Summary
CRISPR-Cas adaptive immune systems protect bacteria and archaea against foreign genetic elements. In Escherichia coli, Cascade (CRISPR-associated complex for antiviral defense) is an RNA-guided surveillance complex that binds foreign DNA and recruits Cas3, a trans-acting nuclease-helicase for target degradation. Here we use single-molecule imaging to visualize Cascade and Cas3 binding to foreign DNA targets. Our analysis reveals two distinct pathways, dictated by the presence or absence of a protospacer adjacent motif (PAM). Binding to a protospacer flanked by a PAM recruits a nuclease-active Cas3 for degradation of short singlestranded regions of target DNA, whereas PAM mutations elicit an alternative pathway that recruits a nuclease-inactive Cas3 through a mechanism that is dependent upon the Cas1 and Cas2 proteins. These findings explain how target recognition by Cascade can elicit distinct outcomes, and supports a model for acquisition of new spacer sequences through a mechanism involving processive, ATP-dependent Cas3 translocation along foreign DNA.
Introduction
Many prokaryotes harbor an RNA-guided adaptive immune system comprised of a genetic locus called CRISPR (clustered regularly interspaced short palindromic repeats) and the CRISPR-associated (Cas) genes (Barrangou and Marraffini, 2014; van der Oost et al., 2014; Westra et al., 2012a). The CRISPR locus was first identified in E. coli as an unusual series of 29-base pair (bp) repeats separated by 32-bp spacer sequences (Ishino et al., 1987). It was later recognized that these spacers were derived from foreign genetic elements, suggesting the CRISPR locus might serve as an RNA-guided immune system (Bolotin et al., 2005; Makarova et al., 2006; Mojica et al., 2005). It is now known that CRISPR-Cas immunity is conferred through integration of short DNA fragments into the CRISPR locus, and these spacer sequences record the history of past infections (Barrangou and Marraffini, 2014; van der Oost et al., 2014; Westra et al., 2012a). The CRISPR locus is transcribed and the resulting transcript is processed into shorter CRISPR-RNAs (crRNAs), each containing a sequence complementary to a previously encountered foreign DNA element.
CRISPR-Cas systems are classified as Type I, II or III, which can be distinguished based on the presence of the signature Cas3, Cas9, or Cas10 genes, respectively (Barrangou and Marraffini, 2014; Westra et al., 2012a). Type I are the most common, and much of our understanding of Type I CRISPR-Cas systems comes from studies of E. coli Cascade (CRISPR-associated complex for antiviral defense), which is comprised of the five proteins Cse1, Cse2, Cas7, Cas5e, and Cas6e. These proteins assemble on a 61-nucleotide (nt) crRNA, yielding a 405-kilodalton (kDa) complex. The crRNA contains the 32-nt spacer sequence, which directs Cascade to sequences (protospacers) in foreign DNA, leading to formation of an R-loop intermediate. Cascade then recruits Cas3, which has an N-terminal histidine-aspartate (HD) nuclease domain and C-terminal superfamily 2 (SF2) helicase domain, to degrade the DNA (Mulepati and Bailey, 2013; Sinkunas et al., 2013).
Cascade must discriminate between spacer sequences found in the bacterial chromosome and those found in foreign DNA. This discrimination is thought to be accomplished through recognition of a tri-nucleotide sequence motif called the PAM (protospacer adjacent motif; 5’-A[A/T]G-3’ for E. coli Cascade), which is adjacent to the protospacer in foreign DNA, but absent in the CRISPR locus. Strict sequence requirements present a potential weakness because mutations in either the PAM or protospacer can allow foreign DNA to escape CRISPR-Cas immunity (Semenova et al., 2011). However, bacteria can rapidly restore immunity using a positive feedback loop to update the CRISPR locus (Datsenko et al., 2012; Fineran et al., 2014). The mechanism of primed spacer acquisition (priming) remains perhaps one of the most poorly understood aspects of CRISPR-Cas immunity (Datsenko et al., 2012; Fineran et al., 2014; Heler et al., 2014). Priming requires Cascade with a crRNA bearing at least partial complementarity to the escape target, suggesting Cascade must be able to locate targets even when they bear mutations sufficient to escape immunity (Datsenko et al., 2012). Priming also requires Cas3 (Datsenko et al., 2012) and the Cas1–Cas2 complex (Nunez et al., 2014), which integrate new sequences into the CRISPR locus (Nunez et al., 2015). It is not known how these complexes elicit the priming response to foreign elements bearing escape mutations.
Here we use single-molecule imaging to visualize individual Cascade complexes as they search for protospacers within the bacteriophage λ genome. Our work reveals PAM-dependent and PAM-independent search pathways. The PAM-dependent pathway is highly efficient, and allows Cascade to recruit Cas3 for strand-specific degradation of the target genome. The PAM-independent pathway is less efficient, but Cascade can still bind tightly to the DNA, ensuring that it can initiate the sequence of molecular events that precede primed spacer acquisition. Through this pathway, Cas3 recruitment becomes strictly dependent upon Cas1–Cas2, and Cas1–Cas2 also attenuate Cas3 nuclease activity and enable Cas3 to rapidly translocate in either direction along the foreign DNA. These results establish Cas1–Cas2 as a trans-acting factor necessary for the recruitment and regulation of Cas3 at escape targets. Based on our findings, we propose a mechanistic framework describing how Cascade, Cas1, Cas2, and Cas3 work together to process and disable foreign genetic elements.
Results
DNA curtain assay for target binding by Cascade
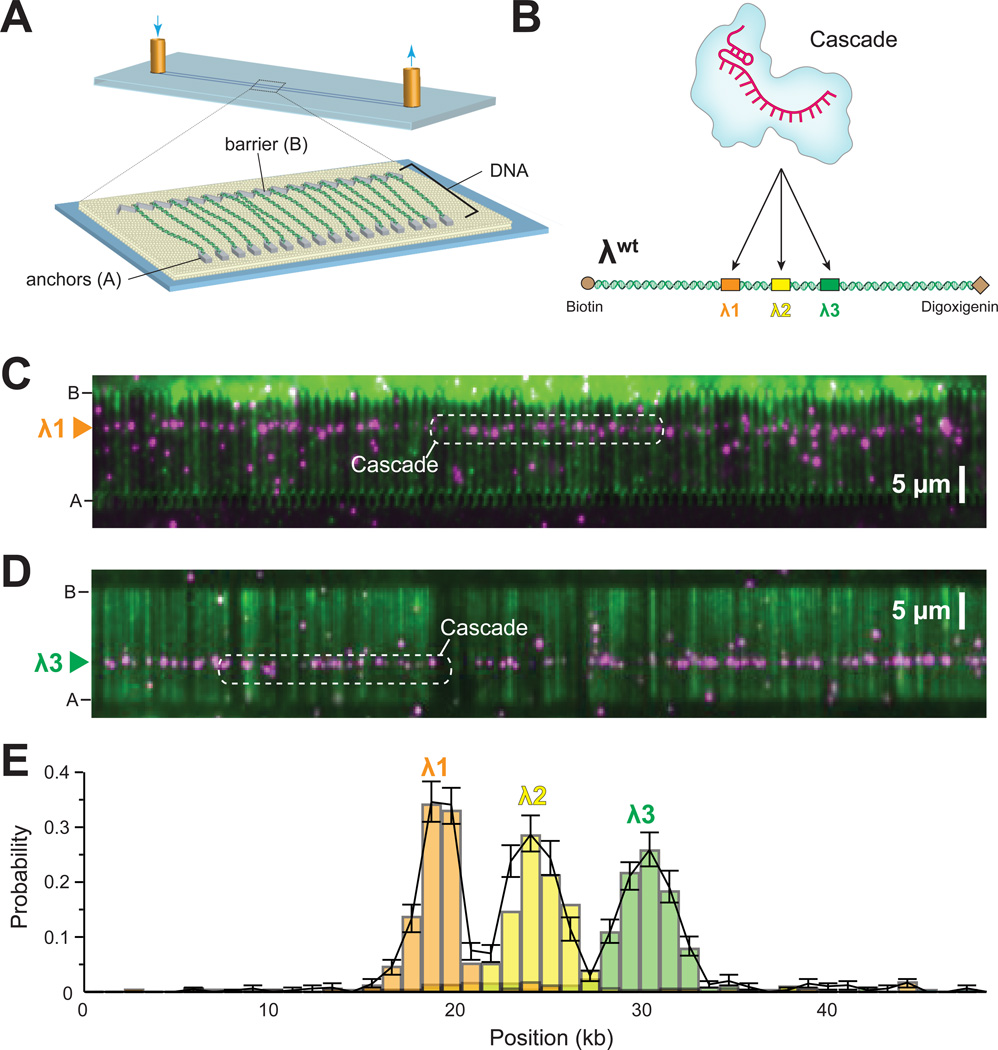
We sought to establish a DNA curtain assay using total internal reflection fluorescence microscopy (TIRFM) for visualizing the behavior of Cascade on individual molecules of wildtype phage λ DNA (λwt) (Figure 1A & Supplemental Information)(Greene et al., 2010). In brief, the surface of a microfluidic sample chamber was coated with a lipid bilayer, and DNA molecules were anchored to the bilayer through a biotin-streptavidin interaction. The DNA was then pushed to the leading edges of nanofabricated barriers to lipid diffusion, and the downstream ends were anchored to pedestals through an antibody-hapten linkage (Gorman et al., 2010; Gorman et al., 2012). Cascade was prepared with one of three crRNAs targeted to different regions of λwt, and then labeled with antiFLAG-quantum dots (QDs) attached to the 3xFLAGtagged Cas6e subunit (Figure 1B). When visualized on DNA curtains, Cascade bound to target sites corresponding to DNA sequences complementary to the three different crRNAs (Figure 1C–E). Cascade remained bound for at least 57 minutes; this lifetime represents a lower limit for the Cascade-protospacer interaction because these measurements are limited by the stability of the 3xFLAG-antiFLAG interaction (Sternberg et al., 2014). Stable binding was not observed for Cascade bearing a control crRNA (P7-crRNA) that was not complementary to λwt.
