ABSTRACT
Macrophages are a target for infection with HIV and represent one of the viral reservoirs that are relatively resistant to current antiretroviral drugs. Here we demonstrate that methylglyoxal-bis-guanylhydrazone (MGBG), a polyamine analog and potent S-adenosylmethionine decarboxylase inhibitor, decreases HIV expression in monocytes and macrophages. MGBG is selectively concentrated by these cells through a mechanism consistent with active transport by the polyamine transporter. Using a macrophage-tropic reporter virus tagged with the enhanced green fluorescent protein, we demonstrate that MGBG decreases the frequency of HIV-infected cells. The effect is dose dependent and correlates with the production of HIV p24 in culture supernatants. This anti-HIV effect was further confirmed using three macrophage-tropic primary HIV isolates. Viral life cycle mapping studies show that MGBG inhibits HIV DNA integration into the cellular DNA in both monocytes and macrophages.
IMPORTANCE Our work demonstrates for the first time the selective concentration of MGBG by monocytes/macrophages, leading to the inhibition of HIV-1 expression and a reduction in proviral load within macrophage cultures. These results suggest that MGBG may be useful in adjunctive macrophage-targeted therapy for HIV infection.
INTRODUCTION
One of the major remaining obstacles in treating human immunodeficiency type 1 (HIV-1)-related diseases is the elimination of viral reservoirs that contribute to persistent infection and chronic immune activation. Although highly active antiretroviral therapy (HAART) is effective at reducing the plasma viral load and slowing down the decline of CD4+ T lymphocytes and dendritic cells, it has failed to eradicate the residual long-lived HIV-infected cellular reservoirs (1–3). At the molecular level, HAART successfully suppresses HIV RNA replication and decreases total viral DNA load in treated individuals. However, it fails to eliminate the integrated proviral DNA (4, 5). Even in HAART-treated patients with an undetectable plasma viral load, discontinuation of treatment is routinely associated with the reappearance of HIV RNA in circulation, confirming the presence of significant HIV proviral reservoirs in HAART-treated individuals (6, 7).
HIV-1 cellular reservoirs include resting CD4+ T lymphocytes and cells of the monocyte and macrophage lineage (1, 8, 9). Research on the HIV reservoirs has largely focused on T cells and more specifically a rare subset of resting memory CD4 T cells that has been reproducibly identified in the blood of patients with low or no detectible plasma viral loads (10). Cells of the monocyte and macrophage lineage may represent another mechanism for viral persistence. These cells are among the cells infected early upon exposure to HIV (11), and the presence of CCR5-utilizing “macrophage-tropic” forms of HIV in the plasma of essentially all newly infected individuals is consistent with the notion that macrophages represent an early target for HIV infection (12–15). In contrast to CD4 T cells which die with effective HIV replication, macrophages are resistant to the cytopathic effect of the virus. Furthermore, macrophages and dendritic cells are capable of transmitting HIV to T cells or other macrophages via cell-cell contact (16). Depending on the origins of the virus, both cis-infection and trans-infection modes have been demonstrated (17–19). Because tissue-resident macrophages are extremely long-lived, the infected macrophages can serve as a long-term source for infectious HIV virions, particularly in patients who develop HIV-associated neurocognitive disorder (HAND) (20–22). For instance, macrophages in the central nervous system (CNS [microglia]) turn over at a rate of less than 1% per year, and they can become superinfected with HIV, leading to the generation of recombinant forms of the virus (23). Therefore, not only are macrophages among the early infected cells in vivo, they also serve as a source of continuous HIV evolution. Prevention of macrophage infection could be an aspect of a successful treatment strategy for both initial and ongoing HIV infection.
The existing approved antiretroviral drugs used in HAART therapies were identified and characterized in T cell cultures. In a recent study, various clinically relevant HIV drugs were found to display significantly lower intracellular concentrations in macrophages versus lymphocytes, leading to a 5 to 200 times difference in antiviral potency between the two cell types (24). In addition, a major site of the macrophage reservoir is within the CNS, and most of the current antiretroviral drugs on the market fail to efficiently cross the blood-brain barrier (25). Thus, the macrophage compartment can serve as a long-term source of HIV that is relatively drug resistant, underscoring the need for additional drugs that specifically target HIV infection in macrophages (26).
HAART has dramatically improved the survival of HIV-positive patients, generally by reversing complications from T cell loss. However, several major diseases such as HAND (27) and atherosclerosis (28) are becoming increasingly prevalent in HAART-treated patients. These diseases result from persistent macrophage activation and currently affect more than half of HIV-infected individuals (29), suggesting a failure in the primarily T cell-targeted approach to treat HIV-associated diseases.
In the course of studying the effects of polyamine analogs on macrophage function, our attention was drawn to methylglyoxal-bis-guanylhydrazone (MGBG) because of its extensive experimental history in both laboratory and clinical studies (30, 31). MGBG is believed to interfere with polyamine metabolism and functions predominantly through the inhibition of S-adenosine-methionine decarboxylase (SAMDC), a rate-limiting enzyme for polyamine biosynthesis, thus resulting in depletion of the intracellular pool of spermine and spermidine (32–35). Polyamines are required for cell proliferation and differentiation in general (36, 37). Since fully differentiated and activated macrophages appear to be targets for HIV infection (38–40) and MGBG was reported to interfere with macrophage activation in vitro (41, 42), we evaluated MGBG for its ability to interfere with HIV infection in primary macrophages.
To test the effect of MGBG on HIV expression in macrophages, we first employed a macrophage-tropic enhanced green fluorescent protein (EGFP)-labeled replication-competent HIV construct that allows convenient tracking of viral expression by flow cytometry. This system has been utilized in several previous studies, the most recent of which showed that cellular activation by osteopontin caused increased expression of HIV in macrophages (43). In the present study, we have shown that MGBG inhibits HIV expression and proviral integration into cellular DNA within macrophages and monocytes. Furthermore, we have extended our finding beyond the EGFP-tagged HIV strain, which is a laboratory strain, and demonstrated the effectiveness of MGBG against multiple primary viral isolates.
MATERIALS AND METHODS
MGBG uptake in cultured cells.
Buffy coats from healthy donors were obtained from the Stanford Blood Center. White blood cells were prepared from buffy coats using red blood cell lysis solution (Miltenyi Biotec, Auburn, CA), and peripheral blood mononuclear cells (PBMCs) were isolated by Percoll or Ficoll-Paque Plus (GE Healthcare Life Sciences, Pittsburgh, PA) gradient centrifugation. Monocytes and T cells were isolated from PMBCs and granulocytes from white blood cells using CD14, CD3, and CD15 microbeads (Miltenyi Biotec, Auburn, CA), respectively. As confirmed by fluorescence-activated cell sorter (FACS) analysis, the purities of all the cell types were 90 to 95%. To generate macrophages, monocytes were cultured for 7 days to allow differentiation.
For drug uptake studies, the freshly isolated primary human blood cells or the in vitro differentiated macrophages were cultured at 1 million per ml in suspension in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 1 mM sodium pyruvate for 24 h with or without drug treatment. For experiments with activated T cells, phytohemagglutinin (PHA) (Sigma-Aldrich, St. Louis, MO) was also added to the cultures at a final concentration of 5 μg/ml. The cells were then harvested and counted manually following trypan blue staining. Cell lysates were prepared in M-PER mammalian protein extraction reagent (Fisher Scientific, Pittsburgh, PA) according to the manufacturer's instructions. Protein and MGBG concentrations in the lysate were measured, respectively, by the Bradford assay (Thermo Fisher Scientific, Waltham, MA) and liquid chromatography-mass spectrometry (LC-MS) at contract laboratories (Seventh Wave, Chesterfield, MO; MPI Research, Mattawan, MI). MGBG levels were normalized to the amount of the protein in each lysate. Calculations of intracellular MGBG concentrations were based on the average of experimentally determined MGBG contents of the cells and the published mean cell volumes of 419 fl for the monocytes (44, 45), 4,990 fl for the macrophages (46), and 800 fl for the activated T cells (47). However, given the inherent individual variations in the cell volumes, the reported intracellular MGBG concentrations should be considered estimates only.
Drug cytotoxicity was assessed using the CellTiter-Glo kit (Promega, Madison, WI). Briefly, the cells were isolated from normal human blood as described above and cultured at 45,000 cells/ml (9,000 cells per well) in triplicate in a 96-well plate in the presence of various concentrations of MGBG. After 24 h, the CellTiter-Glo reagent was added, resulting in cell lysis and generation of a luminescent signal proportional to the amount of ATP present in each well. The data were normalized to the average signals from the untreated cells. The absence of drug-associated cytotoxicity at 24 h was also confirmed by trypan blue exclusion.
Cell culture and HIV-1 infection.
The HIV-1 reporter virus construct pSF162R3 Nef+ plasmid (48) was obtained from Amanda Brown from Johns Hopkins University, School of Medicine. This virus is replication competent and expresses EGFP in conjunction with HIV nef expression and has been used to track HIV expression in macrophage cultures (49). The plasmid was transformed into Max Efficiency Stbl2 competent cells (Life Technologies, Grand Island, NY); large-scale plasmids were prepared using the PureYield plasmid MaxiPrep system (Promega, Madison, WI). The viral stocks were generated by transient transfection into 293T/17 cells (ATCC, Manassas, VA) using SuperFect transfection reagents (Qiagen, Valencia, CA) according to the manufacturer's protocol. PBMCs were isolated from buffy coats of healthy donors by Ficoll-Paque Plus gradient centrifugation. Monocytes were enriched by plating onto a tissue culture plate for at least 4 h before nonadherent cells were washed away. The adherent cells were cultured in 50% Myelocult (Stemcell Technologies, Vancouver, Canada), 25% Iscove's modified Dulbecco's medium (IMDM) (containing 10% FBS), 25% HS27 human fibroblast conditioned IMDM, and 1 ng/ml each of macrophage colony-stimulating factor (M-CSF) and interleukin-3 (IL-3) (Sigma-Aldrich, St. Louis, MO) for 6 to 7 days to allow differentiation into macrophages with or without drug treatment. Differentiated macrophages were infected with viral supernatant at 150 to 180 ng p24 per million cells (0.45 to 0.75 μg per well, 6 to 8 M cells per well) for 6 h in the presence or absence of drugs. Subsequently, the viral inoculate was removed, and the cells were washed and cultured in the same medium for 7 days with or without drug treatment. Medium was exchanged every 3 to 4 days, and clarified culture supernatants were collected and stored at −80°C for subsequent p24 antigen quantitation. Cells were inspected for EGFP expression before harvesting using a fluorescence microscope.
