Abstract
R-Ras small GTPase enhances cell spreading and motility via RalBP1/RLIP76, an R-Ras effector that links GTP-R-Ras to activation of Arf6 and Rac1 GTPases. Here, we report that RLIP76 performs these functions by binding cytohesin-2/ARNO, an Arf GTPase guanine exchange factor, and connecting it to R-Ras at recycling endosomes. RLIP76 formed a complex with R-Ras and ARNO by binding ARNO via its N-terminus (residues 1-180) and R-Ras via residues 180-192. This complex was present in Rab11-positive recycling endosomes and the presence of ARNO in recycling endosomes required RLIP76, and was not supported by RLIP76(Δ1-180) or RLIP76(Δ180-192). Spreading and migration required RLIP76(1-180), and RLIP76(Δ1-180) blocked ARNO recruitment to recycling endosomes, and spreading. Arf6 activation with an ArfGAP inhibitor overcame the spreading defects in RLIP76-depleted cells or cells expressing RLIP76(Δ1-180). Similarly, RLIP76(Δ1-180) or RLIP76(Δ180-192) suppressed Arf6 activation. Together these results demonstrate that RLIP76 acts as a scaffold at recycling endosomes by binding activated R-Ras, recruiting ARNO to activate Arf6, thereby contributing to cell spreading and migration.
Keywords: Ras, migration, spreading, recycling endosome, small GTPase
Introduction
Migrating cells rely on the concerted actions of small GTPases to regulate cytoskeletal dynamics, to establish and maintain polarity, and to modulate the formation and release of ECM attachments[1, 2]. Formation of new integrin attachments at the cell front stimulates activation of Arf6 and Rac1 GTPases, which cause localized actin polymerization to create lamellipodia[3–6]. Similarly, integrin-mediated adhesion of detached cells to ECM substrates leads to rapid un-polarized Arf6 and Rac1 activation, resulting in lamellipodial extension around the cell perimeter in a process known as cell spreading[7]. Spreading, lamellipodial extension, and cell migration are thus closely related Arf6- and Rac1-mediated events. We have shown that the Ras family GTPase R-Ras participates in Arf6- and Rac1-dependent cell spreading and motility[8].
R-Ras has unique functions distinct from other Ras family GTPases, such as stimulating integrins and formation of attachment structures[9–13], and adhesion-mediated Rac1 activation and cell migration[14, 15]. R-Ras has an almost identical effector-binding region to Ras[16, 17], and couples to common Ras effectors including Raf-1 (albeit at much lower affinity), RalGDS, RapL/NORE1 and PI3-K[12]. However, until recently no unique R-Ras effectors had been described to account for its functions distinct from Ras and Rap1. We identified RLIP76 (Ral-interacting protein of 76 kDa, also Ral-binding protein 1 or RalBP1) as a requisite R-Ras-specific effector in R-Ras-dependent cell spreading and migration[8, 18].
We previously showed that the role of RLIP76 in R-Ras-dependent spreading and migration is to regulate activation of Rac1 and Arf6. The N-terminal domain of RLIP76 interacts with ARNO (cytohesin 2), a Sec7-domain-containing ArfGEF which activates both Arf1 and Arf6 and promotes Arf6-dependent activation of Rac1[19–21]. Furthermore, ARNO over-expression rescued the spreading defect in RLIP76-depleted cells[8]. The sub-cellular localization of GTPases and of the GEFs and GAPs which control their activities plays an important role in spatial segregation of signaling by small G proteins such as Arf6[22, 23]. We hypothesized that RLIP76 interaction with ARNO may contribute to its effects on R-Ras-dependent Arf6 activation leading to cell spreading and migration. In this study we describe an Arf6 activation complex consisting of activated R-Ras, RLIP76 and ARNO, localized to recycling endosomes. We present a model for a molecular mechanism of R-Ras stimulation of Arf6 leading to cell spreading and motility.
Materials and Methods
Antibodies and reagents
Monoclonal anti-Rac1 antibody (23A8) was obtained from Millipore. α–human RalBP1 (RLIP76), α–R-Ras, α-Arf6, α-GFP, α-FLAG, α-myc and α-ARNO antibodies were from Santa Cruz Biotechnology, Inc. Anti-HA antibody was from Covance. Restriction endonucleases were from New England Biolabs.
Cell lines and transfections
RLIP76 knockout mice have been described previously(Awasthi et al., 2005). C57Bl/6 mice were from Jackson Laboratories (Bar Harbor, ME). Embryonic fibroblasts were derived from 8-week-old C57Bl/6 (wild type) and RLIP76-null female mice. At day E13, uteri containing embryos were removed and placed in ice-cold PBS. The embryos were separated from the uteri and then dissected, removing the head, organs, and appendages from the fetuses. The carcasses were placed in trypsin for 1 hour, then centrifuged to remove larger particles. The cell suspensions were transferred to culture plates for propagation. Cell culture and transfections were as described[24]. All animal experimental procedures were approved by the Temple University Institutional Animal Care and Use Committee.
