Abstract
Background
High-density lipoprotein cholesterol (HDL-C) exerts many anti-atherogenic properties including its role in reverse cholesterol transport (RCT). Scavenger receptor class B member 1 (SCARB1) plays a key role in RCT by selective uptake of HDL cholesteryl esters. We aimed to explore the genetic contribution of SCARB1 to affecting lipid levels in African Blacks from Nigeria.
Methods
We resequenced 13 exons and exon-intron boundaries of SCARB1 in 95 individuals with extreme HDL-C levels using Sanger method. Then, we genotyped 147 selected variants (78 sequence variants, 69 HapMap tagSNPs, and 2 previously reported relevant variants) in the entire sample of 788 African Blacks using either the iPLEX Gold or TaqMan methods. A total of 137 successfully genotyped variants were further evaluated for association with major lipid traits.
Results
The initial gene-based analysis demonstrated evidence of association with HDL-C and apolipoprotein A-I (ApoA-I). The follow-up single-site analysis revealed nominal evidence of novel associations of nine common variants with HDL-C and/or ApoA-I (P < 0.05). The strongest association was between rs11057851 and HDL-C (P = 0.0043), which remained significant after controlling for multiple testing using false discovery rate. Rare variant association testing revealed a group of 23 rare variants (frequencies ≤1 %) associated with HDL-C (P = 0.0478). Haplotype analysis identified four SCARB1 regions associated with HDL-C (global P < 0.05).
Conclusions
To our knowledge, this is the first report of a comprehensive association study of SCARB1 variations with lipid traits in an African Black population. Our results showed the consistent association of SCARB1 variants with HDL-C across various association analyses, supporting the role of SCARB1 in lipoprotein-lipid regulatory mechanism.
Electronic supplementary material
The online version of this article (doi:10.1186/s12881-015-0250-6) contains supplementary material, which is available to authorized users.
Keywords: African continental ancestry group; Candidate gene association study; Genetic variation; Haplotypes; Lipids; SCARB1 protein, human; Sequence analysis, DNA
Background
Abnormal lipid and lipoprotein levels are a major risk factor for coronary heart disease (CHD) [1], the leading cause of death worldwide [2]. Elevated low-density lipoprotein cholesterol (LDL-C) levels and decreased high-density lipoprotein cholesterol (HDL-C) levels are correlated with the development of CHD. There is a strong genetic basis for lipoprotein-lipid levels with heritability estimates of 40–80 % [3]. A large number of genes and genetic variants associated with lipid traits have been discovered in genome-wide association studies (GWAS) [4–6]. Most of the common variants (minor allele frequency [MAF] ≥5 %) identified by GWAS have modest effects on lipid levels, and have overall a small contribution to total genetic variance of lipid traits (~25–30 % of the heritability) [4–8]. A portion of the missing heritability of lipid traits could be explained by low frequency (LoF)/rare variants (MAF <5 %) as suggested by recent studies [9–11].
HDL, the smallest and densest (d = 1.063–1.21 g/mL) class of lipoprotein particles, has a variety of antiatherogenic properties [12]. One of the HDL properties to protect against CHD is mediated by reverse cholesterol transport (RCT) from peripheral tissues back to the liver [13]. Scavenger receptor class B member 1 (SCARB1, protein; SCARB1, gene) serves as a HDL-C receptor in RCT that mediates selective uptake of HDL-C cholesteryl esters (CE) by the liver and free cholesterol efflux from cells to HDL-C [14]. SCARB1 is also implicated in the metabolism of apolipoprotein B (ApoB)-containing particles [15–21].
The SCARB1 gene (Entrez Gene ID: 949) is located on human chromosome 12, and is abundantly expressed in liver and steroidogenic tissues [22, 23]. The role of SCARB1 in HDL-C and ApoB-containing lipoproteins metabolism has been established in animal studies. The disruption of SCARB1 is associated with increased HDL-C levels and decreased CE uptake [24–26]. Whereas the overexpression of SCARB1 reduces levels of HDL-C, ApoA-I, very low-density lipoprotein cholesterol (VLDL-C), LDL-C, and ApoB [15–17, 19] and promotes the hepatic uptake of CE as well as the biliary secretion of HDL-C [15, 27]. The SCARB1 expression is also significantly associated with hepatic VLDL-triglycerides (TG) and VLDL-ApoB production. Hepatic VLDL cholesterol production together with VLDL clearance is enhanced in response to SCARB1 overexpression [21]. In contrast, reduced hepatic VLDL-TG and VLDL-ApoB production is associated with SCARB1 knockout status [18, 20, 21].
In humans, three SCARB1 mutations (rs397514572 [p.Ser112Phe], rs187831231 [p.Thr175Ala], and rs387906791 [p.Pro297Ser]; MIM: 601040) have been reported to be associated with significantly increased HDL-C levels [28, 29]. Moreover, several genetic studies have demonstrated the association of common SCARB1 variation with lipoprotein-lipid levels [5, 28–39] and subclinical atherosclerosis [40].
To our knowledge, no genetic study has exclusively investigated the association between SCARB1 and lipid traits in native African populations to date. The objective of this study was to resequence all 13 exons and exon-intron boundaries of SCARB1 in 95 African Blacks from Nigeria with extreme HDL-C levels for variant discovery and then to genotype selected variants in the entire sample of 788 African Blacks, followed by genotype-phenotype association analyses with five major lipid and apolipoprotein (Apo) traits (HDL-C, LDL-C, TG, ApoA-I and ApoB). Because our initial gene-based analysis demonstrated evidence of association with HDL-C and ApoA-I, our subsequent analyses focused on these two traits.
Methods
Study population
The present study was carried out on 788 African Black subjects from Benin City, Nigeria, who were recruited as part of a population-based epidemiological study on CHD risk factors. Detailed information on the study design and population description is provided elsewhere [41]. In brief, 788 recruited subjects were healthy civil servants (37.18 % females) from three government ministries of the Edo state in Benin City, Nigeria, aged between 19 and 70 years, including 464 junior staff (non-professional staff with salary grades 1–6), and 324 senior staff (professional and administrative staff with salary grades 7–16). The summary features, including biometric and quantitative data of the entire sample of 788 subjects are given in Table 1 and Additional file 1: Table S1.
Table 1.
Characteristics and lipid profile of 95 individuals with extremea HDL-C levels and of the entire sample of 788 African Blacks
| 95 Individuals with Extremea HDL-C Levels | The Entire Sampleb | |||
|---|---|---|---|---|
| Variables | High HDL-C Group | Low HDL-C Group | P d | |
| (HDL-C rangec: 68.30–99.00 mg/dL) | (HDL-C rangec: 10.30–35.00 mg/dL) | |||
| N (Females, n) | 48 (24) | 47 (24) | 1.00 | 788 (293) |
| Age, years | 41.29 ± 8.72 | 40.87 ± 7.12 | 0.80 | 40.95 ± 8.39 |
| BMI, kg/m2 | 22.06 ± 4.70 | 23.91 ± 5.51 | 0.08 | 22.87 ± 4.04 |
| Total Cholesterol, mg/dL | 201.00 ± 39.68 | 141.68 ± 31.03 | 2.40E-12 | 172.01 ± 38.47 |
| LDL-Cholesterol, mg/dL | 112.55 ± 39.75 | 95.04 ± 28.28 | 0.02 | 109.25 ± 34.40 |
| HDL-Cholesterol, mg/dL | 76.05 ± 7.53 | 25.51 ± 5.66 | 2.20E-16 | 47.88 ± 12.87 |
| Triglycerides, mg/dL | 61.98 ± 19.85 | 95.79 ± 73.21 | 0.004 | 72.96 ± 39.32 |
| Apolipoprotein A-I, mg/dL | 166.04 ± 28.19 | 103.84 ± 27.23 | 2.20E-16 | 137.03 ± 28.46 |
| Apolipoprotein B, mg/dL | 66.00 ± 20.22 | 69.64 ± 21.46 | 0.40 | 66.98 ± 22.19 |
BMI body mass index, HDL-C/HDL-Cholesterol high-density lipoprotein cholesterol, LDL-Cholesterol low-density lipoprotein cholesterol
Values are presented as unadjusted means ± standard deviation (SD), unless otherwise mentioned
aDistribution of HDL-C was adjusted for sex and age: HDL-C levels ≥90th % tile defined as the “High HDL-C group”, and HDL-C levels ≤10th % tile defined as the “Low HDL-C group”
bAll data were unadjusted and included individuals with missing values or outliers (values beyond mean ± 3.5 SD)
cUnadjusted range values
dUnadjusted P-values were calculated with t-test or χ2 test depending on types of variables
For resequencing, 95 individuals with extreme HDL-C levels (within the upper and lower 10th percentiles of HDL-C distribution) were chosen from the entire sample of 788 African Blacks. Resequencing sample comprised of 48 individuals with high HDL-C levels (≥90th percentile, range 68.30–99.00 mg/dL; Table 1) and 47 individuals with low HDL-C levels (≤10th percentile, range 10.30–35.00 mg/dL; Table 1). The University of Pittsburgh Institutional Review Board approved the study protocol. All participants gave their informed consent.
Lipid and apolipoprotein measurements
At least 8-hour fasting blood samples were collected from all participants. Serum specimens were separated by centrifugation of blood samples and then stored at −70 °C for 6–12 months until ready for testing. Lipid and apolipoprotein measurements included total cholesterol, HDL-C, TG, ApoA-I, and ApoB and were done with standard assays at the Heinz Nutrition Laboratory, University of Pittsburgh under the Centers for Disease Control Lipid Standardization Program [41]. LDL-C was calculated with the Friedewald equation [42] when TG levels were less than 400 mg/dL.
PCR and sequencing
Genomic DNA was isolated from clotted blood using the standard DNA extraction procedure. All 13 SCARB1 exons (isoform 1, NM_005505), exon-intron boundaries, and 1 kb of each of 5′ and 3′ flanking regions on chromosome 12 (hg19, chr12: 125,262,175-125,348,519) were polymerase chain reaction (PCR) amplified and sequenced. Specific primers were designed using the Primer3 software (Whitehead Institute for Biomedical Research, http://bioinfo.ut.ee/primer3-0.4.0/) to cover 13 target regions, resulting in 14 amplicons, including two overlapping amplicons for the largest last exon 13. PCR reaction and cycling conditions are available upon request. The primer sequences and amplicon sizes are given in Additional file 2: Table S2.
Automated DNA sequencing of PCR products was performed in a commercial lab (Beckman Coulter Genomics, Danvers, MA, USA) using Sanger method and ABI 3730XL DNA Analyzers (Applied Biosystems, Waltham, MA, USA). Variant analysis was performed using Variant Reporter (version 1.0, Applied Biosystems, Waltham, MA, USA) and Sequencher (version 4.8, Gene Codes Corporation, Ann Arbor, MI, USA) software in our laboratory.
Variant selection for genotyping
Of 83 variants identified in the discovery step (see Additional file 3: Table S3, Additional file 4: Table S4, Additional file 5: Figure S1, and Additional file 6: Figure S2), 78 (28 with MAF ≥5 % and 50 with MAF <5 %) were selected based on the pairwise linkage disequilibrium (LD) and Tagger analysis using an r2 threshold of 0.90 (5 were excluded due to high LD) in Haploview (Broad Institute of MIT and Harvard, https://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/haploview) [43] for follow-up genotyping in the entire sample (n = 788). Since our sequencing was focused primarily on coding regions, in addition we selected 69 HapMap tag single nucleotide polymorphisms [SNPs] (out of total 108 HapMap tagSNPs; see Additional file 7: Table S5 and Additional file 8: Figure S3) based on Tagger analysis (MAF ≥5 % and r2 ≥ 0.80) of HapMap data (Release #27) from the Yoruba people of Ibadan, Nigeria (YRI), in order to cover the entire gene for common genetic variation information. Moreover, we selected two SCARB1 variants previously reported to be significantly associated with lipid traits in the literature (Additional file 9: Table S6). Conclusively, a total of 149 variants, comprising of 78 sequence variants, 69 common HapMap-YRI tagSNPs, and two relevant associated variants, were selected for follow-up genotyping.
Genotyping
Genotyping of selected variants in the total sample of 788 individuals was performed by using either iPLEX Gold (Sequenom, Inc., San Diego, CA, USA) or TaqMan (Applied Biosystems, Waltham, MA, USA) methods and following the manufacturers’ protocols.
Out of 149 selected variants, two failed assay designs and nine failed genotyping runs (see details in Additional file 3: Table S3, Additional file 7: Table S5, and Additional file 9: Table S6). Quality control (QC) measures for successfully genotyped variants were as follow: a genotype call rate of ≥90 %, a discrepancy rate of <1 in 10 % replicates, and no deviation from Hardy-Weinberg equilibrium [HWE] (P >3.62 × 10−4 after Bonferroni correction). Ultimately, a total of 137 QC-passed genotyped variants were included in genetic association analyses (see Additional file 9: Table S6, Additional file 10: Table S7, Additional file 11: Figure S4, and Additional file 12: Figure S5).
Statistical analysis
We used the Haploview program to determine allele frequencies, to test HWE for genotype distribution, and to evaluate the LD and pairwise correlations (r2) between variants [43].
The values of each lipid phenotype outside the mean ± 3.5 standard deviation (SD) were excluded from downstream gene-based, single-site, and haplotype analyses. However, the extreme phenotypic values associated with rare variants (MAF ≤1 %) were maintained during rare variant analysis, as was the case for the p70201/chr12:125279319 variant (see study workflow in Fig. 1). Values of the five lipid and apolipoprotein traits—HDL-C, LDL-C, TG, ApoA-I, and ApoB—were transformed using the Box-Cox transformation. For each trait, we used stepwise regression method to select the most parsimonious set of covariates from the following list: sex, age, body mass index, waist, current smoking (yes/no), minutes of walking or biking to work each day (jobmin), and occupational status (staff: junior [non-professional staff]/senior [professional and administrative staff]). Genetic association analyses, including gene-based, single-site, LoF/rare variant, and haplotype association tests, were performed using linear regression models that included significant covariates for each variable (Additional file 13: Table S8).
Fig. 1.
Summary of the study design and flow. Chart presents an overview of the study design and flow, including sequencing and genotyping stages and analysis approaches. ApoA-I, apolipoprotein A-I; ApoB, apolipoprotein B; HDL-C, high-density lipoprotein cholesterol; LD, linkage disequilibrium; LDL-C, low-density lipoprotein cholesterol; LoF, low-frequency; MAF, minor allele frequency; SD, standard deviation; SKAT-O, an optimal sequence kernel association test; SNP, single nucleotide polymorphism; TG, triglycerides; YRI, Yoruba people of Ibadan from Nigeria
The gene-based association analysis was conducted under linear additive model for the combined evaluation of common and LoF/rare variants (n = 136, excluding p70201/chr12:125279319; see details above in paragraph two of this section) for five major lipid traits using the versatile gene-based association study [VEGAS] (http://gump.qimr.edu.au/VEGAS/) software [44]. The significance threshold for the gene-based test was set at P-value of 0.05.
Following gene-based analysis, which primarily implicated SCARB1 in regulation of HDL-C and ApoA-I levels, we further elucidated the association of SCARB1 variants with these two traits using additional tests. In single-site association analysis, P-values for each trait were adjusted for multiple testing using Benjamini-Hochberg procedure [45] to determine the false discovery rate [FDR] (q-value). For common variants (MAF ≥5 %), a nominal P-value of <0.05 was considered to be suggestive evidence of association, and an FDR cut-off of 0.20 was used to define statistical significance. For LoF/rare variants (MAF <5 %), the single-site association results were interpreted separately because of inadequate power of our study to detect individual statistical significance for these variants.
We conducted an optimal sequence kernel association test (SKAT-O) [46] to evaluate the association between a total of 43 LoF/rare variants (MAF <5 %) and the two lipid traits (HDL-C and ApoA-I) by using three different MAF thresholds: <5 % (n = 43), ≤2 % (n = 26), and ≤1 % (n = 23). A significant SKAT-O test was set at a P-value of <0.05.
Haplotype association analysis was performed using the generalized linear model. We applied a fixed sliding window approach that included four variants per window and sliding for one variant at a time. For each window, a global P-value was used to assess the association between the haplotypes with frequency >1 % and a given trait. A global P-value threshold of 0.05 was used to define significant haplotype association.
All analyses, except for VEGAS, were performed using the R statistical software (http://www.r-project.org/) and relevant R packages (i.e., Haplo.Stats for haplotype analysis and SKAT for SKAT-O analysis).
Results
Identification and distribution of SCARB1 sequence variants in 95 individuals with extreme HDL-C levels
Resequencing of SCARB1 exons and exon-intron boundaries plus flanking regions in 95 African Blacks with extreme HDL-C levels identified 83 variants, of which 51 had MAF <5 % (Additional file 3: Table S3 and Additional file 5: Figure S1). The majority of 83 variants (n = 73) were previously identified (dbSNP build 139: GRCh37.p10). Most variants (n = 80) were singlenucleotide variations [SNVs] (67 transitions and 13 transversions); the rest (n = 3) were short insertion and deletion variations (indels).
Tagger analysis using an r2 cutoff of 0.9 identified 28 bins for 32 common variants (MAF ≥5 %), of which three included more than one variant (r2 ranging from 0.95 to 1.0) (Additional file 6: Figure S2). One of these three bins contained two variants (rs204901986 and rs34339961) in complete LD (r2 = 1.0). Of 51 LoF/rare variants (MAF between 1 and 5 %, n = 31; MAF ≤1 %, n = 20), 17 were present only in the high HDL-C group (MAF ranging between 0.010 and 0.042) and eight were observed only in the low HDL-C group (MAF ranging between 0.011 and 0.033). In the high HDL-C group, 29 of 48 (~60 %) individuals cumulatively carried at least one LoF/rare variant, ranging from 1 to 7 variants. Similarly, in the low HDL-C group, 27 of 47 (~57 %) individuals carried at least one LoF/rare variant, ranging from 1 to 9 variants.
Most variants (n = 60) from our sequencing were located in intronic regions, of which two (rs113910315, MAF = 0.005 and rs10396210, MAF = 0.138) were within splice sites (defined as ± 20 bp from the start or end of an exon). The former splice site variant was observed only in the low HDL-C group.
