Abstract
Although considerable progress has been made toward understanding glioblastoma biology through large-scale genetic and protein expression analyses, little is known about the underlying metabolic alterations promoting their aggressive phenotype. We conducted global metabolomic profiling on patient-derived glioma specimens and identified specific metabolic programs differentiating low- and high-grade tumors, with the metabolic signature of glioblastoma reflecting accelerated anabolic metabolism. When coupled with transcriptional profiles, we identified the metabolic phenotype of the mesenchymal subtype to consist of accumulation of the glycolytic intermediate phosphoenolpyruvate and decreased pyruvate kinase activity. Unbiased hierarchical clustering of metabolomic profiles identified three subclasses, which we term energetic, anabolic, and phospholipid catabolism with prognostic relevance. These studies represent the first global metabolomic profiling of glioma, offering a previously undescribed window into their metabolic heterogeneity, and provide the requisite framework for strategies designed to target metabolism in this rapidly fatal malignancy.
Introduction
The World Health Organization (WHO) classifies glioma into grades 1 to 4, based on abundance of atypical cells, mitoses, endothelial proliferation, and necrosis. Tumor grade plays a central role in prognosis and guides clinical management. For example, patients with grade 1 tumors are typically cured following surgical resection, whereas patients diagnosed with grade 4 tumors, termed glioblastoma, have a median survival of approximately 1 year despite aggressive multimodality treatment consisting of surgery, radiotherapy, and chemotherapy. Although grade 2 gliomas typically have a better prognosis than higher grader tumors and are often categorized as benign, this is somewhat of a misnomer, as these tumors are rarely cured and typically transform to higher grade tumors (1).
Considerable progress has been made in understanding the underlying biology of gliomas. For example, common molecular alterations identified in low-grade oligodendrogliomas and astrocytomas are allelic loss of 1p and 19q and mutations in p53, respectively, whereas grade 3 and 4 tumors typically are driven by alterations in phosphoinositide 3-kinase (PI3K), EGF receptors (EGFR), VEGF, and PTEN signaling (1). Furthermore, through whole genome sequencing, recent data presented by The Cancer Genome Atlas Research Network both reinforced these previously identified mutations and highlighted tumor heterogeneity, identifying aberrant signaling through the RTK/RAS/PI3K, p53, and retinoblastoma (RB) pathways being central for glioblastoma development (2).
Despite these advancements in our understanding of the upstream events signaling tumorigenesis, relationships between underlying metabolic alterations and mechanisms promoting the observed aggressive phenotype in these tumors remain unclear. The seminal observation made by Otto Warburg nearly a century ago (3, 4), described aerobic glycolysis, that is, a high fermentative metabolism of glucose resulting in production and release of lactic acid, even in the presence of adequate oxygen. We have proposed that acid production provides a competitive advantage for invasive cancers (5), yet a definitive explanation for why tumor cells metabolize glucose through the seemingly inefficient process of aerobic glycolysis continues to be elusive. Nonetheless, its clear relevance to cancer biology is evident with the widespread application of 2[18F]fluoro-2-deoxy-D-glucose–positron emission tomography (18-FDG-PET) imaging, which can predict histologic grade in glioma with relatively high accuracy. Grade 2 tumors typically show low specific uptake values (SUV), whereas high-grade tumors (grade 3 and 4) show high SUVs (6). Hence, there are grade-associated changes in glioma metabolism, yet these have not been extensively characterized.
Metabolomics is the global quantitative assessment of endogenous metabolites within a biologic system, taking into account genetic regulation, altered kinetic activity of enzymes, and changes in metabolic reactions. Thus, compared with genomics and proteomics, metabolomics reflects changes in phenotype, and therefore function (7, 8). On the basis of the clear metabolic shift between low-grade and high-grade tumors, we evaluated global metabolomic profiles in grade 2, 3, and 4 gliomas to comprehensively evaluate metabolic underpinnings of grade-specific changes in glioma beyond that of glucose consumption. Furthermore, metabolomic signatures were coupled with gene expression profiles to evaluate for subtype specific changes in tumor metabolism and provide insight into upstream signaling networks that may be driving the observed metabolic alterations.
Materials and Methods
Tumor samples and patient characteristics
A summary of all glioma cases studied is included in Supplementary Table S1. All surgeries were conducted at the H. Lee Moffitt Cancer Center and Research Institute (Tampa, FL), and tissue was obtained from the Moffitt Cancer Center Tissue Core Facility. All of the grade 3 and 4 tumors used in this analysis were newly diagnosed malignancies. Tumors were fresh-frozen and their integrity and histology confirmed by a staff pathologist before aliquoting samples. Clinical outcomes were obtained through the Cancer Center Tumor Registry. Institutional Review Board/Human Subjects approval was obtained for this retrospective study.
Sample preparation and metabolic profiling
Metabolomic studies were conducted at Metabolon Inc. using a nontargeted platform that enables relative quantitative analysis of a broad spectrum of molecules with a high degree of confidence (9). The metabolic profiling analysis combined 3 independent platforms, ultrahigh performance liquid chromatography/tandem mass spectrometry (UHPLC/MS-MS2) optimized for basic species, UHPLC/MS-MS2 optimized for acidic species, and gas chromatography/mass spectrometry (GC/MS). Samples were processed essentially as described previously (9, 10). Tissue samples were homogenized in a minimum volume of water and 100 μL withdrawn for subsequent analyses. Using an automated liquid handler (Hamilton LabStar), protein was precipitated from the homogenate with methanol that contained 4 standards to report on extraction efficiency. The resulting supernatant was split into equal aliquots for analysis on the 3 platforms. Experimental samples and controls were randomized across platform run days. For UHPLC/MS/MS2 analysis, aliquots were separated using a Waters Acquity UPLC (Waters) and analyzed using an LTQ mass spectrometer (Thermo Fisher Scientific, Inc.), which consisted of an electrospray ionization (ESI) source and linear ion-trap (LIT) mass analyzer. The MS instrument scanned 99 to 1,000 m/z and alternated between MS and MS2 scans using dynamic exclusion with approximately 6 scans per second. Derivatized samples for GC/MS were separated on a 5% phenyldimethyl silicone column with helium as the carrier gas and a temperature ramp from 60°C to 340°C and then analyzed on a Thermo-Finnigan Trace DSQ MS (Thermo Fisher Scientific, Inc.) operated at unit mass resolving power with electron impact ionization and a 50 to 750 atomic mass unit scan range. Metabolites were identified by automated comparison of the ion features in the experimental samples with a reference library of chemical standard entries that included retention time, molecular weight (m/z), preferred adducts, and in-source fragments as well as associated MS spectra, and were curated by visual inspection for quality control using software developed at Metabolon (11). Metabolomic subtypes were generated using unsupervised hierarchical clustering on GeneCluster 3.0. Metabolite concentrations (excluding xenobiotics) were log transformed and clustering was conducted with uncentered correlation and single linkage metrics, and visualized using Java TreeView.