Figure 1. Programed target binding by E. coli Cascade.
(A) Overview of DNA curtains. (B) Schematic of E. coli Cascade programed with a crRNA targeting one of three different binding sites (designated λ1, λ2, and λ3) on λwt. (C) Wide-field TIRFM image showing QD-tagged Cascade (magenta) bound to DNA (green) at λ1. (D) Wide-field image showing Cascade bound at λ3. (E) Binding distribution for Cascade targeted to each of the three protospacers; error bars in this and all subsequent binding distributions represent 95% confidence intervals obtained through bootstrap analysis.
PAM-dependent target recognition
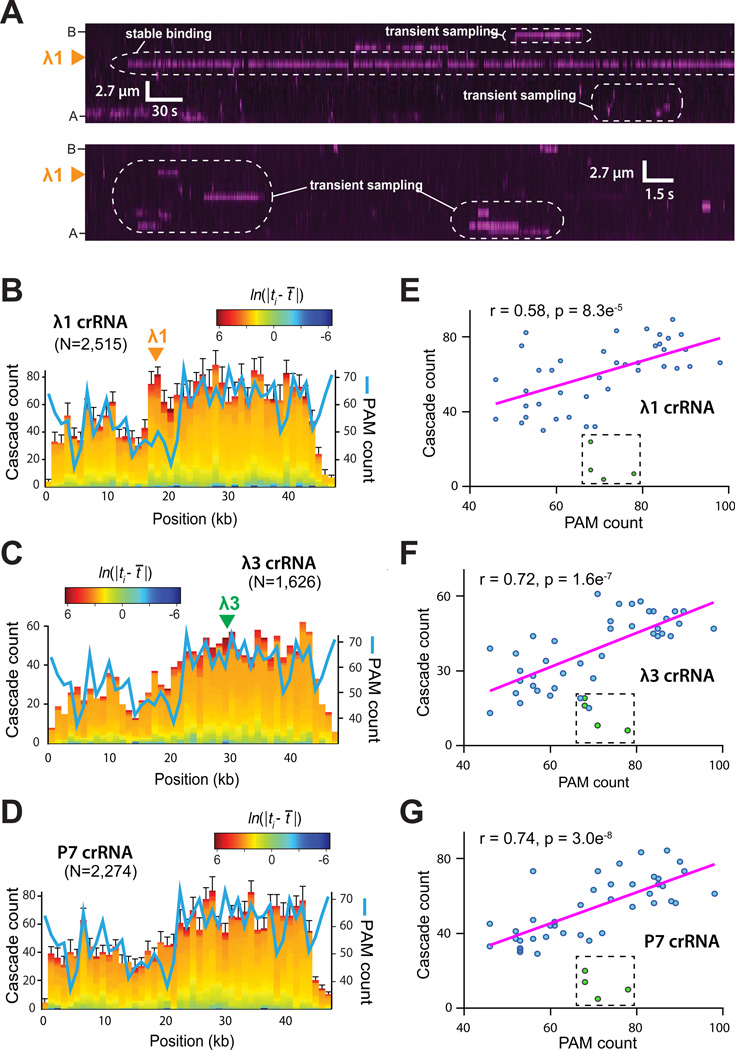
We next sought to determine how Cascade locates protospacers by visualizing reactions in realtime. Most Cascade (>75%) appeared immediately at the protospacer without exhibiting any evidence of microscopically detectable motion along the DNA (Figure 2A). This finding leads us to conclude that Cascade located the protospacer through a pathway that was dominated by 3D diffusion at the microscopic scale. Based on our optical resolution limits these experiments provide an upper limit on any potential 1D diffusion by Cascade of no more than ~250-bp, although we do not exclude that possibility that Cascade may diffuse shorter distances along the DNA (Gorman et al., 2010; Gorman et al., 2012). The remaining fraction of Cascade molecules (<25%) underwent optically detectable 1D diffusion; we did not pursue a detailed analysis of this behavior because it coincided with a loss of binding specificity and appeared to arise from Cascade aggregates (not shown).
Figure 2. Cascade searches for PAMs while interrogating foreign DNA.
(A) Kymographs highlighting examples of Cascade binding events over two different time regimes (see scale bars). Examples of transient sampling and stable recognition are highlighted. Distribution of PAMs (blue line) and transient binding events for Cascade programed with (B) the λ1-crRNA, (C) the λ3-crRNA, or (D) a P7-crRNA. Count refers to number of occurrences within 1-kbp of DNA. The locations of the λ1 and λ3 target sites are indicated, and the heat map color-coding reflects the binding dwell time (ti) relative to the mean dwell time (t̄). Correlation of PAMs with the transient binding events for Cascade programed with (E) the λ1-crRNA, (F) the λ3-crRNA, or (G) P7-crRNA, as indicated. Outlying data points (colored green and boxed) reflect underrepresented binding events at PAM sites near the ends of the DNA; detection of binding at these sites is hindered by the chromium barriers. See also Figure S1.
Analysis of the 3D events revealed long-lived binding to the protospacers, as well as transient binding events all along the λwt DNA (Figure 2A). The λwt genome contains a total of 3,151 PAM sites (5’-A[A/T]G-3’), corresponding to ~1 PAM per 15.4-bp, which are asymmetrically distributed across the phage genome. Cascade did not randomly sample the DNA, instead the transient binding events were correlated with the PAM distribution (Figure 2E–G), as we have reported for S. pyogenes Cas9 (Sternberg et al., 2014). Control reactions using Cascade programed with P7-crRNA revealed a similar pattern of transient binding (Figure 2B–G), and we could detect no binding activity for Cascade lacking Cse1 (not shown), which is the subunit responsible for PAM recognition (Sashital et al., 2012). Cascade programed with either λ1-crRNA or λ3-crRNA displayed many reversible binding events at their targets, which are revealed by the ~50% increased prevalence of longer-lived intermediates at both of these target sites relative to non-target sites (Figure 2B–C & Figure S1A), and also by the peak in binding at λ1 for the λ1-crRNA, which is observable due to the overall lower density of PAM sites in this region of DNA (Figure 2B–C). This category of long-lived, but reversible binding events at the protospacers likely represents abortive engagement, suggesting Cascade must often make multiple attempts before stably engaging the protospacer, similar to what we have observed for Cas9 (Sternberg et al., 2014).
The transient binding events exhibited double-exponential decays similar to S. pyogenes Cas9 (Sternberg et al., 2014), with lifetimes of ~3 seconds and ~25 seconds (Figure S1A–D), indicating that at least two intermediates exist on the pathway towards target recognition. The lifetimes of these intermediates were not appreciably affected by either salt concentration or temperature (Figure S1D), similar to findings for Cas9 (Sternberg et al., 2014). These characteristics, more commonly attributed to site-specific association, provide further evidence that the initial observed interactions are based upon sequence-dependent association with PAM sites rather than nonspecific interactions with the DNA phosphate backbone.