For assays with primary HIV isolates, monocytes were isolated from PMBCs by adhesion to FBS-coated flasks for 2 days followed by repeated washing. Trypsin-EDTA-harvested monocytes were pooled from 6 healthy donors and infected with the specified viral isolates for 2 h at a multiplicity of infection (MOI) of 0.005 in suspension. Unabsorbed virus were washed out, and the cells were plated at 2 × 104 cells (200 μl) per well in a 96-well plate. After overnight culture, the medium was replaced and drugs were added to the cells at various concentrations in triplicate. After being cultured for another 7 days, the supernatant from each well was collected for p24 antigen quantitation. Three primary HIV isolates (Ba-L, 33931N, and 873) were used in the study. In parallel, the effect of the drugs on cell viability was determined using the CellTiter-Glo assay.
For the HIV receptor CD4 and coreceptor CCR5 expression study, monocytes were prepared by overnight adhesion on plastic, and the cells from 6 healthy donors were pooled. They were then cultured with or without MGBG treatment for 7 days in the same medium used for the EGFP-tagged HIV infection study before being harvested for FACS analysis.
Flow cytometry analysis. (i) Studies with EGFP-tagged HIV.
Differentiated macrophages were washed and harvested by scraping. Cells were stained using the LIVE/DEAD fixable red dead cell staining kit (Life technologies, Grand Island, NY) according to the manufacturer's manual, followed by fixation in 3% formaldehyde. The samples were analyzed on a FACScan instrument and the percentage of EGFP+ cells were determined after gating out the debris by FSC/SCC and the dead cells by the LIVE/DEAD stain (see Fig. S1 in the supplemental material). Since macrophages display strong and highly variable autofluorescence in all 3 channels (FL1 to -3), instrument compensation was focused on the cell populations that were relatively dim (as an example, see the lower left population, donor 3, in Fig. S5 in the supplemental material) and thus might not be optimal for other cell populations.
(ii) CCR5 and CD4 study.
Macrophages were double stained using fluorescein isothiocyanate (FITC)-CCR5/peridinin chlorophyll protein (PerCP)-CD14 or FITC-CD4/PerCP-CD14 antibodies or matching isotypes (Biolegend, San Diego, CA). Debris and the main subpopulations of the cells were gated using forward and side scatter (FSC and SSC, respectively) plots.
Quantitative real-time PCR analysis.
Before DNA extraction, the cells were treated as follows to remove the contaminating plasmid DNA used for viral packaging. First, the uninfected cells were mock infected by incubation with the viral supernatant on ice for 2 h in order to estimate the baseline of residual plasmid DNA in the final preparation. After being washed twice with medium without supplements, the mock-infected cells as well as other samples were treated with an excess amount of DNase I (75 to 100 U in 1 ml Hanks' balanced salt solution) [HBSS] (for each digestion) at room temperature for 1 h. The cells were then washed again before total DNA isolation using the Puregene kit (Qiagen, Valencia, CA). The yield and purity of the DNA samples were assessed by spectrometry using a NanoDrop instrument (Thermo Scientific, Lafayette, CO).
The proviral DNA load was determined essentially as described previously (50). Briefly, total DNA samples (1 to 2 μg each) were separated by agarose gel electrophoresis. The ∼20-kb band which represented the high-molecular-weight (HMW) genomic DNA was excised using a razor blade. Subsequently, the GeneJet gel extraction kit (Thermo Scientific, Lafayette, CO) was used to purify the DNA from the gel slices. Each sample was eluted in 20 to 30 μl Tris-HCl buffer at pH 8.
For quantitative PCR (qPCR) assays, the primers and TaqMan probes were synthesized according to previously published sequences for the HIV gag region gene and β-globin gene, a single-copy conserved gene that serves as the control (51). Each 20-μl qPCR mixture contained 10 μl of the PerfeCTa qPCR master mix (Quant BioSciences, Gaithersburg, MD), 10 ng of total DNA or 4 μl of the HMW genomic DNA as the template, 0.05 μM globin primers, 0.1 μM Gag primers, and 0.025 μM VIC- or 6-carboxyfluorescein (FAM)-labeled probes. Cycling was performed in an Agilent Mx3000P qPCR system (Agilent, Santa Clara, CA) using the following parameters: 95°C for 5 min followed by 40 cycles of 95°C for 15 s and 60°C for 45 s. The assay conditions were preoptimized to give 95 to 100% amplification efficiency and zero nonspecific amplifications in the absence of the template (data not shown). All of the reactions were set up in triplicate using white 0.2-ml qPCR tubes (BrandTech Scientific, Essex, CT). Standards from 3 to 30,000 copies for both HIV and the globin gene were created using genomic DNA from the 8E5 cell line, which harbors a single copy of integrated HIV per genome (52).
For data analysis, the threshold cycle (CT) numbers were determined using the instrument's built-in software. Standard curves were created by plotting the CT number versus the log of the initial copy number of the 8E5 genome in each dilution. The copy numbers of β-globin and HIV gag DNA for all of the test samples were interpolated from the respective linear regression curves using GraphPad Prism 5 (GraphPad Software, San Diego, CA). Typically the standard curves had an R2 value of greater than 0.99, and the coefficient of variation of the CT values between the triplicates was ≤1%. The copy number of the β-globin gene was used to normalize the HIV DNA copy number in each sample. The effect of the drug treatment was expressed as a percentage of that in the untreated control sample.
HIV p24 antigen quantitation.
For studies with the EGFP-tagged HIV, HIV p24 antigen in the diluted culture supernatant was quantified using a p24 antigen enzyme-linked immunosorbent assay (ELISA) kit (Zeptometrix Corp., Buffalo, NY) according to the manufacturer's protocol. For assays with the primary HIV isolates, the PerkinElmer Alliance HIV-1 P24 antigen ELISA kit was used.
Statistical analysis.
All experiments included at least three donors unless otherwise indicated. The linear trend in the relationship between the response (EGFP, p24, or proviral DNA, all expressed as percentage of the untreated response) and dose of MGBG was computed separately, and repeated-measures analyses were conducted using SAS v9.4 (SAS Institute Inc., Cary, NC). Linear trend was based on the Greenhouse-Geiser sphericity adjusted within-subject effect of the MGBG dose, which accounts for variability in variances of the differences between the doses, which are assumed to be constant.
RESULTS
Selective uptake of MGBG in cultured primary human blood cells.
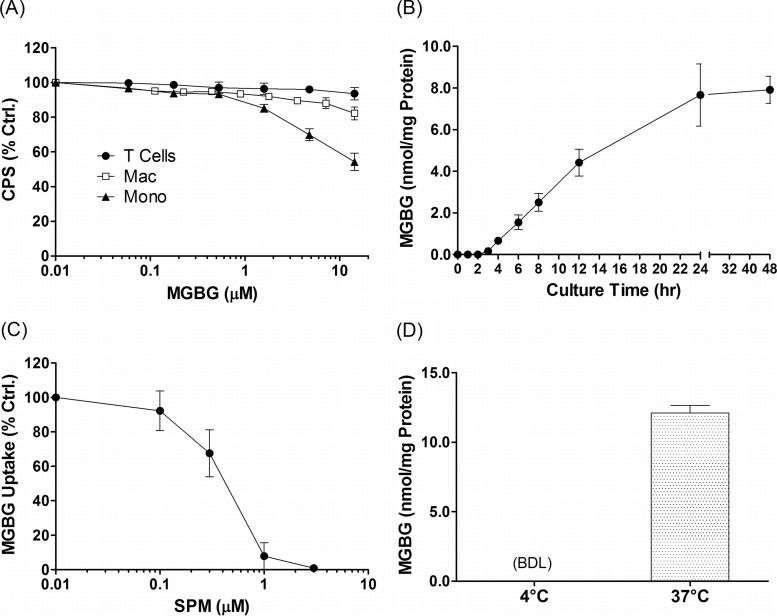
Historically, MGBG had been developed as a cytotoxic agent for treatment of cancer. In order to test the effects of MGBG on HIV in macrophage cultures, we first determined its in vitro cytotoxicity using the CellTiter-Glo assay, which measures the level of ATP in metabolically active cells. The results showed that MGBG had various levels of cytotoxic effect toward monocytes and macrophages, while resting T cells were completely insensitive to the drug (Fig. 1A). Nevertheless, even monocytes, the most sensitive cell type, showed no significant cell death at MGBG concentrations of 1.4 μM or less after a 24-h exposure. Interestingly, the fully differentiated macrophages were much less sensitive than freshly isolated monocytes, with no significant toxicity detected at close to 10 μM MGBG. Based on these results, we performed all subsequent studies at these noncytotoxic doses.
FIG 1.
Kinetics and mechanism of MGBG uptake in cultured primary human monocytes. (A) Effect of MGBG on cell viability. Purified human CD14+ monocytes (Mono), in vitro differentiated macrophages (Mac), and CD3+ cells were cultured in 96-well plates with various concentrations of MGBG in triplicate for 24 h, and the relative number of viable cells in each well was measured using the CellTiter-Glo kit. The signals were normalized against that of the untreated (control [Ctrl.]) cells. (B) Time course of MGBG uptake in cultured monocytes. Human monocytes were purified and cultured with 1.4 μM MGBG, and cells were collected at various time points for quantification of both MGBG and total cellular proteins. The amount of MGBG was normalized against the total amount of proteins in the lysates. The analytical limit of detection for MGBG is 10 nM or ∼55 pmol/mg protein. (C) Competition of MGBG uptake in monocytes by spermine (SPM). Purified human CD14+ cells were cultured in the presence of 1.4 μM MGBG and increasing concentrations of SPM for 24 h before being harvested for MGBG and protein measurement. Data are expressed as a percentage of the MGBG concentration in the absence of SPM. (D) Temperature effect on MGBG uptake in monocytes. Purified human monocytes were cultured with 1.4 μM MGBG at 4 or 37°C for 24 h before being collected for MGBG and protein measurement. BDL, below the analytical detection limit. Data represent the mean results from at least 3 independent donors, with error bars showing the standard error of the mean (SEM) for panels A to C. Data are presented as the mean and SEM from duplicates of a single donor for panel D.
Initially, a time course experiment was performed to determine the kinetics of MGBG uptake in freshly isolated human monocytes. The cells were exposed to 1.4 μM MGBG for various lengths of time before being harvested for protein and MGBG quantification. As shown in Fig. 1B, MGBG was rapidly taken up by the monocytes and was clearly detectable within 3 to 4 h. The drug level reached an apparent steady state by 24 h, with no further increase upon longer incubation (Fig. 1B). Next, we explored the potential mechanism of MGBG uptake in these cells. Natural polyamines such as spermidine and spermine are normally taken up via the cell membrane-associated polyamine transporter in an ATP-dependent manner. MGBG is known to compete with spermidine for uptake in established cell lines (53). To confirm that such a mechanism also operates in monocytes/macrophages, the effect of exogenously added spermine on MGBG uptake was examined. As expected, spermine blocked MGBG uptake in the cultured monocytes in a dose-dependent manner (Fig. 1C), consistent with the involvement of the polyamine transporter in MGBG uptake.
