Abstract
Allelic variation at 4 loci in the human olfactory receptor gene OR7D4 is associated with perceptual variation in the sex steroid-derived odorants, androstenone, and androstadienone. Androstadienone has been linked with chemosensory identification whereas androstenone makes pork from uncastrated pigs distasteful (“boar taint”). In a sample of 2224 individuals from 43 populations, we identified 45 OR7D4 single nucleotide polymorphisms. Coalescent modeling of frequency-site-spectrum-based statistics identified significant deviation from neutrality in human OR7D4; individual populations with statistically significant deviations from neutrality include Gujarati, Beijing Han, Great Britain, Iberia, and Puerto Rico. Analysis of molecular variation values indicated statistically significant population differentiation driven mainly by the 4 alleles associated with androstenone perception variation; however, fixation values were low suggesting that genetic structure may not have played a strong role in creating these group divisions. We also studied OR7D4 in the genomes of extinct members of the human lineage: Altai Neandertal and Denisovan. No variants were identified in Altai but 2 were in Denisova, one of which is shared by modern humans and one of which is novel. A functional test of modern human and a synthesized mutant Denisova OR7D4 indicated no statistically significant difference in responses to androstenone between the 2 species. Our results suggest non-neutral evolution for an olfactory receptor gene.
Key words: Altai Neandertal, androstenone perception, Denisova, human variation, OR7D4, selection
Introduction
Olfactory receptor genes were originally identified in rats (Buck and Axel 1991); in humans, these genes are unusually diverse and rapidly evolving (Olender et al. 2004, 2012; Mainland et al. 2014). Olfactory receptors are proteins expressed in the cell membranes of olfactory sensory neurons that detect airborne odor molecules (odorants); each receptor detects more than one odor and each odor activates more than one receptor (Buck and Axel 1991). Most odorant-olfactory receptor gene relationships have yet to be identified in humans but roughly 40 are known (Mainland et al. 2014).
Comparative rates of change in olfactory receptor genes indicate that humans have not experienced more pseudogenization compared with other primates (Matsui et al. 2010). Analysis of population differences in human olfactory haplotypes suggests high diversity, perhaps reflecting local adaptation (Menashe et al. 2002). Substantial variation exists at the individual level: the difference in number of expressed olfactory receptors between any pair of individuals is at least 14% (Zhang et al. 2007; Verbeurgt et al. 2014) but a rate of 30% has also been reported (Mainland et al. 2014). In addition, variation in olfactory receptor gene sequences among individuals is very high, second only to that seen in immune system genes (Menashe et al. 2003).
The evolutionary source of variation in human olfactory receptors is not fully understood but several studies have found evidence for positive selection acting on some human olfactory receptor genes (Gilad et al. 2000, 2003, 2005; Menashe et al. 2002; Gilad and Lancet 2003; Gimelbrant et al. 2004; Alonso et al. 2008; Moreno-Estrada et al. 2008). The underlying adaptive context of possible selective pressures acting on olfactory receptor genes is not known but may be linked to food (Matsui et al. 2010; Jaeger et al. 2013; McRae et al. 2013) and health (Spehr et al. 2003; Griffin et al. 2009; Neuhaus et al. 2009; Pluznick et al. 2009; Gu et al. 2014; Busse et al. 2014). In addition, copy number variants (deletions and duplications of large genetic segments across genomes) have affected the olfactory receptor gene family significantly more often than other genomic coding regions (Hasin et al. 2008; Waszak et al. 2010; Veerappa et al. 2013). The phenotypic impact of copy number variants on odorant perception is currently unknown.
This article is focused on genetic variation in OR7D4, which has been associated with an olfactory phenotype (Keller et al. 2007). The OR7D4 receptor recognizes androstenone. Androstenone is biologically significant in mammal social behavior as a sex steroid but the role of androstenone as a chemical signal in humans (Loos et al. 2014; Zhou et al. 2014) is contested (Wyatt 2015). Androstenone is the primary odorant in boar taint, the smell of meat from adult non-castrated male pigs and boars. The odor is unpleasant to the majority of humans, with the exception of those individuals carrying mutations that render the receptor less functional or nonfunctional (Lunde et al. 2012). Phenotypic variation in the detection and perception of androstenone in humans has been associated with 2 single nucleotide polymorphisms (SNPs) in OR7D4 that are in complete linkage disequilibrium (rs61729907 and rs5020278) (Keller et al. 2007; Knaapila et al. 2012a, 2012b; Lunde et al. 2012). The ancestral genotype of the linked SNPs has a phenotype that is sensitive to androstenone whereas the derived genotype is less sensitive to the odorant. Two additional SNPs also have been shown to decrease (rs61732668) and increase (rs5020280) sensitivity to androstenone.
In this study, we address the following question: Is there a significant pattern in the geographic distribution of variant alleles in OR7D4 and, if so, what evolutionary explanation best describes it? To answer this question, we generated new sequence data and extracted variant calls from the 1000 Genomes data set to analyze geographic variation and identify possible evolutionary adaptive trends. We also extracted variant calls from the published genomes of Denisova and Altai Neandertal. Finally, we created a mutant Denisova OR7D4 to test its response to androstenone.
