Abstract
Proteus mirabilis is a Gram-negative bacterium which is well-known for its ability to robustly swarm across surfaces in a striking bulls’-eye pattern. Clinically, this organism is most frequently a pathogen of the urinary tract, particularly in patients undergoing long-term catheterization. This review covers P. mirabilis with a focus on urinary tract infections (UTI), including disease models, vaccine development efforts, and clinical perspectives. Flagella-mediated motility, both swimming and swarming, is a central facet of this organism. The regulation of this complex process and its contribution to virulence is discussed, along with the type VI-secretion system-dependent intra-strain competition which occurs during swarming. P. mirabilis uses a diverse set of virulence factors to access and colonize the host urinary tract, including urease and stone formation, fimbriae and other adhesins, iron and zinc acquisition, proteases and toxins, biofilm formation, and regulation of pathogenesis. While significant advances in this field have been made, challenges remain to combatting complicated UTI and deciphering P. mirabilis pathogenesis.
Proteus mirabilis is well-known in clinical laboratories and microbiology survey courses as the species that swarms across agar surfaces, overtaking any other species present in the process. Urease production and robust swarming motility are the two hallmarks of this organism. This species can be identified as a Gram-negative rod that is motile, urease-positive, lactose-negative, indole-negative, and produces hydrogen sulfide (1). It is a member of the same bacterial family (Enterobacteriaceae) as E. coli.
Disease
P. mirabilis is capable of causing symptomatic infections of the urinary tract including cystitis and pyelonephritis and is present in cases of asymptomatic bacteriuria, particularly in the elderly and patients with type 2 diabetes (2, 3). These infections can also cause bacteremia and progress to potentially life-threatening urosepsis. Additionally, P. mirabilis infections can cause the formation of urinary stones (urolithiasis).
P. mirabilis is often isolated from the gastrointestinal tract, although whether it is a commensal, a pathogen, or a transient organism, is somewhat controversial (4). It is thought that the majority of P. mirabilis urinary tract infections (UTI) result from ascension of bacteria from the gastrointestinal tract while others are due to person-to-person transmission, particularly in healthcare settings (1). This is supported by evidence that some patients with P. mirabilis UTI have the same strain of P. mirabilis in their stool, while others have no P. mirabilis in their stools (5). In addition to urinary tract infection, this species can also cause infection in the respiratory tract, eye, ear, nose, skin, throat, burns, and wounds and has been implicated in neonatal meningoencephalitis, empyema, and osteomyelitis (1, 6). Several studies have linked P. mirabilis to rheumatoid arthritis, although others have failed to find an association (reviewed in (7) and (8)). It is thought that antibodies against hemolysin and urease enzymes are subsequently able to recognize self antigens targeted in rheumatoid arthritis patients (8).
Incidence
P. mirabilis causes between 1-10% of all urinary tract infections, varying with the geographic location of the study, the types of samples collected, and the characteristics of the patients examined. In the most recent large North American study, this species caused 4% of almost 3,000 UTI cases (9). In 2006, UTIs in the United States were the cause of 11 million physician visits and cost $3.5 billion dollars (10). This organism is more common in complicated urinary tract infections (such as patients with spinal cord injury or anatomical abnormality) and especially contributes to catheter-associated UTI (CAUTI), causing 10-44% of long-term CAUTIs at a cost of $43-256 million in the US annually (6, 11, 12). The wide range of P. mirabilis CAUTI likely reflects differences in the population surveyed and the types of samples collected. The highest incidence of P. mirabilis CAUTI occurs in elderly patients during long-term catheterization. P. mirabilis is also a common agent of Gram-negative bacteremia, particularly in patients with concurrent UTI; in recent studies, this species was found in 5-20% of these cases and as high as a 50% mortality rate in geriatric patients (13-16).
VIRULENCE FACTORS
P. mirabilis virulence has primarily been tested using mouse or rat models of infection. Two models of ascending UTI are employed. Independent challenge and co-challenge experiments insert bacteria directly into the bladder using a urethral catheter. In an independent challenge, each strain is tested for the ability to cause infection in the absence of other bacteria, while during a co-challenge experiment two different strains of bacteria are mixed prior to catheterization and must compete to colonize the urinary tract. A third model investigates a hematogenous route of infection, in which bacteria are injected intravenously and the ability of the bacteria to colonize the kidneys is examined.
Throughout this section, genes will be referenced by their PMI gene designations in the sequenced and annotated P. mirabilis genome, strain HI4320 (17), although other wild-type isolates have been studied.
Urease
The cytoplasmic nickel metalloenzyme urease acts by hydrolyzing urea into ammonia and carbon dioxide. The resulting ammonia is the preferred nitrogen source for many species of bacteria, and may be assimilated into biomolecules via glutamine synthetase (GlnA) or glutamate dehydrogenase (GdhA). Further general information on urease is reviewed in (18) and P. mirabilis urease is reviewed in (19). A direct result of urease activity and ammonia generation is an increase in local pH. In the urinary tract alkaline pH leads to precipitation of calcium and magnesium ions and the formation of urinary stones composed of magnesium ammonium phosphate (struvite) and calcium phosphate (apatite) (20). In P. mirabilis, urease is encoded by the ureDABCEFG operon (PMI3682-88). A regulator, UreR (PMI3681), is encoded on the reverse strand adjacent to this operon (21). UreR is an AraC-type DNA binding protein which positively induces urease expression in the presence of urea; a ureR mutant lacks measurable urease activity (22). UreR also positively regulates its own expression when bound to the intergenic region between ureR and ureD, and ureR transcription is repressed by the global regulator H-NS (23). Urease expression is induced in the urinary tract of experimentally-infected mice (24, 25).
VIRULENCE
Urease mediates virulence via the production of urinary stones. These stones can block urinary flow and cause tissue damage; they can also become quite large (> 1 cm2) (26, 27). The precipitated minerals may mix with bacteria adherent to a urinary catheter, forming a crystalline biofilm and eventually blocking urine flow through the catheter (28, 29). Similar intracellular crystals have been visualized in cultured urinary epithelial cells that have been experimentally infected with invasive P. mirabilis (30). Although other urease-positive bacterial species are associated with catheter-associated UTI, only P. mirabilis has a positive association with catheter obstruction (31). Bacteria can become embedded in these stones, which may protect pathogens from antibiotics or the immune system (32) (Fig. 1). Furthermore, urinary stones can act as a focal point for other species of bacteria to establish UTI (19). A ureC urease mutant is incapable of forming stones, and this has a direct impact on the ability of P. mirabilis to cause UTI. When this urease mutant was tested in independent challenge using a mouse model of ascending UTI, there was a highly-significant decrease in bacterial numbers compared to the wild-type parent in the bladder, kidneys, and urine (33). The effect was especially pronounced in the kidneys, where no mutant bacteria were detectable in most mice 48 hours post-infection. The murine 50% infective dose for the urease mutant (2.7 × 109 CFU) is 1000-fold higher than the wild type (2.2 × 106 CFU) (34). From two days to two weeks post-inoculation, kidneys from mice infected with the urease mutant displayed less pathology (acute inflammation, epithelial necrosis) compared to wild-type infection. Likewise, a ureR mutant tested in cochallenge with the parent strain was unrecoverable in most mice (22). Urease activity during UTI may be influenced by polymicrobial infection; experimental co-infection of mice with P. mirabilis and urease-positive Providencia stuartii resulted in increased urolithiasis and bacteremia despite similar bacterial loads compared to monospecies infection (35). Indeed, in vitro co-culture of P. mirabilis and P. stuartii in human urine resulted in enhanced total urease activity compared to either species alone (35).
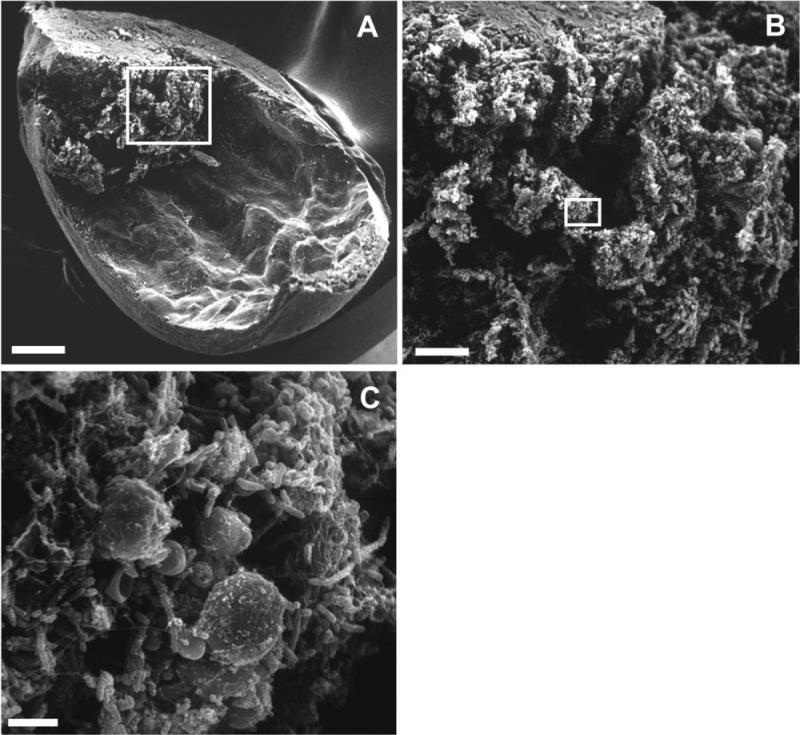
Figure 1.
P. mirabilis in urease-induced bladder stone. A, One-quarter bladder of experimentally-infected mouse (bar, 500 μm). B, Higher magnification of the area indicated in panel A (bar, 100 μm). C, Higher magnification of the area indicated in panel B with individual bacteria visible (bar, 5 μm). Modified with permission from Infection and Immunity, volume 70, page 392, 2002 (32).
Due to the prominent role of urease in P. mirabilis virulence, this enzyme is an active target of investigation to identify clinically useful inhibitors (36). Since the ability of P. mirabilis to generate urinary stones and crystalline biofilms is dependent upon alkaline pH, another approach to prevent catheter blockage is to acidify the urine. Similarly, mineral nucleation can be inhibited by reducing mineral concentration in the urine, i.e., by increasing fluid intake (37). These efforts are aimed at increasing the urinary nucleation pH (the pH at which minerals will precipitate from the urine); a lower nucleation pH is associated with increased stone formation. Preliminary results with patients consuming lemon juice are promising, with the result of increased nucleation pH (38). However, the effect of such treatments on catheter blockage has not yet been reported.
Flagella
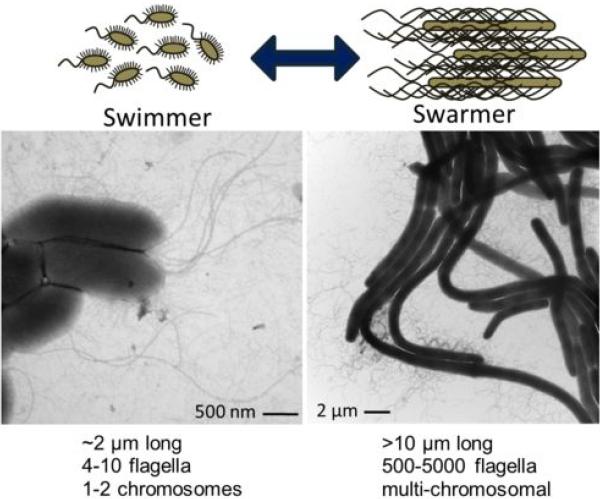
Like many bacteria, P. mirabilis uses flagella to swim through liquids and toward chemical gradients (reviewed in (39)). In liquid culture, P. mirabilis has a short rod shape and typically possesses a few peritrichous flagella. However, on rich solid media, P. mirabilis differentiates into very long (typically 20-80 μm, although cells longer than 100 μm occur), nonseptate polyploid cells with hundreds to thousands of flagella. These swarmer cells move as a population across surfaces, and will be discussed later in this chapter. Although the flagella produced by this organism are generally similar to flagella produced by other bacteria, there are two unusual characteristics of P. mirabilis flagella. First, all genes encoding flagellar components, including the class I flagellar master regulatory genes flhDC (PMI1671-72), are found within a single 54 kb locus in the chromosome (PMI1617-72). This is in contrast to most other flagella-producing bacteria, which have flagellar operons in disparate loci. Second, P. mirabilis encodes two flagellins, FlaA (PMI1620) and FlaB (PMI1619) (also known as FliC1 and FliC2, respectively) (40), which comprise the whip structure of the flagellum.
ANTIGENIC VARIATION
FlaA is the major flagellin. Despite the proximity of flaA and flaB, flaA is transcribed as a monocistronic message and flaB message is generally undetectable (41). Recombination between flaA and flaB can occur. This phenomenon was discovered when flaA mutants were often found to revert to a motile phenotype; these revertants produced antigenically distinct flagella that were the product of recombination resulting in a hybrid flaAB gene (41, 42). Later studies revealed that wild-type populations of P. mirabilis are heterogeneous, with a portion of the cells possessing hybrid flaAB genes (42, 43). RNA experiments suggested that 1.0-1.5% of the total flagellin message in wild-type populations is flaAB (43). Recombination occurs between homologous regions of flaA and flaB, and leads to deletion of the intervening sequence. Hybrid flagellins have been detected in bacteria excreted in the urine of mice experimentally infected with P. mirabilis. There may be a selective advantage to particular recombination events, as the types of rearrangements typically found depend on the bacterial environment (broth, swarm agar, or murine urinary tract) (42). The recombinant flagella are functional and may serve as a method of immune evasion during UTI or to provide motility under adverse conditions. Indeed, bacteria possessing specific hybrid FlaAB flagella are more motile under conditions of high salinity (255-425 mM NaCl) or extreme pH (5.2 or 8.2) compared to bacteria with wild-type FlaA flagella; conversely, the wild-type flagellum confers a motility advantage in low salinity (85 mM NaCl) (44). Additionally, immune serum from mice experimentally infected with P. mirabilis reacts with both FlaA and FlaAB flagellins (45). A possible third flagellin, designated FlaC or FliC3, was identified by DNA-DNA hybridization (40); however, fliC3 was not annotated in the P. mirabilis HI4320 genome sequence and is not readily identifiable by nucleotide BLAST using queries that correspond to the probes used in the initial discovery of fliC3.
REGULATION OF FLAGELLA
As has been characterized for other bacteria, P. mirabilis flagellar genes are transcribed in a three-tier hierarchy (reviewed in (46)). Regulation of flagella is mediated through the class 1 flagellar master regulator genes flhDC. The regulator functions as the heteromer FlhD2C2 or FlhD4C2 (47, 48). There are multiple inputs to regulation of flhDC as well as post-translational modification of FlhD4C2 and downstream regulation of the class II and class III flagellar genes. Perturbations in flagellar expression may lead to different outcomes with regard to swimming versus swarming, and will be discussed in detail below.
Expression of flagellar genes is regulated during experimental UTI. Within 24 h post-infection, flagella are repressed compared to an in vitro mid-logarithmic phase broth culture (25). However, by seven days post-infection, this repression is relieved (25) (Fig. 2). It is very likely that flagella are produced at some point during P. mirabilis-mediated UTI, as experimentally-infected mice produce antibodies that recognize flagella (45, 49). Flagella are also repressed during uropathogenic E. coli (UPEC)-mediated UTI (50), but are transiently expressed around 4-8 hours post-infection (51). This time coincides with bacterial ascension from the bladder to the kidneys. It is possible that P. mirabilis flagella undergo similar transient expression during early stages of ascending UTI.
Figure 2.
Adherence and motility genes are inversely regulated during UTI. Each line represents fold-change of a specific flagellar (left panel) or fimbrial (right panel) gene in vivo relative to mid-logarithmic phase culture in vitro. Genes in the mrp operon are highly induced early during infection, but expression falls by seven days postinfection. Flagellar genes are initially repressed, but expression increases late in infection. Modified with permission from Infection and Immunity, volume 79, page 2625 (25).
ROLE OF FLAGELLA IN VIRULENCE
There are differing conclusions in the literature about the role of motility in P. mirabilis virulence. In one study, the flagellum was found to contribute to ascending UTI (52). In that report, an hpmA (hemolysin, PMI2057) mutant was compared in independent challenge of CBA/J mice to the wild-type parent and an isogenic hpmA flaD (PMI1621) double mutant. FlaD is the capping protein of the flagellum; this mutant produces unassembled flagellin and is nonmotile. At one week post-infection, the hpmA flaD mutant was recovered from the urine, bladder, or kidneys in numbers 100-fold lower than either the wild type or the hpmA single mutant. Functional flagella contribute to bacterial spread during UTI; immobilizing antibodies prevented the ability of P. mirabilis to traverse from one kidney to the other via the ureters using a rat model (53). However, another group has reported that nonmotile P. mirabilis strains also cause UTI (54, 55). In the first of these studies, both ascending UTI and hematogenous routes of infection were used to assess the ability of motile and nonmotile P. mirabilis isolates to colonize the bladders or kidneys of outbred mice at seven days post-infection (54). The nonmotile strain was as infective as the motile strains. In the second study, an isogenic mutant missing the 3’ portion of flaA and the 5’ portion of flaB was compared to the wild-type parent in the ascending UTI outbred mouse model (55). Again, at seven days post-infection, there was not a significant difference between wild type and mutant in colonization of bladder or kidneys. Taken together, it is likely that flagella contribute to P. mirabilis UTI, but the effect may be modest and dependent on the strains of bacteria and animal models used. The specific contribution of swarming to virulence will be considered later in this chapter.
Signature-tagged mutagenesis (STM) studies have also identified mutants with diminished or absent production of flagella that were less competitive in the mouse UTI model (56, 57). In addition, STM identified a chemotaxis mutant (cheW), suggesting that not just motility but also the ability to move toward one or more unidentified signals contributes to bacterial fitness during UTI (56). Although eight likely methyl-accepting chemotaxis proteins (MCP) were identified in the HI4320 genome (two in the flagellar locus and six encoded elsewhere (17)), it is worth noting that the molecules sensed by these MCP have not been elucidated for P. mirabilis.
Swarming
When P. mirabilis is added to an agar surface, the bacteria grow in place for a time (which varies by medium, humidity, and temperature), differentiate into swarmer cells, and move forward as a population. The ability of P. mirabilis to swarm as an organized group across solid surfaces was first noted by Hauser in 1885 (58). DNA replication without septation occurs during swarm cell formation, which results in very long, polyploid cells. At defined intervals, the bacteria stop moving and revert to a shorter morphotype (consolidate). After a period, the bacteria redifferentiate into swarmer cells. These bacteria move across many media surfaces in a repeated process of swarming and consolidation, resulting in a characteristic bull's-eye pattern (Fig. 3). In fact, these bacteria are named for the Greek god Proteus, who was able to “change shape at will to avoid questioning” (58). Swarmer cells are phenotypically distinct from swimmer or vegetative cells, and are characterized by great length (typically 20-80 μm) and hyperflagellation (Fig. 4). The shift to the swarmer form is accompanied by changes in lipopolysaccharide (LPS), peptidoglycan, and membrane fatty acid composition (59, 60). Swarm cells move together as a group, forming rafts of parallel cells (61). A capsular polysaccharide termed colony migration factor and an uncharacterized slime are associated with swarming cells and may be used to aid motility across surfaces (62-64). Numerous genes are differentially regulated during swarming, including genes that are not required for swarming (65, 66). Swarming motility has also been observed for other bacterial species, both Gram-negative and Gram-positive (reviewed in (67)). Specifically, P. mirabilis shares common features with swarming by E. coli (68) and Salmonella (69, 70); however, P. mirabilis swarming is famously robust compared to these species and will occur on most laboratory media unless inhibitors are used. Swarming does not typically occur on chemically-defined minimal media (71), and may also be controlled in the laboratory by reducing the concentration of salt to ≤0.5 g/L or by adding inhibitors (e.g., glycerol, p-nitrophenyl glycerin, or 4% agar) (72-76), although the success of these techniques may depend on incubation time, temperature, humidity, and strain of P. mirabilis. Mathematical models have been used to represent P. mirabilis swarming (77-80); these models recapitulate the terracing that occurs during swarm-consolidation cycles and are beginning to address issues such as water channeling.
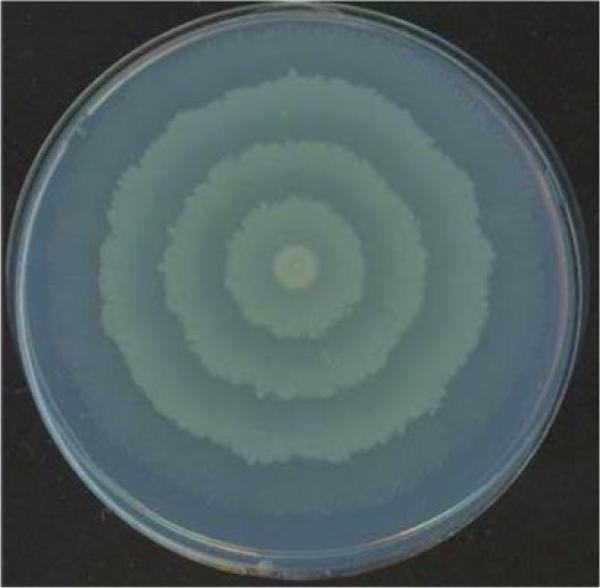
Figure 3.
Swarming colony of P. mirabilis.
Figure 4.
P. mirabilis switches between swimming and swarming forms. On the left is a transmission electron micrograph (TEM) of broth-cultured, vegetative cells displaying peritrichous flagella. On the right is a TEM of differentiated swarm cells. Bundles of flagella are visible.
CONTRIBUTION OF FLAGELLA TO SWARMING
Transcription of flagella is greatly increased during the initial transition from swimming to swarming, and remains high (though cyclical) throughout the swarm cycle. When transcription of swarm and consolidate cells was measured by microarray, flaA (flagellin) was the third and sixth most highly-expressed transcript, respectively (65). Flagella are not only required for swarming motility, but they are also linked to swarmer cell differentiation. For example, a flaD mutant, which produces flagellin but does not assemble flagella, does not swarm and fails to elongate or undergo polyploidy (40). Interaction with a surface is important to swarmer cell development. Elongation of cells can be triggered by culture in viscous fluids, such as by addition of polyvinylpyrollidone (71) or by impeding flagellar rotation through addition of anti-flagellin antibodies (81). It is believed that solid or viscous surfaces are sensed by restricted rotation of flagella, which transmit the signal to transcribe genes associated with swarming (71, 82, 83).
Transposon mutagenesis has been used to identify genes that contribute to swarming, with many variable phenotypes noted (swarming null or crippled swarming, failure to elongate or constitutive elongation, positive or negative for swimming motility or chemotaxis) (73, 84-86) (Table 1). As a caveat, not all of the transposon mutants described in this chapter were complemented, and there could be polar effects. Most mutations of flagellar genes lead to a motility, swarm, and elongation null phenotype (81, 83), which is in keeping with the hypothesis that swarming differentiation requires surface sensing by flagella. However, mutants that were defective in flagellar production or assembly were not universally non-motile, and had distinct phenotypes (81, 87). A flaA mutant was motility and swarming null and failed to elongate under swarming conditions; however, this mutant occasionally reverted to being motility and swarming positive (87). This was later shown to be due to recombination with the flaA and flaB flagellin genes (42). Another swarming-null, motility-positive transposon insertion in flgN (flagella filament assembly, PMI1657) was identified by Gygi et al (85). This mutation resulted in the secretion of unassembled flagellin. The few mature flagella assembled by the flgN mutant were apparently sufficient to mediate swimming motility but did not allow the hyperflagellation required for swarming motility to occur (85). Negative feedback occurs on flhDC when flagellar assembly is blocked in an flhA mutant; restoration of elongation but not flagellation occurs when flhDC is supplied in trans (88).
Table 1.
