Abstract
Intramuscular administration of plasmid DNA (pDNA) with non-ionic Pluronic block copolymers increases gene expression in injected muscles and lymphoid organs. We studied the role of immune cells in muscle transfection upon inflammation. Local inflammation in murine hind limb ischemia model (MHLIM) drastically increased DNA, RNA and expressed protein levels in ischemic muscles injected with pDNA/Pluronic. The systemic inflammation (MHLIM or peritonitis) also increased expression of pDNA/Pluronic in the muscles. When pDNA/Pluronic was injected in ischemic muscles the reporter gene, Green Fluorescent Protein (GFP) co-localized with desmin+ muscle fibers and CD11b+ macrophages (MØs), suggesting transfection of MØs along with the muscle cells. P85 enhanced (~4 orders) transfection of MØs with pDNA in vitro. Moreover, adoptively transferred MØs were shown to pass the transgene to inflamed muscle cells in MHLIM. Using a co-culture of myotubes (MTs) and transfected MØs expressing a reporter gene under constitutive (cmv-luciferase) or muscle specific (desmin-luciferase) promoter we demonstrated that P85 enhances horizontal gene transfer from MØ to MTs. Therefore, MØs can play an important role in muscle transfection with pDNA/Pluronic during inflammation, with both inflammation and Pluronic contributing to the increased gene expression. pDNA/Pluronic has potential for therapeutic gene delivery in muscle pathologies that involve inflammation.
Keywords: Inflammation, gene delivery, ischemia, macrophage, plasmid DNA, Pluronic block copolymer, skeletal muscle transfection
Introduction
Direct intramuscular (i.m.) injection of naked plasmid DNA (pDNA) for skeletal muscle transfection was pioneered by J.A. Wolff in 1990 (1). As of today i.m. injection of pDNA makes up around 18% of worldwide gene therapy human clinical trials (2). These trials are related to DNA vaccines, Duchene muscular dystrophy, hind limb ischemia and cardiac ischemia (3–5). The i.m. injection of pDNA has shown excellent safety profiles, but low gene expression levels, which limits its use in various therapeutic applications. To overcome this limitation, we and others have proposed a very simple and inexpensive approach of co-administering naked pDNA with non-ionic Pluronic block copolymers (“Pluronics” or “poloxamers”) that were shown to drastically increase the levels and duration of muscle gene expression (6, 7). Pluronics consist of non-ionic ethylene oxide (EO) and propylene oxide (PO) blocks arranged in a basic triblock A-B-A structure: EOx-POy-EOx. They do not form complexes with the pDNA (8). So far the mechanisms by which Pluronic increases the transfection with naked pDNA in skeletal muscles remained not well understood.
It has been known that skeletal muscles have a remarkable ability to regenerate after tissue injury, which coincides with the inflammatory events and presence of immune cells, in particular, macrophages (MØs). These cells play a key role in the process of skeletal muscle regeneration (9–13). Due to constant persistence throughout the inflammatory response, MØs help muscle membrane repair in a cell-to-cell contact dependent manner (14). Similarly, we know that direct i.m. injection of pDNA also triggers an inflammation response due to first, the physical injury caused by the needle, and second, the exposure to unmethylated CpG islands in bacterially derived pDNA. A previous study reported an increase in muscle transfection of pDNA during hind limb ischemia (15). It was already noted before that Pluronic effects pDNA expression in the muscle were lacking in immune-deficient athymic mice, which implied the role of immune cells in muscle transfection (8). Moreover, co-administration of Pluronic with pDNA in the muscle greatly increased expression of the transgene along with the cDNA and mRNA levels in distal lymphoid organs, such as draining lymph nodes and spleen, with the transgene being co-localized there with antigen presenting cells (APCs) (16).
Most disease pathologies involve recruitment of inflammatory cells to the tissues and secretion of pro-inflammatory factors either locally or systemically. Since the therapeutic genes in the pDNA constructs are driven by constitutive cytomegalovirus (cmv) promoter with multiple NF-κb binding sites (17), inflammation may influence the gene expression. However, many studies involving i.m. injections of pDNA have been conducted in healthy muscles even when the ultimate goal was to use this method for therapeutic protein expression in disease-affected tissues. Therefore, studies of the gene expression using relatively new platforms such as Pluronic block copolymers in various disease models will further help understanding the limits and opportunities for application of these platforms in gene therapy.
The aim of this study was to evaluate the effect of local and systemic inflammation on muscle gene expression after i.m. delivery of naked pDNA with or without Pluronics. To induce local inflammation, we used a murine hind limb ischemia model (MHLIM) and for systemic inflammation both MHLIM and peritonitis. In previous work, we ranked the potency of various Pluronics on the expression of a plasmid containing a luciferase reporter gene under the control of cmv promoter, and found that Pluronic P85 (P85) and SP1017 were the most effective and safe (16). Here we examine the effects of these copolymers on the MØ ability to take up and express pDNA and horizontally transfer the transgene to the muscle cells both during in vitro coculture and adoptive transfer to healthy and disease affected animals.
Materials and Methods
Plasmids
The gWIZ™ high expression vectors encoding the reporter genes, luciferase (gWIZ™ Luc) and green fluorescent protein (GFP; gWIZ™ GFP), both under control of an optimized human cmv promoter followed by intron A from the cmv immediate-early gene were used throughout the study (Gene Therapy Systems, San Diego, CA). The pDRIVE5Lucia-mDesmin and pDRIVE5GFP-mDesmin (InvivoGen, San Diego, CA) encodes luciferase and GFP reporter proteins respectively, transcribed under the control of murine desmin promoter for muscle-specific expression. All plasmids were expanded in E. coli DH5α and isolated using Qiagen’s EndoFree Plasmid Giga Kit (Qiagen, Valencia, CA) according to the supplier’s protocol, reconstituted in phosphate buffered saline (PBS) and stored at −20°C until use.
Block copolymers
Pluronic L61 (batch # WPNT-511B), P85 (batch # WPNT-511B), F127 (batch # WPNT-511B), were a gift of the BASF Co. (Mount Olive, NJ). A mixed composition of L61 (0.25% w/v) and F127 (2% w/v), SP1017 (2.25% w/v) was obtained from Supratek Pharma Inc. (Montreal, Canada) or prepared using the corresponding copolymers.
pDNA/Pluronic formulations
The pDNA formulations were prepared as described (16) and used immediately for i.m. injections.
Cells
RAW264.7 immortalized mouse MØs cell line and C2C12 immortalized myoblasts (MBs) cell line were purchased from ATCC and cultured at 37°C and 5% CO2. To obtain terminally differentiated skeletal myotubes (MTs), C2C12 MBs were plated in 96-well plates (50,000 cells/well) in complete media (CM) containing Dulbecco’s minimal essential media (DMEM) and 10% fetal bovine serum (FBS). After 24 h or ~100% confluence, CM was replaced with 200 µl/well differentiation media (DM) containing DMEM and 2% horse serum (HS) to facilitate fusion of MBs (18). DM was changed every 12 h thereafter until day 5–10 when healthy long differentiated MTs were formed.
Animals
All animal experiments were carried out with approval of the University of Nebraska Medical Center (UNMC) and University of North Carolina at Chapel Hill (UNC-CH) Institutional Animal Care and Use Committee and in accordance with the NIH Guide for Laboratory Animal Use. Female Balb/c mice (6- to 8-weeks-old, Charles River Laboratories, Wilmington, MA) were used throughout this study. The animals were kept in groups of five and fed ad libitum.
Inflammation models and scheme of experiments
MHLIM was generated by surgical procedure as described (19, 20). Briefly, under general anesthesia, the femoral artery (FA) of right hind limb was completely excised after ligation at its proximal origin as a branch of the external iliac artery and before the point distally where it bifurcates into the saphenous and popliteal arteries. Saphenous artery and saphenous vein were also excised. An interval of 10 days was allowed for postoperative recovery before DNA administration in ischemic tibialis anterior (TA) muscle by direct i.m. injections. In some groups MHLIM surgery was performed on ipsilateral hind limb and test articles were simultaneously injected in contralateral TA muscles of the same mouse. Peritonitis was induced by intraperitoneal (i.p.) injections of filter-sterilized λ-carageenean (CGN; 1 mg/200 µl PBS) on the 1st and 2nd day and followed by test articles injections in TA muscles on the 3rd day. In some groups pDNA and Pluronic were in administered in separate legs in both healthy and peritonitis model.
pDNA injections
Animals were anesthetized by i.p. injection of mixed solution of ketamine (100 mg/kg) and xylazine (25 mg/kg) (Sigma, St. Louis, MO). Single injection of pDNA in 50 µl of Hank’s balanced salt solution (HBSS) alone or 50 µl of the block copolymer solution in HBSS was administered directly into right TA muscle of the mice using 28G 1 cc sterile syringe (BD Bioscience, Franklin Lakes, NJ).
Luciferase activity in vivo
Unless indicated otherwise mice were euthanized at the time points indicated in the figure legends and tissues were processed. The luciferase activity in 10 µl tissue homogenates was quantified using a TD20/20 or Glomax 20/20 luminometer (Promega, Fitchburg, WI) for an integration period of 20 s and 10 s respectively and normalized per mg of tissue as described before (16). Alternatively, luciferase activity was measured in live animals using in vivo imaging system IVIS-200 (Xenogen Corporation, Alameda, CA) 5 min after i.p injection of D-luciferin and the imaging data were quantified as described before (16).
Polymerase Chain Reaction (PCR)
PCR and Reverse transcription PCR (RT-PCR) were performed as described in (16).
Percent White Blood Cell (WBC) count in blood samples
Fresh serial blood samples from the tail vein (30–40 µl), collected into EDTA coated capillaries by tail milking method, were analyzed in VetScanHM5 hematology analyzer (Abaxis Ltd, Union City, CA) and % WBC counts were plotted on a time scale (days) after ischemia surgery.
Tissue Histology
Muscle tissues [quadriceps (Q), adductor (A), gastrocnemius (G) and TA)] at specified time points were removed en bloc and processed as described earlier (16).
Adoptive transfer of GFP transfected MØs in ischemic mice
RAW 264.7 MØs were plated in vitro in T75 flask in macrophage colony stimulating factor (MCSF) supplemented CM for 24 h and transfected with gWIZ™ GFP pDNA using genePORTER300 transfection reagent as per supplier’s protocol (extrapolated from 60 mm2 dish to T75 flask based on surface area). After another 24 h, 5×106 MØs were injected to ischemic mice by intrajugular vein injection (i.j.v.) 48 h post ischemic surgery. Ipsilateral ischemic and contralateral healthy muscle tissues were isolated 3 days after adoptive transfer of MØs (5 days post-surgery), embedded and frozen. 10 µm tissue sections were processed for immunohistochemistry (IHC) as described in (16).
