Abstract
Fragile X syndrome, the most common cause of inherited intellectual disability, is caused by a trinucleotide CGG expansion in the 5′-untranslated region of the FMR1 gene, which leads to the loss of expression of the fragile X mental retardation protein (FMRP). FMRP, an RNA-binding protein that regulates the translation of specific mRNAs, has been shown to bind a subset of its mRNA targets by recognizing G quadruplex structures. It has been suggested that FMRP controls the local protein synthesis of several protein components of the Post Synaptic Density (PSD) in response to specific cellular needs. We have previously shown that the interactions between FMRP and mRNAs of the PSD scaffold proteins PSD-95 and Shank1 are mediated via stable G-quadruplex structures formed within the 3′-untranslated regions of these mRNAs. In this study we used biophysical methods to show that a comparable G quadruplex structure forms in the 3′-untranslated region of the glutamate receptor subunit NR2B mRNA encoding for a subunit of N-methyl-D-aspartate (NMDA) receptors that is recognized specifically by FMRP, suggesting a common theme for FMRP recognition of its dendritic mRNA targets.
INTRODUCTION
Fragile X Syndrome (FXS), an inherited developmental disorder, is caused by the trinucleotide CGG expansion and silencing of the FMR1 gene that codes for the fragile X mental retardation protein (FMRP). Loss of FMRP results in the disruption of the molecular composition of the Post Synaptic Density (PSD), affecting normal dendritic spine development and synaptic function 1, 2, 3. FMRP is an RNA-binding protein whose function is strongly implicated in mRNA translation regulation mechanisms, and whose absence severely affects the spatiotemporal dynamics of mRNA in neurons 4, 5. It is suggested that FMRP locally controls the synthesis of various protein components of PSD, by acting as a switch that suppresses/allows their mRNA translation depending on the current cellular requirements 6, 7. This translational switch is believed to be perpetually disabled in FXS patients where FMRP is absent, leading to an abnormal dendritic spine phenotype 7. Dendritic spines are important excitatory synaptic networks and are crucial for proper communication among neurons 1, 8. There are several confirmed mRNA targets of FMRP that are encoding for important scaffold proteins in PSD and whose translational disruption has been linked to FXS phenotype. Using HITS-CLIP to identify FMRP target mRNAs in brain in vivo, several PSD components were identified: PSD-95 and Shank1-3, as well as mRNAs encoding the NMDA receptor subunits, NR1, NR2A, NR2B and NR3A9, 10. These PSD scaffold proteins play essential roles in the majority of synaptic functions, being responsible for trafficking, anchoring and clustering of glutamate receptors and adhesion molecules, in addition to their role of linking the postsynaptic receptors to their downstream signalling proteins 11. For instance, Mudashetty et al. (2011) 7 showed that there is a dysregulated translation of PSD-95 when FMRP is absent, while Schutt et al. (2009) 12 identified disrupted levels of Shank1, SAPAP1-3, PSD-95 and of glutamate receptor subunits NR1 and NR2B in FMRP knock-out mice mimicking the disease. Given that these scaffold proteins are essential members of the PSD network, any disruption in their expression levels would lead to an abnormal synapse development and function. Schutt et al. (2009) 12 examined whether the changes in the scaffold protein levels affect the abundance of glutamate receptors in the PSD, given the role of these scaffold proteins in anchoring neurotransmitter receptors in the post synaptic membrane 12. They found elevated levels of NR1 in both the neocortex and hippocampus, of NR2B in the hippocampus and of GluR1 in the neocortex of FMRP-deficient mice as compared with wild type mice. In contrast, only a slight increase of the glutamate receptor subunits NR2A, GluR2/3, and GluR4 was found in the PSD fractions of the FMRP-deficient mice. Moreover, this study has also shown that FMRP associates in vivo with mRNAs encoding for scaffold proteins and glutamate receptor units (such as PSD-95, SAPAP1, SAPAP2, SAPAP3, Shank1, NR1 and NR2B) and concluded that the observed elevated protein levels in the FMRP-deficient mouse brain result from their dysregulated translation. The exact details of the mechanisms by which FMRP controls the translation of its mRNA targets are not known. It has been shown that the arginine-glycine-glycine (RGG) domain of FMRP has high affinity for specific G quadruplex structures of neuronal mRNA targets 13, 14, 15. G quadruplex structures are formed when four guanine nucleotides connected through Hoogsteen hydrogen bonding assemble into a square planar arrangement 16, 17. DNA G quadruplexes require the presence of potassium ions for folding, while RNA G quadruplexes of identical sequence can fold even in the absence of these ions, but have low stability 18. Previously, we have directly shown that the interactions between FMRP and mRNAs of the scaffold PSD-95 and Shank1 proteins are mediated via stable G-quadruplex structures formed within the 3′-UTRs of these mRNAs 19, 20. In this work, we used biophysical methods to show that a comparable G quadruplex structure forms in the glutamate receptor subunit NR2B mRNA that is coding for a subunit of N-methyl-D-aspartate (NMDA) receptors, a class of ligand-gated ions channels acting as excitatory amino acid receptors 21. Our results indicate that this G quadruplex structure is recognized specifically by FMRP, suggesting a common theme for FMRP recognition of its dendritic mRNA targets.
METHODS
RNA and peptides synthesis
NR2B mRNA (5′ GGGUACGGGAGGGUAAGGC UGUGGGUCGCGUG 3′) and the mutant NR2B mRNA (5′ GGGUACGCGACCCUAAGGCUGUG GGUCGCGUG 3′) were transcribed using synthetic DNA templates (TriLink BioTechnologies, Inc.) and expressed by T7 RNA polymerase driven in vitro transcription reactions. The RNA samples were purified by 20% polyacrylamide, 8 M urea gel electrophoresis and electroelution and were subsequently dialyzed against 10 mM cacodylic acid, pH 6.5. The 2-aminopurine (2AP) fluorescently labelled NR2B mRNA (5′ GGGU(2AP)CGGGAGGGUAAGGCUGUGGGUCGCGUG 3′) was chemically synthesized by Dharmacon, Inc.
