Abstract
Placental growth factor (PlGF) plays an important role in various pathological conditions and diseases such as inflammation, cancer, atherosclerosis and sickle cell disease (SCD). Abnormally high PlGF levels in SCD patients are associated with increased inflammation and pulmonary hypertension (PHT) and reactive airway disease; however, the transcriptional and post-transcriptional mechanisms regulating PlGF expression are not well defined. Herein, we show that treatment of human erythroid cells and colony forming units with erythropoietin (EPO) increased PlGF expression. Our studies showed EPO-mediated activation of HIF-1α led to subsequent binding of HIF-1α to hypoxia response elements (HREs) within the PlGF promoter, as demonstrated by luciferase transcription reporter assays and ChIP analysis of the endogenous gene. Additionally, we showed miR-214 post-transcriptionally regulated the expression of PlGF as demonstrated by luciferase reporter assays using wild-type (wt) and mutant PlGF-3′-UTR constructs. Furthermore, synthesis of miR-214, located in an intron of DNM3 (dynamin 3), was transcriptionally regulated by transcription factors, peroxisome proliferator-activated receptor-α (PPARα) and hypoxia-inducible factor-1α (HIF-1α). These results were corroborated in vivo wherein plasma from SCD patients and lung tissues from sickle mice showed an inverse correlation between PlGF and miR-214 levels. Finally, we observed that miR-214 expression could be induced by fenofibrate, a Food and Drug Administration (FDA) approved PPARα agonist, thus revealing a potential therapeutic approach for reduction in PlGF levels by increasing miR-214 transcription. This strategy has potential clinical implications for several pathological conditions including SCD.
Keywords: dynamin 3 opposite strand (DNM3os), fenofibrate, hypoxia-inducible factor-1α (HIF-1α), microRNA (miRNA), peroxisome proliferator-activated receptor-α (PPARα), placenta growth factor
INTRODUCTION
Placental growth factor (PlGF), an angiogenic hormone and a member of the vascular endothelial growth factor (VEGF) family, plays an important role in various diseases and pathological conditions such as cancer and inflammation[1,2], myocardial angiogenesis and atherosclerosis [3,4], sepsis [5], allergic asthma [6,7], diabetic wound healing [8], pre-eclampsia [9] and sickle cell disease (SCD) [10].
PlGF is produced by placental trophoblasts and umbilical vein endothelial cells [11], non-placental tissues [12] and erythroid cells [10,13]. Its expression is induced by erythropoietin (EPO) in a cluster of differentiation (CD)34+ progenitor cells of bone marrow [10]. The human PlGF gene encodes four isoforms, PlGF-1–4 by alternative mRNA splicing [14]. PlGF-mediated signalling events begin after binding to its cognate VEGF receptor-1(VEGF-R1) [15–17]. Plasma levels of PlGF in SCD patients are high, when compared with healthy control subjects, which correlates to the frequency of vaso-occlusive pain episodes [10]. Thus, the abnormal levels of systemic PlGF, associated with SCD, could be a consequence of increased erythropoiesis, hypoxia and higher levels of EPO, all resulting from haemolytic anaemia observed in these patients [10,13,18].
PlGF activates endothelial cells and monocytes, increasing the expression of cytochemokines, endothelin-1 (ET-1) and endothelin-B receptor (ET-BR) via activation of the transcription factor-1, hypoxia-inducible factor-1α (HIF-1α), independently of hypoxia [10,19,20]. Moreover, overexpression of PlGF in wild–type (wt) mice, to the levels seen in sickle mice, leads to increased ET-1 production and increased right ventricular pressure; both are markers of pulmonary hypertension (PHT) [21] and these findings were corroborated in SCD patients [21]. Thus, therapeutic strategies that block PlGF-mediated signalling or its production may be beneficial to SCA (sickle cell anaemia) patients. Previous studies have established the role of transcription factors, metal regulatory transcription factor-(MTF-1) [10,12,22,23] and glial cell missing-1 (GCM-1) in the expression of PlGF [24,25]. However, processes that regulate PlGF expression at the transcriptional and the post-transcriptional levels are poorly understood and may provide an alternative approach for therapeutic strategies.
Levels of EPO, a major hormone regulator of erythropoiesis, are elevated in SCD patients [25]. EPO is constitutively secreted by the kidneys, but its production can be further stimulated by tissue hypoxia or anaemia. For example, any reduction in erythrocytes via bleeding or anaemia leads to reduced oxygen delivery to the major organs including the kidneys, thus stimulating EPO production. In SCD, increased EPO secretion [25] occurs during augmented erythropoiesis as a compensatory response to the severe haemolytic anaemia associated with this condition. Increased levels of serum EPO in mice are protective from PHT development in some studies [26], whereas other studies reported higher levels of EPO were instead associated with higher tricuspid regurgitation velocity (TRV) and development of PHT in SCD patients [27].
Since erythroid cells produce PlGF and increased EPO is needed for stimulating compensatory erythropoiesis in severe anaemia associated with SCD, we examined the effect of EPO on the expression of PlGF in erythroid cells. In the present report, we show that EPO induced expression of PlGF in K562 human erythroleukaemia cell line, used as a model for erythroid cells. In this system, PlGF is transcriptionally regulated by HIF-1α and post-transcriptionally regulated by miR-214; the latter targets the 3′-UTR of PlGF mRNA. Moreover, we show miR-214, intronically located in dynamin- 3 (DNM3) and synthesized from the DNM3 opposite strand (DNM3os) transcription unit [28] is also trans-activated by peroxisome proliferator receptor-α (PPARα) and HIF-1α. We observed that agonists of PPARα induced transcription of DNM3os, premiR-214 and miR-214, culminating in reduced expression of PlGF. Indeed, plasma levels of miR214 were reduced in SCD patients compared with controls and in lung tissues of Berkeley SS mice (BK-SS) compared with C57BL/6NJ control mice. These findings were consistent with the regulatory model showing that lowered miR-214 levels were permissive for increased PlGF expression. A consequence of higher basal PlGF levels would subsequently lead to increased expression of PlGF induced genes such as ET-1 [19] and plasminogen activator inhibitor-1 (PAI-1) [29], both of which have been implicated in development of PHT in SCD individuals.
MATERIALS AND METHODS
Cell culture and reagents
K562, a human erythroleukaemia cell line, was cultured in Iscove’s modified DMEM (Dulbecco’s Modified Eagle’s medium; IMDM), supplemented with 10 %heat-inactivated FBS, 100 μg/ml streptomycin and 100 units/ml penicillin, [30]. Cells were kept in serum-free medium overnight prior to treatment with EPO (3 units/ml). EPO was a kind gift from Dr Vinod Pullarkat (City of Hope, Duarte, California). LY294002, PD98059, SB203580, SP600125 and R59949 were purchased from EMD Millipore and used at concentrations as previously described [31]. Drugs were used at the indicated final concentrations and were obtained from vendors as noted: fenofibrate (100 μM) and GW6471 (5 μM) were purchased from Sigma–Aldrich; Ro31 (33 μM) and Ro32 (31 μM) were purchased from Tocris. Cells were pre-incubated with drugs for 30 min prior to treatment with EPO. Primary antibodies for HIF-1α (1:250) and PPARα (1:250) were purchased from Santa Cruz Biotechnology and Abcam respectively. Horseradish peroxidase (HRP)-conjugated antibody for β-actin and secondary antibodies for HIF-1α and PPARα were purchased from Sigma–Aldrich. The miR-214 mimics, anti-miR-214 inhibitors and appropriate controls were purchased from Shanghai GenePharma as well as from Exiqon, designated as miR-214 inhibitor and locked nucleic acid (LNA)-miR214 inhibitor respectively.
Luciferase vectors
An ~2.6-kb region of the PlGF promoter, spanning nts − 2622/+ 60 relative to the transcription start site, was PCR-amplified from human genomic bacterial artificial chromosome (BAC) clone RP11-668 L1 as template (BACPAC Resource Center) and inserted into the pGL3-Basic vector (Promega) using the In-Fusion HD cloning kit (Clontech Laboratories).
An ~2100-bp segment containing the DNM3os promoter (nts − 2100/+ 10) was PCR-amplified from human genomic BAC clone RP11-455 O13 as template and inserted into the NheI restriction site of pGL3-Basic luciferase reporter plasmid following manufacturer’s instructions (Promega). The 3′-UTR region of the PlGF mRNA, fused downstream of the luciferase ORF in pMirTarget-luciferase was purchased from Origene. Mutagenesis of the HIF-1α-binding sites within the PlGF promoter, the PPARα- and HIF-1α-binding sites within the DNM3os promoter and the miR-214-binding sites within the 3′-UTR of PlGF were performed using the Q5 mutagenesis kit (New England Biolabs), using corresponding templates and PCR primers listed in Table 1. All constructs and subsequent mutations were verified by DNA sequencing (Retrogen).
Table 1.
Primers used in the present study
| Gene | Method | Forward primer | Reverse primer |
|---|---|---|---|
| PlGF mRNA | qRT-PCR | TGTTCAGCCCATCCTGTGTC | ACAGTGCAGATTCTCATCGCC |
| GAPDH mRNA | qRT-PCR | AACCTGCCAAGTACGATGACATC | GTAGCCCAGGATGCCCTTGA |
| DNM3os RNA | qRT-PCR | GCCACAGCCGAACTGTACATC | CAGTCATTTCATCTTCAGCCCA |
| premiR-199a2 | qRT-PCR | AGGAAGCTTCTGGAGATCC | TGCTCTCCCTTGCCCAG |
| premiR-214 | qRT-PCR | GGCCTGGCTGGACAGAG | GGCTGGGTTGTCATGTGAC |
| mPlGF | qRT-PCR | AGGTCCTTTGTCCCTCTCTG | TGTATCGGTCAAAGTCCACGG |
| mpremiR-214 | qRT-PCR | GCTGGACAGAGTTGTCATGTGTC | TGTGACTGCCTGTCTGTGCC |
| mGAPDH | qRT-PCR | TTGCAGTGGCAAAGTGGAGA | GTCTCGCTCCTGGAAGATGG |
| PlGF promoter (HRE at − 937/− 933) | ChIP | GGACACAGAAGGCAG | TCTGTCCGCTGTGTAT |
| Bcl-XL promoter | ChIP | CCGGGTCGCATGATCCCTC | CCACCTACATTCAAATCCGCCT |
| G6PD promoter | ChIP | GGACAGTAGGGGCGGGG | GCTCTGCATCCCCAATTCCG |
| DNM3OS HRE mutant | SDM | CTCTCCAGCGgtatAGCGCATACC | ACCGCTAGAATCGCCTGC |
| DNM3OS PP1 mutant | SDM | CTTTTCCATAtgaatGCCCACTTCCTGCC | CTGTAAAAAAAAAAGAGTCAAGATG |
| DNM3OS PP2 mutant | SDM | CTTCTCTAGCtgaatTCAGACAGTAAATTTGTACTTCTAC | TAATTTGAAGATAGAGGGTAGATG |
| DNm3OS PP3 mutant | SDM | AGCATTCAGAtgaatAGCCAAAATAGCTGAGGG | GTGCTATAGTTTGACCTAAAG |
| PlGF HRE1 mutant | SDM | ACAGGCAGGCataCAGACTCACAG | CTCTGCGTGTGTGTGTCTG |
| PlGF HRE2 Mutant | SDM | GAGAGAGAGGataAGTCTTCACCCACAGAGACAGACAC | TGCGTTTGTGTGTGCCCT |
| PlGF–miR-214-binding site 1 mutant | SDM | CAGTGACTCCaaaTGGTACCTGC | AAGAGTGTGACGGTAATAAATAC |
| PlGF–miR-214-binding site 2 mutant | SDM | ACTGTTTCCCaaaTGAATGCCTCG | TGGCTAATAAATAGAGGGC |
| miR-214 mimic | ACAGCAGGCACAGACAGGCAGU | ||
| miR-214 inhibitor | ACUGCCUGUCGUGCCUGCUGU | ||
| miR-214 probe | Northern | ACTGCCTGTCTGTGCCTGCTGT | |
| 5S rRNA probe | Northern | CAGGCCCGACCCTGCTTAGCTTCC GAGATCAGACGAGAT |
SDM, site-directed mutagenesis.