Cytotoxicity evaluation
Cytotoxicity was determined using the CellTiter® 96 Aqueous Cell Proliferation Assay (MTS assay) (Promega, Fitchburg, WI) as per manufacturer recommendations. Briefly, cells (20,000 MØs, 16,000 MBs or day 7 MTs per well) were seeded in a 96-well plate 24 h before the experiment. Cells treated with Pluronic solutions in 100 µl of serum free media (SFM) for 2, 4, 6 and 8 h were rinsed thrice with SFM and cultured in CM. After another 24 h, cells were incubated with a mixture of 100 µl SFM + 20 µl MTS assay reagent for 1 h at 37°C at 5% CO2 and finally, calorimetric readout at 490 nm was done using Spectramax M5 plate reader (Molecular Devices, Sunnyvale, CA). The results were expressed as the mean percentage cell viability relative to untreated cells ± SEM (n=6).
In vitro luciferase gene expression
To study the effects of P85 on gene expression, 500,000 MØs were plated in 24-well plates in antibiotic free media. 24 h later, cells were treated with mixture of gWIZ™ Luc in different concentrations of P85 in SFM for 2h, and cultured in CM. After another 24 h, luciferase activity was measured in cell lysates as described in (16). Luciferase activity in coculture experiments (below) was measured in 10 µl cell lysates (cmv-coculture model) or 10 µl supernatant media (desmin-coculture model) using luciferase assay reagent or Quantiluc® respectively. The data was integrated over 10 s. Total protein content was measured using Pierce™ BCA Protein Assay Kit (Thermo-Fisher Scientific Inc, Waltham, MA) and data was expressed as RLU/mg protein.
Gene expression in coculture
RAW 264.7 MØs were transfected with either gWIZ™ Luc or pDRIVE5Lucia-mDesmin plasmids using genePORTER® 3000 transfection reagent (Genlantis, San Diego, CA) as per supplier’s recommended protocol. Heparin sulfate washing disintegrates the polyion complexes of DNA and removes cell surface bound DNA lipoplexes or polyplexes (21–23). Therefore, to remove membrane bound complexes after transfection prior to coculture, transfected MØs were briefly rinsed with 1 mg/ml heparin sulfate solution in PBS, followed by two times rinse with PBS and this entire washing procedure was repeated twice. MØs were then resuspended in CM and 50,000 MØs were added on top of 50,000 MBs or MTs in each well. Exactly after 2 h when MØs adhere to MBs or MTs, the coculture was rinsed with SFM (thrice) to remove traces of serum and exposed to SFM with/without P85 (0.01%, 0.1%, 0.3% and 1.0% w/v) for another 2 h, rinsed again with SFM (thrice) and cultured in 280 µl CM. After 24 h, gene expression was analyzed as described above. To study time-course of gene expression in coculture, the media was replaced every 12 h and samples were collected at the defined time points. For confocal imaging, gWIZ™ GFP or pDRIVE5GFP-mDesmin plasmid were used. At different time points, post coculture samples were fixed and labeled as explained in immunocytochemistry (ICC) (see in Ref. (24)).
Western blotting
Cell lysates prepared using mammalian protein extraction reagent (M-PER®; Pierce Biotechnology, Rockford, IL) were mixed with 2× Laemmli sample buffer (Bio-Rad Laboratories, Hercules, CA), boiled for 5 min and separated on a precast 12% SDS-PAGE gel (Bio-Rad Laboratories, Hercules, CA). Proteins were transferred on PVDF membranes, blocked with 5% w/v skim milk and probed overnight (o/n) with antibodies against β-actin (Abcam, Cambridge, MA) and desmin (Abcam, Cambridge, MA) at 4°C with gentle shaking. Membranes were washed with tris-buffered-saline (TBS) containing 0.1% w/v tween and probed with goat anti-rabbit IgG-HRP (Santa Cruz Biotechnology, Dallas, TX) and donkey anti-goat IgG-HRP (Abcam, Cambridge, MA) for 1 h at room temperature (RT) with gentle shaking before visualizing the protein bands using Pierce™ ECL Western Blotting substrate (Thermo Fisher Scientific, Waltham, MA).
Statistical analysis
As specified in legends to figures statistical comparisons were made using either unpaired Student’s t-test with Welch’s correction, or one-way ANOVA with Bonferroni correction for multiple comparison, multiple t-tests with Holm-Sidak correction for multiple comparisons.
RESULTS
Pluronic enhances gene expression during local inflammation in MHLIM
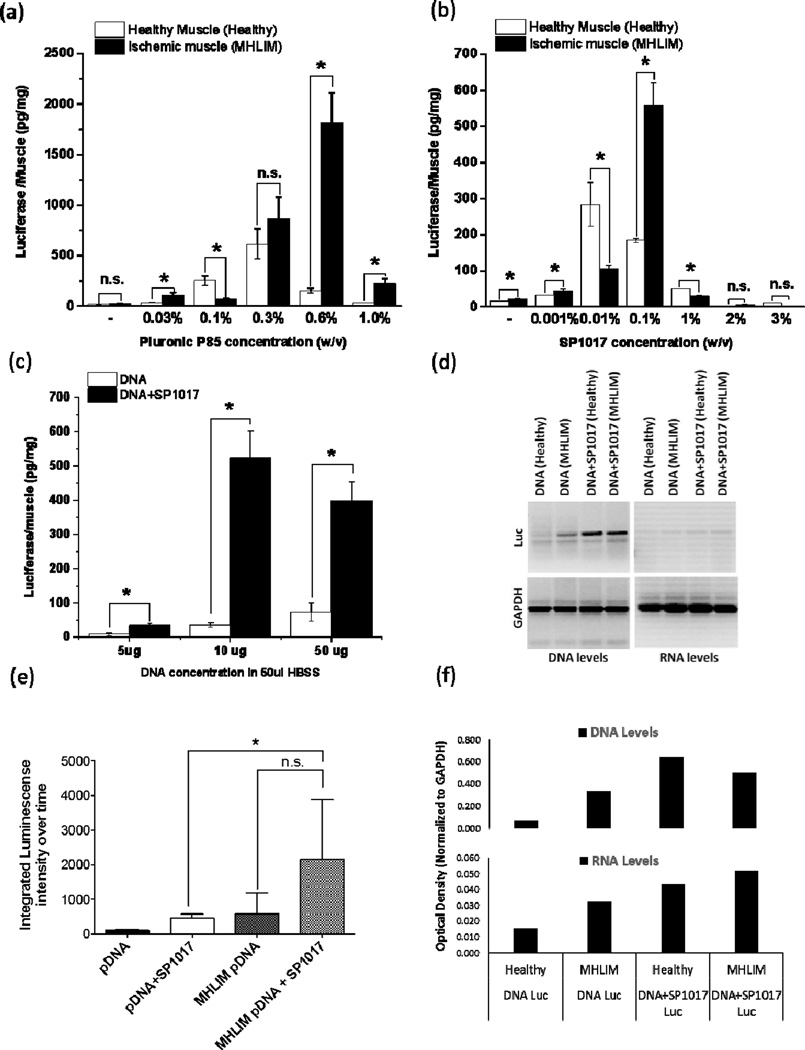
It was reported that co-injection of Pluronics with pDNA into the TA muscle of healthy animals increases and prolongs the gene expression compared to the naked pDNA (16). Here, to determine the effect of copolymers on the gene expression in the inflamed muscle of the disease-affected animal, we established and characterized MHLIM using previously described procedures (19, 20). The ischemia and tissue inflammation in this model were validated by histological examination of various ischemic muscle sections (see Fig. 1 in Ref. (24)). This model shows a typical response consistent with inflammatory cellular infiltrate. The pDNA, with or without Pluronic (P85 or SP1017), was injected into ischemic TA muscle of MHLIM on day 10 after surgery (“ischemic muscle” injections). In parallel experiments, same pDNA formulations were injected in the healthy mice (“healthy muscle” injections). The expression of luciferase was determined in the muscle homogenates on day 4 after injection, as previously described (8). The results suggested that P85 and SP1017 increased the luciferase expression in a dose-dependent manner, revealing typical bell-shaped dependencies of the expression vs. copolymer concentration, (Figs. 1a, b) in both ischemic and healthy muscles. However, there was a shift in the optimal concentration of Pluronic in the ischemic muscles, where the maximal expression was observed at higher concentration of the copolymers compared to the expression in the healthy muscles (0.6% vs. 0.3% w/v for P85 and 0.1% vs. 0.01% w/v for SP1017). At the maximum, in ischemic muscles, the expression levels of pDNA/copolymer mixtures were increased compared to the naked pDNA by ca. 72 fold for pDNA/P85 and 26-fold for pDNA/SP1017. Moreover, the maximal gene expression in ischemic muscles exceeded that in healthy muscles by ca. 8- and 3-fold for pDNA/P85 and pDNA/SP1017 respectively. Interestingly, the expression of naked pDNA also somewhat increased in ischemic muscles compared to that in healthy muscles.
Figure 1. Effect of co-formulation of pDNA with Pluronic on the transgene expression (a-c, e), DNA and RNA levels (d, f) in healthy and ischemic skeletal muscles.
Luciferase expression (a-c, e) or DNA and RNA levels (d, f) in ischemic TA muscles of MHLIM (a-f) or normal TA muscles of healthy animals (a, b, d, e, f) after single injections of the pDNA, pDNA/P85, or pDNA/SP1017. (a-f) All injected solutions contained 10 µg gWIZ™ Luc pDNA in 50 µl HBSS with or without copolymers except (c) where the pDNA concentration was varied. (c-f) SP1017 concentration in injected solution was 0.1 % w/v. (a-d, f) Measurements were done on day 4 by tissue sampling. (e) The AUC 0 to 60 days of the time course of luciferase expression determined in live animals by quantifying the bioluminescence imaging data (see Fig. 2 in Ref. (24)) (a-c, e) Data are mean ± SEM (a-c) n = 5, pairwise comparisons using unpaired Student’s t-test with Welch’s correction * p < 0.05, n.s. – not significant, (e) n = 3–4, the AUCs for each individual animal have been calculated and analyzed using one-way ANOVA with Bonferroni correction for multiple comparisons (f) Data are relative optical density of luciferase DNA or RNA levels presented in (d).