For statistical analyses and data display purposes, any missing values were assumed to be below the limits of detection, and these values were imputed with the compound minimum (minimum value imputation). Statistical analysis of log-transformed data was conducted using “R” (http://cran.r-project.org/), which is a freely available, open-source software package. Welch t tests were conducted to compare data between experimental groups. Multiple comparisons were accounted for by estimating the false discovery rate (FDR) using q values (12). Random forest (RF) analysis (13) was carried out on untransformed data. When data from the glioblastoma grade categories were used in comparisons for classification by RF, the number of in-bag samples was set to 50% of smallest subgroup to account for unbalanced group sizes, with 50,000 trees. RF analysis was conducted using the R-package “randomForest” (14).
Expression transcriptional subtyping
Total RNA was extracted from snap-frozen malignant glioma tissue using magnetic binding beads for cDNA and Qiagen RNeasy kits for cRNA purification. The final in vitro transcription incorporated biotin moieties that were later labeled with phycoerythrin. Samples were fragmented to improve hybridization sensitivity and consistency. The labeled molecules were biotinylated-cRNA. GeneChipmicroarrays were loaded with the fragmented target sample/hybridization buffer mix using standard techniques. Arrays were hybridized for 18 hours at 45°C with vigorous mixing. Unbound sample was removed and staining was accomplished through the binding of streptavidin-conjugated phycoerythrin to the hybridized target. Excess label was removed. Washing and staining steps were carried out by the Affymetrix FS450 fluidics station using standard protocols. Arrays were scanned using a GeneChip Scanner 3000 7G with a 48 array autoloader. Samples were hybridized to a Affymetrix-based chip designed by Merck. A total of 35 probesets from the Merck chip that most closely matched the signature genes described by Phillips and colleagues (15) were clustered using GeneCluster 3.0 and visualized by Java Tree-View 1.1.6. The 3 distinct clusters were evaluated for sample membership with respect to 3 glioma subtypes; defined as proliferative, mesenchymal, or proneural. Selection of differentially expressed genes was done using significance analysis of microarrays (SAM; 16). Principal component analysis (PCA) was conducted in the Evince software package. GeneGO Meta-Core was used to identify significant biologic pathways.
Pyruvate kinase activity assay
Pyruvate kinase (PK) activity was measured using a commercially available kit according to the manufacturer recommendations (BioVision). Briefly, tissues were homogenized and extracted with assay buffer. Three microgram of protein was used to ensure that the reading was within the linear range of the standard curve. For the colorimetric assay, optical density (OD) was measured at 570 nm at T1 to read A1 and measured again at T2 after incubating the reaction at 25°C for 10 and 20 minutes. PK activity was calculated by applying the ΔA (A2 – A1) to the standard curve to yield nmol of pyruvate generated between T1 and T2 by PK in the reaction wells and expressed in mU/mL.
Western blot analysis
Western blot analyses were conducted as previously described (17) using antibodies against PKM2 (Cell Signaling; 1:3,000) and β-actin (Sigma-Aldrich; 1:20,000). Blots were quantified using ImageJ (NIH), and the PKM2 expression of individual samples were normalized to loading control (β-actin).
Statistics
Statistics involved with metabolomic profiling are as described earlier. Estimates of overall survival were evaluated using the Kaplan–Meier product limit method and compared using a Wilcoxon log-rank test with SAS version 9.1.3 (SAS Institute). Student t test was used for box-plot comparisons.
Results
Global metabolic profiles distinguish glioma tumor grades
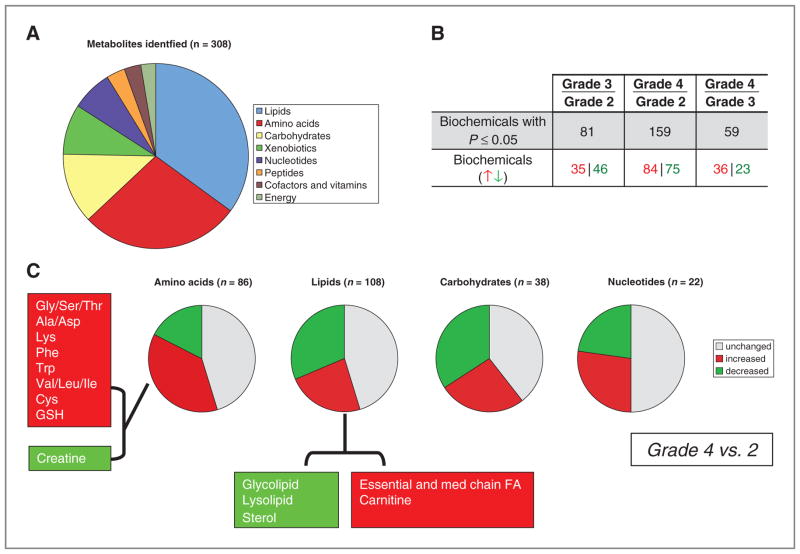
Global metabolomic profiling was conducted using a combination of high-throughput LC- and GC–based MS on a total of 69 fresh-frozen glioma specimens surgically resected at the H. Lee Moffitt Cancer Center (18 grade 2, 18 grade 3, and 33 grade 4; specific histologies are provided in Supplementary Table S1). From a metabolomic library consisting of more than 2,000 purified standards, a total of 308 named biochemicals were detected. The distribution of metabolic pathways identified is presented in Fig. 1A, with a majority of metabolites involved in lipid, amino acid, and carbohydrate metabolism. Following log transformation and imputation with minimum observed values for each compound, Welch 2-sample t tests were used to identify biochemicals that differed significantly between histologic grades. Summaries of the numbers of biochemicals that achieve statistical significance (P ≤ 0.05) are provided in Fig. 1B, with the largest number of significant metabolic changes observed between grade 2 and grade 4 gliomas. A summary of the metabolic pathways differentiating grade 4 and 2 tumors is presented in Fig. 1C. There was a significant increase in several metabolites involved in amino acid metabolism in grade 4 tumors, including glutathione and tryptophan. In addition, a decrease in creatine was also shown, which is consistent with magnetic resonance spectroscopy data for these highly proliferating tumors (18). There seemed to be a significant change in lipid metabolism in grade 4 tumors, with notable decreases in glycolipids, lysoplipids (which include derivatives of phosphocholine and phosphoethanolamine), and sterols, and increases in essential and medium chain fatty acids and metabolites associated with carnitine metabolism. Although grade 4 tumors seem to show a global decrease in carbohydrate metabolites, significant increases in key metabolic intermediaries phosphoenolpyruvate (PEP) and 3-phosphoglycerate (3-PG) were observed. In addition, a signature of increasingly altered nucleotide metabolism was also evident in grade 4 tumors, primarily involving pyrimidine catabolism.
