Abstract
Endovascular infections caused by Staphylococcus aureus involve interactions with fibronectin present as extracellular matrix or surface ligand on host cells. We examined the expression, structure, and binding activity of the two major S. aureus fibronectin-binding proteins (FnBPA, FnBPB) in 10 distinct, methicillin-resistant clinical isolates from patients with either persistent or resolving bacteremia. The persistent bacteremia isolates (n = 5) formed significantly stronger bonds with immobilized fibronectin as determined by dynamic binding measurements performed with atomic force microscopy. Several notable differences were also observed when the results were grouped by clonal complex 5 (CC5) strains (n = 5) versus CC45 strains (n = 5). Fibronectin-binding receptors on CC5 formed stronger bonds with immobilized fibronectin (P < 0.001). The fnbA gene was expressed at higher levels in CC45, whereas fnbB was found in only CC5 isolates. The fnbB gene was not sequenced because all CC45 isolates lacked this gene. Instead, comparisons were made for fnbA, which was present in all 10 isolates. Sequencing of fnbA revealed discrete differences within high-affinity, fibronectin-binding repeats (FnBRs) of FnBPA that included (i) 5-amino-acid polymorphisms in FnBR-9, FnBR-10, and FnBR-11 involving charged or polar side chains, (ii) an extra, 38-amino-acid repeat inserted between FnBR-9 and FnBR-10 exclusively seen in CC45 isolates, and (iii) CC5 isolates had the SVDFEED epitope in FnBR-11 (a sequence shown to be essential for fibronectin binding), while this sequence was replaced in all CC45 isolates with GIDFVED (a motif known to favor host cell invasion at the cost of reduced fibronectin binding). These complementary sequence and binding data suggest that differences in fnbA and fnbB, particularly polymorphisms and duplications in FnBPA, give S. aureus two distinct advantages in human endovascular infections: (i) FnBPs similar to that of CC5 enhance ligand binding and foster initiation of disease, and (ii) CC45-like FnBPs promote cell invasion, a key attribute in persistent endovascular infections.
INTRODUCTION
Staphylococcus aureus interaction with host fibronectin (Fn), present as either an extracellular matrix ligand or surface protein on host cells (e.g., endothelial cells), is often cited as a key event in the pathogenesis of endovascular infections (e.g., infective endocarditis) (1–5). Most clinical S. aureus strains express two distinct fibronectin-binding proteins (FnBPs), FnBPA and FnBPB. FnBPA has been shown to be an important virulence factor (5–7) that contributes to pathogenesis through various mechanisms, including both binding to and invasion of endothelial cells (3, 8, 9); the latter event has in turn been linked to S. aureus persistence on cardiac valves in experimental endocarditis (7). The structure of FnBPA contains 11 nonidentical repeats (Fn-binding repeats [FnBRs]) that each bind Fn with different affinities (10). FnBR-1, -4, -5, -9, -10, and -11 exhibit the highest affinities for the Fn ligand (11). Although less is known about the clinical importance of FnBPB, studies suggest that it, too, plays a role in binding to Fn as it has the same number (six) of high-affinity Fn-binding sites as FnBPA (11).
In previous work, we showed that amino acid polymorphisms in FnBR-5 and FnBR-9 of FnBPA are linked to S. aureus cardiac device infections (CDIs) in U.S. patients (12). This was recently corroborated by another study of European patients with S. aureus CDIs (13). In the present study, we examine whether such polymorphisms in FnBPs are associated with another common, but distinct, S. aureus syndrome, persistent bacteremia (despite appropriate antibiotics). In this syndrome, the presence of an indwelling cardiac device was not a prerequisite for patient selection. Ten isolates of methicillin-resistant S. aureus from a multinational, antibiotic trial of S. aureus bacteremia were selected based on (i) the persistence of bacteremia in response to seemingly appropriate antibiotic treatment (based on in vitro susceptibility profiles) and (ii) distinct genetic lineages. In terms of the former, five isolates each came from patients with either persistent bacteremia (PB) or resolving bacteremia (RB) upon appropriate antibiotic therapy (14). Five isolates each were typed as clonal complex 5 (CC5) or CC45. We focused on these CCs because CC5 is a leading cause of hospital-acquired methicillin-resistant S. aureus (MRSA) associated with complicated infections (15–17), whereas CC45 is a predominant colonizer of healthy individuals, and is also found in bloodstream infections associated with suboptimal clinical outcomes in Europe (Berlin-IV) and the United States (USA600) (18, 19). By strategically selecting samples from opposite ends of a continuum of clinical readouts and clonotypes (PB/RB and CC5/45, respectively), we postulated that this would provide the best opportunity to test linkage between FnBPs and differential therapeutic outcomes in non-CDI S. aureus bacteremia.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
As noted above, 10 MRSA strains in the present study were obtained from a multinational trial comparing daptomycin versus standard antibiotic therapy in S. aureus bacteremia and right-sided infective endocarditis (14). These isolates were classified as CC5 or CC45 by multilocus sequence typing (Table 1). Persistent bacteremia (PB) isolates were obtained from patients demonstrating ≥7 days of positive blood cultures despite appropriate antibiotic therapy based on in vitro susceptibility profiles (20). Resolving bacteremia (RB) isolates were from patients in whom the initial bacteremia resolved rapidly (within 2 to 4 days of antibiotic therapy) (21, 22) (see Table 1). All bacterial testing was performed on the initial blood isolate only. Unless otherwise specified, bacteria were grown overnight in tryptic soy broth (TSB) at 37°C after thawing from −80°C storage.
TABLE 1.
S. aureus clinical strains used in this studya
| Strain | CC type | PB or RB detected in human patients |
|---|---|---|
| 300-087 | 45 | RB |
| 300-103 | 45 | RB |
| 301-188 | 45 | RB |
| 300-169 | 45 | PB |
| 324-136 | 45 | PB |
| 010-016 | 5 | RB |
| 088-237 | 5 | RB |
| 077-107 | 5 | PB |
| 088-180 | 5 | PB |
| 300-246 | 5 | PB |
Data for the persistent (PB) and resolving (RB) bacteremia strains examined in this study are shown. PB isolates were obtained from patients demonstrating ≥7 days of positive blood cultures despite appropriate antibiotic therapy based on in vitro susceptibility profiles (20). RB isolates were obtained from patients in whom the initial bacteremia resolved within 2 to 4 days of antibiotic therapy (21, 22). CC, clonal complex.
Transcription of fnbA Northern blot analysis.
The relative expression of fnbA (gene encoding FnBPA) among the strain cohorts was assessed using Northern blot analysis as described earlier (23) with the exception that 10 μg (rather than 5 μg) was separated by electrophoresis. Cells were harvested by centrifugation (3,200 rpm) at 3 or 6 h postinoculation corresponding to the early and late exponential phases of growth at 37°C. These time points encompass the expected maximal expression window for fnb genes cited in the literature (24). Digoxigenin (DIG)-labeled fnbA DNA probes were synthesized by using a PCR-based DIG probe synthesis kit (Roche Applied Science, Indianapolis, IN), a primer pair (forward primer, 5′-TGGATTTGATTCCTCAGAGG-3′; reverse primer, 5′-CGGAAATGAGAAAAATGGTCCG-3′) that spans the region most dissimilar to fnbB, and a gel-purified fnbA fragment excised from a cloned plasmid as the template.
qRT-PCR of fnbA and fnbB.
