New human genetic findings paired with biochemical studies are challenging current thinking on the function of chromatin remodeling complexes.
Keywords: ATP-dependent chromatin remodelers, human cancer
Abstract
Over the past 4 years, nearly 100 exome sequencing studies have revealed the high frequency of mutations in the genes encoding the subunits of ATP-dependent chromatin remodelers in human cancer. Most of these mutations are within the genes encoding subunits of the BAF (Brg/Brahma-associated factors) or mSWI/SNF complex, which is one of two dozen predicted ATP-dependent chromatin remodeling complexes in mammals. Considering BAF complexes as a single entity, the 15 subunits encoded by 29 genes are mutated in >20% of human cancer, across a broad range of tumor types. These observations demonstrate that there is little redundancy in the oncogenic function of BAF complexes with the other remodeling complexes, underscoring their unique roles. Several important conclusions emerge from these genomic data: specific subunits appear to be mutated in specific cancers, highlighting tissue-specific protective roles; mutations can function as tumor suppressors or oncogenes; mutations can be homozygous or, more commonly, heterozygous, implying their dosage-sensitive roles in an unknown yet fundamental process used to suppress the genesis of cancer. These new human genetic findings paired with biochemical studies are challenging old ideas on how chromatin remodeling complexes function, generating new hypotheses with respect to their normal and oncogenic mechanisms and highlighting potential avenues for therapeutic intervention in human cancer.
Following the 13-year effort to sequence the full human genome, sequencing of disease genomes, notably of human cancer, began to surface. With the advent of next-generation sequencing platforms readily accessible to the academic community, study after study reporting the sequencing of human cancer genomes began appearing almost weekly, starting in 2010. One of the most surprising results from these genome– and exome-wide sequencing efforts was that chromatin regulation and epigenetic centered processes, which had previously been only loosely connected to oncogenesis, were tightly linked to the development of cancer. Previous studies over the years involving drugs such as histone deacetylase and histone methyl transferase inhibitors, hydroxamic acids, sirtuins, and others had suggested the important role of epigenetic modulation in cancer—indeed, nearly all cancers display epigenetic changes, and most cancer mutations, in either a direct or an indirect manner, affect the epigenome (1, 2)—yet the extent of involvement was not fully appreciated. Strikingly, among the most frequent mutations uncovered in human cancer sequencing efforts were mutations in genes encoding the subunits of adenosine triphosphate (ATP)–dependent chromatin remodeling complexes, most notably the mammalian SWI/SNF or BAF (Brg/Brahma-associated factors) complexes. This is the core focus of this review.
With more than 20% of human cancers bearing a mutation to one subunit of a 15-subunit complex (3, 4), mechanistic studies that translate the meaning of these genetic perturbations are critical for understanding oncogenic function. The sheer heterogeneity of perturbations unveiled by sequencing presents a substantial challenge and motivates multidirectional mechanistic hypotheses: mutations can be heterozygous or homozygous, somatic or germline, and result in deletion point mutation, and even translocation resulting in fused protein segments to stable subunits. Collectively, this yields a complex picture of BAF complex compromise, which appears to drive both loss-of-function and gain-of-function oncogenic mechanisms.
Here, we review the evolution and function of SWI/SNF complexes, the landmark studies pertaining to their roles in both development and malignancy, as well as subunit-specific perturbation in cancer, and posit new directions for future investigation and therapeutic approaches.
ATP-DEPENDENT CHROMATIN REMODELING: DISCOVERY AND EVOLUTION
ATP-dependent chromatin remodeling is one of several mechanisms that permit the compaction and decompaction of DNA in the nucleus while retaining the capacity for replication, selective gene expression, and DNA repair and recombination. This mechanism was first discovered in yeast, ironically in screens for signal transduction molecules involved in responses to mating factor that lead to mating type switching (hence the name Switch or SWI) (5). Three of these genes were independently discovered in screens for signaling molecules leading to sucrose fermentation in response to nutrient switching in yeast (sucrose nonfermenting or SNF) (6). Genetic reversion of these signaling phenotypes was achieved by second mutations in histone genes, showing that specific SWI and SNF genes in yeast actually regulated chromatin remodeling rather than acting as participants in a signaling mechanism. Biochemical characterization then led to the discovery that these genes encoded proteins that associated with one another; hence, the term “SWI/SNF complex” was used to characterize this chromatin regulatory entity (7).
Within a few years, similar complexes were discovered in Drosophila from genetic screens designed to detect genes opposing Polycomb-mediated repression of homeotic genes (8, 9). Notably, the ATPase subunit, of the complex in flies, Brm, was found to be homologous to SWI2 or SNF2. Significantly, revertants to mutations in Brm have never been found in histone genes, as is the case in yeast. Thus, it appeared that for the Drosophila SWI/SNF or BAP (Brm-associated protein) (10) complex, the primary targets were not histones, but rather opposing repressive complexes known as Polycomb complexes; however, because of the large number of histone genes, a dominant mutation in a histone may appear genetically invisible. Evidence indicating that Polycomb complexes are an important primary target of mammalian SWI/SNF or BAF complexes has emerged in more recent years from the observation that mutation of the ATPase Brg1 (Smarca4) of BAF complexes leads to H3K27Me3 accumulation and repression of many genes in embryonic stem (ES) cells (11). Additional evidence that BAF opposes Polycomb has come from studies of human malignancy, as will be discussed below (see “BAF Complexes as Tumor Suppressors: Key Mechanistic Themes”). However, genes with related functions (such as to oppose Polycomb-mediated repression) were also discovered in histone modification genes, notably in the MLL genes that are subunits of the COMPASS complex and provide the activating modification H3K4Me3 (2, 12).
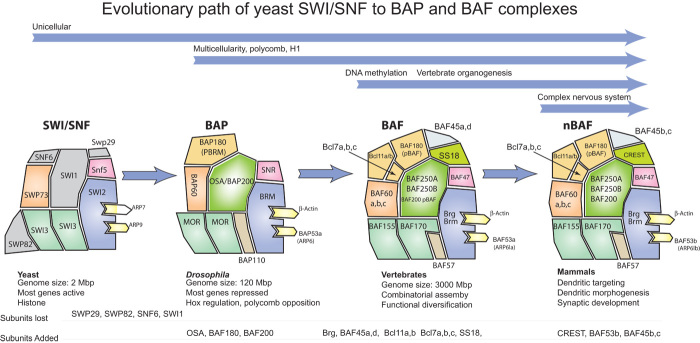
Early sequencing efforts led to the realization that ATP-dependent chromatin remodeling was a mechanism likely to be used extensively. In mammals, 29 genes are predicted to encode ATPases similar to the yeast SWI2/SNF2 protein. Genetically, these ATPases are nearly all nonredundant, indicating that they play specific biologic roles and their mutations give rise to specific phenotypes and diverse human diseases, as will be discussed below. Biochemical studies in yeast, flies, and mammals have heralded a complete or near-complete characterization of all the subunits of several of the ATP-dependent chromatin remodeling complexes. Perhaps the most well studied of the complexes, the SWI/SNF complex, has evolved extensively in the past 500 million years from yeast to mammals (Fig. 1). The steps in this evolutionary process illustrate well how this fundamental mechanism of chromatin regulation has adapted to accommodate changes in strategies of gene regulation.
Fig. 1. Evolution of the yeast SWI/SNF complexes to the fly BAP and vertebrate BAF complexes.
The figure depicts the subunit structure of these related complexes over the last 500 million years of evolutionary history. Colors are used to indicate homology. The development of multicellularity and the need to repress most genes is coupled with the appearance of polycomb-mediated repression, histone H1, and major changes in the subunit structure of SWI/SNF in its transition to BAP complexes in flies. The emergence of vertebrates, appearance of a much larger genome, DNA methylation, and vertebrate complexity is accompanied by another transition in subunit structure and combinatorial assembly. Finally, with the emergence of a complex nervous system, four new neuron-specific subunits enter the complex and are essential for dendritic morphogenesis, synaptogenesis, and connectivity within the nervous system.
SWI/SNF, BAP, and BAF complexes through evolution
A great deal of information can be gained from an examination of the evolution of the subunit composition of the SWI/SNF or BAF complexes (Fig. 1). A clear transition occurs with the appearance of multicellularity, which can be appreciated by the additional complexity in multicellular organisms. Four subunits are lost from the yeast complex, and three different ones are added in flies (and probably also worms, although the complex in worms has not been purified). These changes in SWI/SNF probably reflect newly emerging strategies of chromatin regulation that must support the fact that whereas most genes are expressed in yeast, most genes are repressed in multicellular organisms. Although Polycomb complexes appeared evolutionarily before the split of plants and animals, they were lost in Saccharomyces cerevisiae and Saccharomyces pombe (13). Genetic studies in flies indicate that the major function of the BAP complex is to oppose Polycomb, whereas in yeast, genetic studies clearly demonstrated that histones and presumably nucleosomes are the major targets for SWI/SNF action. Several other strategic changes to chromatin occur in multicellular organisms, including the appearance of histone H1, which may also create chromatin structures that require BAF complexes for conversion to more accessible DNA. With the evolutionary emergence of vertebrates and DNA methylation, nine additional essential subunits are added, which do not have homologs in yeast or flies. In vertebrates, the complexes become polymorphic and combinatorially assembled, allowing them to take on highly specialized functions (14–18). Finally, very late in vertebrate evolution, four additional neuron-specific subunits emerge that are essential for the formation and wiring of our complex nervous systems (19, 20). Thus, the evolution of these complexes might be illustrative of the way that functionalities of the complex are gained and lost with each major change in gene expression and epigenetic strategies over the past half-billion years.
However, through 500 million years of evolution, several subunits, which are remnants of the yeast SWI/SNF complex remain constant. First is the structure of the ATPase subunit and the so-called helicase domains of Brg and Brm, which show a remarkable degree of conservation between them, as with all of the 29 members of this family of chromatin remodeling complexes. This suggests that fundamental mechanisms are conserved among the entire group of SWI2-like remodelers. In addition, the BAF155 and BAF170 subunits are clearly homologs of SWI3, BAF47 is clearly a homolog of SNF5, and SWP73 is a homolog of BAF60. Whereas the mammalian and Drosophila complexes contain β-actin, actin is only found in the INO80 and SWR1 complexes and not in the yeast SWI/SNF or RSC complexes (21–23). The yeast SWI/SNF complex has two Arp subunits; however, they do not polymerize, nor do they have ATPase activity. In contrast, β-actin has both of these critical activities. Thus, in this respect, mammalian BAF complexes resemble yeast INO80 and SWR1, which play critical roles as guardians of the genome against the accumulation of mutations (24) and are probably most analogous to the p53 function in vertebrates.
Early functional discoveries
Until several years ago, the mechanism of action of ATP-dependent chromatin remodeling complexes was thought to be fairly simple (25–28). They appeared to be recruited to DNA by sequence-specific transcription factors or general factors, and they subsequently attacked nucleosomes to facilitate the binding of proteins to DNA (29). Nucleosomes were thought to be the primary target of the complexes. Using in vitro transcription on nucleosomal templates, the complexes could phase or position nucleosomes, exchange nucleosomes, induce nucleosome mobility, evict nucleosomes, or relax torsional stress possibly by their direct actions on nucleosomes (28). However, doubts surrounding these mechanisms began to arise when it was discovered that the deletion of subunits of the mammalian complexes that gave the strongest phenotypes in mice were not required for in vitro chromatin remodeling (30, 31). Initially, this was thought to be due to a role for these subunits in specific targeting to regions of the genome. However, this explanation now seems unlikely because mutations in the subunits involved had phenotypes nearly identical to rapid conditional deletion of the Brg and/or Brm ATPase (32–35), indicating that individual subunit function was central to the fundamental mechanism of the complexes. In addition, these essential subunits are dedicated to the BAF complex, as indicated by the fact that they comigrate with the 2-MDa complex and can be completely depleted using high-affinity antibodies to the other subunits (19, 36). These doubts about the fundamental mechanism of chromatin remodelers were further reinforced when several unexpectedly instructive functions of these complexes were discovered in the conversion of fibroblasts to induced pluripotent stem cells (37) and the conversion of human fibroblasts to neurons (31, 38). Indeed, a large number of highly instructive functions have been described in flies and worms, including instructive functions in the targeting of dendritic trees to their proper termini (39) and cell type specification (40–42). More recently, the model in which a transcription factor recruits complexes has been questioned by genomic studies in ES cells (11). Also, the ability of oncogenic BAF complexes to activate genes in heterochromatin where there is no detectable transcription factor binding strongly indicates that transcription factors are not necessary for recruitment of these complexes (see “BAF Complexes as Oncogenes: The SS18-SSX Fusion in Synovial Sarcoma”) (36). Finally, and most compellingly, recent exome sequencing studies conducted on a number of human diseases have shown that the subunits most commonly mutated in human disease were not those required for in vitro chromatin remodeling (4, 43–45).
These recent findings underscore the fact that the fundamental mechanism of ATP-dependent chromatin remodeling complexes remains, in large part, unclear and that new techniques must be developed for the assessment of the contributions of long-range topologic effects, the complexity of posttranscriptional histone modifications, and the tissue-specific chromatin states, to name a few of the critical aspects of chromatin that cannot be detected using previously standard in vitro chromatin remodeling methods.
WHAT ARE PROTEIN COMPLEXES AND HOW DOES NATURE USE THEM?