Of the total eight coding variants observed, four were common variants (rs2070242 [p.Ser4Ser], rs10396208 [p.Cys21Cys], and rs5888 [p.Ala350Ala], and rs701103 [p.Gly499Arg]—3′ untranslated region [UTR] in isoform 1 and exon 13 in isoform 2), and the remaining four were LoF/rare variants (rs4238001 [p.Gly2Ser], rs5891 [p.Val135Ile], rs5892 [p.Phe301Phe], and rs141545424 [p.Gly501Gly]). Of note, two LoF/rare coding variants, (rs5891 [p.Val135Ile] and rs141545424 [p.Gly501Gly]), were found only in the high HDL-C group.
Fifteen variants were located in either UTRs (n = 5) or flanking regions (n = 10). One 3′ UTR variant (rs150512235, MAF = 0.006) was very close to a predicted microRNA-145 (miR-145) target site (TargetScanHuman version 6.2, http://www.targetscan.org/). One 5′ flanking variant (rs181338950, MAF = 0.048) was located in the putative promoter region [47].
All 10 novel variants (9 SNVs and 1 insertion) identified in this study have been submitted to dbSNP database ([batch ID: SCARB1_AB]:
http://www.ncbi.nlm.nih.gov/SNP/snp_viewTable.cgi?handle=KAMBOH) and were non-coding with MAF <5 % (ranging between 0.005 and 0.011; Additional file 4: Table S4). Of these novel variants, six and four were present only in the high and low HDL-C groups, respectively.
Genotyping of SCARB1 variants in the entire sample of 788 individuals
Since our sequencing was focused primarily on coding regions, we selected additional HapMap tagSNPs from the HapMap-YRI data in order to cover the entire SCARB1 gene for common genetic variation in SCARB1. Altogether we selected 149 variants for genotyping in our entire African Black sample as follows: 78 variants (28 common variants and 50 LoF/rare variants) discovered in the sequencing step (Additional file 3: Table S3, Additional file 5: Figure S1, and Additional file 6: Figure S2), 69 common HapMap-YRI tagSNPs (Additional file 7: Table S5), and two additional variants with reported association in the literature (Additional file 9: Table S6).
Of these 149 variants, 11 (10 from sequencing, including one promoter [rs181338950], one coding (rs4238001 [p.Gly2Ser]), and one novel [p87459/chr12:125262061], and 1 from HapMap tagSNPs [rs4765180]) failed genotyping, and one (rs866793 from HapMap tagSNPs) failed QC measures. Thus, a total of 137 variants (Additional file 9: Table S6 and Additional file 11: Figure S4) that passed QC were advanced into association analyses with five lipoprotein-lipid traits.
The majority of 137 genotyped variants (n = 120) were located in introns, 11 were in exons, and six were in 3′ flanking region (Table 2 and Additional file 12: Figure S5). Ninety-four of 137 variants had MAF ≥5 %, including four coding variants, one UTR variant, two deletions, and one splice site variant. The remaining 43 variants had MAF <5 % (MAF between 1 and 5 %, n = 20; MAF ≤1 %, n = 23), including three coding variants, three UTR variants, one insertion, and one splice variant.
Table 2.
Distribution of 137 SCARB1 genotyped variants
| Total | MAF ≥5 % | MAF between 1-5 % | MAF ≤1 % | |
|---|---|---|---|---|
| N (%) | n (%) | n (%) | n (%) | |
| Total variants | 137 (100.00) | 94 (68.61) | 20 (14.60) | 23 (16.79) |
| By known/novela | ||||
| Known | 128 (93.43) | 94 (68.61) | 20 (14.60) | 14 (10.22) |
| Single-nucleotide variation | 126 | 92 | 20 | 14 |
| Short indels | 2 | 2 | ||
| Novel | 9 (6.57) | 9 (6.57) | ||
| Single-nucleotide variation | 8 | 8 | ||
| Short indels | 1 | 1 | ||
| By location | ||||
| Exons-codingc | 7 | 4c | 1 | 2 |
| Exons-UTRs | 4 | 1 | 1 | 2 |
| Introns | 118 | 85 | 16 | 17 |
| Introns-splice sitesb | 2 | 1 | 1 | |
| 3′ flanking | 6 | 3 | 2 | 1 |
| By amino acid change | ||||
| Non-synonymousc | 2 | 1c | 1 | |
| Synonymous | 5 | 3 | 1 | 1 |
Indels insertion and deletion variations, MAF minor allele frequency, UTR untranslated region
The list of 137 genotyped variants is shown in Additional file 9: Table S6
The list of 10 novel variants is shown in Additional file 4: Table S4
adbSNP build 139: GRCh37.p10. All 10 novel variants identified in this study have been submitted to dbSNP (batch ID: SCARB1_AB): http://www.ncbi.nlm.nih.gov/SNP/snp_viewTable.cgi?handle=KAMBOH
bSplice site, defined as ± 20 bp from the start or end of an exon
cIncluding rs701103 (p.Gly499Arg; MAF = 0.2451) that is located in exon 13-3′ UTR and translated only in isoform 2
Of the 10 novel variants discovered in the sequencing step, nine (8 SNVs and 1 insertion) with MAF <1 % were successfully genotyped (Additional file 4: Table S4). There was one individual with plasma HDL-C levels above the mean + 3.5 SD carrying one novel variant—p70201/chr12:125279319 (MAF = 0.0010). Although this extreme HDL-C value was excluded as outlier from the gene-based, single-site, and haplotype analyses, it was included in the SKAT-O rare variant analysis considering a possible large effect size of this variant (Fig. 1).
Gene-based association analyses
Gene-based tests revealed a nominally significant association (P = 0.0421; Table 3) of SCARB1 variants with HDL-C levels (best SNP: rs141545424 [p.Gly501Gly], exon 12, MAF = 0.0007, P = 0.0016). Additionally, a trend for association (P = 0.1016) was also observed for ApoA-I levels (best SNP: rs7134858, intron 6, MAF = 0.1560, P = 0.0052).
Table 3.
Gene-based association analysis results
| Trait | Variants | Test Statistics | P | Best SNP | ||
|---|---|---|---|---|---|---|
| (n) | SNP Namea-SNP IDb | MAF | P | |||
| HDL-C | 136 | 207.5483 | 0.0421 | p82264-rs141545424 | 0.0007 | 0.0016 |
| LDL-C | 136 | 134.1860 | 0.4640 | p32777-rs11057841 | 0.2805 | 0.0047 |
| TG | 136 | 118.1598 | 0.6700 | p86316-rs701104 | 0.0487 | 0.0357 |
| ApoA-I | 136 | 183.5565 | 0.1016 | p55963-rs7134858 | 0.1560 | 0.0052 |
| ApoB | 136 | 143.7284 | 0.3760 | p22116-rs12370382 | 0.0645 | 0.0153 |
ApoA-I apolipoprotein A-I, ApoB apolipoprotein B, HDL-C high-density lipoprotein cholesterol, LDL-C low-density lipoprotein cholesterol, MAF minor allele frequency, SNP single nucleotide polymorphism, TG triglycerides
All results were adjusted for covariates: sex, age, body mass index, waist, current smoking (yes/no), minutes of walking or biking to work each day (jobmin), and occupational status [staff: junior (non-professional staff)/senior (professional and administrative staff)]
Nominally significant gene-based P-values (P < 0.05) are shown in bold
aRefSeq of SCARB1: hg19, NM_005505 (CHIP Bioinformatics)
bdbSNP build 139: GRCh37.p10
Since the gene-based tests showed evidence of associations with HDL-C and ApoA-I, we primarily focused on these two traits to further examine the SCARB1 variants in the entire sample of 788 African Blacks.
Single-site association analyses of common SCARB1 variants
Of 94 common SCARB1 variants with MAF ≥5 %, 10 showed nominal associations (P < 0.05) with HDL-C and/or ApoA-I (Table 4; see results for each trait in Additional file 14: Table S9 and Additional file 15: Table S10), of which three (rs11057851, rs4765615, and rs838895) exhibited associations with both HDL-C and ApoA-I.
Table 4.
Nominally significant single-site associations (P < 0.05) of common SCARB1 variants
| SNP Namea | SNP IDb | Chr12 Positionc | Location | Amino Acid Change | RegDB Scored | Major/Minor Alleles | MAF | β | SE | R2 (%) | P | FDR | Secondary Trait (Effect) | Top 3 Variants |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HDL-C | ||||||||||||||
| p20207 | rs11057853 | 125329313 | Intron 1 | 5 | G/A | 0.4484 | 0.4082 | 0.1925 | 1.0650 | 0.0343 | 0.4235 | |||
| p20741 | rs11057851 | 125328779 | Intron 1 | 5 | C/T | 0.3237 | −0.5924 | 0.2067 | 1.3010 | 0.0043 | 0.1465 | ApoA-I (↓) | Top 1 | |
| p45516 | rs1902569 | 125304004 | Intron 1 | 5 | G/A | 0.1544 | 0.5447 | 0.2629 | 0.6390 | 0.0386 | 0.4375 | |||
| p49690 | rs4765615 | 125299830 | Intron 2 | 5 | G/A | 0.4426 | −0.4646 | 0.1866 | 0.9330 | 0.0130 | 0.2526 | ApoA-I (↓) | ||
| p79828 | rs838895 | 125269692 | Intron 11 | 5 | C/G | 0.3171 | 0.4961 | 0.2059 | 0.8220 | 0.0162 | 0.2756 | ApoA-I (↑) | ||
| ApoA-I | ||||||||||||||
| p20741 | rs11057851 | 125328779 | Intron 1 | 5 | C/T | 0.3237 | −1.2331 | 0.5117 | 0.8600 | 0.0162 | 0.3186 | HDL-C (↓) | ||
| p49690 | rs4765615 | 125299830 | Intron 2 | 5 | G/A | 0.4426 | −0.9139 | 0.4614 | 0.6770 | 0.0480 | 0.5022 | HDL-C (↓) | ||
| p55963 | rs7134858 | 125293557 | Intron 6 | 6 | C/T | 0.1560 | 1.7537 | 0.6260 | 1.0710 | 0.0052 | 0.2918 | Top 2 | ||
| p63483 | rs838912 | 125286037 | Intron 7 | 7 | G/A | 0.0867 | 1.8700 | 0.8230 | 0.6880 | 0.0234 | 0.3972 | |||
| p64772 | rs5888 | 125284748 | Exon 8 | Ala350Ala | 3a | C/T | 0.0961 | 2.0962 | 0.7888 | 0.9460 | 0.0080 | 0.2918 | Top 3 | |
| p79721 | rs838896 | 125269799 | Intron 11 | 5 | G/C | 0.3104 | 1.1147 | 0.5056 | 0.7270 | 0.0278 | 0.4197 | |||
| p79828 | rs838895 | 125269692 | Intron 11 | 5 | C/G | 0.3171 | 1.2206 | 0.5074 | 0.7800 | 0.0164 | 0.3186 | HDL-C (↑) | ||
| p83884 | rs701106 | 125265636 | Intron 12 | 5 | C/T | 0.2597 | 1.2967 | 0.5352 | 0.7770 | 0.0156 | 0.3186 |
ApoA-I apolipoprotein A-I, FDR false discovery rate, HDL-C high-density lipoprotein cholesterol, MAF minor allele frequency, RegDB RegulomeDB, SE standard error, SNP single nucleotide polymorphism, UTR untranslated region, R2, the proportion of the phenotypic variance explained by the variant; ↓, decreased; ↑, increased
Alleles on reverse strand. HDL-C and ApoA-I variables were in mg/dL and Box-Cox transformed
Results were adjusted for covariates: sex, age, waist, current smoking (yes/no), and minutes of walking or biking to work each day (jobmin) for HDL-C; sex and age for ApoA-I
The most significant P-value for each trait is shown in bold, see the single-site association (−log10 P) plot and pairwise correlations (r2) in Fig. 2
FDR that reached a threshold of <0.20 is shown in bold
a, cRefSeq of SCARB1: hg19, NM_005505 (CHIP Bioinformatics)
bdbSNP build 139: GRCh37.p10
dDetailed RegulomeDB (version 1.0) scoring scheme is described in Additional file 17: Table S12 or at http://regulome.stanford.edu/help, see functional assignments in Additional file 18: Table S13
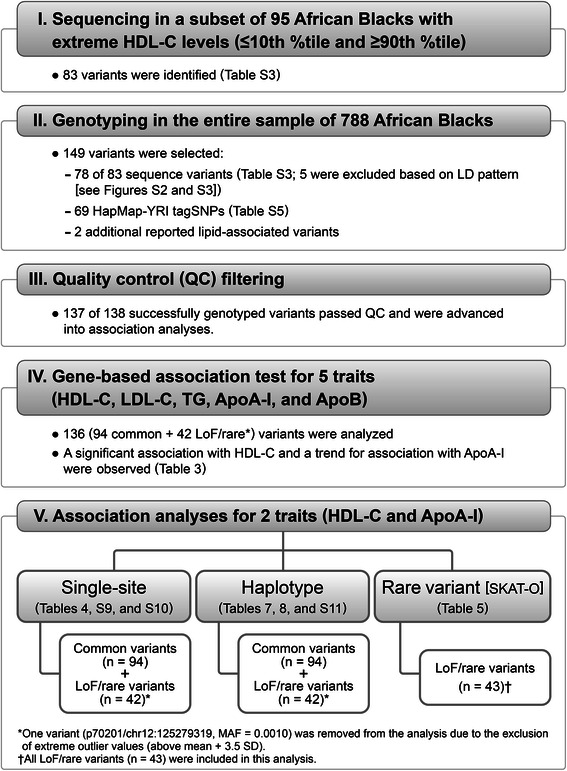
The most significant association was found between rs11057851 and HDL-C (β = −0.5924, P = 0.0043, FDR = 0.1465). The second best association was between rs7134858 and ApoA-I (β = 1.7537, P = 0.0052, FDR = 0.2918), followed by the association of rs5888 (p.Ala350Ala) with ApoA-I (β = 2.0962, P = 0.0080, FDR = 0.2918).
Of 10 variants that showed nominal associations, high LD (r2 > 0.80) was observed for two pairs of variants (Fig. 2), between rs8388912 and rs5888 (p.Ala350Ala; r2 = 0.86), and between rs838896 and rs838895 (r2 = 0.84).
Fig. 2.
Single-site P-values of 94 SCARB1 common variants for HDL-C and ApoA-I. Top: The -log10 P-values are presented in the Y-axis. A total of 94 genotyped variants with MAF ≥5 % are shown on SCARB1 gene (5′ → 3′; RefSeq: hg19, NM_005505) in the X-axis. The dash line indicates the nominal significance threshold (P = 0.05). Middle: Gene structure of SCARB1. Bottom: Linkage disequilibrium (LD) plot of 10 SCARB1 variants with P-values <0.05. Shades and values (r2 × 100) in each square of LD plot indicate pairwise correlations: black indicating r2 = 1, white indicating r2 = 0, and shade intensity indicating r2 between 0 and 1. Marker names are shown as “SNP name-SNP ID”. SNP ID is based on dbSNP build 139. ApoA-I, apolipoprotein A-I; FDR, false discovery rate; HDL-C, high-density lipoprotein cholesterol; LD, linkage disequilibrium; MAF, minor allele frequency; SNP, single nucleotide polymorphism; UTR, untranslated region
Association analyses of low-frequency/rare SCARB1 variants
The LoF/rare variants (n = 43) were categorized into three groups based on their frequencies for association analysis with HDL-C and ApoA-I using SKAT-O: MAF <5 % (n = 43), MAF ≤2 % (n = 26), and MAF ≤1 % (n = 23). Although no association between LoF/rare variants and ApoA-I was detected, the group of 23 variants with MAF ≤1 % yielded nominal association with HDL-C levels (P = 0.0478; Table 5).
Table 5.
Association results for low-frequency and rare SCARB1 variants (MAF <5 %)
| MAF | No of Variants | No of Samples with/without Variants | HDL-C | ApoA-I | ||
|---|---|---|---|---|---|---|
| Stat | P | Stat | P | |||
| ≤0.01 | 23a | 93/694 | 126653.8207 | 0.0478 | 60151.0985 | 0.3707 |
| ≤0.02 | 26 | 134/653 | 123009.0805 | 0.1324 | 48439.6697 | 0.5166 |
| <0.05 | 43 | 442/346 | 135697.1974 | 0.0737 | 298813.0544 | 0.1517 |
ApoA-I apolipoprotein A-I, HDL-C high-density lipoprotein cholesterol, MAF minor allele frequency, SD standard deviation, SNP single nucleotide polymorphism
Results were adjusted for covariates: sex, age, waist, current smoking (yes/no), and minutes of walking or biking to work each day (jobmin) for HDL-C; sex and age for ApoA-I
Nominally significant P-values (P < 0.05) are shown in bold
aIncluding p70201/chr12:125279319 that was observed in one individual with an outlier value (above the mean + 3.5 SD). See details in Result Section 3.5
We then individually examined the association of 23 variants with MAF ≤1 % with HDL-C and ApoA-I. Six of these rare variants showed association with either HDL-C levels or both HDL-C and ApoA-I levels (Table 6). While three of them are known variants (rs115604379, rs377124254, and rs141545424 [p.Gly501Gly]), the other three are novel (p52919/chr12:125296601, p54611/chr12:125294909, and p54856/chr12:125294664). Moreover, four of these six rare variants (rs377124254, rs141545424 [p.Gly501Gly], p54611/chr12:125294909, and p54856/chr12:125294664) were present in individuals with extreme phenotypic values (above or below the 3rd percentile). Two of these variants (rs377124254: β = 11.5518, P = 0.0016; rs141545424 [p.Gly501Gly]: β = 11.585, P = 0.0016) were found in a single subject who had very high HDL-C level. Whereas the other two were observed in one individual each, who had extremely low HDL-C levels (p54611/chr12:125294909: β = −9.5243, P = 0.0097; p54856/chr12:125294664: β = −8.4305, P = 0.0215) and ApoA-I levels (p54611/chr12:125294909: β = −19.3821, P = 0.0344; p54856/chr12:125294664: β = −24.0757, P = 0.0082). This rare variant group also included a novel variant (p70201/chr12:125279319) that was observed in one individual with an unusually high plasma HDL-C level (above the mean + 3.5 SD).