The FMRP RGG box peptide and the HCV peptide derived from the HCV core protein, were chemically synthesized by the Peptide Synthesis Unit at the University of Pittsburgh Center for Biotechnology and Bioengineering.
Native gel electrophoresis
Prior to their use in the native gels, the RNA samples (10 μM) were annealed by boiling for 5 minutes in the presence of various KCl concentrations, followed by incubation at room temperature for 10 minutes. For the electromobility shift assay experiments the NR2B mRNA (10 μM) was incubated with increasing ratios (1:1, 1:2 and 1:3) of the FMRP RGG box for an additional 20 minutes. The 20% native gels in in 0.5 X Tris/Borate/EDTA buffer were run at 4°C, 85 V for 6 h and visualized by UV shadowing at 254 nm using an AlphaImager (AlphaInnotech, Inc.).
UV spectroscopy thermal denaturation
The UV spectroscopy thermal denaturation experiments were performed on a Cary 3E UV-VIS Spectrophotometer (Varian, Inc.) equipped with a peltier cell. 200 μL samples containing 10 μM RNA in 10 mM cacodylic acid buffer pH 6.5 and in the presence of 5–75 mM KCl were annealed as described above and thermally denatured by varying the temperature in the range 20°C–95°C, at a rate of 0.2 °C/minute and monitoring the absorbance changes at 295 nm, wavelength sensitive to G quadruplex denaturation 22. In order to prevent sample evaporation, a layer of mineral oil was added to the cuvettes.
To study if an intermolecular or intramolecular G quadruplex is formed within NR2B mRNA, the UV spectroscopy thermal denaturation experiments were performed at variable RNA concentrations ranging from 10–40 μM and a fixed KCl concentration of 75 mM in 10 mM cacodylic acid buffer, pH 6.5. In the case of G quadruplex structure formation between n number of RNA strands, the melting temperature (Tm) depends on the total RNA concentration (equation [1]), whereas the melting temperature of an intramolecular G quadruplex structure (n=1) is independent of the RNA concentration (equation [2]) 23:
| [1] |
| [2] |
where R is the gas constant and ΔH°νH and ΔS°νH are the Van’t Hoff thermodynamic parameters. The thermodynamic parameters of the G quadruplex structures were obtained by fitting their UV thermal denaturation curves to equation 3, which assumes a two-state model 23:
| [3] |
where AU and AF represent the absorbance of the unfolded and native G quadruplex RNA structure, respectively, and R is the universal gas constant.
To calculate the number of K+ ions bound specifically by the G quadruplex structure, a simple model for folded to unfolded G quadruplex was assumed in which Δn K+ ions are lost upon heat treatment and G quadruplex unfolding. Δn is equal to the slope of the plot of ΔG° as a function of logarithm of K+ concentration:
| [4] |
where and is the slope of the plot of ΔG° as a function of the logarithm of K+ ion concentration 23.
Circular Dichroism (CD) spectroscopy
All experiments were performed on a Jasco J-810 spectropolarimeter at 25°C, using a 1 mm path-length quartz cuvette (Starna Cells). 200 μL volumes of 10 μM samples of annealed RNA were prepared in 10 mM cacodylic acid buffer, pH 6.5. The G quadruplex formation was monitored between 200–350 nm by titrating KCl in the range 5–150 mM, and averaging a series of 7 scans with a 1 s response time and a 2 nm bandwidth. The spectra were corrected by subtracting the cacodylic acid buffer contributions.
1H Nuclear Magnetic Resonance (NMR) spectroscopy
The G quadruplex formation was monitored by acquiring one dimensional (1D) 1H NMR spectra of the NR2B mRNA at 25°C on a 500 MHz Bruker AVANCE spectrometer. A 350 μM RNA sample was prepared in 10 mM cacodylic acid buffer, pH 6.5 in a 90%H2O/10%D2O ratio and KCl was titrated in the range 5–100 mM. The water suppression was accomplished using the Watergate pulse sequence 24. Similar experiments were performed for the NR2B mutant mRNA to show that when G nucleotides predicted to be engaged in G quadruplex formation were mutated the structure no longer formed.
Fluorescence spectroscopy
Steady-state fluorescence spectroscopy experiments of the NR2B 5 2AP mRNA (5′ GGGU(2AP)CGGGAGGGUAAGGCUGUGGGUCGCGUG 3′) were performed on a Horiba Scientific Fluoromax-4 and accompanying software fitted with a 150 W ozone-free xenon arclamp. Experiments were performed in a 150 μL sample volume, 3 mm path-length quartz cuvette (Starna Cells). The excitation wavelength was set to 310 nm, the emission spectrum was recorded in the range of 330 – 450 nm, and the bandpass for excitation and emission monochromators were both set to 5 nm.
FMRP ISO1 and FMRP S500D were titrated (50 nM) into a fixed concentration of NR2B 2AP mRNA (200 nM) and the quenching of the fluorescence signal was recorded as a result of protein-RNA interactions (each point was corrected for tryptophan fluorescence contributions originating from the protein). 1 μM of bovine serum albumin (BSA) was added in the RNA sample prior to FMRP titration to prevent non-specific binding. The binding dissociation constant (Kd) was determined by fitting the binding curves to the equation:
| [5] |
where represents the ratio of the steady state fluorescence intensity of the bound and free mRNA, [RNA]t is the total concentration of mRNA, and [P]t is the total peptide/protein concentration 23. These experiments were performed in triplicate for each FMRP ISO1 and FMRP S500D, and the reported errors represent the standard deviations of the dissociation constants determined from independent fits to the three measurements. Control experiments were performed in identical conditions, titrating 50 nM increments of BSA into a fixed concentration of NR2B 2AP mRNA (200 nM).