Materials
Modern human data
We used 24 world populations from 1000 Genomes (n = 2081) (1000 Genomes Project Consortium 2012; Table 1). Some 1000 Genomes populations are genetically admixed (e.g., Puerto Ricans, Peruvians in Lima) or recent immigrants to an area (e.g., Gujarati in Houston Texas, Mexicans in LA) but many are from (comparatively) nonadmixed groups (e.g., Esan, Mende, Punjabi). We also sequenced 19 populations from Coriell Institute for Medical Research (n = 143) not included in 1000 Genomes; these are (comparatively) nonadmixed groups (e.g., Atayal and Ami hunter-gatherers in Taiwan, Mbuti and Biaka hunter-gatherers in Central Africa). Only samples that did not overlap with 1000 Genomes data were used. These samples were included to provide Native American and Melanesian data and additional sets of indigenous populations.
Table 1.
Materials (n = 2224)
| Africa | Central Africaa | Biakaa | 5 |
|---|---|---|---|
| Mbutia | 4 | ||
| Sub-Saharan | Sub-Saharan Africana | 9 | |
| Esanb | 99 | ||
| Gambianb | 113 | ||
| Kenyanb | 99 | ||
| Mendeb | 85 | ||
| Yorubanb | 108 | ||
| North Africa | North Africaa | 7 | |
| Middle East | Ashkenazima | 10 | |
| Asia | South Asia | Bengalib | 86 |
| Telugub | 102 | ||
| Punjabib | 96 | ||
| Gujarati | 103 | ||
| Tamil, Sri Lankab | 102 | ||
| East Asia | Amia | 10 | |
| Atayala | 9 | ||
| Daib | 93 | ||
| Han-Beijingb | 103 | ||
| Han-Southernb | 105 | ||
| Japaneseb | 104 | ||
| Kinhb | 99 | ||
| Oceania | Melanesia | Boungainvillea | 5 |
| Europe | Basquea | 10 | |
| Czecha | 10 | ||
| Finlandb | 99 | ||
| Great Britainb | 91 | ||
| Hungariana | 9 | ||
| Iberianb | 107 | ||
| Icelandb | 12 | ||
| Northern Europeana | 10 | ||
| Russiana | 9 | ||
| Toscanib | 107 | ||
| Americas | Indigenous America | Karitianaa | 5 |
| Mayaa | 4 | ||
| Pimaa | 5 | ||
| Quechuana | 4 | ||
| Suruia | 5 | ||
| Admixed America | Columbiansb | 94 | |
| Mexicans | 64 | ||
| Puerto Ricanb | 104 | ||
| Barbadoan | 96 | ||
| Peruviansb | 85 |
aCoriell samples that do not overlap with 1000 Genomes.
b1000 Genomes samples.
Hominin data
Denisova and Altai Neandertal are Pleistocene Eurasian hominin species known from specimens excavated from Denisova Cave in southern Siberia in 2010 (Krause et al. 2010; Reich et al. 2010; Prüfer et al., 2014; Table 1). Altai is related to Neandertals from Croatia, Germany, and the Caucasus (Prüfer et al., 2014). Denisova may have interbred with the ancestors of modern-day Melanesians during the Denisova migration from Africa to Siberia (Reich et al. 2010; Prüfer et al., 2014). Analysis of genomic data indicates that Denisova and Altai branched away from modern humans between 550000 and 765000 years ago and from each other between 445000 and 473000 years ago (Prüfer et al., 2014). Denisova inhabited the cave between 30000 and 50000 years ago. Individuals genetically identified as Neandertals were found as close as 100 km away from Denisova Cave. Finally, indirect evidence (i.e., stone tools) places modern humans in the area at 40000 years ago. Thus, there is a clear possibility that three hominin species were contemporary to Altai at the same time (Krause et al. 2010).
Methods
Modern human DNA sequencing (Coriell samples)
Human genomic DNA sequences were amplified using Qiagen HotStar Taq with primers upstream (5′-AAGTGATGACAAGCTGAGCTGC-3′) and downstream (5′-CCACAACATTTGCCTTAGGGGTA-3′) of the OR7D4 open reading frame. PCR products were purified using Sephacryl S-400 (GE HealthCare) then sequenced with an ABI Biosystems 3100 or 3730 Genetic Analyzer.
Hominin variant calling
The Denisovan genome (Reich et al. 2010) was generated from DNA extracted from a finger bone and has an average coverage of 30×. The Altai Neandertal genome was generated from DNA extracted from a toe bone and has an average coverage of 50×. Contamination of Denisova and Altai Neandertal with modern human DNA is estimated to be less than 1%. We identified variants in the published genomes of Altai Neandertal and Denisova from VCF files aligned to NCBI Build Human Genome Build GRCh37/hg19. We verified these variants using the genome browser at UC Santa Cruz (UCSC Genome Browser 2002) by creating a custom track linked to BAM format files aligned to NCBI Build Human Genome Build GRCh37/hg19 and viewing the data relative to the modern human genome. VCF and BAM files are located at http://cdna.eva.mpg.de/neandertal/altai/ in the “AltaiNeandertal” and “Denisova folders” (at the Max Planck Institute for Evolutionary Anthropology in Leipzig).