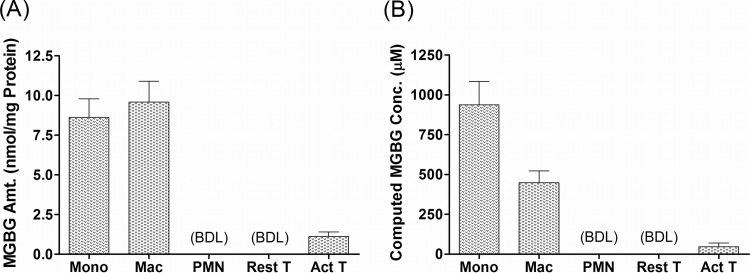
To determine if MGBG uptake was cell type specific, we quantified its level in primary monocytes, macrophages, resting T cells, PHA-activated T cells, and granulocytes. The results are shown in Fig. 2. After exposure to 1.4 μM MGBG for 24 h, monocytes and macrophages took up a very high level of the compound, with estimated intracellular concentrations reaching 0.94 mM in monocytes and 0.45 mM in macrophages. These concentrations translate into enrichment factors of ∼670- and 320-fold in monocytes and macrophages, respectively. Note that the level of intracellular MGBG in these cells is similar when normalized to the amount of cellular proteins in the lysates, but the concentration is roughly 2 times higher in the monocytes than that in the macrophages, probably due to the larger size of the latter cell type. This may help to explain the differential sensitivities of these cell populations to the drug (Fig. 1A). In sharp contrast to monocytes and macrophages, resting T cells and granulocytes had no detectable intracellular MGBG, whereas there was a low level of uptake in PHA-activated T cells, which is consistent with previous reports (54, 55). Overall, monocytes/macrophages appear to be the major blood cell population targeted by MGBG and can apparently concentrate the drug by several hundred-fold without significant cytotoxicity under the conditions tested.
FIG 2.
Cell-type-specific uptake of MGBG in primary human blood cells. Different cell types prepared from human PBMCs or white blood cells were cultured with 1.4 μM MGBG for 24 h. Cell lysates were prepared from the cell for quantification of MGBG and cellular proteins. MGBG uptake was either normalized to the amount of cellular proteins (A) or converted to intracellular MGBG concentrations by using published mean cell volumes (B). PMN, polymorphonuclear leukocytes. The mass peaks for MGBG in the resting T cells and granulocytes are below the analytical detection limit (BDL), which is 10 nM in the lysates, ∼120 pmol/mg protein for the resting T cells, and 70 pmol/mg protein for the granulocytes. The graphs show means and SEMs (n ≥ 3).
Since MGBG is taken up via the polyamine transport system, which is energy and temperature dependent, we also tested MGBG uptake in monocytes at 4°C versus 37°C. The results confirmed that MGBG uptake in monocytes is temperature dependent (Fig. 1D). The MGBG concentration was below the detection level in monocytes after a 24-h incubation with the drug at 4°C.
MGBG inhibits HIV-1 expression in cultured macrophages.
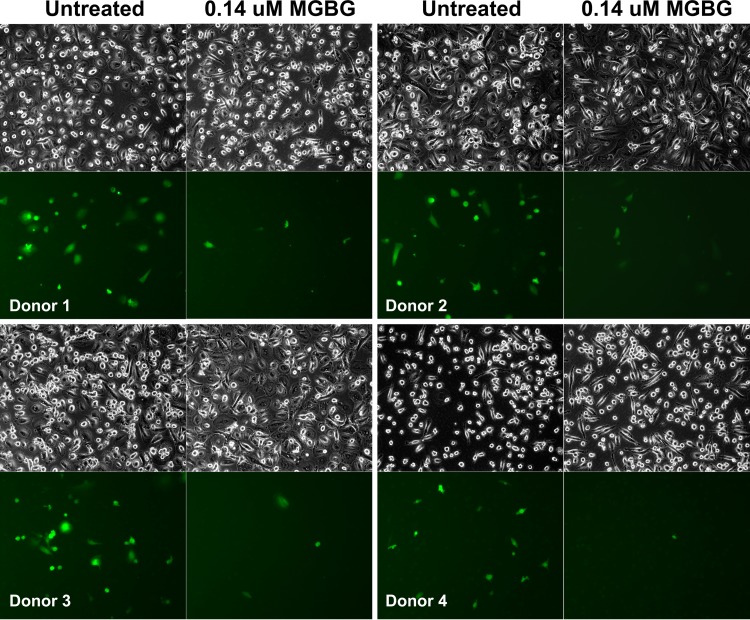
Using the EGFP-tagged macrophage-tropic HIV construct developed by Cheng-Meyer (48), we tested the effect of MGBG on macrophage infection by HIV. When examined under a fluorescence microscope, we observed a significant reduction in the proportion of EGFP-HIV+ macrophages following treatment with 0.14 μM MGBG (Fig. 3, bottom rows). The phase-contrast images of the same cells confirmed that MGBG treatment in this setting showed no obvious cytotoxicity (Fig. 3, top rows). These results were consistently seen in different donors (i.e., inhibition of HIV expression was observed in all cases).
FIG 3.
Inhibition of HIV expression by a noncytotoxic dose of MGBG in human macrophages. Human blood monocytes were isolated from 3 healthy donors and allowed to differentiate for 7 days in the culture. The cells were then infected with EGFP-tagged HIV for 6 h and cultured for another week. MGBG was present throughout the process. EGFP+ cells were visualized under a fluorescence microscope; all cells in the corresponding fields were also imaged by phase-contrast microscopy.
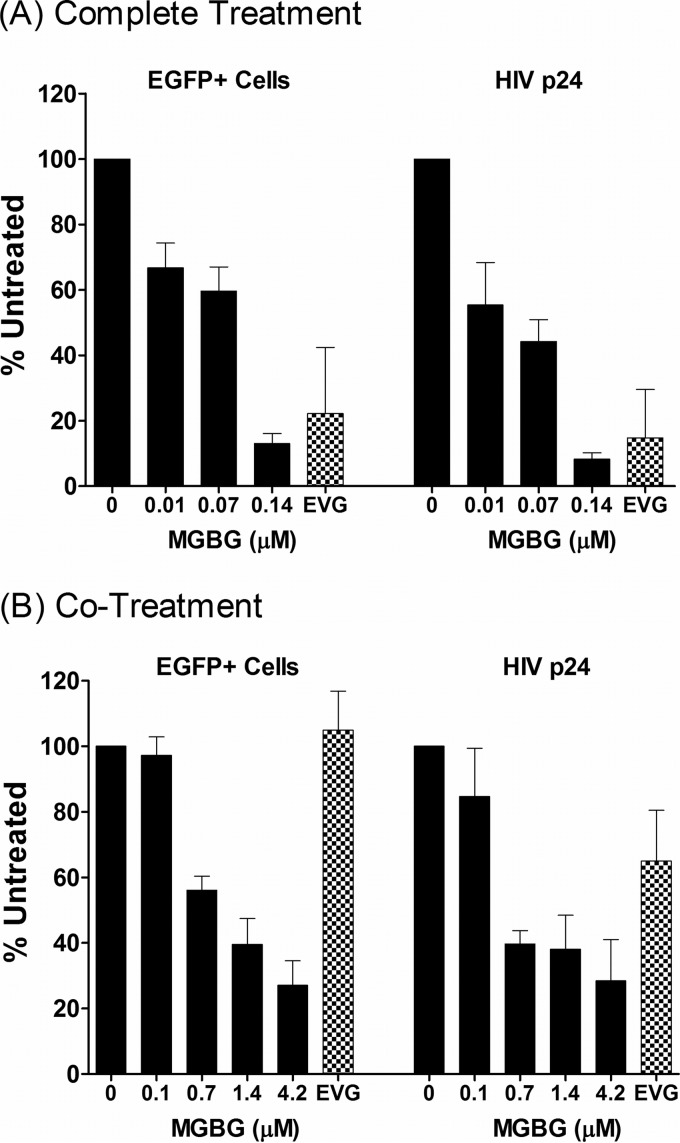
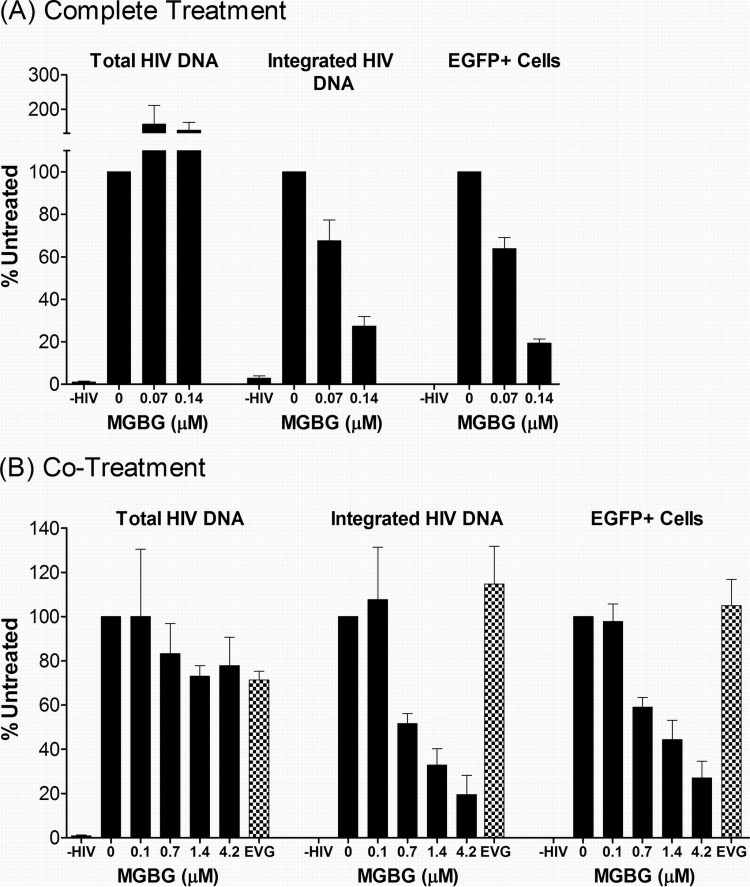
This phenomenon was further investigated using two different treatment modalities. In the “complete treatment” mode, MGBG was present throughout the experimental regimen: 6- to 7-day monocyte-to-macrophage differentiation, 6-h infection with the viral supernatant, and 7-day-postinfection culturing. In the “cotreatment” mode, MGBG was added to the fully differentiated macrophages only during the 6-h infection period. Figure 4 shows the summary of anti-HIV studies performed on cells from multiple donors. In both treatment modalities, a statistically significant linear trend in the association between MGBG dose and effect on EGFP was observed for both complete (P = 0.007) and cotreatment (P = 0.009) modes, as were the associations for p24 (P = 0.0002 and P = 0.0009 for complete treatment and cotreatment, respectively). However, after adjustment for deviations from sphericity (based on Greenhouse-Geiser corrections), the association for EGFP was attenuated (P = 0.10 and P = 0.11 for complete treatment and cotreatment, respectively), and a linear effect was observed for p24 only (P = 0.008 and P = 0.02 for complete treatment and cotreatment, respectively). In these experiments, elvitegravir (EVG), an approved HIV drug which is a potent inhibitor of the viral integrase with a 50% inhibitory concentration (IC50) of 1 nM or less in cultured T cells, was used as a positive control. In the complete treatment mode, 0.14 μM MGBG caused ∼90% inhibition of HIV expression, as measured by both FACS analysis of EGFP expression and ELISA quantification of supernatant HIV p24 antigen 1 week post-initial infection (Fig. 4A; see Fig. S2 in the supplemental material). In comparison, EVG at a concentration that exceeds 20 times the IC50 in cell culture (56) caused an 80% reduction in HIV-expressing macrophages.