RNA-based affinity pull down assay
A biotinylated control HCV RNA probe was denatured at 95°C for 3 minutes and cooled quickly in a dry ice-ethanol bath. The biotinylated NR2B mRNA and PSD-95 mRNA probes were denatured at 95°C for 5 minutes and cooled at room temperature for 15 minutes. 5 μM of either probe was incubated with E17 mouse brain lysate for 20 minutes at room temperature and NeutrAvidin agarose (Thermo Scientific, Inc.) pre-blocked with BSA was used to precipitate the probes. The pull-downs were performed with RIPA buffer that contained 150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 1% NP-40, 0.5% deoxycholate and 0.1% SDS. After extensive washing, the proteins were detected by immunoblot against FMRP (1:1250, Sigma) and SMN (1:500, BD Transduction Laboratories & San Jose) using an Odyssey Infrared Imaging System.
RESULTS
NR2B mRNA forms an intramolecular G quadruplex structure with parallel topology
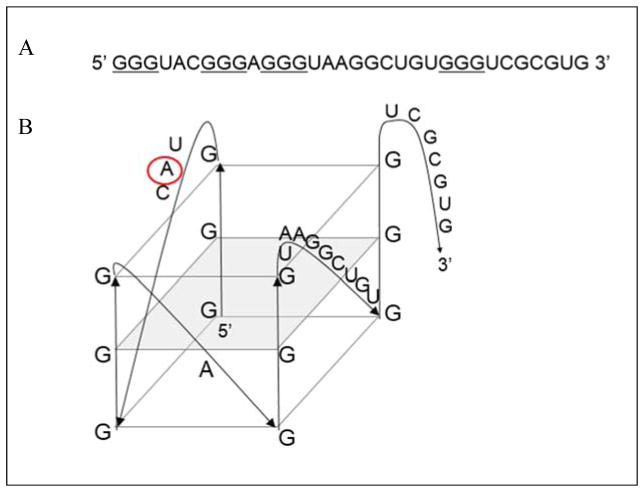
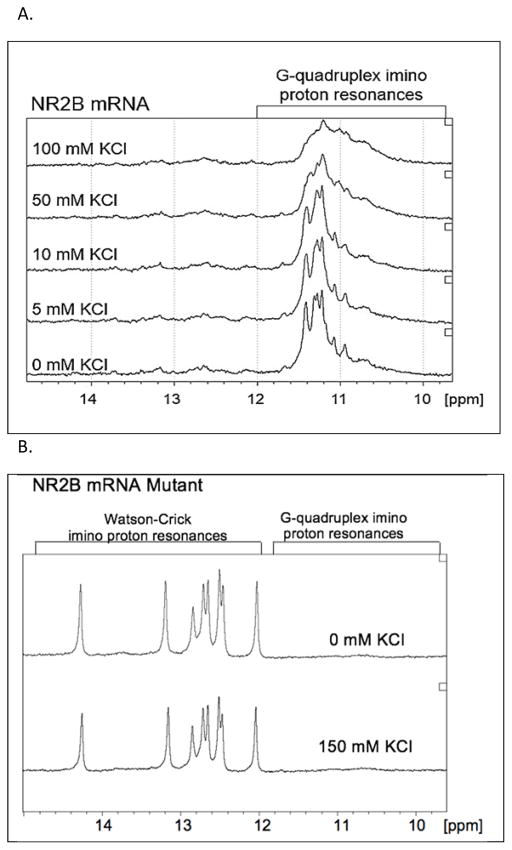
It has been shown that for a subset of mRNAs, FMRP-facilitated translational control is carried on in regions located within their 3′-UTR 14. We have previously demonstrated the presence of G quadruplex structures in the 3′-UTR regions of several FMRP mRNA targets such as Semaphorin 3F, PSD-95, Shank1, that are bound specifically and with high affinity by both FMRP and its phosphorylated mimic FMRP S500D 19, 20, 25. Given that NR2B mRNA has been found to be associated with FMRP in whole mouse brain homogenates 12 and directly crosslinked in vivo 9 we analyzed its sequence for the potential to adopt G quadruplex structures. We used an online G quadruplex prediction software QGRS Mapper (http://bioinformatics.ramapo.edu/QGRS/analyze.php) which predicted the existence of a high score G quadruplex in the NR2B mRNA 3′-UTR (NM_000834.3, position 4659) (Figure 1, the Gs predicted to engage in G quadruplex formation are underlined). To determine if such a structure forms in the NR2B mRNA 3′-UTR we have produced by in vitro transcription reaction a 32 nucleotide (nt) sequence containing this G rich segment. The formation of G quadruplex structures in this RNA was tested first by 1D 1H NMR spectroscopy in the presence of increasing concentrations of K+ ions. This technique is commonly used to detect G quadruplex structures in DNA/RNA since the guanine imino protons engaged in G quadruplex formation have a signature chemical shift between 10 and 12 ppm 23, 26, 27. We observed a strong signal in the region 10–12 ppm for the NR2B mRNA even in the absence of K+, indicating G quadruplex formation (Figure 2A, bottom spectrum). Additionally, low intensity and broad resonances were observed in the region 12–14.5 ppm, indicative of the presence of a minor alternate structure involving Watson-Crick base pairs. No significant changes in imino proton resonances were observed after titration of 5 and 10 mM K+. However, the broadening of the spectra was observed after the addition of 50 and 100 mM KCl, suggesting that additional G quadruplex conformations might be supported by an increased amount of salt. Additionally, at the concentrations used in NMR spectroscopy experiments, the G quadruplex structures might form higher order complexes stabilized by the K+ ions. The analysis of the 32-nt NR2B mRNA sequence revealed the possibility of formation of alternate G quadruplex structures containing two plane G quartets, that were also predicted by the QGRS Mapper, but with lower probability scores than the highest score three-plane G quadruplex (Figure 1).
