Abstract
Host–pathogen interactions provide valuable systems for the study of evolutionary genetics and natural selection. The sequestration of essential iron has emerged as a crucial innate defense system termed nutritional immunity, leading pathogens to evolve mechanisms of ‘iron piracy’ to scavenge this metal from host proteins. This battle for iron carries numerous consequences not only for host–pathogen evolution but also microbial community interactions. Here we highlight recent and potential future areas of investigation on the evolutionary implications of microbial iron piracy in relation to molecular arms races, host range, competition, and virulence. Applying evolutionary genetic approaches to the study of microbial iron acquisition could also provide new inroads for understanding and combating infectious disease.
Keywords: evolution, arms race, iron, microbe, pathogen, immunity
Trends
The battle between microbes and their hosts for nutrient iron is emerging as a new front of evolutionary genetic conflict.
Molecular arms races can emerge between host iron-binding proteins and microbial ‘iron piracy’ factors that steal this nutrient for growth. Such rapid evolution may also contribute to the host range of pathogenic microbes.
Iron acquisition plays an important role in evolutionary interactions between microbes, both in the environment and within the host. Competition for iron can prevent infection by pathogens, while genetic changes in iron acquisition systems can enhance microbial virulence.
Evolutionary conflicts for nutrient iron are revealing potential new genetic mechanisms of disease resistance as well as avenues for therapeutic development.
An Evolving View of Host–Microbe Interactions
The outcome of an infection can have profound consequences for both host and pathogen populations. Intense selective pressures make host–pathogen interactions an attractive biological model to study evolutionary genetics over relatively short intervals of time. To date, much work has focused on rapid evolution involving canonical host immune defenses or antibiotic resistance 1, 2. However, we now know that hosts possess numerous additional means to restrict pathogens, including factors engaged in other core physiological functions. Nutrient iron sequestration provides one such alternative mode of host defense against bacteria and eukaryotic pathogens [3]. Iron is an essential micronutrient for microbes, as well as their hosts, due to its ability to readily shift between ferrous (Fe2+) and ferric (Fe3+) oxidative states for redox catalysis or electron transport. This ability to readily accept and donate electrons also makes iron highly volatile, necessitating a well-coordinated iron transport and storage system in metazoans to prevent the production of toxic free radicals [4]. The sequestration of free iron by host proteins simultaneously prevents acquisition by microbes, a protective effect termed nutritional immunity (see Glossary) 5, 6. While appreciation has grown for the role of nutrient metals in infection, these ‘battles for iron’ and other trace metals provide intriguing cases for investigation from an evolutionary perspective. Here we discuss emerging questions on the control of iron in microbial infection and highlight recent and potential future insights regarding the evolution of molecular arms races, host range, microbial competition, and pathogen virulence.
The Battle for Iron
A potential role for iron in immunity became apparent following an elegant series of experiments by Arthur Schade and Leona Caroline in the early 1940s [7]. While attempting to develop a vaccine against Shigella, the researchers observed that addition of raw egg white to their culture media severely inhibited the growth of diverse bacteria as well as fungi. The antiseptic properties of egg white have in fact been recognized since the days of Shakespeare, where it was applied to wounds during Act III of King Lear. While various nutrient supplements failed to reverse the antimicrobial effect of egg whites, incinerated yeast extract did, suggesting that the limiting component was elemental in nature. Of 31 individual elements tested, supplementation with iron alone was sufficient to restore microbial growth in the presence of egg white. Adding to the fortuitous nature of their discovery, the authors posited that an iron-binding component present in the egg white prevented acquisition of this nutrient by microbes, which could have important implications for immunity. Two years later the scientists reported similar activity present in human blood serum [8]. The factor responsible for this activity in both cases was later revealed to be the protein transferrin, which plays a central role in animal iron metabolism by binding and transporting this metal to target cells 9, 10.
In the decades following Schade and Caroline's initial discoveries, Eugene Weinberg proposed that withholding iron from microbial pathogens provided an important cornerstone of host defense, which he termed nutritional immunity [11]. Weinberg's theory explained previous observations that human iron overload disorders such as hereditary hemochromatosis and thalassemia render affected individuals highly susceptible to bacterial and fungal infections. The theory of nutritional immunity was also consistent with George Cartwright's earlier observations that infection induces an acute reduction in circulating iron levels 12, 13, 14. Subsequent microbiology and molecular genetic studies established that nutritional immunity plays a pivotal role in defense against an array of pathogens, including bacteria, fungi, and parasites 3, 15. Owing to the iron-binding properties of proteins, such as transferrin, circulating levels of free iron in the body are orders of magnitude below the requirements for optimal microbial growth.
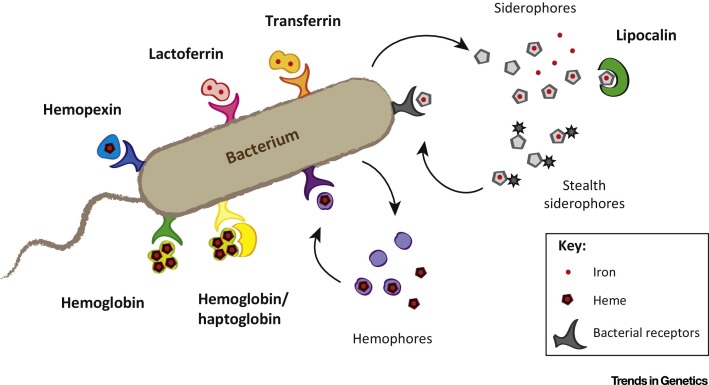
Microbes respond to iron starvation by actively scavenging this nutrient from host proteins to meet their metabolic requirements (Figure 1 ) [16]. One of the most common microbial iron acquisition strategies involves the secretion of siderophores, small molecule chelators, which possess an affinity for iron unmatched even by proteins such as transferrin 17, 18. Microbes then recover iron–siderophore complexes via cell surface receptors. Obviating the need for siderophores, several microbes also express receptors that directly recognize and extract iron from host proteins including transferrin and lactoferrin 19, 20, 21, 22, 23. Additional mechanisms involve the acquisition of heme, the iron-containing porphyrin cofactor, from abundant host proteins such as hemoglobin 24, 25, 26. Ferric reductases are an important class of iron acquisition systems in fungal pathogens, which convert transferrin or lactoferrin-bound ferric iron into a soluble ferrous form [27]. The identification of iron acquisition genes as pathogen virulence factors further underscores the role of iron in infection, as well as the potential for evolutionary conflicts to arise in the struggle for this precious nutrient.