Many chromatin regulators are subunits within large complexes, yet there seems to be little agreement on what the term “complex” implies and the difference between a “subunit” and an “associated protein.” In this review, we use the term “subunit” to imply two critical features. The first is stability of an interaction within the complex such that specific structural features are imparted to the surface of the complex. Here, the criteria that were first used came from early characterizations of the ribosome (46), showing stability at near-denaturing conditions with urea or other denaturing reagents. Generally, the stability of the subunit within the complex to 2 M urea is taken as an arbitrary criterion (36, 47). Stability is critical because it gives rise to specific surfaces that can have new biologic meanings, much in the same way that letters are assembled into words (48). The second is dedication of the subunit to the complex. We use this criterion to indicate that the protein does not exhibit binding interactions or functions outside the complex and performs its function only within one or more complexes of the same class. Biochemically, this means that the subunit comigrates with the complex on density sedimentation (gradient centrifugation) assays and can be entirely depleted from the nuclear extract with high-affinity antibodies to other complex subunits. Genetically, dedication is often reflected in a similar phenotype, but for a variety of reasons, dedicated subunits can also have different phenotypes from the phenotypes of null mutations in other subunits, and indeed, this is often the case if they are members of a highly homologous family. For example, BAF53a deletion has a phenotype that is slightly more severe than that of the ATPase Brg (33), whereas BAF53b has an entirely different phenotype because it is a subunit of the neuron-specific nBAF complex (49). Yet both are dedicated to their complexes and resist dissociation in 5 M urea. These are important considerations to be taken into account for the functional assessment of tissue- and cancer-specific complexes.
Almost all chromatin regulators are present as members of complexes, which is decidedly different from signaling proteins, which are often monomeric. Why does nature use proteins in complexes rather than leave them to perform their functions as monomers? Most likely, protein complexes have evolved to allow an ordered series of biochemical reactions and interactions to occur on the surface of the complex. The same thing might be accomplished if all the proteins were monomers, but the reactions would be very slow and perhaps the biological functions would not be performed with sufficient efficiency. The best example is, again, the ribosome: protein translation is an ordered series of events involving many subunits working simultaneously and in tandem. Almost certainly, chromatin regulation by the large ATP-dependent chromatin remodelers and perhaps other chromatin regulators involves an ordered series of specific biochemical reactions that must be spatially coordinated. Thus, a critical question in the study of ATP-dependent chromatin remodeling is the nature of the spatial relationships within the complex and the order of reactions occurring on the surfaces (as well as within the hydrophobic core) of the complexes. Even for the simplest and most well studied ATP-dependent chromatin regulator, the yeast SWI/SNF complex, these key spatially-based processes remain poorly understood.
GENOME-WIDE MAPPING OF BAF COMPLEXES
Murine genetics in concert with genome-wide binding studies have been informative for understanding features of the mechanism of action of these complexes. The complexes bind over the genome in rather large peaks covering about 2 to 4 kb (50). There are generally about 20,000 to 40,000 sites on chromatin in murine ES cells, lymphocytes, neurons, and some cell lines (50, 51). The number of peaks obtained varies, likely due to the quality of the antibodies and their signal-to-noise ratios. Because the complexes are so frequently inactivated in long-term cell lines, one must be aware that the location and number of binding sites in these lines might reflect a dysfunctional (albeit mechanistically informative) complex. There are about 300,000 BAF complexes per cell in murine ES cells, and it seems that about half of these are chromatin-associated, meaning that each BAF peak must reflect the binding of about 5 or 10 complexes with an average footprint of about 2 to 4 kb (47, 50). This is consistent with the size of the 2-MDa complex, which is about 12 times the molecular weight of a nucleosome (though often misillustrated in reviews as being smaller than a nucleosome).
The localization of BAF complexes over the genome coincides with the major fate-determining factors of the cell type. For example, in ES cells, a BAF complex of specialized subunit composition not found in other cell types to date (esBAF) is found to co-bind strongly with each of the pluripotency factors (50). This observation is consistent with the findings of Rada-Iglesias and colleagues (52), who have shown that BAF complex binding is highly predictive of human enhancers. In addition, in murine ES cells, esBAF co-binds with Polycomb repressive complexes (Kadoch et al., unpublished data), which might reflect their opposition in much the same way the histone acetylases and deacetylase co-bind over the genome, reflecting their constant opposition and the highly labile nature of this histone modification (53).
BIOLOGIC SPECIFICITY GENERATED BY COMBINATORIAL SUBUNIT ASSEMBLY: LESSONS FROM DEVELOPMENT
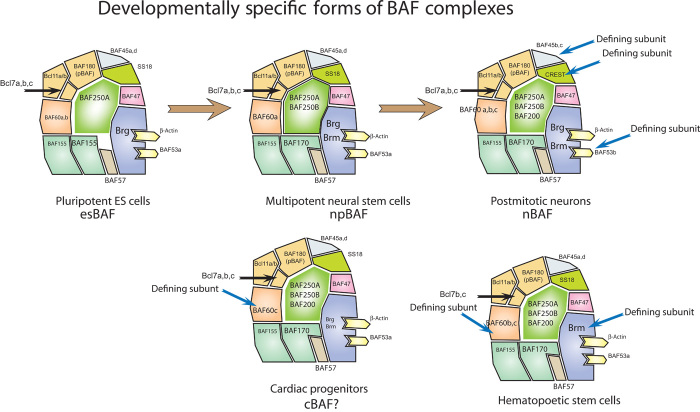
During the 1990s, as signaling pathways were being defined for a variety of specific cell surface receptors, the signaling community surprisingly found that only a few dozen ubiquitous signaling pathways seem to account for nearly all biologic and developmental responses (54). The community studying signal transduction expected specific pathways for specific receptors, and the general question of the biochemical basis of signaling specificity arose. Although still a matter of debate, much of the answer to this question now appears to be that genetic loci are made accessible for the binding of transcription factors at the terminus of a signaling pathway before or concurrently with the use of the signaling pathway. This result indicated that the nucleus and cell membrane somehow anticipated the next step in development. Almost certainly, the development of highly specific regions of the genome that are open to incoming signaling pathways is brought about through the actions of lineage-specifying transcription factors and specific cell membrane receptors, but other components must be present that are capable of reversing long-standing patterns of epigenetic repression. A biochemical search for proteins binding to accessible NFAT (nuclear factor of activated T cells) sites, which is the terminus of the ubiquitous Ca2+/calcineurin/NFAT signaling pathway, revealed that they bound a complex resembling yeast SWI/SNF, which had been discovered earlier in yeast. However, as the subunits were purified and cloned, it became clear that the mammalian complex (which we initially called the mSWI/SNF complex) was not as close to the yeast complex as we initially thought (4, 14–17, 19). The complexes were assembled combinatorially from the products of at least 29 genes encoded by 15 gene families (36, 48), again highlighting the immense combinatorial diversity and possibly functional diversity encoded within these complexes in much the same way that letters are assembled into words (48). In addition, some of the subunits were found to be tissue-specific, such as those found only in BAF complexes of mature neurons (Fig. 2). For example, BAF53b (an actin-like molecule), BAF45b [a double plant homeodomain (PHD) finger protein], and CREST (SS18L1) (a Ca2+-responsive regulator) subunits were found only in postmitotic neurons (19, 20, 55, 56). These subunits appeared during evolution as a more complex brain with greater diversity of neurons came onto the evolutionary scene (Fig. 1). Forcing the assembly of this neuron-specific nBAF complex in human fibroblasts can convert them to neurons (31, 38), and knocking out the neuron-specific nBAF subunits leads to profound defects in critical postmitotic neuronal function such as dendritic outgrowth and synaptogenesis (49, 57). Following these reports, exome sequencing studies of human neurologic diseases revealed that individuals with language acquisition deficiencies with or without mental retardation (Coffin-Siris syndrome and Nicolaides-Baraitser syndrome) generally have mutations in one of several subunits of the BAF complex (43, 44). These discoveries have led to a search for tissue-specific and/or instructive roles for other chromatin remodeling and histone modification complexes. These studies and the finding that minor changes in subunit composition could result in malignancy (see below) put forward the understanding that biologic specificity likely emerged from subunit composition as the means of diversification of function or dysfunction (36, 48). With the view that cancer is development gone wrong, BAF250B (ARID1B) (one of three genes that occupy this position in BAF complexes) was recently found by trio and quad sequencing (mother, father, normal sib, and affected sib) to be the most frequently mutated gene in human neurodevelopmental disorders (58). Here also, the (heterozygous) mutations are generally dominant, implying that a dosage-sensitive process also underlies the developmental roles as well as the role in oncogenesis.
Fig. 2. BAF complexes change their subunit composition during development.
Murine ES cells have a specific subunit composition (esBAF) that is apparently not found in other cell types to date. Overexpression of the esBAF subunits can facilitate induction of pluripotent cells from fibroblasts. In neural stem cells that line the developing neural tube, a second form of BAF complexes is found that again is distinguished, but not by subunits expressed only in neural progenitors, but rather an assembly that appears to be found only in neural progenitors. In postmitotic neurons, three subunits found only in neurons are present in complexes immediately after mitotic exit. These include BAF53b, BAF45b, and CREST. In cardiac progenitors, complexes are distinguished by the expression of BAF60c, whereas in hematopoietic stem cells, complexes contain Brm, but not Brg, and BAF60b and BAF60c, but not BAF60a, and Bcl7b and Bcl7c, but not Bcl7a.
DOMAIN SPECIFICITY: MECHANISMS OF LOCALIZATION OF ATP-DEPENDENT CHROMATIN REMODELERS OVER THE GENOME
Early studies indicated that SWI/SNF complexes are targeted to their sites of action by simply being recruited by a DNA binding protein, such as SWI5, which normally is located in the cytoplasm and moves into the nucleus upon treatment with mating factor (29). In this way, SWI5 is both a signaling protein and a transcription factor. From these studies, it was concluded that in yeast, the SWI5 transcription factor recruits the SWI/SNF complex to its site on the HO endonuclease gene. After recruitment, the complex stays in this location, whereas the transcription factor can move to another location. This mechanism provided an easy understanding of the origin of specificity of binding and is well suited to yeast, where most DNA is accessible and few genes are epigenetically suppressed.
In multicellular organisms, BAP or BAF complexes have enhanced abilities to bind histone modifications and hence might be guided by locus recognition, driven by specific histone modifications and regional architecture, rather than by simple sequence specificity or recruitment by transcription factors. The mammalian BAF (mSWI/SNF) and pBAF complexes collectively contain eight bromodomains (six on PBRM1, one on either BRG or BRM, and one on BRD7), two PHD finger proteins (BAF45 subunits), two chromodomains (BAF155 and BAF170), and between seven and nine DNA binding domains that can bind architectural features such as cruciform DNA structures, AT-rich sequences, or HMG recognition features (15–17, 19). Although a formal calculation of binding affinities has not been performed, the combined affinities would probably outweigh the binding energy provided by a transcription factor or even a group of them. An epigenetic locus recognition mechanism would provide a means by which complexes in multicellular organisms could be targeted to the loci with specific features determined by previous developmental events, and hence provide access to specific groups of genes bearing histone modifications that record their developmental history. In the future, it will be critical to test this hypothesis using a thoughtful combination of in vitro biophysical and in vivo methods.
PRE-GENOMICS: EARLY LINKS TO HUMAN CANCER
Early studies found that malignant cell lines often had lost critical subunits of mSWI/SNF or BAF complexes, indicating that these complexes were likely to be tumor suppressors. In addition, they were found to bind the RB protein and repressed E2F function (59, 60). Definitive evidence that they were tumor suppressors came from the work of Versteege and colleagues, who found that BAF47 (INI1, hSNF5, SMARCB1) was uniformly lost in a rare childhood cancer known as malignant rhabdoid tumor (MRT). In these tumors, most often, germline loss of one allele is followed by loss of the second allele in the tumor tissue (rhabdoid predisposition syndrome), indicating that mutations in BAF47 behave as classic tumor suppressors with genetics similar to that of retinoblastoma (61). Additional early studies by Reisman and colleagues (62) discovered cell lines with inactivation of both Brg and Brm, such as the SW13 line. These studies were confirmed by Wong and colleagues (63), who found that several breast cancer cell lines had lost both Brg and Brm at the protein level but seemed to have mutations in only one of the two homologous subunits. The observation that both Brg and Brm could be disrupted and yet cells remain viable and active was unexpected because several biochemical studies using in vitro transcription on nucleosomal templates indicated that these ATPases might be required for RNA polymerase binding, and conflicting reports in yeast indicated that they might or might not be subunits of RNA polymerase II. These early studies were highly limited because sequencing was labor-intensive. Thus, before the dawn of exome sequencing, the common view was that tumor suppression might require loss of function of both alleles of both of the homologous ATPases or loss of both alleles of subunits that were encoded by single genes, such as BAF47.