Transient transfections and luciferase assays
K562 cells (106) were transfected with luciferase vectors (1 μg), shRNA constructs (1 μg) or miR-214 mimics (90 pmoles), as indicated, using Hi-Perfect transfection reagent (Qiagen), as per manufacturer’s protocols. Transfected cells were kept overnight in complete medium, followed by replacement with serum-free medium and incubation for 3 h prior to treatment with EPO. Cells transfected with luciferase reporter vectors were treated with EPO and lysed in 1× Reporter lysis buffer (Promega) followed by luciferase assay with Dual-Glo luciferase reagent (Promega). Luciferase values were normalized to Renilla luciferase values for transfection efficiency.
Westerns blots
For Western blots, 2 × 106 K562 cells were treated with indicated reagents, lysed using radioimmunoprecipitation assay (RIPA) buffer and probed with indicated antibodies [31]. Twenty-five micrograms of protein from each lysate was run on a SDS/PAGE (10 %gel) followed by transfer to PVDF membrane. Membranes were incubated overnight at 4 °C with primary antibodies to either PPARα (1:250) or HIF-1α (1:250). Membranes were washed and incubated with the appropriate secondary antibodies (1:10000), followed by development using the Clarity chemiluminescent kit (Bio-Rad). The membranes were stripped and re-probed with an antibody for β-actin (1:20000), as a loading control. Quantification of images was performed using the ImageJ software suite.
RNA extraction and quantitative real-time PCR
Cells following treatment were lysed in TRIzol reagent (Life Technologies) and total RNA isolated following the manufacturer’s protocol. 100 ng of RNA were transcribed using the One-Step SYBR PrimeTime RT-PCR kit (Clontech Laboratories) and the primers listed in Table 1, following the manufacturer’s protocols [31].
Isolation and quantification of miRNAs
miRNA-binding sites in the 3′-UTR of PlGF were identified using the Microcosm Targets web tool (URL: http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/). K562 cells were treated with EPO and miRNAs were purified utilizing the mirVana isolation kit (Life Technologies) [32]. Individual miRNA expression levels were determined by quantitative real-time (qRT)-PCR using TaqMan MicroRNA assay kits for indicated miRNAs (Invitrogen), according to the manufacturer’s instructions. miRNA expression levels were normalized to the reference gene, RNU6B (U6 snRNA) [32] using the comparative threshold cycle method (ΔCt). miRNAs were also quantified by Northern blot analysis. Briefly, 35 μg of total RNA from each sample was run on a 15 μ non-denaturing PAGE. The RNA was transferred to Biodyne B nylon membrane (Pall Corporation) followed by cross-linking under UV light and pre-hybridized for 30 min using the UltraHyb hybridization buffer (Ambion) at 60°C. The membrane was then incubated with biotin-labelled hybridization probes for miR-214 and 5S rRNA, purchased from Valugene in Ultrahyb hybridization buffer (Ambion), at 60°C overnight. The membranes were washed twice in washing buffer (Thermo Scientific) at 60°C followed by blocking with 5 μ non-fat milk in 1× PBS at room temperature. Streptavidin-HRP (1:250) was added to the membrane, incubated at room temperature for 3 h and followed by two washes with 1× washing buffer (LightShift Chemiluminescent EMSA kit, Thermo Scientific/Pierce). The membranes were developed and resulting images quantified using the ImageJ analysis software.
ChIP analysis
K562 cells (5 × 106) were incubated in serum-free medium overnight, prior to treatment with EPO as described above. Cells were lysed and sonicated as previously described [33]. Chromatin was immunoprecipitated with HIF-1α antibody (5 μg; Santa Cruz Biotechnology). The DNA was subjected to PCR amplification for 30 cycles under the following conditions: 95 °C for 30 s, 60 °C for 1 min, 72 °C for 120 s, using primers listed in Table 1. The PCR products were electrophoresed on a 2 %agarose gel. Images were quantified using the ImageJ analysis software.
Isolation of erythroid colony forming units from human blood
Five millilitres of blood was obtained from normal volunteers, utilizing informed consent, as approved by University of Southern California Institutional Review Board. Samples were collected in heparinized tubes and diluted with an equal volume of IMDM/2 μ FBS. Six millilitres of the cell suspension was layered on 3 ml of Ficoll–Paque (GE HealthCare) for isolation of the mononuclear cell fraction. Cells were carefully removed from the interface between the plasma/Ficoll–Paque medium and centrifuged at 400 g for 30 min without brake. Cells were washed twice with IMDM/2 μ FBS and re-spun at 300 g for 10 min with the brake ‘on’. Cells (106) were suspended in methyl-cellulose (1 %) medium supplemented with FBS (30 %), interleukin (IL)-3 (10 ng/ml) and stem cell factor (SCF) [10]. Cells in the methyl-cellulose medium were incubated for 15 days in a CO2 incubator at 37 °C [34]. CFU-E (erythroid colony forming units) clusters were isolated and washed with IMDM medium supplemented with FBS and kept for 2 days in the absence of EPO, followed by serum-free medium for 3 h and EPO treatment.
Quantification of secreted PlGF from erythroid cells
K562 cells were transfected with miR-214 mimic (90 pmoles) and the negative control mimic (NC mimic, 90 pmoles). Transfected cells were incubated in complete medium for 24 h followed by serum-free medium for 3 h and treated with either EPO (3 units/ml) or fenofibrate (0.1 mM) for 18 h. The culture supernatants were collected and an aliquot (100 μl) was assayed for secreted PlGF by ELISA (Peprotech). The cells were washed with PBS and lysed with RIPA buffer and assayed for protein content using the Bradford method.
Human subjects and mice
All blood samples were obtained from children with homozygous SCD at steady state (absence of sickle cell related acute event for 3 weeks prior to blood draw) at their elective clinical visit for routine check-up and clinical blood draws. Permissions were obtained with the informed consent of the patient or parent/legal guardian using Institutional Review Board approved protocols through the Repository for Non-Malignant Hematology Specimens at Cincinnati Children’s Hospital Medical Center. The plasma from Sickle cell homozygous (SS) and their unaffected sibling control subjects were collected and stored at − 80 °C until assayed. BK-SS mice originally obtained from Jackson Laboratories were bred up to the sixth generation against a C57BL/6NJ background [35]. Animal protocols were approved by the Institutional Animal Care Use Committee at Cincinnati Children’s Hospital Medical Center. In-house bred C57BL/6NJ mice served as controls. Blood and lungs were collected after animals were exsanguinated and stored at − 80 °C for assay of miRNAs and PlGF levels. Plasma EPO levels were assayed using an ELISA kit (R & D Systems).
Statistical analysis
Data are presented as means ± S.E.M. Control and EPO-treated samples were compared using a Student’s t test. One-way ANOVA followed by Tukey–Kramer test were used for multiple comparisons using the Instat-2 Software (Graph Pad). P < 0.05 was considered statistically significant. *P<0.05, **P < 0.01, ***P < 0.001 and ns (not significant) with P > 0.05.
RESULTS
EPO augments expression of PlGF via activation of PI-3 kinase, p38 MAP kinase and MAP kinase in K562 erythroid cells
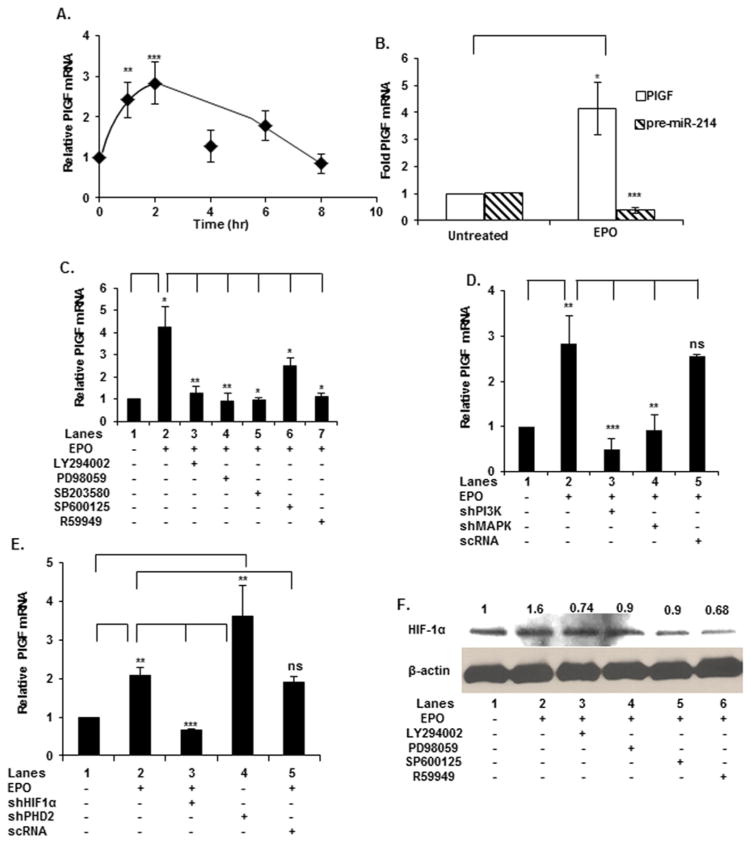
Treatment of K562 cells with EPO showed a time-dependent increase in PlGF mRNA expression with a 3-fold increase observed at 2 h (Figure 1A). Similarly, treatment of human CFU-E cells with EPO increased expression of PlGF mRNA by ~4-fold (Figure 1B). Since EPO treatment of both K562 and CFU-Es behaved similarly with regard to PlGF induction, we used K562 cells as the model system for our continued studies.
Figure 1. EPO-induced PlGF mRNA expression involved signalling via the PI3 kinase, MAP kinase, JNK and HIF-1α pathways.
