Abstract
Background and Aims
Genetic studies of alcohol dependence (AD) have identified several candidate loci and genes, but most observed effects are small and difficult to reproduce. A plausible explanation for inconsistent findings may be a violation of the assumption that genetic factors contributing to each of the seven DSM-IV criteria point to a single underlying dimension of risk. Given that recent twin studies suggest that the genetic architecture of AD is complex and likely involves multiple discrete genetic factors, the current study employed common single nucleotide polymorphisms in two multivariate genetic models to examine the assumption that the genetic risk underlying DSM-IV AD is unitary.
Design, setting, participants, measurements
AD symptoms and genome-wide SNP data from 2596 individuals of European descent from the Study of Addiction: Genetics and Environment were analyzed using Genomic-relatedness-matrix restricted maximum likelihood. DSM-IV AD symptom covariance was described using two multivariate genetic factor models.
Findings
Common SNPs explained 30% (s.e.=0.136, p=0.012) of the variance in AD diagnosis. Additive genetic effects varied across AD symptoms. The Common Pathway Model approach suggested that symptoms could be described by a single latent variable that had a SNP-heritability of 31% (0.130, p=0.008). Likewise, the Exploratory Genetic Factor Model approach suggested that the genetic variance/covariance across symptoms could be represented by a single genetic factor that accounted for at least 60% of the genetic variance in any one symptom.
Conclusion
Additive genetic effects on DSM-IV alcohol dependence criteria overlap. The assumption of common genetic effects across alcohol dependence symptoms appears to be a valid assumption.
Keywords: Alcoholism, Genetics, Alcohol Dependence, GCTA, Diagnostic criteria, DSM-IV
INTRODUCTION
Alcohol dependence (AD) is a multifactorial disease defined by uncontrolled drinking and multiple physiological and psychological problems. Based on the Diagnostic and Statistical Manual of Mental Disorders (Version IV)(1), AD is characterized by seven symptoms which include, tolerance, withdrawal, and using alcohol in larger amounts or for longer periods than intended, to name a few. DSM-IV symptoms are hypothesized to index vulnerability in biological systems that influence AD. Consequently, DSM criteria are used in genetic research and are now complemented by other measures that indicate other aspects of problematic drinking (e.g., factor scores based on drinking behavior in the past year)(2).
To date, a number of genome-wide studies (GWAS) have identified genetic variants associated with AD(3–21). Studies suggest that associated variants are of small effect(17) and little overall heritability is explained by the sum of genome-wide significant SNPs(22, 23). Results from GWAS of AD follow a similar pattern to GWAS of other complex disorders, such as nicotine dependence(24, 25), major depression(26), and schizophrenia(27, 28). One possible cause of small effect sizes observed in GWAS of complex disorders in general and of AD in particular, is a violation of the assumption that genetic factors contributing to each indicator of DSM-IV AD point to a single underlying dimension of risk. Twin studies have demonstrated the role of additive genetic and non-shared environmental factors in the etiology of AD(29); however, studies of individual symptoms suggest varying genetic effects(30, 31). To date, a single twin study has explored the possibility of multiple genetic factors for DSM-IV AD symptoms(31), and another multivariate twin study has examined the shared variance across several alcohol related items (i.e., social and occupational problems, withdrawal, tolerance, compulsive drinking, and impairment in major life activities)(32). Using same-sex adult twins from the Virginia Adult Twin Study of Psychiatric and Substance Use Disorders (VATSPUD), Kendler et al.(31) reported that DSM-IV AD may not reflect a single dimension of genetic liability. Using a slightly different approach with twins from the Minnesota Center for Twin and Family Research, McGue et al.,(32) examined the genetic contribution to five factor-analytically derived measures of behavioral disinhibition (including an alcohol dependence composite indicated by the aforementioned indicators(33)). Their AD composite had a twin-based heritability of 70%, but the estimated contribution of genotyped SNPs was 8%. Kendler et al.’s(31) study, which highlighted up to three weakly correlated genetic factors, highlights a potentially serious problem for molecular genetic studies using outcomes derived from multiple indicators. To the extent that the genetic liability across different indicators of AD is not completely overlapping, collapsing, or averaging across symptoms minimizes the likelihood of identifying relevant quantitative trait loci for AD. More specifically, the clustering of weak-moderately correlated symptoms for a diagnostic outcome may result in imprecision of the phenotype. On the contrary, continuous indicators (e.g., factor scores based on the shared variance across items) would be less influenced by etiological differences across indicators and provide greater power to detect those mechanisms.
Based on our review of the literature, there are no multivariate candidate gene or GWAS studies that have attempted to test the assumption that DSM-IV AD symptoms are under genetic influence and have largely overlapping effects. In this report, we investigated the polygenic nature of AD using common genome-wide SNPs to quantify additive genetic effects on DSM-IV AD diagnosis and AD symptoms. Further, we investigated the extent to which the covariation between DSM-IV AD indicators could be accounted for by one or more independent genetic factors or, more stringently, the extent to which a latent variable (i.e., AD factor) indicated by DSM-IV AD symptoms is determined by additive genetic and environmental effects.
METHODS AND MATERIALS
Sample
Data are from the Study of Addiction: Genetics and Environment (SAGE)(34). Analyses focused on 2596 unrelated individuals (44% male; mean age=38.58 years [standard deviation (SD)=9.80]) to account for any bias that might occur due to cryptic relatedness. European ancestry was confirmed using principal component analysis (while including HapMap control subjects) on the entire set of SAGE participants (n=4121). Subjects of European ancestry were identified and extracted and individuals more related than second cousins were removed by imposing a relatedness cut-off of 0.05(35). Additional details on SAGE are available at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1.
Assessments
SAGE collected DSM-IV symptoms (coded as present or absent) for AD using the Semi-Structured Assessment for the Genetics of Alcoholism(36, 37). Responses were limited to individuals who have been exposed to alcohol (and possibly other drugs). Analyses were limited to AD diagnosis, individual symptoms, and alcohol dependence factor scores (described below).