Figure 1.
Nutritional Immunity and Microbial Iron Piracy. Illustration highlighting major components of bacterial iron acquisition, including surface receptors as well as secreted siderophores and hemophores. Host nutritional immunity proteins are denoted in bold.
New Perspectives on Ancient Evolutionary Arms Races
Novel mutations that alter host–pathogen interactions can provide a substantial fitness advantage and spread in a population through positive selection. Recurrent bouts of positive selection at such interfaces can give rise to so-called ‘molecular arms races’, in which the host and pathogen must continually adapt to maintain comparative fitness [1]. Genes subject to such evolutionary conflicts are often characterized by an increased rate of nonsynonymous to synonymous substitutions (termed dN/dS or ω), reflecting recurrent selection for novel amino acid substitutions that alter protein interaction surfaces. Instances of such molecular arms races also exemplify Leigh Van Valen's Red Queen Hypothesis, which proposed that antagonistic coevolution leads to a perpetual cycle of adaptation in which neither opponent gains a permanent advantage [28]. Several core components of the vertebrate immune system have subsequently been shown to engage in such conflicts, some of which are able to dictate the outcome of an infection 29, 30, 31, 32, 33, 34, 35.
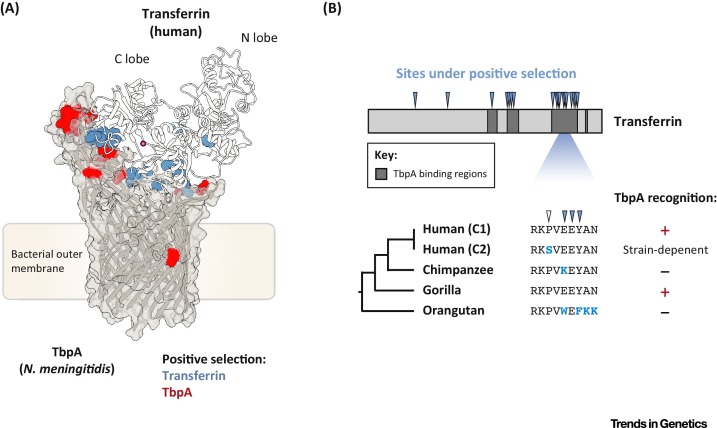
Our recent work highlighted the battle for iron as a new interface for Red Queen evolutionary conflicts [36]. As described earlier, transferrin was among the first vertebrate proteins to be implicated in nutritional immunity and is also a frequent target of iron acquisition by microbes. Reasoning that transferrin could be a focal point for genetic conflicts with pathogens, we performed phylogenetic analyses of transferrin gene divergence in the primate lineage. Not only has transferrin been subject to strong positive selection in primates but also rapidly evolving sites almost entirely overlap with the binding interface of a bacterial surface receptor, transferrin-binding protein A (TbpA), an important virulence factor in several human pathogens including Neisseria meningitidis, Neisseria gonorrhoeae, Haemophilus influenzae, and Moraxella catarrhalis, as well as a number of agricultural pathogens (Figure 2A) 21, 37, 38, 39, 40, 41. Single amino acid substitutions at rapidly evolving sites in transferrin were sufficient to control TbpA-binding specificities between related primates as well as for an abundant human transferrin variant, termed C2 (Figure 2B). Genetic signatures of positive selection at the transferrin–TbpA binding interface suggest that this interaction has been a key determinant of infection during millions of years of primate divergence. More broadly, these results demonstrate that nutritional immunity, similar to more established immune pathways, has strongly impacted host fitness during our long and intertwined history with microbes.
Figure 2.
Evolutionary Conflict at the Transferrin–Transferrin-binding protein A (TbpA) interface. (A) Cocrystal structure (Protein Data Bank: 3V8X) of human transferrin bound to TbpA from Neisseria meningitidis. Side chains of rapidly evolving amino acid positions in primate transferrin are shown in blue, with rapidly evolving TbpA sites among human pathogens shown in red (as described in [36]). (B) Schematic highlighting rapidly evolving regions in primate transferrin. Sites subject to positive selection are denoted with blue arrows; a variable site in humans (the C2 variant) is marked by a white arrow. Divergent amino acids among humans and other primates are shown in blue, and the ability of human-adapted TbpA to recognize each transferrin ortholog is shown on the right. The human transferrin C2 variant is recognized by TbpA from some but not all pathogens.
Evidence for an evolutionary arms race between transferrin and TbpA raises the question as to whether other host nutritional immunity factors may be subject to similar conflicts. Many pathogens encode receptors for other host iron-binding proteins including lactoferrin, a transferrin paralog expressed in milk, saliva, tears, mucus, and the secondary granules of neutrophils 37, 42, 43, 44. The evolution of lactoferrin introduces a fascinating twist; in addition to sequestering iron, lactoferrin has acquired mutations to generate antimicrobial peptide (AMP) domains that bind and disrupt pathogen membranes 45, 46. Many pathogens in turn encode factors that either scavenge lactoferrin-bound iron or inhibit associated AMP activity 47, 48, 49. How these distinct functions have shaped lactoferrin evolution or potential arms races with pathogens remains to be determined.
Genetic conflicts in nutritional immunity may also unfold by means other than simple point mutation and selection at protein interaction sites. For example, the vertebrate protein lipocalin 2 (also known as siderocalin or NGAL) is a potent innate immunity factor that functions in part through binding and sequestration of siderophores, preventing their uptake by microbes 50, 51. Some pathogens evade this defense through production of modified ‘stealth siderophores’, which are not recognized by lipocalin 2 52, 53. Whether lipocalin 2 in turn has undergone adaptation resulting in enhanced or modified siderophore recognition is unknown. Understanding the extent to which molecular arms races have influenced other nutritional immunity factors beyond transferrin could reveal additional modes of adaptation underlying host–pathogen evolutionary conflicts.
