end of the genome (Gorchakov et al., 2004). However, some of the adaptive AU additions functioned in vertebrate but not mosquito cells, suggesting host-specific factor involvement. Thus, taken together, these studies begin to illuminate the complexities of the interactions between viral/host factors and the 5′ cis-acting elements during RNA synthesis.
3′ Elements
The 3′ end of the genome contains the largest UTR, in the range of several hundred nucleotides depending on the virus species (Fig. 3). The final 19–20 nt before the poly(A) tail comprise the 3′ CSE, which is highly conserved in sequence across the genus. As the 3′ end of the genome represents the initiation site for minus-strand synthesis, these elements are presumed to contain the core minus-strand promoter. Nevertheless, recent studies have reported that in the absence of viral RNA cellular RNAs may be utilized as a template for the synthesis of dsRNA (Nikonov et al., 2013). Mutational analysis of the 3′ UTR found alterations throughout that reduced RNA synthesis, with residues in the CSE found to be most critical and other portions being more tolerant of changes (Kuhn et al., 1990). The functions of the 3′ elements in minus-strand synthesis were tested in vitro by introducing point mutations or small deletions into the 3′ CSE and poly(A) tail of synthetic templates (Hardy & Rice, 2005). The 3′ 13 nt of the CSE and a poly(A) tail of at least 11 residues were found to be most critical for minus-strand synthesis. These analyses were extended to determine the initiation site of minus-strand synthesis (Hardy, 2006). WT templates resulted in initiation at the 3′ end of the CSE, whilst alteration of the CSE or the poly(A) tail in many cases shifted the initiation site to within the poly(A) tail. These data suggest that polyadenylation of alphavirus plus-strand RNAs is not templated in contrast to previous reports (Sawicki & Gomatos, 1976). The observation that nsP4 possesses a TATase activity suggests that a poly(A) tract can be added in a template-independent manner (Rubach et al., 2009; Tomar et al., 2006). However, it is also possible that both a templated and non-templated mechanism of polyadenylation and terminal addition are at play during an alphavirus infection, allowing repair and restoration of infectivity of damaged or defective viral genomes. Further work by Raju and colleagues has found that, despite the essential nature of the 3′ UTR, genomes lacking significant portions of this element are capable of producing progeny by a novel repair mechanism (George & Raju, 2000; James et al., 2007; Raju et al., 1999). Synthetic genomes with a set of deletions to the 3′ CSE, UTR and/or poly(A) tail were introduced into cells, and viable progeny were recovered from many of these. Sequencing revealed recovered genomes had regenerated 3′ elements of non-WT AU-rich sequences as well as a poly(A) tail and these viruses were capable of establishing infection in neonatal mice. It is currently unclear precisely how these additions to the genomic RNA occur, but nsP4-mediated terminal addition combined with slippage during template copying is a likely mechanism.
Junction region elements
Between the non-structural and structural ORFs is a third UTR, or junction region (Fig. 3). The promoter for subgenomic synthesis, active solely in the minus-strand, lies partially in this region and partially in the nsP4-coding region. The 5′ UTR of the subgenomic RNA is also derived from this sequence. The minimal promoter for subgenome synthesis was identified by sequence comparisons and confirmed by studies using defective interfering SINV RNAs as a backbone for analysing the junction region (Levis et al., 1990; Ou et al., 1982a). Levis and colleagues found that truncation of the promoter beyond nucleotides − 19 to +5 from the transcription start site abolished activity. Additional enhancer elements and contextual effect were identified by studies using dual-promoter viruses (Raju & Huang, 1991). The full, optimal promoter was mapped to − 98 to +14 and the specific requirements within this region were identified (Wielgosz et al., 2001). Similar to the 5′ and 3′ elements, some these requirements were distinct between vertebrate and arthropod host cells. Additionally, the functional conservation of these elements was tested by assaying the capacity of the SINV replicase to utilize a suite of different alphavirus junction regions and most functioned well, indicating a lack of divergence in subgenome initiation (Hertz & Huang, 1992).
Other functions of genomic RNA elements
Cis-acting elements also perform functions in the evasion of host antiviral responses, translation of alphavirus RNAs, packaging of genome into progeny virions and evasion of RNA degradation by host cells. Many of these cis-acting elements are predicted or have been shown to have secondary structures. Nevertheless, in several instances, e.g. the U-rich element (URE) of SINV, significant secondary RNA structures are absent, indicating a role for the primary sequence of the element. The functions of these cis-acting elements are reported below and their relative positions are shown in Fig. 3.
Recently, secondary structures present in the 5′ UTR have been shown to prevent the recognition of the viral RNAs (Hyde et al., 2014). These RNA structures were identified as a pathogenicity determinant as they served to mask the viral type 0 cap structure, which differs from the cellular type 1 cap structure and is recognized by the host protein IFIT1 (Daffis et al., 2010; Reynaud et al., 2015). It has been known for some time that a polymorphism at position 5 of the SINV genome and position 3 of the VEEV genome that increases the base-paired length of the 5′ stem–loop correlates with increased pathogenicity of these viruses (Kinney et al., 1993; Kuhn et al., 1992). Increasing the base-pairing at the very 5′ end of the genome appears to ‘hide’ the cap from detection by IFIT1, leading to avoidance of host detection; the subsequent activation of innate immunity results in enhanced translation and replication of the alphaviral genome in vertebrate cells.
For many alphaviruses, including VEEV and SINV, the presence of a stem–loop RNA secondary structure immediately adjacent to the opal stop codon has been reported to influence readthrough, thereby generating the P1234 polyprotein(Firth et al., 2011). Indeed, mutational analysis indicated that the presence of the stem–loop structure enhanced readthrough as much as 10-fold (Firth et al., 2011). A similar more complex arrangement of secondary structures including putative pseudoknot-like structures can be observed proximal to the frameshift site of the 6K/TF structural protein (Chung et al., 2010; Firth et al., 2008, 2011). For some alphaviruses, such as SINV, VEEV and Barmah forest virus; a stem–loop structure 3′ adjacent to the UUUUUUA (U6A) motif enhances the ribosomal frameshifting, resulting in the production of TF. Nevertheless, this structural element is not ubiquitous amongst the genus as SFV lacks an identifiable local secondary structure proximal to the U6A motif (Chung et al., 2010).
The translation of the structural protein ORF in the subgenomic RNA is enhanced by a stem–loop in the capsid-coding region known as the translational enhancer (Frolov & Schlesinger, 1996). Later characterization of the translational enhancer revealed the presence of a secondary structure downstream of the initiating methionine of the SINV subgenomic RNA. Functional characterization of the downstream loop (DLP) indicated that it imparts resistance to the shutoff of host translation mediated by the phosphorylation of eukaryotic initiation factor eIF2α (Ventoso et al., 2006). To date, the existence of a DLP element has been reported for SINV and SFV; however, this feature is apparently absent from other members of the genus, notably VEEV and CHIKV. The mechanism by which the DLP enhances translation is not clear, but it bestows resistance to low eIF2 levels in infected vertebrate cells allowing translation to occur in cells in which the RNA-dependent protein kinase is active, thus overcoming a primary antiviral response by the cell (McInerney et al., 2005; Ventoso et al., 2006).