Genotyping
Blood samples were genotyped using the ILLUMINA Human 1M platform, which included 1,040,106 SNPs. Further details are available at: http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/document.cgi?study_id=phs000092.v1.p1&phv=22928&phd=2274&pha=&pht=116&phvf=&phdf=20&phaf=&phtf=&dssp=1&consent=&temp=1.
Quality Control
Markers with an allele frequency >1%, a call rate ≥98%, and a Hardy–Weinberg Equilibrium (HWE) p-value greater than 0.0001 were retained for analysis; 819,554 (796,125 autosomal) SNPs. Autosomal SNPs were used for all analyses and to estimate the genetic relatedness between all possible pairs of individuals using Genome Complex Trait Analysis (GCTA)(38–40).
Estimation of variance
The h2SNP of AD diagnosis and AD symptoms was determined using Genomic-relatedness-matrix restricted maximum likelihood (GREML), which was implemented in GCTA. Specifically, we used genetic relationship matrices (GRMs) to predict phenotypic similarity for each pair of individuals. This approach is analogous to quantitative genetic methods for estimating heritability, such as the twin method. SNP-heritability estimates were transformed on the liability scale to account for the fact that the proportion of dependent cases in SAGE is larger than the prevalence in the general population. Likewise, the proportion of subjects (i.e., cases/controls) endorsing AD symptoms is larger than the prevalence in the general population. Transformation of h2SNP was achieved by using population levels of AD and AD symptoms based on data from the National Epidemiological Survey on Alcohol and Related Conditions (NESARC)(41). All estimates were derived while controlling for the following covariates: gender, age, study of origin, and the first five ancestral principal components (APCs; derived using GCTA).
Estimation of the covariance explained by SNPs
We used two multivariate genetic approaches to determine the extent to which the same genes contribute to the phenotypic correlation between AD symptoms. The first approach, which is analogous to the Common Pathway Model (CPM)(42, 43), tests for additive genetic influences on a latent variable based on the phenotypic covariation between symptoms. The second approach, referred to as Exploratory Genetic Factor Analysis (EGFA), uses common factor analysis to identify the number of latent genetic variables that can capture the genetic variance/covariance between symptoms. It should be noted that, similar to exploratory factor models, EGFA makes no assumption about factor structure. As such, we present it as multivariate extension of GCTA. On the contrary, the CPM assumes that the covariation between symptoms is a consequence of their relationship with the latent variable.
CPM Approach
We used the same procedures as in Palmer et al., (2015) to identify phenotypic factor(s) for AD symptoms and to estimate the additive genetic variance of the factor(s). Briefly, exploratory and confirmatory factor analyses (EFA and CFA, respectively) were fitted in MPlus and the identified factor scores were extracted and analyzed using the same univariate GCTA steps described above.
EGFA Approach
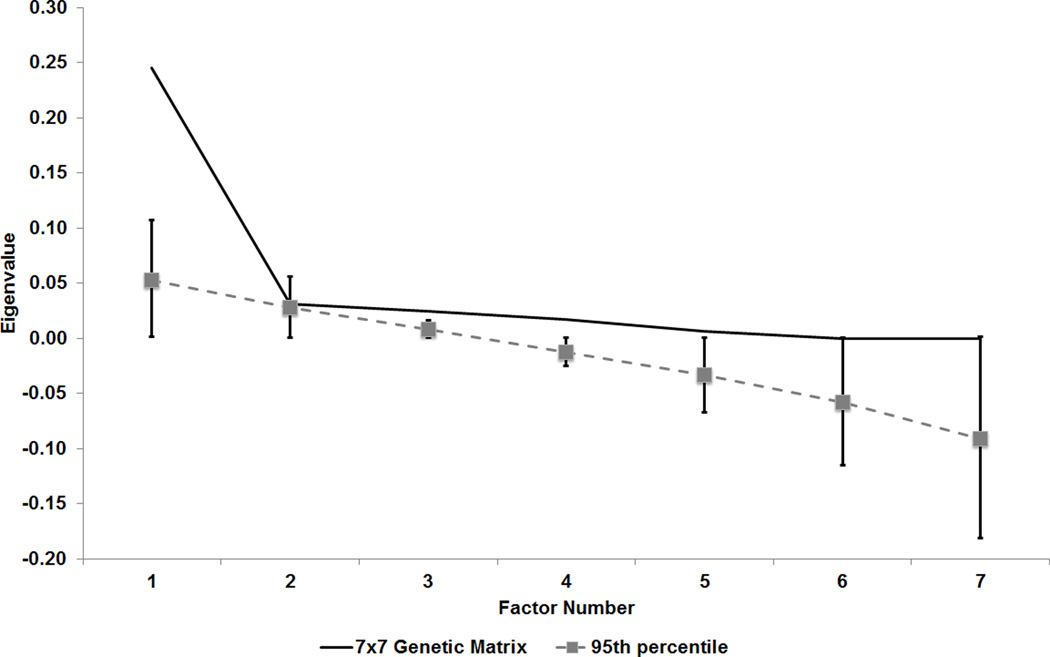
Given GCTA’s [version 1.24] inability to model more than two outcome variables, we devised a multi-stage approach to examine the additive genetic covariance across the seven symptoms simultaneously. We also report the standardized covariance (rG-SNP), which represents the additive-genetic covariance between traits standardized by the geometric mean of the individual trait genetic variances. First, GCTA was used to estimate the extent to which phenotypic covariance could be explained by genetic variance (while controlling for the aforementioned covariates). Second, the identified bivariate SNP-genetic covariance estimates were used to construct a 7 ×7 genetic variance/covariance matrix. Third, given that k-by-k covariance matrices from bivariate estimates are not guaranteed to be positive definite, we first computed eigenvalues for confirmation and, if necessary, determined the nearest positive definite variance/covariance matrix from the approximated matrix using the algorithm of Higham(44) (see nearPD in R [v3.02](45)). Finally, we used factor analysis in R to examine the multivariate genetic relationship between AD symptoms. The number of genetic factors to be retained was determined using the Monte-Carlo-based approach, Parallel Analysis (implemented in R using the package nFactors, repeated 1000 times), which has been shown to perform better than other factor retention methods (e.g., Bartlett’s chi-square, Kaiser’s eigenvalue >1) in a variety of sample conditions(46). A factor was retained if the eigenvalue of the genetic variance/covariance matrix was greater than the 95th percentile of the eigenvalues of the “parallel factor” (i.e., factor of the same rank determined from the randomly generated data)(47, 48).