Cloning and sequencing
The Denisova OR7D4 was created by mutating the 204th amino acid in human OR7D4 from alanine to threonine (A204T) using chimeric PCR. Forward and reverse PCR primers containing the desired mutation were designed to have a 56°C or 58°C annealing temperature, obtained from Integrated DNA Technologies, and diluted to 5 μM. The respective forward and reverse primer sequences were 5′-TGTGGCCACGACACTGCTGGG-3′ and 5′-CCCAGCAGTGTCGTGGCCACA-3′ for the A204T mutant. Chimeric PCR was performed using Phusion polymerase and OR7D4 in a pCI vector as a template, with separate reactions using the forward primers paired with a 3′ pCI primer and reverse primers paired with a 5′ pCI primer. The reaction was started at 98°C for 30 s, then run for 25 cycles of the following conditions: denaturation at 98°C for 5 s, primer annealing at 55°C for 15 s, and elongation at 72°C for 30 s. The reaction was then held at 72°C for 5min. The products resulting from the forward and reverse primers were combined for each mutant and diluted 10× with distilled water (Gibco). A second PCR was performed using Phusion polymerase, 5′ and 3′ pCI primers, and the combined and diluted products for each desired mutant as the template. The same PCR conditions were used.
The products were purified using the QIAquick PCR Purification Kit (Qiagen), cut with MluI and NotI restriction enzymes, run on a 1.1% agarose gel with GelRed, and extracted using the QIAquick Gel Extraction Kit (Qiagen). The products were then ligated into Rho-tagged pCI cut with MluI and NotI using T4 ligase (New England Biolabs), and used to transform competent ampicillin-resistant Escherichia coli. These were plated on LB-ampicillin plates and incubated at 37°C overnight, then grown in 4mL of 2XYT-ampicillin (100mg/mL) medium overnight at 37°C. The Denville Miniprep Purification Kit was used to lyse the bacteria and purify the plasmid DNA. The concentration of the products was determined using an Eppendorf Biophotometer, and then adjusted to 100ng/μL using tris–ethylenediaminetetraacetic acid buffer. The products were amplified using HotstarTaq PCR and purified using Sephacryl (GE Healthcare). The products were sequenced using BigDye Terminator Sequencing Kit (Applied Biosystems), purified using Sephadex (GE Healthcare), and sequenced with 3130 Genetic Analyzer (Applied Biosystems).
Luciferase assay
Hana 3A cells were plated on Poly-D-lysine coated 96-well plates (Corning #3843) and cotransfected using 20 μL Lipofectamine2000 (Invitrogen) per plate with the prepared OR7D4 mutant (5ng/well); the receptor transporting protein 1S (RTP1S, 5ng/well) and the type 3 muscarinic acetylcholine receptor as previously described (Zhuang et al. 2009). The cells were also transfected with elements from the Dual-Glo Luciferase Assay System (Promega): the cAMP response element (CRE) coupled with a luciferase reporter gene (10ng/well); and the constitutively active SV40 promoter region coupled with the Renilla luciferase (RL) reporter (5ng/well). Each OR was transfected into 3 columns (24 wells) of the 96-well plate. About 18–24h after transfection, the medium was replaced with 0, 0.1, 0.316, 1, 3.16, 10, or 31.6 μM of androstenone or decenal (a negative control) in CD293 (Gibco). The cells were incubated at 37°C for 3.5h, then stimulated with 10 μL of Dual-Glo luciferase buffer (Promega) per well, then read using BMG Labtech POLARStar Optima plate reader. Promega Stop-and-Glo buffer and RL substrate were then added and the plates were read again. The degree of activation was quantified for each well in Microsoft Excel using the formula (L − 400)/(RL − 400), where L is the plate reader output after luciferase activation and RL is the output after addition of RL substrate.
Data analyses
PHASE (Stephens et al. 2001; Stephens and Scheet 2005) was used to reconstruct ancestral haplotypes for the Coriell populations (1000 Genomes data were already phased). To identify signatures of selection in human OR7D4, we tested the null-hypothesis that all mutations are selectively neutral (Kimura 1983) via Ka/Ks ratio and an analysis of expected and observed amounts of mutation and genetic drift using Fu and Li’s D (with Pan troglodytes and Gorilla gorilla as outgroups) (Fu and Li 1993) in DnaSP (Librado and Rozas 2009). We visualized the geographical distribution of allele frequencies in the statistical package R (R Development Core Team 2008) using the “Rworldmap” package (South 2011). Using the geographic distribution of alleles and geographic proximity of populations, we created groups and tested the null hypothesis that the geographic distribution of phased haplotypes do not show geographical genetic structure using ARLEQUIN (20000 permutations) (Excoffier and Lischer 2010). Finally, we used fixation index values to determine if genetic structure had an impact on selection.
Results
Variation data
There were 45 SNPs (33 missense, 17 of which were singletons; 12 synonymous, 5 of which were singletons) among populations in the coding region of human OR7D4 (Supplementary Table S1). Full allele frequency data for this sample are found in Supplementary Table S2 (global minor allele frequency data from NCBI are reported in Supplementary Table S1); data for 50 haplotypes (25 of which are singletons) are found in Supplementary Table S3. The most common missense SNPs were the linked pair associated with androstenone perception variation (rs61729907, rs5020278); these were found at an average frequency of 16% across our sample with the highest frequencies in Eurasia—up to 40% in some populations. The mutation rs61732668 results in a severely impaired receptor and phenotype that is insensitive to androstenone (or sensitive only in very high concentrations). The mutation was found in most African populations (excepting Mbuti and North Africa) with several having a frequency of 20%. The mutation was also found in some new world populations at a rate of 2% (Puerto Rico, Columbia, and Peru) and 12% (Afro-Caribbean population in Barbados). The mutation rs5020280 results in a phenotype of increased sensitivity to androstenone and was found in all African populations (excepting Biaka and Sub-Saharan Africa) at a rate between 1% and 10%. New world populations with the mutation included Puerto Rico, Afro-Caribbean, and Mexicans in LA at a rate of less than 1%—the same rates were found in Kihn, Beijing Han, and Tuscans. The most common synonymous SNP is rs8109935.