EXOME SEQUENCING STUDIES IN HUMAN CANCER REVEAL EXTENSIVE AND DIVERSE ROLES FOR CHROMATIN REMODELING
When exon sequencing studies of human cancer began in 2010, a more complicated genetic picture emerged. The remainder of this review focuses in large part on the nature of these mutations and their possible mechanisms. Initial reports showed that about 57% and 46% of ovarian clear cell carcinomas and endometriosis-associated ovarian carcinomas, respectively, contained inactivating mutations, distributed along the length of the BAF250A (ARID1A) subunit (45, 64). Nearly all of these cases had mutations in only a single allele, indicating that BAF250A might be a dominant tumor suppressor subunit within the complex. These initial studies were confirmed repeatedly, indicating that somehow the loss of a single allele of a single subunit contributed to the pathogenesis of ovarian cancer (65). About this same time, studies of an unusual intellectual disability condition (Coffin-Siris syndrome) characterized by difficulty with language acquisition and mental retardation found mutations in BAF250B (ARID1B), which were also heterozygous in most cases. Because the issue of tumor heterogeneity can often inaccurately skew the determination of whether one or both alleles are inactivated, the discovery of the heterozygous BAF250B mutations in patients with these neurologic conditions was conceptually informative, lending further grounds to consider heterozygous mutations as causative candidates and prompting studies to identify the mechanism of allelic dominance. Thus, it appears that a fundamental mechanism for the prevention of cancer and the development of the human nervous system relies on two functional copies of the BAF250A and B family, respectively. Because BAF250 is not necessary for in vitro chromatin remodeling, it appears that this critical, dosage-sensitive function is independent of the limited set of chromatin regulatory events that can be monitored by in vitro chromatin remodeling experiments on nucleosomal templates, strongly suggesting the need for new assays for the function of ATP-dependent chromatin regulatory complexes.
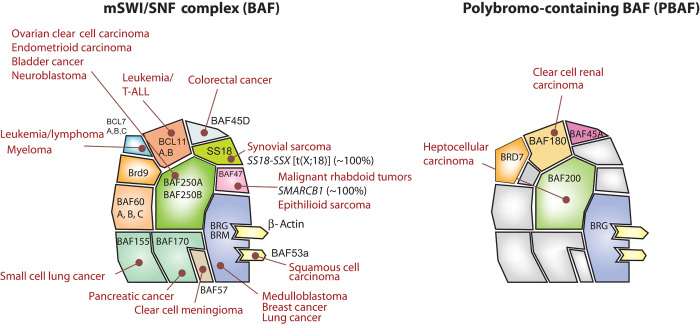
Additional exome sequencing studies revealed that specific cancers have mutations in different subunits of BAF complexes (Fig. 3). Many subunits do not have an elevated mutational rate in any tumor sequenced thus far and hence do not appear to contribute to tumor suppression. In some cases, this is because they are simply not expressed in dividing cells, as is true for the postmitotic, neuron-specific subunits, BAF53b, BAF45b, BAF45c, and CREST (SS18L1). However, in other cases, certain subunits that are ubiquitously expressed also appear to make no detectable genetic contribution to human cancer, but are physically located next to subunits that make extensive contributions. Indeed, this is the case with BAF53A, which is bound directly to BRG1. BRG1 is highly mutated in cancer, whereas BAF53A (ACTL6A) is not. However, the null phenotype for BAF53a is stronger than Brg, which might make it genetically less visible. Again, many of the subunits most highly mutated in cancer and human neurologic disease are not required for in vitro chromatin remodeling. The extensive genomic observations described above have placed a set of three constraints on the fundamental mechanism underlying tumor suppression by BAF complexes. The first is that the mechanism is dosage-sensitive, giving rise to genetic dominance. Second, the fundamental mechanism cannot be detected in an in vitro chromatin remodeling assay. Third, whereas this fundamental tumor-suppressive mechanism might be general, it manifests itself in a highly tumor-specific and apparently tissue-specific manner. For example, the ATPase BRG is mutated in more than 90% of small cell ovarian cancers (66, 67) but in less than 5% of small cell lung cancers. Thus, heterozygous mutations in BRG (SMARCA4) cause small cell ovarian cancer, but likely just bear passenger mutations related to dysfunction of genome maintenance mechanisms in certain other tumors in which they are less prevalent. These biologic observations are critical for designing approaches to understand the fundamental mechanism(s) of tumor suppression.
Fig. 3. BAF subunits are frequently mutated in human cancer.
Left: The subunit composition of the BAF complex is shown with the cancers containing frequent mutations in specific BAF subunits. The general pattern that emerges is that specific subunits protect against cancer in specific tissues. Right: The polybromo containing the pBAF complex with known specific subunits: BAF180 (polybromo PBRM1), BAF200 (ARID2), BAF45a (PHF10), and BRD7.
BAF COMPLEXES AS TUMOR SUPPRESSORS: KEY MECHANISTIC THEMES
One of the most well-studied mechanisms by which ATP-dependent chromatin remodeling can assume tumor suppressor functions comes from the rare childhood cancer MRT caused by biallelic inactivation of BAF47 (hSNF5, INI1) in virtually 100% of the tumors (61, 68). These tumors show a classic loss of heterozygosity in the tumor and hence are recessive tumor suppressors. Wilson and colleagues (69) have shown that the underlying mechanism is rooted in the ability of BAF complexes to regulate the placement and function of Polycomb repressive complexes. The conditional deletion of BAF47 in mice leads to T cell lymphomas with a short latency that is unprecedented for the deletion of a single gene (70). This short latency time recalls the fact that human rhabdoid sarcomas often occur in the first 2 years of life, an observation that indicates that the mouse model, even though yielding a different type of tumor, likely has a similar pathogenesis. BAF complexes in the mutant cells are unable to remove Polycomb complexes and their histone modification, H3K27Me3, from the Ink4a (Cdkn2a) locus, which normally suppresses proliferation (69). The genomes of MRTs are devoid of mutations outside of SMARCB1 loss and are estimated to have the lowest mutational burden of any human tumor sequenced to date (71). Thus, the tumors seem to be induced exclusively by changes in epigenetic regulation (except for the initiating mutations in BAF47). The mechanism by which loss of BAF47 leads to a failure to remove Polycomb is unclear and suggests that one of the important roles of BAF complexes is to oppose Polycomb as indicated from studies in flies and mammals (8, 11).
The advent of exome sequencing across a diverse range of human cancers has led to the realization that BAF complexes are one of the most significant tumor suppressors in humans, with a cumulative incidence of mutation of more than 20% of human cancers sequenced to date (Fig. 3). However, the pathogenesis of this much larger group of tumors is almost certainly different from that of MRTs. Most cancers bearing BAF subunit mutations are found in older age groups, as is cancer in general, and do not occur selectively in young children. In addition, many, if not most, of the cancers have a mutation in only one allele of the affected subunit, making them dominant tumor suppressors, rather than recessive tumor suppressors as in the case of MRTs. Thus, the mechanism underlying tumor suppression by BAF subunits must relate to a defect that demonstrates dosage sensitivity, providing a ripe foundation for the assessment of possible mechanisms. Mutations that produce neurologic disease are also genetically dominant, suggesting that a single fundamental process, albeit context-dependent, is at fault in both neurologic disease and cancer caused by heterozygous mutations in BAF subunits.
The most commonly affected BAF subunit in cancer to date is ARID1A (BAF250A), which has now been found to be mutated in a wide variety of tumors including endometrial carcinoma (65), colon and rectal cancers (72), pancreatic cancer (73), transitional cell carcinoma of the bladder (74), gastric cancer (75, 76), cholangiocarcinomas (77), and childhood neuroblastoma (78). BAF250a and BAF250b are dedicated subunits that appear to have no life outside of BAF complexes, which is the case with all other subunits except BAF53a and β-actin, which have a fraction as monomers. BAF250A mutations occur throughout the length of the gene, without clustering to any one domain, and most appear to be frameshift mutations predicted to lead to a loss of function or alternative splicing. Missense mutations are also present, but only a few are repeated and hence they are not particularly informative with respect to function. Chandler and colleagues (79) have created mice with a point mutation in the Arid domain and showed that the Arid domain in mice is critical for the function of the protein, yet mutations in the human gene are not especially localized to the Arid domain. Thus far, the Arid domain mutant mice do not have a higher incidence of cancer. Several frameshift mutations occur near the end of the protein, suggesting a critical function for the C terminus of the protein. The function of this subunit is unclear, but one report indicated that it could behave as a ubiquitin ligase (80). A murine mutation near the N terminus of BAF250a leads to alternative splicing and the production of a protein that induces hematopoietic proliferation in a non–cell-autonomous manner (81).
SMARCA4 (or BAF190, BRG1) is rather frequently mutated in cancer, and here, the clustering of the mutations is informative. Most of the missense mutations are scattered within the highly conserved ATPase domain and group in interesting ways. A highly penetrant ATPase domain mutation made many years ago, K785R (14), is present in several cancers and suggest that at least some of these could be functioning as dominant negative mutations. Similar mutations have been reported in the homologous Brm protein (SMARCA2); these mutations are less frequent in human cancer, yet far more common in neurologic disease. These human genetic observations therefore indicate that in Brg mutant cancers, targeting the synthetic lethal protein Brm (82) (identified in several studies; see “Paralogous subunit compensation as unique synthetic lethalities”) might lead to neurologic defects. In addition, the combined loss of Brg and Brm has been reported in many cancer cell lines, indicating that resistance might quickly develop when Brm is targeted in a Brg mutant.
Tumors with mutations in BAF57 (SMARCE1) provide an illustrative example of both the remarkable tissue specificity and domain specificity of the tumorigenic actions of BAF complexes. For most cancers, rates of mutation of BAF57 are extremely low (yet breast cancers have about a 10 to 14% incidence of amplification). However, in non–NF2-driven multiple spinal meningiomas, nearly 100% of the tumors have mutations in this gene (83). These tumors are more aggressive than normal meningiomas, which are localized and can be cured by surgical resection. Clear cell meningioma spreads through the spinal cord and central nervous system (CNS), giving rise to a lethal metastatic-like state. However, they rarely metastasize to other tissues. This subunit has the capacity to bind cruciform structures in DNA and might be a source of sequence-specific architectural DNA binding; however, it is not known if this occurs in vivo. The DNA binding domain is a nonspecific HMG domain, and introducing a point mutation into this domain gives rise to a mouse that suppresses the endogenous alleles and a phenotype very much like the loss of the Brg ATPase (84). In addition, mutations in SMARCE1 detected in patients with Coffin-Siris syndrome are also exclusive to the HMG domain (85).
BAF subunits less frequently mutated in cancer
BAF170 (SMARCC2) and BAF155 (SMARCC1) are mutated infrequently in cancers. BAF170 is mutated in gastric and colorectal cancers with microsatellite instability (86). The homologous subunit BAF155 is mutated in about 10% of small cell lung cancers (4). Both BAF155 and BAF170 are true homologs of yeast SWI3 (>20% identical over the entire length of the protein). These two members will dimerize in solution and appear to make hetero- or homodimers within the complexes in mammalian cells. Only BAF155 (and not BAF170) is expressed in pluripotent cells and is part of the specialized esBAF complex, which is essential for pluripotency in murine ES cells. The mutations seen in these proteins are scattered over the body of the gene and appear to generally inactivate the protein.
The BAF45 subunit is encoded by a family of four genes (BAF45a, b, c, and d), which have radically different expression patterns. For example, BAF45b is expressed only in postmitotic neurons. In general, these genes are relatively rarely mutated, but BAF45d is amplified in about 17% of breast cancers. As yet, it is not known if this is due to a carrier effect (that is, coamplification of a nearby essential oncogenic gene) or if its amplification could actually play a role in breast cancer progression. This family was discovered during the purification of BAF complexes from the developing CNS (19), and the remarkable neural specificity of BAF45b helped in understanding how combinatorial assembly could generate functionally distinct complexes (19). These proteins contain two PHD fingers of unclear specificity, which might help localize the complex to specific genetic loci, bearing specific histone marks. The BAF60 family (SMARCD1, 2, and 3) is also less frequently mutated in human cancers. SMARCD1 appears to be biallelically mutated in breast cancer, albeit less frequently (87).
BAF COMPLEXES AS ONCOGENES: THE SS18-SSX FUSION IN SYNOVIAL SARCOMA
As with all genetic discoveries that have poured out of laboratories in recent years—from thousands of genes associated with rare Mendelian disorders and common disease to the hundreds that have been implicated in human cancer—driver, causative function is often challenging to assign, especially in the setting of tumors with numerous mutations. However, in certain tumors, specific genes are mutated in 100% of the cancers in 100% of the cells, which provides definitive evidence that these mutations cause the development and maintenance of the tumor. An example is human synovial sarcoma (88, 89), which has provided ground for the discovery of a mechanism underpinning perturbation to the SS18 subunit of BAF complexes by the t(X;18) translocation hallmark to human synovial sarcoma. This mechanism has been highly informative for understanding the emergence of specificity of actions of mSWI/SNF or BAF complexes.
Human synovial sarcoma is a soft-tissue tumor in which 100% of tumors have a precise translocation involving the SS18 subunit of BAF complexes (90). The t(X;18) chromosomal translocation is the hallmark, diagnostic feature of the disease which results in the direct fusion of 78 amino acids of the C terminus of SSX to the SS18 C terminus. This well established driver, the SS18-SSX oncogenic fusion, overtakes complexes, outcompeting wild-type SS18 (from the one remaining normal allele) and causing the displacement of BAF47, likely an adjacent or nearby subunit in the complex (Fig. 4). The SS18-SSX containing complexes are then retargeted to oncogenic loci such as SOX2 and PAX6, activating these genes by displacement of PRC2 complexes and their H3K27me3 repressive marks. Both the biochemical changes to subunit composition in the complex and the targeting on chromatin can be reversed by removing the SS18-SSX fusion [that is, via short hairpin RNA (shRNA) or CRISPR] or altering the stoichiometric balance of SS18-SSX relative to wild-type SS18 (that is, via wild-type SS18 overexpression) assembling into complexes. It should be noted that this mechanism is quite distinct from that of rhabdoid sarcomas. Although both tumors lose BAF47 from the complex, the dominance of the SS18-SSX fusion in synovial sarcoma arises from the ability of the SS18-SSX fusion protein to target the BAF complex to specific oncogenic loci, inducing proliferation (36).
