Abstract
Background and Aims A worldwide increase in tree decline and mortality has been linked to climate change and, where these represent foundation species, this can have important implications for ecosystem functions. This study tests a combined approach of phylogeographic analysis and species distribution modelling to provide a climate change context for an observed decline in crown health and an increase in mortality in Eucalyptus wandoo, an endemic tree of south-western Australia.
Methods Phylogeographic analyses were undertaken using restriction fragment length polymorphism analysis of chloroplast DNA in 26 populations across the species distribution. Parsimony analysis of haplotype relationships was conducted, a haplotype network was prepared, and haplotype and nucleotide diversity were calculated. Species distribution modelling was undertaken using Maxent models based on extant species occurrences and projected to climate models of the last glacial maximum (LGM).
Key Results A structured pattern of diversity was identified, with the presence of two groups that followed a climatic gradient from mesic to semi-arid regions. Most populations were represented by a single haplotype, but many haplotypes were shared among populations, with some having widespread distributions. A putative refugial area with high haplotype diversity was identified at the centre of the species distribution. Species distribution modelling showed high climatic suitability at the LGM and high climatic stability in the central region where higher genetic diversity was found, and low suitability elsewhere, consistent with a pattern of range contraction.
Conclusions Combination of phylogeography and paleo-distribution modelling can provide an evolutionary context for climate-driven tree decline, as both can be used to cross-validate evidence for refugia and contraction under harsh climatic conditions. This approach identified a central refugial area in the test species E. wandoo, with more recent expansion into peripheral areas from where it had contracted at the LGM. This signature of contraction from lower rainfall areas is consistent with current observations of decline on the semi-arid margin of the range, and indicates low capacity to tolerate forecast climatic change. Identification of a paleo-historical context for current tree decline enables conservation interventions to focus on maintaining genetic diversity, which provides the evolutionary potential for adaptation to climate change.
Keywords: Climate change, Eucalyptus wandoo, Myrtaceae, evolution, forest decline, haplotypes, last glacial maximum, LGM, phylogeography, refugia, species distribution modelling, tree decline.
INTRODUCTION
In the last few decades, a worldwide increase in tree decline and mortality has been linked to climate change, including increasingly frequent extreme weather events, such as droughts and heatwaves (e.g. Breshears et al., 2005; Allen et al., 2010), and long-term global increases in temperature and decreases in rainfall (Carnicer et al., 2011). Climate change interacts with, and amplifies, the impact of other disturbance factors affecting forest ecosystems, such as insect and pathogen outbreaks, landscape fragmentation and fire (Dale et al., 2001; Breshears et al., 2005). As such, and because climate change projections indicate an intensification of recent trends, it is thought that forest ecosystems could be increasingly vulnerable to climate-induced decline (Anderegg et al., 2013). Given that trees can be foundation species, with a major influence on other biota, community structure and composition, and ecosystem function (Manning et al., 2006; Anderegg et al., 2013), understanding their response to current and future climatic patterns is important for climate change adaptation strategies that maintain ecosystem function and facilitate species persistence (Moritz and Agudo, 2013).
Phylogeography can provide important insights into the effects of paleo-historical climate change on the current geographic distribution and genetic structure of species (Avise, 2000; Byrne, 2007), and this historical context is helpful to understand species responses to future climate change (Forester et al., 2013). The effects of historical climatic oscillations on biodiversity were particularly pronounced from the middle Pleistocene through to the last glacial maximum (LGM; approx. 21 000 years BP). While in the northern hemisphere significant areas were affected by glaciation, the southern hemisphere experienced cyclic conditions, varying from warm, wet environments during the interglacial periods to cold, dry environments during glacial periods. Responses to these climatic oscillations, such as repeated contraction and expansion, or persistence in localized refugia, have left signatures in the genetic composition of current populations that can be used to deduce the influence of the historical processes on their evolution (Avise, 2000; Hewitt, 2004; Byrne, 2008).
Species distribution modelling (SDM) is a useful approach to validate and/or complement biogeographic inferences derived from molecular markers (Scoble and Lowe, 2010; Svenning et al., 2011; Forester et al., 2013), and has been increasingly used to support evolutionary and phylogeographic studies (e.g. Schorr et al., 2013). SDM describe the species ecological niches by quantifying the relationship between empirical observations of species distribution and environmental data across space and/or time (Guisan and Zimmermann, 2000). By assuming that the climatic niche of a species remains unchanged over a time period of interest (Wiens and Graham, 2005), its past distribution can be reconstructed by projecting SDM to earlier time periods using paleoclimatic data (Svenning et al., 2011). Yet, a number of factors can affect SDM and result in poor projections, including uncertainty of climatic models, changes in ecological niche, decoupling of regional and local climates, and occurrence of non-analogue environments in the LGM (Worth et al., 2014).
A combined approach using phylogeography and SDM can provide a helpful framework where current tree decline phenomena can be investigated. In this study, phylogeographic analysis of diversity in the chloroplast genome and SDM were used to assess the impacts of past and future climate changes on the genetic structure and geographic distribution of Eucalyptus wandoo (Myrtaceae), an endemic tree species of the inland woodlands of the south-western Australian global biodiversity hotspot (Hopper and Gioia, 2004). Recent and ongoing climate change has been implicated in the decline of E. wandoo crown health and increased mortality, which have been more pronounced in the low rainfall areas of the species distribution range (Hooper and Sivasithamparam, 2005; Brouwers et al., 2013; Poot and Veneklaas, 2013).
Given these patterns of decline in E. wandoo, it is hypothesized that the species may have a long-term response to historical climate change that is different from that observed in phylogeographic studies of other widespread south-western Australian Eucalyptus species (e.g. E. loxophleba, E. marginata, E. kochii, E. horistes and E. gomphocephala; Byrne and Macdonald, 2000; Byrne et al., 2003; Byrne and Hines, 2004; Wheeler and Byrne, 2006; Nevill et al., 2014). These studies have identified a high diversity and differentiation in chloroplast DNA (cpDNA) and the presence of highly localized haplotypes throughout the species distribution ranges, which indicates persistence in localized refugia during the LGM. It is hypothesized that E. wandoo may have a different genetic signature in chloroplast diversity, with signals of contraction during times of aridity and expansion during more mesic conditions, and a geographic pattern that is consistent with modelling of the species distribution at the LGM. Insight into the evolutionary history of E. wandoo will enable understanding of the species response to ongoing and future climate change and provide a context for the management of factors influencing its current decline.
MATERIALS AND METHODS
Study area and study species
Eucalyptus wandoo Blakely has a broad distribution in south-western Australia, from the high (800–1200 mm mean annual rainfall) to the transitional rainfall (300–800 mm) zones (Fig. 1). Nevertheless, the species has been largely cleared for agriculture, with <5 % of the pre-European extent (approximately the 1830s) of E. wandoo remaining, mostly in isolated remnants affected by multiple disturbance factors (e.g. fragmentation and salinity; Yates et al., 2000). The species occurs in lowland areas across broad valleys and grows on loamy or sandy soils. Two subspecies are recognized; subspecies pulverea is restricted to the northern boundary of the species distribution and is distinguished from subspecies wandoo by its pruinose branchlets and powdery bark (Brooker and Hopper, 1991).
Fig. 1.
