Abstract
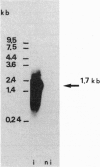
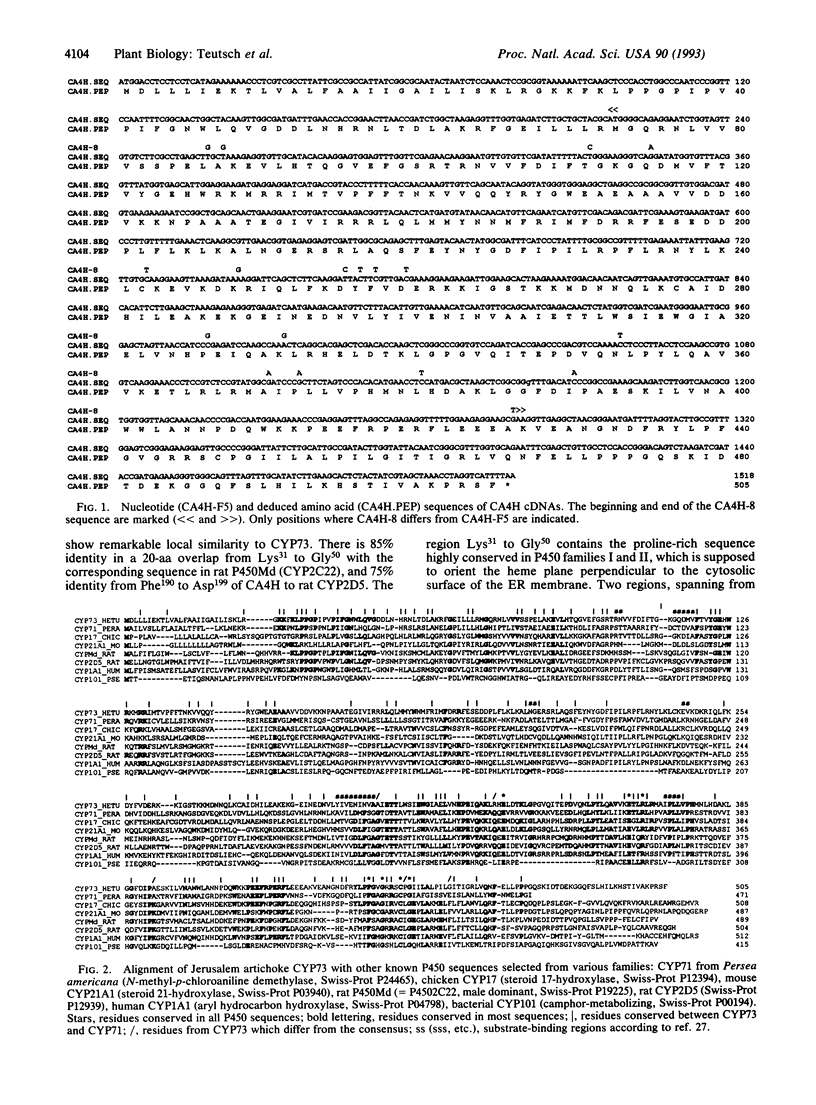
Cinnamate 4-hydroxylase [CA4H; trans-cinnamate,NADPH:oxygen oxidoreductase (4-hydroxylating), EC 1.14.13.11] is a cytochrome P450 that catalyzes the first oxygenation step of the general phenylpropanoid metabolism in higher plants. The compounds formed are essential for lignification and defense against predators and pathogens. We recently reported the purification of this enzyme from Mn(2+)-induced Jerusalem artichoke (Helianthus tuberosus L.) tuber tissues. Highly selective polyclonal antibodies raised against the purified protein were used to screen a lambda gt11 cDNA expression library from wound-induced Jerusalem artichoke, allowing isolation of a 1130-base-pair insert. Typical P450 domains were identified in this incomplete sequence, which was used as a probe for the isolation of a 1.7-kilobase clone in a lambda gt10 library. A full-length open reading frame of 1515 base pairs, encoding a P450 protein of 505 residues (M(r) = 57,927), was sequenced. The N terminus, essentially composed of hydrophobic residues, matches perfectly the microsequenced N terminus of the purified protein. The calculated pI is 9.78, in agreement with the chromatographic behavior and two-dimensional electrophoretic analysis of CA4H. Synthesis of the corresponding mRNA is induced in wounded plant tissues, in correlation with CA4H enzymatic activity. This P450 protein exhibits the most similarity (28% amino acid identity) with avocado CYP71, but also good similarity with CYP17 and CYP21, or with CYP1 and CYP2 families. According to current criteria, it qualifies as a member of a new P450 family.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bozak K. R., Yu H., Sirevåg R., Christoffersen R. E. Sequence analysis of ripening-related cytochrome P-450 cDNAs from avocado fruit. Proc Natl Acad Sci U S A. 1990 May;87(10):3904–3908. doi: 10.1073/pnas.87.10.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Oba K., Uritani I. Properties of a Mixed Function Oxygenase Catalyzing Ipomeamarone 15-Hydroxylation in Microsomes from Cut-Injured and Ceratocystis fimbriata-Infected Sweet Potato Root Tissues. Plant Physiol. 1982 Aug;70(2):573–578. doi: 10.1104/pp.70.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriac B., Werck-Reichhart D., Teutsch H., Durst F. Purification and immunocharacterization of a plant cytochrome P450: the cinnamic acid 4-hydroxylase. Arch Biochem Biophys. 1991 Jul;288(1):302–309. doi: 10.1016/0003-9861(91)90199-s. [DOI] [PubMed] [Google Scholar]
- Gonzalez F. J., Nebert D. W. Evolution of the P450 gene superfamily: animal-plant 'warfare', molecular drive and human genetic differences in drug oxidation. Trends Genet. 1990 Jun;6(6):182–186. doi: 10.1016/0168-9525(90)90174-5. [DOI] [PubMed] [Google Scholar]
- Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992 Jan 5;267(1):83–90. [PubMed] [Google Scholar]
- Hagmann M. L., Heller W., Grisebach H. Induction and characterization of a microsomal flavonoid 3'-hydroxylase from parsley cell cultures. Eur J Biochem. 1983 Aug 15;134(3):547–554. doi: 10.1111/j.1432-1033.1983.tb07601.x. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989 Apr;5(2):151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- Janin J., Wodak S. Conformation of amino acid side-chains in proteins. J Mol Biol. 1978 Nov 5;125(3):357–386. doi: 10.1016/0022-2836(78)90408-4. [DOI] [PubMed] [Google Scholar]