Figure 1.
(A) 32 nt guanine rich sequence located in the 3′-UTR of NR2B mRNA (NM_000834.3, position 4659) predicted to form G quadruplex (guanine triplets proposed to be involved in the structure are underlined). (B) Arrangement of the predicted G quadruplex structure in the NR2B mRNA. QGRS Mapper software was used for the prediction (http://bioinformatics.ramapo.edu/QGRS/analyze.php)42. Adenine replaced by the 2-amino purine fluorescent analog is circled in red.
Figure 2.
(A)1H NMR spectra of 350 μM NR2B mRNA at various concentrations of K+ ions in 10 mM cacodylic acid buffer, pH 6.5, at 25°C. (B) 1H NMR spectra of 350 μM NR2B mRNA mutant at 0 mM and at 150 mM KCl in 10 mM cacodylic acid buffer, pH 6.5, at 25°C.
A mutant NR2B mRNA was also synthesized in which G nucleotides predicted to be engaged in G quadruplex formation were replaced by Cs and its secondary structure was analyzed by 1H NMR spectroscopy. As seen in figure 2B, which shows the imino proton resonance region of the mutant NR2B mRNA in the absence and presence of 150 mM KCl, the G quadruplex imino proton resonances are no longer present in the 10–12 ppm region. This result confirms that the mutation of the G nucleotides abolishes the G quadruplex structure formation. Intense and sharp resonances are present in the region 12–14.5 ppm, corresponding to imino protons involved in Watson-Crick base pairs, consistent with the predicted structure of the NR2B mutant that contains an extended hairpin stem (supplemental figure 1).
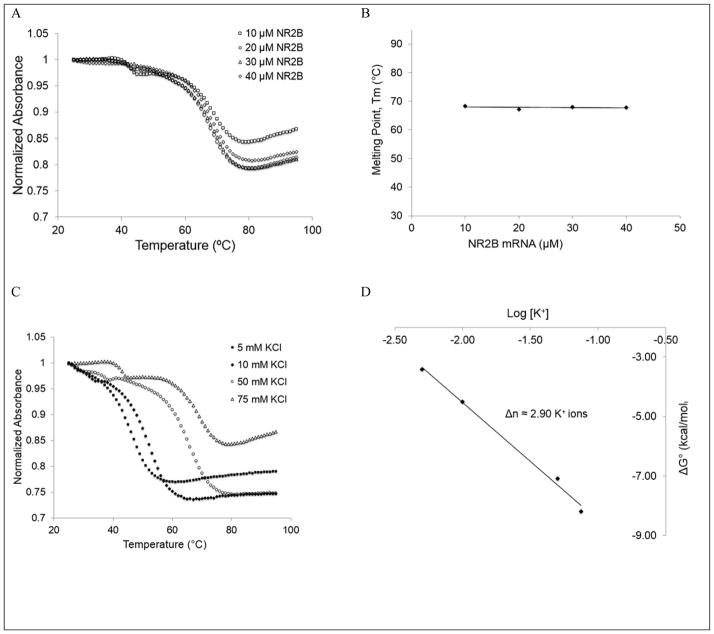
Next, we performed 20% non-denaturing polyacrylamide gel electrophoresis of the 32-nt NR2B mRNA in 0.5 X TBE buffer to obtain additional information about its G quadruplex structure (supplemental figure 2). In the absence of K+, an more intense upper band is present as well as a faint lower band that indicates the presence of at least two conformations, one of which is dominant (supplemental figure 2, lane 1). Upon titrating KCl in the range 5 to 150 mM, the intensity of the upper band decreases with the concomitant increase of the lower band intensity, indicating that in the presence K+ ions the second conformation becomes stabilized (supplemental figure 2, lanes 2–7). Additionally, very faint upper bands appears in the presence of higher KCl concentrations (supplemental figure 2, lanes 3–7, arrow), corresponding to higher molecular weight complexes that could be formed by the stacking of G quadruplex structures. These results are consistent with the NMR spectroscopy results in the presence of higher concentrations of salt, indicating the presence of multiple G quadruplex conformations and/or possible aggregation. It is known that the stability of G quadruplexes highly depends on their architecture, with more G quartet planes stacked within the G quadruplex increasing the overall thermodynamic stability of the structure 28. In our proposed 32-nt NR2B mRNA fragment, there is only one three-plane intramolecular G quadruplex that could be possibly formed due to existence of only four guanine triplets (underlined in figure 1A), and other G quadruplex structures would involve less stable two-plane arrangements. Another possibility could be that guanine residues engage in an intermolecular arrangement that will result in a G quadruplex formed from different strands. To examine if intramolecular or intermolecular G quadruplex conformations are formed by the 32-nt NR2B mRNA fragment, we performed UV thermal denaturation experiments at a fixed KCl concentration (75 mM KCl) and variable RNA concentration in the range 10–40 μM, varying the temperature from 25–95°C and monitoring the absorbance changes at 295 nm, wavelength sensitive to G quadruplex denaturation 29. For intramolecular species the melting temperature is independent of the RNA concentration (materials and methods, equations 1 and 2). A clearly defined transition with a melting temperature, Tm, of 68°C was observed at all RNA concentrations investigated (figure 3A and 3B), indicating that the G quadruplex conformation giving rise to it is intramolecular. A second minor hypochromic transition was also observed with a Tm, of ~42°C, which based on its lower melting temperature, we assign the unfolding of alternate G quadruplex structures containing only two G quadruplex planes. Upon establishing that the G quadruplex conformation giving rise to the main hypochromic transition in the UV thermal denaturation curve of NR2B mRNA is intramolecular we fitted it with equation 3 (materials and methods) that assumes a two state model 23, to determine the thermodynamic parameters of G quadruplex formation in the presence of 75 mM KCl (Supplemental figure 3). The enthalpy of formation of a single G-quartet plane has been reported to range between −18 kcal/mol and −25 kcal/mol 30. Thus, the value obtained for the enthalpy of G quadruplex formation in NR2B mRNA for the main transition (ΔH =−64.6±0.1 kcal/mol) is consistent with the presence of three G quartet planes.