PAM-independent target recognition
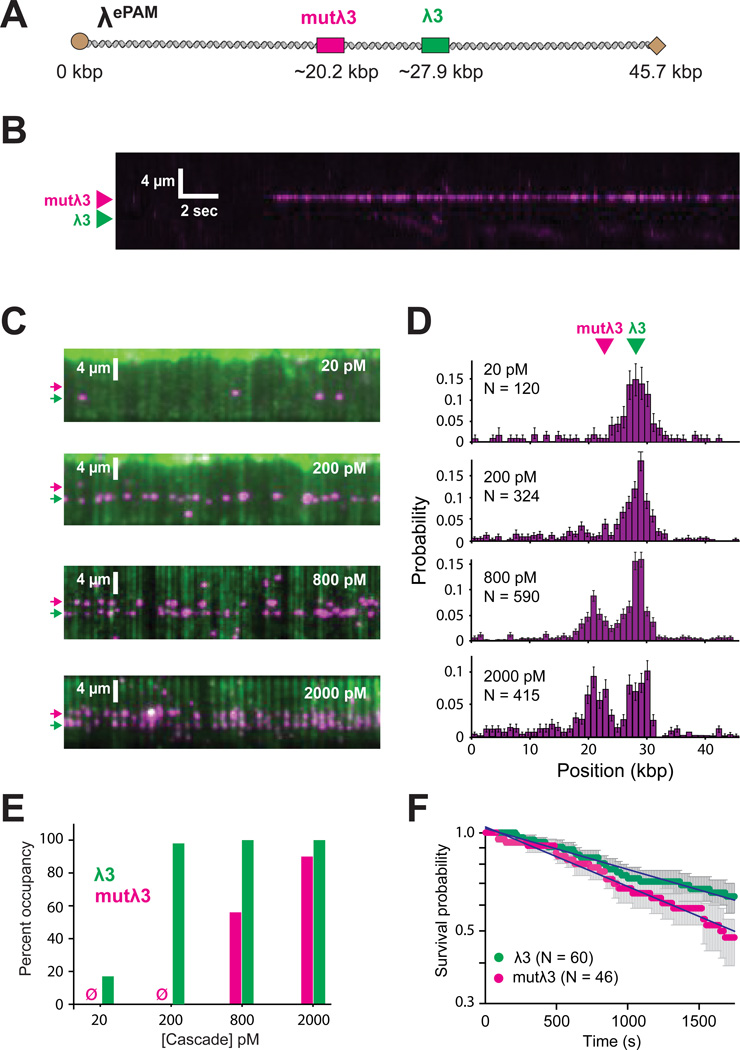
We next sought to determine whether and how Cascade locates targets that lack a canonical PAM. For this, we generated a new phage construct (λePAM) bearing two duplicate targets (Figure 3A). One of the protospacers (λ3) was adjacent to a cognate PAM [5’-ATG-3’], whereas second protospacer (mutλ3) was adjacent to a mutated PAM [5-ATT-3’]. This escape PAM (ePAM) was chosen because it enables an invading DNA to escape the CRISPR-Cas machinery, but still elicits a rapid priming response (Datsenko et al., 2012; Fineran et al., 2014). Surprisingly, Cascade could still bind both protospacers, and binding of mutλ3 still occurred through 3D diffusion (Figure 3B), but recognition of mutλ3 was much less efficient than recognition of λ3 (Figure 3C–E). This difference was evidenced by the ~10-fold higher Cascade concentration necessary to achieve similar levels of occupancy at both protospacers. Despite the large difference in initial recognition, the lifetimes of Cascade at λ3 and mutλ3 were comparable (57 min and 40 min, respectively; Figure 3F). We conclude that PAMs increase the efficiency of target recognition, but that Cascade is still capable of protospacer recognition and high-affinity binding in the absence of a cognate PAM, and this conclusion is consistent with previous studies (Szczelkun et al., 2014).
Figure 3. Recognition of escape PAM mutants.
(A) Schematic of λePAM bearing two identical protospacers, one with a cognate PAM (λ3) and the other with an escape PAM (mutλ3). (B) Kymograph highlighting example of Cascade binding to the mutλ3 through 3D diffusion. (C) Wide-field images showing binding to each of the two targets at different Cascade concentrations following a 10 minute incubation. Arrowheads indicate the locations of the λ3 (green) and mutλ3 (magenta) targets. (D) Binding distributions showing relative occupancy at each Cascade concentration. (E) Quantification of percent occupancy; Ø indicates no detectable binding. (F) Survival probability plots for Cascade bound to the two targets; error bars in this and all subsequent survival probability plots represent 70% confidence intervals obtained through bootstrap analysis.
Cas3 recruitment leads to disruption of the target DNA duplex
We next sought to visualize Cascade-dependent recruitment of Cas3. Cas3 interacts with Cse1, and the displaced ssDNA strand that is generated by R-loop formation (Hochstrasser et al., 2014; Mulepati and Bailey, 2013; Sinkunas et al., 2013). Upon recruitment, Cas3 first nicks the DNA, and is thought to then translocate in the 3’→5’ direction along the non-target strand while unwinding and degrading duplex DNA through an ATP-, Mg2+-, and Co2+-dependent mechanism (Mulepati and Bailey, 2011, 2013; Sinkunas et al., 2013). Cas3 degrades both target DNA strands in bulk biochemical assays (Mulepati and Bailey, 2013; Sinkunas et al., 2013). However, these measurements use relatively high concentrations of Cas3 (50 nM – 1 µM) (Hochstrasser et al., 2014; Mulepati and Bailey, 2011, 2013; Sinkunas et al., 2011; Sinkunas et al., 2013), suggesting that DNA degradation may be due to the action of multiple Cas3 molecules, only the first of which is directly recruited by Cascade. Given these considerations, it is plausible that the initial Cascade-recruited molecule of Cas3 only introduces a small nick or ssDNA gap in the target DNA (Mulepati and Bailey, 2013).
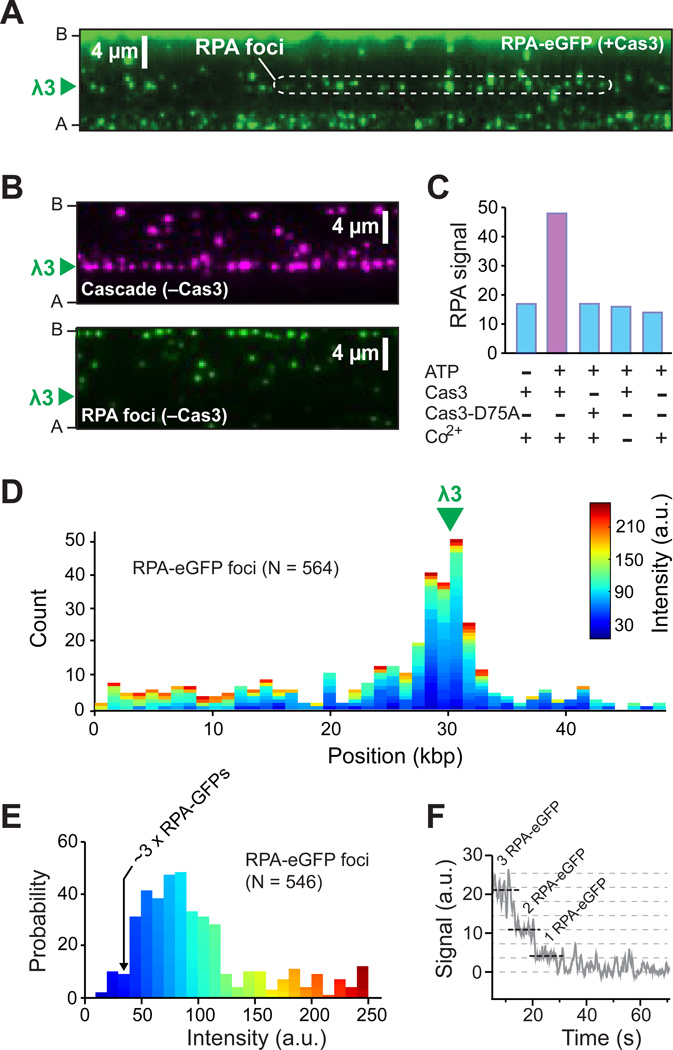
We reasoned that if Cas3 was initially generating ssDNA after loading at Cascade, then this might be revealed in reactions with low concentrations of Cas3 (4 nM) followed by the addition of eGFP-tagged replication protein A (RPA), which binds ssDNA. When RPA-eGFP was added after Cascade and Cas3, bright eGFP foci were detected at the λ3 protospacer (Figure 4A). Formation of RPA-eGFP foci was dependent upon Cascade, Cas3, ATP and Co2+ and the conditions under which we detected RPA-eGFP foci paralleled the conditions necessary for plasmid degradation in bulk biochemical assays (Figure 4B & Figure S2C). Furthermore, RPA-eGFP foci were not observed for a Cas3 nuclease mutant (D75A) (Figure 4C & Figure S2A–C). Notably, the DNA in the single-molecule assays was not liberated from the flowcell surface and there was no evidence for long tracts of RPA-eGFP, indicating that Cas3 only generated a small ssDNA gap. To estimate the size of the ssDNA gaps we measured the intensity of the RPA-eGFP foci (Figure 4D–E), and then used photobleaching steps to roughly estimate the number of RPA-eGFP molecules present (Figure 4F & Supplemental Information). We estimate that the average focus contained ~8–10 molecules of RPA-eGFP, corresponding to ~240–300 nts of ssDNA. These results suggest that the first Cas3 molecule recruited by Cascade makes a short ssDNA gap adjacent to the protospacer.
