Abstract
FLT3 internal tandem duplication (ITD) mutations resulting in constitutive kinase activity are common in acute myeloid leukemia (AML) and carry a poor prognosis. Several agents targeting FLT3 have been developed, but their limited clinical activity suggests that the inhibition of other factors contributing to the malignant phenotype is required. We examined gene expression data sets as well as primary specimens and found that the expression of GLI2, a major effector of the Hedgehog (Hh) signaling pathway, was increased in FLT3-ITD compared to wild type FLT3 AML. To examine the functional role of the Hh pathway, we studied mice in which Flt3-ITD expression results in an indolent myeloproliferative state and found that constitutive Hh signaling accelerated the development of AML by enhancing STAT5 signaling and the proliferation of bone marrow myeloid progenitors. Furthermore, combined FLT3 and Hh pathway inhibition limited leukemic growth in vitro and in vivo, and this approach may serve as a therapeutic strategy for FLT3-ITD AML.
Introduction
Mutations involving internal tandem duplications (ITD) of the FMS-like tyrosine kinase 3 (FLT3) juxtamembrane domain result in constitutive receptor tyrosine kinase (RTK) activity and are clinically associated with high disease burden at diagnosis, short remissions, and poor overall survival (1). FLT3-ITD mutations are typically found in AML along with a wide range of other genetic alterations, suggesting that additional cellular events are required for its full pathogenic effects (2, 3). In transgenic mouse models, the expression of Flt3-ITD as a single genetic lesion within the hematopoietic system causes a gradual expansion of myeloid cells resembling an indolent myeloproliferative neoplasm (MPN), but other genetic events are required for the full development of AML (4–11). Unfortunately, most of these cooperating genetic lesions cannot be therapeutically targeted and therefore, the identification of other cellular pathways activated in FLT3-mutant cells may lead to improved treatment strategies.
The highly conserved Hedgehog (Hh) signaling pathway is required for normal embryonic development and is active in a wide variety of cancers by promoting tumor cell proliferation and survival (12). Signaling is initiated by binding of one of the three mammalian Hh ligands to the cell surface receptor Patched (PTCH1), a 12-pass transmembrane receptor that inhibits the 7-pass transmembrane protein Smoothened (SMO). After Hh ligand binding, SMO is de-repressed and ultimately modulates the three GLI transcriptional regulators and expression of target genes such as CyclinD1 and N-Myc to regulate cell proliferation, survival, and differentiation. Although the pathway is silenced in most post-natal tissues, it is reactivated in several solid tumors, and the clinical activity of the SMO antagonist vismodegib in advanced basal cell carcinoma (aBCC) has confirmed that Hh signaling represents a bona fide anti-cancer target (13).
Aberrant Hh signaling has also been implicated in hematologic malignancies of lymphoid origin, including multiple myeloma and acute lymphoblastic leukemia (14, 15), but its role in myeloid malignancies is less clear. Multiple studies have found that the modulation of Smo activity impacts tumor growth in mouse models of BCR-ABL driven chronic myeloid leukemia (CML) but does not affect the development or propagation of AML induced by the MLL-AF9 fusion gene (16–18). Because crosstalk between Hh and RTK signaling occurs in several systems (19–21) and Hh pathway activation accentuates the oncogenic effects of the BCR-ABL tyrosine kinase, we examined the impact of Hh signaling in FLT3-ITD AML. We found that the expression of GLI2 was increased in FLT3-ITD compared to wild type FLT3 AML cases. Constitutive activation of Hh signaling in the hematopoietic system of mice expressing Flt3-ITD enhanced STAT5 activity, myeloid progenitor expansion, and the development of AML. Furthermore, combined blockade of Hh and FLT3 signaling inhibited leukemic growth in vitro and in vivo. Therefore, aberrant Hh pathway activation promotes the development of AML, and combinatorial therapy with FLT3 and Hh pathway inhibitors may improve the treatment of FLT3-mutated disease.
Results
The expression of the Hh pathway effector GLI2 is increased in FLT3-ITD AML
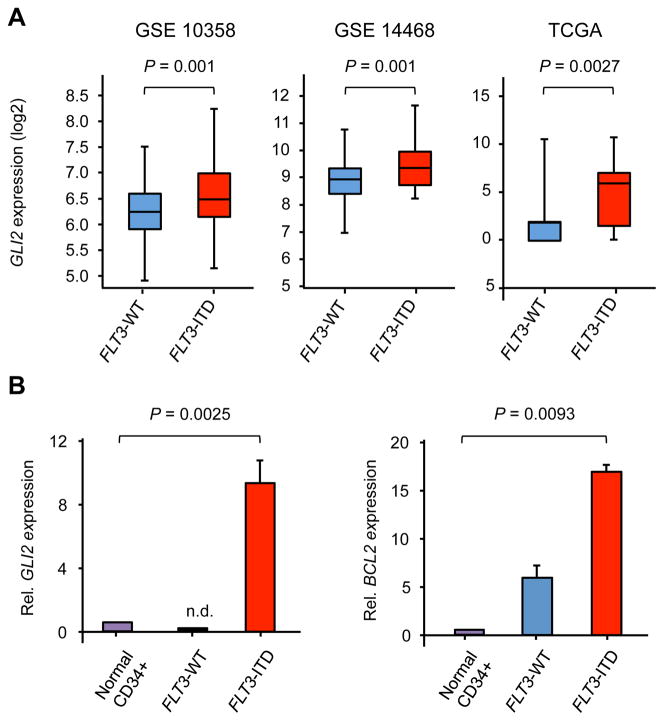
To identify cellular pathways active in FLT3-mutant AML, we initially examined several gene expression profiling (GEP) datasets. Early GEP studies distinguished subgroups of AML on the basis of hierarchical clustering, and a landmark report identified two subgroups enriched in FLT3-ITD mutations (22). Within one of these FLT3-ITD subgroups (cluster 2), the most significantly expressed gene by significance analysis of microarrays (SAM) was GLI2, a critical effector of Hh signaling required for responsiveness to Hh ligand during embryonic development (23–25). We examined three additional AML data sets and similarly found that GLI2 expression was higher in FLT3-ITD compared to wild type FLT3 AML (Figure 1A) (3, 26, 27). Moreover, within the Cancer Genome Atlas (TCGA), the expression of GLI2 in FLT3-ITD cases correlated with the major Hh target gene GLI1, indicative of active Hh signaling (Figure S1A). GLI2 and its target gene BCL2 were also overexpressed in primary FLT3-ITD clinical specimens compared to both wild type FLT3 AML and normal CD34+ hematopoietic stem and progenitor cells (HSPCs) (Figure 1B), whereas GLI1 was highly expressed in both FLT3-ITD and wild type AML (Figure S1B). We also examined FLT3-ITD specimens within TCGA’s data set and found that higher GLI2 (P=0.046), but not GLI1 expression was associated with a shorter median overall survival (Figures S1C and S1D). Therefore, the inferior survival of FLT3-ITD AML cases overexpressing GLI2 suggests that the Hh pathway contributes to the pathogenic impact of this common genetic abnormality.
Figure 1. GLI2 expression is increased in human FLT3-ITD AML.
(A) GLI2 expression in human AML data sets (probe GLI2 – 228537_at) or TCGA. (B) Relative GLI2 and BCL2 expression in primary AML samples compared to normal CD34+ HSPCs (n=5 for HSPCs, wild type FLT3, and FLT3-ITD specimens). Data represent mean ± standard deviation (SD). n.d=not detected.
Hh pathway activation drives the progression of indolent myeloproliferative disease
To examine the functional impact of Hh signaling in FLT3-ITD AML we crossed transgenic Flt3-ITD mice in which an 18 base pair ITD mutation has been knocked into the Flt3 juxtamembrane domain with mice expressing the constitutively active SMO mutant, SmoM2, fused to yellow fluorescent protein (YFP) from the Rosa26 locus (28, 29). Conditional expression of both FLT3-ITD and SmoM2 within the hematopoietic system was induced by Mx1-Cre and poly(I:C) treatment and confirmed by PCR and the detection of YFP in peripheral blood cells (Figures S2A–C) (30). Furthermore, we detected the expression of Gli2 in Mx1-Cre;SmoM2 (SmoM2), Mx1-Cre;Flt3-ITD (Flt3/ITD), and Mx1-Cre;SmoM2;Flt3-ITD (Flt3/ITD-SmoM2) mice, as well as Gli1, a major Hh pathway target, in SmoM2 and Flt3/ITD-SmoM2 mice (Figure S2D). In contrast, Gli1 and Gli2 were not expressed in animals lacking Mx1-cre. The expression of Gli2 in the absence of Gli1 by Flt3/ITD bone marrow cells suggests that GLI2 does not primarily drive pathway activation but allows cells to be responsive to Hh ligands, similar to what happens in the developing neural tube and genital tubercle (31, 32). We also found that recombinant Sonic Hh ligand (SHH) could induce Gli1 expression in bone marrow cells of Flt3/ITD but not wild type mice (Figure S2E).