We further determined the pDNA dose response on the gene expression by injecting different doses (5 µg, 10 µg and 50 µg) of naked pDNA or pDNA/Pluronic (SP1017, 0.1% w/v) into ischemic muscle. The results suggested that the optimal dose of pDNA/SP1017 was 10 µg in 50 µl HBSS (Fig. 1c), and this dose was same as the one previously reported for healthy animals (8). At the lower dose of 5 µg, the levels of gene expression declined. At the higher dose of 50 µg, the gene expression leveled off and was not significantly different from that observed at 10 µg pDNA. Therefore, 10 µg pDNA was selected for further injections.
Next, the naked pDNA or pDNA/Pluronic was injected into ischemic and healthy muscles and the levels of luciferase pDNA and mRNA were determined 4 days after injection as previously described (16). The results suggested that Pluronic (SP1017, 0.1% w/v) increased pDNA and mRNA levels in healthy as well as in ischemic muscles compared to the animals injected with pDNA alone (Fig. 1d, f). Interestingly, the levels of DNA and mRNA in MHLIM injected with pDNA alone were considerably higher compared to these levels in the healthy animals (Fig. 1d, f).
Our previous studies showed that Pluronic prolongs transgene expression in the muscle of healthy Balb/c mice up to several weeks (6). Here we determined the time course of the luciferase expression in the ischemic TA muscle of MHLIM by quantifying luciferase bioluminescence in live animals (Fig. 1e) and (Fig. 2 in Ref. (24)). The areas under the curve (AUC) for the 60 days period were determined for each animal in all groups using the trapezoidal rule. When comparing the ischemic and healthy muscles injected with the naked pDNA, the former displayed much higher gene expression at every time point. Moreover, the gene expression was further significantly enhanced and prolonged when pDNA was formulated with SP1017, resulting in increased exposure of the animal to the transgene by nearly an order of magnitude for at least 60 days (Fig. 2 in Ref. (24)). Thus, the AUC values in the MHLIM injected with pDNA/SP1017 and naked pDNA were ~2146 and ~570 day*photons/s*106, respectively, suggesting ~ 3.8-fold increase in the transgene expression as a result of the pDNA formulation with SP1017. For comparison, in healthy animals the corresponding AUC values for pDNA/SP1017 and naked pDNA were ~449 and ~107 day*photons/s*106 respectively. Statistical comparison of the AUC values for the pDNA/SP1017 groups in the MHLIM and healthy animals suggest a significant difference between these groups (Fig. 1e).
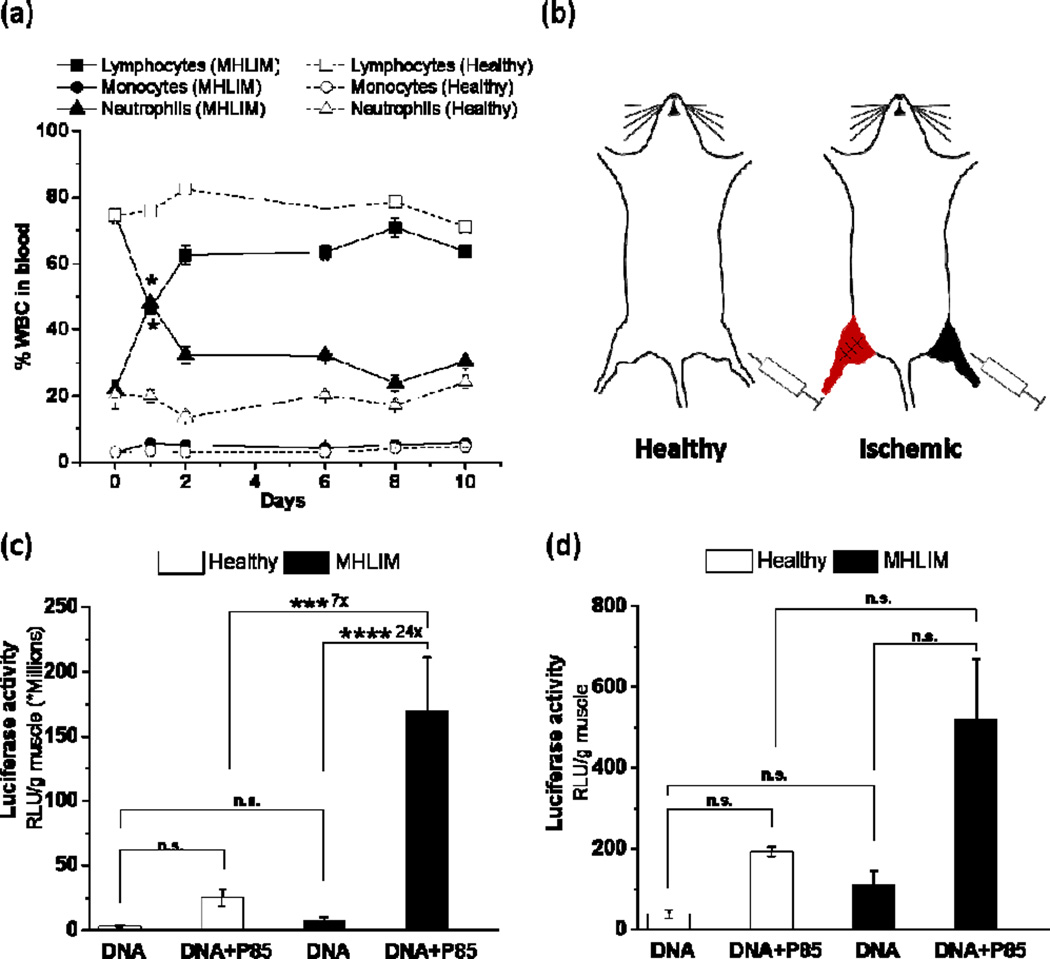
Figure 2. Effect of skeletal muscle ischemia on transgene expression in contralateral muscles.
(a) Changes in percent WBC counts in serial blood samples at different time points after ischemia surgery in MHLIM (solid lines, filled symbols) in comparison to healthy mice (dotted lines, open symbols). Day 0 corresponds to surgery. (b) Test articles were injected in TA muscles in healthy mice or in contralateral TA muscles simultaneously with the ipsilateral ischemia surgery in MHLIM. (c, d) Luciferase gene expression in the (c) TA muscles and (d) draining lymph nodes of MHLIM (black) and healthy mice (white) 3 days after single administration of 10 µg gWIZ™ Luc with or without 0.3 % w/v P85. Data are mean ± SEM (n = 3 (a) or n = 10 (c, d)), * p < 0.05, ***p<0.005, ****p<0.001, n.s. – non significant. (a) Statistical comparisons were made for Lymphocytes and Neutrophils in MHLIM and healthy mice using unpaired Student’s t-Test with Welch’s correction for 1 day time point; (c, d) comparisons were made using one-way ANOVA with Bonferroni correction for multiple comparison.
The GFP reporter gene expression in ischemic TA muscles, harvested on day 4 after gWIZ™ GFP pDNA injection, was mainly seen in the muscle fibers located in and around the site of the injection, and co-localized with desmin muscle marker (see Fig. 3a in Ref. (24)). Also, a substantial GFP expression was observed in small punctate cells, uniformly spread throughout ischemic tissue section, and co-localized with CD11b MØ marker (see Fig. 3b in Ref. (24)). Interestingly, the number of GFP expressing CD11b+ MØs in ischemic muscles appeared to be much higher than previously reported by us in non-inflamed muscles of the same mouse strain (16). Moreover, we also observed a higher number of GFP expressing muscle fibers in ischemic muscles injected with pDNA/P85 compared to pDNA alone (see Fig. 4 and Fig. 5 in Ref. (24)).
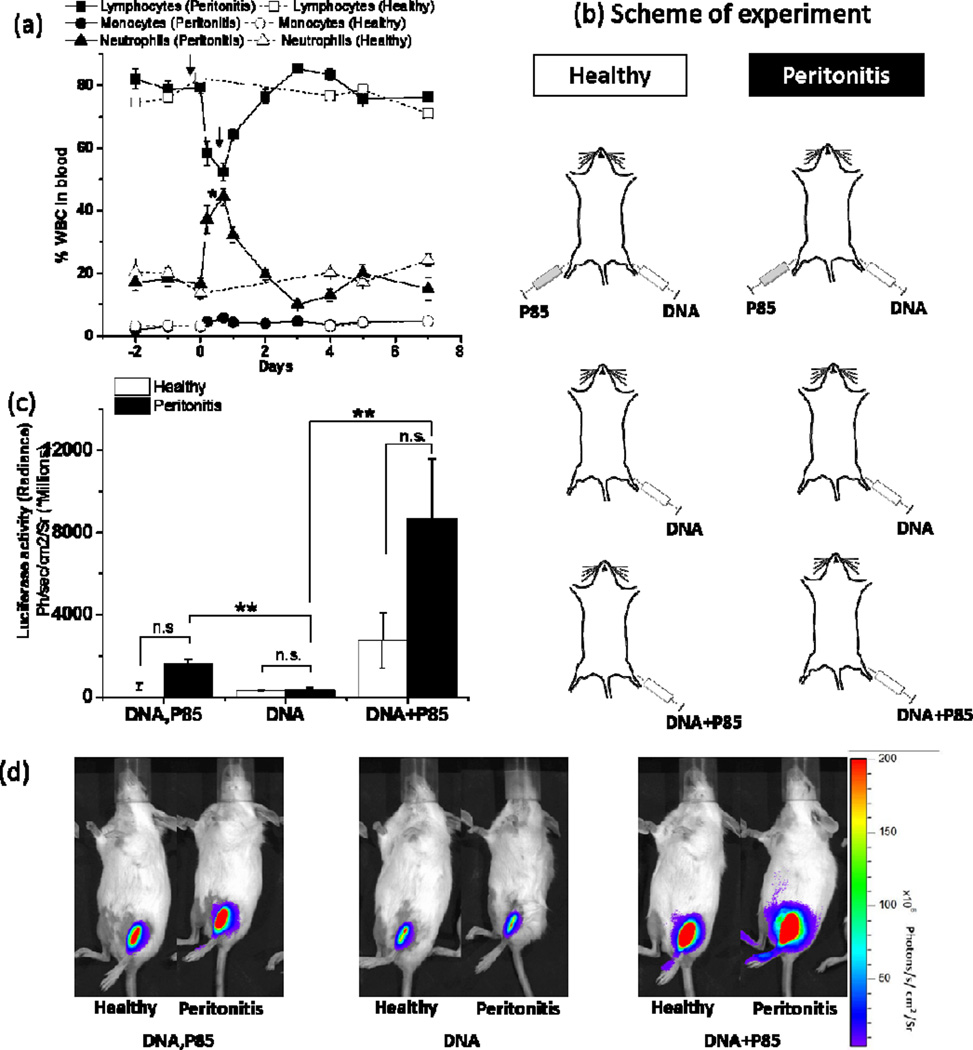
Figure 3. Effect of peritonitis on transgene expression in skeletal muscles.
