Abstract
Discriminating between rare benign and pathogenic variation is a key challenge in clinical genetics, particularly as increasing numbers of non-synonymous SNPs are identified in resequencing studies. Here, we describe an approach for the functional annotation of non-synonymous variants that identifies functionally important, disease-causing residues across protein families using multiple sequence alignment. We applied the methodology to long QT syndrome (LQT) genes, which cause sudden death, and their paralogues, which largely cause neurological disease, and accurately classified known LQT disease-causing variants (positive predictive value=98.4%) with a better performance than established bioinformatic methods. The analysis also identified 1078 new putative disease loci, which we incorporated along with known variants into a comprehensive and freely accessible long QT resource (http://cardiodb.org/Paralogue_Annotation/), based on newly created Locus Reference Genomic sequences. We propose that paralogous annotation is widely applicable for Mendelian human disease genes.
Keywords: Variant annotation, Paralogue, Non-synonymous, Long QT Syndrome, Inherited Heart Disease
Inherited long QT syndrome (LQT [MIM607542]) is a life-threatening Mendelian disease caused by genetic variants in ion channel genes(Campuzano, et al., 2010; Tester and Ackerman, 2011). While clinical guidelines(Ackerman, et al., 2011) indicate that genetic testing of patients with LQT should be performed, it is often difficult to reach a conclusive genetic diagnosis when a new or rare sequence change is detected(Cooper and Shendure, 2011). This relates to uncertainty concerning the significance of novel non-synonymous SNPs (nsSNPs), non-uniform annotation of genomic coordinates of known disease variants, and inconclusive genotype-phenotype relationships in existent databases(Cooper and Shendure, 2011; Dalgleish, et al., 2010).
In one study it was noted that variation at an equivalent residue in KCNQ1 (expressed in the heart [MIM 607542]) and KCNQ4 (expressed in the inner ear [MIM 603537]) causes LQT and autosomal dominant deafness respectively(Kubisch, et al., 1999) [MIM 192500; 600101]. To date pathogenic variation in gene paralogues represents an unexploited resource for the functional annotation of nsSNPs in disease genes. We hypothesized that systematic analysis of disease-causing amino acid substitutions in LQT paralogues could be used to annotate new pathogenic residues in LQT genes for clinical research and molecular diagnostics.
Here we assessed whether known disease-causing variants in LQT paralogues predict pathogenic variation in LQT genes. Potential paralogues were identified using the defined functional gene families of the IUPHAR Ion Channel Database(Sharman, et al., 2011) and BLAST(Altschul, et al., 1990), and disease-causing nsSNPs in these genes were retrieved from HGMD professional 2011.1(Stenson, et al., 2003) and locus specific databases (LSDBs) where available (http://www.LOVD.nl/SCN4A, http://www.LOVD.nl/CACNA1F, http://grenada.lumc.nl/LOVD2/FHM/). Of the 13 genes known to cause LQT(Hedley, et al., 2009; Yang, et al., 2010), eight have one or more paralogues (n=87) that contain variants causing autosomal dominant disease (Supp. Table S1). No disease-causing nsSNPs were found in paralogues of five LQT genes (LQT9-13: CAV3, SCN4B, AKAP9, SNTA1, KCNJ5; [MIM 601253,608256,604001,601017,600734]). Typically LQT paralogues represent ion channels and cause diseases such as familial epilepsy, ataxia and deafness that are attributable to perturbed neuronal ion channel function.
We constructed multiple sequence alignments for these eight LQT genes and their paralogues using the M-Coffee algorithm(Wallace, et al., 2006) (Supp. Table S2, in html format). As well as producing high-quality alignments, M-Coffee has the advantage of reporting a consensus score for each residue. This may be considered a measure of reliability of the alignment in any given region, so that mappings in a region of low consensus may be disregarded if appropriate. Paralogues without disease-causing nsSNPs were included to achieve the best possible alignments. Using these multiple sequence alignments, each paralogue protein residue with a known disease-causing substitution was mapped onto the equivalent amino acid of the LQT protein. In total, known disease-causing amino acid substitutions in LQT paralogues mapped to 1277 residues across the eight LQT proteins (Table 1).
Table 1.
