Summary
Aminoacyl-phosphatidylglycerol synthases (aaPGSs) are membrane proteins that utilize aminoacylated tRNAs to modify membrane lipids with amino acids. Aminoacylation of membrane lipids alters the biochemical properties of the cytoplasmic membrane, and enables bacteria to adapt to changes in environmental conditions. aaPGSs utilize alanine, lysine, and arginine as modifying amino acids, and the primary lipid recipients have heretofore been defined as phosphatidylglycerol (PG) and cardiolipin. Here we identify a new pathway for lipid aminoacylation, conserved in many Actinobacteria, which results in formation of Ala-PG and a novel alanylated lipid, Ala-diacylglycerol (Ala-DAG). Ala-DAG formation in Corynebacterium glutamicum is dependent on the activity of an aaPGS homolog, while formation of Ala-PG requires the same enzyme acting in concert with a putative esterase encoded upstream. The presence of alanylated lipids is sufficient to enhance the bacterial fitness of C. glutamicum cultured in the presence of certain antimicrobial agents, and elucidation of this system expands the known repertoire of membrane lipids acting as substrates for amino acid modification in bacterial cells.
Keywords: phospholipids, multiple peptide resistance factor (MprF), Actinobacteria, diacylglycerol, phosphatidylglycerol, aminoacyl-tRNA synthetase, transfer RNA (tRNA), amino acid, lipids, membrane proteins
Introduction
Aminoacyl-phosphatidylglycerol synthases (aaPGSs) are integral membrane proteins responsible for the synthesis of aminoacyl-phosphatidylglycerol (aa-PG) in the cytoplasmic membranes of bacteria. The hydrophilic domain of aaPGS, located in the cytoplasm, bears the active site of the transferase activity and utilizes aminoacyl-tRNAs (aa-tRNAs) as amino acid (aa) donors, and phosphatidylglycerol (PG) as an aa acceptor (Peschel et al., 2001). A separate domain, integral to the membrane, is required for translocation of newly formed aminoacylated phospholipids from the inner leaflet of the membrane to the outer leaflet (Ernst et al., 2009, Ernst & Peschel, 2011, Slavetinsky et al., 2013, Ernst et al., 2015). Initially discovered in Staphylococcus aureus, the gene mprF encodes a LysPGS, which attaches lysine to PG, increasing the organism’s virulence, as well as resistance to various classes of antibacterial agents (i.e., cationic antimicrobial peptides (Peschel et al., 2001), beta-lactams (Komatsuzawa et al., 2001), glycopeptides, and lipopeptides (Hachmann et al., 2009)). Investigation of aaPGS homologs in other species allowed for discovery of enzymes that exhibit different aa-tRNA specificities. For example, two aaPGS paralogs in C. perfringens, LysPGS and AlaPGS, are responsible for the synthesis of Lys-PG and Ala-PG, respectively (Roy & Ibba, 2008b). Investigation of an aaPGS homolog from Enterococcus faecium revealed a triple specific enzyme (i.e., RakPGS) that enables formation of Lys-PG, Ala-PG and Arg-PG. The function of a second paralog in this species remains to be determined (Roy & Ibba, 2009). To date, the specificity of a total of 23 aaPGSs has been characterized either directly in vitro, or by analyzing aminoacylated lipids produced in vivo (Arendt et al., 2012). Lys-tRNALys, Ala-tRNAAla, and Arg-tRNAArg are currently the only known aa donors for aa-PG synthesis, while PG is the only known naturally occurring aa acceptor, with the exception of di-phosphatidylglycerol (i.e., cardiolipin), which acts as an aa acceptor (in addition to PG) in Listeria monocytogenes (Thedieck et al., 2006). Besides the different aa and lipid substrate specificities, some more distant aaPGS homologs exhibit functions that are quite different than those of traditional aaPGSs. One example is the protein VlmA that is found in various Actinobacteria, and which was the focus of a recent study carried out in Streptomyces viridifaciens. VlmA was found to be unable to aminoacylate any membrane lipids. Instead, this protein was shown to utilize Ser-tRNASer for serine addition to an intermediate in the pathway for biosynthesis of the antibiotic valanimycin (Garg et al., 2008). Thus, the aaPGS family of proteins is a heterogeneous family of tRNA-dependent transferases that exhibit diverse substrate specificities. Despite recent progress, no relationship between the structure and function of aaPGS homologs has been established, and it is not possible to predict the function of individual enzymes based solely on primary sequence (Arendt et al., 2012). Moreover, it is not known whether the aaPGS family of proteins contains additional enzymes with functions that are distinct from those described to date.
A recent analysis to identify potential functional associations between aaPGSs and other genes revealed three conserved families of proteins encoded at aaPGS loci. Each of the three families, α/β-hydrolases (Pfam: PF12697), esterases (Pfam: PF00756), and VirJ proteins (Pfam: PF06057), represents a distinct class of putative hydrolytic enzymes distributed within specific bacterial phyla (Smith et al., 2013). Enzymes belonging to the VirJ family of proteins are found exclusively in Gram-negative bacteria, whereas proteins from the α/β-hydrolases family are found in both Gram-positive and Gram-negative species. Proteins from the esterase group, on the other hand, are found mostly in Actinobacteria and Cyanobacteria. Two of these putative hydrolytic enzymes, AhyD from E. faecium and PA0919 from P. aeruginosa, representing the α/β-hydrolase class of enzymes and the VirJ proteins, respectively, were recently the focus of independent functional studies (Smith et al., 2013, Arendt et al., 2013). Both of these proteins were shown to be aa-PG hydrolases that modulate overall levels of aa-PG in the bacterial membrane, and that are necessary for providing optimal resistance to antibiotics and other stress conditions. The function of proteins belonging to the third group of hydrolytic enzymes (i.e., the esterases) has not yet been determined.
During the course of this study, we performed an analysis on aaPGS sequences and identified a new type of aaPGS homolog found exclusively in Actinobacteria. We studied the role of one representative protein (Cg1103) from Corynebacterium glutamicum, a Gram-positive, non-motile, soil dwelling bacterium, and showed that this enzyme displays a tRNA-dependent alanyl-diacylglycerol synthase (AlaDAGS) activity, which synthesizes Ala-DAG using Ala-tRNAAla as an aa donor and diacylglycerol (DAG) as an aa acceptor. Although Cg1103 was unable to synthesize Ala-PG in vitro or in vivo, we found that Ala-PG synthesis could be reconstituted by co-expression of Cg1103 with PesT, a conserved putative esterase (Cg1104) encoded upstream and adjacent to cg1103. These results suggest that the cg1103/pesT system generates two distinct alanylated lipids, expanding the known range of lipid components utilized for aa modification. Growth curve analysis demonstrated that expression of cg1103/pesT moderately enhances the fitness of C. glutamicum grown in the presence of lactic acid and the antimicrobial agents protamine sulfate and polymyxin B.
Results
aaPGS homologs consist of seven types of proteins
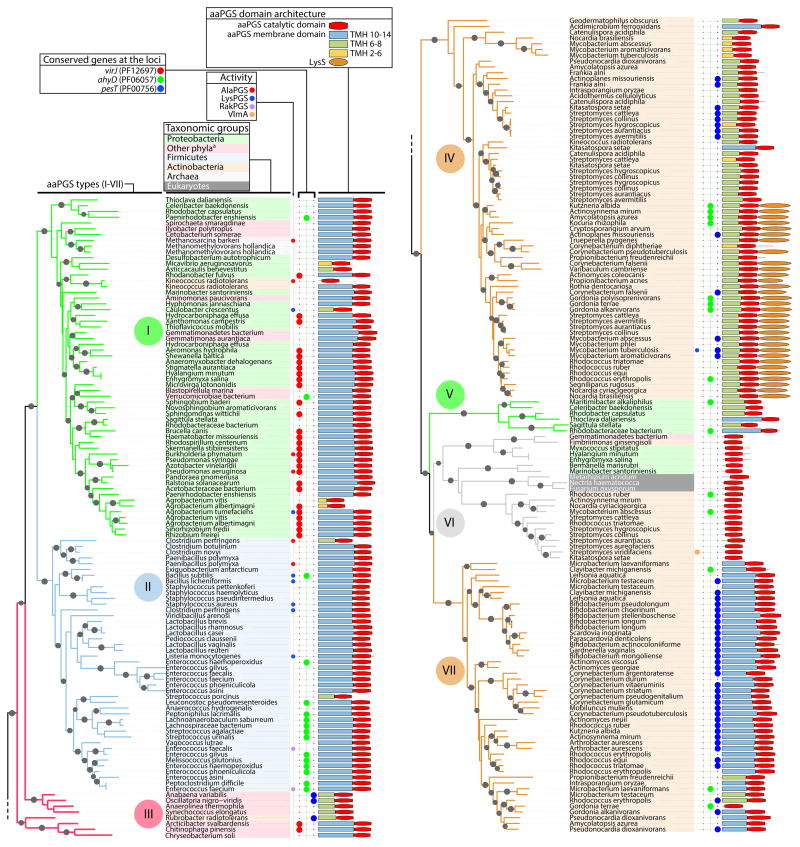
To determine whether aaPGSs can be categorized into distinct structural types, and whether, in light of recent functional studies, these structural types can be correlated to function, 257 representative sequences of aaPGSs were aligned and compared. The set of sequences used in this analysis originated from 174 bacterial species, each containing one or more aaPGS homolog. A phylogenetic analysis was performed on 160 degapped positions of the aminoacyl-transferase domain (in the C-terminus) of the proteins. Based on the topology of the resulting phylogenetic tree, aaPGS homologs can be delineated into seven main types of proteins (I–VII, with nodes exhibiting local support values >0.75; Fig. 1). To understand whether the seven types correlate to functions observed in aaPGS homologs, additional information such as substrate specificities of characterized enzymes, origin of the sequences, and information regarding the architecture of the proteins and loci, was added to the analysis. This method revealed that each of the aaPGS structural types are confined to specific bacterial phyla, suggesting that the dichotomy between types may reflect phylogenetic origin rather than functional differences. Type I and type III enzymes are found mostly in Proteobacteria, type II are found in Firmicutes, type IV and type VII are found in Actinobacteria, and type VI are found in Actinobacteria and some Proteobacteria.