Fig. 4. BAF complexes can be oncogenes or tumor suppressors.
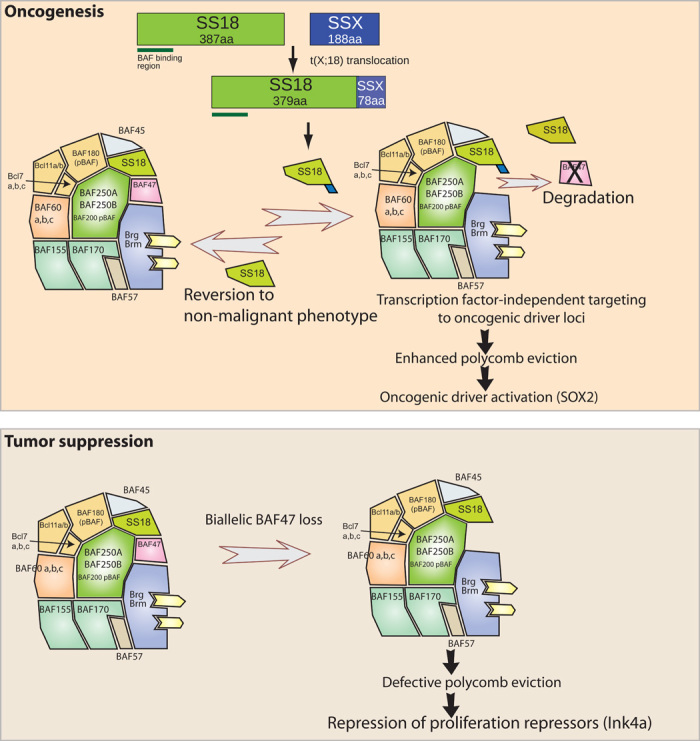
Top: The fusion of the SS18 gene to the SSX gene adds 78 amino acids (aa) of SSX to SS18, giving a fusion protein that evicts wild-type SS18 as well as BAF47 (hSNF5). The resulting oncogenic BAF complex is then targeted to new loci over the genome, such as Sox2, through a transcription factor–independent mechanism to genes that are drivers of proliferation. At these genes, it robustly evicts polycomb by unknown mechanisms leading to activation of genes such as Sox2 that can drive proliferation. Bottom: In the rare rhabdoid sarcoma of young children, the biallelic loss of BAF47 leads to a complex with defective ability to evict polycomb at loci such as Ink4a that repress proliferation. Cells are then transformed without additional mutations. Note that these mechanisms are brought about by a gain of ability to evict polycomb versus a loss of ability to evict polycomb. Both mechanisms are probably distinct from the more common cancers produced in older individuals with mutations in other subunits that compromise the ability to allow TopoII function, as shown in Fig. 6.
THE PBAF COMPLEX IN CANCER
The pBAF complex was discovered by biochemical purification of proteins required for ligand-activated transcription in vitro (91). The name pBAF came from the discovery that this complex contained polybromo (PBRM1 or BAF180) and BAF200 (Arid2), as well as Brg (but not Brm) and several other subunits of the BAF complex. The BAF180 protein contains six bromodomains that are similar to the single bromodomain found in Brg1. BAF180 is mutated or deleted in more than 50% of clear cell renal cell carcinoma (ccRCC) (92). Here again, the tumors do not occur in childhood, and to date, there are no specific pathologic features of this tumor subgroup. Although a number of alleles are predicted not to produce protein, many of the missense mutations occur in the bromodomains. It seems that the bromodomains are not redundant for the tumor suppressor function in that mutation of any single bromodomain in one allele is sufficient to contribute to cancer formation. We assume that the bromodomains bind to acetylated histones, and they do indeed do this in vitro, but their in vivo binding specificity has not yet been determined.
ccRCC is most often associated with a mutation in the VHL (von Hippel–Lindau) gene, which encodes an E3 ubiquitin ligase, either as a point mutation or as part of a large deletion of chromosomal arm 3p. This chromosomal arm contains BAF180, BAF155, the Set domain–containing protein 2 (SetD2, a H3K36 methyltransferase), and the BAP1 tumor suppressor, which is a deubiquitinase. Thus, a large number of tumor suppressors reside in this region. However, it appears that missense mutation of either VHL, BAP1, or BAF180 can contribute to ccRCC independently, suggesting an interesting interplay of these genes in the pathogenesis of this tumor. Mutations in PBRM1 correlate with mutations in VHL, the most commonly mutated gene in ccRCC, and have a tendency toward mutual exclusivity with BAP1 mutations (93). Somewhat surprisingly, the other subunits of the pBAF complex are not mutated with high frequency in ccRCC.
BAF180 (pBRM) and BAF200 (Arid2), which are present in the same pBAF complex, are both frequently mutated in liver cancers (94), and about 18% of hepatitis-associated hepatocellular carcinomas have mutations in BAF200 (94). In other exome sequencing studies, PBRM1, ARID2, and BAP1 were found to be inactivated in a large proportion of specific liver cancers [LCB (liver cancer displaying biliary phenotype)]. This finding suggests that they might work together cooperatively to contribute to the pathogenesis of this tumor (95).
POTENTIAL COOPERATIVE ONCOGENIC INTERACTIONS BETWEEN MUTATIONS IN BAF SUBUNITS AND OTHER GENES
The advent of exome sequencing made it possible to not only identify recessive tumor suppressors but also identify mutations in genes that tend to co-occur with them, pointing toward potential mechanisms and therapeutic approaches. When BAF250A (ARID1A) mutations were first identified in ovarian cancer, it was immediately clear that cooperating mutations in phosphatidylinositol 3-kinase (PI3K) were frequent. This trend became even more clear as additional mutations were found in Brg and other subunits. The association seems invariably to be an activating mutation in PI3K with a loss of function in one of the BAF subunits, most commonly BAF250a or BAF250b. This is intriguing for a number of reasons. First, activation of PI3K should deplete the supply of PIP2 (phosphatidylinositol 4,5-bisphosphate) by phosphorylation on the 3′ position of the inositol ring, giving rise to phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3]. PIP2 binds the BAF complex to one site on the Brg subunit, near the ATPase domain (96). As yet, the significance of this binding is unclear, but it may regulate the activity of BAF complexes and their association with chromatin (47), and hence, PI3K activation could further reduce the activity of the BAF complex. However, the two mutations could cooperate by a completely different mechanism. For example, if the activation of PI3K leads to increased cell division in a cell lacking a BAF subunit, one would expect the consequent defect due to failure of TopoII function (97) (see below) to be sensitized. Activated PI3K would drive more “at-risk” cell divisions, leading to more mutations and advancement along the pathway to cancer. Another possibility would be that loss of BAF function leads to more point mutations and a greater likelihood of activation of PI3K. These possibilities can at least in part be resolved by understanding the time that the mutations occur during the course to cancer using allele frequency calculations.
Recently, Chandler and colleagues (35) demonstrated that mice with one allele of BAF250a deleted do not get cancer, but when bred to a mouse with a mutation in PI3K, they develop ovarian clear cell cancer, establishing this as an excellent model for these cancers. They found that PI3K leads to the dissociation of BAF complexes from the interleukin-6 (IL-6) promoter, relieving its repression and leading to high levels of IL-6 production in the murine cancers. Treating the mice with PI3K inhibitors leads to reduction in tumor growth. IL-6 is also produced from human cancers and appears to contribute to their growth. The mechanism by which PI3K inactivates BAF on the IL-6 promoter is unclear.
Another cooperating mutation is the activation of β-catenin. This co-occurrence is common in medulloblastomas (98). β-catenin binds Brg and regulates its ability to activate genes (99). Hence, one could propose a transcriptional mechanism for this cooperation. On the other hand, as with activation of PI3K, activation of β-catenin would increase the number of cell divisions at risk for TopoII dysfunction during anaphase, thereby generating more mutations and moving the cell further along on the pathway to cancer. Future studies will be necessary to resolve these questions.
OTHER ATP-DEPENDENT CHROMATIN REMODELING COMPLEXES AND CANCER
The ATRX gene contains the SNF2/SWI2-related “helicase” domains and has ATP-dependent chromatin remodeling activity. ATRX ranks 17th on Davoli et al.’s list of tumor suppressors (100), taking into account the nature of the mutations as well as gene length, ratios of silent to nonsilent mutations, and other features likely to predict tumor suppressor activity. It is mutated in about 40% of low-grade gliomas, 20% of pancreatic cancers, 10% of small cell lung cancers, and 9% of uterine endometrial cancers. ATRX cooperates with DAXX to insert the histone variant H3.3 into chromatin and hence appears to function within a histone chaperon complex (101). Mutations in ATRX cause a syndrome characterized by anemia (actually α-thalassemia) and mental retardation, and are encoded on the X chromosome (102). However, since its discovery, it has been implicated in telomere function, DNA methylation, and sister chromatid cohesion and congression. The 280-kD protein includes an unusual N-terminal PHD designated the ATRX-DNMT3-DNMT3L (ADD) domain, owing to its similarity to a protein region found in the DNA methyltransferases (DNMTs). However, the relationship of this domain to its role in DNA methylation is unclear. Remarkably, in several tumors, notably glioma, it is mutated along with DAXX and histone H3.3, identifying a genetic pathway contributing to these tumors (103). Patients with somatic null mutations in ATRX do not have a predisposition to cancer (102), indicating that other genes must contribute to the pathogenesis of cancer in those cases. Almost certainly, these genes are DAXX and H3.3. ATRX is located in the heterochromatin, and null mutations in SUV39H1 and SUV39H2, which place the H3K9Me3 mark, prevent its localization to the heterochromatin (104). Although human genetics strongly implicate a pathway contributing to cancer, the role of defective H3.3 placement in cancer is still unclear.
Other ATP-dependent chromatin regulators also appear to function as tumor suppressors in humans. Of note is CHD1, which is mutated in about 8 to 10% of cases of stomach cancer and colorectal cancer and is 22nd on Davoli et al.’s list of tumor suppressors (100). By contrast, BAF250a is 4th, BAF180 is 6th, BAF200 is 15th, and BAF250b (ARID1B) is 43rd. CHD1 is located on human chromosome 5q21, which is frequently deleted in prostate cancer and is required for androgen receptor–driven gene expression and the androgen-dependent translocation of the ERG gene (105–107). The observation (from cBio portal) that it is mutated in only 1 to 2% of prostatic cancers suggests that other genes located within the deleted region might contribute to its role in tumor suppression. Furthermore, unlike SMARCA4 (BRG1 or BAF190), there are no recurrent missense mutations in the ATPase domain of CHD1 in cancer that would indicate that this particular function is critical. A knockdown was reported to interfere with murine ES cell formation and to be required for pluripotency, but this has not been confirmed yet with a null mutation.
The other chromatin regulator commonly mutated in cancer is CHD8, which ranks 90th among the genes contributing to tumor suppression in a recent analysis provided by Davoli and colleagues. CHD8 is mutated in about 10% of pancreatic, colorectal, and uterine cancers. In Drosophila, CHD8 is called Kismet, and its deletion leads to the accumulation of H3K27Me3 over the fly genome (108), much in the same way that deletion of BAP or BAF subunits leads to enhanced H3K27Me3 at many loci over the mammalian genome. Another possible mechanism by which CHD8 could contribute to cancer is through binding and inhibition of β-catenin in mammalian cells. CHD8-like Brg and Brm show repeated missense mutations within the ATPase domain, which are likely to have an effect on the protein. Notably, like BAF complexes, CHD8 is implicated in neurologic disease and is the most frequently mutated gene in autism spectrum disorders where loss-of-function mutations in one allele are most common.
In yeast, the ATP-dependent chromatin remodelers INO80 and SWR1 are major guardians of the genome (24) and prevent the accumulation of mutations, much as p53 does in mammalian cells. They have a critical role in double-strand break repair and also function in checkpoint pathways and nucleotide excision repair. Curiously, these complexes are similar to mammalian BAF complexes in that they contain β-actin and similar actin-related proteins (ARPs), which are most homologous to BAF53a and BAF53b. Thus, they may be structurally and functionally related to vertebrate BAF complexes. Although their importance in genome maintenance is clear, their mammalian homologs are not commonly mutated in human cancer. This may reflect the fact that BAF subunits are genetically dominant and dosage-sensitive and, hence, represent a significant organismic susceptibility. Genes that are frequently mutated in a disease are not necessarily more important, but rather they cause human disease and are found frequently in exome sequencing studies because of their genetic dominance and dosage sensitivity.