Functional classification of residues in long QT proteins.
| Residues with published variants in LQT proteins |
Residues with disease variants mapped from paralogues |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein | LQT | ODP | Benign | PB | Conflict | UN | Total | LQT | ODP | Benign | PB | Conflict | UN | Total | |
| KCNQ1 | LQT1 | 178 | 8 | 12 | 5 | 15 | 458 | 676 | 36 | 1 | 1 | 0 | 2 | 34 | 74 |
|
| |||||||||||||||
| KCNH2 | LQT2 | 249 | 4 | 32 | 4 | 14 | 856 | 1159 | 27 | 0 | 0 | 0 | 0 | 37 | 64 |
|
| |||||||||||||||
| SCN5A | LQT3 | 328 | 23 | 34 | 6 | 30 | 1595 | 2016 | 95 | 4 | 0 | 1 | 2 | 303 | 405 |
|
| |||||||||||||||
| ANK2 | LQT4 | 9 | 5 | 31 | 18 | 1 | 3860 | 3924 | 0 | 0 | 0 | 0 | 0 | 6 | 6 |
|
| |||||||||||||||
| KCNE1 | LQT5 | 22 | 0 | 5 | 2 | 4 | 96 | 129 | 3 | 0 | 1 | 2 | 0 | 10 | 16 |
|
| |||||||||||||||
| KCNE2 | LQT6 | 12 | 1 | 2 | 0 | 1 | 107 | 123 | 2 | 0 | 1 | 0 | 1 | 24 | 28 |
|
| |||||||||||||||
| KCNJ2 | LQT7 | 33 | 1 | 3 | 1 | 0 | 389 | 427 | 17 | 0 | 0 | 0 | 0 | 78 | 95 |
|
| |||||||||||||||
| CACNA1C | LQT8 | 8 | 0 | 3 | 17 | 0 | 2110 | 2138 | 2 | 0 | 0 | 1 | 0 | 586 | 589 |
|
| |||||||||||||||
| Totals | 839 | 42 | 122 | 53 | 65 | 9471 | 10592 | 182 | 5 | 3 | 4 | 5 | 1078 | 1277 | |
| Percentage | 7.9% | 0.4% | 1.2% | 0.5% | 0.6% | 89.4% | 14.3% | 0.4% | 0.2% | 0.3% | 0.4% | 84.4% | |||
Left panel: for each protein, residues are categorised according to the phenotype associated with variants at that residue: variation at the residue causes definite long QT, short QT or Brugada syndrome (LQT); causes other disease phenotype (ODP); is benign (Benign) or is probably benign (PB). A number of residues have conflicting reports of pathogenicity in the literature (Conflict) and many residues have no reported variation and are unannotated (UN). Right panel: residues identified by mapping of disease-causing variants from paralogues are significantly enriched for known disease-causing variants (p=4.8×10−7, Fisher’s exact test), and annotate 1078 novel putative disease-causing loci.
We next annotated all known disease-causing amino acid substitutions in these eight LQT proteins using HGMD professional 2011.1(Stenson, et al., 2003), dbSNP build 132(Sherry, et al., 2001), LSDBs (Gene Connection for the Heart, http://www.fsm.it/cardmoc/, Human Variome project in China, http://www.genomed.org/LOVD/) and additional variants retrieved from the published literature(Kapa, et al., 2009). All online databases were accessed on 31st July 2011. A single canonical isoform for each LQT gene was used for this analysis, using new Locus Reference Genomic (LRG) transcripts. The LRG accession numbers for these isoforms are shown in Supp. Table S1.
Variants were classified according to reported clinical phenotypes. All variants reported as definite causes of long QT, short QT (SQT [MIM 609620]), Brugada syndrome (BrS [MIM 601144]), or variants thereof were grouped together under the label “LQT”. LQT, SQT and BrS are caused by variants in the same set of genes, but differ in whether variants lead to gain or loss of function. They were grouped together so that any deleterious altered function (gain or loss) in a paralogue could be mapped to the LQT gene, independent of direction of effect. Variants causing other inherited diseases were categorized as Other Disease Phenotype (ODP). Disease associations (e.g. from genome-wide association studies) were excluded. ODP variants included those reported as “possible” causes of LQT, BrS or SQT, or as “definite” causes of an intermediate phenotype such as cardiac arrhythmia. Many or most of these variants may in fact cause LQT, but have been annotated distinctly to allow for comparison of only the most robustly phenotyped variants.
Published variants described as benign in LSDBs or literature case series, or reported in dbSNP with no disease phenotype and a population frequency of >1% in any population (which we consider incompatible with the population frequency of LQT), were categorized as Benign polymorphisms. All other missense mutations in dbSNP with no reported cardiac phenotype were classified as Probably Benign (PB): as many variants have been reported in dbSNP without associated phenotype data, and LQT is not always highly penetrant, there may be some false negatives in this dataset. Instances where the same variant or residue was classified as disease-causing and benign in different reports were labeled as conflicts.