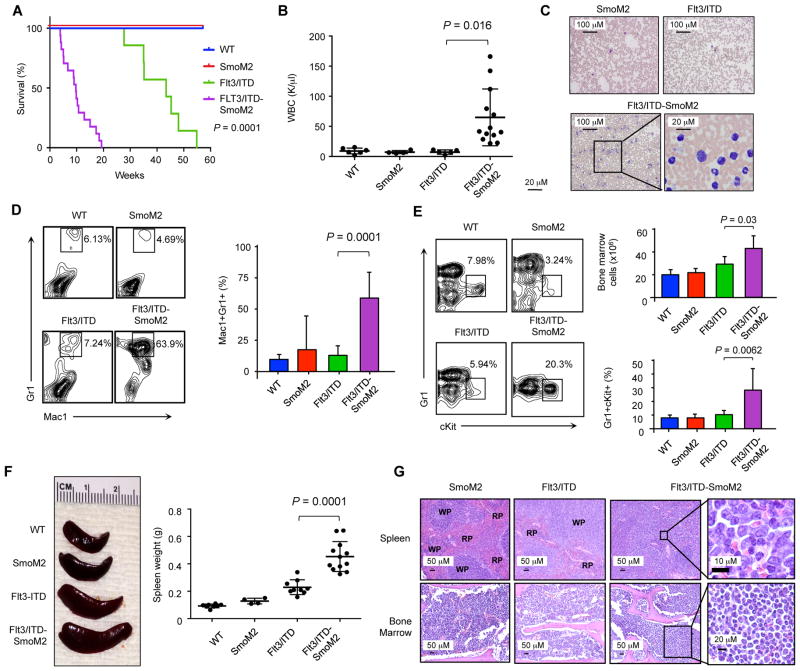
FLT3-ITD mutations primarily occur in de novo AML and are associated with rapidly proliferating disease, but similar to previous findings, heterozygous Flt3-ITD expression in mice resulted in gradually increasing peripheral WBC counts resembling a chronic MPN, with a median lifespan of 40 weeks; the expression of SmoM2 alone did not alter peripheral blood counts or survival compared to wild type controls (Figure 2A) (28). In contrast, the median survival of Flt3/ITD-SmoM2 mice was significantly shorter than Flt3/ITD animals at 12 weeks (P=0.0001) and was associated with elevated peripheral WBC counts and the generation of a new population of Mac1+Gr1int leukemic blasts approximately 3 weeks after poly(I:C) administration (Figures 2B–D). Within the bone marrow, Flt3/ITD-SmoM2 animals demonstrated significantly increased cellularity compared to Flt3/ITD mice, due to the accumulation of an abnormal population of immature c-Kit+Gr-1int myeloid cells (Figure 2E; P=0.03). These cells also infiltrated the spleen, resulting in splenomegaly, as well as non-hematopoietic organs such as the liver and lungs (Figures 2F, 2G, S2F, and Supplemental Table 1). Infiltration within the bone marrow also resulted in the reduction of red blood cell and platelet counts (Figure 2G and S2G). The accelerated death of Flt3/ITD-SmoM2 mice along with increased numbers of myeloid blasts in the bone marrow and spleen, infiltration of myeloid cells into non-hematopoietic organs, and increased number of myeloid cells in the peripheral blood indicate that constitutively active Hh signaling induces disease progression in Flt3/ITD animals to a MPN-like myeloid leukemia based on established criteria (33).
Figure 2. Flt3/ITD-SmoM2 mice develop rapidly fatal AML.
(A) Kaplan Meier survival curve of transgenic animals after poly(I:C) induction. Statistical significance determined by log-rank (Mantel-Cox) test comparing Flt3/ITD and Flt3/ITD-SmoM2 animals. (B) White blood cell counts at 3 months after the completion of poly(I:C) treatment. Data represent mean ± SD. (C) Wright-Giemsa staining of peripheral blood smears. (D) FACS analysis of peripheral blood cells. Bar graph represents percentage of Mac1+Gr1+ cells (n=5–7 per genotype). Data represent mean ± SD. (E) FACS analysis of bone marrow cells. Bar graph depicts bone marrow cellularity and percentage of Gr1+cKit+ cells (n=3–7 per genotype). Data represent mean ± SD. (F) Spleen weights and representative spleen sizes. Data represent mean ± SD. (G) Hematoxylin and eosin staining of bone marrow and spleen sections. RP: red pulp, WP: white pulp.
Myeloid progenitor compartments are expanded in Flt3/ITD-SmoM2 mice
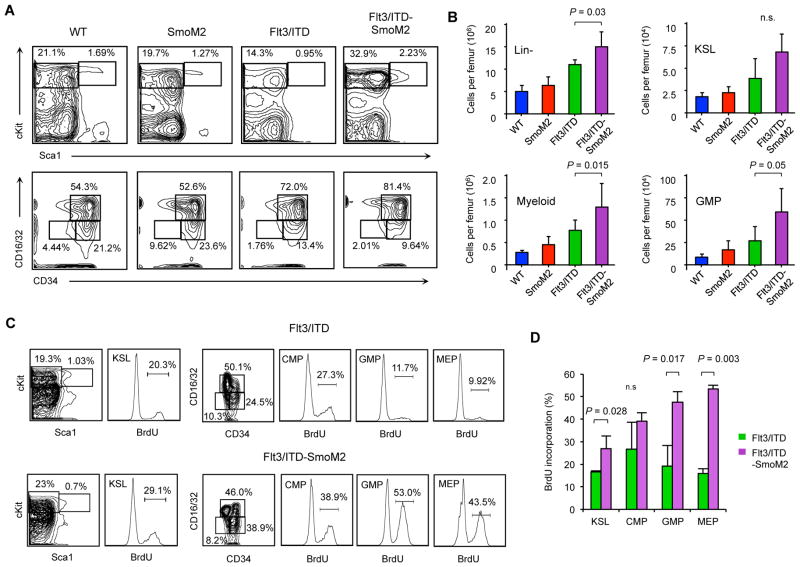
Compared to wild type mice, Flt3/ITD mice display increased numbers of lineage negative (Lin−) multipotent bone marrow cKit+Sca1+ (KSL) HSPCs as well as committed myeloid progenitors (28). In Flt3/ITD-SmoM2 mice, we found that Lin− cells were increased compared to Flt3/ITD mice, but the frequency and total number of KSL cells, long-term hematopoietic stem cells (LT-HSC; KSL CD150+CD48−CD34−), short-term hematopoietic stem cells (ST-HSC; KSL CD34+Flk2−) and multipotent progenitors (MPP; KSL CD34+Flk2+) did not change (Figure 3A, 3B, and S3A). In contrast, cKit+Sca1−Lin− myeloid progenitors were increased by 2-fold in Flt3/ITD-SmoM2 mice, primarily due to the expansion of granulocyte/monocyte progenitors (GMP; cKit+Sca1−Lin−CD34+Flk2+) but not common myeloid (CMP; cKit+Sca1−Lin−CD34+Flk2−) or megakaryocyte/erythrocyte progenitors (MEP; cKit+Sca1−Lin−CD34−Flk2−). Therefore, peripheral blood leukocytosis and the increased frequency of leukemic blasts in Flt3/ITD-SmoM2 mice arise from the relative expansion of the GMP compartment.
Figure 3. Myeloid progenitors are expanded and more proliferative in Flt3/ITD-SmoM2 mice.
(A) Representative FACS plots depicting relative percentages of stem and progenitor KSL (Lin−Sca1+cKit+), myeloid progenitor (Lin−Sca1−cKit+), GMP (Lin−Sca1−cKit+CD34+CD16/32+), CMP (Lin− Sca1−cKit+CD34+CD16/32−), and MEP (Lin−Sca1−cKit+CD34−CD16/32−) cells in bone marrow of transgenic animals. (B) Total number of cells of each specific type per femur (n=3–7 animals per genotype). Data represent mean ± SD. n.s.=non-significant. (C) Representative FACS plots of cells with BrdU incorporation in KSL, CMP, GMP, and MEP compartments. (D) Frequency of BrdU positive cells from each population (n=5). Data represent mean ± SD. n.s.=non-significant.
The proliferation of myeloid progenitors is increased in Flt3/ITD-SmoM2 mice
Both increased FLT3 signaling and Hh pathway activity enhance the proliferation and survival of myeloid leukemia cells (16, 34). To determine the impact of Hh signaling on FLT3-ITD cells, we studied the proliferation of bone marrow cells by quantifying BrdU incorporation. Compared to Flt3/ITD mice, BrdU incorporation in bone marrow cells of Flt3/ITD-SmoM2 mice was similar for most primitive HSPC populations (Figure S3B), but it was increased by greater than 2-fold in GMPs, which were markedly expanded in the bone marrow of Flt3/ITD-SmoM2 mice (Figures 3C and 3D). BrdU incorporation was also increased in KSL cells and MEPs, although total numbers of these cells were not increased in Flt3/ITD-SmoM2 mice. Therefore, Hh pathway activity increases the number of GMPs by increasing their proliferative potential.