RESULTS
Symptom levels & phenotypic covariance in SAGE
Approximately 45% (n=1185) of subjects met diagnostic criteria for DSM-IV AD. Table 1 presents the prevalence of endorsement of each AD symptom, stratified by individuals with and without a diagnosis of AD and across the total sample. As expected, the prevalence of symptoms in SAGE exceeded the levels observed in NESARC. Across the entire sample, symptoms frequently co-occurred. Phenotypic tetrachoric correlations ranged from 0.744 to 0.890.
Table 1.
Prevalence and associations among DSM-IV alcohol dependence (AD) symptoms
| Prevalence % (N) | Tetrachoric Correlations | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Symptom | without AD | with AD | Total Sample |
Sx 1 | Sx 2 | Sx 3 | Sx 4 | Sx 5 | Sx 6 | Sx 7 |
| Sx 1: Tolerance | 15.66 (221) | 88.01 (1042) | 48.67 (1263) | 1.000 | ||||||
| Sx 2: Withdrawal | 0.21 (3) | 51.73 (613) | 23.74 (616) | 0.764 | 1.000 | |||||
| Sx 3: Using longer than intended | 32.95 (464) | 95.77 (1133) | 61.64 (1597) | 0.754 | 0.744 | 1.000 | ||||
| Sx 4: Unsuccessful attempts to cut down | 8.79 (124) | 83.21 (986) | 42.76 (1110) | 0.759 | 0.825 | 0.808 | 1.000 | |||
| Sx 5: Great time spent using/recovering | 0.85 (12) | 58.40 (692) | 27.12 (704) | 0.765 | 0.842 | 0.768 | 0.785 | 1.000 | ||
| Sx 6: Social/Occupation activities foregone | 0.14 (2) | 58.28 (690) | 26.67 (692) | 0.775 | 0.854 | 0.763 | 0.836 | 0.890 | 1.000 | |
| Sx 7: Continued use despite problems | 13.69 (193) | 89.11 (1056) | 48.13 (1249) | 0.772 | 0.844 | 0.809 | 0.797 | 0.812 | 0.853 | 1.000 |
Note: Table presents level of alcohol dependence (AD) symptom endorsement among alcohol users in SAGE stratified by individuals with and without an alcohol dependence diagnosis and for the total sample. The table also shows the tetrachoric correlations across AD symptoms.
Estimation of variance explained by the SNPs
Common SNPs explained 30% (s.e.=0.136) of the variation in AD diagnosis. We found modest differences in h2SNP across the seven AD symptoms with estimates ranging from 7% to 32% (Table 2). Among the significant estimates, “Using longer than intended” had the highest h2SNP (32%) followed by “Tolerance” (24%). Notably, with the exception of “Great time spent using/recovering”, common SNPs captured more than one-third of the additive genetic effects observed using the twin-study approach.
Table 2.
Univariate SNP heritability (h2SNP) and standard errors (s.e.)
| Phenotype | Population prevalence* |
Current Study | Kendler Study** | |
|---|---|---|---|---|
| h2SNP | s.e. | Twin heritability | ||
| Alcohol Dependence Diagnosis | 0.08 | 0.300a | 0.136 | N/A |
| DSM-IV Alcohol Dependence Symptoms | ||||
| Tolerance | 0.08 | 0.242a | 0.129 | 0.440 |
| Withdrawal | 0.08 | 0.281 | 0.174 | 0.490 |
| Using longer than intended | 0.13 | 0.324a | 0.158 | 0.380 |
| Unsuccessful attempts to cut down | 0.12 | 0.197 | 0.146 | 0.360 |
| Great time spent using/recovering | 0.02 | 0.072 | 0.104 | 0.590 |
| Social/Occupation activities foregone | 0.01 | 0.199a | 0.091 | 0.530 |
| Continued use despite problems | 0.05 | 0.237a | 0.109 | 0.450 |
Table describes proportion of phenotypic variance explained by all autosomal SNPs (h2SNP) while controlling for all covariates.
Notations - a = p-value < 0.05.
- Disease prevalence obtained from NESARC (Saha et al., 2006) was used to transform the estimate of the variance explained on the observed scale to that of the underlying scale.
- Heritability estimates derived from Kendler et al. 2012. N/A – Not applicable.
Due to differences in length between chromosomes, we also examined whether the proportion of variance attributable to individual chromosomes was related to chromosomal length (see Supplemental Table S1; Supplemental Figure S1). There was no indication that larger chromosomes accounted for more phenotypic variation than smaller chromosomes. Further, no single chromosome emerged as a disproportionate source of variance across all AD symptoms although large standard errors preclude strong conclusions. We also conducted an independent test for the effects of stratification across chromosomes by examining the total variance explained by considering 22 GRMs (one per chromosome) simultaneously versus the sum of the genetic variance (VG) from 22 models fitted on each chromosome, such that any difference in magnitude of VG across the two approaches is attributable to the fact that the model that estimated the effects across all chromosomes simultaneously accounts for the correlation between markers across chromosomes. In many instances, the sum of VG from the 22 models fitted on each chromosome separately (Supplemental Table S1 row – “Total (ignoring shared effects)” was slightly greater than the total variance explained when fitting a model across all chromosomes simultaneously (Total) (mean difference between estimates across all phenotypes=0.135 [SD=0.039]), suggesting the possibility of shared effects across chromosomes on AD and AD symptoms. Such shared effects across chromosomes could be due to sampling error, but could also denote assortative mating, which creates LD between physically unlinked causal variants, or residual population stratification not removed by controlling for ethnic principal components.
Analysis of the genetic covariance across DSM-IV AD symptoms
CPM Approach
EFA and CFA of the phenotypic covariance across DSM items revealed a latent variable (referred to as AD factor) with loadings in excess of 0.84 (Table 3). Overall, the CPM model described the phenotypic relationships between the symptoms very well, as all models resulted in a Root Mean Square Error of Approximation value less than 0.05(49). Likewise, the Comparative Fit Index and the Tucker Lewis indices were almost one, indicating reasonably good fit(50). The total h2SNP of the AD factor (h2SNP=0.307 (0.130), p=0.008) and the contribution of individual chromosomes to the AD factor were similar to those observed for AD diagnosis (Supplemental Table S1).