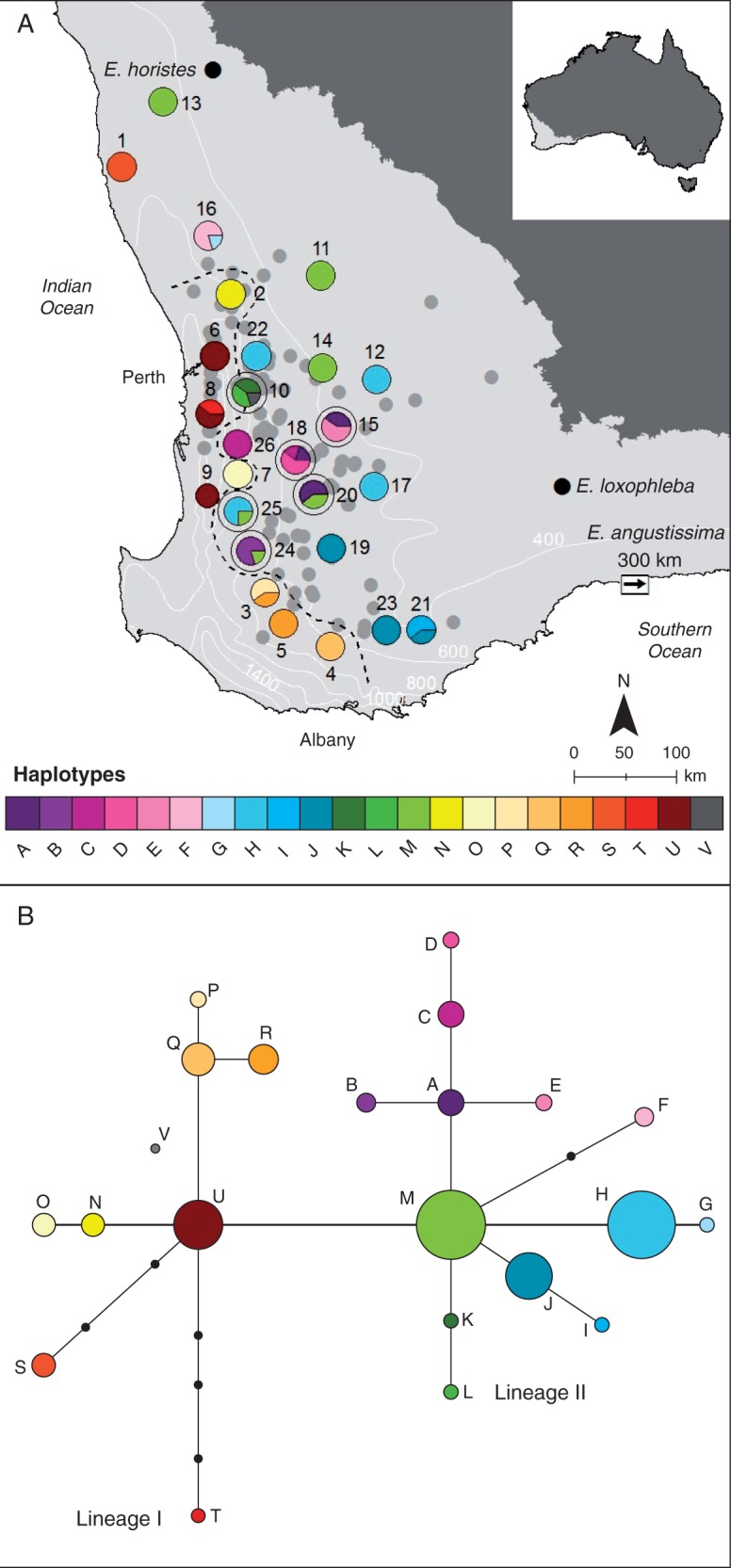
Location of the sampled populations of Eucalyptus wandoo in south-western Australia and their haplotype composition. (A) Sampled populations of E. wandoo are represented with large circles that are coloured according to their haplotype composition. Populations with high haplotype diversity within the refugium area are marked with an outer circle. Species records obtained from NatureMap are represented by light grey circles. Locations of the outgroup species samples are represented with black dots (except for E. angustissima, which is located 300 km east from the arrow, on the coast). The dashed line represents the phylogeographic break between the two lineages. Rainfall isohyets lines (200 mm) are represented in white. (B) Haplotype network for E. wandoo. Haplotypes are identified with the letters A–V, according to Table 2. Interior haplotypes not detected in the samples are represented by small black circles. Each line represents a single polymorphism.
South-western Australia experiences a mediterranean climate, with 80 % of the rainfall occurring in winter and only 4 % in summer. The region has undergone significant climatic changes since the 1970s, with an increase of 0·6 °C in mean annual temperature and a decline of 17 % in mean winter rainfall (Bates et al., 2008; CSIRO and Bureau of Meteorology, 2014). Further drying and warming is projected, with a forecast decrease of 5–60 % in annual rainfall and an increase of 1–5·5 °C in annual temperature by 2070 (Bates et al., 2008).
Genotyping
Phylogeographic analyses were undertaken using restriction fragment length polymorphism (RFLP) technology, as this is known to detect intraspecific polymorphism and phylogeographic patterns in the chloroplast genome of Western Australian species, including eucalypts (Byrne, 2007, 2008), and enables comparison with other species from the study area previously analysed using the same method. A recent comparative study in Calothamnus quadrifidus has shown congruence between cpRFLP and sequence data sets, demonstrating the validity of RFLP methodology (Nistelberger et al., 2014).
Previous studies on Western Australian eucalypts have shown that cpDNA diversity is generally maintained between populations (Byrne, 2007). Therefore, leaves were collected from five healthy and well-spaced (50–100 m) trees from 26 provenances covering the distribution range of E. wandoo (Fig. 1; Table 1). The majority of the populations sampled were from the subspecies wandoo, while two populations (Mt Lesueur and Dookanooka Nature Reserve) were from the restricted northern subspecies pulverea. A sample from each of three related species, E. angustissima (Point Malcolm), E. loxophleba (Lake King) and E. loxophleba (Bowgada), were included as outgroups. Although occasional interseries hybrids have been noted between E. wandoo and E. loxophleba, these are rare (Brooker and Hopper, 1991) due to the differences in the ecological niche of the two species. The aridity index of each provenance location (ratio between annual rainfall and annual potential evapotranspiration) was calculated, as it is regarded as a useful proxy for potential water availability (Gao and Giorgi, 2008).
Table 1.
Name, location, annual potential evapotranspiration (mm), annual rainfall (mm), aridity index and lineage identifier of the 26 Eucalyptus wandoo populations that were used in the chloroplast DNA analysis
| Population name (ID) | Latitude (S) | Longitude (E) | Annual potential evapotranspiration (mm) | Annual rainfall (mm) | Aridity index | Lineage |
|---|---|---|---|---|---|---|
| Mt Lesueur NP (1) | 30°09′47″ | 115°11′48″ | 1650 | 625 | 0·38 | I |
| Bindoon east (2) | 31°19′42″ | 116°15′47″ | 1605 | 654 | 0·41 | I |
| Boyup Brook south (3) | 33°59′16″ | 116°30′51″ | 1299 | 568 | 0·44 | I |
| Rocky Gully east (4) | 34°28′41″ | 117°11′56″ | 1217 | 575 | 0·47 | I |
| Frankland west (5) | 34°15′43″ | 116°42′16″ | 1251 | 611 | 0·49 | I |
| John Forrest NP (6) | 31°53′15″ | 116°04′35″ | 1605 | 830 | 0·52 | I |
| Gorrie Rd (7) | 32°53′49″ | 116°16′41″ | 1570 | 832 | 0·53 | I |
| Serpentine NP (8) | 32°22′55″ | 116°00′45″ | 1535 | 1016 | 0·66 | I |
| Harvey Dam (9) | 33°05′13″ | 115°56′47″ | 1454 | 963 | 0·66 | I |
| Running Brook Rd (10) | 32°11′47″ | 116°23′36″ | 1542 | 795 | 0·52 | I and II |
| Namalcatchem NR (11) | 31°11′18″ | 117°11′53″ | 1642 | 348 | 0·21 | II |
| Boolanelling NR (12) | 32°07′06″ | 117°44′55″ | 1525 | 333 | 0·22 | II |
| Dookanooka NR (13) | 29°36′04″ | 115°38′28″ | 1746 | 438 | 0·25 | II |
| Quairading-York roadside (14) | 32°00′26″ | 117°11′27″ | 1549 | 400 | 0·26 | II |
| Tutanning NR (15) | 32°31′47″ | 117°19′16″ | 1473 | 397 | 0·27 | II |
| Moora south (16) | 30°48′13″ | 116°02′60″ | 1670 | 463 | 0·28 | II |
| Dongolocking NR (17) | 33°03′56″ | 117°41′52″ | 1385 | 398 | 0·29 | II |
| Dryandra woodlands NP (18) | 32°49′11″ | 116°52′56″ | 1451 | 473 | 0·33 | II |
| Wingedyne NR (19) | 33°36′24″ | 117°14′12″ | 1342 | 442 | 0·33 | II |
| Highbury SF (20) | 33°07′34″ | 117°03′51″ | 1398 | 461 | 0·33 | II |
| Stirling Range NP east (21) | 34°20′39″ | 118°09′53″ | 1247 | 412 | 0·33 | II |
| Talbot Block north (22) | 31°53′04″ | 116°30′29″ | 1352 | 647 | 0·38 | II |
| Stirling Range NP west (23) | 34°20′36″ | 117°47′53″ | 1573 | 599 | 0·38 | II |
| Boyup Brook north (24) | 33°36′38″ | 116°23′04″ | 1223 | 470 | 0·48 | II |
| Collie east (25) | 33°14′19″ | 116°15′53″ | 1405 | 825 | 0·59 | II |
| Boddington west (26) | 32°43′45″ | 116°17′50″ | 1442 | 928 | 0·64 | II |
The two populations of subspecies pulverea are indicated in italic.