(a) Changes in percent WBC counts in serial blood samples at different time points after induction of peritonitis (solid lines, filled symbols) in comparison to healthy mice (dotted lines, open symbols, same group as in Fig. 2). Arrows indicate time points of CGN injections to induce peritonitis. Day 0 corresponds to the first injection. (b) Healthy mice or mice with peritonitis were injected in TA muscles with 10 µg gWIZ™ Luc pDNA alone, pDNA coformulated with 0.3 % w/v P85 or simultaneously injected in ipsilateral and contralateral TA muscles with pDNA and 0.3 % w/v P85. (c) Luciferase gene expression in pDNA injected muscle of healthy (white bars) and peritonitis (black bars) mice was quantified using by quantifying the bioluminescence imaging data 4 days after pDNA administration. (d) Representative IVIS images used for quantification of gene expression data 4 days after pDNA administration. (a, c) Data are mean ± SEM (n=3), * p < 0.05, ** p < 0.01, n.s. - non significant. (a) Statistical comparisons were made for Lymphocytes and Neutrophils in healthy and peritonitis mice groups using unpaired Student’s t-Test with Welch’s correction for 1 day time point; (c) comparisons are made using one-way ANOVA with Bonferroni correction for multiple comparisons.
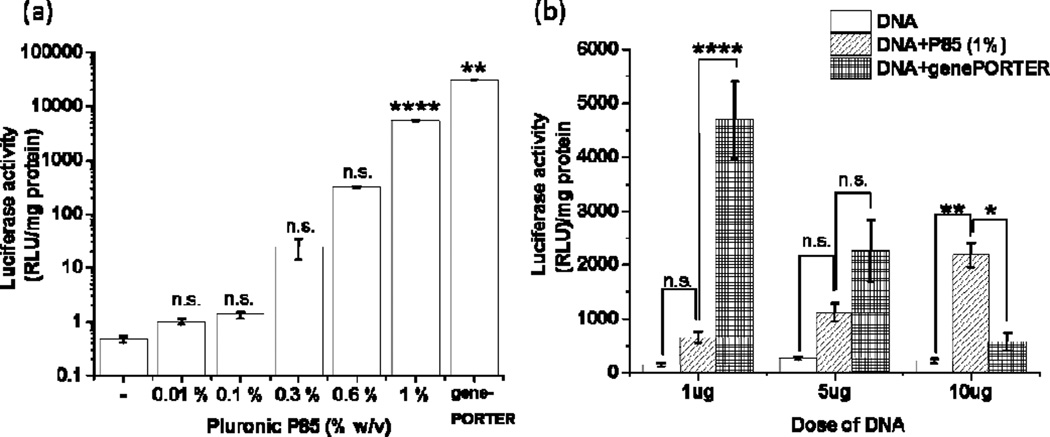
Figure 4. Effect of Pluronic on the gene expression in RAW 264.7 MØs.
(a, b) Cells were exposed to gWIZ™ Luc pDNA with or without P85 in SFM (a) for 2 h using 1 µg pDNA and various concentrations of P85 or (b) for 4 h using different amounts of pDNA (1 µg, 5 µg and 10 µg) and 1.0% w/v P85. (a, b) Cells transfected using genePORTER® 3000 transfection reagent are presented as positive control. Data were acquired using Glomax 20/20 luminometer or M5 Molecular Devices microplate reader. Data are mean ± SEM (n=4), * p<0.05, ** p<0.01, **** p<0.001 n.s. – non significant. Statistical comparisons were made for (a) treated vs. untreated groups or (b) otherwise indicated groups by one-way ANOVA with Bonferroni correction for multiple comparisons.
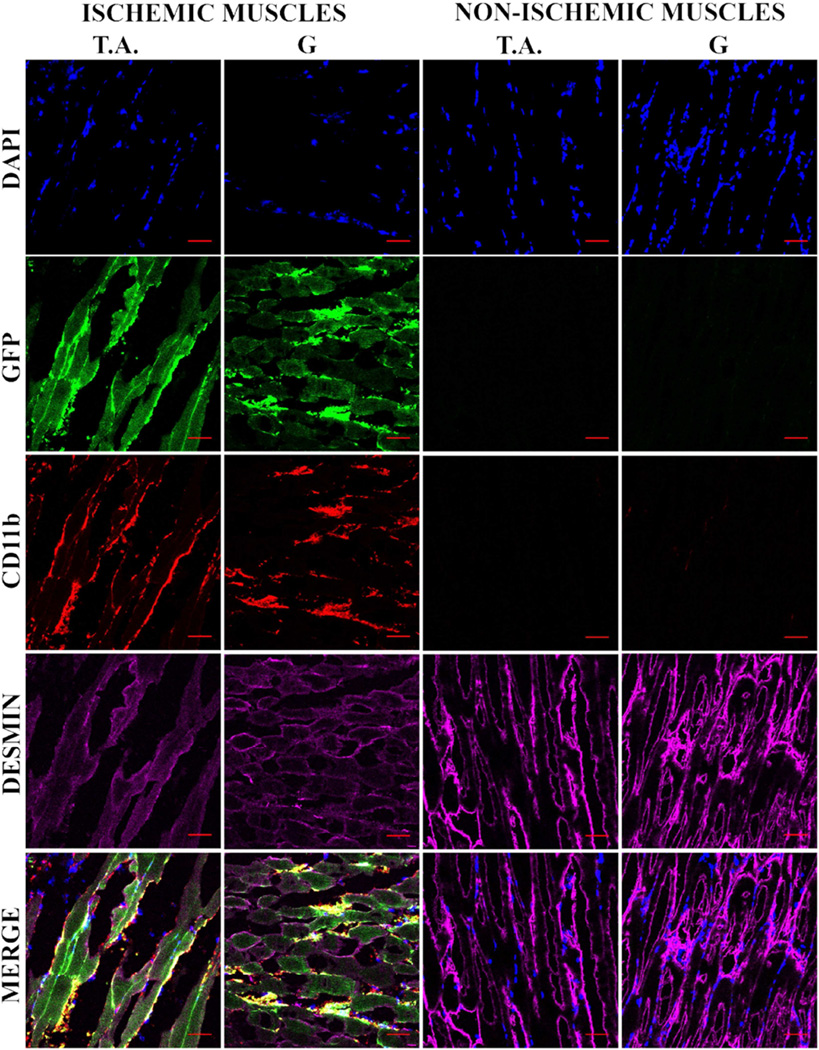
Figure 5. In vivo transfection of muscle cells upon adoptive transfer of GFP transfected MØs.
RAW 264.7 MØs transfected ex vivo with gWIZ™ GFP pDNA and administered i.j.v in MHLIM Balb/c mice (24 h after transfection, 48 h post ischemia surgery). Tissues (TA and G) were isolated 3 days thereafter and 10 µm thick sections of frozen tissues were processed for IHC. The color staining corresponds to nuclear DAPI (blue), GFP (green), CD11b (red) and desmin (magenta). The bottom panels present digitally superimposed images of preceding panels to visualize the co-localization (yellow or white). The images (20 ×) are representative of 3 sections per muscle and 3 mice per group. Scale bar = 50 µm.
Pluronic enhances gene expression in muscle during systemic inflammation
MHLIM was accompanied by systemic inflammation as evidenced by increase in neutrophils count in blood (Fig. 2a). Therefore, we examined the effect of inflammation during MHLIM on the transgene expression in the distal muscle. In this experiment the MHLIM was developed as described before, but the site of pDNA injection was spatially separated from the inflammation site (Fig. 2b). Naked pDNA alone or pDNA/Pluronic were injected in contralateral TA muscle simultaneously with the ischemia surgery performed on ipsilateral hind limb, and gene expression levels in muscles and draining lymph nodes were quantified in tissue homogenates at day 3. Consistent with our previous studies (8, 16) in healthy mice P85 increased gene expression compared to pDNA alone. The ischemia surgery in MHLIM resulted in a further increase in the expression, which was statistically significant in contralateral muscle (Fig. 2c) and displayed a trend in lymph nodes (Fig. 2d). Importantly, the effect of inflammation on gene expression was only observed with pDNA/P85 and not with pDNA alone. These effects appeared to be dependent on the type of the copolymer used since they were not statistically significant for the pDNA/SP1017 although revealed similar trends, especially in the contralateral muscles (see Fig. 6 in Ref. (24)).