Complementary DNAs
pEGFP-C1 was from CLONTECH Laboratories, Inc. (Mountain View, CA). pEGFP-C1-Rac1 WT was a gift from Miguel del Pozo (Fundación Centro Nacional de Investigaciones Cardiovasculares, Madrid, Spain). pEGFP-R-Ras(G38V) was as described[24]. pEGFP-Rab11 (wt) was a gift from Eugene Tkatchenko (University of California, San Diego, La Jolla, CA). FLAG-ARNO was a gift from Lorraine Santy (Penn State University, State College, PA). Human RLIP76 cDNA containing a 5′ influenza HA tag and shRNA-insensitive (mismatch) mutations, RLIP76 truncation plasmids, and R-Ras(43N) were as described[8, 19]. HA-RLIP76(Δ180-192) was generated by PCR from the same template using the following primer set: 5'-GGAGCCAGAGGTGCCTCTGGTTGCTGCTCAAAGTCACAATCGCGTTTTGGAAACTCCTTTGGCTGATGC-3' and 5'-CATCAGCCAAAGGAGTTTCCAAAACGCGATTGTGACTTTGAGCAGCAACCAGAGGCACCTCTGGCTCC-3'. For shRNA-mediated RLIP76 knockdown, complementary single-strand oligonucleotides with overhangs (5'-GATCCCCGTAGAGAGGACCATGATGTTTCAAGAGAACATCATGGTCCTCTCTACTTTTTA-3' and 5'-AGCTTAAAAAGTAGAGAGGACCATGATGTTCTCTTGAAACATCATGGTCCTCTCTACGGG-3') targeting human and mouse RLIP76 (IDT Technologies) were dissolved in TE buffer (10 mM Tris-Cl, pH 7.5, 0.1 mM EDTA). Sequence-scrambled complementary oligonucleotides were also obtained. Double-stranded oligonucleotides were generated by annealing in annealing buffer (100 mM KOAc, 30 mM HEPES-KOH, pH 7.4, and 2 mM MgOAc), and duplexes were ligated directly into HinDIII/BglII-digested pSUPER.retro.puro vector (OligoEngine).
Cell spreading and migration assays and microscopy
Cell spreading and confocal microscopy were performed as described previously[8, 24].
Arf6 activation assays and immunoprecipitations
Adhesion-induced Arf6 activation and co-immunoprecipitation assays were performed as described previously[8, 21].
Statistical analysis
One-way ANOVA followed by Fisher PLSD analysis was used for all statistical data analysis, using StatView (SAS). A 5% probability was considered significant. All experiments in this study were performed at least in triplicate, except where indicated otherwise, and for microscopy results representative images are shown.
Results
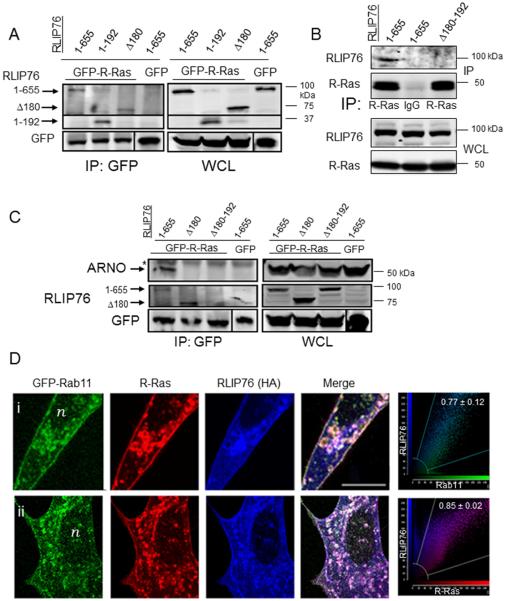
We recently identified the RLIP76 N-terminus (aa 1-180) as the ARNO interaction domain [19]. We sought to map the R-Ras interaction site in RLIP76 and determine whether R-Ras and ARNO can interact in a tri-molecular complex with RLIP76. RLIP76(Δ180), which does not interact with ARNO [19], retained R-Ras binding (Fig. 1A); however, a larger RLIP76 N-terminal fragment (1-192) also bound activated R-Ras; suggesting that aa 180-192 are important for R-Ras interaction. We mutated this region to a randomized sequence in the full-length protein (RLIP76(Δ180-192), which disrupted RLIP76 interaction with R-Ras(Fig. 1B), demonstrating that the 180-192 region of RLIP76 is responsible for R-Ras interaction. RLIP76(Δ180) and RLIP76(Δ180-192) also disrupted ARNO/R-Ras interaction (Fig. 1C). Thus, whereas ARNO interacts with RLIP76 in a region between aa 1-180, activated R-Ras binding to RLIP76 requires the region between aa 180-192. Together, these findings demonstrate that activated R-Ras, RLIP76 and ARNO can form a tri-molecular complex in cells, RLIP76 interacts with R-Ras and ARNO through distinct sites, and ARNO association with R-Ras requires RLIP76. Thus, RLIP76 acts as an adapter protein to link activated R-Ras to ARNO in cells.