Figure 1.
Global metabolomic profiling in glioma identifies grade specific metabolic changes. A, a combination of high-throughput LC- and GC–based MS was conducted on a total of 69 fresh-frozen glioma specimens (grades and histologies are provided in Supplementary Table S1). From a metabolomic library consisting of more than 2,000 purified standards, a total of 308 named biochemicals were detected, and the metabolic pathways that the identified metabolites reside in are presented. B, following log transformation and imputation with minimum observed values for each compound, Welch 2-sample t tests were used to identify biochemicals that differed significantly between histologic grades. Summaries of the numbers of biochemicals that achieve statistical significance (P ≤ 0.05) are provided. C, a summary of the metabolic pathways differentiating grade 4 and 2 tumors, with red and green indicating a statistically significant increase and decrease in identified metabolic pathways, respectively. Gly, glycine; Ser, serine; Thr, threonine; Ala, alanine; Asp, aspartate; Lys, lysine; Phe, phenylalanine, Trp, tryptophan; Val, valine; Leu, leucine; Ile, isoleucine; Cys, cysteine; GSH, glutathione; FA, fatty acids.
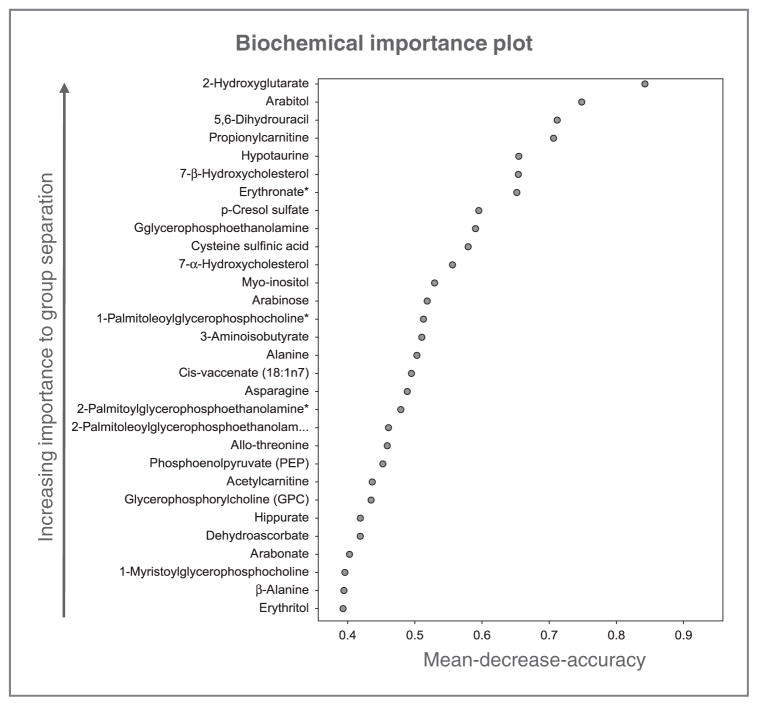
RF classifier models were developed to determine the capacity of global metabolic profiles to differentiate between tumor grades and to identify biochemicals important to the classification. The RF analyses yielded an overall predictive accuracy of 71% for classifying samples among groups (Supplementary Fig. S1). By this method, grade 3 tumors were poorly distinguished from both grade 2 and 4 tumors, suggesting progressive and overlapping metabolic changes during glioma tumorigenesis, whereas grade 2 and 4 tumors were best distinguished in the RF, reinforcing the important role altered metabolism plays in the aggressive phenotype associated with glioblastoma. In addition to producing a metric of predictive accuracy, RF analyses produced a prioritized list of biochemicals ranked in order of their importance to the classification scheme. To provide insight into grade-associated differences in global metabolism, the top 30 biochemicals for the RF classification scheme are provided in Fig. 2. In this analysis, 2-hydroxyglutarate (2-HG) emerged as the top-ranked biochemical for tumor grade classification. Several recent seminal studies have identified mutations in the metabolic enzyme isocitrate dehydrogenase 1 (IDH1) unique to low-grade glioma, resulting in structural changes allowing a new ability of the enzyme to catalyze the NADPH-dependent reduction of α-ketoglutarate to 2-HG (19–22). In addition to validating our described methodologies, identifying a significant accumulation of 2-HG in low-grade glioma specimens relative to its global metabolic profile further supports its potential role as an oncometabolite in this tumor.
Figure 2.
RF analysis identifies key metabolites differentiating glioma grade. RF analysis was conducted to determine the capacity of global metabolic profiles to classify samples between tumor grades and to identify biochemicals important to the classification; the top 30 biochemicals for the RF classification scheme are provided.