Genes that contribute to swarming in P. mirabilis.
| Motility | ||||||
|---|---|---|---|---|---|---|
| Name | ORF | Function | Swim | Swarm | Elong | Ref |
| Flageila | ||||||
| flhDC | PMI1671-2 | flagellar transcriptional activator (cl. I) | - | - | - | (81, 88) |
| fliF | PMI1630 | flagellar MS-ring protein (class II) | - | - | consta | (57, 81) |
| fliG | PMI1631 | flagellar motor switch protein (cl. II) | - | - | const | (81) |
| fliL | PMI1636 | flagellar basal body-associated protein (class II) | less | - | const | (81, 87) |
| fliM | PMI1637 | flagellar motor switch protein (cl. II) | slight | - | - | (81) |
| fliP | PMI1640 | flagellar biosynthetic protein (cl. II) | + | - | - | (81) |
| fliQ | PMI1641 | flagellar biosynthetic protein (cl. II) | - | - | - | (81) |
| flgH | PMI1648 | flagellar L-ring (class II) | + | cr | const | (87) |
| flgE | PMI1651 | flagellar hook protein (class II) | NR | - | NR | (57) |
| flgB | PMI1654 | flagellar basal body rod protein (cl. II) | - | - | - | (81) |
| flgN | PMI1657 | flagella filament assembly (cl. II) | + | - | NR | (85) |
| flhA | PMI1659 | flagellar assembly (class II) | - | - | - | (81, 83) |
| flaA | PMI1620 | flagellin (class III) | -b | -b | - | (40, 41) |
| flaD | PMI1621 | flagellar capping protein (cl. III) | - | - | - | (40) |
| flgK | PMI1645 | flagellar hook-associated protein 1 (III) | - | - | - | (81) |
| flgL | PMI1644 | flagellar hook-associated protein 3 (III) | - | - | - | (81) |
| cheW | PMI1667 | chemotaxis; required for CheA function (class III) | NR | - | NR | (56) |
| Flagellar regulation | ||||||
| lon | PMI0117 | ATP-dependent Lon protease | + | ++ | const | (94) |
| lrp | PMI0696 | leucine-responsive regulator | less | - | - | (95) |
| rsbA | PMI1729 | phosphotransfer intermediate protein | + | ++ | + | (105, 106) |
| rcsBC | PMI1730-1 | capsular synthesis two-component system | + | ++ | + | (92, 106) |
| umoA | PMI3115 | upregulator of flagellar master operon | less | cr | + | (91) |
| umoB | PMI3018 | upregulator of flagellar master operon | less | cr | - | (91) |
| umoC | PMI1939 | upregulator of flagellar master operon | less | cr | + | (91) |
| umoD | PMI0876 | upregulator of flagellar master operon | less | cr | - | (91) |
| rppA | PMI1697 | two-component system sensor kinase | NR | ++ | NR | (108) |
| disA | PMI1209 | amino acid decarboxylase | ++ | ++ | + | (97) |
| LPS | ||||||
| rfaD | PMI3176 | ADP-L-glycero-D-manno-heptose-6-epimerase | + | cr | - | (87) |
| galU | PMI1490 | UTP--glucose-1-phosphate uridylyltransferase | + | cr | - | (87, 111) |
| ugd | PMI3189 | UDP-glucose 6-dehydrogenase | NR | - | - | (111) |
| waaL | PMI3163 | O-antigen ligase | + | - | - | (113) |
| wzz/cld | PMI2183 | O-antigen chain length determinant | + | - | - | (113) |
| Metabolism | ||||||
| cyaA | PMI3333 | adenylate cyclase | NR | cr | NR | (57) |
| aceE | PMI2046 | pyruvate dehydrogenase E1 component | NR | cr | NR | (57) |
| sdhC | PMI0565 | succinate dehydrogenase cytochrome b556 subunit | NR | cr | NR | (57) |
| fumC | PMI1296 | fumarate hydratase, class II | NR | cr | NR | (134) |
| sdhB | PMI0568 | succinate dehydrogenase iron-sulfur protein | NR | cr | NR | (134) |
| gnd | PMI0655 | 6-phosphogluconate dehydrogenase, decarboxylating | NR | cr | NR | (134) |
| talB | PMI0006 | transaldolase B | NR | cr | NR | (134) |
| pfkA | PMI3203 | 6-phosphofructokinase | NR | cr | NR | (134) |
| tpiA | PMI3205 | triosephosphate isomerase | NR | cr | NR | (134) |
| Amino acids | ||||||
| serC | PMI0711 | phosphoserine aminotransferase | NR | cr | NR | (56) |
| cysJ | PMI2250 | sulfite reductase [NADPH] flavoprotein alpha-component | NR | crc | + | (65) |
| dppA | PMI2847 | dipeptide ABC transporter, substrate-binding protein | NR | crc | + | (65) |
| oppB | PMI1474 | oligopeptide ABC transporter, permease protein | NR | crc | + | (65) |
| glnA | PMI2882 | glutamine synthetase | less | - | NR | (120) |
| hisG | PMI0665 | ATP phosphoribosyltransferase | + | cr | + | (120) |
| Other | ||||||
| gidA | PMI3055 | tRNA uridine 5-carboxymethylaminomethyl modification enzyme | + | cr | - | (87) |
| pepQ | PMI3551 | proline peptidase | + | cr | - | (87) |
| dapE | PMI1556 | N-succinyl-diaminopimelate deacylase | + | cr | - | (87) |
| cmfA/cpsF | PMI3190 | colony migration factor (capsular polysaccharide) | + | cr | + | (57, 63) |
| ppaA | PMI3600 | P-type ATPase zinc transporter | + | cr | slight | (116, 117) |
| znuC | PMI1151 | high-affinity zinc uptake system ATP-binding protein | slight | cr | slight | (118) |
| nrpG | PMI2605 | 4′-phosphopantetheinyl transferase | NR | cr | NR | (56, 119) |
| hemY | PMI3329 | porphyrin biosynthesis protein | NR | cr | NR | (56) |
| speB | PMI2093 | agmatinase | + | cr | delay | (120, 121) |
| ccmA | PMI1961 | putative membrane protein, curved cell morphology | + | - | + | (115) |
| dsbA | PMI2828 | thiol:disulfide interchange protein | NR | cr | NR | (56) |
| surA | PMI2332 | peptidyl-prolyl cis-trans isomerase | + | - | NR | (56, 61) |
| mrcA | PMI3021 | penicillin-binding protein 1A | NR | cr | NR | (56) |
| parE | PMIP32 | plasmid stabilization (toxin-antitoxin) | NR | cr | NR | (56) |
| sufI | PMI2342 | putative multicopper oxidase (suppressor of ftsI) | NR | - | NR | (57) |
| hexA | PMI1764 | LysR-family transcriptional regulator | NR | crd | + | (65) |
| lrhA | PMI0629 | LysR-family transcriptional regulator | NR | crd | + | (65) |
| PMI1874 | two-component system sensor kinase | NR | cr | NR | (56) | |
| PMI1046 | putative polysaccharide deacetylase | NR | cr | NR | (56) | |
| PMI3692 | putative lipoprotein | NR | cr | NR | (56) | |
| Phenotypes observed by overexpression | ||||||
| rsmA/csrA | PMI0377 | carbon storage regulator | NR | - | - | (100) |
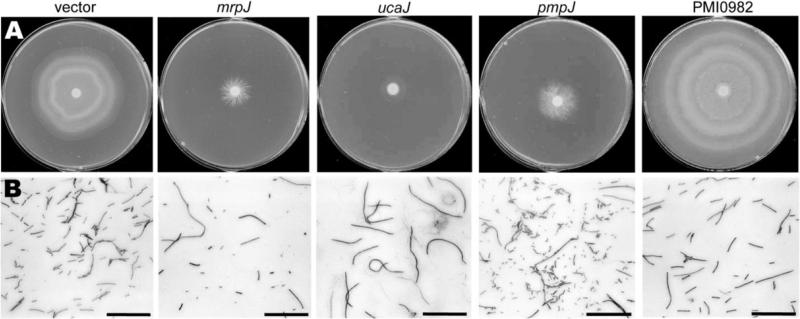
| mrpJ d | PMI0271 | fimbrial operon regulator | - | - | aberr | (162, 203) |
| wosA | PMI0608 | regulator of swarming motility | ++ | ++ | const | (99) |
const, constitutive; cr, crippled; ++, hyperswarm/hypermotility; aberr, aberrant; NR, not reported
elogates on nonswarm agar but not in broth
reverts to wild-type
phenotype detected on MinA-T agar
12 of 14 additional mrpJ homologs also repress motility when overexpressed
Constitutive elongation flagellar mutants
In contrast to the previously described flagellar mutants, fliL (PMI1636, hook basal body) and fliG (PMI1631, flagellar motor switch) mutants exhibited a constitutively elongated (pseudoswarmer) phenotype (81, 87). The fliL phenotype is especially interesting because mutations in other genes in its operon (fliLMNOPQR) lead to a failure to elongate (81). A nonpolar fliL mutant has relatively few flagella and low levels of flaA transcription despite its elongated phenotype (89). However, transcription of the class I master regulator flhDC and the class II flagellar cascade sigma factor fliA genes is increased in this strain compared to its wild-type parent. Complementation with fliL rescues the elongation phenotype but does not restore flagellin expression. Although FliM levels are not disrupted in the fliL mutant, complementation with both fliL and fliM is necessary to restore swarming motility, suggesting that there is an element of fliM DNA that contributes to fliL function (89). The fliL mutant also has increased expression by vegetative cells of virulence factors (zapA and hpmA) which are induced during wild-type swarming (81). RNA-seq analysis of fliL mutant pseudoswarmer cells found that the umoA regulator of swarm cell differentiation was induced (90), similar to the induction that occurs in wild-type cells during swarming (91). Although the mechanism of FliL-mediated swarming differentiation has not been elucidated, this protein has been proposed to sense the torque generated on the basal body when flagella encounter viscosity, or to count the rate of proton flow through the motor (81).
REGULATION OF flhDC
Cyclic regulation of flagellin contributes to the swarm-consolidation pattern of P. mirabilis, and perturbation of this regulation results in aberrant swarming. Artificial overexpression of the flagellar master regulator genes flhDC (PMI1671-2) results in earlier and faster swarming (88). A transposon screen by Clemmer and Rather identified two similar mutants with insertions in the flhDC promoter, which had constitutively high levels of expression of these genes during swarming (92). These mutants initiate swarming sooner and swarm at a higher velocity, but lack the consolidation phase and thus do not form a bull's-eye pattern when swarming. Likewise, factors that regulate flhDC expression, transcript stability, or posttranslational modification also influence swarming behavior. As might be expected, the FlhD and FlhC proteins have high turnover and are rapidly degraded during swarming, with a half-life of approximately two minutes (93). Three regulators that are known to repress flhDC in E. coli have little or no effect on swarming when mutated in P. mirabilis: ompR, lrhA and hdfR (65, 92). Known regulators of FlhD4C2 in P. mirabilis are summarized in Table 1 and discussed in detail below.
Lon protease
A mutation in the gene encoding Lon protease (PMI0117) results in cells that initiate swarming normally but migrate faster than the wild type; complementation with lon restored the wild-type phenotype (94). The lon mutant bacteria also had a tendency to differentiate into swarm cells (elongation, increased flagellation) under non-permissive conditions such as during broth culture. This regulation is likely due to Lon-dependent degradation of FlhD (94). Expression of lon is regulated during swarming (65, 94).
Lrp
The global regulator Lrp (PMI0696) is required to initiate cell elongation and swarming. The lrp mutant swarming phenotype can be complemented by artificial overexpression of flhDC, although this strain is not complemented for expression of hemolysin during swarming (95). Notably, Lrp accumulates at a higher rate in P. mirabilis compared to E. coli or Vibrio cholerae during culture in rich media (a condition that is permissive for swarming) and is subject to only weak auto-repression, unlike E. coli Lrp (96).
DisA
The DisA decarboxylase (PMI1209, decarboxylase inhibitor of swarming) is induced during swarming (97). Mutation of disA leads to increased flhDC transcript and correspondingly enhanced swarming and swimming motility. Overexpression of disA blocks flagellar class 2 (fliA) and class 3 (flaA) gene expression, but does not significantly alter flhDC transcription. DisA is predicted to be an amino acid decarboxylase. Addition of the decarboxylated amino acid phenethylamine reduced both swarming and disA expression (97, 98). Phenethylamine also inhibited transcription of class 2 and 3 flagellar genes yet had a minimal effect on flhDC. FlhC levels are not affected by disA overexpression, suggesting that DisA activity does not destabilize this protein. It has been proposed that the DisA decarboxylation product interferes with FlhD4C2 assembly or DNA binding (97).
WosA
The wosA gene (PMI0608, wild-type onset with superswarming) encodes a predicted membrane protein that induces flhDC expression and hyperswarming when overexpressed (99). It also causes constitutive swarm cell differentiation in liquid media. Inhibition of flagellar rotation, either in a fliL mutant or in viscous broth, increases wosA expression; this regulation is only partially dependent on the presence of flagella. Transcription is also increased over time during both broth and agar culture. However, a wosA mutant only has a modest decrease in swarming compared to wild type, suggesting that wosA is one of several inputs to flhDC regulation (99).
RsmA/CsrA
Overexpression of another gene, rsmA (PMI0377, repressor of secondary metabolites, also called csrA) results in repression of swarming motility and differentiation (100). The predicted protein is 96% identical to CsrA, a positive regulator of flhDC in E. coli (101). RsmA and CsrA appear to have opposite effects on flhDC regulation, and this warrants further investigation. However, attempts to mutate rsmA in P. mirabilis were unsuccessful. Expression of P. mirabilis rsmA complemented an E. coli csrA mutant with regard to glycogen storage, although the effect of rsmA on flhDC expression in E. coli was not reported (100). Overexpression of rsmA decreased the half-life of hemolysin mRNA, indicating that RsmA, like CsrA, regulates by affecting mRNA stability.
Umo proteins
A screen to identify genetic regions that restore swarming motility to the motile but non-swarming flgN mutant (described above) revealed four additional genes that positively regulate flhDC (91). These four genes were designated umoA, umoB, umoC, and umoD for upregulator of the master operon (PMI3115, PMI3018, PMI1939, and PMI0876, respectively). Expression of umoA and umoD is induced during swarming and these genes are subject to negative feedback when flagellar assembly is blocked and positive feedback by flhDC (91). Despite the plethora of sequenced genomes available since the discovery of the umo genes, umoA remains unique to species within Proteus and Morganella. The UmoB homolog IgaA has been recognized as a regulator of the Rcs phophorelay (102, 103), and UmoB homologs are widespread in the Enterobacteriaceae. The Rcs phophorelay (reviewed in (104)) has been implicated in swarming by P. mirabilis and is discussed below. In P. mirabilis, UmoB is a negative regulator of swarming inhibitor disA (discussed above) (98).
TWO-COMPONENT SYSTEMS THAT REGULATE flhDC
RsbA-RcsBC
Generation of hyperswarming or precocious (early) swarming mutants has been used to identify repressors that contribute to swarming. RsbA (regulator of swarming behavior, PMI1729; also called rcsD) has twice been identified as a repressor of swarming (105, 106). The rsbA gene encodes a phosphotransfer intermediate that is part of the RcsBCD phophorelay system. An rsbA mutant has a similar hyperswarming phenotype to that seen during flagellin overexpression (105, 106). Interestingly, overexpression of rsbA also results in precocious swarming (106). Liaw and colleagues examined the ability of transposon mutants to swarm in the presence of the swarming inhibitor p-nitrophenylglycerol (PNPG) (105) and identified a mutant with an insertion in rsbA. During swarming, this mutant expresses higher levels of flagellin, as well as other swarming co-regulated virulence factors including hemolysin, protease, and urease (105). An RsbA-mediated pathway may involve sensing of saturated fatty acids to determine a tendency toward swarming or biofilm formation (107). That is, in the presence of specific fatty acids (myristic acid, lauric acid, palmitic acid), swarming behavior is inhibited while biofilm formation and extracellular polysaccharide production is enhanced. The rsbA mutant is unresponsive to these fatty acids (i.e., hyperswarms) and is deficient in biofilm formation under permissive conditions. To further investigate the contribution of the RcsBCD phosphorelay to swarming, Clemmer and Rather constructed an rcsB (PMI1730, response regulator) mutant; this strain also had a hyperswarming phenotype (92). Thus, the RcsBCD system is likely a repressor of swarming behavior.
RppAB
The rppAB genes (PMI1696-7) form a two-component system that regulates multiple cellular functions in P. mirabilis including LPS biosynthesis and repression of flagella. An rppA mutant was identified in a transposon screen for hyperswarming mutants (108). The rppA mutant also had increased expression of virulence factors associated with swarming (HpmA) and an altered LPS profile which conferred increased susceptibility to polymyxin B. RppA regulates pmrI (PMI1045, also called arnA), which is predicted to be a bacterial UDP-glucuronic acid decarboxylase and contributes to LPS modification (109). Expression of rppA is induced in the presence of polymyxin B and repressed when 10 mM Mg2+ is added (108). This profile is similar to what is observed for the PhoPQ two-component system of Salmonella enterica (reviewed in (110)). Although RppAB has some homology to PhoPQ, another two-component system encoded by P. mirabilis has greater similarity (phoPQ, PMI0884-5) (17). The function of this second locus has not yet been reported.
NON-FLAGELLAR LOCI THAT CONTRIBUTE TO SWARMING
LPS
Several elongation-negative mutants with defects in LPS synthesis have been identified through transposon mutagenesis screens (PMI3176 rfaD, PMI1490 galU, PMI3189 ugd, PMI3163 waaL/rfaL, an O-acetyltransferase, and a probable O-antigen chain-length determinant mutant) (87, 111-113). Swarming is connected to LPS O-antigen but not to the related enterobacterial common antigen (ECA), as a wzyE (PMI3326) mutant swarms (113). Notably, mutation of rcsB or rcsC, and to a lesser extent rcsF, suppresses the waaL swarming deficient mutant (103, 113), showing that although O-antigen itself is not necessary for swarming, there is a regulatory link between LPS biosynthesis and swarming differentiation. UmoB and UmoD have been proposed to activate the Rcs system in an O-antigen dependent manner that is distinct from the canonical Rcs surface-sensed activation by RcsF (91, 103). Mutations in LPS also confer sensitivity to the cationic antimicrobial peptides (CAP) such as polymyxin B; P. mirabilis mutants defective in CAP resistance are either swarming negative or swarm poorly (111, 112). Mutations in at least two of these genes, ugd and galU, activate the alternative sigma factor RpoE, which leads to flhDC repression (111).
Capsule and cell morphology
A mutation in cmfA (colony migration factor, PMI3190, also called cpsF) results in a capsular polysaccharide defect. This mutant is able to elongate and become hyperflagellated, but exhibits reduced swarm velocity (63). The structure of this polysaccharide has been determined to be a tetrasaccharide repeat for one strain of P. mirabilis (64), and this polysaccharide contributes to virulence (114). Another transposon mutant with a motility and elongation-positive but swarming-deficient phenotype was localized to ccmA (curved cell morphology, PMI1961) (115). Although ccmA mutant cells become hyperflagellated and elongate under permissive conditions, the cells are curved and uneven in width. A second ccmA mutant in which 80% of the gene was deleted resulted in a less severe but still distinctive curved cell phenotype. Overexpression of ccmA results in cells with an ellipsoidal or spherical shape. CcmA is predicted to be an integral membrane protein, and its expression increases during swarming differentiation (65, 115). CcmA has been proposed to help maintain linearity during swarm cell elongation, perhaps by organizing peptidoglycan assembly (115). Immunoblotting with CcmA antibodies indicated that there are two forms of CcmA produced in wild-type cells; the larger size matched the predicted full-length protein (CcmA1), and the shorter protein corresponded to an alternative methionine at position 59 of the full-length protein (CcmA2) (115).
Zinc and iron acquisition
A study by Lai et al described an aberrant swarm mutant with a disruption in a gene encoding a zinc-transporting membrane P-type ATPase (PMI3600 ppaA) (86, 116, 117). This mutant swarms at a lower velocity and does not fully elongate during swarming differentiation, yet swarms for longer intervals and has aberrant consolidation terracing. Despite producing lower levels of flagellin transcript and protein as well as repressing the motility regulator lrp, it has normal swimming motility. In wild-type P. mirabilis, ppaA expression is induced during swarming (116). Another zinc uptake mutant, znuC (PMI1151) was subsequently found to display aberrant swarming; furthermore, wild-type P. mirabilis has a similarly altered swarming pattern in the presence of the zinc chelator TPEN (118). Mutation of a nonribosomal peptide system involved in iron acquisition also leads to aberrant swarming (56, 119).
Multiple genes that contribute to swarming were identified in two STM screens (56, 57); however, the roles of these genes in swarming have not been elucidated. Two other elongation-negative transposon mutants had defects in cellular division (PMI3055 gidA) and a proline peptidase (PMI3551 pepQ) (87), although their roles in swarming differentiation have not been described further. These genes are listed in Table 1.
EXTRACELLULAR CONTRIBUTORS TO SWARMING
Glutamine
Glutamine is required for swarmer cell differentiation (71, 120). When Allison et al added a variety of components to a defined minimal growth medium that is normally nonpermissive for swarming, only glutamine triggered swarming behavior(71). Addition of the other 19 amino acids mixed together did not allow swarming within 24 hours. However, swarming occurred more rapidly and the characteristic bull's-eye pattern only developed when all 20 amino acids were present. Swarmer cell differentiation was not blocked in a glutamine transport mutant, but was inhibited by the addition of glutamine analog γ-glutamyl hydroxamate (71). Furthermore, glutamine acts as a chemoattractant for swarmer cells but not swimmer cells. In contrast, glycine, histidine, glutamate, alanine, aspartate, aspargine, tyrosine, and valine were chemoattractants solely for swimmer cells; only methionine and cysteine were chemoattractants for both cell types (71). A glutamine synthetase mutant (glnA PMI2882) is completely unable to swarm, even on rich media (120). Swarming is restored when exogenous L-glutamine, but not D-glutamine, is supplied.
Putrescine
Putrescine has been implicated as a trigger for differentiation in P. mirabilis (121). This molecule is continually produced and accumulates in the media during growth and is a component of the outer membrane in some Gram-negative bacteria, including P. mirabilis (122). In an effort to investigate cell-cell signaling by P. mirabilis, a lacZ fusion transposon screen was used to identify genes responsive to signals in spent culture supernatants (123). One mutant, with an insertion in speA (PMI2094, arginine decarboxylase), was repressed in the presence of spent wild-type supernatant (121). However, spent mutant supernatant did not repress speA. SpeA converts L-arginine to agmatine; the gene is next to speB (PMI2093), which converts agmatine to putrescine. Sturgill and Rather found that addition of putrescine (down to 25 μM), but not agmatine, repressed speA::lacZ expression. The speA mutant also displayed an aberrant swarm pattern comprised of very small, irregular swarm rings. An independent speB mutation had the same phenotype; taken together with the putrescine complementation of speA::lacZ repression, this suggested that the speA::lacZ phenotype was due to a polar effect on speB. Normal swarming was restored when extracellular putrescine was added (121), but not when added to a plaP (putrescine importer PMI0843) speA double mutant (124). Likewise, the speB mutants swarmed when inoculated onto agar in the vicinity of an undefined nonswarming P. mirabilis mutant that still produced extracellular signal (121). Swarming by a speB mutant can also be restored by the addition of ornithine, which can be converted to putrescine by the alternate SpeF pathway (120). Thus putrescine, which accumulates with increasing cell density, may be a signal that initiates swarming. Indeed, uptake of extracellular putrescine appears to be necessary for normal swarming, as the plaP putrescine importer mutant does not swarm as robustly as its wild-type parent (124). The effect could be mediated by putrescine forming a complex with LPS or capsular polysaccharide (125).
Arginine, histidine, malate, and ornithine
Previous approaches to defining the signals to initiate swarming involved adding substances to minimal chemically-defined media, or screening mutated bacteria for the loss of swarming ability. A new study was able to expand the list of factors that contribute to swarming by using a rich medium (LB) with low (10mM) NaCl (120). On this medium, P. mirabilis HI4320 does not swarm when incubated at 37 °C. Under these conditions, addition of 20 mM L-glutamine, L-arginine, DL-histidine, malate, or DL-ornithine promoted swarming. Fumarate and agmatine promoted swarming to a lesser extent. None of these substances enhanced swimming motility, nor did they cause aberrant cell elongation in broth culture. A panel of clinical isolates responded to these swarming cues, although there was some variation in the response to each specific substance. Two of the stronger cues (ornithine and arginine) and the weaker cue agmatine are part of the putrescine biosynthetic pathway. However, as putrescine itself was not sufficient as a swarming cue on low salt LB, and ornithine and arginine stimulated swarming in different ways and induced unique responses when pH or media were changed, these substances most likely stimulate swarming through distinct pathways (120).
Specific mutants were used to further assess the roles of the five swarming cues (L-glutamine, L-arginine, DL-histidine, malate, or DL-ornithine) (120). All five of these cues are present in human urine. Wild-type P. mirabilis cannot swarm well on urine solidified with agar; this is likely due to increased pH and crystal formation, both due to urease activity. However, a ureC urease mutant swarms on urine agar, and swarming is further promoted by all five swarming cues. Mutation of glutamine (glnA) or histidine (hisG) biosynthetic pathways led to abolished or reduced swarming, respectively; swarming was restored with exogenous supply of the corresponding amino acid. Conversely, mutation of gdhA (glutamate dehydrogenase, PMI3008) had no effect on swarming behavior (120). This finding is intriguing when compared with transcription during UTI, where gdhA is induced and glnA repressed (this is discussed later) (25).