Figure 3.
UV spectroscopy thermal denaturation data of NR2B mRNA. (A) NR2B mRNA at 75 mM KCl as a function of RNA concentration, indicating an intramolecular structure formation. (B) G-quadruplex melting temperature at 75 mM KCl as a function of the RNA concentration. (C) 10 μM NR2B mRNA as a function of KCl concentration, showing an increase in Tm due to K+ driven stability. (D) Plot of NR2B mRNA ΔG° as a function of logarithm of K+ ion concentration, where Δn represents the number of K+ ions released upon G quadruplex unfolding.
To determine the number of K+ ions coordinated with this G quadruplex structure we performed similar UV thermal denaturation experiments, keeping the RNA concentration constant (10 μM) and varying the KCl concentration in the range 5–75 mM. The minor transition with the lower melting temperature was not well defined at low KCl concentrations, precluding its further analysis (Figure 3C). Thus, only the major transition was fitted with equation 3 (materials and methods), determining the free energy of G quadruplex formation at each KCl concentration which was plotted as a function of the log[K+] to calculate the number of K+ ions coordinated within the main G quadruplex conformation (materials and methods, equation 4). On average ~ 2.9 K+ ions coordinate the main G quadruplex structure within the NR2B mRNA (Figure 3D).
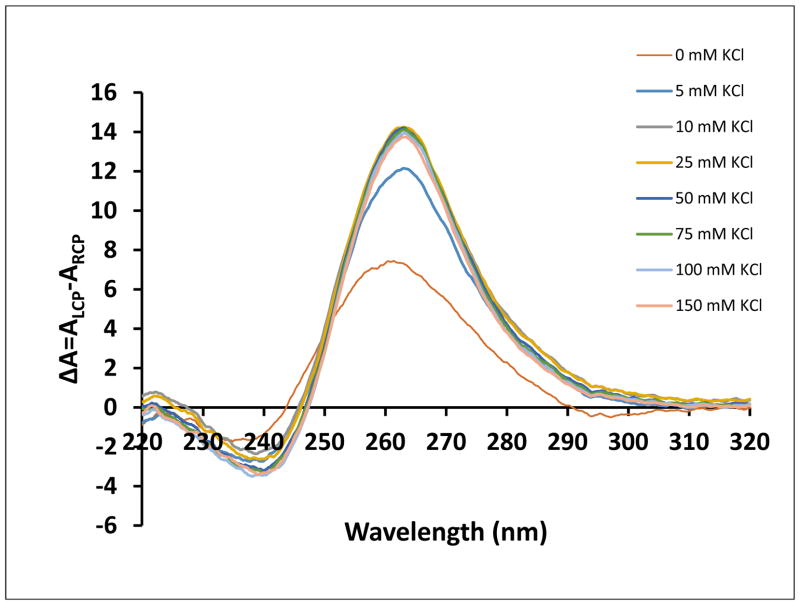
To examine the fold of the NR2B mRNA G quadruplex structure we employed circular dichroism (CD) spectroscopy, technique widely used for G quadruplex analysis, due to unique signature responses of different G quadruplex folding arrangements. For parallel G-quadruplex structures, a positive band at around 265 nm and a negative band at around 240 nm have been observed, whereas the signatures of an antiparallel G quadruplex structure are a negative band at around 260 nm and a positive band at around 295 nm 31–35. We performed the analysis in K+ concentrations ranging from 0–150 mM and observed a positive band at 265 nm and a negative one at 240 nm, even in the absence of KCl, indicating that NR2B mRNA folds into a parallel G quadruplex (Figure 4). The intensities of the bands increased as K+ was titrated, implying that salt is necessary to improve stability. However, no significant intensity changes are observed at KCl concentrations higher than 10 mM, consistent with the 1H NMR spectroscopy results.
Figure 4.
CD spectra of NR2B mRNA in increasing salt concentration at 25°C. 10 μM RNA sample was suspended in 10 mM cacodylic acid buffer pH 6.5 and KCl was titrated in the range 5–150 mM. The spectra were corrected by subtracting 10 mM cacodylic acid buffer contributions.
In summary, our biophysical analysis determined that the G rich stretch within the NR2B mRNA 3′-UTR folds into parallel, G quadruplex structures, the most stable of which is intramolecular, contains three G quartet planes and is coordinated by ~ 3 K+ ion equivalents (figure 1B).