The expression levels of fnbA and fnbB (gene for FnBPB) were determined at two growth phases (early and late exponential) by quantitative reverse transcription-PCR (qRT-PCR), as described previously (19). Reaction mixtures were prepared using 100 nM concentrations of the following primers: fnbA-F, 5′-CCTGGGTTTGTATCTTC-3′; fnbA-R, 5′-CCACACAGCTATAGATGG-3′; fnbB-F, 5′-CAACCAGTCGTTAAGCTCTGTGAC-3′; and fnbB-R, 5′-GCTGACATCATCAAGCTTTGC-3′. gyrB was used to normalize for transcript quantification, since it is not affected by cell density or quorum sensing (19). qRT-PCR experiments were performed using two biological replicates, each tested in triplicate.
DNA sequence analysis of Fn-binding region (FnBR) of the fnbA locus.
The PCR product was used as the template for sequencing reactions. This template was generated from genomic DNA extracted from S. aureus cultures, with fnbA-F and fnbA-R primers (see Table S1 in the supplemental material). Genomic DNA was extracted and processed as described previously (25). PCRs were performed using high-fidelity Platinum Taq DNA polymerase (Invitrogen) in a 50-μl reaction mixture. The PCR condition was comprised of an initial denaturation step (2 min at 95°C), followed by 35 cycles of amplification (denaturation for 30 s at 95°C, annealing for 30 s at 60°C, and extension for 4 min at 68°C), with a final extension of 10 min. A total of 5 μl of the PCR product was visualized in an ethidium bromide-stained 2% agarose gel to determine its purity and concentration. The remainder was purified with QIAquick columns (Qiagen), eluted in 30 μl of EB buffer (Qiagen), and stored at −20°C. Sequencing reactions were performed using four pairs of sequencing primers to cover all 11 Fn-binding domains as shown in Table S1 in the supplemental material. Finally, sequence alignment was performed with Vector NTI. DNA contigs were aligned with Vector NTI contig express, and the consensus sequence was translated to an amino acid sequence. The generated amino acid sequence was subsequently aligned by the CLUSTAL W method and compared to the reference amino acid sequence obtained from the reference strain, S. aureus NCTC 8325.
Confirmation of fnbB gene and surface expression of FnBPA and FnBPB.
The prevalence of the fnbB gene on these 10 isolates was determined by using standard PCR techniques as described above and the primer pair fnbB-F (5′-CTTTACCTTGTTCCACTGGTTTAGAAG-3′) and fnbB-R (5′-GGGAGTAACAGCTAATGGTCG-3′). The surface expression of FnBPs on all ten S. aureus isolates was confirmed by Western ligand blotting, as described previously (26).
AFM.
The strength of the interaction between Fn and FnBPs among the strain cohorts was assessed by using atomic force microscopy (AFM; a Veeco/Digital Instruments Bioscope AFM and a NanoSCOPE IV controller) as described previously (27, 28). AFM allows dynamic measurement of the strength of a binding reaction directly on single cells or molecules, as opposed to more indirect, static measures of adhesion (e.g., counting numbers of cells on a Fn-coated slide). Briefly, bacteria were harvested at the exponential growth phase (optical density at 600 nm = 0.51 ± 0.02) and then plated onto Fn-coated slides (BD Biosciences). An Fn-coated cantilever tip (radius ∼20 nm) was brought into contact with individual bacterial cells and then retracted from contact with the outer cell surface (see Fig. S1 in the supplemental material). Each approach-retraction cycle took ∼1 s. Hundreds of force spectra were collected and analyzed for each isolate (13,751 total force spectra). Data acquisition was confined to a narrow time-window after harvesting (<30 min) to minimize data collection on quiescent bacteria. As a key control experiment, force spectra were also collected on the nonpathogenic, Gram-positive organism Lactococcus lactis that was genetically engineered to express fnbA and fnbB, as previously detailed (6). Native L. lactis is an ideal control organism because its outer envelope is similar to that of S. aureus (both are Gram positive), but it lacks Fn-binding proteins such as FnBPA and FnBPB.
Nucleotide sequence accession numbers.
The nucleotide sequences reported here have been deposited in the GenBank database under accession numbers KF956095 to KF956099.
RESULTS
Dynamic binding reactions probed with AFM.
Single-cell AFM experiments were carried out for each of the 10 S. aureus strains shown in Table 1. Figure 1 shows a few representative force spectra, from thousands of force curves that were obtained by “tapping” a Fn-coated tip on single cells from the CC5 and CC45 clonotype groups. Binding events of Fn on single S. aureus cells resulted in a distinct, nonlinear response as the Fn-coated tip was pulled away from the bacterial cell surface. We confirmed that these interactions were, in fact, specifically due to the bacterial FnBPs, as the interaction spectra for L. lactis engineered to express FnBPA and FnBPB (see Fig. S2 in the supplemental material) parallel those of the S. aureus strains. As previously described by us (28), the shapes of these “unbinding” events originate from the unraveling and eventual unbinding (or “rupture”) of Fn-FnBP complexes.
FIG 1.
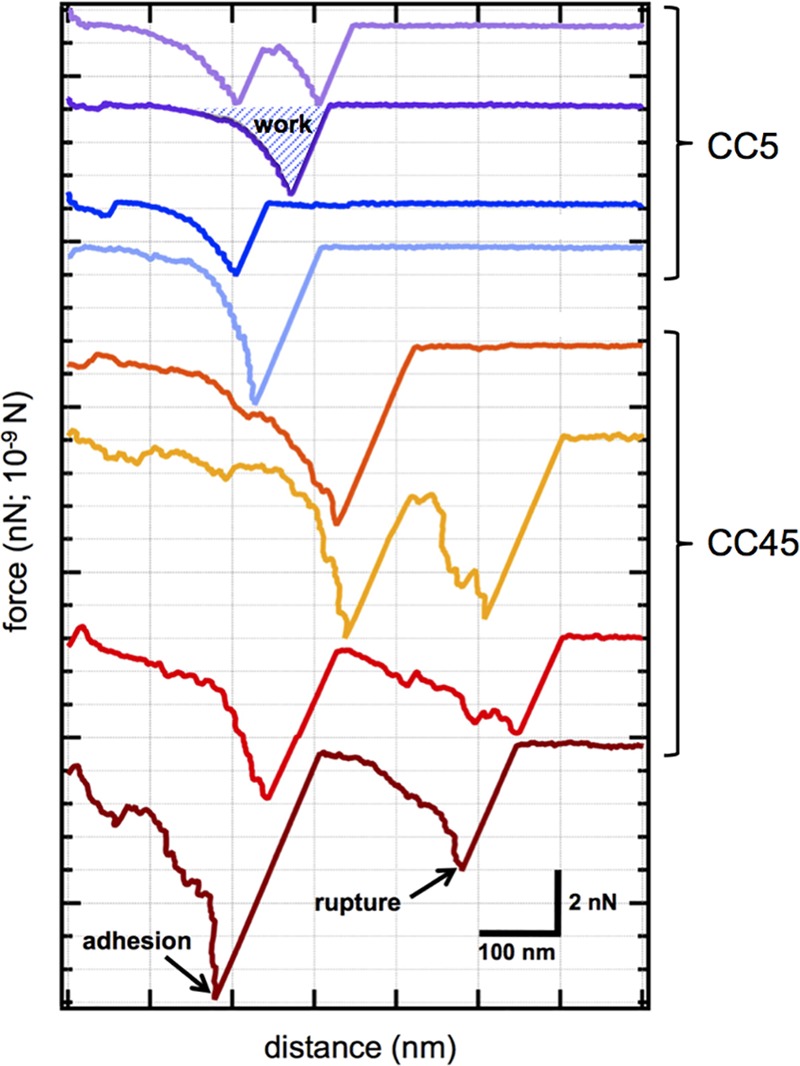
AFM binding force profiles. Examples of AFM force spectra from CC5 (blue and purple curves) and CC45 (red and orange curves) strains as probed with a fibronectin-coated tip. These curves were randomly selected from thousands of force spectra and illustrate the range of binding phenomena that were observed with AFM. From left to right on the x axis, the tip was pulled from contact with the exterior cell wall of individual living cells. For each spectrum, we determined the adhesion force (the maximum force value in a spectrum) and the rupture force (the force associated with the final event before complete separation). For a spectrum with more than one local minima, the adhesion and rupture forces could differ (e.g., see the dark red curve). The energy or work of binding was determined by integrating force with respect to distance for individual spectra (e.g., see the purple curve). Cell samples were immersed in saline buffer during the experiments. Scale bars correspond to 2 nN in force and 100 nm in extension distance.
