Abstract
To maximize the potential of genome-wide association studies, many researchers are performing secondary analyses to identify sets of genes jointly associated with the trait of interest. Although methods for gene-set analyses (GSA), also called pathway analyses, have been around for more than a decade, the field is still evolving. There are numerous algorithms available for testing the cumulative effect of multiple SNPs, yet no real consensus in the field about the best way to perform a GSA. This paper provides an overview of the factors that can affect the results of a GSA, the lessons learned from past studies, and suggestions for how to make analysis choices that are most appropriate for different types of data.
Keywords: genome-wide association studies, polygenic effects, gene set analysis, complex traits
INTRODUCTION
With the success of genome-wide association studies (GWAS), a gap has emerged between researchers’ ability to identify genetic variants associated with complex traits and the ability to interpret the biological significance of those variants. GWAS have provided two important pieces of the puzzle of complex disease genetics. First, these studies have identified a large number of genetic variants significantly associated with human disease. These disease-associated variants have provided candidate genes for further study and hypotheses about disease mechanisms. Second, GWAS have been able to confirm the polygenic nature of complex diseases, particularly for psychiatric disorders. For instance, studies have found that the cumulative effect of a large number of weakly associated SNPs, most of which are not statistically significant on their own, can predict disease status or symptoms [Wray et al., 2014]. This cumulative effect, referred to as a polygenic risk score, indicates that even very small genetic effects can contribute to disease risk when taken together.
Despite these successes, insights about specific biological mechanisms responsible for disease risk have been elusive. Individual genetic associations explain only a very small fraction of disease risk. And while polygenic risk scores may explain a greater portion of disease risk, they are not easily interpreted in terms of biological mechanisms. The need for methods that can provide biological context for genetic associations has led to a surge of interest in gene set analyses, which test for association between biologically meaningful sets of genes and a phenotype.
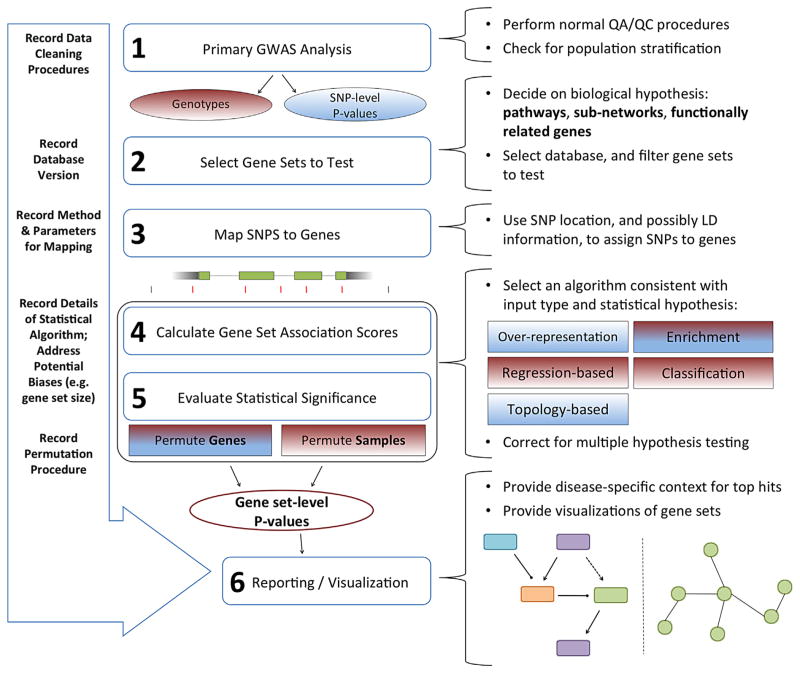
A wide variety of methods are available for testing gene set associations. As a result, a number of important decisions must be made when planning a gene set analysis (Fig. 1).
FIG. 1.
A gene set analysis workflow, including the possible choices that must be made at each step of the analysis. The data type requirements for the various statistical methods are indicated by color. For instance, regression-based methods and permutation procedures that randomize samples require genotypes as inputs. On the other hand, over-representation methods utilize summary statistics. Some methods (multicolored) are not restricted to one type of input data.
WHY GENE SET ANALYSIS?
Gene set analyses have the potential to provide a number of benefits when used as a tool for secondary analysis of a GWAS data set. First, because of the polygenic nature of complex diseases, testing for association with sets of functionally related variants can provide biological context for multiple genetic risk factors and can provide insights into disease mechanisms and possible treatment targets. Second, given the small effect sizes of most reported associations with common variants, examining the cumulative effect of multiple variants can improve the power to detect genetic risk factors for complex diseases. And third, testing for associations at the pathway level may also account for the genetic heterogeneity within affected populations. Since genetic heterogeneity within a study population will lead to a mixture of small genetic effects, detecting their cumulative effect may be possible with GSA methods if enough small effects are present within the same gene set.
Despite these potential benefits, considerable care is critical when interpreting the results of a gene set analysis. Because results can be highly dependent on the definitions of the gene sets and statistical methods used, GSAs should generally be viewed as exploratory analyses. Significantly associated gene sets can provide functional context for individual SNP or gene associations. However, care should be taken not to assign undue meaning to individual genes within a statistically identified gene set if those genes do not show at least weak association on their own. Following this sentiment, it has been suggested that it is not appropriate to apply gene set analyses to a dataset with no indication of any SNP effects (e.g., no deviation from diagonal on a Q-Q plot) [Sedeño- Cortés and Pavlidis, 2014].
Step 1: Primary GWAS Analysis and Data Cleaning
Gene set analyses are typically performed as a secondary analysis of GWAS data, and therefore can make use of either genotype data or summary statistics (e.g., SNP P-values). To avoid false positive associations at the gene set level, before conducting a GSA all standard quality control and data cleaning procedures should be applied to the GWAS data, including correcting summary statistics for population stratification.
Recommendations:
Always follow best practices for GWAS QA/QC and data cleaning prior to downstream analyses like GSA.