The metabolic signature of glioblastoma reflects accelerated anabolic metabolism
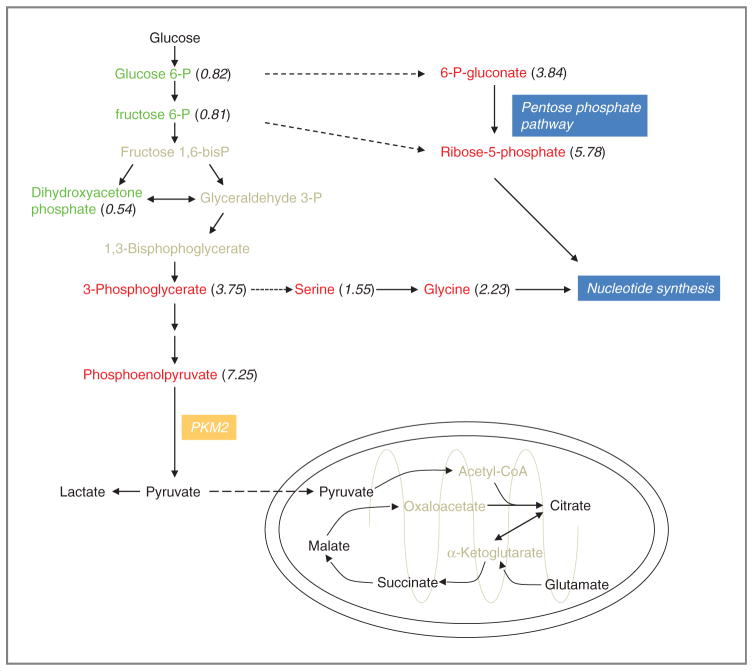
As the majority of our understanding of aberrant metabolism associated with tumorigenesis involves altered carbohydrate metabolism, we focused on alterations in glucose metabolism as a function of tumor grade. An evaluation of metabolites involved in glycolysis and the oxidative energy metabolism of the tricarboxylic acid (TCA) cycle/oxidative phosphorylation (23), provided in Fig. 3, identified accumulation of glycolytic intermediates as a key metabolic alteration in high-grade glioma. As glucose is taken up by cells, it is phosphorylated by hexokinase to glucose-6-phosphate (G6P), which is metabolized via either the Embden–Meyerhof (E–M; glycolysis or conversion of glucose to pyruvate) or the pentose phosphate pathways. In our analysis, progressively higher levels of triose phosphate glycolytic intermediates, including 3-PG and the penultimate intermediate, PEP, are more than 7-fold higher in grade 4 tumors over grade 2 and were among the top biochemicals in the RF importance plot for classification by tumor grade. To more definitively determine if these concerted changes represented a biologically relevant alteration in the metabolic pathway rather than independent events, we evaluated the concordance of 3-PG and PEP within individual samples. This analysis resulted in a Pearson’s r correlation coefficient of 0.96 (Supplementary Fig. S2), supporting the conclusion that increased levels of these 2 metabolites represent a clear metabolic shift in high-grade glioma and the implicit strength of global pathway analysis offered by metabolomics. Although absolute flux of metabolites cannot be definitively made from single steady-state measurements, the accumulation of PEP and 3-PG in grade 4 tumors does suggest the potential for diversion of glycolytic carbon from the E–M mediated generation of ATP into alternate pathways for macromolecule biosynthesis. Two important glycolytic shunts are through the pentose phosphate pathway and serine metabolism. The pentose phosphate shunt, by production of NADPH and ribose-5-phosphate (R5P) contributes to nucleotide biosynthesis, generates reducing equivalents essential to anabolic metabolism, and modulates DNA methylation (24–28). Accordingly, the metabolomic signature of grade 4 tumors consisted of an accumulation of key metabolites in these pathways (Fig. 3), including 6PG (3.84-fold increase), R5P (5.78-fold increase), serine (1.55-fold increase), and glycine (2.23-fold increase).
Figure 3.
The metabolic signature of glioblastoma reflects accelerated anabolic metabolism. Schematic of metabolites involved in carbohydrate metabolism, comparing grade 4 with grade 2 glioma. Metabolites in red reflect an increase accumulation in grade 4 tumors; green, decrease; black, no change; and gray, not identified in this analysis. Ratios were generated (in parenthesis) by normalizing the individual metabolite concentration to the median concentration of the respective metabolite obtained from all samples.
The metabolic phenotype of the mesenchymal subtype involves PEP accumulation and decreased PK activity
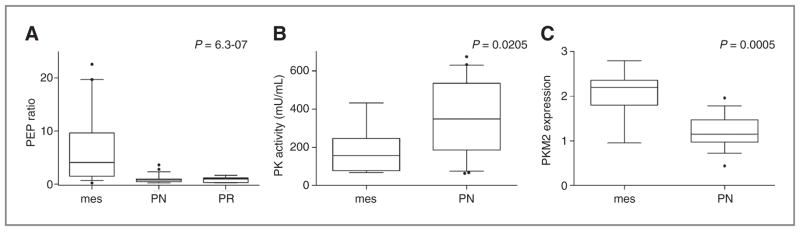
Although malignant glioma is defined as grade 3 or 4 based on histopathologic criteria, clear molecular heterogeneity has been uncovered within these tumor grades. Phillips and colleagues recently described 3 specific subtypes of malignant glioma based on expression profiles associated with distinct molecular signatures and clinical outcome, termed proneural (PN), proliferative (PR), and mesenchymal (15). To determine if metabolic phenotypes were associated with specific molecular subtypes in malignant glioma, global expression profiling followed by subtype designation was conducted on glioma samples. A majority of the grade 3 tumors were PN (67%), whereas the distribution of glioblastoma subtypes were 50% mesenchymal, 20% PR, and 30% PN, which was consistent with previous reports (15; Supplementary Fig. S3). Coupling metabolomic data with expression profiles revealed that the accumulation of PEP strongly correlated with the mesenchymal subtype (P = 6.3 × 10−7; Fig. 4A). Notably, 3 of the 4 grade 3 tumors that were molecularly classified as mesenchymal had significantly elevated PEP. In all, any tumor with a 4-fold or greater accumulation of PEP was invariably mesenchymal (n = 10).