Fig. 1. Phylogenetic analysis of aaPGS homologs.
Phylogenetic tree was constructed based on alignment of 160 residues in the C-terminal domain of 253 aaPGS homolog sequences using the program FastTree. Organism names are shown on a color background delineating the major taxonomic groups. The functions of characterized homologs (LysPGS, AlaPGS, RakPGS, VlmA), occurrence of adjacently encoded hydrolytic enzymes (VirJ, AhyD, PesT), and aaPGS domain architectures are indicated for each organism. When present, the number of predicted transmembrane helices (TMH 2–14) in the membrane domain is color-coded. Sequence identification (GI) numbers are shown in Fig. S3. aBacteria from these phyla belong to one of the following: Fusobacteria, Gemmatimonadetes, Planctomycetes, Bacteroidetes, Chloroflexi, Cyanobacteria, Spirochaetes, Synergistetes, Verrucomicrobia.
Type I and II homologs include numerous aaPGSs that have been well characterized including LysPGSs, AlaPGSs, and the enterococcal triple specific RakPGS. The actinobacterial type IV homologs exhibit a putative membrane domain that is shorter than that of type I and type II proteins, while some type IV proteins exhibit an additional C-terminal domain containing a class II lysyl-tRNA synthetase (Fig. 1). Characterization of one type IV homolog, the protein LysX from Mycobacterium tuberculosis, showed that this enzyme is responsible for synthesis of Lys-PG (Maloney et al., 2009). Type VI homologs include the VlmA protein, which catalyzes a tRNA-dependent aa addition during biosynthesis of valanimycin (Garg et al., 2008). This activity correlates with the lack of a membrane domain, suggesting that type VI proteins are probably not associated with the membrane. There is currently no evidence regarding the function of type III, V, and VII homologs. Type III and type V homologs, which are found primarily in Gram-negative species, tend to display shorter transmembrane domains than the other types. Type VII enzymes, on the other hand, display (like most type I and type II aaPGSs) a membrane domain composed of 14 predicted transmembrane helices.
Some bacterial species exhibit multiple aaPGS homologs with distinct functions (i.e., paralogs). In general, paralogs arise through a gene duplication event and acquire distinct functions that are mechanistically related during the course of evolution (Koonin, 2005). To gain additional clues into the functional roles of aaPGSs, we considered the co-occurrence of multiple paralogs in individual species (Table 1 and Fig. S1). Multiple aaPGS paralogs are encountered most frequently in actinobacterial species (Fig. S1). For example, certain Actinobacteria contain up to five distinct aaPGS paralogs, whereas Firmicutes display a maximum of two paralogs in a given species. About 50% of the actinobacterial species included in our dataset displayed two or more aaPGS paralogs, whereas only 20% of the species belonging to other phyla exhibited two paralogs. Table 1 summarizes the co-occurrence of aaPGS paralogs in relation to the seven main types of aaPGSs. In some species, all the aaPGS paralogs belong to a single type (e.g., both the AlaPGS and LysPGS from C. perfringens are type II enzymes). In other cases, multiple types of aaPGSs co-occur in a single species (e.g., Corynebacterium pseudotuberculosis contains a type IV and a type VII homolog), suggesting that these types may exhibit distinct functionalities.
TABLE 1.
Co-occurrence of aaPGS paralogs according to type defined in Fig. 1
| Paralog type | I | II | III | IV | V | VI | VII |
|---|---|---|---|---|---|---|---|
| I | + | + | + | + | |||
| II | + | ||||||
| III | |||||||
| IV | + | + | + | + | |||
| V | + | ||||||
| VI | + | + | + | ||||
| VII | + | + | + |
The co-occurrence of aaPGS paralogs was analyzed according to the seven aaPGS types defined in Fig. 1. For instance, some organisms harboring a type VI homolog also harbor a type I, type IV, and/or type VII aaPGS homolog. A comprehensive depiction of distribution and co-occurrence of each type is shown in Fig. S1.
Our sequence analysis also included the three families of putative hydrolytic enzymes that are encoded within the aaPGS loci in many bacterial species (Smith et al., 2013). While enzymes belonging to the α/β-hydrolase and VirJ families of proteins have been shown to be aa-PG hydrolases (Smith et al., 2013, Arendt et al., 2013), the function of proteins belonging to the esterase family (PF00756) is still unknown. To determine whether the putative esterases correlate with a certain type of aaPGS homolog, the distribution of these enzymes was considered. The putative esterases were found to be primarily associated with type VII and type IV aaPGS homologs. Based on the observations that type VII and type IV homologs sometimes co-occur in a single species (Fig. S1), and that some type IV homologs exhibit a LysPGS activity, we hypothesized that type VII paralogs might bear an alternate functionality. We selected the type VII enzyme Cg1103 from C. glutamicum (accession number CAF19673) for further analysis. The genomic context of the cg1103 gene, along with the putative esterase pesT, is depicted in Fig. S2.
aaPGS from C. glutamicum aminoacylates a lipid other than PG
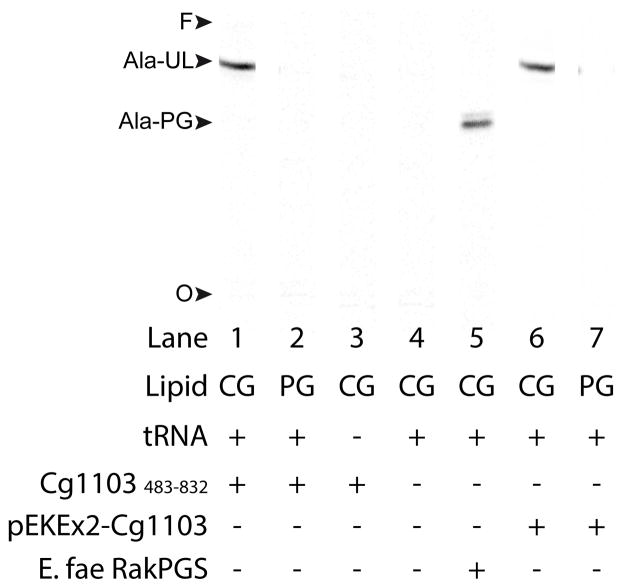
To determine whether type VII homologs exhibit aaPGS activity, the gene cg1103 from C. glutamicum was cloned and expressed in Escherichia coli, which does not harbor an endogenous aaPGS activity. Cg1103 was expressed as a full-length protein, or as an N-terminal 6xHis-tagged variant deprived of the membrane domain and encompassing only the last 350 residues of the C-terminus (residues 483–832). The membrane domains of several aaPGSs have been shown to be dispensable for reconstituting enzymatic activity in vitro (Roy & Ibba, 2008b), and so the latter construct (Cg1103483–832) was included in our studies to facilitate expression of this large membrane protein. Indeed, we were able to isolate heterologously expressed Cg1103483–832 as a soluble, pure protein after a single step of affinity chromatography. Cg1103483–832 was tested for tRNA-dependent transfer of aa using [14C]Lys-tRNALys or [14C]Ala-tRNAAla as aa donors, and commercial egg PG, or total lipids extracted from C. glutamicum, as aa acceptors. Cg1103483–832 was unable to modify any of the lipid sources with lysine (data not shown). However, Cg1103483–832 was capable of alanylation of an unknown lipid (UL) when C. glutamicum lipid extract was used as the aa acceptor (Fig. 2, lane 1). The alanylated lipid (Ala-UL) exhibited a chromatographic migration pattern different from that of an Ala-PG control, formed using the RakPGS from E. faecium and C. glutamicum lipids as aa acceptors (lane 5). Ala-UL was not observed when commercial PG was substituted as a lipid source (lane 2), or when tRNA or enzyme was eliminated from the reaction (lane 3, 4). Membrane extracts from the E. coli strain expressing full-length Cg1103 did not yield activity with any of the substrates tested, suggesting there may have been a problem with expression of this construct (data not shown).