FIG 4.
Dose-dependent inhibition of HIV expression in human macrophages following MGBG treatment. In vitro differentiated human macrophages were infected with EGFP-tagged HIV for 6 h and cultured for another week. MGBG was either present continuously (the “complete treatment” mode) or only during the 6-h acute infection period (the “cotreatment” mode). EGFP expression in the cells was determined by FACS, and the p24 antigen in the culture supernatant was quantified by ELISA. Due to individual variations, the data were normalized against an HIV-infected, nontreated sample in each experiment. The graphs show the means and SEMs of the normalized results from multiple donors. Noninfected cells were included as a negative control for assay specificity. A 20 nM concentration of elvitegravir (EVG) was included in these experiments as a positive control. The results for the complete treatment mode were from 5 donors, and those for the cotreatment mode were from 3 donors. The average percentage of HIV-infected, untreated EGFP cells was around 5%, and the average HIV p24 concentration of HIV-infected, untreated cells was about 10 ng/ml. A statistically significant linear trend in the association between MGBG dose and effect on p24 was observed for both complete (P = 0.008) and cotreatment (P = 0.02) modes. However, the association for EGFP was attenuated after Greenhouse-Geiser correction for sphericity. Detailed FACS data for EGFP expression are shown in Fig. S2 and S3 in the supplemental material.
In the complete treatment mode, MGBG could affect multiple biological processes, including monocyte differentiation as well as HIV infection or expression in macrophages. To test whether pretreatment with MGBG was required for the observed inhibition of HIV in macrophages, cotreatment mode studies were conducted. Fully differentiated macrophages were exposed to MGBG and EGFP-HIV supernatant simultaneously for only 6 h. Considering the short exposure time to the drug, higher concentrations of MGBG were employed in these experiments. Importantly, these concentrations showed no cytotoxicity toward macrophages (Fig. 1A) (data not shown) and are potentially achievable in vivo based on pharmacokinetic (PK) studies (Jeremy Blitzer, Philip Needleman, and John McKearn, unpublished data). As shown in Fig. 4B (see Fig. S3 in the supplemental material), MGBG caused sustained inhibition of HIV expression in macrophages in a dose-dependent manner. The results from flow cytometry, which measured EGFP HIV-expressing cells, matched the concurrently measured HIV p24 production in the culture supernatants. These data were qualitatively similar to those observed in the complete treatment mode, although the cotreatment mode was less effective (0.7 μM versus 0.07 μM for ∼50% inhibition). In contrast to MGBG, EVG showed no significant effect on HIV expression in macrophages when present only during the 6-h infection period.
The decrease in p24 level could be due to the presence of fewer infected cells in the entire culture (Fig. 3 to 4) and/or reduced HIV gene expression in each of the infected cells. The latter possibility would be expected to result in a decrease in the mean fluorescence intensity of the EGFP+ cells. In both treatment modes, the mean fluorescence intensities were similar regardless of MGBG treatment (data not shown), which is consistent with the notion that exposure to MGBG resulted in fewer HIV-expressing cells while having little effect on the expression level on a per cell basis.
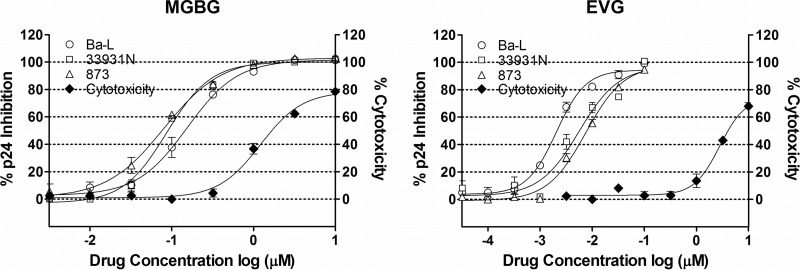
To further confirm that MGBG inhibits HIV in macrophages, we tested the effect of MGBG on multiple primary HIV isolates using a standard antiviral assay (57). Partially differentiated macrophages were infected with the HIV isolates Ba-L, 33931N, and 873 for 2 h at a low MOI (0.005). Subsequently the cells were cultured for 7 days with or without drug treatment, and culture supernatant was collected for p24 measurement. EVG was included as the positive control. As shown in Fig. 5, MGBG was active against all three CCR5-trophic HIV isolates tested, with IC50s of 0.08 to 0.15 μM. To rule out that the effects on HIV were due to cell killing, cytotoxicity was measured in parallel. As summarized in Table 1, the IC50 of MGBG against HIV was significantly (8 to 15 times) below the 50% cytotoxic concentration (CC50), indicating that the activity against HIV was distinct from cytotoxicity.
FIG 5.
Inhibition of primary HIV isolates in MGBG-treated macrophages. Partially differentiated human macrophages were infected with primary HIV isolate Ba-L, 33931N, or 873 for 2 h, followed by drug treatment for 7 days. Culture supernatant was collected for p24 measurement. EVG was used as a positive control. Drug cytotoxicity was assessed in parallel. The results were normalized against an HIV-infected, nontreated sample in each experiment. IC50s were obtained using the Prism software. The HIV p24 concentrations for the infected, untreated controls were between 2.4 and 4 ng/ml.
TABLE 1.
Drug IC50s for primary HIV isolates and cytotoxicity in macrophagesa
| Drug | IC50 (μM) for isolate: |
CC50 (μM) | ||
|---|---|---|---|---|
| Ba-L | 33931N | 873 | ||
| MGBG | 0.155 | 0.093 | 0.079 | 1.22 |
| EVG | 0.002 | 0.005 | 0.008 | 2.79 |
The activity of MGBG on primary HIV isolates was determined using a standard antiviral assay as described in the text. Cytotoxicity (CC50) was determined in parallel. The results were analyzed with the Prism software to obtain the IC50 values.
MGBG treatment inhibits integration of HIV DNA into cultured macrophages.
To understand how MGBG affects the replication cycle of HIV within the cells, we measured the intracellular HIV-1 DNA, both total and integrated forms, by real-time quantitative PCR. The results showed that surprisingly MGBG treatment did not decrease the levels of total HIV DNA in either mode. In fact, the level of total HIV DNA might even be increased in the MGBG-treated cells in the complete treatment mode (Fig. 6A, left set of bars). These results imply that MGBG blocks neither viral penetration into cells nor subsequent reverse transcription and replication of the viral genome. The drug did, however, significantly reduce the integrated proviral form of HIV in the cells. As shown in Fig. 6A, middle set of bars, a dose-dependent reduction of proviral DNA was observed in MGBG-treated cells in the complete treatment mode, with nearly 70% inhibition at 0.14 μM MGBG, which translated to nearly 85% when taking into consideration the total HIV DNA level. At higher MGBG doses than those shown, cellular toxicity in this culture system precluded the interpretation of studies attempting to completely block HIV integration. In the cotreatment mode, a 6-h exposure to MGBG also led to a dose-dependent reduction of proviral DNA load, with ∼50% inhibition at 0.7 μM, and the effect on HIV was not accompanied by any detectable cytotoxicity even at 4.2 μM (Fig. 6B, middle set of bars) (data not shown). In this experimental setting, MGBG also did not decrease the level of total HIV DNA in the cells (Fig. 6B, left set of bars). In contrast to MGBG, the potent HIV integrase inhibitor EVG showed no reduction of the proviral DNA load in the same time frame (6 h). A statistically significant linear trend (P = 0.045) in the association between MGBG dose and integrated HIV DNA was observed for the complete mode. (For the cotreatment mode, the sphericity-adjusted P value is 0.073.) In all experiments, a good correlation between proviral load, EGFP expression, and p24 level was observed in MGBG-treated cells (Fig. 4 and 6; see Fig. S2 to S5 in the supplemental material), suggesting that the primary effect of the compound on HIV is the inhibition of a step or process that results in viral DNA integration.
FIG 6.
Inhibition of HIV DNA integration in MGBG-treated cells. Human macrophages were infected with EGFP-tagged HIV and treated with MGBG in either the “complete treatment” mode or “cotreatment” mode as indicated. At the end of the experiments, a fraction of the cells were used for FACS analysis of EGFP expression, while the rest were used for DNA extraction. Real-time PCR was used to quantify HIV DNA in the total or high-molecular-weight DNA. The copy numbers of β-globin in the samples were simultaneously determined to calculate the load or relative burden of the HIV DNA. Noninfected cells were included as a control for the specificity of the assays. The data for the complete treatment and cotreatment modes were from 7 and 3 donors, respectively. Due to individual variations, all data were normalized against an HIV-infected, nontreated sample in each experiment. The graphs show the means and SEMs of the normalized results from multiple donors. The P values for a linear trend in the association between MGBG dose and effect on proviral DNA were 0.045 and 0.073 for the complete and cotreatment modes, respectively. Detailed FACS data for EGFP expression are shown in Fig. S4 and S5 in the supplemental material.
HIV integration is inhibited by MGBG in both monocytes and macrophages.
