Abstract
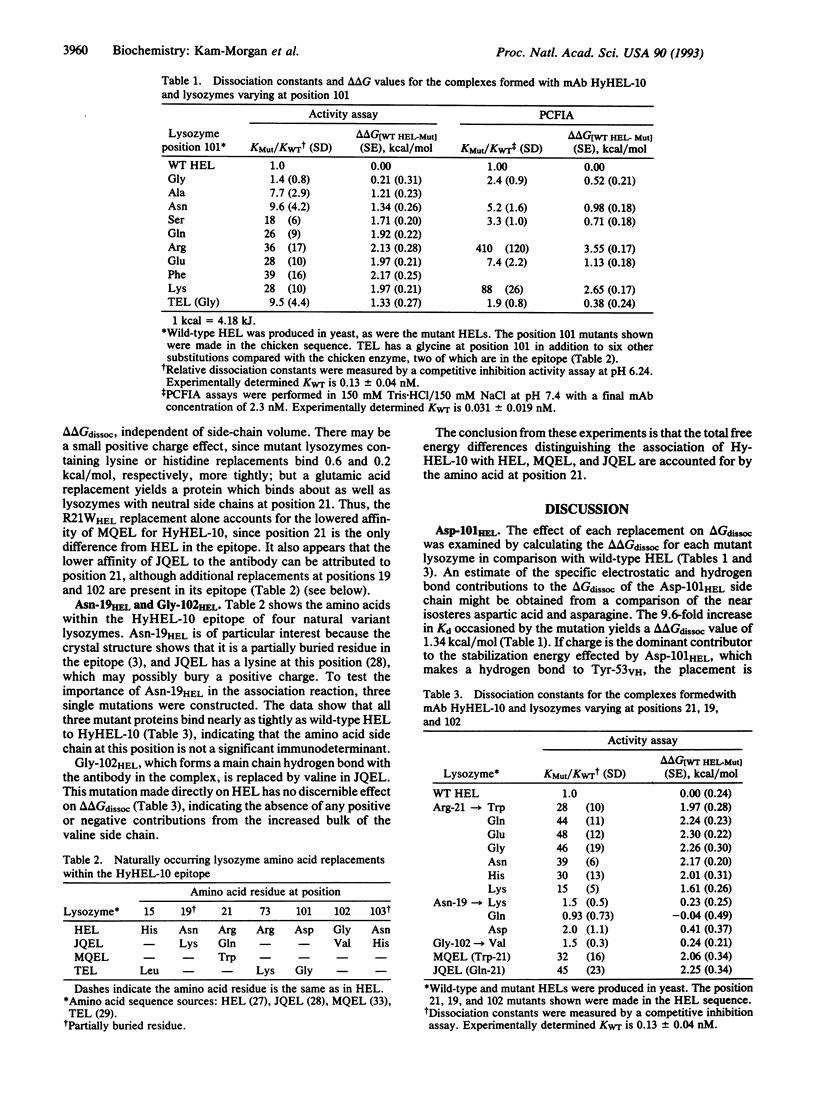
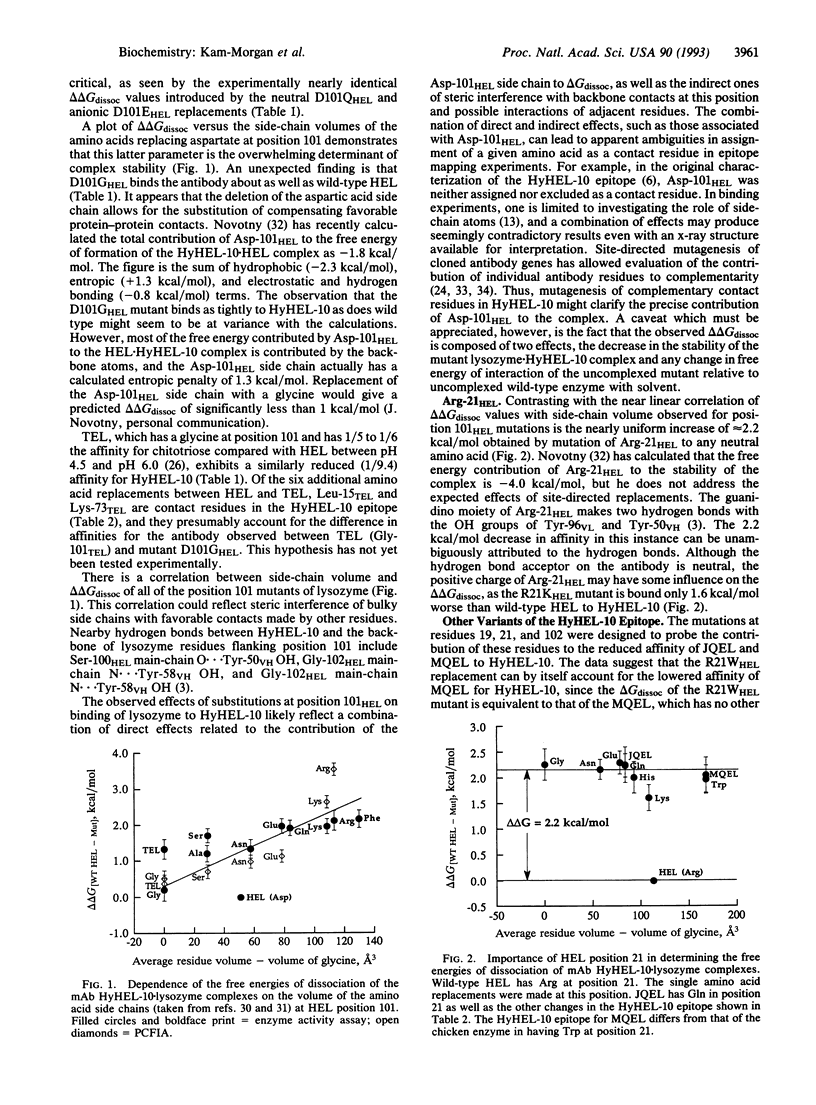
The complex formed between hen egg white lysozyme (HEL) and the monoclonal antibody HyHEL-10 Fab fragment has an interface composed of van der Waals interactions, hydrogen bonds, and a single ion pair. The antibody overlaps part of the active site cleft. Putative critical residues within the epitope region of HEL, identified from the x-ray crystallographic structure of the complex, were replaced by site-directed mutagenesis to probe their relative importance in determining affinity of the antibody for HEL. Twenty single mutations of HEL at three contact residues (Arg-21HEL, Asp-101HEL, and Gly-102HEL) and at a partially buried residue (Asn-19HEL) in the epitope were made, and the effects on the free energies of dissociation were measured. A correlation between increased amino acid side-chain volume and reduced affinity for HELs with mutations at position 101 was observed. The D101GHEL mutant is bound to HyHEL-10 as tightly as wild-type enzyme, but the delta delta Gdissoc is increased by about 2.2 kcal (9.2 kJ)/mol for the larger residues in this position. HEL variants with lysine or histidine replacements for arginine at position 21 are bound 1.4-2.7 times more tightly than those with neutral or negatively charged amino acids in this position. These exhibit 1/40 the affinity for HyHEL-10 Fab compared with wild type. There is no side-chain volume correlation with delta delta Gdissoc at position 21. Although Gly-102HEL and Asn-19HEL are in the epitope, replacements at these positions have no effect on the affinity of HEL for the antibody.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexenko A. P., Izotova L., Strongin AYa Reconstruction of an epitope capable of binding murine monoclonal antibodies NK2 within the sequence of human leukocyte interferon alpha F by site-directed mutagenesis. Biochem Biophys Res Commun. 1990 Jun 29;169(3):1061–1067. doi: 10.1016/0006-291x(90)92002-h. [DOI] [PubMed] [Google Scholar]
- Amit A. G., Mariuzza R. A., Phillips S. E., Poljak R. J. Three-dimensional structure of an antigen-antibody complex at 2.8 A resolution. Science. 1986 Aug 15;233(4765):747–753. doi: 10.1126/science.2426778. [DOI] [PubMed] [Google Scholar]
- Arnheim N., Millett F., Raftery M. A. Nuclear magnetic resonance and ultraviolet difference spectral studies of the binding properties of turkey egg white lysozyme. Consequences of the replacement of Asp 101 by glycine. Arch Biochem Biophys. 1974 Nov;165(1):281–287. doi: 10.1016/0003-9861(74)90166-0. [DOI] [PubMed] [Google Scholar]