SPECIFIC MECHANISMS OF TUMOR SUPPRESSION AND THERAPEUTIC TARGETING
BAF-Polycomb antagonism
Early developmental studies in Drosophila foreshadowed the opposing, antagonistic functions of Trithorax (of which BAF is a member) and Polycomb complexes in the regulation of mammalian gene expression and, more recently, cancer. The antagonistic nature of BAF and Polycomb complexes has emerged as a likely culprit in causing extensive, misplaced repression (or in some cases, activation) genome-wide (Fig. 5). The clearest example of this came from studies by Roberts and colleagues, who discovered that BAF47-deficient MRTs displayed marked increases in the H3K27me3 repressive mark, a mark known to be placed only by PRC2 complexes. Given the important tumor-suppressive function of p16-INK4A, several groups honed in on this locus to reveal the extensive repression over this site in the setting of BAF47 (SNF5) loss (69, 109). From studies in ES cells, we know that BAF complexes oppose PRC complexes at most sites, with the exception of genes such as the Hox loci at which they appear to act synergistically to enable the placement of the H3K27Me3 mark (11). This is in marked contrast to Drosophila, where BAF opposes Polycomb at the Hox genes (8). As yet, it is unclear whether humans follow the strategy observed in Drosophila or mice. Hence, in MRTs, the loss of BAF47 results in altered balance of the activity of BAF and Polycomb complexes, skewing toward Polycomb placement of repressive marks (Fig. 5).
Fig. 5. Mutations in BAF complexes and polycomb complexes affect the balance between these two major genomic chromatin regulators.
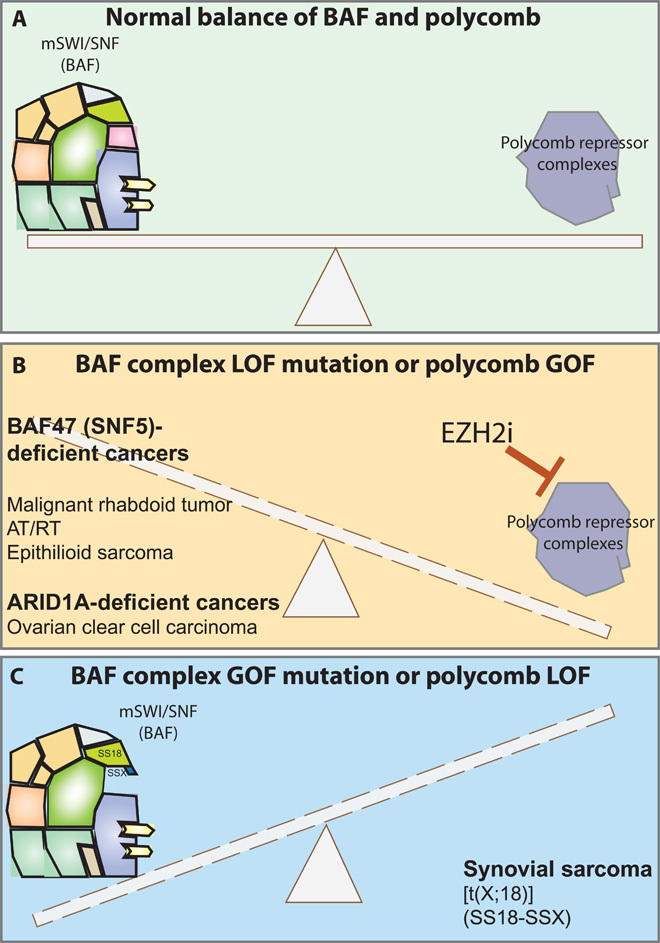
(A) In cells without defined mutations in genes encoding BAF complex subunits, BAF and polycomb complexes oppose one another to facilitate the coordinate regulation of gene expression. (B) Upon loss-of-function (LOF) mutations, such as biallelic inactivation of hSNF5 (BAF47) in MRTs, BAF complexes lose the ability to oppose polycomb, resulting in higher overall levels and repressive histone mark placement genome-wide. (C) In specific gain-of-function (GOF) settings, such as human synovial sarcomas that contain the SS18-SSX oncogenic fusion protein, BAF complexes appear to oppose polycomb complexes at key oncogenic loci.
In contrast, in human SS tumors, oncogenic, SS18-SSX fusion–bearing complexes appear to have gained the ability to oppose Polycomb complexes (Figs. 4 and 5), at least at genes critical for the proliferative maintenance of these tumors such as SOX2 (36). Although the specific mechanism of this unique gain-of-function phenotype still remains unclear, it is clear that the 78 amino acids confer an advantage to these complexes, which enhances the displacement of Polycomb. This oncogenic eviction of Polycomb can be reversed by stoichiometrically altering the balance of SS18-SSX versus wild-type SS18 within BAF complexes, thereby reversing the complexes to an induced wild-type state, bearing normal subunit composition. Hence, an exciting therapeutic opportunity emerges from these findings, which is distinct from the reverse direction of opposition seen in MRT complexes (36).
The unbalanced state produced by either loss-of-function BAF subunit mutations or gain-of-function PRC2 mutations might be an effective site for therapeutic intervention (Fig. 5). Several studies have indicated that EZH2 inhibition to a level that removes nearly all H3K27Me3 over the genome leads to death of cells having a BAF47 mutation (110, 111). Although one would anticipate substantial toxicity associated with removal of PRC2 function, mice appeared to tolerate the treatment, and their tumors underwent regression with EZH2 inhibition. Also, a diffuse large cell lymphoma with activating mutations in Ezh2 (Y641F/N) appears to be sensitive to Ezh2 inhibition (112).
Given that therapies based on polycomb repression are already being advanced as possible treatments of several BAF mutation–driven cancers, it has become important to understand the underlying mechanism of opposition between these complexes. Studies using in vitro transcription on nucleosomal templates are insufficient because they do not reproduce the domains of polycomb-mediated repression found over genes such as Sox2 in most somatic cell types. In addition, these in vitro techniques cannot detect the effects of long-range interactions, tissue-specific epigenetic changes, occurrence of histone H1, and a wide variety of other critical aspects of chromatin regulation. Hence, a major goal for the future will be to generate systems in which one lets the cell assemble chromatin with all its complexity and then challenges this locus with specific chromatin modifiers. This objective led us to develop a system for studying the complex chromatin regulation in different cell types using a mouse with an indicator and an array of regulatory sites knocked into the Oct4 gene (113). This system allows one to study the dynamic behavior of the locus, the rate of heterochromatin propagation, the dissolution and assembly of heterochromatin, and the in vivo synergy with other chromatin modifiers (114) in any tissue of the developing or adult animal. Understanding the dynamics and mechanisms underlying BAF-polycomb opposition might also be fruitfully approached with this system (Kadoch et al., unpublished results).
Synergy between TopoII and BAF complexes
Recent studies have indicated that BAF complexes help TopoII untangle DNA during replication, perhaps providing an explanation for BAF’s tumor-suppressive functions (Fig. 6). At each cell division, hundreds or even thousands of catenes must be relieved by passing one strand of DNA through another (115). This risky task is carried out by TopoIIα and TopoIIβ by first cleaving one strand with a 4–base pair overlap and then forming a covalent bond between the enzyme and DNA. Because the enzyme is a dimer, it can hold a first strand in place while creating a channel for the passage of the second strand. Afterward, the two ends are ligated together by TopoII, relieving the tangle and allowing cell division to proceed (116, 117). Deletion or shRNA depletion of the oncogenic BAF subunits leads to the formation of anaphase bridges, indicating a failure to untangle DNA at anaphase, whereas deletion of non-oncogenic subunits did not have this effect (97). The deletion of oncogenic BAF subunits surprisingly leads to arrest at G2/M with high levels of histone H3 S-10 phosphorylation, an indicator of incomplete mitosis. The deletion of the oncogenic subunits gives rise to the decatenation checkpoint, which arrests cells until the tangled DNA can be decatenated. Consistent with this, the arrest can be overcome by a mutation in TopoIIa, which prevents its phosphorylation and the sensing mechanism that tells the cell that its DNA is tangled. BAF complexes bind robustly to endogenous TopoII, and peptides from TopoII have been recovered in each of five different proteomic analysis of endogenous, untagged BAF complexes from different tissues. The role of BAF seems to allow TopoII to bind to DNA. TopoII’s interaction with DNA is so transient that it is difficult to detect by ordinary Chip-seq. However, by arresting the progression of the mechanism of TopoII using etoposide, it was possible to capture the enzyme covalently attached to DNA before the religation step could be completed. Using this approach, the Brg (Smarca4) ATPase was found to be essential for the binding of TopoII to 11,000 of 16,000 sites over the genome (97). TopoII was found to bind to the BAF250a subunit, which is the one most frequently mutated in human cancer.
Fig. 6. Mechanism of synergy between BAF and TopoII and its loss in common cancers bearing BAF subunit mutations.
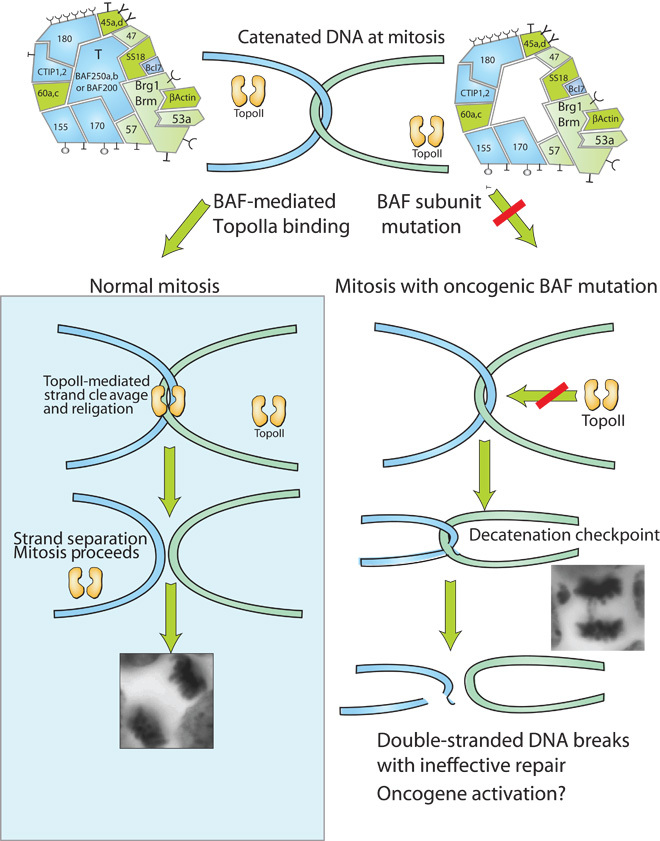
On the left is the normal mechanism of TopoII function. On the right is the mechanism when an oncogenic BAF subunit is mutated. BAF is necessary for binding of TopoIIa to DNA at 11,000 of 16,000 sites over the genome. When TopoII fails, it leads to the inability to resolve tangled DNA and the production of anaphase bridges, as shown in the inset. The mechanism by which DNA is repaired after possibly being cleaved in the cytoplasm by cytoplasmic DNase is unclear, but might be error-prone and lead to an accumulation of mutations.
These studies suggest that mutations in BAF subunits lead to the inability to resolve tangles at anaphase, with subsequent strand breakage and attempted repair of these breaks in the cytoplasm of the separating cells. This could generate potential oncogenic driving mutations that would contribute to the pathogenesis of the cancer. As mentioned above, this potential mechanism of tumor suppression must be examined in the light of the results of exome sequencing studies. First, the effect of BAF deletion on TopoII must be shown to be genetically dominant. Thus, we would expect that loss of a single allele of an oncogenic BAF subunit would lead to impairment of TopoII function. Second, the ultimate effect of TopoII dysfunction would be expected to result in tissue-specific lesions in DNA. This could happen if TopoII used different cleavage sites in different tissues, as would be consistent with the highly tissue-specific nature of deoxyribonuclease (DNase) sensitivity over the genome (118). Finally, we would expect that the effect of TopoII would not be seen in an in vitro transcription reaction using nucleosomal templates. Indeed, this is the case because the minimal set of BAF subunits necessary for in vitro transcription does not include TopoII, and the subunit to which TopoII binds, BAF250, is not necessary for transcription on nucleosomal templates. In yeast, TopoII function was shown to require nucleosome-free DNA, and the loss of the snf5 gene interfered with the binding of TopoIIa (119).
Because BAF subunit mutations prevent TopoII from contacting DNA, these studies predict that cancers with BAF subunit mutations should be resistant to TopoII inhibitors. This has not been shown to date, but could be helpful in guiding the use of these highly toxic inhibitors.
In other studies, the presence of a Brg (Smarca4) mutation has been shown to predict sensitivity to a combination of EZH2 and TopoIIa inhibition in a group of small cell lung cancers (120). The underlying mechanism is unclear, but it is possible that EZH2 inhibition blocks Polycomb placement, allowing the binding of TopoIIa to DNA and, hence, sensitivity to these inhibitors in the face of a BAF subunit mutation. However, the speculation that polycomb and its repressive marks block the binding of TopoIIa has not been tested to date.
Paralogous subunit compensation as unique synthetic lethalities
Another potential therapeutic approach arises from the combinatorial assembly of these complexes, specifically, at the mutually exclusive ATPase position, containing either Brg or Brm (16, 17, 19, 48). For example, loss or mutation of the Brg ATPase appears to make tumors containing these mutations highly susceptible to loss of the alternative ATPase, Brm (82). In fact, in the largest shRNA-based screen (Achilles, Broad Institute), this was the most highly ranked synthetic lethal relationship in human cancer. Similarly, in an shRNA-based screen for genes that were synthetically lethal with a mutation in BAF250a, the loss of BAF250b was detected (120). These studies indicate that when one subunit is mutated, the tumor becomes dependent on the other subunit that can occupy this position in the complex. Developmentally, BAF250a is not redundant with BAF250b and Brg is not redundant with Brm. For example, BAF250b is the most commonly mutated gene discovered in a recent exome sequencing study of human developmental diseases (58), and Brm is mutated in and causes a number of human neurodevelopmental diseases (43). Thus, therapeutic targeting of Brm or BAF250b in human cancer may have inherent toxicity. In addition, many tumors show genetic inactivation of one subunit and epigenetic inactivation of the homolog. For example, SW13 and several other cell lines contain a mutation in Brg, but do not express Brm, presumably by epigenetic inactivation (59, 62). In addition, a number of tumors have lost both BAF250a and BAF250b. Again, the mechanism appears to be genetic inactivation of one subunit and epigenetic suppression of the homologous subunit. It is not clear what allows the cell to compensate for the loss of both Brg and Brm or BAF250a and BAF250b, but this mechanism might be an additional susceptibility that would provide more lasting therapeutic effect than inhibition of one paralogous subunit but not the other.