(A) K562 cells were treated with EPO for indicated time periods followed by isolation of total RNA for qRT-PCR analysis of PlGF mRNA. (B) CFU-E cells were treated with EPO for 2 h followed by isolation of RNA and qRT-PCR analysis. (C) K562 cells were pre-treated for 30 min with the indicated inhibitors followed by EPO-treatment for 2 h. (D) K562 cells were transfected, as indicated, with 1 μg of shRNAs for PI-3 kinase, MAP kinase and scRNA followed by EPO treatment for 2 h. (E) Cells were transfected, as indicated, with 1 μg of shRNAs for the transcription factor HIF-1α, scRNA and PHD2 followed by treatment with EPO for 2 h. In panels (A–D) total RNA was subjected to quantitative qRT-PCR for PlGF mRNA. PlGF mRNA expression was normalized to GAPDH mRNA levels. Data are expressed as means ± S.D. of three independent experiments. (F) Western Blot analysis of HIF-1α protein in K562 cells treated with the indicated pharmacological inhibitors followed by EPO treatment for 2 h. All Western blots were stripped and reprobed with β-actin to normalize protein loading. Data are representative of three independent experiments. ***P < 0.001, **P < 0.005, *P < 0.01, ns, P > 0.05.
Next, we examined the cellular signalling pathways required for induction of EPO-mediated PlGF expression, utilizing established pharmacological inhibitors for known signalling kinases [36]. An inhibitor screen was performed using inhibitors for phosphoinositide (PI)-3 kinase (LY294002, 15 μM), p38 mitogen-activated protein (MAP) kinase (SB203580, 1 μM) and extracellular signal-regulated kinase (ERK)/MAP kinase (PD98059, 10 μM); all drugs completely antagonized EPO-mediated PlGF induction (Figure 1C). An inhibitor of Jun-N-terminal kinase (JNK), SP600125 (100 nM), was tested and observed to reduce EPO-induction of PlGF expression by ~40 μ (Figure 1C). An inhibitor for the transcription factor, HIF-1α, R59949 (30 μM) also attenuated PlGF mRNA expression (Figure 1C, lane 7). Pharmacological inhibitors may have non-specific effects; therefore we validated our results by utilizing kinase specific shRNA. As shown in Figure 1(D), shRNAs for PI-3 kinase and MAP kinase (ERK-1 and ERK-2) inhibited by >90 μ the EPO-induced expression of PlGF mRNA. It is pertinent to note that shRNAs for PI-3 kinase and MAP kinase (ERK-2) were effective in attenuating expression of these proteins by 50 μ as shown in the Supplementary Figures S1(A) and S1(B). Taken together, these data showed EPO-mediated signalling for PlGF expression involved PI3 kinase as well as previously shown p38 MAP kinase, MAP kinase and JNK pathways [30].
EPO-mediated expression of PlGF involves activation of HIF-1α
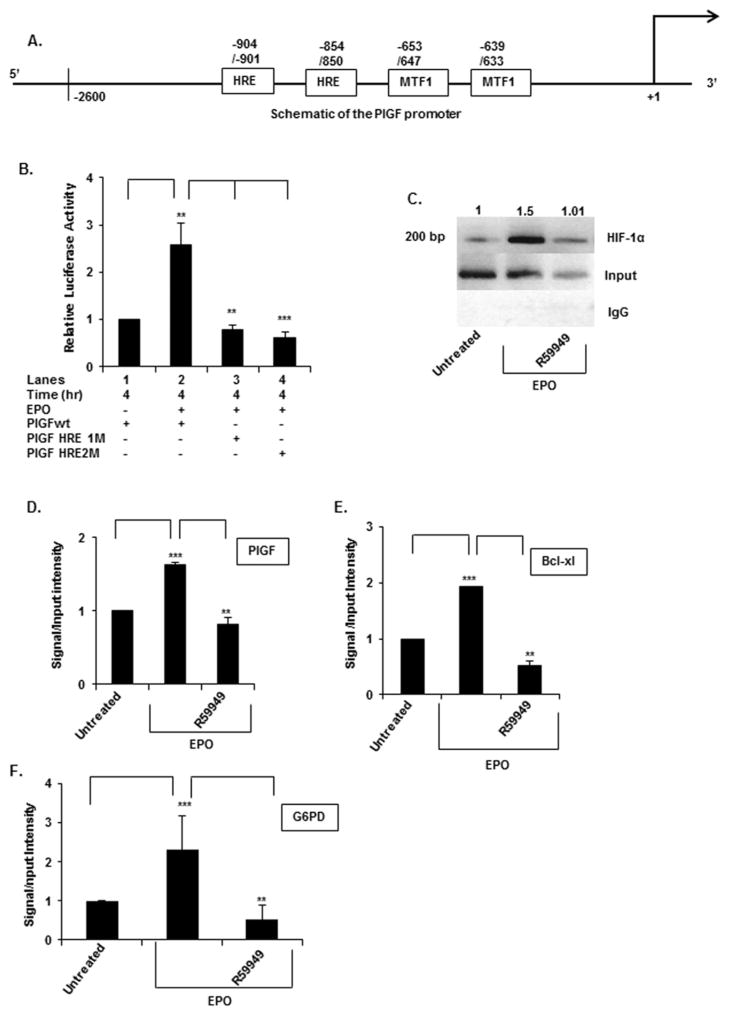
In silico analysis of the PlGF promoter showed the presence of two canonical-binding sites for HIF-1α [hypoxia response elements (HREs)], at nt positions − 854/− 850 and − 904/− 900 as illustrated in the schematic diagram of Figure 2(A). Additionally, the PlGF promoter has two binding sites for metal transcription factor, MTF-1 [10], at positions − 639/− 633 and − 647/− 653 (Figure 2A). We examined whether EPO induction of PlGF required participation of HIF-1α.
Figure 2. PlGF promoter activity requires the activity of HIF-1α-binding sites (HREs).
(A) Schematic diagram of the PlGF promoter (nt positions − 2622/+ 10, relative to transcription start site) showing two HRE and two MTF-1-binding sites. (B) K562 cells were transfected with the wt PlGF promoter-luc, PlGF promoter with mutations in the HRE site at positions − 854/− 850 (HRE1M) and at positions − 904/− 901 (HRE2M). K562 cells were co-transfected with Renilla luciferase plasmid for 24 h. Cells were treated with EPO for 4 h. Luciferase activity was normalized to Renilla luciferase activity to correct for transfection efficiency and the data are expressed as relative expression compared with either the luciferase activity of untreated cells or the wt construct, as indicated in the figure. Data are means ± S.D. of three independent experiments. (C) ChIP analysis of K562 cells treated without and with EPO for 2 h, for assay of HIF-1α binding to the PlGF promoter. HIF-1α antibody (top row) or control rabbit IgG (bottom row) were used for immuno-precipitation of soluble chromatin. The middle panel represents the amplification of input DNA before immunoprecipitation. (D–F) Quantitative analysis of PlGF (D) (ChIP, panel C), the Bcl-XL (E) and the G6PD (F) promoters by qPCR of CHIP utilizing HIF-1α antibody. Primers used to amplify the PCR products in the PlGF, Bcl-XL and the G6PD promoter are indicated in Table 1. Data are representative of three independent experiments. ***P < 0.01, **P < 0.01, *P < 0.05, ns, P > 0.05.
Knockdown of HIF-1α in K562 cells with transfected shRNA vector was performed prior to EPO-treatment for 2 h. This resulted in ~90 μ reduction in PlGF mRNA level relative to control (Figure 1E, lane 3 compared with lane 2). Western blot analysis showed HIF-1α shRNA was ~80 μ effective in reducing EPO mediated HIF-1α expression (Supplementary Figure S1C). To further elucidate the role of HIF-1α in EPO-mediated PlGF expression, we utilized a shRNA for prolyl hydroxylase-2 (PHD2). Under basal conditions, in ambient oxygen, PHD2 catalyses hydroxylation of specific proline residues of HIF-1α, marking it for ubiquitination and subsequent degradation [37]. In the absence of EPO, PHD2 shRNA increased expression of PlGF mRNA by ~3.5-fold, when compared with the untreated cells (Figure 1E, lane 4 compared with lane 1). PHD-2 shRNA was ~50 μ effective in reducing PHD2 protein (Supplementary Figure S1D). Cells transfected with a scrambled non-specific shRNA (scRNA) showed no effect on PlGF mRNA levels (Figure 1E, lane 5 compared with lane 2).
We further examined HIF-1α expression in order to ascertain whether EPO contributed to transcription of HIF-1α itself or regulated it via a post-translational mechanism. As shown in Figure 1(F), EPO increased expression of HIF-1α protein from basal levels, which was significantly inhibited by the inhibitors of PI-3 Kinase (LY294002), MAP kinase (PD 98059) and JNK kinase (SP600125). R59949, an inhibitor of HIF-1α, also significantly inhibited EPO-mediated HIF-1α protein levels (Figure 1F).
In order to determine whether increased HIF-1α levels mediated by EPO resulted from decreased proteasomal turnover, we treated cells with cycloheximide to specifically examine its turnover in the presence and absence of EPO.
Under these conditions, there was measurable HIF-1α turnover in the presence of EPO compared with control; however, in both conditions the t1/2 was observed to be considerably longer than 6 h (Supplementary Figure S1E). Given the 2 h EPO incubation period used in these studies, the amount of protein turnover would be limited. Thus, we concluded that the EPO-dependent net increase in HIF-1α was due to increased rates of HIF-1α mRNA translation [38,39]. This interpretation is consistent with observed effects of specific protein kinase inhibitors on HIF-1α levels in response to EPO described above. Specific and general stimulation of mRNA translation is known to be mediated through PI-3 kinase acting through mammalian target of rapamycin C (mTORC) [40,41].
EPO-mediated expression of PlGF in erythroid cells involves HRE elements in its promoter
We examined the role of HIF-1α in the transcription of PlGF using a luciferase reporter driven by the PlGF promoter. For these studies, the 2.6-kb PlGF-promoter region were used to test the response of the luciferase reporter to EPO. As described above, the presumptive HRE elements found in the PlGF promoter were individually mutated to establish their role(s) in response to EPO treatment. The HRE sites were mutated as follows: HRE-1 (RCGTG to RCATA, bases − 854/− 850) and HRE-2 (RGCAC to RGATA, bases − 904/− 900), as illustrated in the gene schematic diagram of Figure 2(A). Transfection of wt PlGF-luc in K562 cells followed by treatment with EPO for 4 h resulted in ~2.5-fold increase in luciferase activity (Figure 2B). Moreover, mutation of either the HRE-1 (Figure 2B, lane 3 compared with lane 2) or the HRE-2 (Figure 2B, lane 4 compared with lane 2) sites completely abrogated EPO induction of the PlGF promoter-luciferase activity. Taken together, these data showed that both the HRE-1 and the HRE-2 sites were equally functional and therefore together played a role in the EPO-mediated transcription of PlGF in erythroid cells.