Table 6.
Characteristics and effects of 6 SCARB1 rare variants of interest
| SNP Namea | SNP IDb | Chr12 Positionc | Location | Amino Acid Change | RegDB Scored | Major/Minor Alleles | MAF | GT | GT Count (Carrier Freq) | Adjusted Mean ± SD (mg/dL) | β | SE | R2 (%) | P | FDR | Second Assoc Trait (Effect) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HDL-C | ||||||||||||||||
| p52919 | 125296601 | Intron 4 | 5 | G/T | 0.0013 | GG | 734 | 47.87 ± 12.71 | −7.4063 | 2.5863 | 1.1050 | 0.0043 | 0.1465 | ApoA-I (↓) | ||
| GT | 2 (0.27) | 24.67 ± 9.26 | ||||||||||||||
| p53372 | rs115604379 | 125296148 | Intron 5 | 5 | C/T | 0.0066 | CC | 729 | 47.68 ± 12.64 | 3.0372 | 1.1642 | 0.9140 | 0.0093 | 0.2190 | ||
| CT | 10 (1.35) | 58.2 ± 13.03 | ||||||||||||||
| p54611 | 125294909 | Intron 5 | 4 | T/C | 0.0007 | TT | 742 | 47.86 ± 12.68 | −9.5243 | 3.6710 | 0.8920 | 0.0097 | 0.2190 | ApoA-I (↓) | ||
| TC | 1 (0.13) | 19.59 ± NA | ||||||||||||||
| p54856 | 125294664 | Intron 6 | 4 | C/T | 0.0007 | CC | 742 | 47.85 ± 12.70 | −8.4305 | 3.6579 | 0.7130 | 0.0215 | 0.3243 | ApoA-I (↓) | ||
| CT | 1 (0.13) | 21.48 ± NA | ||||||||||||||
| p77620 | rs377124254 | 125271900 | Intron 10 | 5 | G/A | 0.0007 | GG | 735 | 47.77 ± 12.67 | 11.5518 | 3.6514 | 1.3500 | 0.0016 | 0.1104 | ||
| GA | 1 (0.14) | 90.2 ± NA | ||||||||||||||
| p82264 | rs141545424 | 125267256 | Exon 12 | Gly501Gly | 5 | C/A | 0.0007 | CC | 739 | 47.77 ± 12.66 | 11.5850 | 3.6469 | 1.3530 | 0.0016 | 0.1104 | |
| CA | 1 (0.14) | 90.31 ± NA | ||||||||||||||
| ApoA-I | ||||||||||||||||
| p52919 | 125296601 | Intron 4 | 5 | G/T | 0.0013 | GG | 741 | 136.81 ± 27.74 | −13.4137 | 6.4689 | 0.5750 | 0.0385 | 0.4359 | HDL-C (↓) | ||
| GT | 2 (0.27) | 97.42 ± 18.38 | ||||||||||||||
| p54611 | 125294909 | Intron 5 | 4 | T/C | 0.0007 | TT | 748 | 136.83 ± 27.66 | −19.2831 | 9.0970 | 0.5980 | 0.0344 | 0.4359 | HDL-C (↓) | ||
| TC | 1 (0.13) | 80.62 ± NA | ||||||||||||||
| p54856 | 125294664 | Intron 6 | 4 | C/T | 0.0007 | CC | 748 | 136.87 ± 27.61 | −24.0757 | 9.0781 | 0.9330 | 0.0082 | 0.2918 | HDL-C (↓) | ||
| CT | 1 (0.13) | 67.98 ± NA | ||||||||||||||
ApoA-I apolipoprotein A-I, FDR false discovery rate, GT genotype, HDL-C high-density lipoprotein cholesterol, MAF minor allele frequency, RegDB RegulomeDB, SD standard deviation, SE standard error, SNP single nucleotide polymorphism; R2, the proportion of the phenotypic variance explained by the variant; ↓, decreased
All alleles were on reverse stand. HDL-C and ApoA-I variables were in mg/dL and Box-Cox transformed
Results were adjusted for covariates: sex, age, waist, current smoking (yes/no), and minutes of walking or biking to work each day (jobmin) for HDL-C; sex and age for ApoA-I.
Detailed single-site association results are shown in Additional file 14: Table S9 and Additional file 15: Table S10.
a, cRefSeq of SCARB1: hg19, NM_005505 (CHIP Bioinformatics)
bdbSNP build 139: GRCh37.p10. All 10 novel variants identified in this study have been submitted to dbSNP (batch ID: SCARB1_AB): http://www.ncbi.nlm.nih.gov/SNP/snp_viewTable.cgi?handle=KAMBOH
dThe RegulomeDB (version 1.0) scoring scheme and functional assignments are described in Additional file 17: Table S12 and Additional file 18: Table S13, respectively
Haplotype association analyses
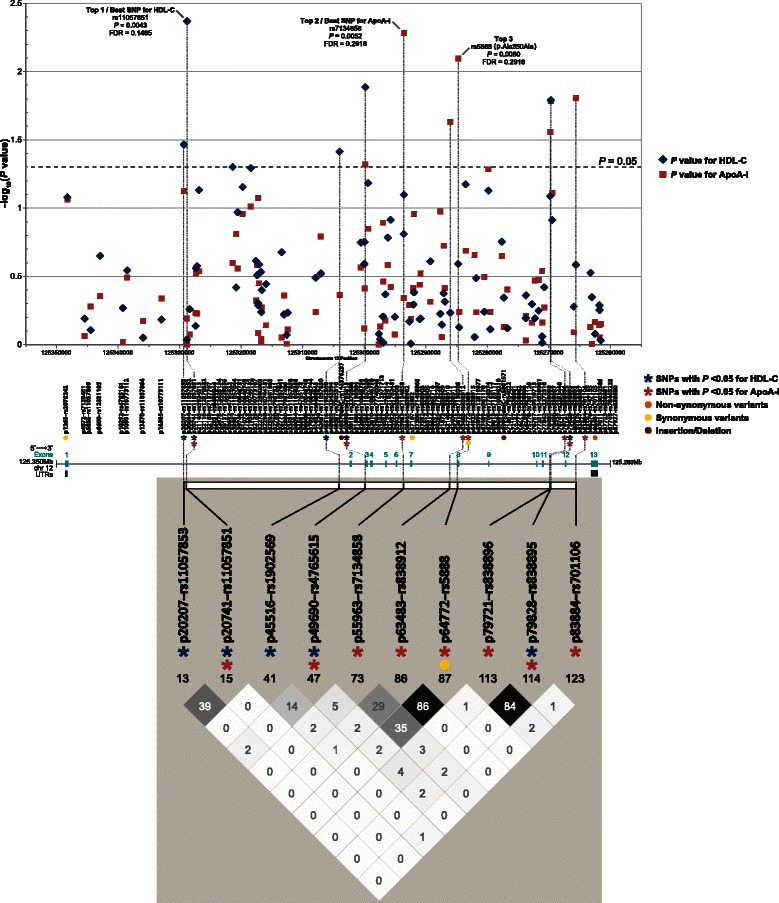
The 4-SNP sliding window haplotype analyses revealed associations of 32 haplotype windows with HDL-C and/or ApoA-I (global P < 0.05; Table 7; see results for each trait in Additional file 16: Table S11), of which five (windows #47, #72, #111, #112, and #123) were associated with both.
Table 7.
Significant haplotype association (global P < 0.05) of 136 SCARB1 genotyped variants with HDL-C and ApoA-I
| Wind # | SNP 1 - SNP 4 | Chr12 Positionc | Location | Amino Acid Change | Major/ Minor Alleles | MAF | β | Single-site P | Haplotype # | Hap Seq | Hap Freq | Coef | SE | t.stat | Hap P | Global P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (SNP Namea-SNP IDb/Chr12 Posc) | ||||||||||||||||
| HDL-C | ||||||||||||||||
| 39 | p41632-rs6488943 | 125307888 | Intron 1 | A/C | 0.2954 | −0.2195 | 0.3244 | h39.1 | CCGG | 0.0315 | 0.4305 | 0.6471 | 0.6654 | 0.5060 | 0.0207 | |
| 39 | p42467-rs11057830 | 125307053 | Intron 1 | C/T | 0.1523 | −0.2810 | 0.3015 | h39.2 | CCGA | 0.2508 | −0.5918 | 0.2725 | −2.1713 | 0.0302 | ||
| 39 | p45516-rs1902569 | 125304004 | Intron 1 | G/A | 0.1544 | 0.5447 | 0.0386 | h39.3 | ATGA | 0.1414 | −0.6841 | 0.3192 | −2.1433 | 0.0324 | ||
| 39 | p45627-rs12297372 | 125303893 | Intron 1 | A/G | 0.0487 | −0.0483 | 0.9156 | h39.4 | ACAA | 0.1514 | 0.1991 | 0.2963 | 0.6720 | 0.5018 | ||
| h39.5 | ACGG | 0.0155 | −1.7144 | 0.9080 | −1.8880 | 0.0594 | ||||||||||
| h39.6 (rare) | **** | 0.0148 | 2.5239 | 1.0902 | 2.3151 | 0.0209 | ||||||||||
| hap.base39 | ACGA | 0.3946 | NA | NA | NA | NA | ||||||||||
| 44 | p48969-rs2343394 | 125300551 | Intron 2 | C/T | 0.1898 | 0.3165 | 0.1788 | h44.1 | TCWG | 0.1855 | 0.5292 | 0.2523 | 2.0977 | 0.0363 | 0.0271 | |
| 44 | p49537-rs7305310 | 125299983 | Intron 2 | C/T | 0.1007 | −0.3396 | 0.2566 | h44.2 | CCDG | 0.2244 | 0.4676 | 0.2429 | 1.9249 | 0.0546 | ||
| 44 | p49570delC-rs145376237 | 125299950 | Intron 2 | W/D | 0.2276 | 0.3121 | 0.1773 | h44.3 | CCWG | 0.0446 | 1.0491 | 0.4882 | 2.1489 | 0.0320 | ||
| 44 | p49690-rs4765615 | 125299830 | Intron 2 | G/A | 0.4426 | −0.4646 | 0.0130 | h44.4 | CTWG | 0.1018 | −0.1197 | 0.3121 | −0.3835 | 0.7015 | ||
| h44.5 (rare) | **** | 0.0089 | −0.9887 | 1.0998 | −0.8990 | 0.3689 | ||||||||||
| hap.base44 | CCWA | 0.4348 | NA | NA | NA | NA | ||||||||||
| 45 | p49537-rs7305310 | 125299983 | Intron 2 | C/T | 0.1007 | −0.3396 | 0.2566 | h45.1 | CDGC | 0.2282 | 0.4661 | 0.2393 | 1.9473 | 0.0519 | 0.0155 | |
| 45 | p49570delC-rs145376237 | 125299950 | Intron 2 | W/D | 0.2276 | 0.3121 | 0.1773 | h45.2 | CWGC | 0.2302 | 0.6926 | 0.2376 | 2.9146 | 0.0037 | ||
| 45 | p49690-rs4765615 | 125299830 | Intron 2 | G/A | 0.4426 | −0.4646 | 0.0130 | h45.3 | TWGC | 0.1020 | −0.0653 | 0.3085 | −0.2115 | 0.8325 | ||
| 45 | p49759-rs146272788 | 125299761 | Intron 2 | C/T | 0.0020 | 2.5988 | 0.2219 | h45.4 (rare) | **** | 0.0030 | 2.0667 | 2.0848 | 0.9913 | 0.3219 | ||
| hap.base45 | CWAC | 0.4366 | NA | NA | NA | NA | ||||||||||
| 46 | p49570delC-rs145376237 | 125299950 | Intron 2 | W/D | 0.2276 | 0.3121 | 0.1773 | h46.1 | DGCG | 0.2228 | 0.4373 | 0.2413 | 1.8123 | 0.0703 | 0.0278 | |
| 46 | p49690-rs4765615 | 125299830 | Intron 2 | G/A | 0.4426 | −0.4646 | 0.0130 | h46.2 | WGCG | 0.3311 | 0.4910 | 0.2105 | 2.3326 | 0.0199 | ||
| 46 | p49759-rs146272788 | 125299761 | Intron 2 | C/T | 0.0020 | 2.5988 | 0.2219 | h46.3 (rare) | **** | 0.0080 | 1.9089 | 1.0569 | 1.8061 | 0.0713 | ||
| 46 | p49978-rs5891 | 125299542 | Exon 3 | Val135lle | G/A | 0.0058 | 1.3374 | 0.2791 | hap.base46 | WACG | 0.4381 | NA | NA | NA | NA | |
| 47 | p49690-rs4765615 | 125299830 | Intron 2 | G/A | 0.4426 | −0.4646 | 0.0130 | h47.1 | ACGG | 0.4346 | −0.4701 | 0.1824 | −2.5777 | 0.0101 | 0.0079 | |
| 47 | p49759-rs146272788 | 125299761 | Intron 2 | C/T | 0.0020 | 2.5988 | 0.2219 | h47.2 (rare) | **** | 0.0101 | 1.4683 | 0.9441 | 1.5552 | 0.1203 | ||
| 47 | p49978-rs5891 | 125299542 | Exon 3 | Val135lle | G/A | 0.0058 | 1.3374 | 0.2791 | hap.base47 | GCGG | 0.5553 | NA | NA | NA | NA | |
| 47 | p50024-rs368880622 | 125299496 | Intron 3 | G/T | 0.0026 | 1.6506 | 0.4362 | |||||||||
| 63 | p53359-rs112371713 | 125296161 | Intron 5 | G/A | 0.1243 | 0.4193 | 0.1651 | h63.1 | ACGA | 0.1237 | 0.3273 | 0.3011 | 1.0871 | 0.2773 | 0.0394 | |
| 63 | p53372-rs115604379 | 125296148 | Intron 5 | C/T | 0.0066 | 3.0372 | 0.0093 | h63.2 | GCGG | 0.0427 | −0.1630 | 0.4738 | −0.3441 | 0.7309 | ||
| 63 | p53790-rs4765614 | 125295730 | Intron 5 | G/A | 0.2653 | −0.3281 | 0.1218 | h63.3 | GCAA | 0.2678 | −0.2408 | 0.2194 | −1.0975 | 0.2728 | ||
| 63 | p54445-rs60910935 | 125295075 | Intron 5 | A/G | 0.0418 | −0.1247 | 0.7963 | h63.4 (rare) | **** | 0.0068 | 2.9428 | 1.2559 | 2.3432 | 0.0194 | ||
| hap.base63 | GCGA | 0.5591 | NA | NA | NA | NA | ||||||||||
| 72 | p55923-rs838900 | 125293597 | Intron 6 | G/A | 0.3921 | 0.2787 | 0.1549 | h72.1 | ACAG | 0.2725 | 0.4039 | 0.2520 | 1.6024 | 0.1095 | 0.0315 | |
| 72 | p55963-rs7134858 | 125293557 | Intron 6 | C/T | 0.1560 | 0.4418 | 0.0799 | h72.2 | ACGG | 0.1086 | −0.1763 | 0.3929 | −0.4486 | 0.6538 | ||
| 72 | p56845-rs838902 | 125292675 | Intron 6 | A/G | 0.4249 | −0.0786 | 0.6801 | h72.3 | GTAG | 0.1284 | 0.3877 | 0.3170 | 1.2228 | 0.2218 | ||
| 72 | p57004-rs187562853 | 125292516 | Intron 6 | G/A | 0.0098 | 1.6474 | 0.0872 | h72.4 | GTGG | 0.0297 | 0.8722 | 0.6546 | 1.3323 | 0.1832 | ||
| h72.5 | GCAG | 0.1716 | −0.4913 | 0.3344 | −1.4690 | 0.1422 | ||||||||||
| h72.6 (rare) | **** | 0.0101 | 1.7731 | 0.9506 | 1.8653 | 0.0625 | ||||||||||
| hap.base72 | GCGG | 0.2791 | NA | NA | NA | NA | ||||||||||
| 111 | p78747-rs2293440 | 125270773 | Intron 11 | T/C | 0.4112 | −0.1684 | 0.3806 | h111.1 | CCCG | 0.0306 | 0.7458 | 0.5599 | 1.3321 | 0.1832 | 0.0040 | |
| 111 | p78791-rs75289200 | 125270729 | Intron 11 | T/C | 0.0321 | 0.7037 | 0.2078 | h111.2 | CTGC | 0.1534 | −0.5556 | 0.2830 | −1.9629 | 0.0500 | ||
| 111 | p79721-rs838896 | 125269799 | Intron 11 | G/C | 0.3104 | 0.3565 | 0.0817 | h111.3 | CTCG | 0.2269 | 0.1234 | 0.2391 | 0.5162 | 0.6058 | ||
| 111 | p79828-rs838895 | 125269692 | Intron 11 | C/G | 0.3171 | 0.4961 | 0.0162 | h111.4 | TTGG | 0.0180 | 2.3022 | 0.7617 | 3.0225 | 0.0026 | ||
| h111.5 | TTCG | 0.0439 | 0.5755 | 0.5317 | 1.0823 | 0.2795 | ||||||||||
| h111.6 | TTCC | 0.0145 | 0.9606 | 0.8068 | 1.1907 | 0.2342 | ||||||||||
| h111.7 (rare) | **** | 0.0033 | 0.7755 | 2.1917 | 0.3538 | 0.7236 | ||||||||||
| hap.base111 | TTGC | 0.5094 | NA | NA | NA | NA | ||||||||||
| 112 | p78791-rs75289200 | 125270729 | Intron 11 | T/C | 0.0321 | 0.7037 | 0.2078 | h112.1 | CCGA | 0.0311 | 0.7440 | 0.5559 | 1.3384 | 0.1812 | 0.0055 | |
| 112 | p79721-rs838896 | 125269799 | Intron 11 | G/C | 0.3104 | 0.3565 | 0.0817 | h112.2 | TGGA | 0.0171 | 2.3734 | 0.7506 | 3.1621 | 0.0016 | ||
| 112 | p79828-rs838895 | 125269692 | Intron 11 | C/G | 0.3171 | 0.4961 | 0.0162 | h112.3 | TGCA | 0.0112 | −1.2672 | 0.9074 | −1.3964 | 0.1630 | ||
| 112 | p80045-rs838893 | 125269475 | Intron 11 | G/A | 0.3244 | 0.3127 | 0.1224 | h112.4 | TCGA | 0.2704 | 0.2488 | 0.2164 | 1.1501 | 0.2505 | ||
| h112.5 | TCCG | 0.0139 | 1.1219 | 0.8186 | 1.3704 | 0.1710 | ||||||||||
| h112.6 (rare) | **** | 0.0068 | 1.6244 | 1.2691 | 1.2800 | 0.2009 | ||||||||||
| hap.base112 | TGCG | 0.6493 | NA | NA | NA | NA | ||||||||||
| 113 | p79721-rs838896 | 125269799 | Intron 11 | G/C | 0.3104 | 0.3565 | 0.0817 | h113.1 | GGAG | 0.0171 | 2.3949 | 0.7509 | 3.1895 | 0.0015 | 0.0048 | |
| 113 | p79828-rs838895 | 125269692 | Intron 11 | C/G | 0.3171 | 0.4961 | 0.0162 | h113.2 | GCAG | 0.0120 | −1.1963 | 0.8784 | −1.3619 | 0.1736 | ||
| 113 | p80045-rs838893 | 125269475 | Intron 11 | G/A | 0.3244 | 0.3127 | 0.1224 | h113.3 | CGAG | 0.2996 | 0.3071 | 0.2067 | 1.4861 | 0.1377 | ||