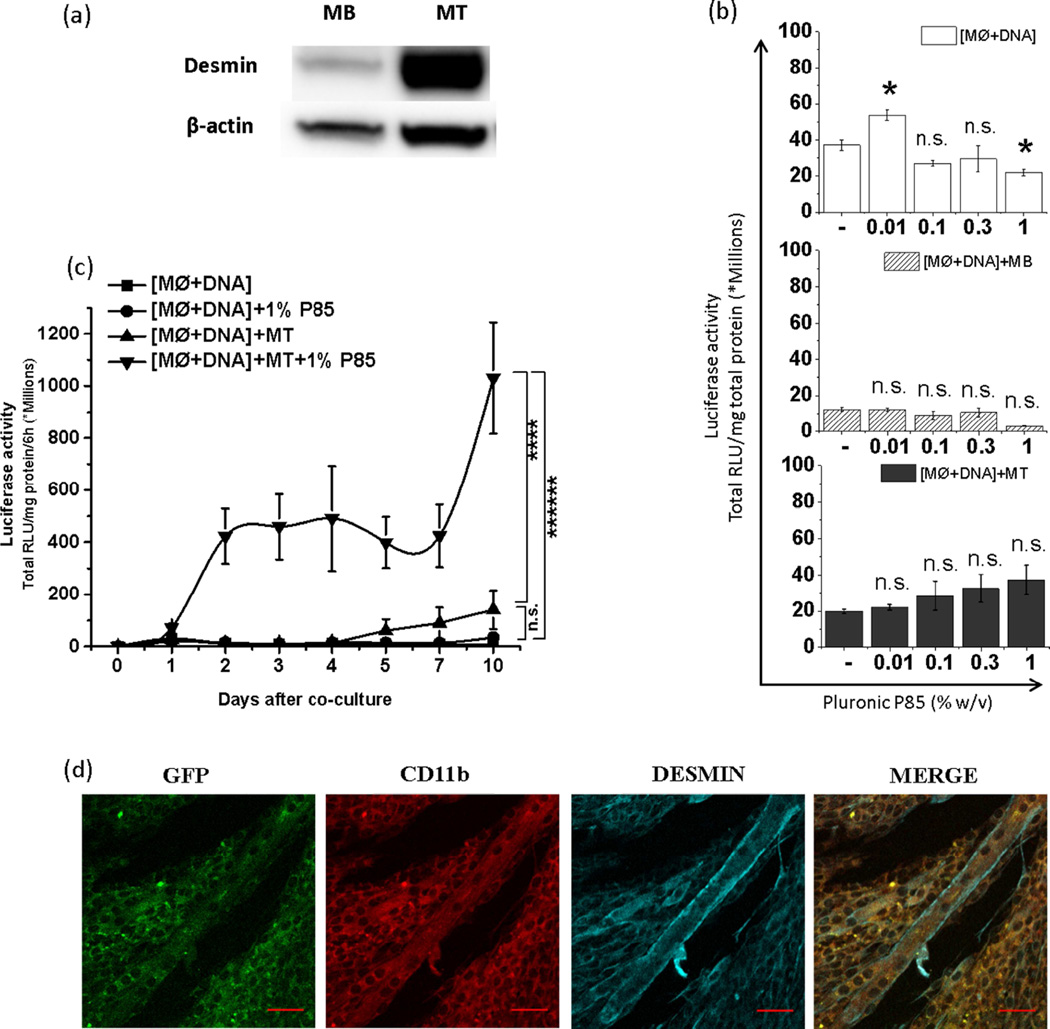
Figure 6. Effect of P85 on horizontal gene transfer from transfected MØs to muscle cells upon co-culture.
(a) Increased desmin expression upon differentiation of MBs to day 7 MTs. (b, c) pDRIVE5Lucia-mDesmin transfected MØs were plated alone, [MØ+DNA], or cocultured with MBs, [MØ+DNA]+MB, or MTs, [MØ+DNA]+MT, and exposed to P85 (0.01, 0.1, 0.3 and 1.0 % (b) or 1 % w/v (c)) for 2 h. Luciferase secreted in cell culture media was quantified after next 24 h (b) or at different time points. In the latter case (c) the media was replaced every 12 h and luciferase secreted in the media over 6 h period was determined daily until day 10. Data represents mean ± SEM, (b) (n=6), (c) (n=12). (b) Statistical comparisons were made using one-way ANOVA with Bonferroni correction for multiple comparisons (c) The AUCs for each individual condition are calculated and compared using one-way ANOVA with Bonferroni correction for multiple comparisons. * p<0.05, n.s. – non significant (d) GFP expression (green) in desmin+ MTs (cyan) was validated 3 days after their coculture with CD11b+ MØs (red) transfected with gWIZ™ GFP pDNA. Transfected MØs were placed on top of MTs. The confocal images were focused at MTs with MØs not visible. The last panels in each row present digitally superimposed images (20 ×) of preceding panels to visualize the co-localization (yellow). Scale bar = 50 µm.
The effect of inflammation on Pluronic-mediated gene expression at a distal site was striking and led to another experiment using peritonitis model to confirm the generality of the observation. Similar to MHLIM, systemic inflammation in peritonitis mouse model was confirmed by quantifying percent WBCs by hematological analysis of serial blood samples from tail vein (Fig. 3a). In this experiment, in addition to pDNA alone and pDNA/P85 groups, we included a group injected with P85 and pDNA in spatially different hind limb muscles (Fig. 3b). Since effect of P85 was more pronounced, we did not use SP1017 in this experiment. Luciferase reporter gene expression was quantified by in vivo imaging after i.p. injection of D-luciferin substrate. In both Pluronic treatment groups the animals with peritonitis showed a trend to increase gene expression in the muscle compared to healthy animals (Fig. 3c). In the peritonitis animals the contralaterally injected P85 significantly increased gene expression of pDNA in ipsilateral muscle, although the effect was numerically greater when P85 and pDNA were codelivered (Figs. 3c, d). Healthy mice injected with naked pDNA in ipsilateral leg and P85 in contralateral leg (DNA, P85) showed gene expression levels similar to naked pDNA alone.
Pluronic enhances transfection of MØs with pDNA in vitro
Since in vivo study suggested colocalization of the reporter gene with MØs (see Fig. 3b in Ref. (24)), we examined whether Pluronics can enhance transfection of MØs with pDNA in vitro. To this end we exposed RAW 264.7 cells for 2 h to pDNA at various concentrations of P85 and quantified luciferase in cell lysates 24 h after the exposure. Addition of P85 to the media, at the concentration as low as 0.01%, resulted in enhancement of the gene expression (Fig. 4a). This effect increased as the copolymer dose increased and reached over 4 orders of magnitude at 1% P85 concentration when the gene expression levels nearly matched that observed using a commercial transfection kit, genePORTER® 3000. Moreover, at the higher dose of pDNA (10 µg) the gene expression with 1.0% w/v P85 exceeded that using genePORTER® 3000 transfection (Fig. 4b). Analysis of pDNA uptake in the cells showed higher pDNA uptake in MØs than muscle cells and marginal increase of pDNA uptake in MØs in the presence of P85 ((see Fig. 7 in Ref. (24)). No such increase in uptake was observed in cultured muscle cells with the addition of P85 (MBs or MTs).
Adoptive gene transfer to muscle by ex vivo transfected MØs
To examine whether MØs can transfer genes to ischemic animals, the RAW 264.7 MØs were transfected ex vivo with gWIZ™ GFP using genePORTER 3000 and adoptively transferred via jugular vein to MHLIM animals 24 h post transfection and 48 h post ischemia surgery. The lower hind limb muscles (ischemic and non-ischemic) were isolated 3 days after MØs administration, sectioned and analyzed by confocal imaging for co-localization of GFP with CD11b+ MØs and desmin+ muscle fibers. Firstly, we found that the ischemic muscles in contrast to non-ischemic muscles stained positive for the MØs cell surface marker Cd11b (Fig. 5). This suggested recruitment of MØs to ischemic muscles and not to non-ischemic muscles of MHLIM. We observed lower levels of muscle-specific marker (desmin) in ischemic muscle compared to non-ischemic muscle, which is typical for ischemia (25, 26). Notably, CD11b staining in the ischemic muscles localized closely to the desmin expressing muscle cells, which suggests that either MØs were in close proximity to muscle fibers or CD11b was transferred from MØs to muscle cells. Secondly, the GFP expression in ischemic tissues co-localized with desmin+ muscle fibers having typical elongated shape (Fig. 5). Larger ischemic muscle fibers clearly expressed GFP protein unlike non-ischemic control muscles. This suggested that MØs can transfer GFP gene or protein to the muscle cells, and along with the muscle cells can collectively represent a reservoir or conduit for the gene expression. This may also imply that cell-to-cell contacts play role in exchange of genes or proteins from MØs across the cell membranes to otherwise hard to transfect skeletal muscle fibers. In the in vitro co-culture of the GFP-transfected MØs with muscle cells we observed unidirectional exchange of CD11b from MØs to the muscle cells followed by expression of the reporter gene in the muscle cells (see Fig. 8 and Fig. 9 in Ref. (24)). No transfer of desmin from muscle cells to MØs was observed. Our data was similar to other reports that describe bi/uni-directional exchange of membrane components as a general cell-to-cell communication amongst various cell types (27–29).
Pluronic enhances horizontal transfer of pDNA from MØs to muscle cells
After demonstrating that Pluronic can enhance the transfection of MØs with pDNA, we further examined whether it could also increase the pDNA transfer from the transfected MØs to muscle cells. To evaluate such possibility, we studied in vitro effect of P85 on the transgene expression in the coculture of transfected MØs with muscle cells. First we transfected the constantly multiplying RAW 264.7 MØs with pDNA [MØ+DNA] and then cocultured them on top of the monolayers of 1) either terminally differentiated C2C12 MTs (depicted herein [MØ+DNA]+MT), or 2) precursor MBs ([MØ+DNA]+MB). Notably, MTs display high level of expression of desmin, which is much lower in MBs (Fig. 6a). Thus we used two different plasmids, one with the constitutive (cmv) promoter, gWIZ™ Luc pDNA (same as the plasmid used above), and the other with the muscle specific-desmin promoter, pDRIVE5Lucia-mDesmin (to quantify the muscle specific gene expression). In both cases, the transfected MØs or the cocultures were exposed to different concentrations of P85 (0.01, 0.1, 0.3, 1.0% w/v) for 2 h and the total gene expression was determined after 24 h and normalized to the total protein. Using the desmin-driven pDNA we observed relatively low levels of normalized gene expression in all cell systems - [MØ+DNA], [MØ+DNA]+MT, and [MØ+DNA]+MB (Fig. 6b). In the transfected MØs and [MØ+DNA]+MB coculture the gene expression was decreased at 1% P85 compared to respective copolymer-free controls. In contrast, [MØ+DNA]+MT coculture after exposure to the copolymer displayed a trend for a monotonous increase in the normalized gene expression, this increase becoming significant at 1% P85. At this copolymer concentration the total gene expression in [MØ+DNA]+MT coculture was much higher than that in [MØ+DNA] and [MØ+DNA]+MB (see Fig. 10 in Ref. (24)).
Therefore, we further examined the gene expression in [MØ+DNA] and [MØ+DNA]+MT coculture at different time points after exposure to 1% P85 (Fig. 6c). In this case starting from day 2 there was a dramatic increase in gene expression in [MØ+DNA]+MT coculture but not in the transfected MØ alone. Compared to the coculture the level of gene expression in MØ+DNA was negligible with or without exposure to P85. The latter is probably explained by the promoter selectivity of the pDRIVE5Lucia-mDesmin plasmid used in this study as well by some toxicity of the 1% P85 to macrophages (see Fig. 11 in Ref. (24)). Notably, when similar co-culture experiments were carried out using constitutive (cmv) promoter driven gWIZ™ Luc pDNA, both the total and normalized gene expression in MØ+DNA groups after 24 h was high ((see Fig. 12 and 13 in Ref. (24)). However, at later time points the normalized gene expression in MØ+DNA sharply decreased while in the coculture it increased, especially in the groups treated by P85 (see Fig. 12 in Ref. (24)). Using plasmids expressing GFP in the [MØ+DNA]+MT coculture groups, we were able to clearly see gene expression in the MTs (Fig. 6d) (see also Fig. 13c in Ref. (24)). This result is particularly amazing given the fact that MTs are most difficult to transfect cells using normal cell transfection means (see Fig. 14 in Ref. (24)).