Fig. 2. tRNA-dependent lipid alanylation by Cg1103.
tRNA-dependent lipid alanylation activity was measured in vitro using either the purified construct Cg1103483–832 (lanes 1–3), or a membrane preparation from C. glutamicum harboring pEKEx2-Cg1103 (lanes 6 and 7). The lipids used as aa acceptors were a total lipid preparation isolated from C. glutamicum (CG), or commercial egg-PG (PG). Lipid products were analyzed by TLC using solvent A for the mobile phase. Activity with a membrane extract isolated from E. coli cells expressing RakPGS from E. faecium is shown as a positive control for [14C]Ala-PG synthesis (lane 5). O: TLC origin, F: solvent front, Ala-UL: alanylated unknown lipid.
To verify that synthesis of Ala-UL by Cg1103483–832 was not due to lack of the membrane domain in the truncated protein, the full-length ORF encoding Cg1103 was cloned into the expression vector pEKEx2-GFP (yielding the construct pEKEx2-Cg1103), and expressed in C. glutamicum. Activity in a membrane extract isolated from this strain was tested using commercial PG, or lipids extracted from C. glutamicum, as potential aa acceptors. A single product with chromatographic mobility identical to that of the Ala-UL formed by the truncated enzyme was observed in reactions containing C. glutamicum lipid extract as aa acceptors (Fig. 2, lane 6). This product was not observed in reactions containing commercial PG as a lipid source (lane 7). These findings show that both Cg1103483–832 and the full-length enzyme are able to utilize [14C]Ala-tRNAAla to form [14C]Ala-UL (but not [14C]Ala-PG) in vitro.
Identification of diacylglycerol as a substrate of Cg1103
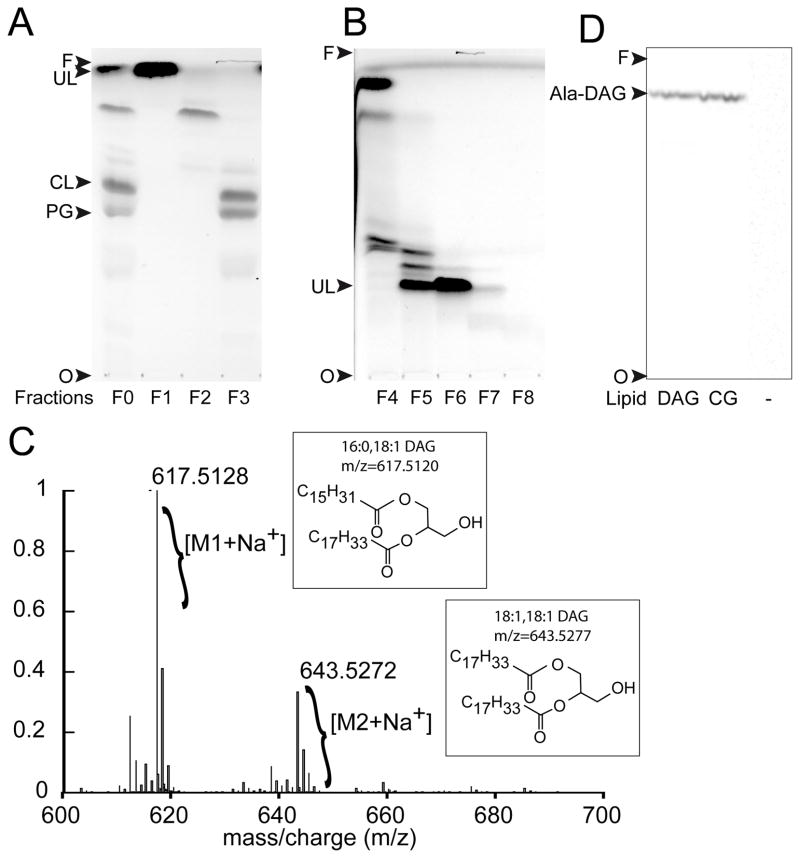
For identification, UL was purified from a lipid extract, prepared from the wild-type strain of C. glutamicum, and characterized by mass spectrometry. Total lipids were extracted using the Bligh and Dyer procedure (Bligh & Dyer, 1959), and non-lipid contaminants were eliminated using the Folch method (Folch et al., 1957). Lipid classes were fractionated by flash chromatography on silica gel to yield three fractions containing non-polar lipids (fraction F1), glycolipids (fraction F2), and phospholipids (fraction F3, Fig. 3A). The presence or absence of UL in each fraction was determined based on the enzymatic activity observed when Cg1103483–832 was added to an aliquot from each fraction (data not shown). UL was only detected in fraction F1 containing the non-polar lipids. A second step of flash chromatography on silica gel was performed to further purify UL, yielding a single fraction containing UL at approximately 90% purity as estimated by TLC analysis (Fig. 3B, Fraction F6).
Fig. 3. Identification of DAG as a lipid substrate of Cg1103.
(A) TLC analysis (using solvent A for the mobile phase) of lipids extracted from the wild-type strain of C. glutamicum, before (F0) and after (F1–F3) fractionation of lipid classes by flash chromatography. TLC spots were isolated and tested for tRNA-dependent lipid alanylation activity to identify the unknown lipid (UL) substrate of Cg1103. (B) Lipids in fraction F1 were further separated using a second step of flash chromatography on silica gel. Resulting fractions (F4–F8) were analyzed by TLC using solvent B for the mobile phase. (C) ESI/TOF mass spectrum of purified UL from fraction F6. (D) tRNA-dependent lipid alanylation activity of Cg1103483–832 was assayed in the presence of commercial 18:1-16:0 DAG, lipids extracted from C. glutamicum (CG), or in the absence of lipids (−). Lipids modified with [14C]Ala were analyzed by TLC (in solvent A), and visualized by phosphorimaging. Ala-DAG: alanyl-diacylglycerol, CL: cardiolipin, PG: phosphatidylglycerol, O: TLC origin, F: solvent front.
Figure 3C shows the ESI/TOF mass spectrum of fraction F6 containing UL. The main peak at a m/z ratio of 617.5128, and the isotopic distribution of the corresponding compound, are characteristic of diacylglycerol (DAG) substituted with palmitoyl and oleoyl fatty acid chains (16:0–18:1 DAG). The second most abundant compound present in the preparation exhibited a m/z value of 643.5272, corresponding to dioleyl-glycerol (18:1,18:1 DAG). The observation of these acyl chains is consistent with those that are found in several species of Corynebacterium (see (Crellin et al., 2013) for review). To confirm that DAG is a substrate of Cg1103, we tested a commercial preparation of 16:0–18:1 DAG as a [14C]-Ala acceptor. Alanylated DAG (Ala-DAG) was formed in vitro and displayed a chromatographic mobility identical to that of the Ala-UL formed when total lipids from C. glutamicum were used as aa acceptors (Fig. 3D). These findings demonstrate that Cg1103 exhibits an alanyl-diacylglycerol synthase (AlaDAGS) activity, which uses DAG and Ala-tRNAAla as substrates to form Ala-DAG.
Expression of Cg1103 in C. glutamicum produces Ala-DAG and Ala-PG
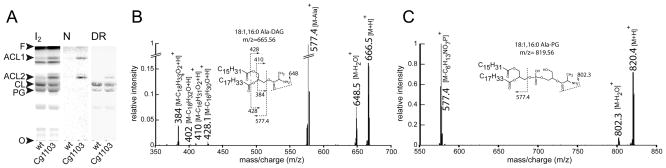
To verify whether Ala-DAG is formed by Cg1103 in vivo, C. glutamicum lipids from the wild-type strain, or from the strain harboring the expression plasmid pEKEx2-Cg1103, were analyzed by TLC. Total lipids were visualized using iodine vapor, and amine-containing lipids (ACL) were revealed by treatment with ninhydrin. No ACLs were detected in the wild-type strain, suggesting that aminoacylated lipids are not abundant in cells grown in the standard laboratory conditions used here. However, two ACLs were detected in the lipid preparation isolated from the strain expressing Cg1103 (Fig. 4A). One ACL (ACL1) exhibited mobility consistent with Ala-DAG, whereas the second one (ACL2) displayed a slower migration pattern similar to Ala-PG. Additional insight into the identities of ACL1 and ACL2 was gained using Dittmer dye (Dittmer & Lester, 1964), which confirmed that ACL2 contains phosphorous, while ACL1 does not (Fig. 4A, lane 4). To verify the identities of ACL1 and ACL2, both lipids were purified from a C. glutamicum total lipid extract isolated from cells expressing Cg1103. Lipid classes were fractionated by flash chromatography on silica gel as described above. Non-polar lipids and ACL1 were sequentially eluted with chloroform, and ACL2 was eluted, along with other phospholipids, using methanol (Figure S4A). Fractions containing each ACL were subjected to a second round of flash chromatography yielding ACL1 at > 90% purity (Fig. S4B, Fraction 3), and ACL2 mixed with a small amount (~10%) of contaminating PG (Fig. S4C, Fraction 3). Both lipids were characterized by ESI/TOF and ESI/MS/MS. Exact mass, isotopic distribution (Fig. S5), and fragmentation patterns of the major peaks confirmed the identities of ACL1 and ACL2 as being Ala-DAG and Ala-PG, respectively (Fig. 4B and 4C). Acyl moieties observed in the purified Ala-DAG and Ala-PG (i.e., oleoyl and palmitoyl) were the same as those found in the non-alanylated DAG characterized above. These findings demonstrate that expression of Cg1103 in wild-type C. glutamicum yields synthesis of Ala-DAG and Ala-PG in vivo. These findings only partially corroborate the in vitro results, which showed that Cg1103 is able to alanylate DAG, but not PG.