Hh signaling impacts malignant hematopoiesis in a cell-intrinsic manner
In some cancers, aberrant Hh pathway activation is mediated by autocrine signaling, in which tumor cells both produce and respond to Hh ligand (14, 35, 36). In other diseases, paracrine signaling has also been described, such that tumor cells secrete Hh ligands and induce pathway activation in non-malignant stromal cells but not tumor cells (37). We initially studied the role of bone marrow stromal cells in the propagation of leukemia from Flt3/ITD-SmoM2 mice by transplanting AML cells into wild-type recipients. However, we could not detect leukemic engraftment 8 weeks after transplantation despite cell doses of up to 10 million cells, similar to previous studies with Flt3-ITD mice (38). Because the Mx1 promoter can be activated by poly(I:C) in multiple cell types, including bone marrow stromal cells (30), we next examined whether the generation of AML by SmoM2 requires cell intrinsic or extrinsic Hh signaling. We transplanted bone marrow from unexcised CD45.2 Flt3/ITD-SmoM2 mice into wild type congenic CD45.1 recipient animals (Figure S4A). After the generation of stable donor blood chimerism, recipient mice were treated with poly(I:C) and developed AML with similar tumor cell phenotype and survival as Flt3/ITD-SmoM2 mice (Figures S4B and 4C). We also detected YFP expression by flow cytometry within CD45.2+ bone marrow hematopoietic cells but not CD45− cells, indicating that stromal cells did not express SmoM2 (Figure S4D). Therefore, Hh signaling enhances AML progression in Flt3/ITD animals in a cell autonomous manner.
Constitutive Smo activity enhances STAT5 signaling in Flt3-ITD cells
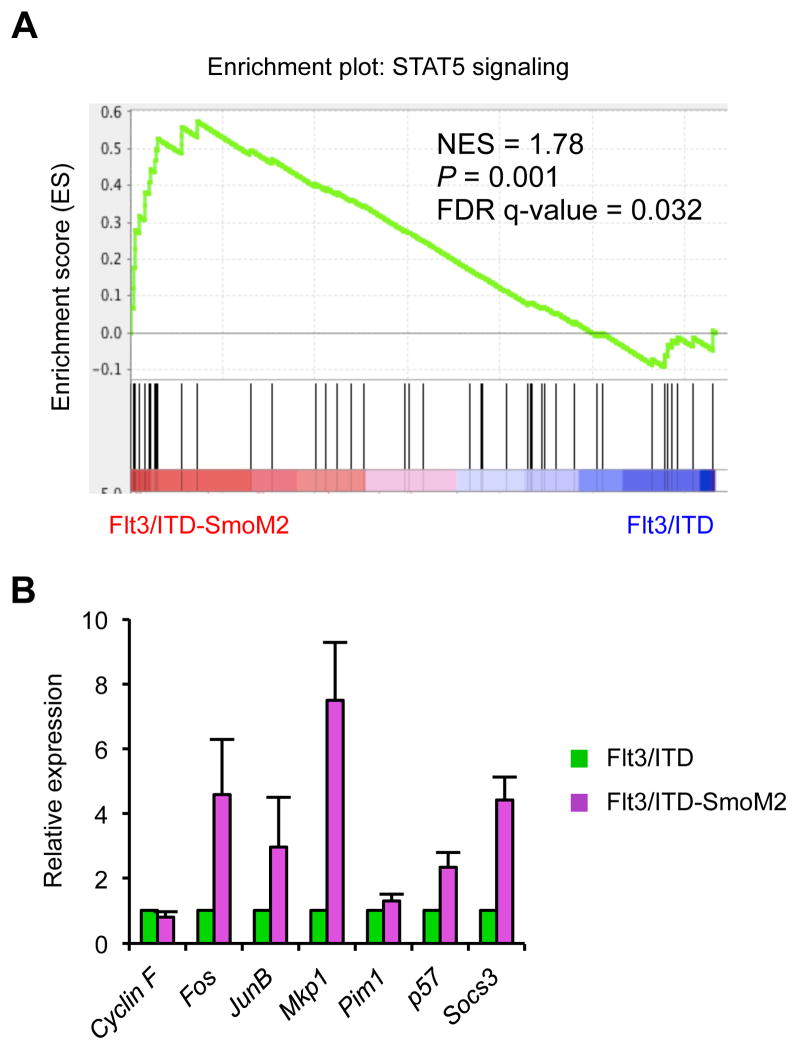
To determine the mechanisms by which Hh pathway activation affects FLT3-ITD signaling, we initially compared the gene expression profiles of isolated KSL and GMP cells from wild type, Flt3/ITD, and Flt3/ITD-SmoM2 mice. We used Gene Set Enrichment Analysis (GSEA) and focused on biological pathways frequently activated during oncogenesis, such as proliferation, survival, and self-renewal (39). Among the top GSEA sets, we identified a gene signature consistent with increased STAT5 signaling in Flt3/ITD compared to wild type animals, as expected from the known role of STAT5 as a major downstream target of FLT3-ITD required for the survival and proliferation of myeloid leukemia cells (Wierenga_STAT5_targets_group2) (Figure S5A). We also used the Analysis of Functional Annotation (AFA) method to further confirm the enrichment of STAT5 signaling functional gene sets (40, 41). Compared to Flt3/ITD mice, the STAT5 signature was further augmented in cells from Flt3/ITD-SmoM2 mice (Figure 4A). To confirm these findings, we quantified the expression of STAT5 target genes and found that several were elevated in GMPs isolated from the bone marrow of Flt3/ITD-SmoM2 mice compared to Flt3/ITD mice (Figure 4B). STAT5 also enhances cell survival, and we found that an anti-apoptotic genetic signature was enriched in Flt3/ITD-SmoM2 compared to Flt3/ITD mice (Anti_apoptosis) (Figure S5B).
Figure 4. STAT5 signaling is enhanced in Flt3/ITD-SmoM2 mice.
(A) GSEA analysis comparing Flt3/ITD and Flt3/ITD-SmoM2 mice. (B) qRT-PCR analysis of STAT5 target genes in sorted GMP cells (n=3). Data represent mean ± SD.
The extent of STAT5 activation corresponds to disease acuity, as evidenced by homozygous Flt3ITD/ITD mice, which experienced a shorter survival than heterozygous Flt3-ITD mice (42, 43). In multiple studies, the transformation of MPN to AML is associated with increased STAT5 signaling driven by loss of the wild-type Flt3 allele to generate hemizygous Flt3ITD/− AML (6–8). We examined both whole bone marrow cells and FACS-isolated GMPs and did not find loss of heterozygosity; both wild type and ITD mutant Flt3 genes were detected in Flt3/ITD-SmoM2 animals (Figure S5C). Therefore, Hh pathway activation in Flt3/ITD-SmoM2 mice promotes increased STAT5 activity and myeloid cell expansion.
Combined FLT3 and Hh pathway inhibition limits AML growth and proliferation in vitro
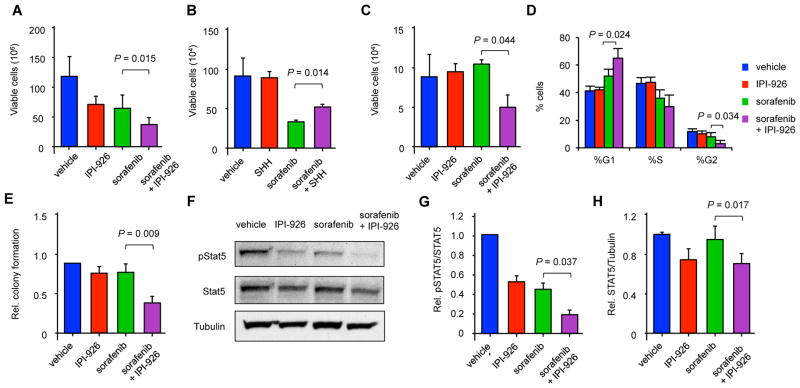
FLT3-ITD mutations carry a relatively poor prognosis in adult AML (1), and several small molecule FLT3 tyrosine kinase inhibitors have undergone clinical testing (44–47). Many of these agents can eliminate leukemic blasts in the peripheral blood, but the clinical responses are transient because tumor cells persist within the bone marrow (48). Seeing that Hh pathway activation enhanced the proliferative effects of FLT3-ITD, we examined whether pathway inhibition could augment the activity of FLT3 antagonists. The MV4-11 and Molm-14 human leukemia cell lines harbor the FLT3-ITD mutation (49), and we found that both expressed Hh signaling pathway components as well as Indian Hh ligand by RT-PCR (Figure S6A). We treated cells with the tyrosine kinase inhibitor sorafenib, which has clinical activity against FLT3-ITD AML (50), and the SMO antagonist IPI-926 to block Hh signaling (51). IPI-926 decreased GLI1 and GLI2 gene expression (Figure S6B) and modestly impacted the growth of either cell line as a single agent (Figures 5A and S6C). However, the anti-proliferative effects of sorafenib against MV4-11 and Molm-14 cells were significantly enhanced by IPI-926 (Figures 5A and S6C, P=0.015 and P=0.006) or knock down of GLI2 by siRNA (Figure S6D). These effects were specific for SMO inhibition, because both a second SMO inhibitor, LDE225, and siRNA against SMO had similar effects to IPI-926 (Figures S6E and S6F). Moreover, further activation of Hh signaling in Molm-14 and MV4-11 cells with recombinant SHH ligand limited the effects of sorafenib (Figures 5B and S6G). Finally, we also found that this combination was active in clinical FLT3-ITD AML bone marrow specimens (Figure 5C).