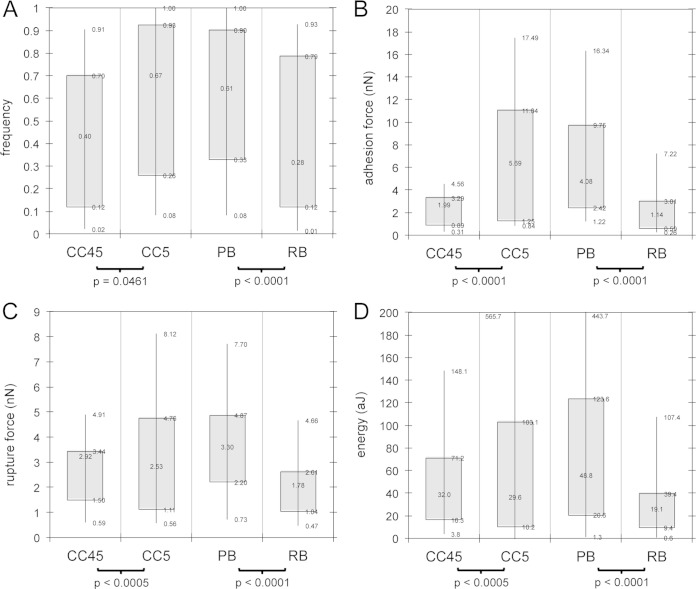
For the hundreds of AFM spectra generated for each S. aureus isolate, we determined the adhesion force, rupture force, and energy of binding (see Fig. 1 for definition of these metrics). These data, along with the occurrence (i.e., frequency) of binding, are summarized in Fig. 2 for (i) CC5 versus CC45 strains and (ii) PB versus RB strains. Differences in the frequency of binding (Fig. 2A) provide a rough estimate of the amount of FnBPs on the S. aureus surface (since the concentration of Fn on the AFM tip should be constant). Higher frequencies of binding were noted for CC5 and PB groups, suggesting the presence of more Fn-binding receptors (or more accessible receptors) on these S. aureus isolates. The “strength” of binding is an additional, and perhaps better, comparator than the frequency of binding. Strength of binding can be assessed as a composite of the adhesion force, rupture force, and energy of binding (labeled in Fig. 1). All three of these parameters were stronger for CC5 and PB groups (Fig. 2B, C, and D).
FIG 2.
Binding metrics on living S. aureus as determined by AFM. Box-and-whisker plots of frequency of binding (A), adhesion force (B), rupture force (C), and energy or work of binding (D) for a fibronectin-coated probe on S. aureus cells grouped according to clonal complex (CC5 versus CC45) and persistence in human infection (PB versus RB) are shown. The frequency of binding (A) is defined as the incidence of observing a binding event (like those shown in Fig. 1), where a value of 0 represents no observed binding, and a value of 1 means that all spectra exhibited a binding force signature. All other attributes (adhesion force, rupture force, and energy/work) are defined in Fig. 1. For panels B, C, and D, force is in nanonewtons (10−9 N) and energy/work is in attojoules (10−18 J). Box ends represent the first and third quartiles. Whisker ends represent the 9th and 91st percentiles. A Student t test was used to determine P values.
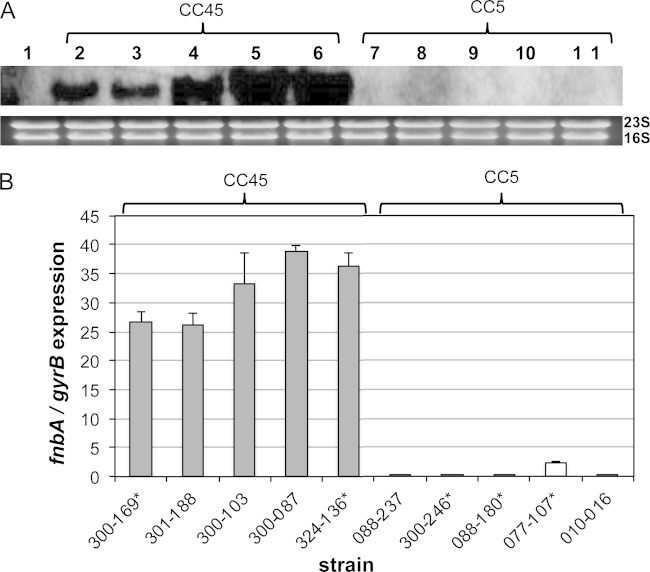
Transcription profiles of fnbA.
All five CC45 strains exhibited robust fnbA expression at 6 h (late-exponential) growth (Fig. 3A). In contrast, the five CC5 strains had relatively low fnbA transcription detectable at this time point by Northern blotting analyses (Fig. 3A). The growth rates for all strains were similar (data not shown), suggesting that the observed differences in fnbA expression were not due to differences in growth dynamics. To exclude any differences in processing (e.g., blotting or film exposure time) that might lead to differences in transcript detection, qRT-PCR of the 6-h samples was carried out, and results were in accordance with the Northern blotting outcomes (Fig. 3B). Of note, these same differences in fnbA expression between the CC5 and CC45 isolates were also observed by qRT-PCR at early exponential growth (3 h) (see Fig. S3 in the supplemental material). When grouped according to clinical outcomes, there were no clear differences in fnbA expression for the PB versus RB strains (see asterisks in Fig. 3B).
FIG 3.