Figure 4. Cas3 generates a ssDNA gap at the λ3 protospacer.
(A) Image showing RPA-eGFP foci at λ3 for reactions with unlabeled Cascade and unlabeled Cas3. (B) Control images showing that RPA-eGFP foci are not present when Cas3 is omitted from the reactions; the upper and lower panels show the same field of view. (C) Requirements for RPA-eGFP foci formation at λ3. (D) Distribution of RPA-eGFP foci in reactions containing both Cascade and Cas3, Count refers to number of occurrences within 1-kbp of DNA. (E) Signal intensities for RPA-eGFP foci. The intensity of a focus comprised of 3 molecules of RPA-eGFP is indicated, and each successive bin corresponds to ~1 additional molecule of RPA-eGFP. The heat-map color-coding in (D) and (E) are the same. (F) Representative stepwise photo-bleaching curve used to estimate the number of RPA-eGFP molecules in each focus. See also Figure S2.
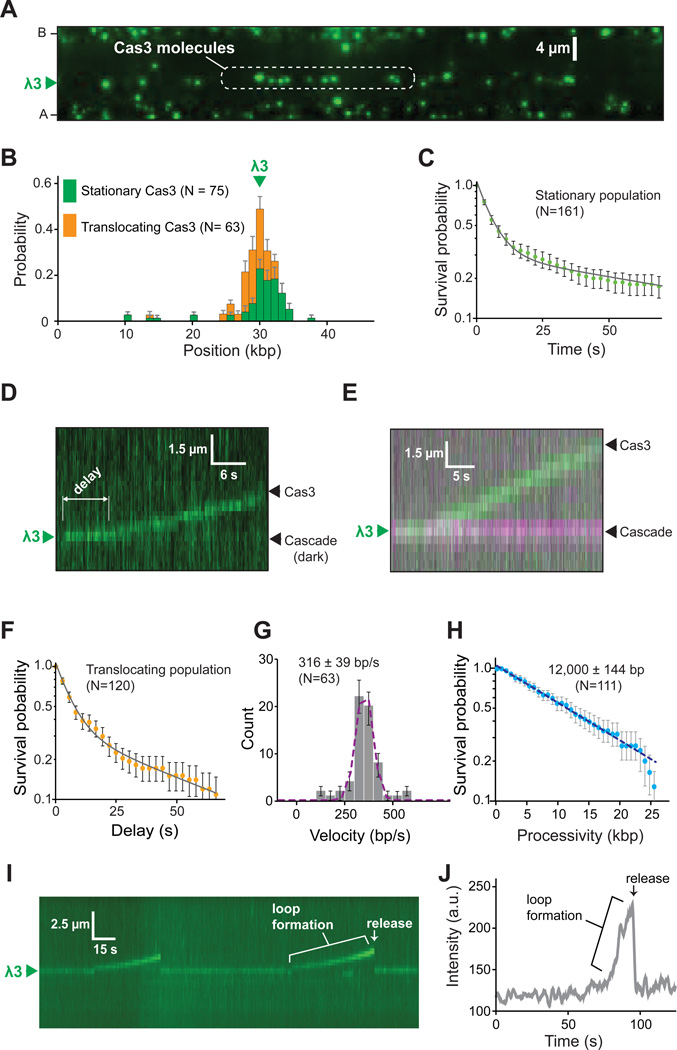
Cas3 recruitment to target-bound Cascade
We next sought to visualize the behavior of fluorescently-tagged Cas3 (Figure S3A & Supplemental Information). We were unable to detect stable binding of Cas3 to Cascade when ATP or Mg2+ were omitted, or when ATP was replaced with ADP or AMP-PNP (not shown). However, Cas3 bound stably to Cascade when ATP and Mg2+ were included in the reactions (Figure 5A & 5B). Cas3 located Cascade through 3D diffusion during initial recruitment (see Figure 5D below). Once bound, ~55% of the Cas3 molecules remained stationary within optical resolution limits (Figure 5B). These seemingly stationary molecules exhibited two distinct lifetimes: one population with a lifetime (τ1) of ~6 seconds; and a second population (τ2) with a lifetime of ≥1 minute (Figure 5C & Figure S3B–C). These findings suggest that Cas3 transiently samples target-bound Cascade before transitioning into a more stably bound state, and that entry into this longer-lived state requires ATP hydrolysis. Interestingly, once a longer-lived Cas3 binding event was observed at a given molecule of Cascade, then that particular Cascade complex appeared incapable of recruiting any additional Cas3 at the protein concentrations used in these assays.
Figure 5. Cascade-mediated recruitment of Cas3.
(A) Image showing that QD-tagged Cas3 is recruited to unlabeled Cascade at λ3. (B) Binding of Cas3 to λ3. The distribution is segregated into the translocation (orange) and stationary (green) Cas3 populations. (C) Survival probabilities of the stationary Cas3 population. (D) Kymograph illustrating the translocation of Cas3 away from λ3 in a reaction with unlabeled Cascade. The delay period prior to the initiation of Cas3 translocation is indicated. (E) Two-color experiment showing that Cas3 (green) translocates away from Cascade (magenta). (F) Survival probability (delay time) of the translocating population of Cas3 prior to moving away from λ3. (G) Cas3 velocity distribution. (H) Cas3 processivity distribution. (I) Kymograph showing an example of Cas3 repeatedly looping the DNA. (J) Intensity profile showing the increase in Cas3 fluorescence signal coinciding with DNA loop formation. See also Figure S3 and Video S1.
Cas3 is a highly processive molecular motor
Many of the Cas3 molecules (~45%) translocated along the DNA (Figure 5B & 5D–E). In these instances, Cas3 was recruited to Cascade at the λ3 protospacer and then moved rapidly away from the protospacer in a direction consistent with 3’→5’ translocation on the non-target strand, as expected from bulk biochemical experiments (Mulepati and Bailey, 2013). There was no evidence that Cas3 translocation could initiate from any other location on the DNA other than the λ3 protospacer, and Cas3 translocation was entirely dependent upon the presence of Cascade. Remarkably, Cascade remained tightly bound to the protospacer even after Cas3 had begun translocating along the DNA (Figure 5E). Moreover, once Cas3 had translocated away from Cascade, then no additional molecules of Cas3 could bind to or translocate away from that particular Cascade complex.