| 113 | p81863-rs185445624 | 125267657 | Intron 11 | G/A | 0.0020 | −0.9612 | 0.6510 | h113.4 | CCGG | 0.0139 | 1.1509 | 0.8168 | 1.4090 | 0.1592 | ||
| h113.5 (rare) | **** | 0.0081 | 1.1622 | 1.0896 | 1.0666 | 0.2865 | ||||||||||
| hap.base113 | GCGG | 0.6493 | NA | NA | NA | NA | ||||||||||
| 114 | p79828-rs838895 | 125269692 | Intron 11 | C/G | 0.3171 | 0.4961 | 0.0162 | h114.1 | GAGC | 0.3173 | 0.3755 | 0.2023 | 1.8559 | 0.0639 | 0.0447 | |
| 114 | p80045-rs838893 | 125269475 | Intron 11 | G/A | 0.3244 | 0.3127 | 0.1224 | h114.2 | CGGT | 0.0306 | −0.8840 | 0.5344 | −1.6541 | 0.0985 | ||
| 114 | p81863-rs185445624 | 125267657 | Intron 11 | G/A | 0.0020 | −0.9612 | 0.6510 | h114.3 | CAGC | 0.0111 | −1.2612 | 0.9170 | −1.3754 | 0.1694 | ||
| 114 | p82019-rs838890 | 125267501 | Intron 11 | C/T | 0.0320 | −1.0051 | 0.0618 | h114.4 (rare) | **** | 0.0086 | 0.9073 | 1.0936 | 0.8296 | 0.4070 | ||
| hap.base114 | CGGC | 0.6325 | NA | NA | NA | NA | ||||||||||
| 117 | p82019-rs838890 | 125267501 | Intron 11 | C/T | 0.0320 | −1.0051 | 0.0618 | h117.1 | CCAG | 0.0238 | −1.0596 | 0.6275 | −1.6884 | 0.0917 | 0.0433 | |
| 117 | p82264-rs141545424 | 125267256 | Exon 12 | Gly501Gly | C/A | 0.0007 | 11.5850 | 0.0016 | h117.2 | TCGG | 0.0311 | −0.9657 | 0.5302 | −1.8215 | 0.0689 | |
| 117 | p82340-rs77483223 | 125267180 | Intron 12 | G/A | 0.0231 | −1.0458 | 0.1012 | h117.3 (rare) | **** | 0.0067 | 1.6191 | 1.2946 | 1.2507 | 0.2114 | ||
| 117 | p82369-rs75446635 | 125267151 | Intron 12 | G/A | 0.0059 | 0.5896 | 0.6322 | hap.base117 | CCGG | 0.9383 | NA | NA | NA | NA | ||
| 118 | p82264-rs141545424 | 125267256 | Exon 12 | Gly501Gly | C/A | 0.0007 | 11.5850 | 0.0016 | h118.1 | CAGT | 0.0238 | −1.0621 | 0.6274 | −1.6929 | 0.0909 | 0.0375 |
| 118 | p82340-rs77483223 | 125267180 | Intron 12 | G/A | 0.0231 | −1.0458 | 0.1012 | h118.2 | CGGC | 0.0307 | −1.0134 | 0.5313 | −1.9073 | 0.0569 | ||
| 118 | p82369-rs75446635 | 125267151 | Intron 12 | G/A | 0.0059 | 0.5896 | 0.6322 | h118.3 (rare) | **** | 0.0067 | 1.6189 | 1.2762 | 1.2685 | 0.2050 | ||
| 118 | p82434-rs838889 | 125267086 | Intron 12 | T/C | 0.0315 | −1.0389 | 0.0526 | hap.base118 | CGGT | 0.9387 | NA | NA | NA | NA | ||
| 123 | p83884-rs701106 | 125265636 | Intron 12 | C/T | 0.2597 | 0.2471 | 0.2601 | h123.1 | TCCT | 0.0256 | −1.2114 | 0.6218 | −1.9483 | 0.0518 | 0.0386 | |
| 123 | p86245-rs188375019 | 125263275 | Intron 12 | C/T | 0.0341 | 0.7447 | 0.1639 | h123.2 | TCCG | 0.2327 | 0.5306 | 0.2403 | 2.2085 | 0.0275 | ||
| 123 | p86276-rs747155 | 125263244 | Intron 12 | C/T | 0.1495 | 0.2793 | 0.2980 | h123.3 | CCTG | 0.1476 | 0.3955 | 0.2811 | 1.4071 | 0.1598 | ||
| 123 | p86316-rs701104 | 125263204 | Intron 12 | G/T | 0.0487 | −0.9838 | 0.0286 | h123.4 | CCCT | 0.0233 | −0.2329 | 0.7038 | −0.3309 | 0.7408 | ||
| h123.5 | CTCG | 0.0330 | 0.8888 | 0.5458 | 1.6283 | 0.1039 | ||||||||||
| h123.6 (rare) | **** | 0.0029 | 1.1191 | 3.2961 | 0.3395 | 0.7343 | ||||||||||
| hap.base123 | CCCG | 0.5348 | NA | NA | NA | NA | ||||||||||
| 124 | p86245-rs188375019 | 125263275 | Intron 12 | C/T | 0.0341 | 0.7447 | 0.1639 | h124.1 | CTGA | 0.1476 | 0.1530 | 0.2692 | 0.5683 | 0.5700 | 0.0368 | |
| 124 | p86276-rs747155 | 125263244 | Intron 12 | C/T | 0.1495 | 0.2793 | 0.2980 | h124.2 | CCTG | 0.0465 | −1.1879 | 0.4699 | −2.5281 | 0.0117 | ||
| 124 | p86316-rs701104 | 125263204 | Intron 12 | G/T | 0.0487 | −0.9838 | 0.0286 | h124.3 | CCGA | 0.0915 | 0.1086 | 0.3376 | 0.3218 | 0.7477 | ||
| 124 | p86481-rs701103 | 125263039 | Exon 13-3' UTR | Gly499Arg (isoform 2) | G/A | 0.2451 | 0.1642 | 0.4492 | h124.4 | TCGG | 0.0337 | 0.7348 | 0.5362 | 1.3702 | 0.1710 | |
| h124.5 (rare) | **** | 0.0045 | 4.0859 | 2.1131 | 1.9336 | 0.0535 | ||||||||||
| hap.base124 | CCGG | 0.6761 | NA | NA | NA | NA | ||||||||||
| 125 | p86276-rs747155 | 125263244 | Intron 12 | C/T | 0.1495 | 0.2793 | 0.2980 | h125.1 | TGAA | 0.1476 | 0.1543 | 0.2689 | 0.5737 | 0.5664 | 0.0307 | |
| 125 | p86316-rs701104 | 125263204 | Intron 12 | G/T | 0.0487 | −0.9838 | 0.0286 | h125.2 | CTGA | 0.0465 | −1.1980 | 0.4691 | −2.5535 | 0.0109 | ||
| 125 | p86481-rs701103 | 125263039 | Exon 13-l3' UTR | Gly499Arg (isoform 2) | G/A | 0.2451 | 0.1642 | 0.4492 | h125.3 | CGAA | 0.0915 | 0.1139 | 0.3375 | 0.3375 | 0.7359 | |
| 125 | p86967-rs187492239 | 125262553 | Exon 13-3' UTR | A/G | 0.0355 | 0.7743 | 0.1412 | h125.4 | CGGG | 0.0352 | 0.7974 | 0.5241 | 1.5216 | 0.1285 | ||
| h125.5 (rare) | **** | 0.0045 | 4.0989 | 2.1134 | 1.9394 | 0.0528 | ||||||||||
| hap.base125 | CGGA | 0.6747 | NA | NA | NA | NA | ||||||||||
| ApoA-I | ||||||||||||||||
| 47 | p49690-rs4765615 | 125299830 | Intron 2 | G/A | 0.4426 | −0.9139 | 0.0480 | h47.1 | ACGG | 0.4351 | −0.8907 | 0.4584 | −1.9432 | 0.0524 | 0.0343 | |
| 47 | p49759-rs146272788 | 125299761 | Intron 2 | C/T | 0.0020 | 1.5883 | 0.7630 | h47.2 (rare) | **** | 0.0106 | 3.5858 | 2.2998 | 1.5592 | 0.1194 | ||
| 47 | p49978-rs5891 | 125299542 | Exon 3 | Val135lle | G/A | 0.0058 | 5.6762 | 0.0628 | hap.base47 | GCGG | 0.5543 | NA | NA | NA | NA | |
| 47 | p50024-rs368880622 | 125299496 | Intron 3 | G/T | 0.0026 | 1.6012 | 0.7255 | |||||||||
| 48 | p49759-rs146272788 | 125299761 | Intron 2 | C/T | 0.0020 | 1.5883 | 0.7630 | h48.1 | CGGT | 0.0206 | 3.3555 | 1.6564 | 2.0258 | 0.0431 | 0.0293 | |
| 48 | p49978-rs5891 | 125299542 | Exon 3 | Val135lle | G/A | 0.0058 | 5.6762 | 0.0628 | h48.2 (rare) | **** | 0.0106 | 4.0750 | 2.3644 | 1.7235 | 0.0852 | |
| 48 | p50024-rs368880622 | 125299496 | Intron 3 | G/T | 0.0026 | 1.6012 | 0.7255 | hap.base48 | CGGC | 0.9688 | NA | NA | NA | NA | ||
| 48 | p50118-rs58710319 | 125299402 | Intron 3 | C/T | 0.0208 | 3.1376 | 0.0571 | |||||||||
| 49 | p49978-rs5891 | 125299542 | Exon 3 | Val135lle | G/A | 0.0058 | 5.6762 | 0.0628 | h49.1 | GGTT | 0.0213 | 3.3792 | 1.6416 | 2.0584 | 0.0399 | 0.0289 |
| 49 | p50024-rs368880622 | 125299496 | Intron 3 | G/T | 0.0026 | 1.6012 | 0.7255 | h49.2 | GGCC | 0.1928 | 0.8864 | 0.5841 | 1.5176 | 0.1295 | ||
| 49 | p50118-rs58710319 | 125299402 | Intron 3 | C/T | 0.0208 | 3.1376 | 0.0571 | h49.3 (rare) | **** | 0.0086 | 4.7388 | 3.1873 | 1.4868 | 0.1375 | ||
| 49 | p50151-rs2278986 | 125299369 | Intron 3 | T/C | 0.1933 | 0.8568 | 0.1419 | hap.base49 | GGCT | 0.7774 | NA | NA | NA | NA | ||
| 70 | p54627-chr12_125294893 | 125294893 | Intron 5 | G/C | 0.0020 | 3.6910 | 0.4850 | h70.1 | GCAC | 0.3873 | 0.8579 | 0.5090 | 1.6854 | 0.0923 | 0.0140 | |
| 70 | p54856-chr12_125294664 | 125294664 | Intron 6 | C/T | 0.0007 | −24.0757 | 0.0082 | h70.2 | GCGT | 0.1568 | 2.0940 | 0.6700 | 3.1254 | 0.0018 | ||
| 70 | p55923-rs838900 | 125293597 | Intron 6 | G/A | 0.3921 | 0.3606 | 0.4549 | h70.3 (rare) | **** | 0.0027 | −2.5567 | 5.2200 | −0.4898 | 0.6244 | ||
| 70 | p55963-rs7134858 | 125293557 | Intron 6 | C/T | 0.1560 | 1.7537 | 0.0052 | hap.base70 | GCGC | 0.4532 | NA | NA | NA | NA | ||
| 71 | p54856-chr12_125294664 | 125294664 | Intron 6 | C/T | 0.0007 | −24.0757 | 0.0082 | h71.1 | CACA | 0.2736 | 0.7883 | 0.6210 | 1.2694 | 0.2047 | 0.0488 | |
| 71 | p55923-rs838900 | 125293597 | Intron 6 | G/A | 0.3921 | 0.3606 | 0.4549 | h71.2 | CACG | 0.1134 | 1.1284 | 0.9724 | 1.1604 | 0.2462 | ||
| 71 | p55963-rs7134858 | 125293557 | Intron 6 | C/T | 0.1560 | 1.7537 | 0.0052 | h71.3 | CGTA | 0.1296 | 2.1103 | 0.7906 | 2.6691 | 0.0078 | ||
| 71 | p56845-rs838902 | 125292675 | Intron 6 | A/G | 0.4249 | −0.3052 | 0.5129 | h71.4 | CGTG | 0.0300 | 2.1358 | 1.6772 | 1.2734 | 0.2032 | ||
| h71.5 | CGCA | 0.1706 | −0.1013 | 0.8355 | −0.1212 | 0.9035 | ||||||||||
| hap.base71 | CGCG | 0.2822 | NA | NA | NA | NA | ||||||||||
| 72 | p55923-rs838900 | 125293597 | Intron 6 | G/A | 0.3921 | 0.3606 | 0.4549 | h72.1 | ACAG | 0.2733 | 0.7471 | 0.6218 | 1.2016 | 0.2299 | 0.0463 | |
| 72 | p55963-rs7134858 | 125293557 | Intron 6 | C/T | 0.1560 | 1.7537 | 0.0052 | h72.2 | ACGG | 0.1057 | 0.7094 | 0.9850 | 0.7202 | 0.4716 | ||
| 72 | p56845-rs838902 | 125292675 | Intron 6 | A/G | 0.4249 | −0.3052 | 0.5129 | h72.3 | GTAG | 0.1297 | 2.0304 | 0.7898 | 2.5707 | 0.0103 | ||
| 72 | p57004-rs187562853 | 125292516 | Intron 6 | G/A | 0.0098 | 3.2853 | 0.1690 | h72.4 | GTGG | 0.0299 | 2.1741 | 1.6857 | 1.2897 | 0.1975 | ||
| h72.5 | GCAG | 0.1712 | −0.3122 | 0.8263 | −0.3778 | 0.7057 | ||||||||||
| h72.6 (rare) | **** | 0.0100 | 3.9105 | 2.4373 | 1.6044 | 0.1090 | ||||||||||
| hap.base72 | GCGG | 0.2801 | NA | NA | NA | NA | ||||||||||
| 78 | p57592-rs838903 | 125291928 | Intron 7 | G/A | 0.3763 | −0.7661 | 0.1109 | h78.1 | GCAC | 0.0559 | 1.8913 | 1.0469 | 1.8067 | 0.0712 | 0.0326 | |
| 78 | p58514-rs838905 | 125291006 | Intron 7 | T/C | 0.4329 | −0.4213 | 0.3646 | h78.2 | GTAC | 0.0367 | 1.0784 | 1.2814 | 0.8415 | 0.4003 | ||
| 78 | p58664-rs865716 | 125290856 | Intron 7 | A/T | 0.2708 | 0.5369 | 0.3008 | h78.3 | GTAT | 0.2557 | 0.3365 | 0.6035 | 0.5576 | 0.5773 | ||
| 78 | p60255-rs3782287 | 125289265 | Intron 7 | C/T | 0.2831 | 0.3715 | 0.4856 | h78.4 | GTTC | 0.2463 | 0.4962 | 0.5864 | 0.8462 | 0.3977 | ||
| h78.5 | GTTT | 0.0238 | 5.5715 | 1.6643 | 3.3477 | 0.0009 | ||||||||||
| h78.6 (rare) | **** | 0.0075 | 0.6333 | 2.9303 | 0.2161 | 0.8289 | ||||||||||
| hap.base78 | ACAC | 0.3740 | NA | NA | NA | NA | ||||||||||
| 79 | p58514-rs838905 | 125291006 | Intron 7 | T/C | 0.4329 | −0.4213 | 0.3646 | h79.1 | CACT | 0.1270 | 0.3290 | 0.8318 | 0.3955 | 0.6926 | 0.0256 | |
| 79 | p58664-rs865716 | 125290856 | Intron 7 | A/T | 0.2708 | 0.5369 | 0.3008 | h79.2 | TACC | 0.0379 | 0.6384 | 1.2921 | 0.4941 | 0.6214 | ||
| 79 | p60255-rs3782287 | 125289265 | Intron 7 | C/T | 0.2831 | 0.3715 | 0.4856 | h79.3 | TATC | 0.2563 | 0.1851 | 0.6336 | 0.2921 | 0.7703 | ||
| 79 | p61872-rs838909 | 125287648 | Intron 7 | C/T | 0.2199 | 0.9232 | 0.1056 | h79.4 | TTCC | 0.1587 | −0.6020 | 0.7769 | −0.7749 | 0.4386 | ||
| h79.5 | TTCT | 0.0880 | 1.8902 | 0.8856 | 2.1342 | 0.0331 | ||||||||||
| h79.6 | TTTC | 0.0238 | 5.1755 | 1.6851 | 3.0714 | 0.0022 | ||||||||||
| h79.7 (rare) | **** | 0.0059 | 1.2466 | 3.1079 | 0.4011 | 0.6885 | ||||||||||
| hap.base79 | CACC | 0.3024 | NA | NA | NA | NA | ||||||||||
| 80 | p58664-rs865716 | 125290856 | Intron 7 | A/T | 0.2708 | 0.5369 | 0.3008 | h80.1 | ACCG | 0.0389 | −0.3521 | 1.2793 | −0.2753 | 0.7832 | 0.0030 | |
| 80 | p60255-rs3782287 | 125289265 | Intron 7 | C/T | 0.2831 | 0.3715 | 0.4856 | h80.2 | ACTG | 0.1274 | −0.1816 | 0.7909 | −0.2297 | 0.8184 | ||
| 80 | p61872-rs838909 | 125287648 | Intron 7 | C/T | 0.2199 | 0.9232 | 0.1056 | h80.3 | ATCG | 0.2611 | −0.1400 | 0.6323 | −0.2213 | 0.8249 | ||
| 80 | p62140-rs838910 | 125287380 | Intron 7 | G/T | 0.3047 | −0.0755 | 0.8821 | h80.4 | TCCG | 0.1549 | −1.3614 | 0.7489 | −1.8178 | 0.0695 | ||
| h80.5 | TCTG | 0.0901 | 2.0511 | 0.8921 | 2.2992 | 0.0218 | ||||||||||
| h80.6 | TTCG | 0.0224 | 4.7307 | 1.8842 | 2.5107 | 0.0123 | ||||||||||
| h80.7 (rare) | **** | 0.0083 | 3.1429 | 3.4362 | 0.9147 | 0.3607 | ||||||||||
| hap.base80 | ACCT | 0.2970 | NA | NA | NA | NA | ||||||||||