Northern blots and qRT-PCR of fnbA. The effect of clonal variation on fnbA expression was assessed by Northern blotting (A) and qRT-PCR (B). (A) Total cellular RNA from S. aureus strains SH1000ΔfnbA (lane 1), CC45 (lanes 2, 3, 4, 5, and 6 for strains 300-169, 301-188, 300-103, 300-087, and 324-136, respectively), and CC5 (lanes 7, 8, 9, 10, and 11 for strains 088-237, 300-246, 088-180, 077-107, and 010-016, respectively). Cells were grown in TSB and isolated at 6 h postinoculation. (B) fnbA transcription by qRT-PCR, normalized by gyrB, at 6 h postinoculation. Isolates marked by an asterisk (*) came from human patients with persistent bacteremia (PB) (strains 300-169, 324-136, 300-246, 088-180, and 077-107), whereas isolates without an asterisk are from human patients with resolving bacteremia (RB). For CC45 versus CC5, a P value of <0.001 was determined with the Student t test.
Transcription profiles of fnbB.
Transcription profiles for fnbB were different than those described above for fnbA. Most CC45 isolates expressed relatively low levels of fnbB (Fig. 4). Four of the five strains of CC5 expressed higher levels of fnbB (Fig. 4). Similar to the results for fnbA, PB and RB strains did not exhibit any clear differences in fnbB expression (see asterisks in Fig. 4).
FIG 4.
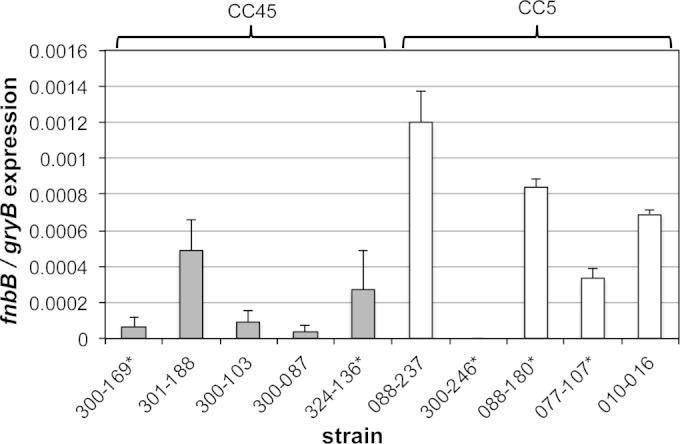
qRT-PCR of fnbB. The effect of clonal variation on fnbB transcription by qRT-PCR, normalized by gyrB, at 6 h postinoculation was evaluated. Isolates with an asterisk (*) were associated with persistent bacteremia (PB) (isolates 300-169, 324-136, 300-246, 088-180, and 077-107) in human patients, whereas isolates without an asterisk are from the RB cohort. For CC45 versus CC5, a P value of 0.10 was determined with Student t test.
fnbA sequence analyses.
We sequenced all 11 Fn-binding sites (i.e., FnBR-1 though FnBR-11) in FnBPA for each of the 10 strains in this study. Figure 5 shows the sequence alignment of three high-affinity domains within FnBPA (FnBR-9, -10, and -11) where we observed significant differences compared to the sequence of FnBPA from the reference strain, 8325, which has a similar genetic background (12). There were three primary differences between the sequences of fnbA in the CC5 versus CC45 isolates (Table 2 and Fig. 5). (i) A unique 38-amino-acid insert between FnBR-9 and FnBR-10 was observed in all CC45, but no CC5, isolates (Fig. 5). (ii) In FnBR-11, the SVDFEED epitope seen in the reference strain (and all CC5 isolates) was replaced with a GIDFVED motif in all CC45 strains (see Fig. 5). (iii) There were two nonsynonymous single nucleotide polymorphisms (SNPs) in FnBR-9 of all CC45 strains, whereas all CC5 strains had four SNPs in FnBR-9, -10, and -11 (Fig. 5).
FIG 5.
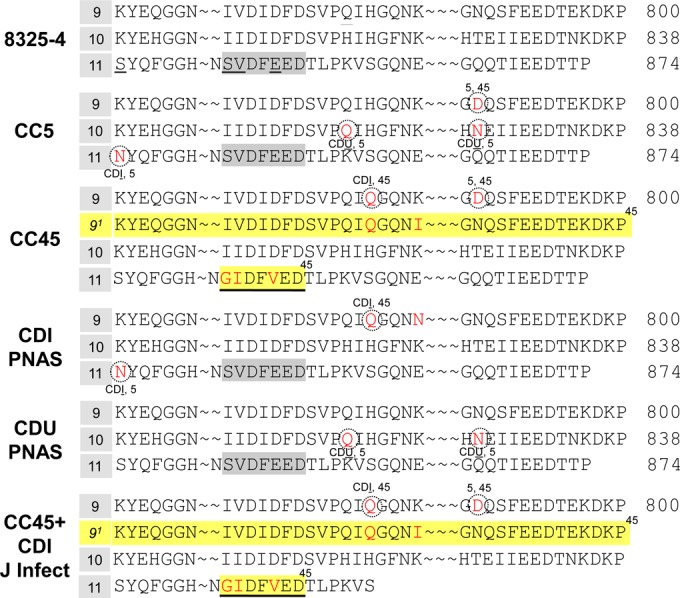
Sequence of FnBPA from CC5 versus CC45. A CLUSTAL W alignment of FnBR-9, FnBR-10, and FnBR-11 of FnBPA in type strain 8325-4, CC5, and CC45 is shown. Also shown are consensus sequences of fnbA for three cohorts of S. aureus obtained from patients with cardiovascular device implants. Two of the cohorts come from patients in the United States, as described by Lower et al. (CDI PNAS and CDU PNAS) (12). The third cohort (CC45+CDI J Infect) comes from a European study lead by Hos et al. (13). Numbering corresponds to the position of amino acids in FnBPA from S. aureus 8325-4. Note (i) the 38-amino-acid insert (yellow highlight) in all CC45 isolates, (ii) the SVDFEED motif in the CC5, CDI, and CDU isolates, whereas the GIDFVED motif is present in all CC45 isolates, and (iii) the nonsynonymous single nucleotide polymorphisms (SNPs) in fnbA (indicated in red).
TABLE 2.
Summary of differences between FnBPA for the type strain of S. aureus and those of various other clinical isolates of S. aureus
| Specimena | Position in FnBPA from S. aureus 8325-4 (Fn-binding repeat no.)b |
Presence of: |
||||||
|---|---|---|---|---|---|---|---|---|
| 782 (9) | 786 (9) | 788 (9) | 818 (10) | 826 (10) | 839 (11) | Extra FnBR between no. 9 and 10 | SVDFEED in no. 11 | |
| Type strain (8325-4) | H | K | N | H | T | S | No | Yes |
| CC5* | H | K | D | Q | N | N | No | Yes |
| CC45* | Q | K | D | H | T | S | Yes | GIDFVED |
| CDI† | Q | N | N | H | T | N | No | Yes |
| CDU† | H | K | N | Q | N | S | No | Yes |
| CDI+CC45‡ | Q | K | D | H | T | S | Yes | GIDFVED |
fnbB sequence analyses.