Fig. 1. ARNO and activated R-Ras associate with RLIP76 in a complex at recycling endosomes.
(A) Co-immunoprecipitations (IP) with GFP antibodies from cells expressing GFP-tagged R-Ras(G38V) and HA-RLIP76 truncated or mutant proteins as indicated. (B) Co-IPs of myc-tagged R-Ras(G38V) with HA-tagged RLIP76 variants with α-R-Ras antibodies or control IgG as indicated. (C) Co-IPs of GFP-R-Ras, HA-RLIP76 variants as indicated, and myc-ARNO, with GFP antibodies. *, Ig heavy chain bands in the IPs. (D) Cells from two independent experiments are shown (I, ii). Secondary antibodies used alone as controls showed no detectable staining, and there were no detectable bleedthrough effects across channels (results not shown). Representative two-color component scatter plots comparing the distribution of correlated pixels for Rab11 and RLIP76 (i) or for R-Ras and RLIP76 (ii) and the calculated Pearson's correlation coefficients are shown on the far right. Bar, 10 μm. n, nucleus.
R-Ras, ARNO and RLIP76 co-localize to recycling endosomes
R-Ras has been localized to Rab11-positive vesicles, presumed to be recycling endosomes (RE)[25, 26]. To investigate a correlation of this property of R-Ras with RLIP76 localization, we assessed the cellular distribution of R-Ras and RLIP76 by confocal microscopy. WT R-Ras localized to Rab11-positive structures, likely RE, and RLIP76 was also partially localized to the Rab11-positive structures, overlapping with R-Ras (Fig. 1D). In addition, both R-Ras and RLIP76 were observed in other unidentified sub-cellular compartments and at the plasma membrane; these observations are also consistent with previous reports for R-Ras[25–27]. RLIP76 staining did not overlap with the distribution of GFP-tagged markers for early endosomes (Rab5)[28], cis-Golgi (GM130)[29], or cis-Golgi-to-ER transport vesicles (p23)[30], (Supplemental Figure 1). Thus, RLIP76 partially co-localizes with R-Ras in recycling endosomes.
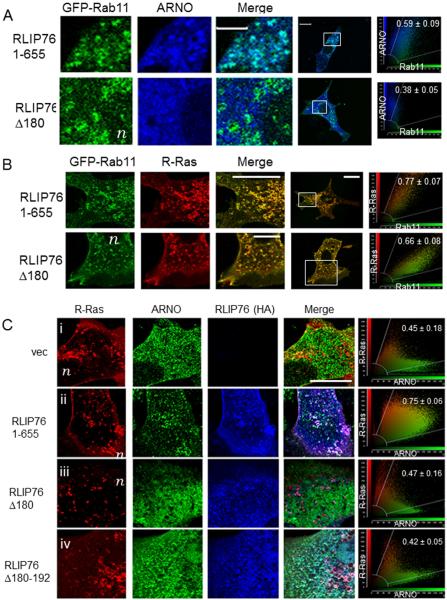
We next investigated whether the R-Ras/RLIP76 complex associates with ARNO at RE. Both ARNO and R-Ras were present in Rab11-positive endosomes (Fig. 2A and B). ARNO was also observed in diffuse patterns throughout the cytosol and at the plasma membrane. Cells transfected with RLIP76(Δ180) exhibited markedly diminished endosomal localization of ARNO (Fig. 2A); however, R-Ras was retained in RE and at the plasma membrane (Fig. 2B). ARNO did not co-localize with R-Ras in RLIP76-null embryonic fibroblasts (Fig. 2Ci). Reconstitution of these cells with full-length RLIP76(1-655) restored ARNO co-localization with R-Ras (Fig. 2Cii), demonstrating that RLIP76 is required for ARNO and R-Ras co-localization. However, reconstitution with RLIP76(Δ180) or RLIP76(Δ180-192) did not restore ARNO to R-Ras-containing RE (Fig. 2Ciii,iv). R-Ras knockdown with shRNA also displaced ARNO from RE, confirming that R-Ras is required for RE targeting of ARNO (Supplemental Figure 2). Thus, ARNO co-localization with R-Ras at RE requires the RLIP76 N-terminus and an R-Ras/RLIP76 complex.
Fig. 2. ARNO co-localizes with R-Ras to recycling endosomes in a RLIP76-dependent manner.