Sequences that direct the genome for packaging have been identified and comparison across the genus shows differences between subgroups (Kim et al., 2011). In the absence of packaging signals both genomic and non-genomic RNAs are packaged – another example of the virus defaulting to a less specific mechanism to rescue infectivity. Viral RNA encapsidation occurs late during infection when the predominant RNA species available for packaging is the subgenome. In spite of this, only genomic RNA is packaged during alphavirus infection with the possibility of one exception; Aura virus is known to package both genomic and subgenomic RNAs indiscriminately (Rümenapf et al., 1994). Nevertheless, generally, the specified packaging of the genomic RNA suggests a highly selective mechanism for alphavirus nucleocapsid assembly.
A number of studies have supported the presence of a packaging signal located within the non-structural coding region of the viral genome (Frolova et al., 1997; Kim et al., 2011; Weiss et al., 1989, 1994). Originally, work by Weiss et al. (1989) demonstrated that a 600 nt fragment derived from within the coding region of nsP1 (nt 721–1306) interacted specifically with the SINV capsid protein (Weiss et al., 1989). Later, this 600-base region was narrowed to a 132 nt fragment; this region, which spanned nt 945–1076 of the SINV genome, was shown to bind capsid protein and a 68-residue capsid-derived peptide (SINV capsid aa 76–132) (Weiss et al., 1994). Nearly a decade later, genetic and biochemical approaches in conjunction with computer-generated predictive models revealed consistent results for the encephalitic viruses. EEEV, WEEV and VEEV contain packaging signals within nsP1 (VEEV nt 856–1150) that consist of four to six predicted stem–loop structures marked by a GGG conserved sequence motif at the base of each loop (Kim et al., 2011). Disruption of the stem–loops or mutation of the conserved GGG sequence reduces packaging efficiency relative to WT virus.
Alternatively, studies conducted in Ross river virus (RRV) revealed three distinct capsid interacting sites, all of which localized to the nsP2-coding region (Frolova et al., 1997). Whilst one capsid interacting site at nt 2761–3062 in RRV appeared noticeably stronger than the others, it is possible that multiple packaging signals exist for RRV. Interestingly, the predominant site of RRV capsid interaction within the nsP2-coding region is consistent with the capsid interaction site proposed for SFV and other members of the SFV clade.
Outside of the 19 nt CSE described above, the alphaviral 3′ UTR exhibits a remarkable level of sequence diversity. A notable feature of the alphaviral 3′ UTRs are the repeat sequence elements (RSEs), which differ in their primary sequence and organization across the members of the genus. The precise functions of the RSEs are largely unclear; however, they are presumed to exhibit secondary structures (Ou et al., 1982b). For CHIKV, the arrangement and overall architecture of the 3′ UTR was found to be largely a consequence of selection within and adaptation to the mosquito vector rather than the mammalian host (Chen et al., 2013).
Elements of the alphaviral 3′ UTR have been identified using SINV that regulate the stability of the viral RNAs during infection (Garneau et al., 2008; Sokoloski et al., 2010). The relative stability of SINV RNAs appears to be dependent on the stage of infection, as the half-lives of incoming viral genomes vary greatly from those synthesized during active infection (Sokoloski et al., 2010, 2015). Late during infection the interaction of elements within the viral 3′ UTR with host factors, specifically the cellular HuR protein, was found to confer resistance to deadenylation in vitro and in tissue culture models of infection (Sokoloski et al., 2010). The region of the 3′ UTR responsible for the repression of deadenylation varied amongst the members of the genus, with SINV and VEEV relying on a URE for HuR binding, whereas other members of the genus relied upon the RSEs for binding. Given the overall diversity of the elements found in the 3′ UTR of the alphaviruses it is highly likely that additional functions, either via host interactions or cis functions, of the viral 3′ UTR elements await discovery.
Outstanding questions in the field
Despite the depth of our understanding of the molecular biology of replication of some alphaviruses there are still many significant knowledge gaps in the field. A major question regarding alphavirus RNA synthetic complexes relates to the precise structural arrangements that underlie the functions observed. The interactions and arrangement of non-structural proteins and non-structural polyproteins within the RNA synthetic complex must change to produce the observed functional shifts, yet these interactions remain largely uncharacterized. Interactions between individual non-structural proteins within the complexes would be informative for determining how the complex is assembled and matures during the virus replication cycle. Determining the spatial arrangement within the RNA synthetic complex would be a large step towards understanding the role each non-structural protein plays in each RNA synthetic activity. Additionally, it is apparent that the non-structural proteins interact with the host cell in ways that do not directly involve RNA synthesis. Multiple populations of nsP3 are observed in infected cells, only a minority of which co-localize with dsRNA, indicating most nsP3 is involved in processes independent of viral RNA synthesis (Frolova et al., 2006). The studies by Foy et al. referred to above demonstrate clearly that nsP3 has cell-type-dependent functions that may be important for virus replication in different tissues and hence for pathogenesis (Foy et al., 2013a, b). Similarly, nsP1 and nsP2 have functions that manipulate the host cell to promote viral spread and inhibit host response (Akhrymuk et al., 2012; Garmashova et al., 2006; Laakkonen et al., 1998). Almost certainly more functions for non-structural proteins remain to be elucidated.
The involvement of host factors in RNA synthesis and regulation of viral RNA function is also an area that is only just beginning to be explored. Whilst numerous host proteins have been found to associate with viral non-structural protein complexes, their specific roles during virus replication remain elusive (Cristea et al., 2006; Frolova et al., 2006; Varjak et al., 2013). Little is known about sites within viral RNAs to which host proteins may bind and what the consequences of their binding might be. Whether there is functional redundancy, either across interactions or host factors, or not is a significant confounding factor for the characterization of alphavirus host interactions. This is undoubtedly an area of research that will be important in understanding the regulation of viral RNA replication and viral gene expression, and will likely enhance our understanding of virus species-specific pathogenesis.
Finally, the vertebrate host system has been used for the majority of the observations reported above. The fact that alphaviruses shut-off host cell macromolecular synthesis in vertebrates, but not in mosquito cells, clearly indicates that the intracellular environments in which the virus replicates are tremendously different. The replication cycle and outcome of infection for the alphaviruses is dependent on the host. Vertebrate hosts exhibit acute infection, whereas invertebrates often develop persistent infection with little incidence of cellular pathogenesis. Hence, there is a critical need to better understand the mechanisms of alphavirus replication in invertebrate systems. Understanding how the viral replication complex forms, what host components are recruited and how RNA synthesis persists in mosquito cells will be invaluable in identifying possible means of antiviral intervention at the level of the vector.
Acknowledgements
We apologize to those individuals whose work we were unable to cite. This work was supported by grants from the National Institutes of Health/National Institute of Allergy and Infectious Diseases (9R01 AI090077 to R. W. H. and F32 AI104217 to K. J. S.).