Quorum sensing
There is no strong evidence that P. mirabilis engages in quorum sensing, despite the coordinated behavior during swarming and the regularity with which P. mirabilis is found as part of multi-species communities. This species produces cyclic dipeptides (cyclo(ΔAla-l-Val) and cyclo(l-Pro-l-Tyr)) that were initially thought to be agonists of the Vibrio fischeri acylhomoserine lactone (AHL)-dependent LuxR quorum-sensing system (126). However, more recent work indicates that these cyclic dipeptides do not affect quorum sensing (127). AHL autoinducers regulate swarming via a LuxI/LuxR-type system in Serratia marcescens (128). Although the P. mirabilis genome sequence did not reveal any putative LuxI or LuxM-type synthases (17), various exogenously-supplied AHLs affect swarming and proteolytic activity (129). P. mirabilis does encode luxS and produces the LuxS-dependent quorum sensing molecule AI-2 during swarming; however, a luxS mutant has neither a swarming nor a virulence defect (130). Despite these observations, the highly ordered P. mirabilis swarm cycle suggests a mechanism for multicellular coordination exists (131).
TRANSCRIPTION AND METABOLISM DURING SWARMING
Despite the vigorous motility displayed during swarming, differentiated P. mirabilis bacteria are less metabolically active than consolidating cells (132). When 27 random DNA probes were hybridized with RNA isolated from different stages of the swarm process, most of the sequences were less transcribed during swarming (66). This finding was later confirmed by microarray analysis of the transcriptome of swarming P. mirabilis, in which there was a general repression of transcription of swarm cells compared to consolidate (541 genes repressed during swarming; 9 induced relative to consolidated cells) (65). Flagellin (flaA) is among the most highly-expressed genes in both swarm and consolidate despite the lack of motion observed in consolidating cells. Consolidating cells are distinct from broth-cultured cells used to inoculate a swarm plate; expression by swarm and consolidate share more in common with each other than with broth-cultured bacteria. During consolidation, bacteria are metabolically active with increased expression of genes involved with central metabolism, respiration, nutrient uptake, and cell wall remodeling. The alternative sigma factor rpoS, associated with stationary phase and stress response, is among the genes induced during consolidation (65). Once initiated, protein synthesis is not required to maintain the swarm; swarming continues even in the presence of chloramphenicol (65, 133). Genes co-regulated with swarming are not necessarily required or involved with the swarming cycle, including virulence genes hpmA and zapA (these virulence genes will be discussed in another section). Investigation of genes regulated during swarming has led to the identification of virulence processes. Mutation of genes involved in peptide uptake (oppB PMI1474 and dppA PMI2847) and amino acid synthesis (cysJ PMI2250) that were regulated during swarming led to minor attenuation of swarming. However, the dppA and cysJ mutants were less fit in animal co-challenge. A similar result was found for the transcriptional regulator hexA (PMI1764) (65).
Swarming occurs under both aerobic and anaerobic conditions (133). STM studies for genes involved in virulence also led to the identification of central metabolism genes (aceE and sdhC) that cause aberrant swarming when mutated, suggesting that a complete (aerobic) TCA cycle contributes to swarming (57). To further investigate how central metabolism affects swarming, selected metabolic genes were mutated and were found to affect swarming in four distinct patterns (134). Two of these classes were characterized by altered distances between rings of swarming and consolidation; TCA cycle mutations (fumC PMI1296 and sdhB PMI0568) had decreased distances, and pentose phosphate pathway mutants (gnd PMI0655 and talB PMI0006) had increased distances. Mutations in glycolysis (pfkA PMI3203 and tpiA PMI3205) resulted in reduced swarming diameter. These mutations were rescued by complementation with the corresponding wild-type alleles or by addition of the missing biochemical intermediate to the growth medium. Specifically, the fumC mutant was rescued by addition of succinate or malate but not fumarate; this distinction indicates fumC is acting as part of the oxidative TCA cycle during swarming and not the reduced branched TCA cycle. Mutations in gluconeogenesis (pckA PMI3015) or the Entner-Doudoroff pathway (edd PMI2760) had no effect on swarming patterns, nor did a mutation in the fumarate reductase subunit gene frdA (PMI3588) which is involved in anaerobic respiration using the branched TCA pathway (134).
Swarming also occurs in the presence of the aerobic respiration poison sodium azide supplied at growth inhibitory concentrations (133, 134). The frdA (branched TCA) mutant also swarms on azide, but the fumC mutant, used in both aerobic and anaerobic TCA cycles, is unable to swarm (134). Thus, an alternative anaerobic electron acceptor has been proposed to provide energy during swarming. A transposon screen for mutants that are able to swarm on LB swarm agar but not in the presence of azide yielded 18 mutants. Not all of the mutated genes were expected to be involved in swarming per se, as mutations leading to increased permeability or susceptibility to azide would also be found. Two genes of particular interest that were identified are hybB (PMI0033 hydrogenase-2), which encodes an anaerobic cytochrome, and PMI2646, which encodes a putative quinine hydroxylase. To further address whether fermentation occurs during swarming, bacteria were inoculated onto swarm agar with the pH indicator phenol red. During aerobic conditions, P. mirabilis produces alkaline conditions; fermentation would have resulted in secretion of acidic byproducts. Swarming under anaerobic conditions, however, results in acidity. Taken together, the authors concluded that anaerobic respiration with a complete oxidative TCA cycle generates the proton motive force required for flagellar rotation (134).
CELLULAR INVASION
P. mirabilis uses its flagella to invade cultured cells derived from the urinary tract, including Vero (green monkey kidney parenchyma) (135, 136), EJ/28 and 5637 (transformed human transitional cell carcinoma of the urinary bladder) (66, 114, 135), NTUB1 (human urothelium) (105), and primary human renal proximal tubular epithelial cells (HRPTEC) (52, 137). In this experimental system, P. mirabilis is highly invasive, in numbers comparable to Salmonella Typhimurium (52) or Salmonella typhi (138). Allison and colleagues found the greatest invasive capability for P. mirabilis coincided with the swarmer cell phase (135). Likewise, a hyperswarming rsbA mutant is more invasive than its wild-type parent (105). Mobley et al found that invasion was greatly impaired when flagella of swarmer cells were immobilized by the addition of antiserum (52). Furthermore, an isogenic flagella mutant (hpmA flaD) was less than 1% as invasive as the parent (hpmA). Centrifugation of the nonmotile bacteria onto the cultured cell monolayer partially restored invasiveness to about 10% of the parental level. Invasion is aided by non-flagellar components including the HpmA hemolysin, autotransporter protein AipA (139) and LPS modification protein PmrI (109). Using a variety of intestinal and urinary epithelial cell lines, Oelschlaeger and Tall were able to inhibit P. mirabilis invasion by blocking bacterial protein, RNA, or DNA synthesis, but not by blocking host cell (eukaryotic) protein synthesis (138). P. mirabilis colocalizes with mucins MUC2 and MUC5AC, and mucin expression has been found to correlate with invasion (140). Cellular invasion could be involved in the transit of P. mirabilis from the kidneys to the bloodstream. Alternately, P. mirabilis could have an intracellular population during bladder or kidney infections. Of course, cultured cells differ in fundamental ways from intact tissues (e.g. types and polarization of proteins, tight junctions). At this time, invasion of urinary cells by P. mirabilis during ascending UTI has not been investigated in much detail.
ROLE OF SWARMING IN VIRULENCE
Virulence factors, including urease, ZapA protease, and hemolysin are induced during swarming compared to broth culture or older bacteria in the interior of a swarm colony (65, 66, 141). Production of both ZapA and HpmA is increased during overexpression of the flagellar regulator umoB, and hpmA transcription is responsive to Lrp (141). The swarming ability of P. mirabilis is especially relevant in catheterized patients, as this organism is able to swarm across catheters made of silicone or latex (61, 142) (Fig. 5). Since expression of several virulence genes is increased during swarming, it is possible that P. mirabilis swarming up catheters is primed to infect the urinary tract. However, the role of swarming during UTI is debated. In one study, outbred mice intravenously infected with P. mirabilis resulted in extensive kidney infection, and long-form, swarmer bacteria were observed in the kidney parenchymal tissue 15 days post-infection (114). Transposon mutants that were motile but nonswarming caused lower rates of lethality and kidney abscesses in this model. Several of these mutants were also less virulent compared to wild type in an ascending UTI model using suckling mice (114). In contrast, swarmer cells were very rarely found in the urinary tracts of CBA/J mice infected via bladder catheterization with GFP-expressing P. mirabilis when examined two or four days post-infection (143). In that study, 7 of 5087 (0.14%) bacteria counted in the bladders and kidneys had an elongated swarm form (> 10 μm); no swarmer cells were observed in the ureters. Combined with the previously described reports in which nonmotile P. mirabilis was fully virulent, this suggests that swarming may not be an important contributor to UTI virulence. However, swarming might only be apparent when a catheter is in place, or during possible invasion of renal cells at later stages of disease progression.
Figure 5.
P. mirabilis swarms across sections of latex catheter. Reproduced with permission from Infection and Immunity, vol 72, page 3942 (61).
It is noteworthy that the putrescine biosynthetic pathway also results in the formation of urea. P. mirabilis encodes genes that may catalyze the ultimate generation of putrescine and urea from ammonia and ATP (17). Because putrescine biosynthesis is required to initiate swarming, and excess urea could potentially drive the putrescine biosynthetic pathway in the opposite direction, it is possible that the abundant urea in the urinary tract represses swarming behavior.
Dienes lines and T6SS
P. mirabilis strains are self-recognizing during swarming; that is, any given pair of isolates, when inoculated on opposite ends of a swarm agar plate, will likely swarm up to but not into each other. When this occurs, a thin clear line of demarcation remains between the two strains, called a Dienes line (144). In contrast, swarms formed by identical strains will merge. Dienes line formation has been used as a method for typing clinical strains of P. mirabilis (145-147). When opposing swarms from two different strains meet, within one to two hours one of the strains will form large, rounded cells (148, 149). Over a period of hours, the rounded cells die and lyse, while the other strain dedifferentiates into vegetative cells. This rounding is not observed when a swarming strain meets nonswarming cells. Dienes line formation is dependent upon cell-cell contact or at least very close cell proximity (<60 μm) between opposing strains (148). When a membrane permeable to most macromolecules but not bacteria is placed between swarming strains, no Dienes line or cell rounding occurs.
The Dienes phenomenon is likely a mechanism of territoriality. When two strains (one marked with red fluorescent protein and one marked with green fluorescent protein) are mixed and placed on swarm agar, one strain dominates the resulting swarm colony, while the other strain forms rounded cells when detected at all. However, in broth culture, neither strain is dominant, and when other strains were tested for biofilm formation, dominance of a given strain did not correlate with Dienes dominance (148).
Recent studies suggest that type VI-mediated secretion of toxic effector proteins is the main mechanism of Dienes line formation (149, 150). Type VI secretion systems (T6SS) comprise a method of protein export that generally requires cell-cell contact, whether between two bacteria or a bacterium and a eukaryotic host cell. They are widespread in Gram-negative bacteria and may be involved in virulence, commensalism, or bacterial competition (reviewed in (151-153)). A core set of structural genes are essential for function and are thought to be involved in the production or function of the secretion apparatus. This includes the hcp/vgrG genes, which respectively encode proteins that form the tube through which export occurs and a needle-like structure used to penetrate the outer membrane of the target cell.
Type VI secretion has been studied in two P. mirabilis isolates, BB2000 and HI4320. Both strains encode the core structural genes essential for T6SS function, as well as multiple putative effector operons associated with the hcp/vgrG genes (17, 154-156). In T6SS characterized in other bacterial species, the multiple hcp/vgrG homologs are thought to act as adapters between the structural components and the effectors, allowing different effector proteins to be secreted (153). In both BB2000 and HI4320, the T6SS is essential for Dienes line formation and identification of self. However, the number of effector operons present and the significance of individual operons in self-recognition varies between the two strains (155, 156).
In BB2000, a transposon screen for mutants that form Dienes lines when in competition with the parent strain (that is, the strains no longer recognize the parent strain as “self”) led to the ids operon (identification of self, idsABCDEF PMI2990-95) (149). The first two genes of this locus, idsA and idsB, have homology to hcp and vgrG, respectively. The idsD and idsE genes display the most variation between P. mirabilis strains, and are believed to be the determinants of self-recognition (149). Expression of the ids operon increases during late logarithmic and early stationary phase. During swarming, expression is highest in the center of the swarm colony where the highest cell density is observed, but expression also occurs within a subset of cells at the leading edge of approaching swarm fronts (150).
A second transposon mutagenesis screen identified two “no-merge” mutants which form Dienes lines with both the wild-type and the idsA-F mutant (155). These mutants mapped to a previously uncharacterized five gene operon, named idr for identity recognition. As in the ids locus, the first two genes in the idr locus (idrA and idrB) have homology to hcp and vgrG, while the remaining three genes encode proteins of unknown function. IdsA, IdsB, IdsD, IdrA, and IdrB were detected using mass spectrometry in culture supernatants of wild-type BB2000 but not in a T6SS mutant, providing further evidence that these proteins are exported from the cell by the T6SS. It is apparent that competition and killing via T6SS effectors is a complex phenomenon in P. mirabilis. For example, wild-type BB2000 can dominate HI4320 in a swarm competition. However, BB2000 Δids mostly dominates HI4320 while HI4320 can dominate BB2000 idrB::tn5, suggesting that the idr operon is more important than the ids operon in BB2000 killing of HI4320 (155).
Unlike in BB2000, mutations in idsB and idsD in HI4320 do not result in a loss of self-recognition (156). However a transposon screen for HI4320 mutants which no longer recognize the wild type as self, and thus form a Dienes line with HI4320, identified hypothetical protein PMI0756 as a putative component of the T6SS. A secondary screen to identify additional mutants that no longer form a Dienes line with the PMI0756 mutant identified two groups of mutants. The first group of mutants clustered in the major T6SS structural operon (PMI0733-0749), while the second group of mutants clustered in the same operon as PMI0756 (PMI0750-0758). The first two genes in this operon are hcp and vgrG homologs, respectively; the other genes in the operon were named pef for primary effector operon. Additional targeted mutagenesis revealed that a mutation in any gene in the pef operon results in loss of self-recognition. Complementation of the pefE mutant with pefE is sufficient to restore immunity from wild-type killing, but is not sufficient to restore killing of non-immune strains. To restore both immunity to wild type and killing of the pefE mutant, complementation with pefEFG is needed (156). It is notable that although the pef and idr operons are both adjacent to the structural T6SS apparatus operon in HI4320 and BB2000, respectively, the organization and sequences of these operons are completely different (aside from the hcp and vgrG homologs at the front of both operons).
Analysis of the HI4320 genome revealed three additional putative hcp-vgrG effector operons in addition to the ids and pef operons. The proteins encoded by the hcp genes are highly similar (with the exception of PMI1332 which appears to be truncated), while the genes predicted to encode VgrG homologs are similar at the N-terminus with decreasing similarity at the C-terminus. All five of the promoters for the hcp-vgrG operons are capable of driving luciferase expression during swarming (156), though they result in different patterns of expression.
Although significant gains have been made in the understanding of Dienes line formation and T6SS competition in P. mirabilis, a number of questions remain. The function of most of the genes in the effector operons remains unclear, and it is not understood how cell death and immunity are mediated. Nor is it clear why P. mirabilis possesses several effector operons, or how they are regulated. Perhaps most importantly, it is unknown when Type VI secretion-mediated killing occurs when P. mirabilis is in its natural environments or if this phenomenon has clinical relevance.
Fimbriae
Although the fimbriae of P. mirabilis are essential virulence factors in urinary tract infections (Table 2), much remains unknown about their expression, physical characteristics, and biological functions. The first P. mirabilis fimbriae were identified by their ability to agglutinate red blood cells and bind uroepithelial cells. Using these methods, the mannose-resistant Proteus-like (MR/P), mannose-resistant Klebsilla-like (MR/K), and urothelial cell adhesin (UCA) fimbriae were described (157, 158). However, as P. mirabilis can simultaneously express multiple types of fimbriae, identification of genes encoding specific fimbriae is often complicated (157). For example, the attempts to identify genes encoding UCA fimbriae led to the discovery of the P. mirabilis fimbriae (PMF) (159), and the genes associated with MR/K hemagglutination remain unidentified.
Table 2.
The fimbriae of P. mirabilis.
| Fimbria | Genes | Classa | Implicated in virulence? | MrpJ homolog |
|---|---|---|---|---|
| MR/P' | PMI0254-PMI0261 | π | ND | PMI0261 |
| MR/P | PMI0262-PMI0271 | π | yesIC,CO | PMI0271 |
| Fimbria 3 | PMI0296-PMI0304 | π | ND | PMI0296 |
| UCA | PMI0532-PMI0536 | γ 1 | yes | PMI0532 |
| Fimbria 5 | PMI1060-PMI1067 | π | ND | PMI1060 |
| Fimbria 6 | PMI1185-PMI1190 | γ 1 | ND | None |
| Fimbria 7 | PMI1193-PMI1197 | γ 1 | ND | None |
| Fimbria 8 | PMI1464-PMI1470 | γ 1 | yes | PMI1470 |
| PMF | PMI1877-PMI1881 | π | yesIC,CO | None |
| Fimbria 10 | PMI2207-PMI2214 | γ 1 | ND | PMI2209, PMI2207 |
| PMP | PMI2216-PMI2224 | π | yes | PMI2224 |
| Fimbria 12 | PMI2533-PMI2539 | γ 2 | ND | None |
| ATF | PMI2728-PMI2733 | γ 1 | noIC,CO | PMI2733 |
| Fimbria 14 | PMI2997-PMI3003 | ND | yesSTM | PMI3003 |
| Fimbria 15 | PMI3086-PMI3093 | π | ND | None |
| Fimbria 16 | PMI3348-PMI3352 | γ 1 | ND | None |
| Fimbria 17 | PMI3435-PMI3440 | γ 1 | ND | None |
The name, genomic location, Greek classification, determination of virulence, and MrpJ homolog of each fimbrial operon in P. mirabilis.
Greek classification was determined as in (161, 185). Fimbria 14 does not have an identified chaperone, and thus cannot be classified in the Greek system. ND, not determined. IC, the mutant was tested in an independent challenge experiment. CO, the mutant was tested in a co-challenge experiment. STM, the mutant was tested in a signature-tagged mutagenesis experiment.
Prior to sequencing the P. mirabilis genome in 2008, five P. mirabilis fimbriae had been discovered. Sequencing revealed an additional 12 chaperone-usher fimbriae encoded in the P. mirabilis genome (17). In comparison, UPEC typically encode nine to 12 fimbrial operons (160). As the name suggests, chaperone-usher fimbriae are defined by the presence of a chaperone and an usher in the fimbrial operon. The chaperone protects the fimbrial subunits during transport from the cytoplasm to the cell surface which allows the fimbrial subunits to fold properly in the periplasm, and the usher, an integral membrane protein, releases the fimbrial subunit from the chaperone and guides it through assembly (161). Ten of the P. mirabilis chaperone-usher operons encode a homolog of MrpJ, a regulatory protein at the beginning or end of the operon, which allows the coordination of adhesion and motility, and may allow fimbrial regulation of other fimbriae (162). The following sections describe the fimbriae of P. mirabilis in order of their discovery, as well as the role of MrpJ homologs in coordinating regulation of fimbriae and flagella.
MANNOSE-RESISTANT PROTEUS-LIKE (MR/P)
Genetic organization
MR/P is the best characterized fimbria in P. mirabilis. The mrp operon (mrpABCDEFGHJ) encodes the major structural subunit (mrpA), four minor subunits (mrpBEFG), a chaperone (mrpD), an usher (mrpC), a tip adhesin (mrpH), and a transcriptional regulator (mrpJ) (163, 164).
The operon is preceded by a σ70 promoter inside an invertible element whose orientation is controlled by MrpI, a recombinase transcribed divergently from the structural operon (164, 165). When the σ70 promoter faces the structural operon, MR/P is “ON”, while when the σ70 promoter faces MrpI, MR/P is “OFF” (165). Transcription of mrpI is unaffected by the orientation of the invertable element, and mrpI is likely transcribed by a promoter outside of the invertable element (164). The MrpI C-terminal domain is homologous to the catalytic domains of the E. coli fimbrial recombinases FimB and FimE, while the N-terminal domain binds DNA (165). Like FimB, MrpI switches the invertible element both from ON to OFF and OFF to ON (165). However, when mrpI transcription decreases, mrpA transcription increases, suggesting MrpI favors switching from ON to OFF (166). In the absence of MrpI, the promoter orientation is fixed, suggesting that MrpI is the sole recombinase for the mrp promoter (165). To facilitate analysis of the role of MR/P fimbriae, two MR/P mutants were engineered. Both of these mutants have an insertion that inactivates mrpI; in the “MR/P L-ON” mutant, the mrp promoter is in the on position, while in the “MR/P L-OFF” mutant, the mrp promoter is in the off position.
Expression
Like many fimbriae, in vitro MR/P expression is encouraged through static culture. The percentage of bacteria with ON or OFF mrp promoters can be determined by a PCR assay, where the percentage of ON mrp promoters correlates to the percentage of cells expressing MR/P (Fig. 6) (165, 166). When a lower oxygen level is maintained in LB under shaking conditions, for example by culturing P. mirabilis in 5% O2, the mrp promoter is up to 70% ON compared to 2% ON in aerated culture. The increased expression of MR/P under static conditions is due to decreased availability of oxygen gas, not increased levels of CO2 (166).
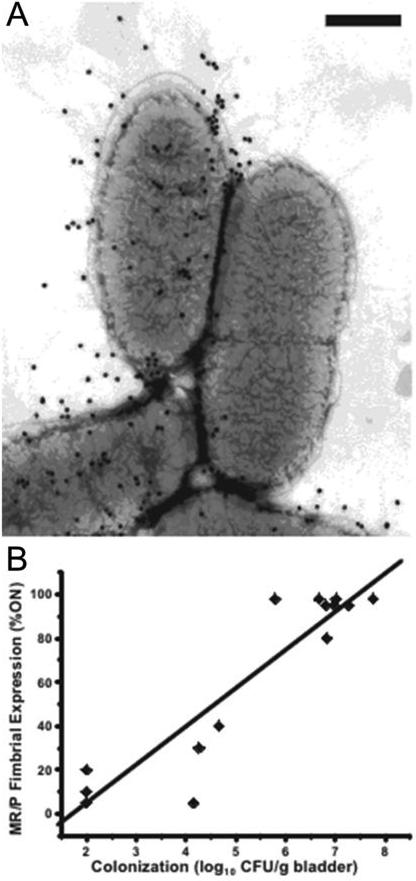
Figure 6.
Expression of MR/P fimbriae is phase-variable and induced during UTI. A, Immunogold electron microscopy of wild-type P. mirabilis HI4320 labeled with gold particles targeting the MrpH tip adhesin. The cell on the left is expressing MR/P fimbriae, and the cell on the right is not. Bar, 500 nm. B, The amount of MR/P fimbriae present positively correlates with murine bladder colonization. Data were obtained seven days post-inoculation. Modified with permission from Journal of Bacteriology vol 191, page 1385 (166).
Oxygen-limiting conditions enrich for expression of MR/P through two mechanisms. First, the expression of MR/P lends a competitive advantage during growth in low oxygen conditions. Although their growth during independent culture is similar, a P. mirabilis L-ON mutant will out-compete a P. mirabilis L-OFF mutant in liquid culture (166, 167). It is hypothesized that this is due to fimbrial expression and flagellar repression driving electron transport and maintaining the proton gradient (166). Secondly, oxygen limitation increases the expression of MR/P. Under limited oxygen conditions, almost twice as much mrpA is transcribed as under atmospheric oxygen, possibly because less MrpI is expressed, leading to less switching of the mrpA promoter from ON to OFF (166).
Assembly
The role of each subunit in the assembly and structure of the MR/P fimbria is partially understood. MrpA has been identified as the major structural subunit by its strong expression and location in the mrp operon (164, 168).
MrpB is homologous to E. coli PapH, which terminates fimbrial growth while anchoring fimbriae to the cell wall and modulating fimbria length (169, 170). P. mirabilis ΔmrpB displays fewer but longer fimbriae on the surface of the cell. In P. mirabilis ΔmrpB, 5% of cells were fimbriated after three 48 hour passages in static culture, while 48% of wild-type cells were fimbriated. MR/P fimbriae from an mrpB mutant are 62-fold longer than wild-type fimbriae (18 μm compared to 0.29 μm) (171). These data suggest MrpB is needed to either initiate and/or terminate assembly of MR/P (171). The mrpEFG genes are predicted to encode minor structural subunits. When MrpG is found in P. mirabilis fimbriae, it is located on short fimbriae, possibly newly synthesized, and where fimbriae meet between aggregating bacteria (172).