Populations are ordered by lineage (refer to Fig. 2) and aridity index (ratio between rainfall and evapotranspiration). Abbreviations in population names represent: Rd, Road; NP, National Park; NR, Nature Reserve; SF, State Forest.
Genomic DNA was extracted from 10–15 g of leaf material (Byrne et al., 1993) with 0·1 m sodium sulphate (Byrne et al., 2001) and 0·05 % bovine serum albumin (BSA) added to the extraction buffer. DNA of each sample was digested with six restriction enzymes (BclI, BglII, DraI, EcoRI, EcoRV and XbaI) and hybridized with heterologous probes covering most of the chloroplast genome, according to the protocol described in Byrne et al. (1993).
Genetic data analyses
Banding patterns obtained for each probe–enzyme polymorphism combination were interpreted as length or restriction site mutations. Mutations identified by consecutive cp probes were counted only once. Nucleotide diversity, the average number of nucleotide differences per site between two sequences (Nei, 1978), and haplotype diversity, Nei’s gene diversity measure (Nei, 1973), were calculated using Arlequin v3.5 (Excoffier et al., 2005), and partitioned within and between populations. The relationship between nucleotide and haplotype diversity was estimated using PERMUT (Pons and Petit, 1996). A network of haplotype relationships was generated in Network v4.612 (http://www.fluxus-engineering.com) using a median-joining algorithm (Bandelt et al., 1999). A parsimony analysis of haplotype relationships characterized by the presence or absence of each polymorphism was undertaken using Phylip v3.68 (Felsenstein, 2008). Regular bootstraps of 1000 data sets were produced (SEQBOOT), 100 trees were prepared using PARS and a majority rule (>50 %) consensus tree was created using CONSENSE (Felsenstein, 2008).
Nucleotide divergence can be used to estimate the time since separation, although there are caveats to the use of a molecular clock given the difficulties in calibration from independent evidence (Wheeler and Byrne, 2006). Nevertheless, similar levels of divergence between lineages in several south-western Australian species from different botanical families provide evidence for consistent influences of major historical events in the region (Wheeler and Byrne, 2006; Byrne, 2007). Estimation of the time of divergence between lineages was made as 0·1 % nucleotide divergence representing a separation of approx. 1 million years (Zurawski et al., 1984), as this has been used for restriction site data from across the chloroplast genome.
Coalescent analyses using mismatch distributions and neutrality tests were completed for the entire data set, as well as within Lineages I and II separately using Arlequin v3.5. Mismatch distribution analyses were conducted by calculating the distribution of the total observed differences between pairs of haplotypes. The parameters of instantaneous demographic expansion and spatial expansion were estimated using a least-squares approach with 1000 bootstrap replicates. Tajima’s D (Tajima, 1989, 1996) and Fu’s Fs (Fu, 1997) were calculated using 1000 coalescent simulations to test for departure from neutral evolution.
Species distribution modelling
A total of 182 presence-only records of E. wandoo were compiled from field work and from the online biodiversity database NatureMap (http://naturemap.dec.wa.gov.au/; accessed February 2014), which includes data from the Western Australian Herbarium. Data were scrutinized for misidentified, suspected inaccurate or duplicate records. The background environment was constrained to the focal biogeographic regions (IBRA v7, http://www.environment.gov.au/topics/land/national-reserve-system/science-maps-and-data/australias-bioregions-ibra) occupied by the species.
Modern climatic data (averaging period 1961–1990) at a spatial resolution of 0·0025 ° (approx. 250 m) were obtained from Yates et al. (2010), who derived 19 bioclimatic variables using minimum monthly temperature, average maximum monthly temperature, average monthly precipitation and average monthly areal potential evapotranspiration data layers provided by the Australian Bureau of Meteorology National Climate Centre. In this study, a sub-set of five bioclimatic variables was used. These variables have been suggested by Yates et al. (2010) as being most relevant to plant distributions in south-western Australia, and include annual precipitation, precipitation of the driest quarter, mean temperature of the warmest quarter, mean temperature of the wettest quarter and isothermality. Projections of these bioclimatic variables for 2070 were also obtained from Yates et al. (2010), using a medium severity climate change scenario that includes the moderate impact model MIROC-M combined with the A1B emission scenario and medium climate sensitivity.
The same five bioclimatic variables for the LGM were obtained at a spatial resolution of 0·05 ° (approx. 5 km) from the WorldClim database (Hijmans et al., 2005), and resampled to a 0·0025 ° resolution via bilinear interpolation. Initially, two general circulation models (GCMs) were considered, CCSM3 (Collins et al., 2006) and MIROC (Hasumi and Emori, 2004), both derived from the Paleoclimate Modelling Inter-comparison Project Phase II (Braconnot et al., 2007). However, only CCSM was used, because MIROC predicts LGM annual mean precipitation values that are higher than modern values (Supplementary Data Table S1), which is highly inconsistent with the palynological evidence that the LGM climate in south Australia was drier than at present (see Dodson, 2001).
Maxent v3.3.3k (Phillips et al., 2006; Phillips and Dudík, 2008) was used to model the distribution range of E. wandoo under modern climate conditions. This model was then used to project the species distribution at the LGM and in 2070. Maxent is one of the most widely used and effective methods for species distribution modelling using presence-only data (Elith et al., 2011). The models were trained on 75 % randomly assigned occurrences, with the remaining 25 % used as an evaluation data set. Furthermore, the default Maxent settings were used, with the exception that model building was restricted to hinge features (Phillips and Dudík, 2008). Because species occurrence data often exhibit a spatial bias in survey effort (Schulman et al., 2007), a sampling effort bias layer (target group method; Phillips et al., 2009) was created using the records density of all plant species belonging to the four main terrestrial botanical families (Myrtaceae, Proteaceae, Fabaceae and Cyperaceae). The variables response curves, their percentage contribution and individual importance on the Jackknife tests were used for selection of the best predictors.
The influence of niche suitability and stability on haplotype diversity was tested using generalized linear models (GLM) in R 3.1.2 (http://www.R-project.org/). Niche stability was calculated between the LGM and the present (NStabLGM–Present = 1 – NSLGM–NSPresent|) and between the present and 2070 (NStabPresent–2070 = 1 – |NSPresent–NS2070|), using Maxent’s niche suitability (NS) values (Gugger et al., 2013). Five explanatory covariates were considered in the GLM: niche stability since the LGM, past and present niche suitability, latitude and longitude. These covariates have low Pearson’s pairwise correlation coefficients, except NSPresent and NSLGM (r = 0·63). The Akaike information criterion (AIC) was used to select the most parsimonious model.
RESULTS
Polymorphism in cpDNA
The analysis of cpDNA variation in Eucalyptus wandoo revealed polymorphisms with all the enzymes used. In total, 27 polymorphisms were detected, 18 length mutations and nine restriction site mutations. The majority of the polymorphisms (18) were located in the large single-copy region of the cp genome, and the rest (9) in the small single-copy region. The arrangement of mutations in individuals resulted in the identification of 22 haplotypes. The most common haplotype (M) was present in six populations and represented 14·7 % of the samples (Table 2). Haplotype H was the second most common haplotype, present in four populations (14·0 % frequency). Haplotypes U, J and A were present in three populations with 10·1, 9·3 and 4·7 % frequencies, respectively. Haplotypes R and C were present in two populations, with 5·4 and 4·7 % frequencies, respectively. The remaining haplotypes (15) were only present in one population, with frequencies ranging from 0·8 to 3·9 %. Diversity within E. wandoo populations was mostly represented by a single haplotype (16 out of 26 populations; frequency = 61·5 %). Eight populations (30·8 %) were represented by two haplotypes, and two populations (7·7 %) by three haplotypes.