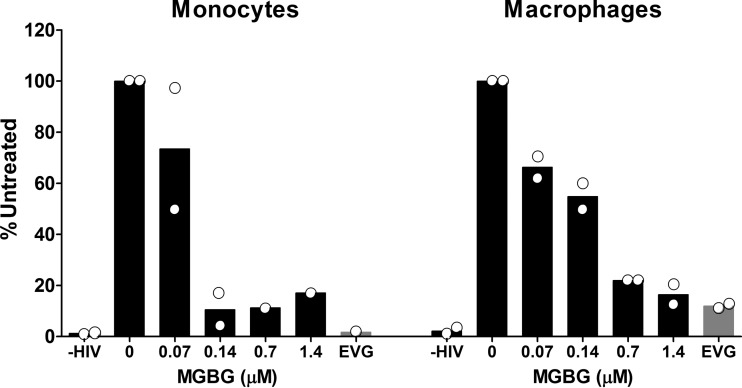
HIV can infect both monocytes and macrophages, although in most experimental systems macrophages are a better target for productive infection (58). To test whether MGBG also interferes with HIV infection of monocytes, experiments using both freshly isolated monocytes and in vitro differentiated macrophages from the same donors were performed. Figure 7 shows the data from 2 donors with both monocyte and macrophage infection, wherein MGBG inhibited HIV integration in both cell types in a dose-dependent manner. In these experiments, MGBG appears to be more potent in monocytes compared to macrophages, which could be due to the higher MGBG intracellular concentrations in monocytes. The average proviral DNA load in untreated monocytes is lower than that in the macrophages (37 versus 121 copies per 1,000 cells, respectively), consistent with other observations that monocytes are not very infectible by HIV, perhaps due to the restriction by SAM domain- and HD domain-containing protein 1 (SAMHD1) (59, 60).
FIG 7.
Inhibition of HIV integration by MGBG in both monocytes and macrophages. Isolated human monocytes were either infected immediately with HIV overnight or allowed to differentiate into macrophages prior to viral infection. Cells were cultured for 2 days after infection before being collected for DNA extraction. Drugs were present both during infection and postinfection. The load of total HIV DNA or integrated proviral DNA was determined by qPCR. The graph shows the means, while the dots indicate the actual values from 2 donors, all normalized against the results for the untreated samples.
Effects of MGBG on HIV receptor CD4 and coreceptor CCR5 expression in macrophages.
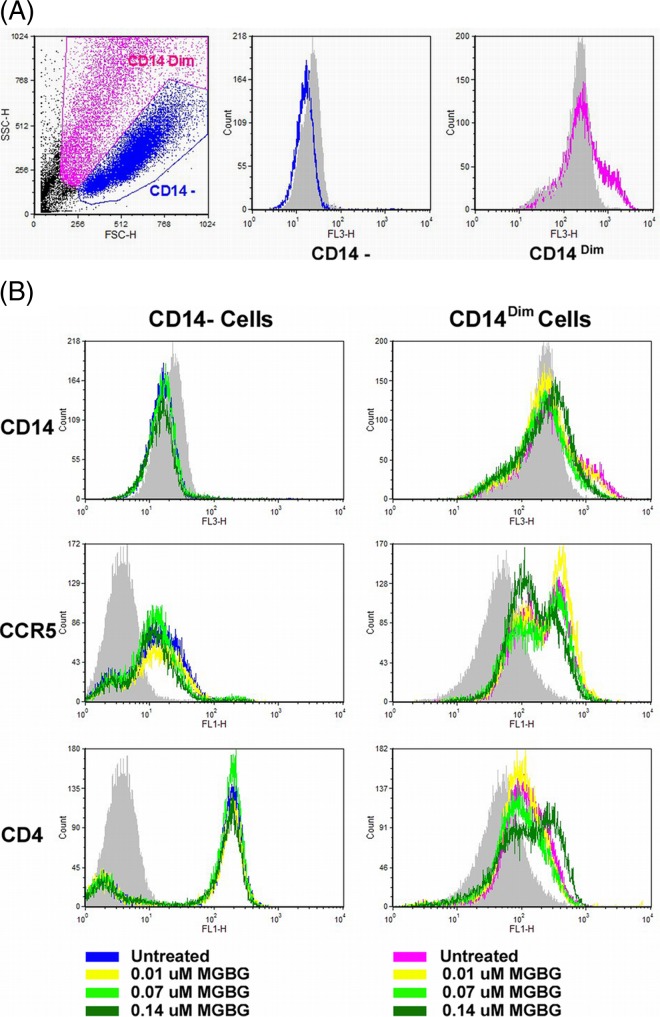
The HIV-1 reporter virus used in this study is a macrophage-trophic strain that uses CCR5 as the coreceptor in combination with the CD4 receptor. In the complete treatment mode, the monocytes were differentiated in the presence of MGBG and the drug was more efficacious. A potential explanation for this is that MGBG might affect the expression of CD4 and/or CCR5. We directly examined CD4 and CCR5 expression in macrophages following MGBG treatment. In this experiment, monocytes were differentiated for 7 days in the absence or presence of various doses of MGBG. The resulting macrophages were analyzed for surface expression of CD4, CCR5, and CD14 by flow cytometry. Like typical macrophages, the cells were very heterogeneous in both size and granularity; and two distinct cell populations were evident in the SSC and FSC plots (Fig. 8A). The levels of CD14 expression also appeared to be slightly different among the two subsets of cells. In the CD14− macrophages, MGBG had no significant effects on the expression of CD4 or CCR5. In the CD14Dim macrophages, MGBG treatment appeared to increase the level of CD4 expression at the higher doses tested, but the level of CCR5 expression was decreased slightly (Fig. 8B).
FIG 8.
Effects of MGBG on CCR5 and CD4 expression in macrophages. Pooled monocytes from 6 healthy donors were differentiated in vitro with or without MGBG treatment for 7 days. Cells were harvested and stained for CCR5, CD4, and CD14 for FACS analysis. (A) Two distinct populations of cells were gated based on forward and side scatters (FSC and SSC, respectively). (B) MGBG treatment showed no apparent effects on CCR5 or CD4 expression in the CD14− macrophages. In the CD14Dim cells, MGBG at 0.14 μM, the highest concentration tested, caused an observable increase in CD4 but a slight decrease in CCR5 expression.
DISCUSSION
Macrophages are one of the HIV reservoirs in vivo, and accumulating evidence suggests that they are involved in the pathogenesis of HIV-1-associated diseases (61–63). Targeting these cells may be a valuable strategy for treating macrophage-driven neurological and cardiovascular diseases that now affect more than 50% of HIV-infected individuals in the United States.
In the present study, we demonstrate that primary monocytes and macrophages take up MGBG in vitro, apparently via the polyamine transporter system. The uptake appears to be cell type specific as several other types of circulating blood cells take up either no drug or drug at a level 10 to 20 times lower than that found in monocytes and macrophages. Besides granulocytes and T cells, MGBG uptake was also tested in isolated B cells. However, due to the low abundance of this cell population in the peripheral blood, cell sorting with the CD20 microbeads was not very effective, resulting in only 30 to 50% purity. A low level of MGBG was observed in these cells (data not shown), which could be from contaminating monocytes. The capacity to rapidly and selectively concentrate in macrophages and monocytes appears to be a unique characteristic of the compound. The magnitude of MGBG accumulation in these cells is likely to be different in vitro versus in vivo. In culture, the intracellular concentrations of MGBG can reach a micromolar concentration of tens to hundreds, depending on the cell type. In vivo, the compound is expected to accumulate to a lesser extent due to the presence of low-level natural polyamines in the blood (64, 65), which can compete with MGBG for cellular uptake. This property, together with the cell type selective uptake, may contribute to the excellent tolerability and safety of the drug when given orally to animals in preclinical testing (Blitzer et al., unpublished).
All experiments reported in this article were performed using noncytotoxic doses of MGBG, and we focused on acute HIV infection rather than addressing macrophages that might be chronically infected with HIV. Most of our studies were performed using the EGFP-tagged HIV strain (48), which allows easy tracking of viral expression by flow cytometry and microscopy. Initially, we tested MGBG in a “complete treatment” mode, wherein the drug was present throughout the experiment. Since MGBG is readily detectable in macrophages after just a few hours of exposure, subsequent studies utilized a “cotreatment” mode wherein MGBG was present only during the 6-h incubation with the virus. Although the complete treatment modality is more efficacious, MGBG in the cotreatment mode also shows significant activity. Despite its highly reproducible activities, MGBG treatment did not completely abolish the expression of the EGFP-tagged HIV in the macrophage culture system. One possible explanation is that a subset of the cells in blood-derived macrophage cultures may express a lower level of the polyamine transporter, which led to a lower level of MGBG accumulation. Another possibility is that the extent of viral spreading might be limited because we infected the cells with a relatively large quantity of the viral supernatant (the equivalent of ∼1 MOI), which resulted in the infection of most if not all of the infectible cells at once. Other possibilities also exist. Of interest, such incomplete inhibition of HIV in macrophage cultures has also been reported for several approved integrase inhibitors (24, 66). In these studies, T cells were completely protected from HIV, whereas residual infection persisted in macrophage cultures despite the high drug concentrations employed.
To confirm that activity of MGBG is not strain specific, we have tested the effect of MGBG on multiple primary HIV isolates using a standard antiviral assay. Unlike the studies with the EGFP-tagged virus performed at high virus input, these experiments used a low MOI in order to capture anti-HIV data that reflected the blockade of viral spreading throughout the macrophage cultures. The new data show an almost complete protection of the cultures in a dose-dependent manner. MGBG was active against all three CCR5-trophic HIV isolates tested, with IC50 values of 0.08 to 0.15 μM (Fig. 5). Importantly, the activity against HIV was clearly separable from drug-associated cytotoxicity measured in parallel.
To test where in the retroviral life cycle MGBG exerts its inhibitory activity, we evaluated the intracellular fate of HIV DNA within infected macrophages by qPCR. Because of the low level of HIV infection in monocytes/macrophages (<1 copy/genome in most experiments [data not shown]), the Alu-PCR assay typically used for quantifying HIV integration was not expected to have sufficient sensitivity in our system. We therefore employed another published method, which separates proviruses integrated into high-molecular-weight genomic DNA from free, unintegrated HIV DNA based on their differential mobilities in low-concentration agarose gel. MGBG treatment does not decrease the level of total intracellular HIV DNA; however, integrated proviral DNA is significantly reduced in a dose-dependent manner. Therefore, MGBG does not appear to interfere with early viral infection events, such as attachment, viral penetration, or reverse transcription of HIV RNA. The primary effect of the drug treatment seems to be the reduction of integrated viral DNA, although the precise molecular mechanism is currently unknown.