Two Denisova variants were identified across multiple sequences (≥75%); one was a missense mutation resulting in an amino acid change in the sixth transmembrane region and the other was a synonymous change also found in modern humans. Nine variants were identified in Altai Neandertal (7 of which are shared with modern humans) but none appeared in more than 3 sequences (≥10%) and are likely to be artifacts of genome degeneration or sequencing errors. Altai Neandertal and Denisova had no variants in common.
Selection
The ratio of substitution rates at nonsynonymous and synonymous sites (Ka/Ks) was 0.392 when humans were compared with 2 apes (chimpanzee and gorilla) and 0.474 when compared only to chimpanzee (Supplementary Table S4). Results of a coalescent model of Fu and Li’s D statistic and Tajima’s D statistic (Supplementary Table S4) suggested some variants may have been selected across human OR7D4. A sliding windows analysis with a window size of 200 and a step size of 50 (Supplementary Table S5) identified androstenone variants as having statistically significant low values (−4.86). One other window had a slightly lower value (−5.07) and may have contributed to the overall low Fu and Li’s D value; the only nonsingleton missense variant in that windows was rs138510982 which causes a mutation in the seventh transmembrane region. A coalescent model of Fu and Li’s D* for specific populations identified 5 populations with statistically significantly low values (Figure 1 and Supplementary Table S4).
Figure 1.
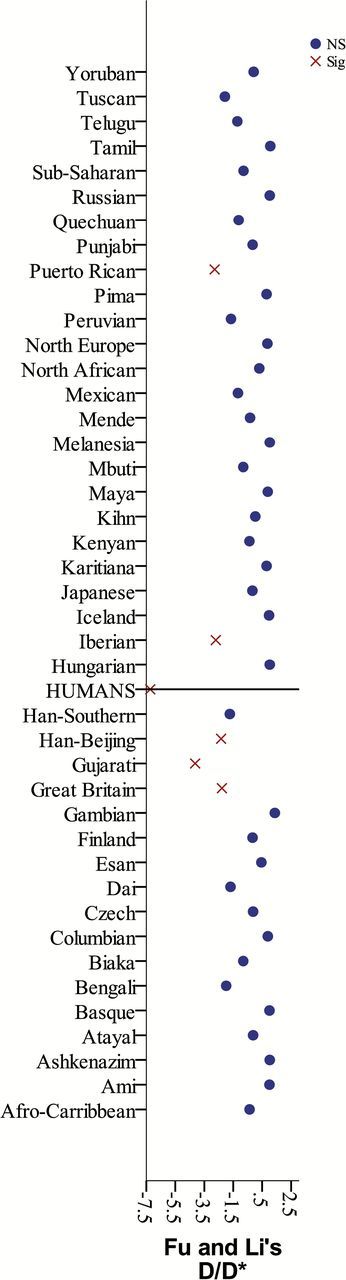
Selection test results for Fu and Li’s D (compared with chimpanzee and gorilla) and D* by population (except Surui, who are monomorphic).
Genetic and geographic variation
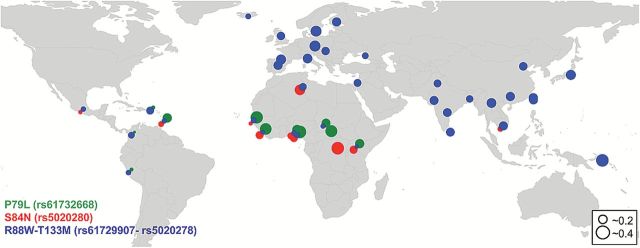
The global distribution of androstenone-related alleles (rs61729907–rs5020278, rs61732668, rs5020280) suggested a geographic pattern of increased frequencies in the linked pair outside of Africa and loss of the other 2 nonlinked alleles (Figure 2); while Gujurati have the nonlinked alleles, those are carried on singleton haplotypes (Supplementary Table S3). Furthermore, African populations are dominated by the nonlinked alleles (rs61732668, rs5020280) rather than the linked alleles (rs61729907–rs5020278), except in the North African and Mbuti populations.
Figure 2.
Geographic distribution of 4 main androstenone variants affecting phenotype.