Step 2: Select Gene Set Definitions
One of the first questions to ask when planning a gene set analysis is: What gene sets will be tested? The investigator has a number of options regarding the source of gene set annotations, and the appropriate selection depends on multiple factors.
Decisions to make at this step:
What is the biological hypothesis you want to investigate?
What data source for gene set annotations should you use?
How many gene sets do you want to test?
Biologically meaningful gene sets can be defined in a variety of ways, which can represent different biological hypotheses [Mooney et al., 2014]. For instance, biological pathways, protein-protein interaction (PPI) networks, and functionally related gene sets (e.g., gene ontology categories) each suggest different types of relationships between the members of a gene set.
Pathway models suggest a common function or end goal for the pathway’s members, and also provide specific information about how the gene members interact to accomplish that end goal (e.g., folate biosynthesis). PPI networks, on the other hand, provide information about biological interactions (e.g., physical interactions) among genes or gene products, but do not imply a common goal or directed action for a set of genes. This difference between pathways and networks is due mainly to the fact that biological interaction data often come from a heterogeneous mixture of sources (e.g., different experiments, different tissues, different animal models). And finally, functionally-related gene sets, such as gene ontology (GO) categories, suggest that member genes share a common function or are involved in a common process, but they do not provide any information about how, or if, the members biologically interact (e.g., different gene products may perform the same function, but in different tissues).
Pathways and functionally-related gene sets are self-contained groups of genes and are therefore ready to use in a GSA with minimal processing (see Step 3). However, because PPI databases contain interactions on a genome-wide scale, the use of network data for GSA requires an additional step. In order to use PPI data for a GSA, subsets of genes (sub-networks) must first be extracted from the global network of all genes. These sub-networks become the gene sets that are later tested for association using methods discussed in Step 4. Sub-networks can be identified in a number of ways, including community detection algorithms, which use topological measures to identify tightly clustered nodes [Xu et al., 2010; Cowley et al., 2012], and heuristic search algorithms [Bakir-Gungor and Sezerman, 2011; Jia et al., 2011; Vandin et al., 2011].
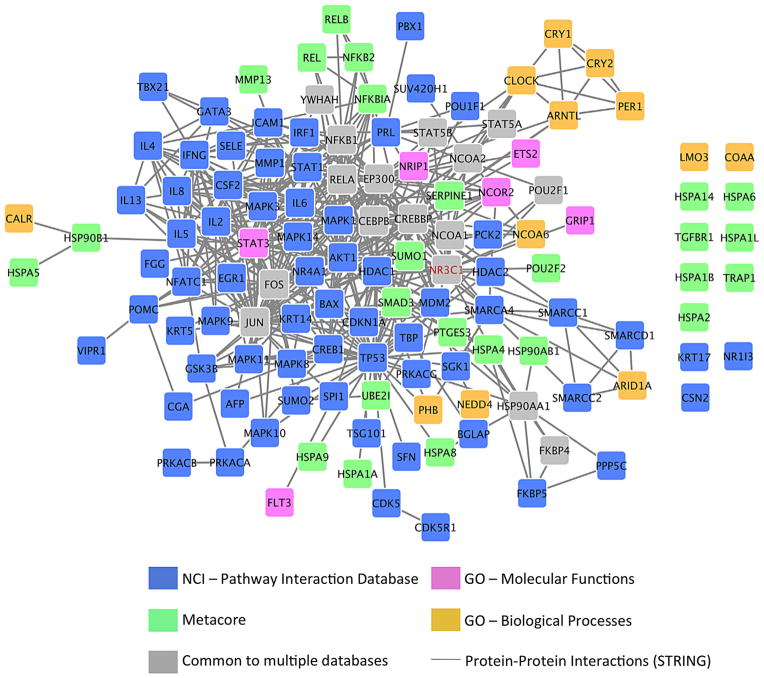
Numerous databases contain gene set annotations or gene interaction data (Table I). The membership of gene sets can vary significantly depending on the data source, even for similar or related biological concepts [Mooney et al., 2014; Belinky et al., 2015]. For example, Figure 2 shows multiple gene sets related to glucocorticoid receptor processes, and illustrates the differences among data sources for this same conceptual domain.
TABLE I.
A Selection of Gene Set Databases
| Database | Canonical pathways | Functionally-related gene sets | Gene/protein interactions | Links/references |
|---|---|---|---|---|
| Pathway Commons | X | pathwaycommons.org, [Cerami et al., 2011] | ||
| PathCards | X | pathcards.genecards.org, [Belinky et al., 2015] | ||
| KEGG | X | genome.jp/kegg, [Kanehisa and Goto, 2000; Kanehisa et al., 2014] | ||
| Reactome | X | reactome.org, [Croft et al., 2014] | ||
| Biocarta | X | biocarta.com | ||
| Panther | X | X | pantherdb.org/data, [Mi et al., 2013] | |
| NCI-PID | X | pid.nci.nih.gov, [Schaefer et al., 2009] | ||
| MSigDB | X | X | broadinstitute.org/gsea/msigdb, [Subramanian et al., 2005] | |
| ConsensusPathDB | X | X | consensuspathdb.org, [Kamburov et al., 2013] | |
| Gene Ontology | X | geneontology.org, [Ashburner et al., 2000; Gene Ontology Consortium, 2010] | ||
| STRING | X | string-db.org, [Franceschini et al., 2013] | ||
| HPRD | X | hprd.org, [Prasad et al., 2009] | ||
| Metacore* | X | X | X | thomsonreuters.com/metacore |
| Ingenuity* | X | X | X | ingenuity.com/products/ipa |
Proprietary database.
FIG. 2.