References
- Aaskov J., Jones A., Choi W., Lowry K., Stewart E. (2011). Lineage replacement accompanying duplication and rapid fixation of an RNA element in the nsP3 gene in a species of alphavirus Virology 410 353–359 10.1016/j.virol.2010.11.025 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T., Kääriäinen L. (1995). Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP Proc Natl Acad Sci U S A 92 507–511 10.1073/pnas.92.2.507 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T., Karlin D.G. (2015). Sequence analysis reveals a conserved extension in the capping enzyme of the alphavirus supergroup, and a homologous domain in nodaviruses Biol Direct 10 16 10.1186/s13062-015-0050-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T., Laakkonen P., Vihinen H., Kääriäinen L. (1997). Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities J Virol 71 392–397 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T., Lampio A., Auvinen P., Kääriäinen L. (1999). Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity EMBO J 18 3164–3172 10.1093/emboj/18.11.3164 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T., Kujala P., Tuittila M., Blom T., Laakkonen P., Hinkkanen A., Auvinen P. (2000). Effects of palmitoylation of replicase protein nsP1 on alphavirus infection J Virol 74 6725–6733 10.1128/JVI.74.15.6725-6733.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhrymuk I., Kulemzin S.V., Frolova E.I. (2012). Evasion of the innate immune response: the Old World alphavirus nsP2 protein induces rapid degradation of Rpb1, a catalytic subunit of RNA polymerase II J Virol 86 7180–7191 10.1128/JVI.00541-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M.D., Buckle A.M., Cordell S.C., Löwe J., Bycroft M. (2003). The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A J Mol Biol 330 503–511 10.1016/S0022-2836(03)00473-X . [DOI] [PubMed] [Google Scholar]
- Atasheva S., Gorchakov R., English R., Frolov I., Frolova E. (2007). Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning J Virol 81 5046–5057 10.1128/JVI.02746-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton D.J., Sawicki S.G., Sawicki D.L. (1991). Solubilization and immunoprecipitation of alphavirus replication complexes J Virol 65 1496–1506 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büchen-Osmond C.E. (2006). In: ICTVdB - The Universal Virus Database, version 4. Online: Columbia University, New York, USA http://www.ncbi.nlm.nih.gov/ICTVdb/Ictv/fs_index.htm . [Google Scholar]
- Burnham A.J., Gong L., Hardy R.W. (2007). Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA Virology 367 212–221 10.1016/j.virol.2007.05.008 . [DOI] [PubMed] [Google Scholar]
- Chen R., Wang E., Tsetsarkin K.A., Weaver S.C. (2013). Chikungunya virus 3′ untranslated region: adaptation to mosquitoes and a population bottleneck as major evolutionary forces PLoS Pathog 9 e1003591 10.1371/journal.ppat.1003591 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R.H., Kuhn R.J., Olson N.H., Rossmann M.G., Choi H.K., Smith T.J., Baker T.S. (1995). Nucleocapsid and glycoprotein organization in an enveloped virus Cell 80 621–630 10.1016/0092-8674(95)90516-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B.Y., Firth A.E., Atkins J.F. (2010). Frameshifting in alphaviruses: a diversity of 3′ stimulatory structures J Mol Biol 397 448–456 10.1016/j.jmb.2010.01.044 . [DOI] [PubMed] [Google Scholar]
- Cristea I.M., Carroll J.-W.N., Rout M.P., Rice C.M., Chait B.T., MacDonald M.R. (2006). Tracking and elucidating alphavirus-host protein interactions J Biol Chem 281 30269–30278 10.1074/jbc.M603980200 . [DOI] [PubMed] [Google Scholar]
- Cristea I.M., Rozjabek H., Molloy K.R., Karki S., White L.L., Rice C.M., Rout M.P., Chait B.T., MacDonald M.R. (2010). Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication J Virol 84 6720–6732 10.1128/JVI.01983-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross R.K. (1983). Identification of a unique guanine-7-methyltransferase in Semliki Forest virus (SFV) infected cell extracts Virology 130 452–463 10.1016/0042-6822(83)90099-5 . [DOI] [PubMed] [Google Scholar]
- Cruz C.C., Suthar M.S., Montgomery S.A., Shabman R., Simmons J., Johnston R.E., Morrison T.E., Heise M.T. (2010). Modulation of type I IFN induction by a virulence determinant within the alphavirus nsP1 protein Virology 399 1–10 10.1016/j.virol.2009.12.031 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., other authors (2010). 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members Nature 468 452–456 10.1038/nature09489 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P.K., Merits A., Lulla A. (2014). Functional cross-talk between distant domains of chikungunya virus non-structural protein 2 is decisive for its RNA-modulating activity J Biol Chem 289 5635–5653 10.1074/jbc.M113.503433 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis N.L., Willis L.V., Smith J.F., Johnston R.E. (1989). In vitro synthesis of infectious venezuelan equine encephalitis virus RNA from a cDNA clone: analysis of a viable deletion mutant Virology 171 189–204 10.1016/0042-6822(89)90526-6 . [DOI] [PubMed] [Google Scholar]
- Dé I., Sawicki S.G., Sawicki D.L. (1996). Sindbis virus RNA-negative mutants that fail to convert from minus-strand to plus-strand synthesis: role of the nsP2 protein J Virol 70 2706–2719 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dé I., Fata-Hartley C., Sawicki S.G., Sawicki D.L. (2003). Functional analysis of nsP3 phosphoprotein mutants of Sindbis virus J Virol 77 13106–13116 10.1128/JVI.77.24.13106-13116.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Hardy W.R., Shirako Y., Strauss J.H. (1990). Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo EMBO J 9 2631–2638 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Rümenapf T., Kuhn R.J., Strauss E.G., Strauss J.H. (1991). Sindbis virus RNA polymerase is degraded by the N-end rule pathway Proc Natl Acad Sci U S A 88 8967–8971 10.1073/pnas.88.20.8967 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C., other authors (2006). Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains J Virol 80 8493–8502 10.1128/JVI.00713-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata C.L., Sawicki S.G., Sawicki D.L. (2002a). Alphavirus minus-strand RNA synthesis: identification of a role for Arg183 of the nsP4 polymerase J Virol 76 8632–8640 10.1128/JVI.76.17.8632-8640.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata C.L., Sawicki S.G., Sawicki D.L. (2002b). Modification of Asn374 of nsP1 suppresses a Sindbis virus nsP4 minus-strand polymerase mutant J Virol 76 8641–8649 10.1128/JVI.76.17.8641-8649.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayzulin R., Frolov I. (2004). Changes of the secondary structure of the 5′ end of the Sindbis virus genome inhibit virus growth in mosquito cells and lead to accumulation of adaptive mutations J Virol 78 4953–4964 10.1128/JVI.78.10.4953-4964.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A.E., Chung B.Y., Fleeton M.N., Atkins J.F. (2008). Discovery of frameshifting in Alphavirus 6K resolves a 20-year enigma Virol J 5 108 10.1186/1743-422X-5-108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A.E., Wills N.M., Gesteland R.F., Atkins J.F. (2011). Stimulation of stop codon readthrough: frequent presence of an extended 3′ RNA structural element Nucleic Acids Res 39 6679–6691 10.1093/nar/gkr224 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester N.L., Palacios G., Tesh R.B., Savji N., Guzman H., Sherman M., Weaver S.C., Lipkin W.I. (2012). Genome-scale phylogeny of the alphavirus genus suggests a marine origin J Virol 86 2729–2738 10.1128/JVI.05591-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy N.J., Akhrymuk M., Akhrymuk I., Atasheva S., Bopda-Waffo A., Frolov I., Frolova E.I. (2013a). Hypervariable domains of nsP3 proteins of New World and Old World alphaviruses mediate formation of distinct, virus-specific protein complexes J Virol 87 1997–2010 10.1128/JVI.02853-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy N.J., Akhrymuk M., Shustov A.V., Frolova E.I., Frolov I. (2013b). Hypervariable domain of nonstructural protein nsP3 of Venezuelan equine encephalitis virus determines cell-specific mode of virus replication J Virol 87 7569–7584 10.1128/JVI.00720-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey T.K., Gard D.L., Strauss J.H. (1979). Biophysical studies on circle formation by Sindbis virus 49 S RNA J Mol Biol 132 1–18 10.1016/0022-2836(79)90493-5 . [DOI] [PubMed] [Google Scholar]