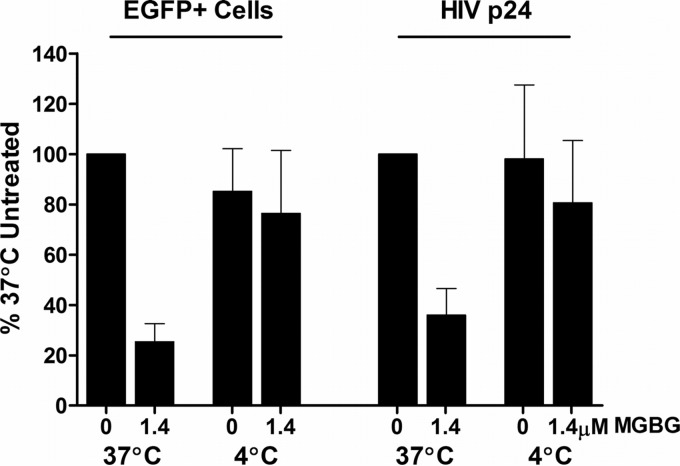
To further confirm that MGBG acts intracellularly post-virion attachment, we compared its effect on HIV infection in macrophages at 37°C versus 4°C in the cotreatment mode. Incubation of macrophages with the viral supernatant and MGBG for 6 h at 37°C inhibited HIV expression as expected. However, the inhibitory activity is abrogated by parallel infection/treatment performed at 4°C (Fig. 9; see Fig. S6 in the supplemental material). At 4°C, HIV virions can still attach to the cells (67), but MGBG does not enter the cells since the uptake of MGBG via the polyamine transporter is energy dependent and thus temperature sensitive (Fig. 1D). When the culture was subsequently brought back to 37°C, the attached virus could enter the cells and continue with the rest of the replication cycle (67). These results suggest that the inhibition of HIV is mediated by intracellular MGBG, and the extracellular MGBG in the media as a polycation had no apparent effect on viral attachment.
FIG 9.
Temperature-sensitive inhibition of HIV-1 expression by MGBG in macrophages. In vitro differentiated human macrophages were infected with HIV-1 and treated with or without 1.4 μM MGBG for 6 h at either 37 or 4°C as indicated. After drug removal, the cells were cultured for 7 more days before harvested for both EGFP and p24 measurements. The experiments were set up in duplicate using cells from a single donor. Detailed FACS data for EGFP expression are shown in Fig. S6 in the supplemental material.
In the macrophage culture system, the complete treatment mode is significantly more effective than the cotreatment mode. The difference in drug potencies between the two modes is likely due to the difference in drug uptake levels. It takes ∼24 h for MGBG to accumulate to a steady-state level in the cells (Fig. 1B). The drug uptake level in the complete treatment mode should be much higher than that in the cotreatment mode. Another interesting observation we made in the course of these studies is that the activity of MGBG is relatively fast acting, with significant efficacy after just a 6-h exposure. By comparison, the potent HIV integrase inhibitor EVG showed little efficacy under the same conditions. The antiviral activity of EVG in T cells has been documented (56), but the kinetics of EVG uptake in macrophages has not been reported. It is possible that EVG has slower uptake kinetics in macrophages than MGBG. Cobicistat, a potent inhibitor of cytochrome P450 3A enzymes and intestinal transport proteins, has been used together with EVG to enhance overall drug absorption in the clinical settings (68, 69).
How MGBG exerts its effects on HIV integration remains to be determined. The activity of MGBG as an inhibitor of SAMDC has been extensively described in the early studies of the compound, primarily in the context of its potential use as an anticancer agent (53, 70). Multiple studies have confirmed that MGBG is indeed a potent inhibitor of SAMDC (32, 33; Blitzer et al., unpublished). Intracellular polyamine depletion, which can result from SAMDC inhibition by MGBG, is also known to interfere with normal chromatin structure (71). A more direct role for MGBG at the level of the chromosome is also a possibility. It has been shown that MGBG can directly interact with DNA (72). Therefore, MGBG interaction with DNA or chromosome, either directly or indirectly, may result in inhibition of HIV integration. Yet another possibility is that MGBG may directly inhibit the HIV integrase in a manner similar to those of other DNA modulators, such as topoisomerase inhibitors, which have activity in HIV integrase assays (73). Although polyamines have been reported to be involved in HIV reverse transcription (74), this step of the viral life cycle is unlikely the target for MGBG in macrophages because we did not detect any decrease in the level of total HIV DNA following MGBG treatment. Alternatively, MGBG could block integrase activity indirectly through its interaction with host proteins. Host cellular proteins such as LEDGF/p75 and TRN-SR2 have been shown to act as cofactors for lentiviral integrase (75). The possibility that MGBG treatment might lead to conformational changes in the viral DNA, which renders it incompetent for integration, cannot be precluded either. Obviously, more work will be needed to address the mechanism.
Macrophage infection by CCR5-trophic HIV isolates potentially could be modulated through CD4 and CCR5 (76). We thus examined the effect of MGBG on CCR5 and CD4 expression in a time frame that mimicked the first stage of the complete treatment mode. As shown in Fig. 8, exposure to MGBG during monocyte differentiation caused an observable increase in CD4 and a slight decrease in CCR5 expression in CD14Dim cells and had no apparent effect in CD14− cells. It has been reported that CD4 level is a limiting factor for HIV infection of macrophages, while changes in CCR5 level have little effect (77). Thus, the inhibitory effect of MGBG on HIV infection is unlikely through CD4 and CCR5 modulation.
An increased total HIV DNA level was seen in the complete treatment mode. The observed increase in CD4 level may contribute to the increased total HIV DNA load. Consistent with this hypothesis, in the cotreatment mode, MGBG showed no significant effect on total HIV DNA level. Alternatively, MGBG-mediated blockage of HIV proviral DNA integration may drive the accumulation of unintegrated DNA. Another possible explanation is that the high level of MOI in this culture system could result in macrophage superinfection by HIV, a process known to occur in vivo (23). The observation that MGBG neither downmodulates the CD4 nor decreases the levels of unintegrated DNA is consistent with the interpretation that the drug does not negatively affect viral adhesion or the subsequent reverse transcription process.
Anti-HIV activities have also been reported with other inhibitors of polyamine biosynthesis. For example, several SAMDC inhibitors, including 5′-5′-deoxyadenosine and 1-aminoxyethylamine, can block HIV replication in vitro (78). SAM486A, a very potent SAMDC inhibitor, apparently suppresses HIV replication by inhibiting HIV Rev function via interference with the hypusination of eukaryotic initiation factor 5A (eIF5A), the Rev cofactor. As a result, expression from the HIV-1 promoter at the level of transcription initiation was blocked (79). As a SAMDC inhibitor, MGBG could theoretically affect eIF5A maturation in a similar fashion. However, the overall data presented—especially the observation that MGBG treatment has no inhibitory effect on total HIV DNA—suggest a novel anti-HIV activity for MGBG that is not related to eIF5A, although the exact mechanism remains to be defined.
The activity of MGBG against HIV may have several therapeutic implications. Macrophages are one of the earliest infected cell types (12–15), with the resultant HIV strains uniformly falling into the CCR5 coreceptor-utilizing “macrophage-tropic” genotype. Unlike T cells, which are prone to the cytopathic effect of HIV, macrophages—especially those in the CNS—are long lived and can be superinfected by HIV in vivo (23). Infected macrophages may in turn efficiently infect T cells and upon HAART discontinuation can serve as a source of recurrent viral spread. Currently, there is no effective agent that completely addresses the macrophage HIV reservoir in the CNS. Virtually all drugs currently approved for treatment of HIV infection are markedly less effective in macrophages than in T cells (24). More importantly, to be active in the CNS, HIV drugs must cross the blood-brain barrier, a property shown by few of the existing drugs (24, 80). In contrast, MGBG is selectively taken up by macrophages, readily crosses the blood-brain barrier, and appears efficacious in vivo, as shown in recent studies in a simian immunodeficiency virus encephalopathy (SIVE) model. In these studies with SIV infection in CD8-depleted macaques, all animals in the vehicle control arm developed AIDS, most with SIVE and myocarditis, whereas almost none of the MGBG treated animals developed AIDS, and none had SIVE or myocarditis (81, 82; Ken Williams, unpublished data). The SIVE study suggests that even though low concentrations of natural polyamines are present in the blood (64, 65), sufficient amounts of MGBG are getting into macrophages in vivo and most importantly into the CNS to mediate significant macrophage-targeted effects. In addition to the activity against HIV described here, MGBG has been shown to inhibit macrophage activation mediated by type I macrophage activators, such as tumor necrosis factor alpha (TNF-α), gamma interferon, and lipopolysaccharide (LPS) (41, 42). Considering the importance of inflammation in the pathogenesis of HAND and atherosclerosis driven in a large part by HIV-infected macrophages, the use of MGBG provides an approach against both HIV infection and the associated pathogenic inflammation. Finally, this property, together with its selective uptake by monocytes and macrophages, suggests that MGBG may be effective against other monocyte/macrophage-driven diseases, such as liver fibrosis (83).
In summary, this study demonstrates that MGBG exhibits selective uptake and is rapidly concentrated in primary human monocytes and macrophages. At nontoxic doses, it inhibits multiple HIV-1 strains in macrophages, most likely through reduced HIV DNA integration. This activity, together with its effects on macrophage activation, suggests that MGBG may be a valuable addition to the treatment of HIV-associated diseases, where chronic macrophage-associated immune activation plays a major role.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to William Rutter for guidance and support throughout the project. In addition, we thank Amanda Brown for kindly providing the reporter virus construct, Stephanie Yu for assistance with the drug uptake experiments, Cheryl Stoddard and Pheroze Joshi for work on primary HIV isolates, Paige Bracci for help with statistical analysis, and Philip Needleman, John McKearn, and Jeremy Blitzer for information on MGBG pharmacology data.
This study was supported in part by the grant Macrophage Targeted Therapy for HAD and HIV Disease (PPG)—5U19MH081835-05.
Xia Jin and Hua Xu are employees of Pathologica; Michael S. McGrath is a consultant and shareholder of the company.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01692-15.