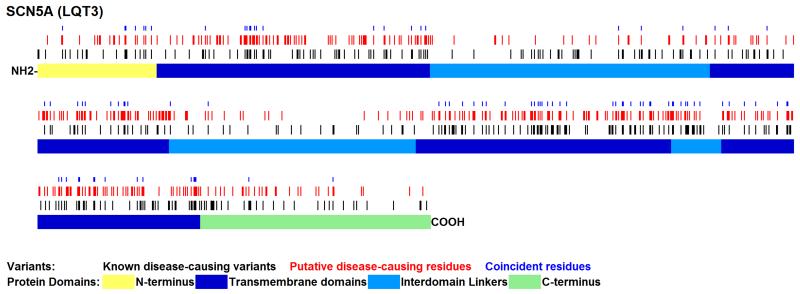
Of the 1277 mappable pathogenic variants from LQT gene paralogues, 185 mapped to LQT residues where variation has previously been unambiguously defined as either pathogenic (n=182, 98.4%) or benign (n=3, 1.6%). This demonstrates that LQT residues at equivalent sites to known pathogenic variation in LQT paralogues are significantly enriched for LQT-causing variants (Table 1; p=4.8×10−7, Fisher’s exact test), as illustrated for SCN5A (Figure 1). Comparing the most robust datasets (known LQT v. known Benign), paralogous annotation has a positive predictive value (PPV) of LQT pathogenicity of 98.4%. When including less reliable annotations, i.e. including ODP as additional true positives and PB as false positives, the PPV of the method remains high (96.4%), and the enrichment significant (p=2.0×10−7). By comparison, SIFT(Kumar, et al., 2009) (version 4.0.3b) and PolyPhen-2(Adzhubei, et al., 2010) (version 2.1.0) have PPVs of 93.6-95.5% and 90.6-95.9% respectively, using the same dataset (Supp. Table S3).
Figure 1. Schematic representation of SCN5A protein showing known disease-causing residues and those predicted to be disease-causing by paralogous annotation.
Previously annotated disease-causing residues are shown in black. Predicted disease-causing residues are shown in red. Coincidences of known and predicted variants are denoted by a blue dot. A schematic of the protein is shown below the variants with color-coding of protein domains. All known variants and novel paralogue mappings for this and for the other seven LQT genes are shown in Supp. Figure S1.
Mendelian pathogenic variants are more frequently located in particular protein domains, and this has been used previously to calculate domain-specific estimates of the probability of pathogenicity of variants in three LQT genes(Kapa, et al., 2009). We annotated each protein with its protein structure, using the domain annotations previously reported in (Kapa, et al., 2009) (for KCNQ1, KCNH2 and SCN5A), or annotations from Swiss-Prot (for KCNE1, KCNE2, KCNJ2 and CACNA1C). ANK2 is not presented due to a paucity of domain features and small number of mapped variants. Expected and observed numbers of variants in each protein region were tabulated, and chi-square tests used to identify genes with non-uniform distributions of variants (Supp. Table S4). We observed that variants in paralogous genes mapped to LQT residues with patterns of domain enrichment similar to those previously reported, and we suggest that the location of mapped paralogue variants may be used to inform domain-specific estimates of pathogenicity for less well-characterized LQT genes (e.g. CACNA1C & KCNJ2).
Paralogous annotation of pathogenic Mendelian variation is widely applicable. By inference the technique can be applied in a reciprocal fashion across the gene families that we have studied, using annotated variants in LQT genes to interpret variation in inherited epilepsy genes (Supp. Table S5). We examined all disease genes in HGMD pro 2011.1(Stenson, et al., 2003), and identified 1824 genes (45.5% of the dataset) with one or more paralogues that contain disease-causing variants (average 3.2 paralogues per gene). This preliminary analysis suggests there are over 150,000 potentially informative annotations from disease-causing variants in paralogues of human disease genes.
The accuracy of the method we describe here depends on reliable phenotype data associated with genetic variants that need to be in an accessible format and readily available (e.g. via the European Genome-Phenome Archive, http://www.ebi.ac.uk/ega/). The correct and standardized annotation of genetic variants in these datasets is fundamental. In the course of this study we identified numerous errors arising from the use of alternate reference sequences (Supp. Table S6). The Human Genome Variation Society recommends the use of a Locus Reference Genomic sequence (LRG) (Dalgleish, et al., 2010) to overcome these deficiencies. Hence, to enable accurate annotation of LQT variants for clinical application we established new LRG coordinates for all 13 LQT genes. For the eight LQT genes studied here, we have collated published variants, submitted annotations to dbSNP and created a comprehensive and freely available resource for LQT research and clinical diagnostics (http://cardiodb.org/Paralogue_Annotation/; Supp. Table S7).