Figure 4.
The mesenchymal subtype of malignant glioma is characterized by PEP accumulation and altered PKM2 expression and activity. Malignant glioma tumors (n = 51) were classified as mesenchymal (mes; n = 20), proneural (PN; n = 22), and proliferative (PR; n = 9) based on their transcriptional profiles (Supplementary Fig. S3). A, box-plots were generated from the PEP ratios of tumor clustering to individual subtypes. Band C, of the initial 51 malignant glioma samples used for global metabolomic and transcriptional profiling, 23 samples had sufficient tissue remaining to evaluate for differential PK activity and PKM2 expression between subtypes (mes, n = 8; PN, n = 15). PK activity was determined using a calorimetric-based assay and PKM2 expression was determined by Western blot analysis, with expression quantified relative to β-actin.
Accumulation of PEP could occur through reduced activity of PK. Isoenzyme variations of PK have been documented to play a role in the diversion of glycolytic metabolites. Specifically, there are 4 PK isoenzymes (M1, M2, L, and R) that differ in their kinetic properties and distribution among cells and tissues. PKM1 is found in the vast majority of cells, whereas PKM2 is abundant during embryogenesis, in selected differentiated tissues, and is the predominant form found in cancer cells (24, 29, 30). PKM2 can be further regulated by tyrosine kinase phosphorylation, which can switch PKM2 from its more active tetrameric form with high affinity toward its substrate PEP, to a less active dimer form, which favors the diversion of trioses toward synthetic processes, such as lipid and amino acid biosynthesis (24, 27, 30, 31). Hence, we determined if PK activity was correlated with increased PEP accumulation associated with the mesenchymal subtype. Of the initial tumor specimens evaluated, 26 had tissue evaluable for enzyme analysis. Of these, only 3 tumors clustered in the proliferative subtype; therefore, further analysis on this subtype was not conducted. As shown in Fig. 4B, the mesenchymal subtype had a significantly decreased PK activity (P = 0.0205) compared with the proneural group, suggesting a mechanism for the observed metabolic phenotype of PEP accumulation. Interestingly, although PK activity was observed to be lower in the mesenchymal subtype, overall expression of PKM2 was significantly increased (P = 0.0005) when compared with malignant gliomas clustering in the proneural subtype (Fig. 4C).
Global metabolism reveals metabolic signatures in malignant glioma
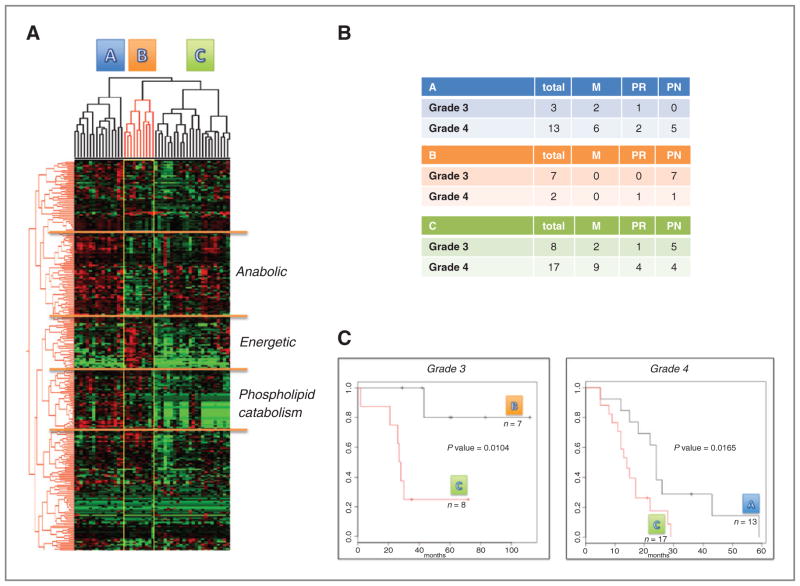
To determine if specific malignant glioma subtypes could be identified on the basis of the global metabolomic profiles, unbiased hierarchical clustering was conducted. Three distinct subgroups (A, B, and C) were identified (Fig. 5A), which were defined by 3 unique metabolomic signatures. Metabolites comprising the individual profiles that showed statistical significance in malignant glioma are provided in Supplementary Table S2. Although each profile consisted of a diverse set of metabolites, we applied the terms energetic, anabolic, and phospholipid catabolism to describe the individual profiles based on the characteristics of specific metabolites found therein. Specifically, subgroup B was defined by a classic glycolytic or energetic profile that consisted of an accumulation of upstream intermediates of glycolysis, including G6P and fructose-6-phosphate. Subgroup A was mutually exclusive to subgroup B, with decreases in these glycolytic metabolites, and elevated levels of metabolites typically associated with diverted glycolytic intermediates and anabolic metabolism, including PEP, 3-PG, and 6-PG. Notably, these also represent key metabolites differentiating grade 4 and 2 tumors, as described in Fig. 3. Other interesting metabolites in this list included those involved in serine, carnitine, tryptophan, and essential and long-chain fatty acid metabolism. Subgroup C had a similar profile as subgroup A, with the additional metabolites of long-chain fatty acid and lysolipid catabolism. Hence, this subtype was denoted as phospholipid catabolism. Both subgroups A and C primarily consisted of grade 4 (n = 30/33) tumors, whereas the majority of tumors that clustered in subgroup B (n = 7/9) were grade 3 gliomas, further supporting that divergence of glycolytic carbons plays an important role in the metabolic switch involved in glioblastoma. Although subgroup B consisted largely of tumors clustering to the PN subtype, these metabolic profiles were otherwise largely independent from the transcriptional signatures (Fig. 5B).
Figure 5.
Global metabolism reveals metabolic signatures in malignant glioma. A, unsupervised, hierarchical clustering was conducted on global metabolomic profiles generated in malignant glioma (n = 51). B, glioma grade and transcriptional subtypes of tumors clustering to the identified metabolomic subtypes. C, Kaplan–Meier estimates for overall survival based on metabolomic subtype.