Fig. 4. ESI/MS/MS analysis of amine-containing lipids in C. glutamicum.
(A) Lipids from wild-type C. glutamicum (wt), and a strain harboring the plasmid pEKEx2-Cg1103, were analyzed by TLC using solvent A as the mobile phase. Total lipids were revealed using iodine vapor (I2), and amine and phosphorus containing lipids were revealed using ninhydrin (N) and Dittmer reagent (DR), respectively. Amine-containing lipids (ACL1 and ACL2) were purified using a two-step flash chromatography procedure as described in methods (see Fig. S4 for fractionation data) and analyzed by mass spectrometry. ESI/MS/MS spectra of fractions containing ACL1 (B) and ACL2 (C) revealed molecular ion and fragmentation peaks consistent with m/z values for Ala-DAG (665.56) and Ala-PG (810.56), respectively. CL: cardiolipin, PG: phosphatidylglycerol, O: TLC origin, F: solvent front.
Ala-PG synthesis is dependent on a putative esterase encoded upstream of cg1103
The ORFs encoding the putative esterase pesT (accession CAF19674) and cg1103 are separated by only 2 nucleotides, and potentially constitute a single operon. PesT is a 426 aa protein, belonging to a family of putative hydrolytic enzymes (Pfam: PF00756) that are frequently encoded at the same loci as type VII aaPGS homologs (Fig. 1). 32 putative esterase sequences, encoded in organisms that also harbor a type VII aaPGS homolog, were subjected to a membrane domain prediction analysis using the program TOPCONS. This analysis revealed that these proteins are made up of two domains. The N-terminal portion contains a membrane domain consisting of four transmembrane helices, and the C-terminal portion, which is more hydrophilic, contains a putative esterase signature exhibiting a conserved serine residue (Pfam: PF00756). In 26 of the proteins, the C-terminal domain of the protein was predicted to be localized to the outer side of the cytoplasmic membrane. These observations suggested that PesT might work in concert with Cg1103 to affect the modification of lipids in C. glutamicum.
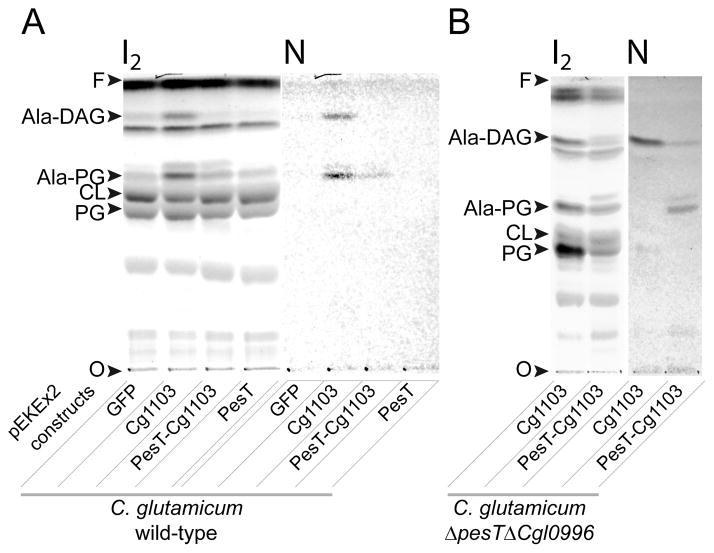
To gain insight into the possible role of pesT in lipid aminoacylation, two constructs were cloned into the pEKEx2-GFP expression vector, and transformed into wild-type C. glutamicum. The first construct included the entire pesT-cg1103 operon, and the second harbored the pesT ORF alone. Figure 5A shows that expression of PesT by itself did not induce synthesis of any ACLs in vivo, whereas expression of PesT and Cg1103 together resulted in synthesis of Ala-PG, but not Ala-DAG. These results, together with the observation that Cg1103 uses DAG, but not PG as a substrate in vitro (Fig. 2), led to the hypothesis that PesT is required for formation of Ala-PG. This hypothesis satisfies the initially confounding observation that expression of Cg1103 in wild-type C. glutamicum, which contains pesT on the chromosome, was capable of synthesizing both Ala-DAG and Ala-PG (Fig. 5A). To further test this hypothesis we constructed a C. glutamicum mutant strain in which the region encompassing pesT and cg1103 was deleted (ΔpesTΔcg1103). The ΔpesTΔcg1103 strain was transformed with the different pEKEx2 constructs harboring GFP (as a negative control), cg1103, or both cg1103 and pesT. Similar to wild-type C. glutamicum, no lipid aminoacylation was measured in strain ΔpesTΔcg1103 transformed with either pEKEx2-GFP or pEKEx2-pesT (data not shown). However, expression of Cg1103 in the double knockout strain resulted in formation of Ala-DAG, while co-expression of Cg1103 and PesT yielded Ala-PG and, to a lesser extent, Ala-DAG (Fig. 5B). These results support the hypothesis that synthesis of Ala-PG is dependent on the presence of both pesT and cg1103, whereas synthesis of Ala-DAG is catalyzed by cg1103 alone.
Fig. 5. Expression of Cg1103 and PesT in C. glutamicum.
TLC analysis of lipids isolated from wild-type C. glutamicum (A), and a deletion strain Δcg1103ΔpesT (B), transformed with the expression vector pEKEx2 harboring GFP (negative control) cg1103, pesT, or both genes. TLCs were developed in solvent A. Total lipids were visualized with iodine vapor (I2), and Ala-DAG and Ala-PG were detected with ninhydrine (N). CL: cardiolipin, PG: phosphatidylglycerol, O: TLC origin, F: solvent front.
cg1103 confers a fitness advantage to C. glutamicum grown in certain conditions of stress
To date, synthesis of aa-PG in some organisms has been linked to an increase in the minimum inhibitory concentrations (MICs) of various drugs such as certain cationic antimicrobial peptides (CAMPs), lipopetides and glycopetides (e.g., vancomycin and daptomycin), and to some osmolytes such as lactic acid (Klein et al., 2009, Dare et al., 2014) (for review see (Roy, 2009)). To determine whether cg1103 or pesT provides C. glutamicum with resistance to some of these compounds, markerless deletion strains lacking the two genes were constructed, and the MICs for various CAMPs and other types of inhibitors were measured. No difference was observed between the MICs for wild-type C. glutamicum and the mutant strains, Δcg1103 and ΔpesT, when tested in the presence of nisin (20 μg ml−1), bacitracin (0.5 μg ml−1), polymyxin B (1.6 μg ml−1), protamine sulfate (40 μg ml−1), daptomycin (3.3 μg ml−1), vancomycin (0.2 μg ml−1), and lactic acid (0.2 % v/v).
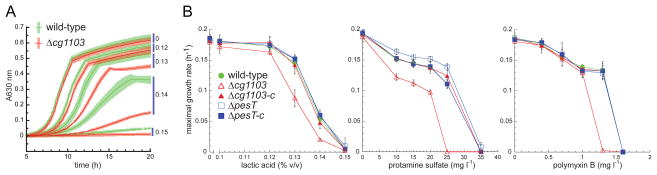
To determine whether lipid alanylation affects C. glutamicum fitness (i.e., the ability of the organism to adapt to conditions in its environment), growth of the wild-type strain was compared to that of the Δcg1103 and ΔpesT strains in the presence or absence of the compounds listed above. Growth was monitored continuously in BHI broth, and the maximum growth rate (μmax), which is commonly used for evaluating bacterial fitness ((Walkiewicz et al., 2012, Andersson & Hughes, 2010)), was calculated using the grofit R package (Kahm et al., 2010). In the absence of drugs, growth of C. glutamicum was not affected by the loss of either cg1103 or pesT in the knockout strains (Fig. 6). Among the compounds tested, only lactic acid, protamine sulfate, and polymyxin B had a markedly higher inhibitory effect on the Δcg1103 strain than on wild-type cells. Inhibition of growth of the Δcg1103 strain and wild-type C. glutamicum, in the presence of lactic acid, is shown as an example in Figure 6A, and the fitness of each strain (i.e., the μmax) is plotted relative to variable concentrations of each of the three compounds in Figure 6B. The fitness advantage of the wild-type strain over the Δcg1103 mutant was observed in a narrow range of concentrations for each compound. For example, upon addition of 0.13 % lactic acid, or 20 mg l−1 of protamine sulfate, the Δcg1103 cell growth rate was ~30% lower than that of the wild-type strain. At higher concentrations of each of these two compounds, no growth was recorded for the Δcg1103 mutant, whereas the wild-type strain exhibited some growth within 20 h. Likewise, for polymyxin B, the Δcg1103 strain was unable to thrive with drug concentrations at or above 1.3 mg l−1 while the wild-type strain continued to grow. The inhibited growth phenotype of the Δcg1103 strain was completely reverted back to wild-type levels in the complementation strain, Δcg1103-c, in which the cg1103 gene was replaced in the genome at the original locus (Fig. 6B).