- Friedman R.M., Levin J.G., Grimley P.M., Berezesky I.K. (1972). Membrane-associated replication complex in arbovirus infection J Virol 10 504–515 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I., Schlesinger S. (1996). Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation J Virol 70 1182–1190 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I., Agapov E., Hoffman T.A., Jr, Prágai B.M., Lippa M., Schlesinger S., Rice C.M. (1999). Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells J Virol 73 3854–3865 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I., Hardy R., Rice C.M. (2001). Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis RNA 7 1638–1651 10.1017/S135583820101010X . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova E., Frolov I., Schlesinger S. (1997). Packaging signals in alphaviruses J Virol 71 248–258 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova E., Gorchakov R., Garmashova N., Atasheva S., Vergara L.A., Frolov I. (2006). Formation of nsP3-specific protein complexes during Sindbis virus replication J Virol 80 4122–4134 10.1128/JVI.80.8.4122-4134.2006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova E.I., Gorchakov R., Pereboeva L., Atasheva S., Frolov I. (2010). Functional Sindbis virus replicative complexes are formed at the plasma membrane J Virol 84 11679–11695 10.1128/JVI.01441-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fros J.J., Domeradzka N.E., Baggen J., Geertsema C., Flipse J., Vlak J.M., Pijlman G.P. (2012). Chikungunya virus nsP3 blocks stress granule assembly by recruitment of G3BP into cytoplasmic foci J Virol 86 10873–10879 10.1128/JVI.01506-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froshauer S., Kartenbeck J., Helenius A. (1988). Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes J Cell Biol 107 2075–2086 10.1083/jcb.107.6.2075 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith S.E., Sheahan B.J., Atkins G.J. (2006). Deletions in the hypervariable domain of the nsP3 gene attenuate Semliki Forest virus virulence J Gen Virol 87 937–947 10.1099/vir.0.81406-0 . [DOI] [PubMed] [Google Scholar]
- Garmashova N., Gorchakov R., Frolova E., Frolov I. (2006). Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription J Virol 80 5686–5696 10.1128/JVI.02739-05 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmashova N., Gorchakov R., Volkova E., Paessler S., Frolova E., Frolov I. (2007). The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff J Virol 81 2472–2484 10.1128/JVI.02073-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau N.L., Sokoloski K.J., Opyrchal M., Neff C.P., Wilusz C.J., Wilusz J. (2008). The 3′ untranslated region of sindbis virus represses deadenylation of viral transcripts in mosquito and mammalian cells J Virol 82 880–892 10.1128/JVI.01205-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J., Raju R. (2000). Alphavirus RNA genome repair and evolution: molecular characterization of infectious sindbis virus isolates lacking a known conserved motif at the 3′ end of the genome J Virol 74 9776–9785 10.1128/JVI.74.20.9776-9785.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomatos P.J., Kääriäinen L., Keränen S., Ranki M., Sawicki D.L. (1980). Semliki Forest virus replication complex capable of synthesizing 42S and 26S nascent RNA chains J Gen Virol 49 61–69 10.1099/0022-1317-49-1-61 . [DOI] [PubMed] [Google Scholar]
- Gomez de Cedrón M., Ehsani N., Mikkola M.L., García J.A., Kääriäinen L. (1999). RNA helicase activity of Semliki Forest virus replicase protein NSP2 FEBS Lett 448 19–22 10.1016/S0014-5793(99)00321-X . [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. (1988). A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination FEBS Lett 235 16–24 10.1016/0014-5793(88)81226-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R., Hardy R., Rice C.M., Frolov I. (2004). Selection of functional 5′ cis-acting elements promoting efficient sindbis virus genome replication J Virol 78 61–75 10.1128/JVI.78.1.61-75.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R., Frolova E., Frolov I. (2005). Inhibition of transcription and translation in Sindbis virus-infected cells J Virol 79 9397–9409 10.1128/JVI.79.15.9397-9409.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R., Frolova E., Sawicki S., Atasheva S., Sawicki D., Frolov I. (2008a). A new role for ns polyprotein cleavage in Sindbis virus replication J Virol 82 6218–6231 10.1128/JVI.02624-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R., Garmashova N., Frolova E., Frolov I. (2008b). Different types of nsP3-containing protein complexes in Sindbis virus-infected cells J Virol 82 10088–10101 10.1128/JVI.01011-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimley P.M., Berezesky I.K., Friedman R.M. (1968). Cytoplasmic structures associated with an arbovirus infection: loci of viral ribonucleic acid synthesis J Virol 2 1326–1338 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui H., Lu C.-W., Adams S., Stollar V., Li M.-L. (2010). hnRNP A1 interacts with the genomic and subgenomic RNA promoters of Sindbis virus and is required for the synthesis of G and SG RNA J Biomed Sci 17 59 10.1186/1423-0127-17-59 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y.S., Grakoui A., Rice C.M., Strauss E.G., Strauss J.H. (1989a). Mapping of RNA– temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4 J Virol 63 1194–1202 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y.S., Strauss E.G., Strauss J.H. (1989b). Mapping of RNA– temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins J Virol 63 3142–3150 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R.W. (2006). The role of the 3′ terminus of the Sindbis virus genome in minus-strand initiation site selection Virology 345 520–531 10.1016/j.virol.2005.10.018 . [DOI] [PubMed] [Google Scholar]
- Hardy R.W., Rice C.M. (2005). Requirements at the 3′ end of the sindbis virus genome for efficient synthesis of minus-strand RNA J Virol 79 4630–4639 10.1128/JVI.79.8.4630-4639.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy W.R., Strauss J.H. (1989). Processing the nonstructural polyproteins of sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans J Virol 63 4653–4664 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy W.R., Hahn Y.S., de Groot R.J., Strauss E.G., Strauss J.H. (1990). Synthesis and processing of the nonstructural polyproteins of several temperature-sensitive mutants of Sindbis virus Virology 177 199–208 10.1016/0042-6822(90)90473-5 . [DOI] [PubMed] [Google Scholar]