- Kalb V. F., Loper J. C. Proteins from eight eukaryotic cytochrome P-450 families share a segmented region of sequence similarity. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7221–7225. doi: 10.1073/pnas.85.19.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochs G., Werck-Reichhart D., Grisebach H. Further characterization of cytochrome P450 involved in phytoalexin synthesis in soybean: cytochrome P450 cinnamate 4-hydroxylase and 3,9-dihydroxypterocarpan 6a-hydroxylase. Arch Biochem Biophys. 1992 Feb 14;293(1):187–194. doi: 10.1016/0003-9861(92)90383-8. [DOI] [PubMed] [Google Scholar]
- Nebert D. W., Nelson D. R., Coon M. J., Estabrook R. W., Feyereisen R., Fujii-Kuriyama Y., Gonzalez F. J., Guengerich F. P., Gunsalus I. C., Johnson E. F. The P450 superfamily: update on new sequences, gene mapping, and recommended nomenclature. DNA Cell Biol. 1991 Jan-Feb;10(1):1–14. doi: 10.1089/dna.1991.10.1. [DOI] [PubMed] [Google Scholar]
- Nelson D. R., Strobel H. W. Evolution of cytochrome P-450 proteins. Mol Biol Evol. 1987 Nov;4(6):572–593. doi: 10.1093/oxfordjournals.molbev.a040471. [DOI] [PubMed] [Google Scholar]
- Nelson D. R., Strobel H. W. On the membrane topology of vertebrate cytochrome P-450 proteins. J Biol Chem. 1988 May 5;263(13):6038–6050. [PubMed] [Google Scholar]
- Nelson D. R., Strobel H. W. Secondary structure prediction of 52 membrane-bound cytochromes P450 shows a strong structural similarity to P450cam. Biochemistry. 1989 Jan 24;28(2):656–660. doi: 10.1021/bi00428a036. [DOI] [PubMed] [Google Scholar]
- O'Keefe D. P., Bozak K. R., Christoffersen R. E., Tepperman J. A., Dean C., Harder P. A. Endogenous and engineered cytochrome P-450 mono-oxygenases in plants. Biochem Soc Trans. 1992 May;20(2):357–361. doi: 10.1042/bst0200357. [DOI] [PubMed] [Google Scholar]
- O'keefe D. P., Leto K. J. Cytochrome P-450 from the Mesocarp of Avocado (Persea americana). Plant Physiol. 1989 Apr;89(4):1141–1149. doi: 10.1104/pp.89.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picado-Leonard J., Miller W. L. Homologous sequences in steroidogenic enzymes, steroid receptors and a steroid binding protein suggest a consensus steroid-binding sequence. Mol Endocrinol. 1988 Nov;2(11):1145–1150. doi: 10.1210/mend-2-11-1145. [DOI] [PubMed] [Google Scholar]
- Poulos T. L., Finzel B. C., Howard A. J. High-resolution crystal structure of cytochrome P450cam. J Mol Biol. 1987 Jun 5;195(3):687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- Raag R., Martinis S. A., Sligar S. G., Poulos T. L. Crystal structure of the cytochrome P-450CAM active site mutant Thr252Ala. Biochemistry. 1991 Dec 3;30(48):11420–11429. doi: 10.1021/bi00112a008. [DOI] [PubMed] [Google Scholar]
- Reichhart D., Salaün J. P., Benveniste I., Durst F. Time Course of Induction of Cytochrome P-450, NADPH-Cytochrome c Reductase, and Cinnamic Acid Hydroxylase by Phenobarbital, Ethanol, Herbicides, and Manganese in Higher Plant Microsomes. Plant Physiol. 1980 Oct;66(4):600–604. doi: 10.1104/pp.66.4.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shen S., Strobel H. W. The role of cytochrome P450 lysine residues in the interaction between cytochrome P450IA1 and NADPH-cytochrome P450 reductase. Arch Biochem Biophys. 1992 Apr;294(1):83–90. doi: 10.1016/0003-9861(92)90140-r. [DOI] [PubMed] [Google Scholar]
- Vetter H. P., Mangold U., Schröder G., Marner F. J., Werck-Reichhart D., Schröder J. Molecular Analysis and Heterologous Expression of an Inducible Cytochrome P-450 Protein from Periwinkle (Catharanthus roseus L.). Plant Physiol. 1992 Oct;100(2):998–1007. doi: 10.1104/pp.100.2.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]