MrpH is a two-domain adhesin, where the predicted MrpH C-terminus is similar to other mrp structural subunits and the larger N-terminal domain likely mediates receptor binding (Fig. 6). This two-domain structure and the presence of a proline at the last residue identify MrpH as an adhesin (163, 173). Less than 1% of mrpH mutant cells express MR/P fimbriae compared to 50% of the wild-type, suggesting that MrpH is essential for stable production of MR/P. Oddly, the mrpH mutant still mediates weak hemagglutination, possibly due to the expression of a different fimbria in the absence of mrpH (163). The N-terminus of MrpH has four conserved cysteine residues, which in other tip adhesins form disulfide bonds that are essential for the proper folding of the adhesin (174, 175). When the MrpH cysteine residues are mutated to serine, the MR/P fimbrial structure is produced and MrpA is found in fimbrial preparations, but there is less pellicle formation and hemagglutination, suggesting that the tip adhesin is inactivated (163). In P. mirabilis and other organisms, periplasmic disulfide bond formation is mediated by DsbA (PMI2828) (176). A dsbA mutant is deficient in MR/P production and in establishing infection in a mouse model of UTI, which is consistent with disulfide bond formation being an essential component of the MR/P structure (56, 166).
Biofilm formation
MR/P contributes to biofilm formation, presumably by mediating attachment to surfaces and autoaggregation (177, 178). P. mirabilis L-ON forms significantly more biofilm during culture in urine than a P. mirabilis L-OFF mutant or the wild type, which form similar levels of biofilms (177). Scanning electron microscopy showed that after two days, wild-type bacteria had adhered as small colonies to a cover slip, while P. mirabilis L-ON formed a full three-dimensional biofilm. By seven days however, the wild-type biofilm had become much thicker than the L-ON or the L-OFF biofilm, suggesting that the ability to turn MR/P on and off may be essential for robust biofilm maintenance (Fig. 7) (177).
Figure 7.
P. mirabilis biofilm formation is MR/P-dependent. P. mirabilis bacteria expressing GFP were grown on a cover glass in urine for 7 days. The resulting biofilm was imaged with confocal microscopy, and the 30 resulting z-stacks were stitched together to form the sagittal view. Wild-type P. mirabilis forms thick, robust biofilms. P. mirabilis MR/P L-ON forms dense, but thin, biofilms while P. mirabilis MR/P L-OFF forms weak biofilms. Reprinted with permission from Infection and Immunity vol 72, page 7299 (177).
Role in infection
MR/P fimbriae are expressed in the urinary tract and contribute to virulence. After transurethral infection with wild-type P. mirabilis, mice with high IgG antibody responses produced serum antibodies to MR/P (179). During infection, the mrp operon genes (including mrpJ), are upregulated 500 to 1300-fold compared to culture in LB, and are the nine most induced genes in vivo (25). The greatest upregulation in vivo occurs early in infection (1-3 days) and then falls later during infection (7 days), although these genes remain induced compared to in vitro culture (Fig 2) (25). Greater than 90% of the mrp promoters are in the ON orientation in the bladder and urine during murine infection (165, 167). Direct observation of P. mirabilis in the bladder, urine, and kidneys of mice revealed MR/P fimbriation in all parts of the urinary tract (177). However, in the kidneys up to 85% of bacteria do not express MR/P (167). Phase variation of the mrp promoter orientation may contribute to evasion of host defenses.
Multiple mutations in the mrp operon have been tested in mouse models of infection. All mutants that do not express MR/P are defective in infection. However, different mutations result in different colonization defects. The P. mirabilis mrpG mutant is inefficient at colonizing the mouse urinary tract in an independent challenge. Fewer mrpG mutant bacteria than wild-type colonize the urine, bladder (> 10,000-fold), and kidney (>250 fold) (172). This large of a defect has not been observed for any other mrp mutant.
An mrpA mutant compared to wild-type P. mirabilis showed decreased colonization of the bladder (28 fold), kidneys (18 fold), and urine (6 fold), as well as reduced pathology in the kidneys of mice in an independent challenge (180). In another study, independent challenge with wild-type P. mirabilis and an mrpH mutant revealed no difference in colonization of the mouse urinary tract. However, in a co-challenge infection, the mrpH mutant was outcompeted by the wild type in the bladder, kidneys, and urine (181).
Similarly, independent challenges between P. mirabilis L-ON, L-OFF, and wild-type strains revealed no differences in the ability of each strain to colonize the urinary tract (181). After a 24-hour co-challenge experiment, there was no difference in the ability of a wild-type, L-ON, or L-OFF P. mirabilis strain to colonize the bladder (177). However, there was a significant difference in the localization of adherent bacteria. The wild-type bacteria largely colonized the surface of uroepithelial umbrella cells and were occasionally attached to exfoliated cells. When examined by electron microscopy, the wild-type bacteria had a fuzzy appearance, indicating fimbriation (177). A P. mirabilis L-ON mutant was similarly able to colonize the intact bladder epithelium. However, the L-ON mutant had a distinct morphology from the wild-type. They were elongated (6-10 μm) and heavily fimbriated (177). The L-OFF mutant colonized a distinct location in the bladder. Instead of colonizing the intact epithelium, the P. mirabilis L-OFF mutant colonized exfoliated cells and areas of the bladder where the uroepithelium was disrupted (177). Exfoliation of umbrella cells exposed the lamina propia, and L-OFF bacteria were able to colonize the underlying basement membrane.
After a seven day co-challenge experiment, wild-type P. mirabilis out-competed the L-OFF mutant in the bladder, kidneys, and urine. However, in a competition between the L-ON mutant and wild-type P. mirabilis, the L-ON mutant dominated the wild type in the bladder, but was similar to the wild type in the urine and the kidneys (167). These data support that MR/P fimbriae are essential for infection of the bladder and contribute to colonization of the kidneys.
MANNOSE-RESISTANT KLEBSIELLA-LIKE (MR/K)
Early characterization of the hemagglutination patterns of P. mirabilis revealed a MR/K hemagglutinin. MR/K hemagglutination is characterized by the ability of bacteria to agglutinate tannic acid treated red blood cells but not un-treated red blood cells (157). Expression of fimbriae 4-5 nm in diameter was found in approximately 60% of strains that showed MR/K hemagglutination patterns, suggesting that there may be more than one fimbria in P. mirabilis that is capable of mediating MR/K hemagglutination (157). The genes that encode the MR/K fimbria remain unidentified.
UROEPITHELIAL CELL ADHESIN (UCA)
Characterization
UCA, also known as the non-agglutinating fimbria (NAF) was identified by its ability to bind the surface of uroepithelial cells. P. mirabilis strains that express UCA bind shed uroepithelial cells from human urine (158). When the uca operon from an adherent P. mirabilis strain was expressed in E. coli K-12, which is not natively adherent to uroepithelial cells, the E. coli became adherent (182). A purified protein preparation of UcaA, the major subunit, also bound uroepithelial cells (158). As the major subunit does not typically mediate adhesion, it is possible that the preparation contained the predicted tip adhesin. Electron microscopy revealed fimbriae 4-6 nM in diameter and 0.09 μm to 0.83 μm in length (158, 183). However, a recent study found that a P. mirabilis ΔucaA mutant is impaired in colonization of the kidneys, but colonizes the bladder as well as wild-type P. mirabilis, suggesting that the UCA fimbriae may be important for kidney rather than bladder colonization (184). The major structural subunit of UCA fimbriae, UcaA, is variable (185), and it is possible that variant UCA fimbriae mediate adherence to different targets. Despite the evidence for a role for UCA in infection, the UCA fimbriae are not differentially regulated in vivo compared to culture in LB (25), and there is considerable variation in the amino acid sequence of UcaA in the seven currently sequenced P. mirabilis genomes (185). The structural proteins in the uca operon are homologous to the F17 family of fimbriae in uropathogenic E. coli (182). F17 fimbriae in enteropathogenic E. coli attach to intestinal epithelium via a terminal N-acetyl-D-glucosamine (186, 187). P. mirabilis is commonly found in the gut, but whether UCA fimbriae mediate the same role as F17 fimbriae in the gut is unknown (188).
Binding
UCA is the only P. mirabilis fimbria with a characterized binding target. Purified UcaA, presumably with the tip adhesin co-purified, binds asialo-GM1, asialo-GM2, and lactosyl-ceramide in in vitro biochemical assays (189). Asialo-GM1 is a Galβ1-3GalNAcβ1-4Galβ1-4Glcβ1-1'ceramide glycolipid commonly found on neuronal and a variety of immune cells. It has been found on the surface of polymorphonuclear leukocytes (PMN) (190), which respond during UTI, but at this time there are no reports of P. mirabilis binding PMN. Asialo-GM1 has also been identified as a potential receptor for Pseudomonas aeruginosa fimbriae in the lung epithelium (191). P. mirabilis expressing UCA can be partially prevented in vitro from binding cultured uroepithelial cells either by use of an anti-UCA antibody (75% blocked) or an anti-asialo-GM1 antibody (50% blocked) (189). The presence of asialo-GM1 on the surface of the bladder has not been shown, and it remains unclear if asialo-GM1 is a valid target of UCA in the urinary tract (189).
PROTEUS MIRABILIS FIMBRIA (PMF)
PMF was discovered due to cross-reactivity with both a ucaA degenerate DNA probe and a UcaA antibody (159). PMF is essential for full infection in a urethral challenge murine model. Infection with wild-type P. mirabilis leads to the production of serum antibodies against PMF (179). A P. mirabilis ΔpmfA mutant colonized the bladder less efficiently than the wild-type parent strain in both independent (83-fold) and co-challenge experiments (700-fold) (192, 193). The pmfA mutant also colonized the kidneys less effectively than the wild type, but only in the more sensitive co-challenge experiment (2630-fold) (192). In a hematogenous challenge, which tests P. mirabilis’ ability to colonize the urinary tract from the bloodstream via the kidneys, fewer mice were colonized by the ΔpmfA mutant than the wild type. Specifically, the wild type colonized 94% of mice, while the ΔpmfA mutant colonized 45% of mice (192). In spite of the critical contribution of PMF toward infectious fitness in animal models, the pmf operon is downregulated in vivo compared to in vitro (25). In E. coli, flagella are down-regulated in vivo compared to in vitro, but are essential to infection. This apparent paradox was explained by an in vivo imaging experiment, which revealed that flagella are temporarly up-regulated in vivo to allow E. coli to ascent the ureters (51). PMF may have a similar short-lived but essential role in causing P. mirabilis urinary tract infections.
A double P. mirabilis ΔmrpA-D ΔpmfA mutant is attenuated at colonizing the urinary tract compared to either the single mrp or pmf mutants. In independent and co-challenge experiments, P. mirabilis ΔmrpA-D ΔpmfA colonized the kidneys less effectively than either a P. mirabilis ΔmrpA-D or P. mirabilis ΔpmfA single mutant. However, P. mirabilis ΔmrpA-D ΔpmfA was less effective at colonizing the bladder than P. mirabilis ΔmrpA-D, but colonized the bladder similarly to P. mirabilis ΔpmfA (194).
AMBIENT TEMPERATURE FIMBRIA (ATF)
When the fimbrial profile of a P. mirabilis mrpA mutant was examined, an additional fimbria was discovered. The fimbria was named the ambient temperature fimbria (ATF) because it is maximally expressed at 25° C (195). The atf operon was initially thought to consist of only a major subunit (atfA), a chaperone (atfB), and an usher (atfC); however, subsequent sequencing of the P. mirabilis genome identified a minor pilin and a tip adhesin encoded in the atf operon (17, 196). A mutant with the atfB gene and parts of the atfA and atfC genes deleted is capable of infecting mice at the same level as wild type, which suggests that ATF may be important for environmental survival, but not for infection of the urinary tract. (197). However, in the absence of MR/P, ATF is upregulated both in vivo and in vitro (167, 177). Bacteria without MR/P are still able to bind the uroepithelium in vivo, but it is unclear if ATF is the only fimbria expressed under the circumstances or if it contributes to adherence (177).
P. MIRABILIS P-LIKE FIMBRIA (PMP)
The P. mirabilis P-like fimbria (PMP) is a prime example of the difficulties encountered when P. mirabilis expresses multiple fimbriae at the same time. PMP was first named in 1995, for a fimbria expressed by P. mirabilis isolated from canine urine (198). However, the authors noted that the PmpA protein sequence had been previously identified in a 1991 paper which determined the N-terminal sequence of MrpA (49, 198). In 1993, the genes which encode MrpA were identified, but the nucleotide sequence encodes a different protein sequence than was initially identified (168). Thus, the first paper describing the isolation of MrpA was likely actually describing the isolation of PmpA.
Although the importance of PMP in human infection is unknown, genome sequencing has revealed the presence of pmpA in all the currently sequenced P. mirabilis genomes, and pmpA was detected in 57 of 58 clinical isolates using a PCR screen (185). Additionally, after infection with wild-type P. mirabilis, mice with the highest bacterial loads over a 6-week infection produced serum antibodies to PMP (49).
FIMBRIA 14
The presence of the fimbria 14 operon was first discovered upon sequencing of the P. mirabilis genome (17). Concurrent with the sequencing of the genome, a random transposon insertion in the fim14 operon was identified which is attenuated in the mouse model compared to the wild-type parent (57). During infection, the fim14 operon is upregulated compared to culture in LB, but it is not highly expressed (25). It is also induced when cultured in vitro at pH 8.0 compared to neutral pH, suggesting fim14 may be responsive to the alkaline pH that is a result of urease activity in urine (185). Oddly, the fim14 operon does not encode an obvious chaperone. Despite the apparent lack of an assembly system for fimbria 14, major structural subunit Fim14A protein has been detected in sheared surface protein preparations from strain HI4320 (185). It is possible that the structural components of Fimbria 14 are transported and assembled on the bacterial surface by a heterologous chaperone-usher pair.
CONSERVATION AND EXPRESSION OF THE 17 FIMBRIAE
Overall, the chaperone-usher fimbriae of P. mirabilis are highly conserved across isolates collected decades apart, from diverse geographical locations and different anatomical sites from either humans or mice. In a comparison between HI4320 and six additional sequenced genomes, 14 of the 17 fimbrial major subunits have ≥95% amino acid identity. UcaA, Fim6A, and Fim3A have greater diversity in amino acid identity. Only Fim3A, which is missing from two isolates, was not found in all sequenced isolates. In a PCR-based screen for the major fimbrial subunits of 58 clinical isolates, only fim3A was not detected in a majority of isolates (185). This high degree of conservation stands in contrast to the patterns detected in E. coli and Salmonella enterica, in which the majority of fimbrial genes are variable, and specific fimbrial operons are associated with isolation from a particular body site (199-201). The lifestyle of P. mirabilis outside of the urinary tract is poorly characterized; however, it is likely that the high degree of fimbrial conservation indicates that possessing a wide array of fimbriae is important for the survival of this species in the environment.
Transcript from all of the 17 major structural genes can be detected in logarithmic-phase aerated cultures. Fimbrial genes were generally induced during stationary-phase and static culture conditions. Mass spectrometry identified proteins from six different fimbriae (MR/P, PMF, UCA, ATF, Fimbria 8, and Fimbria 14) in sheared surface protein preparations of stationary-phase, aerated cultures (185). Again, this is in contrast to UPEC, where a single fimbria dominates under specific culture conditions (202).
MrpJ REGULATES THE TRANSITION BETWEEN SWIMMING AND SWARMING
To survive in both the urinary tract and the environment, P. mirabilis must coordinate adhesion to surfaces with swimming and swarming. Fimbriae are essential for adhesion while flagella are essential for swimming and swarming. P. mirabilis uses the product of a gene in the fimbrial operon to directly repress flagellar synthesis (162). Sequencing of the mrp operon revealed mrpJ, which encodes a helix-turn-helix xenobiotic response element (HTH-XRE) transcriptional regulator downstream of the mrpH gene (181). When mrpJ is expressed in vitro at a level similar to the amount during experimental UTI, swarming, swimming, and FlaA levels are repressed (162). MrpJ binds to the flhDC promoter and represses transcription of flhDC, which results in repression of the downstream target of FlhD4C2, flagellin (flaA) (162, 203). During experimental murine infection, mrpJ transcription is highly induced within one day post-infection and flagellar genes are repressed (Fig. 2) (25). At seven days post-infection, mrp gene expression falls (though it is still induced compared to broth culture) and flagellar gene expression rises. MrpJ is required for P. mirabilis virulence; during a co-challenge experiment, an mrpJ mutant was outcompeted in both the bladder (>100,000-fold) and kidneys (>10-fold) by the wild-type parent strain (203). It is unclear whether this reduction in virulence is due to changes in levels of FlaA and MrpA, the loss of coordination of motility and attachment, or loss of regulation of other virulence factors.
The P. mirabilis genome sequence revealed a surprising 14 additional MrpJ paralogs (17). Ten paralogs are located within fimbrial operons (Table 2), while four are orphans. Overexpression of 10 of the 14 MrpJ paralogs repressed swimming and swarming motility (162). Interestingly, one orphan paralog (PMI3508) repressed swimming but not swarming (162). MrpJ and its paralogs use multiple mechanisms and result in distinct phenotypes. For example, overexpression of mrpJ results in a radial swarm pattern with both swimming and swarming cells at the edge of the swarm front, while ucaJ overexpression completely represses FlaA and swarming, but does not affect differentiation into elongated swarmer cells. On the other hand, overexpressing pmpJ does not affect FlaA levels, but results in disorganized swarming, with both swimming and swarming cells at the edge of the swarm front (162) (Fig. 8). Overexpression of ucaJ results in the fimbrial structures on the surface of vegetative cells changing from short, thick fimbriae to long, thin, fimbriae, suggesting that MrpJ and its paralogs might also coordinate expression of fimbriae (162).
Figure 8.
Overexpression of mrpJ and its paralogs results in distinct phenotypes. A, Swarming assays of P. mirabilis with an empty vector or expressing mrpJ or an mrpJ paralog. B, Gram-stained bacteria from the edge of the swarm front. The reference bar is 50 μm. Modified with permission from Molecular Microbiology, vol 69, page 551 (162).
Alignment of the 15 MrpJ sequences revealed a 10-26 amino acid N-terminal region followed by a predicted HTH motif and a 10-30 amino acid C-terminus. The HTH has a core region of nine conserved amino acids (SQQQFSRYE) (162). Single mutation of 8 of the 9 residues to alanine results in increased swimming motility when mrpJ is overexpressed. Insertion of 15 nucleotide linkers into the HTH domain also resulted in increased swimming motility, suggesting that this region is essential for function. Deletion of the C-terminal 27 amino acids, 25% of the protein, had no effect on MrpJ function (162). To completely abrogate MrpJ function, the C-terminal 45 amino acids had to be deleted, suggesting the C-terminus may not be important for MrpJ regulation of flagella (162).
Interestingly, although there is no sequence homology, MrpJ can be functionally complemented by PapX, a transcriptional regulator from the E. coli pap fimbrial operon (203). In E. coli, PapX directly binds the flhDC promoter to repress flagellar gene expression when P fimbriae are expressed (204, 205). Repression of flagella by components of fimbrial operons is emerging as a common regulatory theme. Searches of the recently annotated urinary pathogen Providencia stuartii 25827 genome, available in GenBank, revealed that mrpJ paralogs are also frequently associated with fimbrial operons in that species. Likewise, thirteen mrpJ paralogs are encoded by Morganella morganii, although only two are associated with fimbrial operons (206). Thus far, mrpJ paralogs have been identified by BLAST homology searches (207) in at least 20 other bacterial species. MrpJ functional homologs have also been discovered in enterohemorrhagic E. coli, and sequence homologs have been reported in Xenorhabdus nematophila and Photorhabdus temperata (208-210).
Autotransporters
The P. mirabilis genome encodes six putative autotransporters, also known as type V secretion proteins (211). Unlike other bacterial secretion systems, autotransporters do not encode specific proteins for their transport to the cell surface, but rather use conserved bacterial systems. Autotransporters contain an N-terminal Sec-pathway-dependent signal sequence, an effector (α) domain, and a transmembrane (β) domain (211). Classical autotransporters encode a full β-barrel domain and can act as monomers, while trimeric autotransporters encode a shortened β-domain which requires a trimer to form a β-barrel (212). P. mirabilis encodes three classical autotransporters and three trimeric autotransporters (17).
CLASSICAL AUTOTRANSPORTERS
Proteus toxic agglutinin (PMI2341, Pta) is the only characterized classical autotransporter from P. mirabilis. As its name implies, Pta acts as both a bacterial autoagglutinin and as a toxin to host cells. The Pta α-domain is a serine protease which, despite remaining attached to the bacterial cell, can mediate cytotoxicity to host cells (213). Although the exact mechanism of Pta cytotoxicity is unknown, intoxication of host cells with Pta results in membrane damage, actin depolymerization, and eventual lysis (213). Pta expression is maximized during culture at 37°C and alkaline pH, conditions which occur during infection (213). During co-infection with wild-type P. mirabilis, a pta mutant is severely defective in colonizing the kidneys and spleen of mice, and has a minor defect in colonizing the bladder (213). The other two classical autotransporters (PMI0844 and PMI2126) are predicted to be proteases (17).
TRIMERIC AUTOTRANSPORTERS
P. mirabilis encodes three putative trimeric autotransporters, of which AipA (adhesion and invasion mediated by the Proteus autotransporter, PMI2122) and TaaP (trimeric autoagglutination autotransporter of Proteus, PMI2575) have been characterized. Both proteins mediate binding to collagen and laminin in vitro (139). AipA mediates binding to HEK293 (kidney), UMUC-3 (bladder), and Vero (monkey kidney) cells in vitro. Co-challenge experiments revealed that a P. mirabilis aipA mutant colonizes the kidneys and the spleen less than the wild type (139). TaaP does not bind any tested cell line, but mediates autoaggregation of P. mirabilis. A P. mirabilis taaP mutant is less successful at colonizing the bladder than the wild type in a co-challenge infection model (139). Also of note, TaaP is encoded within the horizontally-transmissible genetic element ICEPm1 (214).
Other adhesins
In addition to the chaperone-usher fimbriae, the P. mirabilis genome sequence revealed genes encoding one or two potential type IV pili. A putative type IV pilus on the surface of P. mirabilis was reported, although the size and expression pattern was consistent with the MR/P fimbria (215). However, P. mirabilis is capable of twitching motility, a flagellar-independent movement which is associated with type IV pili (216, 217). More work is needed to determine if type IV pili are expressed and functional in P. mirabilis. Additionally, P. mirabilis encodes two genes that are homologous to the attachment and invasion locus genes of Yersinia enterocolitica (17). These genes are not upregulated in the murine urinary tract and have not been further characterized (25).
Hemolysin
In addition to the autotransporter toxin Pta, P. mirabilis encodes a Serratia-type calcium-independent hemolysin which can lyse nucleated cells in addition to red blood cells (218, 219). The hemolysin is encoded by hpmA, while a second gene, hpmB, encodes a membrane transporter; both genes are highly conserved across P. mirabilis isolates (220). The levels of hemolysin in P. mirabilis correlate with its ability to invade cultured kidney cells, and an isogenic P. mirabilis ΔhpmA mutant is minimally invasive in cultured cells (136, 137). However, the contribution of HpmA to UTI remains unclear. In an independent challenge, P. mirabilis hpmA mutant bacteria colonize as well as the wild type, and hpmA is expressed at background levels in a murine infection model (25, 52, 219). In contrast, when wild-type or hpmA mutant P. mirabilis were tested in an IV injection mouse model, the P. mirabilis hpmA mutant had a 6-fold higher LD50 than wild-type P. mirabilis, suggesting that hemolysin might be important during septic infection (219). A truncated hemolysin crystal structure has been solved, and a CXXC disulfide bond motif contributes to activity and stability of the protein (221). Co-challenge experiments with P. mirabilis hpmA mutant and wild-type P. mirabilis coupled with examination of tissue pathology are needed to further clarify the role of hemolysin in P. mirabilis infection.
Proteases
ZapA
For P. mirabilis to survive in the urinary tract, it must overcome host defenses, including antibodies and antimicrobial peptides. To accomplish this, P. mirabilis encodes an array of proteases (17). ZapA (mirabilysin) is a metalloprotease capable of mediating the degradation of numerous host proteins in vitro (222, 223). It was originally thought that zapABCD (PMI0276-0279) formed a single operon, with zapBCD encoding an ABC transporter that could export ZapA (222). However, Northern blot analysis detected only a single zapA transcript corresponding to the size of the zapA gene, and a putative promoter and ribosome binding site have been identified upstream of zapBCD (224). Transposon mutations in zapBCD affect production of ZapA activity, suggesting that ZapBCD is involved in the export of ZapA (222).