FMRP recognizes specifically the NR2B mRNA G quadruplex structure
Previously it has been shown that the FMRP interactions with NR2B mRNA are located within the mRNA’s 3′-UTR 11. Thus, we investigated if the 32-nt G quadruplex forming NR2B mRNA fragment is sufficient for FMRP recognition, by using electromobility shift assay to analyze its interactions with the FMRP RGG box, the protein domain shown to recognize G quadruplex structures in other mRNA targets13, 14, 15 (supplemental figure 4). The bands corresponding to the free NR2B mRNA disappear in the presence of increasing FMRP RGG box ratios, indicating the formation of a complex with the peptide (supplemental figure 4, lanes 2–4). Because the FMRP RGG box confers the complex formed with the small size NR2B mRNA an overall positive charge, it does not give rise to a well-defined shifted band. The upper bands that we attribute to stacks of G quadruplex structures are also shifted in the presence of the FMRP RGG box, and in this case due to the larger sixe of the higher molecular weight RNA, the bands corresponding to complexes formed with the peptide are distinct.
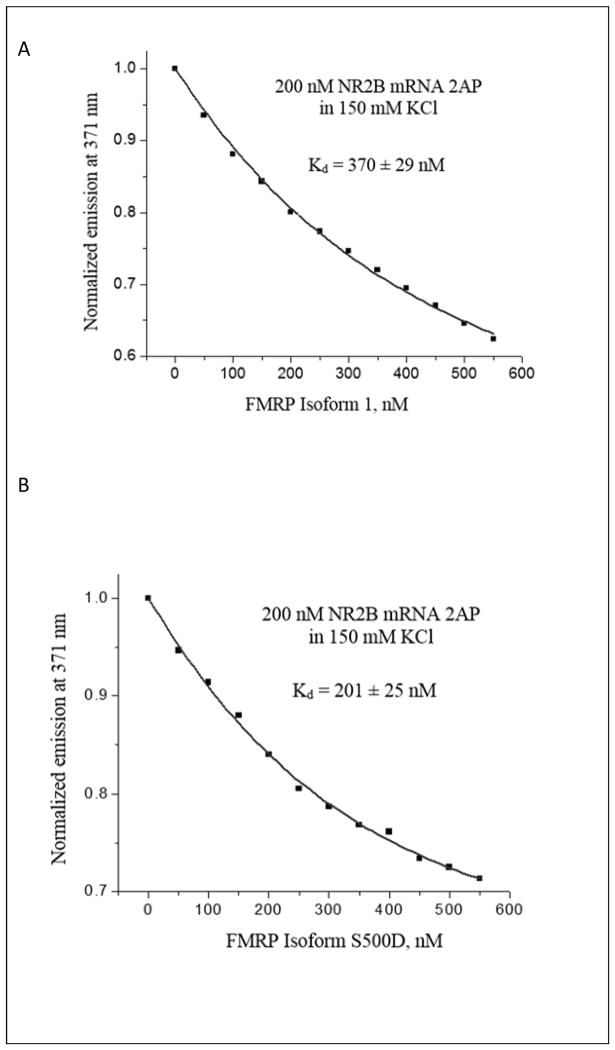
Next, we analyzed the interactions of NR2B mRNA with the full-length FMRP (isoform 1), as well as with its phosphorylated mimic FMRP S500D 36 by fluorescence spectroscopy. The FMRP S500D has been included since it has been shown that the phosphorylation status of FMRP plays a role in mRNA translation control, where in phosphorylated state FMRP acts as a translation inhibitor, whereas upon its dephosphorylation triggered by synaptic input, it supports the mRNA translation 7. FMRP and FMRP S500D were in vitro expressed and purified according to the protocol previously established in our laboratory 37. We have designed a fluorescently labelled NR2B 2AP mRNA, in which the adenine at position 5 was replaced with 2-aminopurine (2AP) (Figure 1B, adenine 5 is circled). 2AP is a highly fluorescent analog of adenine whose steady-state fluorescence is sensitive to changes in its microenvironment 38, 39. A 200 nM NR2B 2AP mRNA sample was prepared in 10 mM cacodylic acid buffer pH 6.5 and 50 nM increments of either of FMRP or FMRP S500D were titrated, monitoring the changes in the steady-state fluorescence of the 2AP reporter (figures 5A and 5B). To demonstrate that the fluorescence quenching observed in the presence of FMRP or FMRP S500D is not due to non-specific interactions, similar control experiments were performed by titrating BSA into the NR2B 2AP mRNA sample. As expected, the steady-state fluorescence of NR2B 2AP mRNA did not change significantly in the presence of BSA (supplemental figure 5). The resulting binding curves were fit with equation 5 (materials and methods), to determine the dissociation constants, Kd, of 370 ± 29 nM for the complex formed by NR2B mRNA with FMRP and of 201 ± 25 nM for the complex formed with FMRP S500D (figures 5A and 5B). These dissociation constant values are within range with those reported for the binding of the full length FMRP or FMRP S500D to G quadruplex structures formed within other mRNA targets such as Shank1 19, FMR1 40, Semaphorin 3F 37. The dissociation constant values for FMRP and FMRP S500D binding to NR2B mRNA yield a free energy of binding of ΔG0b = −8.7 ± 0.1 kcal/mol and of ΔG0b = −9.1 ± 0.1 kcal/mol, respectively, hence a difference of only 0.4 kcal/mol in binding free energy for FMRP S500D, versus unphosphorylated FMRP. This finding suggests that a difference in binding G quadruplex target mRNA binding does not play the predominant role in mediating the function of the unphosphorylated versus phosphorylated FMRP with respect to translation regulation.
Figure 5.
Binding of (A) FMRP Isoform 1 and (B) FMRP S500D to 200 nM NR2B 2AP mRNA in 150 mM KCl and in the presence of 5-fold excess of BSA. FMRP Isoforms were titrated in 50 nM increments. Kd values were calculated for FMRP Isoform 1 (370 ± 29 nM) and FMRP S500D (201 ± 25 nM).