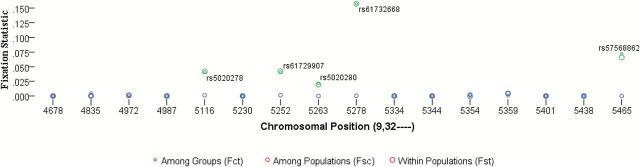
We used geographic regions within continents as well as the distribution of androstenone alleles to create groups of populations in order to test genetic structure using analysis of molecular variation (AMOVA). AMOVA results (Supplementary Table S6) indicated significant molecular variation among-groups, 6.62% (F CT: 0.07, P = 0.00), within-populations, 92.59% (F ST: 0.07, P = 0.00), and among-populations within groups 0.79% (F SC: 0.01; P = 0.00). The majority of the variation is within populations. Fixation for nonsingleton missense variants are visualized in Figure 3 (negative values set to 0); there were 5 variants with statistically significant fixation values. About 3 of the 5 variants with statistically significant values (rs5020280, rs61729907, and rs5020278) shared the same F ST and F CT fixation values (with no statistically significant fixation for F SC); these variants included the 2 linked androstenone SNPs and the 1 variant that increases sensitivity (rs5020280). The among-group and within-populations fixation value for the allele that decreases androstenone sensitivity (rs61732668) were the only high values across the table; this variant was predominant in African populations and new world populations with African admixture. The final variant, rs57568862, had the highest among-populations value and a within-populations value similar to the linked pair; this variant appeared only in Yoruba, Sub-Saharan Africa, and Afro-Caribbean from Barbados.
Figure 3.
Nonsingleton missense variant fixation values from AMOVA (negative values of FST were set to 0).
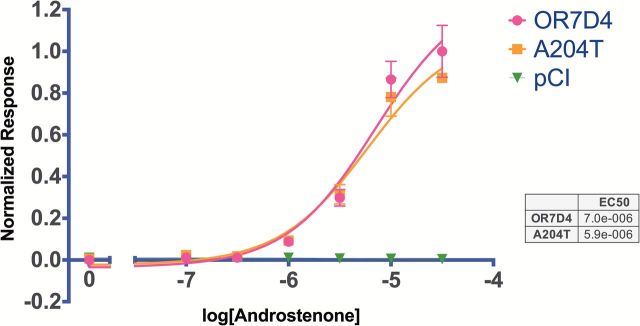
The reconstruction of Denisovan OR7D4 in vitro resulted in functional data not statistically different from human OR7D4: the Denisova receptor responded to androstenone in a similar fashion to the modern human OR7D4 wild type receptor (Figure 4).
Figure 4.
Denisova and Human OR7D4 functional response data: normalized in vitro dose response of human OR7D4 and the Denisova variant A204T to androstenone, compared with vector pCI with no insert, as measured in a luciferase assay.
Discussion
There is tremendous functional variability in the human sense of smell (Mainland et al. 2014) which may be attributable to population specific selective pressure to local environments (Menashe et al. 2002; Gilad and Lancet 2003) or food (Jaeger et al. 2013). Significant population differentiation is a major indicator of local adaptation but it may be confounded by demography especially for species with relatively low effective population size, such as humans. The variation present in OR7D4 alters perception of the odorant androstenone, rendering those with mutations less sensitive to the smell and rating it positively (Keller et al. 2007; Alonso et al. 2008). We analyzed the geographic distribution of genetic variants across OR7D4 in 43 human populations and identified possible evolutionary adaptive trends.
Tests for selection included the Ka/Ks test and coalescent modelling of allele frequency spectra tests. The low Ka/Ks ratio indicates there have been more synonymous than nonsynonymous changes which suggests purifying selection or conservation of the ancestral state. But, a low ratio also could suggest an evolutionary history of both positive and purifying selection acting within different areas of the gene or at different points in its evolutionary history. Indeed, the results of the coalescent modeling of Tajima’s D and Fu and Li’s D and Fu and Li’s D* indicated a signature of positive selection acting on OR7D4 whereas the sliding window analysis indicated that the high frequency alleles associated with phenotypic variation in androstenone perception were the targets of that selection. The population-specific tests gave mixed results: five populations had statistically significant and negative D* but not statistically significant Tajima’s D.
Fixation values aid interpretation of the discrepancy between the Ka/Ks and frequency spectra tests results. Under a model of purifying selection, the expectation is decreasing levels of diversity in the targeted gene. In humans, there are more missense mutations than synonymous mutations and less functioning alleles are at higher frequencies in various world populations in comparison to normal functioning alleles. Indeed, the highest statistically significant F-value from the AMOVA (rs61732668, 0.157) is one that impairs receptor functioning and sensitivity to androstenone; high F ST values relative to other SNPs are interpreted as evidence for natural selection (Weir and Cockerham 1984). Thus, the high frequency of variants that impair receptor function, high fixation values for deleterious alleles, and selection test results all suggest the interpretation of the Ka/Ks ratio as maintenance of the ancestral state via purifying selection is not likely.
Another possible explanation for the signature of positive selection is population structure (e.g., demographic expansion). The geographic distribution of alleles associated with androstenone perception suggested a typical pattern—more variation in African populations (Tishkoff et al. 2002; Campbell et al. 2008; Jorde et al. 2000; Li et al. 2008; Tishkoff et al. 2009) and low variation in Native American populations (Wang et al. 2007). An unexpected pattern identified was that Africans had higher frequencies of variation in the 2 unlinked androstenone alleles whereas Eurasians had higher frequencies of the linked pair. Indeed, the only populations with statistically significant selection statistics were Eurasian except Puerto Ricans; however, Puerto Ricans are an admixed new world population with a large contribution of their genetic makeup from Europe. One estimate for genome-wide European ancestry in Puerto Ricans is 67% (18% African and 15% Native American) (Tang et al. 2007) whereas another (using only 1000 Genomes data) estimate is 52% European (27% African, 11% Native American, 9% southwest Asian) (Elhaik et al. 2014). Furthermore and excluding singleton haplotypes, 1 of the 2 haplotypes (Haplotype 6) containing the linked pair is found exclusively in Eurasian populations and Puerto Ricans; the other haplotype containing the linked pair is found in all world populations (excepting indigenous Americans and Mbuti and Biaka) and is at the highest frequency in Europe and Asia (19%), the lowest frequency in Africa (6%), and a low frequency in admixed new world populations (9%). Of the two unlinked alleles, rs61732668, is found only in African and New World admixed populations and rs50202280 exhibits the same pattern except it is also found in 1 Han from Beijing and 3 Kihn. With the exception of rs61732668, fixation values (excepting rs61732668) were low compared with the average human F ST of ≈0.11 (Akey et al. 2002; Weir et al. 2005; Barreiro et al. 2008). The low fixation values suggest that the statistically significant selection test results are not likely to have been influenced by population structure (Akey et al. 2002).