Discussion
Administration of pDNA to skeletal muscle is a strategy to deliver various genes for prophylactic and therapeutic purposes in healthy and diseased muscles (30–32). Currently, the application of direct i.m. pDNA injections is hindered by low levels of gene expression. Improvements in this area include electroporation-facilitated gene transfer known for its simplicity and use in human clinical trials (33). However, electroporation is associated with the risk of extensive and irreversible muscle damage induced by electrical pulses (34). This has limited its application in therapeutics, especially, when the goal is to rescue the muscle fibers during muscular pathologies, such as hind limb ischemia, polymyositis, muscular dystrophies, etc. Most of these pathologies involve inflammation and recruitment of inflammatory cells, which further contribute to tissue degeneration and muscle damage (35, 36). We have reported that coadministration of pDNA with non-ionic amphiphilic Pluronic block copolymers leads to drastic increases in the level and duration of transgene expression in the healthy muscles (6, 37). In contrast to electroporation, muscle tissue did not appear to be affected by pDNA/Pluronic mixture as observed by histological analysis (16) and animals injected with these formulations did not show any signs of discomfort. Furthermore, Pluronic was coadministered with pDNA in the muscle during electroporation, where it alleviated the pulse-induced tissue damage and further increased the transgene expression (38).
Here, we report supplementary effects of Pluronic and inflammation caused by muscle ischemia on the transgene expression in the muscle. The greatest expression was observed in MHLIM when pDNA/Pluronic was injected directly in the ischemic muscle, although a significant increase compared to expression in healthy animals was also seen when the site of injection was distal from the ischemia site. The latter suggests that systemic inflammation contributes to the pDNA/Pluronic gene delivery in the muscle. This conclusion was also reinforced by an observed trend for increase in muscle transgene expression with pDNA/Pluronic during systemic inflammation in the peritonitis model. Interestingly, the effects of Pluronic in peritonitis animals were higher when Pluronic was co-delivered with pDNA in the same muscle than upon their separate administration in different muscles. Still in the latter case gene expression in peritonitis model was increased possibly suggesting systemic distribution of Pluronic after intramuscular injection. In the absence of Pluronic inflammation had little if any effect on pDNA expression during peritonitis and relatively small effect in MHLIM. Notably, other studies also reported that inflammation could enhance muscle gene expression upon direct i.m. injection of naked pDNA (15, 39).
Previously published data suggested that Pluronics do not enhance muscle transfection in immune-deficient athymic nude mice (8, 40), which implied a mutually inclusive role of both immune cells and copolymers in the muscle transfection. We have also reported that 1) transgenes can be transferred from in vitro transfected MØs to the surrounding cells (e.g. neurons) upon coculture and 2) that after adoptive transfer, transfected MØs can deliver transgenes to the distal sites of inflammation, specifically to the brain in the mouse model of Parkinsonism (41, 42). These studies suggested that transfected MØs can repackage pDNA and deliver across cell membranes to neuron cells via exosomes and other extracellular vesicles. The present study reinforces the idea that Pluronic may assist muscle transfection through the effects on immune cells and specifically MØs that are recruited in the inflamed muscles, pick up the pDNA and express the transgene. MØs are known to internalize bacterial derived pDNA by a very specific transport mechanism resulting in gene expression both in vitro (43) and in vivo (44). Here we demonstrate that treatment with Pluronic increases the transfection of MØs with naked pDNA and boosts gene expression up to 4 orders of magnitude. Moreover, after adoptive transfer in MHLIM the transfected MØs are shown to deliver transgene to the ischemic but not the healthy muscles. These observations suggested that MØs can transfer transgene across the muscle cell membrane, and led to specific experiments using in vitro coculture models.
In these experiments we demonstrated that MØs can horizontally transfer pDNA to muscle cells and this process is greatly enhanced in the presence of Pluronic. Specifically we demonstrated the transfer of pDNA from MØs to MTs in vitro. Since MØs were transfected with pDNA bearing the muscle-specific promoter the enhanced gene expression in MTs could only result from pDNA transfer and not from the protein transfer. Using horizontal gene transfer with Pluronic we were able to efficiently transfect MTs that are terminally differentiated cells and therefore notoriously difficult to transfect in vitro. That is why researchers use transiently transfected mitotically active MBs and differentiate them to form transfected MTs. This procedure limits the skeletal muscle biology studies to early stages of muscle development. Our finding provides a novel approach for gene delivery to MTs at later stages (1–3 weeks) after differentiation and can find application in research labs.
Interestingly, we observed a unidirectional exchange of MØs cell surface marker (CD11b) to muscle cells but not vice versa (see Fig. 8 in Ref. (24)), which may explain the mechanism of delivering pDNA/protein across the membranes resulting in muscle cell transfection. Exchange of plasma membrane fragments, including cell-surface proteins and lipids, in conjugates formed between immune cells and their cellular partners is a field of intense investigation (45). This phenomenon may be in common with the evolutionary process, by which MØs recruited to the site of muscle damage deliver growth factors required for muscle regeneration. Recent studies have also reported a novel mechanism of vesicular exchange of protein/genetic material between APCs during cell-to-cell communication (46), which justifies our observations. It has been shown that immune cells can deliver RNA containing exosomes to an adjacent cell as soon as 10 minutes of establishing cell-to-cell contact (47). We have also reported that exosomes isolated from the media over transfected MØs can further transfect neuronal cells (41, 42).
Skeletal muscles are known for complex muscle biology, and recent studies have shown that MØs play a significant role in helping muscle regeneration and preventing muscle atrophy (9, 48–50) via cell-to-cell contacts (14). The role of stem and hematopoietic cells in muscle regeneration has been also extensively studied (51, 52). Evidence points to involvement of cell-to-cell fusion in muscle regeneration, whereby cells with stem cells’ properties fuse with damaged muscle fibers or satellite cells to form new muscle fibers. Technologies to deplete and repopulate specific cells including transgenic mice were developed and showed that the stem cells with myeloid cell properties are responsible for muscle regeneration (13, 53, 54). Similar mechanisms were reported for regeneration of liver by MØs fusion (55–57). Therefore, we posit a mechanism in which MØs are recruited to the sites of naked pDNA injections as a result of physiological insult caused by needle injury. These MØs phagocytose extracellular naked pDNA and then fuse with the existing muscles as a part of regeneration process thus, delivering pDNA into otherwise hard to transfect skeletal muscles. Based on our observations Pluronic can enhance both the transfection of the MØs with pDNA and the horizontal gene transfer from the transfected MØs to muscle fibers. Polyethylene glycol (PEG) mediated cell-to-cell fusion has been extensively studied since 1970s to generate somatic cell hybrids for cloning, gene mapping, gene expression and antibody production (12, 58, 59). The proposed model involves exclusion of water molecules between lipid bilayers and changing the orientation of molecular dipoles that results in membrane fusion (60). Since Pluronics contain PEG as well as polypropylene glycol chains capable of incorporating into lipid membranes (61), we believe that they can enhance the MØs to muscle cell fusion events eventually delivering pDNA to muscle fibers and increasing transfection and gene expression both in healthy and inflamed muscles.
It is known that after direct i.m. injection in skeletal muscle naked pDNA is rapidly degraded in extracellular matrix (62, 63). Therefore, is noteworthy that we observed a block copolymer-dependent increase in pDNA copies in both healthy and ischemic muscles. Unlike cationic lipids or polycations, Pluronics are non-ionic molecules and do not form complexes with pDNA that can protect it from the degradation in the extracellular matrix similar to lipoplexes/polyplexes (16, 64). However, Pluronics can act as adjuvants that activate NF-κB cell signaling and increase pDNA uptake (65), pDNA nuclear import (66) and transcriptional activation of gene expression of pDNA delivered into cells in vitro with polyplexes (7, 67). It is well-known that inflammation results in increased blood neutrophils counts and secretion of pro-inflammatory cytokines like TNF alpha and IL-1β (68). The overall activation of inflammatory signaling can also influence cmv promoter driven gene expression and may explain somewhat increased gene expression of naked pDNA in non-ischemic muscle of MHLIM. It is important to point out in this regard that the pDNA/Pluronic expression in normal muscles was shown to be promoter selective and dependent on the NF-κB signaling pathway (8, 69). Moreover, systemic inflammation caused by ischemia resulted in increased number of circulatory MØs, which is consistent with previous studies (70, 71). This may result in increased MØs recruitment at the site of pDNA injection due to inflammatory response triggered by physical injury and exposure to bacterial CpG motif and ultimately increase muscle transfection with pDNA/Pluronic. We also observed increased transgene expression in draining lymph nodes, which was in agreement with our earlier studies using i.m. pDNA/Pluronic suggesting co-localization of transgene expression with APCs in lymphoid organs (16).
Altogether, MØs have gained recent attention for their promiscuity or plasticity to perform central roles in development, homeostasis and disease and have emerged as novel therapeutics target. Here we resolved a key issue of low muscle transfection in ischemic muscles by simple mixing of naked pDNA with Pluronic and showed how MØs assist gene transfer to otherwise hard to transfect skeletal muscle fibers. Since Pluronics are listed in U.S. Pharmacopoeia and have been demonstrated to be safe in clinical trials (72) such formulations have promise for human use.