Table 2.
Frequency and distribution of haplotypes in Eucalyptus wandoo, E. angustissima, E. loxophleba and E. horistes
| Haplotype | Frequency (%) | Mutations | Population ID (no. of individuals) |
|---|---|---|---|
| A | 4·65 | 8, 23, 46, 47, 48 | 18 (1), 20 (3), 15 (2) |
| B | 3·10 | 8, 15, 23, 46, 47, 48 | 24 (4) |
| C | 4·65 | 4, 8, 23, 46, 47, 48 | 18 (1), 26 (5) |
| D | 2·33 | 4, 8, 11, 23, 46, 47, 48 | 18 (3) |
| E | 2·33 | 8, 10, 23, 46, 47, 48 | 15 (3) |
| F | 3·10 | 1, 23, 28, 46, 47, 48 | 16 (4) |
| G | 0·78 | 21, 23, 25, 46, 47, 48 | 16 (1) |
| H | 13·95 | 21, 23, 46, 47, 48 | 12 (5), 17 (5), 22 (5), 25 (3) |
| I | 2·33 | 13, 20, 23, 46, 47, 48 | 21 (3) |
| J | 9·30 | 20, 23, 46, 47, 48 | 21 (2), 23 (5), 19 (5) |
| K | 1·55 | 23, 46, 47, 48, 51 | 10 (2) |
| L | 1·55 | 5, 23, 46, 47, 48, 51 | 10 (2) |
| M | 14·73 | 23, 46, 47, 48 | 24 (1), 13 (5), 20 (2), 11 (5), 14 (5), 25 (1) |
| N | 3·88 | 14, 46, 47, 48 | 2 (5) |
| O | 3·88 | 14, 26, 46, 47, 48 | 7 (5) |
| P | 2·33 | 10, 22, 46, 47, 48 | 3 (3) |
| Q | 3·88 | 22, 46, 47, 48 | 4 (5) |
| R | 5·43 | 7, 22, 46, 47, 48 | 3 (2), 5t (5) |
| S | 3·88 | 16, 24, 27, 46, 47, 48 | 1 (5) |
| T | 1·55 | 3, 6, 9, 12, 19, 46, 47, 48 | 8 (2) |
| U | 10·08 | 46, 47, 48 | 6 (5), 9 (5), 8 (3) |
| V | 0·78 | 50, 46, 47, 48 | 10 (1) |
| – | 31, 36, 40, 43, 49 | E. angustissima (1) | |
| – | 18, 29, 30, 33, 36, 40, 41, 45 | E. loxophleba (1) | |
| – | 2, 17, 32, 34, 35, 37, 38, 39, 42, 44 | E. horistes (1) |
Populations are identified by their ID number followed in parentheses by the number of samples for each population with that haplotype.
Haplotype relationships
The network of haplotype relationships showed a structured pattern of diversity, with haplotypes separated into two groups centred on haplotypes U and M (referred to as lineages for ease of identification). Lineage I consisted of nine haplotypes (N–V) representing ten populations from the western margin of the species distribution (Fig. 1). Lineage II was formed by 13 haplotypes (A–M) present in 17 populations in the eastern inland areas of the distribution (Fig. 1). Both lineages showed a star-shaped pattern of haplotype relationships, but the level of structure within the lineages differed. The haplotypes in Lineage I showed little structure, with only two sets of related haplotypes (N/O and P/Q/R) (Fig. 1). Lineage II showed greater structure, with several pairs of related haplotypes (G/H, I/J and K/L) and one group of five haplotypes (A/B/C/D/E) (Fig. 1). Lineage II contained three out of the four most widespread haplotypes present in E. wandoo. There was no genetic distinction between the two identified subspecies, as haplotypes in the two populations at Mt Lesueur and Dookanooka (S and M) were not closely related, and Dookanooka had the common haplotype (M) that was present in many of the subspecies wandoo populations.
The phylogenetic parsimony analysis produced a single tree with a length of 55 and a consistency index of 0·93, with only one case of homoplasy found (mutation 10; Fig. 2 ). The phylogeny clearly distinguished E. wandoo from the outgroup species E. angustissima, E. loxophleba and E. horistes. There were no shared haplotypes with the E. loxophleba sample. In addition, comparison of haplotypes identified here with those identified in a study across the range of E. loxophleba (Byrne and Hines, 2004) also showed no shared haplotypes, indicating that any introgression is minimal. Within E. wandoo, the phylogeny showed the same structured pattern of diversity as the network with haplotypes separated into two main groups, although group 1 was identified as a polytomy and group 2 as a derived lineage within the polytomy.
Fig. 2.
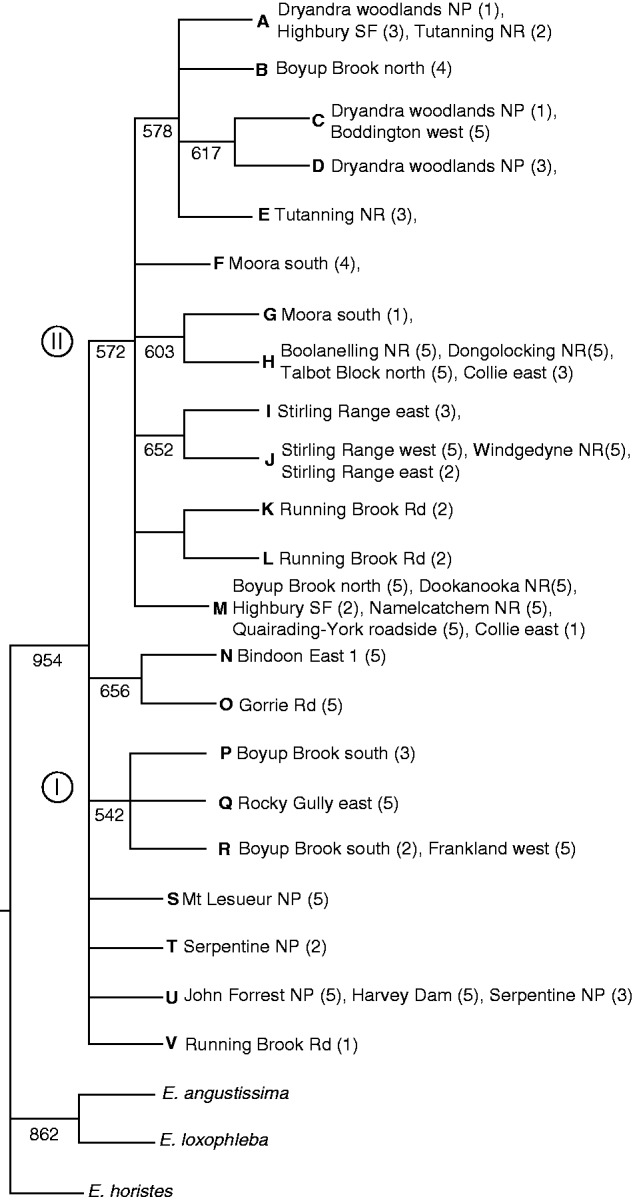
Phylogenetic parsimony tree of haplotype relationships in Eucalyptus wandoo. The consensus tree is based on 1000 bootstrap replications. Haplotypes are identified with the letters A–V, according to Table 2. Numbers below lines represent bootstrap values. Numbers in parentheses indicate sample numbers of each provenance. Roman numerals refer to the lineage number.
Geographic distribution of haplotypes
Populations with haplotypes in Lineage I were distributed in areas with a mean annual rainfall of 742 mm and an aridity index of 0·51, whereas populations with haplotypes in Lineage II occupied areas with lower mean annual rainfall (502 mm) and aridity index (0·34).