Fig. 6. Fitness of C. glutamicum in the presence of selected antimicrobial compounds.
(A) Effect of different concentrations of lactic acid on the growth of wild-type C. glutamicum (green) and the Δcg1103 mutant strain (red). Percent of lactic acid (v/v) is indicated on the right. For each time point, the average value (n=4) of the A630 is indicated by a dark line, and the standard deviation is represented by a light colored area. (B) Derived dose-response curves from growth experiments performed in the presence of lactic acid, protamine sulfate, or polymyxin B. μmax values were calculated for each growth curve using the grofit R package. Averages from four experiments are shown (n=4), and error bars represent standard deviations. Δcg1103-c and ΔpesT-c represent complementation strains in which the deleted genes have been replaced back into the original loci. A growth rate of 0 indicates that no growth was recorded after an incubation period of 20 h.
While a difference was observed in the growth of the Δcg1103 strain (relative to wild-type) in the presence of the three compounds, no similar effect was observed with the ΔpesT strain (Fig. 6B). Both the wild-type strain and the ΔpesT strain grew equally well in the presence of all of the compounds tested. Altogether, these results suggest that Ala-DAG alone is able to confer a fitness advantage to C. glutamicum in the presence of certain inhibitors, and the physiological relevance of Ala-PG is not immediately apparent based on these experiments.
Discussion
An AlaDAGS and a putative esterase are responsible for synthesis of two alanylated lipids in C. glutamicum
aaPGSs are a family of enzymes capable of using various aa-tRNAs as aa donors for aminoacylation of phospholipids. The most common lipid that acts as an aa acceptor is PG, while di-PG (i.e., cardiolipin) has also been reported to be an efficient substrate for LysPGS in L. monocytogenes (Thedieck et al., 2006). Here we identify a novel pathway for membrane lipid aminoacylation. This pathway, which is present in various Actinobacteria species, involves an aaPGS homolog (Cg1103), and a putative esterase (PesT) that is encoded upstream. In vitro and in vivo data demonstrated that Cg1103 is able to alanylate the membrane lipid DAG, while synthesis of a second aminoacylated lipid, Ala-PG, was found to be dependent on co-expression of Cg1103 and PesT.
Additional work is required to clarify the precise role of PesT in this process. One possibility is that Cg1103 alone synthesizes Ala-DAG, but the presence of PesT modulates the specificity of Cg1103 to allow formation of Ala-PG. In this scenario, PesT may or may not exhibit an Ala-DAG hydrolytic activity. In an alternative model, Cg1103 synthesizes Ala-DAG, and PesT supports a transferase activity (instead of a hydrolytic one) that transfers the Ala born by DAG, to PG, to form Ala-PG. This latter scenario is supported by the predicted membrane topology of PesT, which places the location of the active site of the protein on the outer surface of the cytoplasmic membrane. The periplasmic localization of the active site of PesT would allow for synthesis of Ala-PG on the outer face of the membrane, using Ala-DAG as an aa donor, without depleting PG from the inner leaflet.
Although the putative esterase PesT exhibits a conserved Ser residue for potential catalysis of a hydrolytic reaction, this residue could instead be involved in an acyltransferase reaction. Esterases and acyltransferases are mechanistically and evolutionary related enzymes, which both frequently use a catalytic Ser in their active sites. Both types of enzymes use the catalytic residue to produce an acyl-enzyme intermediate, which is released by a second nucleophilic attack of either a water molecule (in the case of esterases), or an acceptor molecule (in the case of transferases). The distinct functions of the two proteins are due to subtle differences in the structures of their active sites that determine which substrate is used for the second nucleophilic attack. These differences cannot be detected based on sequence comparison alone (Jiang et al., 2011). If PesT supports a transferase activity, DAG, which has the particularity of flipping spontaneously from one lipid leaflet to the other, would act as an Ala carrier for the synthesis of Ala-PG in the periplasm. Similar strategies, where functional groups are transferred from carrier lipids to acceptor groups in the periplasm, are used for synthesis of peptidoglycan, acylation of lipoproteins, and decoration of oligosaccharides and lipoteichoic acids (for review see (Zhang & Rock, 2008)).
Ala-DAG provides a fitness advantage to C. glutamicum and increases the range of aminoacylated components available for membrane remodeling
The observations presented above show that lipid alanylation confers a modest fitness advantage to C. glutamicum cultured in the presence of protamine sulfate or polymyxin B. This effect was dependent on cg1103, but not pesT, suggesting that Ala-DAG is sufficient to provide a fitness advantage against these compounds. Synthesis of aa-PG is a common strategy used by bacteria to reduce the net negative charge of the cytoplasmic membrane, thus decreasing its affinity for positively charged antimicrobial compounds (Peschel et al., 2001, Klein et al., 2009). Studies have shown that PG alanylation or lysylation is correlated in some species with resistance to protamine sulfate (Klein et al., 2009, Samant et al., 2009) and polymyxin B (Sohlenkamp et al., 2007, Maloney et al., 2009, Maloney et al., 2011). Both of these compounds are CAMPs with a broad-spectrum antibiotic activity affecting both Gram-negative and Gram-positive species. Therefore, it is not surprising that we observe an Ala-DAG dependent increase in the fitness of C. glutamicum in the presence of these CAMPs, since Ala-DAG is expected to alter the net charge of the cellular membrane similarly to Ala-PG.
In Pseudomonas aeruginosa, Ala-PG formation confers enhanced resistance to lactic acid (Klein et al., 2009). In the current study, we similarly observed an increase in the fitness of C. glutamicum cells grown in the presence of this compound. This phenotype was dependent on cg1103, but not pesT, suggesting that Ala-DAG alone is sufficient to enhance fitness. Lactic acid (in its protonated form) can cross the lipid bilayer by passive diffusion, without the aid of transport proteins (Finkelstein, 1976, Rubin et al., 1982). PG aminoacylation has been shown to impact the biophysical properties of lipid bilayers by altering the molecular packing and interactions between lipid head groups, which in turn affects the passive diffusion of solutes through the membrane (Tocanne et al., 1974, Haest et al., 1972, Sacre et al., 1977). For now, it is unclear whether similar alterations to the biophysical properties of the bacterial membrane, resulting from Ala-DAG synthesis, are responsible for the increased fitness of C. glutamicum grown in the presence of lactic acid.
The effects we observed on the fitness of C. glutamicum cells capable of producing alanylated lipids were modest. This may be due to low levels of Ala-PG and Ala-DAG present in the membranes of cells grown using the culture conditions described here. Indeed, we were unable to detect any amine-containing lipids (by TLC) in wild-type C. glutamicum cells grown in BHI, and the levels of these components in cells grown in the presence of inhibitory compounds was not determined. Different culture conditions may result in increased synthesis of alanylated lipids, which may significantly alter the associated resistance phenotypes.
A system producing two distinct aminoacylated lipids (e.g., Ala-DAG and Ala-PG) may provide an advantage over a system utilizing only PG. For example, a dual system may confer an advantage, in peculiar metabolic conditions, during which PG may not be abundant (e.g., (Zavaleta-Pastor et al., 2010, Lopez-Lara et al., 2003)). Some organisms are able to, during phosphorus limiting conditions, replace constitutive phospholipids in the membrane with phosphorus-free lipids. During this adaptation, liberated phosphate can then be re-routed for synthesis of essential molecules such as nucleic acids. Ala-DAG and Ala-PG may also differentially affect the basic biophysical properties of the cell wall, and be expressed in variable amounts (even when conditions are not limiting) to best suit a given cellular environment. Since no alanylated lipids were detected in C. glutamicum cells cultured in rich media, it is apparent that they do not always constitute major components of the cytoplasmic membrane, but may be produced by a system that is induced only in peculiar metabolic or stress conditions. Investigations into the regulatory networks controlling levels of alanylated lipids in C. glutamicum will provide much needed insight into the role of this system in general membrane homeostasis, as well as bacterial fitness and adaptability.
Experimental Procedures
Bacterial strains and growth conditions
C. glutamicum (ATCC 13032) was obtained from the American Type Culture Collection, and grown in BHI broth at 30 °C with aeration by shaking. Alternatively, cells were grown on plates of the same medium containing 1.5% (w/v) agar and supplemented with 25 mg l−1 of kanamycin, as needed. Electrocompetent cells of C. glutamicum were prepared and used according to established methods (Ming et al., 2010).