Table 3.
CPM Approach: Exploratory and confirmatory factor models of alcohol dependence symptoms
| Exploratory Factor Analysis | Confirmatory Factor Analysis | ||
|---|---|---|---|
| Parameters | Sample 1- EFA |
Sample 2- CFA |
Full sample |
| Fit statistics | |||
| χ2 | 39.149a | 39.392a | 64.380a |
| df | 8 | 14 | 14 |
| RMSEA | 0.037 | 0.037 | 0.037 |
| CFI | 0.999 | 0.999 | 0.999 |
| TLI | 0.998 | 0.998 | 0.998 |
| Factor loadings | |||
| Sx 1: Tolerance | 0.832a | 0.858a | 0.844a |
| Sx 2: Withdrawal | 0.904a | 0.923a | 0.913a |
| Sx 3: Using longer than intended | 0.867a | 0.869a | 0.868a |
| Sx 4: Unsuccessful attempts to cut down | 0.882a | 0.903a | 0.892a |
| Sx 5: Great time spent using/recovering | 0.926a | 0.904a | 0.915a |
| Sx 6: Social/Occupation activities foregone | 0.944a | 0.938a | 0.941a |
| Sx 7: Continued use despite problems | 0.917a | 0.904a | 0.910a |
Abbreviations: CFI - Comparative fit index, TLI - Tucker Lewis index, RMSEA - Root mean square error of approximation.
Notations: a - p-value < 0.05.
Note that samples 1 and 2 are random halves of the total SAGE sample and do not overlap.
EGFA Approach
The pattern of SNP-correlations between several individual symptoms of AD (Table 4) suggested shared and unique genetic influences across AD symptoms. For example, the bivariate analyses indicated a strong (rG-SNP>0.60) genetic correlation between “tolerance” and “use longer than intended” (rG-SNP=0.609 (s.e.=0.259)). Analysis of the genetic covariance across AD symptoms indicated a single common genetic factor. The nearest positive definite genetic variance/covariance matrix used for the analysis was very similar to the matrix obtained from the observed variance/covariance estimates (i.e., the mean of the differences observed between all of the cells of the approximated and nearest-positive-definite genetic covariance matrix was 1.29E-3 [SD=1.27E-3]). Parallel analysis indicated that only the first eigenvalue from the 7×7 genetic variance/covariance matrix exceeded the 95th percentile of the randomly generated data (Figure 1). This indicated that only the first factor should be retained. The common genetic factor accounted for more than a third of the genetic effects on any one symptom (Table 5). Given the high overlap between “unsuccessful attempts to cut down” and other items, we also conducted a post-hoc analysis using a covariance matrix that excluded this symptom; these analyses also suggested a common genetic factor (Supplemental Figure S2).
Table 4.
EGFA Approach: Inter-item SNP-Correlations (SE)
| DSM-IV Symptom of Alcohol Dependence | Sx 1 | Sx 2 | Sx 3 | Sx 4 | Sx 5 | Sx 6 | Sx 7 |
|---|---|---|---|---|---|---|---|
| Sx 1: Tolerance | 1.000 | ||||||
| Sx 2: Withdrawal | 0.437 (0.330) | 1.000 | |||||
| Sx 3: Using longer than intended | 0.609 (0.259) | 0.637 (0.315) | 1.000 | ||||
| Sx 4: Unsuccessful attempts to cut down | 0.995 (0.301)a | 0.941 (0.328)a | 1.000 (0.360) | 1.000 | |||
| Sx 5: Great time spent using/recovering | 0.473 (0.485) | 0.657 (0.437) | 0.917 (0.604) | 0.873 (0.527) | 1.000 | ||
| Sx 6: Social/Occupation activities foregone | 0.530 (0.269) | 0.872 (0.217)a | 0.655 (0.265)a | 0.977 (0.289)a | 0.479 (0.393) | 1.000 | |
| Sx 7: Continued use despite problems | 0.744 (0.223)a | 0.619 (0.266) | 0.760 (0.214)a | 0.858 (0.256)a | 0.944 (0.109)a | 0.816 (0.196)a | 1.000 |
Table displays SNP correlation estimates between DSM-IV symptoms with adjustment for covariates.
Notations - a = p-value < 0.05.
Figure 1.
Table 5.
GFA Approach: Genetic variance in each DSM-IV symptom explained by common genetic factor
| Factor Loadings | % Total Genetic Variance Explained |
|
|---|---|---|
| Tolerance | 0.846 | 0.716 |
| Withdrawal | 0.816 | 0.666 |
| Using longer than intended | 0.838 | 0.702 |
| Unsuccessful attempts to cut down | 0.998 | 0.996 |
| Great time spent using/recovering | 0.692 | 0.479 |
| Social/Occupation activities foregone | 0.829 | 0.687 |
| Continued use despite problems | 0.824 | 0.679 |
Table shows the loadings and communality estimates based on the first genetic factor identified using the genetic variance/covariance matrix.
DISCUSSION
This is the first study to examine the genetic covariance of DSM-IV indicators of AD using common genome-wide SNPs. We estimated the genetic variation in AD symptoms as well as the degree of genetic overlap between symptoms. In addition, we estimate the genetic contribution to AD diagnosis and an AD factor. We have shown that common SNPs account for approximately one half (h2SNP=0.30) of the genetic variance of AD observed in twin and adoption studies (twin-based h2=50–70%(29)), with similar magnitudes of effect when using the AD factor. Further, the genetic contribution to the observed factor score coupled with the ability to reduce the genetic variance/covariance matrix into a single factor suggests common mechanisms acting on individual symptoms of DSM-IV AD.