Figure 5. IPI-926 and sorafenib inhibit leukemic cell growth in vitro.
(A) Viable cell counts of MV4-11 cells treated with IPI-926 and sorafenib for 3 days (n=4). Data represent mean ± SD. (B) Viable cell counts of MV4-11 cell lines treated with recombinant SHH ligand and sorafenib for 3 days (n=3). Data represent mean ± SD. (C) Viable cell counts of clinical FLT3-ITD AML specimens treated with IPI-926 and sorafenib for 3 days (n=4 individual patients). Data represent mean ± SD. (D) Cell cycle distribution of MV4-11 cells treated with IPI-926 and sorafenib after 3 days (n=3). Data represent mean ± SD. (E) Relative colony formation of MV4-11 cells in methylcellulose after treatment with IPI-926 and sorafenib for 3 days compared to vehicle control (n=4). Data represent mean ± SD. (F) Western blot of pSTAT5, total STAT5, and tubulin from MV4-11 cells treated for 3 days. (G) Relative ratio of pSTAT5 to total STAT5 (n=4). Data represent mean ± SD. (H) Relative levels of total STAT5 normalized to tubulin (n=4). Data represent mean ± SD.
Leukemic progression within Flt3/ITD-SmoM2 mice is associated with the increased proliferation of GMPs (Figures 3C and 5D), therefore, we examined the combined effects of sorafenib and IPI-926 on cell cycle progression and the survival of MV4-11 cells. Sorafenib induced an increase in G1 and G2/M cell cycle arrest compared to vehicle treated cells, and the addition of IPI-926 further enhanced this effect (Figure 5D; P=0.024 and P=0.034). Furthermore, this combination inhibited in vitro clonogenic leukemia growth more effectively than either sorafenib or IPI-926 alone (Figures 5E and S6H). At the concentrations used, sorafenib induced a modest amount of apoptosis measured by Annexin V binding, but this effect was not significantly increased with the addition of IPI-926 or in wild type FLT3 HL60 cells that lack GLI2 expression (Figures S6I–K). Therefore, dual FLT3 and SMO inhibition primarily impacts AML proliferation.
Because we found that the enhanced proliferation of GMPs in Flt3/ITD-SmoM2 mice is associated with increased STAT5 activity (Figure 4A), we studied the effects of sorafenib and IPI-926 on STAT5 activity in MV4-11 cells. Sorafenib alone decreased the phosphorylation of STAT5 (pSTAT5), similar to previous reports (52), and it inhibited GLI1 and GLI2 expression (Figure 5F, 5G, and S7A). IPI-926 alone also decreased the ratio of pSTAT5 to total STAT5 as well as decreased the overall expression of total STAT5 (Figure 5G and 5H). Treatment with the combination of sorafenib and IPI-926 further decreased overall pSTAT5 compared to either drug alone, because of both lower expression of total STAT5 as well as a decreased proportion of STAT5 that was phosphorylated (Figure 5F). SHH increased pSTAT5 and limited the effects of sorafenib but did not impact pAKT or pERK, which can also be increased by FLT3 signaling (Figure S7B). To further demonstrate that the effects of combined treatment with sorafenib and IPI-926 were primarily mediated by STAT5, we studied BAF3 cells engineered to express FLT3/ITD (53). In these cells, Interleukin-3 (IL-3) can rescue the effects of FLT3 inhibitors through the induction of STAT5 activation, and we found that it also decreased the effects of sorafenib and IPI-926 on both pSTAT5 and cell growth (Figure S7C and S7D). Therefore, the inhibition of the Hh signaling pathway specifically enhances the anti-leukemic effects of FLT3 tyrosine kinase inhibition associated with the inhibition of STAT5.
Sorafenib and IPI-926 cooperate to inhibit leukemic cell growth in vivo
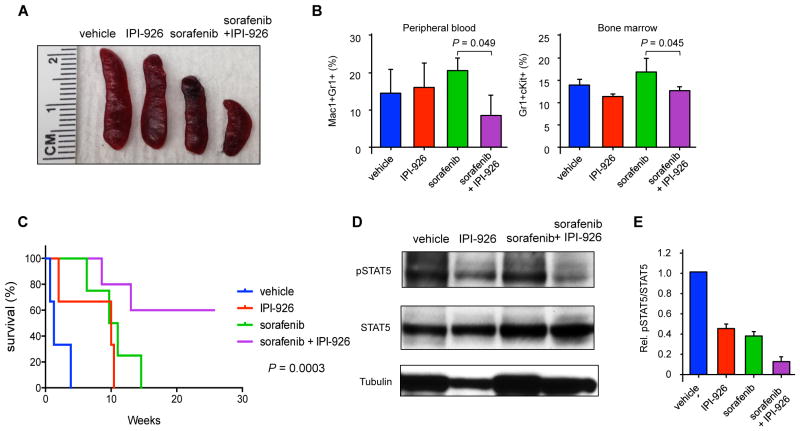
Given the impact of sorafenib and IPI-926 on AML cell lines in vitro, we examined this combination in vivo against Flt3/ITD-SmoM2 derived AML. Wild type recipient mice were engrafted with bone marrow from Flt3/ITD-SmoM2 mice, treated with poly(I:C), and after the development of AML, treated with sorafenib and/or IPI-926 daily for 16 days. Although mutated SmoM2 is relatively resistant to the naturally occurring Smo antagonist cyclopamine, IPI-926 inhibits SmoM2 in vitro and reduced the expression of the Hh target genes Gli1, Gli2, and Ptch1 by bone marrow cells in SmoM2 mice after drug treatment (Figure S8). The combination of sorafenib and IPI-926 reduced splenomegaly compared to either drug alone (Figure 6A). Furthermore, this combination produced a significant reduction in the percentage of Mac1+Gr1+ leukemic cells in the peripheral blood and Gr1+cKit+ cells in the bone marrow (Figure 6B, P=0.049 and P=0.045). Both sorafenib and IPI-926 treatment as single agents increased survival compared to vehicle-treated controls, but the combination further extended survival, with a proportion of the mice failing to succumb to disease despite no further treatment beyond the initial 16-day period (Figure 6C). Similar to our in vitro studies, treatment with IPI-926 and sorafenib decreased both total STAT5 and active pSTAT5 compared to treatment with sorafenib alone (Figures 6D and 6E).
Figure 6. Combined IPI-926 and sorafenib treatment improves survival.
(A) Representative spleen sizes of animals after 16 days of drug treatment. (B) Frequency of abnormal Gr1+Mac1+ and cKit+Gr1+ cells in the peripheral blood and bone marrow, respectively, of animals treated with drugs for 16 days (n=3 per group). Data represented as mean ± SD. (C) Kaplan Meier survival curve of animals treated with IPI-926 and sorafenib (n=3–5 per group). Statistical significance determined by log-rank (Mantel-Cox) test. (D) pSTAT5 and total STAT5 in bone marrow cells harvested after 24 hours of drug treatment. (E) Ratio of pSTAT5 to total STAT5, normalized to vehicle-treated cells (n=3). Data represent mean ± SD.
Discussion
Most newly diagnosed cases of FLT3-mutated AML present with rapidly proliferating disease, but Flt3-ITD expression in mice causes a MPN marked by the gradual elevation of peripheral WBC counts (28). We found that constitutively active SMO cooperates with aberrant FLT3 activity to generate AML, and combined treatment with FLT3 and Hh pathway inhibitors decreased leukemic growth in vitro and in vivo. We also identified increased GLI2 expression in FLT3-ITD clinical AML specimens, suggesting that these findings are clinically relevant. Mutations in Hh pathway components are rare in AML (3), but a recent report also demonstrated that increased wild type spleen tyrosine kinase (SYK) activity can induce the transformation of Flt3-mutant AML (54). Therefore, mutated FLT3 may remain dependent on the activity of non-mutated cellular pathways for AML pathogenesis.
There are limitations to our study. SMO antagonists are clinically effective in aBCC in which Hh signaling is aberrantly activated by PTCH1 and SMO mutations. We used a Smo mutant to constitutively activate the pathway in vivo and found that SMO inhibitors were active when combined with FLT3 inhibition. However, alterations in PTCH1 or SMO are uncommon in most human cancers, including AML (3, 12), and it is possible that SMO inhibition will not be effective in the absence of these pathway-activating mutations. We also used sorafenib to study FLT3 inhibition, but several new FLT3 antagonists, such as quizartinib, are in clinical development (57). Therefore, it is also possible that Hh pathway antagonists will not enhance the activity of these agents because our observed effects were due to the inhibition of other kinases that can be targeted by sorafenib (58), and further studies with next generation FLT3 and Hh pathway inhibitors are needed.