Cas3 exhibited a short delay prior to moving away from Cascade (Figure 5D); analysis of these delay times revealed two lifetimes that were similar to the τ1 and τ2 lifetimes for the stationary Cas3 population, suggesting that the observed intermediates reflected the same underlying molecular processes (Figure 5C, 5F & Figure S3B–C). Cas3 traveled at a mean velocity of ~316 bp/s for 12,000 bp before stalling or dissociating from the DNA (Figure 5G & 5H), and >99% of molecules exhibited unidirectional movement (Figure 5D & 5E, Supplemental Video S1, & see below). Three key observations suggested that Cas3 was not extensively degrading the DNA during translocation. First, there was no evidence that the translocating population of Cas3 caused double-strand breaks. Second, we saw no evidence for long ssDNA tracts when reactions were chased with RPA-eGFP. Finally, if Cas3 had generated tracts of ssDNA long enough to be optically detected, then Cascade would also appear to move in the same direction because of the change in persistence length that accompanies the conversion of dsDNA to ssDNA, but Cascade always remained stationary at the protospacer. We conclude that Cas3 is a highly processive molecular motor that first generates a small ssDNA gap and then translocates in 3’→5’ direction along the non-target DNA strand away from Cascade.
Evidence for looped DNA intermediates
Surprisingly, in addition to our observation that Cas3 recruitment and translocation did not coincide with ejection of Cascade from the DNA, inspection of the Cas3 translocation trajectories revealed evidence that the contacts between Cas3 and Cascade were not immediately broken. In many instances (14%) Cas3 began to translocate along the DNA, but then returned almost instantaneously to the original binding site (Figure 5I). This behavior coincided with an increase in Cas3 fluorescence, suggesting that the molecules were pulled closer to the surface of the flowcell because of increased tension on the DNA. These observations are most consistent with looped DNA intermediates where Cas3 maintains contact with Cascade while simultaneously translocating for a short distance along the flanking duplex DNA (Figure S3D). We conclude that Cas3 can initially remain bound to Cascade as it begins translocating along the DNA, and that a subset of these molecules generate optically detectable DNA loops.
PAM is essential for Cascade-mediated recruitment of Cas3
We next sought to determine whether Cascade could recruit Cas3 to mutλ3, and if so, whether the properties of Cas3 differ in the absence of a cognate PAM. Interestingly, Cas3 did not colocalize with Cascade at mutλ3 (Figure S4A), and we were unable to detect even transient binding of Cas3 to Cascade at the mutλ3 protospacer. We were also unable to detect RPA-eGFP foci at mutλ3 (Figure S4B–E), and Cas3 did not cleave plasmid substrates bearing the mutλ3 protospacer (see below). We conclude that Cascade cannot recruit Cas3 to DNA in the absence of a cognate PAM, in agreement with previous bulk biochemical experiments (Hochstrasser et al., 2014; Mulepati and Bailey, 2013).
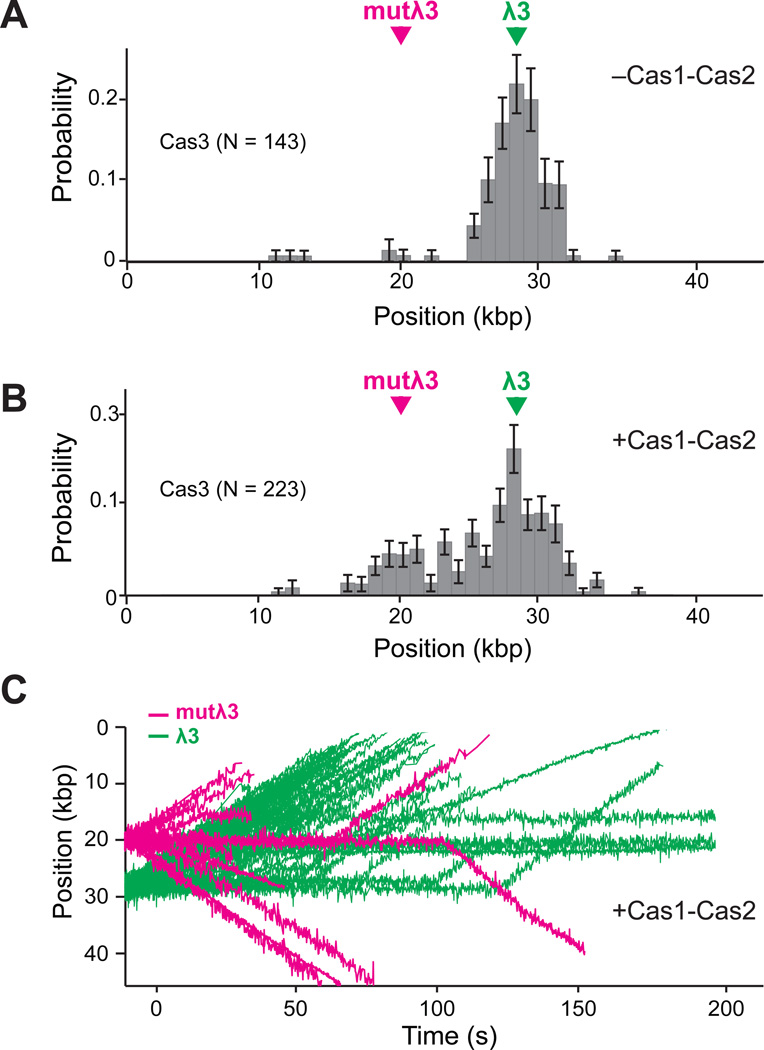
PAM-independent recruitment of Cas3 by Cas1–Cas2
Cas3 is required for primed sequence acquisition (Datsenko et al., 2012), suggesting that alternative pathways must exist to recruit Cas3 to escape targets. Cas1 and Cas2 are universally conserved across CRISPR types and are also necessary for primed sequence acquisition, suggesting the possibility that these proteins may work in concert with Cascade to promote the recruitment of Cas3 to escape targets. Therefore, we next asked whether the Cas1–Cas2 complex might affect target recognition, target processing, or both, in reactions with Cas3. Attempts to generate fluorescently tagged Cas1 or Cas2 yielded inactive proteins, therefore these experiments utilized wild-type (unlabeled) Cas1–Cas2.
Remarkably, the addition of Cas1–Cas2 enabled the recruitment of Cas3 to mutλ3 and also ~3-fold enhanced recruitment of Cas3 to λ3 (Figure 6A–B, Supplemental Video S1 & S2). The velocity and processivity of Cas3 were not altered by Cas1–Cas2 (Figure S5A–B). However, Cas3 recruited to the escape target behaved markedly different from Cas3 that was recruited to cognate protospacer. Most strikingly, Cas3 targeted to mutλ3 could rapidly translocate in either direction away from Cascade (Figure 6C & Supplemental Video S3). Moreover, Cas3 exhibited only a ~6 second delay prior to moving away from mutλ3, but there was no evidence for the second longer-lived intermediate (τ2) that was always observed at λ3 (Figure S3B–C & Figure S5C). There was also no evidence for ssDNA gaps at mutλ3 in the presence of Cas1–Cas2 (Figure S5D), and bulk biochemical assays with Cascade, Cas1–Cas2, and Cas3 revealed no nicking or cleavage of plasmids with the mutλ3 protospacer (Figure S6A–B), even though Cascade was capable of binding the mutλ3 protospacer in bulk assays (Figure S6C). Finally, there was no evidence for Cas3-mediated DNA looping at mutλ3 in reactions with Cas1–Cas2 (Figure 6C). Together, these results show that Cas1–Cas2 are necessary to recruit Cas3 to mutλ3, and attenuate the nuclease activity of Cas3 at these escape targets, enabling Cas3 to translocate away from Cascade in either direction along the foreign DNA.
Figure 6. Cas1–Cas2 mediated recruitment of Cas3 to escape targets.
(A) Binding distribution of Cas3 on λePAM in the absence of Cas1–Cas2. (B) Cas3 binding distribution histogram on λePAM in the presence of Cas1–Cas2. (C) Overlaid trajectories showing examples of Cas3 translocation events originating from either the λ3 protospacer (green) or the mutλ3 protospacer (magenta). Of the trajectories originating from mutλ3, 59% of the Cas3 molecules move towards the downstream anchor points, and the remaining 41% travel in the opposite direction. See also Figures S4 and S4, and Videos S2, S3, and S4.
Cas1–Cas2 also appeared to affect the behavior of Cas3 at the λ3 protospacer. Specifically, Cas1–Cas2 partially attenuated Cas3 nuclease activity in bulk biochemical assays (Figure S6A–B), and the presence of Cas1–Cas2 also enabled iterative Cas3 binding and translocation events from the same Cascade complex bound to the λ3 protospacer (Supplemental Video 4). This observation was in stark contrast to reactions done in the absence of Cas1–Cas2, where we never detected evidence of multiple Cas3 recruitment events to the same Cascade complex. These findings suggest that Cas1–Cas2 not only enhances the recruitment of Cas3 to Cascade bound at λ3, but may also enable iterative Cas3 loading events.