Only 4 of the 10 isolates contained the fnbB gene. We did not perform comparative clonotype-specific sequence analysis of the fnbB genes because this gene was found only in the CC5 isolates. This explains the observed virtual absent expression of fnbB in the CC45 isolates, as determined by RT-PCR (see above).
Western blots of FnBPs.
Western ligand blots were performed to confirm the presence of FnBPs on the outer cell wall of the S. aureus isolates. As shown in Fig. S4 in the supplemental material, all 10 isolates expressed FnBPs. It was not possible to resolve FnBPA versus FnBPB due to similar electrophoretic mobility on the SDS-PAGE gels. Nonetheless, these results confirm that FnBPA and/or FnBPB are present on the outer cell walls of all 10 isolates.
Virulence gene profiles.
S. aureus has a large repertoire of virulence factors associated with induction and progression of disease. For the 10 S. aureus isolates examined herein, Table 3 shows the occurrence of 38 different virulence genes (toxins and adhesins), which were assayed as part of a much larger phase III drug trial (29). Of the 12 adhesin genes (bbp, clfA, clfB, cna, ebpS, fnbA, fnbB, map [or eap], sdrC, sdrD, sdrE, and spa), only one of the Fn-binding genes (fnbB) exhibited statistical difference in occurrence between the CC5 and CC45 isolates.
TABLE 3.
Differences in virulence gene profiles of CC5 and CC45 S. aureus isolates
| Virulence factor genea | No. of isolatesb |
Pc | |
|---|---|---|---|
| CC5 (n = 5) | CC45 (n = 5) | ||
| agrI | 0 | 5 | 0.008 |
| agrII | 5 | 0 | 0.008 |
| fnbA | 5 | 5 | |
| fnbB | 4 | 0 | 0.048 |
| SCCmec IV | 0 | 5 | 0.008 |
| sec | 0 | 5 | 0.008 |
Data for genes, other than fnbA and fnbB, were obtained from Lalani et al. (29).
No significant differences were observed among CC5 and CC45 isolates for the following genes: agrIII, agrIV, bbp, chp, clfA, clfB, cna, ebpS, efb, eta, etb, fnbA, hlg, icaA, map (or eap), SCCmec I to III, SCCmec O, sdrC, sdrD, sdrE, sea, seb, sed, see, seg, seh, sei, sej, spa, tst, and V8.
As determined by the Fisher exact test.
DISCUSSION
In this study, we focused on the two major Fn-binding adhesins in S. aureus (FnBPA and FnBPB) since they play critical roles in the pathogenesis of endovascular infection. FnBPs have been shown to promote bacterial adherence to and invasion of vascular endothelium in vitro (3, 21, 30–32). Moreover, in vivo evidence supports the role of FnBPA as a virulence factor in experimental infective endocarditis (6, 33). For instance, fnbA transposon mutagenesis variants impaired in their ability to adhere to Fn in vitro showed a reduced capacity to induce experimental infective endocarditis in rats (34). Studies with nonpathogenic L. lactis, as an adoptive transfer recipient of the S. aureus fnbA gene, yielded constructs with an enhanced ability to persist and proliferate within damaged endocardium in vivo (6). Finally, investigations with the same L. lactis recipient demonstrated that FnBPA facilitates bacterial invasion of mammalian cells (8, 30).
Amino acid polymorphisms in S. aureus FnBPA have recently been linked to S. aureus cardiac device infections (CDIs) (12). For example, bacteremia isolates of S. aureus from U.S. patients with CDIs have signature, nonsynonymous SNPs in FnBPA (E652D, H782Q, and K786N) versus bacteremia isolates from patients with cardiac implants in place that are uninfected (CDU). In addition, the CDI isolates formed a more resilient bond with Fn, as determined by AFM and computational simulations (12, 26). This link between CDI and polymorphisms in high-affinity Fn-binding repeats of FnBPA in the U.S. patient cohort has since been externally validated using an independent cohort of European S. aureus isolates (13).
Here, we attempted to extend these observations to another well-defined and problematic clinical S. aureus endovascular syndrome, persistent bacteremia. To do this, we examined MRSA strains from a multinational trial focused on antibiotic therapy in S. aureus bacteremia and infective endocarditis where the presence of a cardiac implant was not a criterion used for patient selection (14). From this drug trial, 10 isolates were selected from distinct ends of the sampling continuum in terms of clinical outcome (persistence versus resolving bacteremia) and clonal complex (CC5 versus CC45) (Table 1). We postulated that this purposeful, rather than random, selection would provide an opportunity to test potential correlations between FnBPs and therapeutic outcomes in non-CDI S. aureus bacteremia.
Because FnBPA/B are adhesins allowing S. aureus to colonize key target tissues (e.g., cardiac values in endocarditis), AFM was used to measure the dynamic binding metrics for each of the 10 bacteremia isolates. Clear differences were observed for the PB versus RB cohorts. PB isolates bound Fn with significantly greater force and energy than RB isolates (see Fig. 2B, C, and D). This supports the notion that Fn binding is one contributor to recalcitrant S. aureus infections. AFM measurements also showed clear differences in Fn binding for the CC5 versus CC45 isolates (Fig. 2). These differences could be due to the quantity of reaction pairs (amount of Fn-binding proteins on S. aureus) or to the quality of the interaction, as determined by the primary sequence of amino acids in the bacterial FnBPs.
We attempted to determine the relative quantity of FnBPA and FnBPB expressed on S. aureus via Northern blots and RT-PCR (Fig. 3 and 4). The CC45 isolates transcribed abundant fnbA but lacked the gene for FnBPB. This strongly suggests that the CC45 isolates use only FnBPA to bind to Fn. In contrast, CC5 strains transcribed both fnbB and relatively low levels of fnbA. It is difficult to compare the magnitudes of expression between fnbA and fnbB because the primers are different. Of note, Western blots did confirm the presence of FnBPs in all 10 strains (see Fig. S4 in the supplemental material); however, because of similar molecular weights and probe cross-reactivity, it was not possible to distinguish between FnBPA versus FnBPB for CC5 isolates that had both fnb genes.
It is tempting to utilize the gene expression profiles as a proxy for protein levels (i.e., CC45 binds via FnBPA, whereas CC5 binds primarily with FnBPB). However, the cellular concentrations of adhesion proteins do not always correlate with abundances of their corresponding mRNAs (35) due to translational regulation, as well as localization and turnover of the protein product. For example, the CC45 isolates with the highest expression of fnbA (e.g., 301-188 and 300-087; see Fig. 3, as well as Fig. S3 in the supplemental material) do not necessarily have the highest yield of FnBP in Western blots (see Fig. S4 in the supplemental material). Likewise, isolates with low expression of fnbA and fnbB (e.g., CC5 strain 300-246; see Fig. 3 and 4) still exhibited Fn-binding events (Fig. 1) and similar Western blot profiles (see Fig. S4 in the supplemental material) as the “high-expression” isolates noted previously (i.e., CC45 strains 301-188 and 300-087). Others have reported similar observations. For example, Green et al. (1) show that a mutant of S. aureus (879R4SSp) with low-level transcription of only one fnb gene (fnbA) still exhibited significant adhesion to Fn-coated surfaces, through robust FnBPA production at the cellular level. This could be the case for our CC5 isolates. Other work has shown that abundances of bacterial proteins and mRNAs in single bacterial cells differ from those measured across populations of the same bacteria (35, 36). This could be true for our experiments where RT-PCR and Western blots assess populations of cells, whereas AFM is conducted at the single-cell level. For CC5 isolates, which have both fnbA and fnbB, it is difficult to assign a particular force signature to FnBPA versus FnBPB because spectra overlap even for a model organism like L. lactis (see Fig. S2 in the supplemental material).