The barrier imposed by nutritional immunity has seemingly produced an even more drastic evasion strategy by one pathogen – giving up iron altogether. Previous work has demonstrated that the bacterial spirochete Borrelia burgdorferi, the causative agent of Lyme disease, lacks a requirement for iron shared by nearly all other organisms [54]. This was an astounding discovery given that iron serves as a cofactor for numerous metalloproteins involved in essential cellular processes including the electron transport chain and DNA metabolism. How then has B. burgdorferi managed such an evolutionary feat? Closer inspection of the B. burgdorferi genome revealed that numerous genes encoding iron-binding proteins have been lost, and remaining enzymes that normally bind iron have undergone modification to bind manganese in its place 54, 55. Beyond these general observations, we are only beginning to unravel the stepwise genetic mechanisms that lead to such major evolutionary innovations 56, 57, 58. Identifying other microbes that have foregone the requirement for iron could provide useful comparison points to understand the mechanics of complex evolutionary transitions.
Because many host nutritional immunity proteins also carry out crucial ‘day jobs’ in metal metabolism or transport, it is conceivable that antagonistic pleiotropy, or an evolutionary trade-off, could arise from an arms race with adverse consequences for the host. Sickle cell anemia in humans provides a quintessential example, whereby hemoglobin mutations confer resistance to malaria infection at the expense of severe anemia in homozygous carriers [59]. Hereditary hemochromatosis (HH) is a condition characterized by increased iron absorption in the gut as well as serum iron overload, leading to iron accumulation in various organs and subsequent tissue damage 9, 60. HH caused by the C282Y mutation in the HFE gene is the most common genetic disorder among those of European descent, carried by approximately 10% of these individuals. Although the molecular mechanisms by which HFE mutations cause HH are still unclear, HFE is expressed on the surface of several cell types where it interacts with the transferrin receptor Tf-R to regulate iron absorption in the gut and release of iron stored in circulating macrophages. The high frequency of the C282Y mutation among Europeans has led to speculation as to the underlying cause for its abundance [61]. In addition to other associated health problems, individuals with HH are highly susceptible to infection by normally noninvasive microbes, such as the bacteria Vibrio vulnificus and Yersinia enterocolitica 62, 63, 64. Ironically, this increased susceptibility to extracellular pathogens may be offset by resistance to others that normally infect macrophages such as Mycobacterium tuberculosis or Salmonella enterica serovar Typhi, which cause tuberculosis and typhoid fever, respectively [65]. While many questions remain regarding the consequences of HH mutations, these studies provide fascinating examples of how the role of iron in infection may contribute to instances of antagonistic pleiotropy in human genetic disorders.
Iron in Host Range and Zoonoses
The term zoonosis refers to an infectious disease of animals that can be transmitted to humans. Because naïve populations typically lack pre-existing genetic resistance to these new pathogens, zoonotic diseases have caused some of the most deadly epidemics in human history, including the Black Death and the 1918 Spanish flu, along with recently emerging pathogens such as Ebola virus and the MERS coronavirus 66, 67. The mechanisms that dictate the host range of pathogens are thus of high interest to evolutionary biologists and infectious disease researchers alike. A number of studies have now implicated nutritional immunity factors in restricting the host range of several bacterial pathogens. For example, bacteria that utilize transferrin receptors possess extremely narrow host ranges, such as the human-specific N. gonorrhoeae and the porcine pathogen Actinobacillus pleuropneumoniae. Consistent with this observation, TbpA and its coreceptor TbpB exclusively recognize their host transferrin proteins when compared with diverse mammals 68, 69, 70. Our recent work further demonstrated that single rapidly evolving sites in transferrin are sufficient to dictate TbpA-binding differences between even our closest relatives, such as chimpanzees [36]. Moreover, expressing or injecting human transferrin in mice can promote infection with human-specific Neisseria 71, 72. These findings suggest that adaptive evolution of nutritional immunity factors may be sufficient to establish a host range barrier against pathogens. The implications for transferrin variation on host range likely extend beyond bacteria as well, given that transferrin receptors have been implicated in iron acquisition and tropism of Trypanosoma brucei, the eukaryotic parasite that causes African sleeping sickness 73, 74.
In addition to transferrin, it is likely that other nutritional immunity proteins contribute to limiting pathogen host range. Elegant work by Pishchany et al. demonstrated that the Gram-positive bacterium Staphylococcus aureus exhibits strong preference for human hemoglobin over mouse hemoglobin, which correlates with bacterial growth in murine models of infection [75]. While hemoglobin evolution has been extensively studied in the context of both environmental adaptation and malaria parasite resistance 59, 76, the implications for this variation on nutritional immunity against bacterial pathogens has not been investigated. It is also notable that pathogens exhibiting restricted host iron requirements, including N. gonorrhoeae and S. aureus, pose urgent public health concerns given their increasing resistance to conventional antibiotics [77]. Applying genetic variants of transferrin to protein-based therapeutics provides one new means of treating infectious disease [78]. Therapeutic strategies building from evolutionary studies of nutritional immunity could therefore provide new weapons against increasingly dire scenarios of resistance to traditional antimicrobial treatments.
Microbial Competition for Iron: The Red Queen Is Back in Black
The biology of competition has been a long-standing area of interest for both evolutionary theorists and microbial ecologists. A recent contribution to the field of microbial evolutionary theory came with the Black Queen Hypothesis [79]. The metaphor stems from the card game Hearts, in which players avoid holding the Queen of Spades or face a significant point penalty. The Black Queen Hypothesis posits that gene loss can be adaptive and proceed by natural selection, allowing individuals to reap public goods provided by other members of the microbial community. Iron acquisition is one such ‘leaky’ biological process that may be particularly prone to Black Queen conflicts. It has been widely observed that bacteria that do not produce siderophores nonetheless express siderophore receptors, allowing them to harvest this resource at the expense of their neighbors [16]. This hypothesis also invokes the long-standing concept of evolutionary ‘cheaters’, which can profoundly influence the stability of microbial populations. Previous studies have demonstrated that bacterial siderophore production follows many predictions of kin selection, whereby relatedness and degrees of competition influence the emergence of cheaters that do not produce siderophores 80, 81. Iron acquisition therefore provides as an informative system in which to study microbial population biology and evolution, including Black Queen dynamics.