Modulation of chromatin remodeling complexes by phosphoinositols
Natural small-molecule control of chromatin remodelers would be another possible avenue of therapeutic intervention. In mammals, PIP2 levels were found to modulate chromatin association of BAF complexes (47), and later in yeast, inositol hexakisphosphate (IP6) and inositol polyphosphate were shown to modulate the activity of the ATP-dependent chromatin remodeling complexes INO80 and SWR1 (23, 121), which as mentioned above are structurally similar to BAF in mammals. PIP2 binds to a single site in the BAF complex near the ATPase domain of Brg (96). However, to date, the function of PIP2 binding has not been clear and could be a biochemical artifact. Because PIP2 is membrane-associated, it would indicate that modulation by PIP2 would have to occur near a membrane such as the nuclear membrane or the more elusive “nuclear matrix,” a poorly characterized nuclear fraction. Activation of cell signaling pathways leads to the movement of BAF into the nuclear matrix fraction (47), but again there has been no functional characterization of the consequences of this localization. The realization of the importance of BAF complexes in cancer gave new impetus to discover and understand pathways modulating the activity of these complexes, particularly the importance of activated PI3K as a cooperating oncogene.
PATHWAYS FOR THERAPEUTIC ADVANCES ARISING FROM NEW MECHANISTIC UNDERSTANDING
Perhaps the clearest example to date for which the understanding of mechanism has opened a path for therapeutic development has been in synovial sarcoma (36). Here, the driving, oncogenic mutation is always the same and always results in the addition of exactly 78 amino acids of SSX to the SS18 gene at precisely the same place (88, 90). The fusion protein is incorporated into complexes and then dramatically retargets the BAF complex to oncogenic drivers. Clearly, the cancer-specific SS18-SSX fusion protein is an excellent target for small-molecule therapeutics in much the same way that Bcr-ABL fusions are in chronic myeloid leukemia (122). One would predict that a molecule that would prevent the dominant entry of the SS18-SSX fusion into the BAF complex would prevent tumor growth. This speculation is supported by the fact that overexpressing the wild-type SS18 protein in synovial sarcoma cells leads to reassembly of the normal complex, arrest of cell proliferation, and cell death (36). The precise amino acids that must be targeted are predicted from the sequences of SSX genes that are permissive to tumor development versus those that are not. Thus, a specific path to a cancer-specific drug has been revealed by the combination of human genetics and biochemistry.
In rhabdoid tumors, the definitive mutation is a loss of function of both alleles of BAF47 (SMARCB1, INI1, hSNF5) (61). Because there is no specific fusion to target or gained interaction identified to date, a cancer-specific treatment seems less likely unless the loss of BAF47 produces specific allosteric effects over the complex that would render it uniquely susceptible to small molecules. However, Roberts and colleagues have shown that these cancers have essentially only the initiating BAF47 loss, which suggests that targeting MRT-specific BAF complexes in this setting might be sufficient to initiate tumor regression. The discovery that nearly all of the effects of BAF47 loss could be explained by accumulation of polycomb and its products over the Ink4a locus indicated that polycomb inhibitors may be effective in these cancers. Although not specific for rhabdoid cancers, EZH2 inhibitors could hold hope for this essentially untreatable disease and are already being clinically exploited.
FUTURE CONSIDERATIONS
The discovery of the extensive role of ATP-dependent chromatin remodeling in cancer has highlighted several important goals for the future. The precise mechanism of tumor suppression used by these complexes must be understood if therapeutic advances are to be made. This fundamental mechanism is characterized by dosage sensitivity, tumor specificity, and the fact that this mechanism cannot be detected using present in vitro assays. These mechanisms are likely to be shared with those used in neural development. Hence, a major goal for the future is to develop new assays that faithfully predict the oncogenic functions of the complexes. The discovery that BAF complexes can also be oncogenes and definitive drivers of cancer has led to the realization of a true cancer-specific avenue toward therapeutic development for synovial sarcoma. The biochemical pathways that are likely to modulate BAF complex function must be understood as potential susceptibility factors for cancer. Finally, the possibility of inhibition of homologous subunits may prove a fruitful approach.
REFERENCES AND NOTES
- 1.Dawson M. A., Kouzarides T., Cancer epigenetics: From mechanism to therapy. Cell 150, 12–27 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Morgan M. A., Shilatifard A., Chromatin signatures of cancer. Genes Dev. 29, 238–249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shain A. H., Pollack J. R., The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLOS One 8, e55119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kadoch C., Hargreaves D. C., Hodges C., Elias L., Ho L., Ranish J., Crabtree G. R., Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592–601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stern M., Jensen R., Herskowitz I., Five SWI genes are required for expression of the HO gene in yeast. J. Mol. Biol 178, 853–868 (1984). [DOI] [PubMed] [Google Scholar]
- 6.Neigeborn L., Carlson M., Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108, 845–858 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson C. L., Herskowitz I., Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 68, 573–583 (1992). [DOI] [PubMed] [Google Scholar]
- 8.Tamkun J. W. Deuring R., Scott M. P., Kissinger M., Pattatucci A. M., Kaufman T. C., Kennison J. A., brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68, 561 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Kennison J. A., Tamkun J. W., Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 85, 8136–8140 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papoulas O., Daubresse G., Armstrong J. A., Jin J., Scott M. P., Tamkun J. W., The HMG-domain protein BAP111 is important for the function of the BRM chromatin-remodeling complex in vivo. Proc. Natl. Acad. Sci. U.S.A. 98, 5728–5733 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho L., Miller E. L., Ronan J. L., Ho W. Q., Jothi R., Crabtree G. R., esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 13, 903–913 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller T., Krogan N. J., Dover J., Erdjument-Bromage H., Tempst P., Johnston M., Greenblatt J. F., Shilatifard A., COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. U.S.A. 98, 12902–12907 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumesic P. A., Homer C. M., Moresco J. J., Pack L. R., Shanle E. K., Coyle S. M., Strahl B. D., Fujimori D. G., Yates J. R. III, Madhani H. D., Product binding enforces the genomic specificity of a yeast polycomb repressive complex. Cell 160, 204–218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khavari P. A., Peterson C. L., Tamkun J. W., Mendel D. B., Crabtree G. R., BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366, 170–174 (1993). [DOI] [PubMed] [Google Scholar]
- 15.Wang W., Chi T., Xue Y., Zhou S., Kuo A., Crabtree G. R., Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl. Acad. Sci. U.S.A. 95, 492–498 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W., Côté J., Xue Y., Zhou S., Khavari P. A., Biggar S. R., Muchardt C., Kalpana G. V., Goff S. P., Yaniv M., Workman J. L., Crabtree G. R., Purification and biochemical heterogeneity of the mammalian SWI–SNF complex. Embo J 15, 5370–5382 (1996). [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W., Xue Y., Zhou S., Kuo A., Cairns B. R., Crabtree G. R., Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10, 2117–2130 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Muchardt C., Yaniv M., A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12, 4279–4290 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lessard J., Wu J. I., Ranish J. A., Wan M., Winslow M. M., Staahl B. T., Wu H., Aebersold R., Graef I. A., Crabtree G. R., An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55, 201–215 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staahl B. T., Tang J., Wu W., Sun A., Gitler A. D., Yoo A. S., Crabtree G. R., Kinetic analysis of npBAF to nBAF switching reveals exchange of SS18 with CREST and integration with neural developmental pathways. J. Neurosci. 33, 10348–10361 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cairns B. R., Lorch Y., Li Y., Zhang M., Lacomis L., Erdjument-Bromage H., Tempst P., Du J., Laurent B., Kornberg R. D., RSC, an essential, abundant chromatin-remodeling complex. Cell 87, 1249–1260 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Shen X., Mizuguchi G., Hamiche A., Wu C., A chromatin remodelling complex involved in transcription and DNA processing. Nature 406, 541–544 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Shen X., Xiao H., Ranallo R., Wu W. H., Wu C., Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science 299, 112–114 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Gerhold C. B., Hauer M. H., Gasser S. M., INO80-C and SWR-C: Guardians of the Genome. J. Mol. Biol. 427, 637–651 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Kwon H., Imbalzano A. N., Khavari P. A., Kingston R. E., Green M. R., Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370, 477–481 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Imbalzano A. N., Kwon H., Green M. R., Kingston R. E., Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370, 481–485 (1994). [DOI] [PubMed] [Google Scholar]
- 27.Clapier C. R., Cairns B. R., The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Narlikar G. J., Sundaramoorthy R., Owen-Hughes T., Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154, 490–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosma M. P., Tanaka T., Nasmyth K., Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle– and developmentally regulated promoter. Cell 97, 299–311 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Bao X., Tang J., Lopez-Pajares V., Tao S., Qu K., Crabtree G. R., Khavari P. A., ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell 12, 193–203 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoo A. S., Staahl B. T., Chen L., Crabtree G. R., MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642–646 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho L., Ronan J. L., Wu J., Staahl B. T., Chen L., Kuo A., Lessard J., Nesvizhskii A. I., Ranish J., Crabtree G. R., An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U.S.A. 106, 5181–5186 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krasteva V., Buscarlet M., Diaz-Tellez A., Bernard M. A., Crabtree G. R., Lessard J. A., The BAF53a subunit of SWI/SNF-like BAF complexes is essential for hemopoietic stem cell function. Blood 120, 4720–4732 (2012). [DOI] [PubMed] [Google Scholar]
- 34.de Bruijn D. R., Peters W. J., Chuva de Sousa Lopes S. M., van Dijk A. H., Willemse M. P., Pfundt R., de Boer P., Geurts van Kessel A., Targeted disruption of the synovial sarcoma-associated SS18 gene causes early embryonic lethality and affects PPARBP expression. Hum. Mol. Genet. 15, 2936–2944 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Chandler R. L., Damrauer J. S., Raab J. R., Schisler J. C., Wilkerson M. D., Didion J. P., Starmer J., Serber D., Yee D., Xiong J., Darr D. B., Pardo-Manuel de Villena F., Kim W. Y., Magnuson T., Coexistent ARID1A–PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nat. Commun. 6, 6118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadoch C., Crabtree G. R., Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 153, 71–85 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singhal N., Graumann J., Wu G., Araúzo-Bravo M. J., Han D. W., Greber B., Gentile L., Mann M., Schöler H. R., Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 141, 943–955 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Yoo A. S., Sun A. X., Li L., Shcheglovitov A., Portmann T., Li Y., Lee-Messer C., Dolmetsch R. E., Tsien R. W., Crabtree G. R., MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tea J. S., Luo L., The chromatin remodeling factor Bap55 functions through the TIP60 complex to regulate olfactory projection neuron dendrite targeting. Neural Dev. 6, 5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lickert H., Takeuchi J. K., Von B. I., Walls J. R., McAuliffe F., Adamson S. L., Henkelman R. M., Wrana J. L., Rossant J., Bruneau B. G., Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432, 107–112 (2004). [DOI] [PubMed] [Google Scholar]