- Benjamin D. C., Berzofsky J. A., East I. J., Gurd F. R., Hannum C., Leach S. J., Margoliash E., Michael J. G., Miller A., Prager E. M. The antigenic structure of proteins: a reappraisal. Annu Rev Immunol. 1984;2:67–101. doi: 10.1146/annurev.iy.02.040184.000435. [DOI] [PubMed] [Google Scholar]
- Benjamin D. C. Molecular approaches to the study of B cell epitopes. Int Rev Immunol. 1991;7(2):149–164. doi: 10.3109/08830189109061772. [DOI] [PubMed] [Google Scholar]
- Bhat T. N., Bentley G. A., Fischmann T. O., Boulot G., Poljak R. J. Small rearrangements in structures of Fv and Fab fragments of antibody D1.3 on antigen binding. Nature. 1990 Oct 4;347(6292):483–485. doi: 10.1038/347483a0. [DOI] [PubMed] [Google Scholar]
- Davies D. R., Sheriff S., Padlan E. A. Antibody-antigen complexes. J Biol Chem. 1988 Aug 5;263(22):10541–10544. [PubMed] [Google Scholar]
- Friguet B., Chaffotte A. F., Djavadi-Ohaniance L., Goldberg M. E. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J Immunol Methods. 1985 Mar 18;77(2):305–319. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- Glockshuber R., Stadlmüller J., Plückthun A. Mapping and modification of an antibody hapten binding site: a site-directed mutagenesis study of McPC603. Biochemistry. 1991 Mar 26;30(12):3049–3054. doi: 10.1021/bi00226a010. [DOI] [PubMed] [Google Scholar]
- Hogan K. T., Clayberger C., Bernhard E. J., Walk S. F., Ridge J. P., Parham P., Krensky A. M., Engelhard V. H. Identification by site-directed mutagenesis of amino acid residues contributing to serologic and CTL-defined epitope differences between HLA-A2.1 and HLA-A2.3. J Immunol. 1988 Oct 1;141(7):2519–2525. [PubMed] [Google Scholar]
- Ibrahimi I. M., Prager E. M., White T. J., Wilson A. C. Amino acid sequence of California quail lysozyme. Effect of evolutionary substitutions on the antigenic structure of lysozyme. Biochemistry. 1979 Jun 26;18(13):2736–2744. doi: 10.1021/bi00580a008. [DOI] [PubMed] [Google Scholar]
- Kaneda M., Kato I., Tominaga N., Titani K., Narita K. The amino acid sequence of quail lysozyme. J Biochem. 1969 Nov;66(5):747–749. [PubMed] [Google Scholar]
- LaRue J. N., Speck J. C., Jr Turkey egg white lysozyme. Preparation of the crystalline enzyme and investigation of the amino acid sequence. J Biol Chem. 1970 Apr 25;245(8):1985–1991. [PubMed] [Google Scholar]
- Lavoie T. B., Drohan W. N., Smith-Gill S. J. Experimental analysis by site-directed mutagenesis of somatic mutation effects on affinity and fine specificity in antibodies specific for lysozyme. J Immunol. 1992 Jan 15;148(2):503–513. [PubMed] [Google Scholar]
- Malcolm B. A., Rosenberg S., Corey M. J., Allen J. S., de Baetselier A., Kirsch J. F. Site-directed mutagenesis of the catalytic residues Asp-52 and Glu-35 of chicken egg white lysozyme. Proc Natl Acad Sci U S A. 1989 Jan;86(1):133–137. doi: 10.1073/pnas.86.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny J. Protein antigenicity: a thermodynamic approach. Mol Immunol. 1991 Mar;28(3):201–207. doi: 10.1016/0161-5890(91)90062-o. [DOI] [PubMed] [Google Scholar]