Cloning and gene expression
Bacterial strains, plasmids, and oligonucleotide sequences are reported in Supplementary Tables S1 and S2. The DNA sequence encoding amino acids 483–832 of Cg1103 (accession number CAF19673) from C. glutamicum was amplified by PCR (using oligonucleotides 998 and 999), and cloned between the NdeI and XhoI restriction sites of a modified version of the pET-28a vector (kanR, Novagen), in which the thrombin cleavage site was replaced with an HRV 3C cleavage site. The construct was over-expressed in E. coli strain C41 as an N-terminally His-tagged protein using an autoinduction medium as described (Roy & Ibba, 2008a). Cg1103 (483–832) was purified by cobalt-affinity chromatography according to standard procedures. After dialysis and concentration, the protein was stored at a concentration of 17 mg ml−1 in a buffer containing 50 mM Tris•HCl, pH 8.1, 100 mM NaCl, 2 mM β-mercaptoethanol (β-ME), and 50% glycerol. For gene expression in C. glutamicum, the shuttle plasmid pEKEx2-GFP (kanR), for expression of GFP in C. glutamicum (Lausberg et al., 2012), was used. The vectors pEKEx2-Cg1103 and pEKEx2-PesT-Cg1103 were obtained by substituting the fragment between the PstI and SalI restriction sites (encoding GFP in pEKEx2-GFP) by the PCR fragments encoding either cg1103 (amplified with oligonucleotides 1004 and 978), or cg1103 and pesT (amplified with oligonucleotides 1000 and 978). A plasmid expressing pesT (pEKEx2-PesT) alone was obtained by inactivating cg1103 in the pEKEx-2-PesT-Cg1103 construct by removing the EcoRI fragment encoding the 258 aa of the C-terminal end of Cg1103. Constructs were electrotransformed into C. glutamicum as described (Ming et al., 2010), and gene expression was achieved by incubation overnight in liquid BHI. Membrane extracts from recombinant C. glutamicum and E. coli strains were prepared as described previously (Roy & Ibba, 2009).
Markerless deletion of cg1103 and pesT from C. glutamicum
Markerless inactivation of C. glutamicum genes was achieved using allelic exchange by homologous recombination in a classic two step approach with the vector pK19mobsacB (Schäfer et al., 1994). Briefly, the upstream and downstream regions (1 kb each) flanking the targeted genes were PCR amplified using C. glutamicum ATCC13032 genomic DNA as template. Resulting PCR products were stitched together by overlap extension during PCR using oligonucleotides as indicated in Figure S2 and Table S2. The resulting cassettes, containing the upstream and downstream regions fused together, were cloned into the suicide vector pK19mobsacB, following digestion with the restriction enzymes HindIII and XbaI. Thus, pesT, cg1103, or both genes were inactivated using the vectors pK19mobsacB-ΔpesT, pK19mobsacB-Δcg1103, or pK19mobsacB-Δcg1103ΔpesT, respectively. The genetic environments, upstream and downstream regions, and location of oligonucleotides are indicated in Fig. S2. Plasmid constructs were transformed into C. glutamicum by electroporation as described (Ming et al., 2010), and cells were plated on BHI supplemented with 9% (w/v) sorbitol and kanamycin (25 mg l−1). Strains containing plasmid integrated at the targeted loci (pop-in) were subjected to one passage in BHI broth supplemented with kanamycin, and two passages without antibiotic, before being plated on BHI containing 10% (w/v) sucrose. Sucrose-resistant and kanamycin-sensitive clones, having lost the pK19mobsacB constructs (pop-out), were identified by replica plating on BHI alone and BHI supplemented with kanamycin. Positive clones were subjected to PCR screening and sequence analysis to verify removal of the targeted genes.
Extraction, fractionation, and purification of lipids from C. glutamicum
For analytical purposes, lipids from as little as 10 OD of C. glutamicum cells (i.e., 2ml of culture exhibiting an OD=5) were prepared and analyzed by TLC using the Bligh-Dyer procedure (Bligh & Dyer, 1959) with modifications as described (Smith et al., 2013). TLC solvent systems consisted of chloroform:methanol:water (14:6:1, solvent A), and hexane:diethylether:acetic acid (7:3:0.2, solvent B). Extraction and purification of DAG, Ala-DAG, and Ala-PG (i.e., also referred to as UL, ACL1, and ACL2, respectively) for mass spectrometry characterization was performed in glassware as follows. Cells from an overnight culture grown in 1 l of BHI (OD~10) were washed in a buffer containing 30 mM Tris•HCl and 3 mM β-ME, followed by resuspension in 40 ml of the same buffer and disruption by sonication. Lipids were extracted according to the Bligh-Dyer procedure (Bligh & Dyer, 1959) by addition of 100 ml of methanol and 50 ml of chloroform. After 1 h of extraction under vigorous agitation, phase separation was carried out by addition of 50 ml of chloroform and 50 ml of water. Lipids in the organic phase were dried using a rotavapor, and hydrosoluble contaminants were removed using the Folch procedure (Folch et al., 1957) by addition of 15 ml of chloroform:methanol:water (8:4:3). Lipids in the organic phase were dried again and resuspended in 1 ml of chloroform. Lipid classes were separated by flash chromatography on 10 ml of silica gel (mesh 220–440, Alfa Aesar) under nitrogen gas as described previously (Christie & Han, 2010). Non-polar and slightly polar lipids were eluted with chloroform. Specifically, DAG was eluted with 40 ml of chloroform, and Ala-DAG was eluted with an additional 80 ml of chloroform. After two successive wash steps (with 40 ml of chloroform followed by 50 ml of acetone) Ala-PG and other phospholipids were eluted using 50 ml of methanol collected in 2 ml fractions (Fig. S4A).
Fractions containing DAG were identified using the tRNA-dependent lipid aminoacylation assay (see below), and those containing Ala-PG and Ala-DAG were revealed by ninhydrine treatment of an aliquot subjected to TLC. A second step of flash chromatography on silica gel was used to remove contaminants. Fractions containing lipids of interest from the first chromatography step were dried and resuspended in 1 ml of the solvent used to equilibrate the matrix in the second column. DAG was loaded onto 6 ml of matrix equilibrated with hexane:diethylether:acetic acid (7:3:0.2) and was eluted in 6 ml fractions by application of the same solvent. Ala-DAG was loaded onto 4.5 ml of silica gel equilibrated with diethylether:acetic acid (10:0.2). After washing with 6 volumes of the same solvent, Ala-DAG was eluted with solvent A (Fig. S4B). Ala-PG was loaded onto 4.5 ml of matrix equilibrated with chloroform:methanol:water (14:3:0.2), and eluted with the same solvent (Fig. S4C).
Lipid analysis using mass spectrometry
Lipid fractions were dried and resuspended in acidified methanol (0.1% formic acid). Mass spectrometry analyses were performed at the University of Florida Department of Chemistry Mass Spectrometry Services (Gainesville, FL). Accurate mass experiments were conducted using an Agilent 6220 electrospray ionization-time of flight (ESI/TOF) mass spectrometer equipped with an electrospray DART source. ESI/MS/MS experiments were performed on a ThermoScientific LCQ Deca ion trap mass spectrometer equipped with an electrospray source. Both devices were operated in positive ion mode. Optimal accurate mass determination conditions were as follows: injection rate 100 μl min−1, capillary voltage 4000 V, source temperature 350°C, and cone voltage 60 V. The TOF analyzer was scanned over a m/z range of 100–1000 with a 1 s integration time. Optimal conditions for MS/MS were: injection rate 5 μl min−1, spray voltage 4.0 kV, and capillary temperature 250°C, with nitrogen as the nebulizer gas (8 psi). Collision-induced dissociation was performed by isolating the m/z of interest and fragmenting at a collision energy required for ~50% beam reduction.
tRNA-Dependent lipid aminoacylation assay
tRNA-dependent lipid aminoacylation was measured according to established methods (Roy & Ibba, 2009). Activity was assayed using purified aaPGSs (or homologs), or enzymes present in a crude membrane extract isolated from C. glutamicum as previously described (Roy & Ibba, 2009). Aminoacylation reaction media contained 200 mM Hepes•NaOH, pH 7.2, 60 mM KCl, 20 mM MgCl2, 4 mM ATP, 2 mg ml−1 total tRNA (from E. coli MRE600, Roche Applied Science), 20 μM [14C]Ala (75 Ci/mole, Perkin Elmer), 0.1 μM alanyl-tRNA synthetase, and either 0.5 mg ml−1 membrane extract, or 1 μM purified Cg1103483–832. Lipids were dried under vacuum and resuspended (by low power sonication) in a buffer containing 100 mM Hepes, 30 mM KCl, and 10 mM MgCl2. Lipids added to the reaction mixture consisted of 2 mg ml−1 egg-PG (Avanti), with or without 0.5 mg ml−1 of DAG (Avanti), or lipids isolated from 5 OD of C. glutamicum cells. Lipids purified by TLC were eluted from plates using chloroform:methanol (1:1). After incubation of the complete reaction mixture for 30 min at 37°C, lipids were extracted using the Bligh and Dyer procedure (22), dried, and resuspended in 20 μl of chloroform:methanol (2:1). Reaction products were subjected to analysis by TLC in chloroform:methanol:water (14:6:1), and [14C]-alanylated lipids were visualized by phosphorimaging.