Early in vitro assays indicated that ZapA degrades mouse immunoglobulins, but recent work suggests that ZapA-mediated degradation of native immunoglobulins is less effective than degradation of the human antimicrobial proteins β-defensin-1 and LL-37 (also known as cathelicidin/CRAMP) (222, 223). Both β-defensin-1 and LL-37 are present in the urinary tract, with increased levels during urinary tract infection (225, 226). In particular, mice deficient in LL-37 are more susceptible to UPEC colonization of the kidney (225). ZapA-mediated degradation of β-defensin-1 and LL-37 decreases their antimicrobial activity (223). Although zapA is expressed at low levels in vivo, a P. mirabilis zapA mutant is less efficient than wild type at colonizing the urine, bladder, and kidneys in independent challenge murine infections (25, 224). ZapA also contributes to acute prostatitis in a rat model of infection, and a zapA mutant is severely impaired in establishing chronic prostatitis in this model (227). In support of a role for ZapA in human infections, ZapA-like protease activity has been detected in the urine of individuals with P. mirabilis urinary tract infections (228). ZapA is induced during swarming differentiation (66, 224), and expression remains high at the edge of an expanding swarm colony (65). ZapA is a possible therapeutic target, and toward this goal, chemical inhibitors have been identified (229).
OTHER PROTEASES
Sequencing revealed four additional putative metalloproteases in the five open reading frames upstream of ZapA (17). The gene immediately upstream of zapA was named zapE, and the three putative metalloproteases appear to be zapE copies. One of the zapE copies (PMI0283) was identified as a virulence determinant using STM, although the mutagenesis employed a transposon which might have affected the production of multiple metalloproteases (57). Additionally, a U32-family peptidase (PMI3442) was identified as important for infection in a separate STM study (230).
Metal Acquisition Systems
IRON
To acquire free iron in the host, pathogenic bacteria encode a number of different iron-acquisition systems, including siderophore-based systems, ferrous iron transport, and metal-type ABC transporters (231, 232). Siderophores are extracellular high affinity ferric iron (Fe3+) chelators which bind and solubilize iron. The siderophore-iron complex is then taken up by a specific outer membrane receptor, a process mediated by the TonB-ExbB-ExbD system (232). Uptake of ferrous iron (Fe2+) is largely mediated by the FeoAB system, which is induced under anerobic, iron-limiting conditions. Ferrous iron is then converted to ferric iron, although the mechanism remains unknown (232). Finally, metal-type ABC transporters are specific for iron, but may not require outer membrane receptors or siderophores. Additionally, bacteria may utilize bound iron in the forms of heme, transferrin, and lactoferrin (232). P. mirabilis is predicted to encode all four types of uptake systems, and can grow on hemoglobin, hemin, and ferric citrate as the sole iron source, but not on transferrin and lactoferrin (233) (Table 3). Additionally, P. mirabilis may use α-keto acids as iron chelators.
Table 3.
Iron-related genes in P. mirabilis.
| Gene Designation(s) |
Proposed function |
PMI number(s) |
Upregulated in iron limitation |
Upregulated in vivo |
Implicated in vivo |
Antigenic in vivo |
Citations |
|---|---|---|---|---|---|---|---|
| Heme uptake | |||||||
| TonB-dependent receptor | PMI0409 | ✓ | ✓ | (45, 238) | |||
| hemin uptake protein | PMI1424 | ✓ | ✓ | (25, 238) | |||
| hmuR1R2STUV | hemin uptake system | PMI1425-1430 | ✓ | ✓ | ✓ | ✓ | (25, 45, 238, 239) |
| Ferrous Iron Uptake | |||||||
| sitDCBA | Iron ABC transporter | PMI1024-1027 | ✓ | ✓ | (25, 238) | ||
| feoAB | ferrus iron transport | PMI2920-2921 | ✓ | (25) | |||
| Ferric citrate transport | |||||||
| exported protease | PMI3704 | ✓ | (238) | ||||
| iron-related ABC transporter | PMI3705 | ✓ | ✓ | (57, 238) | |||
| iron receptor | PMI3706-3707 | ✓ | (238) | ||||
| extracytoplasmic function family σ factor | PMI3708 | ✓ | (238) | ||||
| TonB-like protein | PMI3709 | ✓ | (238) | ||||
| Siderophore production | |||||||
| TonB-dependent receptor | PMI2596 | ✓ | ✓ | ✓ | (45, 238) | ||
| nrpXYRSUTABG | Nrp siderophore | PMI2597-2605 | ✓ | ✓ | ✓ | (25, 56, 238) | |
| pbtIABCDEFGH | proteobactin | PMI0231-0239 | ✓ | ✓ | (57, 238) | ||
| Other TonB-dependent receptors | |||||||
| irgA | TonB-dependent enterobactin receptor | PMI0842 | ✓ | ✓ | ✓ | ✓ | (25, 45, 238) |
| ireA | ferric siderophore receptor | PMI1945 | ✓ | ✓ | ✓ | (25, 45, 238) | |
| TonB-dependent receptor | PMI0363 | ✓ | ✓ | (25, 238) | |||
| TonB-dependent receptor | PMI1548-1551 | ✓ | ✓ | (25, 238) | |||
| hasR | TonB-dependent receptor | PMI3120-3121 | ✓ | (56) | |||
| ABC-transport system | |||||||
| iron-related ABC transporter | PMI0331 | ✓ | (25, 238) | ||||
| iron-related ABC transporter | PMI2957-2960 | ✓ | ✓ | ✓ | (25, 56, 238) | ||
| Iron-related ABC transporter | PMI0229-0230 | (238) | |||||
| Iron Metabolism | |||||||
| iron utilization protein | PMI1437 | ✓ | ✓ | (25, 238) | |||
| Iron Sulfur Cluster Formation/Uptake | |||||||
| iron sulfur cluster | PMI1411-1416 | ✓ | ✓ | (25, 238) | |||
| iron sulfur cluster | PMI3253 | ✓ | ✓ | (25, 238) | |||
| iron sulfur cluster | PMI0176-0172 | ✓ | ✓ | (25, 238) | |||
Iron-related genes from P. mirabils were identified by homology to other iron-related genes (17, 238). Genes identified as iron-related by homology but not identified using one of the four conditions shown were excluded. A checkmark indicates that one or more of the genes in the row were identified using the condition specified.
Prior to sequencing of the P. mirabilis genome, α-keto acids were the only known iron scavenging system in P. mirabilis. Initial studies show that α-keto acids, produced by an amino acid deanimase (Aad, PMI2834), could chelate iron from solutions (234, 235). Alpha-keto acids are also produced by Providencia, Morganella, and Salmonella species (234, 236). In Salmonella typhimurium, α-keto acids chelate iron in a TonB-dependent manner, but are less effective than siderophores (236, 237). In both Salmonella and P. mirabilis, α-keto acid production is not directly regulated by iron limitation (235, 236). Overall, α-keto acids appear to participate in iron chelation, but are probably not essential for growth during iron limitation.
P. mirabilis encodes two siderophore systems, the novel non-ribosomal peptide synthetase-independent siderophore system (proteobactin), and the non-ribosomal peptide siderophore system (Nrp). Proteobactin is predicted to be a hydroxycarboxylate siderophore (238). The proteobactin locus involves three transcripts: one encodes an ABC transporter (PMI0229-030); the second transcript includes a single gene pbtI (PMI0231), which encodes an enzyme thought to catalyze synthesis of a proteobactin precursor; and the third transcript encompasses pbtABCDEFGH (PMI0232-0239), which encodes the remaining components necessary for siderophore synthesis and utilization (238).
The P. mirabilis Nrp system was identified in a swarming-defective P. mirabilis mutant (119). It was predicted to encode a siderophore based on its homology to the genes encoding the Yersinia pestis siderophore yersiniabactin (17). However, the nrp locus (PMI2596 and operon nrpXYRSUTABG PMI2597-2605) is missing genes essential for yersiniabactin production and encodes genes not found in the yersiniabactin operon (238). Since P. mirabilis is unable to produce or utilize yersiniabactin, the nrp operon is likely involved in the production and utilization of a siderophore distinct from yersiniabactin.
The Nrp and proteobactin systems are both involved in iron chelation, but only Nrp has been implicated in urinary tract infection (238). Wild-type P. mirabilis can chelate iron in the chrome azul S (CAS) assay (238). Single P. mirabilis nrp or pbt mutants have no defect in iron chelation, while a double mutant is incapable of iron chelation in the CAS assay (Fig. 9) (238). In an independent challenge experiment, neither a single P. mirabilis pbt, nrp, nor a double nrp pbt mutant had a defect in colonization compared to wild type. However, during a co-challenge experiment, the P. mirabilis nrp mutant was impaired in colonization of the bladder and kidneys (238). Interestingly, the P. mirabilis nrp pbt mutant was only impaired in colonization of the kidneys (238). These results suggest that the Nrp system is more important for colonizing the urinary tract than proteobactin.
Figure 9.
P. mirabilis iron chelation is Nrp and proteobactin dependent. A, agar; and B, solution chrome azurol S (CAS) assays of uropathogenic E. coli CFT073 and P. mirabilis HI4320; a color change from blue to orange indicates iron chelation. In B), P. mirabilis supernatants from log-phase cultures grown in MOPS defined media either with 0.1 mM FeCl3·6H2O (black bars) or without supplementation (white bars) were concentrated 50-fold before being used in a liquid CAS assay (E. coli supernatants were not concentrated). Single P. mirabilis nrpR and pbtA mutants are not impaired in iron chelation, but the double P. mirabilis nrpR pbtA mutant is. Reprinted with permission from Molecular Microbiology, vol 78, page 149 (238).
P. mirabilis can grow on hemin, and a putative heme uptake system is upregulated during iron limitation and in vivo. PMI1426 (hmuR2), which encodes a putative outer membrane receptor in the heme uptake system, binds heme, is implicated in infection, and has been identified as an antigen in vivo (25, 233, 238, 239).
Additionally, P. mirabilis encodes genes predicted to be involved in uptake of ferrous iron, ferric iron, and ferric citrate; iron sulfur cluster formation and uptake; as well as a number of predicted TonB-dependent receptors and iron-related ABC-transport systems (17, 238). Several of these genes have been identified as being iron regulated in vitro, or contribute to UTI by being positively regulated in vivo, expressed on the bacterial cell surface in vivo, or important for full colonization of the urinary tract (Table 2) (25, 45, 56, 57, 238). However, the functions of these systems have not been directly tested.
ZINC
P. mirabilis encodes a functional zinc uptake system, znuACB (118). Zinc levels are thought to be limited in the urinary tract and zinc uptake provides a competitive advantage to other pathogenic bacteria, including uropathogenic E. coli (240). In P. mirabilis, znuCB is upregulated in vivo and a P. mirabilis znuC mutant is out-competed by wild-type P. mirabilis during co-challenge infection (25, 118). Zinc is predicted to be needed for other virulence factors, such as flagella and zapA. The master flagellar regulator, FlhD4C2, has a putative zinc binding site and a P. mirabilis znuC mutant expresses less flagellin, suggesting that zinc uptake is essential for flagellar synthesis (48, 119). As previously discussed, the metalloprotease ZapA provides a competitive advantage during infection, and its activity is thought to be zinc-dependent (223). Without a functional zinc-uptake system, P. mirabilis may improperly regulate expression of flagellin, ZapA, and possibly other factors, leading to weakened colonization of the urinary tract.
Phosphate Transport
P. mirabilis encodes a high-affinity inorganic phosphate (Pi) transporter very similar to the E. coli Pst operon (241). The P. mirabilis pst operon includes genes (PMI2893-PMI2896) encoding four proteins associated with phosphate transport (PstSCAB) and a negative transcriptional regulator (PMI2897) of the phosphate regulon, PhoU. When the pst operon is upregulated, so are other members of the phosphate regulon, including phoA (PMI2500), the gene encoding alkaline phosphatase Thus, alkaline phosphatase activity can be measured as an indirect output of the pst operon and of phosphate limitation (241). When alkaline phosphatase activity is measured in wild-type P. mirabilis, there is very little activity in LB media and very high activity in phosphate-limited media. In human urine, there is intermediate induction of alkaline phosphatase, suggesting that urine is somewhat phosphate limited (241).
Two mutations in the pst operon were identified using STM as being essential for full infection of the murine urinary tract (56). Additional experiments showed that individual transposon insertions in pstA and pstS were outcompeted in the urine, bladder, and kidneys during co-challenge infection with wild type, which could be complemented by plasmid-encoded expression of the pst operon (56, 241). A general growth defect was not responsible for the phenotype observed in mice, as the pst mutants out-competed wild type during co-culture in human urine (241). However, both pst mutants were impaired in biofilm formation. While wild-type P. mirabilis formed biofilms with a dense 3D structure reminiscent of towering mushrooms with liquid channels throughout, the pst mutant biofilms lacked these distinctive structures (242). This may suggest a role for biofilms during urinary tract infection independent of catheter encrustation, or the pst operon may contribute to urinary tract infection through a yet to be characterized mechanism.
Virulence factors identified by STM
Three studies used an unbiased signature-tagged mutagenesis screen to identify genes important for infection in the murine urinary tract. These screens identified genes important for many of the virulence factors described above, including urease, motility, iron acquisition, fimbriae, phosphate transport, ZapE, and TaaP (56, 57, 230). Approximately 20% of the genes identified are involved in metabolism, including genes involved with glycolysis (aceE PMI2046), the TCA cycle (sdhC PMI0565), and the Entner-Doudoroff pathway (edd PMI2760). Genes associated with amino acid metabolism and transport were identified, including D-methionine transporter metN (PMI2259), dihydrouridine synthase dusB (PMI3623), carbamoyl phosphate synthetase carA (PMI0020), and L-serine deaminase sdaA (PMI1607) (56, 57). Genes involved in GTP synthesis (guaB PMI1546), adenylate cyclase (cyaA PMI3333) and a predicted oxidoreductase with similarities to formate dehydrogenase (cbbBc PMI2378) were also identified (57). A gene recognized to regulate flhDC in E. coli (hdfR, PMI3295) was identified, although this mutation resulted in no apparent motility defect in P. mirabilis (56). A second transcriptional regulator (nhaR PMI0012) was also identified (57). Two genes likely involved in plasmid maintenance and that localized to the P. mirabilis plasmid pHI4320 were identified in these screens (parE (PMIP32) and pilX (PMIP08-09)), suggesting that this plasmid plays a role in virulence (56). The other genes identified largely encode hypothetical proteins, and the mechanism of their contribution to virulence has yet to be deduced.
Genome organization
Following the first completely sequenced P. mirabilis genome, UTI isolate HI4320 (17), three new P. mirabilis genome sequences have been published: laboratory strain BB2000 (154), diarrhea isolate C05028 (243), and blood isolate PR03 (244). There are three more strains that have been partially sequenced and assembled (type strain ATCC 29906, urine isolate WGLW4, and mouse stool isolate WGLW6). The genome size of these seven strains ranges from 3.82 to 4.06 Mb, and they have GC content ranging from 38.5-39.0%. Strain HI3420 also possesses a plasmid, pHI4320. STM studies indicate the plasmid contributes to virulence during UTI (56). The BB2000 genome does not contain any plasmids; the plasmid status of the other partially assembled genomes has not been specified. Most of the notable features of the HI3420 genome have already been discussed in this chapter, including the 17 chaperone-usher fimbrial operons, contiguous flagellar locus, and other virulence factors. A type III secretion system is encoded by all currently-sequenced P. mirabilis genomes, and although there is evidence for transcription of the genes within this locus, it does not appear to contribute to UTI (245). Comparison of ten uropathogenic E. coli genomes led to the identification of 131 genes that are specific to UPEC compared to three fecal E. coli isolates (246); P. mirabilis encodes homologs of 25 of these genes (17). This species also encodes a pathogenicity island, ICEPm1, which is commonly found in P. mirabilis, Providencia stuartii, and Morganella morganii (214). Virulence genes, including the taaP autotransporter and the nrp siderophore system, are encoded in ICEPm1. This element can be transmitted to other P. mirabilis clinical isolates (247).
Gene expression during UTI
Microarray analysis has been used to examine the transcriptome of P. mirabilis isolated from the urine of experimentally-infected mice (25). Compared to mid-logarithmic phase broth culture, 471 genes were induced and 82 were repressed in vivo. Many of the upregulated genes encode virulence factors that have been already discussed (urease, MR/P fimbriae, iron uptake). These findings were generally similar to the transcriptome of uropathogenic E. coli in vivo (induction of fimbriae, iron and peptide uptake systems, pyruvate catabolism, osmoprotection; repressed flagella) (50).
There were several differences in metabolic gene expression between UPEC and P. mirabilis, most notably with respect to nitrogen assimilation: in P. mirabilis, gdhA (glutamate dehydrogenase, PMI3008) is induced and glnA (glutamine synthetase, PMI2882) is repressed (25); the opposite is true for UPEC. Since the glutamine synthetase system primarily operates during nitrogen starvation, it appears that the carbon-nitrogen balance in P. mirabilis is altered compared to E. coli. Genes in the TCA cycle corresponding to the steps from acetyl coenzyme A to α-ketoglutarate synthesis are also induced in vivo by P. mirabilis. The depletion of α-ketoglutarate pools from the TCA cycle due to GDH-mediated ammonia uptake was proposed to be responsible for the induction of this portion of the TCA cycle. Although urease seemed the likely candidate responsible for this pattern, culture of P. mirabilis in the presence of urease or exogenous ammonium did not recapitulate in vivo gene expression. Instead, the pattern was restored using citrate as a sole carbon source. A gdhA mutant is also not as fit during experimental murine co-challenge (25).
Biofilm formation
CAUTI are generally initiated by biofilm formation on the urinary catheter. After catheter placement, host-derived factors from the urine and urogenital tract are deposited on the catheter, creating a coating that facilitates bacterial attachment. A number of studies have reported on the contribution of specific factors to P. mirabilis biofilm formation in vitro, some of which have been described in detail above (urease, MR/P fimbriae, Pst, and RsbA; see also the review by Jacobsen and Shirtliff (248)). Capsular polysaccharides purified from P. mirabilis have been reported to increase crystalline biofilm formation in artificial urine (249). More recently, P. mirabilis transposon mutants were screened within an artificial catheterized bladder model to find factors that contribute to catheter biofilms (250). In this assay, nirB nitrate reductase (PMI1479), bcr putative multidrug efflux pump (PMI0829), and two genes of unknown function (PMI1608 and PMI3402) were identified. Mutations in nirB and bcr resulted in decreased biomass and calcium associated with the catheter compared to wild type, but similar levels of un-attached biomass and of adherence to silicone surfaces (250).
CLINICAL ASPECTS OF P. MIRABILIS UTI
Prevention
The main risk factors for P. mirabilis infection are an anatomical or functional abnormality in the urinary tract or the presence of a catheter. The first cannot be easily prevented. The presence of a catheter is associated with a 3-10% risk of bacterial infection per day (11). Catheterization should be avoided whenever possible, and when it cannot be avoided, duration of catheterization should be limited. The preferred technique when a catheter is needed is clean, intermittent catheterization (11). It is recommended that catheters are changed regularly when they cannot be removed, but there is no strong clinical evidence to support this recommendation (11). However, as catheterization is indeed necessary in some patients, development of bacterial growth-resistant catheters and of a P. mirabilis vaccine is ongoing (see the section on vaccine development).
Treatment
The currently recommended treatment for acute uncomplicated cystitis is a 3 day course of double-strength trimethoprim-sulfamethoxazole (SXT) if the local SXT resistance rate is less than 10-20% (9, 251). However, this organism has a wide spectrum of local SXT resistance rates ranging from 16-83% (9, 252) (see “Antibiotic Resistance”, below). If the SXT resistance rate is above 10-20%, the recommended alternative antibiotic therapy for uncomplicated cystitis may include fluroquinolones, nitrofurantoin, or fosfomycin, all of which have some reported resistance (9, 251, 253). There are not specific recommendations for complicated- or catheter-associated urinary tract infections. Instead, antibiotic treatment is based on the specific situation and the clinician's discretion.
When P. mirabilis infections are found in connection with a urinary stone, extra precautions need to be taken because bacteria are found inside the stone. The stone needs to be completely eradicated with shockwave lithotripsy or otherwise removed, and urine should be checked monthly for three months after antibiotic treatment is completed (254). Even in the absence of an obvious stone, follow-up urine cultures are recommended in P. mirabilis UTI to ensure that the infection is cleared from the urinary tract and to prevent stones from occurring.
Antibiotic Resistance
Worldwide, P. mirabilis has developed resistance to several classes of antibiotics, complicating treatment. In addition to the described resistance to SXT, resistance to β-lactams (both penicillins and cephalosporins), fluroquinolones, nitrofurantoin, fosfomycin, aminoglycosides, tetracyclines, and sulfonamides has been reported (252, 253, 255). In particular, most isolates are resistant to tetracycline (1). The tendency of this organism to become encased in urinary stones or within crystalline biofilms on urinary catheters can shield bacteria and thus lead to treatment failures (see the urease section, above). P. mirabilis is also highly resistant to antimicrobial peptides including polymyxin B, protegrin, LL-37, and defensin (112, 223). This resistance relies on LPS modifications and extracellular proteases such as ZapA (112, 223, 256). The issues of increasing antibiotic resistance by this organism are otherwise largely similar to those for UPEC, which is covered in detail elsewhere in this book.
Progress toward a P. mirabilis vaccine
The difficulty in treating P. mirabilis and the increase in antibiotic-resistant infection speaks to the importance of developing a P. mirabilis vaccine. Vaccinating the general population is unlikely to be necessary as P. mirabilis only causes 3-5% of all UTI. This vaccine would be beneficial, however, for populations more prone to P. mirabilis UTI, including individuals with functional or structural urinary tract abnormalities, individuals starting long-term catheterization, and patients suffering from recurrent UTI (257). Vaccination of these patients could drastically decrease the incidence of P. mirabilis UTI.
An initial P. mirabilis infection only modestly protects against subsequent homologous and heterologous challenge despite the fact that infection generates a specific adaptive immune response (45, 257). Although this is also true for uropathogenic E. coli, vaccines are available that partially protect humans from recurrent E. coli UTI (258). This suggests that a vaccine is a viable option for P. mirabilis. A successful vaccine will activate a strong adaptive immune response, specifically, the production of mucosal IgA in the urinary tract against P. mirabilis antigens exposed to the immune system.
A number of P. mirabilis vaccines have been tested in a murine urinary tract infection model. The vaccines tested have varied the route of inoculation, the antigen, and the method of antigen display. Although the protocol varies between studies, an initial vaccination is generally followed by 3-5 booster doses, given 7-10 days apart. The post-vaccination challenge occurs seven days after the last booster was administered (257, 259).
Initial P. mirabilis studies largely vaccinated via the transurethral and subcutaneous routes (260-262). However, in order to generate a stronger mucosal IgA response, oral and intranasal vaccines were introduced. Intranasal vaccination provides the best protection and generates a strong mucosal antibody response in both the urine and the vagina (257).
Complex antigens, such as formalin-killed whole P. mirabilis and P. mirabilis outer membranes provide some protection against homologous and heterologous challenge and generate a specific immune response (257, 261). However, as they are difficult to manipulate and failed to initiate an acceptable immune response, other vaccine types were tried.
Several P. mirabilis proteins have been tested in subunit vaccines, where a purified protein, sometimes conjugated to an adjuvant, is used to elicit an immune response. Fimbrial proteins (MrpA, UcaA, PmfA, and MrpH) have been the primary target of these vaccine efforts, though the Pta toxin has also been tested. Although it is difficult to compare results from different vaccine trials, MrpH seems to elicit the strongest response. A truncated MrpH-maltose binding protein fusion conjugated to cholera toxin prevented infection in 75% of mice (257). All these antigens provide some protection, with protection in the kidneys generally stronger than in the bladder (257, 263, 264). Additionally, all tested antigens induce specific antibody responses in either the blood or the urine of the host (257, 263, 264). However, none of the studies to date can correlate the induction of a specific antibody response with the level of protection, as has been seen with an E. coli FimH vaccine (257, 263-265).
Recent studies have also tried recombinant-vector vaccines, where the P. mirabilis antigen is expressed by different, non-pathogenic bacteria. An advantage of a recombinant-vector vaccine is that it can multiply in the host, increasing the exposure to the immune system, but there is no possibility of reversion as in an attenuated vaccine. Two recombinant-vector vaccines have been tested as a delivery vectors for MrpA: Salmonella typhimurium and Lactococcus lactis. Although the S. typhimurium vaccine protected better than the L. lactis vaccine, it is unclear whether the live vaccine approach provided greater protection than the recombinant MrpA vaccine (266, 267).
A defined multi-antigen vaccine has yet to be tested. In particular, because the P. mirabilis fimbriae may be redundant in their ability to adhere to the surfaces of the urinary tract (and at least one fimbria, MR/P, is phase-variable), a vaccine with proteins from multiple fimbriae should be tested. Recent work identifying antigenic proteins in the outer membrane of P. mirabilis during infection identified several other potential vaccine targets, including iron acquisition genes essential for UTI (45). Perhaps a combination of antigens including fimbrial and non-fimbrial targets will prove most effective.