Discussion
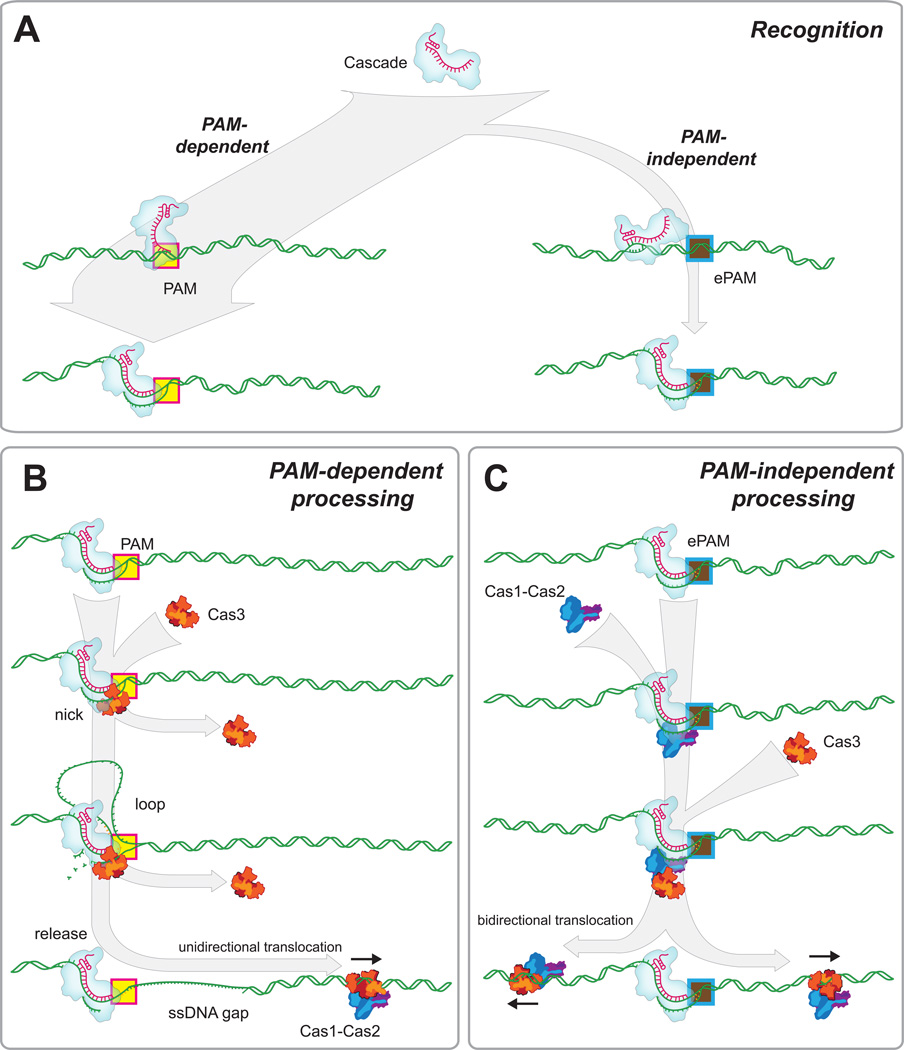
CRISPR-Cas immunity involves the complex interplay among multiple macromolecular components, with the potential for overlapping or convergent pathways. Our work reveals two distinct pathways for target recognition and processing, and shows that the choice of pathway is dictated by the presence or absence of a PAM sequence adjacent to the targeted protospacer (Figure 7).
Figure 7. Model for foreign DNA recognition and processing by Cascade, Cas1, Cas2 and Cas3.
(A) The predominant mechanism for protospacer recognition is through the PAM-dependent pathway. (B) PAM-dependent processing involves the recruitment of Cas3 to the protospacer by Cascade. Cas3 nicks the R-loop and generates an ssDNA gap; Cas3 can dissociate at either of these two steps. Cas3 then breaks free from Cascade and travels unidirectionally along the DNA. (C) PAM-independent processing requires Cas1–Cas2 to recruit Cas3. Cas3 is loaded onto the DNA in one of two possible orientations through a mechanism that attenuates Cas3 nuclease activity. Cas3 then travels in either direction along the DNA as part of a spacer acquisition complex. See also Figure S6.
A conserved mechanism for PAM-dependent target recognition
Our results support a model in which an initial search for PAM sequences is the predominant mode of DNA surveillance by E. coli Cascade (Figure 7A). Once a PAM is identified, Cascade interrogates the flanking DNA for sequence complementarity to the crRNA via directional unwinding of the DNA beginning at the PAM, and identification of a matching protospacer leads to stable capture and R-loop formation (Rutkauskas et al., 2015). This PAM-dependent search process is strikingly similar to that of S. pyogenes Cas9, the crRNA-guided surveillance complex in Type II CRISPR-Cas systems, which also initiates the search by looking for PAMs (Sternberg et al., 2014). In addition, the Type IF CRISPR-Cas system of Pseudomonas aeruginosa also searches for PAM sequences before probing the flanking DNA for sequence complementarity to the crRNA (Rollins et al., 2015). The Type II CRISPR-Cas systems require only a single polypeptide for target recognition and cleavage, whereas Type I CRISPR-Cas systems require large multimeric complexes for target recognition, and a separate trans-acting protein (Cas3) for DNA cleavage. Cas9 and Cse1 share no amino acid sequence homology, and the Cas9 PAM (5’-NGG-3’) and the Cascade PAM (5’-A[A/T]G-3’) are located on opposite ends of the protospacer and on different DNA strands (Jinek et al., 2012; Sashital et al., 2012). Given these differences, there was no reason to assume that S. pyogenes Cas9 and E. coli Cascade would search for target sites using the same general mechanism. The similarities between Cascade and Cas9 suggest that an initial search for PAMs may be a broadly conserved mechanism for DNA surveillance among the Type I and Type II CRISPR-Cas systems.
Facilitated diffusion versus reduced complexity
It is often assumed that site-specific DNA binding proteins accelerate target searches relative to 3D diffusion by facilitated diffusion, which reduces the dimensionality of the search process through 1D sliding, hopping and/or intersegmental transfer (von Hippel and Berg, 1989). However, there is little evidence supporting this general assumption (Halford, 2009). The Cascade target search is remarkably similar to that of Cas9’s, which also exhibits no evidence of 1D sliding (Sternberg et al., 2014). Instead, we find that Cascade and Cas9 both appear to optimize their target searches by reducing the complexity of the sequence space that is sampled while surveying DNA. They accomplish this task by first looking for a small portion of the overall binding site, the PAM, before probing the flanking DNA for sequences complementary to the crRNA, which provides an additional layer of discrimination enabling Cascade to sample and reject incorrect targets (Rutkauskas et al., 2015; Sternberg et al., 2014). The effectiveness of this strategy can be illustrated by considering that based on sequence composition alone Cascade can avoid ~90% of the λ genome just by utilizing the PAM as an initial recognition signal, while kinetically ignoring other sequences. The finding that much higher Cascade concentrations are necessary to achieve similar occupancy at protospacers with an escape PAM compared to those with a cognate PAM also reflects the effectiveness of reducing search complexity.