Gene sets related to glucocorticoid receptor processes. Gene sets from NCI’s Pathway Interaction Database, the proprietary Metacore database, and the Gene Ontology database were overlaid onto an interaction network from the STRING protein-protein interaction database (only high confidence interactions are shown, STRING combined score ≥0.9). Colored genes are unique to a particular database, while gray genes are shared between two or more databases (only NR3C1 is common to all four databases). There are clear differences in membership between gene sets from different data sources. These differences may be due to an attempt to model distinct processes, but are also indicative of incomplete annotation. For instance, some genes are unique to a single database even when there is evidence of interaction with multiple genes from another database (e.g., ARID1A is not part of the NCI-PID gene set, but is connected to four of its member genes).
Gene sets from multiple databases can be combined to improve genome coverage and to take advantage of knowledge from a variety of sources. Some databases, such as Pathway Commons, ConsensusPathDB, and PathCards already integrate data from multiple sources [Cerami et al., 2011; Kamburov et al., 2013; Belinky et al., 2015]. However, because gene sets from publicly available databases do not represent all possible biological processes and because they are constructed from heterogeneous data sources (i.e., a wide variety of experiments conducted under a variety of conditions), manual curation of gene sets (via systematic literature review for example) can be beneficial. Manual curation may be particularly useful for defining gene sets that are relevant to a particular biological context, such as a disease state or tissue type. An example of the result of this type of manual curation is the neurodevelopmental network described in (Poelmans et al., 2011).
Gene set annotations are dynamic, not static and therefore change over time, as genes are further characterized and new evidence about gene function is revealed. It is important that the source of the gene sets chosen for an analysis is consistent with the study’s biological hypothesis, and that any reported results include adequate information about the gene set membership to ensure comparability with other studies.
Once the type and source of the gene sets has been selected, a decision must be made regarding the scope of the analysis. The choice here is between an analysis which focuses on a few candidate gene sets that are hypothesized to play an important role in the disease under study, or a hypothesis-free global analysis, which tests a large number of gene sets, usually from a repository such as those listed in Table I.
An analysis that tests an entire database of gene sets is more common, but this approach may not always be appropriate, particularly for small data sets. Testing a large number of gene sets can reduce statistical power, given the need to correct for testing multiple hypotheses. Permutation methods for evaluating the statistical significance of a gene set association will be discussed below. It should be noted that most of the proposed permutation methods do not adjust for the number of gene sets tested, and therefore standard multiple hypothesis testing corrections such as a False Discovery Rate (FDR) correction are necessary. However, some GSA methods, such as ALIGATOR and INRICH [Holmans et al., 2009; Lee et al., 2012], incorporate two stages of permutation, one to produce an empirical P-value and another to correct for multiple testing.
Recommendations:
Choose a source for gene set annotations that are consistent with your biological hypothesis.
Combine annotations from multiple databases to take advantage of different sources of knowledge about gene function.
Use up-to-date annotations and record information about the data sources (e.g., database versions) to allow replication by other researchers.
Step 3: Prepare Your Data
Decisions to make at this step:
How to filter, or clean, the list of gene sets?
How to map SNPs to genes?
Should imputed genotypes be used?
Several data preprocessing steps are necessary to integrate the genotype data and the gene set annotations. First, it is often necessary to filter the gene sets to remove those with only a few genes and those with a very large number of genes. This step is important because of known biases related to the size (number of genes) of a gene set [Holmans, 2010; Wang et al., 2010; Ramanan et al., 2012]. Although the limits are arbitrary, it is common to limit the size of gene sets to between 10 and 200 genes. Removing very large gene sets, which may encompass multiple cellular processes, also has the benefit of improving the specificity and interpretability of results.
Because gene set analyses attempt to summarize the effects of multiple SNPs in order to create a single gene set-level association measure, it is necessary to map SNPs to genes. The most straightforward way to do this is based on SNP location. For example, a common method is to assign a SNP to a gene if the SNP lies within the gene boundaries or within a fixed window upstream or downstream of the gene, which is meant to cover regulatory regions [Holmans et al., 2009; Chen LS et al., 2010a; Fridley and Biernacka, 2011).
Linkage disequilibrium (LD) can also be used to map SNPs to genes. In this case, SNPs are mapped to a gene if they are correlated with other SNPs located within the gene boundaries. Methods that use LD to map SNPs to genes have the advantage of not losing all the information contained in intergenic SNPs. However, using LD to map SNPs to genes can create problems when a SNP is correlated with, and therefore assigned to, multiple genes. If those genes are in the same gene set, this situation can lead to “multiple-counting“ of a single SNP and can erroneously inflate a gene set’s association measure [Sedeño-Cortés and Pavlidis, 2014].
The ProxyGeneLD and INRICH methods both account for LD during the process of mapping SNPs. These methods also avoid the multiple-counting problem by merging or removing genes that are highly correlated with other genes in the same gene set [Hong et al., 2009; Lee et al. 2012].
Imputing SNPs that are not directly genotyped by a SNP array is now common practice in GWAS because it can improve the power to detect significant associations. Imputation to a common set of SNPs can clearly improve the comparability of studies and can facilitate meta-analyses. However, GSA presents unique challenges compared to single-SNP analyses, and the effects of using imputed genotypes for GSA are not clear. The use of imputed genotypes increases the number of SNPs included in the analysis, and therefore may increase the number of genes represented. For GSA methods that utilize genotypes, rather than summary statistics, the increased number of SNPs can increase computational burden. Furthermore, given that missing genotypes are imputed using the information from multiple neighboring SNPs (haplotypes), methods that use the genotypes of multiple SNPs to model gene-level or gene set-level effects will not benefit from the addition of imputed SNPs. It should also be noted that significantly increasing the number of SNPs in a gene set might adversely affect model fit in studies with small sample size.
GSA methods that use a single SNP to summarize a gene (e.g., assigning the minimum P-value of all SNPs assigned to the gene) may potentially benefit from genotype imputation, since some gene-level p-values may become more significant. However, the increased representation of genes may mean that the number of non-significant genes included in the analysis is increased, potentially affecting enrichment results. For example, one study found that the use of imputed genotypes increased the representation of smaller genes, and that these smaller genes were less likely to contain significantly associated SNPs [Hong et al., 2009].