The Hh pathway interacts with several other cellular pathways in a wide range of tumor types (12), and we have identified a functionally relevant interaction between Hh and FLT3 signaling in AML. In several solid tumors, mutant RAS or the activation of RTKs, such as the Epidermal Growth Factor Receptor, results in aberrant RAS/RAF/MEK and PI3K/AKT signaling and the direct activation of GLI1 (56, 59, 60). Although FLT3-ITD is a constitutively active RTK, Gli1 expression was low in bone marrow cells from Flt3/ITD mice at baseline but induced by SHh ligand. In contrast, Gli1 levels did not change in wild type cells, and the differential expression of Gli2 may have restricted Hh ligand responsiveness to Flt3/ITD cells. Therefore, Flt3-ITD may directly or indirectly induce Gli2, which in turn allows canonical signaling by Hh ligand or directly impacts AML cell proliferation. Both wild type and mutant FLT3 cells expressed more GLI1 than normal HSPCs, suggesting that GLI1 activity may be broadly required in AML. Furthermore, because GLI2 was primarily increased in FLT3-ITD cases and RAS mutations are also common in AML (3), GLI1 activation may broadly occur in AML through Hh ligand mediated canonical signaling or direct activation by mutant RAS. Sorafenib alone decreased the growth of FLT3-ITD cells as well as GLI1 and GLI2 expression, therefore, it is also possible that Hh signaling can be at least partially inhibited by FLT3 inhibition alone.
Despite the biologic diversity of mutations that enhance leukemic progression of Flt3/ITD mice, a common mechanism for leukemic progression is the loss of the wild type Flt3 allele by acquired segmental uniparental disomy, resulting in increased tyrosine kinase activity and activation of the major downstream target STAT5 (42, 43). Although loss of heterozygosity of the Flt3 allele did not occur in the Flt3/ITD-SmoM2 mice, we found that STAT5 activity was increased. STAT5 is one of the main downstream targets of aberrant FLT3-ITD signaling, and increased STAT5 activity is associated with shortened overall survival in patients with FLT3-mutated AML (61, 62). The treatment of both human and mouse AML cells with sorafenib and IPI-926 decreased active pSTAT5 by inhibiting both its phosphorylation by FLT3-ITD and total STAT5 expression in response to SMO inhibition. The modulation of STAT5 activity by the Hh signaling pathway has also been described in the development of the mammary gland and brain (63–65), and chromatin immunoprecipitation studies have identified the STAT5a gene within GLI1-bound regions in both medulloblastoma and normal granule neuron precursor cells (65). Because STAT5 signaling is important in the pathogenesis of CML as well as other forms of leukemia (66, 67), it is possible that this interaction may also explain the impact of the Hh pathway on BCR-ABL activity (16, 17).
FLT3 mutations are the most common genetic alterations identified in AML, and several inhibitors of FLT3 kinase activity have undergone clinical testing. Although these inhibitors can eliminate leukemic blasts in the peripheral blood, bone marrow responses are much less frequent (48). The failure of these agents to produce complete remissions may be due in part to the lack of in vivo potency and complete and sustained FLT3 inhibition (68). We found that IPI-926 enhanced the effects of sorafenib on the proliferation and survival of mutant FLT3 AML cells both in vitro and in vivo. Other recent data demonstrated that inhibition of GLI1 and SMO can enhance the activity of the conventional cytotoxic agent cytarabine in AML, suggesting a broad role for the Hh pathway in therapeutic resistance (69). Therefore, SMO antagonists may improve the clinical utility of FLT3 inhibitors for the treatment of AML through multiple mechanisms.
Materials and Methods
Study design
The overall study objective was to determine the role of the Hh signaling pathway in FLT3-ITD AML by analyzing the development of leukemia in a transgenic mouse model with or without the constitutive activation of SMO. Sample size calculations were not performed for initial studies because these were undertaken to identify an interaction between the FLT3 and Hh pathways. Subsequent studies were stopped when double transgenic mice became symptomatic, and littermate controls were analyzed at the same time points. Controlled laboratory experiments also included the analysis of AML cell lines treated with FLT3 and Hh antagonists. Studies were carried out at least in triplicate and no data were excluded in the final analysis. No blinding occurred throughout the study.
Clinical specimens, cell lines, and drug treatment
Bone marrow specimens were obtained from patients with newly diagnosed AML or normal donors granting informed consent as approved by the Johns Hopkins Medical Institutes Institutional Review Board. CD34+ cells were isolated using magnetic microbeads (Miltenyi). Clinical specimens, human FLT3-ITD MV4-11 and Molm-14, and wild type FLT3 HL60 AML cell lines were cultured in RPMI medium supplemented with 10% FBS, 0.5% L-glutamine and 0.5% penicillin/streptomycin at 37°C. Similar culture conditions were used for the BaF3/ITD cell line (53). Treatments consisted of IPI-926 (2.5 μM, Infinity Pharmaceuticals), SHH ligand (10 ng/ml, StemRD), LDE225 (5 μM, LC laboratories), IL-3 (10 ng/ml, Peprotech) and/or sorafenib (LC laboratories) for 3 days. Sorafenib was used at concentrations of 1 nM for MV4-11 cells and 3 nM for clinical specimens, Molm-14, and BaF3/ITD cells. Viable cells were quantified with a hemocytometer and Trypan blue dye exclusion, or in some cases, incubated (500 cells/ml) in methylcellulose at 37°C for 7 days followed by the quantification of tumor cell colonies (>40 cells/colony) using an inverted microscope (Nikon). Cell viability was also assessed using the Cell Proliferation I kit (Roche) and an iMark Microplate reader (BioRad). Cell cycle analysis was carried out by incubating cells in PBS containing 0.6% NP40 (Sigma), 40 μg propidium iodide, and 2 μg/ml RNase (Roche) followed by flow cytometry analysis with a FACS Calibur (BD Biosciences).
Mice
All animal procedures were approved by the Institutional Animal Care and Use Committee. Flt3/ITD mice harboring an ITD mutation knocked into the endogenous Flt3 locus have been previously described (28). SmoM2 and Mx1-Cre mice were obtained from the Jackson Laboratory. Genotyping was determined by PCR (Table S2), and mice harboring alleles heterozygous for SmoM2, Flt3-ITD, and/or Mx1-Cre were obtained. Littermates with single copies of the SmoM2 or Flt3-ITD alleles were used as controls, whereas wild type controls carried the Mx1-Cre allele alone. After weaning and genotyping, mice were treated with 5 doses of poly(I:C) (300 μg, Sigma) i.p. every other day, and successful transgene excision was confirmed 4 weeks after the last poly(I:C) injection by PCR of peripheral blood DNA with primers for the loxP sites (Table S2). Peripheral blood mononuclear cells were also analyzed for YFP expression by flow cytometry (FACSCalibur, BD Biosciences). The development of AML was assessed by the presence of Mac1+Gr1+ cells in the peripheral blood and Gr1+cKit+ cells in the bone marrow by flow cytometry. Mice were sacrificed after the developed lethargy, hunching, and weight loss, and bone marrow was analyzed by flow cytometry. Formalin-fixed tissues were stained with hematoxylin and eosin and blood smears with Wright-Giemsa stain. Complete peripheral blood counts were quantified using a Hemavet 950 analyzer (Drew Scientific).
In vivo drug treatment
IPI-926 was formulated in a 5% β-cyclodextrin (Sigma) solution with deionized H2O, and mice were given 20 mg/kg of IPI-926 or the same volume of 5% β-cyclodextrin solution once a day for 16 days by oral gavage. Sorafenib was formulated in 100% DMSO (Sigma), and mice were given 10 mg/kg of sorafenib or vehicle once a day for 16 days by oral gavage. The vehicle solution for sorafenib consisted of 30% (w/v) Cremophor EL, 30% (w/v) PEG 400, 10% ethanol, and 10% glucose (all Sigma-Aldrich). Some mice were also given the combined treatment of sorafenib and IPI-926. The vehicle treated group received both 5% β-cyclodextrin solution as well as the vehicle solution for sorafenib.
Antibodies and FACS
Monoclonal antibodies against Mac1, Gr1, B220, and CD3 (all from eBioscience) were used for peripheral blood analysis. Bone marrow lineage staining was carried out with biotinylated antibodies against Gr1, Ter119, B220, and CD3 (all from eBioscience). Bone marrow cells were also stained with antibodies against cKit, Sca1, CD16/32, CD34, FLT3, CD150, CD48, and IL7R (all from eBioscience). Flow cytometry and BrdU staining were carried out as previously described (70). Apoptosis was determined by AnnexinV binding (BD Biosciences). Flow cytometry was performed on FACSCalibur or LSRII flow cytometers (BD Biosciences) and analyzed with FlowJo software. Cell sorting was carried out on a FACSAria II (BD Biosciences).
Quantitative Real-time PCR analysis
Total RNA was extracted with the RNeasy Plus Mini kit (Qiagen) and reverse transcribed with Superscript III reverse transcriptase (Invitrogen). qRT-PCR was carried out with Taqman and SYBR Green mastermixes (ABI) according to manufacturer’s protocols (Table S2).