NIH 3T3 cells were transfected with FLAG-ARNO, Rab11-GFP and HA-tagged RLIP76 constructs as indicated. (A) Rab11, green; ARNO, blue. (B) Rab11, green; R-Ras, red. (C) RLIP76-null MEFs transfected with FLAG-ARNO, Rab11-GFP and HA-tagged RLIP76 constructs as indicated. Merged images and lower magnification images showing the selected regions are to the right. Representative two-color component scatter plots comparing the distribution of correlated pixels for ARNO and Rab11 (A), R-Ras and Rab11 (B), or R-Ras and ARNO (C) and the calculated Pearson's correlation coefficients are shown to the far right. Bars, 10 μm. n, nucleus.
An RLIP76/ARNO/R-Ras complex is required for cell spreading via Arf6 activation
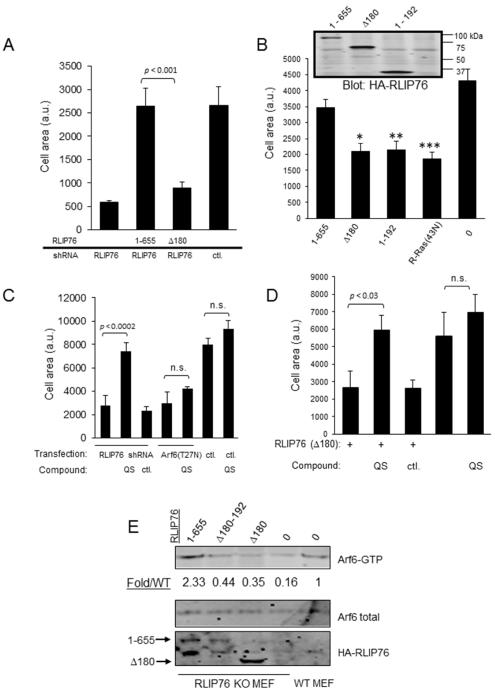
Following our previous finding that ARNO-RLIP76 interaction is required for RLIP76-dependent cell spreading[19], we investigated a role of the RLIP76/ARNO/R-Ras complex in spreading. Reconstitution of RLIP76-depleted cells with full-length RLIP76 restored spreading, whereas reconstitution with RLIP76(Δ180) (Fig. 3A), or ectopic over-expression in WT cells of either RLIP76(Δ180) or RLIP76(1-192), or dominant negative R-Ras[8], inhibited spreading (Fig. 3B), demonstrating that RLIP76/ARNO/R-Ras interaction is necessary for cell spreading.
Fig. 3. The RLIP76 N-terminus is required for efficient cell spreading upstream of Arf6.
(A – D). Cells were transfected as indicated and seeded on fibronectin-coated surfaces for spreading assays. HA-RLIP76 expression levels are shown in the inset panel in (B). (0) = vector control. *, p < 0.02; **, p < 0.004; ***, p < 0.001 (n=3). The average surface areas of GFP-positive cells are shown + SEM (n=3). (C,D) Cells were treated with 1 μM of either QS11 (QS) ArfGAP1 inhibitor or QS11-NC inactive control compound (ctl.), and assayed for spreading. (E) Arf6 activation in cells transfected with the indicated RLIP76 consdtructs, shown as fold increase in the ratio of GTP-Arf6 to total Arf6, normalized to WT (Fold/WT). Representative of four independent experiments.
To investigate the role of Arf6 activation in RLIP76- and ARNO-dependent cell spreading, we used a small molecule inhibitor of ARFGAP1, QS11, which has been extensively used to stimulate Arf6 activation[31–33]. QS11, but not the negative control analog[32], overcame the spreading defect in RLIP76 knockdown cells (Fig. 3C). Ectopic expression of dominant negative Arf6(T27N) blocked spreading, as we have observed previously[8]. QS11 treatment had no effect in cells expressing this Arf6 mutant (Fig. 3C). We next used QS11 to attempt to bypass the requirement for RLIP76/ARNO interaction in spreading. QS11 completely restored spreading in cells over-expressing RLIP76(Δ180) (Fig. 3D), indicating that Arf6 activation downstream of RLIP76/ARNO interaction is sufficient for cell spreading on integrin ligands. Adhesion-induced Arf6 activation was suppressed in RLIP76-null cells, and was restored by reconstitution with full-length RLIP76(1-655), consistent with our previous findings[8, 19]. In contrast, reconstitution with RLIP76(Δ180) or RLIP76(Δ180-192) failed to restore Arf6 activation (Fig. 3E). Thus, the adapter function of RLIP76, linking ARNO to R-Ras in a complex in cells, is necessary for adhesion-induced Arf6 activation leading to cell spreading.