- 41.de la Serna I. L., Carlson K. A., Imbalzano A. N., Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27, 187–190 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Sawa H., Kouike H., Okano H., Components of the SWI/SNF complex are required for asymmetric cell division in C. elegans. Mol. Cell 6, 617–624 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Santen G. W., Aten E., Sun Y., Almomani R., Gilissen C., Nielsen M., Kant S. G., Snoeck I. N., Peeters E. A., Hilhorst-Hofstee Y., Wessels M. W., den Hollander N. S., Ruivenkamp C. A., van Ommen G. J., Breuning M. H., den Dunnen J. T., van Haeringen A., Kriek M., Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat. Genet. 44, 379–380 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Tsurusaki Y., Okamoto N., Ohashi H., Kosho T., Imai Y., Hibi-Ko Y., Kaname T., Naritomi K., Kawame H., Wakui K., Fukushima Y., Homma T., Kato M., Hiraki Y., Yamagata T., Yano S., Mizuno S., Sakazume S., Ishii T., Nagai T., Shiina M., Ogata K., Ohta T., Niikawa N., Miyatake S., Okada I., Mizuguchi T., Doi H., Saitsu H., Miyake N., Matsumoto N., Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat. Genet. 44, 376–378 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Jones S., Wang T. L., Shih I. M., Mao T. L., Nakayama K., Roden R., Glas R., Slamon D., Diaz L. A. Jr, Vogelstein B., Kinzler K. W., Velculescu V. E., Papadopoulos N., Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petermann M., Pavlovec A., Dissociation of rat liver ribosomes to active subunits by urea. Biochemistry 10, 2770–2775 (1971). [DOI] [PubMed] [Google Scholar]
- 47.Zhao K., Wang W., Rando O. J., Xue Y., Swiderek K., Kuo A., Crabtree G. R., Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95, 625–636 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Wu J. I., Lessard J., Crabtree G. R., Understanding the words of chromatin regulation. Cell 136, 200–206 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu J. I., Lessard J., Olave I. A., Qiu Z., Ghosh A., Graef I. A., Crabtree G. R., Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56, 94–108 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Ho L., Jothi R., Ronan J. L., Cui K., Zhao K., Crabtree G. R., An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. U.S.A. 106, 5187–5191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Euskirchen G. M., Auerbach R. K., Davidov E., Gianoulis T. A., Zhong G., Rozowsky J., Bhardwaj N., Gerstein M. B., Snyder M., Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLOS Genet. 7, e1002008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A., Wysocka J., A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z., Zang C., Cui K., Schones D. E., Barski A., Peng W., Zhao K., Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crabtree G. R., Generic signals and specific outcomes: Signaling through Ca2+, calcineurin, and NF-AT. Cell 96, 611–614 (1999). [DOI] [PubMed] [Google Scholar]
- 55.Olave I., Wang W., Xue Y., Kuo A., Crabtree G. R., Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 16, 2509–2517 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aizawa H., Hu S. C., Bobb K., Balakrishnan K., Ince G., Gurevich I., Cowan M., Ghosh A., Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science 303, 197–202 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Vogel-Ciernia A., Matheos D. P., Barrett R. M., Kramár E. A., Azzawi S., Chen Y., Magnan C. N., Zeller M., Sylvain A., Haettig J., Jia Y., Tran A., Dang R., Post R. J., Chabrier M., Babayan A. H., Wu J. I., Crabtree G. R., Baldi P., Baram T. Z., Lynch G., Wood M. A., The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat. Neurosci. 16, 552–561 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Developmentalstudgroup , Large-scale discovery of novel genetic causes of developmental disorders. Nature 519, 223–228 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunaief J. L., Strober B. E., Guha S., Khavari P. A., Alin K., Luban J., Begemann M., Crabtree G. R., Goff S. P., The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79, 119–130 (1994). [DOI] [PubMed] [Google Scholar]
- 60.Trouche D., Le Chalony C., Muchardt C., Yaniv M., Kouzarides T., RB and hbrm cooperate to repress the activation functions of E2F1. Proc. Natl. Acad. Sci. U.S.A. 94, 11268–11273 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Versteege I., Sévenet N., Lange J., Rousseau-Merck M. F., Ambros P., Handgretinger R., Aurias A., Delattre O., Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394, 203–206 (1998). [DOI] [PubMed] [Google Scholar]
- 62.Reisman D. N., Strobeck M. W., Betz B. L., Sciariotta J., Funkhouser W. Jr, Murchardt C., Yaniv M., Sherman L. S., Knudsen E. S., Weissman B. E., Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: Differential effects on RB-mediated growth arrest versus CD44 expression. Oncogene 21, 1196–1207 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Wong A. K., Shanahan F., Chen Y., Lian L., Ha P., Hendricks K., Ghaffari S., Iliev D., Penn B., Woodland A. M., Smith R., Salada G., Carillo A., Laity K., Gupte J., Swedlund B., Tavtigian S. V., Teng D. H., Lees E., BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 60, 6171–6177 (2000). [PubMed] [Google Scholar]
- 64.Wiegand K. C., Shah S. P., Al-Agha O. M., Zhao Y., Tse K., Zeng T., Senz J., McConechy M. K., Anglesio M. S., Kalloger S. E., Yang W., Heravi-Moussavi A., Giuliany R., Chow C., Fee J., Zayed A., Prentice L., Melnyk N., Turashvili G., Delaney A. D., Madore J., Yip S., McPherson A. W., Ha G., Bell L., Fereday S., Tam A., Galletta L., Tonin P. N., Provencher D., Miller D., Jones S. J., Moore R. A., Morin G. B., Oloumi A., Boyd N., Aparicio S. A., Shih I. M., Mes-Masson A. M., Bowtell D. D., Hirst M., Gilks B., Marra M. A., Huntsman D. G., ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 363, 1532–1543 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cancer Genome Atlas Research Network Kandoth C., Schultz N., Cherniack A. D.,Akbani R., Liu Y., Shen H., Robertson A. G., Pashtan I., Shen R., Benz C. C., Yau C., Laird P. W., Ding L., Zhang W., Mills G. B., Kucherlapati R., Mardis E. R., Levine D. A., Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Witkowski L., Carrot-Zhang J., Albrecht S., Fahiminiya S., Hamel N., Tomiak E., Grynspan D., Saloustros E., Nadaf J., Rivera B., Gilpin C., Castellsagué E., Silva-Smith R., Plourde F., Wu M., Saskin A., Arseneault M., Karabakhtsian R. G., Reilly E. A., Ueland F. R., Margiolaki A., Pavlakis K., Castellino S. M., Lamovec J., Mackay H. J., Roth L. M., Ulbright T. M., Bender T. A., Georgoulias V., Longy M., Berchuck A., Tischkowitz M., Nagel I., Siebert R., Stewart C. J., Arseneau J., McCluggage W. G., Clarke B. A., Riazalhosseini Y., Hasselblatt M., Majewski J., Foulkes W. D., Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat. Genet. 46, 438–443 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Ramos P., Karnezis A. N., Craig D. W., Sekulic A., Russell M. L., Hendricks W. P., Corneveaux J. J., Barrett M. T., Shumansky K., Yang Y., Shah S. P., Prentice L. M., Marra M. A., Kiefer J., Zismann V. L., McEachron T. A., Salhia B., Prat J., D’Angelo E., Clarke B. A., Pressey J. G., Farley J. H., Anthony S. P., Roden R. B., Cunliffe H. E., Huntsman D. G., Trent J. M., Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat. Genet. 46, 427–429 (M2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roberts C. W., Galusha S. A., McMenamin M. E., Fletcher C. D., Orkin S. H., Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc. Natl. Acad. Sci. U.S.A. 97, 13796–13800 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson B. G., Wang X., Shen X., McKenna E. S., Lemieux M. E., Cho Y. J., Koellhoffer E. C., Pomeroy S. L., Orkin S. H., Roberts C. W., Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18, 316–328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X., Werneck M. B., Wilson B. G., Kim H. J., Kluk M. J., Thom C. S., Wischhusen J. W., Evans J. A., Jesneck J. L., Nguyen P., Sansam C. G., Cantor H., Roberts C. W., TCR-dependent transformation of mature memory phenotype T cells in mice. J. Clin. Invest. 121, 3834–3845 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lawrence M. S., Stojanov P., Mermel C. H., Robinson J. T., Garraway L. A., Golub T. R., Meyerson M., Gabriel S. B., Lander E. S., Getz G., Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cancer Genome Atlas Network , Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Biankin A. V., Waddell N., Kassahn K. S., Gingras M. C., Muthuswamy L. B., Johns A. L., Miller D. K., Wilson P. J., Patch A. M., Wu J., Chang D. K., Cowley M. J., Gardiner B. B., Song S., Harliwong I., Idrisoglu S., Nourse C., Nourbakhsh E., Manning S., Wani S., Gongora M., Pajic M., Scarlett C. J., Gill A. J., Pinho A. V., Rooman I., Anderson M., Holmes O., Leonard C., Taylor D., Wood S., Xu Q., Nones K., Fink J. L., Christ A., Bruxner T., Cloonan N., Kolle G., Newell F., Pinese M., Mead R. S., Humphris J. L., Kaplan W., Jones M. D., Colvin E. K., Nagrial A. M., Humphrey E. S., Chou A., Chin V. T., Chantrill L. A., Mawson A., Samra J. S., Kench J. G., Lovell J. A., Daly R. J., Merrett N. D., Toon C., Epari K., Nguyen N. Q., Barbour A., Zeps N. Australian Pancreatic Cancer Genome Initiative Kakkar N., Zhao F., Wu Y. Q., Wang M., Muzny D. M., Fisher W. E., Brunicardi F. C., Hodges S. E., Reid J. G., Drummond J., Chang K., Han Y., Lewis L. R., Dinh H., Buhay C. J., Beck T., Timms L., Sam M., Begley K., Brown A., Pai D., Panchal A., Buchner N., De Borja R., Denroche R. E., Yung C. K., Serra S., Onetto N., Mukhopadhyay D., Tsao M. S., Shaw P. A., Petersen G. M., Gallinger S., Hruban R. H., Maitra A., Iacobuzio-Donahue C. A., Schulick R. D., Wolfgang C. L., Morgan R. A., Lawlor R. T., Capelli P., Corbo V., Scardoni M., Tortora G., Tempero M. A., Mann K. M., Jenkins N. A., Perez-Mancera P. A., Adams D. J., Largaespada D. A., Wessels L. F., Rust A. G., Stein L. D., Tuveson D. A., Copeland N. G., Musgrove E. A., Scarpa A., Eshleman J. R., Hudson T. J., Sutherland R. L., Wheeler D. A., Pearson J. V., McPherson J. D., Gibbs R. A., Grimmond S. M., Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gui Y., Guo G., Huang Y., Hu X., Tang A., Gao S., Wu R., Chen C., Li X., Zhou L., He M., Li Z., Sun X., Jia W., Chen J., Yang S., Zhou F., Zhao X., Wan S., Ye R., Liang C., Liu Z., Huang P., Liu C., Jiang H., Wang Y., Zheng H., Sun L., Liu X., Jiang Z., Feng D., Chen J., Wu S., Zou J., Zhang Z., Yang R., Zhao J., Xu C., Yin W., Guan Z., Ye J., Zhang H., Li J., Kristiansen K., Nickerson M. L., Theodorescu D., Li Y., Zhang X., Li S., Wang J., Yang H., Wang J., Cai Z., Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 43, 875–878 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang K., Kan J., Yuen S. T., Shi S. T., Chu K. M., Law S., Chan T. L., Kan Z., Chan A. S., Tsui W. Y., Lee S. P., Ho S. L., Chan A. K., Cheng G. H., Roberts P. C., Rejto P. A., Gibson N. W., Pocalyko D. J., Mao M., Xu J., Leung S. Y., Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat. Genet. 43, 1219–1223 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Wang K., Yuen S. T., Xu J., Lee S. P., Yan H. H., Shi S. T., Siu H. C., Deng S., Chu K. M., Law S., Chan K. H., Chan A. S., Tsui W. Y., Ho S. L., Chan A. K., Man J. L., Foglizzo V., Ng M. K., Chan A. S., Ching Y. P., Cheng G. H., Xie T., Fernandez J., Li V. S., Clevers H., Rejto P. A., Mao M., Leung S. Y., Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 46, 573–582 (2014). [DOI] [PubMed] [Google Scholar]
- 77.Chan-On W., Nairismägi M. L., Ong C. K., Lim W. K., Dima S., Pairojkul C., Lim K. H., McPherson J. R., Cutcutache I., Heng H. L., Ooi L., Chung A., Chow P., Cheow P. C., Lee S. Y., Choo S. P., Tan I. B., Duda D., Nastase A., Myint S. S., Wong B. H., Gan A., Rajasegaran V., Ng C. C., Nagarajan S., Jusakul A., Zhang S., Vohra P., Yu W., Huang D., Sithithaworn P., Yongvanit P., Wongkham S., Khuntikeo N., Bhudhisawasdi V., Popescu I., Rozen S. G., Tan P., Teh B. T., Exome sequencing identifies distinct mutational patterns in liver fluke–related and non-infection-related bile duct cancers. Nat. Genet. 45, 1474–1478 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Sausen S. M., Leary R. J., Jones S., Wu J., Reynolds C. P., Liu X., Blackford A., Parmigiani G., Diaz L. A. Jr, Papadopoulos N., Vogelstein B., Kinzler K. W., Velculescu V. E., Hogarty M. D., Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat. Genet. 45, 12–17 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chandler R. L., Brennan J., Schisler J. C., Serber D., Patterson C., Magnuson T., ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol. Cell. Biol. 33, 265–280 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li X. S., Trojer P., Matsumura T., Treisman J. E., Tanese N., Mammalian SWI/SNF-A subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol. Cell. Biol. 30, 1673–1688 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krosl J., Mamo A., Chagraoui J., Wilhelm B. T., Girard S., Louis I., Lessard J., Perreault C., Sauvageau G., A mutant allele of the Swi/Snf member BAF250a determines the pool size of fetal liver hemopoietic stem cell populations. Blood 116, 1678–1684 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hoffman G. R., Rahal R., Buxton F., Xiang K., McAllister G., Frias E., Bagdasarian L., Huber J., Lindeman A., Chen D., Romero R., Ramadan N., Phadke T., Haas K., Jaskelioff M., Wilson B. G., Meyer M. J., Saenz-Vash V., Zhai H., Myer V. E., Porter J. A., Keen N., McLaughlin M. E., Mickanin C., Roberts C. W., Stegmeier F., Jagani Z., Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc. Natl. Acad. Sci. U.S.A. 111, 3128–3133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith M. J., O’Sullivan J., Bhaskar S. S., Hadfield K. D., Poke G., Caird J., Sharif S., Eccles D., Fitzpatrick D., Rawluk D., du Plessis D., Newman W. G., Evans D. G., Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat. Genet. 45, 295–298 (2013). [DOI] [PubMed] [Google Scholar]