Acknowledgments
This study was supported in parts by the National Institutes of Health grants R01 CA116591 (AVK), the Department of Defense grant W81XWH-09-1-0386 (AVK), Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant P20GM103480, The Carolina Partnership, a strategic partnership between the UNC Eshelman School of Pharmacy, The University Cancer Research Fund through the Lineberger Comprehensive Cancer Center, and the American Heart Association fellowship 0610065Z (ZG). The assistance of the following individuals at UNMC is also gratefully acknowledged: Maram Reddy Prasanna K Reddy (assistance in establishing MHLIM); Janice A. Taylor and James R. Talaska (confocal microscopy); Margaret A. Jennings (histology and IHC); Terence A. Lawson (in vivo imaging); and Dominic Moore (statistical analysis). These studies were carried out using the UNMC Confocal Laser Scanning Microscope and Flow Cytometry Research Cores and UNC-CH Microscopy services laboratories, Department of Pathology and Laboratory Medicine core facilities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, et al. Direct gene transfer into mouse muscle in vivo. Science. 1990;247(4949 Pt 1):1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 2.Ginn SL, Alexander IE, Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2012 - an update. J Gene Med. 2013;15(2):65–77. doi: 10.1002/jgm.2698. [DOI] [PubMed] [Google Scholar]
- 3.Hammer SM, Sobieszczyk ME, Janes H, Karuna ST, Mulligan MJ, Grove D, et al. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N Engl J Med. 2013;369(22):2083–2092. doi: 10.1056/NEJMoa1310566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romero NB, Braun S, Benveniste O, Leturcq F, Hogrel JY, Morris GE, et al. Phase I study of dystrophin plasmid-based gene therapy in Duchenne/Becker muscular dystrophy. Hum Gene Ther. 2004;15(11):1065–1076. doi: 10.1089/hum.2004.15.1065. [DOI] [PubMed] [Google Scholar]
- 5.Morishita R, Makino H, Aoki M, Hashiya N, Yamasaki K, Azuma J, et al. Phase I/IIa clinical trial of therapeutic angiogenesis using hepatocyte growth factor gene transfer to treat critical limb ischemia. Arterioscler Thromb Vasc Biol. 2011;31(3):713–720. doi: 10.1161/ATVBAHA.110.219550. [DOI] [PubMed] [Google Scholar]
- 6.Lemieux P, Guerin N, Paradis G, Proulx R, Chistyakova L, Kabanov A, et al. A combination of poloxamers increases gene expression of plasmid DNA in skeletal muscle. Gene therapy. 2000;7(11):986–991. doi: 10.1038/sj.gt.3301189. [DOI] [PubMed] [Google Scholar]
- 7.Kabanov A, Zhu J, Alakhov V. Pluronic Block Copolymers for Gene Delivery. Adv Genet. 2005;53PA:231–261. doi: 10.1016/S0065-2660(05)53009-8. [DOI] [PubMed] [Google Scholar]
- 8.Yang Z, Zhu J, Sriadibhatla S, Gebhart C, Alakhov V, Kabanov A. Promoter- and strain-selective enhancement of gene expression in a mouse skeletal muscle by a polymer excipient Pluronic P85. J Control Release. 2005;108(2–3):496–512. doi: 10.1016/j.jconrel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Tidball JG, Wehling-Henricks M. Macrophages promote muscle membrane repair and muscle fibre growth and regeneration during modified muscle loading in mice in vivo. J Physiol. 2007;578(Pt 1):327–336. doi: 10.1113/jphysiol.2006.118265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kharraz Y, Guerra J, Mann CJ, Serrano AL, Munoz-Canoves P. Macrophage plasticity and the role of inflammation in skeletal muscle repair. Mediators Inflamm. 2013;2013:491497. doi: 10.1155/2013/491497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rigamonti E, Zordan P, Sciorati C, Rovere-Querini P, Brunelli S. Macrophage plasticity in skeletal muscle repair. Biomed Res Int. 2014;2014:560629. doi: 10.1155/2014/560629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnold L, Henry A, Poron F, Baba-Amer Y, van Rooijen N, Plonquet A, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204(5):1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Summan M, Warren GL, Mercer RR, Chapman R, Hulderman T, Van Rooijen N, et al. Macrophages and skeletal muscle regeneration: a clodronate-containing liposome depletion study. Am J Physiol Regul Integr Comp Physiol. 2006;290(6):R1488–R1495. doi: 10.1152/ajpregu.00465.2005. [DOI] [PubMed] [Google Scholar]
- 14.Sonnet C, Lafuste P, Arnold L, Brigitte M, Poron F, Authier FJ, et al. Human macrophages rescue myoblasts and myotubes from apoptosis through a set of adhesion molecular systems. J Cell Sci. 2006;119(Pt 12):2497–2507. doi: 10.1242/jcs.02988. [DOI] [PubMed] [Google Scholar]
- 15.Takeshita S, Isshiki T, Sato T. Increased expression of direct gene transfer into skeletal muscles observed after acute ischemic injury in rats. Lab Invest. 1996;74(6):1061–1065. [PubMed] [Google Scholar]
- 16.Gaymalov ZZ, Yang Z, Pisarev VM, Alakhov VY, Kabanov AV. The effect of the nonionic block copolymer pluronic P85 on gene expression in mouse muscle and antigen-presenting cells. Biomaterials. 2009;30(6):1232–1245. doi: 10.1016/j.biomaterials.2008.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kako K, Nishikawa M, Yoshida H, Takakura Y. Effects of inflammatory response on in vivo transgene expression by plasmid DNA in mice. J Pharm Sci. 2008;97(8):3074–3083. doi: 10.1002/jps.21254. [DOI] [PubMed] [Google Scholar]
- 18.Kee HJ, Kim JR, Joung H, Choe N, Lee SE, Eom GH, et al. Ret finger protein inhibits muscle differentiation by modulating serum response factor and enhancer of polycomb1. Cell Death Differ. 2012;19(1):121–131. doi: 10.1038/cdd.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couffinhal T, Silver M, Zheng LP, Kearney M, Witzenbichler B, Isner JM. Mouse model of angiogenesis. The American journal of pathology. 1998;152(6):1667–1679. [PMC free article] [PubMed] [Google Scholar]
- 20.Couffinhal T, Silver M, Kearney M, Sullivan A, Witzenbichler B, Magner M, et al. Impaired collateral vessel development associated with reduced expression of vascular endothelial growth factor in ApoE−/− mice. Circulation. 1999;99(24):3188–3198. doi: 10.1161/01.cir.99.24.3188. [DOI] [PubMed] [Google Scholar]
- 21.Iwasa A, Akita H, Khalil I, Kogure K, Futaki S, Harashima H. Cellular uptake and subsequent intracellular trafficking of R8-liposomes introduced at low temperature. Biochim Biophys Acta. 2006;1758(6):713–720. doi: 10.1016/j.bbamem.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 22.Naik RJ, Chandra P, Mann A, Ganguli M. Exogenous and cell surface glycosaminoglycans alter DNA delivery efficiency of arginine and lysine homopeptides in distinctly different ways. J Biol Chem. 2011;286(21):18982–18993. doi: 10.1074/jbc.M111.227793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mislick KA, Baldeschwieler JD. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc Natl Acad Sci U S A. 1996;93(22):12349–12354. doi: 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahajan V, Gaymalov ZZ, Alakhova D, Gupta R, Zucker IH, Kabanov AV. Supporting Data of Horizontal Gene Transfer from Macrophages to Ischemic Muscles upon Delivery of Naked DNA with Pluronic Block Copolymers. Data in Brief. doi: 10.1016/j.biomaterials.2015.10.002. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouyang J, Guzman M, Desoto-Lapaix F, Pincus MR, Wieczorek R. Utility of desmin and a Masson's trichrome method to detect early acute myocardial infarction in autopsy tissues. Int J Clin Exp Pathol. 2009;3(1):98–105. [PMC free article] [PubMed] [Google Scholar]
- 26.Hein S, Scheffold T, Schaper J. Ischemia induces early changes to cytoskeletal and contractile proteins in diseased human myocardium. J Thorac Cardiovasc Surg. 1995;110(1):89–98. doi: 10.1016/S0022-5223(05)80013-3. [DOI] [PubMed] [Google Scholar]
- 27.Strassburg S, Hodson NW, Hill PI, Richardson SM, Hoyland JA. Bi-directional exchange of membrane components occurs during co-culture of mesenchymal stem cells and nucleus pulposus cells. PLoS One. 2012;7(3):e33739. doi: 10.1371/journal.pone.0033739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, Wall D. Cell contact-dependent outer membrane exchange in myxobacteria: genetic determinants and mechanism. PLoS Genet. 2012;8(4):e1002626. doi: 10.1371/journal.pgen.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niu X, Gupta K, Yang JT, Shamblott MJ, Levchenko A. Physical transfer of membrane and cytoplasmic components as a general mechanism of cell-cell communication. J Cell Sci. 2009;122(Pt 5):600–610. doi: 10.1242/jcs.031427. [DOI] [PubMed] [Google Scholar]
- 30.Davis HL, Demeneix BA, Quantin B, Coulombe J, Whalen RG. Plasmid DNA is superior to viral vectors for direct gene transfer into adult mouse skeletal muscle. Hum Gene Ther. 1993;4(6):733–740. doi: 10.1089/hum.1993.4.6-733. [DOI] [PubMed] [Google Scholar]
- 31.Kessler PD, Podsakoff GM, Chen X, McQuiston SA, Colosi PC, Matelis LA, et al. Gene delivery to skeletal muscle results in sustained expression and systemic delivery of a therapeutic protein. Proc Natl Acad Sci U S A. 1996;93(24):14082–14087. doi: 10.1073/pnas.93.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis HL, Michel ML, Whalen RG. DNA-based immunization induces continuous secretion of hepatitis B surface antigen and high levels of circulating antibody. Hum Mol Genet. 1993;2(11):1847–1851. doi: 10.1093/hmg/2.11.1847. [DOI] [PubMed] [Google Scholar]
- 33.Widera G, Austin M, Rabussay D, Goldbeck C, Barnett SW, Chen M, et al. Increased DNA vaccine delivery and immunogenicity by electroporation in vivo. J Immunol. 2000;164(9):4635–4640. doi: 10.4049/jimmunol.164.9.4635. [DOI] [PubMed] [Google Scholar]
- 34.Bhatt DL, Gaylor DC, Lee RC. Rhabdomyolysis due to pulsed electric fields. Plastic and reconstructive surgery. 1990;86(1):1–11. doi: 10.1097/00006534-199007000-00001. [DOI] [PubMed] [Google Scholar]