In summary, we applied systematic multiple sequence alignment of LQT gene paralogues to predict sites of disease-causing variation. This identified novel putative disease-causing variants in ~10% of previously un-annotated LQT residues that adds significantly to the functional annotation of LQT genes. We generated a comprehensive LQT resource based on new LRGs to disseminate these findings, along with existing annotations, for the wider scientific and clinical community that will become increasingly powerful with cumulative annotations from resequencing studies(Cooper and Shendure, 2011). The technique we describe here is widely transferable to human disease genes and, we believe, important for clinical interpretation of novel missense mutations.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge funding from the Wellcome Trust, the British Heart Foundation, the Medical Research Council (UK), and the National institute for Health Research, Royal Brompton and Harefield Cardiovascular Biomedical Research Unit.
Grant Sponsors
Wellcome Trust (087183/Z/08/Z [to JSW])
British Heart Foundation (SP/10/10/28431 [to SAC & EB])
Medical Research Council (UK)
Royal Brompton & Harefield Cardiovascular Biomedical Research Unit, National Institute for Health Research, UK
Footnotes
WEB RESOURCES
Long QT paralogue annotations and compendium of known nsSNPs in LQT genes, http://cardiodb.org/Paralogue_Annotation/
dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/
Locus Reference Genomic sequences, http://www.lrg-sequence.org/
ACCESSION NUMBERS
Published variants in LQT genes collated in this paper have been submitted to dbSNP with batch ID 1056584, submitter handle RBH_CV_BRU.
REFERENCES
- Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, Camm AJ, Ellinor PT, Gollob M, Hamilton R, Hershberger RE, Judge DP, Le Marec H, McKenna WJ, Schulze-Bahr E, Semsarian C, Towbin JA, Watkins H, Wilde A, Wolpert C, Zipes DP. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Europace. 2011;13:1077–109. doi: 10.1093/europace/eur245. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Campuzano O, Beltran-Alvarez P, Iglesias A, Scornik F, Perez G, Brugada R. Genetics and cardiac channelopathies. Genet Med. 2010;12:260–7. doi: 10.1097/GIM.0b013e3181d81636. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–40. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- Dalgleish R, Flicek P, Cunningham F, Astashyn A, Tully RE, Proctor G, Chen Y, McLaren WM, Larsson P, Vaughan BW, Beroud C, Dobson G, Lehvaslaiho H, Taschner PE, den Dunnen JT, Devereau A, Birney E, Brookes AJ, Maglott DR. Locus Reference Genomic sequences: an improved basis for describing human DNA variants. Genome Med. 2010;2:24. doi: 10.1186/gm145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley PL, Jorgensen P, Schlamowitz S, Wangari R, Moolman-Smook J, Brink PA, Kanters JK, Corfield VA, Christiansen M. The genetic basis of long QT and short QT syndromes: a mutation update. Hum Mutat. 2009;30:1486–511. doi: 10.1002/humu.21106. [DOI] [PubMed] [Google Scholar]
- Kapa S, Tester DJ, Salisbury BA, Harris-Kerr C, Pungliya MS, Alders M, Wilde AA, Ackerman MJ. Genetic testing for long-QT syndrome: distinguishing pathogenic mutations from benign variants. Circulation. 2009;120:1752–60. doi: 10.1161/CIRCULATIONAHA.109.863076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubisch C, Schroeder BC, Friedrich T, Lutjohann B, El-Amraoui A, Marlin S, Petit C, Jentsch TJ. KCNQ4, a novel potassium channel expressed in sensory outer hair cells, is mutated in dominant deafness. Cell. 1999;96:437–46. doi: 10.1016/s0092-8674(00)80556-5. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Sharman JL, Mpamhanga CP, Spedding M, Germain P, Staels B, Dacquet C, Laudet V, Harmar AJ. IUPHAR-DB: new receptors and tools for easy searching and visualization of pharmacological data. Nucleic Acids Res. 2011;39:D534–8. doi: 10.1093/nar/gkq1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–81. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- Tester DJ, Ackerman MJ. Genetic testing for potentially lethal, highly treatable inherited cardiomyopathies/channelopathies in clinical practice. Circulation. 2011;123:1021–37. doi: 10.1161/CIRCULATIONAHA.109.914838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace IM, O'Sullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–9. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liang B, Liu J, Li J, Grunnet M, Olesen SP, Rasmussen HB, Ellinor PT, Gao L, Lin X, Li L, Wang L, Xiao J, Liu Y, Zhang S, Liang D, Peng L, Jespersen T, Chen YH. Identification of a Kir3.4 mutation in congenital long QT syndrome. Am J Hum Genet. 2010;86:872–80. doi: 10.1016/j.ajhg.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.