The findings from this study are of particular importance to understanding the complex nature of the genetic liability to AD. This study demonstrates that common variants capture a significant proportion of the genetic liability to AD diagnosis, as well as specific aspects of AD indicated by DSM-IV symptoms, and the shared variance across DSM-IV symptoms. The h2SNP estimates of both AD diagnosis and the AD factor score were similar to the recent report on alcohol abuse/dependence by Vrieze and colleagues(23), but were greater than the effects observed in a recent report by McGue and colleagues(32), possibly due to either sample differences or differences in phenotype scoring. The h2SNP of AD symptoms was at least half of the total heritability observed in a recent multivariate twin study using Caucasian twins from VATSPUD(31). In that study, Kendler and colleagues estimated the additive genetic effect of ‘Withdrawal’ at 0.49. In the current study, the h2SNP of “Withdrawal” was marginally significant at 0.28. One possible explanation for the difference between this study and Kendler et al.’s(31) is that around half of “causal variants” underlying Withdrawal are in poor LD with common SNPs captured by the Illumina 1M array (e.g., because they are rare). It is also possible that twin-based heritability estimates are inflated by the joint effects of non-additive and common environmental effects(51).
Contrary to the multivariate genetic analysis by Kendler et al.(31), findings from the CPM and EGFA approaches show greater overlap of genetic effects on AD symptoms. Kendler et al.’s best-fitting model consisted of three underlying dimensions of genetic risk and no symptom-specific genetic factors. Our findings were not consistent with the “Loss of control/social dysfunction”, “Tolerance”, and “Withdrawal and Continued Use” factors observed by Kendler and colleagues. For reasons similar to those explained above, it is not necessarily surprising that the genetic factor structures estimated by these two methods differ. As with univariate heritability estimates, genetic correlations estimated from twins can include non-additive and common environmental effects; rG-SNP from GCTA is minimally influenced by these factors. Furthermore, it is possible that rare causal variants, whose effects are not well estimated by GCTA but are included in twin estimates, are more symptom specific. Finally, the differences between these two multivariate studies might also arise from ascertainment differences. Unlike SAGE, which is a case-control study made up of alcoholic probands recruited from treatment facilities, the VATSPUD is a population-based study of common psychiatric disorders. Consequently, the AD symptomology and the covariance among symptoms may differ between the samples because of differences in (1) presentation of AD symptoms, and (2) inclusion/exclusion criteria for psychopathology that are often comorbid with AD and reflect vulnerability to different AD symptoms. For example, research has shown that adults with a diagnosis of ADHD are more sensitive to the disinhibiting effects of alcohol(52), suggesting that innate differences in inhibitory mechanisms and possibly attentional process are important to physiological responses to alcohol. To the extent that psychiatric disorders that predicate/moderate alcohol involvement(53, 54) are important risk indicators, differences in ascertainment between studies could affect the covariance among symptoms. Overall, while the results from this selected sample of cases and controls may not generalize to differently ascertained subjects, the evidence of a common genetic architecture across DSM-IV alcohol symptoms provides support for larger mega-case-control GWAS studies of AD diagnosis.
Implications
These findings and the approach herein are of potential value to future genetic studies of AD, especially as the field of psychiatric genetics shifts toward more dimensional conceptualizations of disorders. A growing body of multivariate typological and developmental research on alcohol use disorders suggests that individuals are differentially at risk for high-risk alcohol use patterns and AD(55). For example, an early attempt to identify homogenous alcoholic groups of individuals by Bucholz et al.(56) revealed four distinct classes made up of non-problem drinkers, mild alcoholics (exhibiting tolerance to alcohol, blackouts, and a persistent desire to quit), moderate alcoholics (exhibiting health and social/occupational alcohol-related problems), and severely affected alcoholics (exhibiting alcohol withdrawal, craving, health and occupation alcohol-related problems, and an inability to quit). Notably, (1) individuals with a DSM-IV AD diagnosis were more likely (>90%) to be from the latter two classes, and (2) with the exception of a diagnosis of major depression, more severe classes of alcohol problems were more likely to have a history of other psychiatric illnesses. Phenotyping (i.e., how we define normal drinking versus problem drinking) remains an important aspect of all genetic research, especially in population-based case-control studies as the model assumes phenotypic homogeneity, as well as genetic homogeneity within groups classified as cases and controls. These assumptions also hold true for dimensional measures based on multiple alcohol indices, including symptom counts, factor scores based on multiple indicators (e.g., DSM-IV symptoms, levels of use), or severity scores (absent, mild, moderate and severe) based on multiple indicators of addiction (as in DSM-5). Like diagnostic measures, dimensional measures reduce multiple testing but have the added benefit of treating the data in such a way that precludes the possibility of assigning the same phenotypic score to individuals with very different phenotypic characteristics (e.g., an alcohol abstainer versus a social drinker versus a social binge drinker, etc.). The presence of a common genetic factor that accounts for over 60% of the genetic variance in any one DSM-IV AD symptom suggests that the degree of genetic overlap among symptoms of AD may be less of a problem for association analyses than initially hypothesized for samples selected for AD. This is further supported by our ability to fit a model with the constraint that the overlap between symptoms purely arises from their relationship with the latent variable. It should be noted however that these data do not say that combinations of symptoms are uninformative, but it appears unlikely that such additional steps will provide additional information in genome-wide association studies on AD, in particular amongst users with a dependence diagnosis.
Strengths and Limitations
Several strengths and limitations of these analyses are worth noting. First, the current study utilized drug addiction samples that comprise SAGE (i.e., data from three separate cohorts for studying the genetics of alcohol (Collaborative Study on the Genetics of Alcoholism (COGA)), nicotine (Collaborative Study on the Genetics of Nicotine Dependence (COGEND)) and cocaine (Family Study of Cocaine Dependence (FSCD)). While we were able to maximize sample size, replication is needed using larger sample sizes. Second, the above analyses corrected the SNP-heritability estimates for ascertainment bias by utilizing prevalence rates from the NESARC survey. All univariate and bivariate GCTA analyses utilized NESARC prevalence rates to transform the estimate of variance explained by the SNPs. In doing so, we derived unbiased estimates of genetic comorbidity for this select set of substance using cases and control. However, it should be noted that large population-based studies of drug users might show different patterns of comorbidity (as observed in Kendler et al.(31)). It should also be noted that the current study does not address the possibility of measurement invariance between the individuals identified as cases and controls. The current study assumes that differences between cases and controls in symptom endorsement rates are the result of threshold differences in the underlying continuum of risk for each symptom. Since GCTA utilizes genetic resemblance among individuals who are distantly related to predict phenotypic similarity, the present study had limited power to conduct univariate and bivariate models on only the alcohol dependent cases. That said, these results may not generalize to other samples, as the samples may differ on other indicators of problematic alcohol consumption that contribute to the threshold difference between “cases” and “controls” (e.g., level of consumption, personality traits, externalizing/internalizing behaviors, and sociodemographic factors)(57). Future research incorporating non-DSM indicators of AD (e.g., level of response to ethanol) into diagnostic criteria is needed to refine the classification of risk for disease and to tease apart the risk continuum. Finally, these findings only generalize to the variance/covariance captured by common SNPs among subjects of European ancestry, and may not generalize to rare variants that may be in low LD with these SNPs or subjects of a different ancestral background.