Finally, to confirm the interactions of the NR2B G quadruplex with endogenous FMRP, we performed a biotin-RNA pull down assay using a biotin labelled 32-nt NR2B mRNA probe that was incubated with lysates from E17 mouse brain and further precipitated with NeutrAvidin agarose. It is clear from the Western blot analysis that FMRP interacts with NR2B mRNA probe (Figure 6, lane 5, upper panel). The presence of several bands suggests that the NR2B mRNA probe pulled down more than one FMRP isoform that exists in the mouse brain lysates. A biotinylated probe derived from PSD-95 mRNA that contains two G quadruplex structures 19 was used as a positive control, and because this probe contains multiple binding sites for FMRP, it gives rise to more intense bands than those corresponding to the NR2B mRNA probe (figure 6, lane 4, upper panel). A biotinylated unrelated probe derived from the hepatitis C virus genomic RNA was used as a negative control, and as expected, it did not pull down FMRP (Figure 6, lane 3, upper panel). Western blot analysis was also performed for the survival of motor neuron (SMN) protein, and interestingly, while as predicted the HCV probe did not pull it down (Figure 6, lane 3, bottom panel), the PSD-95 probe pulled it down (Figure 6, lane 4, bottom panel), and a faint band seems also to be present for the NR2B probe (Figure 6, lane 5, bottom panel). The SMN protein is not an RNA binding protein, however it has been shown to directly interact with FMRP41, so it is possible that it is detected not due to its direct interactions with the RNA probes, but rather indirectly, through interactions with FMRP.
Figure 6.
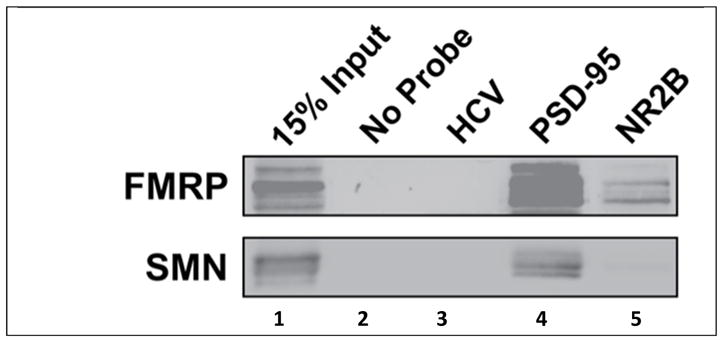
Western blot analysis of FMRP interaction with G quadruplex within NR2B mRNA. The 5′-biotin labelled NR2B mRNA, PSD-95 mRNA and HCV RNA probes were incubated with E17 mouse brain lysate. The probes were precipitated with NeutrAvidin agarose beads and co-purified FMRP and SMN proteins were assessed by immunoblotting. Lane 1: 15% of brain lysate input showing existence of FMRP; Lane 2: No detection of FMRP or SMN when the probe is not present; Lane 3: No detection of FMRP and SMN using HCV probe; Lane 4: FMRP detected with PSD-95 mRNA probe. Lane 5: FMRP detected with NR2B mRNA probe.
DISSCUSION
In this study we have shown for the first time that a main G quadruplex structure forms within 3′-UTR of the NR2B mRNA, which is of parallel type, consists of three G quartet planes and is coordinated by ~ 3 K+ ions. We have also proven that both FMRP and its phosphorylated mimic FMRP S500D bind with nM affinity to this G quadruplex, which also interacts with endogenous FMRP from E17 mouse brain lysates. Thus, our results indicate that from the entire NR2B mRNA that has been shown to co-IP with FMRP 12, only a 32-nt G quadruplex located in its 3′-UTR is sufficient to act as a recruiter of FMRP. NR2B mRNA joins other dendritic mRNAs, such as PSD-95 and Shank1 19, 20 that have been shown to adopt one or more G quadruplex structures in their 3′-UTR recognized specifically by FMRP, suggesting a common mechanism of recognition. In the case of PSD95 mRNA these G quadruplex structures seem to be implicated together with FMRP and the microRNA mir-125a in mediating the translation of their mRNA. Further investigations are underway in our laboratories to determine if there are microRNAs that, similar to the case of PSD-95 mRNA, work in conjunction with FMRP to control the translation of NR2B mRNA.
Supplementary Material
Acknowledgments
This work was supported by the NIH grants 9R15HD078017-03A1 to M.R.M. and 1R21NS089080 to G.J.B.