As all 10 isolates had fnbA, we pursued the idea that Fn binding was affected by the primary sequence of amino acids in FnBPA (i.e., the quality of the interaction). There were three key differences in the primary sequence of the FnBPA proteins, as deduced by the sequences of the fnbA gene, in CC5 versus CC45 cohorts (Table 2; Fig. 5). First, compared to the reference type strain (8325-4), both CC5 and CC45 isolates exhibited amino acid polymorphisms in several high-affinity Fn-binding repeat regions (e.g., FnBR-9, -10, and -11). In some instances (e.g., N788D), the same polymorphism was present in both CC5 and CC45 isolates. Other SNPs were specific to only CC5 (e.g., H818Q, T826N, and S839N) or only CC45 (e.g., H782Q) isolates. Second, the SVDFEED epitope in FnBR-11 of the reference type strain and CC5 isolates was replaced with the GIDFVED motif in all CC45 isolates. Others have shown that the SVDFEED epitope is essential for Fn binding, whereas the GIDFVED motif allows S. aureus to evade the immune system, but at a cost of reduced Fn binding (37, 38). Finally, all CC45 isolates demonstrated an extra, large Fn-binding repeat insert that was absent in CC5 strains (see FnBR-91 in Fig. 5).
There were both similarities and differences between these FnBPA alterations and previously reported perturbations in S. aureus strains obtained from CDI patients (12, 13, 26). In terms of similarities, polymorphisms were observed in three high-affinity Fn-binding repeats (FnBR-9, -10, and -11) for our bacteremia isolates. Such polymorphisms in high-affinity FnBRs (e.g., FnBR-9) were also observed for CDI isolates from both the United States (12) and Europe (13) (Table 2, Fig. 5). Another similarity is the GIDFVED motif, as well as the extra 38-amino-acid repeat insert, which was observed for all of our CC45 strains (Table 2, Fig. 5). This parallels the results of Hos et al. (13) for all European CDI isolates that were the CC45 clonotype (n = 34). Conversely, all 46 of the CDI and CDU isolates from U.S. patients (12), including eight CC5 and two CC45 isolates, contained the wild-type SVDFEED epitope in FnBR-11, similar to the CC5 isolates examined here (see Table 2 and Fig. 5).
As noted above, the SVDFEED epitope in FnBR-11 is important for binding to Fn (37, 38). Indeed, AFM measurements on single cells showed that CC5 isolates bind more strongly (Fig. 2) than the CC45 isolates, which have the GIDFVED replacement in FnBR-11 (Fig. 5). Hos et al. (13), using a microtiter assay, showed a similar trend. Their CC45-CDI isolates with GIDFVED bound to a lesser extent to Fn-coated surfaces than non-CC45-CDI isolates featuring the SVDFEED epitope in FnBR-11.
Rice et al. (37) and Gomes et al. (38) suggest that the phenomenon of GIDFVED-for-SVDFEED replacement evolved as a possible strategy for immune evasion. The GIDFVED epitope may not promote Fn binding, but it does seem to enhance pathogen invasion of host cells, thereby allowing S. aureus to potentially evade the immune response encountered during successive cycles of colonization, transmission and infection (37, 38). Perhaps this is the case for our CC45 isolates. Indeed, Seidl et al. (21) point out that the same isolates used in the present study could initiate a “brisk” inflammatory response in vivo in terms of the host immune system attempts to eliminate these pathogens.
Another intriguing finding of our bacteremia strains is the presence of an extra Fn-binding repeat insert in all CC45 isolates (not found in CC5 isolates) (Fig. 5). Hos et al. first reported a similar duplication in CC45 S. aureus associated with CDI (13). This insert occurs between two repeats that are known to have high affinity for Fn (39). Perhaps this insert plays a role in invasion as it is associated with the GIDFVED motif. Alternatively, the CC45 isolates may have evolved this additional high-affinity FnBR to offset the reduced Fn binding attributed to the GIDFVED motif in its FnBR-11. From a biophysical perspective, the corollary is that the sum of multiple weak bonds in parallel is as effective as one strong bond (e.g., two, identical parallel bonds each with a strength of F/2 are just as strong as one bond with a strength of F) (28, 40).
Together, the present study and other recent investigations (12, 13, 26) support the hypothesis that S. aureus endovascular syndromes are correlatable to amino acid polymorphisms and/or duplications within high-affinity Fn-binding regions of FnBPs. These changes in FnBPs may indeed lead to stronger binding reactions and enhanced capacity to initiate an infection (12). Alternatively, the polymorphisms could induce mechanical or conformational changes in the associated ligand, as suggested by Buck et al. (28), which in turn could impact that bacterium's capacity to bind, invade, and persist in host cells (13). For example, Fn bound on the surface of S. aureus has been shown to form multimers that promote interaction with integrins and subsequent invasion of host cells (8, 30, 41). Shinji et al. (42) suggest that “if FnBPA molecules were distributed in clusters on the bacterial surface … integrins would easily be activated by FnBPA because the cluster of FnBPAs could efficiently organize integrins.” Interestingly, we previously discovered clusters of FnBPs on the surface of individual S. aureus by probing living cells with a Braille-like, force-avidity mapping technique (40).
Ligand binding and host cell invasion are both critical factors affecting the course of distinct S. aureus infections. While blood cultures taken from staphylococcemic patients usually yield only a monoclonal culture of S. aureus, it is possible that in vivo infections can be, in fact, polyclonal. For example, an infection could be initiated by a strain maximized for Fn binding, i.e., one that has polymorphisms H782Q and K786N in FnBR-9 as well as SVDFEED in FnBR-11. The same infection could progress by virtue of another clonotype strain within a mixed culture that is better adapted for invasion (e.g., a CC45 strain with GIDFVED in FnBR-11). Alternatively, a single clone could genetically evolve in vivo during the course of infection (12). This could be important in the circumstance of the PB outcome, as virulence genes such as fnbA or fnbB can undergo relatively rapid molecular evolution within a population of S. aureus cells (43). For our S. aureus strains, the PB outcome appears to have two distinct, clonotype-specific pathogenesis pathways: (i) CC5 via enhanced binding to target tissues and proliferation therein or (ii) CC45 via sacrificing Fn binding for augmentation of intracellular persistence via unique SNPs or amino acid motif duplications. This dual pathway for PB is illustrated in Fig. S5 in the supplemental material showing AFM data pooled for both clonotypes (CC5/45) and clinical outcomes (PB/RB).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants R01-HL119648 (S.K.L.), R01-AI-039108-17 (A.S.B.), and R01-AI068804 (V.G.F.) from the National Institutes of Health. N.N.C.-I. was supported by a grant from the American Heart Association (14POST20460073). S.K.L. acknowledges the support of J. Tak.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01074-15.