The distribution of haplotypes in Lineage I was highly structured, with most haplotypes specific to populations. Only two haplotypes were present in more than one population. In Lineage II there were some haplotypes specific to populations, mainly in the centre of the distribution, but five of the haplotypes were present in multiple populations and had widespread distributions, particularly Haplotype M to the north, Haplotype H to the east and Haplotype J to the south. An area of high diversity was identified that included six populations (populations 10, 15, 18, 20, 24 and 25) located east of Perth, in the central western area of Lineage II (Fig. 1). All these populations had two or three haplotypes, and both widespread (M) and restricted haplotypes (H, A, C) were present (Fig. 1).
At the species level, tests of neutrality were negative, Fu’s Fs was significant (P = 0·014) and Tajima’s D was just non-significant (P = 0·068). Tests of mismatch distribution were not significantly different from that expected under a sudden expansion or a spatial expansion model (Table 3). Similarly, Fu’s Fs and Tajima’s D were negative but not significant for both lineages, and tests of mismatch distribution were not significant for either sudden expansion or spatial expansion models for both lineages (Table 3).
Table 3.
Nucleotide and haplotype diversity, and parameters for tests of neutrality and mismatch distributions for Eucalyptus wandoo at the species level and for each lineage, including: haplotype diversity between populations (GST), nucleotide diversity between populations (NST), Harpenders raggedness index under a spatial expansion model (Hspat) and Harpenders raggedness index under a spatial expansion model (Hsudd)
| All populations | Lineage I | Lineage II | |
|---|---|---|---|
| Nucleotide diversity | 0·091 (0·054) | 0·081 (0·050) | 0·063 (0·040) |
| Haplotype diversity | 0·929 (0·009) | 0·862 (0·027) | 0·870 (0·019) |
| NST | 0·830 (0·047) | 0·857 (0·098) | 0·811 (0·041) |
| GST | 0·765 (0·060) | 0·887 (0·077) | 0·708 (0·086) |
| P NST > GST | 0·525 | 0·619 | 0·478 |
| Tajima’s D | –1·315 (0·068) | –0·714 (0·262) | –0·747 (0·223) |
| Fu’s Fs | –7·793 (0·014) | –0·560 (0·421) | –3·624 (0·08) |
| Hspat | 0·029 (0·560) | 0·023 (0·807) | 0·051 (0·266) |
| Hsudd | 0·029 (0·560) | 0·023 (0·811) | 0·051 (0·271) |
Standard errors (for nucleotide diversity, haplotype diversity, GST and NST) and probability of significance (for D, Fs, Hspat, Hsudd) are presented in parentheses.
Nucleotide and haplotype diversity
Nucleotide diversity and haplotypes diversity are presented in Table 3. Nucleotide diversity was higher in Lineage I than in Lineage II, although haplotype diversity was similar between lineages. Nucleotide diversity among populations (NST) and haplotype diversity among populations (GST) were 0·830 and 0·765, respectively, and were higher for Lineage I than for Lineage II. Nucleotide diversity was not significantly higher than haplotype diversity overall or in either lineage. Nucleotide divergence between Lineage I and II was 0·053 %, and the two lineages are estimated to have been separated during the later Pleistocene, approx. 530 000 years ago, later than in other species where divergence was estimated during the middle Pleistocene or earlier (700 000 to 1 million years ago, Byrne et al., 2002, 2003, Byrne and Hines, 2004; 1·76 mya, Nistelberger et al., 2014).
Nucleotide diversity averaged 0·051 % over all pairs of individuals in the area of high diversity in Lineage II, whereas in the rest of Lineage II distribution, nucleotide diversity averaged 0·021 %. In the high diversity area, total haplotype diversity was 0·896, haplotype diversity within populations was 0·601 and the proportion of haplotype diversity between populations was 0·329. For the rest of the Lineage II area, total haplotype diversity and diversity within populations was lower (0·788 and 0·091, respectively) and diversity between populations was higher (0·884).
Species distribution modelling
The model explaining the current distribution of E. wandoo (Fig. 3A) had high predictive performance [area under the curve (AUC) = 0·87] and included all five bioclimatic variables considered: mean temperature of the wettest (37·7 % variation explained) and warmest quarter (16·3 %), isothermality (22·0 %), annual precipitation (13·0 %) and precipitation of the driest quarter (11·0 %). Although mean temperature of the warmest quarter and precipitation of the driest quarter had a high Pearson pairwise correlation coefficient (r = –0·82), they were both introduced in the model because they were both important and not as strongly correlated during the LGM (r = –0·73). The realized current distribution of the species was generally well predicted by the model, with the exception of the far northern, eastern and southern range boundaries, which were scored with a low presence probability value, despite the presence of species records in those areas.
Fig. 3.
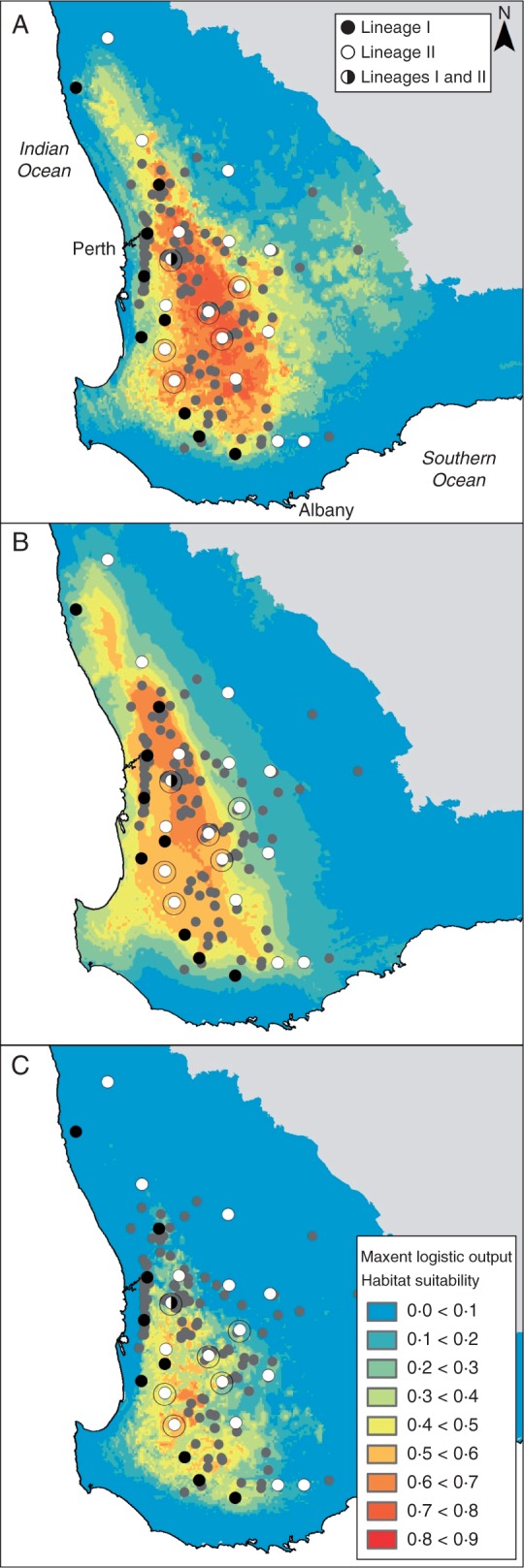
Distribution models of Eucalyptus wandoo for (A) the present, (B) the last glacial maximum (approx. 21 000 years) and (C) 2070 periods. Warmer colours indicate a higher probability of species presence, whereas colder colours indicate a lower probability of species presence (see key). Records of E. wandoo obtained from NatureMap are represented by light grey circles, and sampled populations are represented by black/white circles as indicated in the key in (A). Populations with high haplotype diversity within the refugium area are marked with an outer circle.
The distribution model of E. wandoo at the LGM was not strikingly different from the current distribution model, yet it showed a lower climatic suitablility in the east (drier), and a higher suitability in the west (more mesic) of the range (Fig. 3B), suggesting range contraction. Most importantly, a central area of high climatic suitability with a north-west–south-east orientation was apparent in the LGM modelled distribution (Fig. 3B). The forecast distribution of E. wandoo for 2070 was consistent with the current and LGM distribution patterns, showing a pattern of decreased niche suitability from the eastern but also northern range boundaries. Under the medium severity climate change scenario, the model predicted the distribution of E. wandoo to become more restricted to the western and southern regions of the current distribution, with possible extirpations from the northern and eastern regions. Despite the location of the refugial populations in the area of high past climatic suitability and stability (Fig. 4A), the GLM did not show any significant effects of these variables, nor of latitude or longitude, on present haplotype diversity. Only current climatic suitability was positively related to haplotype diversity when included in a model containing only this variable (β = 0·57; P = 0·042; AIC = 11·23).