Recommendations:
Filter out very small and very large gene sets.
Take care in mapping SNPs to genes to avoid issues related to correlated genes.
Do not impute genotypes, except to improve cross-study comparability.
Steps 4 and 5: Select a Gene Set Analysis Method/Evaluate Statistical Significance
Decisions to make at this step:
What statistical hypothesis do you want to test?
Should genotypes or summary statistics be used?
How should statistical significance be determined?
The statistical tests employed in pathway analyses can be categorized as either competitive or self-contained, depending on the test’s null hypothesis [Goeman and Buhlmann, 2007]. A competitive test compares the proportion of association signal within the target gene set to the proportion of association signal outside of the target gene set. The null hypothesis for a competitive test is that there is no difference between the target gene set and random gene sets of the same size in terms of association to the trait of interest. However, this type of test does not tell you how strongly the gene set itself is associated to the trait. Methods that use a competitive test must have data (i.e., genotypes or P-values) for all genes, not only those within the target gene set.
In contrast, a self-contained test does not require data for any genes outside of the target gene set, since it is concerned only with the association signal within a single gene set. In this case, the test tells you how strong the association is with the trait of interest, but not how important the gene set is compared to other gene sets. The null hypothesis for a self-contained test is simply that none of the genes in the gene set are associated with the trait of interest [Wu et al., 2010].
Most GSA methods use a permutation test to evaluate the statistical significance of pathway-level association measures. Permutation tests can also correct for known biases, such as gene size. However, which permutation method is the most appropriate is still a matter of debate and numerous different approaches have been proposed [Efron and Tibshirani, 2007; Holmans et al., 2009; Yaspan et al., 2011; Cabrera et al., 2012; Jia et al., 2012].
In general, there are two types of permutation tests used in gene set analyses, those that permute samples (randomly assigning case/ control status) and those that permute genes (creating random gene sets). In either case, the association measure for a target gene set is calculated using one of a variety of methods (discussed below). This association measure is then compared to a null distribution of association measures created through repeated permutation of the data.
The null hypotheses of these two types of permutation procedures relate back to the null hypotheses of competitive and self-contained tests. Permuting samples is consistent with the self-contained null hypothesis, as no data on genes outside the target gene set is needed. And permuting genes is consistent with the competitive null hypothesis, since the target gene set is compared to a collection of random gene sets [Goeman and Buhlmann, 2007; Khatri et al., 2012]. This is not to say that competitive methods cannot use sample-permutation procedures, or self-contained methods cannot use gene-permutation procedures. However, algorithms that take this approach become, in a sense, hybrids somewhere between competitive and self-contained tests. For instance, it is important to realize that a self-contained test statistic that is adjusted using a gene-permutation procedure is no longer strictly “self-contained“ since it has been adjusted relative to other gene sets. Similarly, when a competitive test statistic is adjusted by sample permutation, it is the self-contained null hypothesis that is ultimately being tested [Goeman and Buhlmann, 2007].
Much of the debate over the procedures used to evaluate statistical significance in GSA has unfolded within the context of gene expression studies, but the issues are relevant to GWAS data as well. Correlation between genes in expression studies is due to local co-regulation and large differences between groups which results in many differentially expressed genes; correlation among genes in GWAS is due to both linkage disequilibrium (LD) and the polygenic nature of complex traits. Therefore, a competitive test can identify gene sets that are enriched above the relatively high background due to a polygenic trait.
Because gene-set analyses originated in the context of gene-expression studies, a number of GSA methods originally designed to analyze expression results have been adapted to GWAS datasets (over-representation analyses and variations of the GSEA method listed in Table II are examples). It is crucial that researchers use such methods only after taking steps to account for the unique challenges presented by GWAS data [Fridley and Biernacka, 2011; Ramanan et al. 2012; Mooney et al., 2014]. For example, the choice of method for aggregating or summarizing SNP-level association measures at the gene level is a critical aspect of many GWAS-based GSA. The size of a gene (i.e., the number of SNPs it contains) and the correlation between SNPs (LD structure) are two potential sources of bias that can significantly influence gene-level statistics. Fortunately, a number of methods have been developed to calculate unbiased gene-level p-values [Saccone et al., 2007; Hong et al., 2009; Segrè et al., 2010; de Leeuw et al., 2015]. Permutation procedures, as stated above, can also correct for gene size and LD structure.
TABLE II.