Microarray and GSEA analysis
Analysis of published data sets (3, 26, 27) involved normalized gene expression data, as originally provided by the authors, downloaded from the NCBI Gene Expression Omnibus (GEO) database (GSE10358, GSE14468), the TCGA Research Network (AML) at http://cancergenome.nih.gov, or cBioPortal at http://www.cbioportal.org/public-portal (71, 72). Log2-transformed GLI2 and GLI1 gene expression was compared between FLT3-ITD and wild type FLT3 tumors using a t-test. KSL and GMP cells were isolated by FACS from wild type, Flt3/ITD, and Flt3/ITD-SmoM2 animals at approximately 3 months of age or when Flt3/ITD-SmoM2 animals were moribund. Total RNA was extracted and hybridized onto the Affymetrix Mouse genome 430 2.0 array (Affymetrix) followed by microarray scanning with the Affymetrix GeneChip Scanner and data analysis with the GeneChip Operating Software (Affymetrix). Gene expression data preprocessing and normalization were performed by the Frozen Robust Multiarray Analysis (fRMA) method (73). Gene set enrichment analysis was performed with JAVA-based Gene Set Enrichment Analysis (GSEA) software provided by the Broad Institute of the Massachusetts Institute of Technology (39). The Analysis of Functional Annotation (AFA) was performed as previously described (40, 41). Gene sets were obtained from Molecular Signatures Database (MSigDB, C2 collection). Anti-apoptosis gene set was obtained from the gene ontology collection (GO:0006916). The curated gene set of STAT5 signature in CD34+ hematopoietic stem and progenitor cells was obtained from MSigDB and was previously published (74).
Western Blot
Cell lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis followed by western blotting using rabbit anti-total STAT5 (#9396), pSTAT5-Y694 (#9351), total STAT3 (#12640), total AKT (#9272), pAKT (#9271), total ERK1/2 (#9102), and pERK (#9101), all from Cell Signaling, or tubulin (#21058, AbCam), followed by a horseradish peroxidase–conjugated goat anti-rabbit secondary antibody (#NA934V, Amersham). Film was developed using Amersham ECL Plus™ Western Blotting Detection Reagents (GE Healthcare). Images were obtained with the GS-900™ calibrated densitometer and quantified with Image Lab software (BioRad).
siRNA
MV4-11 and MOLM-14 cell lines were transfected with ON-TARGET plus SMARTpool siRNAs targeting human SMO and GLI2 (Dharmacon), using an Amaxa Nucleofector (Lonza). Successful knockdown of the genes was determined by qRT-PCR 24 hours after transfection, after which cells were treated with sorafenib.
Statistics
Log-rank (Mantel-Cox) test P-values for the Kaplan-Meier survival curves were generated using GraphPad Prism. All other P values were calculated with an unpaired two-tailed Student’s t test, unless otherwise specified. All experiments were performed independently at least three times. Pearson’s R value for correlation and P value was generated in GraphPad Prism. P values <0.05 were considered statistically significant. Original data for patient samples and animal experiments are provided in Table S3.
Supplementary Material
Summary.
Activation of the Hedgehog pathway drives FLT3-mutated leukemia, and dual pathway inhibition effectively inhibits tumor growth.
Acknowledgments
We thank all patients and donors who contributed peripheral blood and/or bone marrow specimens for these studies.
Funding: This study was supported by research grants from the NIH (R01CA127574, R01CA174951, R01CA090668, R21CA155733, P30DK090868, P30CA006973, and UL1TR001079) to W.M., D.S., D.L.H., and L.M. Support was also provided by the Leukemia and Lymphoma Society, the Gabrielle’s Angel Foundation for Cancer Research, the Edward P. Evans Foundation, and the Petre Foundation.
Footnotes
Author contributions: Y.L. and W.M. designed the studies, analyzed the data, and wrote the manuscript. Y.L., L.G., L.L., Q.W., H.M., E.C., D.L.H., S.F., C.D.P., and A.A.M. carried out the studies. K.M., D.N.W., and A.A.M. contributed to the design and performance of some of the studies. L.G. and L.M. carried out the bioinformatics studies. B.D.S. and M.L. provided the clinical specimens. L.G., B.D.S., A.A.M., and D.S. contributed to the study design and writing of the manuscript.
Competing interests: K.M. is an employee of Infinity Pharmaceuticals. The other authors declare that they have no competing interests.
Data and materials availability: Oligonucleotide microarray data have been deposited in the Gene Expression Omnibus under accession number GSE67134. IPI-926 was obtained from Infinity Pharmaceuticals through a materials transfer agreement with Johns Hopkins University.
References and Notes
- 1.Frohling S, Schlenk RF, Breitruck J, Benner A, Kreitmeier S, Tobis K, Dohner H, Dohner K. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: a study of the AML Study Group Ulm. Blood. 2002;100:4372–4380. doi: 10.1182/blood-2002-05-1440. [DOI] [PubMed] [Google Scholar]
- 2.Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, Van Vlierberghe P, Dolgalev I, Thomas S, Aminova O, Huberman K, Cheng J, Viale A, Socci ND, Heguy A, Cherry A, Vance G, Higgins RR, Ketterling RP, Gallagher RE, Litzow M, van den Brink MRM, Lazarus HM, Rowe JM, Luger S, Ferrando A, Paietta E, Tallman MS, Melnick A, Abdel-Wahab O, Levine RL. Prognostic Relevance of Integrated Genetic Profiling in Acute Myeloid Leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stubbs MC, Kim YM, Krivtsov AV, Wright RD, Feng Z, Agarwal J, Kung AL, Armstrong SA. MLL-AF9 and FLT3 cooperation in acute myelogenous leukemia: development of a model for rapid therapeutic assessment. Leukemia. 2008;22:66–77. doi: 10.1038/sj.leu.2404951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim HG, Kojima K, Swindle CS, Cotta CV, Huo Y, Reddy V, Klug CA. FLT3-ITD cooperates with inv(16) to promote progression to acute myeloid leukemia. Blood. 2008;111:1567–1574. doi: 10.1182/blood-2006-06-030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zorko NA, Bernot KM, Whitman SP, Siebenaler RF, Ahmed EH, Marcucci GG, Yanes DA, McConnell KK, Mao C, Kalu C, Zhang X, Jarjoura D, Dorrance AM, Heerema NA, Lee BH, Huang G, Marcucci G, Caligiuri MA. Mll partial tandem duplication and Flt3 internal tandem duplication in a double knock-in mouse recapitulates features of counterpart human acute myeloid leukemias. Blood. 2012;120:1130–1136. doi: 10.1182/blood-2012-03-415067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenblatt S, Li L, Slape C, Nguyen B, Novak R, Duffield A, Huso D, Desiderio S, Borowitz MJ, Aplan P, Small D. Knock-in of a FLT3/ITD mutation cooperates with a NUP98-HOXD13 fusion to generate acute myeloid leukemia in a mouse model. Blood. 2012;119:2883–2894. doi: 10.1182/blood-2011-10-382283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reckzeh K, Bereshchenko O, Mead A, Rehn M, Kharazi S, Jacobsen SE, Nerlov C, Cammenga J. Molecular and cellular effects of oncogene cooperation in a genetically accurate AML mouse model. Leukemia. 2012;26:1527–1536. doi: 10.1038/leu.2012.37. [DOI] [PubMed] [Google Scholar]
- 9.Mallardo M, Caronno A, Pruneri G, Raviele PR, Viale A, Pelicci PG, Colombo E. NPMc+ and FLT3_ITD mutations cooperate in inducing acute leukaemia in a novel mouse model. Leukemia. 2013;27:2248–2251. doi: 10.1038/leu.2013.114. [DOI] [PubMed] [Google Scholar]