References
- 1.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 3.Muddashetty Ravi S, VCN Fragile X Syndrome: A Disorder of Synaptic Protein Synthesis Dynamics. Journal of the Indian Institute of Science. 2012;92:452. [Google Scholar]
- 4.Devys D, Lutz Y, Rouyer N, Bellocq JP, Mandel JL. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 5.Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A Direct Role for FMRP in Activity-Dependent Dendritic mRNA Transport Links Filopodial-Spine Morphogenesis to Fragile X Syndrome. Dev Cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeiffer BE, Huber KM. The State of Synapses in Fragile X Syndrome. Neuroscientist. 2009;15:549–567. doi: 10.1177/1073858409333075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible Inhibition of PSD-95 mRNA Translation by miR-125a, FMRP Phosphorylation, and mGluR Signaling. Molecular Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nimchinsky EA, Sabatini BL, Svoboda K. Structure and Function of Dendritic Spines. Annual Review of Physiology. 2002;64:313–353. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- 9.Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darnell JC, Klann E. The Translation of Translational Control by FMRP: Therapeutic Targets for FXS. Nat Neurosci. 2013;11:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verpelli C, Schmeisser MJ, Sala C, Boeckers TM. Scaffold Proteins at the Postsynaptic Density. In: Kreutz MR, Sala C, editors. In Synaptic Plasticity. Springer; Vienna: 2012. pp. 29–61. [DOI] [PubMed] [Google Scholar]
- 12.Schütt J, Falley K, Richter D, Kreienkamp H-J, Kindler S. Fragile X mental retardation protein regulates the levels of scaffold proteins and glutamate receptors in postsynaptic densities. J Biol Chem. 2009;284:25479–25487. doi: 10.1074/jbc.M109.042663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 15.Menon L, Mihailescu M-R. Interactions of the G quartet forming semaphorin 3F RNA with the RGG box domain of the fragile X protein family. Nucleic Acids Res. 2007;35:5379–5392. doi: 10.1093/nar/gkm581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hud NV, Smith FW, Anet FA, Feigon J. The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by 1H NMR. Biochemistry. 1996;35:15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 17.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 18.Joachimi A, Benz A, Hartig JS. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorganic & Med Chem. 2009;17:6811–6815. doi: 10.1016/j.bmc.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 19.Stefanovic S, Bassell GJ, Mihailescu MR. G quadruplex RNA structures in PSD-95 mRNA: potential regulators of miR-125a seed binding site accessibility. RNA. 2015;21:48–60. doi: 10.1261/rna.046722.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Gaetano CM, Williams KR, Bassell GJ, Mihailescu MR. FMRP interacts with G-quadruplex structures in the 3′-UTR of its dendritic target Shank1 mRNA. RNA Biol. 2014;11:1364–1374. doi: 10.1080/15476286.2014.996464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z, Zhao W, Xu T, Pei D, Peng Y. Alterations of NMDA receptor subunits NR1, NR2A and NR2B mRNA expression and their relationship to apoptosis following transient forebrain ischemia. Brain Res. 2010;1361:133–139. doi: 10.1016/j.brainres.2010.09.035. [DOI] [PubMed] [Google Scholar]
- 22.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Letters. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 23.Menon L, Mader SA, Mihailescu MR. Fragile X mental retardation protein interactions with the microtubule associated protein 1B RNA. RNA. 2008;14:1644–1655. doi: 10.1261/rna.1100708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piotto M, Saudek V, Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 25.Menon L, Mihailescu M-R. Interactions of the G quartet forming semaphorin 3F RNA with the RGG box domain of the fragile X protein family. Nucleic Acids Res. 2007;35:5379–5392. doi: 10.1093/nar/gkm581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fürtig B, Richter C, Wöhnert J, Schwalbe H. NMR Spectroscopy of RNA. ChemBioChem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 27.Nambiar M, Goldsmith G, Moorthy BT, Lieber MR, Joshi MV, Choudhary B, Hosur RV, Raghavan SC. Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14;18) translocation in follicular lymphoma. Nucleic Acids Res. 2011;39:936–948. doi: 10.1093/nar/gkq824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petraccone L, Erra E, Duro I, Esposito V, Randazzo A, Mayol L, Mattia CA, Barone G, Giancola C. Relative stability of quadruplexes containing different number of G-tetrads. Nucleosides Nucleotides Nucleic Acids. 2005;24:757–760. doi: 10.1081/ncn-200060080. [DOI] [PubMed] [Google Scholar]
- 29.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Letters. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 30.Hardin CC, Perry AG, White K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers. 2000;56:147–194. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 31.Miyoshi D, Nakao A, Sugimoto N. Structural transition from antiparallel to parallel G-quadruplex of d(G4T4G4) induced by Ca2+ Nucleic Acids Res. 2003;31:1156–1163. doi: 10.1093/nar/gkg211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kypr J, Kejnovska I, Renciuk D, Vorlickova M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paramasivan S, Rujan I, Bolton PH. Circular dichroism of quadruplex DNAs: Applications to structure, cation effects and ligand binding. Methods. 2007;43:324–331. doi: 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Randazzo A, Spada GP, da Silva MW. Circular dichroism of quadruplex structures. Top Curr Chem. 2013;330:67–86. doi: 10.1007/128_2012_331. [DOI] [PubMed] [Google Scholar]
- 35.Vorlíčková M, Kejnovská I, Sagi J, Renčiuk D, Bednářová K, Motlová J, Kypr J. Circular dichroism and guanine quadruplexes. Methods. 2012;57:64–75. doi: 10.1016/j.ymeth.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 36.Tarrant MK, Cole PA. The Chemical Biology of Protein Phosphorylation. Annual Review of Biochemistry. 2009;78:797–825. doi: 10.1146/annurev.biochem.78.070907.103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans TL, Mihailescu M-R. Recombinant Bacterial Expression and Purification of Human Fragile X Mental Retardation Protein Isoform 1. Protein Expr Purif. 2010;74:242–247. doi: 10.1016/j.pep.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bharill S, Sarkar P, Ballin JD, Gryczynski I, Wilson GM, Gryczynski Z. Fluorescence intensity decays of 2-aminopurine solutions: lifetime distribution approach. Anal Biochem. 2008;377:141–149. doi: 10.1016/j.ab.2008.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serrano-Andrés L, Merchán M, Borin AC. Adenine and 2-aminopurine: Paradigms of modern theoretical photochemistry. PNAS. 2006;103:8691–8696. doi: 10.1073/pnas.0602991103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blice-Baum AC, Mihailescu MR. Biophysical characterization of G-quadruplex forming FMR1 mRNA and of its interactions with different fragile X mental retardation protein isoforms. RNA. 2014;20:103–114. doi: 10.1261/rna.041442.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piazzon N, Rage F, Schlotter F, Moine H, Branlant C, Massenet S. In vitro and in cellulo evidences for association of the survival of motor neuron complex with the fragile X mental retardation protein. J Biol Chem. 2008;283:5598–610. doi: 10.1074/jbc.M707304200. [DOI] [PubMed] [Google Scholar]
- 42.Kikin O, D’Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34(Web Server issue):W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathews DH. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA. 2004;10:1178–1190. doi: 10.1261/rna.7650904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.