REFERENCES
- 1.Le Douce V, Herbein G, Rohr O, Schwartz C. 2010. Molecular mechanisms of HIV-1 persistence in the monocyte-macrophage lineage. Retrovirology 7:32. doi: 10.1186/1742-4690-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geeraert L, Kraus G, Pomerantz RJ. 2008. Hide-and-seek: the challenge of viral persistence in HIV-1 infection. Annu Rev Med 59:487–501. doi: 10.1146/annurev.med.59.062806.123001. [DOI] [PubMed] [Google Scholar]
- 3.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. 2009. The challenge of finding a cure for HIV infection. Science 323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 4.Riva E, Antonelli G, Solmone MC, Turriziani O, Narciso P, Tozzi V, Dianzani F. 2000. Significant reduction in HIV-1 plasma viral load but not in proviral infected cells during sub-optimal antiretroviral therapy. J Biol Regul Homeost Agents 14:1–3. [PubMed] [Google Scholar]
- 5.Ibáñez A, Puig T, Elias J, Clotet B, Ruiz L, Martínez MA. 1999. Quantification of integrated and total HIV-1 DNA after long-term highly active antiretroviral therapy in HIV-1-infected patients. AIDS 13:1045–1049. doi: 10.1097/00002030-199906180-00007. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Chung C, Hu BS, He T, Guo Y, Kim AJ, Skulsky E, Jin X, Hurley A, Ramratnam B, Markowitz M, Ho DD. 2000. Genetic characterization of rebounding HIV-1 after cessation of highly active antiretroviral therapy. J Clin Invest 106:839–845. doi: 10.1172/JCI10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrigan PR, Whaley M, Montaner JS. 1999. Rate of HIV-1 RNA rebound upon stopping antiretroviral therapy. AIDS 13:F59–F62. doi: 10.1097/00002030-199905280-00001. [DOI] [PubMed] [Google Scholar]
- 8.Alexaki A, Liu Y, Wigdahl B. 2008. Cellular reservoirs of HIV-1 and their role in viral persistence. Curr HIV Res 6:388–400. doi: 10.2174/157016208785861195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koppensteiner H, Brack-Werner R, Schindler M. 2012. Macrophages and their relevance in human immunodeficiency virus type I infection. Retrovirology 9:82. doi: 10.1186/1742-4690-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siliciano RF, Greene WC. 2011. HIV latency. Cold Spring Harb Perspect Med 1:a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiramizu B, Gartner S, Williams A, Shikuma C, Ratto-Kim S, Watters M, Aguon J, Valcour V. 2005. Circulating proviral HIV DNA and HIV-associated dementia. AIDS 19:45–52. doi: 10.1097/00002030-200501030-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP Jr, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. 2013. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A 110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen R, Richter HE, Clements RH, Novak L, Huff K, Bimczok D, Sankaran-Walters S, Dandekar S, Clapham PR, Smythies LE, Smith PD. 2009. Macrophages in vaginal but not intestinal mucosa are monocyte-like and permissive to human immunodeficiency virus type 1 infection. J Virol 83:3258–3267. doi: 10.1128/JVI.01796-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman CM, Wu L. 2009. HIV interactions with monocytes and dendritic cells: viral latency and reservoirs. Retrovirology 6:51. doi: 10.1186/1742-4690-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philpott SM. 2003. HIV-1 coreceptor usage, transmission, and disease progression. Curr HIV Res 1:217–227. doi: 10.2174/1570162033485357. [DOI] [PubMed] [Google Scholar]
- 16.Waki K, Freed EO. 2010. Macrophages and cell-cell spread of HIV-1. Viruses 2:1603–1620. doi: 10.3390/v2081603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald D, Wu L, Bohks SM, KewalRamani VN, Unutmaz D, Hope TJ. 2003. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 300:1295–1297. doi: 10.1126/science.1084238. [DOI] [PubMed] [Google Scholar]
- 18.Granelli-Piperno A, Delgado E, Finkel V, Paxton W, Steinman RM. 1998. Immature dendritic cells selectively replicate macrophagetropic (M-tropic) human immunodeficiency virus type 1, while mature cells efficiently transmit both M- and T-tropic virus to T cells. J Virol 72:2733–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharova N, Swingler C, Sharkey M, Stevenson M. 2005. Macrophages archive HIV-1 virions for dissemination in trans. EMBO J 24:2481–2489. doi: 10.1038/sj.emboj.7600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho DD, Rota TR, Hirsch MS. 1986. Infection of monocyte/macrophages by human T lymphotropic virus type III. J Clin Invest 77:1712–1715. doi: 10.1172/JCI112491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson JK, Cross GD, Callaway CS, McDougal JS. 1986. In vitro infection of human monocytes with human T lymphotropic virus type III/lymphadenopathy-associated virus (HTLV-III/LAV). J Immunol 137:323–329. [PubMed] [Google Scholar]
- 22.Williams KC, Corey S, Westmoreland SV, Pauley D, Knight H, deBakker C, Alvarez X, Lackner AA. 2001. Perivascular macrophages are the primary cell type productively infected by simian immunodeficiency virus in the brains of macaques: implications for the neuropathogenesis of AIDS. J Exp Med 193:905–915. doi: 10.1084/jem.193.8.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamers SL, Salemi M, Galligan D, de Oliveira T, Fogel GB, Granier SC, Zhao L, Brown JN, Morris A, Masliah E, McGrath MS. 2009. Extensive HIV-1 intra-host recombination is common in tissues with abnormal histopathology. PLoS One 4:e5065. doi: 10.1371/journal.pone.0005065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavegnano C, Detorio MA, Bassit L, Hurwitz SJ, North TW, Schinazi RF. 2013. Cellular pharmacology and potency of HIV-1 nucleoside analogs in primary human macrophages. Antimicrob Agents Chemother 57:1262–1269. doi: 10.1128/AAC.02012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams KC, Hickey WF. 2002. Central nervous system damage, monocytes and macrophages, and neurological disorders in AIDS. Annu Rev Neurosci 25:537–562. doi: 10.1146/annurev.neuro.25.112701.142822. [DOI] [PubMed] [Google Scholar]
- 26.Gavegnano C, Schinazi RF. 2009. Antiretroviral therapy in macrophages: implication for HIV eradication. Antivir Chem Chemother 20:63–78. doi: 10.3851/IMP1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garden GA. 2002. Microglia in human immunodeficiency virus-associated neurodegeneration. Glia 40:240–251. doi: 10.1002/glia.10155. [DOI] [PubMed] [Google Scholar]
- 28.Crowe SM, Westhorpe CL, Mukhamedova N, Jaworowski A, Sviridov D, Bukrinsky M. 2010. The macrophage: the intersection between HIV infection and atherosclerosis. J Leukoc Biol 87:589–598. doi: 10.1189/jlb.0809580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gannon P, Khan MZ, Kolson DL. 2011. Current understanding of HIV-associated neurocognitive disorders pathogenesis. Curr Opin Neurol 24:275–283. doi: 10.1097/WCO.0b013e32834695fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tulpule A, Espina BM, Pedro Santabarbara AB, Palmer M, Schiflett J, Boswell W, Smith S, Levine AM. 2004. Treatment of AIDS related non-Hodgkin's lymphoma with combination mitoguazone dihydrochloride and low dose CHOP chemotherapy: results of a phase II study. Invest New Drugs 22:63–68. doi: 10.1023/B:DRUG.0000006175.32100.2c. [DOI] [PubMed] [Google Scholar]
- 31.Herr HW, Kleinert EL, Conti PS, Burchenal JH, Whitmore WF Jr. 1984. Effects of alpha-difluoromethylornithine and methylglyoxal bis(guanylhydrazone) on the growth of experimental renal adenocarcinoma in mice. Cancer Res 44:4382–4385. [PubMed] [Google Scholar]
- 32.Williams-Ashman HG, Schenone A. 1972. Methyl glyoxal bis(guanylhydrazone) as a potent inhibitor of mammalian and yeast S-adenosylmethionine decarboxylases. Biochem Biophys Res Commun 46:288–295. doi: 10.1016/0006-291X(72)90661-4. [DOI] [PubMed] [Google Scholar]
- 33.Corti A, Dave C, Williams-Ashman HG, Mihich E, Schenone A. 1974. Specific inhibition of the enzymic decarboxylation of S-adenosylmethionine by methylglyoxal bis(guanylhydrazone) and related substances. Biochem J 139:351–357. doi: 10.1042/bj1390351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pegg AE. 1973. Inhibition of spermidine formation in rat liver and kidney by methylglyoxal bis(guanylhydrazone). Biochem J 132:537–540. doi: 10.1042/bj1320537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabor CW, Tabor H. 1976. 1,4-Diaminobutane (putrescine), spermidine, and spermine. Annu Rev Biochem 45:285–306. doi: 10.1146/annurev.bi.45.070176.001441. [DOI] [PubMed] [Google Scholar]
- 36.Pegg AE, McCann PP. 1982. Polyamine metabolism and function. Am J Physiol 243:C212–C221. [DOI] [PubMed] [Google Scholar]
- 37.Jänne J, Hölttä E, Kallio A, Käpyaho K. 1983. Role of polyamines and their antimetabolites in clinical medicine. Spec Top Endocrinol Metab 5:227–293. [PubMed] [Google Scholar]
- 38.Ellery PJ, Tippett E, Chiu YL, Paukovics G, Cameron PU, Solomon A, Lewin SR, Gorry PR, Jaworowski A, Greene WC, Sonza S, Crowe SM. 2007. The CD16+ monocyte subset is more permissive to infection and preferentially harbors HIV-1 in vivo. J Immunol 178:6581–6589. doi: 10.4049/jimmunol.178.10.6581. [DOI] [PubMed] [Google Scholar]
- 39.Ziegler-Heitbrock L. 2007. The CD14+ CD16+ blood monocytes: their role in infection and inflammation. J Leukoc Biol 81:584–592. [DOI] [PubMed] [Google Scholar]
- 40.Fischer-Smith T, Croul S, Sverstiuk AE, Capini C, L'Heureux D, Régulier EG, Richardson MW, Amini S, Morgello S, Khalili K, Rappaport J. 2001. CNS invasion by CD14+/CD16+ peripheral blood-derived monocytes in HIV dementia: perivascular accumulation and reservoir of HIV infection. J Neurovirol 7:528–541. doi: 10.1080/135502801753248114. [DOI] [PubMed] [Google Scholar]
- 41.Messina L, Spampinato G, Arcidiacono A, Malaquarnera L, Paqano M, Kaminska B, Kaczmarek L, Messina A. 1992. Polyamine involvement in functional activation of human macrophages. J Leukoc Biol 52:585–587. [DOI] [PubMed] [Google Scholar]
- 42.Kaczmarek L, Kaminska B, Messina L, Spampinato G, Arcidiacono A, Malaquarnera L, Messina A. 1992. Inhibitors of polyamine biosynthesis block tumor necrosis factor-induced activation of macrophages. Cancer Res 52:1891–1894. [PubMed] [Google Scholar]
- 43.Brown A, Islam T, Adams R, Nerle S, Kamara M, Eger C, Marder K, Cohen B, Schifitto G, McArthur JC, Sacktor N, Pardo CA. 2011. Osteopontin enhances HIV replication and is increased in the brain and cerebrospinal fluid of HIV-infected individuals. J Neurovirol 17:382–392. doi: 10.1007/s13365-011-0035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nibbering PH, Zomerdijk TP, Corsel-Van-Tilburg AJ, Van-Furth R. 1990. Mean cell volume of human blood leucocytes and resident and activated murine macrophages. J Immunol Methods 129:143–145. doi: 10.1016/0022-1759(90)90432-U. [DOI] [PubMed] [Google Scholar]