We then investigated whether these identified metabolic subtypes were clinically relevant by evaluating the outcome of patients from individual subgroups. As grade is a clear prognostic factor in malignant glioma, grade 3 and grade 4 tumors were evaluated independently. Our initial evaluation compared grade 3 tumors with subgroups B and C (n = 7 and 8, respectively). Subgroup A was not analyzed because of low incidence (n = 3). We hypothesized that grade 3 patients with subgroup C metabolic profiles would have a worse outcome, because it is more consistent with the profile observed in grade 4 tumors. Consistent with our predictions, patients with grade 3 tumors that clustered in subgroup B had a significantly improved overall survival (Fig. 5C; P = 0.0104), with median survival not being reached, compared with a median survival of approximately 28 months for grade 3 tumors clustered in subgroup C. We next compared grade 4 tumors that clustered in subgroup A (n = 13) with C (n = 17). Interestingly, in contrast to transcriptional subtypes, metabolomic subtypes identified prognostic relevance in grade 4 tumors, with tumors clustering in subgroup A showing a significantly improved median survival of 24.4 months versus 14.7 months in tumors clustering to subgroup C (P = 0.0165). Notably, subgroup C had the worst outcome, independent of grade.
We then investigated whether specific signaling networks driving individual metabolomic subtypes could be identified. Using transcriptional profiles from individual tumors, glioma grade–specific comparisons were made similar to those conducted in the survival analysis, including comparisons between grade 3 tumors clustering in subgroups B and C and grade 4 tumors clustering between A and C. Interestingly, statistical analysis of microarrays (SAM) analysis of global transcriptional profiles for all of these groupings identified no apparent genes that were significantly differentially expressed within a reasonable FDR (5%). As a proof-of-concept, we conducted similar analyses using SAM comparing transcriptional profiles between grade 2 and grade 4 tumors. These studies identified several thousand probesets at a low FDR (specifically 1,970 genes at a 2-fold or more change and a 0% FDR), which encompassed several well-known pathways differentiating grade 2 and 4 tumors, including anti-apoptotic, signal transducer and activator of transcription (STAT), extracellular signal–regulated kinase (ERK), endoplasmic reticulum (ER) stress, WNT, and hedgehog signaling pathways.
Further analysis was conducted to examine the relationship between the 3 identified metabolic profiles (anabolic, energetic, and phospholipid catabolism) and the existing transcriptional subtypes. By using PCA on each metabolic profile and plotting the first 2 components, there was no discernable separation between transcriptional subtypes or grade (Supplementary Fig. S4). To determine if any genes were correlated to metabolic profiles, we examined the correlation between the first PC of each metabolic profile and individual gene expression. Only weak correlation was observed (max R2; anabolic = 0.53; energetic = 0.53; phospholipid catabolism = 0.31). Finally, we asked if there were gene expression differences among the 3 metabolic subtypes identified. Using the Kruskal–Wallis test for differences among the 3 subtypes and applying a FDR filter of 5%, we identified 2,510 probesets (representing 1,605 genes) significantly different (Supplementary Table S3). Post hoc tests for pair-wise differences indicate that 85% of the differences are driven by cluster B (Supplementary Table S3). Mapping these genes onto biologic pathways using GeneGO MetaCore, we identified several significant pathways including cell adhesion and cytoskeleton remodeling (see Supplementary Table S4 for full list of pathways identified). Taken together, this cross-platform analysis supports the concept that metabolomic profiles may provide a unique insight into the underlying biology of brain tumors beyond that recognized from traditional transcriptional signatures.
Discussion
Here, we describe, for the first time, global metabolomic signatures in glioma, which provide insight into their underlying biology that seems to have prognostic significance. An enhanced biosynthetic capacity from divergence of glycolytic carbons has been proposed as an important metabolic phenotype associated with tumorigenesis to provide the requisite genome, proteins, and lipids for these rapidly dividing cells, however, this study represents one of the first to provide support for this adaptive process in human tumors. One important mechanism tumors have adapted to allow for glycolytic shunting involves modulation of PKM2 activity. By converting from its highly active tetramer form that favors the conversion of PEP to pyruvate, to its less active dimer, upstream intermediates including PEP and 3-PG accumulate, increasing substrate availability for alternate pathways important for rapidly dividing cells (24, 27, 28, 31). Here, we identify some of the highest ranking metabolites differentiating grade 4 from grade 2 gliomas to be accumulation of PEP and 3-PG. Interestingly, within high-grade gliomas, integrative metabolomic, genomic, and enzyme affinity assays identified PEP accumulation and decreased PK activity highly correlated with the mesenchymal subtype, representing a particularly aggressive subgroup in this malignancy. It should be noted that extrapolations of absolute flux cannot be made from single steady-state measurements, although inferences about relative flux can be made using the cross-over theorem. Notably, this is complicated by the observation that the rate of feeding carbons into these pathways is highly variable, as measured with FDG-PET, and correlates with grade and survival. Nonetheless, our kinetic measurements revealed decreased PK activity in the “shunted” phenotype, suggesting that these metabolic profiles can be generated, in part, by modulation of PK activity. Another limitation is that, although tissue obtained in this study was rapidly frozen following excision, the resulting ischemia and hypoxia contributing to anaerobic metabolism within a specimen is difficult to control in the context of a resection. Therefore, the concentrations of specific metabolites may be affected by the unknown state of metabolic degradation during this period of ischemia and/or hypoxia. However, since comparisons made herein are between glioma grades, including the identified anabolic phenotype and altered phospholipid metabolism, we postulate that these same inconsistencies involving anaerobic glycolysis associated with surgery-related ischemia occur at a similar rate between the low- and high-grade lesions, essentially normalizing this effect. This limitation extends to the inability in determining grade-specific differences in pyruvate metabolism into lactate, which may not be accurately recapitulated in these studies based on the accelerated anaerobic metabolism following resection.