Phylogenetic analysis of aaPGS homologs
Protein sequences were retrieved from the non-redundant database at the National Center for Biotechnology Information (NIH, Bethesda). The database was searched using the BLAST program (with default parameters) (Altschul et al., 1997) with the LysPGS from B. subtilis being used as a query sequence. Redundant sequences were removed using a Perl script to eliminate sequences displaying >90% identity with another sequence from the same species. In this process, species exhibiting paralogous aaPGSs were preferentially kept in the dataset. Sequences were aligned using the ClustalO program with default parameters (Sievers et al., 2011); columns of the alignment containing gaps and non-homogenous columns (exhibiting an entropy >2.2) were removed from the alignment with BioEdit software (Hall, 1999). The resulting 160-column alignment was used to construct a maximum likelihood phylogenetic tree using the FastTree program (Price et al., 2009) with default parameters (JTT evolutionary model, discrete gamma model with 20 rate categories). Transmembrane helices were predicted using the TOPCONS program (Bernsel et al., 2009), and phylogenetic tree graphics were generated using iTol (Letunic & Bork, 2007).
Determination of the maximal growth rate (μmax) of C. glutamicum
Growth kinetics were performed in 96-well plates with each well containing 10 μl of inhibitor solution, or inhibitor solvent (water) alone as a negative control, and 140 μl of BHI inoculated with 14,000 cfu of C. glutamicum (1 OD = 185,000 cfu μl−1). Plates were sealed with gas permeable film and incubated in an Elx800 Microplate Reader (BioTek) at 30°C with constant, linear agitation. The A630 was measured at 6 min intervals over 20 h. The maximal growth rates were determined with the R package grofit using the model-free spline method. Each growth condition was tested in duplicate in two sets of independent experiments (n=4).
Determination of MICs
MICs were measured in BHI broth by microdilution of inhibitory compounds (diluted serially 1:1) using a final C. glutamicum inoculum of 105 cfu ml−1. The MIC was defined as the lowest drug concentration preventing visible turbidity after 20 h of incubation at 30°C.
Supplementary Material
Fig. S1. Phylogenetic analysis of aaPGS homologs, and paralogy relationships. The seven types of aaPGS homologs are indicated (I–VII). Paralogs in a single species are indicated by connecting lines. Connections and names of each organism are indicated by colors depicting distinct taxonomic groups. Connecting lines are shaded according to the number of paralogs present in a given organism. aBacteria from these phyla belong to one of the following: Fusobacteria, Gemmatimonadetes, Planctomycetes, Bacteroidetes, Chloroflexi, Cyanobacteria, Spirochaetes, Synergistetes, Verrucomicrobia.
Fig. S2: Genetic context of pesT-cg1103 in C. glutamicum, and plasmid constructs used for markerless deletion and protein expression. The upstream and downstream regions used for markerless deletion of pesT, cg1103, or both genes (using the suicide vector pK19mobsacB) are indicated. The regions amplified for expression of pesT, cg1103, or both genes (using the expression vector pEKEx2) are indicated. Plasmids and oligonucleotides are detailed in Tables S1 and S2. pEKEx2-PesT was obtained by inactivation of cg1103 through removal of the EcoRI fragment from the pEKEx2-PesT-Cg1103 construct.
Fig. S3. Phylogenetic analysis of aaPGS homologs. Taxon names and GI numbers (NCBI) of protein sequences used to build the tree shown in Fig. 1 are indicated. Taxonomic groups and aaPGS homolog types (I–VII) are color-coded. aBacteria from these phyla belong to one of the following: Fusobacteria, Gemmatimonadetes, Planctomycetes, Bacteroidetes, Chloroflexi, Cyanobacteria, Spirochaetes, Synergistetes, Verrucomicrobia.
Fig. S4. Purification of amine-containing lipids from C. glutamicum expressing Cg1103. Amine-containing lipids (ACL1 and ACL2) were purified from a lipid extract isolated from C. glutamicum harboring the expression vector pEKEx2-Cg1103. (A) TLC analysis of total lipids (F0) and fractions collected during the first chromatographic step (F1–9). Total lipids were detected with iodine vapor (I2), and amine-containing lipids were revealed with ninhydrin (N). Fractions enriched in ACL1 (F2 and F3) and ACL2 (F8) were subjected to a second chromatographic step to remove contaminants. F1–F4 were eluted with chloroform, F5 with acetone, and F6–F9 with methanol. TLC analysis of ACL1 (B) and ACL2 (C) containing fractions collected during the second chromatographic step. The F3 fractions were selected (for both ACL1 and ACL2) for analysis by ESI/MS/MS. CL: cardiolipin, PG: phosphatidylglycerol, O: TLC origin, F: solvent front.
Fig. S5. ESI/TOF/MS spectra of amine-containing lipids from C. glutamicum. ACL1 and ACL2 were purified from C. glutamicum as described in Experimental Procedures (see also Fig. S4). ESI/TOF/MS spectra of ACL1 (A) and ACL2 (B). Upper panels show the full spectra, while lower panels depict detail images of the m/z ranges of interest.
Table S1. Strains and plasmids used in this study
Table S2. Oligonucleotides used in this study
Acknowledgments
This work was supported by National Institute of General Medical Sciences Grant 109404. Mass spectrometry analyses were performed by the University of Florida Department of Chemistry Mass Spectrometry Services (Gainesville, FL) and supported by CRIF grant 0541761 from the National Science Foundation.
We would like to acknowledge Dr. Roland Freudl (IBG1, Jülich, Germany) for providing the plasmids pEKEx2-GFP and pK19mobsacB.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- Arendt W, Groenewold MK, Hebecker S, Dickschat JS, Moser J. Identification and characterization of a periplasmic aminoacyl-phosphatidylglycerol hydrolase responsible for Pseudomonas aeruginosa lipid homeostasis. J Biol Chem. 2013;288:24717–24730. doi: 10.1074/jbc.M113.482935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt W, Hebecker S, Jager S, Nimtz M, Moser J. Resistance phenotypes mediated by aminoacyl-phosphatidylglycerol synthases. J Bacteriol. 2012;194:1401–1416. doi: 10.1128/JB.06576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsel A, Viklund H, Hennerdal A, Elofsson A. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37:W465–468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Christie WW, Han X. Lipid Analysis - Isolation, Separation, Identification and Lipidomic Analysis. 4. Oily Press; Bridgwater, U.K: 2010. [Google Scholar]
- Crellin PK, Luo CY, Morita Y. Metabolism of Plasma Membrane Lipids in Mycobacteria and Corynebacteria. Lipid Metabolism. 2013;Chapter 6 [Google Scholar]
- Dare K, Shepherd J, Roy H, Seveau S, Ibba M. LysPGS formation in Listeria monocytogenes has broad roles in maintaining membrane integrity beyond antimicrobial peptide resistance. Virulence. 2014;5 doi: 10.4161/viru.28359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer JC, Lester RL. A simple, specific spray for the detection of phospholipids on thin-layer chromatograms. J Lipid Res. 1964;5:126–127. [PubMed] [Google Scholar]
- Ernst CM, Kuhn S, Slavetinsky CJ, Krismer B, Heilbronner S, Gekeler C, Kraus D, Wagner S, Peschel A. The lipid-modifying multiple Peptide resistance factor is an oligomer consisting of distinct interacting synthase and flippase subunits. MBio. 2015:6. doi: 10.1128/mBio.02340-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst CM, Peschel A. Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol Microbiol. 2011;80:290–299. doi: 10.1111/j.1365-2958.2011.07576.x. [DOI] [PubMed] [Google Scholar]
- Ernst CM, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 2009;5:e1000660. doi: 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein A. Water and nonelectrolyte permeability of lipid bilayer membranes. J Gen Physiol. 1976;68:127–135. doi: 10.1085/jgp.68.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Garg RP, Qian XL, Alemany LB, Moran S, Parry RJ. Investigations of valanimycin biosynthesis: elucidation of the role of seryl-tRNA. Proc Natl Acad Sci U S A. 2008;105:6543–6547. doi: 10.1073/pnas.0708957105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachmann AB, Angert ER, Helmann JD. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob Agents Chemother. 2009;53:1598–1609. doi: 10.1128/AAC.01329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haest CW, de Gier J, den Kamp JO, Bartels P, van Deenen LL. Changes in permeability of Staphylococcus aureus and derived liposomes with varying lipid composition. Biochim Biophys Acta. 1972;255:720–733. doi: 10.1016/0005-2736(72)90385-9. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Jiang Y, Morley KL, Schrag JD, Kazlauskas RJ. Different active-site loop orientation in serine hydrolases versus acyltransferases. Chembiochem. 2011;12:768–776. doi: 10.1002/cbic.201000693. [DOI] [PubMed] [Google Scholar]
- Kahm M, Hasenbrick G, Lichtenberg-Fraté H, Ludwig J, Kschischo M. Grofit: fitting biological growth curves with R. Journal of Statistical Software. 2010;33:1–21. [Google Scholar]