| 81 | p60255-rs3782287 | 125289265 | Intron 7 | C/T | 0.2831 | 0.3715 | 0.4856 | h81.1 | CCGC | 0.1740 | −1.5355 | 0.7276 | −2.1103 | 0.0352 | 0.0050 | |
| 81 | p61872-rs838909 | 125287648 | Intron 7 | C/T | 0.2199 | 0.9232 | 0.1056 | h81.2 | CCGT | 0.0215 | −0.5623 | 1.6155 | −0.3481 | 0.7279 | ||
| 81 | p62140-rs838910 | 125287380 | Intron 7 | G/T | 0.3047 | −0.0755 | 0.8821 | h81.3 | CCTC | 0.0352 | 3.6130 | 1.4518 | 2.4886 | 0.0130 | ||
| 81 | p62409-rs838911 | 125287111 | Intron 7 | C/T | 0.4211 | −0.6245 | 0.1888 | h81.4 | CCTT | 0.2683 | −0.7498 | 0.6337 | −1.1832 | 0.2371 | ||
| h81.5 | CTGC | 0.0886 | 1.4787 | 0.9259 | 1.5970 | 0.1107 | ||||||||||
| h81.6 | CTGT | 0.1287 | −0.2477 | 0.7967 | −0.3109 | 0.7560 | ||||||||||
| h81.7 (rare) | **** | 0.0017 | 4.9120 | 8.4190 | 0.5834 | 0.5598 | ||||||||||
| hap.base81 | TCGC | 0.2819 | NA | NA | NA | NA | ||||||||||
| 82 | p61872-rs838909 | 125287648 | Intron 7 | C/T | 0.2199 | 0.9232 | 0.1056 | h82.1 | CGTT | 0.0214 | 0.3707 | 1.6055 | 0.2309 | 0.8175 | 0.0137 | |
| 82 | p62140-rs838910 | 125287380 | Intron 7 | G/T | 0.3047 | −0.0755 | 0.8821 | h82.2 | CTCT | 0.0364 | 3.8641 | 1.3703 | 2.8199 | 0.0049 | ||
| 82 | p62409-rs838911 | 125287111 | Intron 7 | C/T | 0.4211 | −0.6245 | 0.1888 | h82.3 | CTTT | 0.2692 | −0.2007 | 0.5674 | −0.3537 | 0.7237 | ||
| 82 | p62615-rs7138386 | 125286905 | Intron 7 | T/C | 0.1137 | −0.6495 | 0.3851 | h82.4 | TGCT | 0.0869 | 2.1488 | 0.8777 | 2.4481 | 0.0146 | ||
| h82.5 | TGTT | 0.0179 | 3.0085 | 1.9599 | 1.5351 | 0.1252 | ||||||||||
| h82.6 | TGTC | 0.1116 | −0.1961 | 0.7815 | −0.2510 | 0.8019 | ||||||||||
| h82.7 (rare) | **** | 0.0020 | −4.7635 | 9.0097 | −0.5287 | 0.5972 | ||||||||||
| hap.base82 | CGCT | 0.4546 | NA | NA | NA | NA | ||||||||||
| 83 | p62140-rs838910 | 125287380 | Intron 7 | G/T | 0.3047 | −0.0755 | 0.8821 | h83.1 | GCTA | 0.0854 | 2.0624 | 0.8886 | 2.3211 | 0.0205 | 0.0187 | |
| 83 | p62409-rs838911 | 125287111 | Intron 7 | C/T | 0.4211 | −0.6245 | 0.1888 | h83.2 | GTTG | 0.0389 | 1.3667 | 1.2527 | 1.0910 | 0.2756 | ||
| 83 | p62615-rs7138386 | 125286905 | Intron 7 | T/C | 0.1137 | −0.6495 | 0.3851 | h83.3 | GTCG | 0.1129 | −0.3143 | 0.7855 | −0.4002 | 0.6891 | ||
| 83 | p63483-rs838912 | 125286037 | Intron 7 | G/A | 0.0867 | 1.8700 | 0.0234 | h83.4 | TCTG | 0.0368 | 3.8488 | 1.3757 | 2.7977 | 0.0053 | ||
| h83.5 | TTTG | 0.2675 | −0.1681 | 0.5759 | −0.2918 | 0.7705 | ||||||||||
| h83.6 (rare) | **** | 0.0031 | −0.5696 | 5.5038 | −0.1035 | 0.9176 | ||||||||||
| hap.base83 | GCTG | 0.4554 | NA | NA | NA | NA | ||||||||||
| 86 | p63483-rs838912 | 125286037 | Intron 7 | G/A | 0.0867 | 1.8700 | 0.0234 | h86.1 | ATCG | 0.0871 | 2.5431 | 0.8550 | 2.9743 | 0.0030 | 0.0290 | |
| 86 | p64772-rs5888 | 125284748 | Exon 8 | Ala350Ala | C/T | 0.0961 | 2.0962 | 0.0080 | h86.2 | GCAG | 0.1457 | 0.3613 | 0.6957 | 0.5194 | 0.6037 | |
| 86 | p64923-rs838915 | 125284597 | Intron 8 | C/A | 0.1435 | −0.3684 | 0.5766 | h86.3 | GCCA | 0.2814 | 1.0972 | 0.5782 | 1.8976 | 0.0581 | ||
| 86 | p65999-rs12819677 | 125283521 | Intron 8 | G/A | 0.2813 | 0.6769 | 0.2052 | h86.4 | GTCG | 0.0116 | 1.6563 | 2.1240 | 0.7798 | 0.4357 | ||
| hap.base86 | GCCG | 0.4736 | NA | NA | NA | NA | ||||||||||
| 95 | p71867-rs7954022 | 125277653 | Intron 9 | C/T | 0.1323 | 0.8502 | 0.2241 | h95.1 | TACT | 0.1311 | 0.8202 | 0.7688 | 1.0669 | 0.2864 | 0.0131 | |
| 95 | p72197-rs838861 | 125277323 | Intron 9 | A/G | 0.3777 | −0.1507 | 0.7464 | h95.2 | CACC | 0.0507 | 0.3188 | 1.2809 | 0.2489 | 0.8035 | ||
| 95 | p72777-rs838862 | 125276743 | Intron 9 | C/T | 0.0887 | 0.7012 | 0.3938 | h95.3 | CGCT | 0.1846 | −0.7832 | 0.6960 | −1.1253 | 0.2608 | ||
| 95 | p75766-rs838866 | 125273754 | Intron 9 | T/C | 0.2116 | −0.0497 | 0.9306 | h95.4 | CGCC | 0.1022 | 0.7176 | 0.8581 | 0.8362 | 0.4033 | ||
| h95.5 | CGTT | 0.0324 | 4.7525 | 1.5071 | 3.1534 | 0.0017 | ||||||||||
| h95.6 | CGTC | 0.0582 | −1.3987 | 1.0854 | −1.2887 | 0.1979 | ||||||||||
| h95.7 (rare) | **** | 0.0009 | 18.2723 | NA | NA | NA | ||||||||||
| hap.base95 | CACT | 0.4399 | NA | NA | NA | NA | ||||||||||
| 96 | p72197-rs838861 | 125277323 | Intron 9 | A/G | 0.3777 | −0.1507 | 0.7464 | h96.1 | ACCT | 0.0443 | 1.0796 | 1.2832 | 0.8413 | 0.4004 | 0.0484 | |
| 96 | p72777-rs838862 | 125276743 | Intron 9 | C/T | 0.0887 | 0.7012 | 0.3938 | h96.2 | GCTC | 0.1849 | −0.7979 | 0.6554 | −1.2176 | 0.2238 | ||
| 96 | p75766-rs838866 | 125273754 | Intron 9 | T/C | 0.2116 | −0.0497 | 0.9306 | h96.3 | GCCT | 0.0727 | −0.3866 | 0.9478 | −0.4079 | 0.6835 | ||
| 96 | p75778-rs7301120 | 125273742 | Intron 9 | C/T | 0.1135 | 0.3767 | 0.6174 | h96.4 | GCCC | 0.0282 | 1.9372 | 1.6107 | 1.2027 | 0.2295 | ||
| h96.5 | GTTC | 0.0319 | 4.2363 | 1.4400 | 2.9419 | 0.0034 | ||||||||||
| h96.6 | GTCC | 0.0595 | −1.3421 | 1.0101 | −1.3286 | 0.1844 | ||||||||||
| h96.7 (rare) | **** | 0.0058 | −3.2342 | 3.8265 | −0.8452 | 0.3983 | ||||||||||
| hap.base96 | ACTC | 0.5728 | NA | NA | NA | NA | ||||||||||
| 97 | p72777-rs838862 | 125276743 | Intron 9 | C/T | 0.0887 | 0.7012 | 0.3938 | h97.1 | CTCT | 0.1997 | −1.0781 | 0.6237 | −1.7287 | 0.0843 | 0.0098 | |
| 97 | p75766-rs838866 | 125273754 | Intron 9 | T/C | 0.2116 | −0.0497 | 0.9306 | h97.2 | CCTT | 0.1141 | 0.2005 | 0.7597 | 0.2639 | 0.7919 | ||
| 97 | p75778-rs7301120 | 125273742 | Intron 9 | C/T | 0.1135 | 0.3767 | 0.6174 | h97.3 | CCCT | 0.0336 | 0.7963 | 1.3894 | 0.5731 | 0.5667 | ||
| 97 | p76757-rs9919713 | 125272763 | Intron 9 | A/T | 0.4390 | −0.1860 | 0.6921 | h97.4 | TTCT | 0.0301 | 4.3773 | 1.4494 | 3.0201 | 0.0026 | ||
| h97.5 | TCCT | 0.0588 | −1.4125 | 1.0117 | −1.3961 | 0.1631 | ||||||||||
| h97.6 (rare) | **** | 0.0050 | −6.5869 | 3.6167 | −1.8213 | 0.0690 | ||||||||||
| hap.base97 | CTCA | 0.5587 | NA | NA | NA | NA | ||||||||||
| 109 | p78402-rs838898 | 125271118 | Intron 10 | G/A | 0.0714 | −0.9806 | 0.2889 | h109.1 | AGCT | 0.0288 | −1.4134 | 1.6436 | −0.8600 | 0.3901 | 0.0195 | |
| 109 | p78430-rs838897 | 125271090 | Intron 10 | C/G | 0.3830 | −0.1887 | 0.6887 | h109.2 | AGTT | 0.0451 | −1.5093 | 1.2496 | −1.2078 | 0.2275 | ||
| 109 | p78747-rs2293440 | 125270773 | Intron 11 | T/C | 0.4112 | −0.2984 | 0.5352 | h109.3 | GGCC | 0.0317 | 3.0784 | 1.3763 | 2.2366 | 0.0256 | ||
| 109 | p78791-rs75289200 | 125270729 | Intron 11 | T/C | 0.0321 | 3.6568 | 0.0086 | h109.4 | GGCT | 0.1633 | −0.4126 | 0.6911 | −0.5971 | 0.5506 | ||
| h109.5 | GGTT | 0.1088 | −1.6537 | 0.8639 | −1.9142 | 0.0560 | ||||||||||
| h109.6 | GCCT | 0.1851 | −1.8104 | 0.7168 | −2.5256 | 0.0118 | ||||||||||
| hap.base109 | GCTT | 0.4363 | NA | NA | NA | NA | ||||||||||
| 110 | p78430-rs838897 | 125271090 | Intron 10 | C/G | 0.3830 | −0.1887 | 0.6887 | h110.1 | GCCC | 0.0305 | 3.0357 | 1.4224 | 2.1342 | 0.0331 | 0.0012 | |
| 110 | p78747-rs2293440 | 125270773 | Intron 11 | T/C | 0.4112 | −0.2984 | 0.5352 | h110.2 | GCTG | 0.0189 | −3.0973 | 2.2833 | −1.3565 | 0.1753 | ||
| 110 | p78791-rs75289200 | 125270729 | Intron 11 | T/C | 0.0321 | 3.6568 | 0.0086 | h110.3 | GCTC | 0.1696 | −0.0290 | 0.6830 | −0.0424 | 0.9662 | ||
| 110 | p79721-rs838896 | 125269799 | Intron 11 | G/C | 0.3104 | 1.1147 | 0.0278 | h110.4 | GTTG | 0.1400 | −2.3158 | 0.7741 | −2.9914 | 0.0029 | ||
| h110.5 | GTTC | 0.0189 | 1.3536 | 2.3385 | 0.5788 | 0.5629 | ||||||||||
| h110.6 | CCTG | 0.1379 | −2.4014 | 0.7888 | −3.0443 | 0.0024 | ||||||||||
| h110.7 | CCTC | 0.0514 | −0.8677 | 1.2628 | −0.6871 | 0.4922 | ||||||||||
| h110.8 | CTTC | 0.0398 | −0.1892 | 1.4963 | −0.1264 | 0.8994 | ||||||||||
| h110.9 (rare) | **** | 0.0012 | 7.8235 | 8.0313 | 0.9741 | 0.3303 | ||||||||||
| hap.base110 | CTTG | 0.3918 | NA | NA | NA | NA | ||||||||||
| 111 | p78747-rs2293440 | 125270773 | Intron 11 | T/C | 0.4112 | −0.2984 | 0.5352 | h111.1 | CCCG | 0.0305 | 3.5704 | 1.4077 | 2.5364 | 0.0114 | 0.0038 | |
| 111 | p78791-rs75289200 | 125270729 | Intron 11 | T/C | 0.0321 | 3.6568 | 0.0086 | h111.2 | CTGC | 0.1514 | −2.1697 | 0.7058 | −3.0742 | 0.0022 | ||
| 111 | p79721-rs838896 | 125269799 | Intron 11 | G/C | 0.3104 | 1.1147 | 0.0278 | h111.3 | CTCG | 0.2233 | 0.3086 | 0.5985 | 0.5157 | 0.6062 | ||
| 111 | p79828-rs838895 | 125269692 | Intron 11 | C/G | 0.3171 | 1.2206 | 0.0164 | h111.4 | TTGG | 0.0173 | 1.0502 | 1.9388 | 0.5417 | 0.5882 | ||
| h111.5 | TTGC | 0.0431 | 0.3464 | 1.3140 | 0.2637 | 0.7921 | ||||||||||
| h111.6 | TTCC | 0.0150 | 0.6429 | 1.9745 | 0.3256 | 0.7448 | ||||||||||
| h111.7 (rare) | **** | 0.0047 | 3.8853 | 4.0634 | 0.9562 | 0.3393 | ||||||||||
| hap.base111 | TTGC | 0.5147 | NA | NA | NA | NA | ||||||||||
| 112 | p78791-rs75289200 | 125270729 | Intron 11 | T/C | 0.0321 | 3.6568 | 0.0086 | h112.1 | CCGA | 0.0309 | 3.7315 | 1.3947 | 2.6755 | 0.0076 | 0.0412 | |
| 112 | p79721-rs838896 | 125269799 | Intron 11 | G/C | 0.3104 | 1.1147 | 0.0278 | h112.2 | TGGA | 0.0179 | 1.8646 | 1.8467 | 1.0097 | 0.3130 | ||
| 112 | p79828-rs838895 | 125269692 | Intron 11 | C/G | 0.3171 | 1.2206 | 0.0164 | h112.3 | TGCA | 0.0109 | −3.3720 | 2.3180 | −1.4547 | 0.1462 | ||
| 112 | p80045-rs838893 | 125269475 | Intron 11 | G/A | 0.3244 | 0.8859 | 0.0774 | h112.4 | TCGA | 0.2661 | 0.7087 | 0.5428 | 1.3056 | 0.1921 | ||
| h112.5 | TCCG | 0.0144 | 1.0316 | 2.0147 | 0.5120 | 0.6088 | ||||||||||
| h112.6 (rare) | **** | 0.0068 | 2.8715 | 3.2105 | 0.8944 | 0.3714 | ||||||||||
| hap.base112 | TGCG | 0.6530 | NA | NA | NA | NA | ||||||||||
| 123 | p83884-rs701106 | 125265636 | Intron 12 | C/T | 0.2597 | 1.2967 | 0.0156 | h123.1 | TCCT | 0.0235 | −1.7638 | 1.7393 | −1.0141 | 0.3109 | 0.0468 | |
| 123 | p86245-rs188375019 | 125263275 | Intron 12 | C/T | 0.0341 | 1.8399 | 0.1674 | h123.2 | TCCG | 0.2351 | 1.8726 | 0.6006 | 3.1179 | 0.0019 | ||
| 123 | p86276-rs747155 | 125263244 | Intron 12 | C/T | 0.1495 | −0.2164 | 0.7433 | h123.3 | CCTG | 0.1485 | 0.3912 | 0.6981 | 0.5604 | 0.5754 | ||
| 123 | p86316-rs701104 | 125263204 | Intron 12 | G/T | 0.0487 | −0.6627 | 0.5579 | h123.4 | CCCT | 0.0238 | 1.6476 | 1.7546 | 0.9390 | 0.3480 | ||
| h123.5 | CTCG | 0.0328 | 2.3144 | 1.3655 | 1.6949 | 0.0905 | ||||||||||
| h123.6 (rare) | **** | 0.0024 | 1.2704 | 8.8153 | 0.1441 | 0.8855 | ||||||||||
| hap.base123 | CCCG | 0.5340 | NA | NA | NA | NA | ||||||||||
ApoA-I apolipoprotein A-I, Coef coefficient, del/D deletion, HDL-C high-density lipoprotein cholesterol, MAF minor allele frequency, NA not analyzed, SE standard error, SNP single nucleotide polymorphism, UTR untranslated region, W wild type allele for deletion on RefSeq
All alleles on the reverse strand. HDL-C and ApoA-I variables were in mg/dL and Box-Cox transformed
Results were adjusted for covariates: sex, age, waist, current smoking (yes/no), and minutes of daily walking or biking to work (jobmin) for HDL-C; sex and age for ApoA-I
SNP 1-SNP 4 for each window are shown as “SNP name-SNP ID/Chromosome 12 Position (for novel variants)”. All 10 novel variants identified in this study have been submitted to dbSNP database (batch ID: SCARB1_AB): http://www.ncbi.nlm.nih.gov/SNP/snp_viewTable.cgi?handle=KAMBOH.