REFERENCES
- 1.Greene C, Vaudaux PE, Francois P, Proctor RA, McDevitt D, Foster TJ. 1996. Low-fibronectin-binding mutant of Staphylococcus aureus 879R4S has Tn918 inserted into its single fnb gene. Microbiology 142:2153–2160. doi: 10.1099/13500872-142-8-2153. [DOI] [PubMed] [Google Scholar]
- 2.Foster TJ, Hook M. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol 6:484–488. doi: 10.1016/S0966-842X(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 3.Peacock SJ, Foster TJ, Cameron BJ, Berendt AR. 1999. Bacterial fibronectin-binding proteins and endothelial cell surface fibronectin mediate adherence of Staphylococcus aureus to resting human endothelial cells. Microbiology 145:3477–3486. doi: 10.1099/00221287-145-12-3477. [DOI] [PubMed] [Google Scholar]
- 4.Wann ER, Gurusiddappa S, Hook M. 2000. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J Biol Chem 275:13863–13871. doi: 10.1074/jbc.275.18.13863. [DOI] [PubMed] [Google Scholar]
- 5.Moreillon P, Que YA, Bayer AS. 2002. Pathogenesis of streptococcal and staphylococcal endocarditis. Infect Dis Clin North Am 16:297–318. doi: 10.1016/S0891-5520(01)00009-5. [DOI] [PubMed] [Google Scholar]
- 6.Que YA, Francois P, Haefliger JA, Entenza JM, Vaudaux P, Moreillon P. 2001. Reassessing the role of Staphylococcus aureus clumping factor and fibronectin-binding protein by expression in Lactococcus lactis. Infect Immun 69:6296–6302. doi: 10.1128/IAI.69.10.6296-6302.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Que YA, Haefliger JA, Piroth L, Francois P, Widmer E, Entenza JM, Sinha B, Herrmann M, Francioli P, Vaudaux P, Moreillon P. 2005. Fibrinogen and fibronectin binding cooperate for valve infection and invasion in Staphylococcus aureus experimental endocarditis. J Exp Med 201:1627–1635. doi: 10.1084/jem.20050125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha B, Francois P, Que YA, Hussain M, Heilmann C, Moreillon P, Lew D, Krause KH, Peters G, Herrmann M. 2000. Heterologously expressed Staphylococcus aureus fibronectin-binding proteins are sufficient for invasion of host cells. Infect Immun 68:6871–6878. doi: 10.1128/IAI.68.12.6871-6878.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massey RC, Kantzanou MN, Fowler T, Day NP, Schofield K, Wann ER, Berendt AR, Hook M, Peacock SJ. 2001. Fibronectin-binding protein A of Staphylococcus aureus has multiple, substituting, binding regions that mediate adherence to fibronectin and invasion of endothelial cells. Cell Microbiol 3:839–851. doi: 10.1046/j.1462-5822.2001.00157.x. [DOI] [PubMed] [Google Scholar]
- 10.Edwards AM, Potts JR, Josefsson E, Massey RC. 2010. Staphylococcus aureus host cell invasion and virulence in sepsis is facilitated by the multiple repeats within FnBPA. PLoS Pathog 6:e1000964. doi: 10.1371/journal.ppat.1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meenan NA, Visai L, Valtulina V, Schwarz-Linek U, Norris NC, Gurusiddappa S, Hook M, Speziale P, Potts JR. 2007. The tandem beta-zipper model defines high-affinity fibronectin-binding repeats within Staphylococcus aureus FnBPA. J Biol Chem 282:25893–25902. doi: 10.1074/jbc.M703063200. [DOI] [PubMed] [Google Scholar]
- 12.Lower SK, Lamlertthon S, Casillas-Ituarte NN, Lins RD, Yongsunthon R, Taylor ES, DiBartola AC, Edmonson C, McIntyre LM, Reller LB, Que YA, Ros R, Lower BH, Fowler VG Jr. 2011. Polymorphisms in fibronectin binding protein A of Staphylococcus aureus are associated with infection of cardiovascular devices. Proc Natl Acad Sci U S A 108:18372–18377. doi: 10.1073/pnas.1109071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hos NJ, Rieg S, Kern WV, Jonas D, Fowler VG Jr, Higgins PG, Seifert H, Kaasch AJ. 2015. Amino acid alterations in fibronectin binding protein A (FnBPA) and bacterial genotype are associated with cardiac device related infection in Staphylococcus aureus bacteraemia. J Infect 70:153–159. doi: 10.1016/j.jinf.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Fowler VGJ, Boucher HW, Corey GR, Abrutyn E, Karchmer AW, Rupp ME, Levine DP, Chambers HF, Tally FP, Vigliani GA, Cabell CH, Link AS, DeMeyer I, Filler SG, Zervos M, Cook P, Parsonnet J, Bernstein JM, Price CS, Forrest GN, Fatkenheuer G, Gareca M, Rehm SJ, Brodt HR, Tice A, Cosgrove SE. 2006. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N Engl J Med 355:653–665. doi: 10.1056/NEJMoa053783. [DOI] [PubMed] [Google Scholar]
- 15.Fowler VGJ, Nelson CL, McIntyre LM, Kreiswirth BN, Monk A, Archer GL, Federspiel J, Naidich S, Remortel B, Rude T, Brown P, Reller LB, Corey GR, Gill SR. 2007. Potential associations between hematogenous complications and bacterial genotype in Staphylococcus aureus infection. J Infect Dis 196:738–747. doi: 10.1086/520088. [DOI] [PubMed] [Google Scholar]
- 16.McCalla C, Smyth DS, Robinson DA, Steenbergen J, Luperchio SA, Moise PA, Fowler VG Jr, Sakoulas G. 2008. Microbiological and genotypic analysis of methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob Agents Chemother 52:3441–3443. doi: 10.1128/AAC.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendes RE, Deshpande LM, Smyth DS, Shopsin B, Farrell DJ, Jones RN. 2012. Characterization of methicillin-resistant Staphylococcus aureus strains recovered from a phase IV clinical trial for linezolid versus vancomycin for treatment of nosocomial pneumonia. J Clin Microbiol 50:3694–3702. doi: 10.1128/JCM.02024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fusco DN, Alexander EL, Weisenberg SA, Mediavilla JR, Kreiswirth BN, Schuetz AN, Jenkins SG, Rhee KY. 2009. Clinical failure of vancomycin in a dialysis patient with methicillin-susceptible vancomycin-heteroresistant S. aureus. Diagn Microbiol Infect Dis 65:180–183. doi: 10.1016/j.diagmicrobio.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seidl K, Chen L, Bayer AS, Hady WA, Kreiswirth BN, Xiong YQ. 2011. Relationship of agr expression and function with virulence and vancomycin treatment outcomes in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 55:5631–5639. doi: 10.1128/AAC.05251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fowler VGJ, Sakoulas G, McIntyre LM, Meka VG, Arbeit RD, Cabell CH, Stryjewski ME, Eliopoulos GM, Reller LB, Corey GR, Jones T, Lucindo N, Yeaman MR, Bayer AS. 2004. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J Infect Dis 190:1140–1149. doi: 10.1086/423145. [DOI] [PubMed] [Google Scholar]