- 84.Chi T. H., Wan M., Lee P. P., Akashi K., Metzger D., Chambon P., Wilson C. B., Crabtree G. R., Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity 19, 169–182 (2003). [DOI] [PubMed] [Google Scholar]
- 85.Kosho T., Okamoto N. Coffin-Siris Syndrome International Collaborators , Genotype-phenotype correlation of Coffin-Siris syndrome caused by mutations in SMARCB1, SMARCA4, SMARCE1, and ARID1A. Am. J. Med. Genet. C Semin. Med. Genet. 166C, 262–275 (2014). [DOI] [PubMed] [Google Scholar]
- 86.Kim S. S., Kim M. S., Yoo N. J., Lee S. H., Frameshift mutations of a chromatin-remodeling gene SMARCC2 in gastric and colorectal cancers with microsatellite instability. APMIS 121, 168–9 (2013). [DOI] [PubMed] [Google Scholar]
- 87.Stephens P. J., Tarpey P. S., Davies H., Van Loo P., Greenman C., Wedge D. C., Nik-Zainal S., Martin S., Varela I., Bignell G. R., Yates L. R., Papaemmanuil E., Beare D., Butler A., Cheverton A., Gamble J., Hinton J., Jia M., Jayakumar A., Jones D., Latimer C., Lau K. W., McLaren S., McBride D. J., Menzies A., Mudie L., Raine K., Rad R., Chapman M. S., Teague J., Easton D., Langerød A. Oslo Breast Cancer Consortium (OSBREAC) Lee M. T., Shen C. Y., Tee B. T., Huimin B. W., Broeks A., Vargas A. C., Turashvili G., Martens J., Fatima A., Miron P., Chin S. F., Thomas G., Boyault S., Mariani O., Lakhani S. R., van de Vijver M., van ’t Veer L., Foekens J., Desmedt C., Sotiriou C., Tutt A., Caldas C., Reis-Filho J. S., Aparicio S. A., Salomon A. V., Børresen-Dale A. L., Richardson A. L., Campbell P. J., Futreal P. A., Stratton M. R., The landscape of cancer genes and mutational processes in breast cancer. Nature 486, 400–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Crew A. J., Clark J., Fisher C., Gill S., Grimer R., Chand A., Shipley J., Gusterson B. A., Cooper C. S., Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. Embo J 14, 2333–2340 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Naka N., Takenaka S., Araki N., Miwa T., Hashimoto N., Yoshioka K., Joyama S., Hamada K., Tsukamoto Y., Tomita Y., Ueda T., Yoshikawa H., Itoh K., Synovial sarcoma is a stem cell malignancy. Stem Cells 28, 1119–1131 (2010). [DOI] [PubMed] [Google Scholar]
- 90.Clark J., Rocques P. J., Crew A. J., Gill S., Shipley J., Chan A. M., Gusterson B. A., Cooper C. S., Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat. Genet. 7, 502–508 (1994). [DOI] [PubMed] [Google Scholar]
- 91.Lemon B., Inouye C., King D. S., Tjian R., Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414, 924–927 (2001). [DOI] [PubMed] [Google Scholar]
- 92.Varela I., Tarpey P., Raine K., Huang D., Ong C. K., Stephens P., Davies H., Jones D., Lin M. L., Teague J., Bignell G., Butler A., Cho J., Dalgliesh G. L., Galappaththige D., Greenman C., Hardy C., Jia M., Latimer C., Lau K. W., Marshall J., McLaren S., Menzies A., Mudie L., Stebbings L., Largaespada D. A., Wessels L. F., Richard S., Kahnoski R. J., Anema J., Tuveson D. A., Perez-Mancera P. A., Mustonen V., Fischer A., Adams D. J., Rust A., Chan-on W., Subimerb C., Dykema K., Furge K., Campbell P. J., Teh B. T., Stratton M. R., Futreal P. A., Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gerlinger M., Horswell S., Larkin J., Rowan A. J., Salm M. P., Varela I., Fisher R., McGranahan N., Matthews N., Santos C. R., Martinez P., Phillimore B., Begum S., Rabinowitz A., Spencer-Dene B., Gulati S., Bates P. A., Stamp G., Pickering L., Gore M., Nicol D. L., Hazell S., Futreal P. A., Stewart A., Swanton C., Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 46, 225–233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li M., Zhao H., Zhang X., Wood L. D., Anders R. A., Choti M. A., Pawlik T. M., Daniel H. D., Kannangai R., Offerhaus G. J., Velculescu V. E., Wang L., Zhou S., Vogelstein B., Hruban R. H., Papadopoulos N., Cai J., Torbenson M. S., Kinzler K. W., Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 43, 828–829 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fujimoto A., Furuta M., Shiraishi Y., Gotoh K., Kawakami Y., Arihiro K., Nakamura T., Ueno M., Ariizumi S., Nguyen H. H., Shigemizu D., Abe T., Boroevich K. A., Nakano K., Sasaki A., Kitada R., Maejima K., Yamamoto Y., Tanaka H., Shibuya T., Shibata T., Ojima H., Shimada K., Hayami S., Shigekawa Y., Aikata H., Ohdan H., Marubashi S., Yamada T., Kubo M., Hirano S., Ishikawa O., Yamamoto M., Yamaue H., Chayama K., Miyano S., Tsunoda T., Nakagawa H., Whole-genome mutational landscape of liver cancers displaying biliary phenotype reveals hepatitis impact and molecular diversity. Nat. Commun. 6, 6120 (2015). [DOI] [PubMed] [Google Scholar]
- 96.Rando O. J., Zhao K., Janmey P., Crabtree G. R., Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc. Natl. Acad. Sci. U.S.A. 99, 2824–2829 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dykhuizen E. C., Hargreaves D. C., Miller E. L., Cui K., Korshunov A., Kool M., Pfister S., Cho Y. J., Zhao K., Crabtree G. R., BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature 497, 624–627 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pugh T. J., Weeraratne S. D., Archer T. C., DA Pomeranz K., Auclair D., Bochicchio J., Carneiro M. O., Carter S. L., Cibulskis K., Erlich R. L., Greulich H., Lawrence M. S., Lennon N. J., McKenna A., Meldrim J., Ramos A. H., Ross M. G., Russ C., Shefler E., Sivachenko A., Sogoloff B., Stojanov P., Tamayo P., Mesirov J. P., Amani V., Teider N., Sengupta S., Francois J. P., Northcott P. A., Taylor M. D., Yu F., Crabtree G. R., Kautzman A. G., Gabriel S. B., Getz G., Jäger N., Jones D. T., Lichter P., Pfister S. M., Roberts T. M., Meyerson M., Pomeroy S. L., Cho Y. J., Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488, 106–110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barker N., Hurlstone A., Musisi H., Miles A., Bienz M., Clevers H., The chromatin remodelling factor Brg-1 interacts with β-catenin to promote target gene activation. Embo J 20, 4935–4943 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Davoli T., Xu A. W., Mengwasser K. E., Sack L. M., Yoon J. C., Park P. J., Elledge S. J., Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Clynes D., Higgs D. R., Gibbons R. J., The chromatin remodeller ATRX: A repeat offender in human disease. Trends Biochem. Sci. 38, 461–466 (2013). [DOI] [PubMed] [Google Scholar]
- 102.Gibbons R. J., Picketts D. J., Villard L., Higgs D. R., Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (ATR-X syndrome). Cell 80, 837–845 (1995). [DOI] [PubMed] [Google Scholar]
- 103.Schwartzentruber J., Korshunov A., Liu X. Y., Jones D. T., Pfaff E., Jacob K., Sturm D., Fontebasso A. M., Quang D. A., Tönjes M., Hovestadt V., Albrecht S., Kool M., Nantel A., Konermann C., Lindroth A., Jäger N., Rausch T., Ryzhova M., Korbel J. O., Hielscher T., Hauser P., Garami M., Klekner A., Bognar L., Ebinger M., Schuhmann M. U., Scheurlen W., Pekrun A., Frühwald M. C., Roggendorf W., Kramm C., Dürken M., Atkinson J., Lepage P., Montpetit A., Zakrzewska M., Zakrzewski K., Liberski P. P., Dong Z., Siegel P., Kulozik A. E., Zapatka M., Guha A., Malkin D., Felsberg J., Reifenberger G., von Deimling A., Ichimura K., Collins V. P., Witt H., Milde T., Witt O., Zhang C., Castelo-Branco P., Lichter P., Faury D., Tabori U., Plass C., Majewski J., Pfister S. M., Jabado N., Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012). [DOI] [PubMed] [Google Scholar]
- 104.Eustermann S., Yang J. C., Law M. J., Amos R., Chapman L. M., Jelinska C., Garrick D., Clynes D., Gibbons R. J., Rhodes D., Higgs D. R., Neuhaus D., Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat. Struct. Mol. Biol. 18, 777–782 (2011). [DOI] [PubMed] [Google Scholar]
- 105.Huang S., Gulzar Z. G., Salari K., Lapointe J., Brooks J. D., Pollack J. R., Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene 31, 4164–4170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu W., Lindberg J., Sui G., Luo J., Egevad L., Li T., Xie C., Wan M., Kim S. T., Wang Z., Turner A. R., Zhang Z., Feng J., Yan Y., Sun J., Bova G. S., Ewing C. M., Yan G., Gielzak M., Cramer S. D., Vessella R. L., Zheng S. L., Grönberg H., Isaacs W. B., Xu J., Identification of novel CHD1-associated collaborative alterations of genomic structure and functional assessment of CHD1 in prostate cancer. Oncogene 31, 3939–3948 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Burkhardt L., Fuchs S., Krohn A., Masser S., Mader M., Kluth M., Bachmann F., Huland H., Steuber T., Graefen M., Schlomm T., Minner S., Sauter G., Sirma H., Simon R., CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res. 73, 2795–2805 (2013). [DOI] [PubMed] [Google Scholar]
- 108.Srinivasan S., Dorighi K. M., Tamkun J. W., Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLOS Genet. 4, e1000217 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim K. H., Roberts C. W., Mechanisms by which SMARCB1 loss drives rhabdoid tumor growth. Cancer genetics 207, 365–372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Knutson S. K., Warholic N. M., Wigle T. J., Klaus C. R., Allain C. J., Raimondi A., Porter S. M., Chesworth R., Moyer M. P., Copeland R. A., Richon V. M., Pollock R. M., Kuntz K. W., Keilhack H., Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl. Acad. Sci. U.S.A. 110, 7922–7927 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bitler B. G., Aird K. M., Garipov A., Li H., Amatangelo M., Kossenkov A. V., Schultz D. C., Liu Q., Shih I. M., Conejo-Garcia J. R., Speicher D. W., Zhang R., Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat. Med. 21, 231–238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Knutson S. K., Kawano S., Minoshima Y., Warholic N. M., Huang K. C., Xiao Y., Kadowaki T., Uesugi M., Kuznetsov G., Kumar N., Wigle T. J., Klaus C. R., Allain C. J., Raimondi A., Waters N. J., Smith J. J., Porter-Scott M., Chesworth R., Moyer M. P., Copeland R. A., Richon V. M., Uenaka T., Pollock R. M., Kuntz K. W., Yokoi A., Keilhack H., Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol. Cancer Ther. 13, 842–854 (2014). [DOI] [PubMed] [Google Scholar]
- 113.Hathaway N. A., Bell O., Hodges C., Miller E. L., Neel D. S., Crabtree G. R., Dynamics and memory of heterochromatin in living cells. Cell 149, 1447–1460 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hodges C., Crabtree G. R., Dynamics of inherently bounded histone modification domains. Proc. Natl. Acad. Sci. U.S.A. 109, 13296–13301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang J. C., Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 (2002). [DOI] [PubMed] [Google Scholar]
- 116.Wu C. C., Li T. K., Farh L., Lin L. Y., Lin T. S., Yu Y. J., Yen T. J., Chiang C. W., Chan N. L., Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science 333, 459–462 (2011). [DOI] [PubMed] [Google Scholar]
- 117.Pommier Y., Leo E., Zhang H., Marchand C., DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Thurman R. E., Rynes E., Humbert R., Vierstra J., Maurano M. T., Haugen E., Sheffield N. C., Stergachis A. B., Wang H., Vernot B., Garg K., John S., Sandstrom R., Bates D., Boatman L., Canfield T. K., Diegel M., Dunn D., Ebersol A. K., Frum T., Giste E., Johnson A. K., Johnson E. M., Kutyavin T., Lajoie B., Lee B. K., Lee K., London D., Lotakis D., Neph S., Neri F., Nguyen E. D., Qu H., Reynolds A. P., Roach V., Safi A., Sanchez M. E., Sanyal A., Shafer A., Simon J. M., Song L., Vong S., Weaver M., Yan Y., Zhang Z., Zhang Z., Lenhard B., Tewari M., Dorschner M. O., Hansen R. S., Navas P. A., Stamatoyannopoulos G., Iyer V. R., Lieb J. D., Sunyaev S. R., Akey J. M., Sabo P. J., Kaul R., Furey T. S., Dekker J., Crawford G. E., Stamatoyannopoulos J. A., The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sperling A. S., Jeong K. S., Kitada T., Grunstein M., Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 108, 12693–12698 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fillmore C. M., Xu C., Desai P. T., Berry J. M., Rowbotham S. P., Lin Y. J., Zhang H., Marquez V. E., Hammerman P. S., Wong K. K., Kim C. F., EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature 520, 239–242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Steger D. J., Haswell E. S., Miller A. L., Wente S. R., O’Shea E. K., Regulation of chromatin remodeling by inositol polyphosphates. Science 299, 114–116 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Druker B. J., Translation of the Philadelphia chromosome into therapy for CML. Blood 112, 4808–4817 (2008). [DOI] [PubMed] [Google Scholar]