- Hefti E., Bishop D.H., Dubin D.T., Stollar V. (1975). 5′ Nucleotide sequence of sindbis viral RNA J Virol 17 149–159 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J., Andino R. (2001). Poliovirus RNA replication requires genome circularization through a protein-protein bridge Mol Cell 7 581–591 10.1016/S1097-2765(01)00205-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz J.M., Huang H.V. (1992). Utilization of heterologous alphavirus junction sequences as promoters by Sindbis virus J Virol 66 857–864 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K.R., Hajjou M., Hu J.Y., Raju R. (1997). RNA–RNA recombination in Sindbis virus: roles of the 3′ conserved motif, poly(A) tail, and nonviral sequences of template RNAs in polymerase recognition and template switching J Virol 71 2693–2704 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde J.L., Gardner C.L., Kimura T., White J.P., Liu G., Trobaugh D.W., Huang C., Tonelli M., Paessler S., other authors (2014). A viral RNA structural element alters host recognition of nonself RNA Science 343 783–787 10.1126/science.1248465 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- James F.D., Hietala K.A., Eldar D., Guess T.E., Cone C., Mundell N.A., Barnett J.V., Raju R. (2007). Efficient replication, and evolution of Sindbis virus genomes with non-canonical 3′A/U-rich elements (NC3ARE) in neonatal mice Virus Genes 35 651–662 10.1007/s11262-007-0130-z . [DOI] [PubMed] [Google Scholar]
- Johansson M.A., Powers A.M., Pesik N., Cohen N.J., Staples J.E. (2014). Nowcasting the spread of chikungunya virus in the Americas PLoS One 9 e104915 10.1371/journal.pone.0104915 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kääriäinen L., Ahola T. (2002). Functions of alphavirus nonstructural proteins in RNA replication Prog Nucleic Acid Res Mol Biol 71 187–222 10.1016/S0079-6603(02)71044-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio K., Hellström K., Balistreri G., Spuul P., Jokitalo E., Ahola T. (2013). Template RNA length determines the size of replication complex spherules for Semliki Forest virus J Virol 87 9125–9134 10.1128/JVI.00660-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karo-Astover L., Sarova O., Merits A., Zusinaite E. (2010). The infection of mammalian and insect cells with SFV bearing nsP1 palmitoylation mutations Virus Res 153 277–287 10.1016/j.virusres.2010.08.019 . [DOI] [PubMed] [Google Scholar]
- Kim K.H., Rümenapf T., Strauss E.G., Strauss J.H. (2004). Regulation of Semliki Forest virus RNA replication: a model for the control of alphavirus pathogenesis in invertebrate hosts Virology 323 153–163 10.1016/j.virol.2004.03.009 . [DOI] [PubMed] [Google Scholar]
- Kim D.Y., Firth A.E., Atasheva S., Frolova E.I., Frolov I. (2011). Conservation of a packaging signal and the viral genome RNA packaging mechanism in alphavirus evolution J Virol 85 8022–8036 10.1128/JVI.00644-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney R.M., Chang G.J., Tsuchiya K.R., Sneider J.M., Roehrig J.T., Woodward T.M., Trent D.W. (1993). Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5′-noncoding region and the E2 envelope glycoprotein J Virol 67 1269–1277 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R.J., Hong Z., Strauss J.H. (1990). Mutagenesis of the 3′ nontranslated region of Sindbis virus RNA J Virol 64 1465–1476 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R.J., Griffin D.E., Zhang H., Niesters H.G., Strauss J.H. (1992). Attenuation of Sindbis virus neurovirulence by using defined mutations in nontranslated regions of the genome RNA J Virol 66 7121–7127 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujala P., Ikäheimonen A., Ehsani N., Vihinen H., Auvinen P., Kääriäinen L. (2001). Biogenesis of the Semliki Forest virus RNA replication complex J Virol 75 3873–3884 10.1128/JVI.75.8.3873-3884.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasegaran-Shylini R., Atasheva S., Gorenstein D.G., Frolov I. (2009a). Structural and functional elements of the promoter encoded by the 5′ untranslated region of the Venezuelan equine encephalitis virus genome J Virol 83 8327–8339 10.1128/JVI.00586-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasegaran-Shylini R., Thiviyanathan V., Gorenstein D.G., Frolov I. (2009b). The 5′UTR-specific mutation in VEEV TC-83 genome has a strong effect on RNA replication and subgenomic RNA synthesis, but not on translation of the encoded proteins Virology 387 211–221 10.1016/j.virol.2009.02.027 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Linn M., Gardner J., Warrilow D., Darnell G.A., McMahon C.R., Field I., Hyatt A.D., Slade R.W., Suhrbier A. (2001). Arbovirus of marine mammals: a new alphavirus isolated from the elephant seal louse Lepidophthirus macrorhini. J Virol 75 4103–4109 10.1128/JVI.75.9.4103-4109.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laakkonen P., Hyvönen M., Peränen J., Kääriäinen L. (1994). Expression of Semliki Forest virus nsP1-specific methyltransferase in insect cells and in Escherichia coli J Virol 68 7418–7425 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laakkonen P., Ahola T., Kääriäinen L. (1996). The effects of palmitoylation on membrane association of Semliki forest virus RNA capping enzyme J Biol Chem 271 28567–28571 10.1074/jbc.271.45.28567 . [DOI] [PubMed] [Google Scholar]
- Laakkonen P., Auvinen P., Kujala P., Kääriäinen L. (1998). Alphavirus replicase protein NSP1 induces filopodia and rearrangement of actin filaments J Virol 72 10265–10269 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampio A., Kilpeläinen I., Pesonen S., Karhi K., Auvinen P., Somerharju P., Kääriäinen L. (2000). Membrane binding mechanism of an RNA virus-capping enzyme J Biol Chem 275 37853–37859 10.1074/jbc.M004865200 . [DOI] [PubMed] [Google Scholar]
- LaStarza M.W., Grakoui A., Rice C.M. (1994a). Deletion and duplication mutations in the C-terminal nonconserved region of Sindbis virus nsP3: effects on phosphorylation and on virus replication in vertebrate and invertebrate cells Virology 202 224–232 10.1006/viro.1994.1338 . [DOI] [PubMed] [Google Scholar]
- LaStarza M.W., Lemm J.A., Rice C.M. (1994b). Genetic analysis of the nsP3 region of Sindbis virus: evidence for roles in minus-strand and subgenomic RNA synthesis J Virol 68 5781–5791 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm J.A., Rice C.M. (1993). Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34 J Virol 67 1905–1915 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm J.A., Rümenapf T., Strauss E.G., Strauss J.H., Rice C.M. (1994). Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis EMBO J 13 2925–2934 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm J.A., Bergqvist A., Read C.M., Rice C.M. (1998). Template-dependent initiation of Sindbis virus RNA replication in vitro J Virol 72 6546–6553 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson R.S., Strauss J.H., Strauss E.G. (1990). Complete sequence of the genomic RNA of O'nyong-nyong virus and its use in the construction of alphavirus phylogenetic trees Virology 175 110–123 10.1016/0042-6822(90)90191-S . [DOI] [PubMed] [Google Scholar]
- Levis R., Schlesinger S., Huang H.V. (1990). Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription J Virol 64 1726–1733 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Rice C.M. (1993). The signal for translational readthrough of a UGA codon in Sindbis virus RNA involves a single cytidine residue immediately downstream of the termination codon J Virol 67 5062–5067 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.-L., Stollar V. (2004). Identification of the amino acid sequence in Sindbis virus nsP4 that binds to the promoter for the synthesis of the subgenomic RNA Proc Natl Acad Sci U S A 101 9429–9434 10.1073/pnas.0400995101 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.-L., Stollar V. (2007). Distinct sites on the Sindbis virus RNA-dependent RNA polymerase for binding to the promoters for the synthesis of genomic and subgenomic RNA J Virol 81 4371–4373 10.1128/JVI.02672-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.P., La Starza M.W., Hardy W.R., Strauss J.H., Rice C.M. (1990). Phosphorylation of Sindbis virus nsP3 in vivo in vitro Virology 179 416–427 10.1016/0042-6822(90)90310-N . [DOI] [PubMed] [Google Scholar]