RLIP76/ARNO/R-Ras signaling is required for migration
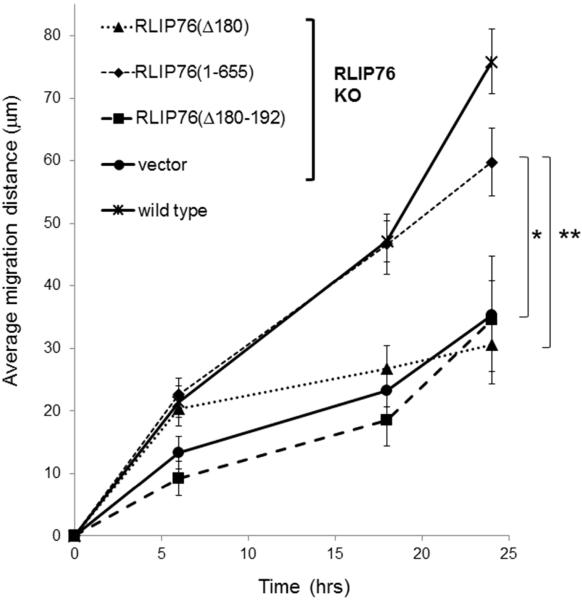
We investigated whether the RLIP76/R-Ras/ARNO complex is important for cell migration. RLIP76-null fibroblasts migrated at a slower rate than wild type cells in scratch wound assays, but migration was restored with full-length RLIP76, as previously observed (Fig. 4)[19]. RLIP76-null cells reconstituted with RLIP76(Δ180) exhibited markedly impaired migration relative to knockout cells reconstituted with full-length RLIP76. Similarly, RLIP76(Δ180-192) did not restore robust migration in these cells (Fig. 4). Thus, the ability of RLIP76 to regulate ARNO interaction, to support ARNO complex formation with R-Ras, ARNO localization, Arf6 activation, and cell spreading are required for robust cell migration in fibroblasts.
Fig. 4. The RLIP76 N-terminus and R-Ras interaction are required for fibroblast migration.
Primary embryonic fibroblasts derived from RLIP76-null or isogenic wild type mice were transfected as indicated plus GFP. Equal numbers of wild type or RLIP76-null (KO) fibroblasts transfected as shown were seeded onto fibronectin-coated surfaces to form confluent monolayers. A single scratch was made through the monolayer with a pipet tip and GFP-positive cells at the wound margins were imaged over time, from which migration distances were calculated. Average migration distances from the starting point are shown ± SEM. *, p < 0.03; **, p < 0.02 (n=5).
Discussion
In this study we present evidence for a signaling pathway through which R-Ras promotes cell spreading and motility. The basis for this pathway is a signaling complex comprised of activated R-Ras anchored to recycling endosomes, recruiting the ArfGEF, ARNO through the RLIP76 linker protein, driving Arf6 activation leading to lamellipodial extension and adoption of a motile phenotype.
RLIP76 can be phosphorylated at Ser and Thr residues in cells[34], the utilized sites are concentrated in the N-terminus[35], and we have identified two Ser residues in this domain required for ARNO interaction[19]. Furthermore, we have identified a region of RLIP76 which is required for R-Ras interaction, but which does not share sequence homology with classical Ras-binding domains (RBDs). These domains typically adopt an ubiquitin-like fold which forms a binding face for the switch I region of Ras[36]. Our previous results indicated that the Ral-binding domain of RLIP76 in the C-terminus, which is likely to adopt a similar conformation as a RBD, was dispensable for R-Ras interaction[8]. How the short segment of RLIP76 (aa 180-192) mediates GTP-dependent R-Ras binding remains to be explained, but suggests a novel mode of effector interaction with a Ras protein.
Activated R-Ras is concentrated in cells at the plasma membrane and in membranes of recycling endosomes (RE)[24–26], also apparent in our cells, but the signaling role of R-Ras in RE is just beginning to be defined. H-Ras and N-Ras also traffic through RE[37]; however, we have shown that R-Ras and H-Ras follow distinct anterograde trafficking pathways from RE to the plasma membrane, suggesting that vesicular R-Ras signaling may be spatially segregated[24]. Our observations here suggest the assembly of an R-Ras signaling complex at the surfaces of RE, in which RLIP76 and ARNO are recruited from the cytosol. ARNO is mostly a cytosolic protein, and perinuclear enrichment has been observed in several cell types; however, the precise localization was not defined[38–40]. Other ARNO adapter proteins, such as CASP, have also been localized to perinuclear tubulo-vesicular structures which may represent RE[41]. Similarly, RLIP76 has been described as both a cytosolic and membrane-associated protein, and has been observed in punctate structures in some cells [42–44]. Thus, R-Ras, RLIP76 and ARNO form a molecular complex recruited to RE membranes, representing a convergence of cytosolic proteins which can engage in signaling localized to those sites.