A final explanation might be that relaxed constraint is operating on OR7D4. Taken as a whole, pseudogenization in the olfactory receptor gene family has been attributed to relaxed functional constraint (Glusman et al. 2001; Gimelbrant et al. 2004). But, there is also evidence of positive selection operating on olfactory receptor genes (Gilad et al. 2000; Menashe et al. 2002; Gilad and Lancet 2003; Gilad et al. 2003, 2005; Gimelbrant et al. 2004; Alonso et al. 2008; Moreno-Estrada et al. 2008). Differentiating positive selection and relaxed constraint might be particularly challenging in olfactory receptor genes due to the rapid evolution of functional sequences and rapid expansion of the olfactory receptor gene family (Vallender et al. 2004).
If selection is operating, why might lower functioning alleles be selected? One environmental source of androstenone is human bodily secretions with more odorant in male sweat and the amount of sweat excreted varying across populations and among individuals; the androstenone-related human steroid androstadienone has been putatively identified as a pheromone conveying masculinity in humans (Zhou et al. 2014) but all roles for these compounds in human chemical communication are contested (Wyatt 2015). This explanation is not very likely. Another source of androstenone is Sus Scrofa, or boar. Boar originated in Asia and Eurasian populations in our study had higher frequencies of the linked alleles that reduce phenotypic sensitivity to boar taint. But, boar bones are not found in high frequency in archaeological sites in Asia until around the Neolithic when they were domesticated. During the Neolithic, pigs were a central resource in the domestication package that spread rapidly throughout Asia into Europe and the Middle East (Larson et al. 2010). Modern Europeans carrying the linked alleles showed minimal (if any) aversion to the heavy concentration of androstenone in non-castrated male pigs (Lunde et al. 2012); perhaps the lack of averseness to boar taint influenced the rapid domestication of these animals during the Neolithic.
Olfaction is a primary sense involved in the detection of environmental and conspecific cues (e.g., recognition of mates, offspring recognition, predators, tainted food, chemical dangers). Mutations often accumulate on redundant genes, including olfactory receptor genes, rendering them nonfunctional over time. This pattern is evident in humans but we also see new functional variants emerge (Mainland et al. 2014). An example in a related family of genes is seen in the pseudogenization and loss of selective constraints on bitter-taste receptors in higher primates alongside evidence for positive selection in TAS2R16 (Wooding et al. 2004, 2006; Kim et al. 2005; Soranzo et al. 2005), which exhibits geographically patterned variation linked to adaptation. Our study is the first analysis of variation in a human olfactory receptor across a large sample of world populations and paleogenomes, and indicates how a range of anthropological, genetic, and neurobiological skills will be necessary to unravel how our olfactory genome evolved.
Supplementary material
Supplementary material can be found at http://www.chemse.oxfordjournals.org/
Funding
This work was partially supported by the National Institutes of Health (DC005782) to H.M.