- Padlan E. A., Silverton E. W., Sheriff S., Cohen G. H., Smith-Gill S. J., Davies D. R. Structure of an antibody-antigen complex: crystal structure of the HyHEL-10 Fab-lysozyme complex. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5938–5942. doi: 10.1073/pnas.86.15.5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHUGAR D. The measurement of lysozyme activity and the ultra-violet inactivation of lysozyme. Biochim Biophys Acta. 1952 Mar;8(3):302–309. doi: 10.1016/0006-3002(52)90045-0. [DOI] [PubMed] [Google Scholar]
- Sharma S., Georges F., Delbaere L. T., Lee J. S., Klevit R. E., Waygood E. B. Epitope mapping by mutagenesis distinguishes between the two tertiary structures of the histidine-containing protein HPr. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4877–4881. doi: 10.1073/pnas.88.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheriff S., Silverton E. W., Padlan E. A., Cohen G. H., Smith-Gill S. J., Finzel B. C., Davies D. R. Three-dimensional structure of an antibody-antigen complex. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8075–8079. doi: 10.1073/pnas.84.22.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Gill S. J., Lavoie T. B., Mainhart C. R. Antigenic regions defined by monoclonal antibodies correspond to structural domains of avian lysozyme. J Immunol. 1984 Jul;133(1):384–393. [PubMed] [Google Scholar]
- Smith A. M., Benjamin D. C. The antigenic surface of staphylococcal nuclease. II. Analysis of the N-1 epitope by site-directed mutagenesis. J Immunol. 1991 Feb 15;146(4):1259–1264. [PubMed] [Google Scholar]
- Smith A. M., Woodward M. P., Hershey C. W., Hershey E. D., Benjamin D. C. The antigenic surface of staphylococcal nuclease. I. Mapping epitopes by site-directed mutagenesis. J Immunol. 1991 Feb 15;146(4):1254–1258. [PubMed] [Google Scholar]
- Stevens F. J. Modification of an ELISA-based procedure for affinity determination: correction necessary for use with bivalent antibody. Mol Immunol. 1987 Oct;24(10):1055–1060. doi: 10.1016/0161-5890(87)90073-3. [DOI] [PubMed] [Google Scholar]
- Tavernier J., Marmenout A., Bauden R., Hauquier G., Van Ostade X., Fiers W. Analysis of the structure-function relationship of tumour necrosis factor. Human/mouse chimeric TNF proteins: general properties and epitope analysis. J Mol Biol. 1990 Jan 20;211(2):493–501. doi: 10.1016/0022-2836(90)90367-U. [DOI] [PubMed] [Google Scholar]
- Zamyatnin A. A. Protein volume in solution. Prog Biophys Mol Biol. 1972;24:107–123. doi: 10.1016/0079-6107(72)90005-3. [DOI] [PubMed] [Google Scholar]