The RLIP76 complex may control Arf6-dependent spreading through exocytic trafficking of specific subsets of migration-related proteins. One such possibility may involve Arf6-dependent recruitment of a WAVE regulatory complex to the plasma membrane, facilitating actin assembly[45]. Both ARNO and Arf6 regulate rapid recycling of β1 integrins, important adhesion receptors mediating cell attachments to the extracellular matrix during cell spreading and migration[39, 46]. Arf6 activated at RE causes exocytic recycling of lipid raft compartments from RE to the plasma membrane, apparently carrying β1 integrins and stimulating Rac activation – the latter is an established function of ARNO[21, 40, 46–48]. Whether the R-Ras/RLIP76/ARNO nexus participates in these particular recycling processes remains to be determined. R-Ras has also been linked recently to β1 integrin trafficking to membrane ruffles via R-Ras association with RE[26]. It is tempting to speculate that RLIP76 engages in the signal crosstalk between R-Ras, Arf6, β1 integrins, and exocytic trafficking to stimulate spreading and migration.
In summary, the RLIP76 N-terminus, through its interaction with R-Ras and ARNO, coordinates a spatially restricted signaling nexus to regulate Arf6 leading to cell spreading and migration. Thus, spatially-restricted small GTPase signaling regulates cytoskeletal dynamics, cell morphology and motility.
Supplementary Material
Highlights.
RLIP76 is a linker protein connecting R-Ras to ARNO, and ArfGEF.
The RLIP76/R-Ras/ARNO complex localizes to recycling endosomes.
Endosomal targeting of the complex promotes spreading and migration through activation of Arf6.
Acknowledgements
The authors thank Tyrina Talesha-Lashay Newkirk and Ankita Patel for technical assistance. This work was supported by grants from the NIH (HL093416 to LG, HL31950 to MG) and USPHS CA 77495 (SA and SS), and American Heart Association Postdoctoral Fellowship 12POST12040257 to SL.
Abbreviations
- Arf
ADP ribosylation factor
- ArfGAP
Arf GTPase activating protein
- ArfGEF
Arf guanine nucleotide exchange factor
- ECM
extracellular matrix
- GTP
guanosine triphosphate
- GTPase
GTP hydrolyzing enzyme
- RE
recycling endosome(s)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- [2].Schwartz MA, Horwitz AR. Integrating adhesion, protrusion, and contraction during cell migration. Cell. 2006;125:1223–1225. doi: 10.1016/j.cell.2006.06.015. [DOI] [PubMed] [Google Scholar]
- [3].Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- [4].Radhakrishna H, Al-Awar O, Khachikian Z, Donaldson JG. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. Journal of Cell Science. 1999;112(Pt 6):855–866. doi: 10.1242/jcs.112.6.855. [DOI] [PubMed] [Google Scholar]
- [5].Song J, Khachikian Z, Radhakrishna H, Donaldson JG. Localization of endogenous ARF6 to sites of cortical actin rearrangement and involvement of ARF6 in cell spreading. J Cell Sci. 1998;111(Pt 15):2257–2267. doi: 10.1242/jcs.111.15.2257. [DOI] [PubMed] [Google Scholar]
- [6].Titus B, Schwartz MA, Theodorescu D. Rho proteins in cell migration and metastasis. Crit Rev Eukaryot Gene Expr. 2005;15:103–114. doi: 10.1615/critreveukaryotgeneexpr.v15.i2.20. [DOI] [PubMed] [Google Scholar]
- [7].Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- [8].Goldfinger LE, Ptak C, Jeffery ED, Shabanowitz J, Hunt DF, Ginsberg MH. RLIP76 (RalBP1) is an R-Ras effector that mediates adhesion-dependent Rac activation and cell migration. Journal of Cell Biology. 2006;174:877–888. doi: 10.1083/jcb.200603111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hughes PE, Oertli B, Han J, Ginsberg MH. R-Ras Regulation of Integrin Function. Methods Enzymol. 2001;333:163–171. doi: 10.1016/s0076-6879(01)33054-9. [DOI] [PubMed] [Google Scholar]
- [10].Hughes PE, Renshaw MW, Pfaff M, Forsyth J, Keivens VM, Schwartz MA, Ginsberg MH. Suppression of integrin activation: A novel function of a Ras/Raf-initiated MAP kinase pathway. Cell. 1997;88:521–530. doi: 10.1016/s0092-8674(00)81892-9. [DOI] [PubMed] [Google Scholar]
- [11].Kwong L, Wozniak MA, Collins AS, Wilson SD, Keely PJ. R-Ras promotes focal adhesion formation through focal adhesion kinase and p130(Cas) by a novel mechanism that differs from integrins. Mol.Cell Biol. 2003;23:933–949. doi: 10.1128/MCB.23.3.933-949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Oertli B, Han J, Marte BM, Sethi T, Downward J, Ginsberg M, Hughes PE. The effector loop and prenylation site of R-Ras are involved in the regulation of integrin function [In Process Citation] Oncogene. 2000;19:4961–4969. doi: 10.1038/sj.onc.1203876. [DOI] [PubMed] [Google Scholar]