Supplementary Material
Acknowledgments
M. J. Ni for assistance and Dr Jianghai Ho for expert advice.
References
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. 2002. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 12(12):1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso S, López S, Izagirre N, de la Rúa C. 2008. Overdominance in the human genome and olfactory receptor activity. Mol Biol Evol. 25(5):997–1001. [DOI] [PubMed] [Google Scholar]
- Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. 2008. Natural selection has driven population differentiation in modern humans. Nat Genet. 40(3):340–345. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. 1991. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 65(1):175–187. [DOI] [PubMed] [Google Scholar]
- Busse D, Kudella P, Grüning NM, Gisselmann G, Ständer S, Luger T, Jacobsen F, Steinsträßer L, Paus R, Gkogkolou P, et al. 2014. A synthetic sandalwood odorant induces wound-healing processes in human keratinocytes via the olfactory receptor OR2AT4. J Invest Dermatol. 134(11):2823–2832. [DOI] [PubMed] [Google Scholar]
- Campbell MC, Tishkoff SA. 2008. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. 9:403–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhaik E, Tatarinova T, Chebotarev D, Piras IS, Maria Calò C, De Montis A, Atzori M, Marini M, Tofanelli S, Francalacci P, et al. ; Genographic Consortium. 2014. Geographic population structure analysis of worldwide human populations infers their biogeographical origins. Nat Commun. 5:3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 10(3):564–567. [DOI] [PubMed] [Google Scholar]
- Fu YX, Li WH. 1993. Statistical tests of neutrality of mutations. Genetics. 133(3):693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Segré D, Skorecki K, Nachman MW, Lancet D, Sharon D. 2000. Dichotomy of single-nucleotide polymorphism haplotypes in olfactory receptor genes and pseudogenes. Nat Genet. 26(2):221–224. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Bustamante CD, Lancet D, Pääbo S. 2003. Natural selection on the olfactory receptor gene family in humans and chimpanzees. Am J Hum Genet. 73(3):489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Lancet D. 2003. Population differences in the human functional olfactory repertoire. Mol Biol Evol. 20(3):307–314. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Man O, Glusman G. 2005. A comparison of the human and chimpanzee olfactory receptor gene repertoires. Genome Res. 15(2):224–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelbrant AA, Skaletsky H, Chess A. 2004. Selective pressures on the olfactory receptor repertoire since the human-chimpanzee divergence. Proc Natl Acad Sci U S A. 101(24):9019–9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G, Yanai I, Rubin I, Lancet D. 2001. The complete human olfactory subgenome. Genome Res. 11(5):685–702. [DOI] [PubMed] [Google Scholar]
- Griffin CA, Kafadar KA, Pavlath GK. 2009. MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev Cell. 17(5):649–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Karp PH, Brody SL, Pierce RA, Welsh MJ, Holtzman MJ, Ben-Shahar Y. 2014. Chemosensory functions for pulmonary neuroendocrine cells. Am J Respir Cell Mol Biol. 50(3):637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin Y, Olender T, Khen M, Gonzaga-Jauregui C, Kim PM, Urban AE, Snyder M, Gerstein MB, Lancet D, Korbel JO. 2008. High-resolution copy-number variation map reflects human olfactory receptor diversity and evolution. PLoS Genet. 4(11):e1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger SR, McRae JF, Bava CM, Beresford MK, Hunter D, Jia Y, Chheang SL, Jin D, Peng M, Gamble JC, et al. 2013. A Mendelian trait for olfactory sensitivity affects odor experience and food selection. Curr Biol. 23(16):1601–1605. [DOI] [PubMed] [Google Scholar]
- Jorde LB, Watkins WS, Bamshad MJ, Dixon ME, Ricker CE, Seielstad MT, Batzer MA. 2000. The distribution of human genetic diversity: a comparison of mitochondrial, autosomal, and Y-chromosome data. Am J Hum Genet. 66(3):979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H. 2007. Genetic variation in a human odorant receptor alters odour perception. Nature. 449(7161):468–472. [DOI] [PubMed] [Google Scholar]
- Kim U, Wooding S, Ricci D, Jorde LB, Drayna D. 2005. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Hum Mutat. 26(3):199–204. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1983. The neutral theory of evolution. Cambridge: Cambridge University Press. [Google Scholar]
- Knaapila A, Hwang LD, Lysenko A, Duke FF, Fesi B, Khoshnevisan A, James RS, Wysocki CJ, Rhyu M, Tordoff MG, et al. 2012a. Genetic analysis of chemosensory traits in human twins. Chem Senses. 37(9):869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaapila A, Zhu G, Medland S, Wysocki C, Montgomery G, Martin N, Wright M, Reed D. 2012b. A genome-wide study on the perception of the odorants androstenone and galaxolide. Chem Senses. 37:541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause J, Fu Q, Good JM, Viola B, Shunkov MV, Derevianko AP, Pääbo S. 2010. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 464(7290):894–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Liu R, Zhao X, Yuan J, Fuller D, Barton L, Dobney K, Fan Q, Gu Z, Liu XH, et al. 2010. Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA. Proc Natl Acad Sci U S A. 107(17):7686–7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 319(5866):1100–1104. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25(11):1451–1452. [DOI] [PubMed] [Google Scholar]
- Loos HM, Doucet S, Soussignan R, Hartmann C, Durand K, Dittrich R, Sagot P, Buettner A, Schaal B. 2014. Responsiveness of human neonates to the odor of 5α-androst-16-en-3-one: a behavioral paradox? Chem Senses. 39(8):693–703. [DOI] [PubMed] [Google Scholar]
- Lunde K, Egelandsdal B, Skuterud E, Mainland JD, Lea T, Hersleth M, Matsunami H. 2012. Genetic variation of an odorant receptor OR7D4 and sensory perception of cooked meat containing androstenone. PLoS One. 7(5):e35259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainland JD, Keller A, Li YR, Zhou T, Trimmer C, Snyder LL, Moberly AH, Adipietro KA, Liu WL, Zhuang H, et al. 2014. The missense of smell: functional variability in the human odorant receptor repertoire. Nat Neurosci. 17(1):114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A, Go Y, Niimura Y. 2010. Degeneration of olfactory receptor gene repertories in primates: no direct link to full trichromatic vision. Mol Biol Evol. 27(5):1192–1200. [DOI] [PubMed] [Google Scholar]