Fig. 4.
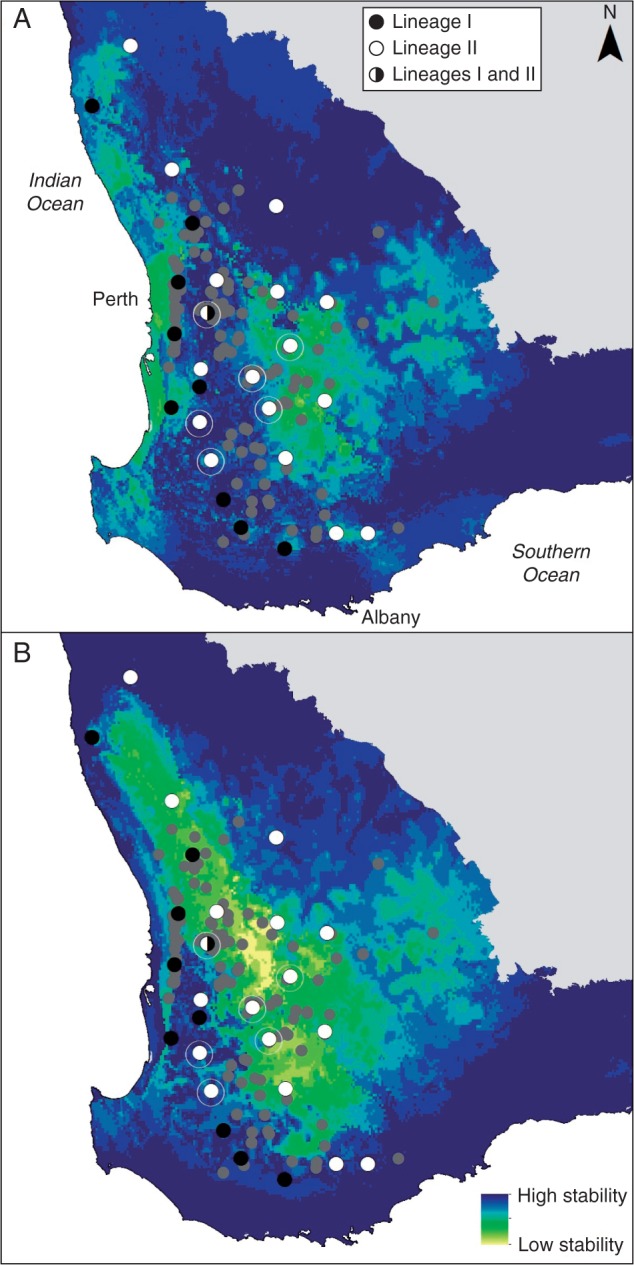
Climatic stability for E. wandoo (A) in the past, between the LGM and present moment, and (B) in the future, projected for 2070. Records of E. wandoo obtained from NatureMap are represented by light grey circles, and sampled populations are represented by black/white circles as indicated in the key in (A). Populations with high haplotype diversity within the refugium area are marked with an outer circle.
DISCUSSION
This study revealed how a combined approach using phylogeographic analysis and SDM can provide a paleo-historical context to understand current climate-driven tree decline phenomena. In the case of E. wandoo, this approach showed a historical signature of long-term persistence in a central-western refugial area, contraction to this area during the LGM, and more recent expansion to the east but also north and south. SDM has also predicted that future climate change will lead to a similar range contraction from the drier eastern and northern regions of the species distribution range. While not evidence of causality, these patterns are consistent with the currently observed mortality and decline of E.wandoo crown health, which are more pronounced in the eastern low rainfall areas (e.g. Brouwers et al., 2013), and consistent with other suggestions that the species is vulnerable to ongoing and future climate change (Hooper and Sivasithamparam, 2005; Poot and Veneklaas, 2013).
Phylogeography, SDM and climate change
While inference of population history from haplotype distributions can be complex (e.g. Ibrahim et al., 1996; Excoffier and Ray, 2008; Worth et al., 2014), the different patterns of haplotype distribution within the E. wandoo lineages suggest the influence of different evolutionary processes in different geographic regions. The highly structured distribution of haplotypes in the western lineage, with most haplotypes specific to populations, is consistent with persistence of the species through multiple climatic cycles. In contrast, the widespread distribution of several haplotypes with internal positions in the network in the northern, eastern and southern peripheral populations of the eastern lineage is consistent with contraction of the species from these areas during arid cycles, and expansion from a refugial area during mesic conditions (Crandall and Templeton, 1993; Templeton et al., 1995). This latter pattern can occur when populations are in equilibrium and have been in their present range for a substantial period of time (Slatkin, 1993).
Coalescence analysis provides inferences of population history (Rosenberg and Nordborg, 2002) and indicated a scenario of both spatial and demographic expansion in E. wandoo following the LGM. The inference of refugia and contraction/expansion was supported by SDM, which showed maintenance of high past climatic suitability and stability in the same area where high haplotype diversity was observed, and lower climatic suitability in the inland peripheral areas at the LGM compared with the current period (although low climatic suitability does not necessarily mean that the species was not able to persist; see Worth et al., 2014). While this pattern could also arise through contraction and persistence in very small populations and subsequent demographic expansion, such a scenario might be expected to lead to population-specific haplotypes (e.g. Byrne and Hines, 2004; Byrne, 2008), which were not observed here.
The E. wandoo western lineage is located along the Darling Scarp and the western edge of the Darling Plateau, which are areas of medium to low current climatic suitability, according to SDM. In this region, E. wandoo has a scattered distribution, occurring in small stands on sloping sides of valleys, in either lateritic or granitic substrates (Boland, 1984). This contrasts with the species main distribution range where it occurs as a dominant species, forming open forests on broad shallow valleys or low ridges, with clay or loam substrates (Boland, 1984). While climatic conditions along the Darling Scarp may not be optimal for E. wandoo, habitat heterogeneity and localized areas maintaining suitable microclimatic conditions may explain the long-term persistence of the species in the area.
The fact that past climate stability did not have a significant influence on haplotype diversity was unexpected; however, it may be explained by the low power of the analysis, due to the relatively low number of sampled populations and low haplotype diversity. Ideally, fossil evidence is required to determine historical distributions (Magri et al., 2006). However, fossil evidence in south-western Australia is very limited and there are no fossil cores from the inland part of the distribution where contraction may have occurred at the LGM (Dodson, 2001).
Phylogeography and SDM can reveal new insights into species responses to climate change, as in this case where the central-western area had not been identified before as having characteristics of refugia. Refugial areas in south-western Australia have been identified in coastal areas, ranges and areas of high topographical complexity (including granite outcrops), and areas of high species richness and endemism (Hopper, 1979; Byrne, 2008; Nistelberger et al., 2014). The putative refugium in E. wandoo does not show these characteristics, but does appear to have been a region of relative climatic stability during the LGM, based on SDM, and possibly during earlier arid cycles within the Pleistocene.
Despite the consistency between phylogeographic data and SDM, and the recognized usefulness of this combined approach (Scoble and Lowe, 2010; Forester et al., 2013), more comprehensive analyses are required to draw more definitive conclusions about species responses to past and future climatic changes. Recognized issues of SDM-based predictions include the uncertainty of climatic models and climatic reconstructions, assumptions that the species fundamental niche do not change over time, difficulties in modelling the occurrence of non-analogue environments in the LGM, and implementing interspecific interaction, dispersal and migration scenarios (Collevatti et al., 2013; Worth et al., 2014).