The link between iron and microbial competition during infection was further illuminated in recent work by Deriu et al., focusing on interactions between pathogenic Salmonella and commensal Escherichia coli in the gut [82]. The Nissle 1917 strain of E. coli was isolated from a soldier during World War I who appeared resistant to an outbreak of dysentery, and has subsequently been applied as a probiotic treatment for gastrointestinal ailments including ulcerative colitis and Crohn's disease 83, 84. The authors demonstrated that Nissle is able to suppress gastroenteritis induced by Salmonella enterica serovar Typhimurium through competition for iron [82]. The ability of Nissle to outcompete S. enterica was also dependent on the presence of the host siderophore-binding protein lipocalin 2, illustrating a complex interplay among host, pathogen, and commensal species. These findings further illustrate how microbial Black Queen dynamics can impact human disease, as well as the potential application of beneficial microbes to combat pathogens through iron sequestration.
The Red Queen and Black Queen Hypotheses highlight distinct modes of evolutionary adaptation ranging from antagonistic arms races to adaptive gene loss [85]. In the case of microbial iron piracy, both processes appear to play roles influencing fundamental functions. In addition to previous examples, loss of iron acquisition genes in B. burgdorferi could be interpreted as a Black Queen process arising within the host cell, whereas rapid evolution of bacterial transferrin receptors reflects a prototypical Red Queen conflict. The Black Queen Hypothesis may also provide a basis for understanding the fitness advantage conferred by bacterial receptors such as TbpA. By forgoing siderophore production and targeting host iron-binding proteins directly, these bacteria use a less leaky system, which may be inherently resistant to cheaters. In turn, dependence on these receptors gives rise to Red Queen conflicts with host proteins, such as transferrin, contributing to narrow host ranges observed for these strains. Future studies of iron acquisition could reveal additional genetic or ecological factors that contribute to these distinct evolutionary outcomes and the implications for infectious disease.
Iron in the Evolution of Virulence
In addition to influencing interactions between microbes, iron can also regulate evolutionary transitions between commensal and pathogenic states. Iron acquisition genes are now established as microbial virulence factors, in the sense that loss of these genes impairs pathogenicity without completely compromising organism viability 3, 86. However, a simple interpretation of iron acquisition dictating pathogenesis breaks down when considering the commensal microbiota. In nearly all cases these organisms require iron for survival, and yet rarely if ever cause disease. The role of iron in microbial virulence is thus more nuanced than it first appears and represents a growing area of research.
New insights on the role of iron in bacterial virulence were recently provided by studies of the opportunistic bacterial pathogen Pseudomonas aeruginosa. While P. aeruginosa is commonly found in the environment and is typically avirulent in immune-competent individuals, it readily colonizes the lungs of patients with cystic fibrosis (CF) where it poses a major source of mortality [87]. P. aeruginosa, like many microbes, possesses multiple distinct iron acquisition systems, including siderophores and a heme uptake system. Recently Marvig et al. investigated the genetic basis of host adaptation in P. aeruginosa strains that had maintained persistent human colonization for over 36 years [88]. Among several potential adaptations that took place during this period, evidence of positive selection was detected in the promoter of phu, the bacterial heme uptake system. Using experimental culture systems, the authors determined that increased expression of the phu system enhanced bacterial growth in a model of the CF lung, and that similar mutations had occurred independently in P. aeruginosa isolates from other patients with CF. These results suggest that a shift to enhanced heme uptake via hemoglobin plays an important role in the transition of P. aeruginosa from an environmental microbe to a dedicated human pathogen.
Iron acquisition can also dictate the ability of a microbe to shift between commensalism and virulence as conditions change, as was recently observed in the yeast Candida albicans. While often present as a commensal member of the gut microbiota, C. albicans is also capable of proliferating in the bloodstream and causing systemic infections in susceptible individuals [89]. The mechanisms by which C. albicans can shift between these distinct lifestyles are still largely mysterious. Chen et al. dissected the evolution of an elaborate transcriptional network in C. albicans whereby incorporation of a novel transcriptional activator, Sef1, into a pre-existing iron-responsive repressor system mediates growth in different host niches [90]. The authors demonstrated that Sef1 promotes iron acquisition and is essential for virulence in vivo. In addition, Sef1 is regulated by and itself regulates Sfu1, a transcriptional repressor. Sfu1 in turn represses iron acquisition and is dispensable for virulence, but is conversely essential for commensal growth in the gastrointestinal tract. Thus, C. albicans has evolved an intricate transcriptional program to regulate iron acquisition during commensal or pathogenic states [91]. Given that iron starvation acts as a trigger for activation of virulence genes in many pathogens [92], it is likely that similar gene regulatory networks also exist in other microbes.
Our understanding of iron in the control of commensal versus pathogenic states is only in its infancy. For example, while transferrin receptors are important virulence factors in pathogenic N. meningitidis and N. gonorrhoeae, these genes are also harbored by commensal Neisseria that inhabit the nasopharynx [93]. Adding to this complexity, Neisseria are naturally competent and horizontal gene transfer readily occurs between pathogenic and commensal strains [94]. Commensal microbes may therefore provide a genetic stockpile of virulence factors available for acquisition by pathogens. As we continue to learn more about the impact of the microbiota on human health and disease, these studies may also shed light on the role of iron in evolutionary interactions between seemingly beneficial or pathogenic inhabitants of our bodies.