PAM-dependent target processing
The PAM-dependent pathway requires only Cascade to recruit Cas3 to protospacers (Figure 7B). Cas3 first transiently samples Cascade before transitioning into a stably bound complex. Formation of this longer-lived species prevents any further Cascade-specific recruitment of Cas3, most likely because the first stably bound Cas3 cleaves the R-loop, which destroys the Cas3 binding site (Mulepati and Bailey, 2013). Consistent with this interpretation, formation of stable Cascade-Cas3 intermediates coincides with the appearance of a ~200–300 nt ssDNA gap adjacent to the protospacer. The first molecule of Cas3 does not appear to induce any damage other than creating this initial ssDNA gap. This finding is notably different from bulk biochemical assays, which reveal more extensive DNA degradation (Mulepati and Bailey, 2013; Sinkunas et al., 2013). This difference may be explained by the potential for recruitment of additional Cas3 molecules in the bulk biochemical assays through a Cascade-independent pathway, as previously suggested (Mulepati and Bailey, 2013). Consistent with this explanation, Cas3 is a potent ssDNA nuclease even in the absence of Cascade (Mulepati and Bailey, 2013; Sinkunas et al., 2011; Sinkunas et al., 2013). Thus, the ssDNA gaps generated by the first molecule of Cas3 likely reflect an early intermediate in the degradation pathway and serve as an entryway for additional ssDNA-specific nucleases, including Cas3 or perhaps other host enzymes. Together, these findings suggest that the early stages of foreign DNA degradation involve the ATP-dependent recruitment of just one molecule of Cas3 through a mechanism that requires Cascade-specific contacts and an intact R-loop. This initial transient binding event exhibits a ~6 second lifetime (τ1) before Cas3 transitions into a more stably bound intermediate. The first stably bound molecule of Cas3 then generates a short ssDNA gap, reflected in the delay time (τ2) prior to moving away from Cascade, and after being released from Cascade this Cas3 molecule can either dissociate into solution or continue traveling along the remaining duplex DNA. Any subsequent recruitment of Cas3 (or other nucleases) occurs through nonspecific interactions with the resultant ssDNA gap.
Cascade remains tightly bound to the DNA even after Cas3 generates an ssDNA gap and moves away from the protospacer. It is possible that continued presence of Cascade may distinguish these Cas3-generated gaps from other ssDNA gaps that can be produced during normal DNA metabolism, and Cascade may perhaps prevent host DNA repair proteins from filling in these gaps before the invading DNA is eventually destroyed.
PAM-dependent Cas3 motor activities
Cas3 is a fast and highly processive molecular motor, which is recruited by Cascade through the PAM-dependent pathway and then translocates along the flanking DNA. This translocation does not coincide with any apparent DNA degradation or persistently unwound DNA. When Cas3 is recruited by Cascade through the PAM-dependent pathway it always moves in the same direction along the DNA, consistent with expectations for 3’→5’ translocation along the nontarget strand. A subset of Cas3 molecules also form optically detectable looped intermediates, and Cas3 likely generates smaller DNA loops that cannot be observed in our experiments, suggesting these looped intermediates may be a common feature of the PAM-dependent pathway (Figure 7B). Interestingly, similar looping behaviors have been reported for many different SF1 and SF2 helicases, including Saccharomyces cerevisiae Pif1 (Zhou et al., 2014), Bacillus. stearothermophilus PcrA (Park et al., 2010), E. coli Rep (Myong et al., 2005), and S. cerevisiae Srs2 (Qiu et al., 2013). The looping behaviors exhibited by these proteins are thought to help establish and maintain a particular structural state of the DNA; for instance, PcrA and Srs2 repeatedly shuttle back and forth while removing proteins from ssDNA proximal to an ssDNA/dsDNA junction to prevent aberrant recombination (Park et al., 2010; Qiu et al., 2013). Similarly, Pif1 repeatedly unwinds G-quadraplexes, ensuring that these structures do not inhibit DNA replication (Zhou et al., 2014). The looping activity observed for Cas3 may reflect attempts to dissociate from Cascade. Alternatively, looping may help keep the ssDNA gap clear of proteins, free of secondary structures or both, until the arrival of additional Cas3 molecules or other accessory nucleases.
PAM-independent target recognition
Like the PAM-dependent search, the PAM-independent pathway also occurs by microscopic 3D diffusion, suggesting that Cascade must test for complementarity to the crRNA by either transiently melting the DNA or by taking advantage of the intrinsic breathing of the DNA duplex (Figure 7A). One primary difference between PAM-dependent and PAM-independent target recognition is that the efficiency of the PAM-independent pathway is comprised, such that a higher concentration of Cascade is required to achieve similar levels of occupancy at both targets. Despite this disparity in apparent association constants, Cascade can still bind tightly to the DNA regardless of whether or not the protospacer has a canonical PAM. In both instances, the lifetime of the target-bound Cascade complexes is significantly longer that the typical doubling time of E. coli, a finding that is in good agreement with the results of magnetic tweezer experiments (Szczelkun et al., 2014). This tight binding would help ensure that even though escape target recognition is inefficient, in the rare instances in which an escape target is captured, Cascade would remain in place long enough to initiate downstream steps necessary for primed sequence acquisition (Figure 7C & see below). Interestingly, not all PAM mutations are equal with respect to Cascade, and the defect in binding with the ATT mutant PAM is more moderate that some other PAM mutations (Szczelkun et al., 2014). Future studies will be essential for testing the effects of other PAM mutations on target binding in these single molecule assays.
Interestingly, recent single-molecule FRET experiments have suggested that Cascade recognizes escape targets with substantially reduced fidelity, and interactions with these targets is characterized by a ~25 second lifetime (Blosser et al., 2015), which is identical to one of the nonspecific lifetimes observed in our experiments (Figure S1). We suggest that these shorterlived complexes found by FRET reflect intermediates that have failed to transition into the more tightly bound complexes observed in our assays.
Importantly, PAM escape mutations reflect only a subset of mutations that can lead to a priming response, with the remainder occurring within the protospacer, but both types of escape mutants lead to similar priming responses (Datsenko et al., 2012; Fineran et al., 2014). We anticipate that Cascade will locate protospacer escape mutants through the normal PAM-dependent search pathway, but may then require Cas1–Cas2 to recruit Cas3 and initiate a priming response from this class of escape mutations.
Cas1–Cas2 recruitment of Cas3 to escape targets
We demonstrate that the Cas1–Cas2 complex serves as a trans-acting factor necessary for the recruitment and regulation of Cas3 at protospacers bearing an escape PAM (Figure 7C). Recruitment may occur through one of two general mechanisms. Cas1–Cas2 may modify the structure of Cascade such that it can now directly recruit Cas3 by the same process as occurs during PAM-dependent recruitment. Alternatively, protein-protein contacts with Cas1–Cas2 may directly recruit Cas3 to the escape target through a mechanism that is distinct from the Cascade-dependent recruitment at cognate protospacers. Importantly, the behavior of Cas3 at the escape targets differs markedly from the behavior of Cas3 at cognate targets. First, Cas3 can translocate in either direction from the escape targets, implying that that Cas3 is loaded onto the flanking phage DNA through a different pathway than is observed at cognate protospacers. Second, there was no evidence that Cas3 generates ssDNA gaps at the escape targets, nor was there any evidence that Cas3 even nicked the DNA when loaded at escape targets, suggesting that the nuclease activity of Cas3 is fully attenuated at escape targets. The inability of Cas3 to cleave the escape target is also consistent with the fact that the vast majority of cells will die when infected with phage bearing an escape mutation, and immunity is only conferred for those rare survivors that successfully update the CRISPR locus (Datsenko et al., 2012). Third, Cas3 loaded at escape targets exhibited only a ~6 second lifetime prior to initiating translocation, but there was no evidence for the longer-lived intermediate (τ2) that we have ascribed to ssDNA degradation. Fourth, there was no evidence for DNA looping when Cas3 initiated translocation from the escape target, suggesting that Cas3 is more readily released from Cascade at the escape target. Together, these observations suggest that Cas1–Cas2 recruits and loads Cas3 onto the DNA flanking the escape targets through a mechanism that is distinct from the Cascade-mediated mechanism that takes place at cognate protospacers.
Primed acquisition of new spacer sequences
Together, our data provide direct support for a model of primed sequence acquisition involving Cas1–Cas2-mediated recruitment of Cas3 to Cascade at escape targets, followed by ATP-dependent translocation of Cas3 along the foreign DNA (Figure 7C). Cas3 can move in either direction away from the escape target, consistent with the expectation that new spacers can be acquired from either side of an escape target (Richter et al., 2014). Translocation of Cas3 away from the escape target does not induce DNA damage, and we speculate that Cas3 may be looking for an as yet unidentified signal (e.g. DNA sequence, partner protein, or both) necessary to activate its nuclease activity, or the nuclease activity of a partner protein, at some distal location. Importantly, although the tagged Cas6e subunits remain bound to the protospacer after Cas3 translocation, we do not know whether the other Cas proteins are also left behind. It is possible that Cas3 takes a subset of Cascade components while translocating along the DNA. In fact, Cas3 is naturally linked with Cse1 in a single polypeptide chain in some systems, suggesting that Cse1 may have additional downstream functions during Cas3 translocation (Westra et al., 2012b). In addition, Cas1–Cas2 are essential to process and insert new spacer sequences into the CRISPR locus (Nunez et al., 2014; Nunez et al., 2015), and one attractive model is that Cas1–Cas2 travel with Cas3 as part of a larger spacer acquisition complex (Figure 7C), which would allow delivery of Cas1–Cas2 to sites distal to an escape target, where they would then be able to process the DNA to promote new spacer acquisition. In support of this model, studies in the closely related Type 1F CRISPR-Cas system from Pectobacterium atrosepticum have shown that Cas3 interacts directly with Cas1 (Richter et al., 2012).