- Klein S, Lorenzo C, Hoffmann S, Walther JM, Storbeck S, Piekarski T, Tindall BJ, Wray V, Nimtz M, Moser J. Adaptation of Pseudomonas aeruginosa to various conditions includes tRNA-dependent formation of alanyl-phosphatidylglycerol. Mol Microbiol. 2009;71:551–565. doi: 10.1111/j.1365-2958.2008.06562.x. [DOI] [PubMed] [Google Scholar]
- Komatsuzawa H, Ohta K, Fujiwara T, Choi GH, Labischinski H, Sugai M. Cloning and sequencing of the gene, fmtC, which affects oxacillin resistance in methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett. 2001;203:49–54. doi: 10.1111/j.1574-6968.2001.tb10819.x. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- Lausberg F, Chattopadhyay AR, Heyer A, Eggeling L, Freudl R. A tetracycline inducible expression vector for Corynebacterium glutamicum allowing tightly regulable gene expression. Plasmid. 2012;68:142–147. doi: 10.1016/j.plasmid.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Lopez-Lara IM, Sohlenkamp C, Geiger O. Membrane lipids in plant-associated bacteria: their biosyntheses and possible functions. Mol Plant Microbe Interact. 2003;16:567–579. doi: 10.1094/MPMI.2003.16.7.567. [DOI] [PubMed] [Google Scholar]
- Maloney E, Lun S, Stankowska D, Guo H, Rajagoapalan M, Bishai WR, Madiraju MV. Alterations in phospholipid catabolism in Mycobacterium tuberculosis lysX mutant. Front Microbiol. 2011;2:19. doi: 10.3389/fmicb.2011.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney E, Stankowska D, Zhang J, Fol M, Cheng QJ, Lun S, Bishai WR, Rajagopalan M, Chatterjee D, Madiraju MV. The two-domain LysX protein of Mycobacterium tuberculosis is required for production of lysinylated phosphatidylglycerol and resistance to cationic antimicrobial peptides. PLoS Pathog. 2009;5:e1000534. doi: 10.1371/journal.ppat.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam Leitch EC, Stewart CS. Escherichia coli O157 and non-O157 isolates are more susceptible to L-lactate than to D-lactate. Appl Environ Microbiol. 2002;68:4676–4678. doi: 10.1128/AEM.68.9.4676-4678.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming L, Tao N, Jun-huan Z, Zhe S, Dong-jun K. High Electroporation Efficiency of Corynebacterium glutamicum with Xenogeneic Plasmid DNA. Bioinformatics and Biomedical Engineering (iCBBE), 2010 4th International Conference on; 2010. pp. 1–4. [Google Scholar]
- Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, van Kessel KP, van Strijp JA. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med. 2001;193:1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy H. Tuning the properties of the bacterial membrane with aminoacylated phosphatidylglycerol. IUBMB Life. 2009;61:940–953. doi: 10.1002/iub.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy H, Ibba M. Monitoring Lys-tRNALys phosphatidylglycerol transferase activity. Methods. 2008a;44:164–169. doi: 10.1016/j.ymeth.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy H, Ibba M. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc Natl Acad Sci U S A. 2008b;105:4667–4672. doi: 10.1073/pnas.0800006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy H, Ibba M. Broad range amino acid specificity of RNA-dependent lipid remodeling by multiple peptide resistance factors. J Biol Chem. 2009;284:29677–29683. doi: 10.1074/jbc.M109.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin HE, Nerad T, Vaughan F. Lactate acid inhibition of Salmonella typhimurium in yogurt. J Dairy Sci. 1982;65:197–203. doi: 10.3168/jds.S0022-0302(82)82177-2. [DOI] [PubMed] [Google Scholar]
- Sacre MM, El Mashak EM, Tocanne JF. A monolayer (pi, deltaV) study of the ionic properties of alanylphosphatidylglycerol: effects of pH and ions. Chem Phys Lipids. 1977;20:305–318. doi: 10.1016/0009-3084(77)90071-8. [DOI] [PubMed] [Google Scholar]
- Samant S, Hsu FF, Neyfakh AA, Lee H. The Bacillus anthracis protein MprF is required for synthesis of lysylphosphatidylglycerols and for resistance to cationic antimicrobial peptides. J Bacteriol. 2009;191:1311–1319. doi: 10.1128/JB.01345-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavetinsky CJ, Peschel A, Ernst CM. Alanyl-phosphatidylglycerol and lysyl-phosphatidylglycerol are translocated by the same MprF flippases and have similar capacities to protect against the antibiotic daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother. 2013;56:3492–3497. doi: 10.1128/AAC.00370-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Harrison JS, Sprague KM, Roy H. A Conserved Hydrolase Responsible for the Cleavage of Aminoacylphosphatidylglycerol in the Membrane of Enterococcus faecium. J Biol Chem. 2013;288:22768–22776. doi: 10.1074/jbc.M113.484402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlenkamp C, Galindo-Lagunas KA, Guan Z, Vinuesa P, Robinson S, Thomas-Oates J, Raetz CR, Geiger O. The lipid lysyl-phosphatidylglycerol is present in membranes of Rhizobium tropici CIAT899 and confers increased resistance to polymyxin B under acidic growth conditions. Mol Plant Microbe Interact. 2007;20:1421–1430. doi: 10.1094/MPMI-20-11-1421. [DOI] [PubMed] [Google Scholar]
- Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, Chakraborty T, Wehland J, Jansch L. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol Microbiol. 2006;62:1325–1339. doi: 10.1111/j.1365-2958.2006.05452.x. [DOI] [PubMed] [Google Scholar]
- Tocanne JF, Ververgaert PH, Verkleij AJ, van Deenen LL. A monolayer and freeze-etching study of charged phospholipids. I. Effects of ions and pH on the ionic properties of phosphatidylglycerol and lysylphosphatidylglycerol. Chem Phys Lipids. 1974;12:201–219. doi: 10.1016/0009-3084(74)90075-9. [DOI] [PubMed] [Google Scholar]
- Walkiewicz K, Benitez Cardenas AS, Sun C, Bacorn C, Saxer G, Shamoo Y. Small changes in enzyme function can lead to surprisingly large fitness effects during adaptive evolution of antibiotic resistance. Proc Natl Acad Sci U S A. 2012;109:21408–21413. doi: 10.1073/pnas.1209335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaleta-Pastor M, Sohlenkamp C, Gao JL, Guan Z, Zaheer R, Finan TM, Raetz CR, Lopez-Lara IM, Geiger O. Sinorhizobium meliloti phospholipase C required for lipid remodeling during phosphorus limitation. Proc Natl Acad Sci U S A. 2010;107:302–307. doi: 10.1073/pnas.0912930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YM, Rock CO. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Phylogenetic analysis of aaPGS homologs, and paralogy relationships. The seven types of aaPGS homologs are indicated (I–VII). Paralogs in a single species are indicated by connecting lines. Connections and names of each organism are indicated by colors depicting distinct taxonomic groups. Connecting lines are shaded according to the number of paralogs present in a given organism. aBacteria from these phyla belong to one of the following: Fusobacteria, Gemmatimonadetes, Planctomycetes, Bacteroidetes, Chloroflexi, Cyanobacteria, Spirochaetes, Synergistetes, Verrucomicrobia.
Fig. S2: Genetic context of pesT-cg1103 in C. glutamicum, and plasmid constructs used for markerless deletion and protein expression. The upstream and downstream regions used for markerless deletion of pesT, cg1103, or both genes (using the suicide vector pK19mobsacB) are indicated. The regions amplified for expression of pesT, cg1103, or both genes (using the expression vector pEKEx2) are indicated. Plasmids and oligonucleotides are detailed in Tables S1 and S2. pEKEx2-PesT was obtained by inactivation of cg1103 through removal of the EcoRI fragment from the pEKEx2-PesT-Cg1103 construct.
Fig. S3. Phylogenetic analysis of aaPGS homologs. Taxon names and GI numbers (NCBI) of protein sequences used to build the tree shown in Fig. 1 are indicated. Taxonomic groups and aaPGS homolog types (I–VII) are color-coded. aBacteria from these phyla belong to one of the following: Fusobacteria, Gemmatimonadetes, Planctomycetes, Bacteroidetes, Chloroflexi, Cyanobacteria, Spirochaetes, Synergistetes, Verrucomicrobia.
Fig. S4. Purification of amine-containing lipids from C. glutamicum expressing Cg1103. Amine-containing lipids (ACL1 and ACL2) were purified from a lipid extract isolated from C. glutamicum harboring the expression vector pEKEx2-Cg1103. (A) TLC analysis of total lipids (F0) and fractions collected during the first chromatographic step (F1–9). Total lipids were detected with iodine vapor (I2), and amine-containing lipids were revealed with ninhydrin (N). Fractions enriched in ACL1 (F2 and F3) and ACL2 (F8) were subjected to a second chromatographic step to remove contaminants. F1–F4 were eluted with chloroform, F5 with acetone, and F6–F9 with methanol. TLC analysis of ACL1 (B) and ACL2 (C) containing fractions collected during the second chromatographic step. The F3 fractions were selected (for both ACL1 and ACL2) for analysis by ESI/MS/MS. CL: cardiolipin, PG: phosphatidylglycerol, O: TLC origin, F: solvent front.
Fig. S5. ESI/TOF/MS spectra of amine-containing lipids from C. glutamicum. ACL1 and ACL2 were purified from C. glutamicum as described in Experimental Procedures (see also Fig. S4). ESI/TOF/MS spectra of ACL1 (A) and ACL2 (B). Upper panels show the full spectra, while lower panels depict detail images of the m/z ranges of interest.
Table S1. Strains and plasmids used in this study
Table S2. Oligonucleotides used in this study