Gene Set Analysis Methods and Software Tools
| Method | Input | References |
|---|---|---|
| Over-representation methods | ||
| WebGestalt | Gene list | Wang et al., [2013] |
| DAVID | Gene list | Huang et al. [2009] |
| Metacore | Gene list | thomsonreuters.com/metacore |
| GeneSetDB | Gene list | [Araki et al. [2012] |
| INRICH | Genomic Regions | Lee et al. [2012] |
| MAGENTA | P-values | Segrè et al. [2010] |
| ALIGATOR | P-values | Holmans et al. [2009] |
| Enrichment methods | ||
| GSEA | Genotypes | Wang et al. [2007] |
| i-GSEA4GWAS | P-values | Zhang et al. [2010] |
| GSA-SNP | P-values | Nam et al. [2010] |
| GSEA-P | Ranked gene list | Subramanian et al. [2007] |
| GSEA-SNP | P-values | Holden et al. [2008] |
| Methods for combining P-values | ||
| Modified fisher’s method | P-values | De la Cruz et al. [2010] |
| Modified fisher’s method | P-values | Luo et al. [2010] |
| Adaptive rank truncated product method | Genotypes or P-values | Yu et al. [2009] |
| FORGE | P-values | Pedroso et al. [2012] |
| Plink set-based test | Genotypes | Purcell et al., [2007] |
| Regression-based methods | ||
| GRASS | Genotypes | Chen LS et al. [2010] |
| MAGMA | Genotypes | de Leeuw et al. [2015] |
| PCgamma | Genotypes | Biernacka et al. [2012] |
| PAGWAS | Genotypes | Evangelou et al. [2014] |
| SGL-BCGD | Genotypes | Silver et al. [2013] |
| Supervised PCA | Genotypes | Chen X et al. [2010] |
| Classification-type methods | ||
| Pathways of distinction analysis | Genotypes | Bruan and Buetow, [2011] |
| Synthetic feature random forest | Genotypes | Pan et al. [2014] |
| Methods that incorporate interaction data (networks) | ||
| PANOGA | P-values | Bakir-Gungor and Sezerman, [2011] |
| dmGWAS | P-values | Jia et al. [2011] |
| HotNet2 | P-values | Vandin et al. [2011] |
| EnrichNet | Gene list | Glaab et al. [2012] |
| NetPEA | Gene list | Liu and Ruan, [2013] |
| NEA | Gene list | Alexeyenko et al. [2012] |
| PANOGA | P-values | Bakir-Gungor et al. [2014] |
| PINA | Gene list | Cowley et al. [2012] |
| Dapple | Gene list | Rossin et al. [2011] |
| DEPICT | P-values | Pers et al. [2015] |
Several reviews of the statistical tests used in gene set analyses are available [Wang et al., 2010; Fridley and Biernacka, 2011; Khatri et al., 2012; Ramanan et al., 2012; Mooney et al., 2014]. Here we will give a brief summary of the various classes of statistical tests that have been proposed for calculating pathway-level association measures. A selection of methods from each class is listed in Table II.
The simplest form of competitive test for GSA is the test for over-representation. In over-representation analyses (also called 2 × 2 table methods), an association measure is calculated for each gene in the dataset and a threshold is used to determine which genes are significantly associated. The proportion of significantly associated genes within a target pathway is compared to the proportion of significantly associated genes among all genes outside of the target pathway. The chi-square or hypergeometric tests are commonly used for tests of over-representation.
A major disadvantage of over-representation analyses is that they require a strict threshold for determining statistical significance [Goeman and Buhlmann, 2007]. This threshold is arbitrary and can influence the results of an analysis. Furthermore, when pathway association measures are based solely on a count of significantly associated genes, information about the strength of association is lost [Khatri et al., 2012]. To overcome these issues, enrichment methods that use all gene-level P-values have been devised. An example of this type of method is the popular gene set enrichment analysis (GSEA) [Subramanian et al., 2005; Subramanian et al., 2007; Wang et al., 2007]. The GSEA algorithm calculates a gene-level P-value for all genes, then ranks the genes based on P-value. The next step is to calculate a running-sum statistic that represents the extent to which the genes in the target set are concentrated at the top of the ranked list. The significance of this statistic is evaluated by comparing it to a null distribution of statistics created by repeatedly permuting the data. A number of modifications of this algorithm have been developed (Table II).
For a self-contained test, the simplest approach is to combine the P-values of all members of a gene set. It is most common to first calculate gene-level P-values, but it is also possible to combine SNP-level P-values. A variety of methods for combining multiple p-values are available, such as Fisher’s method [De la Cruz et al., 2010; Luo et al., 2010], the gamma method [Biernacka et al., 2012], and the adaptive rank truncated product method [Yu et al., 2009].
Regression-based methods, which use genotypes to model the effects of multiple SNPs have also been proposed. Often these methods are combined with some form of feature selection, such as principle component analysis, to select those SNPs that are most informative [Chen LS et al., 2010; Chen X et al., 2010; Biernacka et al., 2012]. Regression-based methods that use multiple SNPs to model gene-level effects have been found to be more powerful than simply selecting the minimum SNP P-value to represent a gene-level statistic [Ballard et al., 2010].
In addition to regression-based methods, classification-type methods, which utilize genotype data to identify gene sets that distinguish cases and controls, have also been proposed. Examples are the Pathways of Distinction Analysis (PoDA) method [Braun and Buetow, 2011], and a random-forest based method that creates synthetic features to represent all SNPs within a particular gene set [Pan et al., 2014].
All the GSA algorithms discussed above treat all genes in a gene set independently and do not account for the relationships between genes. Topology-based GSA methods are fundamentally different because the relationships between genes are used to assign different levels of “importance“ to genes in the set. For example, a gene that interacts with only one other member of the gene set will be weighted less than a gene that interacts with most other members. Because topology-based methods require information about interactions between gene set members, it may be necessary to integrate gene set membership information with interaction data from a separate source. For example, since GO categories do not define interactions between genes, it would be necessary to gather interactions information from another sources, such as a PPI database.
Although most topology-based methods were developed for gene expression data sets, some require only gene-level P-values and therefore can be applied to GWAS data sets as well [Bakir-Gungor and Sezerman, 2011; Jia et al., 2011; Vandin et al., 2011]. For a review of topology-based GSA methods see [Mitrea et al., 2013].
The computational burden of a statistical algorithm can be an important factor in a GSA analysis and depends on the data used as input (e.g., genotypes vs. summary statistics), the complexity of the algorithm, and the permutation procedure used to evaluate statistical significance. The most computationally efficient methods are those that use summary statistics and a permutation procedure that randomizes genes. Permuting samples requires the re-calculation of SNP-level P-values for each permutation, and therefore requires access to the genotype data (or a reference set of genotype data [Evangelou et al., 2014]) as well as greater computational resources.
However, the computational burden of an algorithm should not be the deciding factor when planning a GSA, since it has been demonstrated that genotype-based method can have greater power to detect pathway-level associations [Ballard et al., 2010; Gui et al., 2011]. Furthermore, it has also been suggested that applying multiple GSA methods to the same dataset may be beneficial, given the expectation that different methods are sensitive to different types of genetic effects [Gui et al., 2011; Varemo et al., 2013; Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium, 2015].
Recommendations:
If computational resources are available, select a method that utilizes genotypes.
Use a permutation procedure that is consistent with your statistical hypothesis, and corrects for the size of a gene set.