Although previously identified malignant glioma subtypes based on transcriptional signatures provide insight into underlying molecular heterogeneity, their biologic relevance still remains unclear. Identifying the accumulation of PEP in mesenchymal malignant glioma represents a previously unrecognized phenotype that may contribute to the aggressive nature of this subtype and be subsequently modulated as a form of metabolism-based cancer therapy. Furthermore, recent findings identified transcriptional networks regulated by C/EBPβ and STAT3 are central to mesechymal transformation in glioblastoma (32); therefore, further work linking these pathways to modulation of PKM2 activity are warranted. In addition, despite lower PK activity in the mesenchymal subtype, there seemed to be higher overall expression of this enzyme when compared with tumors clustering into the proneural subtype, suggesting the potential for alternate functions of this enzyme. This is supported by recent work of Yang and colleagues showing the nonmetabolic role of PKM2, involving translocation to the nucleus and EGFR-promoted β-catenin transactivation (33). Another possibility may involve PKM2 phosphorylation, and subsequent inactivation in this subtype (31), which was not evaluated in this study.
One important pathway in which cells divert carbon from glycolysis is through the pentose phosphate pathway. This shunting allows for both the generation of 6-phosphogluconate and R5P, which is used for nucleotide biosynthesis, and generates sufficient reducing potential for detoxification of reactive oxidative species (23, 26, 28). Here, we observed accumulation of both these intermediates, along with a 3.2-fold increase in reduced glutathione in grade 4 glioma, supporting this potential. In contrast, several pentitols, polyols derived from pentose phosphate pathway intermediates, including ribitol and arabitol, as well as the precursor arabinose, showed significantly decreasing levels with increasing tumor grade. Moreover, arabitol and arabinose, as well as additional, related small sugar derivatives were among the top 30 biochemicals in the RF-generated importance plot. The prevalence of this metabolite class among top RF biochemicals suggests that they provide robust indication of tumor grade progression. Although the metabolic pathways for many of these biochemicals in humans are not well understood, because several of these biochemicals are found elevated in certain human metabolic disorders with pentose phosphate enzyme deficiencies, this pattern of decreased levels for these pentose phosphate pathway side-products may serve as a further indication of an activated pentose phosphate pathway and heightened anabolic metabolism.
Another avenue for diversion of glycolytic flux is through de novo synthesis of serine and glycine, which represent precursors for a variety of biosynthetic pathways and epigenetic modulation through DNA methylation. The importance of this shift has been highlighted in 2 recent reports, identifying amplification in the gene phosphoglycerate dehydrogenase (PHGDH), the enzyme whose substrate is 3-PG, in a subset melanomas and breast cancers (25, 34, 35). Our study identified accumulation of both serine and glycine in grade 4 tumors, suggesting activity of this pathway may also be relevant in glioblastoma biology. In addition to serine and glycine, other exogenous essential and nonessential amino acids are required to maintain such processes as protein synthesis, anapleurosis, and nucleotide biosynthesis during tumorigenesis. Accordingly, in our study, the accumulation of amino acid metabolites played a significant role in distinguishing grade 2 and 4 tumors, with 54% of the 86 amino acid metabolites identified showing statistically significant differences. One of the amino acids that have been most extensively implicated in tumorigenesis is glutamine, which contributes toward the core metabolic needs of proliferating tumor cells, including providing bioenergetics, relieving oxidative stress, and complementing glucose metabolism through macromolecule production (36). Interestingly, increased accumulation of metabolites associated with glutamine metabolism was not associated with higher grade glioma. Although glutamine may certainly still be an essential component of glioma metabolism, our findings suggest its metabolism does not seem to be altered between tumor grades.
This report has identified 3 unique subtypes in malignant glioma based on their metabolomic signatures. Through unbiased, hierarchical clustering of the glioma metabolome, metabolites associated with divergence of glycolytic flux, characterized by accumulation of PEP and 3-PG, seemed to play an important role in the biology of a subset of these tumors. These analyses identified a unique, particularly aggressive metabolic subgroup defined by high PEP combined with a signature suggestive of lipid catabolism; mainly consisting of decreased accumulation of several glycerophosphocholines (GPCs) with no associated increase in phosphocholine. Several of these were among the top 30 biochemicals in the RF-generated importance plot. Interestingly, altered phospholipid catabolism has been previously described in glioma, with higher phosphocholine (PCHo)/GPC ratios found in the 1H spectra of human glioblastoma when compared with lower grade tumors (37). Altered phospholipid catabolism has also been observed in breast, prostate, and ovarian cancer (38–41). Furthermore, recent studies suggest that in addition to serving as a “passive” structural building block, phospholipids also have the capacity of “actively” regulating cellular function (42).
In addition to insight into the underlying biology of glioma, the identified metabolic subtypes seem to also provide information on the aggressiveness of an individual tumor. Although limited by the number of samples analyzed and inability to account for known prognostic factors in glioma, including RPA: Recursive partitioning analysis class, 1p/19q status, and promoter methylation of MGMT: O(6)-methylguanine-DNA methyl transferase, these profiles do suggest potential prognostic relevance with potential application for future trial stratification. Additional work will be required to confirm these findings in an expanded cohort accounting for these know prognostic factors. In addition, although names of specific signatures were coined (e.g., energetic, anabolic, and phospholipid catabolism), it is important to note that these were based on selected metabolites from an extensive list (Supplementary Table S2) that clustered individual subtypes. Continued investigations will be required to determine the relative importance of these other metabolites in subtype designation and their overall influence on malignant glioma metabolism. Surprisingly, these identified metabolomic subgroups seemed to be independent from both previously recognized malignant glioma subgroups and transcriptional profiles. This suggests that other global processes as genomic and/or epigenetic regulation, including EGFR amplification or mutation and PTEN activation, may be driving the observed metabolomic subtypes and that integrating these platforms may provide further insight into the signaling processes driving the observed metabolic phenotype.
Although these findings still require validation in an independent dataset, understanding the glioma metabolome offers the potential for several levels of clinical application. In addition to serving as a prognostic factor, subtype designation may allow to personalize therapy toward an individual tumor’s metabolic phenotype. For example, therapies designed to target the energetic subtype may involve Akt inhibitors, hexokinase inhibitors (43), or the metabolic modulator dichloroacetate (44), whereas a therapeutic regimen designed to target the anabolic phenotype of glioblastoma may involve agents designed to modulate PEP accumulation or shunting into the PPP, including such agents as transketolase inhibitors (43).