Early models suggested Cascade might diffuse away from the escape target (Datsenko et al., 2012). However, this model was later disfavored because the distribution of new spacers acquired from a circular plasmid was inconsistent with expectations for a diffusion-based mechanism, which would predict a strong bias toward acquisition of new spacer sequences near the original protospacer (Savitskaya et al., 2013). The high processivity of E. coli Cas3 (~12-kbp) explains why assays using relatively small plasmids (~5-kb) fail to yield a biased distribution of newly acquired spacer sequences as predicted by the original sliding hypothesis (Heler et al., 2014; Savitskaya et al., 2013). Interestingly, the Type 1F CRISPR-Cas system from P. atrosepticum does exhibit a biased distribution of newly acquired spacers in response to an escape mutation (Richter et al., 2014). Assuming that priming occurs through a similar mechanism for the Type 1F and Type 1E CRISPR-Cas systems, our model predicts that P. atrosepticum Cas3 is less processive that E. coli Cas3, explaining why spacer acquisition bias can be observed in plasmid assays for P. atrosepticum.
Our data demonstrate that the first Cas3 molecule recruited to cognate protospacers through the PAM-dependent pathway can translocate rapidly away from Cascade before the DNA is destroyed. Moreover, the nuclease activity of Cas3 was partially attenuated by Cas1–Cas2 at cognate protospacers, allowing iterative Cas3 firing events presumably before the eventual destruction of the R-loop. Together, these observations suggest that priming might take place even when there is no escape mutation present in the invading DNA (Figure 7B). The ability to occasionally acquire new spacers in the absence of an escape mutation may allow microbes to routinely update the CRISPR locus even before foreign genetic elements have the opportunity to evade the CRIPSR/Cas immune response by acquiring new mutations.
Experimental Procedures
Single-molecule assays
DNA curtains were fabricated by electron-beam lithography as previously described (Greene et al., 2010; Sternberg et al., 2014). A lipid bilayer was then deposited on the surface of the sample chamber, the anchor points were coated with anti-digoxigenin antibodies, and the DNA was anchored to the bilayer through a biotin-streptavidin linkage. The DNA was then aligned along the leading edges of the Cr diffusion barriers and coupled to the antibody-coated anchors through the application of hydrodynamic force. Cascade single-molecule binding assays were conducted in reaction buffer containing 40 mM Tris-HCl [pH 7.4], 1 mM MgCl2, 25 mM KCl, 1 mg/mL BSA, 0.8% glucose, YOYO-1, and a glucose oxidase-catalase oxygen scavenging system. The Cas6e subunit of Cascade was expressed with an N-terminal 3xFLAG tag, and the Cascade complex was labeled with antiFLAG-coated QDs (Invitrogen) for 10 minutes on ice prior to use. In experiments with Cas3, the YOYO-1 dye was omitted, and the reaction buffer was supplemented to contain 2 mM MgCl2, 1 mM ATP, and 20 µM CoCl2. Cas3 was labeled by incubation with streptavidin coated QDs (Invitrogen) on ice for 20 minutes prior to injection onto the flowcell at 4 nM final concentration. RPA-eGFP labeling of ssDNA gaps was always performed at the end of the Cas3 experiments as a check for activity. In these assays, Cas3 was flushed from the sample chamber, followed by delivery of buffer containing 100 nM RPA-eGFP. Buffer flow was then terminated and RPA-eGFP was incubated with the DNA for 10 minutes prior to imaging. Buffer conditions for experiments containing Cas1–Cas2 were identical to those above. Cas1 (8 nM) and Cas2 (16 nM) were pre-incubated on ice for 20 minutes and then mixed with Cas3 (4 nM) for an additional 5 minutes before being delivered to the flowcell. All single molecule experiments were conducted at 25°C, unless otherwise indicated, and all data were collected and analyzed as previously described (Sternberg et al., 2014).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Alison Marie Smith and Kaihong Zhou for technical assistance, and members of the Doudna and Greene laboratories for helpful discussions and critical reading of the manuscript. S.H.S. acknowledges support from the NSF and National Defense Science & Engineering Graduate Research Fellowship programs. Funding was provided by the NIH (GM074739 to E.C.G., and GM108888 to B.W.), an HHMI Early Career Scientist Award (to E.C.G.) and the NSF (MCB-1154511 to E.C.G. and MCB-1244557 to J.A.D.). J.A.D. is an HHMI Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
S.R. and S.H.S. conceived the project with initial input from B.W. S.R. conducted single-molecule experiments and data analysis, and designed all biochemical assays. S.H.S. purified proteins and assisted with experimental design and bulk biochemical experiments. M.M. and B.G. conducted bulk biochemical assays and purified proteins. C.K.G., P.B., and B.W. assisted with preparation and biochemical analysis of Cascade and Cas3. All authors discussed the data and co-wrote the manuscript.
References
- Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blosser TR, Loeff L, Westra ER, Vlot M, Kunne T, Sobota M, Dekker C, Brouns SJ, Joo C. Two Distinct DNA Binding Modes Guide Dual Roles of a CRISPR-Cas Protein Complex. Mol Cell. 2015;58:60–70. doi: 10.1016/j.molcel.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- Fineran PC, Gerritzen MJ, Suarez-Diez M, Kunne T, Boekhorst J, van Hijum SA, Staals RH, Brouns SJ. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci U S A. 2014;111:E1629–E1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17:932–938. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J, Wang F, Redding S, Plys AJ, Fazio T, Wind S, Alani EE, Greene EC. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc Natl Acad Sci U S A. 2012;109:E3074–E3083. doi: 10.1073/pnas.1211364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene EC, Wind S, Fazio T, Gorman J, Visnapuu ML. DNA curtains for high-throughput single-molecule optical imaging. Methods Enzymol. 2010;472:293–315. doi: 10.1016/S0076-6879(10)72006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford SE. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem Soc Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- Heler R, Marraffini LA, Bikard D. Adapting to new threats: the generation of memory by CRISPR-Cas immune systems. Mol Microbiol. 2014;93:1–9. doi: 10.1111/mmi.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna JA. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci U S A. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Mulepati S, Bailey S. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3) J Biol Chem. 2011;286:31896–31903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J Biol Chem. 2013;288:22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Lee AS, Engelman A, Doudna JA. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, Ha T. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Antony E, Doganay S, Koh HR, Lohman TM, Myong S. Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat Commun. 2013;4:2281. doi: 10.1038/ncomms3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Dy RL, McKenzie RE, Watson BN, Taylor C, Chang JT, McNeil MB, Staals RH, Fineran PC. Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014;42:8516–8526. doi: 10.1093/nar/gku527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Gristwood T, Clulow JS, Fineran PC. In vivo protein interactions and complex formation in the Pectobacterium atrosepticum subtype I-F CRISPR/Cas System. PLoS One. 2012;7:e49549. doi: 10.1371/journal.pone.0049549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins MF, Schuman JT, Paulus K, Bukhari HS, Wiedenheft B. Mechanism of foreign DNA recognition by a CRISPR RNA-guided surveillance complex from Pseudomonas aeruginosa. Nucleic Acids Res. 2015;43:2216–2222. doi: 10.1093/nar/gkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkauskas M, Sinkunas T, Songailiene I, Tikhomirova MS, Siksnys V, Seidel R. Directional R-Loop Formation by the CRISPR-Cas Surveillance Complex Cascade Provides Efficient Off-Target Site Rejection. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.067. [DOI] [PubMed] [Google Scholar]
- Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitskaya E, Semenova E, Dedkov V, Metlitskaya A, Severinov K. High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA Biol. 2013;10:716–725. doi: 10.4161/rna.24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci U S A. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012a;46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- Westra ER, van Erp PB, Kunne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012b;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Zhang J, Bochman ML, Zakian VA, Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife. 2014;3:e02190. doi: 10.7554/eLife.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.