Concluding Remarks and Future Directions
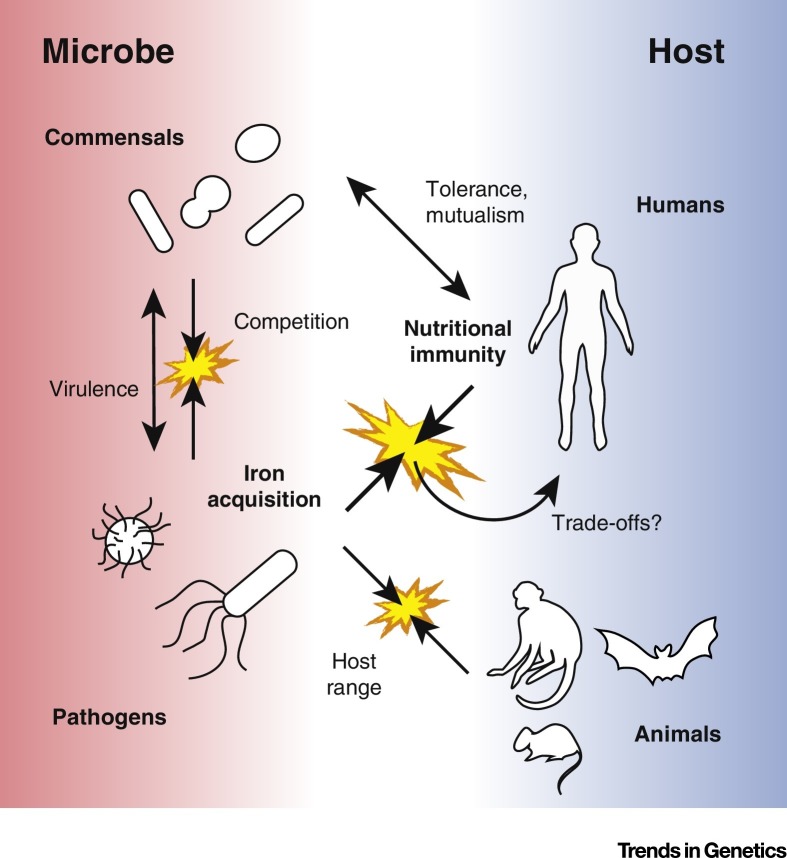
Studies of microbial iron acquisition are proving a useful resource for investigating diverse evolutionary genetic phenomena (Figure 3 ). As new insights emerge, additional questions continue to arise (see Outstanding Questions). For example, while the majority of studies addressing nutritional immunity involve iron, recent work has begun to reveal the contribution of other essential metals including manganese and zinc in this process 95, 96, 97, 98. In addition, while previous findings have demonstrated that iron plays an important role in microbial competition, we understand comparatively little regarding its importance in the evolution of cooperative microbial communities. Finally, we have focused largely here on the role of bacteria in the evolutionary battle for iron, while fungi and parasites are undoubtedly also subject to similar conflicts 74, 99, 100. The potential for adaptive evolution to dictate the outcome of microbial infection further provides an impetus to harness these studies for novel therapeutics. The application of evolutionary insights has led to promising genetic and chemical strategies for combatting human disease, most notably in the case of HIV 66, 101, 102, 103. Future studies promise to reveal additional genetic innovations favoring or countering microbial piracy of iron and other critical nutrients.
Outstanding Questions.
In addition to transferrin, what other nutritional immunity factors are subject to molecular arms races with pathogens?
Do evolutionary conflicts exist for other nutrient metals such as manganese or zinc and their respective binding proteins?
How can we leverage evolution-guided studies of nutritional immunity to combat infectious disease?
Figure 3.
Implications for Iron Piracy in Host–Microbe Evolution. Overview depicting the roles of iron piracy and nutritional immunity in diverse evolutionary processes involving microbes and animal hosts.
Acknowledgments
We thank members of the Elde laboratory and Nicola Barber for helpful comments and suggestions on the manuscript. This work has been supported by awards from the National Institutes of Health to N.C.E. (R00GM090042) and M.F.B. (F32GM108288, K99GM115822). N.C.E. is a Pew Scholar in the Biomedical Sciences and the Mario R. Capecchi Endowed Chair in Genetics.
Glossary
- Antagonistic pleiotropy
also termed an evolutionary ‘trade-off’, in which a single gene controls multiple traits with opposing beneficial and deleterious effects.
- Black Queen Hypothesis
describes the process by which gene loss can progress via natural selection, particularly in cases where individuals reduce investment in costly metabolic functions provided by other members of a microbial community.
- Microbiota
the collection of microorganisms inhabiting a particular environment, such as the human body.
- Nutritional immunity
a host immune defense mechanism by which essential nutrients, such as iron, are withheld in order to limit microbial growth and prevent infection.
- Positive selection
the process by which new, beneficial genetic variation accrues in a population.
- Red Queen Hypothesis
posits that antagonistic coevolution (e.g., between predators and prey or pathogens and hosts) produces a state in which constant adaptation is required to maintain comparative evolutionary fitness.
- Siderophore
a diverse class of small molecule iron chelators, which are secreted by microbes and then internalized via surface receptors to mediate iron acquisition.
- Transferrin
a serum glycoprotein in animals containing two iron-binding ‘lobe’ domains that deliver ferric iron to host cells via receptor-mediated endocytosis, as well as withholding iron from microbes.
- Virulence factor
a gene or molecule that contributes to microbial infection, while not necessarily required for viability in nonpathogenic settings.
- Zoonosis
an infectious disease naturally transmitted from animals to humans.