Ideally, evidence from ecophysiological and genomic analyses, including field trial survival and growth responses and physiological data on drought tolerance thresholds, are required to understand fully species response to climate change. Combined ecophysiological and genomic approaches have identified both adaptation and plasticity across a climatic gradient in eastern Australia in E. tricarpa (McLean et al., 2014; Steane et al., 2014), and similar patterns of adaptation and plasticity identified in E. loxophleba in south-western Australia (D. A. Steane, unpubl. data) are consistent with previous phylogeographic inference of persistence in localized refugia throughout the range (Byrne and Hines, 2004; Byrne, 2008). In contrast, a similar approach identified cryptic lineages with variation not associated with climate in E. salubris (Steane et al., 2015). The recent decline observed in E. wandoo and an evolutionary pattern of contraction and expansion suggest that the adaptive and/or plastic capacity of this species may not be as strong as in other eucalypts.
Evolutionary history, tree decline and conservation
Insights derived from phylogeographic analysis and SDM, while not evidence of causality, provide evidence of species responses to climate change. In E. wandoo they provide additional lines of evidence consistent with recent reports of tree decline, such as a comparison of crown health that found a decline in dry areas and most sensitivity to climate change in the eastern dry, warm end of the range (Brouwers et al., 2013). The observed phenomenon of decline in the last three decades as the climate became drier and warmer is consistent with the species’ inferred response to historical climate change. This emphasizes the need for management and conservation strategies to focus on the western areas of the current distribution where the species has persisted with larger population size through Pleistocene climatic oscillations, since these areas maintain genetic diversity that provides the evolutionary potential for adaptation to climate change.
Other disturbance factors and local environmental conditions are likely to interact with climate change, and possibly exacerbate the impacts in E. wandoo. Effects of landscape fragmentation on remnant vegetation are greatest in regions with high maximum temperatures and declining rainfall (Mantyka-Pringle et al., 2012). Thus, it is likely that the current decline of E. wandoo observed in the highly fragmented inland part of the distribution is impacted by this synergistic effect, as remnant woodlands with small size or a relative large perimeter/area ratio are more prone to crown decline (Brouwers et al., 2013). Furthermore, species with shallower root systems may be affected by soil water deficits, and this has also been demonstrated in E. wandoo, as most water in clay sub-soils becomes unavailable as upper soil layers dry out (Poot and Veneklaas, 2013).
Climate-driven tree decline due to drought and heat stress has been observed worldwide and is expected to increase with future climate change (Anderegg et al., 2013). This study has shown that a combined approach using phylogeography and SDM can provide an evolutionary context to climate-driven tree decline phenomena and a supportive framework where this can be investigated. Indeed, as both methods can cross-validate evidence for refugia and contraction under harsh climatic conditions, the combined approach offers insight into species response to past environmental changes, and sheds light on their trajectory into the future. Consistent patterns across genetic, spatial modelling, observational and/or ecophysiological data can trigger the development of tailored research questions to understand the patterns and mechanisms of tree mortality, and the development of targeted conservation measures and climate change adaptation strategies aiming at reducing and mitigating the ecological, societal and climaticological impacts of forest decline. While a combined approach using phylogeograhy and SDM has been recognized as a useful tool to understand species response to past and future climate change (e.g. Scoble and Lowe, 2010), this study is, to our knowledge, the first one to suggest its use in the context of the global issue of tree decline.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of Table S1: differences in annual mean temperature between current and LGM climate, and proportion of annual mean precipitation between LGM and current climate for the two global circulation models MIROC and CCSM.
ACKNOWLEDGEMENTS
We thank Bronwyn MacDonald for assistance, and Jerome Chopard for adjusting the code script for HAPLO. This work was supported by an Australian Research Council Linkage grant LP0347692. Scholarship support for E.D. was provided by the Department of Parks and Wildlife, and the Cooperative Research Centre for Plant-based Management of Dryland Salinity.
LITERATURE CITED
- Allen CD, Macalady AK, Chenchouni H, et al. 2010. A global overview of drought and heat-induced tree mortality reveals emerging climate change risks for forests. Forest Ecology and Management 259: 660–684. [Google Scholar]
- Anderegg WRL, Kane JM, Anderegg LDL. 2013. Consequences of widespread tree mortality triggered by drought and temperature stress. Nature Climate Change 3: 30–36. [Google Scholar]
- Avise JC. 2000. Phylogeography: the history and formation of species . Cambridge, MA: Harvard University Press. [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37–48. [DOI] [PubMed] [Google Scholar]
- Bates B, Hope P, Ryan B, Smith I, Charles S. 2008. Key findings from the Indian Ocean Climate Initiative and their impact on policy development in Australia. Climatic Change 89: 339–354. [Google Scholar]
- Boland DJ. 1984. Forest trees of Australia . Melbourne, Australia: Nelson and CSIRO. [Google Scholar]
- Braconnot P, Otto-Bliesner B, Harrison S, et al. 2007. Results of PMIP2 coupled simulations of the Mid-Holocene and Last Glacial Maximum – Part 1: experiments and large-scale features. Climate of the Past 3: 261–277. [Google Scholar]
- Breshears DD, Cobb NS, Rich PM, et al. 2005. Regional vegetation die-off in response to global-change-type drought. Proceedings of the National Academy of Sciences, USA 102: 15144–15148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker MJH, Hopper SD. 1991. A taxonomic revision of Eucalyptus wandoo, E. redunca, and allied species (Eucalytus series Levispermae Maiden-Myrtaceae) in Western Australia. Nuytsia 8: 1–189. [Google Scholar]
- Brouwers NC, Mercer J, Lyons T, Poot P, Veneklaas E, Hardy G. 2013. Climate and landscape drivers of tree decline in a Mediterranean ecoregion. Ecology and Evolution 3: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M. 2007. Phylogeography provides an evolutionary context for the conservation of a diverse and ancient flora. Australian Journal of Botany 55: 316–325. [Google Scholar]
- Byrne M. 2008. Evidence for multiple refugia at different time scales during Pleistocene climatic oscillations in southern Australia inferred from phylogeography. Quaternary Science Reviews 27: 2576–2585. [Google Scholar]
- Byrne M, Hines B. 2004. Phylogeographical analysis of cpDNA variation in Eucalyptus loxophleba (Myrtaceae). Australian Journal of Botany 52: 459–470. [Google Scholar]
- Byrne M, Macdonald B. 2000. Phylogeography and conservation of three oil mallee taxa, Eucalyptus kochii ssp. kochii, ssp. plenissima and E. horistes. Australian Journal of Botany 48: 305–312. [Google Scholar]
- Byrne M, Moran GF, Tibbits WN. 1993. Restriction map and maternal inheritance of chloroplast DNA in Eucalyptus nitens. Journal of Heredity 84: 218–220. [Google Scholar]
- Byrne M, Macdonald B, Francki M. 2001. Incorporation of sodium sulfite into extraction protocol minimizes degradation of Acacia DNA. Biotechniques 30: 742–748. [DOI] [PubMed] [Google Scholar]
- Byrne M, Macdonald B, Coates D. 2002. Phylogeographical patterns in chloroplast DNA variation within the Acacia acuminata (Leguminosae: Mimosoideae) complex in Western Australia. Journal of Evolutionary Biology 15: 576–587. [Google Scholar]
- Byrne M, Macdonald B, Brand J. 2003. Phylogeography and divergence in the chloroplast genome of Western Australian sandalwood (Santalum spicatum). Heredity 91: 389–395. [DOI] [PubMed] [Google Scholar]
- Carnicer J, Coll M, Ninyerola M, Pons X, Sánchez G, Peñuelas J. 2011. Widespread crown condition decline, food web disruption, and amplified tree mortality with increased climate change-type drought. Proceedings of the National Academy of Sciences, USA 108: 1474–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collevatti RG, Terribile LC, de Oliveira G, et al. 2013. Drawbacks to palaeodistribution modelling: the case of South American seasonally dry forests. Journal of Biogeography 40: 345–358. [Google Scholar]
- Collins WD, Bitz CM, Blackmon ML, et al. 2006. The Community Climate System Model Version 3 (CCSM3). Journal of Climate 19: 2122–2143. [Google Scholar]