- McRae JF, Jaeger Sara R, Bava Christina M, Beresford Michelle K, Hunter D, Jia Y, Chheang Sok L, Jin D, Peng M, Gamble Joanna C, et al. 2013. Identification of regions associated with variation in sensitivity to food-related odors in the human genome. Curr Biol. 23:1596–1600. [DOI] [PubMed] [Google Scholar]
- Menashe I, Man O, Lancet D, Gilad Y. 2002. Population differences in haplotype structure within a human olfactory receptor gene cluster. Hum Mol Genet. 11(12):1381–1390. [DOI] [PubMed] [Google Scholar]
- Menashe I, Man O, Lancet D, Gilad Y. 2003. Different noses for different people. Nat Genet. 34(2):143–144. [DOI] [PubMed] [Google Scholar]
- Moreno-Estrada A, Casals F, Ramírez-Soriano A, Oliva B, Calafell F, Bertranpetit J, Bosch E. 2008. Signatures of selection in the human olfactory receptor OR5I1 gene. Mol Biol Evol. 25(1):144–154. [DOI] [PubMed] [Google Scholar]
- Neuhaus EM, Zhang W, Gelis L, Deng Y, Noldus J, Hatt H. 2009. Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J Biol Chem. 284(24):16218–16225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olender T, Feldmesser E, Atarot T, Eisenstein M, Lancet D. 2004. The olfactory receptor universe—from whole genome analysis to structure and evolution. Genet Mol Res. 3(4):545–553. [PubMed] [Google Scholar]
- Olender T, Waszak SM, Viavant M, Khen M, Ben-Asher E, Reyes A, Nativ N, Wysocki CJ, Ge D, Lancet D. 2012. Personal receptor repertoires: olfaction as a model. BMC Genomics. 13:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluznick JL, Zou DJ, Zhang X, Yan Q, Rodriguez-Gil DJ, Eisner C, Wells E, Greer CA, Wang T, Firestein S, et al. 2009. Functional expression of the olfactory signaling system in the kidney. Proc Natl Acad Sci U S A. 106(6):2059–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 505(7481):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. (2008). R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; ISBN 3-900051-07-0 URL http://www.R-project.org. [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 468(7327):1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soranzo N, Bufe B, Sabeti PC, Wilson JF, Weale ME, Marguerie R, Meyerhof W, Goldstein DB. 2005. Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr Biol. 15(14):1257–1265. [DOI] [PubMed] [Google Scholar]
- South A. 2011. rworldmap: A new R package for mapping global data. R Journal. 3: 35–43. [Google Scholar]
- Spehr M, Gisselmann G, Poplawski A, Riffell JA, Wetzel CH, Zimmer RK, Hatt H. 2003. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 299(5615):2054–2058. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. 2001. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 68(4):978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Scheet P. 2005. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 76(3):449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Choudhry S, Mei R, Morgan M, Rodriguez-Cintron W, Burchard EG, Risch NJ. 2007. Recent genetic selection in the ancestral admixture of Puerto Ricans. Am J Hum Genet. 81(3):626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature. 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, et al. 2009. The genetic structure and history of Africans and African Americans. Science. 324(5930):1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM. 2002. Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet. 3(8):611–621. [DOI] [PubMed] [Google Scholar]
- UCSC Genome Browser. Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res. 12(6):996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallender EJ, Lahn BT. 2004. Positive selection on the human genome. Hum Mol Genet. 13 Spec No 2:R245–R254. [DOI] [PubMed] [Google Scholar]
- Veerappa AM, Vishweswaraiah S, Lingaiah K, Murthy M, Manjegowda DS, Nayaka R, Ramachandra NB. 2013. Unravelling the complexity of human olfactory receptor repertoire by copy number analysis across population using high resolution arrays. PLoS One. 8(7):e66843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeurgt C, Wilkin F, Tarabichi M, Gregoire F, Dumont JE, Chatelain P. 2014. Profiling of olfactory receptor gene expression in whole human olfactory mucosa. PLoS One. 9(5):e96333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Lewis CM, Jakobsson M, Ramachandran S, Ray N, Bedoya G, Rojas W, Parra MV, Molina JA, Gallo C, et al. 2007. Genetic variation and population structure in native Americans. PLoS Genet. 3(11):e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszak SM, Hasin Y, Zichner T, Olender T, Keydar I, Khen M, Stütz AM, Schlattl A, Lancet D, Korbel JO. 2010. Systematic inference of copy-number genotypes from personal genome sequencing data reveals extensive olfactory receptor gene content diversity. PLoS Comput Biol. 6(11):e1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG. 2005. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 15(11):1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Soc Study Hum Evol. 38(6):1358–1370. [DOI] [PubMed] [Google Scholar]
- Wooding S, Kim UK, Bamshad MJ, Larsen J, Jorde LB, Drayna D. 2004. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet. 74(4):637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S, Bufe B, Grassi C, Howard MT, Stone AC, Vazquez M, Dunn DM, Meyerhof W, Weiss RB, Bamshad MJ. 2006. Independent evolution of bitter-taste sensitivity in humans and chimpanzees. Nature. 440(7086):930–934. [DOI] [PubMed] [Google Scholar]
- Wyatt TD. 2015. The search for human pheromones: the lost decades and the necessity of returning to first principles. Proc Biol Sci. 282(1804):20142994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang H, Chien MS, Matsunami H. 2009. Dynamic functional evolution of an odorant receptor for sex-steroid-derived odors in primates. Proc Natl Acad Sci U S A. 106(50):21247–21251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, De la Cruz O, Pinto JM, Nicolae D, Firestein S, Gilad Y. 2007. Characterizing the expression of the human olfactory receptor gene family using a novel DNA microarray. Genome Biol. 8(5):R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Yang X, Chen K, Cai P, He S, Jiang Y. 2014. Chemosensory communication of gender through two human steroids in a sexually dimorphic manner. Curr Biol. 24(10):1091–1095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.