- Li M.-L., Wang H., Stollar V. (2010). In vitro synthesis of Sindbis virus genomic and subgenomic RNAs: influence of nsP4 mutations and nucleoside triphosphate concentrations J Virol 84 2732–2739 10.1128/JVI.01561-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.-Y., Shih S.-R., Pan M., Li C., Lue C.-F., Stollar V., Li M.-L. (2009). hnRNP A1 interacts with the 5′ untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication J Virol 83 6106–6114 10.1128/JVI.02476-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulla A., Lulla V., Tints K., Ahola T., Merits A. (2006). Molecular determinants of substrate specificity for Semliki Forest virus nonstructural protease J Virol 80 5413–5422 10.1128/JVI.00229-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulla V., Sawicki D.L., Sawicki S.G., Lulla A., Merits A., Ahola T. (2008). Molecular defects caused by temperature-sensitive mutations in Semliki Forest virus nsP1.J Virol 82 9236–9244 10.1128/JVI.00711-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulla A., Lulla V., Merits A. (2012). Macromolecular assembly-driven processing of the 2/3 cleavage site in the alphavirus replicase polyprotein J Virol 86 553–565 10.1128/JVI.05195-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulla V., Karo-Astover L., Rausalu K., Merits A., Lulla A. (2013). Presentation overrides specificity: probing the plasticity of alphaviral proteolytic activity through mutational analysis J Virol 87 10207–10220 10.1128/JVI.01485-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malet H., Coutard B., Jamal S., Dutartre H., Papageorgiou N., Neuvonen M., Ahola T., Forrester N., Gould E.A., other authors (2009). The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket J Virol 83 6534–6545 10.1128/JVI.00189-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.L., McMillan F.M. (2002). SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold Curr Opin Struct Biol 12 783–793 10.1016/S0959-440X(02)00391-3 . [DOI] [PubMed] [Google Scholar]
- Martinez M.G., Snapp E.L., Perumal G.S., Macaluso F.P., Kielian M. (2014). Imaging the alphavirus exit pathway J Virol 88 6922–6933 10.1128/JVI.00592-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney G.M., Kedersha N.L., Kaufman R.J., Anderson P., Liljeström P. (2005). Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation Mol Biol Cell 16 3753–3763 10.1091/mbc.E05-02-0124 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merits A., Vasiljeva L., Ahola T., Kääriäinen L., Auvinen P. (2001). Proteolytic processing of Semliki Forest virus-specific non-structural polyprotein by nsP2 protease J Gen Virol 82 765–773 . [DOI] [PubMed] [Google Scholar]
- Mi S., Stollar V. (1990). Both amino acid changes in nsP1 of Sindbis virusLM21 contribute to and are required for efficient expression of the mutant phenotype Virology 178 429–434 10.1016/0042-6822(90)90340-W . [DOI] [PubMed] [Google Scholar]
- Mi S., Stollar V. (1991). Expression of Sindbis virus nsP1 and methyltransferase activity in Escherichia coli Virology 184 423–427 10.1016/0042-6822(91)90862-6 . [DOI] [PubMed] [Google Scholar]
- Michel G., Petrakova O., Atasheva S., Frolov I. (2007). Adaptation of Venezuelan equine encephalitis virus lacking 51-nt conserved sequence element to replication in mammalian and mosquito cells Virology 362 475–487 10.1016/j.virol.2007.01.009 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasar F., Palacios G., Gorchakov R.V., Guzman H., Da Rosa A.P., Savji N., Popov V.L., Sherman M.B., Lipkin W.I., other authors (2012). Eilat virus, a unique alphavirus with host range restricted to insects by RNA replication Proc Natl Acad Sci U S A 109 14622–14627 10.1073/pnas.1204787109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasar F., Haddow A.D., Tesh R.B., Weaver S.C. (2014). Eilat virus displays a narrow mosquito vector range Parasit Vectors 7 595 10.1186/s13071-014-0595-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasar F., Gorchakov R.V., Tesh R.B., Weaver S.C. (2015). Eilat virus host range restriction is present at multiple levels of the virus life cycle J Virol 89 1404–1418 10.1128/JVI.01856-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuvonen M., Kazlauskas A., Martikainen M., Hinkkanen A., Ahola T., Saksela K. (2011). SH3 domain-mediated recruitment of host cell amphiphysins by alphavirus nsP3 promotes viral RNA replication PLoS Pathog 7 e1002383 10.1371/journal.ppat.1002383 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickens D.G., Hardy R.W. (2008). Structural and functional analyses of stem-loop 1 of the Sindbis virus genome Virology 370 158–172 10.1016/j.virol.2007.08.006 . [DOI] [PubMed] [Google Scholar]
- Niesters H.G., Strauss J.H. (1990a). Defined mutations in the 5′ nontranslated sequence of Sindbis virus RNA J Virol 64 4162–4168 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesters H.G., Strauss J.H. (1990b). Mutagenesis of the conserved 51-nucleotide region of Sindbis virus J Virol 64 1639–1647 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonov A., Mölder T., Sikut R., Kiiver K., Männik A., Toots U., Lulla A., Lulla V., Utt A., other authors (2013). RIG-I and MDA-5 detection of viral RNA-dependent RNA polymerase activity restricts positive-strand RNA virus replication PLoS Pathog 9 e1003610 10.1371/journal.ppat.1003610 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly E.K., Kao C.C. (1998). Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure Virology 252 287–303 10.1006/viro.1998.9463 . [DOI] [PubMed] [Google Scholar]
- Oberste M.S., Parker M.D., Smith J.F. (1996). Complete sequence of Venezuelan equine encephalitis virus subtype IE reveals conserved and hypervariable domains within the C terminus of nsP3 Virology 219 314–320 10.1006/viro.1996.0254 . [DOI] [PubMed] [Google Scholar]
- Ou J.H., Strauss E.G., Strauss J.H. (1981). Comparative studies of the 3′-terminal sequences of several alpha virus RNAs Virology 109 281–289 10.1016/0042-6822(81)90499-2 . [DOI] [PubMed] [Google Scholar]
- Ou J.H., Rice C.M., Dalgarno L., Strauss E.G., Strauss J.H. (1982a). Sequence studies of several alphavirus genomic RNAs in the region containing the start of the subgenomic RNA Proc Natl Acad Sci U S A 79 5235–5239 10.1073/pnas.79.17.5235 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J.H., Trent D.W., Strauss J.H. (1982b). The 3′-non-coding regions of alphavirus RNAs contain repeating sequences J Mol Biol 156 719–730 10.1016/0022-2836(82)90138-3 . [DOI] [PubMed] [Google Scholar]
- Pardigon N., Strauss J.H. (1992). Cellular proteins bind to the 3′ end of Sindbis virus minus-strand RNA J Virol 66 1007–1015 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardigon N., Strauss J.H. (1996). Mosquito homolog of the La autoantigen binds to Sindbis virus RNA J Virol 70 1173–1181 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Griffin D.E. (2009). The nsP3 macro domain is important for Sindbis virus replication in neurons and neurovirulence in mice Virology 388 305–314 10.1016/j.virol.2009.03.031 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson J.R., Fuji R.N. (1998). Evolutionary conservation of histone macroH2A subtypes and domains Nucleic Acids Res 26 2837–2842 10.1093/nar/26.12.2837 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peränen J., Takkinen K., Kalkkinen N., Kääriäinen L. (1988). Semliki Forest virus-specific non-structural protein nsP3 is a phosphoprotein J Gen Virol 69 2165–2178 10.1099/0022-1317-69-9-2165 . [DOI] [PubMed] [Google Scholar]
- Peränen J., Laakkonen P., Hyvönen M., Kääriäinen L. (1995). The alphavirus replicase protein nsP1 is membrane-associated and has affinity to endocytic organelles Virology 208 610–620 10.1006/viro.1995.1192 . [DOI] [PubMed] [Google Scholar]