Nominally significant P-values (P < 0.05) for SNPs with MAF ≥5 % in single-site analysis are shown in bold
Haplotype sequences corresponding to SNP 1-SNP 4 in the 5′ to 3′ direction, respectively
Haplotype association results for all haplotype windows are shown in Additional file 16: Table S11, see haplotype association plots in Fig. 3
a, cRefSeq of SCARB1: hg19, NM_005505 (CHIP Bioinformatics)
bdbSNP build 139: GRCh37.p10
Overall, a total of 21 haplotype windows showed significant associations with ApoA-I, of which 10 contained seven variants associated with ApoA-I in single-site analysis. Haplotype window #110 spanning introns 10–11 showed the best association signal (global P = 0.0012) and contained the rs838896 variant with a nominal evidence of association with ApoA-I (P = 0.0278) in single-site analysis.
A total of 16 haplotype windows yielded significant associations with HDL-C, of which seven contained three HDL-C-associated variants detected in single-site analysis. The most significant association was found with window #111 (global P = 0.0040) spanning intron 11, which contained the rs838895 variant nominally associated with HDL-C (P = 0.0162) in single-site analysis.
We observed nine regions (5 regions for ApoA-I and 4 regions for HDL-C) harboring consecutive significant haplotype windows (global P < 0.05; ranging from 2 to 6 windows per region; Table 8 and Fig. 3). Seven of those regions contained at least one of the six variants that exhibited nominal associations (P < 0.05) with HDL-C and/or ApoA-I (rs4765615, rs7134858, rs838912, rs838896, rs838895, and rs701106) in single-site analysis.
Table 8.
Significantly associated haplotype regions (global P < 0.05) with HDL-C and ApoA-I
| Region # | Trait | Consecutive Significantly Associated Haplotype Windows (global P < 0.05) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Haplotype Windows # | Chr12 Positiona | The Composited Variants in the Region, 5′ to 3′ Direction | Most Relevant Haplotype | ||||||
| (Location) | |||||||||
| Start (5′) | End (3′) | SNP Nameb-SNP IDc/Chr12 Positiona | Major/Minor Alleles | Haplotype # | Sequence | β (Min-Max) | |||
| 1 | HDL-C | 44 | 125300551 | 125299542 | p48969-rs2343394 | C/T | h44.3 | CCWGCGG | 0.4910–1.0491 |
| 45 | (intron 2) | (exon 3) | p49537-rs7305310 | C/T | h45.2 | ||||
| 46 | p49570delC-rs145376237 | W/D | h46.2 | ||||||
| 47 | p49690-rs4765615 | G/A | hap.base47 | ||||||
| p49759-rs146272788 | C/T | hap.base44 | CCWACGG | −0.4701 | |||||
| p49978-rs5891 (p.Val135Ile) | G/A | hap.base45 | |||||||
| p50024-rs368880622 | G/T | hap.base46 | |||||||
| h47.1 | |||||||||
| 2 | ApoA-I | 47 | 125299830 | 125299369 | p49690-rs4765615 | G/A | h47.1 | ACGGTT | (−0.8907)–3.3792 |
| 48 | (intron 2) | (intron 3) | p49759-rs146272788 | C/T | h48.1 | ||||
| 49 | p49978-rs5891 (p.Val135Ile) | G/A | h49.1 | ||||||
| p50024-rs368880622 | G/T | ||||||||
| p50118-rs58710319 | C/T | ||||||||
| p50151-rs2278986 | T/C | ||||||||
| 3 | ApoA-I | 70 | 125294893 | 125292516 | p54627-chr12_125294893 | G/C | h70.2 | GCGTAG | 2.0304–2.1103 |
| 71 | (intron 5) | (intron 6) | p54856-chr12_125294664d | C/T | h71.3 | ||||
| 72 | p55923-rs838900 | G/A | h72.3 | ||||||
| p55963-rs7134858 | C/T | ||||||||
| p56845-rs838902 | A/G | ||||||||
| p57004-rs187562853 | G/A | ||||||||
| 4 | ApoA-I | 78 | 125291928 | 125286037 | p57592-rs838903 | G/A | h78.5 | GTTTCGCTG | 4.7307–5.5715 |
| 79 | (intron 7) | (intron 7) | p58514-rs838905 | T/C | h79.6 | ||||
| 80 | p58664-rs865716 | A/T | h80.6 | ||||||
| 81 | p60255-rs3782287 | C/T | hap.base81 | ||||||
| 82 | p61872-rs838909 | C/T | hap.base82 | ||||||
| 83 | p62140-rs838910 | G/T | hap.base83 | ||||||
| p62409-rs838911 | C/T | h78.2 | GTACCTCTG | 0.6384–3.8641 | |||||
| p62615-rs7138386 | T/C | h79.2 | |||||||
| p63483-rs838912 | G/A | hap.base80 | |||||||
| h81.3 | |||||||||
| h82.2 | |||||||||
| h83.4 | |||||||||
| 5 | ApoA-I | 95 | 125277653 | 125272763 | p71867-rs7954022 | C/T | h95.5 | CGTTCT | 4.2363-4.7525 |
| 96 | (intron 9) | (intron 9) | p72197-rs838861 | A/G | h96.5 | ||||
| 97 | p72777-rs838862 | C/T | h97.4 | ||||||
| p75766-rs838866 | T/C | ||||||||
| p75778-rs7301120 | C/T | ||||||||
| p76757-rs9919713 | A/T | ||||||||
| 6* | ApoA-I | 109 | 125271118 | 125269475 | p78402-rs838898 | G/A | h109.6 | GCCTGCA | (−3.3720)─(−1.8104) |
| 110 | (intron 10) | (intron 11) | p78430-rs838897 | C/G | h110.6 | ||||
| 111 | p78747-rs2293440 | T/C | h111.2 | ||||||
| 112 | p78791-rs75289200 | T/C | h112.3 | ||||||
| p79721-rs838896 | G/C | ||||||||
| p79828-rs838895 | C/G | ||||||||
| p80045-rs838893 | G/A | ||||||||
| 7* | HDL-C | 111 | 125270773 | 125267501 | p78747-rs2293440 | T/C | h111.4 | TTGGAGC | 0.3755–2.3949 |
| 112 | (intron 11) | (intron 11) | p78791-rs75289200 | T/C | h112.2 | ||||
| 113 | p79721-rs838896 | G/C | h113.1 | ||||||
| 114 | p79828-rs838895 | C/G | h114.1 | ||||||
| p80045-rs838893 | G/A | ||||||||
| p81863-rs185445624 | G/A | ||||||||
| p82019-rs838890 | C/T | ||||||||
| 8 | HDL-C | 117 | 125267501 | 125267086 | p82019-rs838890 | C/T | h117.2 | TCGGC | (−1.0134)–(−0.9657) |
| 118 | (intron 11) | (intron 12) | p82264-rs141545424 (p.Gly501Gly)d | C/A | h118.2 | ||||
| p82340-rs77483223 | G/A | ||||||||
| p82369-rs75446635 | G/A | ||||||||
| p82434-rs838889 | T/C | ||||||||
| 9 | HDL-C | 123 | 125265636 | 125262553 | p83884-rs701106 | C/T | h123.4 | CCCTGA | (−1.180)–(−0.2329) |
| 124 | (intron 12) | (exon 13-3′ UTR) | p86245-rs188375019 | C/T | h124.2 | ||||
| 125 | p86276-rs747155 | C/T | h125.2 | ||||||
| p86316-rs701104 | G/T | ||||||||
| p86481-rs701103 (p.Gly499Arg, isoform 2) | G/A | ||||||||
| p86967-rs187492239 | A/G | ||||||||
ApoA-I apolipoprotein A-I, del/D deletion, HDL-C high-density lipoprotein cholesterol, SNP single nucleotide polymorphism, UTR untranslated region, W wild type allele for deletion on the RefSeq
All alleles on the reverse strand. HDL-C and ApoA-I variables were in mg/dL and Box-Cox transformed
Results were adjusted for covariates: sex, age, waist, current smoking (yes/no), and minutes of daily walking or biking to work (jobmin) for HDL-C; sex and age for ApoA-I
All nine haplotype regions are shown in Fig. 3
Detailed single-site associations are shown in Additional file 14: Table S9 and Additional file 15: Table S10
Detailed haplotype associations are shown in Table 7 and Additional file 16: Table S11
Regions with asterisk (*) indicate regions that included the haplotype window exhibiting the most significant association signal (the smallest global P) for the associated trait
For each region, the most significant associated haplotype window is shown in bold
SNPs with significant evidence of association with the same trait in both single-site and haplotype analyses (single-site P < 0.05 and global P < 0.05) are shown in bold
SNPs with significant evidence of association with different trait in single-site and haplotype analyses (single-site P < 0.05 and global P < 0.05) are shown in italic bold
a, bRefSeq of SCARB1: hg19, NM_005505 (CHIP Bioinformatics)
cdbSNP build 139: GRCh37.p10
dRare variants of interest with potential effects on lipid traits; see details in Table 6
Fig. 3.
Haplotype association plots for HDL-C and ApoA-I. Top: The -log10 P-values are presented in the Y-axis. A total of 136 genotyped variants are shown in order on SCARB1 gene (5′ → 3′; RefSeq: hg19, NM_005505) in the X-axis. Middle: gene structure of SCARB1. Marker names are shown as “SNP name-SNP ID/chromosome 12 position (for novel variants)”. Bottom: linkage disequilibrium (LD) plot of 136 variants. SNPs with MAF ≥5 % are shown in bold. SNP ID is based on dbSNP build 139. All 10 novel variants identified in this study have been submitted to dbSNP (batch ID: SCARB1_AB): http://www.ncbi.nlm.nih.gov/SNP/snp_viewTable.cgi?handle=KAMBOH. The dash line indicates the significance threshold (global P = 0.05). Significantly associated haplotype regions are highlighted. The degree of shades and values (r2 × 100) in each square of LD plot represent the pairwise correlations between 136 genotyped variants: black indicating r2 = 1, white indicating r2 = 0, and shade intensity indicating r2 between 0 and 1. ApoA-I, apolipoprotein A-I; HDL-C, high-density lipoprotein cholesterol; LD, linkage disequilibrium; MAF, minor allele frequency; SNP, single nucleotide polymorphism; UTR, untranslated region
Functional evaluation of identified variants
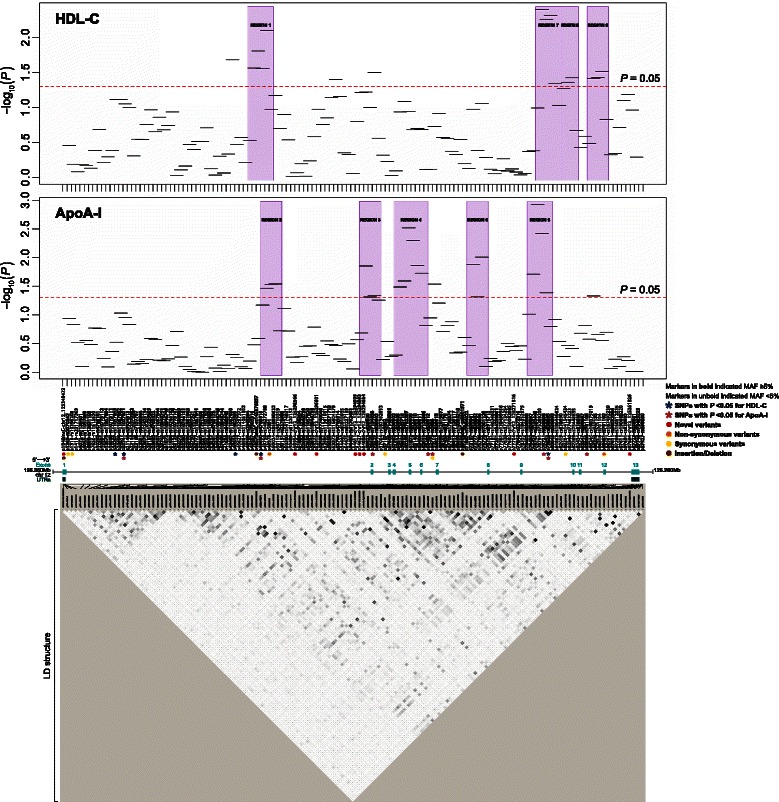
In order to examine the possible regulatory function of all 153 SCARB1 variants (83 variants identified by our sequencing, 68 common HapMap tagSNPs [excluding rs4765180 due to genotyping failure; see Additional file 7: Table S5], and two relevant variants from the literature), we used the RegulomeDB database (version 1.0, Stanford University, http://www.regulomedb.org/) [48]. Although most of 153 variants (n = 132) revealed scores ranging from 1 to 6, only 11 were supported by strong evidence for regulatory function (scores of 1f -2b): one promoter, one 5′ UTR, two coding (rs2070242 [p.Ser4Ser] and rs10396208 [p.Cys21Cys]), five intronic, one 3′ UTR, and one 3′ flanking variants. Summary and detailed regulatory functions are provided in Additional file 17: Table S12 and Additional file 18: Table S13.
Of 10 variants associated with HDL-C and/or ApoA-I, only one ApoA-I associated variant (rs5888 [p.Ala350Ala] in exon 8) showed suggestive evidence of regulatory function with a score of 3a (Table 4).
Of 10 novel variants, one insertion variant (p1048insC/chr12:125348472) located in 5′ UTR-exon 1 had a strong potential for regulatory function with a score of 2a (Additional file 4: Table S4).
Comparison of SCARB1 single-site and haplotype association analysis results between African Blacks (this study) and US Non-Hispanic Whites (previous study [49])
We compared SCARB1 single-site and haplotype association results in African Blacks reported in this study to those in US Non-Hispanic Whites (NHWs) reported in our previously published study [49]. In the sequencing stage, the number of variants identified in African Blacks (n = 83) was greater than that in US NHWs (n = 44). Notably, most (~90 %) of the 22 sequence variants that were shared between the two populations differed in minor alleles and/or MAFs. Although our major findings included the associations with HDL-C and ApoA-I in African Blacks, we also sought to replicate four associations observed with ApoB levels in US NHWs [49] (Table 9); the association between rs11057820 and ApoB (P < 0.05) that we previously reported in US NHWs [49] was also observed in African Blacks (US NHWs [G allele]: β = 0.8700, P = 0.0436; African Blacks [A allele]: β = 1.8661, P = 0.0292). In addition, we observed two variants (rs4765615 and rs701106) exhibiting nominal associations (P < 0.05) in both populations, albeit with different lipid traits (US NHWs| rs4765615 [G allele]: β = 1.2493, P = 0.0059 for ApoB; rs701106 [T allele]: β = 0.0394, P = 0.0066 for HDL-C; African Blacks| rs4765615 [A allele]: β = −0.4646, P = 0.013 for HDL-C and β = −0.9139, P = 0.048 for ApoA-I; rs701106 [T allele]: β = 1.2967, P = 0.0156 for ApoA-I). Moreover, we noticed that two regions associated with HDL-C or ApoA-I (global P < 0.05; Table 10) in African Blacks spanning intron 2 and intron 3 overlapped with the ApoB-associated region (Region I in Fig. 4) previously reported in US NHWs [49]. Three haplotype regions associated with HDL-C (global P < 0.05) spanning intron 11 and exon 13-3′ UTR in African Blacks also overlapped with a large HDL-C-associated region (Region II in Fig. 4) previously reported in US NHWs [49].
Table 9.