References
- 1.Daugherty M.D., Malik H.S. Rules of engagement: molecular insights from host–virus arms races. Annu. Rev. Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 2.Palmer A.C., Kishony R. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nat. Rev. Genet. 2013;14:243–248. doi: 10.1038/nrg3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cassat J.E., Skaar E.P. Iron in infection and immunity. Cell Host Microbe. 2013;13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews N.C. Disorders of iron metabolism. N. Engl. J. Med. 1999;341:1986–1995. doi: 10.1056/NEJM199912233412607. [DOI] [PubMed] [Google Scholar]
- 5.Hood M.I., Skaar E.P. Nutritional immunity: transition metals at the pathogen–host interface. Nat. Rev. Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potrykus J. Conflicting interests in the pathogen–host tug of war: fungal micronutrient scavenging versus mammalian nutritional immunity. PLoS Pathog. 2014;10:e1003910. doi: 10.1371/journal.ppat.1003910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schade A.L., Caroline L. Raw hen egg white and the role of iron in growth inhibition of Shigella dysenteriae, Staphylococcus aureus, Escherichia coli and Saccharomyces cerevisiae. Science. 1944;100:14–15. doi: 10.1126/science.100.2584.14. [DOI] [PubMed] [Google Scholar]
- 8.Schade A.L., Caroline L. An iron-binding component in human blood plasma. Science. 1946;104:340–341. doi: 10.1126/science.104.2702.340. [DOI] [PubMed] [Google Scholar]
- 9.Ganz T., Nemeth E. Hepcidin and disorders of iron metabolism. Annu. Rev. Med. 2011;62:347–360. doi: 10.1146/annurev-med-050109-142444. [DOI] [PubMed] [Google Scholar]
- 10.Gkouvatsos K. Regulation of iron transport and the role of transferrin. Biochim. Biophys. Acta. 2012;1820:188–202. doi: 10.1016/j.bbagen.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 11.Weinberg E.D. Nutritional immunity. Host's attempt to withhold iron from microbial invaders. JAMA. 1975;231:39–41. doi: 10.1001/jama.231.1.39. [DOI] [PubMed] [Google Scholar]
- 12.Weinberg E.D. Iron and infection. Microbiol. Rev. 1978;42:45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinberg E.D. Iron withholding: a defense against infection and neoplasia. Physiol. Rev. 1984;64:65–102. doi: 10.1152/physrev.1984.64.1.65. [DOI] [PubMed] [Google Scholar]
- 14.Cartwright G.E. The anemia associated with chronic infection. Science. 1946;103:72–73. doi: 10.1126/science.103.2664.72. [DOI] [PubMed] [Google Scholar]
- 15.Skaar E.P. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010;6:e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ratledge C., Dover L.G. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 17.Francis J. Mycobactin, a growth factor for Mycobacterium johnei. I. Isolation from Mycobacterium phlei. Biochem. J. 1953;55:596–607. doi: 10.1042/bj0550596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holden V.I., Bachman M.A. Diverging roles of bacterial siderophores during infection. Metallomics. 2015;7:986–995. doi: 10.1039/c4mt00333k. [DOI] [PubMed] [Google Scholar]
- 19.Schryvers A.B., Morris L.J. Identification and characterization of the human lactoferrin-binding protein from Neisseria meningitidis. Infect. Immun. 1988;56:1144–1149. doi: 10.1128/iai.56.5.1144-1149.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee B.C., Schryvers A.B. Specificity of the lactoferrin and transferrin receptors in Neisseria gonorrhoeae. Mol. Microbiol. 1988;2:827–829. doi: 10.1111/j.1365-2958.1988.tb00095.x. [DOI] [PubMed] [Google Scholar]
- 21.Cornelissen C.N. Gonococcal transferrin-binding protein 1 is required for transferrin utilization and is homologous to TonB-dependent outer membrane receptors. J. Bacteriol. 1992;174:5788–5797. doi: 10.1128/jb.174.18.5788-5797.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmon D. A novel heterodimeric transferrin receptor encoded by a pair of VSG expression site-associated genes in T. brucei. Cell. 1994;78:75–86. doi: 10.1016/0092-8674(94)90574-6. [DOI] [PubMed] [Google Scholar]
- 23.Steverding D. The transferrin receptor of Trypanosoma brucei. Parasitol. Int. 2000;48:191–198. doi: 10.1016/s1383-5769(99)00018-5. [DOI] [PubMed] [Google Scholar]
- 24.Lewis L.A., Dyer D.W. Identification of an iron-regulated outer membrane protein of Neisseria meningitidis involved in the utilization of hemoglobin complexed to haptoglobin. J. Bacteriol. 1995;177:1299–1306. doi: 10.1128/jb.177.5.1299-1306.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torres V.J. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol. 2006;188:8421–8429. doi: 10.1128/JB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabian M. Heme transfer to the bacterial cell envelope occurs via a secreted hemophore in the Gram-positive pathogen Bacillus anthracis. J. Biol. Chem. 2009;284:32138–32146. doi: 10.1074/jbc.M109.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saikia S. Role of ferric reductases in iron acquisition and virulence in the fungal pathogen Cryptococcus neoformans. Infect. Immun. 2014;82:839–850. doi: 10.1128/IAI.01357-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Valen L. A new evolutionary law. Evol. Theory. 1973;1:1–30. [Google Scholar]
- 29.Sawyer S.L. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:e275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawyer S.L. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elde N.C. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2008;457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell P.S. Evolution-guided identification of antiviral specificity determinants in the broadly acting interferon-induced innate immunity factor MxA. Cell Host Microbe. 2012;12:598–604. doi: 10.1016/j.chom.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel M.R. Convergent evolution of escape from hepaciviral antagonism in primates. PLoS Biol. 2012;10:e1001282. doi: 10.1371/journal.pbio.1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demogines A. Dual host–virus arms races shape an essential housekeeping protein. PLoS Biol. 2013;11:e1001571. doi: 10.1371/journal.pbio.1001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sironi M. Evolutionary insights into host–pathogen interactions from mammalian sequence data. Nat. Rev. Genet. 2015;16:224–236. doi: 10.1038/nrg3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barber M.F., Elde N.C. Escape from bacterial iron piracy through rapid evolution of transferrin. Science. 2014;346:1362–1366. doi: 10.1126/science.1259329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schryvers A.B. Characterization of the human transferrin and lactoferrin receptors in Haemophilus influenzae. Mol. Microbiol. 1988;2:467–472. doi: 10.1111/j.1365-2958.1988.tb00052.x. [DOI] [PubMed] [Google Scholar]