Apply multiple GSA methods to capture different genetic effects and identify robust gene set associations.
Step 6: Reporting and Visualizing Results
Given that the results of GSA are highly dependent on the source of gene set annotations and the statistical algorithm used, it is often difficult to compare results across studies. However, when reporting results of a GSA, a number of steps can be taken to improve the interpretability and comparability of findings.
Providing detailed information about the gene sets tested (e.g., database versions and dates accessed) and the statistical algorithm used will allow other researchers to attempt to replicate findings. This information will also provide important context for understanding discordance among studies. For example, similar biological concepts may be represented by very different gene sets depending on the database used (Fig. 2). It is also important to address any potential sources of bias (such as gene set size, or correlation between genes) when reporting results.
One of the goals of a GSA is to provide functional context for multiple genetic associations. Therefore, claims of significant gene set associations should be accompanied by evidence supporting a role for the gene set in the disease being studied. This disease-specific context is important for the interpretation of results, given that gene set annotations are often incomplete and are of varying quality.
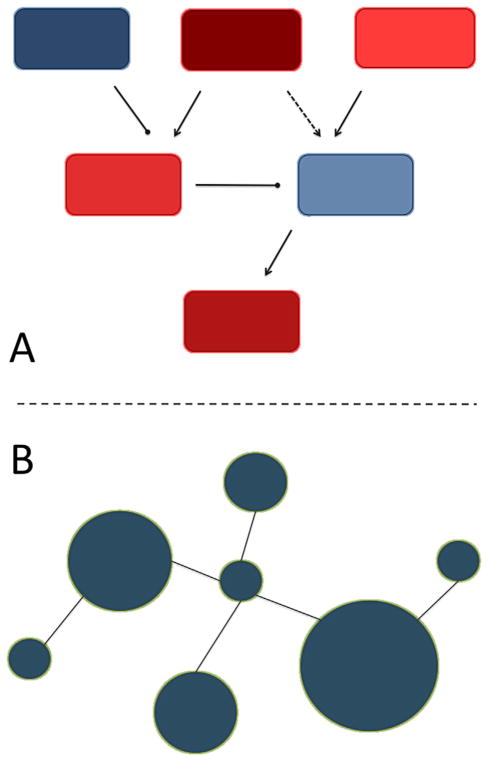
Finally, visualizations, which show the relationships between genes in a gene set as well as each gene’s contribution to the overall association (i.e., gene-level association effects), can be an important part of reporting findings from a GSA (Fig. 3). Tools such as Dapple [Rossin et al., 2011] can provide information about relationships among associated genes. It should be noted that depending on the source of the gene set definitions, these types of visualizations may require the integration of interaction data from a distinct data source (e.g., a PPI database) separate from the source used to define the gene set.
FIG. 3.
Visualizations of gene set analysis results. Depending on the source of the gene sets, gene interaction information can be obtained from pathway maps or PPI databases. A: A signaling pathway map with genes colored to show gene-level association measures. Dark blue indicates a weak association and dark red indicates a strong association. B: A gene set overlaid onto a PPI network to show known interactions between genes. Here the strength of gene-level associations is indicated by node size.
Recommendations:
Provide details about the gene sets tested (e.g., database version), and the statistical algorithm used, to ensure comparability of results.
Address any possible sources of bias.
Provide disease-relevant biological context to aid the interpretation of significant gene set associations.
Provide visualizations of associated gene sets.
DISCUSSION
Gene set analyses have become a popular approach for secondary analyses of GWAS data sets, and have been used successfully to gain additional insights into disease mechanisms and to provide functional context for individual SNP associations. A diverse set of methods for performing GSA has been proposed, and the increased application of these methods has exposed a number of factors that can have an important effect on GSA results. Researchers have also identified a variety of circumstances that can lead to faulty findings, and have proposed ways to avoid misleading results.
In this tutorial we have given an overview of the steps taken during a GSA, including the choices that must be made at each step. We have also attempted to summarize the lessons learned from the numerous applications of GSA methods to GWAS datasets in recent years. We believe the guide presented above will not only allow researchers to make decisions appropriate for the available data and the biological hypotheses of interest when planning a GSA, but will also improve the interpretability and comparability of GSA results.
Acknowledgments
Grant sponsor: NIMH; Grant number: R01MH099064; Grant sponsor: NIH/NCATS; Grant number: UL1TR000128.
The authors thank Joel Nigg and Shannon McWeeney for helpful discussion. Work on this project was supported by NIMH (R01MH099064) and NIH/NCATS (UL1TR000128).
Footnotes
Conflict of interest: None.