- [13].Sethi T, Ginsberg MH, Downward J, Hughes PE. The small GTP-binding protein R-Ras can influence integrin activation by antagonizing a Ras/Raf initiated integrin suppression pathway. Molecular Biology of the Cell. 10(1999):1799–1809. doi: 10.1091/mbc.10.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Holly SP, Larson MK, Parise LV. The unique N-terminus of R-ras is required for Rac activation and precise regulation of cell migration. Molecular Biology of the Cell. 2005;16:2458–2469. doi: 10.1091/mbc.E03-12-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wozniak MA, Kwong L, Chodniewicz D, Klemke RL, Keely PJ. R-Ras controls membrane protrusion and cell migration through the spatial regulation of Rac and Rho. Molecular Biology of the Cell. 2005;16:84–96. doi: 10.1091/mbc.E04-04-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lowe DG, Capon DJ, Delwart E, Sakaguchi AY, Naylor SL, Goeddel DV. Structure of the human and murine R-ras genes, novel genes closely related to ras proto-oncogenes. Cell. 1987;48:137–146. doi: 10.1016/0092-8674(87)90364-3. [DOI] [PubMed] [Google Scholar]
- [17].Self AJ, Paterson HF, Hall A. Different structural organization of Ras and Rho effector domains. Oncogene. 1993;8:655–661. [PubMed] [Google Scholar]
- [18].Goldfinger LE, Ptak C, Jeffery ED, Shabanowitz J, Han J, Haling JR, Sherman NE, Fox JW, Hunt DF, Ginsberg MH. An experimentally derived database of candidate Ras-interacting proteins. J.Proteome.Res. 2007;6:1806–1811. doi: 10.1021/pr060630l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lee S, Wurtzel JG, Goldfinger LE. The RLIP76 N-terminus binds ARNO to regulate PI 3-kinase, Arf6 and Rac signaling, cell spreading and migration. Biochem Biophys Res Commun. 2014;454:560–565. doi: 10.1016/j.bbrc.2014.10.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Al-Awar O, Radhakrishna H, Powell NN, Donaldson JG. Separation of membrane trafficking and actin remodeling functions of ARF6 with an effector domain mutant. Mol.Cell Biol. 2000;20:5998–6007. doi: 10.1128/mcb.20.16.5998-6007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Santy LC, Casanova JE. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. Journal of Cell Biology. 2001;154:599–610. doi: 10.1083/jcb.200104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Casanova JE. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- [23].Henis YI, Hancock JF, Prior IA. Ras acylation, compartmentalization and signaling nanoclusters (Review) Mol Membr Biol. 2009;26:80–92. doi: 10.1080/09687680802649582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wurtzel JG, Kumar P, Goldfinger LE. Palmitoylation regulates vesicular trafficking of R-Ras to membrane ruffles and effects on ruffling and cell spreading. Small GTPases. 2012;3:139–153. doi: 10.4161/sgtp.21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Takaya A, Kamio T, Masuda M, Mochizuki N, Sawa H, Sato M, Nagashima K, Mizutani A, Matsuno A, Kiyokawa E, Matsuda M. R-Ras regulates exocytosis by Rgl2/Rlf-mediated activation of RalA on endosomes. Molecular Biology of the Cell. 2007;18:1850–1860. doi: 10.1091/mbc.E06-08-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Conklin MW, Ada-Nguema A, Parsons M, Riching KM, Keely PJ. R-Ras regulates beta1-integrin trafficking via effects on membrane ruffling and endocytosis. BMC Cell Biol. 2010;11:14. doi: 10.1186/1471-2121-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kashatus DF, Lim KH, Brady DC, Pershing NL, Cox AD, Counter CM. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol. 2011;13:1108–1115. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Barbieri MA, Hoffenberg S, Roberts R, Mukhopadhyay A, Pomrehn A, Dickey BF, Stahl PD. Evidence for a symmetrical requirement for Rab5-GTP in in vitro endosome-endosome fusion. J Biol Chem. 1998;273:25850–25855. doi: 10.1074/jbc.273.40.25850. [DOI] [PubMed] [Google Scholar]
- [29].Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Blum R, Pfeiffer F, Feick P, Nastainczyk W, Kohler B, Schafer KH, Schulz I. Intracellular localization and in vivo trafficking of p24A and p23. J Cell Sci. 1999;112(Pt 4):537–548. doi: 10.1242/jcs.112.4.537. [DOI] [PubMed] [Google Scholar]