Prevention of urinary catheter biofilm formation
Many efforts have been made to design catheters that are resistant to bacterial colonization, with the goals of preventing catheter blockage and reducing UTI in catheterized patients. Catheters made from latex, silicone, polyurethane, and composite biomaterials have been tested, but no single catheter material is sufficient to prevent bacterial colonization (reviewed in (6)). Several groups have attempted to prevent biofilm formation on catheters by applying or embedding an antimicrobial solution on the catheter surface or in the catheter material. Antibiotic coatings, such as nitrofurazone or a combination of broad-spectrum antibiotics, have significantly reduced the number of CAUTIs in small clinical trials (6, 268). However, given the ubiquity of catheterization, this approach will likely result in antibiotic resistance, a difficulty which has not been resolved. A similar approach involving coating catheters with antiseptics, specifically various silver compounds, has been studied extensively. However, the combination of poorly designed clinical trials and conflicting results have made it difficult to determine the efficacy of these treatments (6, 268).
Several promising techniques have been tested in vitro but have not yet been tested in clinical trials. These include the use of catheters impregnated with combinations of chlorhexidine, silver sulfadiazine, and triclosan; the use of urease inhibitors to prevent crystalline biofilm formation; and the treatment of catheters with a cocktail of bacteriophages (6, 268). Most of the techniques tested have been chemical, not mechanical, which makes the promising in vitro results using low energy acoustic waves to prevent bacterial attachment and the development of a urinary catheter that uses inflation-generated catheter strain to clear P. mirabilis crystalline biofilms intriguing alternative approaches (268, 269).
Although complete prevention of biofilms on catheters is ideal, the most serious complications of CAUTI generally occur when the catheter becomes blocked by the presence of crystalline biofilms that are the result of urease activity. A sensor which detects the initial stages of crystalline biofilm formation early enough to allow the catheter to be changed prior to catheter blockage would help mitigate the effects of biofilm formation on catheters. An early prototype of this sensor detected crystalline biofilm formation around 12 days prior to blockage in a clinical trial (270). A more recent sensor, which is amenable to mass production, detected P. mirabilis crystalline biofilm formation 17 to 24 hours in advance (271). This technology could significantly decrease the morbidity and costs associated with CAUTI.
CONCLUSIONS
Significant recent advances have been made in our understanding of the lifestyle of this pathogen both with regard to swarming and to pathogenesis during UTI. However, there are still substantial gaps in our knowledge. What triggers the cyclic switch between swarming and consolidation? How important are motility and swarming to virulence, and at which points during virulence is motility involved? Why does this species encode such an extensive array of adherence factors, which of these contribute to UTI, and what are their host targets? Is invasion of either bladder or renal cells a component of pathogenesis? Very little investigation of the immune response during P. mirabilis UTI has occurred; is the response similar to the response for uropathogenic E. coli, or are there differences? Studies in this area could aid vaccine design. CAUTI are often polymicrobial. How is UTI affected by the interaction of P. mirabilis with other species? Finally, numerous genes have been identified by transposon mutagenesis, STM, and microarray analysis as being important for or regulated during swarming or UTI. However, the mechanism of most of these contributions is yet to be determined.
ACKNOWLEDGMENTS
This work was supported by grant AI083743 from the National Institutes of Health (M.M.P.).
REFERENCES
- 1.O'Hara CM, Brenner FW, Miller JM. Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin Microbiol Rev. 2000;13:534–546. doi: 10.1128/cmr.13.4.534-546.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matthews SJ, Lancaster JW. Urinary tract infections in the elderly population. Am J Geriatr Pharmacother. 2011;9:286–309. doi: 10.1016/j.amjopharm.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Papazafiropoulou A, Daniil I, Sotiropoulos A, Balampani E, Kokolaki A, Bousboulas S, Konstantopoulou S, Skliros E, Petropoulou D, Pappas S. Prevalence of asymptomatic bacteriuria in type 2 diabetic subjects with and without microalbuminuria. BMC Res Notes. 2010;3:169. doi: 10.1186/1756-0500-3-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janda JMA, Abbott SL. The Enterobacteria. 2 ed. ASM Press; Washington, D.C.: 2006. [Google Scholar]
- 5.Mathur S, Sabbuba NA, Suller MT, Stickler DJ, Feneley RC. Genotyping of urinary and fecal Proteus mirabilis isolates from individuals with long-term urinary catheters. Eur J Clin Microbiol Infect Dis. 2005;24:643–644. doi: 10.1007/s10096-005-0003-0. [DOI] [PubMed] [Google Scholar]
- 6.Jacobsen SM, Stickler DJ, Mobley HLT, Shirtliff ME. Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin Microbiol Rev. 2008;21:26–59. doi: 10.1128/CMR.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaston H. Proteus--is it a likely aetiological factor in chronic polyarthritis? Ann Rheum Dis. 1995;54:157–158. doi: 10.1136/ard.54.3.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rashid T, Ebringer A. Rheumatoid arthritis is linked to Proteus--the evidence. Clin Rheumatol. 2007;26:1036–1043. doi: 10.1007/s10067-006-0491-z. [DOI] [PubMed] [Google Scholar]
- 9.Karlowsky JA, Lagacé-Wiens PR, Simner PJ, DeCorby MR, Adam HJ, Walkty A, Hoban DJ, Zhanel GG. Antimicrobial resistance in urinary tract pathogens in Canada from 2007 to 2009: CANWARD surveillance study. Antimicrob Agents Chemother. 2011;55:3169–3175. doi: 10.1128/AAC.00066-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielubowicz GR, Mobley HLT. Host-pathogen interactions in urinary tract infection. Nat Rev Urol. 2010;7:430–441. doi: 10.1038/nrurol.2010.101. [DOI] [PubMed] [Google Scholar]
- 11.Nicolle LE. Catheter-related urinary tract infection. Drugs Aging. 2005;22:627–639. doi: 10.2165/00002512-200522080-00001. [DOI] [PubMed] [Google Scholar]
- 12.Hung EW, Darouiche RO, Trautner BW. Proteus bacteriuria is associated with significant morbidity in spinal cord injury. Spinal Cord. 2007;45:616–620. doi: 10.1038/sj.sc.3102004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams-Sapper S, Sergeevna-Selezneva J, Tartof S, Raphael E, Diep BA, Perdreau-Remington F, Riley LW. Globally dispersed mobile drug-resistance genes in gram-negative bacterial isolates from patients with bloodstream infections in a US urban general hospital. J Med Microbiol. 2012;61:968–974. doi: 10.1099/jmm.0.041970-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mylotte JM. Nursing home-acquired bloodstream infection. Infect Control Hosp Epidemiol. 2005;26:833–837. doi: 10.1086/502502. [DOI] [PubMed] [Google Scholar]
- 15.Sader HS, Flamm RK, Jones RN. Frequency of occurrence and antimicrobial susceptibility of Gram-negative bacteremia isolates in patients with urinary tract infection: results from United States and European hospitals (2009-2011). J Chemother. 2014;26:133–138. doi: 10.1179/1973947813Y.0000000121. [DOI] [PubMed] [Google Scholar]
- 16.Lubart E, Segal R, Haimov E, Dan M, Baumoehl Y, Leibovitz A. Bacteremia in a multilevel geriatric hospital. J Am Med Dir Assoc. 2011;12:204–207. doi: 10.1016/j.jamda.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 17.Pearson MM, Sebaihia M, Churcher C, Quail MA, Seshasayee AS, Luscombe NM, Abdellah Z, Arrosmith C, Atkin B, Chillingworth T, Hauser H, Jagels K, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Walker D, Whithead S, Thomson NR, Rather PN, Parkhill J, Mobley HLT. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J Bacteriol. 2008;190:4027–4037. doi: 10.1128/JB.01981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mobley HLT. Urease. In: Mobley HLT, Mendz GL, Hazell SL, editors. Helicobacter pylori: physiology and genetics. ASM Press; Washington, DC: 2001. doi:NBK2417 [bookaccession] [PubMed] [Google Scholar]
- 19.Mobley HLT. Virulence of Proteus mirabilis. In: Mobley HL, Warren JW, editors. Urinary Tract Infections: Molecular Pathogenesis and Clinical Management. ASM Press; Washington, D.C.: 1996. pp. 245–269. [Google Scholar]
- 20.Griffith DP, Musher DM, Itin C. Urease. The primary cause of infection-induced urinary stones. Invest Urol. 1976;13:346–350. [PubMed] [Google Scholar]
- 21.Nicholson EB, Concaugh EA, Foxall PA, Island MD, Mobley HLT. Proteus mirabilis urease: transcriptional regulation by UreR. J Bacteriol. 1993;175:465–473. doi: 10.1128/jb.175.2.465-473.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dattelbaum JD, Lockatell CV, Johnson DE, Mobley HLT. UreR, the transcriptional activator of the Proteus mirabilis urease gene cluster, is required for urease activity and virulence in experimental urinary tract infections. Infection and Immunity. 2003;71:1026. doi: 10.1128/IAI.71.2.1026-1030.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poore CA, Mobley HLT. Differential regulation of the Proteus mirabilis urease gene cluster by UreR and H-NS. Microbiology. 2003;149:3383–3394. doi: 10.1099/mic.0.26624-0. [DOI] [PubMed] [Google Scholar]
- 24.Zhao H, Thompson RB, Lockatell V, Johnson DE, Mobley HLT. Use of green fluorescent protein to assess urease gene expression by uropathogenic Proteus mirabilis during experimental ascending urinary tract infection. Infect Immun. 1998;66:330–335. doi: 10.1128/iai.66.1.330-335.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson MM, Yep A, Smith SN, Mobley HLT. Transcriptome of Proteus mirabilis in the murine urinary tract: virulence and nitrogen assimilation gene expression. Infect Immun. 2011;79:2619–2631. doi: 10.1128/IAI.05152-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munns J, Amawi F. A large urinary bladder stone: an unusual cause of rectal prolapse. Arch Dis Child. 2010;95:1026. doi: 10.1136/adc.2010.198440. [DOI] [PubMed] [Google Scholar]
- 27.Chew R, Thomas S, Mantha ML, Killen JP, Cho Y, Baer RA. Large urate cystolith associated with Proteus urinary tract infection. Kidney Int. 2012;81:802. doi: 10.1038/ki.2011.506. [DOI] [PubMed] [Google Scholar]
- 28.Jones BV, Mahenthiralingam E, Sabbuba NA, Stickler DJ. Role of swarming in the formation of crystalline Proteus mirabilis biofilms on urinary catheters. J Med Microbiol. 2005;54:807–813. doi: 10.1099/jmm.0.46123-0. [DOI] [PubMed] [Google Scholar]
- 29.Stickler DJ. Bacterial biofilms in patients with indwelling urinary catheters. Nat Clin Pract Urol. 2008;5:598–608. doi: 10.1038/ncpuro1231. [DOI] [PubMed] [Google Scholar]
- 30.Torzewska A, Budzyńska A, Białczak-Kokot M, Różalski A. In vitro studies of epithelium-associated crystallization caused by uropathogens during urinary calculi development. Microb Pathog. 2014;71-72C:25–31. doi: 10.1016/j.micpath.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Mobley HLT, Warren JW. Urease-positive bacteriuria and obstruction of long-term urinary catheters. J Clin Microbiol. 1987;25:2216–2217. doi: 10.1128/jcm.25.11.2216-2217.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Zhao H, Lockatell CV, Drachenberg CB, Johnson DE, Mobley HLT. Visualization of Proteus mirabilis within the matrix of urease-induced bladder stones during experimental urinary tract infection. Infection and Immunity. 2002;70:389–394. doi: 10.1128/IAI.70.1.389-394.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones BD, Lockatell CV, Johnson DE, Warren JW, Mobley HLT. Construction of a urease-negative mutant of Proteus mirabilis: analysis of virulence in a mouse model of ascending urinary tract infection. Infection and Immunity. 1990;58:1120–1123. doi: 10.1128/iai.58.4.1120-1123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson DE, Russell RG, Lockatell CV, Zulty JC, Warren JW, Mobley HLT. Contribution of Proteus mirabilis urease to persistence, urolithiasis, and acute pyelonephritis in a mouse model of ascending urinary tract infection. Infection and Immunity. 1993;61:2748–2754. doi: 10.1128/iai.61.7.2748-2754.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Armbruster CE, Smith SN, Yep A, Mobley HLT. Increased incidence of urolithiasis and bacteremia during Proteus mirabilis and Providencia stuartii coinfection due to synergistic induction of urease activity. J Infect Dis. 2014;209:1524–1532. doi: 10.1093/infdis/jit663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Follmer C. Ureases as a target for the treatment of gastric and urinary infections. J Clin Pathol. 2010;63:424–430. doi: 10.1136/jcp.2009.072595. [DOI] [PubMed] [Google Scholar]
- 37.Suller MT, Anthony VJ, Mathur S, Feneley RC, Greenman J, Stickler DJ. Factors modulating the pH at which calcium and magnesium phosphates precipitate from human urine. Urol Res. 2005;33:254–260. doi: 10.1007/s00240-004-0458-y. [DOI] [PubMed] [Google Scholar]
- 38.Khan A, Housami F, Melotti R, Timoney A, Stickler D. Strategy to control catheter encrustation with citrated drinks: a randomized crossover study. J Urol. 2010;183:1390–1394. doi: 10.1016/j.juro.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 39.Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 40.Belas R, Flaherty D. Sequence and genetic analysis of multiple flagellin-encoding genes from Proteus mirabilis. Gene. 1994;148:33–41. doi: 10.1016/0378-1119(94)90230-5. [DOI] [PubMed] [Google Scholar]
- 41.Belas R. Expression of multiple flagellin-encoding genes of Proteus mirabilis. J Bacteriol. 1994;176:7169–7181. doi: 10.1128/jb.176.23.7169-7181.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy CA, Belas R. Genomic rearrangements in the flagellin genes of Proteus mirabilis. Mol Microbiol. 1999;31:679–690. doi: 10.1046/j.1365-2958.1999.01209.x. [DOI] [PubMed] [Google Scholar]
- 43.Manos J, Belas R. Transcription of Proteus mirabilis flaAB. Microbiology. 2004;150:2857–2863. doi: 10.1099/mic.0.27275-0. [DOI] [PubMed] [Google Scholar]
- 44.Manos J, Artimovich E, Belas R. Enhanced motility of a Proteus mirabilis strain expressing hybrid FlaAB flagella. Microbiology. 2004;150:1291–1299. doi: 10.1099/mic.0.26727-0. [DOI] [PubMed] [Google Scholar]
- 45.Nielubowicz GR, Smith SN, Mobley HLT. Outer membrane antigens of the uropathogen Proteus mirabilis recognized by the humoral response during experimental murine urinary tract infection. Infect Immun. 2008;76:4222–4231. doi: 10.1128/IAI.00533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Claret L, Hughes C. Functions of the subunits in the FlhD(2)C(2) transcriptional master regulator of bacterial flagellum biogenesis and swarming. J Mol Biol. 2000;303:467–478. doi: 10.1006/jmbi.2000.4149. [DOI] [PubMed] [Google Scholar]
- 48.Wang S, Fleming RT, Westbrook EM, Matsumura P, McKay DB. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J Mol Biol. 2006;355:798–808. doi: 10.1016/j.jmb.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 49.Bahrani FK, Johnson DE, Robbins D, Mobley HLT. Proteus mirabilis flagella and MR/P fimbriae: isolation, purification, N-terminal analysis, and serum antibody response following experimental urinary tract infection. Infect Immun. 1991;59:3574–3580. doi: 10.1128/iai.59.10.3574-3580.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, Welch RA, Mobley HLT. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infection and Immunity. 2004;72:6373. doi: 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lane MC, Alteri CJ, Smith SN, Mobley HLT. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A. 2007;104:16669–16674. doi: 10.1073/pnas.0607898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mobley HLT, Belas R, Lockatell V, Chippendale G, Trifillis AL, Johnson DE, Warren JW. Construction of a flagellum-negative mutant of Proteus mirabilis: effect on internalization by human renal epithelial cells and virulence in a mouse model of ascending urinary tract infection. Infection and Immunity. 1996;64:5332–5340. doi: 10.1128/iai.64.12.5332-5340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pazin GJ, Braude AI. Immobilizing antibodies in urine. II. Prevention of ascending spread of Proteus mirabilis. Invest Urol. 1974;12:129–133. [PubMed] [Google Scholar]
- 54.Zunino P, Piccini C, Legnani-Fajardo C. Flagellate and non-flagellate Proteus mirabilis in the development of experimental urinary tract infection. Microb Pathog. 1994;16:379–385. doi: 10.1006/mpat.1994.1038. [DOI] [PubMed] [Google Scholar]
- 55.Legnani-Fajardo C, Zunino P, Piccini C, Allen A, Maskell D. Defined mutants of Proteus mirabilis lacking flagella cause ascending urinary tract infection in mice. Microb Pathog. 1996;21:395–405. doi: 10.1006/mpat.1996.0070. [DOI] [PubMed] [Google Scholar]
- 56.Burall LS, Harro JM, Li X, Lockatell CV, Himpsl SD, Hebel JR, Johnson DE, Mobley HLT. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infection and Immunity. 2004;72:2922–2938. doi: 10.1128/IAI.72.5.2922-2938.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Himpsl SD, Lockatell CV, Hebel JR, Johnson DE, Mobley HLT. Identification of virulence determinants in uropathogenic Proteus mirabilis using signature-tagged mutagenesis. J Med Microbiol. 2008;57:1068–1078. doi: 10.1099/jmm.0.2008/002071-0. [DOI] [PubMed] [Google Scholar]
- 58.Williams FD, Schwarzhoff RH. Nature of the swarming phenomenon in Proteus. Annu Rev Microbiol. 1978;32:101–122. doi: 10.1146/annurev.mi.32.100178.000533. [DOI] [PubMed] [Google Scholar]
- 59.Gué M, Dupont V, Dufour A, Sire O. Bacterial swarming: a biochemical time-resolved FTIR-ATR study of Proteus mirabilis swarm-cell differentiation. Biochemistry. 2001;40:11938–11945. doi: 10.1021/bi010434m. [DOI] [PubMed] [Google Scholar]
- 60.Strating H, Vandenende C, Clarke AJ. Changes in peptidoglycan structure and metabolism during differentiation of Proteus mirabilis into swarmer cells. Can J Microbiol. 2012;58:1183–1194. doi: 10.1139/w2012-102. [DOI] [PubMed] [Google Scholar]
- 61.Jones BV, Young R, Mahenthiralingam E, Stickler DJ. Ultrastructure of Proteus mirabilis swarmer cell rafts and role of swarming in catheter-associated urinary tract infection. Infect Immun. 2004;72:3941–3950. doi: 10.1128/IAI.72.7.3941-3950.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stahl SJ, Stewart KR, Williams FD. Extracellular slime associated with Proteus mirabilis during swarming. J Bacteriol. 1983;154:930–937. doi: 10.1128/jb.154.2.930-937.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gygi D, Rahman MM, Lai HC, Carlson R, Guard-Petter J, Hughes C. A cell-surface polysaccharide that facilitates rapid population migration by differentiated swarm cells of Proteus mirabilis. Mol Microbiol. 1995;17:1167–1175. doi: 10.1111/j.1365-2958.1995.mmi_17061167.x. [DOI] [PubMed] [Google Scholar]
- 64.Rahman MM, Guard-Petter J, Asokan K, Hughes C, Carlson RW. The structure of the colony migration factor from pathogenic Proteus mirabilis: a capsular polysaccharide that facilitates swarming. J Biol Chem. 1999;274:22993–22998. doi: 10.1074/jbc.274.33.22993. [DOI] [PubMed] [Google Scholar]
- 65.Pearson MM, Rasko DA, Smith SN, Mobley HLT. Transcriptome of swarming Proteus mirabilis. Infect Immun. 2010;78:2834–2845. doi: 10.1128/IAI.01222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allison C, Lai HC, Hughes C. Co-ordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol Microbiol. 1992;6:1583–1591. doi: 10.1111/j.1365-2958.1992.tb00883.x. [DOI] [PubMed] [Google Scholar]
- 67.Harshey RM. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol. 2003;57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 68.Inoue T, Shingaki R, Hirose S, Waki K, Mori H, Fukui K. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J Bacteriol. 2007;189:950–957. doi: 10.1128/JB.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Q, Frye JG, McClelland M, Harshey RM. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol Microbiol. 2004;52:169–187. doi: 10.1111/j.1365-2958.2003.03977.x. [DOI] [PubMed] [Google Scholar]
- 70.Kim W, Surette MG. Metabolic differentiation in actively swarming Salmonella. Mol Microbiol. 2004;54:702–714. doi: 10.1111/j.1365-2958.2004.04295.x. [DOI] [PubMed] [Google Scholar]
- 71.Allison C, Lai HC, Gygi D, Hughes C. Cell differentiation of Proteus mirabilis is initiated by glutamine, a specific chemoattractant for swarming cells. Mol Microbiol. 1993;8:53–60. doi: 10.1111/j.1365-2958.1993.tb01202.x. [DOI] [PubMed] [Google Scholar]
- 72.Senior BW. p-nitrophenylglycerol--a superior antiswarming agent for isolating and identifying pathogens from clinical material. J Med Microbiol. 1978;11:59–61. doi: 10.1099/00222615-11-1-59. [DOI] [PubMed] [Google Scholar]
- 73.Belas R, Erskine D, Flaherty D. Transposon mutagenesis in Proteus mirabilis. J Bacteriol. 1991;173:6289–6293. doi: 10.1128/jb.173.19.6289-6293.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu MC, Lin SB, Chien HF, Wang WB, Yuan YH, Hsueh PR, Liaw SJ. 10'(Z),13'(E)-heptadecadienylhydroquinone inhibits swarming and virulence factors and increases polymyxin B susceptibility in Proteus mirabilis. PLoS One. 2012;7:e45563. doi: 10.1371/journal.pone.0045563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang WB, Lai HC, Hsueh PR, Chiou RY, Lin SB, Liaw SJ. Inhibition of swarming and virulence factor expression in Proteus mirabilis by resveratrol. J Med Microbiol. 2006;55:1313–1321. doi: 10.1099/jmm.0.46661-0. [DOI] [PubMed] [Google Scholar]
- 76.Hernandez E, Ramisse F, Cavalho JD. Abolition of swarming of Proteus. J Clin Microbiol. 1999;37:3435. doi: 10.1128/jcm.37.10.3435-3435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ayati BP. A structured-population model of Proteus mirabilis swarm-colony development. J Math Biol. 2006;52:93–114. doi: 10.1007/s00285-005-0345-3. [DOI] [PubMed] [Google Scholar]
- 78.Esipov SE, Shapiro JA. Kinetic model of Proteus mirabilis swarm colony development. Journal of Mathematical Biology. 1998;36:249–268. [Google Scholar]
- 79.Frénod E, Sire O. An explanatory model to validate the way water activity rules periodic terrace generation in Proteus mirabilis swarm. J Math Biol. 2009;59:439–466. doi: 10.1007/s00285-008-0235-6. [DOI] [PubMed] [Google Scholar]
- 80.Xue C, Budrene EO, Othmer HG. Radial and spiral stream formation in Proteus mirabilis colonies. PLoS Comput Biol. 2011;7:e1002332. doi: 10.1371/journal.pcbi.1002332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Belas R, Suvanasuthi R. The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J Bacteriol. 2005;187:6789–6803. doi: 10.1128/JB.187.19.6789-6803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Belas R. Proteus mirabilis swarmer cell differentiation and urinary tract infection. In: Mobley HL, Warren JW, editors. Urinary Tract Infections: Molecular Pathogenesis and Clinical Management. ASM Press; Washington, D.C.: 1996. pp. 271–298. [Google Scholar]
- 83.Gygi D, Bailey MJ, Allison C, Hughes C. Requirement for FlhA in flagella assembly and swarm-cell differentiation by Proteus mirabilis. Mol Microbiol. 1995;15:761–769. doi: 10.1111/j.1365-2958.1995.tb02383.x. [DOI] [PubMed] [Google Scholar]