Summary
In summary, the current study demonstrates shared additive genetic effects on AD symptoms using common variants. Moreover, there are common additive genetic factors acting upon AD symptoms. While these results are tentative, they are important because they suggest that a substantial portion of the variance in AD is captured by common SNPs. More importantly, they suggest shared genetic effects across AD symptoms that are also reflected in the shared phenotypic variance across AD symptoms. Higher-powered whole genome studies that capitalize on larger sample sizes may uncover more variants significantly associated with AD, but increased coverage of rare variants may also yield additional variants. Keeping in mind that genetics represents one of many causal factors for AD, future studies are likely to benefit from the use of AD phenotypes that reflect the shared variance across AD indicators.
Supplementary Material
SUPPORTING INFORMATION LEGENDS:
Supplemental Table S1. Word Document.
Table describing the proportion of phenotypic variation attributable to the genetic variation on individual autosomal chromosomes (i.e., chromosomes 1–22).
Supplemental Figure S1. File format – TIFF
SNP heritability (controlling for five APCs) of AD, AD Factor Score and AD symptoms across chromosomes. The relative contribution of genetic variation from each chromosome is presented for each phenotype reported in the study. With the exception of chromosome 18, variation in almost every phenotype can be attributed to genetic variation along each chromosome. Care should be taking when trying to interpret the stacked histogram bars, which do not reflect the total genetic variance. The vertical scale serves as a guide to measure the observed effect of variation relative to each dependence symptom. Actual values are presented in Supplemental Table T1.
Supplemental Figure S2. File format – TIFF.
Supplemental Figure S2. Post-hoc EGFA Approach – Parallel Analysis using 6×6 genetic covariance matrix.
Observed eigenvalues (solid line) based on the bended 6×6 genetic variance/covariance matrix are compared to 95 percentile of the eigenvalue distribution (square shapes [standard error]) derived from 1000 randomly generated datasets. Factors are retained if the observed eigenvalue exceeds the parallel analysis eigenvalue. In other words, all factors left of where the solid lines first intersect with the dashed line are retained.
Acknowledgements
Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01HG004422). SAGE is one of the genome-wide as association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract "High throughput genotyping for studying the gene tic contributions to human disease" (HHSN268200782096C). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/projects/gap/cgibin/study.cgi?study_id=phs000092.v1.p1 through dbGaP accession number phs000092.v1.p1.
Role of Funding Sources
Funding for this study was provided by AA021113 (Palmer), AA11998 (Heath), NIMH K01 MH085812 & NIMH 1R01MH100141 (Keller), and DA023134 (Knopik). NIAAA, NIMH, and NIDA had no role in the study design, analysis, or interpretation of the data, writing the manuscript, or the decision to submit the paper for publication. The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs.
Footnotes
Declaration of Interests
Conflict of Interest
All of the listed authors declare that they have no conflicts of interests.
Contributors
Author Rohan Palmer conducted literature searches and provided summaries of previous research studies. Drs. Rohan Palmer and Leslie Brick conducted the statistical analyses under the guidance of Drs. Valerie Knopik and Matthew Keller. Dr. McGeary provided clinical expertise on the phenotype and Dr. Heath provided guidance on integration of results into the larger literature. Author Rohan Palmer wrote the first draft of the manuscript and all authors contributed to and have approved the final manuscript.
References
- 1.Diagnostic and statistical manual of mental disorders. Washington, DC: American Psychiatric Association; 2000. [Google Scholar]
- 2.Agrawal A, Grant JD, Littlefield A, Waldron M, Pergadia ML, Lynskey MT, et al. Developing a quantitative measure of alcohol consumption for genomic studies on prospective cohorts. J Stud Alcohol Drugs. 2009;70:157–168. doi: 10.15288/jsad.2009.70.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biernacka JM, Geske JR, Schneekloth TD, Frye MA, Cunningham JM, Choi DS, et al. Replication of genome wide association studies of alcohol dependence: support for association with variation in ADH1C. PLoS One. 8:e58798. doi: 10.1371/journal.pone.0058798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panhuysen CI, Kranzler HR, Yu Y, Weiss RD, Brady K, Poling J, et al. Confirmation and Generalization of an Alcohol-Dependence Locus on Chromosome 10q. Neuropsychopharmacology. 2010;35:1325–1332. doi: 10.1038/npp.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Treutlein J, Rietschel M. Genome-wide association studies of alcohol dependence and substance use disorders. Curr Psychiatry Rep. 2011;13:147–155. doi: 10.1007/s11920-011-0176-4. [DOI] [PubMed] [Google Scholar]
- 6.Zuo L, Zhang XY, Wang F, Li CS, Lu L, Ye L, et al. Genome-wide significant association signals in IPO11-HTR1A region specific for alcohol and nicotine codependence. Alcohol Clin Exp Res. 2013;37:730–739. doi: 10.1111/acer.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang JC, Foroud T, Hinrichs AL, Le NX, Bertelsen S, Budde JP, et al. A genome-wide association study of alcohol-dependence symptom counts in extended pedigrees identifies C15orf53. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin P, Hartz SM, Wang JC, Agrawal A, Zhang TX, McKenna N, et al. Copy number variations in 6q14.1 and 5q13.2 are associated with alcohol dependence. Alcohol Clin Exp Res. 2012;36:1512–1518. doi: 10.1111/j.1530-0277.2012.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuo L, Zhang F, Zhang H, Zhang XY, Wang F, Li CS, et al. Genome-wide search for replicable risk gene regions in alcohol and nicotine co-dependence. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:437–444. doi: 10.1002/ajmg.b.32047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agrawal A, Wetherill L, Bucholz KK, Kramer J, Kuperman S, Lynskey MT, et al. Genetic influences on craving for alcohol. Addict Behav. 2013;38:1501–1508. doi: 10.1016/j.addbeh.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang KS, Liu X, Zhang Q, Wu LY, Zeng M. Genome-wide association study identifies 5q21 and 9p24.1 (KDM4C) loci associated with alcohol withdrawal symptoms. J Neural Transm. 2012;119:425–433. doi: 10.1007/s00702-011-0729-z. [DOI] [PubMed] [Google Scholar]