References
- Alexeyenko A, Lee W, Pernemalm M, Guegan J, Dessen P, Lazar V, Lehtio J, Pawitan Y. Network enrichment analysis: Extension of gene-set enrichment analysis to gene networks. BMC Bioinformatics. 2012;11(13):226. doi: 10.1186/1471-2105-13-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H, Knapp C, Tsai P, Print C. GeneSetDB: A comprehensive meta-database, statistical, and visualisation framework for gene set analysis. FEBS Open Bio. 2012;2:76–82. doi: 10.1016/j.fob.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakir-Gungor B, Sezerman OU. A new methodology to associate SNPs with human diseases according to their pathway related context. PLoS One. 2011;6(10):e26277. doi: 10.1371/journal.pone.0026277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakir-Gungor B, Egemen E, Sezerman OU. PANOGA: A web server for identification of SNP-targeted pathways from genome-wide association study data. Bioinformatics. 2014;30(9):1287–1289. doi: 10.1093/bioinformatics/btt743. [DOI] [PubMed] [Google Scholar]
- Ballard DH, Cho J, Zhao H. Comparisons of multi-marker association methods to detect association between a candidate region and disease. Genet Epidemiol. 2010;34(3):201–212. doi: 10.1002/gepi.20448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinky F, Nativ N, Stelzer G, Zimmerman S, Stein Iny, Lancet M. PathCards: Multi-source consolidation of human biological pathways. Database (Oxford) 2015 doi: 10.1093/database/bav006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernacka JM, Jenkins GD, Wang L, Moyer AM, Fridley BL. Use of the gamma method for self-contained gene-set analysis of SNP data. Eur J Hum Genet. 2012;20(5):565–571. doi: 10.1038/ejhg.2011.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun R, Buetow K. Pathways of distinction analysis: A new technique for multi-SNP analysis of GWAS data. PLoS Genet. 2011;7(6):e1002101. doi: 10.1371/journal.pgen.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera CP, Navarro P, Huffman JE, Wright AF, Hayward C, Campbell H, Wilson JF, Rudan I, Hastie ND, Vitart V, Haley CS. Uncovering networks from genome-wide association studies via circular genomic permutation. G3 (Bethesda) 2012;2(9):1067–1075. doi: 10.1534/g3.112.002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39(Database issue):D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LS, Hutter CM, Potter JD, Liu Y, Prentice RL, Peters U, Hsu L. Insights into colon cancer etiology via a regularized approach to gene set analysis of GWAS data. Am J Hum Genet. 2010;86(6):860–871. doi: 10.1016/j.ajhg.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang L, Hu B, Guo M, Barnard J, Zhu X. Pathway-based analysis for genome-wide association studies using supervised principal components. Genet Epidemiol. 2010;34(7):716–724. doi: 10.1002/gepi.20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley MJ, Pinese M, Kassahn KS, Waddell N, Pearson JV, Grimmond SM, Biankin AV, Hautaniemi S, Wu J. PINA v2.0: Mining interactome modules. Nucleic Acids Res. 2012;40(Database issue):D862–D865. doi: 10.1093/nar/gkr967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, Jassal B, Jupe S, Matthews L, May B, Palatnik S, Rothfels K, Shamovsky V, Song H, Williams M, Birney E, Hermjakob H, Stein L, D’Eustachio P. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42(1):D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Cruz O, Wen X, Ke B, Song M, Nicolae DL. Gene, region and pathway level analyses in whole-genome studies. Genet Epidemiol. 2010;34(3):222–231. doi: 10.1002/gepi.20452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4):e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Tibshirani R. On testing the significance of sets of genes. Ann Appl Stat. 2007;1:107. [Google Scholar]
- Evangelou M, Dudbridge F, Wernisch L. Two novel pathway analysis methods based on a hierarchical model. Bioinformatics. 2014;30(5):690–697. doi: 10.1093/bioinformatics/btt583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelou M, Smyth DJ, Fortune MD, Burren OS, Walker NM, Guo H, Onengut-Gumuscu S, Chen WM, Concannon P, Rich SS, Todd JA, Wallace C. A method for gene-based pathway analysis using genomewide association study summary statistics reveals nine new type 1 diabetes associations. Genet Epidemiol. 2014;38(8):661–670. doi: 10.1002/gepi.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridley BL, Biernacka JM. Gene set analysis of SNP data: Benefits, challenges, and future directions. Eur J Hum Genet. 2011;19(8):837–843. doi: 10.1038/ejhg.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. The Gene Ontology in 2010: Extensions and refinements. Nucleic Acids Res. 2010;38(Database issue):D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaab E, Baudot A, Krasnogor N, Schneider R, Valencia A. Enrich-Net: Network-based gene set enrichment analysis. Bioinformatics. 2012;28(18):i451–i457. doi: 10.1093/bioinformatics/bts389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman JJ, Buhlmann P. Analyzing gene expression data in terms of gene sets: Methodological issues. Bioinformatics. 2007;23(8):980–987. doi: 10.1093/bioinformatics/btm051. [DOI] [PubMed] [Google Scholar]
- Gui H, Li M, Sham PC, Cherny SS. Comparisons of seven algorithms for pathway analysis using the WTCCC Crohn’s Disease dataset. BMC Res Notes. 2011;4:386. doi: 10.1186/1756-0500-4-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M, Deng S, Wojnowski L, Kulle B. GSEA-SNP: Applying gene set enrichment analysis to SNP data from genome-wide association studies. Bioinformatics. 2008;24(23):2784–2785. doi: 10.1093/bioinformatics/btn516. [DOI] [PubMed] [Google Scholar]
- Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P, Owen MJ, O’Donovan MC, Craddock N Wellcome Trust Case-Control Consortium. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet. 2009;85(1):13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P. Statistical methods for pathway analysis of genome-wide data for association with complex genetic traits. Adv Genet. 2010;72:141–179. doi: 10.1016/B978-0-12-380862-2.00007-2. [DOI] [PubMed] [Google Scholar]
- Hong MG, Pawitan Y, Magnusson PK, Prince JA. Strategies and issues in the detection of pathway enrichment in genome-wide association studies. Hum Genet. 2009;126(2):289–301. doi: 10.1007/s00439-009-0676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA. Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics. 2009 doi: 10.1002/0471250953.bi1311s27. [DOI] [PubMed] [Google Scholar]
- Jia P, Zheng S, Long J, Zheng W, Zhao Z. DmGWAS: Dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;27(1):95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Wang L, Fanous AH, Chen X, Kendler KS, Zhao Z International Schizophrenia Consortium. A bias-reducing pathway enrichment analysis of genome-wide association data confirmed association of the MHC region with schizophrenia. J Med Genet. 2012;49(2):96–103. doi: 10.1136/jmedgenet-2011-100397. [DOI] [PubMed] [Google Scholar]
- Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41(Database issue):D793–D800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014;42(1):D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: Current approaches and outstanding challenges. PLoS Comput Biol. 2012;8(2):e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PH, O’Dushlaine C, Thomas B, Purcell SM. INRICH: Interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28(13):1797–1799. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Ruan J. Network-based pathway enrichment analysis. Proceedings. 2013:218–221. doi: 10.1109/BIBM.2013.6732493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Peng G, Zhu Y, Dong H, Amos CI, Xiong M. Genome-wide gene and pathway analysis. Eur J Hum Genet. 18(9):1045–1053. doi: 10.1038/ejhg.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41(Database issue):D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea C, Taghavi Z, Bokanizad B, Hanoudi S, Tagett R, Donato M, Voichiţa C, Drăghici S. Methods and approaches in the topology-based analysis of biological pathways. Front Physiol. 2013;4:278. doi: 10.3389/fphys.2013.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney MA, Nigg JT, McWeeney SK, Wilmot B. Functional and genomic context in pathway analysis of GWAS data. Trends Genet. 2014;30(9):390–400. doi: 10.1016/j.tig.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam D, Kim J, Kim SY, Kim S. GSA-SNP: A general approach for gene set analysis of polymorphisms. Nucleic Acids Res. 2010:W749–W754. doi: 10.1093/nar/gkq428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18(2):199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Hu T, Malley JD, Andrew AS, Karagas MR, Moore JH. A system-level pathway-phenotype association analysis using synthetic feature random forest. Genet Epidemiol. 2014;38(3):209–219. doi: 10.1002/gepi.21794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroso I, Lourdusamy A, Rietschel M, Nothen MM, Cichon S, McGuffin P, Al-Chalabi A, Barnes MR, Breen G. Common genetic variants and gene-expression changes associated with bipolar disorder are over-represented in brain signaling pathway genes. Biol Psychiatry. 2012;72(4):311–317. doi: 10.1016/j.biopsych.2011.12.031. [DOI] [PubMed] [Google Scholar]
- Pers TH, Karjalainen JM, Chan Y, Westra HJ, Wood AR, Yang J, Lui JC, Vedantam S, Gustafsson S, Esko T, Frayling T, Speliotes EK, Boehnke, Raychaudhuri M, Fehrmann S, Hirschhorn RS, Franke JN Genetic Investigation, Consortium (GIANT) Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelmans G, Pauls DL, Buitelaar JK, Franke B. Integrated genome-wide association study findings: Identification of a neurodevelopmental network for attention deficit hyperactivity disorder. Am J Psychiatry. 2011;168(4):365–377. doi: 10.1176/appi.ajp.2010.10070948. [DOI] [PubMed] [Google Scholar]
- Prasad TSK, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Kishore Harrys, Ahmed S, Kashyap M, Mohmood MK, Ramachandra R, Krishna YL, Rahiman V, Mohan BA, Ranganathan S, Ramabadran P, Chaerkady S, Pandey R. Human protein reference database-2009 update. Nucleic Acids Res. 2009;37(Database issue):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan VK, Shen L, Moore JH, Saykin AJ. Pathway analysis of genomic data: Concepts, methods, and prospects for future development. Trends Genet. 2012;28(7):323–332. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, Cotsapas C, Daly MJ International Inflammatory Bowel Disease Genetics Constortium. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7(1):e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, Swan GE, Goate AM, Rutter J, Bertelsen S, Fox L, Fugman D, Martin NG, Montgomery GW, Wang JC, Ballinger DG, Rice JP, Bierut LJ. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: The pathway interaction database. Nucleic Acids Res. 2009;37(Database issue):D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedeño-Cortés AE, Pavlidis P. Pitfalls in the application of gene-set analysis to genetics studies. Trends Genet. 2014;30(12):513–514. doi: 10.1016/j.tig.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segré AV, Groop L, Mootha VK, Daly MJ, Altshuler D Consortium DIAGRAM, investigators MAGIC. Common inherited variation in mitochondrial genes is not enriched for associations with type two diabetes or related glycemic traits. PLoS Genet. 2010;6(8):e1001058. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver M, Chen P, Li R, Cheng CY, Wong TY, Tai ES, Teo YY, Montana G. Pathways-driven sparse regression identifies pathways and genes associated with high-density lipoprotein cholesterol in two Asian cohorts. PLoS Genet. 2013;9(11):e1003939. doi: 10.1371/journal.pgen.1003939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: A desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23(23):3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- Vandin F, Upfal E, Raphael BJ. Algorithms for detecting significantly mutated pathways in cancer. J Comput Biol. 2011;18(3):507–522. doi: 10.1089/cmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- Varemo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41(8):4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81(6):1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010;11(12):843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- Wray NR, Lee SH, Mehta D, Vinkhuyzen AA, Dudbridge F, Middeldorp CM. Research review: Polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry. 2014;55(10):1068–1087. doi: 10.1111/jcpp.12295. [DOI] [PubMed] [Google Scholar]
- Wu D, Lim E, Vaillant F, Asselin-Labat ML, Visvader JE, Smyth GK. ROAST: Rotation gene set tests for complex microarray experiments. Bioinformatics. 2010;26(17):2176–2182. doi: 10.1093/bioinformatics/btq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Bennett L, Papageorgiou LG, Tsoka S. Module detection in complex networks using integer optimisation. Algorithms Mol Biol. 2010;5:36. doi: 10.1186/1748-7188-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaspan BL, Bush WS, Torstenson ES, Ma D, Pericak-Vance MA, Ritchie MD, Sutcliffe JS, Haines JL. Genetic analysis of biological pathway data through genomic randomization. Hum Genet. 2011;129(5):563–571. doi: 10.1007/s00439-011-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Li Q, Bergen AW, Pfeiffer RM, Rosenberg PS, Caporaso N, Kraft P, Chatterjee N. Pathway analysis by adaptive combination of P-values. Genet Epidemiol. 2009;33(8):700–709. doi: 10.1002/gepi.20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Cui S, Chang S, Zhang L, Wang J. I-GSEA4GWAS: A web server for identification of pathways/gene sets associated with traits by applying an improved gene set enrichment analysis to genome-wide association study. Nucleic Acids Res. 2010;38:W90–W95. doi: 10.1093/nar/gkq324. [DOI] [PMC free article] [PubMed] [Google Scholar]