- 84.Belas R, Erskine D, Flaherty D. Proteus mirabilis mutants defective in swarmer cell differentiation and multicellular behavior. J Bacteriol. 1991;173:6279–6288. doi: 10.1128/jb.173.19.6279-6288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gygi D, Fraser G, Dufour A, Hughes C. A motile but non-swarming mutant of Proteus mirabilis lacks FlgN, a facilitator of flagella filament assembly. Mol Microbiol. 1997;25:597–604. doi: 10.1046/j.1365-2958.1997.5021862.x. [DOI] [PubMed] [Google Scholar]
- 86.Allison C, Hughes C. Closely linked genetic loci required for swarm cell differentiation and multicellular migration by Proteus mirabilis. Mol Microbiol. 1991;5:1975–1982. doi: 10.1111/j.1365-2958.1991.tb00819.x. [DOI] [PubMed] [Google Scholar]
- 87.Belas R, Goldman M, Ashliman K. Genetic analysis of Proteus mirabilis mutants defective in swarmer cell elongation. J Bacteriol. 1995;177:823–828. doi: 10.1128/jb.177.3.823-828.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Furness RB, Fraser GM, Hay NA, Hughes C. Negative feedback from a Proteus class II flagellum export defect to the flhDC master operon controlling cell division and flagellum assembly. J Bacteriol. 1997;179:5585–5588. doi: 10.1128/jb.179.17.5585-5588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee YY, Patellis J, Belas R. Activity of Proteus mirabilis FliL is viscosity dependent and requires extragenic DNA. J Bacteriol. 2013;195:823–832. doi: 10.1128/JB.02024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cusick K, Lee YY, Youchak B, Belas R. Perturbation of FliL interferes with Proteus mirabilis swarmer cell gene expression and differentiation. J Bacteriol. 2012;194:437–447. doi: 10.1128/JB.05998-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dufour A, Furness RB, Hughes C. Novel genes that upregulate the Proteus mirabilis flhDC master operon controlling flagellar biogenesis and swarming. Mol Microbiol. 1998;29:741–751. doi: 10.1046/j.1365-2958.1998.00967.x. [DOI] [PubMed] [Google Scholar]
- 92.Clemmer KM, Rather PN. Regulation of flhDC expression in Proteus mirabilis. Res Microbiol. 2007;158:295–302. doi: 10.1016/j.resmic.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 93.Claret L, Hughes C. Rapid turnover of FlhD and FlhC, the flagellar regulon transcriptional activator proteins, during Proteus swarming. J Bacteriol. 2000;182:833–836. doi: 10.1128/jb.182.3.833-836.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Clemmer KM, Rather PN. The Lon protease regulates swarming motility and virulence gene expression in Proteus mirabilis. J Med Microbiol. 2008;57:931–937. doi: 10.1099/jmm.0.47778-0. [DOI] [PubMed] [Google Scholar]
- 95.Hay NA, Tipper DJ, Gygi D, Hughes C. A nonswarming mutant of Proteus mirabilis lacks the Lrp global transcriptional regulator. J Bacteriol. 1997;179:4741–4746. doi: 10.1128/jb.179.15.4741-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lintner RE, Mishra PK, Srivastava P, Martinez-Vaz BM, Khodursky AB, Blumenthal RM. Limited functional conservation of a global regulator among related bacterial genera: Lrp in Escherichia, Proteus and Vibrio. BMC Microbiol. 2008;8:60. doi: 10.1186/1471-2180-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stevenson LG, Rather PN. A novel gene involved in regulating the flagellar gene cascade in Proteus mirabilis. J Bacteriol. 2006;188:7830–7839. doi: 10.1128/JB.00979-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Szostek BA, Rather PN. Regulation of the swarming inhibitor disA in Proteus mirabilis. J Bacteriol. 2013;195:3237–3243. doi: 10.1128/JB.00039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hatt JK, Rather PN. Characterization of a novel gene, wosA, regulating FlhDC expression in Proteus mirabilis. J Bacteriol. 2008;190:1946–1955. doi: 10.1128/JB.01010-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liaw SJ, Lai HC, Ho SW, Luh KT, Wang WB. Role of RsmA in the regulation of swarming motility and virulence factor expression in Proteus mirabilis. J Med Microbiol. 2003;52:19–28. doi: 10.1099/jmm.0.05024-0. [DOI] [PubMed] [Google Scholar]
- 101.Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol. 2001;40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- 102.Cano DA, Dominguez-Bernal G, Tierrez A, Garcia-Del Portillo F, Casadesus J. Regulation of capsule synthesis and cell motility in Salmonella enterica by the essential gene igaA. Genetics. 2002;162:1513–1523. doi: 10.1093/genetics/162.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Morgenstein RM, Rather PN. Role of the Umo proteins and the Rcs phosphorelay in the swarming motility of the wild type and an O-antigen (waaL) mutant of Proteus mirabilis. J Bacteriol. 2012;194:669–676. doi: 10.1128/JB.06047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang YH, Ferrieres L, Clarke DJ. The role of the Rcs phosphorelay in Enterobacteriaceae. Res Microbiol. 2006;157:206–212. doi: 10.1016/j.resmic.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 105.Liaw SJ, Lai HC, Ho SW, Luh KT, Wang WB. Characterisation of p-nitrophenylglycerol-resistant Proteus mirabilis super-swarming mutants. J Med Microbiol. 2001;50:1039–1048. doi: 10.1099/0022-1317-50-12-1039. [DOI] [PubMed] [Google Scholar]
- 106.Belas R, Schneider R, Melch M. Characterization of Proteus mirabilis precocious swarming mutants: identification of rsbA, encoding a regulator of swarming behavior. J Bacteriol. 1998;180:6126–6139. doi: 10.1128/jb.180.23.6126-6139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liaw SJ, Lai HC, Wang WB. Modulation of swarming and virulence by fatty acids through the RsbA protein in Proteus mirabilis. Infect Immun. 2004;72:6836–6845. doi: 10.1128/IAI.72.12.6836-6845.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang WB, Chen IC, Jiang SS, Chen HR, Hsu CY, Hsueh PR, Hsu WB, Liaw SJ. Role of RppA in the regulation of polymyxin b susceptibility, swarming, and virulence factor expression in Proteus mirabilis. Infect Immun. 2008;76:2051–2062. doi: 10.1128/IAI.01557-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang SS, Liu MC, Teng LJ, Wang WB, Hsueh PR, Liaw SJ. Proteus mirabilis pmrI, an RppA-regulated gene necessary for polymyxin B resistance, biofilm formation, and urothelial cell invasion. Antimicrob Agents Chemother. 2010;54:1564–1571. doi: 10.1128/AAC.01219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kato A, Groisman EA. The PhoQ/PhoP regulatory network of Salmonella enterica. Adv Exp Med Biol. 2008;631:7–21. doi: 10.1007/978-0-387-78885-2_2. [DOI] [PubMed] [Google Scholar]
- 111.Jiang SS, Lin TY, Wang WB, Liu MC, Hsueh PR, Liaw SJ. Characterization of UDP-glucose dehydrogenase and UDP-glucose pyrophosphorylase mutants of Proteus mirabilis: defectiveness in polymyxin B resistance, swarming, and virulence. Antimicrob Agents Chemother. 2010;54:2000–2009. doi: 10.1128/AAC.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McCoy AJ, Liu H, Falla TJ, Gunn JS. Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob Agents Chemother. 2001;45:2030–2037. doi: 10.1128/AAC.45.7.2030-2037.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Morgenstein RM, Clemmer KM, Rather PN. Loss of the waaL O-antigen ligase prevents surface activation of the flagellar gene cascade in Proteus mirabilis. J Bacteriol. 2010;192:3213–3221. doi: 10.1128/JB.00196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Allison C, Emody L, Coleman N, Hughes C. The role of swarm cell differentiation and multicellular migration in the uropathogenicity of Proteus mirabilis. J Infect Dis. 1994;169:1155–1158. doi: 10.1093/infdis/169.5.1155. [DOI] [PubMed] [Google Scholar]
- 115.Hay NA, Tipper DJ, Gygi D, Hughes C. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J Bacteriol. 1999;181:2008–2016. doi: 10.1128/jb.181.7.2008-2016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lai HC, Gygi D, Fraser GM, Hughes C. A swarming-defective mutant of Proteus mirabilis lacking a putative cation-transporting membrane P-type ATPase. Microbiology. 1998;144(Pt 7):1957–1961. doi: 10.1099/00221287-144-7-1957. [DOI] [PubMed] [Google Scholar]
- 117.Rensing C, Mitra B, Rosen BP. A Zn(II)-translocating P-type ATPase from Proteus mirabilis. Biochem Cell Biol. 1998;76:787–790. [PubMed] [Google Scholar]
- 118.Nielubowicz GR, Smith SN, Mobley HLT. Zinc uptake contributes to motility and provides a competitive advantage to Proteus mirabilis during experimental urinary tract infection. Infect Immun. 2010;78:2823–2833. doi: 10.1128/IAI.01220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gaisser S, Hughes C. A locus coding for putative non-ribosomal peptide/polyketide synthase functions is mutated in a swarming-defective Proteus mirabilis strain. Mol Gen Genet. 1997;253:415–427. doi: 10.1007/s004380050339. [DOI] [PubMed] [Google Scholar]
- 120.Armbruster CE, Hodges SA, Mobley HLT. Initiation of swarming motility by Proteus mirabilis occurs in response to specific cues present in urine and requires excess L-glutamine. J Bacteriol. 2013;195:1305–1319. doi: 10.1128/JB.02136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sturgill G, Rather PN. Evidence that putrescine acts as an extracellular signal required for swarming in Proteus mirabilis. Mol Microbiol. 2004;51:437–446. doi: 10.1046/j.1365-2958.2003.03835.x. [DOI] [PubMed] [Google Scholar]
- 122.Vinogradov E, Perry MB. Structural analysis of the core region of lipopolysaccharides from Proteus mirabilis serotypes O6, O48 and O57. Eur J Biochem. 2000;267:2439–2446. doi: 10.1046/j.1432-1327.2000.01262.x. [DOI] [PubMed] [Google Scholar]
- 123.Sturgill GM, Siddiqui S, Ding X, Pecora ND, Rather PN. Isolation of lacZ fusions to Proteus mirabilis genes regulated by intercellular signaling: potential role for the sugar phosphotransferase (Pts) system in regulation. FEMS Microbiol Lett. 2002;217:43–50. doi: 10.1111/j.1574-6968.2002.tb11454.x. [DOI] [PubMed] [Google Scholar]
- 124.Kurihara S, Sakai Y, Suzuki H, Muth A, Phanstiel Ot, Rather PN. Putrescine importer PlaP contributes to swarming motility and urothelial cell invasion in Proteus mirabilis. J Biol Chem. 2013;288:15668–15676. doi: 10.1074/jbc.M113.454090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rather PN. Swarmer cell differentiation in Proteus mirabilis. Environ Microbiol. 2005;7:1065–1073. doi: 10.1111/j.1462-2920.2005.00806.x. [DOI] [PubMed] [Google Scholar]
- 126.Holden MT, Ram Chhabra S, de Nys R, Stead P, Bainton NJ, Hill PJ, Manefield M, Kumar N, Labatte M, England D, Rice S, Givskov M, Salmond GP, Stewart GS, Bycroft BW, Kjelleberg S, Williams P. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol Microbiol. 1999;33:1254–1266. doi: 10.1046/j.1365-2958.1999.01577.x. [DOI] [PubMed] [Google Scholar]
- 127.Campbell J, Lin Q, Geske GD, Blackwell HE. New and unexpected insights into the modulation of LuxR-type quorum sensing by cyclic dipeptides. ACS Chem Biol. 2009;4:1051–1059. doi: 10.1021/cb900165y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Eberl L, Winson MK, Sternberg C, Stewart GS, Christiansen G, Chhabra SR, Bycroft B, Williams P, Molin S, Givskov M. Involvement of N-acyl-L-hormoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol Microbiol. 1996;20:127–136. doi: 10.1111/j.1365-2958.1996.tb02495.x. [DOI] [PubMed] [Google Scholar]
- 129.Stankowska D, Kwinkowski M, Kaca W. Quantification of Proteus mirabilis virulence factors and modulation by acylated homoserine lactones. J Microbiol Immunol Infect. 2008;41:243–253. [PubMed] [Google Scholar]
- 130.Schneider R, Lockatell CV, Johnson D, Belas R. Detection and mutation of a luxS-encoded autoinducer in Proteus mirabilis. Microbiology. 2002;148:773–782. doi: 10.1099/00221287-148-3-773. [DOI] [PubMed] [Google Scholar]
- 131.Rauprich O, Matsushita M, Weijer CJ, Siegert F, Esipov SE, Shapiro JA. Periodic phenomena in Proteus mirabilis swarm colony development. J Bacteriol. 1996;178:6525–6538. doi: 10.1128/jb.178.22.6525-6538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Armitage JP. Changes in metabolic activity of Proteus mirabilis during swarming. J Gen Microbiol. 1981;125:445–450. doi: 10.1099/00221287-125-2-445. [DOI] [PubMed] [Google Scholar]
- 133.Falkinham JO, 3rd, Hoffman PS. Unique developmental characteristics of the swarm and short cells of Proteus vulgaris and Proteus mirabilis. J Bacteriol. 1984;158:1037–1040. doi: 10.1128/jb.158.3.1037-1040.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Alteri CJ, Himpsl SD, Engstrom MD, Mobley HLT. Anaerobic respiration using a complete oxidative TCA cycle drives multicellular swarming in Proteus mirabilis. MBio. 2012;3 doi: 10.1128/mBio.00365-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Allison C, Coleman N, Jones PL, Hughes C. Ability of Proteus mirabilis to invade human urothelial cells is coupled to motility and swarming differentiation. Infect Immun. 1992;60:4740–4746. doi: 10.1128/iai.60.11.4740-4746.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Peerbooms PG, Verweij AM, MacLaren DM. Vero cell invasiveness of Proteus mirabilis. Infect Immun. 1984;43:1068–1071. doi: 10.1128/iai.43.3.1068-1071.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chippendale GR, Warren JW, Trifillis AL, Mobley HLT. Internalization of Proteus mirabilis by human renal epithelial cells. Infect Immun. 1994;62:3115–3121. doi: 10.1128/iai.62.8.3115-3121.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Oelschlaeger TA, Tall BD. Uptake pathways of clinical isolates of Proteus mirabilis into human epithelial cell lines. Microb Pathog. 1996;21:1–16. doi: 10.1006/mpat.1996.0037. [DOI] [PubMed] [Google Scholar]
- 139.Alamuri P, Lower M, Hiss JA, Himpsl SD, Schneider G, Mobley HLT. Adhesion, invasion, and agglutination mediated by two trimeric autotransporters in the human uropathogen Proteus mirabilis. Infect Immun. 2010;78:4882–4894. doi: 10.1128/IAI.00718-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mathoera RB, Kok DJ, Verduin CM, Nijman RJ. Pathological and therapeutic significance of cellular invasion by Proteus mirabilis in an enterocystoplasty infection stone model. Infect Immun. 2002;70:7022–7032. doi: 10.1128/IAI.70.12.7022-7032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fraser GM, Claret L, Furness R, Gupta S, Hughes C. Swarming-coupled expression of the Proteus mirabilis hpmBA haemolysin operon. Microbiology. 2002;148:2191–2201. doi: 10.1099/00221287-148-7-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sabbuba N, Hughes G, Stickler DJ. The migration of Proteus mirabilis and other urinary tract pathogens over Foley catheters. BJU Int. 2002;89:55–60. doi: 10.1046/j.1464-4096.2001.01721.x. [DOI] [PubMed] [Google Scholar]
- 143.Jansen AM, Lockatell CV, Johnson DE, Mobley HLT. Visualization of Proteus mirabilis morphotypes in the urinary tract: the elongated swarmer cell is rarely observed in ascending urinary tract infection. Infect Immun. 2003;71:3607–3613. doi: 10.1128/IAI.71.6.3607-3613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dienes L. Reproductive processes in Proteus cultures. Proc Soc Exp Biol Med. 1946;63:265–270. doi: 10.3181/00379727-63-15570. [DOI] [PubMed] [Google Scholar]
- 145.De Louvois J. Serotyping and the Dienes reaction on Proteus mirabilis from hospital infections. J Clin Pathol. 1969;22:263–268. doi: 10.1136/jcp.22.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pfaller MA, Mujeeb I, Hollis RJ, Jones RN, Doern GV. Evaluation of the discriminatory powers of the Dienes test and ribotyping as typing methods for Proteus mirabilis. J Clin Microbiol. 2000;38:1077–1080. doi: 10.1128/jcm.38.3.1077-1080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Senior BW, Larsson P. A highly discriminatory multi-typing scheme for Proteus mirabilis and Proteus vulgaris. J Med Microbiol. 1983;16:193–202. doi: 10.1099/00222615-16-2-193. [DOI] [PubMed] [Google Scholar]
- 148.Budding AE, Ingham CJ, Bitter W, Vandenbroucke-Grauls CM, Schneeberger PM. The Dienes phenomenon: competition and territoriality in swarming Proteus mirabilis. J Bacteriol. 2009;191:3892–3900. doi: 10.1128/JB.00975-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gibbs KA, Urbanowski ML, Greenberg EP. Genetic determinants of self identity and social recognition in bacteria. Science. 2008;321:256–259. doi: 10.1126/science.1160033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gibbs KA, Wenren LM, Greenberg EP. Identity gene expression in Proteus mirabilis. J Bacteriol. 2011;193:3286–3292. doi: 10.1128/JB.01167-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Silverman JM, Brunet YR, Cascales E, Mougous JD. Structure and regulation of the type VI secretion system. Annu Rev Microbiol. 2012;66:453–472. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jani AJ, Cotter PA. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe. 2010;8:2–6. doi: 10.1016/j.chom.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sullivan NL, Septer AN, Fields AT, Wenren LM, Gibbs KA. The complete genome sequence of Proteus mirabilis strain BB2000 reveals differences from the P. mirabilis reference strain. Genome Announc. 2013;1 doi: 10.1128/genomeA.00024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wenren LM, Sullivan NL, Cardarelli L, Septer AN, Gibbs KA. Two independent pathways for self-recognition in Proteus mirabilis are linked by type VI-dependent export. MBio. 2013;4 doi: 10.1128/mBio.00374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Alteri CJ, Himpsl SD, Pickens SR, Lindner JR, Zora JS, Miller JE, Arno PD, Straight SW, Mobley HLT. Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS Pathog. 2013;9:e1003608. doi: 10.1371/journal.ppat.1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Old DC, Adegbola RA. Haemagglutinins and fimbriae of Morganella, Proteus and Providencia. Journal of Medical Microbiology. 1982;15:551. doi: 10.1099/00222615-15-4-551. [DOI] [PubMed] [Google Scholar]
- 158.Wray SK, Hull SI, Cook RG, Barrish J, Hull RA. Identification and characterization of a uroepithelial cell adhesin from a uropathogenic isolate of Proteus mirabilis. Infect Immun. 1986;54:43–49. doi: 10.1128/iai.54.1.43-49.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bahrani FK, Cook S, Hull RA, Massad G, Mobley HLT. Proteus mirabilis fimbriae: N-terminal amino acid sequence of a major fimbrial subunit and nucleotide sequences of the genes from two strains. Infection and Immunity. 1993;61:884–891. doi: 10.1128/iai.61.3.884-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Welch RA, Burland V, Plunkett G, 3rd, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HLT, Donnenberg MS, Blattner FR. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nuccio SP, Baumler AJ. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev. 2007;71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pearson MM, Mobley HLT. Repression of motility during fimbrial expression: identification of 14 mrpJ gene paralogues in Proteus mirabilis. Mol Microbiol. 2008;69:548–558. doi: 10.1111/j.1365-2958.2008.06307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Li X, Johnson DE, Mobley HLT. Requirement of MrpH for mannose-resistant Proteus-like fimbria-mediated hemagglutination by Proteus mirabilis. Infection and Immunity. 1999;67:2822–2833. doi: 10.1128/iai.67.6.2822-2833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bahrani FK, Mobley HLT. Proteus mirabilis MR/P fimbrial operon: genetic organization, nucleotide sequence, and conditions for expression. J Bacteriol. 1994;176:3412–3419. doi: 10.1128/jb.176.11.3412-3419.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhao H, Li X, Johnson DE, Blomfield I, Mobley HLT. In vivo phase variation of MR/P fimbrial gene expression in Proteus mirabilis infecting the urinary tract. Molecular Microbiology. 1997;23:1009–1019. doi: 10.1046/j.1365-2958.1997.2791645.x. [DOI] [PubMed] [Google Scholar]
- 166.Lane MC, Li X, Pearson MM, Simms AN, Mobley HLT. Oxygen-limiting conditions enrich for fimbriate cells of uropathogenic Proteus mirabilis and Escherichia coli. J Bacteriol. 2009;191:1382–1392. doi: 10.1128/JB.01550-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li X, Lockatell CV, Johnson DE, Mobley HLT. Identification of MrpI as the sole recombinase that regulates the phase variation of MR/P fimbria, a bladder colonization factor of uropathogenic Proteus mirabilis. MolMicrobiol. 2002;45:865–874. doi: 10.1046/j.1365-2958.2002.03067.x. [DOI] [PubMed] [Google Scholar]
- 168.Bahrani FK, Mobley HLT. Proteus mirabilis MR/P fimbriae: molecular cloning, expression, and nucleotide sequence of the major fimbrial subunit gene. J Bacteriol. 1993;175:457–464. doi: 10.1128/jb.175.2.457-464.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Båga M, Norgren M, Normark S. Biogenesis of E. coli Pap pili: PapH, a minor pilin subunit involved in cell anchoring and length modulation. Cell. 1987;49:241–251. doi: 10.1016/0092-8674(87)90565-4. [DOI] [PubMed] [Google Scholar]
- 170.Verger D, Miller E, Remaut H, Waksman G, Hultgren S. Molecular mechanism of P pilus termination in uropathogenic Escherichia coli. EMBO Rep. 2006;7:1228–1232. doi: 10.1038/sj.embor.7400833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li X, Mobley HLT. MrpB functions as the terminator for assembly of Proteus mirabilis mannose-resistant Proteus-like fimbriae. Infect Immun. 1998;66:1759–1763. doi: 10.1128/iai.66.4.1759-1763.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li X, Zhao H, Geymonat L, Bahrani F, Johnson DE, Mobley HLT. Proteus mirabilis mannose-resistant, Proteus-like fimbriae: MrpG is located at the fimbrial tip and is required for fimbrial assembly. Infect Immun. 1997;65:1327–1334. doi: 10.1128/iai.65.4.1327-1334.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 174.Kuehn MJ, Normark S, Hultgren SJ. Immunoglobulin-like PapD chaperone caps and uncaps interactive surfaces of nascently translocated pilus subunits. Proc Natl Acad Sci U S A. 1991;88:10586–10590. doi: 10.1073/pnas.88.23.10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Carnoy C, Moseley SL. Mutational analysis of receptor binding mediated by the Dr family of Escherichia coli adhesins. Mol Microbiol. 1997;23:365–379. doi: 10.1046/j.1365-2958.1997.2231590.x. [DOI] [PubMed] [Google Scholar]
- 176.Heras B, Shouldice SR, Totsika M, Scanlon MJ, Schembri MA, Martin JL. DSB proteins and bacterial pathogenicity. Nat Rev Microbiol. 2009;7:215–225. doi: 10.1038/nrmicro2087. [DOI] [PubMed] [Google Scholar]
- 177.Jansen AM, Lockatell V, Johnson DE, Mobley HLT. Mannose-resistant Proteus-like fimbriae are produced by most Proteus mirabilis strains infecting the urinary tract, dictate the in vivo localization of bacteria, and contribute to biofilm formation. Infection and Immunity. 2004;72:7294–7305. doi: 10.1128/IAI.72.12.7294-7305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rocha SP, Elias WP, Cianciarullo AM, Menezes MA, Nara JM, Piazza RM, Silva MR, Moreira CG, Pelayo JS. Aggregative adherence of uropathogenic Proteus mirabilis to cultured epithelial cells. FEMS Immunol Med Microbiol. 2007;51:319–326. doi: 10.1111/j.1574-695X.2007.00308.x. [DOI] [PubMed] [Google Scholar]