- Powers A.M., Brault A.C., Shirako Y., Strauss E.G., Kang W., Strauss J.H., Weaver S.C. (2001). Evolutionary relationships and systematics of the alphaviruses J Virol 75 10118–10131 10.1128/JVI.75.21.10118-10131.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R., Huang H.V. (1991). Analysis of Sindbis virus promoter recognition in vivo, using novel vectors with two subgenomic mRNA promoters J Virol 65 2501–2510 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R., Hajjou M., Hill K.R., Botta V., Botta S. (1999). In vivo addition of poly(A) tail and AU-rich sequences to the 3′ terminus of the Sindbis virus RNA genome: a novel 3′-end repair pathway J Virol 73 2410–2419 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud J.M., Kim D.Y., Atasheva S., Rasalouskaya A., White J.P., Diamond M.S., Weaver S.C., Frolova E.I., Frolov I. (2015). IFIT1 differentially interferes with translation and replication of alphavirus genomes and promotes induction of type I interferon PLoS Pathog 11 e1004863 10.1371/journal.ppat.1004863 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikkonen M. (1996). Functional significance of the nuclear-targeting and NTP-binding motifs of Semliki Forest virus nonstructural protein nsP2 Virology 218 352–361 10.1006/viro.1996.0204 . [DOI] [PubMed] [Google Scholar]
- Rikkonen M., Peränen J., Kääriäinen L. (1994). ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsP2 J Virol 68 5804–5810 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozanov M.N., Koonin E.V., Gorbalenya A.E. (1992). Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses J Gen Virol 73 2129–2134 10.1099/0022-1317-73-8-2129 . [DOI] [PubMed] [Google Scholar]
- Rubach J.K., Wasik B.R., Rupp J.C., Kuhn R.J., Hardy R.W., Smith J.L. (2009). Characterization of purified Sindbis virus nsP4 RNA-dependent RNA polymerase activity in vitro Virology 384 201–208 10.1016/j.virol.2008.10.030 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rümenapf T., Strauss E.G., Strauss J.H. (1994). Subgenomic mRNA of Aura alphavirus is packaged into virions J Virol 68 56–62 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp J.C., Jundt N., Hardy R.W. (2011). Requirement for the amino-terminal domain of sindbis virus nsP4 during virus infection J Virol 85 3449–3460 10.1128/JVI.02058-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A.T., White M.A., Watowich S.J. (2006). The crystal structure of the Venezuelan equine encephalitis alphavirus nsP2 protease Structure 14 1449–1458 10.1016/j.str.2006.07.010 . [DOI] [PubMed] [Google Scholar]
- Salonen A., Vasiljeva L., Merits A., Magden J., Jokitalo E., Kääriäinen L. (2003). Properly folded nonstructural polyprotein directs the semliki forest virus replication complex to the endosomal compartment J Virol 77 1691–1702 10.1128/JVI.77.3.1691-1702.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A., Ahola T., Kääriäinen L. (2005). Viral RNA replication in association with cellular membranes Curr Top Microbiol Immunol 285 139–173 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D.L., Gomatos P.J. (1976). Replication of semliki forest virus: polyadenylate in plus-strand RNA and polyuridylate in minus-strand RNA J Virol 20 446–464 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D.L., Sawicki S.G. (1980). Short-lived minus-strand polymerase for Semliki Forest virus J Virol 34 108–118 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D.L., Sawicki S.G. (1985). Functional analysis of the A complementation group mutants of Sindbis HR virus Virology 144 20–34 10.1016/0042-6822(85)90301-0 . [DOI] [PubMed] [Google Scholar]
- Sawicki D.L., Sawicki S.G. (1993). A second nonstructural protein functions in the regulation of alphavirus negative-strand RNA synthesis J Virol 67 3605–3610 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D.L., Sawicki S.G. (1994). Alphavirus positive and negative strand RNA synthesis and the role of polyproteins in formation of viral replication complexes Arch Virol Suppl 9 393–405 . [DOI] [PubMed] [Google Scholar]
- Sawicki D.L., Kääriäinen L., Lambek C., Gomatos P.J. (1978). Mechanism for control of synthesis of Semliki Forest virus 26S and 42S RNA J Virol 25 19–27 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D.L., Silverman R.H., Williams B.R., Sawicki S.G. (2003). Alphavirus minus-strand synthesis and persistence in mouse embryo fibroblasts derived from mice lacking RNase L and protein kinase R J Virol 77 1801–1811 10.1128/JVI.77.3.1801-1811.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D.L., Perri S., Polo J.M., Sawicki S.G. (2006). Role for nsP2 proteins in the cessation of alphavirus minus-strand synthesis by host cells J Virol 80 360–371 10.1128/JVI.80.1.360-371.2006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton-Shaw K.D., Ledermann J.P., Borland E.M., Stovall J.L., Mossel E.C., Singh A.J., Wilusz J., Powers A.M. (2013). O'nyong nyong virus molecular determinants of unique vector specificity reside in non-structural protein 3 PLoS Negl Trop Dis 7 e1931 10.1371/journal.pntd.0001931 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter I., Berger A. (1967). On the size of the active site in proteases I. Papain. Biochem Biophys Res Commun 27 157–162 10.1016/S0006-291X(67)80055-X . [DOI] [PubMed] [Google Scholar]
- Scheidel L.M., Stollar V. (1991). Mutations that confer resistance to mycophenolic acid and ribavirin on Sindbis virus map to the nonstructural protein nsP1 Virology 181 490–499 10.1016/0042-6822(91)90881-B . [DOI] [PubMed] [Google Scholar]
- Schluckebier G., O'Gara M., Saenger W., Cheng X. (1995). Universal catalytic domain structure of AdoMet-dependent methyltransferases J Mol Biol 247 16–20 10.1006/jmbi.1994.0117 . [DOI] [PubMed] [Google Scholar]
- Scholte F.E., Tas A., Albulescu I.C., Zˇusinaite E., Merits A., Snijder E.J., van Hemert M.J. (2015). Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication J Virol 89 4457–4469 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin G., Yost S.A., Miller M.T., Elrod E.J., Grakoui A., Marcotrigiano J. (2012). Structural and functional insights into alphavirus polyprotein processing and pathogenesis Proc Natl Acad Sci U S A 109 16534–16539 10.1073/pnas.1210418109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirako Y., Strauss J.H. (1990). Cleavage between nsP1 and nsP2 initiates the processing pathway of Sindbis virus nonstructural polyprotein P123 Virology 177 54–64 10.1016/0042-6822(90)90459-5 . [DOI] [PubMed] [Google Scholar]
- Shirako Y., Strauss J.H. (1994). Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis J Virol 68 1874–1885 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirako Y., Strauss J.H. (1998). Requirement for an aromatic amino acid or histidine at the N terminus of Sindbis virus RNA polymerase J Virol 72 2310–2315 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirako Y., Strauss E.G., Strauss J.H. (2003). Modification of the 5′ terminus of Sindbis virus genomic RNA allows nsP4 RNA polymerases with nonaromatic amino acids at the N terminus to function in RNA replication J Virol 77 2301–2309 10.1128/JVI.77.4.2301-2309.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons D.T., Strauss J.H. (1972a). Replication of Sindbis virus. I. Relative size and genetic content of 26 s and 49 s RNA J Mol Biol 71 599–613 10.1016/S0022-2836(72)80026-3 . [DOI] [PubMed] [Google Scholar]
- Simmons D.T., Strauss J.H. (1972b). Replication of Sindbis virus. II. Multiple forms of double-stranded RNA isolated from infected cells J Mol Biol 71 615–631 . [DOI] [PubMed] [Google Scholar]