- 21.Seidl K, Bayer AS, McKinnell JA, Ellison S, Filler SG, Xiong YQ. 2011. In vitro endothelial cell damage is positively correlated with enhanced virulence and poor vancomycin responsiveness in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Cell Microbiol 13:1530–1541. doi: 10.1111/j.1462-5822.2011.01639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seidl K, Bayer AS, Fowler VGJ, McKinnell JA, Abdel Hady W, Sakoulas G, Yeaman MR, Xiong YQ. 2011. Combinatorial phenotypic signatures distinguish persistent from resolving methicillin-resistant Staphylococcus aureus bacteremia isolates. Antimicrob Agents Chemother 55:575–582. doi: 10.1128/AAC.01028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma-Kuinkel BK, Mann EE, Ahn JS, Kuechenmeister LJ, Dunman PM, Bayles KW. 2009. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J Bacteriol 191:4767–4775. doi: 10.1128/JB.00348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong Y, Bayer AS, Yeaman MR, van Wamel W, Manna AC, Cheung AL. 2004. Impacts of sarA and agr in Staphylococcus aureus strain Newman on fibronectin-binding protein A gene expression and fibronectin adherence capacity in vitro and in experimental infective endocarditis. Infect Immun 72:1832–1836. doi: 10.1128/IAI.72.3.1832-1836.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell SJ, Deshmukh HS, Nelson CL, Bae IG, Stryjewski ME, Federspiel JJ, Tonthat GT, Rude TH, Barriere SL, Corey R, Fowler VG Jr. 2008. Genotypic characteristics of Staphylococcus aureus isolates from a multinational trial of complicated skin and skin structure infections. J Clin Microbiol 46:678–684. doi: 10.1128/JCM.01822-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casillas-Ituarte NN, Lower BH, Lamlertthon S, Fowler VGJ, Lower SK. 2012. Dissociation rate constants of human fibronectin binding to fibronectin-binding proteins on living Staphylococcus aureus isolated from clinical patients. J Biol Chem 287:6693–6701. doi: 10.1074/jbc.M111.285692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yongsunthon R, Lower SK. 2006. Force spectroscopy of bonds that form between a Staphylococcus bacterium and silica or polystyrene substrates. J Electron Spec Related Phenomena 150:228–234. doi: 10.1016/j.elspec.2005.06.012. [DOI] [Google Scholar]
- 28.Buck AW, Fowler VGJ, Yongsunthon R, Liu J, DiBartola AC, Que YA, Moreillon P, Lower SK. 2010. Bonds between fibronectin and fibronectin-binding proteins on Staphylococcus aureus and Lactococcus lactis. Langmuir 26:10764–10770. doi: 10.1021/la100549u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lalani T, Federspiel JJ, Boucher HW, Rude TH, Bae IG, Rybak MJ, Tonthat GT, Corey GR, Stryjewski ME, Sakoulas G, Chu VH, Alder J, Steenbergen JN, Luperchio SA, Campion M, Woods CW, Fowler VG. 2008. Associations between the genotypes of Staphylococcus aureus bloodstream isolates and clinical characteristics and outcomes of bacteremic patients. J Clin Microbiol 46:2890–2896. doi: 10.1128/JCM.00905-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinha B, Francois PP, Nusse O, Foti M, Hartford OM, Vaudaux P, Foster TJ, Lew DP, Herrmann M, Krause KH. 1999. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin alpha5beta1. Cell Microbiol 1:101–117. doi: 10.1046/j.1462-5822.1999.00011.x. [DOI] [PubMed] [Google Scholar]
- 31.Peacock SJ, Lina G, Etienne J, Foster TJ. 1999. Staphylococcus schleiferi subsp. schleiferi expresses a fibronectin-binding protein. Infect Immun 67:4272–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Belkum A, Kools-Sijmons M, Verbrugh H. 2002. Attachment of Staphylococcus aureus to eukaryotic cells and experimental pitfalls in staphylococcal adherence assays: a critical appraisal. J Microbiol Methods 48:19–42. doi: 10.1016/S0167-7012(01)00342-6. [DOI] [PubMed] [Google Scholar]
- 33.Joh D, Wann ER, Kreikemeyer B, Speziale P, Hook M. 1999. Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol 18:211–223. doi: 10.1016/S0945-053X(99)00025-6. [DOI] [PubMed] [Google Scholar]
- 34.Kuypers JM, Proctor RA. 1989. Reduced adherence to traumatized rat heart valves by a low-fibronectin-binding mutant of Staphylococcus aureus. Infect Immun 57:2306–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogel C, Marcotte EM. 2012. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Gene 13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taniguchi Y, Choi PJ, Li G, Chen H, Babu M, Hearn J, Emili A, Xie XS. 2010. Quantifying Escherichia coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice K, Huesca M, Vaz D, McGavin MJ. 2001. Variance in fibronectin binding and fnb locus polymorphisms in Staphylococcus aureus: identification of antigenic variation in a fibronectin binding protein adhesin of the epidemic CMRSA-1 strain of methicillin-resistant S. aureus. Infect Immun 69:3791–3799. doi: 10.1128/IAI.69.6.3791-3799.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomes AR, Vinga S, Zavolan M, de Lencastre H. 2005. Analysis of the genetic variability of virulence-related loci in epidemic clones of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 49:366–379. doi: 10.1128/AAC.49.1.366-379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Norris NC, Bingham RJ, Harris G, Speakman A, Jones RPO, Leech A, Turkenburg JP, Potts JR. 2011. Structural and functional analysis of the tandem beta-zipper interaction of a streptococcal protein with human fibronectin. J Biol Chem 286:38311–38320. doi: 10.1074/jbc.M111.276592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lower SK, Yongsunthon R, Casillas-Ituarte NN, Taylor ES, DiBartola AC, Lower BH, Beveridge TJ, Buck AW, Fowler VG. 2010. A tactile response in Staphylococcus aureus. Biophys J 99:2803–2811. doi: 10.1016/j.bpj.2010.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shinji H, Seki K, Tajima A, Uchida A, Masuda S. 2003. Fibronectin bound to the surface of Staphylococcus aureus induces association of very late antigen 5 and intracellular signaling factors with macrophage cytoskeleton. Infect Immun 71:140–146. doi: 10.1128/IAI.71.1.140-146.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shinji H, Yosizawa Y, Tajima A, Iwase T, Sugimoto S, Seki K, Mizunoe Y. 2011. Role of fibronectin-binding proteins A and B in in vitro cellular infections and in vivo septic infections by Staphylococcus aureus. Infect Immun 79:2215–2223. doi: 10.1128/IAI.00133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamers RP, Stinnett JW, Muthukrishnan G, Parkinson CL, Cole AM. 2011. Evolutionary analyses of Staphylococcus aureus identify genetic relationships between nasal carriage and clinical isolates. PLoS One 6:e16426. doi: 10.1371/journal.pone.0016426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.