- 12.Edwards AC, Aliev F, Bierut LJ, Bucholz KK, Edenberg H, Hesselbrock V, et al. Genome-wide association study of comorbid depressive syndrome and alcohol dependence. Psychiatr Genet. 2012;22:31–41. doi: 10.1097/YPG.0b013e32834acd07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Contini V, Bertuzzi GP, Polina ER, Hunemeier T, Hendler EM, Hutz MH, et al. A haplotype analysis is consistent with the role of functional HTR1B variants in alcohol dependence. Drug Alcohol Depend. 2012;122:100–104. doi: 10.1016/j.drugalcdep.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 14.Frank J, Cichon S, Treutlein J, Ridinger M, Mattheisen M, Hoffmann P, et al. Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addict Biol. 2012;17:171–180. doi: 10.1111/j.1369-1600.2011.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lydall GJ, Bass NJ, McQuillin A, Lawrence J, Anjorin A, Kandaswamy R, et al. Confirmation of prior evidence of genetic susceptibility to alcoholism in a genome-wide association study of comorbid alcoholism and bipolar disorder. Psychiatr Genet. 2011;21:294–306. doi: 10.1097/YPG.0b013e32834915c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bae JS, Jung MH, Lee BC, Cheong HS, Park BL, Kim LH, et al. The genetic effect of copy number variations on the risk of alcoholism in a Korean population. Alcohol Clin Exp Res. 2012;36:35–42. doi: 10.1111/j.1530-0277.2011.01578.x. [DOI] [PubMed] [Google Scholar]
- 17.Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, et al. A Quantitative-Trait Genome-Wide Association Study of Alcoholism Risk in the Community: Findings and Implications. Biological Psychiatry. 2011;70:513–518. doi: 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zlojutro M, Manz N, Rangaswamy M, Xuei X, Flury-Wetherill L, Koller D, et al. Genome-wide association study of theta band event-related oscillations identifies serotonin receptor gene HTR7 influencing risk of alcohol dependence. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2011;156:44–58. doi: 10.1002/ajmg.b.31136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, et al. A genome-wide association study of alcohol dependence. Proceedings of the National Academy of Sciences. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, et al. Genome-Wide Association Study of Alcohol Dependence Implicates a Region on Chromosome 11. Alcoholism: Clinical and Experimental Research. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lind PA, Macgregor S, Vink JM, Pergadia ML, Hansell NK, de Moor MHM, et al. A Genomewide Association Study of Nicotine and Alcohol Dependence in Australian and Dutch Populations. Twin Research and Human Genetics. 2010;13:10–29. doi: 10.1375/twin.13.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vrieze SI, Feng S, Miller MB, Hicks BM, Pankratz N, Abecasis GR, et al. Rare Nonsynonymous Exonic Variants in Addiction and Behavioral Disinhibition. Biological psychiatry. 2013 doi: 10.1016/j.biopsych.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vrieze SI, McGue M, Miller MB, Hicks BM, Iacono WG. Three mutually informative ways to understand the genetic relationships among behavioral disinhibition, alcohol use, drug use, nicotine use/dependence, and their co-occurrence: twin biometry, GCTA, and genome-wide scoring. Behav Genet. 2013;43:97–107. doi: 10.1007/s10519-013-9584-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang KS, Liu X, Zhang Q, Zeng M. ANAPC1 and SLCO3A1 are associated with nicotine dependence: meta-analysis of genome-wide association studies. Drug and alcohol dependence. 2012;124:325–332. doi: 10.1016/j.drugalcdep.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Loukola A, Wedenoja J, Keskitalo-Vuokko K, Broms U, Korhonen T, Ripatti S, et al. Genome-wide association study on detailed profiles of smoking behavior and nicotine dependence in a twin sample. Molecular psychiatry. 2013 doi: 10.1038/mp.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Molecular psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nature genetics. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nature genetics. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agrawal A, Lynskey MT. Are there genetic influences on addiction: evidence from family, adoption and twin studies. Addiction. 2008;103:1069–1081. doi: 10.1111/j.1360-0443.2008.02213.x. [DOI] [PubMed] [Google Scholar]
- 30.Slutske WS, True WR, Scherrer JF, Heath AC, Bucholz KK, Eisen SA, et al. The heritability of alcoholism symptoms: "indicators of genetic and environmental influence in alcohol-dependent individuals" revisited. Alcohol Clin Exp Res. 1999;23:759–769. doi: 10.1111/j.1530-0277.1999.tb04181.x. [DOI] [PubMed] [Google Scholar]