- 45.Chapman EH, Kurec AS, Davey FR. 1981. Cell volumes of normal and malignant mononuclear cells. J Clin Pathol 34:1083–1090. doi: 10.1136/jcp.34.10.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krombach F, Münzing S, Allmeling AM, Gerlach JT, Behr J, Dörger M. 1997. Cell size of alveolar macrophages: an interspecies comparison. Environ Health Perspect 105(Suppl 5):S1261–S1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.GE Healthcare Life Sciences. 2010. Application note 28-9650-52 AC. Perfusion culture of T lymphocytes in the WAVE bioreactor system 2/10 (software version 2.61). GE Healthcare Life Sciences, Pittsburgh, PA. [Google Scholar]
- 48.Brown A, Gartner S, Kawano T, Benoit N, Cheng-Mayer C. 2005. HLA-A2 down-regulation on primary human macrophages infected with an M-tropic EGFP-tagged HIV-1 reporter virus. J Leukoc Biol 78:675–685. doi: 10.1189/jlb.0505237. [DOI] [PubMed] [Google Scholar]
- 49.Brown A, Zhang H, Lopez P, Pardo CA, Gartner S. 2006. In vitro modeling of the HIV-macrophage reservoir. J Leukoc Biol 80:1127–1135. doi: 10.1189/jlb.0206126. [DOI] [PubMed] [Google Scholar]
- 50.Koelsch KK, Liu L, Haubrich R, May S, Havlir D, Günthard HF, Ignacio CC, Campos-Soto P, Little SJ, Shafer R, Robbins GK, D'Aquila RT, Kawano Y, Young K, Dao P, Spina CA, Richman DD, Wong JK. 2008. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J Infect Dis 197:411–419. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- 51.Shiramizu B, Ananworanich J, Chalermchai T, Siangphoe U, Troelstrup D, Shikuma C, De Grutolla V, Sithinamsuwan P, Praihirunkit P, Rattanamanee S, Valcour V, SEARCH 001.1 Study Group . 2012. Failure to clear intra-monocyte HIV infection linked to persistent neuropsychological testing impairment after first-line combined antiretroviral therapy. J Neurovirol 18:69–73. doi: 10.1007/s13365-011-0068-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Folks TM, Powell D, Lightfoote M, Koenig S, Fauci AS, Benn S, Rabson A, Daugherty D, Gendelman HE, Hoggan MD, Venkatesan S, Martin MA. 1986. Biological and biochemical characterization of a cloned Leu-3− cell surviving infection with the acquired immune deficiency syndrome retrovirus. J Exp Med 164:280–290. doi: 10.1084/jem.164.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Regenass U, Caravatti G, Mett H, Stanek J, Schneider P, Müller M, Matter A, Vertino P, Porter CW. 1992. New S-adenosylmethionine decarboxylase inhibitors with potent antitumor activity. Cancer Res 52:4712–4718. [PubMed] [Google Scholar]
- 54.Jänne J, Morris DR. 1984. Inhibition of S-adenosylmethionine decarboxylase and diamine oxidase activities by analogues of methylglyoxal bis(guanylhydrazone) and their cellular uptake during lymphocyte activation. Biochem J 218:947–951. doi: 10.1042/bj2180947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kakinuma Y, Hoshino K, Igarashi K. 1988. Characterization of the inducible polyamine transporter in bovine lymphocytes. Eur J Biochem 176:409–414. doi: 10.1111/j.1432-1033.1988.tb14297.x. [DOI] [PubMed] [Google Scholar]
- 56.Shimura K, Kodama E, Sakagami Y, Matsuzaki Y, Watanabe W, Yamataka K, Watanabe Y, Ohata Y, Doi S, Sato M, Kano M, Ikeda S, Matsuoka M. 2008. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137). J Virol 82:764–774. doi: 10.1128/JVI.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stoddart CA, Galkina SA, Joshi P, Kosikova G, Long BR, Maidji E, Moreno ME, Rivera JM, Sanford UR, Sloan B, Cieplak W, Wrin T, Chan-Hui PY. 2014. Efficacy of broadly neutralizing monoclonal antibody PG16 in HIV-infected humanized mice. Virology 462-463:115–125. doi: 10.1016/j.virol.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bergamaschi A, Pancino G. 2010. Host hindrance to HIV-1 replication in monocytes and macrophages. Retrovirology 7:31. doi: 10.1186/1742-4690-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arfi V, Rivière L, Jarrosson-Wuillème L, Goujon C, Rigal D, Darlix JL, Cimarelli A. 2008. Characterization of the early steps of infection of primary blood monocytes by human immunodeficiency virus type 1. J Virol 82:6557–6565. doi: 10.1128/JVI.02321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ryoo J, Choi J, Oh C, Kim S, Seo M, Kim SY, Seo D, Kim J, White TE, Brandariz-Nuñez A, Diaz-Griffero F, Yun CH, Hollenbaugh JA, Kim B, Baek D, Ahn K. 2014. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med 20:936–941. doi: 10.1038/nm.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nath A, Schiess N, Venkatesan A, Rumbaugh J, Sacktor N, McArthur J. 2008. Evolution of HIV dementia with HIV infection. Int Rev Psychiatry 20:25–31. doi: 10.1080/09540260701861930. [DOI] [PubMed] [Google Scholar]
- 62.Gras G, Kaul M. 2010. Molecular mechanisms of neuroinvasion by monocytes-macrophages in HIV-1 infection. Retrovirology 7:30. doi: 10.1186/1742-4690-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian S, Tawakol A, Burdo TH, Abbara S, Wei J, Vijayakumar J, Corsini E, Abdelbaky A, Zanni MV, Hoffmann U, Williams KC, Lo J, Grinspoon SK. 2012. Arterial inflammation in patients with HIV. JAMA 308:379–386. doi: 10.1001/jama.2012.6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rudman D, Kutner MH, Chawla RK, Goldsmith MA, Blackston RD, Bain R. 1979. Serum and urine polyamines in normal and in short children. J Clin Invest 64:1661–1668. doi: 10.1172/JCI109628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shipe JR Jr, Hunt DF, Savory J. 1979. Plasma polyamines determined by negative-ion chemical ionization/mass spectrometry. Clin Chem 25:1564–1571. [PubMed] [Google Scholar]
- 66.Métifiot M, Marchand C, Maddali K, Pommier Y. 2010. Resistance to integrase inhibitors. Viruses 2:1347–1366. doi: 10.3390/v2071347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Henderson HI, Hope TJ. 2006. The temperature arrested intermediate of virus-cell fusion is a functional step in HIV infection. Virol J 3:36. doi: 10.1186/1743-422X-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arribas JR, Eron J. 2013. Advances in antiretroviral therapy. Curr Opin HIV AIDS 8:341–349. doi: 10.1097/COH.0b013e328361fabd. [DOI] [PubMed] [Google Scholar]
- 69.Custodio JM, Wang H, Hao J, Lepist EI, Ray AS, Andrews J, Ling KH, Cheng A, Kearney BP, Ramanathan S. 2014. Pharmacokinetics of cobicistat boosted-elvitegravir administered in combination with rosuvastatin. J Clin Pharmacol 54:649–656. doi: 10.1002/jcph.256. [DOI] [PubMed] [Google Scholar]
- 70.Hibasami H, Tsukada T, Maekawa S, Nakashima K. 1986. Antitumor effect of methylglyoxal bis(butylamidinohydrazone), a new inhibitor of S-adenosylmethionine decarboxylase, against human erythroid leukemia K 562 cells. Cancer Lett 30:17–23. doi: 10.1016/0304-3835(86)90127-8. [DOI] [PubMed] [Google Scholar]
- 71.Snyder RD. 1989. Polyamine depletion is associated with altered chromatin structure in HeLa cells. Biochem J 260:697–704. doi: 10.1042/bj2600697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sartorelli AC, Iannotti AT, Booth BA, Schneider FH, Bertino JR, Johns DG. 1965. Complex formation with DNA and inhibition of nucleic acid synthesis by methylglyoxal bis(guanylhydrazone). Biochim Biophys Acta 103:174–176. doi: 10.1016/0005-2787(65)90550-2. [DOI] [PubMed] [Google Scholar]
- 73.Fesen MR, Kohn KW, Leteurtre F, Pommier Y. 1993. Inhibitors of human immunodeficiency virus integrase. Proc Natl Acad Sci U S A 90:2399–2403. doi: 10.1073/pnas.90.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bakhanashvili M, Rahav G. 2006. The incorporation of nucleoside analogs by human immunodeficiency virus type 1 reverse transcriptase decreases in the presence of polyamines. FEBS Lett 580:5363–5370. doi: 10.1016/j.febslet.2006.08.074. [DOI] [PubMed] [Google Scholar]
- 75.Taltynov O, Desimmie BA, Demeulemeester J, Christ F, Debyser Z. 2012. Cellular cofactors of lentiviral integrase: from target validation to drug discovery. Mol Biol Int 2012:863405. doi: 10.1155/2012/863405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pesenti E, Pastore C, Lillo F, Siccardi AG, Vercelli D, Lopalco L. 1999. Role of CD4 and CCR5 levels in the susceptibility of primary macrophages to infection by CCR5-dependent HIV type 1 isolates. AIDS Res Hum Retroviruses 15:983–987. doi: 10.1089/088922299310494. [DOI] [PubMed] [Google Scholar]
- 77.Bannert N, Schenten D, Craig S, Sodroski J. 2000. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J Virol 74:10984–10993. doi: 10.1128/JVI.74.23.10984-10993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiang PK, McCann PP, Lane JR, Pankaskie MC, Burke DS, Mayers DL. 1996. Antihuman immunodeficiency virus (HIV-1) activities of inhibitors of polyamine pathways. J Biomed Sci 3:78–81. doi: 10.1007/BF02255534. [DOI] [PubMed] [Google Scholar]
- 79.Schäfer B, Hauber I, Bunk A, Heukeshoven J, Düsedau A, Bevec D, Hauber J. 2006. Inhibition of multidrug-resistant HIV-1 by interference with cellular S-adenosylmethionine decarboxylase activity. J Infect Dis 194:740–750. doi: 10.1086/507043. [DOI] [PubMed] [Google Scholar]
- 80.Lindl KA, Marks DR, Kolson DL, Jordan-Sciutto KL. 2010. HIV-associated neurocognitive disorder: pathogenesis and therapeutic opportunities. J Neuroimmune Pharmacol 5:294–309. doi: 10.1007/s11481-010-9205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McGrath M, Jin X, Burdo T, Williams K. 2012. Macrophage targeted therapy for AIDS: regulation of monocyte differentiation with PA300, a novel oral form of MGBG that reverses CNS and myocardial disease in a SIV model, abstr ST106. Annu Int Meet Inst Human Virol. Institute of Human Virology, Baltimore, MD. [Google Scholar]
- 82.Burdo T, Blitzer J, McKearn J, Miller A, McGrath M, Williams K. 2012. Treatment of SIV-infected monkeys with a polyamine biosynthesis inhibitor, PA300 that decreases monocyte activation and infection, blocks AIDS and SIV encephalitis. CROI 2012: 19th Conf Retroviruses Opportunistic Infect. CROI, Seattle, WA. [Google Scholar]
- 83.Heymann F, Trautwein C, Tacke F. 2009. Monocytes and macrophages as cellular targets in liver fibrosis. Inflamm Allergy Drug Targets 8:307–318. doi: 10.2174/187152809789352230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.