In conclusion, metabolomics provides a unique window into the phenotype of glioma that, when integrated with other platforms, may provide a more comprehensive understanding of the complex biology associated with glioma tumorigenesis and malignant transformation. These findings underscore a previously unrecognized metabolic heterogeneity in glioma with both biologic and clinical relevance. A richer understanding of aberrant metabolism will provide a framework for the design and implementation of a personalized approach to malignant glioma therapy through metabolic modulation.
Supplementary Material
Acknowledgments
The authors thank Michelle Fournier, Cynthia Shen, and Vonetta Williams for their assistance in tissue and data acquisition.
Grant Support
This research was supported by the U.S. Army Medical Research and Materiel Command, National Functional Genomics Center project, under award number W81XWH-08-2-0101. P. Chinnaiyan is currently funded by The Ben and Catherine Ivy Foundation and The American Cancer Society (RSG-11-029-01-CSM).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
E. Kensicki is a paid employee of Metabolon, Inc. Opinions, interpretations, conclusions, and recommendations are those of the author(s) and are not necessarily endorsed by the U.S. Army. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: P. Chinnaiyan, R. Gillies
Development of methodology: P. Chinnaiyan, G. Bloom, B. Sarcar, S. Kahali
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): P. Chinnaiyan, E. Kensicki, B. Sarcar
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): P. Chinnaiyan, E. Kensicki, G. Bloom, A. Prabhu, B. Sarcar, S. Kahali, S. Eschrich, X. Qu, P. Forsyth, R. Gillies
Writing, review, and/or revision of the manuscript: P. Chinnaiyan, E. Kensicki, G. Bloom, S. Kahali, S. Eschrich, P. Forsyth, R. Gillies
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): P. Chinnaiyan
Study supervision: P. Chinnaiyan, S. Kahali
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warburg O, Posener K, Negelein E. Uber den Stoffwechsel der Carcinomzelle. Biochem Zeitschr. 1924;152:309–44. [Google Scholar]
- 4.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–30. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gatenby RA, Gawlinski ET, Gmitro AF, Kaylor B, Gillies RJ. Acid-mediated tumor invasion: a multidisciplinary study. Cancer Res. 2006;66:5216–23. doi: 10.1158/0008-5472.CAN-05-4193. [DOI] [PubMed] [Google Scholar]
- 6.Padma MV, Said S, Jacobs M, Hwang DR, Dunigan K, Satter M, et al. Prediction of pathology and survival by FDG PET in gliomas. J Neurooncol. 2003;64:227–37. doi: 10.1023/a:1025665820001. [DOI] [PubMed] [Google Scholar]
- 7.Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat Rev Cancer. 2004;4:551–61. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- 8.Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15:431–40. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–67. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 10.Ohta T, Masutomi N, Tsutsui N, Sakairi T, Mitchell M, Milburn MV, et al. Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicol Pathol. 2009;37:521–35. doi: 10.1177/0192623309336152. [DOI] [PubMed] [Google Scholar]
- 11.Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform. 2010;2:9. doi: 10.1186/1758-2946-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 14.Liaw A, Wiener M. Classification and regression by random forest. R News. 2002;2:18–22. [Google Scholar]
- 15.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 16.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahali S, Sarcar B, Fang B, Williams ES, Koomen JM, Tofilon PJ, et al. Activation of the unfolded protein response contributes toward the antitumor activity of vorinostat. Neoplasia. 2010;12:80–6. doi: 10.1593/neo.91422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Florian CL, Preece NE, Bhakoo KK, Williams SR, Noble M. Characteristic metabolic profiles revealed by 1H NMR spectroscopy for three types of human brain and nervous system tumours. NMR Biomed. 1995;8:253–64. doi: 10.1002/nbm.1940080605. [DOI] [PubMed] [Google Scholar]
- 19.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A. 2011;108:3270–5. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dang CV. PKM2 tyrosine phosphorylation and glutamine metabolism signal a different view of the Warburg effect. Sci Signal. 2009;2:pe75. doi: 10.1126/scisignal.297pe75. [DOI] [PubMed] [Google Scholar]
- 25.DeBerardinis RJ. Serine metabolism: some tumors take the road less traveled. Cell Metab. 2011;14:285–6. doi: 10.1016/j.cmet.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–8. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 28.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to antioxidant responses. Science. 2011;334:1278–83. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–3. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 30.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–6. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 31.Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2009;463:318–25. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;478:118–22. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–74. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–50. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–50. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Usenius JP, Vainio P, Hernesniemi J, Kauppinen RA. Choline-containing compounds in human astrocytomas studied by 1H NMR spectroscopy in vivo and in vitro. J Neurochem. 1994;63:1538–43. doi: 10.1046/j.1471-4159.1994.63041538.x. [DOI] [PubMed] [Google Scholar]
- 38.Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res. 2004;64:4270–6. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]
- 39.Keshari KR, Tsachres H, Iman R, Delos Santos L, Tabatabai ZL, Shinohara K, et al. Correlation of phospholipid metabolites with prostate cancer pathologic grade, proliferative status and surgical stage—impact of tissue environment. NMR Biomed. 2011;24:691–9. doi: 10.1002/nbm.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iorio E, Mezzanzanica D, Alberti P, Spadaro F, Ramoni C, D’Ascenzo S, et al. Alterations of choline phospholipid metabolism in ovarian tumor progression. Cancer Res. 2005;65:9369–76. doi: 10.1158/0008-5472.CAN-05-1146. [DOI] [PubMed] [Google Scholar]
- 41.Morse DL, Carroll D, Day S, Gray H, Sadarangani P, Murthi S, et al. Characterization of breast cancers and therapy response by MRS and quantitative gene expression profiling in the choline pathway. NMR Biomed. 2009;22:114–27. doi: 10.1002/nbm.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Podo F. Tumour phospholipid metabolism. NMR Biomed. 1999;12:413–39. doi: 10.1002/(sici)1099-1492(199911)12:7<413::aid-nbm587>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 43.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–84. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 44.Michelakis ED, Sutendra G, Dromparis P, Webster L, Haromy A, Niven E, et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med. 2010;2:31ra4. doi: 10.1126/scitranslmed.3000677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.