- 31.Kendler KS, Aggen SH, Prescott CA, Crabbe J, Neale MC. Evidence for multiple genetic factors underlying the DSM-IV criteria for alcohol dependence. Molecular psychiatry. 2012;17:1306–1315. doi: 10.1038/mp.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGue M, Zhang Y, Miller MB, Basu S, Vrieze S, Hicks B, et al. A genome-wide association study of behavioral disinhibition. Behav Genet. 2013;43:363–373. doi: 10.1007/s10519-013-9606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hicks BM, Schalet BD, Malone SM, Iacono WG, McGue M. Psychometric and genetic architecture of substance use disorder and behavioral disinhibition measures for gene association studies. Behav Genet. 2011;41:459–475. doi: 10.1007/s10519-010-9417-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, et al. A genome-wide association study of alcohol dependence. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer RH, Brick L, Nugent NR, Bidwell LC, McGeary JE, Knopik VS, et al. Examining the role of common genetic variants on alcohol, tobacco, cannabis and illicit drug dependence: genetics of vulnerability to drug dependence. Addiction. 2015;110:530–537. doi: 10.1111/add.12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- 37.Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--a comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. American journal of human genetics. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saha TD, Chou SP, Grant BF. Toward an alcohol use disorder continuum using item response theory: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Psychol Med. 2006;36:931–941. doi: 10.1017/S003329170600746X. [DOI] [PubMed] [Google Scholar]
- 42.Palmer RH, Button TM, Rhee SH, Corley RP, Young SE, Stallings MC, et al. Genetic etiology of the common liability to drug dependence: evidence of common and specific mechanisms for DSM-IV dependence symptoms. Drug Alcohol Depend. 2012;123(Suppl 1):S24–S32. doi: 10.1016/j.drugalcdep.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neale MC, LR C. Methodology for Genetic Studies of Twins and Families. Dordrecht, the Netherlands: Kluwer Academic Publishers; 1992. [Google Scholar]
- 44.Higham N. Computing the nearest correlation matrix - a problem from finance. IMA Journal of Numerical Analysis 22. 2002;22:329–343. [Google Scholar]
- 45.Statistical R. C. T. R. A. l. a. e. f. s. c. R. F. f. Austria: Computing V.; URL http://www.R-project.org/. [Google Scholar]
- 46.Zwick WR, Velicer WF. Comparison of Five Rules for Determine the Number of Components to Retain. Psychological Bulletin. 1986;99:432–442. [Google Scholar]
- 47.Hayton JC, Allen DG, Scarpello V. Factor retention decisions in exploratory factor analysis: A tutorial on parrallel analysis. Organizational Research Methods. 2004;7:191–205. [Google Scholar]
- 48.Glorfeld LW. An improvement on Horn’s parallel analysis methodology for selecting the correct number of factors to retain. Educational and Psychological Measurement. 1995;55:377–393. [Google Scholar]
- 49.MacCallum RC, Browne MW, Sugawara HM. Power analysis and determination of sample size for covariance structure modeling. Psychological Methods. 1996;1:130–149. [Google Scholar]
- 50.Hu L, Bentler PM. Cutoff criteria for fit indexes in covariance structure analysis: Conventional criteria versus new alternatives. Structural Equation Modeling. 1999;6:1–55. [Google Scholar]
- 51.Coventry WL, Keller MC. Estimating the extent of parameter bias in the classical twin design: a comparison of parameter estimates from extended twin-family and classical twin designs. Twin Res Hum Genet. 2005;8:214–223. doi: 10.1375/1832427054253121. [DOI] [PubMed] [Google Scholar]
- 52.Weafer J, Fillmore MT, Milich R. Increased sensitivity to the disinhibiting effects of alcohol in adults with ADHD. Exp Clin Psychopharmacol. 2009;17:113–121. doi: 10.1037/a0015418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palmer RH, Knopik VS, Rhee SH, Hopfer CJ, Corley RC, Young SE, et al. Prospective effects of adolescent indicators of behavioral disinhibition on DSM-IV alcohol, tobacco, and illicit drug dependence in young adulthood. Addict Behav. 2013;38:2415–2421. doi: 10.1016/j.addbeh.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grucza RA, Robert Cloninger C, Bucholz KK, Constantino JN, Schuckit MI, Dick DM, et al. Novelty seeking as a moderator of familial risk for alcohol dependence. Alcoholism, clinical and experimental research. 2006;30:1176–1183. doi: 10.1111/j.1530-0277.2006.00133.x. [DOI] [PubMed] [Google Scholar]
- 55.Saunders JB, Schuckit MA, Sirovatka PJ, Regier DA. Diagnostic Issues in Substance Use Disorders: Refining the Research Agenda for DSM-V. American Psychiatric Publishing. 2007:352. [Google Scholar]
- 56.Bucholz KK, Heath AC, Reich T, Hesselbrock VM, Kramer JR, Nurnberger JI, Jr, et al. Can we subtype alcoholism? A latent class analysis of data from relatives of alcoholics in a multicenter family study of alcoholism. Alcoholism, clinical and experimental research. 1996;20:1462–1471. doi: 10.1111/j.1530-0277.1996.tb01150.x. [DOI] [PubMed] [Google Scholar]
- 57.Maggs JL, Patrick ME, Feinstein L. Childhood and adolescent predictors of alcohol use and problems in adolescence and adulthood in the National Child Development Study. Addiction. 2008;103(Suppl 1):7–22. doi: 10.1111/j.1360-0443.2008.02173.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORTING INFORMATION LEGENDS:
Supplemental Table S1. Word Document.
Table describing the proportion of phenotypic variation attributable to the genetic variation on individual autosomal chromosomes (i.e., chromosomes 1–22).
Supplemental Figure S1. File format – TIFF
SNP heritability (controlling for five APCs) of AD, AD Factor Score and AD symptoms across chromosomes. The relative contribution of genetic variation from each chromosome is presented for each phenotype reported in the study. With the exception of chromosome 18, variation in almost every phenotype can be attributed to genetic variation along each chromosome. Care should be taking when trying to interpret the stacked histogram bars, which do not reflect the total genetic variance. The vertical scale serves as a guide to measure the observed effect of variation relative to each dependence symptom. Actual values are presented in Supplemental Table T1.
Supplemental Figure S2. File format – TIFF.
Supplemental Figure S2. Post-hoc EGFA Approach – Parallel Analysis using 6×6 genetic covariance matrix.
Observed eigenvalues (solid line) based on the bended 6×6 genetic variance/covariance matrix are compared to 95 percentile of the eigenvalue distribution (square shapes [standard error]) derived from 1000 randomly generated datasets. Factors are retained if the observed eigenvalue exceeds the parallel analysis eigenvalue. In other words, all factors left of where the solid lines first intersect with the dashed line are retained.