- 35.Porter JD, Khanna S, Kaminski HJ, Rao JS, Merriam AP, Richmonds CR, et al. A chronic inflammatory response dominates the skeletal muscle molecular signature in dystrophin-deficient mdx mice. Hum Mol Genet. 2002;11(3):263–272. doi: 10.1093/hmg/11.3.263. [DOI] [PubMed] [Google Scholar]
- 36.Gute DC, Ishida T, Yarimizu K, Korthuis RJ. Inflammatory responses to ischemia and reperfusion in skeletal muscle. Mol Cell Biochem. 1998;179(1–2):169–187. doi: 10.1023/a:1006832207864. [DOI] [PubMed] [Google Scholar]
- 37.Kabanov AV, Batrakova EV, Sriadibhatla S, Yang Z, Kelly D, Alakhov VY. Polymer genomics: shifting the gene and drug delivery paradigms. J Contr Rel. 2005;101(1–3):259–271. doi: 10.1016/j.jconrel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 38.Riera M, Chillon M, Aran JM, Cruzado JM, Torras J, Grinyo JM, et al. Intramuscular SP1017-formulated DNA electrotransfer enhances transgene expression and distributes hHGF to different rat tissues. J Gene Med. 2004;6(1):111–118. doi: 10.1002/jgm.463. [DOI] [PubMed] [Google Scholar]
- 39.Babiuk S, Baca-Estrada ME, Foldvari M, Middleton DM, Rabussay D, Widera G, et al. Increased gene expression and inflammatory cell infiltration caused by electroporation are both important for improving the efficacy of DNA vaccines. J Biotechnol. 2004;110(1):1–10. doi: 10.1016/j.jbiotec.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 40.Kabanov AV, Batrakova EV, Alakhov VY. Pluronic block copolymers as novel polymer therapeutics for drug and gene delivery. J Control Release. 2002;82(2–3):189–212. doi: 10.1016/s0168-3659(02)00009-3. [DOI] [PubMed] [Google Scholar]
- 41.Haney MJ, Zhao Y, Harrison EB, Mahajan V, Ahmed S, He Z, et al. Specific transfection of inflamed brain by macrophages: a new therapeutic strategy for neurodegenerative diseases. PLoS One. 2013;8(4):e61852. doi: 10.1371/journal.pone.0061852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Y, Haney MJ, Gupta R, Bohnsack JP, He Z, Kabanov AV, et al. GDNF-transfected macrophages produce potent neuroprotective effects in Parkinson's disease mouse model. PLoS One. 2014;9(9):e106867. doi: 10.1371/journal.pone.0106867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stacey KJ, Sweet MJ, Hume DA. Macrophages ingest and are activated by bacterial DNA. J Immunol. 1996;157(5):2116–2122. [PubMed] [Google Scholar]
- 44.Meuli M, Liu Y, Liggitt D, Kashani-Sabet M, Knauer S, Meuli-Simmen C, et al. Efficient gene expression in skin wound sites following local plasmid injection. J Invest Dermatol. 2001;116(1):131–135. doi: 10.1046/j.1523-1747.2001.00139.x. [DOI] [PubMed] [Google Scholar]
- 45.Daubeuf S, Lindorfer MA, Taylor RP, Joly E, Hudrisier D. The direction of plasma membrane exchange between lymphocytes and accessory cells by trogocytosis is influenced by the nature of the accessory cell. J Immunol. 2010;184(4):1897–1908. doi: 10.4049/jimmunol.0901570. [DOI] [PubMed] [Google Scholar]
- 46.Gutierrez-Vazquez C, Villarroya-Beltri C, Mittelbrunn M, Sanchez-Madrid F. Transfer of extracellular vesicles during immune cell-cell interactions. Immunol Rev. 2013;251(1):125–142. doi: 10.1111/imr.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dumont N, Frenette J. Macrophages protect against muscle atrophy and promote muscle recovery in vivo and in vitro: a mechanism partly dependent on the insulin-like growth factor-1 signaling molecule. The American journal of pathology. 2010;176(5):2228–2235. doi: 10.2353/ajpath.2010.090884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robertson TA, Maley MA, Grounds MD, Papadimitriou JM. The role of macrophages in skeletal muscle regeneration with particular reference to chemotaxis. Exp Cell Res. 1993;207(2):321–331. doi: 10.1006/excr.1993.1199. [DOI] [PubMed] [Google Scholar]
- 50.Godwin JW, Pinto AR, Rosenthal NA. Macrophages are required for adult salamander limb regeneration. Proc Natl Acad Sci U S A. 2013;110(23):9415–9420. doi: 10.1073/pnas.1300290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mesika A, Grigoreva I, Zohar M, Reich Z. A regulated, NFkappaB-assisted import of plasmid DNA into mammalian cell nuclei. Mol Ther. 2001;3(5 Pt 1):653–657. doi: 10.1006/mthe.2001.0312. [DOI] [PubMed] [Google Scholar]
- 52.Ferrari G, Cusella-De Angelis G, Coletta M, Paolucci E, Stornaiuolo A, Cossu G, et al. Muscle regeneration by bone marrow-derived myogenic progenitors. Science. 1998;279(5356):1528–1530. doi: 10.1126/science.279.5356.1528. [DOI] [PubMed] [Google Scholar]
- 53.Saclier M, Cuvellier S, Magnan M, Mounier R, Chazaud B. Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. FEBS J. 2013;280(17):4118–4130. doi: 10.1111/febs.12166. [DOI] [PubMed] [Google Scholar]
- 54.Camargo FD, Green R, Capetanaki Y, Jackson KA, Goodell MA. Single hematopoietic stem cells generate skeletal muscle through myeloid intermediates. Nat Med. 2003;9(12):1520–1527. doi: 10.1038/nm963. [DOI] [PubMed] [Google Scholar]
- 55.Camargo FD, Finegold M, Goodell MA. Hematopoietic myelomonocytic cells are the major source of hepatocyte fusion partners. J Clin Invest. 2004;113(9):1266–1270. doi: 10.1172/JCI21301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willenbring H, Bailey AS, Foster M, Akkari Y, Dorrell C, Olson S, et al. Myelomonocytic cells are sufficient for therapeutic cell fusion in liver. Nat Med. 2004;10(7):744–748. doi: 10.1038/nm1062. [DOI] [PubMed] [Google Scholar]
- 57.Forbes SJ. Myelomonocytic cells are sufficient for therapeutic cell fusion in the liver. J Hepatol. 2005;42(2):285–286. doi: 10.1016/j.jhep.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 58.Kirk S, Oldham J, Kambadur R, Sharma M, Dobbie P, Bass J. Myostatin regulation during skeletal muscle regeneration. J Cell Physiol. 2000;184(3):356–363. doi: 10.1002/1097-4652(200009)184:3<356::AID-JCP10>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 59.Marsala M, Hefferan MP, Kakinohana O, Nakamura S, Marsala J, Tomori Z. Measurement of peripheral muscle resistance in rats with chronic ischemia-induced paraplegia or morphine-induced rigidity using a semi-automated computer-controlled muscle resistance meter. J Neurotrauma. 2005;22(11):1348–1361. doi: 10.1089/neu.2005.22.1348. [DOI] [PubMed] [Google Scholar]
- 60.Pedrazzoli F, Chrysantzas I, Dezzani L, Rosti V, Vincitorio M, Sitar G. Cell fusion in tumor progression: the isolation of cell fusion products by physical methods. Cancer Cell Int. 2011;11:32. doi: 10.1186/1475-2867-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao Y, Alakhova DY, Kim JO, Bronich TK, Kabanov AV. A simple way to enhance Doxil(R) therapy: drug release from liposomes at the tumor site by amphiphilic block copolymer. J Control Release. 2013;168(1):61–69. doi: 10.1016/j.jconrel.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dupuis M, Denis-Mize K, Woo C, Goldbeck C, Selby MJ, Chen M, et al. Distribution of DNA vaccines determines their immunogenicity after intramuscular injection in mice. J Immunol. 2000;165(5):2850–2858. doi: 10.4049/jimmunol.165.5.2850. [DOI] [PubMed] [Google Scholar]
- 63.Bureau MF, Naimi S, Torero Ibad R, Seguin J, Georger C, Arnould E, et al. Intramuscular plasmid DNA electrotransfer: biodistribution and degradation. Biochim Biophys Acta. 2004;1676(2):138–148. doi: 10.1016/j.bbaexp.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 64.Batrakova EV, Kabanov AV. Pluronic block copolymers: evolution of drug delivery concept from inert nanocarriers to biological response modifiers. J Control Release. 2008;130(2):98–106. doi: 10.1016/j.jconrel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Astafieva I, Maksimova I, Lukanidin E, Alakhov V, Kabanov A. Enhancement of the polycation-mediated DNA uptake and cell transfection with Pluronic P85 block copolymer. FEBS letters. 1996;389(3):278–280. doi: 10.1016/0014-5793(96)00607-2. [DOI] [PubMed] [Google Scholar]
- 66.Yang Z, Sahay G, Sriadibhatla S, Kabanov AV. Amphiphilic block copolymers enhance cellular uptake and nuclear entry of polyplex-delivered DNA. Bioconjug Chem. 2008;19(10):1987–1994. doi: 10.1021/bc800144a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sriadibhatla S, Yang Z, Gebhart C, Alakhov VY, Kabanov A. Transcriptional activation of gene expression by pluronic block copolymers in stably and transiently transfected cells. Mol Ther. 2006;13(4):804–813. doi: 10.1016/j.ymthe.2005.07.701. [DOI] [PubMed] [Google Scholar]
- 68.Bianco-Batlles MD, Sosunov A, Polin RA, Ten VS. Systemic inflammation following hind-limb ischemia-reperfusion affects brain in neonatal mice. Dev Neurosci. 2008;30(6):367–373. doi: 10.1159/000164686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lavigne MD, Pohlschmidt M, Novo JF, Higgins B, Alakhov V, Lochmuller H, et al. Promoter dependence of plasmid-pluronics targeted alpha galactosidase A expression in skeletal muscle of Fabry mice. Mol Ther. 2005;12(5):985–990. doi: 10.1016/j.ymthe.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 70.Capoccia BJ, Gregory AD, Link DC. Recruitment of the inflammatory subset of monocytes to sites of ischemia induces angiogenesis in a monocyte chemoattractant protein-1-dependent fashion. J Leukoc Biol. 2008;84(3):760–768. doi: 10.1189/jlb.1107756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cochain C, Rodero MP, Vilar J, Recalde A, Richart AL, Loinard C, et al. Regulation of monocyte subset systemic levels by distinct chemokine receptors controls post-ischaemic neovascularization. Cardiovasc Res. 2010;88(1):186–195. doi: 10.1093/cvr/cvq153. [DOI] [PubMed] [Google Scholar]
- 72.Danson S, Ferry D, Alakhov V, Margison J, Kerr D, Jowle D, et al. Phase I dose escalation and pharmacokinetic study of pluronic polymer-bound doxorubicin (SP1049C) in patients with advanced cancer. British journal of cancer. 2004;90(11):2085–2091. doi: 10.1038/sj.bjc.6601856. [DOI] [PMC free article] [PubMed] [Google Scholar]