Results for 7 SCARB1 lipid-associated variants in US Non-Hispanic Whites (previous studya) and in African Blacks (this study)
| SNP Nameb | SNP IDc | Chr12 Positiond | Location | RegDB Scoree | Alleles | US Non-Hispanic Whitesa (n = 623) | African Blacks (n = 788) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MA, MAF | β | P | MA, MAF | β | P | Other Assoc Trait(s)f | ||||||
| (SE) | (SE) | |||||||||||
| HDL-C | ||||||||||||
| p28957 | rs11057844 | 125320563 | Intron 1 | 5 | G/A | A, 0.1839 | −0.0395 | 0.0035 | A, 0.2362 | 0.3671 | 0.1075 | |
| (0.0135) | (0.2278) | |||||||||||
| p83884 | rs701106 | 125265636 | Intron 12 | 5 | C/T | T, 0.1527 | 0.0394 | 0.0066 | T, 0.2597 | 0.2471 | 0.2601 | ApoA-I |
| (0.0144) | (0.2192) | |||||||||||
| p87927 | rs838880 | 125261593 | 3′ flanking | 5 | G/A | G, 0.3237 | 0.0257 | 0.0250 | A, 0.2414 | 0.0198 | 0.9314 | |
| (0.0114) | (0.2302) | |||||||||||
| ApoB | ||||||||||||
| p48969 | rs2343394 | 125300551 | Intron 2 | 5 | C/T | T, 0.2850 | 1.2544 | 0.0082 | T, 0.1898 | 0.0383 | 0.9544 | |
| (0.4721) | (0.6696) | |||||||||||
| p49690 | rs4765615 | 125299830 | Intron 2 | 5 | G/A | G, 0.4497 | 1.2493 | 0.0059 | A, 0.4426 | 0.7771 | 0.1338 | HDL-C, ApoA-I |
| (0.4518) | (0.5178) | |||||||||||
| p50151 | rs2278986 | 125299369 | Intron 3 | 5 | T/C | C, 0.2890 | 1.1926 | 0.0122 | C, 0.1933 | 0.1308 | 0.8434 | |
| (0.4735) | (0.6619) | |||||||||||
| p52556 | rs11057820 | 125296964 | Intron 4 | 5 | G/A | G, 0.4871 | 0.8700 | 0.0436 | A, 0.1000 | 1.8661 | 0.0292 | |
| (0.4300) | (0.8542) | |||||||||||
ApoB apolipoprotein B, HDL-C high-density lipoprotein cholesterol, MA minor allele, MAF minor allele frequency, RegDB RegulomeDB, SE standard error, SNP single nucleotide polymorphism
All alleles on the reverse strand
HDL-C and ApoB values for US Non-Hispanic Whites were in mg/dL, Box-Cox transformed, and adjusted for covariates: sex, age, body mass index, and smoking (past/current/never) for HDL-C; age and smoking for ApoB
HDL-C and ApoB values for African Blacks were in mg/dL, Box-Cox transformed, and adjusted for covariates: sex, age, waist, current smoking (yes/no), and daily walking or biking to work (jobmin) for HDL-C; body mass index and staff status for ApoB
Nominally significant P-values (P < 0.05) are shown in bold
aData from Niemsiri V, et al. Circ Cardiovasc Genet 2014, 7(6):838–847 (Ref [49])
b, dRefSeq of SCARB1: hg19, NM_005505 (CHIP Bioinformatics)
cdbSNP version 139: GRCh37.p10
eThe RegulomeDB (version 1.0) scoring scheme is described at the footnote of Additional file 17: Table S12 or at http://regulome.stanford.edu/help
fEvidence is based on SNPs with MAF ≥5 % exhibiting nominally significant association with either HDL-C or ApoA-I (P < 0.05; Additional file 14: Table S9 and Additional file 15: Table S10) in single-site association results in the current study
Table 10.
Significant lipid-associated regions (global P < 0.05) that were observed in US Non-Hispanic Whites (previous studya) and African Blacks (this study)
| Region # | Consecutive Haplotype Windows in 623 US Non-Hispanic Whitesa | Consecutive Haplotype Windows in 788 African Blacks | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Chr12 Positionb (Location) | Length (bp) | The Composited Variants, 5′ to 3′ Direction | Trait | Chr12 Positionb (Location) | Length (bp) | The Composited Variants, 5′ to 3′ Direction | |||||
| Start (5′) | End (3′) | SNP Namec-SNP IDd | Major/Minor Alleles | Start (5′) | End (3′) | SNP Namec-SNP IDd | Major/Minor Alleles | |||||
| I | ApoB | 125300551 | 125299369 | 1183 | p48969-rs2343394 | C/T | HDL-C | 125300551 | 125299496 | 1056 | p48969-rs2343394 | C/T |
| (intron 2) | (intron 3) | p49518-rs144194221 | G/A | (intron 2) | (intron 3) | p49537-rs7305310 | C/T | |||||
| p49690-rs4765615 | A/G | p49570delC-rs145376237 | W/D | |||||||||
| p49978-rs5891 | G/A | p49690-rs4765615 | G/A | |||||||||
| (p.Val135Ile) | ||||||||||||
| p50151-rs2278986 | T/C | p49759-rs146272788 | C/T | |||||||||
| p49978-rs5891 | G/A | |||||||||||
| (p.Val135Ile) | ||||||||||||
| p50024-rs368880622 | G/T | |||||||||||
| ApoA-I | 125299830 | 125299369 | 462 | p49690-rs4765615 | G/A | |||||||
| (intron 2) | (intron 3) | p49759-rs146272788 | C/T | |||||||||
| p49978-rs5891 | G/A | |||||||||||
| (p.Val135Ile) | ||||||||||||
| p50024-rs368880622 | G/T | |||||||||||
| p50118-rs58710319 | C/T | |||||||||||
| p50151-rs2278986 | T/C | |||||||||||
| II | HDL-C | 125269692 | 125262516 | 7177 | p79828-rs838895 | C/G | HDL-C | 125269692 | 125267501 | 2192 | p79828-rs838895 | C/G |
| (intron 11) | (exon 13- 3′ UTR) | p80045-rs838893 | G/A | (intron 11) | (intron 11) | p80045-rs838893 | G/A | |||||
| p83088-rs797729 | A/G | p81863-rs185445624 | G/A | |||||||||
| p83884-rs701106 | C/T | p82019-rs838890 | C/T | |||||||||
| p86436-rs10396214 | C/T | HDL-C | 125267501 | 125267086 | 416 | p82019-rs838890 | C/T | |||||
| (p.Arg484Trp, isoform 2) | ||||||||||||
| p87004-rs184715678 | C/A | (intron 11) | (intron 12) | p82264-rs141545424 | C/A | |||||||
| (p.Gly501Gly) | ||||||||||||
| p82340-rs77483223 | G/A | |||||||||||
| p82369-rs75446635 | G/A | |||||||||||
| p82434-rs838889 | T/C | |||||||||||
| HDL-C | 125265636 | 125262553 | 3084 | p83884-rs701106 | C/T | |||||||
| (intron 12) | (exon 13- 3′ UTR) | p86245-rs188375019 | C/T | |||||||||
| p86276-rs747155 | C/T | |||||||||||
| p86316-rs701104 | G/T | |||||||||||
| p86481-rs701103 | G/A | |||||||||||
| (p.Gly499Arg, isoform 2) | ||||||||||||
| p86967-rs187492239 | A/G | |||||||||||
ApoA-I apolipoprotein A-I, ApoB apolipoprotein B, del/D deletion, HDL-C high-density lipoprotein cholesterol, SNP single nucleotide polymorphism, UTR untranslated region, W wild type allele for deletion on RefSeq
All alleles on the reverse strand
Results for a US Non-Hispanic White sample were Box-Cox transformed, and adjusted for covariates: sex, age, body mass index, and smoking (past/current/never) for HDL-C; age and smoking for ApoB
Results for an African Black sample were Box-Cox transformed, and adjusted for covariates: sex, age, waist, current smoking (yes/no), and minutes of walking or biking to work each day (jobmin) for HDL-C; sex and age for ApoA-I
Location of each region on SCARB1 gene is shown in Fig. 4
SNPs with significant evidence with the same trait in both single-site and haplotype associations (single-site P and global P < 0.05) observed in each population are shown in bold
SNPs with significant evidence with the different trait in single-site and haplotype associations (single-site P and global P < 0.05) in each population are shown in italic bold
aData from Niemsiri V, et al. Circ Cardiovasc Genet 2014, 7(6):838–847 (Ref [49])
b, cRefSeq of SCARB1: hg19, NM_005505 (CHIP Bioinformatics)
ddbSNP version 139: GRCh37.p10
Fig. 4.
Lipid-associated SCARB1 common variants and haplotype regions identified in US Non-Hispanic Whites (previous study; Ref [49]) and African Blacks (this study). Lipid-associated variants with MAF ≥5 % with P-values <0.05 and haplotype regions with global P-values < 0.05 that were previously identified in US Non-Hispanic Whites (US NHWs; n = 623) are shown in top panel and those identified in African Blacks (n = 788) are shown in bottom panel (see details in Table 9 and Table 10). SCARB1 variants and haplotype regions are shown on SCARB1 gene (5′ → 3′; RefSeq: hg19, NM_005505). All SNP IDs are based on dbSNP build 139. Regions I and II that are defined based on consecutive haplotype windows with evidence of lipid-association in US NHWs (global P < 0.05; see details in Ref [49]) also show some significant associations in African Blacks (global P < 0.05; see details in Table 7 and Table 8). ApoA-I, apolipoprotein A-I; ApoB, apolipoprotein B; HDL-C, high-density lipoprotein cholesterol; MAF, minor allele frequency; NHW, Non-Hispanic White; SNP, single nucleotide polymorphism; UTR, untranslated region
Discussion
Our sequencing identified 83 variants, of which 78 were selected for follow-up genotyping in the total sample of 788 African Blacks. Additional 69 tagSNPs from the HapMap-YRI data along with two previously reported lipid-associated SCARB1 variants were also genotyped in the total sample. Of 149 genotyped SCARB1 variants, 137 that passed QC were examined for association with major lipid traits (Table 2). The initial gene-based analyses revealed a nominal association with HDL-C (P = 0.0421) as well as a trend for association with ApoA-I (P = 0.1016; Table 3). Consistent with the gene-based results, single-site association analyses also revealed 10 common variants nominally associated (P < 0.05) with HDL-C (n = 5) and/or ApoA-I (n = 8; Table 4 and Fig. 2). The best association signal was between rs11057851 in intron 1 and HDL-C (P = 0.0043, FDR = 0.1465) followed by two associations with ApoA-I including rs7134858 in intron 6 (P = 0.0052, FDR = 0.2918) and rs5888 (p.Ala350Ala) in exon 8 (P = 0.0080, FDR = 0.2918). Moreover, three variants (rs11057851, rs4765615, and rs838895) exhibited evidence of associations (P < 0.05) with both HDL-C and ApoA-I. These findings are supported by the fact that SCARB1 appears to influence ApoA-I in addition to HDL-C [15, 17]. In our data, there was a moderate correlation between ApoA-I and HDL-C levels (r2 = 0.61).
Except for previously reported association of rs5888 (p.Ala350Ala) with lipid traits (HDL-C or LDL-C) in non-African populations [30–34, 36, 37, 39], the remaining nine associations observed in this study with the lipid traits (HDL-C and/or ApoA-I levels) in general population are novel and await replication in independent African or African-derived populations. Two of these nine SNPs have previously been shown to have differential effects on cholesterol levels in response to statin (rs4765615) [50] or on HDL-C/TG levels in response to estradiol in post-menopausal women (rs838895) [51]. Another variant (rs838896) was found to be associated with decreased SCARB1 expression in liver [51]. Although the latter SNP was not associated with a low RegulomeDB score (<3), we cannot rule out the possibility that it might be affecting the SCARB1 expression in a tissue-specific manner.
The haplotype analysis revealed evidence of significant association (global P < 0.05) of 32 haplotype windows with HDL-C (n = 16) and/or ApoA-I (n = 21; Table 7) and nine regions harboring consecutive overlapping haplotype windows significantly associated with either HDL-C (4 regions) or ApoA-I (5 regions; Table 8 and Fig. 3). In addition, six variants with nominal association (P < 0.05) in single-site analysis were contained in seven of these nine significantly associated regions, indicating the presence of functional variants in these regions. Our findings demonstrate that haplotype analysis may provide more information than single-site analysis.
Our comparison of the single-site and haplotype association results between in African Blacks (this study) and US NHWs (previous study [49]) has revealed three variants (rs11057820, rs4765615, and rs701106; Table 9) and two regions (Regions I and II; Table 10 and Fig. 4) showing evidence of lipid-associations in both ethnic groups. However, there were differences in associated traits, and/or associated alleles or their directional effects between the two ethnic groups, which reflects the genetic heterogeneity of complex phenotypes like lipid traits among diverse populations. This phenomenon can be explained by different ancestry backgrounds associated with differences in LD structure and genetic architecture, as well as by differences in SNP-SNP, gene-gene, and gene-environment interactions. Nonetheless, the lipid associations observed across different ethnic populations provide convincing evidence that causal/functional variants are present in SCARB1 gene that deserves comprehensive sequencing and functional studies in order to confirm and further characterize the effects of its variants on lipid metabolism.
Rare variant analysis showed significant evidence of association between a group of 23 rare variants (MAF ≤1 %) and HDL-C (P = 0.0478; Table 5). Single-site analysis of these rare variants revealed six (including three novel ones) with effects on HDL-C, of which three also had effects on ApoA-I (Table 6). In addition, four of these six rare variants appeared to be carried by individuals with extreme HDL-C and/or ApoA-I levels (above or under the 3rd percentile). This HDL-C-associated rare variant group also included a novel variant (p70201/chr12:125279319) that was observed in one individual with an unusually high plasma HDL-C level (above the mean + 3.5 SD). Our findings suggest that these rare variants might have functional relevance, thus screening of additional large African samples for these rare variants may help to establish their role in HDL-C and ApoA-I metabolism.
To date, there has been limited information concerning possible functional effects of lipid-associated SCARB1 variants, particularly for those located in non-coding regions. In fact, most of common and rare HDL-C/ApoA-I-associated variants observed in the current study are non-coding and do not show strong evidence of regulatory function based on RegulomeDB database. Nonetheless, three of these HDL-C/ApoA-I-associated SCARB1 variants (rs5888 [p.Ala350Ala], rs838885, and rs838886) have been previously demonstrated to influence the SCARB1 expression [51–53]. Therefore, additional functional studies are needed and may help to determine the functional nature of the SCARB1-associated variants and those in LD with them.
Our study has revealed a number of novel findings, although we also acknowledge some limitations. SCARB1 is a large gene and we sequenced only its coding regions and exon-intron junctions and also our sequencing sample size was small. Thus, we may have missed some functional LoF/rare variants due to small sample size and those located in uncovered intronic regions. Moreover, consistent with generally small effect sizes of lipid-associated variants reported in the literature, most of our single-site associations reached nominal significance (P < 0.05) but did not survive multiple testing corrections. Only the top variant (rs11057851) associated with HDL-C yielded an FDR cut-off of <0.20 (FDR = 0.1465; Table 4). Therefore, future larger studies in independent African or African-derived populations are necessary to validate all nominal associations observed in this study.
Conclusions
In conclusion, we report the first comprehensive association study of SCARB1 variants with lipid traits in a native African population, which revealed a number of novel associations in single-site and haplotype analyses. In addition, resequencing allowed us to identify 10 novel rare variants, of which four were in the group of 23 rare variants that has showed association with HDL-C levels. The SCARB1 associated common and rare variants observed in our study explained ~11.09 % of the variation in HDL-C levels and ~8.63 % of the variation in ApoA-I levels. Our findings indicate the genetic contribution of SCARB1, both common and LoF/rare variants, to inter-individual lipid variation in the general African Black population, which warrants further follow-up in independent studies. Insights into the HDL-C and related lipid traits may also lead to new potential targets for CHD treatment.
Acknowledgements
This study was supported by the National Heart, Lung and Blood Institute [NHLBI] (grant numbers HL044413 to C.H.B. and HL084613 to M.I.K.).
Abbreviations
- ApoA-I
Apolipoprotein A-I
- ApoB
Apolipoprotein B
- BP
Base pair
- CE
Cholesteryl esters
- CHD
Coronary heart disease
- FDR
False discovery rate
- GWAS
Genome-wide association studies
- HDL-C
High-density lipoprotein cholesterol
- HWE
Hardy-Weinberg equilibrium
- indel
Insertion and deletion variation
- KB
Kilobase pair
- LD
Linkage disequilibrium
- LDL-C
Low-density lipoprotein cholesterol
- LoF
Low-frequency
- MAF
Minor allele frequency
- NHW
Non-Hispanic White
- PCR
Polymerase chain reaction
- QC
Quality controls
- RCT
Reverse cholesterol transport
- SCARB1
Scavenger receptor class B member 1
- SD
Standard deviation
- SKAT-O
An optimal sequence kernel association test
- SNP
Single nucleotide polymorphism
- SNV
Singlenucleotide variation
- TG
Triglycerides
- UTR
Untranslated region
- VEGAS
Versatile gene-based association study
- VLDL-C
Very low-density lipoprotein cholesterol
- YRI
Yoruba people of Ibadan from Nigeria
Additional files
Characteristics and lipid profile of the entire sample of 788 African Blacks stratified by sex. (PDF 63 kb)
Primer sequences for 14 polymerase chain reaction (PCR) fragments and the sizes of 13 SCARB1 exons. (PDF 96 kb)
Characteristics of 83 SCARB1 sequence variants identified in 95 African Blacks with extreme HDL-C levels. (PDF 146 kb)
Characteristics of 10 SCARB1 novel variants identified by sequencing. (PDF 81 kb)
Linkage disequilibrium (LD) plot of 83 SCARB1 sequence variants. Of 83 sequence variants (see the list in Additional file 3: Table S3), 78 were selected for genotyping. An enlarged view of the part of LD plot (A) shows the pairwise correlations (r2) between four variants including the two variants (shown in bold) in the same bin in our data, of which one selected for genotyping. This bin was not identified by Tagger analysis of common SCARB1 variants in the HapMap-YRI data (see Additional file 7: Table S5 and Additional file 8: Figure S3). The degree of shades and values (r2 × 100) in each square of LD plot represent the pairwise correlations: black indicating r2 = 1, white indicating r2 = 0, and shade intensity indicating r2 between 0 and 1. LD, linkage disequilibrium; MAF, minor allele frequency; YRI, Yoruba people of Ibadan from Nigeria. (PDF 920 kb)
Linkage disequilibrium (LD) plot of 32 SCARB1 common sequence variants. Enlarged view of the parts of the LD plot (A, B, and C) show three LD bins (identified by Tagger analysis of variants with minor allele frequency (MAF) ≥5 % using an r2 cutoff of 0.90) containing more than one variant (r2 ranging between 0.95 and 1.0). The degree of shades and values (r2 × 100) in each square of LD plot represent the pairwise correlations: black indicating r2 = 1, white indicating r2 = 0, and shade intensity indicating r2 between 0 and 1. LD, linkage disequilibrium; MAF, minor allele frequency. (PDF 370 kb)
List of 77 SCARB1 HapMap-YRI tagSNPs. (PDF 110 kb)
Linkage disequilibrium (LD) plot of 108 SCARB1 common HapMap-YRI tagSNPs. The list of 77 common HapMap-YRI tagSNPs identified by Tagger analysis of variants with minor allele frequency ≥5 % using an r2 cutoff of 0.80 is shown in Additional file 7: Table S5. The degree of shades and values (r2 × 100) in each square of LD plot represent the pairwise correlations: black indicating r2 = 1, white indicating r2 = 0, and shade intensity indicating r2 between 0 and 1.LD, linkage disequilibrium; SNP, single nucleotide polymorphism; YRI, Yoruba people of Ibadan from Nigeria. (TIFF 2642 kb)
Characteristics of 138 SCARB1 variants genotyped in the entire sample of 788 African Blacks. (PDF 212 kb)
List of 87 SCARB1 genotyped common tagSNPs. (PDF 108 kb)
Linkage disequilibrium (LD) plot of 137 SCARB1 genotyped variants. The list of 87 genotyped common tagSNPs identified by Tagger analysis for variants with minor allele frequency ≥5 % using an r2 cutoff of 0.90 is shown in Additional file 10: Table S7. The degree of shades and values (r2 × 100) in each square of LD plot represent the pairwise correlations: black indicating r2 = 1, white indicating r2 = 0, and shade intensity indicating r2 between 0 and 1.LD, linkage disequilibrium; SNP, single nucleotide polymorphism. (TIFF 3095 kb)
Location and minor allele frequency (MAF) distributions of 137 SCARB1 genotyped variants. Details for each variant are shown in Additional file 9: Table S6. MAF, minor allele frequency; UTR, untranslated region. (TIFF 324 kb)
Covariates used for the statistical analysis of lipid variables. (PDF 78 kb)
Single-site association results for 136 SCARB1 genotyped variants with HDL-C. (PDF 689 kb)
Single-site association results for 136 SCARB1 genotyped variants with ApoA-I. (PDF 686 kb)
Haplotype association results for 136 SCARB1 genotyped variants for HDL-C and ApoA-I. (PDF 257 kb)
Summary of RegulomeDB scores of 153 SCARB1 variants. (PDF 95 kb)
RegulomeDB scores and functional assignments of 153 SCARB1 variants. (PDF 149 kb)
Footnotes
M. Ilyas Kamboh and F. Yesim Demirci contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceive and design the experiments: FYD, MIK Perform the experiments: VN, FYD Analyze the data: VN, XW, DP, ZHR, MMB, FYD, MIK Contribute reagents/materials/analysis tools: CHB, MIK Write the paper: VN, FYD, MIK Provide critical revisions: XW, DP, ZHR, CHB, MMB Interpret the results: VN, XW, DP, ZHR, CHB, MMB, FYD, MIK All authors read and approved the final manuscript.