The requirement for HIF-1α in PlGF induction by EPO was further supported by ChIP analysis of the endogenous gene. As shown in Figure 2(C), HIF-1α binding to the PlGF promoter was stimulated in EPO treated K562 cells by ~1.5-fold (Figure 2C), compared with untreated cells normalized to input DNA. The expected ChIP PCR product size using primers listed in Table 1 was 200 bp, corresponding to the PlGF 2.6-kb promoter region containing HRE-1 and HRE-2 (Figure 2C). Control ChIP reactions with non-specific antibody did not enrich recovery of the promoter segment containing the HRE elements (Figure 2C, bottom panel). As an alternative approach to support the requirement for HIF-1α, pre-incubation of K562 cells with R59949 (an inhibitor of HIF-1α) was observed to attenuate EPO-mediated HIF-1α binding to the PlGF promoter (Figure 2C). HIF-1α bound to the PlGF promoter in the chromatin was quantified using qPCR (Figure 2D). As a positive control for the ChIP result, we examined the expression of the B-cell lymphoma-extra large (Bcl-XL) gene, which is regulated by HIF-1α and induced by EPO in erythroid cells [42]. The qPCR analysis for Bcl-XL showed an increase of ~2-fold following EPO treatment that was abrogated by pre-treatment with HIF-1α inhibitor R59949 (Figure 2E). These data supported the participation of HIF-1α in transcription of PlGF. We also examined the expression of the glucose-6-phosphate dehydrogenase gene, which is also regulated by HIF-1α [43]. The qPCR result for this gene showed an increase of ~2-fold following EPO treatment which was also inhibited by R59949 (Figure 2F).
miR-214 targets the 3′-UTR of PlGF mRNA
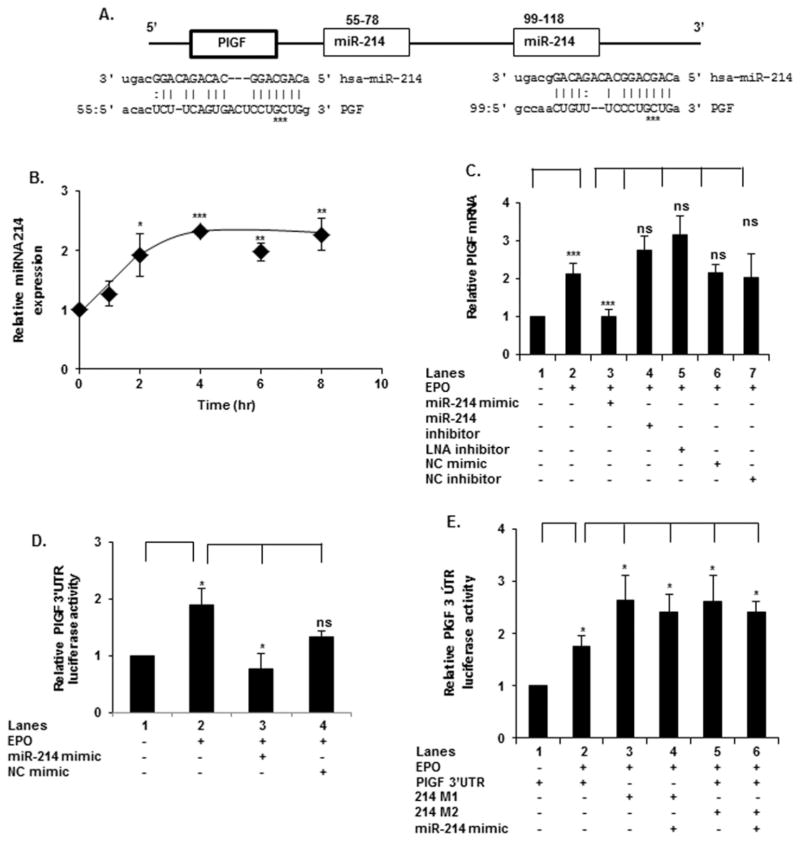
In silico analysis of the 3′-UTR of human PlGF mRNA showed the presence of two predicted complementary-binding sites for miR-214 at nt + 55/+ 78 and + 99/+ 118, beyond the translation stop codon (Figure 3A). These potential miR-214 target sites in PlGF mRNA are highly conserved among several mammalian species (result not shown). In order to correlate miR-214 expression to PlGF mRNA levels, we first examined EPO-mediated regulation of mature miR-214 levels. EPO treatment led to an increase in miR-214 in a time-dependent manner achieving a maximum by 2 h, which was maintained through 8 h of incubation with EPO (Figure 3B).
Figure 3. miR-214 post-transcriptionally regulates EPO-mediated PlGF expression.
(A) Schematic diagram of the PlGF 3′-UTR, depicting two binding sites for miR-214. Mutations in the miR-214-binding sites are indicated by asterisks. (B) K562 cells were treated with EPO for indicated time periods followed by qRT-PCR for miR-214 expression. miR-214 levels were normalized to RNU6 RNA levels. Data are represented as relative expression of miR-214, following EPO treatment, compared with untreated samples. (C) K562 cells were transfected with indicated oligonucleotide miR-214 mimic, miR-214 inhibitor (antagomir) or LNA miR-214 inhibitor, followed by EPO treatment for 2 h. (D) Cells were co-transfected with a PlGF 3′-UTR luciferase plasmid and the miR-214 mimic followed by treatment with EPO for 4 h. (E) K562 cells were transfected with either the wt PlGF 3′-UTR luciferase plasmid or the PlGF 3′-UTR plasmid with miR-214-binding site mutations at positions + 55/+ 78 (214M1) or at positions + 99/+ 118 (214M2), as indicated, along with Renilla luciferase plasmid. Data are means ± S.D. of three independent experiments. ***P < 0.001, **P < 0.01, *P < 0.05, ns, P > 0.05.
In order to establish a possible role of miR-214 in the post-transcriptional regulation of PlGF, we transfected K562 cells with a miR-214 mimic and observed a complete reduction in EPO-mediated PlGF mRNA expression to basal levels (Figure 3C, lane 3 compared with lane 2). Transfection with miR-214 mimic inhibitor did not alter expression of PlGF mRNA (Figure 3C, lane 4 compared with lane 2). Moreover, transfection with LNA miR-214 inhibitor also did not significantly change PlGF mRNA levels (Figure 3C, lane 5 compared with lane 2). Control transfection with a non-specific mimic (NC) had no effect on PlGF mRNA level (Figure 3C, lane 6 and 7 compared with lane 2). We conclude from these results that PlGF has functional miR-214-binding sites in its mRNA 3′-UTR.
The effect of miR-214 on PlGF synthesis was further analysed using a luciferase translation reporter assay. Transfection of K562 cells with the pMIR–PlGF-3′-UTR-luc (wt) reporter followed by treatment with EPO for 4 h resulted in a 2-fold increase in luciferase activity (Figure 3D, lane 2 compared with lane 1); this indicated that the PlGF 3′-UTR participated in post-transcriptional regulation of PlGF. Co-transfection of the wt-PlGF-3′-UTR with miR-214 mimic (Figure 3D, lane 3 compared with lane 2), reduced EPO-induced luciferase activity to basal levels; the reporter activity was not affected by the NC miRNA (Figure 3D, lane 4 compared with lane 2), demonstrating the specificity of this interaction. Next, the specificity of miR-214 interaction with PlGF 3′-UTR in the translation reporter for post-transcriptional regulation of PlGF mRNA was examined. For this, we introduced three base substitutions within both predicted miR-214-binding sites in the PlGF 3′-UTR (from CCTGCTGG to CCAAATGG) of the luciferase translation reporter. These base substitutions will abolish possible base pairing between miR-214 and the PlGF 3′-UTR sequence, as shown in the schematic diagram (Figure 3A). The mutant luciferase translation reporters were assayed for luciferase production in EPO-treated K562 cells. The data showed mutation of miR-214-binding site 1, (positions + 55/+ 78; denoted 214M1, Figure 3E, lane 3) or miR-214-binding site 2 (positions + 99/+ 118; denoted 214M2, Figure 3E, lane 5) individually were refractory to the effect of elevated endogenous miR-214 at >2 h post EPO addition. Furthermore, luciferase expression for 214M1 and 214M2 mutants was significantly elevated compared with the activity of wt 3′-UTR (Figure 3E, lanes 5 and 6 compared with lane 2), which would be expected in the absence of post-transcriptional regulation of the mutant reporter mRNA. Thus, we interpreted this result to indicate that both miR-214 sites were required for reducing reporter activity and by extension, PlGF expression itself in vivo. In addition, co-transfection of the mutant luciferase reporters with miR-214 mimic to augment endogenous miR-214 levels had no effect on reporter activity (Figure 3E, lane 4 compared with lane 3 and lane 6 compared with lane 5) consistent with the predicted interaction between miR-214 and the PlGF 3′-UTR.
miR-214 located in an intron of host gene DNM3 is co-transcriptionally regulated by PPARα
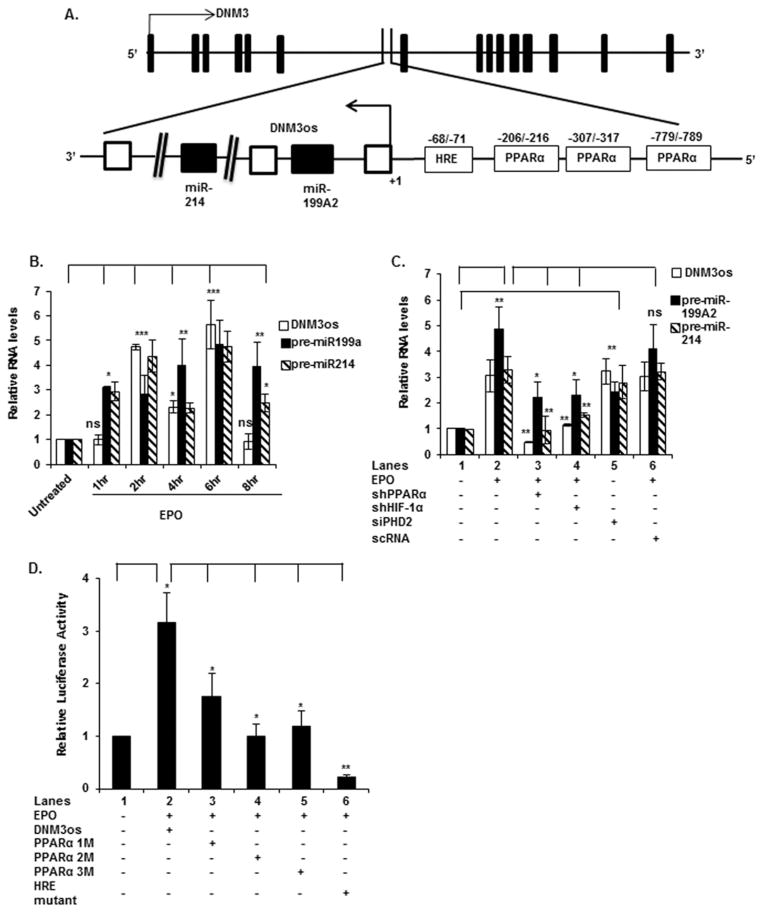
Recent studies from our laboratory [44] and others [45] show that the miR-199a2/214 cluster is located within DNM3os (chr1:172109461-172113934), an opposite strand locus, within an intron of DNM3 (chr1:171810621-172381856). The former encodes a long non-coding transcript, as depicted in the schematic diagram of Figure 4(A). We have shown that induction of DNM3os by PlGF takes place in human endothelial cells; furthermore, this gene could be independently induced by PPARα in this cell type [44]. Accordingly, we examined EPO-mediated transcription of DNM3os RNA and concomitantly miR-214/miR-199a2 in erythroid cells (K562). We also examined whether PPARα-induced DNM3os transcription in these cells. As shown in Figure 4(B), EPO treatment of K562 cells showed a time-dependent increase in DNM3os, premiR-199a2 and premiR-214 RNA expression. The EPO-mediated expression of premiR-199a2/-214 and DNM3os RNAs increased biphasically, showing separate maxima of expression after 2 and 6 h of EPO treatment (Figure 4B). However, CFU-E cells treated with EPO showed reduced expression of premiR-214 (Figure 1B).