- Crandall KA, Templeton AR. 1993. Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics 134: 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CSIRO, Bureau of Meteorology. 2014. State of the climate 2014. CSIRO and Bureau of Meteorology http://www.bom.gov.au/state-of-the-climate/. [Google Scholar]
- Dale VH, Joyce LA, McNulty S, et al. 2001. Climate change and forest disturbances. Bioscience 51: 723–734. [Google Scholar]
- Dodson JR. 2001. Holocene vegetation change in the mediterranean-type climate regions of Australia. Holocene 11: 673–680. [Google Scholar]
- Elith J, Phillips SJ, Hastie T, Dudík M, Chee YE, Yates CJ. 2011. A statistical explanation of MaxEnt for ecologists. Diversity and Distributions 17: 43–57. [Google Scholar]
- Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Ray N. 2008. Surfing during population expansions promotes genetic revolutions and structuration. Trends in Ecology and Evolution 23: 347–351. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 2008. PHYLIP (Phylogeny inference package), version 3.68. Seattle, USA: Department of Genetics, The University of Washington. [Google Scholar]
- Forester BR, DeChaine EG, Bunn AG. 2013. Integrating ensemble species distribution modelling and statistical phylogeography to inform projections of climate change impacts on species distributions. Diversity and Distributions 19: 1480–1495. [Google Scholar]
- Fu Y-X. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Giorgi F. 2008. Increased aridity in the Mediterranean region under greenhouse gas forcing estimated from high resolution simulations with a regional climate model. Global and Planetary Change 62: 195–209. [Google Scholar]
- Gugger PF, Ikegami M, Sork VL. 2013. Influence of late Quaternary climate change on present patterns of genetic variation in valley oak, Quercus lobata Née. Molecular Ecology 22: 3598–3612. [DOI] [PubMed] [Google Scholar]
- Guisan A, Zimmermann NE. 2000. Predictive habitat distribution models in ecology. Ecological Modelling 135: 147–186. [Google Scholar]
- Hasumi H, Emori S. 2004. K-1 coupled GCM (MIROC) description. Tokyo, Japan: Center for Climate System Research, University of Tokyo, National Institute for Environmental Studies, Frontier Research Center from Global Change. [Google Scholar]
- Hewitt GM. 2004. Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society B: Biological Sciences 359: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25: 1965–1978. [Google Scholar]
- Hooper RJ, Sivasithamparam K. 2005. Characterization of damage and biotic factors associated with the decline of Eucalyptus wandoo in southwest Western Australia. Canadian Journal of Forest Research 35: 2589–2602. [Google Scholar]
- Hopper SD. 1979. Biogeographical aspects of speciation in the southwest Australian flora. Annual Review of Ecology and Systematics 10: 399–422. [Google Scholar]
- Hopper SD, Gioia P. 2004. The Southwest Australian Floristic Region: conservation of a global hotspot of biodiversity. Annual Review of Ecology and Systematics 35: 623–650. [Google Scholar]
- Ibrahim KM, Nichols RA, Hewitt GM. 1996. Spatial patterns of genetic variation generated by different forms of dispersal during range expansion. Heredity 77: 282–291. [Google Scholar]
- Magri D, Vendramin GG, Comps B, et al. 2006. A new scenario for the Quaternary history of European beech populations: palaeobotanical evidence and genetic consequences. New Phytologist 171: 199–221. [DOI] [PubMed] [Google Scholar]
- Manning AD, Fischer J, Lindenmayer DB. 2006. Scattered trees are keystone structures – implications for conservation. Biological Conservation 132: 311–321. [Google Scholar]
- Mantyka-Pringle CS, Martin TG, Rhode JR. 2012. Interactions between climate and habitat loss effects on biodiversity: a systematic review and meta-analysis. Global Change Biology 18: 1289–1252. [Google Scholar]
- McLean EH, Prober SM, Stock WD, et al. 2014. Plasticity of functional traits varies clinally along a rainfall gradient in Eucalyptus tricarpa. Plant, Cell and Environment 37: 1440–1451. [DOI] [PubMed] [Google Scholar]
- Moritz C, Agudo R. 2013. The future of species under climate change: resilience or decline? Science 341: 504–508. [DOI] [PubMed] [Google Scholar]
- Nei M. 1973. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences, USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevill PG, Bradbury D, Williams A, Tomlinson S, Krauss SL. 2014. Genetic and palaeo-climatic evidence for widespread persistence of the coastal tree species Eucalyptus gomphocephala (Myrtaceae) during the Last Glacial Maximum. Annals of Botany 113: 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nistelberger H, Gibson N, Macdonald B, Tapper SL, Byrne M. 2014. Phylogeographic evidence for two mesic refugia in a biodiversity hotspot. Heredity 113: 454–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips SJ, Dudík M. 2008. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31: 161–175. [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE. 2006. Maximum entropy modeling of species geographic distributions. Ecological Modelling 190: 231–259. [Google Scholar]
- Phillips SJ, Dudík M, Elith J, et al. 2009. Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data. Ecological Applications 19: 181–197. [DOI] [PubMed] [Google Scholar]
- Pons O, Petit RJ. 1996. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144: 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot P, Veneklaas E. 2013. Species distribution and crown decline are associated with contrasting water relations in four common sympatric eucalypt species in southwestern Australia. Plant and Soil 364: 409–423. [Google Scholar]
- Rosenberg NA, Nordborg M. 2002. Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Nature Reviews Genetics 3: 380–390. [DOI] [PubMed] [Google Scholar]
- Schorr G, Pearman PB, Guisan A, Kadereit JW. 2013. Combining palaeodistribution modelling and phylogeographical approaches for identifying glacial refugia in Alpine Primula. Journal of Biogeography 40: 1947–1960. [Google Scholar]
- Schulman L, Toivonen T, Ruokolainen K. 2007. Analysing botanical collecting effort in Amazonia and correcting for it in species range estimation. Journal of Biogeography 34: 1388–1399. [Google Scholar]
- Scoble J, Lowe AJ. 2010. A case for incorporating phylogeography and landscape genetics into species distribution modelling approaches to improve climate adaptation and conservation planning. Diversity and Distributions 16: 343–353. [Google Scholar]
- Slatkin M. 1993. Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47: 264–279. [DOI] [PubMed] [Google Scholar]
- Steane DA, Potts BM, McLean E, et al. 2014. Genome-wide scans detect adaptation to aridity in a widespread forest tree species. Molecular Ecology 23: 2500–2513. [DOI] [PubMed] [Google Scholar]
- Svenning J-C, Fløjgaard C, Marske KA, Nógues-Bravo D, Normand S. 2011. Applications of species distribution modeling to paleobiology. Quaternary Science Reviews 30: 2930–2947. [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1996. The amount of DNA polymorphism maintained in a finite population when the neutral mutation rate varies among sites. Genetics 143: 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Routman E, Phillips CA. 1995. Separating population structure from population history: a cladistic analysis of the geographical distribution of mitochondrial DNA haplotypes in the tiger salamander, Ambystoma tigrinum. Genetics 140: 767–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MA, Byrne M. 2006. Congruence between phylogeographic patterns in cpDNA variation in Eucalyptus marginata (Myrtaceae) and geomorphology of the Darling Plateau, south-west of Western Australia. Australian Journal of Botany 54: 17–26. [Google Scholar]
- Wiens JJ, Graham CH. 2005. Niche conservatism: integrating evolution, ecology, and conservation biology. Annual Review of Ecology, Evolution, and Systematics 36: 519–539. [Google Scholar]
- Worth JRP, Williamson GJ, Sakaguchi S, Nevill PG, Jordan GJ. 2014. Environmental niche modelling fails to predict Last Glacial Maximum refugia: niche shifts, microrefugia or incorrect palaeoclimate estimates? Global Ecology and Biogeography 23: 1186–1197. [Google Scholar]
- Yates CJ, Hobbs RJ, True DT. 2000. The distribution and status of eucalypt woodlands in Western Australia. In: Hobbs RJ, Yates C, eds. Temperate eucalypt woodlands in Australia: biology, conservation, management and restoration. Chipping Norton, NSW, Australia: Surrey Beatty & Sons Pty., Ltd, 86–106. [Google Scholar]
- Yates CJ, McNeill A, Elith J, Midgley GF. 2010. Assessing the impacts of climate change and land transformation on Banksia in the South West Australian Floristic Region. Diversity and Distributions 16: 187–201. [Google Scholar]
- Zurawski G, Clegg MT, Brown AHD. 1984. The nature of nucleotide sequence divergence between barley and maize chloroplast DNA. Genetics 106: 735–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