- [31].Kanamarlapudi V, Thompson A, Kelly E, Lopez Bernal A. ARF6 activated by the LHCG receptor through the cytohesin family of guanine nucleotide exchange factors mediates the receptor internalization and signaling. J Biol Chem. 2012;287:20443–20455. doi: 10.1074/jbc.M112.362087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang Q, Major MB, Takanashi S, Camp ND, Nishiya N, Peters EC, Ginsberg MH, Jian X, Randazzo PA, Schultz PG, Moon RT, Ding S. Small-molecule synergist of the Wnt/beta-catenin signaling pathway. Proc Natl Acad Sci U S A. 2007;104:7444–7448. doi: 10.1073/pnas.0702136104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhu W, London NR, Gibson CC, Davis CT, Tong Z, Sorensen LK, Shi DS, Guo J, Smith MC, Grossmann AH, Thomas KR, Li DY. Interleukin receptor activates a MYD88-ARNO-ARF6 cascade to disrupt vascular stability. Nature. 2012;492:252–255. doi: 10.1038/nature11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jilkina O, Bhullar RP. A serine kinase associates with the RAL GTPase and phosphorylates RAL-interacting protein 1. Biochim Biophys Acta. 2006;1763:948–957. doi: 10.1016/j.bbamcr.2006.07.007. [DOI] [PubMed] [Google Scholar]
- [35].Herlevsen MC, Theodorescu D. Mass spectroscopic phosphoprotein mapping of Ral binding protein 1 (RalBP1/Rip1/RLIP76) Biochem Biophys Res Commun. 2007;362:56–62. doi: 10.1016/j.bbrc.2007.07.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Herrmann C. Ras-effector interactions: after one decade. Curr.Opin.Struct.Biol. 2003;13:122–129. doi: 10.1016/s0959-440x(02)00007-6. [DOI] [PubMed] [Google Scholar]
- [37].Misaki R, Morimatsu M, Uemura T, Waguri S, Miyoshi E, Taniguchi N, Matsuda M, Taguchi T. Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. J Cell Biol. 2010;191:23–29. doi: 10.1083/jcb.200911143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell. 2007;18:2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Oh SJ, Santy LC. Differential effects of cytohesins 2 and 3 on beta1 integrin recycling. J Biol Chem. 2010;285:14610–14616. doi: 10.1074/jbc.M109.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Santy LC, Ravichandran KS, Casanova JE. The DOCK180/Elmo Complex Couples ARNO-Mediated Arf6 Activation to the Downstream Activation of Rac1. Current Biology. 2005;15:1749–1754. doi: 10.1016/j.cub.2005.08.052. [DOI] [PubMed] [Google Scholar]
- [41].Mansour M, Lee SY, Pohajdak B. The N-terminal coiled coil domain of the cytohesin/ARNO family of guanine nucleotide exchange factors interacts with the scaffolding protein CASP. J Biol Chem. 2002;277:32302–32309. doi: 10.1074/jbc.M202898200. [DOI] [PubMed] [Google Scholar]
- [42].Margutti P, Matarrese P, Conti F, Colasanti T, Delunardo F, Capozzi A, Garofalo T, Profumo E, Rigano R, Siracusano A, Alessandri C, Salvati B, Valesini G, Malorni W, Sorice M, Ortona E. Autoantibodies to the C-terminal subunit of RLIP76 induce oxidative stress and endothelial cell apoptosis in immune-mediated vascular diseases and atherosclerosis. Blood. 2008;111:4559–4570. doi: 10.1182/blood-2007-05-092825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Matsubara K, Hinoi T, Koyama S, Kikuchi A. The post-translational modifications of Ral and Rac1 are important for the action of Ral-binding protein 1, a putative effector protein of Ral. FEBS Letters. 1997;410:169–174. doi: 10.1016/s0014-5793(97)00633-9. [DOI] [PubMed] [Google Scholar]
- [44].Yadav S, Singhal SS, Singhal J, Wickramarachchi D, Knutson E, Albrecht TB, Awasthi YC, Awasthi S. Identification of membrane-anchoring domains of RLIP76 using deletion mutant analyses. Biochemistry. 2004;43:16243–16253. doi: 10.1021/bi0482811. [DOI] [PubMed] [Google Scholar]
- [45].Humphreys D, Davidson AC, Hume PJ, Makin LE, Koronakis V. Arf6 coordinates actin assembly through the WAVE complex, a mechanism usurped by Salmonella to invade host cells. Proc Natl Acad Sci U S A. 2013;110:16880–16885. doi: 10.1073/pnas.1311680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Balasubramanian N, Meier JA, Scott DW, Norambuena A, White MA, Schwartz MA. RalA-exocyst complex regulates integrin-dependent membrane raft exocytosis and growth signaling. Curr Biol. 2010;20:75–79. doi: 10.1016/j.cub.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Balasubramanian N, Scott DW, Castle JD, Casanova JE, Schwartz MA. Arf6 and microtubules in adhesion-dependent trafficking of lipid rafts. Nat Cell Biol. 2007;9:1381–1391. doi: 10.1038/ncb1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hall B, McLean MA, Davis K, Casanova JE, Sligar SG, Schwartz MA. A fluorescence resonance energy transfer activation sensor for Arf6. Anal Biochem. 2008;374:243–249. doi: 10.1016/j.ab.2007.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.