- 38.Cornelissen C.N. The transferrin receptor expressed by gonococcal strain FA1090 is required for the experimental infection of human male volunteers. Mol. Microbiol. 1998;27:611–616. doi: 10.1046/j.1365-2958.1998.00710.x. [DOI] [PubMed] [Google Scholar]
- 39.Yu R.H., Schryvers A.B. The interaction between human transferrin and transferrin binding protein 2 from Moraxella (Branhamella) catarrhalis differs from that of other human pathogens. Microb. Pathog. 1993;15:433–445. doi: 10.1006/mpat.1993.1092. [DOI] [PubMed] [Google Scholar]
- 40.Moraes T.F. Insights into the bacterial transferrin receptor: the structure of transferrin-binding protein B from Actinobacillus pleuropneumoniae. Mol. Cell. 2009;35:523–533. doi: 10.1016/j.molcel.2009.06.029. [DOI] [PubMed] [Google Scholar]
- 41.Noinaj N. Structural basis for iron piracy by pathogenic Neisseria. Nature. 2012;483:53–58. doi: 10.1038/nature10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.García-Montoya I.A. Lactoferrin a multiple bioactive protein: an overview. Biochim. Biophys. Acta. 2012;1820:226–236. doi: 10.1016/j.bbagen.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beddek A.J., Schryvers A.B. The lactoferrin receptor complex in Gram negative bacteria. Biometals. 2010;23:377–386. doi: 10.1007/s10534-010-9299-z. [DOI] [PubMed] [Google Scholar]
- 44.Noinaj N. Structural insight into the lactoferrin receptors from pathogenic Neisseria. J. Struct. Biol. 2013;184:83–92. doi: 10.1016/j.jsb.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamauchi K. Antibacterial activity of lactoferrin and a pepsin-derived lactoferrin peptide fragment. Infect. Immun. 1993;61:719–728. doi: 10.1128/iai.61.2.719-728.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haney E.F. Novel lactoferrampin antimicrobial peptides derived from human lactoferrin. Biochimie. 2009;91:141–154. doi: 10.1016/j.biochi.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 47.Hammerschmidt S. Identification of pneumococcal surface protein A as a lactoferrin-binding protein of Streptococcus pneumoniae. Infect. Immun. 1999;67:1683–1687. doi: 10.1128/iai.67.4.1683-1687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Senkovich O. Structure of a complex of human lactoferrin N-lobe with pneumococcal surface protein A provides insight into microbial defense mechanism. J. Mol. Biol. 2007;370:701–713. doi: 10.1016/j.jmb.2007.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deka R.K. Crystal structure of the Tp34 (TP0971) lipoprotein of Treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin. J. Biol. Chem. 2007;282:5944–5958. doi: 10.1074/jbc.M610215200. [DOI] [PubMed] [Google Scholar]
- 50.Flo T.H. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 51.Bachman M.A. Interaction of lipocalin 2, transferrin, and siderophores determines the replicative niche of Klebsiella pneumoniae during pneumonia. MBio. 2012;3:e00224–e311. doi: 10.1128/mBio.00224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hantke K. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abergel R.J. Anthrax pathogen evades the mammalian immune system through stealth siderophore production. Proc. Natl. Acad. Sci. U.S.A. 2006;103:18499–18503. doi: 10.1073/pnas.0607055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Posey J.E., Gherardini F.C. Lack of a role for iron in the Lyme disease pathogen. Science. 2000;288:1651–1653. doi: 10.1126/science.288.5471.1651. [DOI] [PubMed] [Google Scholar]
- 55.Aguirre J.D. A manganese-rich environment supports superoxide dismutase activity in a Lyme disease pathogen, Borrelia burgdorferi. J. Biol. Chem. 2013;288:8468–8478. doi: 10.1074/jbc.M112.433540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blount Z.D. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489:513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leiby N., Marx C.J. Metabolic erosion primarily through mutation accumulation, and not tradeoffs, drives limited evolution of substrate specificity in Escherichia coli. PLoS Biol. 2014;12:e1001789. doi: 10.1371/journal.pbio.1001789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chou H.H. Fast growth increases the selective advantage of a mutation arising recurrently during evolution under metal limitation. PLoS Genet. 2009;5:e1000652. doi: 10.1371/journal.pgen.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ashley-Koch A. Sickle hemoglobin (HbS) allele and sickle cell disease: a HuGE review. Am. J. Epidemiol. 2000;151:839–845. doi: 10.1093/oxfordjournals.aje.a010288. [DOI] [PubMed] [Google Scholar]
- 60.Vujić M. Molecular basis of HFE-hemochromatosis. Front. Pharmacol. 2014;5:42. doi: 10.3389/fphar.2014.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinberg E.D. Survival advantage of the hemochromatosis C282Y mutation. Perspect. Biol. Med. 2008;51:98–102. doi: 10.1353/pbm.2008.0001. [DOI] [PubMed] [Google Scholar]
- 62.Wright A.C. Role of iron in the pathogenesis of Vibrio vulnificus infections. Infect. Immun. 1981;34:503–507. doi: 10.1128/iai.34.2.503-507.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quenee L.E. Hereditary hemochromatosis restores the virulence of plague vaccine strains. J. Infect. Dis. 2012;206:1050–1058. doi: 10.1093/infdis/jis433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weinberg E.D. Microbial pathogens with impaired ability to acquire host iron. Biometals. 2000;13:85–89. doi: 10.1023/a:1009293500209. [DOI] [PubMed] [Google Scholar]
- 65.Olakanmi O. Hereditary hemochromatosis results in decreased iron acquisition and growth by Mycobacterium tuberculosis within human macrophages. J. Leukoc. Biol. 2007;81:195–204. doi: 10.1189/jlb.0606405. [DOI] [PubMed] [Google Scholar]
- 66.Sawyer S.L., Elde N.C. A cross-species view on viruses. Curr. Opin. Virol. 2012;2:561–568. doi: 10.1016/j.coviro.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karlsson E.K. Natural selection and infectious disease in human populations. Nat. Rev. Genet. 2014;15:379–393. doi: 10.1038/nrg3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schryvers A.B., Gonzalez G.C. Receptors for transferrin in pathogenic bacteria are specific for the host's protein. Can. J. Microbiol. 1990;36:145–147. doi: 10.1139/m90-026. [DOI] [PubMed] [Google Scholar]