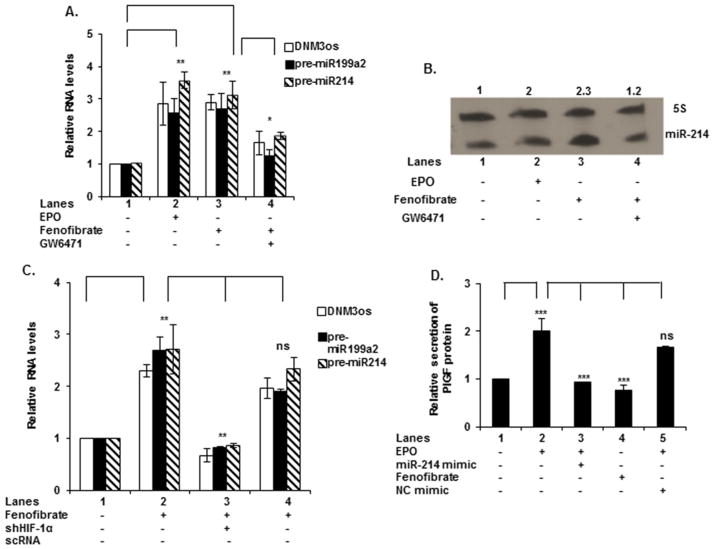
Figure 4. EPO-induced expression of DNM3os RNA, premiR-214 and premiR-199a2 requires the activity of HIF-1α and PPARα.
(A) Schematic diagram of the DNM3os promoter, depicting three binding sites for PPARα and one site for HIF-1α (HRE). Base positions are indicated relative to transcription start site. (B) K562 cells were treated with EPO for the indicated time periods. (C) Cells were transfected with shRNAs for PPARα and HIF-1α followed by EPO treatment for 2 h or transfected with shRNA for PHD2. For (B and C) RNA was subjected to quantitative qRT-PCR utilizing primers listed in Table 1. DNM3os, premiR-199a2 and premiR-214 RNA expression was normalized to GAPDH mRNA levels. qRT-PCR data represent relative levels of DNM3os RNA and premiR-199a2/-214 expression following treatment with EPO compared with untreated cells. (D) Cells were transfected with either the wt DNM3os promoter luciferase construct or the DNM3os promoter constructs with mutations in the PPARα-binding sites at positions − 216/− 206 (PPAR1M), − 317/− 307 (PPAR2M) and − 789/− 779 (PPAR3M) or the HRE site at positions − 71/− 68 (HRE Mut) and treated with EPO for 4 h. Data are expressed as means ± S.D. of three independent experiments. ***P < 0.001, **P < 0.01, *P < 0.05, ns, P>0.05.
Next we examined the transcriptional regulation of miR-214. Although this study focused on expression of miR-214, the DNM3os transcription unit includes the gene for miR-199a2 [44]. In silico analysis of the ~2-kb 5′-flanking region of DNM3os showed three PPARα-binding sites (bases − 216/− 206, bases − 317/− 307 and bases − 789/− 779) and one HRE site (bases − 68 to − 71) proximal to the transcription start site (Figure 4A).
Thus, we examined these cis elements for their roles in the transcriptional regulation of DNM3os as well as premiR-214, in response to EPO treatment. As shown in Figure 4(C), transfection of K562 cells with shRNAs for PPARα and HIF-1α reduced expression of DNM3os RNA and premiR-199a2/-214 by ~90 μ (Figure 4C, lanes 3 and 4 compared with lane 2), whereas a non-specific control (control shRNA) had no significant effect (Figure 4C, lane 6 compared with lane 2). The activity of these shRNAs on corresponding protein synthesis was confirmed by Western blot (Supplementary Figure S1E). PPARα shRNA was ~40 μ effective in reducing PPARα protein expression (Supplementary Figure S1E). In the absence of EPO, PHD-2 shRNA, increased the expression of DNM3os, premiR-199a2 and premiR-214 (Figure 4C, lane 5 compared with lane 1). These results demonstrated that transcription of DNM3os was dependent on PPARα and HIF-1α.
To further determine the contribution of PPARα and HIF-1α in the expression of miR-214 and DNM3os, we mutated the PPARα and HIF-1α-binding sites of the DNM3os promoter. K562 cells transfected with the wt DNM3os promoter-luciferase reporter showed a ~3-fold increase in luciferase activity in response to EPO treatment (Figure 4D, lane 2 compared with lane 1). Mutation of PPARα site-1 (Figure 4D, lane 3 compared with lane 2) partially reduced the activity of DNM3os promoter in the luciferase reporter in response to EPO. By contrast, mutations in PPARα site- 2 (Figure 4D, lane 4 compared with lane 2) and PPARα site-3 (Figure 4D, lane 5 compared with lane 2) significantly abrogated reporter activity in response to EPO. Mutation of the single HRE site (nts − 68/− 71) in the DNM3os promoter (Figure 4A) exhibited the highest reduction in reporter activity when compared with the wt DNM3os promoter (Figure 4D, lane 6 compared with lane 2). These results implicated both PPARα and HIF-1α as positive regulators of DNM3os and premiR-214 transcription, with the latter providing the largest contribution to DNM3os transcription.
Fenofibrate augments expression of miR-214 and concomitantly attenuates expression of PlGF
Since PPARα and HIF-1α were involved in transcription of DNM3os, we examined the effect of fenofibrate, a PPARα agonist, on the expression of DNM3os RNA, premiR-199a2 and pre-miR-214. Fenofibrate is a Food and Drug Administration (FDA) approved drug from the fibrate class, which is widely utilized for the treatment of dyslipidemia [46]. As shown in Figure 5(A) (lanes 2 and 3 compared with lane 1), both EPO and fenofibrate increased by ~3-fold the expression levels of DNM3os, premiR-199a2 and premiR-214 RNA. A PPARα antagonist (GW6471, 15 μM) attenuated by >80 μ the expression of DNM3os RNA, premiR-199a2/premiR-214 (Figure 5A, lane 4 compared with lane 3). These results were also reflected in a Northern blot analysis for miR-214, wherein fenofibrate treatment of K562 cells, in the absence of EPO, induced mature miR-214 expression by ~2.3-fold, whereas addition of the PPARα antagonist (GW6417) inhibited the expression of miR-214 (Figure 5B). By comparison, treatment with EPO alone resulted in a 2-fold increase in the expression of mature miR-214 (Figure 5B). Thus, both EPO and fenofibrate treatment augmented miR-214 expression.
Figure 5. Fenofibrate-induced expression of DNM3os RNA, premir-199a2 and premir-214 and PlGF secretion.
(A) K562 cells were pre-treated with GW6471 (15 μM) for 30 min, where indicated, followed by treatment with either fenofibrate (100 μM) or EPO for 2 h. (B) Northern blot for expression of miR-214. Cells were treated with EPO, fenofibrate or GW6471 for 2 h. Total RNA was isolated and subjected to electrophoresis for Northern blots as described in Materials and Methods. miR-214 levels were normalized to that of 5S RNA levels. (C) K562 cells were transfected with shRNA for HIF-1α prior to treatment with fenofibrate for 2 h. In (A and C) total RNA was used for qRT-PCR. qRT-PCR data represent relative expression levels of DNM3os, premiR-199a2 and premiR-214 RNA following treatment compared with untreated cells. (D) Secretion of PlGF protein from K562 cells in response to overnight transfection with miR-214 mimic, followed by treatment with either EPO or fenofibrate for 24 h. Cell culture supernatants were harvested and secreted PlGF protein was assayed by ELISA; data are normalized to total protein content. Data are representative of three independent experiments. ***P < 0.001, **P < 0.01, *P < 0.05, ns, P>0.05.
To ascertain the role of HIF-1α in the transcription of DNM3os and synthesis of premiR-199a2/214, K562 cells were transfected with shRNA for HIF-1α. As shown in Figure 5(C), fenofibrate-induced synthesis of DNM3os, premiR-199a2 and premiR-214 could be reduced to basal levels by HIF-1α shRNA (Figure 5C, lane 3 compared with lane 2). There was no significant effect on DNM3os RNA levels upon transfection with a non-specific control shRNA (Figure 5C, lane 4 compared with lane 2). These data indicated that both PPARα and HIF-1α co-activated DNM3os transcription.
Next, we examined the effect of EPO and fenofibrate on PlGF synthesis and secretion from erythroid cells. EPO treatment resulted in ~2-fold increase in the release of PlGF from K562 cells (Figure 5D, lane 2 compared with lane 1) and was antagonized by transfection with miR-214 mimic, which reduced PlGF secretion to basal level, as expected from its post-transcriptional effect on PlGF mRNA (Figure 5D, lane 3 compared with lane 2). Under the same conditions, transfection with a non-specific mimic did not significantly reduce PlGF secretion (Figure 5D, lane 5 compared with lane 2). Moreover, PlGF secretion was also significantly reduced by fenofibrate (Figure 5D, lane 4 compared with lane 2). Taken together, these data showed that both PPARα and HIF-1α regulated the transcription of miR-214 and fenofibrate augmented expression of miR-214 and concomitantly reduced PlGF mRNA and protein expression.
EPO-mediated signalling involves phosphorylation of PPARα
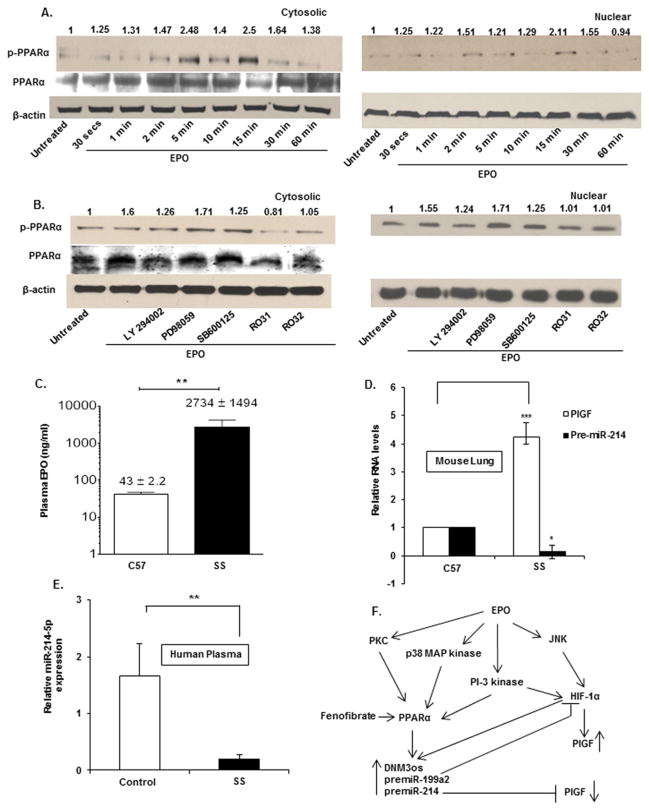
As PPARα increased transcription of DNM3os and miR-214, we examined the mechanisms by which PPARα may be activated in response to EPO. We observed EPO treatment did not significantly increase PPARα mRNA levels (result not shown), therefore any change in PPARα activity could be achieved by a post-translational mechanism. The activity of PPARα is known to be modulated by phosphorylation [47]. Thus, we asked if the phosphorylation state of PPARα changed during EPO treatment. K562 cells were treated with EPO and analysed for the appearance of phosphorylated PPARα over a 30 min time course. Under these conditions, we observed that PPARα phosphorylation increased by ~2.5-fold between 5 and 15 min in both cytosolic and nuclear fractions (Figure 6A). Accordingly, whereas the amount of cytosolic PPARα protein did not change between 2 and 60 min (Figure 6A, middle panel), unphosphorylated PPARα protein was not observed in the nuclear fraction of EPO treated cells during the induction time period (result not shown).