- 10.Rau R, Magoon D, Greenblatt S, Li L, Annesley C, Duffield AS, Huso D, McIntyre E, Clohessy JG, Reschke M, Pandolfi PP, Small D, Brown P. NPMc+ cooperates with Flt3/ITD mutations to cause acute leukemia recapitulating human disease. Exp Hematol. 2014;42:101–113 e105. doi: 10.1016/j.exphem.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kats LM, Reschke M, Taulli R, Pozdnyakova O, Burgess K, Bhargava P, Straley K, Karnik R, Meissner A, Small D, Su SM, Yen K, Zhang J, Pandolfi PP. Proto-Oncogenic Role of Mutant IDH2 in Leukemia Initiation and Maintenance. Cell Stem Cell. 2014;14:329–341. doi: 10.1016/j.stem.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMillan R, Matsui W. Molecular pathways: the Hedgehog signaling pathway in cancer. Clin Cancer Res. 2012;18:4883–4888. doi: 10.1158/1078-0432.CCR-11-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Von Hoff D, Lorusso P, Rudin C, Reddy J, Yauch R, Tibes R, Weiss G, Borad M, Hann C, Brahmer J, Mackey H, Lum B, Darbonne W, Marsters J, de Sauvage F, Low J. Inhibition of the Hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med. 2009 doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 14.Peacock CD, Wang Q, Gesell GS, Corcoran-Schwartz IM, Jones E, Kim J, Devereux WL, Rhodes JT, Huff CA, Beachy PA, Watkins DN, Matsui W. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc Natl Acad Sci USA. 2007;104:4048–4053. doi: 10.1073/pnas.0611682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin TL, Wang QH, Brown P, Peacock C, Merchant AA, Brennan S, Jones E, McGovern K, Watkins DN, Sakamoto KM, Matsui W. Self-renewal of acute lymphocytic leukemia cells is limited by the Hedgehog pathway inhibitors cyclopamine and IPI-926. PLoS ONE. 2010;5:e15262. doi: 10.1371/journal.pone.0015262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dierks C, Beigi R, Guo GR, Zirlik K, Stegert MR, Manley P, Trussell C, Schmitt-Graeff A, Landwerlin K, Veelken H, Warmuth M. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell. 2008;14:238–249. doi: 10.1016/j.ccr.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Zhao C, Chen A, Jamieson CH, Fereshteh M, Abrahamsson A, Blum J, Kwon HY, Kim J, Chute JP, Rizzieri D, Munchhof M, VanArsdale T, Beachy PA, Reya T. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458:776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann I, Stover EH, Cullen DE, Mao J, Morgan KJ, Lee BH, Kharas MG, Miller PG, Cornejo MG, Okabe R, Armstrong SA, Ghilardi N, Gould S, de Sauvage FJ, Mcmahon AP, Gilliland DG. Hedgehog signaling is dispensable for adult murine hematopoietic stem cell function and hematopoiesis. Cell Stem Cell. 2009;4:559–567. doi: 10.1016/j.stem.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kessaris N, Jamen F, Rubin LL, Richardson WD. Cooperation between sonic hedgehog and fibroblast growth factor/MAPK signalling pathways in neocortical precursors. Development. 2004;131:1289–1298. doi: 10.1242/dev.01027. [DOI] [PubMed] [Google Scholar]
- 20.Götschel F, Berg D, Gruber W, Bender C, Eberl M, Friedel M, Sonntag J, Rüngeler E, Hache H, Wierling C, Nietfeld W, Lehrach H, Frischauf A, Schwartz-Albiez R, Aberger F, Korf U. Synergism between Hedgehog-GLI and EGFR signaling in Hedgehog-responsive human medulloblastoma cells induces downregulation of canonical Hedgehog-target genes and stabilized expression of GLI1. PLoS ONE. 2013;8:e65403. doi: 10.1371/journal.pone.0065403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnidar H, Eberl M, Klingler S, Mangelberger D, Kasper M, Hauser-Kronberger C, Regl G, Kroismayr R, Moriggl R, Sibilia M, Aberger F. Epidermal growth factor receptor signaling synergizes with Hedgehog/GLI in oncogenic transformation via activation of the MEK/ERK/JUN pathway. Cancer Res. 2009;69:1284–1292. doi: 10.1158/0008-5472.CAN-08-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valk PJM, Verhaak RGW, Beijen MA, Erpelinck CAJ, van Doorn-Khosrovani SBvW, Boer JM, Beverloo HB, Moorhouse MJ, van der Spek PJ, Löwenberg B, Delwel R. Prognostically Useful Gene-Expression Profiles in Acute Myeloid Leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 23.Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci USA. 2009;106:21666–21671. doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regl G, Neill GW, Eichberger T, Kasper M, Ikram MS, Koller J, Hintner H, Quinn AG, Frischauf AM, Aberger F. Human GLI2 and GLI1 are part of a positive feedback mechanism in Basal Cell Carcinoma. Oncogene. 2002;21:5529–5539. doi: 10.1038/sj.onc.1205748. [DOI] [PubMed] [Google Scholar]
- 26.Tomasson MH, Xiang Z, Walgren R, Zhao Y, Kasai Y, Miner T, Ries RE, Lubman O, Fremont DH, McLellan MD. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008;111:4797–4808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taskesen E, Bullinger L, Corbacioglu A, Sanders MA, Erpelinck CA, Wouters BJ, van der Poel-van SC, Damm F, Krauter J, Ganser A. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117:2469–2475. doi: 10.1182/blood-2010-09-307280. [DOI] [PubMed] [Google Scholar]
- 28.Li L, Piloto O, Nguyen HB, Greenberg K, Takamiya K, Racke F, Huso D, Small D. Knock-in of an internal tandem duplication mutation into murine FLT3 confers myeloproliferative disease in a mouse model. Blood. 2008;111:3849–3858. doi: 10.1182/blood-2007-08-109942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D, McMahon AP. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006;66:10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 31.Ding Q, Motoyama J, Gasca S, Mo R, Sasaki H, Rossant J, Hui CC. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development. 1998;125:2533–2543. doi: 10.1242/dev.125.14.2533. [DOI] [PubMed] [Google Scholar]
- 32.Miyagawa S, Matsumaru D, Murashima A, Omori A, Satoh Y, Haraguchi R, Motoyama J, Iguchi T, Nakagata N, Hui C-c, Yamada G. The Role of Sonic Hedgehog-Gli2 Pathway in the Masculinization of External Genitalia. Endocrinology. 2011;152:2894–2903. doi: 10.1210/en.2011-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kogan SC, Ward JM, Anver MR, Berman JJ, Brayton C, Cardiff RD, Carter JS, de Coronado S, Downing JR, Fredrickson TN, Haines DC, Harris AW, Harris NL, Hiai H, Jaffe ES, MacLennan IC, Pandolfi PP, Pattengale PK, Perkins AS, Simpson RM, Tuttle MS, Wong JF, Morse HC., 3rd Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- 34.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 35.Varnat F, Duquet A, Malerba M, Zbinden M, Mas C, Gervaz P, Ruiz i Altaba A. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol Med. 2009;1 doi: 10.1002/emmm.200900039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park KS, Martelotto LG, Peifer M, Sos ML, Karnezis AN, Mahjoub MR, Bernard K, Conklin JF, Szczepny A, Yuan J, Guo R, Ospina B, Falzon J, Bennett S, Brown TJ, Markovic A, Devereux WL, Ocasio CA, Chen JK, Stearns T, Thomas RK, Dorsch M, Buonamici S, Watkins DN, Peacock CD, Sage J. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat Med. 2011;17:1504–1508. doi: 10.1038/nm.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, Marshall D, Fu L, Januario T, Kallop D, Nannini-Pepe M, Kotkow K, Marsters JC, Rubin LL, de Sauvage FJ. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 38.Chu SH, Heiser D, Li L, Kaplan I, Collector M, Huso D, Sharkis Saul J, Civin C, Small D. FLT3-ITD Knockin Impairs Hematopoietic Stem Cell Quiescence/Homeostasis, Leading to Myeloproliferative Neoplasm. Cell Stem Cell. 2012;11:346–358. doi: 10.1016/j.stem.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kortenhorst MS, Wissing MD, Rodriguez R, Kachhap SK, Jans JJ, Van der Groep P, Verheul HM, Gupta A, Aiyetan PO, van der Wall E, Carducci MA, Van Diest PJ, Marchionni L. Analysis of the genomic response of human prostate cancer cells to histone deacetylase inhibitors. Epigenetics. 2013;8:907–920. doi: 10.4161/epi.25574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross AE, Marchionni L, Vuica-Ross M, Cheadle C, Fan J, Berman DM, Schaeffer EM. Gene expression pathways of high grade localized prostate cancer. Prostate. 2011;71:1568–1577. doi: 10.1002/pros.21373. [DOI] [PubMed] [Google Scholar]
- 42.Li L, Bailey E, Greenblatt S, Huso D, Small D. Loss of the wild-type allele contributes to myeloid expansion and disease aggressiveness in FLT3/ITD knockin mice. Blood. 2011;118:4935–4945. doi: 10.1182/blood-2011-01-328096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kharazi S, Mead AJ, Mansour A, Hultquist A, Boiers C, Luc S, Buza-Vidas N, Ma Z, Ferry H, Atkinson D, Reckzeh K, Masson K, Cammenga J, Ronnstrand L, Arai F, Suda T, Nerlov C, Sitnicka E, Jacobsen SE. Impact of gene dosage, loss of wild-type allele, and FLT3 ligand on Flt3-ITD-induced myeloproliferation. Blood. 2011;118:3613–3621. doi: 10.1182/blood-2010-06-289207. [DOI] [PubMed] [Google Scholar]
- 44.Smith BD, Levis M, Beran M, Giles F, Kantarjian H, Berg K, Murphy KM, Dauses T, Allebach J, Small D. Single-agent CEP-701, a novel FLT3 inhibitor, shows biologic and clinical activity in patients with relapsed or refractory acute myeloid leukemia. Blood. 2004;103:3669–3676. doi: 10.1182/blood-2003-11-3775. [DOI] [PubMed] [Google Scholar]