Contributor Information
Vipavee Niemsiri, Email: vin4@pitt.edu.
Xingbin Wang, Email: xbw1@pitt.edu.
Dilek Pirim, Email: dip16@pitt.edu.
Zaheda H. Radwan, Email: zhr2@pitt.edu
Clareann H. Bunker, Email: bunkerc@pitt.edu
M. Michael Barmada, Email: barmada@pitt.edu.
M. Ilyas Kamboh, Phone: (+1) 412-624-3066, Email: kamboh@pitt.edu.
F. Yesim Demirci, Phone: (+1) 412-624-3066, Email: fyd1@pitt.edu.
References
- 1.National Cholesterol Education Program Third report of the national cholesterol education program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (adult treatment panel III) final report. Circulation. 2002;106(25):3143–3421. [PubMed] [Google Scholar]
- 2.World Health Organization. The 10 leading causes of death in the world, 2000 and 2012. In: The top 10 causes of death. Media centre, World Health Organization, 2014. Available from http://www.who.int/mediacentre/factsheets/fs310/en/. (accessed 30 April 2015).
- 3.O’Connell DL, Heller RF, Roberts DC, Allen JR, Knapp JC, Steele PL, Silove D. Twin study of genetic and environmental effects on lipid levels. Genet Epidemiol. 1988;5(5):323–341. doi: 10.1002/gepi.1370050504. [DOI] [PubMed] [Google Scholar]
- 4.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41(1):56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Consortium GLG, Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khetarpal SA, Rader DJ. Genetics of lipid traits: Genome-wide approaches yield new biology and clues to causality in coronary artery disease. Biochim Biophys Acta. 2014;1842(10):2010–2020. doi: 10.1016/j.bbadis.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Kuivenhoven JA, Hegele RA. Mining the genome for lipid genes. Biochim Biophys Acta. 2014;1842(10):1993–2009. doi: 10.1016/j.bbadis.2014.04.028. [DOI] [PubMed] [Google Scholar]
- 9.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305(5685):869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 10.Frikke-Schmidt R, Nordestgaard BG, Jensen GB, Tybjaerg-Hansen A. Genetic variation in ABC transporter A1 contributes to HDL cholesterol in the general population. J Clin Invest. 2004;114(9):1343–1353. doi: 10.1172/JCI200420361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peloso GM, Auer PL, Bis JC, Voorman A, Morrison AC, Stitziel NO, Brody JA, Khetarpal SA, Crosby JR, Fornage M, et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014;94(2):223–232. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rye KA, Barter PJ. Cardioprotective functions of HDLs. J Lipid Res. 2014;55(2):168–179. doi: 10.1194/jlr.R039297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenson RS, Brewer HB, Jr, Davidson WS, Fayad ZA, Fuster V, Goldstein J, Hellerstein M, Jiang XC, Phillips MC, Rader DJ, et al. Cholesterol efflux and atheroprotection: advancing the concept of reverse cholesterol transport. Circulation. 2012;125(15):1905–1919. doi: 10.1161/CIRCULATIONAHA.111.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trigatti BL, Krieger M, Rigotti A. Influence of the HDL receptor SR-BI on lipoprotein metabolism and atherosclerosis. Arterioscler Thromb Vasc Biol. 2003;23(10):1732–1738. doi: 10.1161/01.ATV.0000091363.28501.84. [DOI] [PubMed] [Google Scholar]
- 15.Kozarsky KF, Donahee MH, Rigotti A, Iqbal SN, Edelman ER, Krieger M. Overexpression of the HDL receptor SR-BI alters plasma HDL and bile cholesterol levels. Nature. 1997;387(6631):414–417. doi: 10.1038/387414a0. [DOI] [PubMed] [Google Scholar]
- 16.Wang N, Arai T, Ji Y, Rinninger F, Tall AR. Liver-specific overexpression of scavenger receptor BI decreases levels of very low density lipoprotein ApoB, low density lipoprotein ApoB, and high density lipoprotein in transgenic mice. J Biol Chem. 1998;273(49):32920–32926. doi: 10.1074/jbc.273.49.32920. [DOI] [PubMed] [Google Scholar]
- 17.Ueda Y, Royer L, Gong E, Zhang J, Cooper PN, Francone O, Rubin EM. Lower plasma levels and accelerated clearance of high density lipoprotein (HDL) and non-HDL cholesterol in scavenger receptor class B type I transgenic mice. J Biol Chem. 1999;274(11):7165–7171. doi: 10.1074/jbc.274.11.7165. [DOI] [PubMed] [Google Scholar]
- 18.Out R, Kruijt JK, Rensen PC, Hildebrand RB, de Vos P, Van Eck M, Van Berkel TJ. Scavenger receptor BI plays a role in facilitating chylomicron metabolism. J Biol Chem. 2004;279(18):18401–18406. doi: 10.1074/jbc.M401170200. [DOI] [PubMed] [Google Scholar]
- 19.Out R, Hoekstra M, de Jager SC, de Vos P, van der Westhuyzen DR, Webb NR, Van Eck M, Biessen EA, Van Berkel TJ. Adenovirus-mediated hepatic overexpression of scavenger receptor class B type I accelerates chylomicron metabolism in C57BL/6 J mice. J Lipid Res. 2005;46(6):1172–1181. doi: 10.1194/jlr.M400361-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Van Eck M, Hoekstra M, Out R, Bos IS, Kruijt JK, Hildebrand RB, Van Berkel TJ. Scavenger receptor BI facilitates the metabolism of VLDL lipoproteins in vivo. J Lipid Res. 2008;49(1):136–146. doi: 10.1194/jlr.M700355-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Wiersma H, Nijstad N, Gautier T, Iqbal J, Kuipers F, Hussain MM, Tietge UJ. Scavenger receptor BI facilitates hepatic very low density lipoprotein production in mice. J Lipid Res. 2010;51(3):544–553. doi: 10.1194/jlr.M000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trigatti B, Rigotti A, Krieger M. The role of the high-density lipoprotein receptor SR-BI in cholesterol metabolism. Curr Opin Lipidol. 2000;11(2):123–131. doi: 10.1097/00041433-200004000-00004. [DOI] [PubMed] [Google Scholar]
- 23.Rhainds D, Brissette L. The role of scavenger receptor class B type I (SR-BI) in lipid trafficking. defining the rules for lipid traders. Int J Biochem Cell Biol. 2004;36(1):39–77. doi: 10.1016/S1357-2725(03)00173-0. [DOI] [PubMed] [Google Scholar]
- 24.Rigotti A, Trigatti BL, Penman M, Rayburn H, Herz J, Krieger M. A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc Natl Acad Sci U S A. 1997;94(23):12610–12615. doi: 10.1073/pnas.94.23.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varban ML, Rinninger F, Wang N, Fairchild-Huntress V, Dunmore JH, Fang Q, Gosselin ML, Dixon KL, Deeds JD, Acton SL, et al. Targeted mutation reveals a central role for SR-BI in hepatic selective uptake of high density lipoprotein cholesterol. Proc Natl Acad Sci U S A. 1998;95(8):4619–4624. doi: 10.1073/pnas.95.8.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trigatti B, Rayburn H, Vinals M, Braun A, Miettinen H, Penman M, Hertz M, Schrenzel M, Amigo L, Rigotti A, et al. Influence of the high density lipoprotein receptor SR-BI on reproductive and cardiovascular pathophysiology. Proc Natl Acad Sci U S A. 1999;96(16):9322–9327. doi: 10.1073/pnas.96.16.9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji Y, Wang N, Ramakrishnan R, Sehayek E, Huszar D, Breslow JL, Tall AR. Hepatic scavenger receptor BI promotes rapid clearance of high density lipoprotein free cholesterol and its transport into bile. J Biol Chem. 1999;274(47):33398–33402. doi: 10.1074/jbc.274.47.33398. [DOI] [PubMed] [Google Scholar]
- 28.Brunham LR, Tietjen I, Bochem AE, Singaraja RR, Franchini PL, Radomski C, Mattice M, Legendre A, Hovingh GK, Kastelein JJ, et al. Novel mutations in scavenger receptor BI associated with high HDL cholesterol in humans. Clin Genet. 2011;79(6):575–581. doi: 10.1111/j.1399-0004.2011.01682.x. [DOI] [PubMed] [Google Scholar]
- 29.Vergeer M, Korporaal SJ, Franssen R, Meurs I, Out R, Hovingh GK, Hoekstra M, Sierts JA, Dallinga-Thie GM, Motazacker MM, et al. Genetic variant of the scavenger receptor BI in humans. N Engl J Med. 2011;364(2):136–145. doi: 10.1056/NEJMoa0907687. [DOI] [PubMed] [Google Scholar]
- 30.Acton S, Osgood D, Donoghue M, Corella D, Pocovi M, Cenarro A, Mozas P, Keilty J, Squazzo S, Woolf EA, et al. Association of polymorphisms at the SR-BI gene locus with plasma lipid levels and body mass index in a white population. Arterioscler Thromb Vasc Biol. 1999;19(7):1734–1743. doi: 10.1161/01.ATV.19.7.1734. [DOI] [PubMed] [Google Scholar]
- 31.Hong SH, Kim YR, Yoon YM, Min WK, Chun SI, Kim JQ. Association between HaeIII polymorphism of scavenger receptor class B type I gene and plasma HDL-cholesterol concentration. Ann Clin Biochem. 2002;39(Pt 5):478–481. doi: 10.1258/000456302320314485. [DOI] [PubMed] [Google Scholar]
- 32.Osgood D, Corella D, Demissie S, Cupples LA, Wilson PW, Meigs JB, Schaefer EJ, Coltell O, Ordovas JM. Genetic variation at the scavenger receptor class B type I gene locus determines plasma lipoprotein concentrations and particle size and interacts with type 2 diabetes: the framingham study. J Clin Endocrinol Metab. 2003;88(6):2869–2879. doi: 10.1210/jc.2002-021664. [DOI] [PubMed] [Google Scholar]
- 33.Tai ES, Adiconis X, Ordovas JM, Carmena-Ramon R, Real J, Corella D, Ascaso J, Carmena R. Polymorphisms at the SRBI locus are associated with lipoprotein levels in subjects with heterozygous familial hypercholesterolemia. Clin Genet. 2003;63(1):53–58. doi: 10.1034/j.1399-0004.2003.630108.x. [DOI] [PubMed] [Google Scholar]
- 34.Morabia A, Ross BM, Costanza MC, Cayanis E, Flaherty MS, Alvin GB, Das K, James R, Yang AS, Evagrafov O, et al. Population-based study of SR-BI genetic variation and lipid profile. Atherosclerosis. 2004;175(1):159–168. doi: 10.1016/j.atherosclerosis.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Richard E, von Muhlen D, Barrett-Connor E, Alcaraz J, Davis R, McCarthy JJ. Modification of the effects of estrogen therapy on HDL cholesterol levels by polymorphisms of the HDL-C receptor, SR-BI: the Rancho Bernardo Study. Atherosclerosis. 2005;180(2):255–262. doi: 10.1016/j.atherosclerosis.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 36.Boekholdt SM, Souverein OW, Tanck MW, Hovingh GK, Kuivenhoven JA, Peters RI, Jansen H, Schiffers PM, van der Wall EE, Doevendans PA, et al. Common variants of multiple genes that control reverse cholesterol transport together explain only a minor part of the variation of HDL cholesterol levels. Clin Genet. 2006;69(3):263–270. doi: 10.1111/j.1399-0004.2006.00578.x. [DOI] [PubMed] [Google Scholar]
- 37.Roberts CG, Shen H, Mitchell BD, Damcott CM, Shuldiner AR, Rodriguez A. Variants in scavenger receptor class B type I gene are associated with HDL cholesterol levels in younger women. Hum Hered. 2007;64(2):107–113. doi: 10.1159/000101962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka T, Delgado-Lista J, Lopez-Miranda J, Perez-Jimenez F, Marin C, Perez-Martinez P, Gomez P, Ordovas JM. Scavenger receptor class B type I (SCARB1) c.1119C > T polymorphism affects postprandial triglyceride metabolism in men. J Nutr. 2007;137(3):578–582. doi: 10.1093/jn/137.3.578. [DOI] [PubMed] [Google Scholar]
- 39.Cerda A, Genvigir FD, Arazi SS, Hirata MH, Dorea EL, Bernik MM, Bertolami MC, Faludi AA, Hirata RD. Influence of SCARB1 polymorphisms on serum lipids of hypercholesterolemic individuals treated with atorvastatin. Clin Chim Acta. 2010;411(9–10):631–637. doi: 10.1016/j.cca.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Manichaikul A, Naj AC, Herrington D, Post W, Rich SS, Rodriguez A. Association of SCARB1 variants with subclinical atherosclerosis and incident cardiovascular disease: the multi-ethnic study of atherosclerosis. Arterioscler Thromb Vasc Biol. 2012;32(8):1991–1999. doi: 10.1161/ATVBAHA.112.249714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bunker CH, Ukoli FA, Okoro FI, Olomu AB, Kriska AM, Huston SL, Markovic N, Kuller LH. Correlates of serum lipids in a lean black population. Atherosclerosis. 1996;123(1–2):215–225. doi: 10.1016/0021-9150(96)05810-8. [DOI] [PubMed] [Google Scholar]
- 42.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18(6):499–502. [PubMed] [Google Scholar]
- 43.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 44.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Investigators AMFS, Hayward NK, Montgomery GW, Visscher PM, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87(1):139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Stat Methodol) 1995;57:289–300. [Google Scholar]
- 46.Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13(4):762–775. doi: 10.1093/biostatistics/kxs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao G, Garcia CK, Wyne KL, Schultz RA, Parker KL, Hobbs HH. Structure and localization of the human gene encoding SR-BI/CLA-1. Evidence for transcriptional control by steroidogenic factor 1. J Biol Chem. 1997;272(52):33068–33076. doi: 10.1074/jbc.272.52.33068. [DOI] [PubMed] [Google Scholar]
- 48.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niemsiri V, Wang X, Pirim D, Radwan ZH, Hokanson JE, Hamman RF, Barmada MM, Demirci FY, Kamboh MI. Impact of genetic variants in human scavenger receptor class B type I (SCARB1) on plasma lipid traits. Circ Cardiovasc Genet. 2014;7(6):838–847. doi: 10.1161/CIRCGENETICS.114.000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peters BJ, Pett H, Klungel OH, Stricker BH, Psaty BM, Glazer NL, Wiggins KL, Bis JC, de Boer A, der Zee AH M-v. Genetic variability within the cholesterol lowering pathway and the effectiveness of statins in reducing the risk of MI. Atherosclerosis. 2011;217(2):458–464. doi: 10.1016/j.atherosclerosis.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiba-Falek O, Nichols M, Suchindran S, Guyton J, Ginsburg GS, Barrett-Connor E, McCarthy JJ. Impact of gene variants on sex-specific regulation of human Scavenger receptor class B type 1 (SR-BI) expression in liver and association with lipid levels in a population-based study. BMC Med Genet. 2010;11:9. doi: 10.1186/1471-2350-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Constantineau J, Greason E, West M, Filbin M, Kieft JS, Carletti MZ, Christenson LK, Rodriguez A. A synonymous variant in scavenger receptor, class B, type I gene is associated with lower SR-BI protein expression and function. Atherosclerosis. 2010;210(1):177–182. doi: 10.1016/j.atherosclerosis.2009.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cerda A, Genvigir FD, Rodrigues AC, Willrich MA, Dorea EL, Bernik MM, Arazi SS, Oliveira R, Hirata MH, Hirata RD. Influence of polymorphisms and cholesterol-lowering treatment on SCARB1 mRNA expression. J Atheroscler Thromb. 2011;18(8):640–651. doi: 10.5551/jat.6544. [DOI] [PubMed] [Google Scholar]