Figure 6. EPO-induced miR-214 expression required the phosphorylation of PPARα.
(A) Time course of PPARα phosphorylation. Cells were treated with EPO for indicated time periods and cell lysates were fractionated into cytosolic and nuclear fractions. Each cell fraction was subjected to Western blot analysis utilizing antibodies to phosphorylated PPARα and unphosphorylated PPARα (middle panel). (B) Effect of pharmacologic inhibitors on PPARα phosphorylation and total PPARα levels. Blots were stripped and reprobed for β-actin to demonstrate equal loading. The data are representative of three independent experiments. (C) The concentrations of EPO in the plasma of sickle mice (SS, n = 4) and control mice (C57, n = 4) were quantified using ELISA. (D) Total RNA was isolated from the lungs of sickle (SS; n = 4) and control mice (C57; n = 4) and subjected to qRT-PCR. PlGF and premiR-214 levels were normalized to GAPDH mRNA levels. (E) The plasma concentrations of miR-214 from sickle patients (SS; n = 13) and corresponding sibling controls (n = 8) were quantified using the TaqMan miRNA assay. Levels of miR-214 were normalized to levels of miR-16 in patient plasma. (F) Schematic diagram of the proposed signalling and regulatory pathways involved in EPO-mediated expression of PlGF mRNA and miR-214. EPO leads to HIF-1α activation and up-regulates the expression of PlGF. EPO signals phosphorylation of PPARα, which transactivates the DNM3os promoter leading to increased expression of miR-199a2 and miR-214. miR-199a2 targets the 3′-UTR of HIF-1α whereas miR-214 targets the 3′-UTR of PlGF. Fenofibrate (agonist) activates PPARα, which in turn increases transcription of DNM3os RNA, co-expressing premiR-199a2 and premiR-214. Increased levels of miR-214 attenuate PlGF levels. ***P < 0.001, **P < 0.01, *P < 0.05, ns, P > 0.05.
In order to gain insight into which protein kinase activities participated in PPARα phosphorylation, we utilized selective pharmacological protein kinase inhibitors which have been previously shown to modulate PPARα phosphorylation [47]. We observed PI-3 kinase, p38 MAP kinase and protein kinase C α (PKCα) inhibitors (Ro31 and Ro32), but not MAP kinase inhibitors, were effective in attenuating phosphorylation of PPARα in the nuclear fraction of K562 cells (Figure 6B), though the PPARα unphosphorylated levels were constant (Figure 6B, middle panel). Taken together, these data showed EPO treatment increased phosphorylation of PPARα and the presumptive pathway involved in its activation involved PI-3 kinase, p38 MAP kinase and members of PKC-family of activators, but not MAP kinase.
PremiR-214 levels inversely correlate with PlGF mRNA levels in lung tissue of Berkeley sickle mice
The present study showed the existence of a regulatory circuit between miR-214 and PlGF in K562 cells. Our previous studies showed significantly elevated levels of circulatory PlGF and ET-1 in BK-SS mice, both of which are associated with right ventricular hypertrophy and pulmonary changes reflecting what is observed in SCD patients [21]. Therefore, this system was revisited to measure plasma levels of EPO in BK-SS mice. As expected, we found that EPO levels were ~64-fold higher in BK-SS mice compared with control mice (Figure 6C), indicating that sickling correlated with increased expression of EPO.
Since PHT is observed in sickle patients, we hypothesized that lungs of BK-SS mice may also express higher levels of PlGF. Indeed, lung tissue from BK-SS mice showed ~4-fold increase in PlGF mRNA expression compared with lung tissue derived from C57BL/6NJ control mice (Figure 6D). Consistent with the increase in PlGF, premiR-214 levels were significantly reduced in BK-SS lung tissue to very low basal expression levels when compared with control mice (Figure 6D).
Reduced miR-214 plasma levels correlate with increased PlGF levels in SCD patients
Plasma levels of PlGF are abnormally high in SCD subjects [10]. Consequently the correlation of reduced miR-214 expression to high PlGF levels we observed in vitro, led us to measure plasma levels of miR-214 in SCD patients and their sibling(s), as controls. As shown in Figure 6(E), this analysis showed miR-214 levels were ~8-fold lower in SCD patients compared with healthy sibling controls. Thus these clinical data coupled with the animal data above, strongly support the role of miR-214 in the regulation of PlGF levels. Lower levels of miR-214, as evident from our data in SCD patients, was probably contributory to increased levels of PlGF in SCD subjects, as was shown in vitro.
DISCUSSION
In the present report, we delineated the transcriptional and post-transcriptional mechanisms that underlie EPO-mediated induction of PlGF in erythroid cells. Using pharmacological inhibitors and RNAi, we showed that the canonical PI-3 kinase pathway, the MAP kinase, p38 MAP kinase and the JNK kinase pathways were all participants in EPO-mediated induction of PlGF. The PlGF promoter has two HREs that serve as binding sites for the transcription factor, HIF-1α, as previously reported [48,49]. A detailed analysis of the PlGF promoter showed a functional requirement for both HRE sites in EPO-mediated PlGF transcription. Moreover, this result was extended wherein we showed in vivo binding of HIF-1α by ChIP analysis to the endogenous PlGF promoter.
miRNAs are known to have an important role in controlling gene expression by affecting the stability and translation of mRNA through base-pairing interactions with the 3′-UTRs of these translation templates [50–53]. In silico analysis of the 3′-UTR of PlGF predicted two potential complementary binding sites for miR-214, at nt positions + 55/+ 78 and + 99/+ 118. We observed miR-214 levels showed an inverse correlation to the levels of PlGF mRNA upon EPO-mediated induction. These results were corroborated wherein overexpression of miR-214 showed attenuation of EPO-induced PlGF promoter luciferase reporter activity, PlGF mRNA expression and resultant PlGF protein secretion. The specificity of miR-214 binding to miRNA response elements (MREs) in the PlGF mRNA was established using a luciferase reporter gene containing the PlGF 3′-UTR. Furthermore, the functionality of these sequences was established by mutation of the binding sites for miR-214 within the 3′-UTR of the luciferase reporter.
The miR-199a2/miR-214 cluster is located within the DNM3os transcription unit and is indispensable for skeletal development and body growth in mammals [54,55]. Results from RNAi of PPARα and HIF-1α activity in these cells indicated that both transcription factors were important for DNM3os transcription and that both PPARα and HIF-1α have major roles in transcription of this locus.
The transactivation of the DNM3os promoter by PPARα and HIF-1α was somewhat surprising because increased miR-214/miR-199a2 expression would be expected to attenuate both the induction of HIF-1α, via regulation through miR-199a2 [44], and subsequently PlGF. We propose that this is actually part of a homoeostatic regulatory loop for the return of PlGF and HIF-1α to basal levels of expression via a post-transcriptional mechanism mediated by miRNAs. Given that PlGF levels are abnormally high in SCD, this implies that this homoeostatic regulatory arm is dysregulated in this chronic illness.
Phosphorylation of PPARα is essential to its function as a transcription factor and is preceded by activation of the PKCα pathway [47]. EPO activated PPARα by inducing its phosphorylation, required activation of PI-3 kinase and the PKC pathway. This would lead to transactivation of the DNM3os promoter, thereby initiating transcription of the DNM3os transcription unit including the miR-199a2/miR-214 cluster. Participation of PI-3 kinase and MAP kinases may also be important for stimulating translation of HIF-1α mRNA as has been previously reported [56,57]. We observed a similar phenomenon based on our observation that there was no net change in HIF-1α mRNA levels, yet EPO treatment resulted in a 2-fold increase in HIF-1α protein. This occurred despite the observation that there was a small increase in HIF-1α turnover during EPO treatment (Supplementary Figure S1F). Modulation of protein synthesis rates in the absence of increases in mRNA levels is a well-established phenomenon [58].
HIF-1α protein levels are primarily regulated by proteasomal degradation under normoxic conditions. Following hypoxia, the proteasomal-dependent pathway is suppressed resulting in increased cytoplasmic levels of HIF-1α leading to nuclear translocation and gene activation events. However, basal HIF-1α levels are also influenced by increased rates of HIF-1α mRNA translation [56]. It has been reported that HIF-1α mRNA is alternatively initiated for protein synthesis by a 5′-cap independent process [57] and by positively acting, trans-acting RNA-binding proteins, poly pyrimidine tract-binding protein (PTB) and human antigen R (HuR) [59,60]. Thus the changes in HIF-1α levels we observed in response to EPO could be due to participation of non-canonical translation pathways leading to the net increase in this key transcription factor [58]. Such changes in HIF-1α levels, not involving the proteasomal pathway, have been reported by others for non-hypoxic stimuli [61].
Plasma levels of PlGF are significantly higher in sickle cell patients compared with healthy control subjects, as a part of the compensatory erythroid hyperplasia response. The higher levels of PlGF in patients with SCD are significantly associated with haemolysis, baseline inflammation and features of PHT [10,21,62]. Recent studies of Kato and co-workers [63] also show correlation of markers of iron overload, i.e. ferritin and transferrin, to increased plasma levels of PlGF in patients with haemolytic anaemia, compared with healthy controls. In cultured endothelial cells and monocytes, PlGF induces HIF-1α-dependent expression of ET-1, PAI-1 and 5-lipoxygenase activating protein [19,29]. The role of miR-214 in regulating expression of PlGF levels in vivo was supported by our findings wherein miR-214 levels were significantly reduced in plasma of SCD patients and lung tissues of BK-SS mice compared with normal subjects and control animals respectively. Thus, the correspondingly high levels of PlGF observed in SCD patients and the sickle mouse model corroborated the in vitro results, further buttressing this novel mechanistic link between PlGF and miR-214. Our studies additionally showed that the miR-199a2/miR-214 cluster, co-synthesized with the non-coding DNM3os transcript, could be induced by the PPARα agonist, fenofibrate. Recent studies show increased expression of miR-199/miR-214 along with DNM3os RNA in biopsies of cardiac tissue from patients with heart failure compared with human control heart tissues [45]. This was accompanied by reduced PPARδ protein levels in cardiac biopsies of heart failure patients due to repression of PPARδ by miR-214 [45].