- 69.Cornelissen C.N. Expression of gonococcal transferrin-binding protein 1 causes Escherichia coli to bind human transferrin. J. Bacteriol. 1993;175:2448–2450. doi: 10.1128/jb.175.8.2448-2450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Litt D.J. Neisseria meningitidis expressing transferrin binding proteins of Actinobacillus pleuropneumoniae can utilize porcine transferrin for growth. Infect. Immun. 2000;68:550–557. doi: 10.1128/iai.68.2.550-557.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oftung F. A mouse model utilising human transferrin to study protection against Neisseria meningitidis serogroup B induced by outer membrane vesicle vaccination. FEMS Immunol. Med. Microbiol. 1999;26:75–82. doi: 10.1111/j.1574-695X.1999.tb01374.x. [DOI] [PubMed] [Google Scholar]
- 72.Zarantonelli M.L. Transgenic mice expressing human transferrin as a model for meningococcal infection. Infect. Immun. 2007;75:5609–5614. doi: 10.1128/IAI.00781-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bitter W. The role of transferrin-receptor variation in the host range of Trypanosoma brucei. Nature. 1998;491:499–502. doi: 10.1038/35166. [DOI] [PubMed] [Google Scholar]
- 74.Steverding D. The significance of transferrin receptor variation in Trypanosoma brucei. Trends Parasitol. 2003;19:125–127. doi: 10.1016/s1471-4922(03)00006-0. [DOI] [PubMed] [Google Scholar]
- 75.Pishchany G. Specificity for human hemoglobin enhances Staphylococcus aureus infection. Cell Host Microbe. 2010;8:544–550. doi: 10.1016/j.chom.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Natarajan C. Epistasis among adaptive mutations in deer mouse hemoglobin. Science. 2013;340:1324–1327. doi: 10.1126/science.1236862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blair J.M.A. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 78.Lin L. Transferrin iron starvation therapy for lethal bacterial and fungal infections. J. Infect. Dis. 2014;210:254–264. doi: 10.1093/infdis/jiu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morris J.J. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio. 2012;3 doi: 10.1128/mBio.00036-12. e00036–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Griffin A.S. Cooperation and competition in pathogenic bacteria. Nature. 2004;430:1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 81.Ghoul M. An experimental test of whether cheating is context dependent. J. Evol. Biol. 2014;27:551–556. doi: 10.1111/jeb.12319. [DOI] [PubMed] [Google Scholar]
- 82.Deriu E. Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nissle A. [Explanations of the significance of colonic dysbacteria and the mechanism of action of E. coli therapy (mutaflor)] Medizinische. 1959;4:1017–1022. (in German) [PubMed] [Google Scholar]
- 84.Jacobi C.A., Malfertheiner P. Escherichia coli Nissle 1917 (Mutaflor): new insights into an old probiotic bacterium. Dig. Dis. 2011;29:600–607. doi: 10.1159/000333307. [DOI] [PubMed] [Google Scholar]
- 85.Morris J.J. Black Queen evolution: the role of leakiness in structuring microbial communities. Trends Genet. 2015;31:475–482. doi: 10.1016/j.tig.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 86.Subashchandrabose S., Mobley H.L.T. Back to the metal age: battle for metals at the host–pathogen interface during urinary tract infection. Metallomics. 2015;7:935–942. doi: 10.1039/c4mt00329b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gellatly S.L., Hancock R.E.W. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog. Dis. 2013;67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 88.Marvig R.L. Within-host evolution of Pseudomonas aeruginosa reveals adaptation toward iron acquisition from hemoglobin. MBio. 2014;5:e00966–e1014. doi: 10.1128/mBio.00966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheng S-C. Interplay between Candida albicans and the mammalian innate host defense. Infect. Immun. 2012;80:1304–1313. doi: 10.1128/IAI.06146-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen C. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe. 2011;10:118–135. doi: 10.1016/j.chom.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Noble S.M. Candida albicans specializations for iron homeostasis: from commensalism to virulence. Curr. Opin. Microbiol. 2013;16:708–715. doi: 10.1016/j.mib.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Troxell B., Hassan H.M. Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell Infect. Microbiol. 2013;3:59. doi: 10.3389/fcimb.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marri P.R. Genome sequencing reveals widespread virulence gene exchange among human Neisseria species. PLoS ONE. 2010;5:e11835. doi: 10.1371/journal.pone.0011835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rotman E., Seifert H.S. The genetics of Neisseria species. Annu. Rev. Genet. 2014;48:405–431. doi: 10.1146/annurev-genet-120213-092007. [DOI] [PubMed] [Google Scholar]
- 95.Liu J.Z. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe. 2012;11:227–239. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gaddy J.A. The host protein calprotectin modulates the Helicobacter pylori cag type IV secretion system via zinc sequestration. PLoS Pathog. 2014;10:e1004450. doi: 10.1371/journal.ppat.1004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Corbin B.D. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 98.Kehl-Fie T.E., Skaar E.P. Nutritional immunity beyond iron: a role for manganese and zinc. Curr. Opin. Chem. Biol. 2010;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kronstad J.W. An encapsulation of iron homeostasis and virulence in Cryptococcus neoformans. Trends Microbiol. 2013;21:457–465. doi: 10.1016/j.tim.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jung W.H., Do E. Iron acquisition in the human fungal pathogen Cryptococcus neoformans. Curr. Opin. Microbiol. 2013;16:686–691. doi: 10.1016/j.mib.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 101.Richardson M.W. Stabilized human TRIM5α protects human T cells from HIV-1 infection. Mol. Ther. 2014;22:1084–1095. doi: 10.1038/mt.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tebas P. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Van Der Ryst E. Maraviroc – a CCR5 antagonist for the treatment of HIV-1 infection. Front. Immunol. 2015;6:277. doi: 10.3389/fimmu.2015.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]