- 45.Stone RM, DeAngelo DJ, Klimek V, Galinsky I, Estey E, Nimer SD, Grandin W, Lebwohl D, Wang Y, Cohen P, Fox EA, Neuberg D, Clark J, Gilliland DG, Griffin JD. Patients with acute myeloid leukemia and an activating mutation in FLT3 respond to a small-molecule FLT3 tyrosine kinase inhibitor, PKC412. Blood. 2005;105:54–60. doi: 10.1182/blood-2004-03-0891. [DOI] [PubMed] [Google Scholar]
- 46.Knapper S, Burnett AK, Littlewood T, Kell WJ, Agrawal S, Chopra R, Clark R, Levis MJ, Small D. A phase 2 trial of the FLT3 inhibitor lestaurtinib (CEP701) as first-line treatment for older patients with acute myeloid leukemia not considered fit for intensive chemotherapy. Blood. 2006;108:3262–3270. doi: 10.1182/blood-2006-04-015560. [DOI] [PubMed] [Google Scholar]
- 47.Fischer T, Stone RM, DeAngelo DJ, Galinsky I, Estey E, Lanza C, Fox E, Ehninger G, Feldman EJ, Schiller GJ, Klimek VM, Nimer SD, Gilliland DG, Dutreix C, Huntsman-Labed A, Virkus J, Giles FJ. Phase IIB Trial of Oral Midostaurin (PKC412), the FMS-Like Tyrosine Kinase 3 Receptor (FLT3) and Multi-Targeted Kinase Inhibitor, in Patients With Acute Myeloid Leukemia and High-Risk Myelodysplastic Syndrome With Either Wild-Type or Mutated FLT3. J Clin Oncol. 2010;28:4339–4345. doi: 10.1200/JCO.2010.28.9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Small D. Targeting FLT3 for the treatment of leukemia. Semin Hematol. 2008;45:S17–21. doi: 10.1053/j.seminhematol.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuo Y, MacLeod R, Uphoff C, Drexler H, Nishizaki C, Katayama Y, Kimura G, Fujii N, Omoto E, Harada M. Two acute monocytic leukemia (AML-M5a) cell lines (MOLM-13 and MOLM-14) with interclonal phenotypic heterogeneity showing MLL-AF9 fusion resulting from an occult chromosome insertion, ins (11; 9)(q23; p22p23) Leukemia. 1997;11:1469–1477. doi: 10.1038/sj.leu.2400768. [DOI] [PubMed] [Google Scholar]
- 50.Zhang W, Konopleva M, Shi YX, McQueen T, Harris D, Ling X, Estrov Z, Quintas-Cardama A, Small D, Cortes J, Andreeff M. Mutant FLT3: a direct target of sorafenib in acute myelogenous leukemia. J Natl Cancer Inst. 2008;100:184–198. doi: 10.1093/jnci/djm328. [DOI] [PubMed] [Google Scholar]
- 51.Tremblay MR, Lescarbeau A, Grogan MJ, Tan E, Lin G, Austad BC, Yu LC, Behnke ML, Nair SJ, Hagel M, White K, Conley J, Manna JD, Alvarez-Diez TM, Hoyt J, Woodward CN, Sydor JR, Pink M, MacDougall J, Campbell MJ, Cushing J, Ferguson J, Curtis MS, McGovern K, Read MA, Palombella VJ, Adams J, Castro AC. Discovery of a potent and orally active hedgehog pathway antagonist (IPI-926) J Med Chem. 2009;52:4400–4418. doi: 10.1021/jm900305z. [DOI] [PubMed] [Google Scholar]
- 52.Auclair D, Miller D, Yatsula V, Pickett W, Carter C, Chang Y, Zhang X, Wilkie D, Burd A, Shi H, Rocks S, Gedrich R, Abriola L, Vasavada H, Lynch M, Dumas J, Trail PA, Wilhelm SM. Antitumor activity of sorafenib in FLT3-driven leukemic cells. Leukemia. 2007;21:439–445. doi: 10.1038/sj.leu.2404508. [DOI] [PubMed] [Google Scholar]
- 53.Tse K, Allebach J, Levis M, Smith B, Bohmer F, Small D. Inhibition of the transforming activity of FLT3 internal tandem duplication mutants from AML patients by a tyrosine kinase inhibitor. Leukemia. 2002;16:2027–2036. doi: 10.1038/sj.leu.2402674. [DOI] [PubMed] [Google Scholar]
- 54.Puissant A, Fenouille N, Alexe G, Pikman Y, Bassil CF, Mehta S, Du J, Kazi JU, Luciano F, Rönnstrand L. SYK is a critical regulator of FLT3 in acute myeloid leukemia. Cancer Cell. 2014;25:226–242. doi: 10.1016/j.ccr.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nolan-Stevaux O, Lau J, Truitt ML, Chu GC, Hebrok M, Fernández-Zapico ME, Hanahan D. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009;23:24–36. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aberger F, Ruiz i Altaba A. Context-dependent signal integration by the GLI code: The oncogenic load, pathways, modifiers and implications for cancer therapy. Semin Cell Dev Biol. 2014;33:93–104. doi: 10.1016/j.semcdb.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levis M. FLT3 mutations in acute myeloid leukemia: what is the best approach in 2013? ASH Education Program Book. 2013;2013:220–226. doi: 10.1182/asheducation-2013.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilhelm S, Carter C, Lynch M, Lowinger T, Dumas J, Smith RA, Schwartz B, Simantov R, Kelley S. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat Rev Drug Discov. 2006;5:835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- 59.Schnidar H, Eberl M, Klingler S, Mangelberger D, Kasper M, Hauser-Kronberger C, Regl G, Kroismayr R, Moriggl R, Sibilia M, Aberger F. Epidermal growth factor receptor signaling synergizes with Hedgehog/GLI in oncogenic transformation via activation of the MEK/ERK/JUN pathway. Cancer Res. 2009;69:1284–1292. doi: 10.1158/0008-5472.CAN-08-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stecca B, Mas C, Clement V, Zbinden M, Correa R, Piguet V, Beermann F, Ruiz i Altaba A. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci USA. 2007;104:5895–5900. doi: 10.1073/pnas.0700776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tse KF, Mukherjee G, Small D. Constitutive activation of FLT3 stimulates multiple intracellular signal transducers and results in transformation. Leukemia. 2000;14:1766–1776. doi: 10.1038/sj.leu.2401905. [DOI] [PubMed] [Google Scholar]
- 62.Brady A, Gibson S, Rybicki L, Hsi E, Saunthararajah Y, Sekeres MA, Tiu R, Copelan E, Kalaycio M, Sobecks R, Bates J, Advani AS. Expression of phosphorylated signal transducer and activator of transcription 5 is associated with an increased risk of death in acute myeloid leukemia. Eur J Haematol. 2012;89:288–293. doi: 10.1111/j.1600-0609.2012.01825.x. [DOI] [PubMed] [Google Scholar]
- 63.Fiaschi M, Rozell B, Bergstrom A, Toftgard R, Kleman MI. Targeted expression of GLI1 in the mammary gland disrupts pregnancy-induced maturation and causes lactation failure. J Biol Chem. 2007;282:36090–36101. doi: 10.1074/jbc.M704280200. [DOI] [PubMed] [Google Scholar]
- 64.Wang J, Pham-Mitchell N, Schindler C, Campbell IL. Dysregulated Sonic hedgehog signaling and medulloblastoma consequent to IFN-alpha-stimulated STAT2-independent production of IFN-gamma in the brain. J Clin Invest. 2003;112:535–543. doi: 10.1172/JCI18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee EY, Ji H, Ouyang Z, Zhou B, Ma W, Vokes SA, McMahon AP, Wong WH, Scott MP. Hedgehog pathway-regulated gene networks in cerebellum development and tumorigenesis. Proc Natl Acad Sci USA. 2010;107:9736–9741. doi: 10.1073/pnas.1004602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moriggl R, Sexl V, Kenner L, Duntsch C, Stangl K, Gingras S, Hoffmeyer A, Bauer A, Piekorz R, Wang D, Bunting KD, Wagner EF, Sonneck K, Valent P, Ihle JN, Beug H. Stat5 tetramer formation is associated with leukemogenesis. Cancer Cell. 2005;7:87–99. doi: 10.1016/j.ccr.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 67.Hoelbl A, Schuster C, Kovacic B, Zhu B, Wickre M, Hoelzl MA, Fajmann S, Grebien F, Warsch W, Stengl G, Hennighausen L, Poli V, Beug H, Moriggl R, Sexl V. Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia. EMBO Mol Med. 2010;2:98–110. doi: 10.1002/emmm.201000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pratz KW, Cortes J, Roboz GJ, Rao N, Arowojolu O, Stine A, Shiotsu Y, Shudo A, Akinaga S, Small D. A pharmacodynamic study of the FLT3 inhibitor KW-2449 yields insight into the basis for clinical response. Blood. 2009;113:3938–3946. doi: 10.1182/blood-2008-09-177030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zahreddine HA, Culjkovic-Kraljacic B, Assouline S, Gendron P, Romeo AA, Morris SJ, Cormack G, Jaquith JB, Cerchietti L, Cocolakis E, Amri A, Bergeron J, Leber B, Becker MW, Pei S, Jordan CT, Miller WH, Borden KL. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature. 2014;511:90–93. doi: 10.1038/nature13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Merchant A, Joseph G, Wang Q, Brennan S, Matsui W. Gli1 regulates the proliferation and differentiation of HSC and myeloid progenitors. Blood. 2010 doi: 10.1182/blood-2009-09-241703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11:242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wierenga AT, Vellenga E, Schuringa JJ. Maximal STAT5-induced proliferation and self-renewal at intermediate STAT5 activity levels. Mol Cell Biol. 2008;28:6668–6680. doi: 10.1128/MCB.01025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.