In conclusion, our data provide evidence for the involvement of PPARα and HIF-1α in EPO-mediated PlGF expression and the role of miR-214 as a post-transcriptional regulator of PlGF mRNA, as illustrated in the schematic diagram shown in Figure 6(F). Moreover, PPARα agonists were shown to augment the expression of miR-214 and concomitantly reduced the expression of PlGF. These studies provide a rational therapeutic approach of utilizing FDA approved fibrates [64] as a potential therapeutic agent for ameliorating the pathological consequences of dysregulated PlGF production in SCD.
Acknowledgments
FUNDING
This work was supported by the National Heart, Lung, and Blood Institute and Analytical–Metabolic Instrumentation Core [grant number R01-HL111372]; and the University of Southern California Research Center for Liver Disease Grant [grant number P30-DK048522].
The content is solely the responsibility of the authors and does not necessarily represent the official views of NHLBI or the National Institute of Health.
Abbreviations
- BAC
bacterial artificial chromosome
- BK-SS
Berkeley sickle mice
- CFU-E
erythroid colony forming units
- DNM3
dynamin 3
- DNM3os
dynamin 3 opposite strand
- EPO
erythropoietin
- ERK
extracellular signal-regulated kinase
- ET-1
endothelin-1
- FDA
Food and Drug Administration
- HIF-1α
hypoxia-inducible factor-1α
- HRE
hypoxia response element
- HRP
horseradish peroxidase
- HuR
Human antigen R
- IMDM
Iscove’s modified DMEM
- JNK
c-Jun N-terminal kinase
- LNA
locked nucleic acid
- MAP
mitogen-activated protein
- MTF-1
metal regulatory transcription factor-1
- NC
negative control
- ns
not significant
- PAI-1
plasminogen activator inhibitor-1
- PHD2
prolyl hydroxylase 2
- PHT
pulmonary hypertension
- PI
phosphoinositide
- PKC
protein kinase C
- PlGF
placental growth factor
- PPARα
peroxisome proliferator-activated receptor-α
- PTB
polypyrimidine tract-binding protein
- qRT
quantitative real-time
- RIPA
radioimmunoprecipitation assay
- SCD
sickle cell disease
- SS
plasma samples from SCD patients
- VEGF
vascular endothelial growth factor
- wt
wild-type
Footnotes
AUTHOR CONTRIBUTION
Caryn Gonsalves performed all in vitro experiments. Chen Li performed Northern blots and analysis of human plasma miRNA levels. Punam Malik and Marthe-Sandrine Mpollo designed human and mouse experiments. Vinod Pullarkat provided the EPO used for experiments. Vijay Kalra and Stanley Tahara designed experiments, supervised analysis and interpreted data. Caryn Gonsalves, Stanley Tahara and Vijay Kalra wrote the manuscript.
References
- 1.Van de Veire S, Stalmans I, Heindryckx F, Oura H, Tijeras-Raballand A, Schmidt T, Loges S, Albrecht I, Jonckx B, Vinckier S, et al. Further pharmacological and genetic evidence for the efficacy of plgf inhibition in cancer and eye disease. Cell. 2010;141:178–190. doi: 10.1016/j.cell.2010.02.039. [DOI] [PubMed] [Google Scholar]
- 2.Kim KJ, Cho CS, Kim WU. Role of placenta growth factor in cancer and inflammation. Exp Mol Med. 2012;44:10–19. doi: 10.3858/emm.2012.44.1.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carnevale D, Lembo G. Placental growth factor and cardiac inflammation. Trends Cardiovas Med. 2012;22:209–212. doi: 10.1016/j.tcm.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 4.Carmeliet P, Moons L, Luttun A, Vincenti V, Compernolle V, De Mol M, Wu Y, Bono F, Devy L, Beck H, et al. Synergism between vascular endothelial growth factor and placental growth factor contributes to angiogenesis and plasma extravasation in pathological conditions. Nat Med. 2001;7:575–583. doi: 10.1038/87904. [DOI] [PubMed] [Google Scholar]
- 5.Yano K, Okada Y, Beldi G, Shih SC, Bodyak N, Okada H, Kang PM, Luscinskas W, Robson SC, Carmeliet P, et al. Elevated levels of placental growth factor represent an adaptive host response in sepsis. J Exp Med. 2008;205:2623–2631. doi: 10.1084/jem.20080398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bobic S, Seys S, De Vooght V, Callebaut I, Hox V, Dooms C, Vinckier S, Jonckx B, Saint-Remy JM, Stassen JM, et al. Placental growth factor contributes to bronchial neutrophilic inflammation and edema in allergic asthma. Am J Resp Cell Mol. 2012;46:781–789. doi: 10.1165/rcmb.2011-0152OC. [DOI] [PubMed] [Google Scholar]
- 7.Oura H, Bertoncini J, Velasco P, Brown LF, Carmeliet P, Detmar M. A critical role of placental growth factor in the induction of inflammation and edema formation. Blood. 2003;101:560–567. doi: 10.1182/blood-2002-05-1516. [DOI] [PubMed] [Google Scholar]
- 8.Cianfarani F, Zambruno G, Brogelli L, Sera F, Lacal PM, Pesce M, Capogrossi MC, Failla CM, Napolitano M, Odorisio T. Placenta growth factor in diabetic wound healing: altered expression and therapeutic potential. Am J Pathol. 2006;169:1167–1182. doi: 10.2353/ajpath.2006.051314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weed S, Bastek JA, Anton L, Elovitz MA, Parry S, Srinivas SK. Examining the correlation between placental and serum placenta growth factor in preeclampsia. Am J Obstet Gynecol. 2012;207:140.e141–140.e146. doi: 10.1016/j.ajog.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Perelman N, Selvaraj SK, Batra S, Luck LR, Erdreich-Epstein A, Coates TD, Kalra VK, Malik P. Placenta growth factor activates monocytes and correlates with sickle cell disease severity. Blood. 2003;102:1506–1514. doi: 10.1182/blood-2002-11-3422. [DOI] [PubMed] [Google Scholar]
- 11.Hauser IA, Johnson DR, Madri JA. Differential induction of VCAM-1 on human iliac venous and arterial endothelial cells and its role in adhesion. J Immunol. 1993;151:5172–5185. [PubMed] [Google Scholar]
- 12.Green CJ, Lichtlen P, Huynh NT, Yanovsky M, Laderoute KR, Schaffner W, Murphy BJ. Placenta growth factor gene expression is induced by hypoxia in fibroblasts: a central role for metal transcription factor-1. Cancer Res. 2001;61:2696–2703. [PubMed] [Google Scholar]
- 13.Tordjman R, Delaire S, Plouët J, Ting S, Gaulard P, Fichelson S, Roméo PH, Lemarchandel V. Erythroblasts are a source of angiogenic factors. Blood. 2001;97:1968–1974. doi: 10.1182/blood.v97.7.1968. [DOI] [PubMed] [Google Scholar]
- 14.De Falco S. The discovery of placenta growth factor and its biological activity. Exp Mol Med. 2012;44:1–9. doi: 10.3858/emm.2012.44.1.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clauss M, Weich H, Breier G, Knies U, Röckl W, Waltenberger J, Risau W. The vascular endothelial growth factor receptor Flt-1 mediates biological activities: implications for a functional role of placenta growth factor in monocyte activation and chemotaxis. J Biol Chem. 1996;271:17629–17634. doi: 10.1074/jbc.271.30.17629. [DOI] [PubMed] [Google Scholar]
- 16.Fischer C, Mazzone M, Jonckx B, Carmeliet P. FLT1 and its ligands VEGFB and PlGF: drug targets for anti-angiogenic therapy? Nat Rev Cancer. 2008;8:942–956. doi: 10.1038/nrc2524. [DOI] [PubMed] [Google Scholar]
- 17.Iyer S, Leonidas DD, Swaminathan GJ, Maglione D, Battisti M, Tucci M, Persico MG, Acharya KR. The crystal structure of human placenta growth factor-1 (PlGF-1), an angiogenic protein, at 2.0 A resolution. J Biol Chem. 2001;276:12153–12161. doi: 10.1074/jbc.M008055200. [DOI] [PubMed] [Google Scholar]
- 18.Duits AJ, Rodriguez T, Schnog JJ. Serum levels of angiogenic factors indicate a pro-angiogenic state in adults with sickle cell disease. Br J Haematol. 2006;134:116–119. doi: 10.1111/j.1365-2141.2006.06103.x. [DOI] [PubMed] [Google Scholar]
- 19.Patel N, Gonsalves CS, Malik P, Kalra VK. Placenta growth factor augments endothelin-1 and endothelin-B receptor expression via hypoxia-inducible factor-1 alpha. Blood. 2008;112:856–865. doi: 10.1182/blood-2007-12-130567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selvaraj SK, Giri RK, Perelman N, Johnson C, Malik P, Kalra VK. Mechanism of monocyte activation and expression of proinflammatory cytochemokines by placenta growth factor. Blood. 2003;102:1515–1524. doi: 10.1182/blood-2002-11-3423. [DOI] [PubMed] [Google Scholar]
- 21.Sundaram N, Tailor A, Mendelsohn L, Wansapura J, Wang X, Higashimoto T, Pauciulo MW, Gottliebson W, Kalra VK, Nichols WC, et al. High levels of placenta growth factor in sickle cell disease promote pulmonary hypertension. Blood. 2010;116:109–112. doi: 10.1182/blood-2009-09-244830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cramer M, Nagy I, Murphy BJ, Gassmann M, Hottiger MO, Georgiev O, Schaffner W. NF-kappaB contributes to transcription of placenta growth factor and interacts with metal responsive transcription factor-1 in hypoxic human cells. Biol Chem. 2005;386:865–872. doi: 10.1515/BC.2005.101. [DOI] [PubMed] [Google Scholar]
- 23.Nishimoto F, Sakata M, Minekawa R, Okamoto Y, Miyake A, Isobe A, Yamamoto T, Takeda T, Ishida E, Sawada K, et al. Metal Transcription Factor-1 is involved in hypoxia-dependent regulation of placenta growth factor in trophoblast-derived cells. Endocrinology. 2009;150:1801–1808. doi: 10.1210/en.2008-0949. [DOI] [PubMed] [Google Scholar]
- 24.Little JA, McGowan VR, Kato GJ, Partovi KS, Feld JJ, Maric I, Martyr S, Taylor JG, Machado RF, Heller T, et al. Combination erythropoietin-hydroxyurea therapy in sickle cell disease: experience from the National Institutes of Health and a literature review. Haematologica. 2006;91:1076–1083. [PMC free article] [PubMed] [Google Scholar]
- 25.Sherwood JB, Goldwasser E, Chilcote R, Carmichael LD, Nagel RL. Sickle cell anemia patients have low erythropoietin levels for their degree of anemia. Blood. 1986;67:46–49. [PubMed] [Google Scholar]
- 26.Satoh K, Kagaya Y, Nakano M, Ito Y, Ohta J, Tada H, Karibe A, Minegishi N, Suzuki N, Yamamoto M, et al. Important role of endogenous erythropoietin system in recruitment of endothelial progenitor cells in hypoxia-induced pulmonary hypertension in mice. Circulation. 2006;113:1442–1450. doi: 10.1161/CIRCULATIONAHA.105.583732. [DOI] [PubMed] [Google Scholar]