- 179.Johnson DE, Bahrani FK, Lockatell CV, Drachenberg CB, Hebel JR, Belas R, Warren JW, Mobley HLT. Serum immunoglobulin response and protection from homologous challenge by Proteus mirabilis in a mouse model of ascending urinary tract infection. Infection and Immunity. 1999;67:6683–6687. doi: 10.1128/iai.67.12.6683-6687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bahrani FK, Massad G, Lockatell CV, Johnson DE, Russell RG, Warren JW, Mobley HLT. Construction of an MR/P fimbrial mutant of Proteus mirabilis: role in virulence in a mouse model of ascending urinary tract infection. Infection and Immunity. 1994;62:3363–3371. doi: 10.1128/iai.62.8.3363-3371.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li X, Johnson DE, Mobley HLT. Requirement of MrpH for Mannose-Resistant Proteus-Like Fimbria-Mediated Hemagglutination by Proteus mirabilis. Inection and Immunity. 1999;67:2822–2833. doi: 10.1128/iai.67.6.2822-2833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cook SW, Mody N, Valle J, Hull R. Molecular cloning of Proteus mirabilis uroepithelial cell adherence (uca) genes. Infect Immun. 1995;63:2082–2086. doi: 10.1128/iai.63.5.2082-2086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tolson DL, Barrigar DL, McLean RJ, Altman E. Expression of a nonagglutinating fimbria by Proteus mirabilis. Infect Immun. 1995;63:1127–1129. doi: 10.1128/iai.63.3.1127-1129.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pellegrino R, Scavone P, Umpiérrez A, Maskell DJ, Zunino P. Proteus mirabilis uroepithelial cell adhesin (UCA) fimbria plays a role in the colonization of the urinary tract. Pathog Dis. 2013;67:104–107. doi: 10.1111/2049-632X.12027. [DOI] [PubMed] [Google Scholar]
- 185.Kuan L, Schaffer JN, Zouzias CD, Pearson MM. Characterization of 17 chaperone-usher fimbriae encoded by Proteus mirabilis reveals strong conservation. J Med Microbiol. 2014 doi: 10.1099/jmm.0.069971-0. doi:10.1099/jmm.0.069971-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Väisänen-Rhen V, Korhonen TK, Finne J. Novel cell-binding activity specific for N-acetyl-D-glucosamine in an Escherichia coli strain. FEBS Lett. 1983;159:233–236. doi: 10.1016/0014-5793(83)80453-0. [DOI] [PubMed] [Google Scholar]
- 187.Saarela S, Westerlund-Wikström B, Rhen M, Korhonen TK. The GafD protein of the G (F17) fimbrial complex confers adhesiveness of Escherichia coli to laminin. Infect Immun. 1996;64:2857–2860. doi: 10.1128/iai.64.7.2857-2860.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dorofeyev AE, Vasilenko IV, Rassokhina OA. Joint extraintestinal manifestations in ulcerative colitis. Dig Dis. 2009;27:502–510. doi: 10.1159/000233289. [DOI] [PubMed] [Google Scholar]
- 189.Lee KK, Harrison BA, Latta R, Altman E. The binding of Proteus mirabilis nonagglutinating fimbriae to ganglio-series asialoglycolipids and lactosyl ceramide. Can J Microbiol. 2000;46:961–966. [PubMed] [Google Scholar]
- 190.Ortaldo JR, Sharrow SO, Timonen T, Herberman RB. Determination of surface antigens on highly purified human NK cells by flow cytometry with monoclonal antibodies. J Immunol. 1981;127:2401–2409. [PubMed] [Google Scholar]
- 191.Saiman L, Prince A. Pseudomonas aeruginosa pili bind to asialoGM1 which is increased on the surface of cystic fibrosis epithelial cells. J Clin Invest. 1993;92:1875–1880. doi: 10.1172/JCI116779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zunino P, Sosa V, Allen AG, Preston A, Schlapp G, Maskell DJ. Proteus mirabilis fimbriae (PMF) are important for both bladder and kidney colonization in mice. Microbiology. 2003;149:3231–3237. doi: 10.1099/mic.0.26534-0. [DOI] [PubMed] [Google Scholar]
- 193.Massad G, Lockatell CV, Johnson DE, Mobley HLT. Proteus mirabilis fimbriae: construction of an isogenic pmfA mutant and analysis of virulence in a CBA mouse model of ascending urinary tract infection. Infection and Immunity. 1994;62:536–542. doi: 10.1128/iai.62.2.536-542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zunino P, Sosa V, Schlapp G, Allen AG, Preston A, Maskell DJ. Mannose-resistant Proteus-like and P. mirabilis fimbriae have specific and additive roles in P. mirabilis urinary tract infections. FEMS Immunol Med Microbiol. 2007;51:125–133. doi: 10.1111/j.1574-695X.2007.00285.x. [DOI] [PubMed] [Google Scholar]
- 195.Massad G, Bahrani FK, Mobley HLT. Proteus mirabilis fimbriae: identification, isolation, and characterization of a new ambient-temperature fimbria. Infection and Immunity. 1994;62:1989–1994. doi: 10.1128/iai.62.5.1989-1994.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Massad G, Fulkerson JF, Jr., Watson DC, Mobley HLT. Proteus mirabilis ambient-temperature fimbriae: cloning and nucleotide sequence of the atf gene cluster. Infect Immun. 1996;64:4390–4395. doi: 10.1128/iai.64.10.4390-4395.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zunino P, Geymonat L, Allen AG, Legnani-Fajardo C, Maskell DJ. Virulence of a Proteus mirabilis ATF isogenic mutant is not impaired in a mouse model of ascending urinary tract infection. FEMS Immunol Med Microbiol. 2000;29:137–143. doi: 10.1111/j.1574-695X.2000.tb01516.x. [DOI] [PubMed] [Google Scholar]
- 198.Bijlsma IG, van Dijk L, Kusters JG, Gaastra W. Nucleotide sequences of two fimbrial major subunit genes, pmpA and ucaA, from canine-uropathogenic Proteus mirabilis strains. Microbiology. 1995;141(Pt 6):1349–1357. doi: 10.1099/13500872-141-6-1349. [DOI] [PubMed] [Google Scholar]
- 199.Spurbeck RR, Stapleton AE, Johnson JR, Walk ST, Hooton TM, Mobley HLT. Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: contribution of Ygi and Yad fimbriae. Infect Immun. 2011;79:4753–4763. doi: 10.1128/IAI.05621-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Townsend SM, Kramer NE, Edwards R, Baker S, Hamlin N, Simmonds M, Stevens K, Maloy S, Parkhill J, Dougan G, Bäumler AJ. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect Immun. 2001;69:2894–2901. doi: 10.1128/IAI.69.5.2894-2901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wurpel DJ, Beatson SA, Totsika M, Petty NK, Schembri MA. Chaperone-usher fimbriae of Escherichia coli. PLoS One. 2013;8:e52835. doi: 10.1371/journal.pone.0052835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Snyder JA, Haugen BJ, Lockatell CV, Maroncle N, Hagan EC, Johnson DE, Welch RA, Mobley HLT. Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect Immun. 2005;73:7588–7596. doi: 10.1128/IAI.73.11.7588-7596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Li X, Rasko DA, Lockatell CV, Johnson DE, Mobley HLT. Repression of bacterial motility by a novel fimbrial gene product. EMBO Journal. 2001;20:4854–4862. doi: 10.1093/emboj/20.17.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Simms AN, Mobley HLT. PapX, a P fimbrial operon-encoded inhibitor of motility in uropathogenic Escherichia coli. Infect Immun. 2008 doi: 10.1128/IAI.00630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Reiss DJ, Mobley HLT. Determination of target sequence bound by PapX, repressor of bacterial motility, in flhD promoter using systematic evolution of ligands by exponential enrichment (SELEX) and high throughput sequencing. J Biol Chem. 2011;286:44726–44738. doi: 10.1074/jbc.M111.290684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chen YT, Peng HL, Shia WC, Hsu FR, Ken CF, Tsao YM, Chen CH, Liu CE, Hsieh MF, Chen HC, Tang CY, Ku TH. Whole-genome sequencing and identification of Morganella morganii KT pathogenicity-related genes. BMC Genomics. 2012;13(Suppl 7):S4. doi: 10.1186/1471-2164-13-S7-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 208.Meslet-Cladiere LM, Pimenta A, Duchaud E, Holland IB, Blight MA. In vivo expression of the mannose-resistant fimbriae of Photorhabdus temperata K122 during insect infection. J Bacteriol. 2004;186:611–622. doi: 10.1128/JB.186.3.611-622.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Allison SE, Silphaduang U, Mascarenhas M, Konczy P, Quan Q, Karmali M, Coombes BK. Novel repressor of Escherichia coli O157:H7 motility encoded in the putative fimbrial cluster OI-1. J Bacteriol. 2012;194:5343–5352. doi: 10.1128/JB.01025-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.He H, Snyder HA, Forst S. Unique organization and regulation of the mrx fimbrial operon in Xenorhabdus nematophila. Microbiology. 2004;150:1439–1446. doi: 10.1099/mic.0.26853-0. [DOI] [PubMed] [Google Scholar]
- 211.Leyton DL, Rossiter AE, Henderson IR. From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat Rev Microbiol. 2012;10:213–225. doi: 10.1038/nrmicro2733. [DOI] [PubMed] [Google Scholar]
- 212.Cotter SE, Surana NK, St Geme JW., 3rd Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 2005;13:199–205. doi: 10.1016/j.tim.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 213.Alamuri P, Mobley HLT. A novel autotransporter of uropathogenic Proteus mirabilis is both a cytotoxin and an agglutinin. Mol Microbiol. 2008;68:997–1017. doi: 10.1111/j.1365-2958.2008.06199.x. [DOI] [PubMed] [Google Scholar]
- 214.Flannery EL, Mody L, Mobley HLT. Identification of a modular pathogenicity island that is widespread among urease-producing uropathogens and shares features with a diverse group of mobile elements. Infect Immun. 2009;77:4887–4894. doi: 10.1128/IAI.00705-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Silverblatt FJ. Host-parasite interaction in the rat renal pelvis: a possible role for pili in the pathogenesis of pyelonephritis. J Exp Med. 1974;140:1696–1711. doi: 10.1084/jem.140.6.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hola V, Peroutkova T, Ruzicka F. Virulence factors in Proteus bacteria from biofilm communities of catheter-associated urinary tract infections. FEMS Immunol Med Microbiol. 2012;65:343–349. doi: 10.1111/j.1574-695X.2012.00976.x. [DOI] [PubMed] [Google Scholar]
- 217.Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 218.Uphoff TS, Welch RA. Nucleotide sequencing of the Proteus mirabilis calcium-independent hemolysin genes (hpmA and hpmB) reveals sequence similarity with the Serratia marcescens hemolysin genes (shlA and shlB). Journal of Bacteriology. 1990;172:1206. doi: 10.1128/jb.172.3.1206-1216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Swihart KG, Welch RA. Cytotoxic activity of the Proteus hemolysin HpmA. Infect Immun. 1990;58:1861–1869. doi: 10.1128/iai.58.6.1861-1869.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Cestari SE, Ludovico MS, Martins FH, da Rocha SP, Elias WP, Pelayo JS. Molecular detection of HpmA and HlyA hemolysin of uropathogenic Proteus mirabilis. Curr Microbiol. 2013;67:703–707. doi: 10.1007/s00284-013-0423-5. [DOI] [PubMed] [Google Scholar]
- 221.Weaver TM, Hocking JM, Bailey LJ, Wawrzyn GT, Howard DR, Sikkink LA, Ramirez-Alvarado M, Thompson JR. Structural and functional studies of truncated hemolysin A from Proteus mirabilis. J Biol Chem. 2009;284:22297–22309. doi: 10.1074/jbc.M109.014431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wassif C, Cheek D, Belas R. Molecular analysis of a metalloprotease from Proteus mirabilis. Journal of Bacteriology. 1995;177:5790. doi: 10.1128/jb.177.20.5790-5798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Belas R, Manos J, Suvanasuthi R. Proteus mirabilis ZapA metalloprotease degrades a broad spectrum of substrates, including antimicrobial peptides. Infect Immun. 2004;72:5159–5167. doi: 10.1128/IAI.72.9.5159-5167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Walker KE, Moghaddame-Jafari S, Lockatell CV, Johnson D, Belas R. ZapA, the IgA-degrading metalloprotease of Proteus mirabilis, is a virulence factor expressed specifically in swarmer cells. Mol Microbiol. 1999;32:825–836. doi: 10.1046/j.1365-2958.1999.01401.x. [DOI] [PubMed] [Google Scholar]
- 225.Chromek M, Slamova Z, Bergman P, Kovacs L, Podracka L, Ehren I, Hokfelt T, Gudmundsson GH, Gallo RL, Agerberth B, Brauner A. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat Med. 2006;12:636–641. doi: 10.1038/nm1407. [DOI] [PubMed] [Google Scholar]
- 226.Ganz T. Defensins in the urinary tract and other tissues. J Infect Dis. 2001;183(Suppl 1):S41–42. doi: 10.1086/318838. [DOI] [PubMed] [Google Scholar]
- 227.Phan V, Belas R, Gilmore BF, Ceri H. ZapA, a virulence factor in a rat model of Proteus mirabilis-induced acute and chronic prostatitis. Infect Immun. 2008;76:4859–4864. doi: 10.1128/IAI.00122-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Senior BW, Loomes LM, Kerr MA. The production and activity in vivo of Proteus mirabilis IgA protease in infections of the urinary tract. J Med Microbiol. 1991;35:203–207. doi: 10.1099/00222615-35-4-203. [DOI] [PubMed] [Google Scholar]
- 229.Carson L, Cathcart GR, Scott CJ, Hollenberg MD, Walker B, Ceri H, Gilmore BF. Comprehensive inhibitor profiling of the Proteus mirabilis metalloprotease virulence factor ZapA (mirabilysin). Biochimie. 2011;93:1824–1827. doi: 10.1016/j.biochi.2011.06.030. [DOI] [PubMed] [Google Scholar]
- 230.Zhao H, Li X, Johnson DE, Mobley HLT. Identification of protease and rpoN-associated genes of uropathogenic Proteus mirabilis by negative selection in a mouse model of ascending urinary tract infection. Microbiology. 1999;145(Pt 1):185–195. doi: 10.1099/13500872-145-1-185. [DOI] [PubMed] [Google Scholar]
- 231.Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Andrews SC, Robinson AK, Rodriguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 233.Piccini CD, Barbe FM, Legnani-Fajardo CL. Identification of iron-regulated outer membrane proteins in uropathogenic Proteus mirabilis and its relationship with heme uptake. FEMS Microbiol Lett. 1998;166:243–248. doi: 10.1111/j.1574-6968.1998.tb13897.x. [DOI] [PubMed] [Google Scholar]
- 234.Drechsel H, Thieken A, Reissbrodt R, Jung G, Winkelmann G. Alpha-keto acids are novel siderophores in the genera Proteus, Providencia, and Morganella and are produced by amino acid deaminases. J Bacteriol. 1993;175:2727–2733. doi: 10.1128/jb.175.9.2727-2733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Massad G, Zhao H, Mobley HLT. Proteus mirabilis amino acid deaminase: cloning, nucleotide sequence, and characterization of aad. J Bacteriol. 1995;177:5878–5883. doi: 10.1128/jb.177.20.5878-5883.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Reissbrodt R, Kingsley R, Rabsch W, Beer W, Roberts M, Williams PH. Iron-regulated excretion of alpha-keto acids by Salmonella typhimurium. J Bacteriol. 1997;179:4538–4544. doi: 10.1128/jb.179.14.4538-4544.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kingsley R, Rabsch W, Roberts M, Reissbrodt R, Williams PH. TonB-dependent iron supply in Salmonella by alpha-ketoacids and alpha-hydroxyacids. FEMS Microbiol Lett. 1996;140:65–70. doi: 10.1111/j.1574-6968.1996.tb08316.x. [DOI] [PubMed] [Google Scholar]
- 238.Himpsl SD, Pearson MM, Arewang CJ, Nusca TD, Sherman DH, Mobley HLT. Proteobactin and a yersiniabactin-related siderophore mediate iron acquisition in Proteus mirabilis. Mol Microbiol. 2010;78:138–157. doi: 10.1111/j.1365-2958.2010.07317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lima A, Zunino P, D'Alessandro B, Piccini C. An iron-regulated outer-membrane protein of Proteus mirabilis is a haem receptor that plays an important role in urinary tract infection and in in vivo growth. J Med Microbiol. 2007;56:1600–1607. doi: 10.1099/jmm.0.47320-0. [DOI] [PubMed] [Google Scholar]
- 240.Sabri M, Houle S, Dozois CM. Roles of the extraintestinal pathogenic Escherichia coli ZnuACB and ZupT zinc transporters during urinary tract infection. Infect Immun. 2009;77:1155–1164. doi: 10.1128/IAI.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Jacobsen SM, Lane MC, Harro JM, Shirtliff ME, Mobley HLT. The high-affinity phosphate transporter Pst is a virulence factor for Proteus mirabilis during complicated urinary tract infection. FEMS Immunol Med Microbiol. 2008;52:180–193. doi: 10.1111/j.1574-695X.2007.00358.x. [DOI] [PubMed] [Google Scholar]
- 242.O'May GA, Jacobsen SM, Longwell M, Stoodley P, Mobley HL, Shirtliff ME. The high-affinity phosphate transporter Pst in Proteus mirabilis HI4320 and its importance in biofilm formation. Microbiology. 2009;155:1523–1535. doi: 10.1099/mic.0.026500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Shi X, Zhu Y, Li Y, Jiang M, Lin Y, Qiu Y, Chen Q, Yuan Y, Ni P, Hu Q, Huang S. Genome sequence of Proteus mirabilis clinical isolate C05028. Genome Announc. 2014;2 doi: 10.1128/genomeA.00167-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Khalid MI, Teh LK, Lee LS, Zakaria ZA, Salleh MZ. Genome sequence of Proteus mirabilis strain PR03, isolated from a local hospital in Malaysia. Genome Announc. 2013;1 doi: 10.1128/genomeA.00327-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Pearson MM, Mobley HLT. The type III secretion system of Proteus mirabilis HI4320 does not contribute to virulence in the mouse model of ascending urinary tract infection. J Med Microbiol. 2007;56:1277–1283. doi: 10.1099/jmm.0.47314-0. [DOI] [PubMed] [Google Scholar]
- 246.Lloyd AL, Rasko DA, Mobley HLT. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J Bacteriol. 2007;189:3532–3546. doi: 10.1128/JB.01744-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Flannery EL, Antczak SM, Mobley HL. Self-transmissibility of the integrative and conjugative element ICEPm1 between clinical isolates requires a functional integrase, relaxase, and type IV secretion system. J Bacteriol. 2011;193:4104–4112. doi: 10.1128/JB.05119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Jacobsen SM, Shirtliff ME. Proteus mirabilis biofilms and catheter-associated urinary tract infections. Virulence. 2011;2:460–465. doi: 10.4161/viru.2.5.17783. [DOI] [PubMed] [Google Scholar]
- 249.Dumanski AJ, Hedelin H, Edin-Liljegren A, Beauchemin D, McLean RJ. Unique ability of the Proteus mirabilis capsule to enhance mineral growth in infectious urinary calculi. Infect Immun. 1994;62:2998–3003. doi: 10.1128/iai.62.7.2998-3003.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Holling N, Lednor D, Tsang S, Bissell A, Campbell L, Nzakizwanayo J, Dedi C, Hawthorne JA, Hanlon G, Ogilvie LA, Salvage JP, Patel BA, Barnes LM, Jones BV. Elucidating the genetic basis of crystalline biofilm formation in Proteus mirabilis. Infect Immun. 2014;82:1616–1626. doi: 10.1128/IAI.01652-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, Moran GJ, Nicolle LE, Raz R, Schaeffer AJ, Soper DE. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: A 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis. 2011;52:e103–120. doi: 10.1093/cid/ciq257. [DOI] [PubMed] [Google Scholar]
- 252.Ma KL, Wang CX. Analysis of the spectrum and antibiotic resistance of uropathogens in vitro: Results based on a retrospective study from a tertiary hospital. Am J Infect Control. 2013 doi: 10.1016/j.ajic.2012.09.015. doi:S0196-6553(12)01257-6 [pii] 10.1016/j.ajic.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 253.Schito GC, Naber KG, Botto H, Palou J, Mazzei T, Gualco L, Marchese A. The ARESC study: an international survey on the antimicrobial resistance of pathogens involved in uncomplicated urinary tract infections. Int J Antimicrob Agents. 2009;34:407–413. doi: 10.1016/j.ijantimicag.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 254.Bichler KH, Eipper E, Naber K, Braun V, Zimmermann R, Lahme S. Urinary infection stones. Int J Antimicrob Agents. 2002;19:488–498. doi: 10.1016/s0924-8579(02)00088-2. [DOI] [PubMed] [Google Scholar]
- 255.Adamus-Bialek W, Zajac E, Parniewski P, Kaca W. Comparison of antibiotic resistance patterns in collections of Escherichia coli and Proteus mirabilis uropathogenic strains. Mol Biol Rep. 2013 doi: 10.1007/s11033-012-2420-3. doi:10.1007/s11033-012-2420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Kaca W, Radziejewska-Lebrecht J, Bhat UR. Effect of polymyxins on the lipopolysaccharide-defective mutants of Proteus mirabilis. Microbios. 1990;61:23–32. [PubMed] [Google Scholar]
- 257.Li X, Lockatell CV, Johnson DE, Lane MC, Warren JW, Mobley HLT. Development of an intranasal vaccine to prevent urinary tract infection by Proteus mirabilis. Infection and Immunity. 2004;72:66–75. doi: 10.1128/IAI.72.1.66-75.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Brumbaugh AR, Mobley HLT. Preventing urinary tract infection: progress toward an effective Escherichia coli vaccine. Expert Rev Vaccines. 2012;11:663–676. doi: 10.1586/erv.12.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Scavone P, Rial A, Umpierrez A, Chabalgoity A, Zunino P. Effects of the administration of cholera toxin as a mucosal adjuvant on the immune and protective response induced by Proteus mirabilis MrpA fimbrial protein in the urinary tract. Microbiol Immunol. 2009;53:233–240. doi: 10.1111/j.1348-0421.2009.00111.x. [DOI] [PubMed] [Google Scholar]
- 260.Jones RJ. Oral vaccination against Proteus mirabilis. Br J Exp Pathol. 1976;57:395–399. [PMC free article] [PubMed] [Google Scholar]
- 261.Moayeri N, Collins CM, O'Hanley P. Efficacy of a Proteus mirabilis outer membrane protein vaccine in preventing experimental Proteus pyelonephritis in a BALB/c mouse model. Infect Immun. 1991;59:3778–3786. doi: 10.1128/iai.59.10.3778-3786.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Pellegrino R, Galvalisi U, Scavone P, Sosa V, Zunino P. Evaluation of Proteus mirabilis structural fimbrial proteins as antigens against urinary tract infections. FEMS Immunol Med Microbiol. 2003;36:103–110. doi: 10.1016/S0928-8244(03)00103-2. [DOI] [PubMed] [Google Scholar]
- 263.Alamuri P, Eaton KA, Himpsl SD, Smith SN, Mobley HLT. Vaccination with Proteus toxic agglutinin, a hemolysin-independent cytotoxin in vivo, protects against Proteus mirabilis urinary tract infection. Infect Immun. 2009;77:632–641. doi: 10.1128/IAI.01050-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Scavone P, Sosa V, Pellegrino R, Galvalisi U, Zunino P. Mucosal vaccination of mice with recombinant Proteus mirabilis structural fimbrial proteins. Microbes Infect. 2004;6:853–860. doi: 10.1016/j.micinf.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 265.Alteri CJ, Hagan EC, Sivick KE, Smith SN, Mobley HLT. Mucosal immunization with iron receptor antigens protects against urinary tract infection. PLoS Pathog. 2009;5:e1000586. doi: 10.1371/journal.ppat.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Scavone P, Miyoshi A, Rial A, Chabalgoity A, Langella P, Azevedo V, Zunino P. Intranasal immunisation with recombinant Lactococcus lactis displaying either anchored or secreted forms of Proteus mirabilis MrpA fimbrial protein confers specific immune response and induces a significant reduction of kidney bacterial colonisation in mice. Microbes Infect. 2007;9:821–828. doi: 10.1016/j.micinf.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 267.Scavone P, Umpierrez A, Maskell DJ, Zunino P. Nasal immunization with attenuated Salmonella Typhimurium expressing an MrpA-TetC fusion protein significantly reduces Proteus mirabilis colonization in the mouse urinary tract. J Med Microbiol. 2011;60:899–904. doi: 10.1099/jmm.0.030460-0. [DOI] [PubMed] [Google Scholar]
- 268.Siddiq DM, Darouiche RO. New strategies to prevent catheter-associated urinary tract infections. Nat Rev Urol. 2012;9:305–314. doi: 10.1038/nrurol.2012.68. [DOI] [PubMed] [Google Scholar]
- 269.Levering V, Wang Q, Shivapooja P, Zhao X, López GP. Soft Robotic Concepts in Catheter Design: an On-Demand Fouling-Release Urinary Catheter. Adv Healthc Mater. 2014 doi: 10.1002/adhm.201400035. doi:10.1002/adhm.201400035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Stickler DJ, Jones SM, Adusei GO, Waters MG, Cloete J, Mathur S, Feneley RC. A clinical assessment of the performance of a sensor to detect crystalline biofilm formation on indwelling bladder catheters. BJU Int. 2006;98:1244–1249. doi: 10.1111/j.1464-410X.2006.06562.x. [DOI] [PubMed] [Google Scholar]
- 271.Malic S, Waters MG, Basil L, Stickler DJ, Williams DW. Development of an “early warning” sensor for encrustation of urinary catheters following Proteus infection. J Biomed Mater Res B Appl Biomater. 2012;100:133–137. doi: 10.1002/jbm.b.31930. [DOI] [PubMed] [Google Scholar]