- Sokoloski K.J., Dickson A.M., Chaskey E.L., Garneau N.L., Wilusz C.J., Wilusz J. (2010). Sindbis virus usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells Cell Host Microbe 8 196–207 10.1016/j.chom.2010.07.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloski K.J., Haist K.C., Morrison T.E., Mukhopadhyay S., Hardy R.W. (2015). Noncapped alphavirus genomic RNAs and their role during infection J Virol 89 6080–6092 10.1128/JVI.00553-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spuul P., Salonen A., Merits A., Jokitalo E., Kääriäinen L., Ahola T. (2007). Role of the amphipathic peptide of Semliki forest virus replicase protein nsP1 in membrane association and virus replication J Virol 81 872–883 10.1128/JVI.01785-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spuul P., Balistreri G., Kääriäinen L., Ahola T. (2010). Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest Virus replication complexes from the plasma membrane to modified lysosomes J Virol 84 7543–7557 10.1128/JVI.00477-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spuul P., Balistreri G., Hellström K., Golubtsov A.V., Jokitalo E., Ahola T. (2011). Assembly of alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells J Virol 85 4739–4751 10.1128/JVI.00085-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoermer Burrack K.A., Hawman D.W., Jupille H.J., Oko L., Minor M., Shives K.D., Gunn B.M., Long K.M., Morrison T.E. (2014). Attenuating mutations in nsP1 reveal tissue-specific mechanisms for control of Ross River virus infection J Virol 88 3719–3732 10.1128/JVI.02609-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J.H., Strauss E.G. (1994). The alphaviruses: gene expression, replication, and evolution Microbiol Rev 58 491–562 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss E.G., Rice C.M., Strauss J.H. (1983). Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon Proc Natl Acad Sci U S A 80 5271–5275 10.1073/pnas.80.17.5271 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss E.G., Rice C.M., Strauss J.H. (1984). Complete nucleotide sequence of the genomic RNA of Sindbis virus Virology 133 92–110 10.1016/0042-6822(84)90428-8 . [DOI] [PubMed] [Google Scholar]
- Suopanki J., Sawicki D.L., Sawicki S.G., Kääriäinen L. (1998). Regulation of alphavirus 26S mRNA transcription by replicase component nsP2 J Gen Virol 79 309–319 . [DOI] [PubMed] [Google Scholar]
- Tomar S., Hardy R.W., Smith J.L., Kuhn R.J. (2006). Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity J Virol 80 9962–9969 10.1128/JVI.01067-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar S., Narwal M., Harms E., Smith J.L., Kuhn R.J. (2011). Heterologous production, purification and characterization of enzymatically active Sindbis virus nonstructural protein nsP1 Protein Expr Purif 79 277–284 10.1016/j.pep.2011.05.022 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuittila M., Hinkkanen A.E. (2003). Amino acid mutations in the replicase protein nsP3 of Semliki Forest virus cumulatively affect neurovirulence J Gen Virol 84 1525–1533 10.1099/vir.0.18936-0 . [DOI] [PubMed] [Google Scholar]
- Tuittila M.T., Santagati M.G., Röyttä M., Määttä J.A., Hinkkanen A.E. (2000). Replicase complex genes of Semliki Forest virus confer lethal neurovirulence J Virol 74 4579–4589 10.1128/JVI.74.10.4579-4589.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjak M., Saul S., Arike L., Lulla A., Peil L., Merits A. (2013). Magnetic fractionation and proteomic dissection of cellular organelles occupied by the late replication complexes of Semliki Forest virus J Virol 87 10295–10312 10.1128/JVI.01105-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L., Merits A., Auvinen P., Kääriäinen L. (2000). Identification of a novel function of the alphavirus capping apparatus RNA 5′-triphosphatase activity of Nsp2.J Biol Chem 275 17281–17287 10.1074/jbc.M910340199 . [DOI] [PubMed] [Google Scholar]
- Vasiljeva L., Merits A., Golubtsov A., Sizemskaja V., Kääriäinen L., Ahola T. (2003). Regulation of the sequential processing of Semliki Forest virus replicase polyprotein J Biol Chem 278 41636–41645 10.1074/jbc.M307481200 . [DOI] [PubMed] [Google Scholar]
- Ventoso I., Sanz M.A., Molina S., Berlanga J.J., Carrasco L., Esteban M. (2006). Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR Genes Dev 20 87–100 10.1101/gad.357006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihinen H., Ahola T., Tuittila M., Merits A., Kääriäinen L. (2001). Elimination of phosphorylation sites of Semliki Forest virus replicase protein nsP3 J Biol Chem 276 5745–5752 10.1074/jbc.M006077200 . [DOI] [PubMed] [Google Scholar]
- Villoing S., Béarzotti M., Chilmonczyk S., Castric J., Brémont M. (2000). Rainbow trout sleeping disease virus is an atypical alphavirus J Virol 74 173–183 10.1128/JVI.74.1.173-183.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.F., Sawicki S.G., Sawicki D.L. (1991). Sindbis virus nsP1 functions in negative-strand RNA synthesis J Virol 65 985–988 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.F., Sawicki S.G., Sawicki D.L. (1994). Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA J Virol 68 6466–6475 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.L., O'Rear J., Stollar V. (1996). Mutagenesis of the Sindbis virus nsP1 protein: effects on methyltransferase activity and viral infectivity Virology 217 527–531 10.1006/viro.1996.0147 . [DOI] [PubMed] [Google Scholar]
- Weaver S.C. (2014). Arrival of chikungunya virus in the new world: prospects for spread and impact on public health PLoS Negl Trop Dis 8 e2921 10.1371/journal.pntd.0002921 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S.C., Lecuit M. (2015). Chikungunya virus and the global spread of a mosquito-borne disease N Engl J Med 372 1231–1239 10.1056/NEJMra1406035 . [DOI] [PubMed] [Google Scholar]
- Weiss B., Nitschko H., Ghattas I., Wright R., Schlesinger S. (1989). Evidence for specificity in the encapsidation of Sindbis virus RNAs J Virol 63 5310–5318 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B., Geigenmüller-Gnirke U., Schlesinger S. (1994). Interactions between Sindbis virus RNAs and a 68 amino acid derivative of the viral capsid protein further defines the capsid binding site Nucleic Acids Res 22 780–786 10.1093/nar/22.5.780 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston J.H., Welsh M.D., McLoughlin M.F., Todd D. (1999). Salmon pancreas disease virus, an alphavirus infecting farmed Atlantic salmon, Salmo salar L Virology 256 188–195 10.1006/viro.1999.9654 . [DOI] [PubMed] [Google Scholar]
- Wielgosz M.M., Huang H.V. (1997). A novel viral RNA species in Sindbis virus-infected cells J Virol 71 9108–9117 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielgosz M.M., Raju R., Huang H.V. (2001). Sequence requirements for Sindbis virus subgenomic mRNA promoter function in cultured cells J Virol 75 3509–3519 10.1128/JVI.75.8.3509-3519.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S., Padmanabhan R. (1999). A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA J Biol Chem 274 33714–33722 10.1074/jbc.274.47.33714 . [DOI] [PubMed] [Google Scholar]
- Zhang W., Mukhopadhyay S., Pletnev S.V., Baker T.S., Kuhn R.J., Rossmann M.G. (2002). Placement of the structural proteins in Sindbis virus J Virol 76 11645–11658 10.1128/JVI.76.22.11645-11658.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Tözsér J., Waugh D.S. (2009). Molecular cloning, overproduction, purification and biochemical characterization of the p39 nsp2 protease domains encoded by three alphaviruses Protein Expr Purif 64 89–97 10.1016/j.pep.2008.10.013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusinaite E., Tints K., Kiiver K., Spuul P., Karo-Astover L., Merits A., Sarand I. (2007). Mutations at the palmitoylation site of non-structural protein nsP1 of Semliki Forest virus attenuate virus replication and cause accumulation of compensatory mutations J Gen Virol 88 1977–1985 10.1099/vir.0.82865-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]