- 27.Gordeuk VR, Campbell A, Rana S, Nouraie M, Niu XM, Minniti CP, Sable C, Darbari D, Dham N, Onyekwere O, et al. Relationship of erythropoietin, fetal hemoglobin, and hydroxyurea treatment to tricuspid regurgitation velocity in children with sickle cell disease. Blood. 2009;114:4639–4644. doi: 10.1182/blood-2009-04-218040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin G, Chen R, Alvero AB, Fu HH, Holmberg J, Glackin C, Rutherford T, Mor G. TWISTing stemness, inflammation and proliferation of epithelial ovarian cancer cells through MIR199A2/214. Oncogene. 2010;29:3545–3553. doi: 10.1038/onc.2010.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel N, Sundaram N, Yang M, Madigan C, Kalra VK, Malik P. Placenta growth factor (PlGF), a novel inducer of plasminogen activator inhibitor-1 (PAI-1) in sickle cell disease (SCD) J Biol Chem. 2010;285:16713–16722. doi: 10.1074/jbc.M110.101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonsalves CS, Crable S, Chandra S, Li W, Kalra VK, Joiner CH. Angiogenic growth factors augment K–Cl cotransporter expression in erythroid cells via hypoxia-inducible factor-1α. Am J Hematol. 2014;89:273–281. doi: 10.1002/ajh.23631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonsalves CS, Kalra VK. Hypoxia-mediated expression of 5-Lipoxygenase-activating protein involves HIF-1{alpha} and NF-{kappa}B and MicroRNAs 135a and 199a-5p. J Immunol. 2010;184:3878–3888. doi: 10.4049/jimmunol.0902594. [DOI] [PubMed] [Google Scholar]
- 32.Patel N, Tahara SM, Malik P, Kalra VK. Involvement of miR-30c and miR-301a in immediate induction of plasminogen activator inhibitor-1by placenta growth factor in human pulmopnary endothelial cells. Biochem J. 2011;434:473–482. doi: 10.1042/BJ20101585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonsalves C, Kalra VK. Endothelin-1 induced macrophage inflammatory protein-1b expression in monocytic cells involves hypoxia-inducible factor-1a and AP-1 and is negatively regulated by microRNA-195. J Immunol. 2010;185:6253–6264. doi: 10.4049/jimmunol.1000660. [DOI] [PubMed] [Google Scholar]
- 34.Miller C, Lai B. Human and mouse hematopoietic colony-forming cell assays. In: Helgason CD, MIller CL, editors. Methods in Molecular Biology: Basic Cell Culture Protocols. Humana Press, Inc; Totowa: pp. 71–89. [DOI] [PubMed] [Google Scholar]
- 35.Pászty C, Brion CM, Manci E, Witkowska HE, Stevens ME, Mohandas N, Rubin EM. Transgenic knockout mice with exclusively human sickle hemoglobin and sickle cell disease. Science. 1997;278:876–878. doi: 10.1126/science.278.5339.876. [DOI] [PubMed] [Google Scholar]
- 36.Hanlon PR, Fu P, Wright GL, Steenbergen C, Arcasoy MO, Murphy E. Mechanisms of erythropoietin-mediated cardioprotection during ischemia-reperfusion injury: role of protein kinase C and phosphatidylinositol 3-kinase signaling. FASEB J. 2005;19:1323–1325. doi: 10.1096/fj.04-3545fje. [DOI] [PubMed] [Google Scholar]
- 37.Semenza GL. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J Appl Physiol. 2000;88:1474–1480. doi: 10.1152/jappl.2000.88.4.1474. [DOI] [PubMed] [Google Scholar]
- 38.Chua YL, Dufour E, Dassa EP, Rustin P, Jacobs HT, Taylor CT, Hagen T. Stabilization of hypoxia-inducible factor-1α protein in hypoxia occurs independently of mitochondrial reactive oxygen species production. J Biol Chem. 2010;285:31277–31284. doi: 10.1074/jbc.M110.158485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang JC, Babak T, Corson TW, Chua G, Khan S, Gallie BL, Hughes TR, Blencowe BJ, Frey BJ, Morris QD. Using expression profiling data to identify human microRNA targets. Nat Methods. 2007;4:1045–1049. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- 40.Caron E, Ghosh S, Matsuoka Y, Ashton-Beaucage D, Therrien M, Lemieux S, Perreault C, Roux PP, Kitano H. A comprehensive map of the mTOR signaling network. Mol Syst Biol. 2010;6:453. doi: 10.1038/msb.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carrière A, Cargnello M, Julien LA, Gao H, Bonneil É, Thibault P, Roux PP. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr Biol. 2008;18:1269–1277. doi: 10.1016/j.cub.2008.07.078. [DOI] [PubMed] [Google Scholar]
- 42.Dolznig H, Habermann B, Stangl K, Deiner EM, Moriggl R, Beug H, Müllner EW. Apoptosis protection by the Epo target Bcl-XL allows factor-independent differentiation of primary erythroblasts. Curr Biol. 2002;12:1076–1085. doi: 10.1016/s0960-9822(02)00930-2. [DOI] [PubMed] [Google Scholar]
- 43.Lord-Dufour S, Copland IB, Levros LC, Post M, Das A, Khosla C, Galipeau J, Rassart E, Annabi B. Evidence for transcriptional regulation of the glucose-6-phosphate transporter by HIF-1α: targeting G6PT with mumbaistatin analogs in hypoxic mesenchymal stromal cells. Stem Cells. 2009;27:489–497. doi: 10.1634/stemcells.2008-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C, Mpollo MSEM, Gonsalves CS, Tahara SM, Malik P, Kalra VK. Peroxisome proliferator-activated receptor-α-mediated transcription of miR-199a2 attenuates endothelin-1 expression via hypoxia-inducible factor-1α. J Biol Chem. 2014;289:36031–36047. doi: 10.1074/jbc.M114.600775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.el Azzouzi H, Leptidis S, Dirkx E, Hoeks J, van Bree B, Brand K, McClellan Elizabeth A, Poels E, Sluimer JC, et al. The hypoxia-inducible microRNA cluster miR-199a~214 targets myocardial pparδ and impairs mitochondrial fatty acid oxidation. Cell Metabolism. 2013;18:341–354. doi: 10.1016/j.cmet.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 46.Delerive P, Martin-Nizard F, Chinetti G, Trottein F, Fruchart JC, Najib J, Duriez P, Staels B. Peroxisome proliferator-activated receptor activators inhibit thrombin-induced endothelin-1 production in human vascular endothelial cells by inhibiting the activator protein-1 signaling pathway. Circul Res. 1999;85:394–402. doi: 10.1161/01.res.85.5.394. [DOI] [PubMed] [Google Scholar]
- 47.Paumelle R, Blanquart C, Briand O, Barbier O, Duhem C, Woerly G, Percevault F, Fruchart JC, Dombrowicz D, Glineur C, Staels B. Acute antiinflammatory properties of statins involve peroxisome proliferator–activated receptor-α via inhibition of the protein kinase C signaling pathway. Circul Res. 2006;98:361–369. doi: 10.1161/01.RES.0000202706.70992.95. [DOI] [PubMed] [Google Scholar]
- 48.Yamakawa M, Liu LX, Date T, Belanger AJ, Vincent KA, Akita GY, Kuriyama T, Cheng SH, Gregory RJ, Jiang C. Hypoxia-inducible factor-1 mediates activation of cultured vascular endothelial cells by inducing multiple angiogenic factors. Circul Res. 2003;93:664–673. doi: 10.1161/01.RES.0000093984.48643.D7. [DOI] [PubMed] [Google Scholar]
- 49.Kelly BD, Hackett SF, Hirota K, Oshima Y, Cai Z, Berg-Dixon S, Rowan A, Yan Z, Campochiaro PA, Semenza GL. Cell type–specific regulation of angiogenic growth factor gene expression and induction of angiogenesis in nonischemic tissue by a constitutively active form of hypoxia-inducible factor 1. Circul Res. 2003;93:1074–1081. doi: 10.1161/01.RES.0000102937.50486.1B. [DOI] [PubMed] [Google Scholar]
- 50.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 51.Bushati N, Cohen SM. microRNA Functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 52.Grimson A, Farh KKH, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe T, Sato T, Amano T, Kawamura Y, Kawamura N, Kawaguchi H, Yamashita N, Kurihara H, Nakaoka T. Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev Dyn. 2008;237:3738–3748. doi: 10.1002/dvdy.21787. [DOI] [PubMed] [Google Scholar]
- 55.Loebel DAF, Tsoi B, Wong N, Tam PPL. A conserved noncoding intronic transcript at the mouse Dnm3 locus. Genomics. 2005;85:782–789. doi: 10.1016/j.ygeno.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 56.Koh MY, Spivak-Kroizman TR, Powis G. HIF-1 regulation: not so easy come, easy go. Trends Biochem Sci. 2008;33:526–534. doi: 10.1016/j.tibs.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 57.Lin W, Wadlington NL, Chen L, Zhuang X, Brorson JR, Kang UJ. Loss of PINK1 attenuates HIF-1α induction by preventing 4E-BP1-dependent switch in protein translation under hypoxia. J Neurosci. 2014;34:3079–3089. doi: 10.1523/JNEUROSCI.2286-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukarotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Galbán S, Kuwano Y, Pullmann R, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1α. Mol Cell Biol. 2008;28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schepens B, Tinton SA, Bruynooghe Y, Beyaert R, Cornelis S. The polypyrimidine tract-binding protein stimulates HIF-1α IRES-mediated translation during hypoxia. Nucleic Acids Res. 2005;33:6884–6894. doi: 10.1093/nar/gki1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Déry MAC, Michaud MD, Richard DE. Hypoxia-inducible factor 1: regulation by hypoxic and non-hypoxic activators. Int J Biochem Cell Biol. 2005;37:535–540. doi: 10.1016/j.biocel.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 62.Brittain JE, Hulkower B, Jones SK, Strayhorn D, De Castro L, Telen MJ, Orringer EP, Hinderliter A, Ataga KI. Placenta growth factor in sickle cell disease: association with hemolysis and inflammation. Blood. 2010;115:2014–2020. doi: 10.1182/blood-2009-04-217950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, Mendelsohn L, Rogers H, Leitman S, Raghavachari N, Yang Y, Yau YY, Tallack M, Perkins A, Taylor JGt, et al. Heme-bound iron activates placenta growth factor in erythroid cells via erythroid Kruppel-like factor. Blood. 2014;124:946–954. doi: 10.1182/blood-2013-11-539718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lalloyer F, Staels B. Fibrates, glitazones, and peroxisome proliferator–activated receptors. Atertio Thromb Vasc Biol. 2010;30:894–899. doi: 10.1161/ATVBAHA.108.179689. [DOI] [PMC free article] [PubMed] [Google Scholar]